Abstract
Bacterial RNA polymerase (RNAP) makes extensive contacts with duplex DNA downstream of the transcription bubble in initiation and elongation complexes. We investigated the role of downstream interactions in formation of catalytically competent transcription initiation complex by measuring initiation activity of stable RNAP complexes with model promoter DNA fragments whose downstream ends extend from +3 to +21 relative to the transcription start site at +1. We found that DNA downstream of position +6 does not play a significant role in transcription initiation when RNAP-promoter interactions upstream of the transcription start site are strong and promoter melting region is AT-rich. Further shortening of downstream DNA dramatically reduces efficiency of transcription initiation. The boundary of minimal downstream DNA duplex needed for efficient transcription initiation shifted further away from the catalytic center upon increasing the GC content of promoter melting region or in the presence of bacterial stringent response regulators DksA and ppGpp. These results indicate that the strength of RNAP-downstream DNA interactions has to reach a certain threshold to retain the catalytically competent conformation of the initiation complex and that establishment of contacts between RNAP and downstream DNA can be coupled with promoter melting. The data further suggest that RNAP interactions with DNA immediately downstream of the transcription bubble are particularly important for initiation of transcription. We hypothesize that these active center-proximal contacts stabilize the DNA template strand in the active center cleft and/or position the RNAP clamp domain to allow RNA synthesis.
Keywords: transcription initiation, transcription regulation, stringent response, DNA-protein interactions, fluorescence spectroscopy
Introduction
Formation of transcription initiation complex between RNA polymerase (RNAP) and promoter DNA is one of the most heavily regulated steps of bacterial gene expression. RNAP holoenzymes containing primary σ factors (σ70 in Escherichia coli) recognize and initiate transcription from most promoters present in a bacterial cell (1). The establishment of initial RNAP contacts with a σ70-dependent promoter is usually initiated through contacts with the –35 promoter element and upstream promoter DNA. These initial interactions are followed by the formation of a catalytically-competent open promoter complex (RPo) in which a stretch of ~13 base pairs of promoter DNA is unwound (forming the so-called “transcription bubble”), and the DNA coding strand is loaded into the active site, providing a template for RNA synthesis (2). The main source of energy driving the DNA strand separation is the strong interaction between σ70 and the non-template strand bases of the −10 element and adjacent downstream nucleotides (2-5). RNAP interaction with the template strand segment of the transcription bubble is considerably weaker (6, 7). RNAP also forms multiple contacts with the duplex DNA downstream of the transcription bubble. These interactions extend from position +3, the first double-stranded position at the downstream edge of the transcription bubble, to as far as position +20 (2). The downstream DNA-RNAP contacts contribute to stabilization of transcription initiation and elongation complexes and may facilitate the nucleation and propagation of promoter DNA melting (8-11). It is unclear, however, whether formation of downstream RNAP-promoter contacts is strictly required for promoter melting (2). RNAP interactions with the downstream DNA are affected by a number of low molecular weight inhibitors and protein factors that target the initiation step of transcription, such as bacterial stringent response regulators DksA and ppGpp (12-16).
Contacts with the downstream duplex DNA are primarily made by the RNAP β’ subunit clamp domain (17-19). The β’-clamp can move relative to the RNAP mainframe assuming conformations that define the “open” and “closed” states of the RNAP active center cleft (17, 20-25). The movement of the β’-clamp and a ratcheting motion of the core and shelf modules of the central part of RNAP relative to each other alter the nucleic acid binding properties of RNAP and stability of transcription complexes (20-25). Structural analysis shows that RNAP interactions with the downstream DNA are essentially the same in bacterial initiation and elongation complexes where RNAP assumes a closed β’-clamp conformation (18,19). Chakraborty et al. proposed that, during the RPo formation, clamp opening allows DNA to be unwound and loaded in the RNAP active-center cleft. In turn, DNA loading and unwinding triggers the β’-clamp closure, leading to formation of stable initiation and elongation complexes (24). Consistent with this view, crystallographic studies of an elongation complex bound to transcription inhibitor Gfh1 or a paused elongation complex reveal open-clamp RNAP structures in which most of downstream contacts are broken and the RNAP active site is blocked by a kinked conformation of the bridge-helix domain (23,25). It was proposed that a similar open-clamp RNAP conformation transiently arises during DNA translocation (23,25).
Considering the extent of RNAP-downstream DNA interactions and their dynamics, it is important to dissect RNAP contacts with specific positions of downstream segment and identify interactions targeted by regulatory factors. Because there is an interplay between the RNAP-downstream DNA interactions and catalytic activity, it is also of interest to determine a minimal set of downstream DNA contacts that is sufficient to support efficient transcription initiation. Here, we elucidated the role of RNAP-promoter downstream contacts in the formation of catalytically-competent transcription initiation complex by the Echerichia coli RNAP σ70 holoenzyme by measuring initiation activity of stable RNAP complexes with model promoter DNA fragments whose downstream ends extend from +3 to +21. Our approach allowed us to observe how the formation of the catalytically competent conformation of transcription initiation complex depends on the establishment of downstream RNAP-promoter interactions. We also investigated the interdependence between the extent of downstream RNAP-promoter contacts and inhibition of transcription initiation by DksA/ppGpp.
Results
RNAP forms stable complexes with N25cons promoter derivatives truncated downstream of the transcription start site
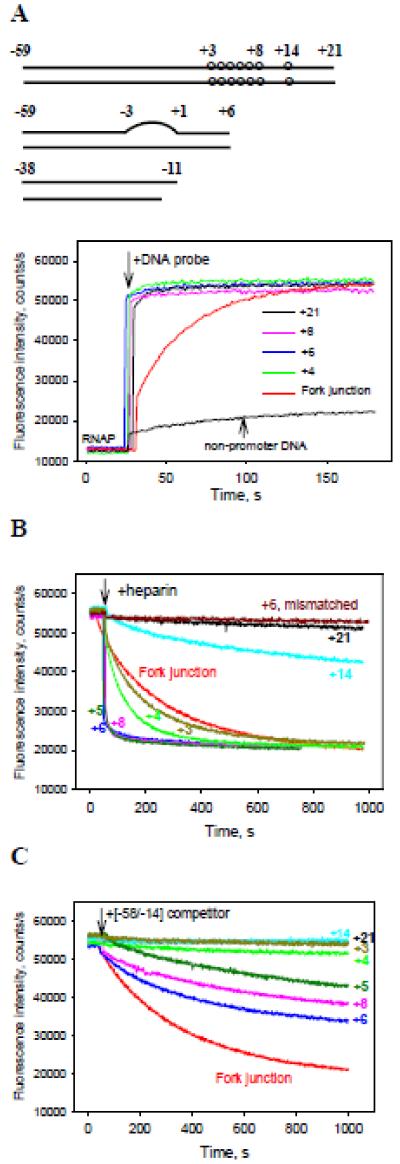
We reasoned that the role of downstream RNAP-promoter interactions in initiation complex formation may be investigated by analyzing properties of E. coli σ70 RNAP holoenzyme complexes with promoter derivatives truncated at different positions downstream of the transcription start site. However, activity studies of such complexes are compromised by the weakened binding of RNAP to promoter fragments with short downstream segments. For example, a certain length of downstream DNA is needed for formation of stable specific complexes on a strong phage T5 N25 promoter (9). Similar results were observed with derivatives of λPr promoter in this work (see below). While the saturation binding of RNAP to truncated T5 N25 fragments could be reached at sufficiently high template concentrations, formation of non-specific complexes made quantitative measurements impossible (data not shown). We therefore sought to maximize the strength of sequence-specific RNAP interactions with promoter elements, such that incomplete or non-specific RNAP binding to promoter DNA variants will not interfere with the analysis. To this end, the N25cons promoter (12) with consensus −35 element, consensus extended −10 elements, and an optimized UP element was chosen for analysis. The parent N25cons probe spans from positions −59 to +21 (probe 1, [−59/+21], Supplemental Table 1). Shorter probes had the same upstream boundary but their downstream edges were at positions +14, +8, +7, +6, +5, +4, or +3 (Supplemental Table 1). Binding of DNA probes to RNAP was detected using the RNAP beacon assay in which the interaction with DNA is revealed by increased fluorescence of RNAP holoenzyme carrying a fluorescent label upon specific interaction of promoter with conserved region 2 of the σ70 subunit (5). Upon addition of 2 nM of each of the probes to 1 nM RNAP beacon, the fluorescent signal increased ~4-fold, reaching peak intensity in a few seconds. Representative traces and a control trace obtained with an upstream fork junction probe 33 that tightly binds to RNAP (26) are shown in Fig. 1A. Further increases in probe concentrations did not enhance signal amplitudes (data not shown) indicating that specific RNAP binding was saturated. The increases in fluorescence intensity observed in these experiments were much higher than that generated by a control non-promoter DNA at similar conditions (Fig. 1A). We also tested several locally mismatched, partially single-stranded (ss) and nicked analogs of the N25cons derivatives (see Table S1, probes 13-21). Binding of these probes to RNAP beacon caused the same enhance in fluorescence intensity as it was observed with probes 1-8. These results demonstrate that RNAP forms high-affinity specific complexes with all tested derivatives of N25cons promoter.
Figure 1. Measuring of RNAP interactions with derivatives of N25cons promoter using the RNAP beacon assay.
(A) Time dependence of the increase in fluorescence upon mixing 1 nM RNAP beacon with 2 nM indicated N25cons derivatives whose downstream ends were located between positions +3 to +21, 2 nM upstream fork junction or 2 nM non-promoter DNA probe.
(B, C) The effect of downstream end position on resistance of RNAP beacon complexes with truncated N25cons derivatives to heparin or [−58/−14] DNA competitor. Time-dependent changes of the fluorescence signal were measured upon the addition of either 20 μg/ml heparin (B) or 4 nM [−58/−14] (C) to RNAP beacon complexes with N25cons derivatives formed as in panel (A).
The structures of the DNA probes are shown in Table. S1 (probes 1-8, 13, 28, 33). The structure of non-promoter DNA probe is shown in Fig. S5. The numbers in the panels indicate positions of downstream edges of the probes, “+6 mismatched” in panel B refers to probe 13 with downstream end at +6 containing a mismatched segment spanning positions −3 to +1.
To determine relative stabilities of RNAP compelx with various N25cons probes, we used RNAP beacon-based competition-binding experiments. Competitive displacement of bound promoter fragments from their complexes with RNAP beacon was initiated by the addition of either heparin, a strong inhibitor of RNAP-DNA interactions, or a less potent inhibitor, DNA probe [−58/−14] (probe 28), which binds RNAP tightly but does not generate the fluorescent signal of the beacon (5). RNAP complexes with probes whose downstream edges extend to positions +21 and +14 were resistant to both competitors (Fig. 1B and C). Moving the edge of the duplex to base pairs +8 or +6 led to complex destabilization, the effect being most evident in the presence of heparin. Further shortening of the downstream DNA segment to positions +5, +4 and +3 increased complex stability (Fig. 1B and 1C). The RNAP beacon complexes with probes 13-21 were found to be heparin-resistant (representative data for mismatched probe 13 with downstream edge at +6 are shown in Fig. 1B). Overall, the data in Fig. 1B and C show that RNAP contacts with the +7 to +21 N25cons segment considerably strengthen the RNAP-promoter binding, as expected (9).
Transcription initiation activity of RNAP complexes with N25cons promoter fragments
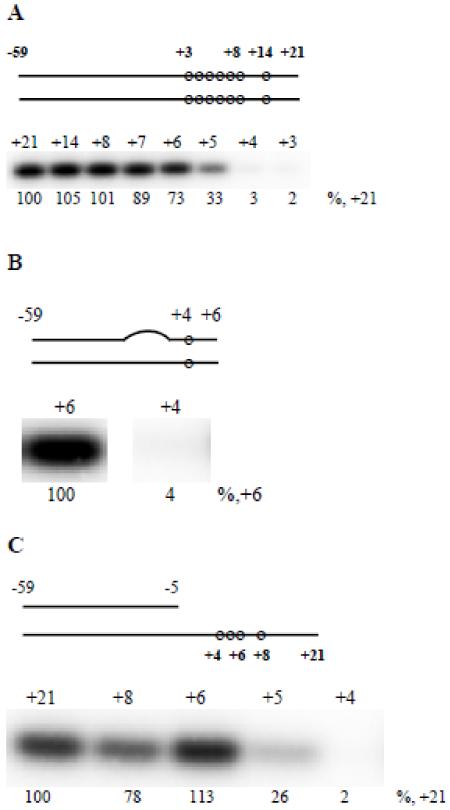
The ability of truncated DNA fragments of N25cons promoter to serve as transcription initiation templates was determined using a standard abortive initiation in vitro transcription assay in which the synthesis of RNA trinucleotide CpApU from CpA dinucleotide primer and radiolabeled UTP was monitored. The activities of complexes formed on probes with downstream edges at +14, +8, and +7 were nearly identical to activity observed on the parent −59/+21 probe (Fig. 2A). Amounts of abortive RNA synthesized from probes with downstream edges at positions +6 and +5 were decreased relative to those produced on parental DNA by 1.5- and 3-fold, respectively (Fig. 2A). Shifting the downstream boundary to positions +4 and +3 led to much more dramatic ~30- and 50-fold drop in transcriptional activity, respectively. Neither raising the CpA or UTP concentrations nor changing the +3 to +6 segment sequence prevented the drop of activity observed upon moving the downstream edge of transcription templates from the +6 to the +4 position (see Fig. S1 and Table S2). These results thus indicate that the downstream promoter segment encompassing nucleotides up to position +5 is required for efficient transcription initiation. RNAP contacts with base pairs at position +6 and +7 further stimulate transcription. In contrast, promoter segment located downstream of +7 is not essential for initiation of RNA synthesis in the context of tested templates.
Figure 2. Abortive transcript synthesis from N25cons derivatives.
Abortive transcript synthesis from: (A) duplex probes 1-8 with downstream edges at +21,+14, +8, +7,+6,+5,+4, and +3. (B) probes 13, 14 with downstream edges at +6 and +4 containing a mismatched segment spanning positions −3 to +1. (C) tailed probes 15-19 bearing a double-stranded DNA segment at positions −59 to −5 followed by single-stranded template strand extensions ending at positions +4, +5, +6, +8, and +21.
Next, we asked which step in the pathway to RPo formation is predominantly affected by shortening of the downstream segment. We hypothesized that templates truncated at +3, +4, and +5 positions may not be efficiently melted by RNAP, which would explain poor levels of transcription activity. To address this, we tested transcription activity from premelted promoter fragments that were similar to truncated ds probes [−59/+4] and [−59/+6] but contained a stretch of DNA with mismatched nucleotides spanning positions −3 to +1, which should facilitate loading of template stand into the RNAP catalytic cleft. As shown in Fig. 2B, the drop in activity levels with these templates was the same as with fully double-stranded probes. Next, we tested a set of tailed N25cons derivatives carrying ds DNA segment at positions −59 to −5 followed by ss template DNA strand extending to positions +4, +5, +6, +8, or +21 (probes 15-19). As can be seen from Fig. 2C, RNAP complexes with tailed probes ending at +4 and +5 were much less active than complexes with probes carrying longer ss DNA tails. The longest tailed probe 15 was about 3-fold less active than the corresponding ds [−59/+21] probe (data not shown). Together the data presented above exclude defects in promoter melting as the main cause for the low transcriptional activity observed with deeply truncated probes.
Experiments with tailed probes that lack non-template strand bases downstream of the −5 position seem to suggest that the template strand of the downstream segment is particularly important for transcription activity. This suggestion was supported by measurements of transcription activity from discontinuous N25cons derivatives that lacked a phosphate group between positions +4/+5 or +6/+7 of the template strand (probes 20 and 21, downstream edges of these probes were extended to +31 to prevent dissociation of short oligos used to construct the probes). The initiation activity from a probe with a break at +4/+5 was 8-fold lower than that from a probe with a break at +6/+7 (Fig. S2), while the initiation activity from a probe with a break at +6/+7 was only about 20% lower than that of ds [−59/+21] probe (data not shown). Overall, the results indicate that RNAP interactions with active center-proximal downstream nucleotides are most essential for retaining efficient initiation activity while RNAP contacts with a distal downstream promoter segment play supplementary role.
In the RNAP holoenzyme, the σ70 subunit region 1.1 (σ1.1) is positioned inside the RNAP main channel from where it is displaced by the downstream promoter segment upon RPo formation (27). Although σ1.1 is not strictly required for in vitro transcription, it modulates the kinetics of RPo formation at some promoters (28,29). σ1.1 is also involved in inhibition of transcription initiation by T7 phage gp2 protein (30, 31). Therefore, it seemed possible that σ1.1 could affect activity of RNAP complexes with truncated promoter fragments. However, the dependence of abortive transcription levels on the length of downstream duplex DNA in complexes with RNAP reconstituted with σ70 lacking region 1.1 was similar to that observed with wild-type σ70 RNAP holoenzyme (Fig. S3 and Fig. 2A, respectively). Thus, the observed dependence of transcription activity on downstream DNA segment length is not determined by σ1.1.
Next, we hypothesized that shortening of downstream DNA segment could affect transcription initiation by shifting the balance between post-and pre-translocated registers of RPo from the former to the latter. This assumption can be experimentally tested since initiation complex in the pre-translocated register will be susceptible to the pyrophosphorolysis reaction (32). To detect pyrophosphorolysis in RNAP complexes with promoter fragments, we used a synthetic chimeric primer in which ATP is fused to rifampicin (Rif-ATP), thus stabilizing the priming nucleotide in the active center (33). Both the forward reaction of Rif-ApU synthesis from Rif-ATP and UTP, and the reverse reaction of UTP formation from Rif-ApU in the presence of pyrophosphate could be readily detected in RNAP complexes with –[−59/+21], [−59/+6], and [−59/+5] probes but not with probe [−59/+4] (Figs. S4A and B). Thus, both catalytic reactions are impaired in complexes with promoter DNA truncated at position +4. Therefore, changing the balance between post-and pre-translocated registers (whether it takes place or not) cannot account for the observed low activity on this template.
Overall, the results above suggest that the active center-proximal downstream interactions play a role in stabilizing a catalytically-competent conformation of promoter complex that is attained after the initial DNA melting step has occurred (See Discussion).
Transcription initiation from N25cons derivatives with GC-rich promoter discriminator segment
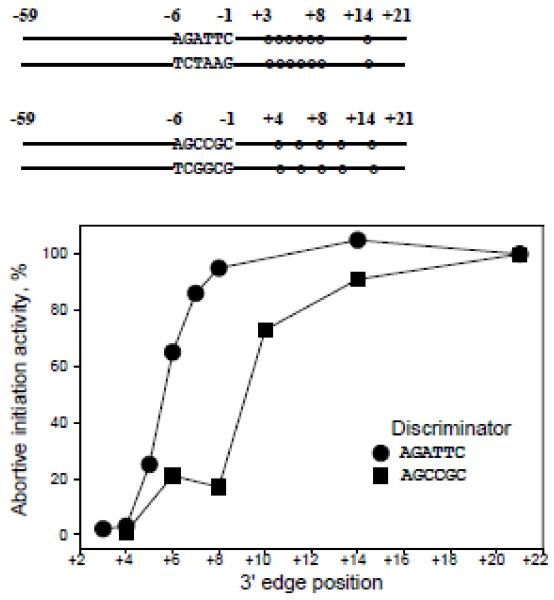
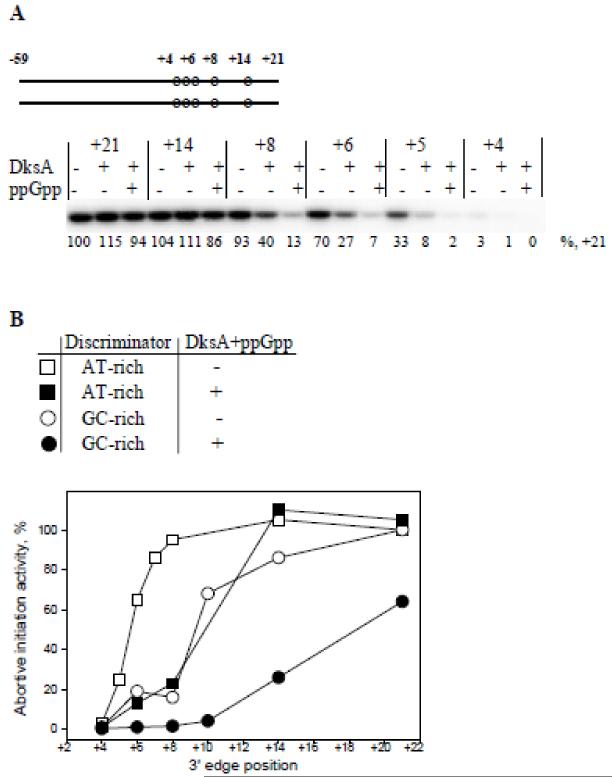
The sequence of N25cons within the DNA-melting region (−11 to +2) has low GC content that favors the separation of DNA strands. Base substitutions increasing the GC content of the region between the −10 element and the transcription start site (termed the “discriminator”) hinder the RPo formation in many promoters (34). We wondered whether increasing the GC content of N25cons discriminator influences the dependence of transcription initiation activity on the length of the downstream DNA segment. Therefore, we measured abortive initiation activity from N25cons derivatives in which the discriminator segment sequence AGATTC was changed to AGCCGC (probes 22-27, Table S1). The RNAP beacon assay experiments showed that probes with increased GC discriminator content specifically bound RNAP with affinity nearly as high as the affinity of original probes 1-8 (Fig. S5). For the longest probes, discriminator sequence change conferred a ~1.5-fold decrease in activity (data not shown). The relative activities from substituted probes are plotted in Fig. 3, along with similar data for original N25cons derivatives (see Fig. 2A). As can be seen, complexes formed on GC rich discriminator probes with downstream edges at +8 and +6 were poorly active (about 20% of activity of parental probe 22), in contrast to high relative activities from similar N25cons derivatives with AT rich discriminator (Fig. 3). As expected, activity from the shortest probe 27 with downstream edge at +4 was very low (2% of longest GC rich probe 22 activity).
Figure 3. Abortive transcript synthesis from N25cons derivatives with GC rich promoter discriminator segment.
Abortive transcript synthesis activities from probes 22-27 containing a GC rich discriminator region are plotted along with similar data for original N25cons derivatives 1-8. The plotted data were averaged from three independent experiments.
The data in Fig. 3 thus demonstrate that the dependence of transcription initiation activity on downstream promoter DNA length is modulated by the GC content of discriminator. A plausible explanation for this observation is that the melting of the N25cons derivatives with low GC discriminator content is driven mainly by RNAP interactions with the DNA strands within the transcription bubble, whereas less energetically favorable melting of probes bearing discriminator with high GC content must be coupled with formation of extended downstream RNAP-promoter contacts. The mechanistic basis for such coupling may be due to a sharp kink in DNA near the transcription start site that is introduced once RNAP contacts with the downstream duplex are established upon the RPo formation and that should stabilize the melted promoter conformation (2,18,19).
RNAP complexes with derivatives of the λPr promoter
To exclude a possibility that a sharp drop in transcription initiation activity observed upon shortening of promoter fragments from +6 to +4 is specific for the N25cons promoter, we analyzed the RNAP-binding kinetics and transcription initiation activities of four λ Pr promoter fragments. The parent probe is a [−62/+21] derivative of λ Pr in which a nonconsensus G at −12 is changed to consensus T to improve the binding. Other probes are derivatives of [−62/+21] truncated at positions +10, +6, and +4 (see Table S1, probes 29-32). Transcription initiation activity observed on the template shortened to +10 was the same as activity from the parental template, whereas complex with template ending at +6 was less active (28% residual activity) (Fig. 4). Upon moving the downstream edge to the +4 position, transcription activity sharply decreased (1% of activity on parental probe), in agreement with data obtained with N25cons-based template series. The longest [−62/+21] probe generated high fluorescent signal upon binding to the RNAP beacon, which suggests that the binding was specific and strong (Fig. S6). However, the fluorescent signals generated by shorter λ Pr probes were considerably lower than that generated by [−62/+21] (Fig. S6), indicating weaker RNAP binding to these probes. Nevertheless, overall trends detected with shortened λ Pr fragments clearly follow results obtained with N25cons and its derivatives.
Figure 4. Abortive transcript synthesis from λPr derivatives.
Abortive transcript synthesis from duplex probes 29-32 with downstream edges at +21,+10,+6, and +4.
Effect of DksA and ppGpp on RNAP binding to model promoter fragments
Transcription factor DksA and alarmone ppGpp synergistically regulate bacterial transcription initiation in response to various cell stresses (e.g. during stringent response) (35). DksA belongs to a class of transcription regulators that bind in the RNAP secondary channel, whereas ppGpp binding site is located between the β’ and ω subunits on the outer surface of RNAP (36-38). Neither of the two regulators interacts directly with DNA. DksA/ppGpp inhibit transcription by destabilizing short-lived RPo formed on sensitive promoters (16,35). It was proposed that by binding to the mobile trigger loop domain in RNAP secondary channel, DksA induces an allosteric change in the position of the RNAP clamp, thus affecting the downstream DNA contacts in RPo (39).
To characterize potential regulatory role of RNAP-downstream promoter interactions in RPo formation, we investigated how DksA/ppGpp affect the properties of RNAP complexes formed on model promoter fragments. We measured the combined effect of 2 μM DksA and 100 μM ppGpp, concentrations that cause maximal effects on in vitro transcription (40). The effect of DksA alone was also determined in some experiments. We found that fast formation of high-affinity complexes between RNAP and N25cons-based promoter fragments was not compromised by DksA/ppGpp, while noticeable decrease in competitor resistance of the preformed complexes was observed (representative data are shown in Fig. S7A, B and C).
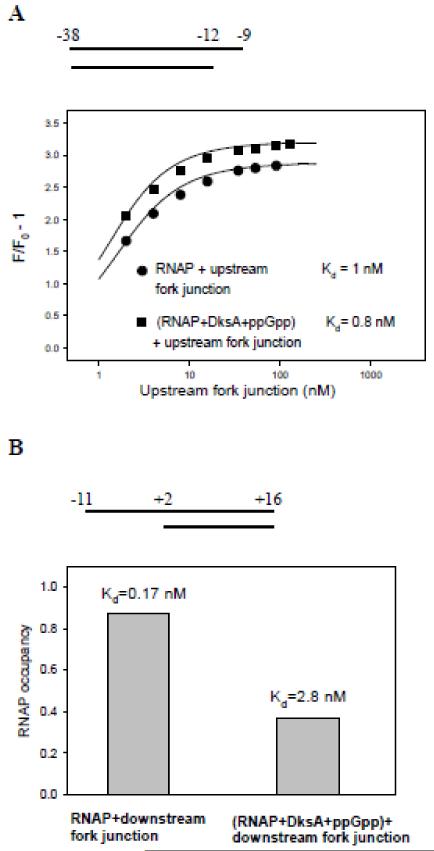
We next studied the effects of DksA/ppGpp on RNAP binding to short promoter fragments (probes 34-37 in Table S1). RNAP binding to the upstream fork junction probe bearing the −35 element (probe 34) was insignificantly improved by DksA/ppGpp (Fig. 5A). However, DksA/ppGpp noticeably affected the rate of RNAP binding to downstream fork junction probe (probe 35), which consisted of ss DNA segment −11/+2 corresponding to the non-template strand part of the transcription bubble followed by downstream ds segment of N25cons from +3 to +16 (Fig. S8A). Because of the very high affinity of this downstream fork junction to RNAP (Kd <0.2 nM (9)), the destabilizing effect of DksA/ppGpp on complex formation could not be quantified directly. Therefore, we performed an RNAP beacon competition binding assay using a similar downstream fork junction probe but carrying a non-consensus T at −8 position (probe 36), which weakened the interaction with RNAP (9). The RNAP occupancies and Kd values were determined in samples containing 1 nM RNAP beacon and 2 nM downstream fork junction probe 36 in the absence and presence of DksA/ppGpp as described in Supplementary Material (see Fig. S9 and accompanying text). The results showed that DksA/ppGpp cause a 16-fold decrease in the apparent affinity of downstream fork junction to RNAP (Fig. 5B). Since DksA/ppGpp did not influence RNAP binding to the ss oligo probe −11/+2 corresponding to the ss DNA part of the fork junctions (Fig. S8B), these experiments indicate that DksA/ppGpp considerably weaken RNAP interaction with the downstream duplex.
Figure 5. The effect of DksA/ppGpp on binding of upstream and downstream fork junction probes to RNAP.
(A) curve 1, titration of RNAP beacon with upstream fork junction (probe 34). Curve 2, same as curve 1, but RNAP beacon was preincubated with 2 μM DksA and 100 μM ppGpp for 2 min prior to the addition of downstream fork junction. The Kd values were determined by fitting the dependence of relative fluorescence signal amplitude (F/Fo) on probe concentration to a chemical equilibrium equation (5).
(B) RNAP occupancy by downstream fork junction 36 was measured in samples containing 1 nM RNAP beacon or RNAP beacon preincubated with 2 μM DksA and 100 μM and 2nM of downstream fork junction probe.
Inhibition of transcription initiation from N25cons derivatives by DksA and ppGpp
DksA/ppGpp did not noticeably affect transcription initiation from longest [−59/+21] and [−59/+14] probes with AT-rich discriminators. Abortive transcript synthesis from shorter [−59/+8], [−59/+6], and [−59/+5] probes decreased ~3 and ~10-fold upon the addition of DksA alone or both DksA and ppGpp, respectively (Fig. 6A). The inhibitory effect was specific since no change in activity was observed in the presence of D74A DksA mutant that binds RNAP without affecting transcription (data not shown) (41). DksA/ppGpp also inhibited abortive initiation from premelted [−59/+6] probe 13 (Fig. S10).
Figure 6. The effect of DksA and ppGpp on abortive transcript synthesis from N25cons derivatives.
(A) Abortive transcript synthesis from probes with AT-rich discriminator (probes 1-3, 5-7). (B) The effects of DksA/ppGpp on abortive transcript synthesis from probes with AT rich and GC rich discriminator sequences. The activity values are presented as percentage of activities of the corresponding parental probes 1 and 22 with edges at +21. The plotted data were averaged from three independent experiments.
A known feature that contributes to susceptibility of promoter complexes to inhibition by DksA/ppGpp is a GC-rich discriminator sequence (35). Consistently, we found that RNAP complexes with probes bearing a GC-rich discriminator region were more susceptible to DksA/ppGpp than RNAP complexes with original N25cons derivatives. Activity of parent GC-rich discriminator containing probe with downstream edge at +21 was only slightly decreased in the presence of DksA/ppGpp (by 1.4-fold), however activities of probes with downstream edges at +14 and +10 decreased 3.3 and 15-fold, respectively (Fig. 6B). The abortive RNA synthesis from shorter probes 25-27 was barely detectable in the presence of DksA/ppGpp (Fig. 6B). Thus, DksA/ppGpp significantly increase the minimal length of downstream DNA segment required for efficient transcription initiation from model templates. This is fully consistent with suppression of downstream RNAP-promoter interactions by DksA/ppGpp (Fig. 5B). These results emphasize the interdependence between establishment of the downstream interactions and RPo formation and suggest that weakening of downstream RNAP-promoter contacts by DksA/ppGpp may be the cause of inhibition of transcription initiation from promoters on which the RPo formation is relatively energetically unfavorable.
Discussion
Structural and biochemical analyses of transcriptional complexes have revealed multiple RNAP interactions with downstream DNA. These interactions are dynamic and can change coordinately with conformational changes in RNAP (17-25). The role of downstream RNAP-DNA contacts in bacterial transcription initiation is not fully understood and is subject to debate (reviewed in Refs. 2, 42). To investigate significance of discrete downstream interactions in formation of catalytically-competent initiation complexes, we here analyzed the ability of model promoter derivatives bearing gradually truncated downstream segments to serve as transcription initiation templates. Interpretation of such activity measurements is potentially hindered by low stability of RNAP complexes with promoter fragments lacking downstream sequences (9). This complication was resolved by using derivatives of an optimized for binding N25cons promoter, which formed specific high-affinity complexes with RNAP (Fig. 1A).
Our results reveal a previously unknown effect of active center proximal downstream DNA interactions on transcription initiation. We found that the efficiency of transcription initiation greatly decreases upon shortening of the downstream promoter segment from +6 to +4. In contrast, promoter segment located downstream to +7 is not essential for initiation in the context of templates based on the N25cons sequence (Fig. 2A), while it increases the strength of RNAP-template binding, as seen from Fig. 1B and C. Similar results were obtained with derivatives of the λ Pr promoter (Fig. 4). These data indicate that active center-distal downstream interactions are dispensable for retaining the catalytically-competent conformation of initiation complex when RNAP-promoter interactions upstream of the start site are optimized.
The low activity of [−59/+4] and [−59/+5] probes can not be bypassed by pre-opening of the transcription start site (Fig. 2B and C). The abortive RNA synthesis from [−59/+4] template was also not improved upon deletion of σ70 subunit region 1.1, which blocks downstream DNA loading into the active center channel (Fig. S3) (27,31,43). Therefore, RNAP interactions with the downstream DNA segment proximal to the transcription start site are likely to be required after promoter melting has occurred. Interestingly, shortening of the downstream DNA segment from position +6 to positions +5, +4 and +3 somewhat increased the complex resistance to competitor challenge (Fig. 1B and C). This result suggests that a short segment of the active center-proximal duplex downstream DNA (up to +6 position) destabilizes the complex. We speculate that establishment of RNAP contacts with the +3/+6 positions might account for this effect by weakening some other interactions within the RNAP-promoter complex. Displacement of σ70 region 1.1 may play a role in this process, however the confirmation of this conjecture requires further studies that are beyond the scope of this work. Measurements of transcription initiation and pyrophosphorolysis from promoter fragments using a chimeric rifampicin-ATP compound as a primer suggest that poor activity of [−59/+4] probe cannot be explained by changes in the translocation equilibrium of complexes on shortened templates (Fig. S4A and B). In principle, correct loading of the template strand of short probes into the active center may be somehow affected by fraying of the downstream probe termini. However, the fact that substitutions of A’s for G and C at positions +3 and +4 did not improve activity of [−59/+4] compared to [−59/+6] (Table S2) argues against such an explanation.
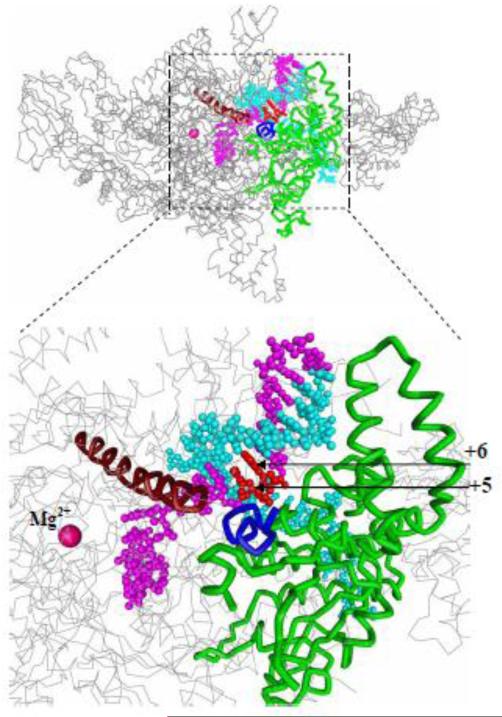
In a bacterial initiation complex structure, downstream DNA nucleotides within +3 to +6 duplex segment are contacted by switch 1 and switch 2 domains, as well as by rudder and N-terminal β’ region which are parts of the β’ clamp (Fig. 7) (19). The “switch region” is located at the base of the clamp and serves as a hinge at which clamp moves from open to closed conformation (17). It has been proposed that direct contacts between the switch region and DNA phosphates might coordinate clamp closure and DNA binding (17,22). Thus, the large increase in the initiation activity observed upon extension from +4 to +6 in the context of [−59/+4] and [−59/+6] probes may be explained by postulating that extension of downstream DNA induces the proper clamp closure required to set up the active center for catalysis. This is consistent with observed inhibitory effect on transcription of the compounds that “freeze” conformation of the clamp (14, 24). The inhibitory effect of the template strand break between +4 and +5 positions (Fig. S2) may thus be a consequence of distortion of the catalytically competent active center conformation caused by the physical discontinuity of the template DNA. On the other hand, the RNAP-promoter contacts downstream from +4 may be required to position the template strand near the transcription start site. Indeed, breaks in the template-strand segment of the transcription bubble and mutations in RNAP that have been shown to affect RNAP contacts with this segment impair the catalytic properties of initiation complexes (19,44-47). These observations suggest that the template strand may be bound in the active center cleft relatively loosely. The proposed effects of downstream contacts on either the active center or template strand conformation are not mutually exclusive.
Figure 7. The template strand +5 and +6 nucleotides interact with switch 1 and β’ clamp.
Light blue, DNA non-template strand; red, +5 and +6 template stand nucleotides; magenta, rest of DNA template strand; green, β’ clamp; brown, bridge helix; dark blue, switch 1; grey, rest of RNAP. The structure of RNAP complex with a downstream promoter fragment is from Ref. 19.
We found that the boundary of a minimal downstream DNA duplex needed for efficient initiation of RNA synthesis shifts further away from the catalytic center under conditions that hamper the RPo formation. This effect was observed when the GC content of the N25cons discriminator sequence was increased or in the presence of bacterial stringent response regulators DksA and ppGpp (Figs. 3 and 6B). The former result suggests that highly energetically unfavorable melting of the GC-rich discriminator is coupled with establishment of extended downstream DNA contacts with RNAP. In contrast, such coupling is not required when the discriminator sequence is AT-rich. As seen in Fig. 6B, the negative effects caused by DksA/ppGpp and by high GC content of promoter discriminator on the efficiency of initiation from promoter fragments are additive.
DNA footprinting data and mutational analysis indicate that DksA disrupts RNAP interactions with the rrnB P1 promoter DNA between positions −6 and +6 (16). This implies that DksA may target RNAP interactions with the promoter melting region and with the downstream promoter duplex. We found that DksA/ppGpp weaken the downstream RNAP-promoter interactions 16-fold but have insignificant effects on RNAP binding to the upstream part of promoter (Fig. 5A and B). Taken together with the fact that the RPo formation can be coupled with establishment of the downstream RNAP-promoter interactions, this suggests that weakening of downstream interactions by DksA/ppGpp may lead to destabilization of initiation complexes formed on promoters whose melting is reversible. We note that our results do not exclude the possibility that DksA/ppGpp may also affect RNAP contacts with the template strand segment of the transcription bubble. Furman et al. reported that deletion of a species-specific insertion in the β’ (called i6) or a partial deletion of the β’ jaw domain make RNAP hypersensitive to DksA (48). Our data suggest that this observation might in part be a consequence of these deletions directly or indirectly weakening RNAP interactions with the downstream duplex DNA (11,49,50).
Overall, the data presented here argue that for transition of RNAP promoter complex to a catalytically competent state, the strength of RNAP-downstream DNA interaction has to reach a certain threshold. The results show that downstream interactions not only contribute to general stability of promoter complexes but also modulate the catalytic efficiency, providing a possible mechanistic basis for transduction of regulatory signals from the downstream part of promoter complex to the RNAP active center.
Materials and methods
Proteins
Wild-type Escherichia coli RNAP core was purchased from Epicenter. Wild-type E. coli σ70 subunit and the σ70 derivative labeled at position 211 with fluorescent label 5-tetramethylrhodamine were prepared as in (5), the σ70 deleted for region 1.1 (residues 1-104) was a gift from S. Wigneshweraraj. RNAP core enzyme carrying 6xHis-tag at the C-terminus of β’ subunit was purified as described (51). The wild type 6xHis-tagged DksA was prepared by cloning dksA gene into pET33b+ vector (Novagen) between NheI and XhoI sites resulting in pET33-NPH-DksA expression plasmid. DksA-D74A mutation was introduced into pET33-NPH-DksA using QuickChange Site-Directed Mutagenesis Kit (Stratagene). The wild type and the mutant 6xHis-tagged DksA proteins were overexpressed in BL21(DE3) E.coli and purified by chelating Ni-NTA agarose followed by size-exclusion chromatography on Superdex 75 HR column as described (40). RNA polymerase holoenzymes were prepared as in (52).
DNA Probes
DNA oligonucleotides were synthesized by Integrated DNA Technologies. Double-stranded and tailed DNA probes were prepared as in (5). The structures of DNA probes used are presented in Supplementary Table S1 and are also schematically depicted in main figures.
In vitro abortive initiation and pyrophosphorolysis
Abortive transcription reactions were performed in a final volume of 10 μl and contained 200 nM E. coli core enzyme (Epicentre) supplemented with 500 nM σ70, 5 μM DksA, 100 μM ppGpp (TriLink Biotechnologies) (where indicated), and 50 nM various DNA templates prepared as described above in standard transcription buffer (30 mM Tris-HCl [pH 7.9], 40 mM KCl, 10 mM MgCl2, 2 mM β-mercaptoethanol). Reactions were mixed as indicated and incubated for 10 min at 37 °C, followed by the addition of CpA RNA dinucleotide primer (200 μM), cold UTP (20 μM), and [α-32P] UTP (3000 Ci/mmol). Where indicated, the chimerical compound Rif-ATP prepared as in (33) or ATP were used as primers in concentration 2 μM. The reactions were incubated for a further 10 min at 37 °C and then terminated by the addition of an equal volume of urea-formamide loading buffer. The reaction products were resolved on a 20% (w/v) polyacrylamide denaturing gel and visualized using a PhosphorImager. To determine relative initiation activities of transcription templates, abortive transcription initiation experiments were repeated at least three times. For most measurements, standard deviations were less than 25%. For shortened templates where very low transcription activity was observed (less than 7% of activity observed on full-sized starting templates), standard deviations were ≤40%.
To obtain the radioactively labeled Rif-ApU dinucleotide for pyrophosphorolysis experiments, the abortive initiation reactions were performed in transcription buffer in a final volume of 50 μl and contained 15 μM E. coli RNAP core enzyme with 10-histidine tagged β’ subunit supplemented with 30 μM σ70, 50 μM Rif-A. After 5 minutes incubation at 37 °C reaction was supplemented with 10 μM N25cons [−59/+21] promoter DNA and further incubated for 10 minutes. Next, 4 μl [α-32P] UTP (3000 Ci/mmol) was added followed by 2 minutes incubation. After that, the transcription complex was incubated for 5 minutes at room temperature with Ni-NTA-agarose beads with occasional mixing. Unbound RNAP and nucleotides were washed out with 1 ml of water and the bound RNAP-Rif-ApU-DNA ternary complex was incubated in 30 μl of water for 5 minutes at 80 °C to release newly synthetized radioactively labeled Rif-ApU. The resulting supernatant with Rif-ApU was treated with DNase I to eliminate the presence of promoter DNA followed by heat inactivation of DNase I. Pyrophosphorolysis of Rif-ApU was performed in 10 μl of transcription buffer for 5 minutes at 37 °C in the presence of 500 μM sodium pyrophosphate (Sigma). The reaction products were resolved on a 20% (w/v) polyacrylamide denaturing gel and visualized using a PhosphorImager.
Fluorometric assays
Fluorescence measurements were performed using a QuantaMaster QM4 spectrofluorometer (PTI) in transcription buffer [40 mM Tris–HCl (pH 8.0), 100 mM NaCl, 5% glycerol, 1 mM DTT and 10 mM MgCl2] containing 0.02% Tween 20 at 30°C. Final assay mixtures (800 μl) contained 1 nM labeled RNAP holoenzyme reconstituted with the σ70 derivative labeled at position 211 with 5-tetramethylrhodamine (RNAP beacon) and DNA probes at various concentrations. The fluorescence intensities were recorded with an excitation wavelength of 550nm and an emission wavelength of 578 nm. Time-dependent fluorescence changes were monitored after manual-mixing of RNAP beacon (800 μl) and a DNA probe (~20 μl) in a cuvette; the mixing dead-time was 15 s. The experimental variation of relative signal amplitude changes among replicate measurements usually did not exceed 10% of the average value.
Supplementary Material
Highlights.
The role of downstream promoter interactions in transcription initiation is unclear.
Downstream promoter interactions can be coupled with formation of open complex.
DksA and ppGpp weaken downstream promoter interactions.
Weakening of downstream promoter interactions can inhibit open complex formation.
Acknowledgements
This work was supported by the National Institutes of Health (R01 GM59295) and by Molecular and Cell Biology Program grant from the Russian Academy of Sciences Presidium (to KS).
Abbreviations used
- RNAP
RNA polymerase
- RPo
open promoter complex
- Kd
dissociation constant
- Rif
rifampicin
- ds
double-stranded
- ss
single-stranded
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 2.Saecker RM, Record MT, Jr, Dehaseth PL. Mechanism of bacterial transcription initiation: RNA polymerase-promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 2011;412:754–771. doi: 10.1016/j.jmb.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marr MT, Roberts JW. Promoter recognition as measured by binding of polymerase to nontemplate strand oligonucleotide. Science. 1997;276:1258–1260. doi: 10.1126/science.276.5316.1258. [DOI] [PubMed] [Google Scholar]
- 4.Haugen SP, Berkmen MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;16:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 5.Mekler V, Pavlova O, Severinov K. The interaction of Escherichia coli RNA polymerase σ70 subunit with promoter elements in the context of free σ70, RNA polymerase holoenzyme and the β’-σ70 complex. J. Biol. Chem. 2011;286:270–279. doi: 10.1074/jbc.M110.174102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo J, Gralla JD. Promoter opening via a DNA fork junction binding activity. Proc. Natl Acad. Sci. USA. 1998;95:11655–11660. doi: 10.1073/pnas.95.20.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mekler V, Severinov K. Cooperativity and interaction energy threshold effects in recognition of the −10 promoter element by bacterial RNA polymerase. Nucleic Acids Res. 2013;41:7276–7285. doi: 10.1093/nar/gkt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saecker RM, Tsodikov OV, McQuade KL, Schlax PE, Jr, Capp MW, Record MT., Jr. Kinetic studies and structural models of the association of E. coli σ70 RNA polymerase with the lambdaP(R) promoter: large scale conformational changes in forming the kinetically significant intermediates. J. Mol. Biol. 2002;319:649–671. doi: 10.1016/S0022-2836(02)00293-0. [DOI] [PubMed] [Google Scholar]
- 9.Mekler V, Minakhin L, Severinov K. A critical role of downstream RNA polymerase-promoter interactions in the formation of initiation complex. J. Biol. Chem. 2011;286:22600–22608. doi: 10.1074/jbc.M111.247080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 11.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J. Biol. Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 12.Mekler V, Minakhin L, Sheppard C, Wigneshweraraj S, Severinov K. Molecular mechanism of transcription inhibition by phage T7 gp2 protein. J. Mol. Biol. 2011;413:1016–1027. doi: 10.1016/j.jmb.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camara B, Liu M, Reynolds J, Shadrin A, Liu B, Kwok K, Simpson P, Weinzierl R, Severinov K, Cota E, Matthews S, Wigneshweraraj SR. T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2247–2252. doi: 10.1073/pnas.0907908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, Irschik H, Arnold E, Ebright RH. The RNA polymerase “switch region” is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babu MM, Teichmann SA. Functional determinants of transcription factors in Escherichia coli: protein families and binding sites. Trends Genet. 2003;19:75–79. doi: 10.1016/S0168-9525(02)00039-2. [DOI] [PubMed] [Google Scholar]
- 16.Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 2009;23:236–248. doi: 10.1101/gad.1745409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 18.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Feng Y, Chatterjee S, Tuske S, Ho MX, Arnold E, Ebright RH. Structural basis of transcription initiation. Science. 2012;338:1076–1080. doi: 10.1126/science.1227786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc. Natl. Acad. Sci. USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landick R. RNA polymerase clamps down. Cell. 2001;105:567–570. doi: 10.1016/s0092-8674(01)00381-6. [DOI] [PubMed] [Google Scholar]
- 22.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 23.Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- 24.Chakraborty A, Wang D, Ebright YW, Korlann Y, Kortkhonjia E, Kim T, Chowdhury S, Wigneshweraraj S, Irschik H, Jansen R, Nixon BT, Knight J, Weiss S, Ebright RH. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weixlbaumer A, Leon K, Landick R, Darst SA. Structural basis of transcriptional pausing in bacteria. Cell. 2013;152:431–441. doi: 10.1016/j.cell.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mekler V, Minakhin L, Kuznedelov K, Mukhamedyarov D, Severinov K. RNA polymerase-promoter interactions determining different stability of the Escherichia coli and Thermus aquaticus transcription initiation complexes. Nucleic Acids Res. 2012;40:11352–11362. doi: 10.1093/nar/gks973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mekler V, Kortkhonjia E, Mukhopadhyay J, Knight J, Revyakin A, Kapanidis AN, Niu W, Ebright YW, Levy R, Ebright RH. Structural organization of bacterial RNA polymerase holoenzyme and the RNA polymerase-promoter open complex. Cell. 2002;108:599–614. doi: 10.1016/s0092-8674(02)00667-0. [DOI] [PubMed] [Google Scholar]
- 28.Hook-Barnard IG, Hinton DM. The promoter spacer influences transcription initiation via sigma70 region 1.1 of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA. 2009;106:737–742. doi: 10.1073/pnas.0808133106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- 30.James E, Liu M, Sheppard C, Mekler V, Camara B, Liu B, Simpson P, Cota E, Severinov K, Matthews S, Wigneshweraraj S. Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2. Mol. Cell. 2012;47:755–766. doi: 10.1016/j.molcel.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bae B, Davis E, Brown D, Campbell E, Wigneshweraraj S, Darst S. Phage T7 Gp2 inhibition of Escherichia coli RNA polymerase involves misappropriation of sigma70 domain 1.1. Proc. Natl. Acad. Sci. USA. 2013;110:19772–19777. doi: 10.1073/pnas.1314576110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borukhov S, Nudler E. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008;16:126–134. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 33.Mustaev A, Zaychikov E, Severinov K, Kashlev M, Polyakov A, Nikiforov V, Goldfarb A. Topology of the RNA polymerase active center probed by chimeric rifampicin-nucleotide compounds. Proc. Natl. Acad. Sci. U.S.A. 1994;91:12036–12040. doi: 10.1073/pnas.91.25.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pemberton IK, Muskhelishvili G, Travers AA, Buckle M. The G+C-rich discriminator of the tyrT promoter antagonises the formation of stable preinitiation complexes. J. Mol. Biol. 2000;299:859–864. doi: 10.1006/jmbi.2000.3780. [DOI] [PubMed] [Google Scholar]
- 35.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev.Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel— structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 37.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol. Cell. 2013;50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol. Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lennon CW, Ross W, Martin-Tumasz S, Toulokhonov I, Vrentas CE, Rutherford ST, Lee JH, Butcher SE, Gourse RL. Direct interactions between the coiled-coil tip of DksA and the trigger loop of RNA polymerase mediate transcriptional regulation. Genes Dev. 2012;26:2634–2646. doi: 10.1101/gad.204693.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 41.Lee J, Lennon C, Ross W, Gourse R. Role of the coiled-coil tip of Escherichia coli DksA in promoter control. J. Mol. Biol. 2012;416:503–517. doi: 10.1016/j.jmb.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brodolin K. Antibiotics trapping transcription initiation intermediates: To melt or to bend, what’s first? Transcription. 2011;2:60–65. doi: 10.4161/trns.2.2.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami KS. The X-ray Crystal Structure of Escherichia Coli RNA Polymerase s70 Holoenzyme. J. Biol. Chem. 2013;288:9126–9134. doi: 10.1074/jbc.M112.430900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pupov D, Miropolskaya N, Sevostyanova A, Bass I, Artsimovitch I, Kulbachinskiy A. Multiple roles of the RNA polymerase b’ SW2 region in transcription initiation, promoter escape, and RNA elongation. Nucleic Acids Res. 2010;38:5784–5796. doi: 10.1093/nar/gkq355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiesler SC, Burrows PC, Buck M. A dual switch controls bacterial enhancer-dependent transcription. Nucleic Acids Res. 2012;40:10878–10892. doi: 10.1093/nar/gks844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Tullius TD, Levin JR. Effects of discontinuities in the DNA template on abortive initiation and promoter escape by Escherichia coli RNA polymerase. J. Biol. Chem. 2007;282:26917–26927. doi: 10.1074/jbc.M702473200. [DOI] [PubMed] [Google Scholar]
- 47.Schroder O, Geiduschek EP, Kassavetis GA. A single-stranded promoter for RNA polymerase III. Proc. Natl. Acad. Sci. U.S.A. 2003;100:934–939. doi: 10.1073/pnas.242735699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Furman R, Tsodikov OV, Wolf YI, Artsimovitch I. An insertion in the catalytic trigger loop gates the secondary channel of RNA polymerase. J. Mol. Biol. 2013;425:82–93. doi: 10.1016/j.jmb.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 50.Drennan A, Kraemer M, Capp M, Gries T, Ruff E, Sheppard C, Wigneshweraraj S, Artsimovitch I, Record MT. Key roles of the downstream mobile jaw of Escherichia coli RNA polymerase in transcription initiation. Biochemistry. 2012;51:9447–9459. doi: 10.1021/bi301260u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kashlev M, Nudler E, Severinov K, Borukhov S, Komissarova N, Goldfarb A. Histidine-tagged RNA polymerase of Escherichia coli and transcription in solid phase. Methods Enzymol. 1996;274:326–334. doi: 10.1016/s0076-6879(96)74028-4. [DOI] [PubMed] [Google Scholar]
- 52.Mukhopadhyay J, Mekler V, Kortkhonjia E, Kapanidis AN, Ebright YW, Ebright RH. Fluorescence resonance energy transfer (FRET) in analysis of transcription-complex structure and function. Methods Enzymol. 2003;371:144–159. doi: 10.1016/S0076-6879(03)71010-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.