Abstract
Structural models of the fibrils formed by the 40-residue amyloid-β (Aβ40) peptide in Alzheimer’s disease typically consist of linear polypeptide segments, oriented approximately perpendicular to the long axis of the fibril, and joined together as parallel in-register β-sheets to form filaments. However, various models differ in the number of filaments that run the length of a fibril, and in the topological arrangement of these filaments. In addition to questions about the structure of Aβ40 monomers in fibrils, there are important unanswered questions about their structure in prefibrillar intermediates, which are of interest because they may represent the most neurotoxic form of Aβ40. To assess different models of fibril structure and to gain insight into the structure of prefibrillar intermediates, the relative solvent accessibility of amino acid residue side chains in fibrillar and prefibrillar Aβ40 preparations was characterized in solution by hydroxyl radical footprinting and structural mass spectrometry. A key to the application of this technology was the development of hydroxyl radical reactivity measures for individual side chains of Aβ40. Combined with mass-per-length measurements performed by dark-field electron microscopy, the results of this study are consistent with the core filament structure represented by two- and three-filament solid state nuclear magnetic resonance-based models of the Aβ40 fibril (such as 2LMN, 2LMO, 2LMP, and 2LMQ), with minor refinements, but they are inconsistent with the more recently proposed 2M4J model. The results also demonstrate that individual Aβ40 fibrils exhibit structural heterogeneity or polymorphism, where regions of two-filament structure alternate with regions of three-filament structure. The footprinting approach utilized in this study will be valuable for characterizing various fibrillar and nonfibrillar forms of the Aβ peptide.
Fibrils composed of amyloid-β (Aβ) peptides are the most abundant component of the senile plaques that are pathognomonic of Alzheimer’s disease (AD), one of the many diseases associated with fibril-forming peptides and/or proteins.1 The molecular pathogenesis of AD is an area of active study, and Aβ fibrils are such a consistent histopathological feature of AD that understanding their structure is likely to be an important step in understanding the pathogenesis of AD.
Early fiber diffraction studies of amyloid fibrils from amyloid-laden tissue revealed a meridial reflection at 4.75 Å, corresponding to the characteristic interstrand distance in a β-sheet, and an equatorial reflection at ∼10 Å, thought to represent the distance between two separate β-sheets.2−4 Solid state nuclear magnetic resonance (SS-NMR) studies have delineated the extent of the two β-sheets and demonstrated that the segments comprising them are arranged in parallel and in register,5,6 findings that have been confirmed by EPR with extrinsic labels,7 one-dimensional infrared (FTIR) spectroscopy,8 and two-dimensional infrared (2D-IR) spectroscopy.9
Several structural models of Aβ fibrils have been proposed to relate the two β-sheets to each other, and to the overall morphological appearance of fibrils observed in transmission electron microscopy (TEM) studies. Two early models based on studies of the 40-residue form of Aβ (i.e., Aβ40) represented fibrils as a pair of “filaments” (defined as linear arrays of peptides linked in β-sheets via hydrogen bonds)10 with axial symmetry. These models [Protein Data Bank (PDB) entries 2LMN and 2LMO, collectively herein termed 2LMN/O]11 differ principally in “stagger”, i.e., the degree to which the two β-sheet-forming segments of a single peptide are displaced from each other along the fibril axis. Additional SS-NMR studies,12 mass-per-length determinations,13 X-ray diffraction studies,14 and computer simulations15 have led to models composed of three filaments and a hollow core structure (PDB entries 2LMP and 2LMQ), again differing principally in stagger (collectively herein termed 2LMP/Q). The possibility that fibrils may have either two or three filaments is variously termed plasticity, polymorphism, or heterogeneity of Aβ40 fibrils.16 A recent study of fibrils grown from “seeds” derived from two different human brains afflicted with AD suggests that structural polymorphism among Aβ40 fibrils is pathogenically significant.17 Of the two cases, the one with an atypical clinical presentation yielded fibrils with a distinct morphology, and a distinct set of low-energy three-filament conformers (PDB entry 2M4J).
While these models are the most detailed to date, they were developed with relatively few experimental constraints compared to the complexity of the system. As a result, the 10 superimposed low-energy conformers for each chain of the fibril models (2LMN/O and 2LMP/Q) display large structural variations between conformers, and the N-terminal regions are not included in the models as they are presumably disordered. In this situation, a topological study of fibril structure by hydroxyl radical footprinting (HRF) and mass spectrometry is useful to test these models and to determine if structural constraints obtained for fibril samples in water are compatible with models that rely on data obtained from samples prepared for SS-NMR.
The fibrils that form in AD are composed of Aβ proteins that vary in length because of imperfect specificity in the γ-secretase that determines their C-terminus, and various N-terminal truncations. It is not known how prevalent each length is in the fibrils of AD, and the relative prevalence of various lengths may well vary between individuals. In any case, Aβ40 is invariably a major if not predominant component of the fibrils in AD, and fibrils made solely from synthetic Aβ40 were the basis for the structural models that we consider herein. Therefore, the fibrils examined by HRF in this work were prepared solely from synthetic Aβ40.
HRF is performed by exposing an aqueous protein sample to hydroxyl radicals generated by various means. In this study, synchrotron X-rays are used to create hydroxyl radicals (•OH) through radiolysis of water. Amino acid side chains are modified by these radicals at rates that are proportional to their solvent accessibility but also a function of side chain reactivity. Modification rates are determined by plotting the extent of side chain modification, quantified by mass spectrometry, versus X-ray exposure time.18 HRF has become a powerful method for developing and refining structural models of macromolecules and their complexes, especially when the relevant functional states are not amenable to crystallographic or NMR-based methods, and when solution state information is desirable.19−22
Two aspects of the HRF technique applied in this study yielded important analytical advantages. The first aspect is that fibrils were digested by pepsin after irradiation to generate a nested set of overlapping segments spanning the entire length of Aβ40. These segments were separated by ultra-high-pressure liquid chromatography to achieve chromatographic separation of the oxidized isomers and provide assessments of solvent accessibility at the level of individual residues, in contrast to the peptide-level resolution typical in most footprinting studies to date.23,24 The availability of overlapping segments also provides multiple readouts for many of the residues, providing greater confidence in the rate measurements. The second aspect is that the results from fibrillar and prefibrillar Aβ40 digested after irradiation were compared to results from Aβ40 digested before irradiation. This “predigested” Aβ40 yielded baseline modification rates for side chains in the context of minimally structured segments while preserving the sequence context of the residues of interest. Rates for predigested Aβ40 were divided by the rates observed for side chains in fibrillar and prefibrillar Aβ40 to obtain a “protection factor” for each residue representing the degree to which the side chain solvent accessibility of a residue is altered upon assembly into prefibrillar and fibrillar Aβ40 states. Protection factors normalize the modification rate data for differences in side chain reactivity, and yield a measure of structurally important differences in solvent accessibility of the Aβ40 states at the single-residue level. These two aspects of the HRF technique applied herein have some precedent in hydrogen–deuterium exchange (HDX) studies.25−28
The HRF results described below provide solvent accessibility data in the form of experimental protection factors for 16 of the 40 residues of Aβ40 in low-molecular weight (LMW) prefibrillar aggregates, and in mature fibrils. Results from Aβ40 fibrils are consistent with the core filament structure identified in both the two- and three-filament structures by SS-NMR. As such, this study represents an important test of the core filament structure proposed by SS-NMR studies. HRF results also provide evidence of ordered structure in two segments of Aβ40 fibrils that remain undefined by SS-NMR, namely, the eight N-terminal residues and the five-residue loop consisting of residues 25–29. The single-residue data from HRF, coupled to the normalization approaches provided by the protection factor assessments, will be useful in characterizing various fibrillar and nonfibrillar forms of the Aβ peptide and in improving our understanding of structure–function relationships in the pathophysiology of Alzheimer’s disease.
Materials and Methods
Materials
Hexafluoroisopropanol (HFIP) was obtained from Fluka and glass distilled prior to use. Formic acid (FA, 98%) was also obtained from Fluka. Ruthenium-tris(2,2′-bipyridyl) dichloride, (Ru(byp)3)Cl2, and ammonium persulfate (APS) were purchased from Sigma (St. Louis, MO). Recombinant HFIP-purified ultrapure Aβ40 (rAβ40) was obtained as a lyophilized powder from rPeptide (Athens, GA). Synthetic Aβ40 was custom synthesized and purified to >95% by the Small-Scale Peptide Synthesis Facility in the Keck Biotechnology Resource Laboratory at Yale University (New Haven, CT).
Seed Formation
Fibril seeds were prepared from rAβ40 without further purification. Lyophilized powder (100–200 μg aliquots) was dissolved in acidified HFIP (40% HFIP in 5 mM HCl) and relyophilized overnight to disaggregate oligomeric and fibrillar forms. It was then reconstituted in 5–10 μL of 1% NH4OH and bath sonicated for ∼30 s (model 1510, Branson, Danbury, CT). This solution was diluted to a concentration of 30 μM in incubation buffer [30 mM HEPES-NaOH (pH 7.4) and 0.01% NaN3] and stored at 37 °C in polypropylene tubes without agitation for at least 3 weeks to allow fibril formation. To remove low-molecular weight forms of Aβ40 after incubation, solutions were washed twice by centrifugation at >10000g for 45 min and replacement of the supernatant with the same buffer. Fibril formation was confirmed by transmission electron microscopy (TEM) with negative staining (ammonium molybdate). Suspended fibrils were bath sonicated for 3 min to break the fibrils into short fragments that subsequently served as seeds.16
Determination of the Purity of Synthetic Aβ40
Synthetic Aβ40 in powder form was dissolved in 0.1% NH4OH and assayed for protein concentration in three ways: a bicinchoninic acid assay, tyrosine absorbance at 280 nm, and mass spectrometric determination of Ala, Val, and Leu in an acid hydrolysate. All three methods showed the material to be 65–70% protein by weight, with the balance presumably water and/or salts. The purity of the protein component was determined by two different high-performance liquid chromatography methods. Method 1 involved a 4.6 mm × 250 mm reversed phase Vydac MS C4 column, mobile phase A composed of 0.1% NH4OH in water, mobile phase B composed of 0.1% NH4OH in acetonitrile (1/200 in volume), a flow rate of 700 μL/min, and detection by absorption at 215 nm. The gradient program was 0% B from 0 to 8 min, with a linear increase to 60% B at 40 min. Method 2 involved a 4.6 mm × 250 mm reversed phase Varian PLRP-S column, the same mobile phases, a flow rate of 500 μL/min, and detection by absorption at 278 nm. The gradient program was a linear increase from 0 to 60% B over 60 min. Only minor peptide impurities were detected as small peaks eluting earlier than the main peak, barely above the baseline noise. Method 1 or 2 suggested that the purity of synthetic Aβ40 in the eluted fraction was >92 or >96%, respectively.
Seeded Fibril Formation
Fibril seeds were prepared from rAβ40 by bath sonication of fibrils formed in a 30 μM pH 7.4 buffer at 37 °C for at least 3 weeks. Synthetic Aβ40 was prepared as a 40 μM solution with 5% seeds and incubated without agitation at 37 °C for at least 1 week. Synthetic Aβ40 was weighed to the nearest microgram on a Cahn microbalance, dissolved in acidified HFIP, lyophilized, and reconstituted at a concentration of 1–2 mM in 0.01% NH4OH. Our experience, based on an infrared spectroscopic study, is that HFIP can be removed from Aβ40 preparations much more easily than TFA, which was why Aβ40 peptides were washed in HFIP and HCl and extensively lyophilized before use. This solution was diluted in incubation buffer containing fibril seeds so that the final concentrations were 40 μM Aβ peptide with 5% seeds. The mixture was incubated without agitation at 37 °C for at least 1 week in polypropylene tubes. Prior to use, fibrils were pelleted by centrifugation at >10000g, washed, and resuspended in measurement buffer [10 mM phosphate (pH 7.2)]. Fibril formation was confirmed for every fibril preparation by TEM.
LMW Aβ40
Synthetic Aβ40 was dissolved in acidified HFIP and lyophilized overnight. Aliquots of the lyophilized powder (100–200 μg) were dissolved in 5–10 μL of 1% NH4OH, diluted to 30 μM in measurement buffer at 4 °C, and kept on ice. Samples studied within 3 h of preparation were deemed “LMW” Aβ40.
Chemical Cross-Linking and Sodium Dodecyl Sulfate–Polyacrylamide Gel Electrophoresis (SDS–PAGE) Analysis
Photoinduced cross-linking of unmodified proteins (PICUP) was performed according to published methods.29−31 In this application, 3 μL of 1 mM (Ru(byp)3)Cl2 and 3 μL of 20 mM APS in 10 mM sodium phosphate (pH 7.4) were added to 54 μL of 30 μM freshly prepared LMW Aβ40. The mixture was irradiated for 1 s using a shuttered 150 W xenon lamp. The reaction was immediately quenched with 10 μL of Tricine sample buffer (Invitrogen) containing 5% β-mercaptoethanol. Cross-linked mixtures were fractionated without boiling by SDS–PAGE using 10–20% Tricine gels (1.0 mm × 10 wells, Invitrogen) and silver stained using a Silver-Xpress silver staining kit (Invitrogen). Bands were quantified by densitometry using ImageJ.
HRF
X-ray exposures were performed at beamline X-28C of the National Synchrotron Light Source with beam currents ranging between 183 and 195 mA using a modified KinTek quench flow apparatus as previously described.32 Samples were exposed for 0, 2.5 5, 10, 15, or 20 ms,33 immediately quenched with methionine amide (10 mM, final concentration) to prevent secondary oxidation, and stored at −80 °C until they were analyzed.34
Proteolysis with Liquid Chromatography (LC) and Mass Spectrometry (MS)
Irradiated fibrils were disrupted with tip sonication (Fisher model 60) in 70% FA. The FA was then removed by three cycles of rotary evaporation and washing with a 1:1 methanol/acetonitrile mixture. Disrupted fibrils and LMW Aβ40 were digested with pepsin at room temperature for 12 h at an enzyme:protein molar ratio of 1:20. Digested Aβ40 was loaded onto a 300 μm (inside diameter) × 2 cm C18 PepMap RP trapping column (Thermo Scientific) using buffer A (100% water and 0.1% FA) at 10 μL/min to preconcentrate the sample and wash away salts. Reverse phase separation was then performed on a 75 μm (inside diameter) × 25 cm C18 PepMap RSLC column with a 2 μm particle size, and 300 Å pore size, using the nano UltiMate 3000 Rapid Separation system (Dionex, Co.) with buffer B (100% acetonitrile and 0.1% FA) increasing at 2% per minute. The extent of oxidation for each amino acid residue was quantified from MS1 spectra of the unmodified proteolytic peptides and their radiolytic products using a Thermo Scientific LTQ FT Ultra mass spectrometer equipped with a nanospray ion source and operating with a needle voltage of 2.4 kV. All MS1 mass spectra were recorded in the positive ion mode at a mass resolution of 100000. The identity of each peptic peptide and specific sites of oxidation were determined using tandem MS and specially optimized liquid chromatographic separation conditions that yielded a base peak resolution of >95%. Collision-induced dissociation was found to be suitable, despite reports of better sequence coverage using electron-transfer dissociation,35 which, in our hands, yielded only 35% coverage. Chromatographic peaks were identified initially using Mascot (Matrix Science, Boston, MA), but all were manually verified.
Calculation of Modification Rates and Protection Factors
The integrated peak areas from selected ion chromatograms of the unmodified peptide (Au) and of a peptide in which a residue is modified (Am) were used to calculate the fraction unmodified: Fu = Au/(Au + Am). A graph of fraction unmodified versus X-ray exposure time was fit to the equation Fu(t) = Fu(0)e–kt, where Fu(0) and Fu(t) are the fractions of unmodified peptide at time zero and time t, respectively, and k is a first-order rate constant as detailed in recent reviews.36,37 Thus, modification rates are obtained from the slope of a logarithmic graph, and each rate relies on multiple measurements. For fragments containing Met residues, the modification rates were determined from doubly modified fragments, because peaks for singly modified fragments containing nonoxidized Met were not apparent. The fragments identified and the amino acid side chains in each fragment for which rates were determined are provided as Supporting Information (Tables S1–S3). Experimental protection factors were calculated by dividing the modification rate for predigested Aβ40 peptides (presumed to be mostly if not fully structure free) by the modification rates for fibrillar and LMW Aβ40, thus normalizing the effects of residue-specific reactivity to provide a pure measure of structure. In addition, any errors in quantification due to variations in ionization and detection efficiency of modified versus unmodified peptide species cancel once protection factors are calculated. Therefore, protection factors provide information that can be compared across experiments and across residues to assess different structure models in an unbiased manner.
Electron Microscopy
Peptide samples (∼20 ng) in 1 μL of buffer were placed onto freshly glow-discharged carbon films on 300 mesh nickel grids for 2 min and blotted with filter paper. A 1% (w/v) solution of ammonium molybdate (Sigma-Aldrich) was adjusted to pH 7.4 with ammonium hydroxide, applied for 2 min, blotted, and air-dried. Images were recorded using a JEOL-1010 transmission electron microscope (JEOL, Tokyo, Japan), operating at 80 kV, equipped with a side-mounted CCD digital camera. All fibril preparations used for HRF studies were examined initially by TEM to verify fibril morphology.
Mass-per-Length Measurements
Approximately 10 ng of seeded Aβ40 fibrils in 1 μL aliquots of buffer was applied to carbon films on 300 mesh nickel grids and mixed with tobacco mosaic virus (TMV) at 5.0 ng/μL. After 2 min, the grids were thoroughly washed in distilled water, blotted, and air-dried. Dark-field TEM images of unstained samples were obtained by shifting the objective aperture, but otherwise using the same electron optics as in bright-field imaging. The MPL of Aβ40 fibrils was determined using the method of Chen et al. with modifications in the size of the integration area as described in Results.38 The intensity in a rectangular area of a digital image containing either Aβ40 fibril or TMV was integrated, and the intensity of a nearby background area was subtracted. The MPL of the Aβ40 fibril was determined by assuming a value of 131.4 kDa/nm for the MPL of TMV and calculating the ratio of background-subtracted signal intensities for Aβ40 fibrils and for TMV.39
Results
Overview of the Footprinting Approach
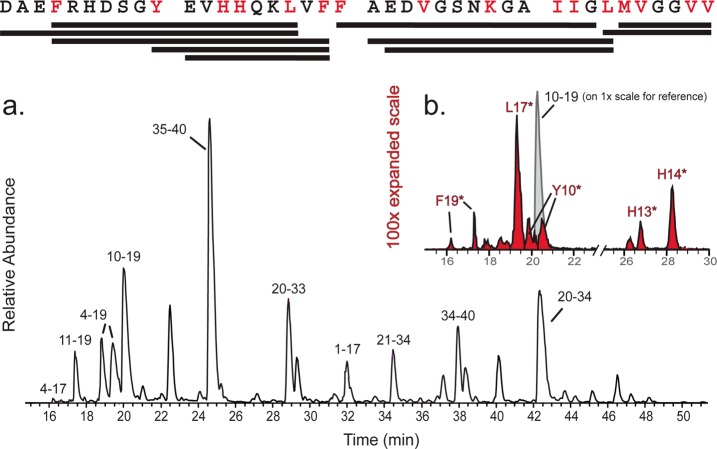
Aβ40 fibrils, LMW Aβ40, and pepsin-digested Aβ40 were exposed to a focused X-ray beam for 0–20 ms to produce hydroxyl radicals from water radiolysis that covalently modified solvent accessible and reactive amino acid side chains. Carefully controlled and reproducible pepsin digestions yielded 10 peptide fragments ranging in length from 5 to 14 residues with overlapping coverage of the entire Aβ40 sequence (Figure 1). Fragments with and without modified residues were chromatographically isolated, identified by tandem mass spectrometry (Figure 1a), and quantified by extraction of peak areas. Figure 1b shows a representative example for the peptide spanning residues 10–19, where retention time differences between peptides oxidized at different locations (e.g., F19, L17, Y10, H13, and H14) allowed the identification and quantification of oxidation products specific to individual residues through retention time-specific extraction of discrete +16 kDa modified species. Residue-specific modification rates were determined by integration of peak areas and plotting those areas as a function of multiple exposure times. Thus, for each structural state investigated, this approach yielded 36 separately measured modification rates from the nested set of pepsin fragments, spanning 16 different residues (Table 1; individual rates provided in Tables S1–S3 of the Supporting Information). Each rate was derived from multiple measurements per peptide (from a peptide-specific dose response) and from multiple peptides (when the particular oxidized species was identified on more than one peptide).
Figure 1.
Aβ40 sequence with the modified amino acids that were detected colored red. Bars under the sequence indicate fragments identified after pepsin digestion. (a) Base peak chromatogram of the unmodified Aβ40 fibril pepsin digest with fragment peaks identified. (b) Selected ion chromatogram (SIC) for unmodified Aβ10–19 (light gray) and for singly oxidized Aβ10–19 (red shading) with the modified residues causing altered retention times identified. Note that the vertical scale for modified Aβ10–19 (red peaks) is 100-fold larger than that for unmodified Aβ10–19 (gray peak), so that the amount of modified polypeptide is a negligible fraction of the amount of unmodified polypeptide.
Table 1. Modification Rates (±standard deviations) for Peptide Fragments, LMW Aβ40, and Aβ40 Fibrils and Protection Factors for LMW Aβ40 and Aβ40 Fibrilsa.
| residue | fragments (s−1) | LMW (s−1) | LMW PF | fibrils (s−1) | fibril PF |
|---|---|---|---|---|---|
| F4 | 5.5 (−) | 3.1 (1.6) | 2 | 1.4 (0.4) | 4 |
| Y10 | 7 (2) | 3.0 (0.5) | 2 | 1.7 (0.4) | 4 |
| HI3 | 4.7 (0.1) | 2.5 (0.6) | 2 | 0.11 (0.04) | 40 |
| HI4 | 5.4 (0.5) | 2.8 (0.7) | 2 | 0.3 (0.1) | 20 |
| L17 | 4.5 (−) | 2.7 (−) | 2 | 0.15 (−) | 30 |
| F19 | 6.2 (−) | 2.7 (0.4) | 2 | 0.08 (0.01) | 80 |
| F20 | 7 (3) | 4.1 (−) | 2 | 0.94 (−) | 7 |
| V24 | 5.0 (0.1) | 4.7 (−) | 1 | 0.31 (−) | 20 |
| K28 | 11 (2) | 1.5 (0.3) | 7 | 0.19 (0.09) | 60 |
| 131 | 7.5 (−) | 5.7 (0.9) | 1 | 0.5 (0.2) | 20 |
| 132 | 7.6 (0.3) | 0.8 (0.5) | 10 | 0.13 (0.04) | 60 |
| L34 | 5 (1) | 0.9 (0.5) | 6 | 0.06 (0.03) | 90 |
| M35 | 15 (6) | 12 (−) | 1 | 2.3 (0.2) | 6 |
| V36/39/40 | 5.4 (0.6) | 0.6 (0.1) | 9 | 0.01 (0.01) | 400 |
A propagation of error analysis is provided in Table S4 of the Supporting Information.
Because it is difficult to be certain that any preparation of Aβ40 is fully unstructured, modification rates were also measured for Aβ40 that had been digested with pepsin prior to irradiation to provide a sample closely representing an unstructured state. The rates obtained from predigested Aβ40 were divided by the rates obtained for LMW and fibrillar Aβ40, which canceled the effects of side chain reactivity and sequence context on modification rates. These calculations yielded two sets of protection factors, one for LMW Aβ40 and one for fibrillar Aβ40 (Table 1). Each protection factor indicates the degree to which the solvent accessible surface area of an amino acid side chain is reduced by protein folding and aggregation.18,40,41
LMW Aβ40
PICUP and SDS–PAGE
The precise physical state of Aβ40 in solution is difficult to ascertain because of its tendency to form polydisperse aggregates and because attempts to characterize these aggregates can alter their populations. TEM suggested that freshly prepared 30 μM solutions of Aβ40 at pH 7.4 contain many oligomeric spherical aggregates (Figure S1a of the Supporting Information). However, TEM involves dehydration and the application of high-ionic strength contrast agents, which could artifactually induce aggregation. PICUP and subsequent SDS–PAGE analysis indicated that LMW samples were approximately 16% dimeric and 7% trimeric, with traces of higher-order species (Figure S1b of the Supporting Information). However, the cross-linking efficiency may be much less than 100%, so this technique could underestimate the population of oligomeric species.
Protection Factors
All seven residues in the N-terminal portion of LMW Aβ40 (F4–F20) examined by HRF have protection factors of 2, suggesting that the potential dimer and trimer structures identified by cross-linking have modest but detectable interactions in this region. Numerous NMR studies of Aβ monomers have suggested that N-terminal residues may form α or 310 helices (Figure S2 of the Supporting Information).42−49 Although most of these studies were performed in the presence of helix-inducing solvents or detergents, an Aβ-binding antibody is known to bind the five N-terminal residues in a helical conformation.50
Several residues in the C-terminal portion of LMW Aβ40 (K28, I32, L34, V36, V39, and V40) have protection factors ranging from 6 to 10. This suggests potential regions in which the dimers and/or trimers may bind tightly or regions in which quaternary interactions may dramatically alter tertiary structure. Fragments containing modified forms of the three valine residues could not be resolved chromatographically, so the rates listed in Table 1 are averages for all three residues. Aside from V39, it is noteworthy that all of the highly protected C-terminal side chains beyond V24 are even-numbered, while the less protected residues (I31 and M35) are odd-numbered. This result is consistent with β-sheet structure alternation in which even-numbered residues are on one side of a β-sheet and more protected from solvent while odd-numbered residues are on the other side and more exposed. An exposed V24 residue is not present in any current monomer structure; further, the high protection factors of C-terminal residues measured in LMW Aβ40 are not consistent with any existing model. However, the relatively high protection factor of 7 for K28 is consistent with an amyloid folding nucleus for the Aβ40 monomer proposed by Lazo et al.51
Fibrillar Aβ40
TEM and MPL Analysis
Most fibrils exhibited semiregular variations in width from ∼4 to ∼8 nm, suggesting a ribbonlike structure (Figure 2a,b). Dark-field MPL determinations using a 40 nm window for intensity integrations indicate that these fibrils consist of a roughly even mixture of two and three filament species with peaks in the histogram of MPL counts at 18.2 and 27.3 kDa/nm [average MPL of 25.2 kDa/nm (Figure 2c)]. To determine whether these results reflect a mixed population of two-filament and three-filament fibrils, or variation in the number of filaments along the length of each fibril, MPL determinations were repeated on the same images using an 80 nm window. This analysis yielded a similar average MPL (27.3 kDa/nm), but only one broad peak in the histogram (Figure 2d). Because the same images were used for both analyses, these results indicate that there is variation in the number of filaments along the length of each fibril, and they confirm previously published results.52
Figure 2.
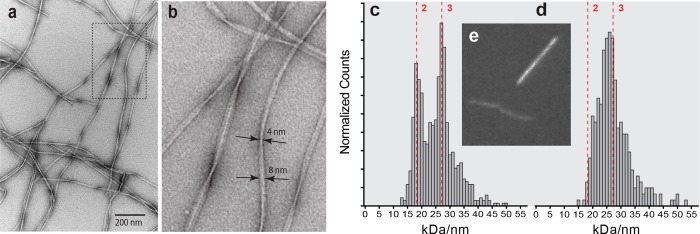
(a) TEM images of Aβ40 fibrils negatively stained with ammonium molybdate (pH 7.4). (b) Five-fold magnification of the portion of panel a outlined in a dotted line, with measurements of a fibril at its widest (8 mm) and narrowest (4 mm) points. (c) MPL histogram (60 nm × 40 nm window, 685 measurements) of unstained TEM dark-field images of seeded Aβ40 fibrils. Dotted red vertical lines and integers represent the x-axis positions corresponding to two and three filaments per fibril. With windows that are 40 nm wide, there is a clear predominance of segments with either two or three filaments. (d) MPL histogram of the same images examined in panel c with a wider window (60 nm × 80 nm, 305 measurements). With the wider window, the same average MPL is obtained, but the two-filament and three-filament regions cannot be resolved. (e) Typical dark-field image with TMV (bright line) that serves as an internal mass standard for determining the MPL of a fibril (dim line).
Protection Factors
In the N-terminal portion of fibrillar Aβ40 from residue 11 to 24, the side chains of odd-numbered residues have protection factors (30–80) much higher than those of the side chains of even-numbered residues (4–20) (Table 1). This pattern is striking for adjacent pairs of identical residues such as H13 and H14 (40 for H13 vs 20 for H14) and F19 and F20 (80 for F19 vs 7 for F20). The opposite is found in the C-terminal portion from residue 30 to 40 (excluding C-terminal valines): odd-numbered residues have protection factors (6–20) lower than those of even-numbered residues (60–90).
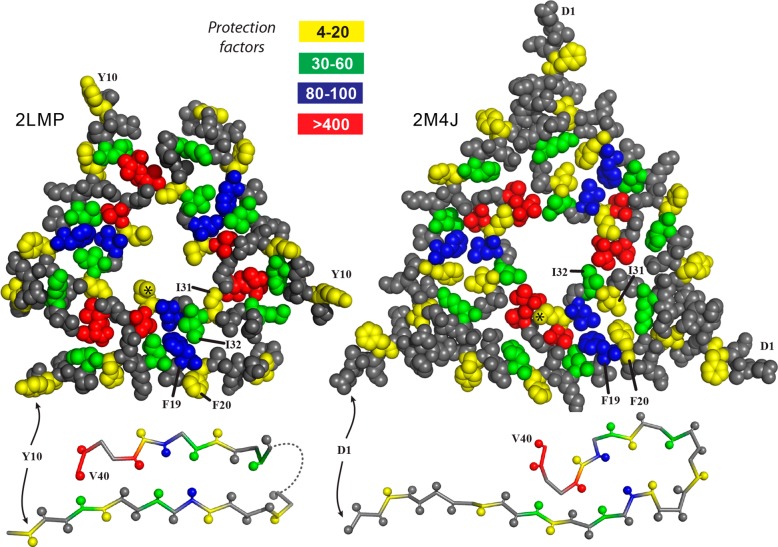
This pattern is consistent with a fibril structure containing two apposing β-sheets. To illustrate, measured protection factors have been mapped onto a schematic representation of the core filament structure common to both the two- and three-filament models (Figure 3, left).11 In addition to positioning all of the relatively unprotected side chains (those with protection factors from 4 to 20) on the periphery, this mapping also illustrates that side chains of F19 (protection factor of 80) and L34 (protection factor of 90) with nearly identical protection factors are in the proximity of each other. Two residues assigned to an unstructured “loop” region (V24 and K28) have relatively high protection factors (20 and 60, respectively), despite a lack of recognized regular secondary structure or order in this region from SS-NMR. However, the high protection factor for K28 is consistent with the SS-NMR detection of a salt bridge between K28 and D23 in Aβ40 samples incubated under conditions conducive to the formation of homogeneous two-filament fibrils.53
Figure 3.
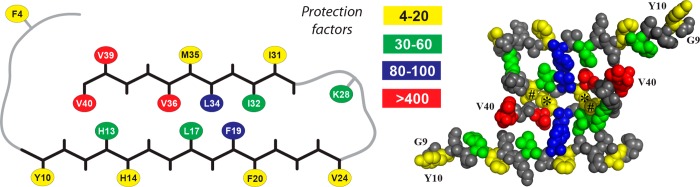
Schematic representation (left) of one Aβ40 molecule from the 2LMO model showing the two β-sheets spanning residues 11–24 and 30–40 (angular black backbone segments) and two structurally uncharacterized segments [residues 1–8 at the N-terminus and residues 25–29 comprising a loop connecting the two sheets (smooth gray backbone segments)]. Residues for which protection factors were determined are color-coded according to the legend. Note that all residues with protection factors of <20 are on the outer surface of this model. Cross section (right) of the 2LMO model with van der Waals spheres for non-hydrogen atoms, illustrating a proposed quaternary relationship between two filaments. The eight N-terminal residues and residues 25–29 have been omitted from this model. The N-termini are marked G9, and the C-termini are marked V40. Note that two residues with low protection factors (I31, marked with number signs, and M35, marked with asterisks) are buried in this model, while residues with high protection factors (V39 and V40) are exposed.
Nevertheless, the two-filament assembly proposed in 2LMO (Figure 3, right) is not consistent with all measured protection factors. Specifically, residues I31 and M35 have protection factors of only 20 and 6, respectively, despite being fully buried in the model. However, the small protection factor for M35 is consistent with the three-filament 2LMP model (Figure 4). Results for Val 36 (protection factor of 400) and L34 (protection factor of 90) are consistent with both models.
Figure 4.
Cross sections of two different three-filament fibril models with van der Waals spheres for non-hydrogen atoms. Below each model is a ball-and-stick rendering illustrating the backbone conformation of one filament in the model. On the left is the 2LMP model with the three Y10 residues at the N-termini labeled. Note that M35 (marked by an asterisk in the lower filament) is exposed to the water-filled central channel, consistent with the low observed protection factor. Also note that residues F20 and I31 appear to be much more solvent accessible than their adjacent residues of the same type (F19 and I32), consistent with the observed protection factors. All other residues for which data are available are either buried or accessible in this model, consistent with the measured protection factors. At the right is the 2M4J model with its three N-terminal “D1” residues marked. Note that the backbone conformation does not correspond to a β-sheet, and there are numerous discrepancies between expected and observed protection factors. For example, F19 and F20 appear to be similarly buried, contrary to the observed protection factors. Also, I31 is buried while I32 is not, the converse of the relationship indicated by the observed protection factors. M35 (marked by an asterisk in the lower filament) appears to be less accessible than V40 in this model, again the converse of the relationship indicated by the observed protection factors.
The three C-terminal valine residues have high experimental protection factors, although V40 is solvent-exposed in the 2LMO model. The high protection factor for this residue may be due to the eight N-terminal residues not included in the 2LMO model, or to quaternary contacts in the three-filament 2LMP and 2LMQ models, where L17 and H13 are in close contact with V36 and V39/40, respectively (Figure 4, left).12 It is important to note that despite an inability to assign individual protection factors to these three residues, the high protection factor assigned collectively implies that all three are protected to some degree individually (because any one of the residues, if unprotected, would dramatically lower the overall protection factor). The high degree of protection for these Val residues is in stark contrast to a much lower protection factor for V24 (protection factor of 20), despite virtually identical modification rates in unstructured fragments (Table 1). Therefore, these residues may be protected by N-terminal contacts where fibrils have a two-filament structure, or by quaternary contacts where the fibrils have a three-filament structure, given that the MPL data described above suggest that these fibrils contain a mixture of two- and three-filament segments.
Many protection factors do not map well onto the three-filament 2M4J model (Figure 4).17 There are three centers of inconsistency between the HRF results and this model in which the C-terminal residues assume an irregular “cotter pin” structure. One inconsistency is that many of the conformers comprising the 2M4J model place the F19 and F20 side chains adjacent to each other, with approximately equal protection from solvent, while the HRF data show these residues exhibit more than 10-fold variation in their protection factors (80 for F19 vs 7 for F20). A second inconsistency is that the model suggests that V40 is less protected than M35, while the HRF data show that V40 is more than 50-fold better protected than M35. Finally, the model suggests that I32 is less protected than I31, whereas the HRF data show 3-fold more protection for I32 than for I31.
Discussion
This study, using a combination of HRF and MPL analysis, unambiguously supports an Aβ40 fibril structure composed of both the two- and three-filament assemblies, and it is consistent with a combination of the SS-NMR models 2LMN/O and 2LMP/Q (Figures 3 and 4). The feature common to both models is a filament structure characterized by two β-sheets, with odd-numbered side chains from the N-terminal sheet apposed to even-numbered side chains from the C-terminal sheet. The models may be brought into better agreement with the HRF data if residues 1–8 (omitted in all the SS-NMR-derived structures) were positioned to protect residue F4 and the C-terminal Val residues. The adoption of regular structure in the loop spanning residues 25–29 would also improve agreement by accounting for the observed protection of V24 and K28. The presence of regular structure in this loop is also supported by limited proteolysis and hydrogen exchange studies,54 as well as by NMR evidence of a salt bridge between the side chains of D23 and K28 in the two-filament 2LMN/O models.51,55
Models other than 2LMN/O/P/Q are more difficult to reconcile with the HRF data. For example, the recently published 2M4J model also has three filaments but is inconsistent with the high degree of protection we observe in the C-terminal Val residues, and the lower level of protection observed for M35. Also, the differences in protection observed in adjacent residue pairs such as I31 and I32, and F19 and F20, are not reflected in 2M4J (Figure 4, right). The models proposed by Lührs et al.56 and Bertini et al.57 are also difficult to reconcile with the HRF results. However, we note that that Lührs et al. were working with Aβ42 and an oxidized M35 residue while Bertini et al. added a Met residue to the N-terminus. Therefore, their samples are not strictly comparable with those used for the study presented here.
Modest revisions of the filament structure and assembly in the two- and three-filament models bring them into agreement with HRF data provided that both forms are present in fibrils as indicated by mass-per-length determinations (Figure 2). We have previously reported that Aβ40 fibrils exhibit “linear heterogeneity”, and the results presented herein confirm this remarkable aspect of fibril structure. In such circumstances, only a modest overall level of protection is expected for residues such as I31 and M35, despite the high degree of protection suggested in the 2LMN/O structures, because they appear to be accessible in the 2LMP/Q structures.
MPL results indicating the presence of linear heterogeneity, with two- and three-filament regions intermixed, recall the cryo-EM results of Sachse et al.,58 who reported an MPL of ∼5 for a fibril that they described as being composed of two double-helical “protofilaments”. They also reported an MPL of 2.5 for a “filamentous substructure” of this fibril. Such a result would arise naturally from fibrils in which two- and three-filament regions alternate along the length of a fibril. We have occasionally observed that fibrils grown at low pH exhibit MPL peaks at three and six filaments per fibril. In these fibrils, two strands are clearly apparent, and one strand occasionally ends before the other (Figure 5). It is valuable to contrast the morphology of fibrils formed at low pH with that of fibrils grown at physiological pH (Figure 2) because it highlights the structural implications of linear heterogeneity in the latter. At low pH, the MPL peaks at three and six filaments per fibril are obtained irrespective of window size; they are readily explained by intertwined three-filament fibrils, and this explanation is supported by TEM results of Figure 5. At physiological pH, however, we have not observed one strand ending before the other by TEM. Instead, the appearance of an MPL peak at both two and three filaments per fibril is resolved using a smaller (40 nm) window in the analysis. Again, we emphasize that both HRF and MPL studies independently lead to the conclusion that regions with two and three filaments are present in individual fibrils.
Figure 5.
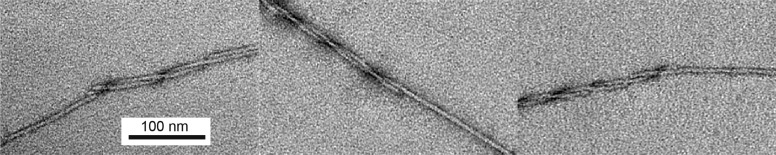
TEM images of Aβ40 fibrils formed at low pH (∼2) and negatively stained with ammonium molybdate (pH 7.4) showing several instances of fibrils that appear to be composed of two strands. In each instance, the strands appear to twist around each other and one terminates before the other. In each case, the strand that continues has a morphology distinctly different from that of the portion with two apparent strands. An MPL analysis of these fibrils exhibited peaks at three and six filaments per fibril (data not shown). It should be noted that the twist pitch of a fibril is sensitive to the type and pH of the negative stain used, suggesting that prominent morphological features observed in TEM images may be artifactual, perhaps due to the high ionic strength of the stain adherent to the fibril that provides contrast. MPL studies, in contrast, are performed on unstained specimens that have been assiduously washed to remove salts.
Our LMW preparation resembles that of Zhang et al.59 in that it consists of a mixed population of dimers and other small oligomers in rapid exchange with a monomeric peptide. Our results for LMW Aβ40 are also similar to their HDX results in suggesting that most residues in the N-terminal half of the peptide are solvent accessible. However, their results for fibrillar Aβ40 also indicated a relatively high level of exchange for residues 35–40, contrary to the high protection factors observed in this study. One way to reconcile these differences is by noting that the fibrils of Zhang et al. were formed in the presence of TFA, whereas we made a vigorous effort to remove TFA before fibril formation. Another way to reconcile the differences is to note that HDX data primarily reflect the stability of the backbone secondary structure. It is possible to have a region that can exchange with solvent in HDX while having an ensemble average structure that is protected from hydroxyl radicals. HDX exchange on a time scale of seconds to minutes can occur, while the millisecond snapshot of the ensemble revealed by HDX shows significant protection.60
The chief point of our results for LMW Aβ40 is that sufficient structure has formed to protect residues such as K28, I32, L34, V36, V39, and V40 despite the LMW Aβ40 being freshly prepared at 4 °C after HFIP dissolution at a concentration of only 30 μM. PICUP results point to the presence of small oligomers that may be involved in this structure. One may speculate that K28 is protected because of salt bridge formation, I32 and L34 are protected by virtue of being on the other side of a β-sheet from I31 and M35, which are not protected, and that V36, V39, and V40 are protected at the center of a protein micelle that has been suggested to be an early aggregated form.61
Given the known polymorphism of fibrils generated under different conditions, it is important to emphasize that the fibrils found in AD are likely to have a high degree of thermodynamic stability because they remain fibrillar while in exchange with nonfibrillar forms of Aβ that are probably present at low nanomolar concentrations. Presumably, the fibrils in AD have had a long time to anneal52 and adopt a stable structure while being incubated at temperatures (e.g., 37 °C) that are significantly higher than the room-temperature conditions commonly used when fibrils are formed in the laboratory. Therefore, it is relevant that the fibrils described herein were generated using seeds of known morphology at neutral pH with relatively low total Aβ40 concentrations (30–40 μM). Fibrils formed in this manner have a self-propagating internal structure and are remarkably stable (they appear to be in exchange with <100 nM nonfibrillar forms of Aβ40).16 In contrast, the thermodynamic stability of fibrils from which the aforementioned models were derived is unknown.
Conclusions
HRF coupled with structural mass spectrometry has provided structural data with single-amino acid resolution, in the form of solvent accessibility protection factors, for 16 of the 40 residues in fibrillar and prefibrillar Aβ40. The HRF-derived protection factors obtained in solution are consistent with the core filament structure proposed by the 2LMP/Q and 2LMN/O models, in which two β-sheets have odd-numbered side chains from the N-terminal sheet apposed to even-numbered side chains from the C-terminal sheet. Linear heterogeneity, such that regions of two- and three-filament assemblies alternate along the length of the fibril, is independently suggested by HRF and MPL techniques. The ability to derive residue-specific solvent accessibility data that might be used to complement structural constraints from SS-NMR studies demonstrates that HRF with single-amino acid resolution may be valuable for the analysis of other fibrillar and oligomeric forms of Aβ40, especially when sample quantities are at a premium.
Acknowledgments
We thank Dr. J. Bohon and Dr. R. D’Mello for assistance with sample irradiation at the Brookhaven National Laboratory (Upton, NY).
Glossary
Abbreviations
- Aβ
amyloid-β peptides
- Aβ40
40-residue amyloid-β peptide
- AD
Alzheimer’s disease
- FA
formic acid
- APS
ammonium persulfate
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- HFIP
hexafluoroisopropanol
- SS-NMR
solid state nuclear magnetic resonance spectroscopy
- EPR
electron paramagnetic resonance spectroscopy
- FTIR
one-dimensional infrared spectroscopy
- 2D-IR
two-dimensional infrared spectroscopy
- TEM
transmission electron microscopy
- HDX
hydrogen–deuterium exchange
- HRF
hydroxyl radical footprinting
- PICUP
photoinduced cross-linking of unmodified proteins
- ASA
solvent accessible surface area
- MPL
mass per length
- TMV
tobacco mosaic virus
- 2LMN, 2LMO, 2LMP, 2LMQ, and 2M4J
Protein Data Bank entries.
Supporting Information Available
Tables of fragment-specific modification rates, model-specific ASA results, and two supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
This work has been supported in part by National Institutes of Health Grants GM76201, NS74178, EB09866, and EB9998 and by grants from the American Health Assistance Foundation, the Glenn Foundation, and the Alzheimer’s Association.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Chiti F.; Dobson C. M. (2006) Protein Misfolding, Functional Amyloid, and Human Disease. Annu. Rev. Biochem. 75, 333–366. [DOI] [PubMed] [Google Scholar]
- Eanes E. D.; Glenner G. G. (1968) X-Ray Diffraction Studies on Amyloid Filaments. J. Histochem. Cytochem. 16, 673–677. [DOI] [PubMed] [Google Scholar]
- Kirschner D. A.; Abraham C.; Selkoe D. J. (1986) X-ray Diffraction from Intraneuronal Paired Helical Filaments and Extraneuronal Amyloid Fibers in Alzheimer-Disease Indicates Cross-β Conformation. Proc. Natl. Acad. Sci. U.S.A. 83, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinchik S. B.; Inouye H.; Szumowski K. E.; Kirschner D. A. (1998) Structural Analysis of Alzheimer’s β(1–40) Amyloid: Protofilament Assembly of Tubular Fibrils. Biophys. J. 74, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antzutkin O. N.; Balbach J. J.; Leapman R. D.; Rizzo N. W.; Reed J.; Tycko R. (2000) Multiple Quantum Solid-State NMR Indicates a Parallel, Not Antiparallel, Organization of β-Sheets in Alzheimer’s β-Amyloid Fibrils. Proc. Natl. Acad. Sci. U.S.A. 97, 13045–13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbach J. J.; Petkova A. T.; Oyler N. A.; Antzutkin O. N.; Gordon D. J.; Meredith S. C.; Tycko R. (2002) Supramolecular Structure in Full-Length Alzheimer’s β-Amyloid Fibrils: Evidence for a Parallel β-Sheet Organization From Solid-State Nuclear Magnetic Resonance. Biophys. J. 83, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torok M.; Milton S.; Kayed R.; Wu P.; McIntire T.; Glabe C. G.; Langen R. (2002) Structural and Dynamic Features of Alzheimer’s Aβ Peptide in Amyloid Fibrils Studied by Site-Directed Spin Labeling. J. Biol. Chem. 277, 40810–40815. [DOI] [PubMed] [Google Scholar]
- Paul C.; Axelsen P. H. (2005) β Sheet Structure in Amyloid β Fibrils and Vibrational Dipolar Coupling. J. Am. Chem. Soc. 127, 5754–5755. [DOI] [PubMed] [Google Scholar]
- Kim Y. S.; Liu L.; Axelsen P. H.; Hochstrasser R. M. (2008) Two-Dimensional Infrared Spectra of Isotopically Diluted Amyloid Fibrils From Aβ40. Proc. Natl. Acad. Sci. U.S.A. 105, 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodali R.; Wetzel R. (2007) Polymorphism in the Intermediates and Products of Amyloid Assembly. Curr. Opin. Struct. Biol. 17, 48–57. [DOI] [PubMed] [Google Scholar]
- Petkova A. T.; Yau W. M.; Tycko R. (2006) Experimental Constraints on Quaternary Structure in Alzheimer’s β-Amyloid Fibrils. Biochemistry 45, 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paravastu A. K.; Leapman R. D.; Yau W. M.; Tycko R. (2008) Molecular Structural Basis for Polymorphism in Alzheimer’s β-Amyloid Fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 18349–18354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.; Thurber K. R.; Shewmaker F.; Wickner R. B.; Tycko R. (2009) Measurement of Amyloid Fibril Mass-Per-Length by Tilted-Beam Transmission Electron Microscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald M.; Box H.; Bian W.; Kendall A.; Tycko R.; Stubbs G. (2012) Fiber Diffraction Data Indicate a Hollow Core for the Alzheimer’s Aβ 3-Fold Symmetric Fibril. J. Mol. Biol. 423, 454–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller Y.; Ma B.; Nussinov R. (2011) The Unique Alzheimer’s β-Amyloid Triangular Fibril Has a Cavity Along the Fibril Axis Under Physiological Conditions. J. Am. Chem. Soc. 133, 2742–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H.; Feingold-Link E.; Sharp K. A.; Rastogi T.; Axelsen P. H. (2010) Intrinsic Linear Heterogeneity of Amyloid β Protein Fibrils Revealed by Higher Resolution Mass-Per-Length Determinations. J. Biol. Chem. 285, 41843–41851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J. X.; Qiang W.; Yau W. M.; Schwieters C. D.; Meredith S. C.; Tycko R. (2013) Molecular Structure of β-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell 154, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselar J. G.; Maleknia S. D.; Sullivan M.; Downard K. M.; Chance M. R. (2002) Hydroxyl Radical Probe of Protein Surfaces Using Synchrotron X-ray Radiolysis and Mass Spectrometry. Int. J. Radiat. Biol. 78, 101–114. [DOI] [PubMed] [Google Scholar]
- Gupta S.; Mangel W. F.; McGrath W. J.; Perek J. L.; Lee D. W.; Takamoto K.; Chance M. R. (2004) DNA Binding Provides a Molecular Strap Activating the Adenovirus Proteinase. Mol. Cell. Proteomics 3, 950–959. [DOI] [PubMed] [Google Scholar]
- Kiselar J. G.; Datt M.; Chance M. R.; Weiss M. A. (2011) Structural Analysis of Proinsulin Hexamer Assembly by Hydroxyl Radical Footprinting and Computational Modeling. J. Biol. Chem. 286, 43710–43716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durer Z. A. O.; Kamal J. K. A.; Benchaar S.; Chance M. R.; Reisler E. (2011) Myosin Binding Surface on Actin Probed by Hydroxyl Radical Footprinting and Site-Directed Labels. J. Mol. Biol. 414, 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. W.; Chance M. R. (2011) Structural Mass Spectrometry of Proteins Using Hydroxyl Radical Based Protein Footprinting. Anal. Chem. 83, 7234–7241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselar J. G.; Janmey P. A.; Almo S. C.; Chance M. R. (2003) Structural Analysis of Gelsolin Using Synchrotron Protein Footprinting. Mol. Cell. Proteomics 2, 1120–1132. [DOI] [PubMed] [Google Scholar]
- Kiselar J. G.; Chance M. R. (2010) Future Directions of Structural Mass Spectrometry Using Hydroxyl Radical Footprinting. J. Mass Spectrom.y 45, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheterpal I.; Wetzel R. (2006) Hydrogen/Deuterium Exchange Mass Spectrometrys: A Window into Amyloid Structure. Acc. Chem. Res. 39, 584–593. [DOI] [PubMed] [Google Scholar]
- Pan J.; Han J.; Borchers C. H.; Konermann L. (2012) Structure and Dynamics of Small Soluble Aβ(1–40) Oligomers Studied by Top-Down Hydrogen Exchange Mass Spectrometry. Biochemistry 51, 3694–3703. [DOI] [PubMed] [Google Scholar]
- Zhang Y.; Rempel D. L.; Zhang J.; Sharma A. K.; Mirica L. M.; Gross M. L. (2013) Pulsed Hydrogen-Deuterium Exchange Mass Spectrometry Probes Conformational Changes in Amyloid β (Aβ) Peptide Aggregation. Proc. Natl. Acad. Sci. U.S.A. 110, 14604–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z. Y.; Walters B. T.; Mayne L.; Englander S. W. (2013) Protein Hydrogen Exchange at Residue Resolution by Proteolytic Fragmentation Mass Spectrometry Analysis. Proc. Natl. Acad. Sci. U.S.A. 110, 16438–16443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fancy D. A.; Kodadek T. (1999) Chemistry for the Analysis of Protein-Protein Interactions: Rapid and Efficient Cross-Linking Triggered by Long Wavelength Light. Proc. Natl. Acad. Sci. U.S.A. 96, 6020–6024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G.; Lomakin A.; Teplow D. B. (2001) Amyloid β-Protein Oligomerization: Prenucleation Interactions Revealed by Photo-Induced Cross-Linking of Unmodified Proteins. J. Biol. Chem. 276, 35176–35184. [DOI] [PubMed] [Google Scholar]
- Bitan G.; Teplow D. B. (2004) Rapid Photochemical Cross-Linking: A New Tool for Studies of Metastable, Amyloidogenic Protein Assemblies. Acc. Chem. Res. 37, 357–364. [DOI] [PubMed] [Google Scholar]
- Gupta S.; Sullivan M.; Toomey J.; Kiselar J.; Chance M. R. (2007) The Beamline X28C of the Center for Synchrotron Biosciences: A National Resource for Biomolecular Structure and Dynamics Experiments Using Synchrotron Footprinting. J. Synchrotron Radiat. 14, 233–243. [DOI] [PubMed] [Google Scholar]
- Maleknia S. D.; Brenowitz M.; Chance M. R. (1999) Millisecond Radiolytic Modification of Peptides by Synchrotron X-rays Identified by Mass Spectrometry. Anal. Chem. 71, 3965–3973. [DOI] [PubMed] [Google Scholar]
- Xu G.; Kiselar J.; He Q.; Chance M. R. (2005) Secondary Reactions and Strategies To Improve Quantitative Protein Footprinting. Anal. Chem. 77, 3029–3037. [DOI] [PubMed] [Google Scholar]
- Srikanth R.; Wilson J.; Bridgewater J. D.; Numbers J. R.; Lim J.; Olbris M. R.; Kettani A.; Vachet R. W. (2007) Improved Sequencing of Oxidized Cysteine and Methionine Containing Peptides Using Electron Transfer Dissociation. J. Am. Soc. Mass Spectrom. 18, 1499–1506. [DOI] [PubMed] [Google Scholar]
- Kiselar J. G.; Maleknia S. D.; Sullivan M.; Downard K. M.; Chance M. R. (2002) Hydroxyl Radical Probe of Protein Surfaces Using Synchrotron X-ray Radiolysis and Mass Spectrometry. Int. J. Radiat. Biol. 78, 101–114. [DOI] [PubMed] [Google Scholar]
- Takamoto K.; Chance M. R. (2006) Radiolytic Protein Footprinting With Mass Spectrometry to Probe the Structure of Macromolecular Complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276. [DOI] [PubMed] [Google Scholar]
- Chen B.; Thurber K. R.; Shewmaker F.; Wickner R. B.; Tycko R. (2009) Measurement of Amyloid Fibril Mass-Per-Length by Tilted-Beam Transmission Electron Microscopy. Proc. Natl. Acad. Sci. U.S.A. 106, 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba K.; Stubbs G. (1986) Structure of Tobacco Mosaic Virus at 3.6 Å Resolution: Implications for Assembly. Science 231, 1401–1406. [DOI] [PubMed] [Google Scholar]
- Guan J. Q.; Vorobiev S.; Almo S. C.; Chance M. R. (2002) Mapping the G-Actin Binding Surface of Cofilin Using Synchrotron Protein Footprinting. Biochemistry 41, 5765–5775. [DOI] [PubMed] [Google Scholar]
- Guan J. Q.; Chance M. R. (2005) Structural Proteomics of Macromolecular Assemblies Using Oxidative Footprinting and Mass Spectrometry. Trends Biochem. Sci. 30, 583–592. [DOI] [PubMed] [Google Scholar]
- Talafous J.; Marcinowski K. J.; Klopman G.; Zagorski M. G. (1994) Solution Structure of Residues 1–28 of the Amyloid β-Peptide. Biochemistry 33, 7788–7796. [DOI] [PubMed] [Google Scholar]
- Sticht H.; Bayer P.; Willbold D.; Dames S.; Hilbich C.; Beyreuther K.; Frank R. W.; Rosch P. (1995) Structure of Amyloid β(1–40)-Peptide of Alzheimers-Disease. Eur. J. Biochem. 233, 293–298. [DOI] [PubMed] [Google Scholar]
- Coles M.; Bicknell W.; Watson A. A.; Fairlie D. P.; Craik D. J. (1998) Solution Structure of Amyloid β-Peptide(1–40) in a Water-Micelle Environment. Is the Membrane-Spanning Domain Where We Think It Is?. Biochemistry 37, 11064–11077. [DOI] [PubMed] [Google Scholar]
- Zhang S.; Iwata K.; Lachenmann M. J.; Peng J. W.; Li S.; Stimson E. R.; Lu Y.; Felix A. M.; Maggio J. E.; Lee J. P. (2000) The Alzheimer’s Peptide Aβ Adopts a Collapsed Coil Structure in Water. J. Struct. Biol. 130, 130–141. [DOI] [PubMed] [Google Scholar]
- Crescenzi O.; Tomaselli S.; Guerrini R.; Salvadori S.; D’Ursi A. M.; Temussi P. A.; Picone D. (2002) Solution Structure of the Alzheimer Amyloid β-Peptide (1–42) in an Apolar Microenvironment: Similarity with a Virus Fusion Domain. Eur. J. Biochem. 269, 5642–5648. [DOI] [PubMed] [Google Scholar]
- Tomaselli S.; Esposito V.; Vangone P.; van Nuland N. A. J.; Bonvin A. M. J. J.; Guerrini R.; Tancredi T.; Temussi P. A.; Picone D. (2006) The α-to-β Conformational Transition of Alzheimer’s Aβ-(1–42) Peptide in Aqueous Media Is Reversible: A Step by Step Conformational Analysis Suggests the Location of β Conformation Seeding. ChemBioChem 7, 257–267. [DOI] [PubMed] [Google Scholar]
- Hoyer W.; Gronwall C.; Jonsson A.; Stahl S.; Hard T. (2008) Stabilization of a β-Hairpin in Monomeric Alzheimer’s Amyloid-β Peptide Inhibits Amyloid Formation. Proc. Natl. Acad. Sci. U.S.A. 105, 5099–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivekanandan S.; Brender J. R.; Lee S. Y.; Ramamoorthy A. (2011) A Partially Folded Structure of Amyloid-β(1–40) in an Aqueous Environment. Biochem. Biophys. Res. Commun. 411, 312–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles L. A.; Crespi G. A. N.; Doughty L.; Parker M. W. (2013) Bapineuzumab Captures the N-Terminus of the Alzheimer’s Disease Amyloid-β Peptide in a Helical Conformation. Sci. Rep. 3, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo N. D.; Grant M. A.; Condron M. C.; Rigby A. C.; Teplow D. B. (2005) On the Nucleation of Amyloid β-Protein Monomer Folding. Protein Sci. 14, 1581–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. Q.; Komatsu H.; Kim Y. S.; Liu L.; Hochstrasser R. M.; Axelsen P. H. (2013) Intrinsic Structural Heterogeneity and Long-Term Maturation of Amyloid β Peptide Fibrils. ACS Chem. Neurosci. 4, 1236–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciarretta K. L.; Gordon D. J.; Petkova A. T.; Tycko R.; Meredith S. C. (2005) Aβ40-Lactam(D23/K28) Models a Conformation Highly Favorable for Nucleation of Amyloid. Biochemistry 44, 6003–6014. [DOI] [PubMed] [Google Scholar]
- Kheterpal I.; Chen M.; Cook K. D.; Wetzel R. (2006) Structural Differences in Aβ Amyloid Protofibrils and Fibrils Mapped by Hydrogen Exchange: Mass Spectrometry With on-Line Proteolytic Fragmentation. J. Mol. Biol. 361, 785–795. [DOI] [PubMed] [Google Scholar]
- Petkova A. T.; Ishii Y.; Balbach J. J.; Antzutkin O. N.; Leapman R. D.; Delaglio F.; Tycko R. (2002) A Structural Model for Alzheimer’s β-Amyloid Fibrils Based on Experimental Constraints From Solid State NMR. Proc. Natl. Acad. Sci. U.S.A. 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luhrs T.; Ritter C.; Adrian M.; Riek-Loher D.; Bohrmann B.; Dobeli H.; Schubert D.; Riek R. (2005) 3D Structure of Alzheimer’s Amyloid-β(1–42) Fibrils. Proc. Natl. Acad. Sci. U.S.A. 102, 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I.; Gonnelli L.; Luchinat C.; Mao J.; Nesi A. (2011) A New Structural Model of Aβ40 Fibrils. J. Am. Chem. Soc. 133, 16013–16022. [DOI] [PubMed] [Google Scholar]
- Sachse C.; Fandrich M.; Grigorieff N. (2008) Paired β-Sheet Structure of an Aβ(1–40) Amyloid Fibril Revealed by Electron Microscopy. Proc. Natl. Acad. Sci. U.S.A. 105, 7462–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A.; Qi W.; Good T. A.; Fernandez E. J. (2009) Structural Differences Between Aβ(1–40) Intermediate Oligomers and Fibrils Elucidated by Proteolytic Fragmentation and Hydrogen/Deuterium Exchange. Biophys. J. 96, 1091–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J. Q.; Takamoto K.; Almo S. C.; Reisler E.; Chance M. R. (2005) Structure and Dynamics of the Actin Filament. Biochemistry 44, 3166–3175. [DOI] [PubMed] [Google Scholar]
- Lomakin A.; Teplow D. B.; Kirschner D. A.; Benedek G. B. (1997) Kinetic Theory of Fibrillogenesis of Amyloid β-Protein. Proc. Natl. Acad. Sci. U.S.A. 94, 7942–7947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.