Abstract
TDP-43 aggregates are the neurohistological landmark of diseases like amyotrophic lateral sclerosis and frontotemporal dementia. Their role in the pathogenesis of these conditions is not yet clear mainly due to the lack of proper models of aggregation that may allow the study of the mechanism of formation, their interactions with other cellular components and their effect on the cell metabolism. In this work, we have used tandem repeats of the prion like Q/N-rich region of TAR DNA-binding protein (TDP-43) fused to additional TDP-43 protein sequences to trigger aggregate formation in neuronal and non-neuronal cell lines. At the functional level, these aggregates are able to sequester endogenous TDP-43 depleting its nuclear levels and inducing loss of function at the pre-mRNA splicing level. No apparent direct cellular toxicity of the aggregates seems to be present beyond the lack of functional TDP-43. To our knowledge, this is the only system that achieves full functional TDP 43 depletion with effects similar to RNAi depletion or gene deletion. As a result, this model will prove useful to investigate the loss-of-function effects mediated by TDP-43 aggregation within cells without affecting the expression of the endogenous gene. We have identified the N-terminus sequence of TDP-43 as the domain that enhances its interaction with the aggregates and its insolubilization. These data show for the first time that cellular TDP-43 aggregation can lead to total loss of function and to defective splicing of TDP-43-dependent splicing events in endogenous genes.
INTRODUCTION
TDP-43 participates in several mechanisms of mRNA metabolism like pre-mRNA splicing, mRNA stability, mRNA transport and miRNA processing (1). In normal conditions, the subcellular localization of the protein is preferentially nuclear, but the presence of a nuclear localization sequence (NLS) and a nuclear export sequence in the N-terminus of the protein allow TAR DNA-binding protein (TDP-43) to shuttle between the nucleus and cytoplasm (2). Downstream of the N-terminus, the first RNA recognition motif (RRM1) is the main responsible for the binding to RNAs containing UG repetitions (3,4). Very recently, our laboratory has observed that RRM1 also plays a key role in recognizing the UG-rich RNA sequences involved in the TDP-43 autoregulation mechanism (5–8). The RRM1 sequence is followed by a second RRM motif, called RRM2. The role of RRM2 in RNA binding is less known. However, it has been shown to bind nucleic acids with low affinity (9), to provide the main structural core of the protein (10) and to participate together with RRM1 in the binding of TDP-43 to UG-rich cross-linking immunoprecipitation sequences (11). Finally, the C-terminal portion of the protein includes a glycine-rich domain that has been involved in most of the protein interactions described up to now (1,12–14). In this region is present a glutamine/asparagine (Q/N) prion-like domain that participates in protein–protein interactions (15,16) and in the TDP-43 aggregation process (16–18).
In recent years, the observation that TDP-43 is the principal component of inclusions (aggregates) present in neurons of patients affected by amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) (19,20) has resulted in further studies on this protein both at the molecular and clinical levels. From a genetic point of view, the link between this protein and the pathology has been reinforced following the discovery of >40 disease-associated mutations localized principally in the C-terminal region of TDP-43 (21–23). However, it is important to always keep in mind that ∼95% of the patients showing TDP-43 inclusions do not carry any type of mutations in this protein. Hence, the pathological mechanisms do not necessarily require a mutation in the protein sequence to trigger aggregation.
In the affected neurons, TDP-43 aggregates localize preferentially in the cell cytoplasm and less frequently in the cell nuclei (that sometimes show complete depletion of this factor). Moreover, these aggregates are distinguished by ubiquitination, abnormal phosphorylation (19,20,24,25) and the presence of cleaved TDP 43 C-terminal fragments (25 and 35 kDa).
Two principal hypotheses have been proposed to account for the neurodegeneration mediated by these pathological TDP-43 aggregates (26). The ‘loss-of-function’ hypothesis suggests that the depletion of nuclear TDP-43 following its aggregation impairs the functions in which TDP-43 is involved (mRNA splicing, mRNA transport, etc.), thus affecting the whole cell metabolism. In this case, the cytoplasmic TDP-43 aggregates act as ‘sinks’ retaining the protein in the cytoplasm and causing loss of function (16,27,28). However, it should be kept in mind that, in the first stages of disease, aggregation may even act in a protective manner. The second hypothesis, known as ‘gain of function’, suggests that cytoplasmic TDP-43 aggregates or the presence of missense mutations could be toxic by ‘themselves’ (29–31). In addition, gain-of-function effects could also originate from abnormal trapping in the aggregates of mRNAs or proteins that usually are partners of TDP-43, and thus affecting also their function.
At the moment, it is not clear which hypothesis is correct or if both mechanisms could equally contribute to disease onset/progression. To address this issue, several cellular models have been described, most of them have been developed by overexpression of different C-terminal regions of TDP-43 (28,32) or by restricting the localization of TDP-43 to the cell cytoplasm (27). Some of these models have been able to reproduce aggregates, but the participation of these inclusions in mechanism of loss- or gain of functions still remains ambiguous.
Previously, we have characterized a particular structural determinant that can trigger TDP-43 loss of function/interaction with hnRNP A2 and whose minimal region corresponds to residues 342–366 of the TDP-43 C-terminal tail (15–17). In addition to these protein–protein interactions, recent studies have supported that this sequence possesses properties similar to those found in yeast prions that in response to stress can recruit the native protein into an inactive aggregate (33). A structural characterization of this Q/N prion-like region has recently shown that this segment is normally in a disordered conformation that can form β-sheet strands spontaneously over time (34), suggesting a possible TDP-43 aggregation pathway during disease. We have used tandem repetitions of the Q/N region of TDP-43 (residues 339–369)-linked enhanced green fluorescent protein (EGFP) to (EGFP-12xQ/N) (16). These aggregates were able to sequester either exogenous or endogenous full-length TDP-43 and recapitulated some properties of the inclusions observed in patients such as ubiquitination and phosphorylation at the C-terminal residues pS409/pS410. However, there was no detectable splicing function deterioration in the presence of these EGFP-12xQ/N-induced TDP-43 aggregates (16), suggesting that the aggregates were not capable of trapping enough endogenous TDP-43 to cause loss of function in the short interval measured in a cell system.
In the present work, we describe a new variant of the previous construct that is based on the TDP-43 molecule itself (or fragments of it) linked to tandem repeats of the 339–369 sequences (TDP-12xQ/N). The advantage of this model over the previous one is that induces in a highly reproducible and efficient manner aggregate formation and TDP-43 nuclear depletion. This led to a clearly visible splicing loss of function both in endogenous and exogenous (transfected) genes. To our knowledge, this is the first model of TDP-43 aggregation capable of displaying this property. As a result, this new model could have an important role in the understanding of the loss-of-function mechanisms that may arise following TDP-43 aggregation. The TDP-43 additional structural determinants for efficient aggregation were mapped to its N-terminus 75 amino acids.
MATERIALS AND METHODS
Expression plasmids
All TDP-43 plasmids prepared to generate the corresponding stable cell lines were based on pCDNA5FRT/TO vector (Invitrogen). To produce the Flag-TDP-12xQ/N construct, the pFlag-TDP-43 wild-type (WT) plasmid (6) was modified by introducing an XhoI restriction site in position 1209 of TDP-43 by site-directed mutagenesis (Stratagene Quick-Change). The oligos used for the mutagenesis were XhoI_S (5′-ggaggctttggctcgagcatggattctaag-3′) and XhoIAS (5′-cttagaatccatgctcgagccaaagcctcc-3′). Once inserted the XhoI site, the Flag-TDP-43 sequence was amplified by PCR using the EcoRV_F (5′-aaattaagatatcatggactacaaagacgatgac-3′) and C-CMV (5′-tattaggacaaggctggtgggcac-3′) primers. The Flag-TDP-43 product was blunted and then cloned in pCDNA5FRT/TO vector previously digested with PmeI enzyme to generate the construct pCDNA5FRT/TO-Flag-TDP-43. Next, the fragment containing the 12xQ/N repetitions was digested from the EGFP-12xQ/N construct (16) using XhoI/BamHI enzymes and cloned in pCDNA5FRT/TO-Flag-TDP-43, generating the final construct pCDNA5FRT/TO Flag-TDP-12xQ/N. Moreover, a small intron from pCI-Neo vector was cloned upstream of the resultant construct pCDNA5FRT/TO Flag-TDP-12xQ/N. The intron was previously cloned in pUC vector and then digested with EcoRI and HindIII restriction enzymes to then be cloned in blunt inside the EcoRV site from pCDNA5FRT/TO Flag-TDP-12xQ/N. In order to facilitate future cloning strategies, a BamHI site contained in the intron was eliminated by site-directed mutagenesis using the forward (5′-ctcccagcggccgctaggggatactctagagtcgacctgcag-3′) and reverse (5′-ctgcaggtcgactctagagtatcccctagcggccgctgggag-3′) primers. All the Flag-TDP-12x-Q/N mutants were generated with the same procedure described before: modifications were performed in pCDNA5FRT/TO-Flag-TDP-43 using site-directed mutagenesis before the introduction of the 12xQ/N repetitions. The primers used to generate each mutant were as follows: Flag-TDP-12xQ/N F4L: forward (5′-aaggggttgggcttggttcgtttt-3′) and reverse (5′-aaaacgaaccaagcccaacccctt-3′); Flag-TDP-12xQ/N ΔRRM1: forward (5′-gtgaaagtgaaaagagcagtcgactgcaaacttcctaattct-3′) and reverse (5′-agaattaggaagtttgcagtcgactgctcttttcactttcac-3′); Flag-TDP-12xQ/N ΔRRM2: forward (5′-caggatgagcctttgagaagctccaatgccgaacctaagcac-3′) and reverse (5′-gtgcttaggttcggcattggagcttctcaaaggctcatcctg-3′); Flag-TDP-12XQ/N ΔN and Flag-TDP-12xQ/N ΔRRM1/2, ΔN: forward (5′-tacaaagacgatgacgacaagcttaactatccaaaagataac-3′) and reverse (5′-gttatcttttggatagttaagcttgtcgtcatcgtctttgta-3′); Flag-TDP-12xQ/N-ΔRRM1/2, ΔCterm: forward (5′-agccaagatgagcctttgagaagctttggctcgagcatggat-3′) and reverse (5′-atccatgctcgagccaaagcttctcaaaggctcatcttggct-3′).
Cell culture and transfection
HEK293 flip-in cell line (Invitrogen) was grown in DMEM–Glutamax-I (GIBCO) supplemented with 10% fetal bovine serum (GIBCO) and Antibiotic-Antimycotic-stabilized suspension (Sigma). Plasmid transfections were carried out using Effectene Transfection reagent (Qiagen) according to the manufacturer's instructions. To generate the stable clones, 0.5 μg of plasmids (Flag-TDP-12x-Q/N and mutant variants) were co-transfected with 0.5 μg pOG44 vector that expresses the Flp-recombinase (Invitrogen). After co-transfection, cells were grown in DMEM–Glutamax-I supplemented with 10% fetal bovine serum, Antibiotic-Antimycotic and after 24 h the stable integration was gradually selected using 100 μg/ml of Hygromycin B (Invivogen). Once the selection was finalized, the induction of Flag-TDP-43-12x-Q/N and mutant proteins was achieved by adding 1 μg/ml of tetracycline (Sigma) at the culture medium.
NSC-34 cells were maintained in DMEM–Glutamax-I (GIBCO) supplemented with 5% fetal bovine serum (Sigma) and Antibiotic-Antimycotic-stabilized suspension (Sigma). The day before the transfection, cells were plated in six-well plates containing 1% fetal bovine serum in order to start the cell differentiation. Cells were transfected with Lipofectamine 2000 (Life Technologies) with 2 μg of each plasmid according to data sheet instructions and always maintained in differentiation conditions. After 48 h post-transfection, cells were fixed in order to perform immunofluorescence analyses.
RT-PCR and splicing assays
The reporter CFTR exon 9 minigene used in the splicing assay, pTB CFTR C155T, has already been described by Pagani et al. (35). When siRNA was used, 1.7 × 105 HEK293 cells were plated in six-well plates (Day 0) and two rounds of TDP-43 siRNA transfections were carried out according to the procedure already described (36) on Days 1 and 2 in order to maximize TDP-43 silencing efficiency. The siRNA target sequence used to silence the endogenous TDP-43 was 5′-aagcaaagccaagaugagccu-3′. After TDP-43 silencing, the transfection of 0.5 μg of the reporter minigene CFTR exon 9 was performed on Day 3 using HiPerFect (Qiagen). Cells were harvested on Day 4 and total RNA was collected using Trizol Reagent (Invitrogen). Reverse transcription was performed using M-MLV Reverse Transcriptase (Invitrogen), according to the manufacturer's protocol. PCR with DNA Polymerase (Roche) was carried out for 20–25 amplification cycles (95°C for 30 s, 55°C for 30 s and 72°C for 30 s). The primers to amplify CFTR exon 9 were the following: forward (5′-caacttcaagctcctaagccactgc-3′) and reverse (5′-taggatccggtcaccaggaagttggttaaatca-3′). The primers used to test the splicing pattern of endogenous gene POLDIP3/SKAR were the following: forward (5′-gcttaatgccagaccgggagttgga-3′) and reverse (5′-tcatcttcatccaggtcatataaatt-3′). Endogenous TDP-43 was not silenced when splicing efficiency was measured following induction of Flag-TDP-43 WT, Flag-TDP-12xQ/N and the additional mutants. All PCR products were analyzed on a 1.8% agarose gels.
Co-immunoprecipitation assays
For co-immunoprecipitation assays, HEK293 flip-in stably expressing the corresponding proteins were induced for 72 h with 1 μg/ml of tetracycline. Cells were collected in RIPA lysis buffer (50 mm Tris/HCl pH 7.4, 150 mm NaCl, 1% NP-40, 0.1% SDS, 1 mm EDTA pH 8, 1 mm PMSF, 0.5% sodium deoxycholate) supplemented with protease inhibitors (Roche, cat. 11836145001) and incubated for 30 min at 4°C. After spin down at 500 g at 4°C, cells were lysed by sonication. The lysates were then incubated with 40 μl of A/G plus Agarose beads (Santa Cruz) to perform a pre-clearing for 1 h at 4°C. In the meantime, an incubation of 40 μl of A/G plus Agarose beads with 5 μg of anti-Flag antibody (Sigma, F1804) was performed in RIPA buffer for 2 h at 4°C. After both incubations, the pre-cleared lysate was incubated with A/G plus Agarose beads/anti-Flag for 3 h at 4°C. Then, the beads were centrifuged and washed with PBS once for 10 min at 4°C. The beads were finally resuspended in 50 μl of resuspension buffer (50 mm Tris/HCl pH 7.4, 5 mm EDTA, 10 mm DTT and 1% SDS) and 20 μl of SDS 5× loading buffer were added. For mass spectrometry detection, immunoprecipitated samples were analyzed by Coomassie blue and then the selected bands were excised from the gel. In case of western blot assays, the analysis of the samples was done using the antibodies anti-Flag (Sigma, F1804), anti-TDP-43 (Protein Tech, 10782-2-AP), anti-HA (Roche, cat. 912CA5) or anti-HSP70 (Sigma, H5147). All Co-IP experiments were performed a minimum of two times and have always given a consistent result.
Immunofluorescence microscopy
For indirect immunofluorescence, all HEK293 stable cell lines were induced with tetracycline for 72 h and the samples were processed as previously described (2). The primary antibodies used were anti-Flag (Sigma, F1804), anti-TDP-43 (Protein Tech, 10782-2-AP) and anti-Myc (Cell Signaling n.c 2272S). The secondary antibodies anti-mouse-AlexaFluor 594 (cat. A21203), anti-rabbit-AlexaFluor 488 (cat. A21200) and the dye TO-PRO3 (cat. T3605) were all purchased from Life Technologies. Cells were analyzed on a Zeiss LSM 510 Meta confocal microscope.
Cell lysate fractionation
To perform cell lysate fractionation in soluble and pellet fractions, 3 × 106 cells were seeded and induced with 1 µg/ml of tetracycline for 72 h. Then, cells were collected and lysed with 1 ml of RIPA + protease inhibitor for 30 min at 4°C. After a centrifugation at 4000 rpm for 20 min, the whole supernatant was furthermore sonicated for 5 min to allow a better lysis. Then, 600 µg of cell lysate were ultracentrifuged in a clean Beckman polycarbonate thick wall Centrifuge tube (rotor type 70.1Ti) for 1 h at 25°C at 33 000 rpm. The supernatants were collected and the pellets washed twice with 100 µl of RIPA buffer. Pellets were finally dissolved in urea buffer (7 M urea, 4% CHAPS, 30 mm Tris, pH 8.5). To analyze each fraction by western blot, 30 μg of protein lysates were run on an SDS–PAGE gel and blotted on a nitrocellulose membrane for standard incubation procedures.
RESULTS
Generation of a cell line stably expressing exogenous TDP-43 aggregates
We have previously developed a cell-based aggregation model by cloning in-tandem 12 copies of the Q/N region from TDP-43 in the C-terminal of EGFP, resulting in the expression of a so-called EGFP-12xQ/N protein (16). In order to generally improve the aggregation process and to further study whether other regions in TDP-43 could contribute to it we considered to include the 12xQ/N repetitions into the C-terminus of the Flag-TDP-43 protein itself (Fig. 1A).
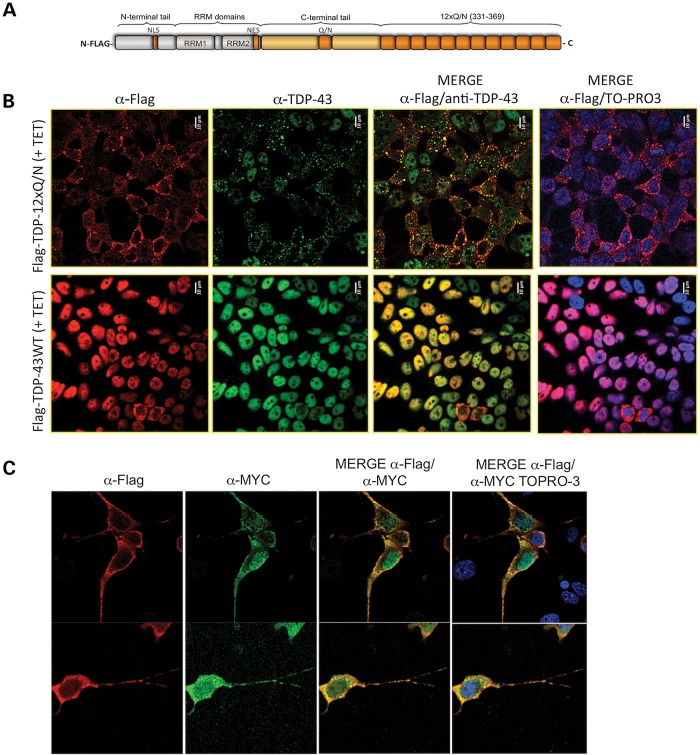
Figure 1.
(A) A schematic representation of the Flag-TDP-12xQ/N construct, describing all its different structural determinants. Residues 403–414 were eliminated due to cloning strategy. (B) An immunofluorescence of the Flag-TDP-43 WT and Flag-TDP-12xQ/N-induced proteins using anti-Flag (red) and anti-TDP-43 (green) antibodies. The cell nuclei were stained with the reagent TOPRO-3. A merge between anti-Flag/anti-TDP-43 antibodies or anti-Flag/anti-TDP-43/TOPRO-3 is also reported. (C) A co-immunofluorescence of differentiated NSC-34 motor neuron cell line co-transfected with constructs expressing Flag-TDP-12xQ/N and Myc-TDP-43 WT. The assay was performed using anti-Flag (red) and anti-Myc (green) antibodies. The cell nuclei were stained with the reagent TOPRO-3. A merge between anti-Flag/anti-Myc antibodies or anti-Flag/anti-Myc/TOPRO-3 is reported.
The resulting construct, Flag-TDP-12xQ/N, was then used to generate a HEK293 human kidney cell line that stably expressed one copy of the construct after tetracycline (TET) addition, in a similar way as previously described for Flag-TDP-43 WT cell line (6). Supplementary Material, Figure S1 shows a western blot using an anti-Flag monoclonal antibody demonstrating that incubation of this cell line with tetracycline for 72 h induced the efficient expression of the Flag-TDP-12xQ/N protein migrating at ∼80 kDa as expected. As a control, we also induced with tetracycline a stable cell line expressing Flag-TDP-43 WT (6).
Immunofluorescence analysis of the induced cells using an anti-Flag antibody showed that the Flag-TDP-43 WT was mainly present in the cell nucleus (Fig. 1B). On the other hand, expression of Flag-TDP-12xQ/N protein induced the formation of inclusions mostly localized in the cytoplasm. However, in some cases, cells displaying inclusions only in the nucleus or both in the nucleus or cytoplasm were observed (Fig. 1B). Interestingly, in many cells no nuclear signal was present, suggesting total absence from the nucleus of TDP-43 species. In fact, staining of these cells using a commercial anti-TDP-43 antibody indicated that several cell nuclei were lacking both endogenous and exogenous TDP-43 proteins, as no positive staining for red (anti-Flag), green (anti-TDP-43) or yellow (merged anti-Flag/anti-TDP-43) was observed (Fig. 1B, MERGE rows).
In addition to the HEK293 cell lines, transient transfection of Flag-TDP-12xQ/N and of a Myc-tagged WT TDP-43 in differentiated motor neuron-like NSC-34 cells was also able to achieve aggregation and nuclear clearance. Interestingly, in this cell line, the induced inclusions were also present along the neuronal neurites (Fig. 1C).
Flag-TDP-12xQ/N expression affects the solubility properties of endogenous TDP-43 in the cellular environment
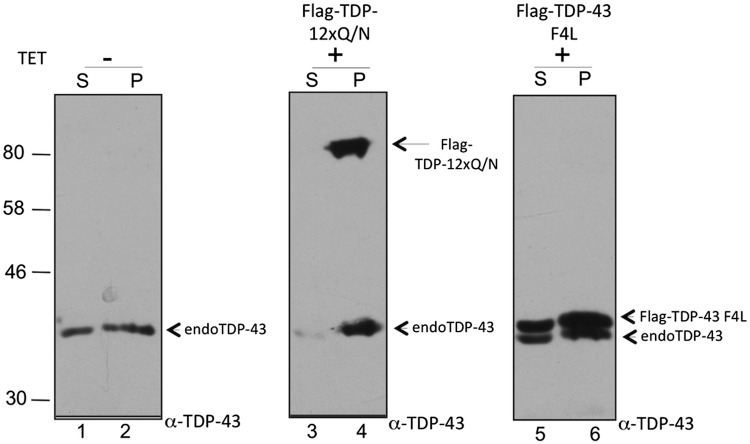
We then proceeded to study the solubility properties of endogenous TDP-43 in the presence or absence of induced aggregates. To do this, total protein extraction was performed using RIPA buffer after 72 h following induction of the Flag-TDP-12xQ/N effector. Subsequently, soluble (S) and insoluble (P) fractions were separated by ultracentrifugation, and the presence of endogenous TDP-43 and exogenous (Flag-TDP-12xQ/N) proteins in each fraction was analyzed by western blot using an anti-TDP-43 antibody.
In Figure 2, it is possible to observe that, in normal conditions, the endogenous TDP-43 is equally distributed in the soluble and insoluble fractions (Fig. 2, lines 1–2). Interestingly, in the presence of Flag-TDP-12xQ/N expression, the endogenous TDP-43 protein shifted almost completely into the insoluble fraction together with this mutant (Fig. 2, lines 3 and 4). Overexpression of TDP-43 alone does not cause aggregation/insolubilization per se in our stable transgenic TDP-43 in the HEK293 cells. This could have partly due to the self-regulation mechanisms that, in a situation of excess protein, shut-off the endogenous gene (6). In order to completely discard the possibility that the shift into the insoluble fraction was produced simply because of the overexpression of Flag-TDP-12xQ/N, we then performed the same experiment using a stable cell line overexpressing Flag-TDP-43 F4L. The advantage of using this particular RNA-binding mutant is that Flag-TDP-43 F4L is unable to downregulate the endogenous TDP-43 levels by the autoregulation mechanism (6). As shown in Figure 2, lanes 5 and 6, following TET induction both this mutant and the endogenous TDP-43 were distributed in the soluble (S) and insoluble (P) fractions in similar amounts as those observed with the endogenous protein alone (Fig. 2, lanes 1 and 2). There may only a slight increase of both proteins in the P fraction, but there is no sequestration of the endogenous TDP-43 in the P fraction as occurred in the presence of Flag-TDP-12xQ/N (Fig. 2, lines 5 and 6).
Figure 2.
A cell lysate fractionation of normal cells, Flag-TDP-12xQ/N HEK293 and Flag-TDP-43 F4L stable cell lines induced or not with tetracycline. The soluble (S) and insoluble (P) cell fractions were separated by ultracentrifugation, and the corresponding Flag-tagged proteins and endogenous TDP-43 were detected by western blot using an anti-TDP-43 antibody.
Flag-TDP-12xQ/N aggregates interact with HSP70
Since we have previously shown that the aggregates induced in our model are ubiquitinated similarly to those found in the brains of ALS patients, it was then of interest to determine whether aggregated TDP-43 was interacting with other cellular factors connected with this modification (37). For this reason, we have explored other protein–protein interactions of transgenic TDP-43 and their eventual presence in the aggregates. For this purpose, the Flag-TDP-43 WT transgenic cell (used as a control) and the corresponding cell line carrying the Flag-TDP-12xQ/N transgene were induced with tetracycline for 72 h. An immunoprecipitation assay was then carried out using an anti-Flag antibody, and the resulting proteins were run in a SDS–PAGE followed by staining with Coomassie Blue (data not shown) followed by mass spectrometry analysis of selected differential bands. Among the proteins that were identified in this manner, one of the most prominent interactors from the sample expressing Flag-TDP-12xQ/N was identified as HSP70, a molecular chaperone that has been observed to participate in the aggregation processes and neurodegeneration (38,39). In keeping with these affinity purification results, in cells overexpressing Flag-TDP-12xQ/N the HSP70 factor was observed redistributed and co-localizing perfectly with the induced aggregates (Supplementary Material, Fig. S2). On the other hand, the subcellular localization of HSP70 in cells expressing Flag-TDP-43 WT consisted in a uniform cellular distribution (Supplementary Material, Fig. S2). The interaction between Flag-TDP-12xQ/N and HSP70 was further confirmed by co-immunoprecipitation analyses. In Supplementary Material, Figure S3, in fact, it is shown that HSP70 can co-precipitate specifically with overexpressed Flag-TDP-12xQ/N, but not with Flag-TDP-43 WT. While this work was in progress, a similar observation was published by other researchers (40,54).
Induced Flag-TDP-12xQ/N aggregates cause TDP-43 loss of function
One very important issue that remained to be evaluated was to determine whether these aggregates were able to induce loss of normal TDP-43 functions within cells.
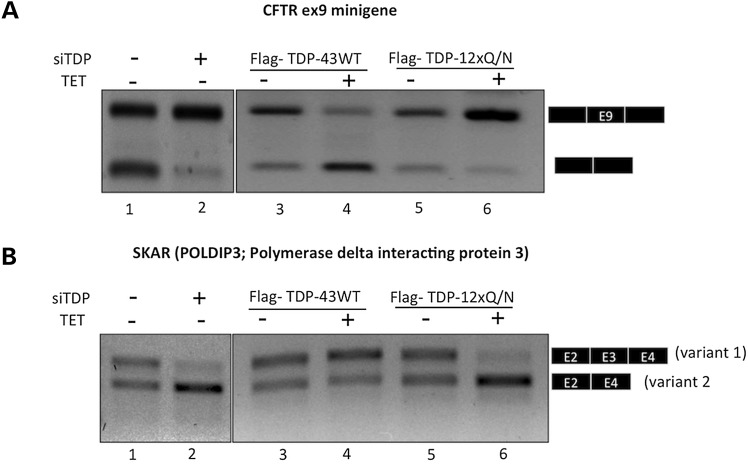
As previously described, loss of TDP-43 function (e.g. by siRNA TDP-43 downregulation) can be easily observed using a minigene system based on the inclusion/skipping of CFTR exon 9 (13,35). Therefore, cells expressing Flag-TDP-12xQ/N and Flag-TDP-43 WT (used as a control) were transfected with this minigene and after 24 h post-transfection the splicing pattern of exon 9 was evaluated by RT-PCR. In order to have a positive control of TDP-43 loss of function, we knocked down TDP-43 by transfecting siTDP-43 against this protein. Knockdown of TDP-43 resulted in the complete inclusion of exon 9 (Fig. 3A, compare lines 1 and 2). In keeping with expectations, the overexpression of Flag-TDP-43 WT repressed the inclusion of exon 9 (Fig. 3A, compare lines 3 and 4), while the overexpression of Flag-TDP-12xQ/N induced the inclusion of this exon in a similar way as the siTDP-43 treatment (Fig. 3A, compare lines 5, 6 with 1, 2). This last observation is compatible with loss of active nuclear TDP-43 following its aggregation in the nucleus/cytoplasm.
Figure 3.
(A) An RT-PCR assay in which it is possible to observe the splicing pattern of CFTR exon 9 minigene in the presence of Flag-TDP-43 WT or Flag-TDP-12xQ/N-induced protein. The corresponding HEK293 stable cell lines were induced or not during 48 h with tetracycline. The cells were transfected with CFTR9 minigene and maintained under induction condition for an additional 24 h. The splicing pattern (exclusion/inclusion) of exon 9 from CFTR9 minigene was evaluated by RT-PCR. In (B) is reported the splicing pattern of the endogenous gene POLDIP3/SKAR (exon 3) following induction of Flag-TDP-43 WT or Flag-TDP-12xQ/N protein. Exon inclusion was evaluated by RT-PCR. In both cases, the result of knocking down the endogenous TDP-43 is reported as a positive TDP-43 loss-of-function control.
Next, we extended our studies to endogenous genes like POLDIP3/SKAR, whose splicing has already been described to be affected by TDP-43 cellular levels (41,42).
Regarding POLDIP3/SKAR, knockdown of TDP-43 by siRNA treatment caused the exclusion of exon 3 from the processed mRNA inducing the appearance of the variant 2 isoform (Fig. 3B, compare lines 1 and 2). As expected, overexpression of Flag-TDP-43 WT promoted the inclusion of POLDIP3 exon3 (Fig. 3B, compare lines 3 and 4). Also in keeping with expectations, the overexpression of Flag-TDP-12xQ/N enhanced the exclusion this exon in the same manner of the siTDP-43 treatment (Fig. 3B, compare lines 5, 6 with 1, 2).
Role of different domains from Flag-TDP-12xQ/N on TDP-43 loss-of-function effect
It was then of interest to use this system to investigate whether other sequences/domains of TDP-43 were contributing together with the 12xQ/N to these phenomena. To understand the contribution of the different domains of TDP-43 to induce aggregation and loss of function, we generated a variety of HEK293 stable cell lines expressing different mutants of Flag-TDP-12xQ/N (Fig. 4A). The expression of these proteins was induced by tetracycline and 72 h later TDP-43 functionality was tested by looking at the splicing of the endogenous POLDIP3/SKAR gene both at the mRNA and protein levels (Fig. 4B and C, respectively, indicated as variant 1 or variant 2).
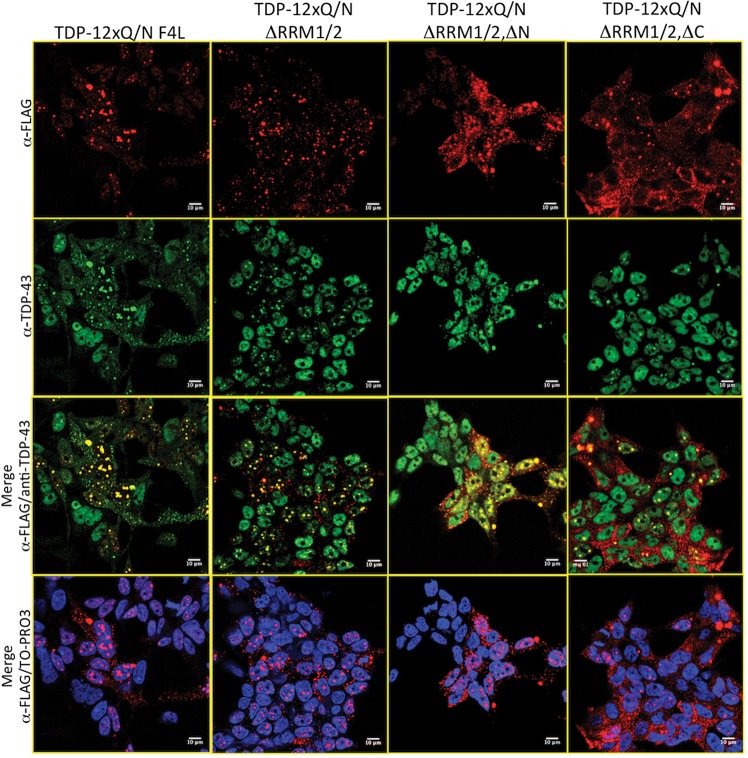
Figure 4.
(A) A schematic representation from constructs that include modifications in Flag-TDP-12xQ/N. TDP-12xQ/N F4/L (F147, 149, 229, 231/L); TDP-12xQ/N ▵RRM1/2 (deleted RMM1 and RMM2); TDP-12xQ/N ▵RRM1/2, ▵N (deleted RMM1, RMM2 and N-terminal portion); TDP-12xQ/N ▵RRM1/2, ▵C (deleted RMM1, RMM2 and C-terminal portion). In (B) is reported an RT-PCR indicating the splicing pattern of endogenous POLDIP3/SKAR gene (exon 3) after the induction or not of Flag-TDP-12xQ/N or its corresponding mutant proteins. (C) A western blot performed with an anti-SKAR antibody using proteins extracted from the same samples used in (B) The red boxes enclose the splicing and protein expression pattern of the mutant that does not induce loss of function.
As observed in Figure 4B, neither point mutations in both RMMs (Flag-TDP-12xQ/N F4L) nor the complete deletion of both RMMs (Flag-TDP-12xQ/N ▵RRM1/2) affected the loss of function caused by the original Flag-TDP-12xQ/N (Fig. 4B, compare lines 5, 6 and 7, 8; with lines 3, 4). All these mutants modified the POLDIP3 splicing pattern by excluding mainly variant 1 and leaving mainly variant 2, indicating that the RNA-binding activity of Flag-TDP-12xQ/N was not responsible of the induced loss of TDP-43 activity. The same effects could also be observed in mutants where each of the two RNA recognition motifs (RRMs) was removed separately (Flag-TDP-12xQ/N ▵RRM1, Flag-TDP-12xQ/N ▵RRM2; Supplementary Material, Fig. S4A).
Next, we evaluated whether the N-terminal or C-terminal domains from Flag-TDP-12xQ/N were necessary to cause the lack of endogenous TDP-43 activity. Thus, we performed N-terminal and C-terminal deletion from the minimal construct. Compared with full-length Flag-TDP-12xQ/N, the deletion of the C-terminal region in Flag-TDP-12xQ/N ▵RRM1/2, ▵C had little effects on its loss-of-function properties and the protein maintained its capacity to induce the generation of variant 2 (loss of function; Fig. 4B, compare lines 11, 12 with lines 3, 4). Interestingly, however, the deletion of the N-terminal portion (residues 1–75) completely abolished the capacity of the resulting protein (Flag-TDP-12xQ/N ▵RRM1/2, ▵N) to cause loss of function (Fig. 4B, compare lines 9, 10, boxed, with 3, 4), leaving the splicing pattern like in an endogenous situation (Fig. 5B, compare line 13 with 2). Such an effect was also observed when the expression of a Flag-TDP-12xQ/N, ▵N (containing the two RRMs) was induced (Supplementary Material, Fig. S5B). The observed changes on POLDIP3 mRNA splicing were further confirmed at the protein level by performing western blot assay using an antibody that detects the two variants of this protein (Fig. 4C). As control, a western blot showing that all the Flag-TDP-12xQ/N proteins were expressed at similar levels is presented in Supplementary Material, Figure S4B.
Figure 5.
An immunofluorescence of HEK293 cells expressing the Flag-TDP-12xQ/N mutants proteins described in Figure 4A. The assay was performed using anti-Flag (red) and anti-TDP-43 (green) antibodies. The cell nuclei were stained with the reagent TOPRO-3. A merge between anti-Flag/anti-TDP-43 antibodies or anti-Flag/anti-TDP-43/TOPRO-3 is also shown.
We then performed an immunofluorescence assay in order to evaluate the capacity of each mutant to produce aggregates. As observed in Figure 5, after 72 h of tetracycline induction, all mutants formed aggregates in a similar way to Flag-TDP-12xQ/N. Most interestingly, also the Flag-TDP-12xQ/N ▵RRM1/2, ▵N mutant lacking residues 1–75 retained its capacity to generate aggregates but not to affect splicing. This suggests that the N-terminal region could be involved not so much in the aggregation process itself but in increasing the efficiency of the aggregates to capture endogenous TDP-43, an aspect that will warrant future in depth studies.
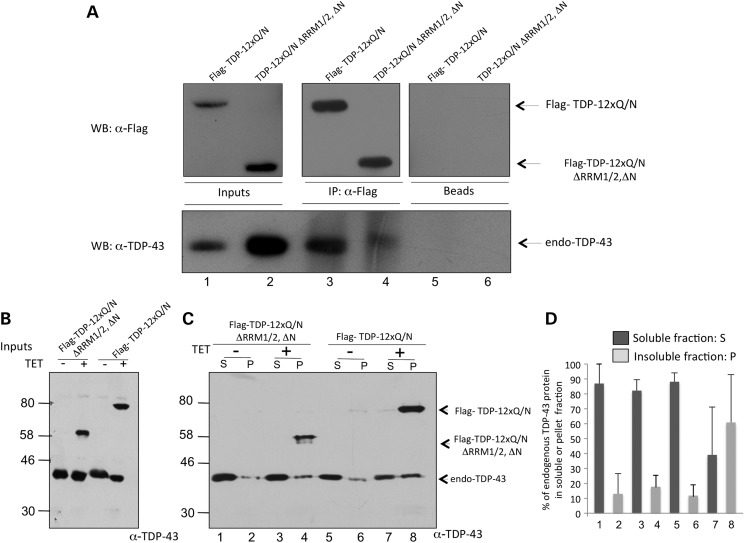
To further validate this possibility, we then performed co-immunoprecipitation and solubility fractionation assays. Figure 6A shows a co-immunopreciptiation in which induced Flag-TDP-12xQ/N and Flag-TDP-12xQ/N ▵RRM1/2, ▵N proteins were immunoprecipitated using an anti-Flag antibody as previously described. Co-precipitation of endogenous TDP-43 with each these Flag-proteins was then evaluated by western blot. As shown in Figure 6A, while Flag-TDP-12xQ/N was able to co-precipitate endogenous TDP-43 the interaction of this protein with Flag-TDP-12xQ/N ▵RRM1/2, ▵N was greatly diminished (Fig. 6A, compare lanes 3 and 4), consistent with the results observed in Figure 4. Interestingly, the same experiment was also repeated for the Flag-TDP-12xQ/N ▵RRM1/2, ▵C construct, and that in the experiments presented in Figure 4 showed an intermediate effect in affecting the splicing profiles (both CFTR reporter and endogenous POLDIP3). The results of the co-immunoprecipitation confirmed that this mutant also had a reduced ability to pull down endogenous TDP-43 with respect to Flag-TDP-12xQ/N (Supplementary Material, Fig. S6). However, although co-IP is not a properly quantitative technique, the level of the remaining interaction was consistently higher than that observed for the Flag-TDP-12xQ/N ▵RRM1/2, ▵N protein, providing a reason why the Flag-TDP-12xQ/N ▵RRM1/2, ▵C mutant still retained an intermediate ability to induce loss of function. Additionally, solubility fractionation was also performed from induced Flag-TDP-12xQ/N and Flag-TDP-12xQ/N ▵RRM1/2, ▵N proteins. In agreement with their ability to form inclusions, both Flag-proteins were present almost completely in the insoluble fraction (Fig. 6C, lines 4 and 8). As expected, endogenous TDP-43 shifted to the insoluble fraction when Flag-TDP-12xQ/N was induced (Fig. 6C, compare lines 7, 8 with 5, 6). However, in the presence of Flag-TDP-12xQ/N ▵RRM1/2, ▵N protein, the endogenous TDP-43 pattern between the S and P fractions remained the same (Fig. 6C, compare lanes 1, 2 with 3, 4). Figure 6B shows the inputs of the solubility experiment. Figure 6D shows the ratio of endogenous TDP-43 between soluble and insoluble fractions as quantified following three independent experiments.
Figure 6.
(A) A western blot from a co-immunoprecipitation between overexpressed Flag-TDP-12xQ/N or Flag-TDP-12xQ/N ▵1/2, ▵N and endogenous TDP-43 protein. After induction of the corresponding HEK293 cell lines, an immunoprecipitation was performed used an anti-Flag antibody. Upper panels show a western blot using an anti-Flag antibody of precipitated Flag-tagged proteins (left panels: inputs used in the assay, middle panel: immunoprecipitated proteins, right panel: negative control). Bottom panel shows a western blot using an anti-TDP-43 antibody of co-precipitated endogenous TDP-43 in each indicated condition. (B) A western blot using an anti-TDP-43 antibody from inputs of HEK293 stable cell lines expressing or not Flag-TDP-12xQ/N F4L or Flag-TDP-12xQ/N ▵RRM1/2, ▵N. (C) A cell lysate fractionation of Flag-TDP-12xQ/N F4L and Flag-TDP-12xQ/N ▵RRM1/2, ▵N stable cell lines. The soluble (S) and insoluble (P) fractions were separated by ultracentrifugation and the corresponding Flag-tagged proteins and endogenous TDP-43 were detected by western blot using an anti-TDP-43 antibody. (C) The distribution of endogenous TDP-43 between the P and S fractions under each condition from three independent experiments.
DISCUSSION
Nuclear factor TDP-43 plays a key role in several neurodegenerative diseases, principally ALS and FTLD (43–45). The most common features of these disorders, in fact, are represented by the presence of abnormal TDP-43 inclusions in the neurons of patients (46).
At the moment, however, it is still unclear how TDP-43 is involved in disease onset and progression. In particular, it is not known whether TDP-43 aggregates are toxic, protective or just a non-pathological epiphenomena (unlikely) since controversial data have been observed from both in vitro (16,29) and in vivo (30,47) models. What is very clear at this moment; however, is that lack of TDP-43 activity can have very serious consequences on the general cellular metabolism. In support of this conclusion, several different works have demonstrated that knockdown of TDP-43 causes in vivo neurodegeneration (48–50).
The Q/N region of TDP-43 has been shown to be involved in aggregate formation and in the interaction of TDP-43 with inclusions (15–17). Our original cellular model, based on the EGFP-12xQ/N construct, was based on 12 tandem repetitions of the core Q/N region (aa 339–369) of TDP-43 (16). Aggregates were preferentially cytoplasmic and had the capacity at least in part to trap exogenous full-length TDP-43 and at least part of the endogenous protein. However, these aggregates were neither toxic nor capable of inducing appreciable loss-of-function effects at the RNA splicing level when induced in different cell lines including primary neuronal cultures (16).
Now, we have generated a new TDP-43 cellular aggregation model that can efficiently induce prominent loss-of-function effects. This model was obtained by cloning 12xQ/N repetitions at the C-terminus of the WT TDP-43 protein. Most importantly, following formation of these aggregates, we were able to observe a clear loss-of-function effect both using a minigene approach and looking at the endogenous gene splicing pattern (POLDIP3/SKAR). This loss of TDP-43 function could be explained because of endogenous TDP-43 was able to interact fully with Flag-TDP-12xQ/N aggregates and to be totally sequestered in the insoluble aggregate. We also observed that the nuclei from some cell populations were almost completely depleted for both endogenous and exogenous TDP-43. The aggregates also presented a mixed distribution as the presence of TDP-43 inclusions was observed in cell cytoplasm, in nuclei or both, indicating that the loss of TDP-43 function was mostly due to the generation of a nonfunctional aggregated form of TDP-43 in any part of the cell.
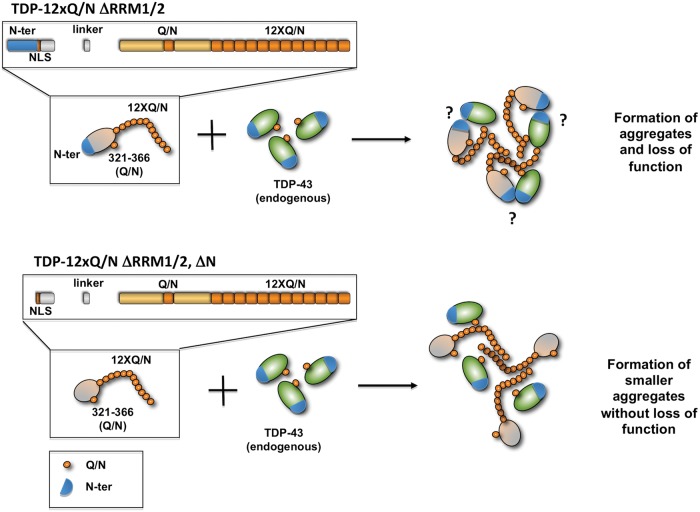
Our new model, schematically represented in Figure 7, also allowed us to study the importance of other TDP-43 domains in the protein aggregation process. First of all, we confirmed previous data indicating that the RNA-binding activity is not necessary for TDP-43 aggregation (16). In addition, we also observed that deletion of the 75 N-terminal residues of Flag-TDP-12xQ/N abolished the ability of the aggregates to efficiently induce loss-of-function effects. This result is particularly interesting as is consistent with previous studies which demonstrated that the N-terminal domain (residues 1–105) is involved in TDP-43 protein oligomerization in vitro (51) and in TDP-43 intermolecular interactions in cell culture (29,52). Most recently, the involvement of the N-terminal domain in TDP-43 aggregation was also proposed in a cellular aggregation model where TDP-43 full-length aggregates were triggered by disrupting the NLS signals (GFP-TDP-43NLSm) as previously described (27). In this case, however, it should be noted that mutating the NLS is just a way to increase the amount of TDP-43 production through its autoregulation process (6,7) by increasing the amount of this protein that is made by the cell and exported to the cytoplasm with minimal return to the nucleus. Furthermore, Zhang et al. (53) found that the deletion of residues 1–10 (GFP-TDP-4310-4014-NLSm) eliminated the interaction with full-length TDP-43 (MYC-TDP-43). A difference between the Zhang et al. model and ours is that, in our case, the elimination of the residues 1–75 from Flag-TDP-12xQ/N did not disrupt aggregate formation, but reduced the efficiency of interaction with endogenous TDP-43. These results indicate that the N-terminal portion of TDP-43 fulfills an important role in an aggregation context by mediating the interaction between endogenous TDP-43 and the aggregates. It should nonetheless be noted that the Co-IP results presented in Figure 6 show that removal of this region does not completely abolish the ability of the mutant to interact with endogenous TDP-43. This further strengthens the concept that TDP-43 oligomerization is mediated by several regions of this protein that co-operate to mediate this self-interaction whose functional significance remains still to be explored.
Figure 7.
A schematic model of how aggregation does not necessarily correlate with loss of function. In particular, loss of TDP-43 function occurs only when the endogenous TDP-43 is able to interact with very high efficiency with Flag-TDP-12xQ/N ▵RRM1/2 aggregates and to be totally sequestered in the insoluble aggregate. The question mark reflects the fact that we still do not know whether the Q/N region and the N-terminus interact with each other or only among themselves. On the other hand, deletion of the 75 N-terminal residues of TDP-43 in the Flag-TDP-12xQ/N ▵RRM1/2, ▵N mutant abolished the ability of the aggregates to efficiently induce loss-of-function effects, being much less able to sequester endogenous TDP-43.
In conclusion, the model developed in this work gives us valuable information on the structural determinants of TDP-43 loss of function through aggregation, thus opening the way to new strategies aimed to overcome the TDP-43 loss of function in neurodegenerative diseases. Most importantly, our data have uncovered an essential difference between aggregation per se and loss of function. In fact, much emphasis has been given to the aggregation process in most models, but not to functional consequences. Our observations have now filled this gap showing that obtaining some aggregation of endogenous TDP-43 is not enough to modify the RNA processing stages in which this protein is involved. This is not surprising as a reduction of TDP-43 returning to the nucleus can be compensated by the self-regulation mechanism that will increase TDP-43 production as necessary. In our case, however, the contribution of the N-terminus is critical to enhance the efficiency of the aggregates to trap endogenous TDP-43 in a nonfunctional insoluble form, and eventually overcome the capacity of the cell to compensate for a drop of TDP-43 nuclear levels.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
The authors are supported by grants from AriSLA (TARMA), Thierry Latran Foundation (REHNPALS) and the EU Joint Programme-Neurodegenerative Diseases JPND (RiMod-FTD, Italy, Ministero della Sanita’). Funding to pay the Open Access publication charges for this article was provided by ARISLA foundation through the TARMA grant.
Supplementary Material
REFERENCES
- 1.Buratti E., Baralle F.E. TDP-43: gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem. Sci. 2012;37:237–247. doi: 10.1016/j.tibs.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Ayala Y.M., Zago P., D'ambrogio A., Xu Y.-F., Petrucelli L., Buratti E., Baralle F.E. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 3.Buratti E., Baralle F.E. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 4.Buratti E., Brindisi A., Pagani F., Baralle F.E. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am. J. Hum. Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avendano-Vazquez S.E., Dhir A., Bembich S., Buratti E., Proudfoot N., Baralle F.E. Autoregulation of TDP-43 mRNA levels involves interplay between transcription, splicing, and alternative polyA site selection. Genes Dev. 2012;26:1679–1684. doi: 10.1101/gad.194829.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayala Y.M., De Conti L., Avendano-Vazquez S.E., Dhir A., Romano M., D'Ambrogio A., Tollervey J., Ule J., Baralle M., Buratti E., et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bembich S., Herzog J.S., De Conti L., Stuani C., Avendano-Vazquez S.E., Buratti E., Baralle M., Baralle F.E. Predominance of spliceosomal complex formation over polyadenylation site selection in TDP-43 autoregulation. Nucleic Acids Res. 2014;42:3362–3371. doi: 10.1093/nar/gkt1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhardwaj A., Myers M.P., Buratti E., Baralle F.E. Characterizing TDP-43 interaction with its RNA targets. Nucleic Acids Res. 2013;41:5062–5074. doi: 10.1093/nar/gkt189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo P.-H., Doudeva L.G., Wang Y.-T., Shen C.-K.J., Yuan H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackness B.C., Tran M.T., McClain S.P., Matthews C.R., Zitzewitz J.A. Folding of the RNA Recognition Motif (RRM) domains of the ALS-linked protein TDP-43 reveals an intermediate state. J. Biol. Chem. 2014;289:8264–8276. doi: 10.1074/jbc.M113.542779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lukavsky P.J., Daujotyte D., Tollervey J.R., Ule J., Stuani C., Buratti E., Baralle F.E., Damberger F.F., Allain F.H. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat. Struct. Mol. Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 12.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 13.D'ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y.M., Baralle F.E. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling S.-C., Albuquerque C.P., Han J.S., Lagier-Tourenne C., Tokunaga S., Zhou H., Cleveland D.W. ALS-associated mutations in TDP-43 increase its stability and promote TDP-43 complexes with FUS/TLS. Proc. Natl. Acad. Sci. USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y.M., Baralle F.E. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Budini M., Buratti E., Stuani C., Guarnaccia C., Romano V., De Conti L., Baralle F.E. Cellular model of TAR DNA-binding protein 43 (TDP-43) aggregation based on its C-terminal Gln/Asn-rich region. J. Biol. Chem. 2012;287:7512–7525. doi: 10.1074/jbc.M111.288720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuentealba R.A., Udan M., Bell S., Wegorzewska I., Shao J., Diamond M.I., Weihl C.C., Baloh R.H. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J. Biol. Chem. 2010;285:26304–26314. doi: 10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Budini M., Romano V., Avendano-Vazquez S.E., Bembich S., Buratti E., Baralle F.E. Role of selected mutations in the Q/N rich region of TDP-43 in EGFP-12xQ/N-induced aggregate formation. Brain Res. 2012;1462:139–150. doi: 10.1016/j.brainres.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 19.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 20.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 21.Lattante S., Rouleau G.A., Kabashi E. TARDBP and FUS mutations associated with amyotrophic lateral sclerosis: summary and update. Hum. Mut. 2013;34:812–826. doi: 10.1002/humu.22319. [DOI] [PubMed] [Google Scholar]
- 22.Buratti E., Baralle F.E. The molecular links between TDP-43 dysfunction and neurodegeneration. Adv. Genet. 2009;66:1–34. doi: 10.1016/S0065-2660(09)66001-6. [DOI] [PubMed] [Google Scholar]
- 23.Mackenzie I., Rademakers R., Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 24.Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T.G., Buratti E., Baralle F., Morita M., et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inukai Y., Nonaka T., Arai T., Yoshida M., Hashizume Y., Beach T.G., Buratti E., Baralle F.E., Akiyama H., Hisanaga S.-I., et al. Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Lett. 2008;582:2899–2904. doi: 10.1016/j.febslet.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 26.Lee E.B., Lee V.M.-Y., Trojanowski J.Q. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. FEBS Lett. 2012;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winton M.J., Igaz L.M., Wong M.M., Kwong L.K., Trojanowski J.Q., Lee V.M. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimoto Y., Ito D., Yagi T., Nihei Y., Tsunoda Y., Suzuki N. Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J. Biol. Chem. 2010;285:608–619. doi: 10.1074/jbc.M109.022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y.-J., Xu Y.-F., Cook C., Gendron T.F., Roettges P., Link C.D., Lin W.-L., Tong J., Castanedes-Casey M., Ash P., et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson B.S., McCaffery J.M., Lindquist S., Gitler A.D. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson B.S., Snead D., Lee J.J., Mccaffery J.M., Shorter J., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. Proc. Natl. Acad. Sci. USA. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Igaz L.M., Kwong L.K., Xu Y., Truax A.C., Uryu K., Neumann M., Clark C.M., Elman L.B., Miller B.L., Grossman M., et al. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 2008;173:182–194. doi: 10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cushman M., Johnson B.S., King O.D., Gitler A.D., Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J. Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mompean M., Buratti E., Guarnaccia C., Brito R.M., Chakrabartty A., Baralle F.E., Laurents D.V. Structural characterization of the minimal segment of TDP-43 competent for aggregation’. Arch. Biochem. Biophys. 2014;545:53–62. doi: 10.1016/j.abb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Pagani F., Buratti E., Stuani C., Baralle F. Missense, nonsense, and neutral mutations define juxtaposed regulatory elements of splicing in cystic fibrosis transmembrane regulator exon 9. J. Biol. Chem. 2003;278:26580. doi: 10.1074/jbc.M212813200. [DOI] [PubMed] [Google Scholar]
- 36.Mercado P.A., Ayala Y.M., Romano M., Buratti E., Baralle F.E. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Prot. Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saibil H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell. Biol. 2013;14:630–642. doi: 10.1038/nrm3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar P., Pradhan K., Karunya R., Ambasta R.K., Querfurth H.W. Cross-functional E3 ligases Parkin and C-terminus Hsp70-interacting protein in neurodegenerative disorders. J. Neurochem. 2012;120:350–370. doi: 10.1111/j.1471-4159.2011.07588.x. [DOI] [PubMed] [Google Scholar]
- 40.Udan-Johns M., Bengoechea R., Bell S., Shao J., Diamond M.I., True H.L., Weihl C.C., Baloh R.H. Prion-like nuclear aggregation of TDP-43 during heat shock is regulated by HSP40/70 chaperones. Hum. Mol. Genet. 2014;23:157–170. doi: 10.1093/hmg/ddt408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fiesel F.C., Weber S.S., Supper J., Zell A., Kahle P.J. TDP-43 regulates global translational yield by splicing of exon junction complex component SKAR. Nucleic Acids Res. 2012;40:2668–2682. doi: 10.1093/nar/gkr1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiga A., Ishihara T., Miyashita A., Kuwabara M., Kato T., Watanabe N., Yamahira A., Kondo C., Yokoseki A., Takahashi M., et al. Alteration of POLDIP3 splicing associated with loss of function of TDP-43 in tissues affected with ALS. PLoS ONE. 2012;7:e43120. doi: 10.1371/journal.pone.0043120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann M. Frontotemporal lobar degeneration and amyotrophic lateral sclerosis: molecular similarities and differences. Rev Neurol. 2013;169:793–798. doi: 10.1016/j.neurol.2013.07.019. [DOI] [PubMed] [Google Scholar]
- 44.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Langenhove T., van der Zee J., Van Broeckhoven C. The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Ann. Med. 2012;44:817–828. doi: 10.3109/07853890.2012.665471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geser F., Martinez-Lage M., Robinson J., Uryu K., Neumann M., Brandmeir N.J., Xie S.X., Kwong L.K., Elman L., McCluskey L., et al. Clinical and pathological continuum of multisystem TDP-43 proteinopathies. Arch. Neurol. 2009;66:180–189. doi: 10.1001/archneurol.2008.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gregory J.M., Barros T.P., Meehan S., Dobson C.M., Luheshi L.M. The aggregation and neurotoxicity of TDP-43 and its ALS-associated 25 kDa fragment are differentially affected by molecular chaperones in Drosophila. PLoS ONE. 2012;7:e31899. doi: 10.1371/journal.pone.0031899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iguchi Y., Katsuno M., Niwa J., Takagi S., Ishigaki S., Ikenaka K., Kawai K., Watanabe H., Yamanaka K., Takahashi R., et al. Loss of TDP-43 causes age-dependent progressive motor neuron degeneration. Brain. 2013;136:1371–1382. doi: 10.1093/brain/awt029. [DOI] [PubMed] [Google Scholar]
- 49.Wu L.S., Cheng W.C., Shen C.K. Targeted depletion of TDP-43 expression in the spinal cord motor neurons leads to the development of amyotrophic lateral sclerosis-like phenotypes in mice. J. Biol. Chem. 2012;287:27335–27344. doi: 10.1074/jbc.M112.359000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feiguin F., Godena V.K., Romano G., D'Ambrogio A., Klima R., Baralle F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 51.Chang C.-K., Wu T.-H., Wu C.-Y., Chiang M.-H., Toh E.K.-W., Hsu Y.-C., Lin K.-F., Liao Y.-H., Huang T.-H., Huang J.J.-T. The N-terminus of TDP-43 promotes its oligomerization and enhances DNA binding affinity. Biochem. Biophys. Res. Commun. 2012;425:219–224. doi: 10.1016/j.bbrc.2012.07.071. [DOI] [PubMed] [Google Scholar]
- 52.Shiina Y., Arima K., Tabunoki H., Satoh J. TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 2010;30:641–652. doi: 10.1007/s10571-009-9489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Y.J., Caulfield T., Xu Y.F., Gendron T.F., Hubbard J., Stetler C., Sasaguri H., Whitelaw E.C., Cai S., Lee W.C., et al. The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet. 2013;22:3112–3122. doi: 10.1093/hmg/ddt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang H.Y., Hou S.C., Way T.D., Wong C.H., Wang I.F. Heat-shock protein dysregulation is associated with functional and pathological TDP-43 aggregation. Nat. Commun. 2013;4:2757. doi: 10.1038/ncomms3757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.