Abstract
Large conductance, voltage- and Ca2+-gated K+ (BKCa) channels play a critical role in smooth muscle contractility and thus represent an emerging therapeutic target for drug development to treat vascular disease, gastrointestinal, bladder and uterine disorders. Several compounds are known to target the ubiquitously expressed BKCa channel-forming α subunit. In contrast, just a few are known to target the BKCa modulatory β1 subunit, which is highly expressed in smooth muscle and scarce in most other tissues. Lack of available high-resolution structural data makes structure-based pharmacophore modeling of β1 subunit-dependent BKCa channel activators a major challenge. Following recent discoveries of novel BKCa channel activators that act via β1 subunit recognition, we performed ligand-based pharmacophore modeling that led to the successful creation and fine-tuning of a pharmacophore over several generations. Initial models were developed using physiologically active cholane steroids (bile acids) as template. However, as more compounds that act on BKCa β1 have been discovered, our model has been refined to improve accuracy. Database searching with our best-performing model has uncovered several novel compounds as candidate BKCa β1 subunit ligands. Eight of the identified compounds were experimentally screened and two proved to be activators of recombinant BKCa β1 complexes. One of these activators, sobetirome, differs substantially in structure from any previously reported activator.
Keywords: BKCa Channel, beta subunit, pharmacophore model
Introduction
Functional BKCa channels result from homomeric assembly of four α (slo, slo1) subunits. In most tissues, however, the native channel complex includes modulatory β subunits that modify ion channel phenotype.[1] Due to the ubiquitous expression of the BKCa channel-forming α subunit, these channels play a significant role in many physiological functions including neuronal excitability, neurotransmission, hormonal secretion and regulation of smooth muscle tone.[2, 3] BKCa α subunits consist of seven transmembrane segments [4] in addition to a long C-terminal tail that recognizes several physiologic ligands, including intracellular Ca2+.[5] In contrast, BKCa β types (β1-4) are small proteins, each made of intracellular N- and C-termini, and two transmembrane helices connected by a large extracellular loop.[6]
Differential expression of β subunit types across tissues allows tissue-specific BKCa current phenotypes that contribute to cell physiology in a rather tissue-specific manner. In particular, when co-expressed with the α subunit, the smooth muscle-abundant β1 subunit increases the channel’s apparent Ca2+ sensitivity and slows down channel gating kinetics.[7] In smooth muscle, an increase in the channel’s apparent Ca2+ sensitivity allows BKCa channels to activate upon increase in intracellular Ca2+ levels near the BKCa channel, which are raised during membrane depolarization and smooth muscle contraction. Thus, BKCa channel activation results in outward K+ currents that tend to hyperpolarize the membrane and oppose depolarization-induced Ca2+ entry. Eventually, smooth muscle contraction is restrained and relaxation is facilitated.[6, 7] Moreover, the critical role of BKCa β1 subunit in smooth muscle physiology has been well documented: for instance, BKCa β1 knock-out mice are characterized by increased arterial tone.[7] Decreased β1 gene expression leads to systemic hypertension in rats [8] and increased phasic contractions in mouse urinary bladder.[9] Considering the key role of BKCa β1 in determining smooth muscle tone, this subunit represents an attractive target for therapeutic developments against a wide clinical spectrum of prevalent human conditions associated with enhanced smooth muscle contraction, including bronchial asthma, systemic hypertension, cerebral vasospasm, erectile dysfunction, premature labor, and some bladder disorders.[10–12]
There are several compounds known to modify BKCa channel function, yet only a few are confirmed to act via the recently identified β1 subunit cholane steroid-recognition site (Figure 1).[13, 14] Cholane steroids such as the bile acid, lithocholic acid (LCA), increase BKCa channel activity through interaction with β1 subunits.[14] This action is β1 subunit-specific and not observed with β2-4 subunit-containing BKCa complexes or with homomeric α channels.[15] Combined results from point mutagenesis, electrophysiology and computational modeling indicate that the cholane steroid-recognition site is located in the β1 subunit second transmembrane domain, with Thr169 playing a critical role in bile acid recognition.[16] Previous structure-activity relationship (SAR) studies indicated that the overall shape of the bile acid, as well as the number of hydroxyls, and placement of hydroxyls and carboxylic acids, were important in bile acid interaction with β1.[14] Among non-steroidal compounds, only one triterpenoid, 3-hydroxyolean-12-en-30-oic acid (3-HENA)[13] has been reported to specifically target β1-containing BKCa complexes. Moreover, although there are several ligands with β1-dependent effects on BKCa channel opening, only LCA and 3-HENA have been reported to interact with a defined recognition site [13] making the other ligands less useful in determination of a pharmacophore.
Figure 1.
BKCa β1 selective activators; lithocholic acid (LCA), 3-hydroxyolean-12-en-30-oic acid (3-HENA), and dehydrosoyasaponin I (DHS-I).
High-resolution structures of the BKCa α gating-ring are available,[17] however structures of neither the complete α subunit nor any β subunit are available. Given this deficit in receptor structural data, the present study focuses on ligand-based pharmacophore model development using BKCa-ligand structures found by our group to selectively target the cholane steroid-recognition site in BKCa β1. As new data and techniques have become available, multiple iterations of pharmacophore modeling have led to a better understanding of how pharmacophore features effect model performance, yet lack of structural diversity amongst known ligands has posed a significant challenge. The best performing model was used to search the PubChem compound database,[18] which led to the confirmation of two novel active compounds that fit structural criteria posed by the β1 steroid-sensing site while offering structural diversity over known ligands. The expanded structural diversity provided by these new BKCa channel activators sets the stage for future pharmacophore model refinements as well as further pharmacological characterization of these new biologically active compounds.
Methods
Pharmacophore Model Development
Initial modeling was performed using the Molecular Operating Environment (MOE)[19] 2006.08 software package. Stochastic conformational searching was performed on LCA (Figure 2 #14) using the MMFF94x forcefield, chosen for its ubiquitous use with small organic molecules, with gas phase solvation and a dielectric of zero. MMFF94x is an unpublished version of the MMFF94 forcefield [20] where conjugated nitrogen atoms are treated as planar instead of tetrahedral. The carboxylic acid of LCA was deprotonated to represent the expected prevalent ionization state of the molecule at physiological pH. Since LCA was reported to be the best activator among the 13 bile acids tested in a previous SAR study,[14] the lowest energy conformation of LCA was used to establish distance constraints (Figure 3A) for crude pharmacophore searching in the National Cancer Institute (NCI) database (http://cactus.nci.nih.gov/ncidb2.2/) using the 3D search feature. Hits selected in this manner proved largely to be unavailable for screening, providing incentive to develop pharmacophore models that could be used to search alternative collections of commercially available compounds.
Figure 2.
Compounds used in pharmacophore model development and testing. Active compounds are indicated with a + before the compound number. Inactive compounds are indicated with a – before the compound number. The most active compounds are denoted with an *. Compounds denoted with a ^ indicate previously unpublished compounds whose synthesis and/or electrophysiology are included in the supplemental material.
Figure 3.
Lithocholic acid is mapped to all generations of pharmacophore models for comparison. Magenta features represent hydrogen bond donors, green features represent hydrophobic centers, teal features represent hydrogen bond acceptors, and the red feature represents a carboxylic acid bioisostere. In the 5th generation model (panel E) the teal feature is a combination of a hydrogen bond acceptor and an anion. In the 6th generation model the teal feature is a hydrogen bond acceptor projection from overlapped hydrogen bond donors in the model training set. A) 1st generation crude model of pharmacophore using atomic distances. B) 2nd generation model developed for conformational database searching. C) 3rd generation model using 5 pharmacophore features. D) 4th generation model generated with more structurally diverse compounds. E) 5th generation model with volume reduced pharmacophore features. F) 6th generation model using ligand projections for a hydrogen bond acceptor (1) and three hydrophobic features (2–4).
A 2nd generation model (Figure 3B) was generated using the Pharmacophore Elucidator feature in MOE 2009.10 to allow for searching of databases other than the NCI database. LCA and deoxycholic acid (#24), previously established activators of β1-containing BKCa channels, were used as positive examples on which to train the model. Three additional bile acids that do not increase channel activity (#16, 17, 18) were used as negative examples.
After the discovery of 3-HENA (#39) as a non-steroidal β1 activator, a 3rd generation model was developed due to the structural diversity offered by 3-HENA [13]. The 3rd generation model (Figure 3C) was generated using LCA and 3-HENA as active compounds, while using two new compounds as inactive compounds to train the model (#8, 46) in addition to #17, which was used as an inactive compound in the previous generation. These compounds were selected to provide more structural diversity over inactive compounds used to train the 2nd generation model.
The 4th generation model (Figure 3D) was obtained using LCA (#24), 3-HENA (#39), and a synthetic bile acid containing a cyano functional group in place of the lateral chain carboxylic acid (#10) as active compounds. Three inactive compounds were selected for structural diversity, which are shown in Figure 2 as compounds #1, 42, 45.
Refinements were made to the 4th generation model to yield a 5th generation model (Figure 3E), as follows. 1) The radii of the pharmacophore features were reduced to be more exclusive, 2) the hydrogen bond acceptor feature was changed to include anionic compounds, 3) feature 2 was moved to align better with the steroid nucleus, and 4) features 1, 3, and 4 were made “essential” features. Thus, any screened compound would contain at least these three of the four features.
Finally, a 6th generation model was developed using newer techniques available in the MOE 2013.08 software that include ligand projection features for hydrogen bond donors and acceptors. The same training set of compounds used to develop models 4 and 5 was used to develop model 6 (Figure 3F).
Model Evaluation
Test set compounds were compiled from various publications [14, 21–23] and previously unpublished data. Electrophysiology data and synthetic schemes for unpublished compounds are provided as supplemental material. A conformational database was generated for test set compounds using LowModeMD conformational searching [24] and the MMFF94x forcefield in MOE 2013.08. Conformations were discarded if their internal energy exceeded the lowest energy conformation by 50 kcal/mol to provide more conformational diversity over the default 7 kcal/mol cutoff. The entire set of compounds used to train any generation of pharmacophore model (compounds #1, 8, 10, 14, 16, 17, 18, 24, 39, 42, 45, and 46) were removed from the standard test set and 37 compounds remained to calculate pharmacophore validation metrics for a common test set to allow comparison of model performance. Sensitivity, specificity, percent yield of actives, enrichment, and accuracy were calculated for each model, as discussed in detail by Triballeau et al.[25] Sensitivity (Equation 1 below) represents the ability of the model to correctly pick out actives, while specificity (Equation 2) indicates the model’s ability to correctly reject inactive compounds. The yield of actives (Equation 3) indicates the proportion of active molecules selected by the model. Enrichment (Equation 4) is a measure of how well the model enriches the hit list with actives compared to the proportion of actives in the whole test set. Accuracy (Equation 5) is a measure of how well the model can discriminate between experimentally active and inactive compounds. A brief description of the calculations is given below:
| 1 |
| 2 |
| 3 |
| 4 |
| 5 |
where TP is the number of true positives, TN is the number of true negatives, FP is the number of false positives, FN is the number of false negatives, n is the number of compounds selected by the model, N is the total number of entries in the database, and A is the total number of active compounds in the database. In this use, a positive compound is one identified to be active by the model, while a negative compound is identified to be inactive by the model. The term true indicates the result from the model agrees with the experimental result, while false indicates a result from the model that disagrees with the experimental result. For example, a true positive is a compound predicted by the model to be active that is experimentally active and a false positive is a compound predicted by the model to be active that is experimentally inactive. For Equations 1, 2, 3, and 5, a value closer to one indicates better performance. For Equation 4, values greater than one indicate performance better than random selection.
Database Searching
Database searching was performed in the 3D compound database, which contains up to 10 conformations per molecule, downloaded from the PubChem FTP site (ftp://ftp.ncbi.nlm.nih.gov/pubchem/Compound_3D/10_conf_per_cmpd/). The compounds were imported into MOE 2012.10 and the 3rd generation model was used to query the database. The number of compounds returned from the database search was reduced by excluding anything with a LogP below 1.7 and above 5.9. These values represent the LogP values of compounds #25 and #39, respectively and were used because they are the highest and lowest LogP values of the most potent activators. The molecular weights of compounds #25 and #39 were used to further reduce the number of compounds by excluding those whose molecular weight fall outside the range of 198.2620 to 455.7030 g/mol. Finally, since the steroid recognition site is located in the transmembrane part of the helix, compounds that would be too polar for membrane insertion were removed. This was accomplished by calculating the number of non-carbon heavy atoms in each molecule. Any compound with more than four non-carbon heavy atoms was removed from the hit list, since all but one known active (#20) contain no more than four non-carbon heavy atoms.
Clustering was performed on the remaining compounds by first calculating MACCS structural keys (a binary description of structure)[26] and similarity was determined using a 65% Tanimoto coefficient (a measure of similarity between two compounds)[27] similarity threshold. The clustering overlap threshold was set at 65% or greater; meaning two compounds would require 65% or greater Tanimoto similarity among their structural neighbor lists to be placed in the same cluster. The formal charge on compounds in each cluster was used to further reduce the number of compounds by eliminating compounds with no formal charge, leaving only compounds with -1 or -2 charges to mimic the charge on the most active compounds (LCA and 3-HENA).
Automated Patch Clamp Procedures on Recombinant BKCa
Cell culture
Recombinant BKCa channel-forming cbv1 (AY330293) and accessory β1 (FJ154955) subunits cloned from rat cerebral artery myocytes [28, 29] were stably expressed in the Human Embryonic Kidney (HEK) 293 cell line (ATCC). Cells were grown in sterile tissue flasks in Dulbecco’s Modified Eagle Medium (DMEM; Cellgro) supplemented with 10% (v/v) heat-inactivated fetal calf serum (FCS; Hyclone) and penicillin:streptomycin (6:100 μg per 1 mL) solution (Cellgro) at 35°C in 5% CO2. Confluent cell cultures were sub-cultured every 2–3 days. Before electrophysiological experiments, cells were treated with trypsin/EDTA (Invitrogen) for 1 min at 35 C, in 5% CO2, harvested, and centrifuged at 1,000 rpm for 5 min. The pellet was washed once with PBS (Cellgro), and the resulting cell suspension was centrifuged again. The pellet was gently re-suspended in the extracellular solution and used for electrophysiological recording.
Solutions
The extracellular solution was buffered to pH 7.4 with an overall osmotic concentration of 298 mOsm and contained (mM): NaCl 140; KCl 4; CaCl2 2; MgCl2 1; D-glucose monohydrate 5; HEPES 10. The seal enhancing solution was buffered to pH 7.4 and contained (mM): NaCl 80; KCl 3; MgCl2 10; CaCl2 35; HEPES 10. The internal Ca2+-free solution was buffered to pH 7.2 and contained (mM): KCl 50, NaF 10; KF 60; EGTA 5; HEDTA 1.6; HEPES 10; MgCl2 2.28. For vehicle- or compound-containing solution internal solution was supplemented by a Ca2+/EGTA stock (1/0.1 M) to render nominal 10 μM Ca2+. The nominal free Ca2+ concentration was calculated with MaxChelator Sliders (C. Patton, Stanford University). All solutions were filtered through 0.45 μM Millex filter unit (Millipore) and stored for no more than 1 week at +4°C.
Compounds
Sobetirome was purchased from APExBIO; PubChem CID 3307939 and 7312086 from MCULE. The remaining compounds were purchased from Cayman Chemical. A 1/9 (v/v) DMSO/EtOH mixture was used as vehicle. The stock solution for each compound was prepared at 333 mM using the aforementioned mixture. On the days of the experiment, the compound stock solution was diluted in the extracellular recording solution to 45 μM. This concentration represents the EC50 for the BKCa channel activator lithocholic acid [12]. The DMSO/ethanol vehicle (≤0.015/≤0.13% final concentrations) in the extracellular solution was used as control.
Patch-clamp recording on Patchliner (Nanion Technologies)
On the day of the experiment, a 500-mL aliquot of the cell suspension in the extracellular solution was placed in the cell-hotel. Cells were added to each chip well and attracted to the holes by suction applied by a pressure controller. Once a stable whole-cell configuration was obtained, the experimental protocol was initiated. K+ currents were measured with a patch–voltage-clamp technique in the whole-cell configuration at room temperature using the Quadro EPC-10 amplifier and associated PatchMaster software (HEKA Elektronik). Currents were low-pass filtered using the internal Bessel filter of the EPC-10 and digitized at 50 kHz. Whole-cell ionic currents were evoked from a holding potential of -80 mV by 200 milliseconds long, 10 mV steps ranging from −150 to +150 mV. After whole-cell currents were recorded in control, vehicle-containing solution, the extracellular solution was switched to the compound solution. Cells were incubated with compound for 20 sec, and the currents were recorded. Between the compounds, the Patchliner pipette was washed with 2% DMSO.
Data analysis
Measurement of the whole-cell BKCa current amplitude at each voltage step was performed using PatchMaster software (HEKA Electronik) 100 msec after the start of the depolarizing step. Macroscopic conductance G/Gmax–V plots were fitted to a Boltzmann function of the type G(V) = Gmax/1 + exp[(−V + V1/2)/k]. Boltzmann fitting routines were run in Origin software (OriginLab) that was also used for the final fitting and plotting of the data. As a measure of channel activity we used Vhalf, i.e., the voltage needed to reach half-maximal macroscopic conductance. Therefore, a decrease in Vhalf indicates an increase in BKCa current while an increase in Vhalf reflects a reduction in BKCa current, independently of changes in ion channel expression. Statistical analysis was conducted by InStat 3.0 (GraphPad) using one-way analysis of variance and Bonferroni’s multiple comparison test. P < 0.05 was considered statistically significant. Data are expressed as mean ± S.E.M., and (n) = number of cells. Each cell was only tested once with control (vehicle) and a single compound.
Results and Discussion
Model Selection
For the 1st generation model, three points were established on the LCA molecule. These points were based in part on a previous study [22]; two points underscored the importance of the lateral chain carboxylic acid and the A ring hydroxyl. A carbon in the steroid nucleus was selected as the third point to represent the concavity of LCA due to a cis A-B ring junction and is shown in Figure 3A. The distances between the points and the ranges used to search the NCI database are shown in Table 1. Since LCA has a LogP of 6.05, a LogP range of 4 to 8 was also used as additional search criteria to narrow the results. Due to the limitations of the browser-based search features of the NCI database and limited availability of compounds in that database, this method was abandoned in favor of more robust three-dimensional model development using MOE. While the 1st generation model provided a starting point, it was inherently limited to 3D distance descriptions of the pharmacophore and did not involve inactive compounds for model development.
Table 1.
Distance constraints used in NCI database searching.
| Pharmacophore Points | Lithocholic Acid Distances | NCI Search Distances |
|---|---|---|
|
| ||
| 1→2 | 4.93Å | 4.60Å→5.80Å |
| 1→3 | 10.48Å | 9.00Å→11.00Å |
| 2→3 | 7.28Å | 6.50Å→8.30Å |
The 2nd generation pharmacophore used LCA and deoxycholic acid as active compounds to develop the model and provided a more robust description of the steroid recognition site in β1 by using four features to describe the pharmacophore in three dimensions (Figure 3B). The set of potential models was analyzed for hydrophobic features that were spaced to convey the “bean” shaped concavity of LCA. Candidate models were also screened using the criteria that the model should contain a hydrogen bond donor feature aligned with the hydroxyl of LCA and a hydrogen bond acceptor feature aligned with the carboxylate of LCA as these two functional groups very likely play a role in the interaction with β1 based on previous SAR, modeling and mutagenesis studies reported by our groups.[13, 16, 22, 30, 31] The top candidate model was tested for accuracy using a test set of compounds with experimentally known activity on β1-containing BKCa channels that excluded the compounds used to train all models so that all models were evaluated using the same set of compounds.
For the 3rd generation model, the top candidate model was selected from the potential models using the same selection criteria as the 2nd generation model. An additional feature, however, was allowed in the 3rd generation model: a carboxylate ion bioisostere feature that allows other functional groups in place of the carboxylate (Figure 3C).
The top candidate pharmacophore model in the 4th generation was selected using the previously mentioned criteria for the 2nd generation model, but the placement of hydrophobic features was more heavily scrutinized than previously. This was accomplished by ensuring any candidate models had hydrophobic features in the steroid nucleus of the active molecules and not on methyl groups (Figure 3D).
For the 6th generation model, the two top models ranked by the model development algorithm were selected to evaluate with the test set. The models differed only in the first feature, where the top model had a hydrogen bond donor projection and the second ranked model had a hydrogen bond acceptor projection. These projections place pharmacophore features on the projected interaction sites for hydrogen bond donor and acceptor atoms in a molecule and could find a partner hydrogen bond acceptor or donator interaction partner common to all activators of the cholane steroid-recognition site. The second model was selected as the 6th generation model after evaluation of performance against the test set compounds (Figure 3E).
Distances between pharmacophore features in models 2–6 are provided in Table 2 and the radius of each feature is provided in Table 3. Because models are generated using conformational databases of active and inactive compounds, distances between similar features are not necessarily the same. The radii of pharmacophore features were generally left at default values except in the 5th generation model where feature radii were intentionally decreased to increase specificity.
Table 2.
Distance (Å) between pharmacophore features in generations 2–6
| Pharmacophore Points | 2nd Generation | 3rd Generation | 4th Generation | 5th Generation | 6th Generation |
|---|---|---|---|---|---|
|
| |||||
| 1→2 | 4.08 | 4.60 | 5.33 | 5.10 | 5.12 |
| 1→3 | 9.58 | 9.63 | 11.95 | 11.95 | 7.68 |
| 1→4 | 8.99 | 9.00 | 8.57 | 8.57 | 13.00 |
| 1→5 | - | 10.18 | - | - | - |
| 2→3 | 7.61 | 7.39 | 9.43 | 9.27 | 2.58 |
| 2→4 | 5.15 | 4.56 | 4.42 | 4.35 | 8.64 |
| 2→5 | - | 7.76 | - | - | - |
| 3→4 | 5.68 | 5.97 | 5.42 | 5.42 | 7.16 |
| 3→5 | - | 0.63 | - | - | - |
| 4→5 | - | 5.96 | - | - | - |
Table 3.
Radii (Å) of pharmacophore features in generations 2–6
| Pharmacophore Point | 2nd Generation | 3rd Generation | 4th Generation | 5th Generation | 6th Generation |
|---|---|---|---|---|---|
|
| |||||
| 1 | 1.4 | 1.4 | 1.6 | 1.0 | 1.0 |
| 2 | 1.4 | 1.4 | 1.4 | 1.3 | 1.4 |
| 3 | 1.4 | 1.4 | 1.6 | 1.3 | 1.4 |
| 4 | 1.4 | 1.4 | 1.4 | 1.3 | 1.4 |
| 5 | - | 1.4 | - | - | - |
Model Performance
The metrics used to evaluate the models are displayed in Table 4. These values are reported because they represent the most commonly reported pharmacophore metrics [25] and because each gives different insight into model performance.
Table 4.
Evaluation metrics for pharmacophore model generations 2–6 using the standard set of test compounds
| 2nd Generation | 3rd Generation | 4th Generation | 5th Generation | 6th Generation | |
|---|---|---|---|---|---|
|
| |||||
| True Positives | 9 | 5 | 10 | 9 | 13 |
| False Positives | 15 | 3 | 15 | 14 | 15 |
| True Negatives | 7 | 19 | 7 | 8 | 7 |
| False Negatives | 6 | 10 | 5 | 6 | 2 |
|
| |||||
| Sensitivity | 0.60 | 0.33 | 0.67 | 0.60 | 0.87 |
| Specificity | 0.32 | 0.86 | 0.32 | 0.36 | 0.32 |
| Yield of Actives | 0.36 | 0.63 | 0.40 | 0.39 | 0.46 |
| Enrichment | 0.89 | 1.54 | 0.99 | 0.97 | 1.15 |
| Accuracy | 0.43 | 0.65 | 0.46 | 0.46 | 0.54 |
The 2nd generation model scored comparably to the 4th and 5th generation models (Table 4) with the standard test set in some calculated metrics, but performed lower in others. Compared to the 4th generation model, this model was 0.07 lower in sensitivity, 0.04 lower in yield of actives, 0.10 lower in enrichment, and 0.03 lower in accuracy. This indicates the inclusion of more recently tested compounds with greater activity in the 4th generation training set led to improvements in model performance.
The 3rd generation model featured a fifth pharmacophore point, which more than doubled the specificity, increased the yield of actives by 0.27, the enrichment by 0.65, and the accuracy by 0.22, but had a sensitivity almost half that of the 2nd generation model with the standard test set. Additionally the 3rd generation model showed the greatest enrichment of any of the models generated. The fifth point was a carboxylate bioisostere intended to improve structural diversity among selected compounds. The low number of true positives and false positives indicate the fifth feature of this model is overly restrictive compared to four feature models. While this fifth feature did improve the model in several metrics, due to the large decrease in sensitivity, the placement and type of feature need to be further refined in future efforts to yield a model that better identifies the active compounds as true positives.
For the 4th generation model, the three most active compounds were used to train the model (#10, 14, 39). The drop in specificity, or true negative rate, of 0.54 compared to the 3rd generation model indicates that the 4th generation model cannot correctly eliminate inactive compounds as well as the 3rd generation. This model outperformed the 3rd generation model only in sensitivity by 0.34, so further refinements were sought to improve upon the 4th generation model, without sacrificing sensitivity as was seen in the 3rd generation model.
In an effort to improve specificity of the 4th generation model, the radii of features were reduced and features were moved to the steroid nucleus to yield the 5th generation model. While the specificity did improve to be better than the 2nd generation model by 0.04, the sensitivity, or true positive rate, declined by 0.07 compared to the 4th generation model to become equivalent to the 2nd generation model. Other metrics remained similar between 4th and 5th generation models.
The 6th generation model, which used the newer model development protocol in MOE 2013.08, provided a significant improvement in sensitivity of 0.20 and 0.54 over the previous 4th and 3rd generation models, respectively. The specificity of the model was on par with the 4th generation model, but 0.04 lower than the 5th generation model and 0.54 lower than the 3rd generation model. Accuracy improved to 0.54, which is 0.08 better than the 4th and 5th generation, but 0.09 lower than the 3rd generation model. Enrichment also improved over 4th and 5th generation by 0.16 and 0.18 respectively, but was 0.39 behind the 3rd generation model.
The major difference in sensitivity and specificity for the 3rd and 6th generation models demonstrates the biggest issue with pharmacophore model development for β1 so far. With current models, any increase in sensitivity caused a decrease in specificity. Since the PubChem database contains over 33 million compounds, the more selective nature of the 3rd generation model helps to reduce the number of compounds returned from the search of a database that large, even though some active compounds will not be returned as hits.
Virtual Screening Results
The 1st generation pharmacophore search of the NCI database yielded several compounds, but due to limitations on availability from NCI none of the compounds were available for screening. Also, the limitations of the web browser-based query in the NCI database with distances assigned to atom types led us to pursue a 3D pharmacophore model with feature annotations. However, the distance constraints established for the NCI search were later used to guide development of potential 2nd generation models. Development of 3D models also allows for searching of databases other than NCI. Further database searching in Hit2Lead.com was performed, but similarity to LCA was used instead of the distance constraints used for NCI. This search yielded 3-HENA as discussed in detail by Bukiya et al, 2013 [13].
Due to the size of the PubChem database, the 3rd generation model was used for virtual screening due to the high specificity of this pharmacophore generation. Initially, there were 317,771 compounds returned by the model. Due to the laborious nature of visually inspecting over 300,000 compounds, filters were applied to the hit list to further reduce the number of compounds returned by the pharmacophore search. The upper and lower limits of the best known activators were used to filter by molecular weight, LogP, and the number of non-carbon heavy atoms. This removed any compounds that were unlikely to be activators based on our current understanding of the pharmacophore by removing compounds that would be too large for the recognition site or too polar for membrane partitioning. Since the inactive compounds have a larger LogP range of 0.47 to 8.0 and a larger molecular weight range of 142.154 to 516.787 compared to a LogP range of 1.7 to 5.9 and a molecular weight range of 192.262 to 455.703 for active compounds, using these filters should remove a larger proportion of inactive than active compounds. Additionally, restricting the number of non-carbon heavy atoms reduced the number of compounds that would not insert well into the membrane to access the steroid recognition site. Finally, compounds lacking a formal negative charge were removed from the hit list. This cutoff was selected due to the -1 charge present in the two most potent compounds: LCA and 3-HENA. However, because the synthetic LCA derivative with a nitrile in place of the carboxylic acid functional group is active, this method could potentially remove some active compounds. The total number of compounds was reduced to 7,193 after all filters were applied.
Structural similarity clustering of the 3rd generation model search results was accomplished using MACCS structural keys to fingerprint molecules and Tanimoto similarity. A total of 953 clusters were produced at a 65% similarity threshold with the largest cluster containing 2,655 compounds.
All clusters were visually inspected to identify promising compounds for screening. The largest cluster included many bile acid derivatives and triterpenoids similar to LCA and 3-HENA, but also contained many compounds with structural diversity compared to current test and training set compounds. Five compounds were selected for screening from this largest cluster, although two originally selected compounds were substituted with close analogs (stereoisomers) when the originally selected compound proved not to be commercially available (Figure 4). Three additional compounds from three other clusters were also selected for screening to enhance structural diversity among these candidate BKCa activators.
Figure 4.
Commercially available compounds selected from PubChem database with compound ID (CID) numbers. Compounds were selected by visual inspection of structural similarity clusters. Compounds above the black line came from the largest cluster of 5429 compounds. Compounds with an asterisk next to the CID were identified by PubChem as similar to a compound originally selected that was no longer commercially available. Common names are given for compounds that have common names.
Experimental Screening Results
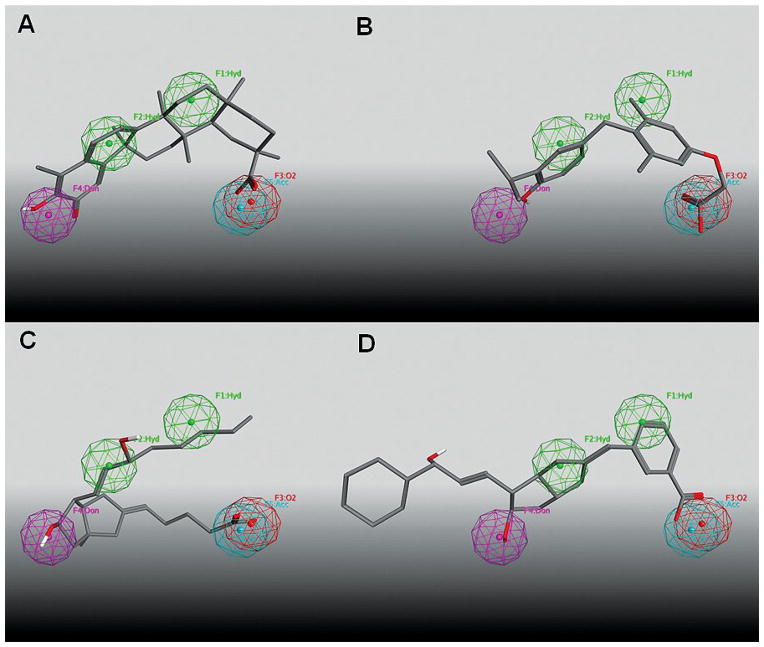
The selected compounds were screened by automated patch-clamp assay for their ability to activate recombinant channels in HEK293 cells expressing both the BKCa channel-forming cbv1 and the β1 accessory subunit (Figure 5). Two of the eight tested compounds, sobetirome and celastrol, reduced Vhalf, the voltage needed to reach half-maximal ionic current. These two compounds therefore activate the channel. Given the small size of the screening set, the 25% hit rate needs further verification, but certainly demonstrates the combined ability of the pharmacophore model and the filtering process to select active compounds and to limit the proportion of compounds screened that prove inactive. The fit of the two active compounds and two of the six inactive compounds (ciprostene and PubChem CID 52224407) on the 3rd generation pharmacophore is shown in Figure 6. All four compounds clearly place appropriate functional groups into the spheres defining the pharmacophore feature locations. Three of the four additionally maintain a concave shape without steric bulk filling the space between the hydrogen bond donor and the carboxylic acid bioisostere. Ciprostene, an inactive compound, fails to exhibit the concave shape that has previously been identified as essential for activity (Figure 6C).[16, 22, 31] The lack of activity observed for PubChem CID 52224407 seems most likely due to the lengthy extension of the molecule outside of the volume defined by the pharmacophore features (Figure 6D). Analysis of the fit of both active and inactive compounds on the pharmacophore model will guide the addition of exclusion volumes that can potentially improve sensitivity without a loss in specificity.
Figure 5.
Two BKCa channel activators were revealed during patch-clamp testing of newly identified compounds. Bar graphs show average Vhalf of BKCa currents obtained from whole-cell recordings to evaluate the activity of the newly identified compounds. Hollow bars represent data obtained in the absence of compounds. Currents were initially tested in Ca2+-free (first hollow bar) versus Ca2+-containing (second hollow bar) intracellular solution: the Vhalf shift towards a lower value in presence of Ca2+ underscores Ca2+-dependent gating, which is a characteristic of BKCa channels. Black bars correspond to the BKCa current Vhalf in presence of tested compounds. * P < 0.05, significantly different from vehicle-containing control.
Figure 6.
Select commercially available compounds selected from PubChem database superposed on the 3rd generation pharmacophore. Panels A–D exhibit sobetirome, celastrol, ciprostene, and PubChem CID 52224407, respectively.
Conclusion
Our pharmacophore model represents a significant step toward the development of a robust tool for the discovery of novel activators of the β1 steroid recognition site. While iterative improvements in pharmacophore modeling and new information have advanced the pharmacophore model over time, further improvements are still needed to address the issues with sensitivity and enrichment without the need for additional filtering. The two active compounds identified in this work represent exciting and structurally novel additions to the set of BKCa channel activators.
Supplementary Material
Highlights.
Six pharmacophore models for the BKCa-β1 site have been evaluated against a common test set.
The top model identified thousands of candidates BKCa activators from the PubChem database.
Two of eight experimentally-screened compounds were confirmed as BKCa activators.
Acknowledgments
Research reported in this publication was supported in part by the National Institutes of Health under Award Numbers R01-HL104631 (AMD) and R37-AA11560 (AMD). Ms. Camisha Terrell (Christian Brothers University, Department of Chemistry) conducted research at UTHSC Department of Pharmacology under the UTHSC-Christian Brothers University undergraduate summer rotation program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Provision of MOE by the Chemical Computing Group (ALP) is also appreciated. Authors deeply thank Nanion Technologies team for help in developing protocols for automated patch-clamp BKCa current recording.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Jacob E. McMillan, Email: jemcmlln@memphis.edu.
Anna N. Bukiya, Email: abukiya@uthsc.edu.
Camisha L. Terrell, Email: cterrell@cbu.edu.
Shivaputra A. Patil, Email: spatil3@uthsc.edu.
Duane D. Miller, Email: dmiller@uthsc.edu.
Alex M. Dopico, Email: adopico@uthsc.edu.
Abby L. Parrill, Email: aparrill@memphis.edu.
References Cited
- 1.Lu R, Alioua A, Kumar Y, Eghbali M, Stefani E, Toro L. MaxiK channel partners: physiological impact. The Journal of physiology. 2006;570:65–72. doi: 10.1113/jphysiol.2005.098913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eichhorn B, Dobrev D. Vascular large conductance calcium-activated potassium channels: functional role and therapeutic potential. Naunyn-Schmiedeberg’s archives of pharmacology. 2007;376:145–55. doi: 10.1007/s00210-007-0193-3. [DOI] [PubMed] [Google Scholar]
- 3.Knaus HG, Eberhart A, Kaczorowski GJ, Garcia ML. Covalent attachment of charybdotoxin to the beta-subunit of the high conductance Ca(2+)-activated K+ channel. Identification of the site of incorporation and implications for channel topology. J Biol Chem. 1994;269:23336–41. [PubMed] [Google Scholar]
- 4.Meera P, Wallner M, Song M, Toro L. Large conductance voltage- and calcium-dependent K+ channel, a distinct member of voltage-dependent ion channels with seven N-terminal transmembrane segments (S0–S6), an extracellular N terminus, and an intracellular (S9–S10) C terminus. Proc Natl Acad Sci U S A. 1997;94:14066–71. doi: 10.1073/pnas.94.25.14066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefani E, Ottolia M, Noceti F, Olcese R, Wallner M, Latorre R, et al. Voltage-controlled gating in a large conductance Ca2+-sensitive K+channel (hslo) Proc Natl Acad Sci U S A. 1997;94:5427–31. doi: 10.1073/pnas.94.10.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaggar JH, Porter VA, Lederer WJ, Nelson MT. Calcium sparks in smooth muscle. Am J Physiol Cell Physiol. 2000;278:C235–56. doi: 10.1152/ajpcell.2000.278.2.C235. [DOI] [PubMed] [Google Scholar]
- 7.Brenner R, Perez GJ, Bonev AD, Eckman DM, Kosek JC, Wiler SW, et al. Vasoregulation by the beta1 subunit of the calcium-activated potassium channel. Nature. 2000;407:870–6. doi: 10.1038/35038011. [DOI] [PubMed] [Google Scholar]
- 8.Chang T, Wu L, Wang R. Altered expression of BK channel beta1 subunit in vascular tissues from spontaneously hypertensive rats. American journal of hypertension. 2006;19:678–85. doi: 10.1016/j.amjhyper.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Petkov GV, Bonev AD, Heppner TJ, Brenner R, Aldrich RW, Nelson MT. Beta1-subunit of the Ca2+-activated K+ channel regulates contractile activity of mouse urinary bladder smooth muscle. The Journal of physiology. 2001;537:443–52. doi: 10.1111/j.1469-7793.2001.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ponte CG, McManus OB, Schmalhofer WA, Shen DM, Dai G, Stevenson A, et al. Selective, direct activation of high-conductance, calcium-activated potassium channels causes smooth muscle relaxation. Mol Pharmacol. 2012;81:567–77. doi: 10.1124/mol.111.075853. [DOI] [PubMed] [Google Scholar]
- 11.Nardi A, Olesen SP. BK channel modulators: a comprehensive overview. Curr Med Chem. 2008;15:1126–46. doi: 10.2174/092986708784221412. [DOI] [PubMed] [Google Scholar]
- 12.Bukiya AN, Liu JX, Toro L, Dopico AM. beta(1) (KCNMB1) subunits mediate lithocholate activation of large-conductance Ca2+-activated K+ channels and dilation in small, resistance-size arteries. Mol Pharmacol. 2007;72:359–69. doi: 10.1124/mol.107.034330. [DOI] [PubMed] [Google Scholar]
- 13.Bukiya AN, McMillan JE, Fedinec AL, Patil SA, Miller DD, Leffler CW, et al. Cerebrovascular dilation via selective targeting of the cholane steroid-recognition site in the BK channel beta1-subunit by a novel nonsteroidal agent. Mol Pharmacol. 2013;83:1030–44. doi: 10.1124/mol.112.083519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dopico AM, Walsh JV, Jr, Singer JJ. Natural bile acids synthetic analogues modulate large conductance Ca2+-activated K+ (BKCa) channel activity in smooth muscle cells. The Journal of general physiology. 2002;119:251–73. doi: 10.1085/jgp.20028537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bukiya AN, Vaithianathan T, Toro L, Dopico AM. Channel beta2–4 subunits fail to substitute for beta1 in sensitizing BK channels to lithocholate. Biochemical and biophysical research communications. 2009;390:995–1000. doi: 10.1016/j.bbrc.2009.10.091. [DOI] [PubMed] [Google Scholar]
- 16.Bukiya AN, Singh AK, Parrill AL, Dopico AM. The steroid interaction site in transmembrane domain 2 of the large conductance, voltage- and calcium-gated potassium (BK) channel accessory beta1 subunit. Proc Natl Acad Sci U S A. 2011;108:20207–12. doi: 10.1073/pnas.1112901108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Yang Y, Ye S, Jiang Y. Structure of the gating ring from the human large-conductance Ca(2+)-gated K(+) channel. Nature. 2010;466:393–7. doi: 10.1038/nature09252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolton EE, Wang Y, Thiessen PA, Bryant SH. Chapter 12 PubChem: Integrated Platform of Small Molecules and Biological Activities. In: Ralph AW, David CS, editors. Annual Reports in Computational Chemistry. Vol. 4. Elsevier; 2008. pp. 217–41. [Google Scholar]
- 19.Molecular Operating Environment (MOE) Chemical Computing Group Inc; Montreal, QC, Canada: 2013. [Google Scholar]
- 20.Halgren TA. Merck molecular force field .1. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem. 1996;17:490–519. [Google Scholar]
- 21.Bukiya AN, Patil SA, Li W, Miller DD, Dopico AM. Calcium- and voltage-gated potassium (BK) channel activators in the 5beta-cholanic acid-3alpha-ol analogue series with modifications in the lateral chain. ChemMedChem. 2012;7:1784–92. doi: 10.1002/cmdc.201200290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bukiya AN, McMillan J, Parrill AL, Dopico AM. Structural determinants of monohydroxylated bile acids to activate beta 1 subunit-containing BK channels. J Lipid Res. 2008;49:2441–51. doi: 10.1194/jlr.M800286-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patil S, Bukiya AN, Li W, Dopico AM, Miller D. Design and synthesis of hydroxy-alkynoic acids and their methyl esters as novel activators of BK channels. Bioorg Med Chem Lett. 2008;18:3427–30. doi: 10.1016/j.bmcl.2008.03.080. [DOI] [PubMed] [Google Scholar]
- 24.Labute P. LowModeMD--implicit low-mode velocity filtering applied to conformational search of macrocycles and protein loops. J Chem Inf Model. 2010;50:792–800. doi: 10.1021/ci900508k. [DOI] [PubMed] [Google Scholar]
- 25.Triballeau NB, HO, Acher F. Pharmacophores and Pharmacophore Searches. Weinheim, Germany: Wiley-VCH; 2006. [Google Scholar]
- 26.Machine Learning: Paradigms and Methods. Cambridge, MA: MIT Press; 1990. [Google Scholar]
- 27.Rogers DJ, Tanimoto TT. A Computer Program for Classifying Plants. Science. 1960;132:1115–8. doi: 10.1126/science.132.3434.1115. [DOI] [PubMed] [Google Scholar]
- 28.Bukiya AN, Liu JX, Dopico AM. The BK channel accessory beta(1) subunit determines alcohol-induced cerebrovascular constriction. FEBS Lett. 2009;583:2779–84. doi: 10.1016/j.febslet.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaggar JH, Li A, Parfenova H, Liu J, Umstot ES, Dopico AM, et al. Heme is a carbon monoxide receptor for large-conductance Ca2+-activated K+ channels. Circ Res. 2005;97:805–12. doi: 10.1161/01.RES.0000186180.47148.7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bukiya AN, Vaithianathan T, Toro L, Dopico AM. The second transmembrane domain of the large conductance, voltage- and calcium-gated potassium channel beta(1) subunit is a lithocholate sensor. FEBS Lett. 2008;582:673–8. doi: 10.1016/j.febslet.2008.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bukiya AN, McMillan J, Fedinec AL, Leffler CW, Parrill AL, Dopico AM. Sodium 3-Hydroxyolean-12-en-30-Oate is a Novel and Selective Activator of beta 1 Subunit-Containing BK Channels and thus Cerebral Artery Dilator. Biophysical journal. 2012;102:133a–4a. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.