Abstract
Cell fate can be controlled through asymmetric division and segregation of protein determinants. But the regulation of this process in the hematopoietic system is poorly understood. Here we show that the dynein binding protein Lis1 (Pafah1b1) is critically required for blood formation and hematopoietic stem cell function. Conditional deletion of Lis1 in the hematopoietic system led to a severe bloodless phenotype, depletion of the stem cell pool and embryonic lethality. Further, the loss of Lis1 accelerated cell differentiation, in part through defects in spindle positioning and inheritance of cell fate determinants. Finally, deletion of Lis1 blocked propagation of myeloid leukemia and led to a marked improvement in animal survival, suggesting that Lis1 is also required for oncogenic growth. These data identify a key role for Lis1 in hematopoietic stem cells, and mark the directed control of asymmetric division as a critical regulator of normal and malignant hematopoietic development.
A key question in biology is how cell fate decisions are regulated and how disruption of this regulation can lead to cancer. One fundamental mechanism that controls fate is asymmetric division, which involves the polarized distribution of determinants within the mother cell and their unequal inheritance by each daughter cell. Such asymmetric division allows one daughter to become differentiated and the other to retain an immature fate; in contrast, symmetric division allows both daughters to adopt equivalent fates. Studies in invertebrates such as Drosophila melanogaster have elucidated the major steps involved in asymmetric division, which include establishment of polarity, localization of fate determinants, and orientation of the mitotic spindle. A key regulator of this process is Lis1, a dynein binding protein that anchors the mitotic spindle to the cellular cortex1,2. By determining the orientation of the spindle, Lis1 ensures that the proper cleavage plane is established during cell division, and thus allows correct inheritance of fate determinants by daughter cells.
While the regulation of asymmetric cell division in invertebrates is well understood, relatively little is known about how it influences hematopoietic development and even less about its role in malignancy. Previous work from our lab and others has shown that hematopoietic stem and progenitor cells can undergo both symmetric and asymmetric division3–5. These findings were supported by more recent studies indicating that genetic modulation of fate determinants4,6–10 can affect hematopoietic stem cell (HSC) function. But how inheritance of fate determinants is controlled during asymmetric division, and whether disruption of this process can affect hematopoietic cell fate and tumorigenesis in vivo, remain unknown. Here we have addressed these questions by focusing on Lis1, and show that its genetic loss triggers an inability to maintain the stem cell state in both normal and malignant hematopoiesis. Conditional deletion of Lis1 in hematopoietic cells leads to a dramatic bloodless phenotype, impaired stem cell function, and depletion of the stem cell pool. Mechanistically, loss of Lis1 in stem cells does not appear to influence proliferation or apoptosis, but leads to accelerated differentiation. At a molecular level, fate determinants such as Numb are properly polarized, but their inheritance is impaired, with more frequent segregation to one daughter driving a rise in asymmetric divisions. We also examined the role of Lis1 in cancer to gain a better understanding of whether and how asymmetric division controls oncogenesis and to define new signals that may be targets of therapy. Using mouse models and patient samples of aggressive leukemias we found that Lis1 is critical for the growth and propagation of blast crisis Chronic Myelogenous Leukemia (bcCML) and therapy-resistant de novo Acute Myelogenous Leukemia (AML). These data show that Lis1 plays a crucial role in the establishment of the hematopoietic system and controls normal and malignant stem cell function.
Results
Loss of Lis1 leads to a bloodless phenotype
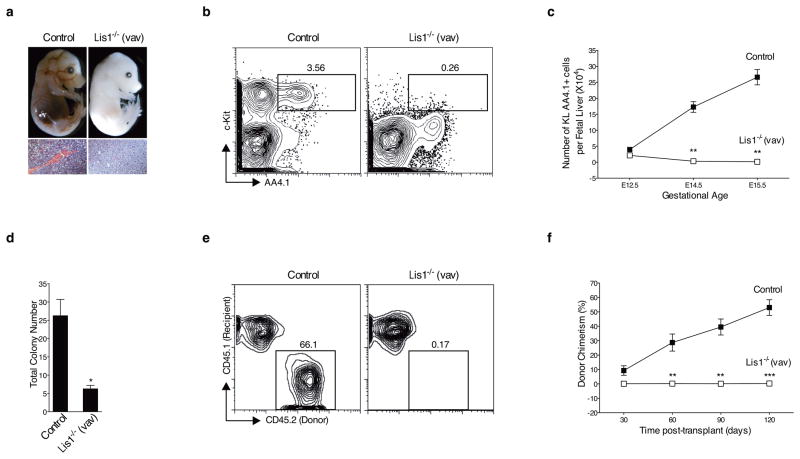
To study the role of Lis1 in the hematopoietic system, we generated mice in which a Lis1 floxed allele11 was conditionally deleted by Cre recombinase under the control of the Vav promoter (Lis1f/f; Vav-Cre)12–14. This approach led to loss of Lis1 expression in hematopoietic cells and enabled assessment of Lis1’s role in establishment of the hematopoietic system (Supplementary Fig. 1). Of 344 viable progeny obtained, none of the 86 expected Lis1−/− mice were born. In a retrospective analysis, we found that loss of Lis1 led to a striking bloodless phenotype, indicative of severe anemia, at E14.5 (Fig. 1a). Subsequently, loss of Lis1 led to lethality between E15.5–E18.5 (Supplementary Table 1). Histologically, Lis1 deletion led to a loss of hematopoietic cells (Fig. 1a) and a ~13.5-fold reduction in the frequency of HSCs (c-Kit+ Lin− AA4.1+ or KL AA4.1+ cells; Fig. 1b) in the fetal liver. Importantly, the 7-fold expansion of HSCs that normally occurs between E12.5–E15.5 and leads to the generation of a functional hematopoietic system (Fig. 1c, solid squares) failed to occur in the absence of Lis1 (Fig. 1c, open squares).
Figure 1. Genetic deletion of Lis1 impairs establishment of the hematopoietic system during embryonic development.
(a) Representative image of Control (Lis1f/+, upper left) and Lis1−/− (Lis1f/f; Vav-Cre, upper right) littermates at E14.5. Hematoxylin and eosin (H&E) stain of fetal liver from Control (bottom left) and Lis1−/− (bottom right) littermates at E14.5. 40X. Scale bars, 55μm. (b) Fetal liver cells from Control (Lis1f/+) and Lis1−/− mice were analyzed for frequency of HSCs (cKit+ Lin− AA4.1+; KL AA4.1+). Dot plots are shown for representative Control (left) and Lis1−/− (right) E14.5 embryos. (c) Absolute number of HSCs (KL AA4.1+) from Control (Lis1f/+ or Lis1f/f; solid squares) or Lis1−/− (open squares) mice at different gestational ages; n=3–5 mice for each genotype for each gestational age; **P=0.0014 for E14.5, **P=0.0018 for E15.5. (d) Number of colonies generated from Control (Lis1f/+ or Lis1f/f) and Lis1−/− fetal liver. Cells are isolated from 3–6 embryos of each genotype; *P=0.0110 (n=3, technical replicates). (e) Representative FACS profile of donor chimerism (4 months) in CD45.1+ recipients transplanted with HSC-enriched cells (Lin− AA4.1+) from either Control (Lis1f/+) or Lis1−/− E14.5 embryos. (f) Average donor chimerism at different times post-transplantation (2–4 donor mice were used for each genotype and 4–6 recipient mice in each cohort). Control: solid squares; Lis1−/−: open squares; **P=0.0088 for 60 days, **P=0.0021 for 90 days and ***P=0.0006 for 120 days. Error bars show standard error of the mean (SEM).
To determine whether the failure of HSC expansion in vivo was linked to functional defects in HSCs we assessed colony formation in methylcellulose cultures. Loss of Lis1 led to a 3-fold reduction in total colony formation; the fact that the colonies formed were similar between wild type and Lis1-deficient cells indicated that differentiation potential was unaffected (Fig. 1d and Supplementary Fig. 2). Further, transplantation of wild type HSC-enriched cells (Lin− AA4.1+) led to ~53% donor chimerism in the recipient mice at 4 months post-transplant, while no chimerism (0%) was detected in mice reconstituted with Lis1-deficient cells (Fig. 1e, f), suggesting that loss of Lis1 affects fetal HSC function. Unsorted whole fetal liver transplants also showed a loss of chimerism, indicating that Lis1 deletion affected functional HSCs and is unlikely to have simply changed their phenotype (data not shown).
Lis1 is required for adult hematopoietic stem cell function
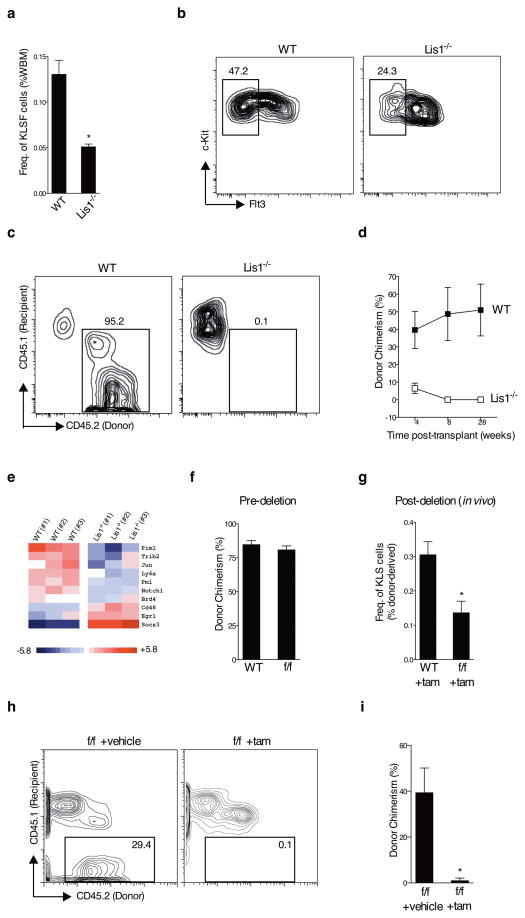
To determine if Lis1 has a conserved functional role in the adult hematopoietic system we crossed the floxed Lis1 mice to mice harboring a Cre-ERT2 allele targeted to the ubiquitously expressed ROSA26 locus15 (denoted hereafter as Lis1f/fcreER mice). Tamoxifen delivery allowed effective temporal control over Lis1 deletion (Supplementary Fig. 3a, b) and a clear reduction in Lis1 mRNA expression in adult bone marrow HSC-enriched cells (cKit+ Lin− Sca1+ or KLS; Supplementary Fig. 3b, c). Consistent with this, Lis1 protein expression was reduced in highly enriched HSCs (KLS CD48−, Supplementary Fig. 3d). Loss of Lis1 led to a significant reduction in the frequency and absolute number of adult HSCs (Fig. 2a, b and Supplementary Fig. 4). HSC defects preceded any changes in differentiated cells, suggesting that HSC maintenance is directly affected by loss of Lis1 (Supplementary Fig. 5). Adult HSC function was also affected by the deletion of Lis1; while transplanted HSCs from control mice led to increasing donor chimerism from 39% to 51%, mice reconstituted with Lis1-deficient cells showed a gradual loss in donor chimerism from ~6.5% to 0% (Fig. 2c, d). Genome wide expression analysis of HSC-enriched cells from control and Lis1 null mice revealed a highly significant loss of core genes that form the stem cell signature16–18 (Supplementary Fig. 6), as well as changes in genes such as Pim1 and Socs3 that play important regulatory roles in hematopoietic stem/progenitor cell maintenance and differentiation (Fig. 2e). That loss of the stem cell signature is a key downstream consequence of Lis1 deletion confirms, through an independent molecular strategy, that Lis1 is critical to maintenance of the stem cell state.
Figure 2. Lis1 is cell-autonomously required for adult hematopoietic stem cell self-renewal.
(a) Average frequency of HSCs (KLSFlt3−, KLSF) in whole bone marrow from tamoxifen-treated control (Lis1+/+; Rosa-creER, indicated as WT) and tamoxifen-treated (Lis1f/f; Rosa-creER (indicated as Lis1−/−) mice; n=4 for control (WT), n=3 for Lis1−/−; *P=0.0140. (b) Representative FACS plots of HSCs (KLSF) from WT and Lis1−/− mice. (c) Repopulation efficiency of Lis1−/− HSCs. Representative FACS plots shows donor chimerism (CD45.2+ cells) in recipients transplanted with HSCs (KLS CD150+ CD48−) from WT or Lis1−/− mice. FACS analysis was performed 28 weeks post-transplantation. (d) Average donor chimerism at different times after transplantation (4–5 mice per cohort). WT is shown with solid squares and Lis1−/− is shown with open squares. (e) Genome wide expression analysis of Lis1-deficient HSC-enriched cells. Heat map of known regulators of stem and progenitor cell activity significantly affected by the loss of Lis1. (f–i) Lis1 chimeras with hematopoietic-specific Lis1 deletion, (f) Donor chimerism prior to tamoxifen (tam) treatment was assessed two months post-transplantation. (WT) indicates control Lis1+/+; Rosa-creER and (f/f) indicates Lis1f/f; Rosa-creER transplanted mice (5 mice in each cohort). (g) Frequency of donor-derived KLS cells in chimeric mice post-deletion. ((WT) +tam) indicates mice that received donor cells from Lis1+/+; Rosa-creER and ((f/f) +tam) indicates mice that received donor cells from Lis1f/f; Rosa-creER mice; n=3 for each cohort, *P=0.0277. (h–i) Repopulation ability of whole bone marrow (WBM) cells isolated from Lis1 chimera mice. (h) Representative FACS plots show donor chimerism (CD45.2+ cells) in recipients that received cells from either control (f/f +vehicle) or (f/f +tam) Lis1 chimeras. (i) Average donor chimerism at 16 weeks post-transplantation (n=3–4 recipients per cohort; *P=0.0369).
Because Cre activation was under the control of a ubiquitous promoter, it was possible that deletion of Lis1 in non-hematopoietic tissues contributed to impaired maintenance of HSCs. To address this, we created chimeras in which only hematopoietic cells harbored the floxed allele and the microenvironment remained wild-type. HSCs from untreated control Lis1+/+creER or Lis1f/fcreER mice were transplanted (Supplementary Fig. 7); following donor repopulation (Fig. 2f), chimeras were treated with tamoxifen to delete Lis1 specifically in the hematopoietic system. This led to a significant reduction in the frequency of HSC-enriched cells (Fig. 2g). Donor-derived whole bone marrow from both control and Lis1-deficient chimeras was re-transplanted to test stem cell function. While the average chimerism from control cells was 53.5%, chimerism from Lis1 null cells was nearly absent (0.3% percent, Fig. 2h, i), roughly recapitulating the phenotype of non-chimeric Lis1 null mice. These data suggest that adult Lis1-deficient HSCs have a cell-autonomous functional defect in vivo.
Lis1 deletion impairs inheritance of fate determinants
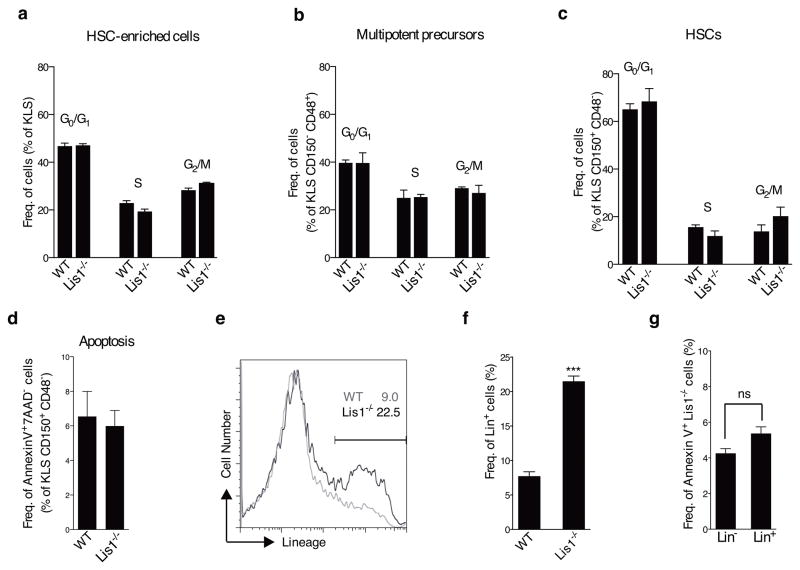
To understand how stem cells are lost in the absence of Lis1, we first examined proliferation and apoptosis. Lis1-deficient HSCs (KLS CD150+ CD48−), HSC-enriched cells (KLS) and multipotent precursors (KLS CD150− CD48+) incorporated BrdU at a rate similar to those from wild type mice and displayed a normal cell cycle distribution (Fig. 3a–c and Supplementary Fig. 8). In contrast, differentiated cells showed decreased proliferation (data not shown), indicating that Lis1 may have context-specific effects at distinct stages of development, consistent with observations in the nervous system2. Lis1-deficient HSCs also had normal frequencies of apoptotic cells (Fig. 3d and Supplementary Fig. 9). The fact that HSC depletion occurred as early as day 3 after Lis1 deletion when no changes in cell survival were observed suggested that apoptosis is unlikely to account for HSC loss. Some increase in necrosis (Supplementary Fig. 9) could contribute to the overall phenotype; however, the fact that it occurs after the HSC depletion is initiated suggested that loss of Lis1 may influence stem cells through other mechanisms.
Figure 3. Lis1 deficiency leads to accelerated differentiation of hematopoietic stem cells.
(a–c) Cell cycle status of hematopoietic cells following Lis1 deletion. Control (Lis1+/+; Rosa-creER; WT) and Lis1f/f; Rosa-creER (Lis1−/−) mice were treated with tamoxifen and cell cycle analyzed after BrdU incorporation. Average frequency of KLS (a), KLS CD48+ CD150− (b), and KLS CD150+ CD48− (c) in G0/G1, S, and G2/M cell cycle phases in control (WT) and Lis1−/− mice. Data shown are from two independent experiments (n=2–3 per cohort). (d) Percentage of HSCs (KLS CD150+ CD48−) undergoing apoptosis (AnnexinV+ 7AAD−) in control (WT) and Lis1−/− mice. Data shown are from three independent experiments (n=2–3 per cohort). (e) Analysis of rate of differentiation of Lis1−/− cells. KLS cells from control (Lis1+/+; Rosa-creER; WT) and Lis1f/f; Rosa-creER (Lis1−/−) mice were treated with tamoxifen in vitro. Representative FACS plot shows frequency of cells expressing lineage markers in WT (shown in gray) and Lis1−/− (shown in black) populations 24 hours post-deletion. (f) Average frequency of cells expressing lineage markers (Lin+) in WT and Lis1−/− cells. Data shown are from three independent experiments; ***P=0.0002. (g) Analysis of apoptosis in Lin− and Lin+ fraction of Lis1−/− cells. Percentage of Annexin V+ cells is shown 24 hours post-deletion. Data shown are from two independent experiments. Error bars show the standard error of mean (SEM).
To test whether depletion of HSCs resulted from defects in maintenance of the undifferentiated state, we tracked the rate of differentiation of Lis1-deficient cells. HSC-enriched cells from either Lis1f/fcreER or control Lis1+/+creER mice were treated with tamoxifen at t=0, and their differentiation monitored (Supplementary Fig. 10). Over 24 hours ~23% of Lis1−/− cells became positive for lineage markers (Lin+) while only ~9% of control cells became positive for lineage markers (Lin+) (Fig. 3e, f). Importantly, the increase in differentiation after Lis1 deletion was not due to preferential death of immature cells (Lin−) (Fig. 3g).
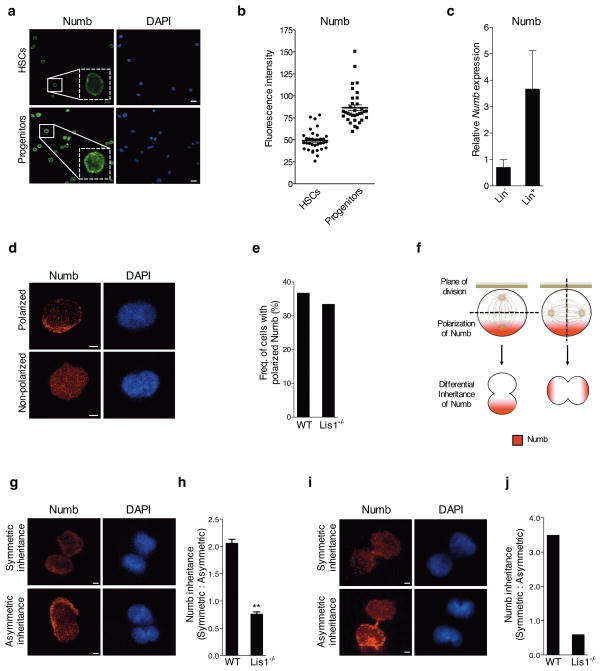
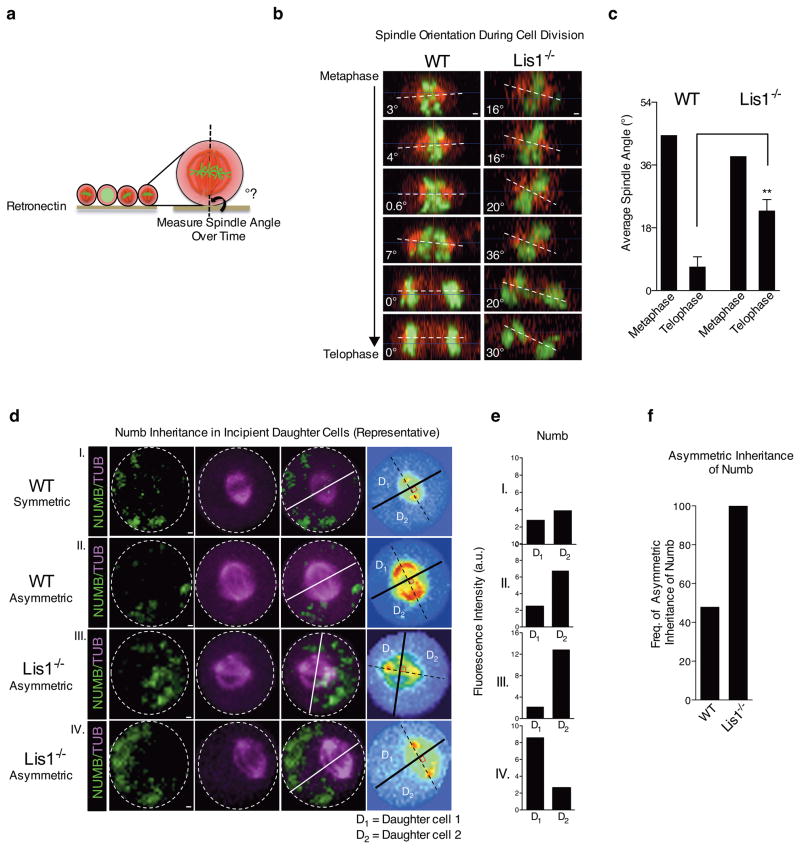
Since accelerated differentiation can be a consequence of defects in asymmetric division, we examined whether the Lis1 loss led to altered polarization of fate determinants within mother cells or altered inheritance of these determinants by daughter cells. Numb is an important fate determinant that marks differentiated cells (Fig. 4a–c) and that can accelerate differentiation upon ectopic expression5; conversely, Numb inhibition can also sustain cells in an undifferentiated state (Supplementary Fig. 11). We thus tracked the polarization and inheritance of Numb in HSC-enriched cells. Numb was distributed evenly in 63% and polarized in 37% of cells. The absence of Lis1 did not affect Numb distribution (Fig. 4d, e and Supplementary Fig. 12a). In cells with polarized Numb, changes in the plane of division can lead to equal or unequal inheritance by the two daughters (Fig. 4f). To analyze changes in Numb inheritance, HSC-enriched cells from control or floxed Lis1 mice were treated with tamoxifen in vitro (Supplementary Fig. 12a). Following Lis1 deletion, cells were stained for Numb and the ratio of symmetric to asymmetric divisions determined. Only cells in late telophase or undergoing cytokinesis were tracked to assess Numb inheritance in incipient daughter cells. Cells either displayed equivalent distribution of low levels of Numb to both daughters (Fig. 4g, symmetric) or unequal distribution, i.e. higher levels of Numb to one daughter and lower levels to the other (Fig. 4g, asymmetric). Control cells displayed two-fold more symmetric inheritance relative to asymmetric inheritance of Numb. In contrast, Lis1 loss led to a complete reversal in the pattern of inheritance, with two-fold more cells undergoing asymmetric divisions (Fig. 4h).
Figure 4. Loss of Lis1 impairs inheritance of fate determinants in hematopoietic development.
(a) Expression of Numb in HSCs and progenitor cells. Representative image with magnified inlay (dotted white box) shows HSCs (KLS CD48− CD150+) and progenitor cells (KLS CD48+) stained with anti-Numb antibody (green) and DAPI (blue), 63X. Scale bars, 10μm. (b) Average fluorescence intensity of Numb in individual HSCs and progenitor cells. Data shown are from two independent experiments (n=34 cells for each genotype); ****P<0.0001. (c) Realtime RT-PCR analysis of Numb expression in Lin− and Lin+ cells (n=2 independent experiments). (d) Representative images of individual HSC-enriched cells with polarized or non-polarized Numb (Numb in red, DAPI in blue, zoomed 63x images, Scale bars, 2.5μm). (e) Frequency of control (WT) or Lis1−/− cells with polarized Numb. Frequencies were determined out of 100 tracked cells for each genotype. (f) Model illustrates how two dividing cells may equivalently polarize Numb (shown in red) to one side of the cell, yet direct the cleavage plane in such a way to ensure either equal or unequal inheritance of Numb into the incipient daughter cells. (g) Representative image of a tracked cell inheriting Numb symmetrically (top) or asymmetrically (bottom) into incipient daughter cells (Numb in red, DAPI in blue, zoomed 63x images, Scale bars, 2.5μm). (h) Relative ratios of symmetric:asymmetric division in vitro. Data shown are from two independent experiments; n=25–27 dividing cells were assessed for each experiment per cohort; **P=0.0032. (i) Representative image of symmetric (top) and asymmetric (bottom) inheritance of Numb by incipient daughter cells in vivo (Numb in red, DAPI in blue, zoomed 63x images, Scale bars, 2.5μm). (j) Relative ratio of symmetric:asymmetric division in vivo (nine dividing cells were assessed for the control (WT) group; eight dividing cells were assessed for the Lis1−/− group. Data analyzed using three independent chimeric mice for each genotype).
We tested if this shift in division pattern occurred after Lis1 deletion in vivo. Due to the limited numbers of telophase HSCs in vivo we focused on lineage-negative cells, a less enriched but nonetheless immature population. We analyzed Numb inheritance in incipient daughters of Lis1 null or control lineage-negative (Lin−) cells (Supplementary Fig. 12b). Consistent with our in vitro analysis, control cells underwent symmetric divisions 3.5 times more frequently, and the loss of Lis1 led to a predominance of asymmetric divisions. This shift led to a seven-fold difference in the ratio of symmetric:asymmetric divisions between control and Lis1-deficient cells (Fig. 4i, j). Because Lis1 deletion affected Numb inheritance but not Numb polarization, these data cumulatively suggest that the absence of Lis1 affects inheritance by changing the cleavage plane (Fig. 4f), thereby generating more cells that have higher levels of Numb.
Lis1 controls spindle orientation in hematopoietic stem cells
To directly test whether loss of Lis1 leads to changes in the cleavage plane and if this results from defects in spindle positioning, we developed a strategy to image spindle orientation during cell division in real time. This imaging approach was a modification of a method previously used to visualize the spindle in epithelial lines19. Cells were infected with H2B-GFP20 to mark separating chromosomes and mCherry-α-tubulin21 to mark the spindle (Supplementary Fig. 13 and Supplementary Video 1). Cells were plated on retronectin and imaged; 4 dimensional movies (x,y,z,t) of dividing cells were visualized from the side to measure the spindle angle relative to the substrate. While a range of spindle angles were seen in metaphase, the spindle always positioned parallel (0–10°) to the substrate in telophase, consistent with previous reports19. This substrate directed re-positioning of the mitotic spindle allowed us to test whether Lis1 controls spindle orientation in hematopoietic cells. Control HSC-enriched cells displayed a range of angles during metaphase, but re-positioned their spindles by telophase (Fig. 5a–c). In contrast, Lis1 deficient cells were unable to correctly position their spindle (Fig. 5a–c). These data suggest that Lis1 loss leads to defects in spindle orientation in HSC-enriched cells.
Figure 5. Loss of Lis1 impairs spindle orientation in hematopoietic development.
(a) Schematic showing how spindle angle is measured relative to the retronectin base. (b) Representative side view images of a control (WT) and Lis1−/− cell undergoing cell division and their spindle angles. Scale bars, 1μm. (c) Average metaphase and telophase spindle angles of control (WT) and Lis1−/− cells relative to substrate; data shown are from three independent experiments; n=5–7 cells per genotype; **P=0.0054. (d) Numb distribution in dividing HSC-enriched cells relative to mitotic spindle orientation. Representative images of control (WT) cells (I and II) or Lis1−/− cells (III and IV) with examples of symmetric (I) or asymmetric (II, III, IV) inheritance of Numb by incipient daughter cells. Numb (green), α-tubulin (magenta); representative videos of a cell undergoing symmetric or asymmetric cell division are shown in Supplementary Videos 2–4. Scale bars, 1μm. On far right panel, each cell is displayed in spectrum color format to facilitate accurate identification of spindle position (dotted black line connecting the two centrosomes highlighted in red) and the cleavage furrow (solid lines; white and black) which partitions the dividing cell into incipient daughter 1 (D1) and daughter 2 (D2). (e) Quantification of fluorescence intensity of Numb in D1 and D2 for each representative control (WT; I and II) or Lis1−/− cell (III, IV) shown in (d). (f) Frequency of cells undergoing asymmetric inheritance of Numb in control (WT) or Lis1−/− cells; data are shown for four independent experiments; n=23 cells for WT and n=9 cells for Lis1−/−. All error bars show the standard error of mean (SEM).
Finally, we tested if the spindle orientation defects driven by Lis1 deficiency leads to the improper inheritance of Numb. Thus we tracked the orientation of the spindle coordinately with Numb inheritance in real time. HSC-enriched cells were infected with mCherry-α-tubulin and Numb-CFP fusion vectors and Numb inheritance tracked relative to the mitotic spindle using time-lapse microscopy (Supplementary Videos 2–4). Of the cells entering mitosis, we focused on those with polarized Numb since changes in the spindle angle would affect whether Numb is inherited asymmetrically or symmetrically only in these cells (i.e. non-polarized cells should invariably undergo symmetric division regardless of spindle orientation). Live imaging of control and mutant primary stem and progenitor cells yielded clear and distinct patterns. While the mitotic spindle was positioned such that Numb was bisected asymmetrically in 56.5% of wild type cells, the mitotic spindle bisected Numb asymmetrically in 100% of the Lis1 null cells (Fig. 5d–f). The functional consequence of spindle positioning on stem cells was tested by ectopic expression of Nde1, a protein that independently controls spindle orientation22. This conferred a partial but significant rescue of the accelerated differentiation seen in Lis1-deficient stem cells (Supplementary Fig. 14). These data suggest that defective spindle positioning in the absence of Lis1 increases asymmetric inheritance of Numb and accelerates differentiation, and identify these changes as a mechanism that underlies, at least in part, the HSC depletion observed.
Although the absence of Lis1 affects spindle orientation, it may also affect other aspects of stem cell function. Because Lis1 is linked to spindle assembly, we tested if Lis1 deficiency affected bipolar spindle formation, spindle morphology and nuclear envelope breakdown and were unable to identify any obvious defects (Supplementary Fig. 15). In addition, no significant changes in mitotic duration were observed in the absence of Lis1 (Supplementary Fig. 15). However, a rise in the number of cells with abnormal mitoses (multipolar or incomplete) did occur (Supplementary Fig. 15). It is thus possible that, in addition to defects in spindle orientation and the inheritance of fate determinants, some loss of cells with abnormal mitoses (possibly linked to late onset necrosis as seen in Supplementary Fig. 9d) contributes to the overall defects observed in the absence of Lis1.
Lis1 is required for mouse and human myeloid leukemia
While proper regulation of the stem cell state is a critical feature of normal development, aberrant adoption of stem cell programs can be a hallmark of oncogenesis23. Whether regulators of spindle orientation and division plane can contribute to cancer is an important question that remains unaddressed. This may be particularly relevant for understanding the regulation of immature cancers and cancer stem cells since a shift toward symmetric renewal divisions could contribute to the failure of differentiation and maintenance of a stem cell state. In the hematopoietic system, acute phase myeloid leukemias such as blast crisis CML (bcCML) and de novo AML display a severe differentiation blockade. We thus used these as models to test whether Lis1 could play a role in blood cancers.
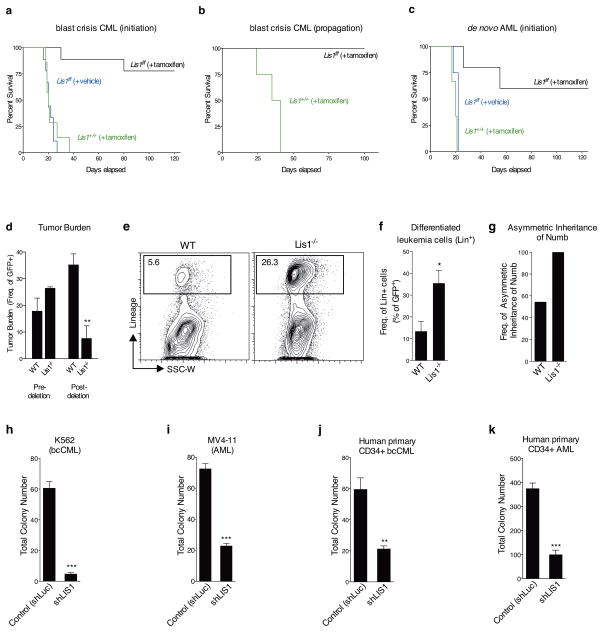
To generate bcCML, HSCs from Lis1+/+creER or Lis1f/fcreER mice were co-infected with BCR-ABL and NUP98-HOXA924–26 and transplanted into sub-lethally irradiated mice, and treated with tamoxifen or corn oil. While all tamoxifen-treated mice transplanted with control BCR-ABL/NUP98-HOXA9-infected cells succumbed to bcCML, only 22% of the tamoxifen-treated mice transplanted with Lis1f/fcreER BCR-ABL/NUP98-HOXA9-infected cells developed leukemia (Fig. 6a). The ability to temporally control Lis1 loss also allowed us to delete Lis1 after disease establishment. Established bcCML cells with a Lis1f/f allele were transplanted into recipients, and treated with tamoxifen seven days later. Whereas all mice transplanted with control leukemia-propagating cells succumbed to leukemia, none of the mice transplanted with cells that conditionally lost Lis1 developed leukemia (Fig. 6b). This indicated that Lis1 is critically important for both the establishment and the continued propagation of bcCML. Importantly, a similar impairment of leukemia growth occurred in de novo AML induced by co-expression of the human mixed-lineage leukemia fusion gene (MLL-AF9) and NRASG12V (ref. 27). While all control mice died of leukemia within 3 weeks, only ~40% of mice transplanted with cells that conditionally lost Lis1 developed AML, and those that did exhibited a longer disease latency (Fig. 6c).
Figure 6. Loss of Lis1 impairs the development and propagation of myeloid leukemia in mouse models and human leukemia cells.
(a) Impact of loss of Lis1 on bcCML initiation. KLS cells from control (Lis1+/+; Rosa-creER) and Lis1f/f; Rosa-creER mice transduced with BCR-ABL-IRES-YFP and NUP98-HOXA9-IRES-GFP were transplanted into recipients, treated with tamoxifen and survival monitored. Data shown are from three independent experiments (n=9 for Lis1f/f +tamoxifen (black), n=9 for Lis1f/f +vehicle (blue) and n=7 for Lis1+/+ +tamoxifen (green)) (b) Impact of loss of Lis1 on propagation of established bcCML. Lin-negative cells from established control Lis1+/+creER or Lis1f/fcreER bcCML were transplanted into secondary recipients. Recipients were administered tamoxifen and survival monitored (n=4 for Lis1+/+, green and n=3 for Lis1f/f, black). (c) Impact of loss of Lis1 on de novo AML. KLS cells were isolated from control (Lis1+/+; Rosa-creER) and Lis1f/f; Rosa-creER mice and co-transduced with MLL-AF9-IRES-GFP and NRASG12V-IRES-YFP, treated with tamoxifen or corn oil and survival was monitored. Data shown are from two experiments (n=5 for Lis1f/f +tamoxifen (black), n=4 for Lis1f/f +vehicle (blue) and n=3 for Lis1+/+ +tamoxifen (green)). (d–f). Growth and cellular behavior of bcCML in vivo. Lineage-negative (Lin−) cells from control Lis1+/+; Rosa-creER or Lis1f/f; Rosa-creER established bcCML were transplanted into secondary recipients and (d) average tumor burden was tracked before (% of peripheral blood) and after (% of spleen cells) tamoxifen delivery; n=4 for control (WT) and n=3 for Lis1−/− mice; **P=0.0065. (e) Representative FACS plots show frequency of bcCML cells expressing lineage markers in control (WT) or Lis1−/− populations 1 day post-tamoxifen treatment. (f) Average frequency of bcCML cells expressing lineage markers (Lin+). Data shown are from two independent experiments (n=4 for WT and n=6 for Lis1−/−, *P=0.0282). (g) Frequency of leukemia cells undergoing asymmetric inheritance of Numb in control (WT) or Lis1−/− cells; n=22 cells for WT and n=7 cells for Lis1−/−. (h–k) Influence of Lis1 loss on human leukemia growth. Human leukemia cells were infected with either control (shLuc) or lentiviral shRNA targeting human LIS1 (shLIS1). Subsequently, infected cells were sorted and plated in methylcellulose. Colony formation was assessed in K562 blast crisis CML cells (h), MV4-11 AML cells (i), Imatinib, Nilotinib and Dastinib-resistant primary human CD34+ bcCML (j) and primary human CD34+ AML (k); One patient sample was tested for each leukemia with n=3 technical replicates, *P<0.05, **P<0.01, ***P<0.001. Error bars represent standard error of the mean (SEM).
To understand the cellular and molecular impact of Lis1 deletion on leukemogenesis, we used the bcCML model. Monitoring GFP+ leukemia cells, we found that deletion of Lis1 well after the tumor burden had begun to climb allowed complete reversion to normal cell counts and resolution of disease (Fig. 6d and Supplementary Fig. 16a, b). At a cellular level, the most notable and immediate impact of Lis1 deletion was a 5-fold increase in the number of differentiated leukemic cells (Fig. 6e, f), accompanied by a rise in Numb expression (Supplementary Fig. 16c, d). In addition, real time imaging revealed that while Numb was bisected asymmetrically in 54.5% of control leukemia cells, it was bisected asymmetrically in 100% of Lis1-null cells (Figure 6g, Supplementary Fig. 17). This suggested that incorrectly directed Numb inheritance could be a possible basis for the increased Numb expression and differentiation observed in Lis1-deficient leukemia cells. In addition, Lis1 loss led to a 1.5 fold reduction in proliferation; thus the differentiation and proliferation defects may act together to lead to the severe defects observed in leukemogenesis. Consistent with our observations in normal hematopoietic stem cells, the loss of Lis1 in leukemic cells did not cause significant defects in apoptosis (data not shown).
To test whether Lis1 is also required for human myeloid leukemia, we deleted LIS1 in leukemic cell lines and primary patient samples. The bcCML cell line K562 and the de novo AML cell line MV4-11 were infected with shRNA targeting LIS1, and colony-forming ability was measured. Knockdown of LIS1 led to a significant reduction in colony-forming ability of both leukemia lines (Fig. 6h, i and Supplementary Fig. 18). LIS1’s role in primary human leukemia was tested by infecting patient derived CD34+ bcCML cells resistant to the tyrosine kinase inhibitors imatinib, nilotinib and dasatinib as well as CD34+ AML patient cells harboring the therapy-resistant MLL-AF9 translocation with shLIS1, and assessing colony formation. As shown (Fig. 6j, k and Supplementary Fig. 18), inhibition of LIS1 expression led to a significant blockade of colony-forming ability in both cancers. To understand the basis of the decrease in colony formation we analyzed the consequence of LIS1 knockdown on growth and differentiation. Inhibition of LIS1 in primary patient AML samples did not affect cell growth in the short term (24–72 hours) but led to accelerated differentiation as indicated by the increased frequency of MAC-1 expressing cells (Supplementary Fig. 19). Because viability of primary myeloid leukemia patient samples decreases significantly after short term culture, longer term analysis of proliferation was carried out in cell lines; inhibition of LIS1 led to a decease in cell numbers over a period of 12 days (data not shown). These data collectively suggest that inhibition of LIS1 increases differentiation in the short term and blocks growth in the longer term (either as a consequence of, or independent of, differentiation), and identifies LIS1 as a critical new regulator of human leukemia propagation.
Discussion
The studies described here show that Lis1 is critically required for development of the hematopoietic system. Its loss leads to a bloodless embryo and severe defects in hematopoietic stem cell maintenance and expansion in both fetal and adult life. Such an impact on fetal hematopoiesis has been previously reported largely for key transcription factors such as Runx1/Aml1, Scl/Tal-1 and Gata228–31. In this context, Lis1’s influence on the hematopoietic system implicates proteins that direct asymmetric division as a new class of regulators of hematopoiesis.
Our data indicate that a predominant genomic consequence of Lis1 deletion is loss of the stem cell core gene signature, suggesting that Lis1 is critical for the maintenance of the stem cell state. How Lis1 deletion leads to a loss of the stem cell state could potentially be explained by defects in the inheritance of cell fate determinants. As depicted in the model (Supplementary Fig. 20), if loss of Lis1 leads to incorrect spindle positioning and thus an increase in asymmetric division, it would generate more cells that inherit high levels of Numb. This would in turn generate an increased number of differentiated cells with each division. Thus if more differentiated cells comprise a greater fraction, and the undifferentiated stem cells comprise a smaller fraction, of the KLS population used in the array analysis, this may lead to the reduction or loss of the stem cell signature observed. It is also possible that Lis1 affects the stem cell core signature through as yet unknown mechanisms that are unrelated to its role in spindle positioning and asymmetric division.
Our results show the stem cell defects that occur in the absence of Lis1 are linked to increased inheritance of Numb and a marked imbalance in asymmetric and symmetric divisions. These findings identify Lis1 as a key component of the molecular machinery that directs asymmetric division in hematopoietic stem cells and provide the first genetic proof for the requirement of a proper balance of asymmetric division and its regulators for hematopoietic development in vivo. Defining the position and orientation of the immature hematopoietic cells within their microenvironment would be an important aspect of future work as environmental cues may be critical for specifying the plane of division of hematopoietic cells through Lis1. While our focus has been on understanding the severe loss of the stem cell pool, we found possibly independent defects in mature erythroid and granulocyte lineages. Interestingly, these findings parallel those of mice lacking the serine-threonine kinase Lkb132,33, which exhibit significant defects in HSCs. Lkb1’s role in asymmetric division34 raises the possibility that Lkb1 and Lis1 may control overlapping, albeit not identical, mechanisms in the hematopoietic system.
Elucidating the basis of the maintenance of the undifferentiated state is important because it may allow us to understand the mechanisms underlying the differentiation blockade seen in cancers such as glioblastoma, breast cancer and leukemia35–37. Emerging studies indicate that the presence and dysregulated expression of fate determinants such as Numb and Musashi may be important elements of the induction of such cancers10,38. Our work now shows that the regulatory mechanisms that direct the inheritance of these determinants are perhaps equally important for the establishment and continued propagation of malignancies. Previous studies have shown that the loss of asymmetric division proteins including Brat, Prospero and Numb can trigger tumor formation in Drosophila neuroblasts39–44. Using mouse models and patient samples of leukemias that are resistant to therapy, our data are the first to link Lis1 and the machinery regulating inheritance with mammalian oncogenesis and thus provide an important clinical complement to studies in Drosophila. This raises the possibility that molecules that can control or modulate the inheritance of fate determinants could serve as a powerful new class of regulators of cancer growth and that further work in this area may define new approaches to therapy.
Methods
Generation and analysis of mice
Hypomorphic conditional knockout mice (Lis1f/f, also Lis1-loxP or Pafah1b1-loxP; Strain: 129-Pafah1b1tm2awb/J)11 were mated with either Rosa26-CreERT2 mice (Strain: B6;129-Gt(ROSA)26Sortm1(cre/Esr1)Tyj)15 or Vav-Cre transgenic mice12. Vav-Cre reporter mice were generated by crossing Vav-Cre mice to Rosa26-stop-tdTomato mice (Stain: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; Stock #: 007909). B6-CD45.1 (Strain: B6.SJL-PtprcaPepcb/BoyJ) mice were used as transplant recipients. All mice were 6–16 weeks of age. Mice were bred and maintained in the animal care facilities at Duke University Medical Center and the University of California, San Diego. Tamoxifen treatment was done as previously described45. In brief, adult mice were administered tamoxifen (Sigma) in corn oil (20 mg/ml) daily by oral gavage at ~114 μg tamoxifen per gram of body weight per day for five consecutive days. For leukemia experiments, all recipient mice weighed ~17.5–20 grams and were administered 2 mg of tamoxifen per day for five consecutive days. Embryos were suspended in phosphate-buffered saline and visualized with a Leica MZ16 FA Fluorescence Stereomicroscope. Embryos were fixed in 4% paraformaldehyde and embedded in paraffin according to standard protocols. Sections (5 μm) were obtained for hematoxylin and eosin staining. All animal experiments were performed according to protocols approved by the Duke University and University of California, San Diego Institutional Animal Care and Use Committees.
Cell isolation and FACS analysis
Cells were suspended in Hanks’ balanced salt solution (HBSS) (Gibco, Life Technologies) containing 5% (vol/vol) fetal bovine serum and 2 mM EDTA and prepared for FACS analysis and sorting as previously described46. The following antibodies were used to define lineage positive cells: 145-2C11 (CD3ε), GK1.5 (CD4), 53-6.7 (CD8), RB6-8C5 (Ly-6G/Gr1), M1/70 (CD11b/Mac-1), TER119 (Ly-76/TER119), 6B2 (CD45R/B220), and MB19-1 (CD19). Red blood cells were lysed using RBC Lysis Buffer (eBioscience) before staining for lineage markers. For fetal liver cell isolation and FACS analysis, single-cell suspensions were prepared by disaggregation and passing through a 74-μm-nylon mesh (Corning). For fetal HSC cell population analysis, the lineage antibody cocktail was used without anti-Mac-1. The following additional antibodies were used to define HSC populations: 2B8 (CD117/c-kit), D7 (Ly-6A/E/Sca-1), AA4.1 (CD93/C1qRp), HM48-1 (CD48/BCM1), TC15-12F12.2 (CD150) and A2F10 (CD135/Flt3). Fetal HSCs were defined as c-Kit+ Lin− AA4.1+ (KL AA4.1+). Adult HSCs were defined as either c-Kit+ Lin− Sca1+ CD48− CD150+ (KLS CD48− CD150+) or c-Kit+ Lin− Sca1+ Flt3− (KLSF). To determine donor-derived chimerism in transplantation-based assays, peripheral blood of recipients were obtained by the submandibular bleeding method and prepared for analysis as previously described10. All antibodies were purchased from BD Pharmigen, eBioscience or BioLegend. Apoptosis assays were performed by staining cells with Annexin-V and 7AAD (BD Pharmingen). Analysis of in vivo BrdU incorporation was performed using the FITC BrdU Flow Kit (BD Pharmingen) after a single intraperitoneal injection of BrdU (2 mg). Analysis and cell sorting were carried out on FACSVantage SE, FACStar, FACSCantoII, FACSDiva and FACSAriaIII machines (all from Becton Dickinson) and data were analyzed with FlowJo software (Tree Star Inc.).
Retro- and Lentiviral constructs and production
MIG-BCR-ABL was a gift from Warren Pear and Ann Marie Pendergast and was cloned into MSCV-IRES-YFP retroviral vector. MSCV-NUP98-HOXA9-IRES-YFP was a gift from Gary Gilliland and was cloned into the MSCV-IRES-GFP vector. MSCV-MLL-AF9-IRES-GFP was generously provided by Scott Armstrong. NRASG12V cDNA was a gift from Christopher Counter and was cloned into MSCV-IRES-YFP retroviral vector. Numb cDNA (p65 isoform, accession number BC033459, NCBI) was either cloned into the MSCV-IRES-GFP vector or fused to CFP in the MSCV-CFP vector following the removal of IRES. The short hairpin RNA construct against Numb (shNumb) was designed and cloned in MSCV/LTRmiR30-PIG (LMP) vector from Open Biosystems according to their instructions. The target sequence is 5′-GGACCTCATAGTTGACCAG-3′ for shNumb. Mouse Nde1 cDNA (accession number BC023267, NCBI) was cloned into MSCV-IRES-GFP. H2B-GFP (pEGFPN1) vector20 was a gift from Geoffrey Wahl and the H2B-GFP chimeric gene was cloned into MSCV-IRES-GFP retroviral vector following the removal of IRES-GFP. mCherry-alpha-tubulin fusion construct21 was generously provided by John Chang and Sarah Russell. Lentiviral short hairpin RNA (shRNA) constructs were cloned in FG12 as described previously47. The target sequences are 5′-AGATGAACTAAATCGAGC-3′ for shLIS1-(592), 5′-TGTCTGCCTCAAGGGATA-3′ for shLIS1-(1191) and 5′-TGCGCTGCTGGTGCCAAC-3′ for luciferase as a negative control. Virus was produced in 293T cells transfected using the FuGENE®6 or X-tremeGENE HP (Roche) with viral constructs along with VSV-G and gag-pol. For lentivirus production Rev was also co-transfected. Viral supernatants were collected for three to five days followed by ultracentrifugal concentration at 50,000× g for 3h.
Cell culture and methylcellulose colony formation
For liquid culture, freshly isolated adult KLS (cKit+ Lin− Sca-1+) cells were plated into a 96-well U bottom plate in X-Vivo15 (with Gentamicin and Phenol Red) (Lonza) supplemented with 50 μM 2-mercaptoethanol, 10% (vol/vol) fetal bovine serum, stem cell factor (SCF; 100 ng/ml, R&D Systems) and thrombopoietin (TPO; 20 ng/ml, R&D Systems). 4-OH tamoxifen (Sigma) was dissolved in ethanol at 1 mg/ml (1000X), and a 1X solution was made immediately before treatment. For certain immunofluorescence experiments, cells were treated for 24 hrs with either 10 μM Cytochalasin B (Sigma) or 10 nM Nocodazole (Sigma). For fetal liver methylcellulose assays, individual fetal livers (FL) from embryonic day 12.5 (E12.5) embryos were dissected in cold phosphate-buffered saline, disaggregated and passed through a 74-μm nylon mesh (Corning) to generate single-cell suspensions. 5,000 FL cells were plated in triplicate in Iscove’s modified medium-based methylcellulose medium (Methocult M3434, StemCell Technologies). Erythroid (BFU-E) hematopoietic progenitors were scored by morphological criteria on day 7 and myeloid (CFU-GM) and multilineage (CFU-GEMM) colonies were scored on day 10.
In vivo transplantation assays
For fetal liver transplants, 5,000 Lin− AA4.1+ fetal liver cells (derived from E14.5 embryos expressing CD45.2) along with 3 ×105 competitive bone marrow cells derived from an unirradiated recipient mouse were transplanted by retro-orbital i.v. injections into lethally irradiated (9.5 Gy) congenic recipient mice (expressing CD45.1). Recipient mice received donor cells derived from one individual embryo of a given genotype. Peripheral blood of recipient mice was collected at 4, 8, 12, and 16 weeks after transplantation. Donor and recipient cells were distinguished by expression of CD45.1 (A20; eBioscience) and CD45.2 (104; eBioscience). For bone marrow transplants, 500 LT-HSCs (cKit+ Lin− Sca-1+ CD150+ CD48−) isolated from bone marrow of mice expressing CD45.2 were transplanted into lethally irradiated (9.8 Gy) congenic recipient mice (expressing CD45.1) along with 3 × 105 Sca1-depleted bone marrow cells derived from an unirradiated recipient mouse. Peripheral blood of recipient mice was collected at 4, 8 and 28 weeks after transplantation. For Lis1 chimera bone marrow transplants, 3 × 105 whole bone marrow cells isolated from Lis1 chimera mice (expressing CD45.2) were transplanted into lethally irradiated (9.8 Gy) recipient mice (expressing CD45.1) along with 3 × 105 Sca1-depleted bone marrow cells derived from an unirradiated recipient mouse. Peripheral blood of recipient mice was collected at 16 weeks after transplantation.
Determining Numb inheritance
For experiments involving fixed cells, cells in late telophase or undergoing cytokinesis were identified by pronounced cytoplasmic cleft by brightfield or visualized by staining cells for alpha-tubulin plus the presence of dual nuclei using DAPI. ImageJ 1.46r was used to determine fluorescence intensity of pixels following Numb staining. The fluorescence intensity of Numb was on average ~2.4-fold higher in the Numbhigh daughter cell relative to the Numblow daughter cell during an asymmetric division. Based on data shown in Fig. 4b, Numb is ~1.8-fold higher in progenitors than in HSCs and thus, incipient daughters that expressed at least a 1.8-fold difference in Numb expression were scored as an asymmetric Numb inheritance. For live imaging experiments, either KLS cells isolated from Lis1f/f; Rosa26-creER/Rosa26-creER and Lis1+/+; Rosa26-creER/Rosa26-creER mice or established WT or Lis1−/− bcCML lineage-negative cells were co-infected with mCherry-α-tubulin and Numb-CFP fusion constructs and doubly-infected cells were subsequently plated in methylcellulose medium (Methocult M3434, StemCell Technologies) and treated with 4-OH tamoxifen (Sigma). Please note that we used Numb-CFP specifically since it allowed clear detection of distinct levels of Numb. In contrast, Numb-YFP led to highly saturated expression of YFP and did not allow easy identification of low and high expressing daughter cells. Dividing cells identified in movie replay were visualized in spectrum color format (where red indicates pronounced α-tubulin expression and centrosome location) to readily identify the centrosomes. Using ImageJ 1.46r software, a line connecting the two centrosomes of a cell was drawn (Line 1; dotted). Subsequently, an additional line (Line 2; solid) was drawn perpendicular to Line 1, which marked the cleavage furrow and partitioned the mother cell into incipient daughter cell 1 (D1) and daughter cell 2 (D2). Using the criteria described above and shown in Fig. 4b, incipient daughters that expressed at least a 1.8-fold difference in Numb expression were scored as an asymmetric Numb inheritance.
Analysis of spindle orientation and mitotic events
KLS cells were isolated and sorted from age-gender matched Lis1f/f; Rosa26-creER/Rosa26-creER and Lis1+/+; Rosa26-creER/Rosa26-creER mice and cultured overnight in X-Vivo™15 media (Lonza) supplemented with 50 μM 2-mercaptoethanol, 10% (vol/vol) fetal bovine serum, SCF (100 ng/ml, R&D Systems) and thrombopoietin (20 ng/ml, R&D Systems). Cells were retrovirally-infected with MSCV-H2B-GFP and mCherry-α-tubulin, harvested 48 hrs after infection and re-sorted for GFP+ mCherry+ KLS cells. Sorted cells were either cultured in 96-well U-bottomed plates (BD Biosciences) for 48 hrs with 4-OH tamoxifen (Sigma) and subsequently placed on chambered coverglass slides (Lab-Tek II®, Thermo Scientific) coated with 0.1 μg/μl Retronectin® (Takara Bio Inc.) in the continual presence of 4-OH tamoxifen or plated in Iscove’s modified medium-based methylcellulose medium (Methocult M3434, StemCell Technologies) supplemented with 4-OH tamoxifen. Images were collected every 3–4 minutes with xyzt acquisition mode using an Axio Observer.Z1 microscope with the LSM 700 scanning module (Zeiss). Cultures were maintained at 37°C, 5% CO2 using a Heating Insert P Lab-Tek S1 with an Incubator PM S1 (Zeiss). Mitotic cells were identified in movie replay. To measure spindle orientation, a concatenation of Z-stack images of each cell at every measured time point from the start of metaphase to early telophase was generated and displayed orthogonally using Zen 2010 software. Subsequently, the angle formed between the substratum plane (Retronectin base) and the virtual line passing through spindle poles was measured using ImageJ 1.44. To quantify mitosis duration, the time between nuclear envelope breakdown and chromatin condensation until the beginning of telophase was determined. For chromosome counts, KLS cells were cultured in X-Vivo™15 media (Lonza) supplemented with 50 μM 2-mercaptoethanol, 10% (vol/vol) fetal bovine serum, SCF (100 ng/ml, R&D Systems) and TPO (20 ng/ml, R&D Systems) for 48 hrs, and then arrested in metaphase by a 2h incubation with 100 ng/ml colcemid (KaryoMAX solution, Gibco). Cells were treated with hypotonic solution (0.56% KCl) for 15 min at 37°C, then fixed with 3:1 methanol:glacial acetic acid and spread on a slide to prepare metaphase spreads. Karyotyping was performed by Cell Line Genetics, Inc.
Generation and analysis of leukemic mice
KLS cells were isolated and sorted from age-gender matched Lis1f/f; Rosa26-creER/Rosa26-creER and Lis1+/+; Rosa26-creER/Rosa26-creER mice and cultured overnight in X-Vivo15 media (Lonza) supplemented with 50 μM 2-mercaptoethanol, 10% (vol/vol) fetal bovine serum, SCF (100 ng/ml, R&D Systems) and TPO (20 ng/ml, R&D Systems). Cells were retrovirally-infected with MSCV-BCR-ABL-IRES-YFP and MSCV-NUP98-HOXA9-IRES-GFP to generate myeloid blast crisis phase CML (bcCML) or MSCV-MLL-AF9-IRES-GFP and MSCV-NRASG12V-IRES-YFP to generate de novo AML. Subsequently, cells were harvested 48 hours after infection. Doubly-infected cells (for bcCML experiments) or unsorted cells (for AML experiments) were transplanted retro-orbitally into cohorts of B6-CD45.1 mice. Before transplantation, for de novo AML transplants, infected cells regardless of donor genotype displayed similar infection efficiency. All recipients were sub-lethally (6–7 Gy) irradiated. For secondary bcCML transplantations, cells recovered from terminally ill primary recipients were sorted for lineage-negative (Lin-), MSCV-BCR-ABL-IRES-YFP and MSCV-NUP98-HOXA9-IRES-GFP and transplanted into secondary recipients. Analysis of diseased mice was conducted as previously described10.
Human leukemia patient samples and cell lines
Patient samples were obtained from: Singapore General Hospital (Singapore), the Fred Hutchinson Cancer Research Center, Duke Adult Bone Marrow Transplant Clinic and Moores UCSD Cancer Center from Institutional Review Board-approved protocols with written informed consent in accordance with the Declaration of Helsinki. For colony-forming experiments, primary bcCML cells were imatinib, nilotinib and dasatinib-resistant and primary AML cells harbored the MLL-AF9 translocation. Leukemia cells were cultured in Iscove’s modified Dulbecco medium (IMDM) with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 μg/ml streptomycin, 55 μM 2-mercaptoethanol and supplemented with SCF, IL-3, IL-6, FLT3L, and TPO. For cell differentiation experiments, leukemia cells were cultured in IMDM with 20% BIT 9500 (StemCell Technologies), 2mM L-Glutamine, 100 IU/ml penicillin, 55 μM 2-mercaptoethanol and supplemented with LDL, SCF and TPO. The human chronic myeloid leukemia cell line K562 (ATCC) was maintained in Roswell Park Memorial Institute medium (RPMI-1640) with 10% FBS, 100 IU/ml penicillin and 100 μg/ml streptomycin. The human acute myeloid leukemia cell line MV4-11 (ATCC) was maintained in IMDM with 10% FBS, 100 IUml−1 penicillin and 100 μg/ml streptomycin. For colony forming assays, human cell lines or sorted hCD34+ cells from primary patient samples were transduced with lentiviral shRNA (cloned in FG12-UbiC-GFP), and GFP-positive cells were sorted at 48 hrs and plated in complete methylcellulose medium (MethoCult Express, StemCell Technologies). All knockdown experiments were conducted with the construct shRNA-LIS1-(592) except those involving human primary CML cells, which were instead transduced with an alternative shLIS1 construct: shRNA-LIS1-(1191). This construct represents an independent hairpin shRNA targeting LIS1 that more effectively knocks down LIS1 in these cells. Colony numbers were counted 10–14 days after plating.
Gene expression microarray and data analysis
Control (Lis1+/+; Rosa-creER) or Lis1f/f; Rosa-creER mice were treated with tamoxifen for five consecutive days. Three days after the final tamoxifen administration, KLS cells were FACS-sorted and total cellular RNAs were purified. RNAs were amplified, labeled, hybridized onto Affymetrix GeneChip Mouse Genome 430 2.0 Arrays and raw hybridization data were collected (Asuragen Inc., Austin, TX). Expression level data were normalized using a multiple-loess algorithm as previously described48. Probes whose expression levels exceed a threshold value in at least one sample were considered detected. The threshold value is found by inspection from the distribution plots of log2 expression levels. Detected probes were sorted according to their q-value, which is the smallest false discovery rate (FDR)49 at which the gene is called significant. An FDR value of α is the expected fraction of false positives among all genes with q ≤α. FDR was evaluated using Significance Analysis of Microarrays and its implementation in the official statistical package samr 50. To prevent unwarranted variances, the percentile of standard deviation values used for the exchangeability factor s0 in the regularized t-statistic was set to 50. The probe list, sorted by q-value in ascending order, was translated into Entrez gene ID’s and parsed so that where several different probes represent the same gene, only the highest-ranking probe was kept for further analysis. The sorted list of genes was subjected to a non-parametric variant of the Gene Set Enrichment Analysis (GSEA)51, in which the p-value of a gene set was defined as the minimal rank-order p-value of a gene in the gene set52 rather than the Kolmogorov-Smirnov statistic as in GSEA. Briefly, let rk be the k-th highest rank among a gene set of size N. The rank-order p-value pk of this gene is the probability that among N randomly chosen ranks without replacement, the k-th highest rank will be at least rk. The p-value of a gene set was defined as the smallest of all pk. Finding the p-value of a gene set of size N requires calculation of N rank-order p-values; however, there is no need to adjust the p-values for the number of genes tested as the tests are highly statistically dependent. A Bonferroni adjustment of gene set p-values for the number of gene sets tested was performed. Gene sets with adjusted p-values ≤ 0.01 were reported. For the analysis of stem cell signature sets published16–18,53–55, all detected genes in the Lis1−/− (Lis1f/fcreER +tamoxifen) to control (Lis1+/+creER +tamoxifen) comparison were sorted according to their q-values as above and gene set enrichment analysis for each signature gene set was performed. Each gene signature’s p-value is Bonferroni-adjusted by a factor of 9 (number of signature gene sets tested). Heatmaps were created using in-house hierarchical clustering software and the colors qualitatively correspond to fold changes.
PCR genotyping and RT-PCR analysis
For genotyping by PCR, the reaction mixture contained MangoMix (Bioline), genomic DNA and 0.5 μM of each primer. PCR conditions for genotyping were as follows: 3 min at 94°C, followed by 35 cycles at 94°C for 30 s, 60°C for 1 min, and 72°C for 1 min. RNA was isolated using RNAqueous-Micro (Ambion) or RNeasy Mini kit (Qiagen). cDNA was prepared from equal amounts of RNAs using Superscript II reverse transcriptase (Invitrogen). Quantitative real-time PCRs were performed using iQ SYBR Green Supermix (Bio-Rad) on a CFX 96 C1000™ Thermal cycler (Bio-Rad). Results were normalized to the level of β2 microglobulin or TATA-binding protein. Mouse Lis1 (Mm01253377_mH) gene levels were analyzed with TaqMan Gene Expression Assays. All primer sequences are listed in Supplementary Table 2.
Immunofluorescence staining
Cells were allowed to settle on poly-L-lysine coated coverslips (BD Biosciences) at 37°C, fixed with 4% paraformaldehyde (USB Corporation) or methanol, permeabilized with 1X Dako wash buffer (Dako) and blocked with 20% normal goat serum (Invitrogen) or donkey serum (Abcam) in 1X Dako wash buffer. Primary antibody incubation was overnight at 4°C. The following primary antibodies were used: rabbit anti-Numb 1:50 or 1:100 (Abcam), goat anti-LIS1 1:500 (Santa Cruz Biotechnology), mouse anti-alpha-tubulin 1:200 (Abcam), rat anti-α-tubulin 1:1000 (Abcam), mouse anti-α-tubulin conjugate FITC 1:200 (Sigma). Secondary antibody incubation was performed for 1 hr at room temperature. DAPI (Molecular Probes) was used to detect DNA. Images were obtained with a Confocal Leica TCS SP5 II (Leica Microsystems) or an Axio Observer.Z1 microscope with the LSM 700 scanning module (Zeiss). ImageJ 1.46r was used to determine fluorescence intensity.
Statistical analysis
Statistical analyses were carried out using Graphpad Prism software version 5.0a or 5.0d (GraphPad Software Inc.). Data are mean ± SEM and ‘center values’ are defined as the median. Chi-square test was used to determine deviation from Mendelian ratios. Two-tailed unpaired Student’s t-tests with Welch’s correction when appropriate were used to determine statistical significance (*P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Supplementary Material
Acknowledgments
We are grateful to Brigid Hogan, John Chang, Arshad Desai, Joseph Gleeson, Ji Eun Lee, MingFu Wu, Maike Sander, and Janel Koop for experimental advice and reagents. We would also like to thank Marcie Kritzik for advice and comments on the manuscript; Mai Nakamura for experimental help; Mike Cook, Lynn Matinek and Beth Harvat, Eric O’Conner and Karl Marquez for cell sorting; Warren Pear and Ann Marie Pendergast for the BCR-ABL construct, Gary Gilliland for the NUP98-HOXA9 construct, Scott Armstrong for the MLL-AF9 construct, Christopher Counter for NRASG12V, Sarah Russell for the mCherry-alpha-tubulin construct, Geoffrey Wahl for the H2B-GFP (pEGFPN1) vector and Dimitris Kioussis for the vav-Cre transgenic line. B.Z. and C.S.K. received support from T32 GM007184-33 and T32 GM007752, respectively. T.I. is a recipient of a California Institute for Regenerative Medicine interdisciplinary stem cell training program fellowship, and T.K. is supported by a postdoctoral fellowship from the Japanese Society for the Promotion of Science. This work was also supported by a Leukemia and Lymphoma Society Scholar Award as well as DK63031, HL097767 and DP1 CA174422 to T.R.
Footnotes
Accession codes
Microarray data reported have been deposited in the ArrayExpress Database (accession number E-MEXP-3855) (European Bioinformatics Institute 2013).
Author Contributions
B.Z. and T.I. planned and designed the research, performed the majority of experiments and helped write the paper. B.Z. and J.B. developed all real time imaging methods for visualizing and tracking spindle orientation in primary hematopoietic cells. A.B., J.B., T.K., J.W., C.S.K., H.Y.K and O.A. provided experimental data and help. D.R., H.E.B., C.C. and V.G.O. provided primary patient samples and experimental advice. R.S. and G.H. carried out all bioinformatics analysis on microarray data. T.R. planned and guided the project, provided experimental advice and wrote the paper.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Siller KH, Doe CQ. Lis1/dynactin regulates metaphase spindle orientation in Drosophila neuroblasts. Dev Biol. 2008;319:1–9. doi: 10.1016/j.ydbio.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yingling J, et al. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suda T, Suda J, Ogawa M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci U S A. 1984;81:2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ting SB, et al. Asymmetric segregation and self-renewal of hematopoietic stem and progenitor cells with endocytic Ap2a2. Blood. 2012;119:2510–2522. doi: 10.1182/blood-2011-11-393272. [DOI] [PubMed] [Google Scholar]
- 5.Wu M, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007;1:541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope KJ, et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell. 2010;7:101–113. doi: 10.1016/j.stem.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Ito K, et al. Regulation of reactive oxygen species by Atm is essential for proper response to DNA double-strand breaks in lymphocytes. J Immunol. 2007;178:103–110. doi: 10.4049/jimmunol.178.1.103. [DOI] [PubMed] [Google Scholar]
- 8.Kharas MG, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16:903–908. doi: 10.1038/nm.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Andres-Aguayo L, et al. Musashi 2 is a regulator of the HSC compartment identified by a retroviral insertion screen and knockout mice. Blood. 2011;118:554–564. doi: 10.1182/blood-2010-12-322081. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirotsune S, et al. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nature Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 12.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 13.Almarza E, et al. Regulatory elements of the vav gene drive transgene expression in hematopoietic stem cells from adult mice. Exp Hematol. 2004;32:360–364. doi: 10.1016/j.exphem.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Ogilvy S, et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94:1855–1863. [PubMed] [Google Scholar]
- 15.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 16.Wong DJ, et al. Module map of stem cell genes guides creation of epithelial cancer stem cells. Cell Stem Cell. 2008;2:333–344. doi: 10.1016/j.stem.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Venezia TA, et al. Molecular signatures of proliferation and quiescence in hematopoietic stem cells. PLoS Biol. 2004;2:e301. doi: 10.1371/journal.pbio.0020301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eppert K, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–1093. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 19.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 21.Day D, et al. A method for prolonged imaging of motile lymphocytes. Immunol Cell Biol. 2009;87:154–158. doi: 10.1038/icb.2008.79. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Walsh CA. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 24.Dash AB, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A. 2002;99:7622–7627. doi: 10.1073/pnas.102583199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayotte N, Roy DC, Yao J, Kroon E, Sauvageau G. Oncogenic interaction between BCR-ABL and NUP98-HOXA9 demonstrated by the use of an in vitro purging culture system. Blood. 2002;100:4177–4184. doi: 10.1182/blood-2002-04-1244. [DOI] [PubMed] [Google Scholar]
- 26.Neering SJ, et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110:2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23:877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsai FY, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226. doi: 10.1038/371221a0. [DOI] [PubMed] [Google Scholar]
- 29.Wang Q, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci U S A. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 31.Porcher C, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 32.Gan B, et al. Lkb1 regulates quiescence and metabolic homeostasis of haematopoietic stem cells. Nature. 2010;468:701–704. doi: 10.1038/nature09595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakada D, Saunders TL, Morrison SJ. Lkb1 regulates cell cycle and energy metabolism in haematopoietic stem cells. Nature. 2010;468:653–658. doi: 10.1038/nature09571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonaccorsi S, et al. The Drosophila Lkb1 kinase is required for spindle formation and asymmetric neuroblast division. Development. 2007;134:2183–2193. doi: 10.1242/dev.02848. [DOI] [PubMed] [Google Scholar]
- 35.Calabretta B, Perrotti D. The biology of CML blast crisis. Blood. 2004;103:4010–4022. doi: 10.1182/blood-2003-12-4111. [DOI] [PubMed] [Google Scholar]
- 36.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nature Rev Cancer. 2007;7:791–799. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 37.Maher EA, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 38.Pece S, et al. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- 40.Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- 41.Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nature Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- 42.Lee CY, et al. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell. 2006;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, et al. Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts. Genes Dev. 2006;20:3453–3463. doi: 10.1101/gad.1487506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang ZJ, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14:135–145. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Domen J, Cheshier SH, Weissman IL. The role of apoptosis in the regulation of hematopoietic stem cells: Overexpression of Bcl-2 increases both their number and repopulation potential. J Exp Med. 2000;191:253–264. doi: 10.1084/jem.191.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasik R, Woelk CH, Corbeil J. Microarray truths and consequences. J Mol Endocrinol. 2004;33:1–9. doi: 10.1677/jme.0.0330001. [DOI] [PubMed] [Google Scholar]
- 49.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- 50.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arnold BC, Balakrishnan N, Nagaraja HN. A First Course in Order Statistics (Wiley Series in Probability and Statistics) 1992 [Google Scholar]
- 53.Metzeler KH, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Somervaille TC, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4:129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yagi T, et al. Identification of a gene expression signature associated with pediatric AML prognosis. Blood. 2003;102:1849–1856. doi: 10.1182/blood-2003-02-0578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.