Abstract
Estrogen receptor coregulator over-expression promotes carcinogenesis and/or progression of endocrine related-cancers in which steroid hormones are powerful mitogenic agents. Recent studies in our laboratory, as well as others, demonstrated that the estrogen receptor coregulator PELP1 is a proto-oncogene. PELP1 interactions with histone demethylase KDM1 play a critical role in its oncogenic functions and PELP1 is a prognostic indicator of decreased survival in breast cancer patients. However, the in vivo significance of PELP1 deregulation during initiation and progression of breast cancer remains unknown. We generated an inducible, mammary gland–specific PELP1-expressing transgenic (Tg) mouse (MMTVrtTA-TetOPELP1). We found more proliferation, extensive side branching and precocious differentiation in PELP1-overexpressing mammary glands than in control glands. Aged MMTVrtTA-TetOPELP1 bitransgenic mice had hyperplasia and preneoplastic changes as early as 12 weeks, and ER-positive mammary tumors occurred at a latency of 14–16 months. Mechanistic studies revealed that PELP1 deregulation altered expression of a number of known ER target genes involved in cellular proliferation (such as cyclin D1, CDKs) and morphogenesis (EGFR, MMPs) and such changes facilitated altered mammary gland morphogenesis and tumor progression. Further, PELP1 was hyper-phosphorylated at its CDK phosphorylation site, suggesting an autocrine loop involving the CDK–cyclin D1–PELP1 axis in promoting mammary tumorigenesis. Treatment of PELP1 Tg mice with a KDM1 inhibitor significantly reduced PELP1 driven hyper branching, reversed alterations in cyclin D1 expression levels and reduced CDK-driven PELP1 phosphorylation. These results further support the hypothesis that PELP1 deregulation has the potential to promote breast tumorigenesis in vivo and represent a novel model for future investigation into molecular mechanisms of PELP1-mediated tumorigenesis.
Keywords: estrogen receptor, ER-coactivator, PELP1, breast cancer, cyclin D1
Introduction
Breast cancer is the most common neoplasm and the second most common cause of cancer death in women, posing a significant public health challenge. Given the hormone-dependent nature of breast cancer and the central function of estrogen receptor-alpha (ER) in estrogen signaling (1), the ER status of breast tumors is an important biomarker for both prognosis and prediction of treatment response (2). A majority of tumors exhibit estrogen dependency (ER-positive), and therefore are suitable candidates for targeted endocrine therapies. Despite the well-documented benefits of endocrine treatment in breast cancer patients, not all ER-positive tumors respond to endocrine manipulation and a substantial number of initially responding tumors later become refractory to treatment due to acquired resistance (3, 4). Such major clinical problems have highlighted a critical need for identifying novel therapeutic targets and additional diagnostic/prognostic biomarkers, and prompted a deeper investigation into the regulation and function of ER by coregulatory proteins.
Transcriptional activity of ER is not only regulated by steroid hormones alone but also by several coregulatory proteins (5, 6), which associate with the ER in response to hormone binding to activate transcription (5). ER coregualtor levels are tightly regulated under normal conditions with deregulation primarily reported in the literature in association with a number of disease states. More than 100 of the nearly 300 distinct coregulators identified were revealed to be over- or under-expressed in human cancers; in breast cancer 38% of the coregulators identified were found to be over-expressed (7). Coregulator over-expression may promote carcinogenesis and/or progression of endocrine related-cancers where steroid hormones are powerful mitogenic agents (8). ER signaling is intact in the therapy resistant tumors, and ER interactions with critical co-regulator proteins containing an LXXLL motif (including PELP1, SRC1, SRC3) appear to mediate ER signaling in these therapy-resistant and ER-positive metastatic tumors (9).
Proline glutamic acid leucine rich protein (PELP1) is a proto-oncogene that interacts with ER through the LXXLL motif and provides breast cancer cells with a distinct growth and survival advantage by functioning as a critical coregulator (10, 11). PELP1 is overexpressed in many hormone-related cancers, promotes E2-mediated cell proliferation (12) and is prognostically linked to shorter breast cancer–specific survival (13), therapy resistance (14), and metastasis (15, 16). Recent studies indicated that PELP1 is needed for optimal epigenetic modifications at ER target genes, that PELP1 interactions with KDM1 play a key role in PELP1 mediated oncogenic functions (17) and that PELP1 deregulation promotes tumor proliferation in xenograft models (16, 17). Collectively, these data provide evidence for PELP1-mediated ER coregulatory functions playing a role in hormonal tumorigenesis by providing cancer cells with a growth and survival advantage. However, the in vivo significance, functional role and mechanism(s) by which PELP1 over-expression promotes initiation and progression of breast cancer have yet to be fully resolved.
In this present study, we generated a pathologically relevant murine breast cancer model by utilizing the tetracycline regulatory system to achieve inducible expression of PELP1 and the MMTV promoter to generate mammary epithelium–specific expression (18). PELP1-expressing mammary glands had more proliferation, extensive side branching, precocious differentiation and mammary tumors. Mechanistic studies revealed that PELP1 deregulation modulates expression of a number of known ER target genes and cancer promoting genes. Our results provide the first direct evidence that defines the in vivo role of PELP1 in oncogenesis and demonstrates that PELP1 is an ER coregulator with tumorigenic potential.
Materials and Methods
Generation of transgenic mice
Full-length T7-His tagged human PELP1 gene was PCR amplified and subcloned into the TMILA-SP plasmid (18) downstream to the human cytomegalovirus minimal promoter combined with tetracycline operator (tetO) sequence and upstream to a luciferase reporter gene translated from an internal ribosome entry site via the In-Fusion™ cloning method. TetO-PELP1 transgenic mice were generated by UTHSC-Houston Transgenic core facilities by pronuclear microinjection into fertilized oocytes and implantation in pseudopregnant C57/Bl6 females. Founder mice harboring the transgene construct were identified by PCR analysis of TetO-PELP1–specific primers and confirmed through Southern blot analysis of genomic DNA isolated from tail biopsies. TetO-PELP1 mice were bred with MMTV-rtTA transgenic mice (18) to generate MMTVrtTA-TetO-PELP1 bitransgenic mice. Offspring were screened for both rtTA and TetO-PELP1 transgenes by PCR and confirmed by using Southern blot analysis of tail DNA. Mice carrying MMTVrtTA-TetO-PELP1 transgene were used to establish two independent bitransgenic founder lines. Transgene induction via doxycycline treatment (MP Biomedicals) began in nulliparous female bitransgenic offspring at eight weeks of age at 200 µg/mL in the drinking water.
Real-time PCR analysis
Mammary gland tissues from nulliparous bitransgenic and wild-type controls were mechanically homogenized in TRIZOL reagent (1 mL TRIZOL for 200 mg of frozen tissue) using the manufacture’s protocol (Invitrogen, Carlsbad, CA). Assessment of RNA integrity was done using a NanoDrop 2000c spectrophotometer and indicated that the RNA extracted from snap frozen mammary glands was in good quality and suitable for real-time RT-PCR. Total RNA (2 µg) was used to generate first-strand complementary DNA with random hexamers and SuperScript® III reverse transcriptase according to supplier’s instructions (Invitrogen, cat no. 18080-051). Mouse-specific primers were obtained from Sigma. Real-Time PCR mixtures contained 25 ng template cDNA, SYBR Green master mix buffer and 300nM forward and reverse primers. The cycling conditions comprised 10-minute polymerase activation at 95C and 40 cycles at 95C for 15 seconds and 60C for one minute. Results and the difference in fold expression were calculated by using ΔΔCT method. Validated primers for each of the analyzed gene were purchased from realtimeprimers.com.
Chromatin immunoprecipitation analysis
The chromatin immunoprecipitation (ChIP) analysis was performed as described previously (19). In brief, frozen-stored Tg mice mammary tissues (n=3) were broken into powder with mortar and pestle, suspended in ChIP lysis buffer, cross-linked using formaldehyde and quenched by glycine. The chromatin was isolated and subjected to immunoprecipitation using the indicated antibodies. Isotype-specific IgG was used as a control. DNA was eluted and re-suspended in 50 µl of TE buffer and used for PCR amplification using published primer sequences (20). Mouse cyclin D1-591-F:ccagcgaggaggaatagatg;R:agcgtccctgtcttctttca; mouse cyclin D1-2707F:tgaaatccgctcagggtaac; R:ggacttggctgtttctgctc
Histology of Whole mounts and side branching quantification
The entire number four inguinal mammary gland was removed, fixed in Carnoy’s solution at room temperature for 48 hours, rehydrated, stained with carmine alum overnight (0.2% carmine0.5% aluminum potassium sulfate), dehydrated through graded series of ethanol, cleared in xylene then stored in methyl salicylate. Images of mammary gland whole mounts from adult nulliparous control (n=3) and bitransgenic animals (n=3 per genotype) were taken for quantitative analysis. Secondary and tertiary branch points within five 5X fields per gland were manually counted. For tumor formation analysis, aging mice were palpated every week and mammary glands containing tumors were isolated and formalin embedded as described above.
Immunohistochemistry analysis
Immunohistochemistry (IHC) and antigen retrieval was done according to a previously established protocol (17) with primary antibodies overnight at 4°C PELP1 (1:1000), T7 (1: 50), CK8 (1:50), phosMAPK (1:50), phos-AKT (1:50), Cyclin D1 (1:50), or Phospho PELP1 (1:50). Sections were then washed three times with 0.05% Tween in PBS, incubated with secondary antibody for 1 hour, washed three times with 0.05% Tween in PBS, visualized by 3,3’-diaminobenzidine (DAB) substrate and counterstained with hematoxylin QS (Vector Lab, Burlingame, CA, USA). The proliferative index was calculated as the percentage of Ki-67–positive cells in 10 randomly selected microscopic fields per slide at 40X. TUNEL analysis was performed by using the In Situ Cell Death Detection Kit (Roche, Indianapolis, IN, USA) as per the manufacturer’s protocol, and 10 randomly selected microscopic fields in each group were used to calculate the relative ratio of TUNEL-positive cells.
Luciferase assay
To monitor luciferase activity in vivo, mammary gland homogenates of aged-matched experimental nulliparous adult female bitransgenic mice treated with or without doxycycline were assessed by using a Dual Luciferase Assay System (Promega, Madison, WI) according to the manufacturer’s instructions. For whole animal in vivo bioluminescence imaging, the mice received 150 mg/kg of D-Luciferin (XenoLight Rediject; Caliper LifeSciences) intraperitoneally, and images were taken with an IVIS imaging system 10 minutes post injection.
Immunoprecipitation and western blot analysis
Mammary gland homogenates of aged-matched experimental nulliparous adult female bitransgenic mice were prepared by using T-PER Tissue Protein Extraction Reagent (Thermo Scientific, Cat no. 78510) following the manufacturer’s protocol. Protein concentrations were determined using the Bradford assay. Equal amounts of total protein extracts of mammary glands were resolved using 8 or 10% SDS-PAGE gels. The proteins were transferred to a nitrocellulose membrane and immunoblotted with primary mouse antibodies against cyclin D1 (1: 500), PTEN (1:500) or Actin (1:1000) and developed using ECL methodology. Using T7-epitope antibody–coupled agarose beads, the PELP1 transgene was immunoprecipitated from homogenates of female integral mammary glands from double and single transgenic animals after six months with or without doxycycline treatment. Western blotting was done with an anti-T7 antibody (EMD BioSciences, Gibbstown, NJ) to identify PELP1. For PELP1-KDM1 immunoprecipitation analysis, mammary gland lysates were prepared using a lysis buffer containing 50mM Tris-Hcl-pH7.5, 0.2% Triton X-100, 0.3% NP-40, 150 mM NaCl, 25 mM NaF, 0.1 mM sodium orthovanadate, along with a phosphatase and protease inhibitor cocktail. Immunoprecipitation was done using PELP1 antibody and western analysis was done using KDM1 antibody (Bethyl Lab, Montgomery, TX).
PCR array analysis
RNA was extracted from replicated sets of sections cut from formalin-fixed paraffin-embedded blocks of mouse mammary gland tissue using the ArrayGrade™ RNA Isolation Kit (SABiosciences), and the changes in RNA were profiled using the RT2 Profiler™ Mouse Breast Cancer Pathway PCR Array (cat no. 330231 Pamm-131ZA). Prior to the RT PCR step, 3 µg of total RNA was treated to digest possible gDNA contamination. Following the elimination step, complementary DNA (cDNA) was synthesized using RT2 First Strand Kit (cat no. 330401) in accordance with manufacture’s protocol. Then, cDNA synthesized from 3 µg total RNA went through another round of amplification before being combined with RT2 Real-Time SYBR Green PCR master mix (cat no. 330500) and loaded onto the array. PCR reactions were executed by utilizing the ABI StepOnePlus™ machine.
Statistical analyses
GraphPad Prism software (GraphPad Software, SanDiego, CA) was used to analyze all data. Results (mean +/− SEM) were analyzed using a two-tailed t test. Survival data are presented as Kaplan-Meier plots and were analyzed by using a log-rank (Mantel-Haenszel) method.
Results
Expression of PELP1 in mammary glands of bitransgenic mice
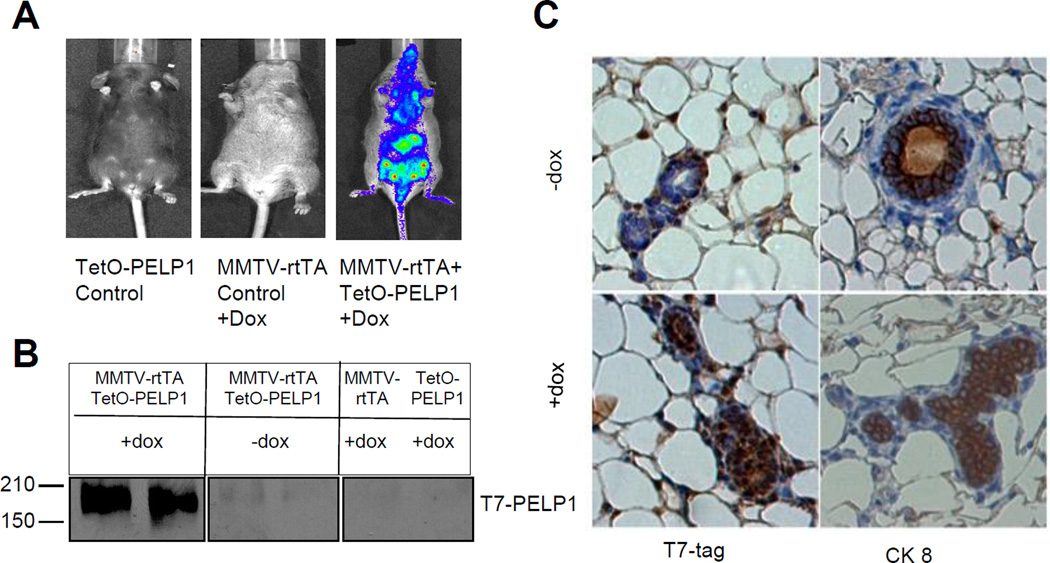
To determine the role of PELP1 in mammary tumorigenesis in vivo, we generated an inducible transgenic mouse model by crossing transgenic mice that express human PELP1 under the control of a tet-responsive promoter (TetO-PELP1) with transgenic mice expressing tetracycline-responsive transactivator protein under the control of the murine mammary tumor virus (MMTV-rtTA(18) (Supplementary Figure S1A). To facilitate transgene detection, this TetO-PELP1 construct encodes a bicistronic mRNA comprising a luciferase reporter gene translated from an internal ribosome entry site downstream of the PELP1-coding sequences as well as T7 epitope tag before ATG. Founder mice harboring the transgene construct were identified using PCR with TetO-PELP1–specific primers and were confirmed by Southern blotting genomic DNA isolated from tail biopsies. Two independent transgenic lines were established (Supplementary Figure S1B and data not shown). The Tg line 1 has 4 copies and the transgenic line 2 has 6 copies of the PELP1 transgene (data not shown). Tg line 2 exhibited poor breeding with small litter size; therefore, we used Tg line 1 for this study and key phenotypes were validated using Tg line 2. PELP1 transgene induction was initiated via doxycycline treatment (200 µg/mL) in the drinking water in nulliparous female bitransgenic offspring at 8 weeks of age and in vivo bioluminescence imaging was performed. Luciferase activity was only observed in bitransgenic animals treated with doxycycline (Fig.1 A and Supplementary Figure S1C). The human PELP1 transcripts in the transgenic mammary glands were 3 to 4 fold higher than the transcripts in mammary glands with endogenous PELP1 (Supplementary Figure S1D). Immunoprecipitation followed by Western analysis revealed that expression of T7-PELP1 transgene only occurred in mammary tissues extracted from the doxycycline-treated bitransgenic mice (Fig. 1B). IHC using the T7-epitope tag also revealed greater PELP1 transgene protein expression in these mice (Fig. 1C, left), confirming our Western data. PELP1-overexpressing cells are of epithelial origin as these cells expressed CK8 (Fig. 1C, right), thus confirming spatial regulation of PELP1 expression in our Tg mice model.
Figure 1.
Mammary-specific transgene expression in MMTV-rtTA-TetO-PELP1 bitransgenic mice. A, Representative images of observed in vivo luminescence from bitransgenic mice fed with doxycycline-added drinking water or normal drinking water. Mice were injected ip of 150 mg/kg D-luciferin and after 10 min imaged on the IVIS imaging system. B, PELP1 protein expression. Mammary gland lysates from mice treated with or without doxycycline were subjected to immunoprecipitation with anti-T7 antibody followed by detection of PELP1 by Western blotting with the anti-T7 antibody. C, Spatial regulation of transgene expression was confirmed by using IHC for the T7 tag of PELP1 and the epithelial surface marker cytokeratin-8 (CK 8).
PELP1 overexpression disrupts mammary gland morphogenesis by inducing hyperbranching and epithelial proliferation
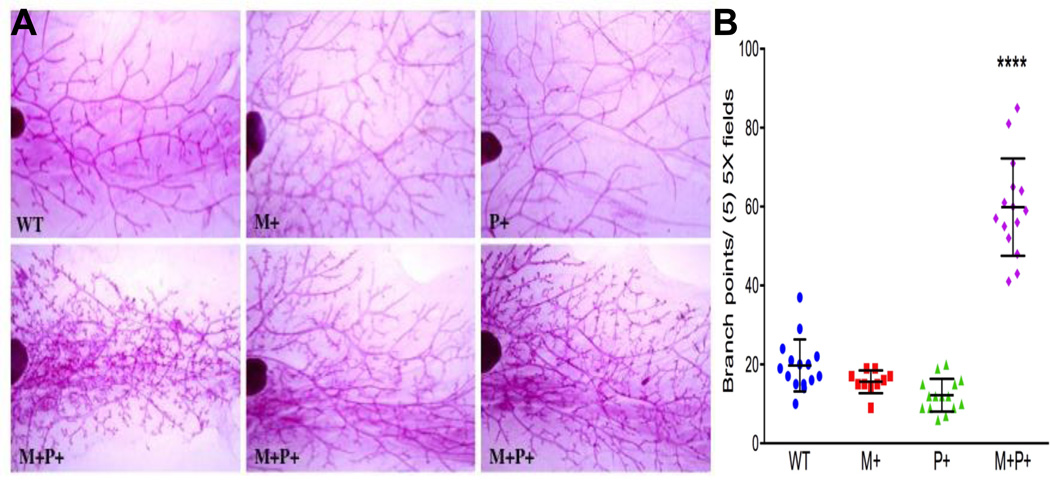
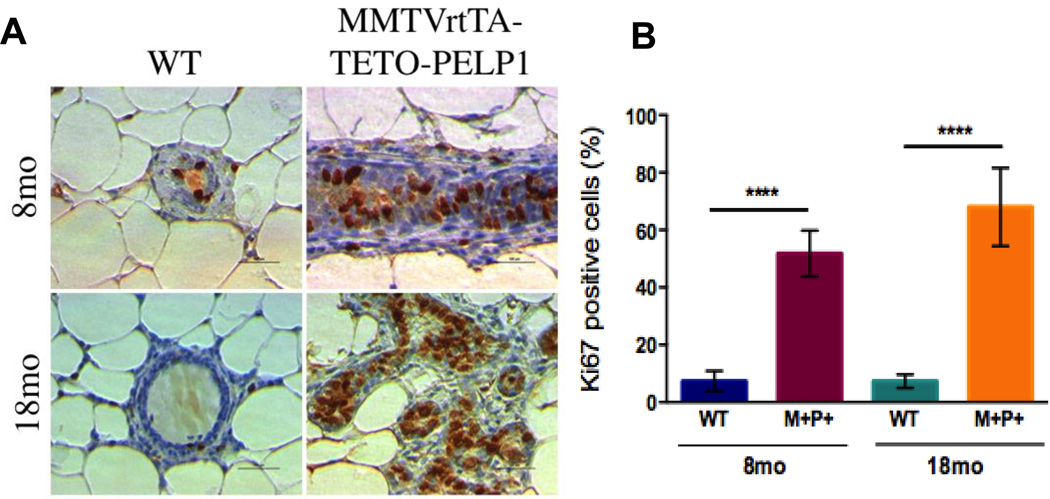
To determine the effect of PELP1 on mammary gland morphology, we examined Carmine-stained whole mount preparations from female nulliparous transgenic animals (Fig. 2A). Differences in the amount of side branching after PELP1 induction was observed as early two months; these differences became significant at 4 months (59.9 +/− 3.2 branch points in transgenic mice vs 15.88 +/− 0.93 control mice) (Fig. 2B). The hyperbranching phenotype in nulliparous mice (quiescent state of mammary epithelium) suggests an active role of PELP1 in proliferation and epithelial remodeling. Since published studies using in vitro models showed that PELP1 plays a role in cell proliferation, we hypothesized that the extensive hyperbranching phenotype mediated by PELP1 overexpression could be due to alterations in proliferation. To test, we analyzed mammary epithelial proliferation and apoptosis. Although TUNEL analysis revealed no change in the number of cells undergoing apoptosis (data not shown) at both 8- and 18-month time points, the Ki-67 labeling index increased significantly (52% and 68%, respectively) (Figure 3A, B). Our data suggest that the increase in mammary epithelial proliferation could in part be contributing to the hyperbranching phenotype in vivo.
Figure 2.
Effect of PELP1 overexpression on mammary gland morphogenesis. The entire number four-inguninal mammary gland was removed, fixed in Carnoy’s solution and stained with carmine alum overnight. A, Representative whole-mount images (magnification 3X) of mammary glands from 6-month-old nulliparous female mice of the indicated genetic controls (top panels) and doxycycline-treated bitransgenic animals (bottom panels). B, Side branching quantification (means +/− sem). All experimental data points were generated from three biological replicates per transgenic line. Statistical significance was determined by using a Student’s t test.****p<0.0001. WT, wild type; M+/−, MMTV-rtTA transgenic mice; P+/−, TetO-PELP1 mice ; and M+P+, MMTV-rtTA-TetO-PELP1 bitransgenic mice.
Figure 3.
Effect of PELP1 overexpression on mammary epithelial proliferation. The entire number nine inguinal mammary gland from wild-type and PELP1 bitransgenic animals at 8 and 18 months of age were removed, formalin-fixed, paraffin-embedded and sectioned every 4 mm. A, mammary epithelial proliferation as reflected by IHC analysis of Ki-67 expression. B Proliferation index was calculated relative to a percentage of total epithelial cell nuclei counts in five 20X fields per slide. All experimental data points were generated from three biological replicates per transgenic line. Statistical significance was determined by using a Student’s t test.****p<0.0001. WT, wild type; and M+P+, MMTV-rtTA-TetO-PELP1 bitransgenic mice.
PELP1 transgene overexpression promotes mammary epithelial hyperplasia and tumor development
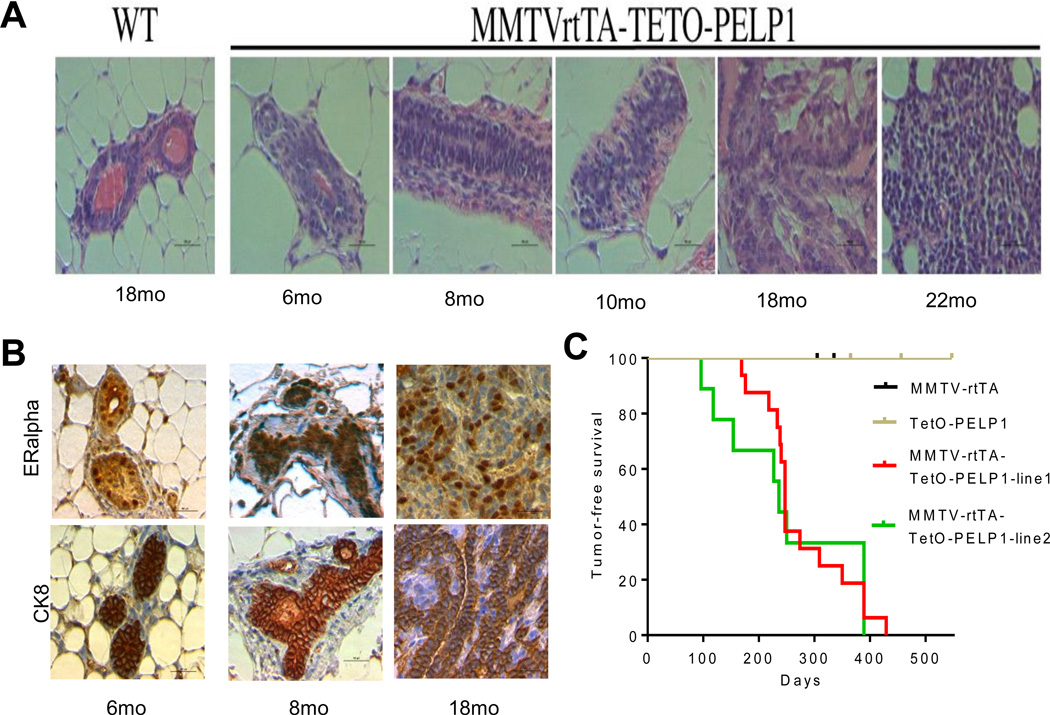
To directly test the tumorigenic potential of deregulated PELP1 expression in vivo, we assessed the premalignant changes within the epithelium and tumor ontogeny. Histological examination of mammary glands in PELP1 bitransgenic mice treated with doxycycline revealed distended primary ducts with areas of hyperplastic growth and structures with multiple cell layers (Fig. 4A). Between 6–10 months of age, precancerous mammary lesions (atypical hyperplasia, mammary intraepithelial neoplasia and carcinoma in situ) were observed. Mammary gland adenocarcinomas were observed in 14 to 18- month-old mice. (Fig. 4A). The presence of ER alpha in PELP1-driven hyperplasia and mammary tumors was analyzed by using IHC. Approximately, 100% of mammary glands and 90% of hyperplasia and mammary tumors from PELP1 transgenic mice had ER alpha expression (Fig 4B). Further analysis revealed that mammary glands, hyperplasia and mammary tumors from PELP1 transgenic mice had low/undetectable levels of ER beta expression (data not shown). Palpable mammary tumor development was first detected between 6 and 8 months of age. The Kaplan–Meier survival curve demonstrates that PELP1 overexpression significantly correlates with shorter mammary tumor–free survival (Fig 4C). Collectively, these results suggest that PELP1 deregulation has potential to promote mammary tumorigenesis in vivo.
Figure 4.
Histopathological analysis of PELP1-mediated mammary tumorigenesis. A, Representative images of histological analysis of H&E–stained mammary gland sections following PELP1 induction over time. B, IHC analysis of MMTVrtTA-TetO-PELP1 on doxycycline with indicated antibodies at specified time points. C, Kaplan-Meier analysis of mammary tumor free survival of bitransgenic mice treated with doxycycline (MMTV-rtTATetO-PELP1-line1 n=16; MMTV-rtTATetO-PELP1-line2 n=9) and genetic controls (dox-treated MMTVrtTA n=8 and TetO-PELP1 n=27). Mice were monitored weekly for a period of 22 months for palpable tumors. p<0.0001.
PELP1 transgene overexpression in mammary gland alters expression of genes involved in cell cycle progression and morphogenesis
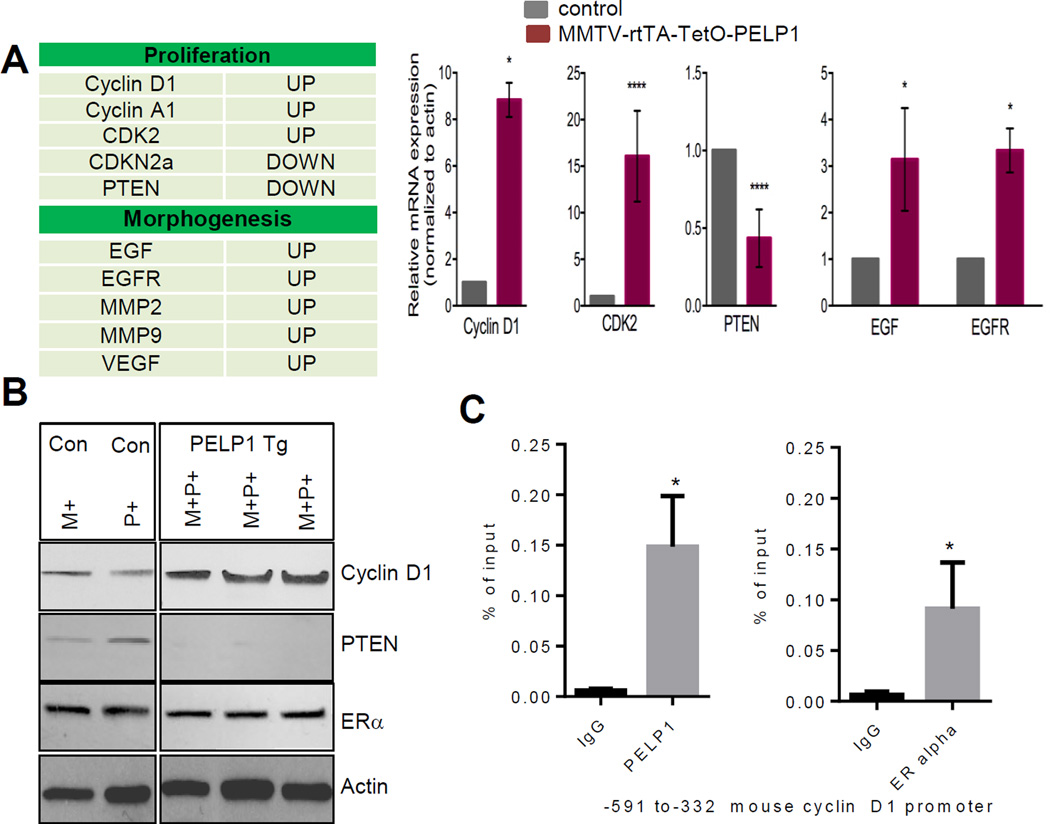
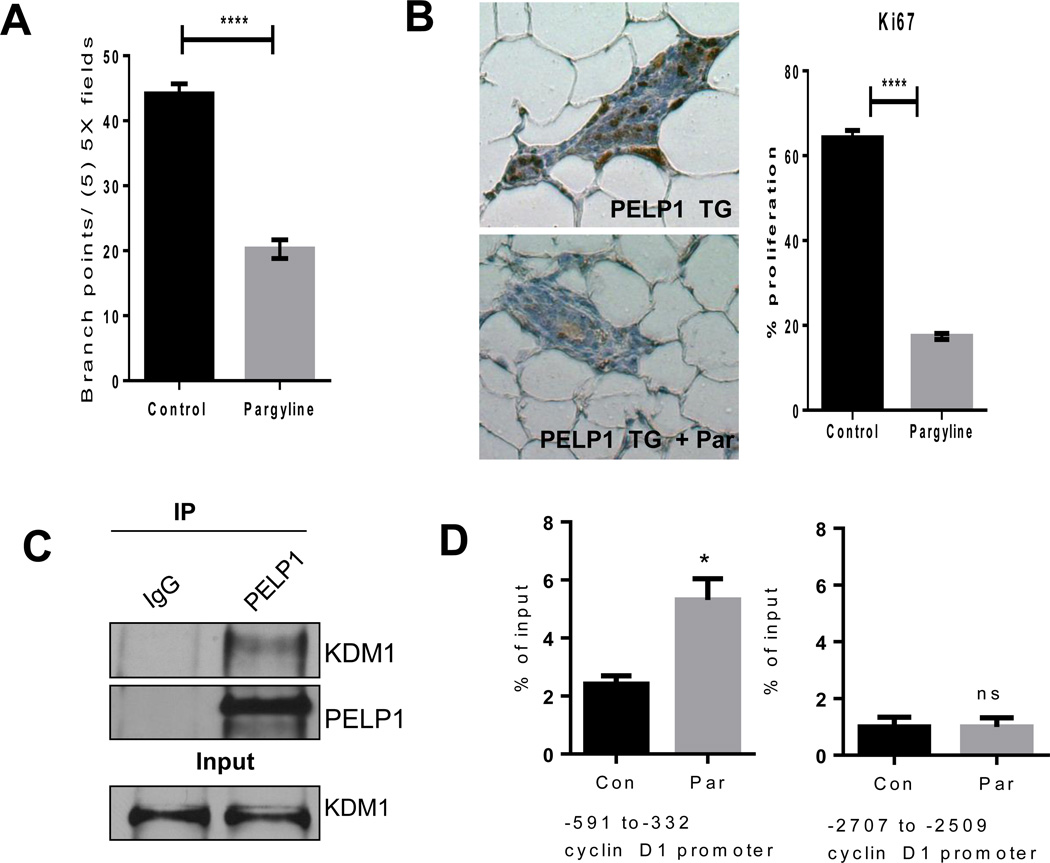
Since PELP1 functions as a coregulator of ER, we hypothesized that PELP1-mediated mammary tumorigenesis could be mediated by alterations of specific genes in the mammary gland. To test this, we investigated the differential expression of breast cancer–focused genes between control and PELP1 transgenic mammary glands using the real-time RT2 Profiler™ PCR array. Out of the 84 mouse breast cancer pathway–focused genes, 67 genes demonstrated at least a three-fold difference (up or down regulation) in expression (Fig. 5A). Of the several genes identified with significantly higher expression, we focused on validating the top candidate genes associated with the cell cycle progression and mammary gland ductal elongation. Specifically, we focused on genes that are shown in previous studies using in vitro model cells to be regulated by PELP1 and ERalpha. The changes in the expression pattern that were detected by PCR array were confirmed by using qRT-PCR (Fig. 5A). Western blot analysis confirmed upregulation of cyclin D1 and down regulation of PTEN in the PELP1 transgene expressing mammary glands (Fig. 5B). Further, IHC analysis revealed greater expression of cyclin D1 in PELP1 transgenic mammary glands than in the control mice, thus validating our qRT-PCR and western data (Supplementary Figure S2A). Earlier studies from our lab suggested that PELP1 recruits to the promoters of a number of ER target genes including cyclin D1 and promotes epigenetic changes. To examine whether PELP1 is recruited to the cyclinD1 gene promoter in mouse mammary glands, we performed ChIP analysis. Results from this experiment showed that both PELP1 and ER alpha are recruited to the proximal region (−591 to −332) of mouse cyclinD1 proximal promoter(20) (Fig. 5C) in PELP1 Tg mouse mammary glands, with no recruitment seen at distal region (-2707-2509) (data not shown). Since PELP1 is a substrate of both CDK4 and CDK2, and since we observed more expression of cyclin D1 and CDK2 in the PELP1 transgene–expressing mammary glands, we examined whether increased expression of cell cycle regulators potentiated an autocrine loop by phosphorylating PELP1. IHC analysis revealed more phosphorylation of PELP1 at ser991, a site phosphorylated by CDKs (Supplementary Figure 2B). Since PELP1 transgene–expressing mammary glands exhibited more EGFR and EGF gene expression, we also analyzed activation of downstream effectors of EGFR pathways such as Phos-AKT and PhosMAPK. Higher levels of phosphorylated MAPK as well as phosphorylated Akt were detected in the PELP1 transgene–expressing mammary glands than in the control mammary glands (Supplementary Figure S3A, B). Earlier studies using in vitro models suggested that PELP1 deregulation promotes changes in the epigenetic mark such as H3K9me2 that contributes to the activation of genes via its association with KDM1 (17), and that PELP1-mediated alterations in H3K9me2 can be reversed by pargyline, an inhibitor of KDM1. Accordingly, treatment of MMTV-rtTA-TetO-PELP1 bitransgenic mice with pargyline (80 mg/Kg/ every other day/ip) for 4 months reversed the PELP1 transgene–mediated effects, including less hyperplasia and hyper-branching were observed and were similar to the control mice (Fig. 6A). Further, pargyline treatment resulted in less expression of Ki67 (Fig. 6B). Immunoprecipitation results showed that PELP1 can form complex with KDM1 in transgenic mammary gland (Fig. 6C). ChIP studies on cyclin D1 promoter showed increased dimethyl modification at H3K9, a mark that is removed by histone demethylase KDM1. These results further supports the hypothesis that PELP1 mediated oncogenic functions may involve KDM1 functions (Fig. 6D). Accordingly, IHC examination revealed more expression of the inhibitory histone methyl mark H3K9me2, lower cyclin D1 levels and less phosphorylation of PELP1 in paragyline treated PELP1 transgene–expressing mammary glands compared to control Tg mammary glands (Supplementary Figure S4A, B). Collectively, the results from these studies revealed that PELP1 overexpression leads to activation of several ER alpha target genes involved in cellular proliferation (cyclin D1 and CDKs) and morphogenesis (EGFR and MMPs), and that PELP1-mediated epigenetic changes may play a role in PELP1-mediated mammary tumor initiation.
Figure 5.
Effect of PELP1 transgene expression on the expression of breast cancer pathway genes in the mammary gland. A, Gene profile of PELP1-mediated alterations in genes in the mammary gland was analyzed by using real-time RT2 ProfilerTM pathway–focused PCR array. Total RNA isolated from 6-month-old control and MMTV-rtTA-TetO-PELP1 mice was used. The list of the top five genes is shown in the table. Validation of five representative genes was done by using real-time RT-PCR. All experimental data points were generated from three biological replicates per transgenic line (left panel). Statistical significance was determined by using a Student’s t test.****p<0.0001; *p<0.05. B, Western blot analyses of the status of cyclin D1, PTEN and ER-alpha in control and MMTV-rtTA-TetO-PELP1 mammary glands (left panel). Actin was used as control. WT, wild type; M+ MMTV-rtTA transgenic mice; P+ TetO-PELP1 mice ; and M+P+, MMTV-rtTA-TetO-PELP1 bitransgenic mice. C, ChIP assay was done using the DNA isolated from the mammary glands of 6 months old doxycycline-treated PELP1 bitransgenic animals with antibodies specific for PELP1, ER or the isotype IgG control. DNA recovered from ChIP or input controls was subjected to real-time quantitative PCR with primers specific to −591 to −332 that represent the proximal promoter regions of mouse cyclin D1 promoter. Statistical significance was determined by using a Student’s t test. *p<0.05.
Figure 6.
Effect of pargyline treatment on PELP1-induced hyperplasia. A, Side branching quantification (means +/− sem) in the mammary glands of 6-month-old MMTV-rtTA-TetO-PELP1 bitransgenic mice treated with PBS or pargyline. All experimental data points were generated from three biological replicates per transgenic line. Statistical significance was determined by using a Student’s t test.****p<0.0001. B, Mammary epithelial proliferation as reflected by IHC analysis of Ki-67 expression in the mammary glands of 6-month-old MMTV-rtTA-TetO-PELP1 bitransgenic mice treated with PBS or pargyline. Proliferation index was calculated relative to a percentage of total epithelial cell nuclei counts in five 20X fields per slide. All experimental data points were generated from three biological replicates per transgenic line. Statistical significance was determined by using a Student’s t test.****p<0.0001. C, Total lysates from the mammary glands of 6-month-old MMTV-rtTA-TetO-PELP1 bitransgenic mice were subjected to immunoprecipitation using the PELP1 antibody and PELP1–KDM1 interaction was verified by western analysis. D, ChIP assay was done using the DNA isolated from the mammary glands of 6-month-old MMTV-rtTA-TetO-PELP1 bitransgenic mice (n=3) treated with PBS or pargyline with antibodies specific for dimethyl H3K9. DNA recovered from ChIP or input controls was subjected to real-time quantitative PCR with primers specific to −591 to −332 and −2707 to −2509 that represent the proximal and distal promoter regions of mouse cyclin D1 promoter. Statistical significance was determined by using a Student’s t test. *p<0.05.
Discussion
Changes in ERalpha-associated coregulator expression have been demonstrated to substantially contribute to ERα activity and often correlate with a poor prognosis for breast cancer patients (21–23). PELP1 is a proto-oncogene that functions as an ER coregulator and exhibits over expression in breast tumors. Despite significant progress in understanding the role of PELP1 in ER signaling using in vitro models, no in vivo models were available to elucidate its significance in the initiation and progression of breast cancer. Thus, we used an engineered mouse model (MMTV-rtTA-TetO-PELP1 bitransgenic mice) to assess the functional role of PELP1 in breast tumor initiation and progression. Our results demonstrated that PELP1 is a critical regulator of ductal morphogenesis and that deregulation of PELP1 expression is sufficient to initiate mammary tumorigenesis.
In our Tg mouse model, PELP1 expression is under control of a doxycycline-regulated promoter. To avoid developmental artifacts, we initiated induction of the PELP1 transgene at 8 weeks of age. We observed a 3 to 4-fold induction of PELP1 transgene over endogenous levels, which is very similar to the levels detected in human breast tumors. Further, transgene expression was maintained in aged PELP1 Tg mice up to 22 months. Mammary glands of MMTV-rtTA-TetO-PELP1 Tg mice had premalignant lesions. Control MMTV-rtTA and TetO-PELP1 mice did not have any signs of premalignant lesions. PELP1 Tg mammary glands and mammary lesions showed high levels of Ki67 expression, which is indicative of proliferation. Many of the premalignant lesions progressed to mammary tumors as the mice aged. Further, PELP1 premalignant lesions and mammary tumors had high levels of cyclin D1 expression. Earlier in vitro studies using breast cancer models showed that cyclin D1 is a PELP1 target gene (24). Our in vivo data also support this observation. Since cyclin D1 is frequently overexpressed in breast cancers and implicated in cancer progression (25), high cyclin D1 level in the PELP1-induced mammary lesions indicated that PELP1-mediated tumorigenesis may involve modulation of the cyclin D1 pathway. We failed to see any significant differences in the tumor latency in multiparous animals compared to nulliparous animals. Since our earlier studies showed that PELP1 deregulation induces local E2 synthesis via aromatase regulation (26) and such findings may explain to some extent the lack of exacerbate phenotype in PELP1 Tg multiparous animals, however, future studies are needed. Our ongoing studies will address the role of local estrogen in PELP1 mediated tumorgenesis and also test the effect of PELP1 overexpression on various stages of development of mammary gland.
Estrogens induce proliferation of ER-positive breast epithelial cells by stimulating G1/S transition (27). Our recent results identified the ER coregulator PELP1 as a novel substrate of CDKs, and PELP1 is sequentially phosphorylated by the CDK4/cyclin D1, CDK2/cyclin E and CDK2/cyclin A complexes (12). PELP1 can couple E2 signaling to the E2F axis, and CDK phosphorylation plays a key role in the PELP1 oncogenic functions (12). EGFs activate the CDK4/cyclin D1 complexes (28) and PELP1 is shown to be phosphorylated at CDK sites when treated with EGF (29). Gene expression analysis revealed more expression in a number of cell cycle regulator genes including CDK2, cyclin A1, and CDK2 with concomitantly less expression of the CDK inhibitor CDKN2a and PTEN in PELP1 Tg mammary glands than in the control glands. Further, PELP1 Tg mammary glands exhibited increased phosphorylation of PELP1 at the CDK site, suggesting functional activation of upregulated CDK pathways. These results also suggest that PELP1 deregulation may enhance tumor growth by promoting a CDK–PELP1 functional autocrine signaling loop.
The steroid hormone estradiol (E2) plays an important role in the development of the mammary gland. ER alpha is the major ER subtype in the mammary epithelium and its importance in mammary gland biology and development has been confirmed by studying ERalpha knockout (KO) mice. These mice display grossly impaired ductal epithelial cell proliferation and branching (30, 31). EGF and EGFR play critical roles in mammary gland branching, and extensive crosstalk in signaling occurs between ER alpha and EGFR signaling (32). Mammary gland branching morphogenesis is also dependent, in part, on the extracellular matrix–degrading enzymes, including matrix metalloproteinases (MMPs) (33). Earlier studies suggested that PELP1 has the potential to regulate expression of several genes involved in the epithelial mesenchymal transition (EMT) including MMPs (34, 35). In our transgenic mice model, induction of PELP1 expression by doxycycline significantly enhanced mammary ductal branching and morphogenesis. Gene expression studies revealed higher expression levels in a number of genes involved in morphogenesis including EGFR, MMP9, MMP2 and Notch in PELP1-overexpressing mammary glands than in control mammary glands. These findings, in part, explain the mammary morphogenesis phenotype observed in PELP1 Tg mice. However, future studies are needed to examine the mechanisms by which PELP1 modulates the expression of genes involved in morphogenesis. Since we analyzed gene expression changes using a focused microarray containing 84 genes, additional studies using whole genome approaches such as RNAseq and CHIPseq are needed to further understand the mechanisms by which PELP1 deregulation promotes branching and morphogenesis phenotypes.
Deregulation of the ER coactivator SRC3 (AIB1) was reported in breast tumors (21, 22). ER coregulatory proteins have been suggested to play roles in the generally observed tissue-specific effects of tamoxifen (36). SRC3 (AIB1) KO mice studies demonstrated that normal expression of the coactivator SRC3 (AIB1) is required for initiation of tumorigenesis by carcinogens and oncogenes (37, 38). Using inducible expression of PELP1 directed into the mammary gland of Tg mice, we provided evidence that PELP1 is another ER coregulator whose deregulation has potential to promote mammary tumor initiation. PELP1 interacts with and functions as a coactivator of a number of transcription factors that play a critical role in mammary gland biology including ER, E2F and STAT3(10, 11). Further, PELP1 interacts with and recruits a number of epigenetic modifiers (such as KDM1) in the target chromatin and thus facilitates activation of number genes involved in proliferation and cancer progression (19). Our recent RNAseq studies identified 318 genes as PELP1 regulated genes and many of them are involved in breast cancer progression (39) and that PELP1 is also involved in splicing of genes. Collectively, multiple roles played by PELP1 in cellular signaling may contribute to important changes in seen the mammary gland of PELP1 transgenic mice.
Approximately 70% of breast tumors are ER-positive at the time of presentation (6, 40, 41). PELP1 expression provides ER-positive breast cancer cells with a distinct growth and survival advantage (12). PELP1 overexpression has been noted in many hormone-related cancers (10, 42), and PELP1 promotes E2-mediated cell proliferation. In the ER-positive/luminal-like group of tumors, PELP1 is prognostically linked to shorter breast cancer–specific survival (13, 43) and to therapy resistance (14, 44). In our studies, PELP1 deregulation promoted hyperplasia and mammary tumors that were predominantly ER positive. Even though hyperplasia was seen as early as 3 months, progression to mammary adenocarcinomas had a long latency of between 14 and 18 months. This long latency in the development of mammary tumors in PELP1 Tg mice could be due to the nature of slow growth characteristics of ER-positive mammary tumors. Similar long latencies were also reported in other TG mouse models such as MMTV AIB1 (21–23). Another possibility is that delay in tumorigenesis in PELP1 Tg mice may indicate need for acquiring additional mutations. This is supported by the fact that some of PELP1 Tg mice developed tumors as early as 8 months. Since coregulators are shared by multiple transcription factors and some of these coactivation functions may promote tumor suppression, and tumor promotion may be delayed until such pathways are inactivated. In support of this, our recent study suggested that PELP1 also functions as co-activator of tumor suppressor p53 and such activation may counteract PELP1 oncogenic functions and could contribute to long latency (45).
Recent evidence suggests that ER coactivators, in addition to ER, are targets of growth factor signaling and growth factor-mediated phosphorylation of ER, and ER-coregulatory proteins have been shown to have a role in tamoxifen resistance (46). ER coregulators are targeted by excessive ER–HER2 crosstalk leading to hormone resistance in a subset of breast tumors (46). PELP1 deregulation and/or its mislocalization in the cytoplasm can contribute to the activation MAPK and AKT pathways by coupling the ER–Src pathways (47). In our transgenic mouse model, wild-type PELP1 was used and it predominantly localized in the nuclear compartment, as we expected. Interestingly, gene expression studies revealed upregulation of EGF and down regulation of PTEN in the TG mice mammary glands than in the control mammary glands. Further, analysis revealed increased activation of MAPK and AKT pathways in PELP1 Tg–expressing mammary glands. We failed to see any activation of Src kinase in the mammary glands (data not shown). Collectively, these results suggest that PELP1 deregulation has the potential to activate MAPK and AKT indirectly by enhancing expression of EGF/EGFR or by downregulating PTEN. Increased activation of MAPK and AKT could have contributed to the increased proliferation and survival of mammary epithelial cells in PELP1 transgene–expressing mammary gland.
Estrogen stimulation induces various histone modifications at ER target gene promoters and is mediated by recruitment of coregulators to the target gene promoters by ER. Histone methylation-dependent mechanism impose ligand dependency for ER-mediated gene activation and may play a vital role in many neoplastic processes, representing a valuable therapeutic target (48). Previous studies showed KDM1 recruitment to a significant fraction of ER target genes, demethylation of H3-K4 and H3-K9 and demethylation of proximal histones enables ER-mediated transcription (49). Emerging evidence also suggests that PELP1 is involved in oncogenesis through its interaction with histones, acetyltransferases, deacetylases and the demethylase KDM1 (9,12). We observed decreased expression of H3k9me2 mark in PELP1-driven hyperplasia and mammary tumors. In our previous studies using preclinical models, we observed pargyline inhibited PELP1-driven tumorigenesis by blocking KDM1 functions (17). Similarly, pargyline treatment delayed the development of hyperplasia in PELP1 Tg mice and reduced the hyperbranching phenotype induced by PELP1 over expression. Since pargyline also has ability to inhibit MAO in addition to inhibition of KDM1, we cannot rule out the possibility that part of the results could be due to inhibition of MAO. In our previous in vitro studies (19), under conditions of KDM1 knockdown, pargyline showed little or no effect on PELP1 mediated functions, suggesting pargyline effects on PELP1 signaling require functional KDM1. Since PELP1 interacts with KDM1 in Tg mouse mammary gland and because pargyline increased dimethyl H3K9 mark at cyclinD1 promoter, we reason that effects observed are due to parglyine inhibition of KDM1. However, future studies are clearly needed using a specific inhibitor of KDM1. These findings underscore the importance of epigenetic modifications in PELP1 oncogenic functions in vivo.
In summary, our data using novel inducible transgenic mouse model provided the first in vivo evidence that overexpression of PELP1 in mammary epithelial cells initiate a pathologic state, leading to breast cancer initiation and progression. The proposed PELP1 Tg model will provide an opportunity to study the biological events that underlie the PELP1 actions in pathophysiological conditions and sets a stage for future investigations to test pharmacologic interventions in conditions of PELP1 deregulation.
Supplementary Material
Acknowledgements
We thank Dr. Lewis A. Chodosh, University of Pennsylvania School of Medicine for providing MMTV-rtTA transgenic mice, Challa Ram Babu for managing mice colony, Dimple Chakravarty for help with vector construction, and Domnique Newallo for initial characterization of the transgene line, UTHSCSA Pathology Core for preparation of blocks, Dr. Martha Hanes for pathological analysis and UTHSC Houston for the generation of transgenic mice.
This study was supported by the NIH/NCI grant CA095681 (RKV) Komen grant KG090447 (RKV), Komen grant KG110812 (RRT) and NIH F31 awardF31CA165814 (CV) and the Cancer Therapy and Research Center at the University of Texas Health Science Center at San Antonio through the NCI Cancer Center Support Grant P30CA054174-17.
Footnotes
Disclosure of Potential Conflicts of Interest
Authors declare no conflicts of interest
Reference List
- 1.Osborne CK. Steroid hormone receptors in breast cancer management. Breast Cancer Res Treat. 1998;51:227–238. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- 2.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–1481. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 3.Normanno N, Di MM, De ME, De LA, de MA, Giordano A, et al. Mechanisms of endocrine resistance and novel therapeutic strategies in breast cancer. Endocr Relat Cancer. 2005;12:721–747. doi: 10.1677/erc.1.00857. [DOI] [PubMed] [Google Scholar]
- 4.Musgrove EA, Sutherland RL. Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer. 2009;9:631–643. doi: 10.1038/nrc2713. [DOI] [PubMed] [Google Scholar]
- 5.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell DP, Norris JD. Connections and regulation of the human estrogen receptor. Science. 2002;296:1642–1644. doi: 10.1126/science.1071884. [DOI] [PubMed] [Google Scholar]
- 7.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–587. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 8.Singh RR, Kumar R. Steroid hormone receptor signaling in tumorigenesis. J Cell Biochem. 2005;96:490–505. doi: 10.1002/jcb.20566. [DOI] [PubMed] [Google Scholar]
- 9.Lonard DM, O’Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vadlamudi RK, Kumar R. Functional and biological properties of the nuclear receptor coregulator PELP1/MNAR. Nucl Recept Signal. 2007;5:e004. doi: 10.1621/nrs.05004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Girard BJ, Daniel AR, Lange CA, Ostrander JH. PELP1: A review of PELP1 interactions, signaling, and biology. Mol Cell Endocrinol. 2013;10 doi: 10.1016/j.mce.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nair BC, Nair SS, Chakravarty D, Challa R, Manavathi B, Yew PR, et al. Cyclin-dependent kinase-mediated phosphorylation plays a critical role in the oncogenic functions of PELP1. Cancer Res. 2010;70:7166–7175. doi: 10.1158/0008-5472.CAN-10-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Habashy HO, Powe DG, Rakha EA, Ball G, Macmillan RD, Green AR, et al. The prognostic significance of PELP1 expression in invasive breast cancer with emphasis on the ER-positive luminal-like subtype. Breast Cancer Res Treat. 2009;120:603–612. doi: 10.1007/s10549-009-0419-9. [DOI] [PubMed] [Google Scholar]
- 14.Vallabhaneni S, Nair BC, Cortez V, Challa R, Chakravarty D, Tekmal RR, et al. Significance of ER-Src axis in hormonal therapy resistance. Breast Cancer Res Treat. 2010;130:377–385. doi: 10.1007/s10549-010-1312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarty D, Nair SS, Santhamma B, Nair BC, Wang L, Bandyopadhyay A, et al. Extranuclear Functions of ER Impact Invasive Migration and Metastasis by Breast Cancer Cells. Cancer Res. 2010;70:4092–4101. doi: 10.1158/0008-5472.CAN-09-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajhans R, Nair S, Holden AH, Kumar R, Tekmal RR, Vadlamudi RK. Oncogenic Potential of the Nuclear Receptor Coregulator Proline-, Glutamic Acid-, Leucine-Rich Protein 1/Modulator of the Nongenomic Actions of the Estrogen Receptor. Cancer Res. 2007;67:5505–5512. doi: 10.1158/0008-5472.CAN-06-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortez V, Mann M, Tekmal S, Suzuki T, Miyata N, Rodriguez-Aguayo C, et al. Targeting PELP1-KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res. 2012;19(14):R108. doi: 10.1186/bcr3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, et al. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB J. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- 19.Nair SS, Nair BC, Cortez V, Chakravarty D, Metzger E, Schule R, et al. PELP1 is a reader of histone H3 methylation that facilitates oestrogen receptor-alpha target gene activation by regulating lysine demethylase 1 specificity. EMBO Rep. 2010;11:438–444. doi: 10.1038/embor.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, et al. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- 21.List HJ, Reiter R, Singh B, Wellstein A, Riegel AT. Expression of the nuclear coactivator AIB1 in normal and malignant breast tissue. Breast Cancer Res Treat. 2001;68:21–28. doi: 10.1023/a:1017910924390. [DOI] [PubMed] [Google Scholar]
- 22.Azorsa DO, Cunliffe HE, Meltzer PS. Association of steroid receptor coactivator AIB1 with estrogen receptor-alpha in breast cancer cells. Breast Cancer Res Treat. 2001;70:89–101. doi: 10.1023/a:1012972808558. [DOI] [PubMed] [Google Scholar]
- 23.Torres-Arzayus MI, Font de MJ, Yuan J, Vazquez F, Bronson R, Rue M, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 24.Balasenthil S, Vadlamudi RK. Functional interactions between the estrogen receptor coactivator PELP1/MNAR and retinoblastoma protein. J Biol Chem. 2003;278:22119–22127. doi: 10.1074/jbc.M212822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 26.Rajhans R, Nair HB, Nair SS, Cortez V, Ikuko K, Kirma NB, et al. Modulation of in situ Estrogen Synthesis by PELP1: Potential ER Autocrine Signaling Loop in Breast Cancer Cells. Mol Endocrinol. 2008;22:649–664. doi: 10.1210/me.2007-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster JS, Henley DC, Bukovsky A, Seth P, Wimalasena J. Multifaceted regulation of cell cycle progression by estrogen: regulation of Cdk inhibitors and Cdc25A independent of cyclin D1-Cdk4 function. Mol Cell Biol. 2001;21:794–810. doi: 10.1128/MCB.21.3.794-810.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paternot S, Dumont JE, Roger PP. Differential utilization of cyclin D1 and cyclin D3 in the distinct mitogenic stimulations by growth factors and TSH of human thyrocytes in primary culture. Mol Endocrinol. 2006;20:3279–3292. doi: 10.1210/me.2005-0515. [DOI] [PubMed] [Google Scholar]
- 29.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 30.Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc Natl Acad Sci U S A. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bocchinfuso WP, Korach KS. Mammary gland development and tumorigenesis in estrogen receptor knockout mice. J Mammary Gland Biol Neoplasia. 1997;2:323–334. doi: 10.1023/a:1026339111278. [DOI] [PubMed] [Google Scholar]
- 32.Sebastian J, Richards RG, Walker MP, Wiesen JF, Werb Z, Derynck R, et al. Activation and function of the epidermal growth factor receptor and erbB-2 during mammary gland morphogenesis. Cell Growth Differ. 1998;9:777–785. [PubMed] [Google Scholar]
- 33.Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast Cancer Res. 2004;6:1–11. doi: 10.1186/bcr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakravarty D, Roy SS, Babu CR, Dandamudi R, Curiel TJ, Vivas-Mejia P, et al. Therapeutic Targeting of PELP1 Prevents Ovarian Cancer Growth and Metastasis. Clin Cancer Res. 2011;17:2250–2259. doi: 10.1158/1078-0432.CCR-10-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy S, Chakravarty D, Cortez V, De MK, Bandyopadhyay A, Ahn JM, et al. Significance of PELP1 in ER-negative breast cancer metastasis. Mol Cancer Res. 2012;10:25–33. doi: 10.1158/1541-7786.MCR-11-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CL, O'Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 37.Kuang SQ, Liao L, Wang S, Medina D, O'Malley BW, Xu J. Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. [DOI] [PubMed] [Google Scholar]
- 38.Kuang SQ, Liao L, Zhang H, Lee AV, O'Malley BW, Xu J. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–1885. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- 39.Mann M, Zou Y, Chen Y, Brann D, Vadlamudi R. PELP1 oncogenic functions involve alternative splicing via PRMT6. Mol Oncol. 2014;8:389–400. doi: 10.1016/j.molonc.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lonard DM, O'Malley BW. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–414. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Ravindranathan P, Ramanan M, Kapur P, Hammes SR, Hsieh JT, et al. Central Role for PELP1 in Nonandrogenic Activation of the Androgen Receptor in Prostate Cancer. Mol Endocrinol. 2012;26:550–561. doi: 10.1210/me.2011-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar R, Zhang H, Holm C, Vadlamudi RK, Landberg G, Rayala SK. Extranuclear coactivator signaling confers insensitivity to tamoxifen. Clin Cancer Res. 2009;15:4123–4130. doi: 10.1158/1078-0432.CCR-08-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakravarty D, Tekmal RR, Vadlamudi RK. PELP1: A novel therapeutic target for hormonal cancers. IUBMB Life. 2010;62:162–169. doi: 10.1002/iub.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nair BC, Krishnan SR, Sareddy GR, Mann M, Xu B, Natarajan M, et al. Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell Death Differ. 2014;21:1409–1418. doi: 10.1038/cdd.2014.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, et al. Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst. 2004;96:926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 47.Vadlamudi RK, Manavathi B, Balasenthil S, Nair SS, Yang Z, Sahin AA, et al. Functional implications of altered subcellular localization of PELP1 in breast cancer cells. Cancer Res. 2005;65:7724–7732. doi: 10.1158/0008-5472.CAN-05-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Bassets I, Kwon YS, Telese F, Prefontaine GG, Hutt KR, Cheng CS, et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.