Abstract
Background
Although maternal atopy is a risk factor for development of peanut allergy, this phenomenon has not been well-characterized experimentally, and the mechanisms underlying offspring risk are unclear.
Objective
To determine if offspring of peanut allergic mothers (O-PAM) are more susceptible to peanut allergy than offspring of naive mothers (O-NM) in a murine model; and if so whether the susceptibility is linked to Th2 biased epigenetic alterations.
Methods
Five-week-old O-PAM and O-NM were intragastrically (i.g.) sensitized to and challenged with peanut. Serum peanut-specific IgE, plasma histamine, anaphylactic reactions, and splenocyte (SPC) and mesenteric lymph node (MLN) cell cytokine production were measured. DNA methylation levels of the IL-4 gene promoter from sensitized offspring SPC and MLN cells and un-sensitized neonatal offspring SPCs were determined by pyrosequencing.
Results
O-PAM exhibited 3-fold higher peanut-specific IgE levels following peanut sensitization as well as 5-fold higher histamine levels and significantly higher anaphylactic symptom scores following challenge than O-NM (p<0.05-0.01). Cultured O-PAM SPCs and MLN produced significantly more Th2 cytokines than O-NM cells (p<0.05-0.01). O-PAM cells exhibited significantly reduced DNA methylation at CpG sites of the IL-4 gene promoter than O-NM cells. DNA methylation levels were inversely correlated with IL-4 and IgE production. O-PAM neonatal splenocyte hypomethylation of the IL-4 gene promoter was also present.
Conclusion
This study is the first to demonstrate that increased susceptibility to peanut allergy in offspring of peanut allergic mothers is associated with epigenetic alternation of the IL-4 gene promoter. This finding may provide an insight into preventing the development of early life allergy.
Clinical Implications
This study is the first to demonstrate that offspring of peanut allergic mothers are predisposed to develop peanut allergy, which is associated with epigenetic alternation of the IL-4 gene promoter. This suggests that an early life intervention that modifies the Th2 biased epigenetic status may reduce the incidence of allergy in high-risk offspring.
Keywords: Peanut allergy, anaphylaxis mice, offspring, IgE, IL-4, DNA methylation
Introduction
Food allergy, a growing public health concern in the U.S. affects up to 8% of children and 4% of adults, and is a major cause of anaphylaxis.(1-3) Among these food allergies, peanut allergy has attracted great public attention because of its prevalence, severity of reactions, and frequent life-long persistence.(4-7) In 1996, Hourihane et al reported that prevalence of peanut and other allergies in the families of people with peanut allergy was increased in successive generations in maternal but not paternal relatives.(8) Additional clinical observational studies also show that maternal peanut allergy and other allergies increase the risk of a child to develop peanut allergy.(9-11) The reason for this is not known, and although allergy associated genes have been identified, none are strongly associated with peanut or other food allergy.(12)
Peanut allergy, like other allergies, is a Th2-mediated immune disorder characterized by increased expression of IL-4, IL-5, and IL-13 in humans (13) and animal models.(14-16) IL-4 is a key Th2 cytokine required for B cells switching to IgE production, mast cell activation and Th2 cell differentiation.(17) Higher IL-4/IFN-γ ratios were found in persistent peanut allergic patients than in patients who naturally outgrew peanut allergy.(18)
Increasing evidence shows that epigenetic mechanisms are involved in regulating T cell differentiation, cytokine expression, allergic sensitization and allergic asthma development.(19-21) CpG DNA methylation is a well known epigenetic modification that affects chromatin remodeling and can drive cytokine expression in the absence of alterations in DNA sequences.(22) DNA hypomethylation activates cytokine expression, whereas hypermethylation represses cytokine expression. (23-25) DNA methylation status is dynamic and is influenced by both external and internal factors. A study investigating the effect of inhalation of diesel exhaust particles and allergen on DNA demethylation status at several CpG sites of the IL-4 promoter showed that the level of DNA CpG-408 demethylation was inversely correlated with increased IgE production in a murine model of asthma.(26) Although it has been suggested that association of maternal atopy with increased offspring susceptibility to food allergy is linked to epigenetic changes,(27;28) direct experimental evidence of such an association is lacking.
We hypothesized that maternal peanut allergy would increase offspring propensity for peanut specific-IgE production, hypersensitivity reactions, Th2 cytokine production, and Th2 cytokine gene promoter hypomethylation. To test this possibility, we used a mouse model to compare IgE levels and anaphylactic reactions following suboptimal active peanut sensitization and challenge in offspring of peanut allergic mothers (O-PAM) to offspring of naive mothers (O-NM). We also determined cytokine production and methylation status of the Th2 signature cytokine IL-4 gene promoter at CpG sites in DNA from offspring mesenteric lymph node cells (MLN), and splenocytes (SPCs).
Methods
Mice and reagents
Six-week (wk) old female and male C3H/HeJ mice purchased from the Jackson Laboratory (Bar Harbor, ME) were maintained on peanut free chow under specific pathogen-free conditions according to standard guidelines for the care and use of animals.(29) Freshly ground, whole roasted peanut and crude peanut extract (CPE) was prepared as described previously (Online Repository Method).(30-32) Sources of reagents used in this study are provided in the Online Repository Methods and Reagents.
Experimental protocols
Maternal protocol
To establish maternal peanut allergy, female C3H/HeJ mice, on a peanut free diet, were orally sensitized with peanut as previously described(15;16) with slight modification. Briefly, 6 week-old female mice were sensitized with peanut (10 mg PN) and cholera toxin (20 μg CT) intragastrically (i.g.) weekly for 5 weeks (wks) and challenged at wk 6 (200 mg PN/mouse). Our sensitization protocol differed from our previous standard protocol in that mice did not receive an additional boost (50 mg PN+10 mg CT). Mice treated with the protocol used in our current study are termed peanut allergic mothers (PAM). As controls, naïve females were PBS (Phosphate Buffered Saline) sham sensitized and challenged; these mice are termed naïve mothers (NM). One-week after peanut challenge, mice in both groups were bred with naive males. All mice were fed peanut-free chow during gestation and lactation.
Determination of peanut protein in milk
During lactation, milk was collected from PAM or NM when their offspring were 10-15 days old with a mouse milking machine modified in our laboratory as described.(33) To establish a positive control, an additional group of PAM lactating mice was administered 10mg of peanut protein intragastrically. Milk was collected 2 hours later. The levels of peanut protein in milk from these three groups were detected using a Veratox® Peanut commercial kit (Neogen Corp, Lansing, Mich) following the manufacturer's instruction and as previously described.(34)
Offspring protocol
Offspring of peanut allergic mothers (O-PAM) and offspring of naive mothers (O-NM), weaned at 4 wks of age, were sensitized one week later by 3 weekly i.g. suboptimal doses of peanut (5mg) and CT (10μg). A third group of age-matched peanut-naive offspring of peanut-free mothers (Naive) served as normal controls (Figure 1). All offspring were orally challenged at week 4 with 200 mg of ground peanut.
Figure 1. Experimental protocols.
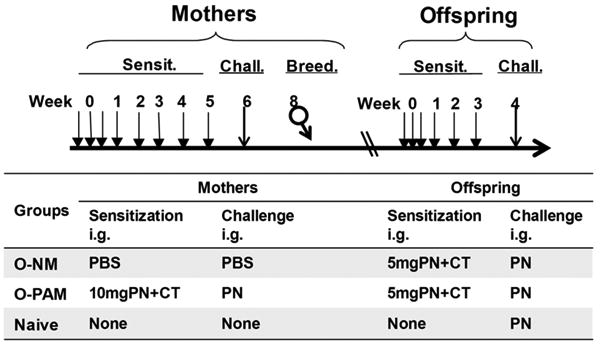
Peanut-sensitized female mice were orally challenged with peanut, and then mated with naïve males one week later. Five-wk-old offspring of peanut allergic mothers (O-PAM) and naïve mothers (O-NM) were sensitized weekly for 3 wks and challenged at week 4. Offspring of PBS challenged naïve mothers served as normal controls. Sensit: Sensitization; Chall: Challenge; Breed: Breeding.
Measurement of peanut-specific antibodies
To monitor antibody responses to peanut sensitization, sera were obtained from blood collected weekly during sensitization and one day prior to challenge by submandibular venipuncture. To measure serum peanut-specific IgE, serum IgG antibodies were first depleted by protein G–Sepharose (BioVisio, Milpitas, CA) centrifugation.(35;36) In brief, equal volumes of protein G–Sepharose and undiluted mouse serum were mixed and incubated at room temperature for 10 minutes, then centrifuged at 1200 rpm for 10 minutes at room temperature. Supernatants were collected and the process was repeated twice. Peanut-specific IgE levels in protein G–depleted sera were measured using previously described ELISA methods.(37;38) Serum peanut-specific IgG1 and IgG2a were determined as previously described.(37;38)
Assessment of hypersensitivity reactions
Anaphylactic symptoms were evaluated 30 minutes following oral challenge utilizing the scoring system described previously (Online Repository Methods).(15) In this scoring system, 0 is no reaction, 1 is mild, 2 is moderate, 3 is severe, 4 very severe, 5 is death.(39) Core body temperatures were measured using a rectal probe (Harvard Apparatus, NJ). To confirm that anaphylactic reactions were not IgG mediated, peanut sensitized mice were pretreated with anti-FcγRIIB/FcγRIII mAb (mAb 2.4G2, 500μg/mouse) or isotype control antibody 24 hours prior to challenge as previously described.(40;41)
Histamine measurement
Histamine levels were measured using an enzyme immunoassay kit (ImmunoTECH Inc., Marseille, France) as described by the manufacturer.
Cell culture and cytokine measurements
Offspring SPCs and MLNs were prepared as previously described,(33) and cultured in 24-well plates (4 × 106/well/ml) in the presence or absence of CPE (200 μg/ml) or Con A (2.5 μg/mi). 72 hrs later, supernatants were collected and cytokine levels were determined by ELISA. SPCs and MLN cells were then collected and preserved in Buffer RTL (Qiagen, MD) for DNA methylation assays.
DNA pyrosequencing methylation analysis
Genomic DNA from offspring MLN cells and SPCs was isolated using an AllPrep mini DNA kit (Qiagen, MD). DNA from the spleens of approximately 1 week old offspring mice (unsensitized) and DNA from maternal peripheral blood leucocytes (PBL) post challenge were extracted using DNeasy Tissue/Blood kit (Qiagen, MD) as previously described.(42) Isolated DNA was then bisulfite converted using an Epitect plus DNA Bisulfite kit (Qiagen, MD) as per manufacturer's instructions. Bisulfite converted DNA was used to amplify promoter regions of the IL-4 gene by PCR, using the primers in Table I. PCR products were subjected to pyrosequencing using the Pyromark Q24 Pyrosequencing system (Qiagen, MD) using the sequencing primers for CpG-408 and CpG-393 in Table I. Bisulfite converted DNA was also used to amplify promoter regions of the IFN-γ and Foxp3 genes by PCR and PCR products were then subjected to pyrosequencing using sequencing primers. Primers, which are listed in Table EI, were chosen based on previous studies.(26;42) Methylation levels of PCR products at each CpG site were determined using PyroMark 24 run software (Qiagen,MD)
Table I. Primers used for PCR amplification and pyrosequencing experiments.
| IL-4 | |
|---|---|
| PCR forward | GTTTTTAAGGGGTTTTTATAGTAGGAAGT |
| PCR reverse | Biotin-AATTACCACTAACTCTCCTCCTACA |
| Pyrosequencing | AGATTTTTTTGATATTATTTTGTTT |
Statistical analysis
Data were analyzed using the SigmaStat statistical software package (Systat, Chicago, IL). Differences between two groups were analyzed by t test if the data were approximately normal or by Mann-Whitney Rank Sum Test if the data were skewed. An ANOVA followed by a Bonferroni t test was performed for all pair-wise comparisons if the data were approximately normal. Differences among more than two groups were analyzed by ANOVA on ranks followed by all pair-wise comparisons (Dunn's method) if the data were not normally distributed. Graphpad Prism 4 was used for non-parametric (Spearman) correlation analysis. P values less than 0.05, based on 2-tailed tests, were considered statistically significant.
Results
Offspring of peanut allergic mothers produce increased peanut-specific IgE following oral peanut sensitization
PAM mothers had increased peanut specific IgE levels following i.g. sensitization and exhibited hypersensitivity reactions following i.g. challenge before breeding (Figure E1 in the Online Repository).
O-PAM and O-NM were sensitized using a suboptimal protocol consisting of 3 weekly 5 mg PN+10 μg CT administrations (half the dose and less than half the duration of our standard sensitization protocol).(15) Based on our preliminary results, sub-optimal offspring sensitization more clearly distinguishes between O-PAM and O-NM sensitivity to allergy. Four weeks after the initial sensitization, O-PAM sera contained approximately 300% more peanut specific-IgE than O-NM sera (4061.2± 2739.6 vs.1280.9±594.3 ng/ml. p<0.01, N=8-11/group, Fig 2A). Sera from normal control mice contained no detectable peanut-specific IgE. Peanut-specific IgG1 and IgG2a levels in sensitized O-PAM and O-NM mice were not significantly different, although they tended to be higher in O-NM than O-PAM (N=8-11/group, Fig 2B&C). Thus, O-PAM produce more IgE than O-NM in response to the same sensitization protocol.
Figure 2. Offspring of peanut allergic mothers have an increased IgE response to peanut sensitization.
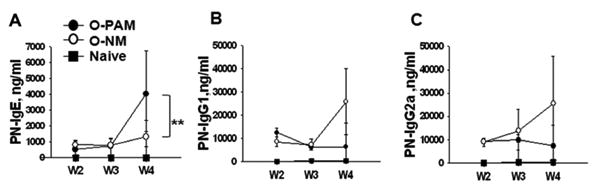
Blood was collected from offspring following peanut sensitization at indicated time-points, and serum peanut-specific IgE levels (A), peanut-specific IgG1 levels (B) and peanut-specific IgG2a levels (C) were determined by ELISA. **p<0.01, vs. O-NM (N=8-11/group). W: week in sensitization protocol.
Offspring of peanut allergic mothers develop more severe anaphylaxis following oral peanut challenge
Only 2 of 11 (18%) peanut challenged O-NM mice developed severe reactions (score 3) and none exhibited very severe or fatal reactions. In contrast, 6 of 8 (75%) O-PAM exhibited severe reactions, including one fatal and one near-fatal reaction. Mean anaphylactic symptom scores of O-PAM were significantly higher than O-NM (O-PAM 3.0±1.2 vs. O-NM 1.8±0.9, p<0.05, N=8-11/group, Fig 3A). Naive control mice showed no sign of anaphylaxis following challenge. O-PAM core body temperatures were significantly below those of O-NM (mean: 32.2 ±3.9° C vs. 36.4 ±1.3° C; p< 0.01; N=8-11/group, Fig 3B) and normal control mice (37.9±0.2 °C, p<0.01, N=8-11/group). Consistent with the physiological changes, O-PAM plasma histamine levels averaged 420% higher than those of O-NM (66175.2±20072.6 vs. 15724.8±9271.1 nM, p< 0.05, N=8-11/group, Fig 3C). In addition, suppression of IgG-mediated anaphylaxis by administration of anti-FcgRIIB/FcgRIII monoclonal antibody (2.4G2) 24 hours prior to challenge in O-PAM had no effect on challenge-induced anaphylactic reactions or decreased core body temperatures (Figure E2 in the Online Repository).
Figure 3. Offspring of peanut allergic mothers develop anaphylaxis following peanut oral challenge.
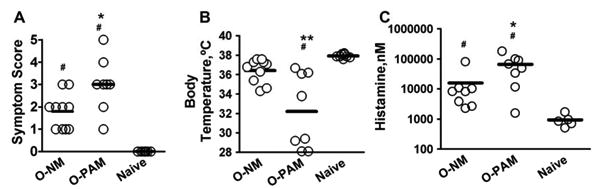
Thirty minutes following challenge, O-NM and O-PAM anaphylactic symptoms were scored (A), core body temperatures were measured (B), and plasma histamine levels were determined (C). Histamine levels are presented in log scale. *p<0.05,**p<0.01, vs. O-NM; # p< 0.05, vs. normal control (N=8-11/group). Each dot represents an individual mouse. Horizontal bar indicates the mean.
Splenocytes and mesenteric lymph node cells from offspring of peanut allergic mothers secrete more IL-4
We compared production of the signature Th2 cytokine, IL-4, by SPCs and MLN cells isolated from peanut-sensitized and challenged O-PAM, O-NM and naïve control mice following in vitro peanut protein stimulation. Peanut stimulated SPCs from O-PAM produced significantly more IL-4 than O-NM SPCs (p<0.05, N=8-11/group, Fig 4A). IL-10 and IFN-γ production did not differ. IL-4 and IL-5 levels were also significantly higher in O-PAM MLN cell cultures than in O-NM cultures following peanut stimulation (p<0.05, N=8-11/group, Fig 4B). O-PAM MLN cells also produced significantly less IL-10 and IFN-γ than O-NM MLN cells (p<0.05, N=8-11/group, Fig 4B). Cytokine production by peanut-stimulated SPCs and MLNs from normal control offspring was negligible, and cytokines were not detected in medium alone cultures (data not shown).
Figure 4. Cytokine secretion by offspring SPC and MLN cultures.
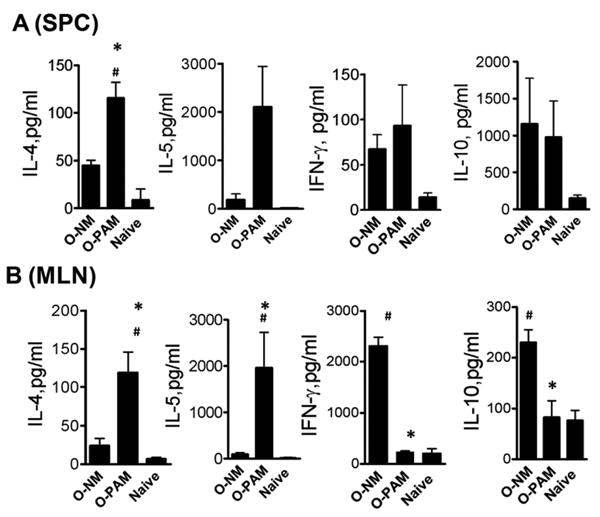
SPC and MLN cells were prepared and cultured for 72 hours in the presence or absence of crude peanut extract. SPC (A), and MLN (B). Culture supernatant IL-4, IFN-g and IL-10 levels were determined. Data are expressed as means ± SDs of each group. *, p <0.05 vs. O-NM; #, p< 0.05 vs. normal control (N=8-11/group).
IL-4 gene promoter DNA methylation was reduced at CpG sites in spleen and mesenteric lymph node cells in offspring of peanut allergic mothers
To determine the differences in DNA methylation, we analyzed both global methylation using a global DNA methylation ELISA kit and site-specific methylation by pyrosequencing. In the global DNA methylation ELISA kit, the methylated fraction of DNA is recognized by anti-5-methylcytosine antibody and quantified through an ELISA-like reaction. We found no significant differences in global DNA methylation of MLN cells between O-PAM, O-NM and normal controls following challenge (Figure E3A in the Online Repository). We next determined the DNA methylation status of the IL-4, Foxp3 and IFN-γ genes promoters in DNA from SPCs and MLNs of O-PAM, O-NM and normal controls. The percent of IL-4 promoter methylation at CpG-408 and CpG-393 in MLN cells and SPCs from O-PAM was significantly less than in DNA from O-NM and normal controls (Fig 5A and B for SPCs, C and D for MLN, all p < 0.05 or p<0.01, N=5-8/group). Thus, O-PAM SPC and MLN DNA have a hypomethylated IL-4 promoter profile. In addition, peripheral blood leukocytes (PBL) from PNA mothers also showed significantly reduced methylation at CpG-408 and CpG-393 sites of the IL-4 promoter as compared to PBL from native mothers (Figure E4 in the Online Repository). However, we did not observe a significant difference in methylation levels at the IFN- γ and Foxp3 gene promoters in MLN cells from O-PAM and O-NM (Figure E3B&C in the Online Repository), indicating that additional factors, such as histone modifications,(43) may contribute to expression of these proteins.
Figure 5. Methylation at the CpG-408 and CpG-393 sites of the IL-4 promoter.
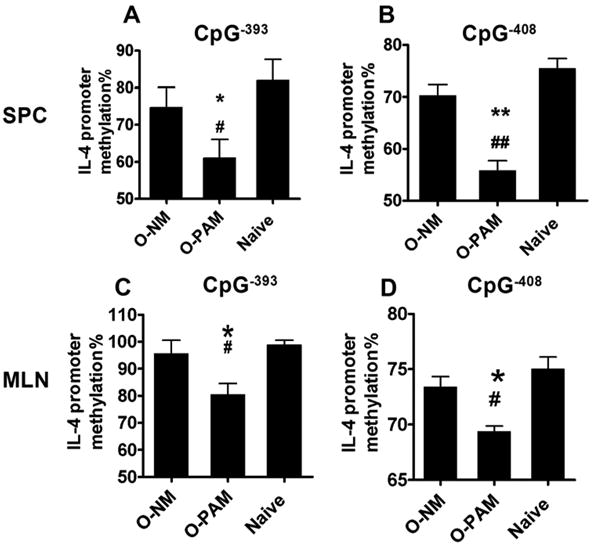
Purified DNA from sensitized and challenged offspring SPCs and MLN cells underwent bisulfite treatment, PCR amplification, and pyrosequencing. Percent of DNA methylation at CpG-408 and CpG-393 sites of the IL-4 promoters in offspring SPCs (A&B) and MLN cells (C&D). Data are expressed as means ± SDs of each group.*p < 0.05; **p < 0.01 vs.O-NM; # p < 0.05; ## p < 0.01 vs. normal control (N=5-8/group).
Offspring spleen and mesenteric lymph node cell DNA methylation levels at CpG-408 of the IL-4 gene promoter inversely correlate with IL-4 and IgE production
Correlation analysis revealed that the percent of methylation at CpG -408 but not CpG -393 of the IL-4 promoter in DNA in SPCs and MLN cells was inversely correlated with in vitro IL-4 production and serum peanut specific IgE levels (p<0.05, N=10-12, Fig 6 A-D).
Figure 6. Correlation between percent DNA methylation of the IL-4 promoter, IL-4 production and peanut-sensitized offspring serum IgE levels (O-PAM+O-NM).
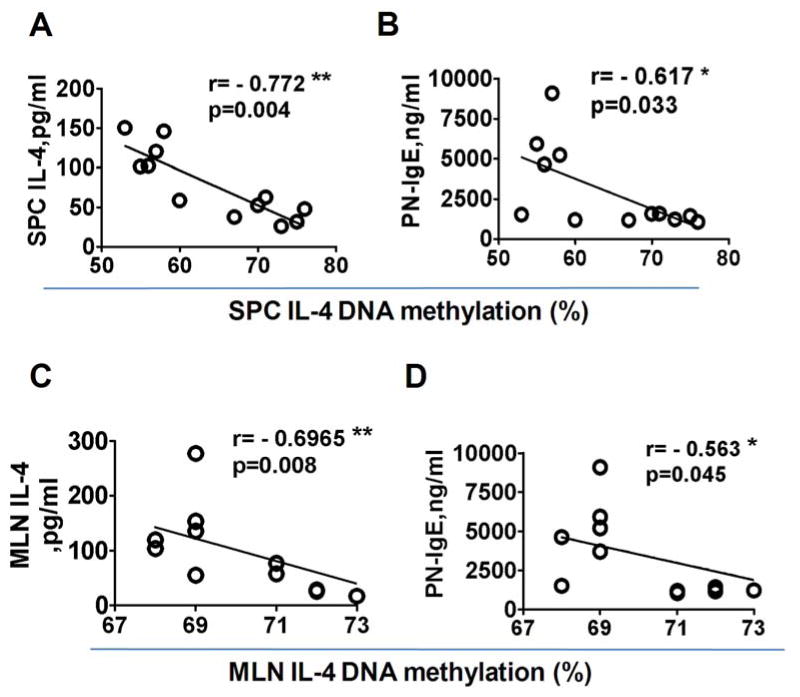
Percent of offspring SPC and MLN DNA methylation at the CpG-408 site of the IL-4 promoter was plotted versus SPC and MLN IL-4 and serum IgE (A-D). Non-parametric (Spearman) correlation was used. *p< 0.05; **, p<0.01(N=10-12). Each dot represents an individual mouse.
Splenocytes from un-sensitized neonatal offspring of peanut allergic mothers produce more ConA stimulated IL-4 and are hypomethylated at CpG sites of the IL-4 gene promoter
We quantitated IL-4 secretion by from neonatal splenocytes from O-PAM and O-NM that had not been immunized with PN and CT. SPCs from both groups did not respond to CPE stimulation (data not shown). However, Con A stimulated IL-4 secretion was higher in O-PAM than in O-NM (Fig 7A, p<0.001, N=5-8/group). In addition, splenocyte DNA from unsensitized neonatal O-PAM was less methylated at the CpG-408 and CpG-393 sites of the IL-4 promoter than splenocyte DNA from unsensitized neonatal O-NM (Fig 7 B&C, p<0.05, N=5-8/group).
Figure 7. Neonatal offspring SPC IL-4 cytokine levels and CpG methylation at CpG-408 and CpG-393 sites of the IL-4 promoter.
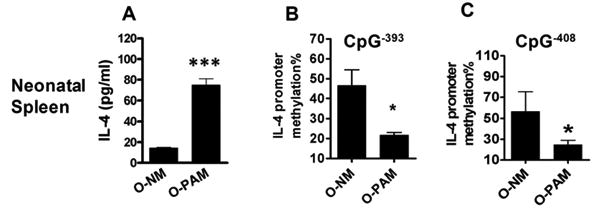
SPCs from unimmunized neonatal offspring were prepared and cultured for 72 hours in the presence of ConA. IL-4 levels in culture supernatants were determined by ELISA. DNA methylation of the IL-4 promoter from unimmunized neonatal SPCs was determined as described in Figure 5. *, p<0.05; ***, p<0.001(N=5-8/group). Data are expressed as means ± SDs of each group.
Discussion
Although epidemiologic studies show that maternal atopy is a significant risk factor for development of peanut allergy,(8;11) the present study provides the first experimental evidence that maternal peanut allergy increases offspring susceptibility to IgE-mediated peanut hypersensitivity. We demonstrated that a suboptimal oral sensitization regimen induces only a low level of IgE production and mild anaphylactic reactions in O-NM, but robust IgE production and severe reactions, including fatal/near fatal reactions in O-PAM. The increased severity of reactions was associated with markedly elevated post-challenge plasma histamine levels. It is well known that IgG as well as IgE induces anaphylactic reactions in mice.(44) In the present study, we demonstrated that the increased peanut hypersensitivity was not IgG-mediated, because peanut specific IgG1 and IgG2a levels in O-PAM and O-NM did not differ, and because FcγRIII blockade did not attenuate O-PAM anaphylactic reactions. This is consistent with previous observations that anaphylactic responses to ingested antigens in the mouse are nearly always IgE-mediated.(45)
Our observations with enteric sensitization of offspring are consistent with the results of a previous study, in which the offspring of peanut allergic mothers and naïve mothers were sensitized epicutaneously with egg white for 4 weeks. (46) It was demonstrated that egg white-specific IgE levels were significantly higher in O-PAM than O-NM, suggesting that maternal atopy is an important, allergen-independent risk factor for offspring IgE hyper-sensitization.
Our data also indicate that maternal peanut allergy did not increase offspring susceptibility to peanut allergy by maternal milk or amniotic fluid transmission of peanut allergen, because our mice were on a peanut free diet and because there was no detectable peanut in milk (Figure E5 in the Online Repository) or amniotic fluid (data not shown) from PAM, while peanut allergens were detectable in the milk of post-partum mice that were fed peanuts (Figure E5 in the Online Repository). The results of our published peanut-egg white experiments, which established that IgE hypersensitivity in O-PAM is not allergen-specific, further excludes a role for neonatal passive sensitization though maternal milk. Thus, alternative mechanisms must be responsible for the ability of maternal atopy to increase susceptibility of offspring to antigen sensitization.
Th2 cytokines play a central role in the pathogenesis of food allergy.(47) IL-4, a signature cytokine of Th2 biased immunity, promotes IgE production and the differentiation of naïve CD4+ T cells into Th2 cells.(48) In this study, PN-restimulated O-PAM MLN cells and SPCs produced significantly more IL-4 than O-NM cells, demonstrating the presence of Th2 biased immunity in O-PAM. In addition, IL-10 and IFN-γ production by MLN cells, but not SPCs from O-PAM was impaired. This might be because the gut mucosal immune response was more affected than the peripheral immune response in these food allergic mice. However, a causative role for IL-4 in impaired IL-10 and IFN-γ production in this model requires further investigation. In addition, we found that increased IL-4 levels from SPCs in response to ConA stimulation were present in O-PAM that had not themselves been immunized with PN + CT, indicating that some inherited epigenetic changes may promote IL-4 transcription and atopy development, regardless of the allergen involved.
Epigenetic mechanisms have been suggested to explain why maternal allergy increases the risk of allergy development by offspring.(28) However, our data provide the initial experimental evidence that maternal peanut allergy induces Th2-promoting epigenetic alterations in offspring. Altered DNA methylation status at CpG sites in the gene regulatory region is an important marker of epigenetic regulation of cytokine expression. Reduced DNA methylation at the IL-4 promoter increases IL-4 expression by increasing the accessibility of transcription factors to the regulatory region.(24;25) In an asthma model, Liu et al(26) investigated CpG sites (CpG-408, CpG-393, CpG-339, and CpG-314 in the IL-4 proximal promoter region, and CpG+101, CpG+113, CpG+185 CpG+225, and CpG+229 in the first intron) relative to the first start codon of the IL-4 gene. In that study, antigen plus diesel exposure induced hypomethylation at CpG-408 of the IL-4 promoter and the degree of methylation at CpG-408 correlated inversely with IgE levels. In the present study, maternal peanut allergy was associated with hypomethylation at CpG-408 and CpG-393 of the IL-4 promoter in SPCs and MLNs in O-PAM following antigen sensitization and challenge, as compared to O-NM and normal controls. The same pattern of hypomethylation of the IL-4 promoter was also present in one-week-old neonatal mice in the O-PAM group prior to sensitization. Methylation at CpG residue (-408), but not CpG-393, correlated inversely with IL-4 and IgE production. These data are the first evidence that maternal peanut allergy induces IL-4 gene epigenetic changes, and that this may play a key role in offspring predisposition to develop peanut allergy. The data also support previous evidence that CpG-408 is a functionally important CpG residue at the IL-4 promoter,(26) due at least partially to its location in the proximal promoter region of the IL-4 gene.(26;42)
In conclusion, we demonstrate that offspring of peanut allergic mothers are more susceptible to development of peanut allergy than offspring of non-peanut allergic mothers. Maternal peanut allergy induces epigenetic alternation of the IL-4 promoter in their offspring, which correlates with a Th2 biased responses (IL-4 and IgE production). These findings may provide insights into developing a preventive intervention by modifying early life epigenetic status.
Supplementary Material
Figure E1: Peanut allergic mothers exhibited hypersensitivity reactions following oral peanut challenge and prior to mating. Blood was collected from peanut allergic mothers (PAM) and naïve mothers (NM) one day prior to challenge. Serum peanut-specific IgE levels (A) were measured by ELISA. Anaphylactic scores (B) and core body temperatures (C) were measured, and plasma histamine levels (D) were determined thirty minutes after challenge. **, p<0.01; ***, p<0.001, vs. NM (N=5/group). Each dot represents an individual mouse. Horizontal bars indicate means. Data shown are representative of two individual experiments.
Figure E2: Following oral challenge, anaphylactic scores and core body temperatures with or without anti-FγRIIB/RIII monoclonal antibody administration in peanut sensitized offspring of peanut allergic mothers. N=5-6. *, p<0.05.Horizontal lines represent the means of each group.
Figure E3: A: Global methylation of MLN cells in O-PAM, O-NM and normal controls following challenge was measured by the global DNA methylation ELISA kit(Epigentek, Farmingdale, NJ). In this ELISA kit, the methylated fraction of DNA is recognized by 5-methylcytosine antibody and quantified through an ELISA-like reaction. (N=6-7/group). Both p>0.05 for O-PAM vs. O-NM and O-PAM vs. Naïve. B:Purified DNA from sensitized and challenged offspring MLN cells underwent bisulfite treatment, PCR amplification, and pyrosequencing. Percent of DNA methylation of the IFN-γ and Foxp3 promoters in offspring MLN cells (B&C). Data are expressed as means ± SDs of each group. (N=5-6/group).
Figure E4: DNA methylation at CpG-408 and CpG-393 sites of the IL-4 promoter from peripheral blood leucocytes (PBL) of peanut allergic mothers (PAM) and naïve mothers (NM) prior to breeding. Purified DNA from mothers PBL prior to breeding underwent bisulfite treatment, PCR amplification, and pyrosequencing. **p < 0.01; ***p < 0.001 vs.NM (N=5).
Figure E5: Breast milk was collected from lactating peanut allergic mothers (PAM) and naïve mothers (NM) when their offspring were 10-15 day-old, using a mouse milking machine modified in our laboratory. An additional group of PAM lactating mice was inoculated intragastrically with10 mg of peanut protein (PAM-PN). Milk was collected 2 hours following peanut protein feeding. Milk was diluted 1:2 in PBS, and peanut protein levels were detected using a commercial kit (Neogen Corp, Lansing, Mich). N=3-4. ***, p<0.001.
Supplement Table I. Primers used for PCR amplification and pyrosequencing experiments
Acknowledgments
This work was supported by the Food Allergy Initiative and National Institute of Health grant no. AT001495-01A1 to X.M.L.
Abbreviations
- Con A
concanavalin A
- CT
cholera toxin
- FBS
fetal bovine serum
- i.g
intragastric gavage, or intragastrically
- MLNs
mesenteric lymph nodes
- SPCs
splenocytes
- PAM
peanut allergic mothers
- NM
naive mothers
- O-PAM
offspring of peanut allergic mothers
- O-NM
offspring of naive mother
- PA
peanut allergy
- PBS
Phosphate buffered saline
- Th
T helper cells
- wk (s)
week (s)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boyce JA, Assa'ad A, Burks AW, Jones SM, Sampson HA, Wood RA, et al. Guidelines for the diagnosis and management of food allergy in the United States: report of the NIAID-sponsored expert panel. J Allergy Clin Immunol. 2010;126(6 Suppl):S1–58. doi: 10.1016/j.jaci.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RS, Springston EE, Warrier MR, Smith B, Kumar R, Pongracic J, et al. The prevalence, severity, and distribution of childhood food allergy in the United States. Pediatrics. 2011;128(1):e9–17. doi: 10.1542/peds.2011-0204. [DOI] [PubMed] [Google Scholar]
- 3.Sampson HA. Anaphylaxis and emergency treatment. Pediatrics. 2003;111(6 Pt 3):1601–8. [PubMed] [Google Scholar]
- 4.Bock SA, Munoz-Furlong A, Sampson HA. Further fatalities caused by anaphylactic reactions to food, 2001-2006. J Allergy Clin Immunol. 2007;119(4):1016–8. doi: 10.1016/j.jaci.2006.12.622. [DOI] [PubMed] [Google Scholar]
- 5.Fleischer DM, Conover-Walker MK, Matsui EC, Wood RA. The natural history of tree nut allergy. J Allergy Clin Immunol. 2005;116(5):1087–93. doi: 10.1016/j.jaci.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Fleischer DM, Conover-Walker MK, Christie L, Burks AW, Wood RA. Peanut allergy: recurrence and its management. J Allergy Clin Immunol. 2004;114(5):1195–201. doi: 10.1016/j.jaci.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 7.Sicherer SH, Munoz-Furlong A, Godbold JH, Sampson HA. US prevalence of self-reported peanut, tree nut, and sesame allergy: 11-year follow-up. J Allergy Clin Immunol. 2010 doi: 10.1016/j.jaci.2010.03.029. [DOI] [PubMed] [Google Scholar]
- 8.Hourihane JO, Dean TP, Warner JO. Peanut allergy in relation to heredity, maternal diet, and other atopic diseases: results of a questionnaire survey, skin prick testing, and food challenges. BMJ. 1996;313(7056):518–21. doi: 10.1136/bmj.313.7056.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greer FR, Sicherer SH, Burks AW. Effects of early nutritional interventions on the development of atopic disease in infants and children: the role of maternal dietary restriction, breastfeeding, timing of introduction of complementary foods, and hydrolyzed formulas. Pediatrics. 2008;121(1):183–91. doi: 10.1542/peds.2007-3022. [DOI] [PubMed] [Google Scholar]
- 10.Lack G. Update on risk factors for food allergy. J Allergy Clin Immunol. 2012;129(5):1187–97. doi: 10.1016/j.jaci.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 11.Lack G, Fox D, Northstone K, Golding J. Factors associated with the development of peanut allergy in childhood. N Engl J Med. 2003;348(11):977–85. doi: 10.1056/NEJMoa013536. [DOI] [PubMed] [Google Scholar]
- 12.Bjorksten B. Genetic and environmental risk factors for the development of food allergy. Curr Opin Allergy Clin Immunol. 2005;5(3):249–53. doi: 10.1097/01.all.0000168790.82206.17. [DOI] [PubMed] [Google Scholar]
- 13.Beyer K, Castro R, Birnbaum A, Benkov K, Pittman N, Sampson HA. Human milk-specific mucosal lymphocytes of the gastrointestinal tract display a TH2 cytokine profile. J Allergy Clin Immunol. 2002;109(4):707–13. doi: 10.1067/mai.2002.122503. [DOI] [PubMed] [Google Scholar]
- 14.Morafo V, Srivastava K, Huang CK, Kleiner G, Lee SY, Sampson HA, et al. Genetic susceptibility to food allergy is linked to differential TH2-TH1 responses in C3H/HeJ and BALB/c mice. J Allergy Clin Immunol. 2003;111(5):1122–8. doi: 10.1067/mai.2003.1463. [DOI] [PubMed] [Google Scholar]
- 15.Qu C, Srivastava K, Ko J, Zhang TF, Sampson HA, Li XM. Induction of tolerance after establishment of peanut allergy by the food allergy herbal formula-2 is associated with up-regulation of interferon-gamma. Clin Exp Allergy. 2007;37(6):846–55. doi: 10.1111/j.1365-2222.2007.02718.x. [DOI] [PubMed] [Google Scholar]
- 16.Srivastava KD, Kattan JD, Zou ZM, Li JH, Zhang L, Wallenstein S, et al. The Chinese herbal medicine formula FAHF-2 completely blocks anaphylactic reactions in a murine model of peanut allergy. J Allergy Clin Immunol. 2005;115(1):171–8. doi: 10.1016/j.jaci.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Steinke JW, Borish L. Th2 cytokines and asthma. Interleukin-4: its role in the pathogenesis of asthma, and targeting it for asthma treatment with interleukin-4 receptor antagonists. Respir Res. 2001;2(2):66–70. doi: 10.1186/rr40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turcanu V, Maleki SJ, Lack G. Characterization of lymphocyte responses to peanuts in normal children, peanut-allergic children, and allergic children who acquired tolerance to peanuts. J Clin Invest. 2003;111(7):1065–72. doi: 10.1172/JCI16142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller RL, Ho SM. Environmental epigenetics and asthma: current concepts and call for studies. Am J Respir Crit Care Med. 2008;177(6):567–73. doi: 10.1164/rccm.200710-1511PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuriakose JS, Miller RL. Environmental epigenetics and allergic diseases: recent advances. Clin Exp Allergy. 2010 doi: 10.1111/j.1365-2222.2010.03599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovinsky-Desir S, Miller RL. Epigenetics, asthma, and allergic diseases: a review of the latest advancements. Curr Allergy Asthma Rep. 2012;12(3):211–20. doi: 10.1007/s11882-012-0257-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Shaw T, Martin P. Epigenetic reprogramming during wound healing: loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10(8):881–6. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9(2):91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 25.Janson PC, Winerdal ME, Winqvist O. At the crossroads of T helper lineage commitment-Epigenetics points the way. Biochim Biophys Acta. 2009;1790(9):906–19. doi: 10.1016/j.bbagen.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Ballaney M, Al-alem U, Quan C, Jin X, Perera F, et al. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci. 2008;102(1):76–81. doi: 10.1093/toxsci/kfm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prescott SL, Clifton V. Asthma and pregnancy: emerging evidence of epigenetic interactions in utero. Curr Opin Allergy Clin Immunol. 2009;9(5):417–26. doi: 10.1097/ACI.0b013e328330634f. [DOI] [PubMed] [Google Scholar]
- 28.Prescott S, Saffery R. The role of epigenetic dysregulation in the epidemic of allergic disease. Clin Epigenetics. 2011;2(2):223–32. doi: 10.1007/s13148-011-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Institute of Laboratory Animal Resources Commission of Life Sciences NRC. Guide for the Care and Use of Laboratory Animals. National Academy Press; 1996. [Google Scholar]
- 30.Beyer K, Morrow E, Li XM, Bardina L, Bannon GA, Burks AW, et al. Effects of cooking methods on peanut allergenicity. J Allergy Clin Immunol. 2001;107(6):1077–81. doi: 10.1067/mai.2001.115480. [DOI] [PubMed] [Google Scholar]
- 31.Srivastava KD, Qu C, Zhang T, Goldfarb J, Sampson HA, Li XM. Food Allergy Herbal Formula-2 silences peanut-induced anaphylaxis for a prolonged posttreatment period via IFN-gamma-producing CD8+ T cells. J Allergy Clin Immunol. 2009;123(2):443–51. doi: 10.1016/j.jaci.2008.12.1107. [DOI] [PubMed] [Google Scholar]
- 32.Srivastava K, Yang N, Chen Y, Lopez-Exposito I, Song Y, Goldfarb J, et al. Efficacy, safety and immunological actions of butanol-extracted Food Allergy Herbal Formula-2 on peanut anaphylaxis. Clin Exp Allergy. 2010 doi: 10.1111/j.1365-2222.2010.03643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lopez-Exposito I, Jarvinen KM, Castillo A, Seppo AE, Song Y, Li XM. Maternal peanut consumption provides protection in offspring against peanut sensitization that is further enhanced when co-administered with bacterial mucosal adjuvant. Food Research International. 2011 doi: 10.1016/j.foodres.2011.04.047. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brough HA, Makinson K, Penagos M, Maleki SJ, Cheng H, Douiri A, et al. Distribution of peanut protein in the home environment. J Allergy Clin Immunol. 2013;132(3):623–9. doi: 10.1016/j.jaci.2013.02.035. [DOI] [PubMed] [Google Scholar]
- 35.Lee SM, Batzer G, Ng N, Lam D, Pattar SS, Patel ND, et al. Regulatory T cells contribute to allergen tolerance induced by daily airway immunostimulant exposures. Am J Respir Cell Mol Biol. 2011;44(3):341–9. doi: 10.1165/rcmb.2010-0001OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson MS, Taylor MD, Balic A, Finney CA, Lamb JR, Maizels RM. Suppression of allergic airway inflammation by helminth-induced regulatory T cells. J Exp Med. 2005;202(9):1199–212. doi: 10.1084/jem.20042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Exposito I, Song Y, Jarvinen KM, Srivastava K, Li XM. Maternal peanut exposure during pregnancy and lactation reduces peanut allergy risk in offspring. J Allergy Clin Immunol. 2009;124(5):1039–46. doi: 10.1016/j.jaci.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y, Fu C, Chiang B. Administration of interleukin-12 exerts a therapeutic instead of a long-term preventive effect on mite Der p I allergen-induced animal model of airway inflammation. Immunology. 1999;97(2):232–40. doi: 10.1046/j.1365-2567.1999.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li XM, Serebrisky D, Lee SY, Huang CK, Bardina L, Schofield BH, et al. A murine model of peanut anaphylaxis: T- and B-cell responses to a major peanut allergen mimic human responses. J Allergy Clin Immunol. 2000;106(1 Pt 1):150–8. doi: 10.1067/mai.2000.107395. [DOI] [PubMed] [Google Scholar]
- 40.Strait RT, Morris SC, Yang M, Qu XW, Finkelman FD. Pathways of anaphylaxis in the mouse. J Allergy Clin Immunol. 2002;109(4):658–68. doi: 10.1067/mai.2002.123302. [DOI] [PubMed] [Google Scholar]
- 41.Tsujimura Y, Obata K, Mukai K, Shindou H, Yoshida M, Nishikado H, et al. Basophils play a pivotal role in immunoglobulin-G-mediated but not immunoglobulin-E-mediated systemic anaphylaxis. Immunity. 2008;28(4):581–9. doi: 10.1016/j.immuni.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 42.Niedzwiecki M, Zhu H, Corson L, Grunig G, Factor PH, Chu S, et al. Prenatal exposure to allergen, DNA methylation, and allergy in grandoffspring mice. Allergy. 2012;67(7):904–10. doi: 10.1111/j.1398-9995.2012.02841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fields PE, Kim ST, Flavell RA. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169(2):647–50. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 44.Finkelman FD. Anaphylaxis: lessons from mouse models. J Allergy Clin Immunol. 2007;120(3):506–15. doi: 10.1016/j.jaci.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 45.Brandt EB, Strait RT, Hershko D, Wang Q, Muntel EE, Scribner TA, et al. Mast cells are required for experimental oral allergen-induced diarrhea. J Clin Invest. 2003;112(11):1666–77. doi: 10.1172/JCI19785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luengo O, Song Y, Srivastava K, Li XM. The Potential of Maternal Dietary Modification for Prevention of Food Allergy. J Aller Ther. 2013;S3:005. doi: 10.4172/2155-6121.S3-005. [DOI] [Google Scholar]
- 47.Schade RP, Ieperen-Van Dijk AG, Van Reijsen FC, Versluis C, Kimpen JL, Knol EF, et al. Differences in antigen-specific T-cell responses between infants with atopic dermatitis with and without cow's milk allergy: relevance of TH2 cytokines. J Allergy Clin Immunol. 2000;106(6):1155–62. doi: 10.1067/mai.2000.110802. [DOI] [PubMed] [Google Scholar]
- 48.Finkelman FD, Katona IM, Urban JFJ, Holmes J, Ohara J, Tung AS, et al. IL-4 is required to generate and sustain in vivo IgE responses. J Immunol. 1988;141:2335–41. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure E1: Peanut allergic mothers exhibited hypersensitivity reactions following oral peanut challenge and prior to mating. Blood was collected from peanut allergic mothers (PAM) and naïve mothers (NM) one day prior to challenge. Serum peanut-specific IgE levels (A) were measured by ELISA. Anaphylactic scores (B) and core body temperatures (C) were measured, and plasma histamine levels (D) were determined thirty minutes after challenge. **, p<0.01; ***, p<0.001, vs. NM (N=5/group). Each dot represents an individual mouse. Horizontal bars indicate means. Data shown are representative of two individual experiments.
Figure E2: Following oral challenge, anaphylactic scores and core body temperatures with or without anti-FγRIIB/RIII monoclonal antibody administration in peanut sensitized offspring of peanut allergic mothers. N=5-6. *, p<0.05.Horizontal lines represent the means of each group.
Figure E3: A: Global methylation of MLN cells in O-PAM, O-NM and normal controls following challenge was measured by the global DNA methylation ELISA kit(Epigentek, Farmingdale, NJ). In this ELISA kit, the methylated fraction of DNA is recognized by 5-methylcytosine antibody and quantified through an ELISA-like reaction. (N=6-7/group). Both p>0.05 for O-PAM vs. O-NM and O-PAM vs. Naïve. B:Purified DNA from sensitized and challenged offspring MLN cells underwent bisulfite treatment, PCR amplification, and pyrosequencing. Percent of DNA methylation of the IFN-γ and Foxp3 promoters in offspring MLN cells (B&C). Data are expressed as means ± SDs of each group. (N=5-6/group).
Figure E4: DNA methylation at CpG-408 and CpG-393 sites of the IL-4 promoter from peripheral blood leucocytes (PBL) of peanut allergic mothers (PAM) and naïve mothers (NM) prior to breeding. Purified DNA from mothers PBL prior to breeding underwent bisulfite treatment, PCR amplification, and pyrosequencing. **p < 0.01; ***p < 0.001 vs.NM (N=5).
Figure E5: Breast milk was collected from lactating peanut allergic mothers (PAM) and naïve mothers (NM) when their offspring were 10-15 day-old, using a mouse milking machine modified in our laboratory. An additional group of PAM lactating mice was inoculated intragastrically with10 mg of peanut protein (PAM-PN). Milk was collected 2 hours following peanut protein feeding. Milk was diluted 1:2 in PBS, and peanut protein levels were detected using a commercial kit (Neogen Corp, Lansing, Mich). N=3-4. ***, p<0.001.
Supplement Table I. Primers used for PCR amplification and pyrosequencing experiments


