Abstract
OxlT, the oxalate/formate exchanger of Oxalobacter formigenes, is a member of the major facilitator superfamily of transporters. In the present work, substrate (oxalate) was found to enhance the reactivity of the cysteine mutant S336C on the cytoplasmic end of helix 11 to methanethiosulfonate ethyl carboxylate. In addition, S336C is found to spontaneously cross-link to S143C in TM5 in either native or reconstituted membranes under conditions that support transport. Continuous wave EPR measurements are consistent with this result and indicate that positions 143 and 336 are in close proximity in the presence of substrate. These two residues are localized within helix interacting GxxxG-like motifs (G140LASG144 and S336DIFG340) at the cytoplasmic poles of TM5 and TM11. Pulse EPR measurements were used to determine distances and distance distributions across the cytoplasmic or periplasmic ends of OxlT and were compared with the predictions of an inside-open homology model. The data indicate that a significant population of transporter is in an outside-open configuration in the presence of substrate; however, each end of the transporter exhibits significant conformational heterogeneity, where both inside-open and outside-open configurations are present. These data indicate that TM5 and TM11, which form part of the transport pathway, transiently close during transport and that there is a conformational equilibrium between inside-open and outside-open states of OxlT in the presence of substrate.
The anaerobic Gram-negative bacterium Oxalobacter formigenes is an important component of the human microbiome that is responsible for degradation of oxalate in the large intestine.1 In the bacterial inner membrane of O. formigenes, OxlT functions as an oxalate-formate exchanger,2 where the high activity of this transporter may play a role as an alternate proton pump.3 OxlT is also a member of the major facilitator superfamily (MFS) of secondary transporters,4 which move diverse substrates such as sugars, ions, amino acids, and neurotransmitters. MFS transporters have a common fold with both an N-terminal and a C-terminal domain of typically six transmembrane helixes (TMs) connected by a central cytoplasmic loop. At the cytoplasmic end of TM2, and frequently at the symmetry-related position in TM8, signature amino acid sequences, Gxxx[D/E][R/K], are found.4 This is consistent with the hypothesis that members of this family arose after a primordial gene duplication and fusion event to generate the observed 2-fold symmetry axis between the N- and C-terminal domains.5−17 The underlying similarity in structure implies that members of the MFS function by a common molecular transport mechanism.
Significant progress has been made in determining the three-dimensional structures of selected MFS members;5−21 however, there is presently limited information on the conformational states and molecular motions that provide the basis for transport. The available crystal structures suggest that the relative positions of helices in the N- and C-terminal domains may change in different states of the transporter, and understanding these states and transitions will be critical to determining mechanisms that drive transport in MFS proteins.22
At the present time, there are no high-resolution structures for OxlT; however, there is an experimentally tested GlpT-based homology model of OxlT, which is shown in Figure 1.23 In the OxlT homology model, the transport pathway is open to the cytoplasm, similar to structures for LacY, GlpT, NarK, GLUT1, PiPT, GlcPSe, and PepT(St).6,7,11,13,15,17,19 In this structure, helices 5 and 11 line the substrate permeation pathway in OxlT,5,23−25 and there is experimental evidence for substrate-dependent conformational changes in TM5 of OxlT at S143C.24 This position lies within a GxxxG motif (G140LASG144), and symmetry arguments suggest that a similar change may occur at a cytoplasmic GxxxG-like motif (S336DIFG340) in TM11.
Figure 1.
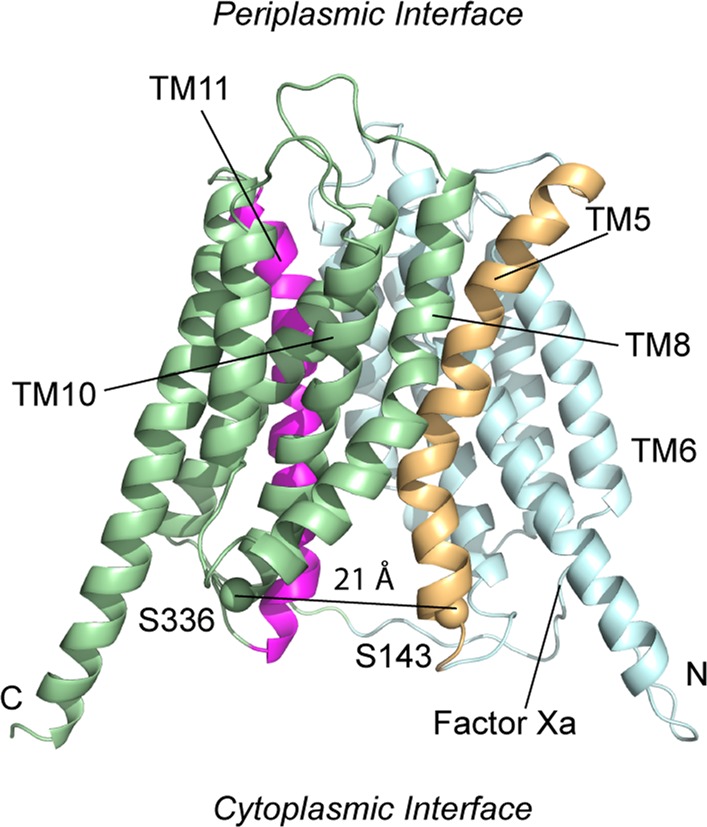
GlpT-based homology model of OxlT23 showing the transport lining helices, TM5 (tan) and TM11 (magenta) (PDB ID: 1ZC7). The approximate location of the Factor Xa cleavage site on the C-terminal end of the loop between TM6 and TM7 is indicated. The transporter is in a cytoplasmic open configuration where the Cα carbons of residues near the ends of TM5 and TM11 (S336 and S143) are separated by 21 Å. Other helices in the N-terminal domain are colored in cyan, and the remaining helices in the C-terminal domain are in light green. Both the C- and N-termini are located on the cytoplasmic side of the membrane.
In the present work, we examine the effect of substrate on covalent modification of S336C, which lies in the loop connecting TM10 and TM11 in the homology model for OxlT, and we demonstrate that there are substrate-induced structural changes at the cytoplasmic end of TM11. In the OxlT homology model, residues S143 and S336 are separated by 21 Å; however, these residues lie within GxxxG and GxxxG-like motifs that are known to mediate helix–helix interactions in membrane proteins.26,27 Evidence for interactions at these sites is obtained using a double-cysteine derivative, S143C/S336C, which is found to readily undergo spontaneous cross-linking. Site-directed spin labeling (SDSL) indicates that spin labels at these sites are separated by a mean distance of 8 Å in the presence of substrate, demonstrating that the cytoplasmic interfaces of these helices come into close proximity during transport. This finding suggests that the pore may be gated at the cytoplasmic side of the transporter by the interaction of TM5 and TM11, perhaps through these GxxxG and GxxxG-like motifs. Using pulse EPR, long-range distance measurements were made, and they demonstrate that OxlT samples both inward-open and outward-open states in the presence of substrate. In contrast to the homology model, a state that is closed to the cytoplasm and open to the periplasmic space is found to be significantly populated.
Experimental Procedures
Construction of Mutants and Protein Expression
A cysteine-less template (C28G/C271A) with a C-terminal nine-histidine tag (His9) in pBluescript II SK+28,29 was the parent for subsequent site-directed mutagenesis. Mutants were generated using QuikChange (Stratagene) and confirmed at the Johns Hopkins DNA sequencing facility. For disulfide trapping/cross-linking, mutations were placed in a cysteine-less variant carrying a tandem Factor Xa cleavage site in the central loop.30 For expression,31 OxlT in pBluescript II SK+ was transformed into Escherichia coli strain XL3 (strain XL1 containing plasmid pMS421 (Specr, LacIq)), grown overnight in a shaker in 25–50 mL of Luria–Bertani (LB) medium in the presence of 100 μg/mL ampicillin and 50 μg/mL spectinomycin, and diluted into 1.5 L of fresh LB broth with antibiotics (as indicated above) to a starting optical density (OD) at 650 nm (OD650) of 0.025–0.03. This diluted sample was grown with shaking until OD650 reached 0.1, at which point 1 mM IPTG was added. After 3 to 4 h (OD650 = 0.35–0.6), the cells were harvested by centrifugation, resuspended in 20 mM potassium phosphate (pH 7.5) as a concentrated stock (OD650 of ca. 35), and frozen at −80 °C until use. No mutational alterations were observed in plasmids sequenced after expression.
Disulfide Trapping and Cross-Linking
For cross-linking and disulfide trapping experiments, cells expressing OxlT were harvested by centrifugation, resuspended in 20 mM Tris·HCl (pH 7.5) at OD650 ∼ 20, and washed by low-speed centrifugation in the same buffer. To prepare membrane vesicles, cells were subjected to high-pressure lysis (18 000 psi) in a French pressure cell. Debris and unbroken cells were removed by low-speed centrifugation; vesicles were collected from the supernatant by high-speed centrifugation (150 000g for 1 h), washed by centrifugation at 150 000g for 1 h with 20 mM Tris·HCl (pH 7.5), resuspended in 20 mM Tris·HCl (pH 7.5) and 20% glycerol at 200–400 μg protein/mL, and frozen at −80 °C until use (<2 weeks).30 For cross-linking, vesicles were diluted in 20 mM Tris·HCl (pH 7.5) to 20 μg protein/mL and treated with 200 μM Cu(II)(1,10-phenanthroline)3 for 10 min at 37 °C or 200 μM of either of two homobifunctional MTS reagents (1,1-methanediyl bismethanethiosulfonate, MTS-1-MTS, and 1,2-ethanediyl bismethanethiosulfonate, MTS-2-MTS) for 10 min at 23 °C. The reactions were then immediately quenched by addition of 5 mM N-ethyl maleimide (NEM) and 10 mM EDTA, and vesicles were washed once by resuspension in 20 mM Tris·HCl (pH 7.5) and pelleted at 290 000g for 26 min. The final pellet was resuspended to 0.2–0.5 μg/μL in 20 mM Tris·HCl (pH 7.5), 0.1 M NaCl, 2 mM CaCl2, and 1% DDM. A solution of 20 mM HEPES (pH 7.5), 500 mM NaCl, 2 mM CaCl2, and 50% glycerol, with or without Factor Xa protease (0.1 mg/mL final concentration) (New England Biolabs), was diluted 1 to 9 into the vesicle suspension, and the mixture was incubated for 6 h on a rotary platform at 4 °C. Control and protease-treated samples were quenched with an equal volume of heated SDS-PAGE loading buffer (12 M urea, 4% SDS, 100 mM Tris·HCl pH 6.8, 20% glycerol, and 0.01% bromophenol blue) before immunoblot analysis using a monoclonal antibody directed against the OxlT C-terminal polyhistidine tag.30 In some experiments and for functional assays following reconstitution, vesicles were solubilized after cross-linking using 100 mM oxalate, 20% glycerol, 20 mM potassium phosphate (pH 7.5), 0.2% E. coli polar lipid (Avanti Polar Lipids, Inc.), and 0.5% DDM.24
Site-Directed Spin Labeling
Cells derived from 1.5 L cultures were processed as described above and then diluted into 20 mL (final) of 200 mM potassium oxalate, 20 mM potassium phosphate (pH 7.5), and 20% glycerol in the presence of 0.25 mM phenylmethylsulfonyl fluoride. The suspension was placed in an ice/water bath, and cells were disrupted by sonication for 5 min using a Sonic Dismembrator model 500 (Fisher Scientific) set to 30% amplitude with an on pulse of 10 s and off pulse of 5 s. After sonication, DDM was added to 0.5%, and lysed material was solubilized for 1 h on a rotary platform in a cold room (4 °C). Solubilized material was clarified by ultracentrifugation at 149 000g for 30 min, after which the extract was incubated with Ni-NTA (Qiagen) for 4–5 h (1 mL of resin/L of cell culture) along with 500 μM TCEP (Sigma-Aldrich). This mixture was then passed into a column, followed by a wash of the retained Ni-NTA resin with 10 bed volumes of 100 μM TCEP, 200 mM potassium oxalate, 0.02% DDM, 20 mM potassium phosphate (pH 7.5), 20% glycerol, and 80 mM imidazole. An additional wash of 5 bed volumes was carried out in 0.02% DDM, 100 mM potassium oxalate, 20 mM potassium phosphate (pH 7.5), and 20% glycerol without TCEP (buffer 2). To spin-label single- and double-cysteine OxlT mutants, 0.5 mM MTSL (Toronto Research Chemicals) was freshly prepared in buffer 2 and added to protein-bound Ni-NTA resin at 130% Ni-NTA bed volume. The reaction was allowed to proceed for 20 min at room temperature in the dark. The column was then washed with 25 bed volumes of buffer 2, after which purified OxlT was eluted by addition of a solution of 20% glycerol, 100 mM potassium oxalate (pH 4.2), and 0.02% DDM. Eluted samples were titrated to pH 7 by addition of a one-ninth volume of 1 M potassium phosphate (pH 8.5). The efficiency of labeling was assessed by monitoring the reactivity of unreacted thiols to 4,4′-dithiodipyridine (Sigma-Aldrich),32 using the OxlT cysteine-less variant as the negative control. For single-cysteine mutants, labeling efficiency was 86 ± 6% (standard deviation) for the six mutants tested (range 79–92%); for the double-cysteine variants, labeling was 88 ± 6% complete for the five variants examined (range 80–95%). In some cases, protein was purified without spin-labeling for functional work. As required, protein was concentrated to 5–13 mg/mL using an Amicon 30 kDa cutoff centrifugal filter device (Millipore).
EPR spectroscopy was performed using solubilized protein purified as described above or with protein that had been reconstituted by detergent dilution (see below) at a protein/lipid ratio of ∼1:18 (w/w).
Reconstitution of OxlT and Assay of Oxalate Transport
Crude detergent extracts or purified material was used for reconstitution by a detergent-dilution protocol, in which proteoliposomes were loaded with 100 mM potassium oxalate and 50 mM potassium phosphate (pH 7) as described.23,24 OxlT function was assessed by following exchange of internal cold oxalate with external 14[C]oxalate (American Radiolabeled Chemicals, St. Louis, MO), using either of two methods. First, and in most cases, 200 μL aliquots of a proteoliposome suspension were spotted onto 0.22 μM GSTF filters (Millipore) on a vacuum manifold, followed by two washes with 5 mL of assay buffer (100 mM potassium sulfate, 50 mM potassium phosphate, pH 7). After disengaging the vacuum, OxlT function was monitored by overlaying trapped proteoliposomes with 0.2 mL of 0.1 mM 14[C]oxalate prepared in the assay buffer; 14[C]oxalate transport was terminated after 1 min by vacuum filtration, immediately followed by two 5 mL washes with assay buffer. To assess the effects of MTSCE (Toronto Research Chemicals), trapped proteoliposomes were overlaid for 5 min with 0.3 mL of MTSCE freshly prepared at the indicated concentrations in assay buffer. MTSCE was removed by vacuum filtration and two washes with assay buffer, after which residual OxlT function was monitored as described above. Similar methods were used to monitor the effect of substrate on MTSCE reactivity with OxlT single-cysteine mutants. In these cases, trapped proteoliposomes were exposed to increasing amounts of external substrate (0–1 mM oxalate) in the presence of an MTSCE concentration initially causing 50% inhibition of OxlT function.24 In the second approach of transport measurement (where better time resolution was required), proteoliposomes were washed twice by centrifugation in ice-cold assay buffer using a Beckman Optima ultracentrifuge (290 000g, 26 min) or in a Beckman L5-50E ultracentrifuge (150 000g, 60 min), and resuspended in 0.45 mL of assay buffer. Subsequently, 0.1 mL aliquots were added to 0.8 mL of assay buffer in a 1.5 mL Eppendorf tube, followed 3 min later with the addition of 0.1 mL of 1 mM 14[C]oxalate to achieve a final concentration of 0.1 mM 14[C]oxalate. At specified times, 0.1 mL portions were removed for vacuum filtration and washing as described above. In experiments requiring disulfide reduction, the proteoliposome suspension was supplemented with 10 mM DTT for 20 min prior to assay.
EPR Spectroscopy
For EPR spectroscopy, 6–10 μL of OxlT at a concentration of approximately 40–120 μM in DDM or lipid bilayers was loaded into borosilicate glass capillary tubes 0.6 mm i.d. × 0.84 mm o.d. (Vitrocom, Mountain Lakes, NJ). EPR spectra were recorded at 23 °C on a Bruker EMX X-band spectrometer equipped with an ER4123D dielectric resonator using 2 mW incident microwave power, 1 G field modulation, and a scan range of 200 G. The spectra were normalized and baseline-corrected. Distances between labels were determined using a Fourier deconvolution approach implemented in the LabVIEW-based program ShortDistances provided by Dr. Christian Altenbach (UCLA).33 Pulse EPR measurements were performed on 25–30 μL samples loaded into 2 mm i.d. × 2.4 mm o.d. quartz capillaries (Fiber Optic Center, Inc., New Bedford, MA) that were flash frozen in a dry ice/isopropanol bath. The DEER data were recorded at 80 K at X-band frequency using a Bruker Elexsys-E580 spectrometer fitted with an ER4118X-MS3 split ring resonator (Bruker Biospin, Billerica, MA). Data were acquired using a four-pulse DEER sequence,34 with a 16 ns π/2 and two 32 ns π observe pulses separated by a 40 ns π pump pulse. The dipolar evolution times were typically 2.0–2.5 μs. The pump frequency was set to the maximum of the central transition of the nitroxide spectrum, and the observe frequency was set to the low-field maximum, typically 65–70 MHz higher. The dipolar evolution data were processed using the model-free approach implemented in the MATLAB-based program DeerAnalysis 2013.35 A background dimensionality of 3 was chosen for the initial background subtraction used to process the data. The validation routine in DeerAnalysis was then used to determine the background subtraction that produced the best fit to the dipolar evolution and to estimate likely errors in the distance distribution.
Results
Cysteine Modification Reveals Substrate-Dependent Structural Changes in TM5 and TM11 of OxlT
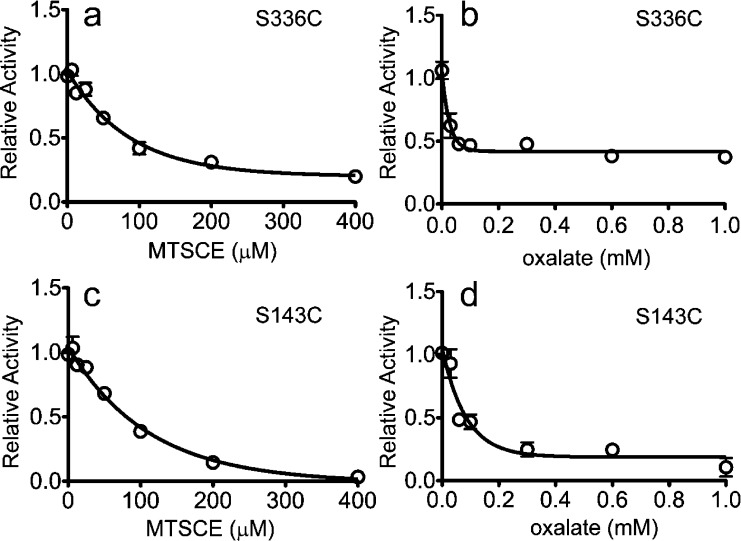
In previous work, a single-cysteine derivative on the cytoplasmic end of TM5, S143C, was found to be more reactive to MTSCE in the presence than in the absence of external oxalate, indicating that a substrate-dependent conformational change occurs at the cytoplasmic end of OxlT.24 In OxlT, the symmetry-related partner to TM5 is TM11 (Figure 1); the cysteine mutant S336C near the cytoplasmic end of TM11 was purified and reconstituted into liposomes, and its reactivity to MTSCE was tested as described previously.24 In this experiment, cysteine derivatization is monitored from the loss in transport activity that takes place when the cysteine reacts with MTSCE. It should be noted that OxlT is symmetrically oriented across proteoliposomes and that MTSCE has access to both interfaces.24 Oxalate-loaded proteoliposomes were initially treated with MTSCE in the absence of external oxalate, and, as seen in Figure 2a, there is a loss of transport activity for S336C as the MTSCE concentration is increased. From Figure 2a, the level of MTSCE producing 50% inhibition was determined, and proteoliposomes (from the same initial stock preparation) were pretreated with this concentration of MTSCE and simultaneously mixed with increasing concentrations of external unlabeled oxalate prior to assaying oxalate transport. As seen in Figure 2b, increasing the concentration of external oxalate reduces the transport activity in the presence of MTSCE, indicating that MTSCE reactivity is enhanced by external cold oxalate. As a positive control, S143C was also examined and was found to respond as seen previously; as shown in Figures 2c,d, the effect of oxalate on transport in the presence of MTSCE indicates that there is an elevated reactivity to MTSCE in the presence of external oxalate. These experiments demonstrate that for both TM5 and TM11 there are substrate-dependent conformational changes during substrate exchange that result in elevated reactivities of residues near the cytoplasmic portions of these helices.
Figure 2.
MTSCE inhibition of transport for S143C and S336C. Purified single-cysteine OxlT mutants were reconstituted into liposomes to form proteoliposomes under conditions where they were loaded with nonradiolabeled (cold) oxalate. The proteoliposomes were washed on a filter, treated with MTSCE, washed again, and assayed for 14[C]oxalate uptake. As MTSCE concentrations are increased, there is a loss in 14[C]oxalate uptake for (a) S336C and (c) S143C in the absence of any external nonradiolabeled oxalate. In a second experiment, the proteoliposomes were washed, incubated with MTSCE sufficient to produce a 50% inhibition in transport and simultaneously with increasing concentrations of external cold oxalate, washed again, and then assayed for 14[C]oxalate transport. The residual 14[C]oxalate transport as a function of external cold oxalate concentration is shown for (b) S336C and (d) S143C. Each data point is the average of three measurements.
The Cytoplasmic Ends of TM5 and TM11 Are Easily Cross-Linked in OxlT
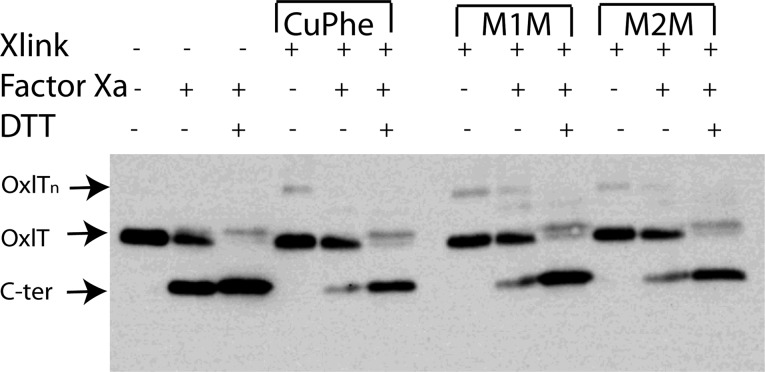
In the homology model for OxlT,23 the Cα atoms of S143 and S336 are separated by ∼21 Å (Figure 1). These sites are also located in GxxxG and GxxxG-like motifs that are often found at the cytoplasmic ends of TM5 and TM11 in MFS transporters (see Supporting Information Table S1). Because the data in Figure 2 indicate that there are conformational changes at the ends of TM5 and TM11 and because GxxxG and GxxxG-like motifs often mediate helix–helix interactions,26 we tested whether S143C and S336C could be cross-linked. In these experiments, we used an OxlT variant having a tandem Factor Xa cleavage site in the central loop that links the N- and C-terminal domains. The cross-linking was carried out in native inner membrane vesicles (see Experimental Procedures) using copper(II) (1,10)-(phenanthroline)3 (a zero-length cross-linking agent), MTS-1-MTS (∼3 Å spacer), and MTS-2-MTS (∼5 Å spacer). The cross-linked protein was examined on SDS-PAGE before and after Factor Xa protease cleavage.30 As shown in Figure 3, cross-linking takes place with all the agents used, suggesting that S143C and S336C interact.
Figure 3.
Western blot demonstrating cross-linking of S143C/S336C in native vesicles. Native membrane vesicles (see Experimental Procedures) containing OxlT with the S143C/S336C mutations and a tandem Factor Xa cleavage site were treated with 200 μM copper(II)(1,10-phenanthroline)3(CuPhe), 200 μM MTS-1-MTS (M1M), or 200 μM MTS-2-MTS (M2M) for 10 min and immediately quenched with 5 mM NEM and 10 mM EDTA. Washed vesicles were processed for western blotting and probed with a C-terminal polyhistidine antibody. Xlink indicates lanes with the addition of the indicated cross-linking agents; Factor Xa indicates lanes where the central loop of OxlT has been cleaved; DTT indicates lanes where DTT is used to reduce disulfides after Factor Xa cleavage. The positions of the covalently intact protein (OxlT), the C-terminal OxlT fragment (C-ter), and OxlT oligomer (OxlTn) are indicated.
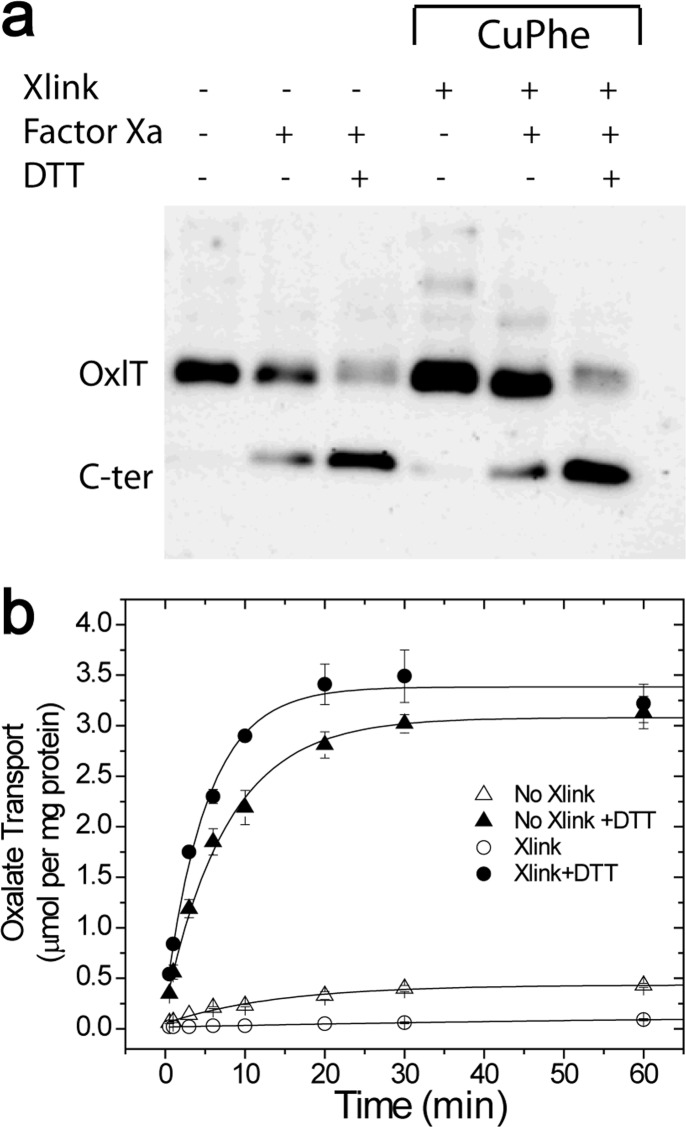
As seen in Figure 3, cross-linking appears to take place between S143C and S336C in native membrane vesicles in the absence of an oxidizing or cross-linking agent. We also determined whether OxlT remains cross-linked in liposomes and whether cross-linking inhibits oxalate transport in proteoliposomes. In this experiment, native membrane vesicles were treated with buffer (control) or freshly prepared copper(II) (1,10-phenanthroline)3, washed, and solubilized in a solution of 100 mM oxalate, 20% glycerol, 20 mM potassium phosphate (pH 7.5), 0.2% E. coli polar lipid, and 0.5% DDM, and solubilized extracts were reconstituted into oxalate-loaded liposomes. An aliquot of proteoliposomes was quenched with NEM and EDTA and processed for protease cleavage while a second aliquot was assayed for transport without quenching. As seen in Figure 4a, a western blot of the cleaved samples indicates significant spontaneous intramolecular cross-linking between S143C and S336C. Furthermore, as seen in Figure 4b, cross-linking inhibits transport and this inhibition may be reversed by externally added DTT (Figure 4b). As expected, the cys-less control does not show evidence of cross-linking when examined on SDS-PAGE or in a transport assay (Figure S1). The results demonstrate that S143C and S336C remain cross-linked in liposomes and suggest that movement at the cytoplasmic ends of helices 5 and 11 is necessary for oxalate transport in OxlT.
Figure 4.
Cross-linking of OxlT S143C/S336C and transport in liposomes. E. coli vesicles containing OxlT with S143C/S336C and a tandem Factor Xa cleavage site were subjected to an oxidizing agent or buffer alone prior to reconstitution into oxalate-loaded vesicles of E. coli polar lipids. (a) Proteoliposomes were immediately quenched with 5 mM NEM after reconstitution and processed for western blotting using a polyhistidine antibody. (b) Another aliquot was assayed for 14[C]oxalate transport in the presence of an oxidizing agent without (○) and with (●) the addition of DTT and in the absence of cross-linking agent without (△) and with (▲) DTT. Each point was measured in triplicate.
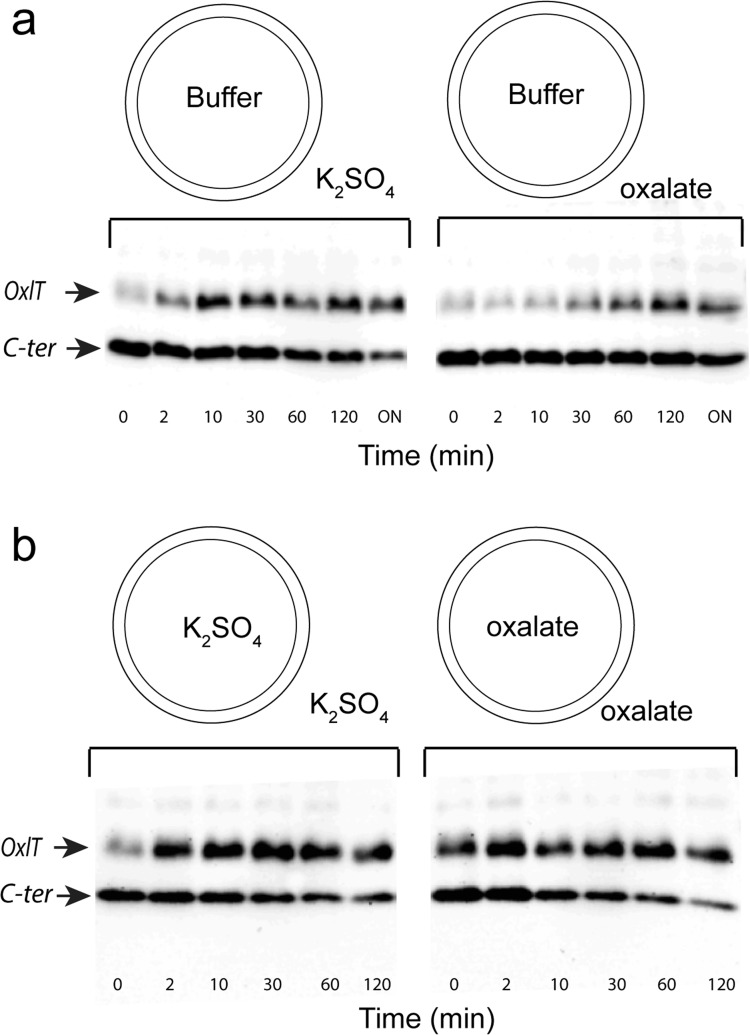
Additional measurements were made on ice (∼4 °C) to examine the effect of substrate on cross-linking. At this lower temperature, conformational exchange between states is slowed, and structures induced by substrate are stabilized. As a result, the effects of substrate on cross-linking should be better resolved. In these measurements, native membrane vesicles were isolated in the presence of potassium sulfate (nonsubstrate) or potassium oxalate (substrate) to create vesicles containing sulfate or oxalate on both sides of the membrane. Vesicles lacking either salt (empty vesicles) were also created in the presence of 20 mM Tris·HCl (see Figure 5, legend). As shown in Figure 5a, addition of substrate (oxalate) to the exterior of empty vesicles inhibits cross-linking compared to the sulfate-treated control. Remarkably, the addition of oxalate to both sides of the membrane (Figure 5b) results in enhanced cross-linking relative to the case where oxalate is added only on the exterior of empty vesicles (Figure 5a). The apparent cross-linking rate is independent of intra- or extra-vesicular placement of sulfate, which is consistent with the idea that sulfate is not a substrate of OxlT. In these native membrane vesicles, OxlT appears to have an inverted orientation,36 and the result indicates that the cytoplasmic ends of helices 5 and 11 are more likely to be in close proximity when oxalate is present on both sides of the membrane than when oxalate is present only on one surface of the transporter. Since OxlT is able to carry out oxalate/oxalate self-exchange and is an obligate exchanger rather than a uniporter,37 conditions that support exchange appear to facilitate proximity of the cytoplasmic ends of TM5 and TM11.
Figure 5.
Effect of oxalate on cross-linking of S143C/S336C FXa. OxlT in native vesicles with S143C/S336C and a tandem Factor Xa cleavage site was treated with the oxidizing agent copper(II) phenanthroline. Cross-linking was followed on ice by quenching the samples at specific times by adding them to tubes preloaded with 5 mM N-ethyl maleimde and 10 mM EDTA, followed by cleavage with Factor Xa. (a) Western blots using a polyhistidine antibody, which show cross-linking of OxlT where buffer (20 mM Tris·HCl (pH 7.5)) was on the vesicle interior and either 10 mM potassium sulfate (left) or 10 mM potassium oxalate (right) were on the vesicle exterior. (b) Buffer on the vesicle interior was replaced with 10 mM potassium sulfate (left) or 10 mM potassium oxalate (right) , and either 10 mM potassium sulfate (left) or 10 mM potassium oxalate (right) are on the exterior. In panel a, ON refers to overnight incubation at 4 °C.
Site-Directed Spin Labeling Indicates That the Cytoplasmic Ends of TM5 and TM11 Are in Close Proximity in the Presence of Substrate
Site-directed spin labeling and EPR spectroscopy were used to test for the proximity of S143C and S336C. The dipolar interaction between spin labels leads to a broadening in the EPR spectrum, and distance measurements may be made in the 8–20 Å range at room temperature.33 The double mutant, S143C/S336C, as well as the corresponding single-cysteine mutants, were labeled with the MTSL spin label (see Experimental Procedures) to attach the R1 side chain (Figure 6). Labeling efficiencies were 85–95% in all cases, as determined from the thiol reactivity to DTDP32 (see Experimental Procedures). Continuous wave (CW) EPR spectra were recorded for S143R1, S336R1, and S143R1/S336R1. As seen in Figure 6a,b, strong dipolar coupling is observed for this pair, which corresponds to distances of 7.8 and 8 Å in bilayers and DDM micelles, respectively. Placing the labeled pair further toward the cytoplasmic surface at I338R1 and L141R1 does not result in any significant dipolar coupling, and the interspin distance exceeds 20 Å in bilayer and DDM preparations (Figure 6c,d).
Figure 6.
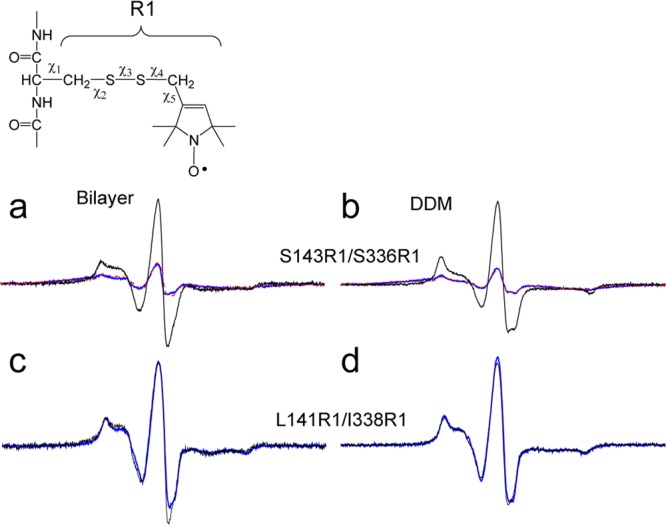
X-band CW EPR line shapes of MTSL-labeled OxlT. The spin-labeled side chain R1 was derivatized to double mutants as well as pairs of the corresponding single-cysteine mutants of OxlT, and X-band EPR spectra were recorded at room temperature. Spectra are shown for the interacting spin pairs (blue traces) and the equivalent spectra without dipolar interactions (black traces). The non-interacting spectra are obtained from the sum of the single-labeled spectra. (a, b) Spectra for S143R1/S336R1 in bilayers and DDM, respectively, where the simulated spectrum obtained from the distance distribution is shown (dashed red trace). The sample in panel a yielded a mean distance of 7.8 Å with a standard deviation of 0.3 Å from three independent experiments. (c, d) Spectra for L141R1/I338R1, which do not show evidence for strong dipolar interaction, for bilayer and DDM environments, respectively. The scans shown are 150 G. Distances were determined using a Fourier deconvolution approach implemented in the LabVIEW program Short Distances (provided by Christian Altenbach, UCLA).
In addition to these spin pairs, six additional spin pairs were produced to examine distances between TM5 and TM11 (see Figures S2 and S3a–d), and the distances measured by CW methods are summarized in Table 1. The spin pair S143R1/S336R1 appears to yield the shortest distance between the cytoplasmic regions of TM5 and TM11, and the spin pair S143R1/I338R1 yields a distance of 17 Å in both DDM and liposomes (Figure S2a,b), consistent with the helical structure in the homology model. As a control, an i, i + 2 pair (S336R1/I338R1) was examined and yielded a distance of 14.3 Å (see Figure S3e), which is close to the distance expected between R1 side chains at i, i + 2 positions in a helix.38 For completeness, we attempted to make measurements on substrate-free OxlT. Dipolar coupling was not observed between S143R1/S336R1 in the absence of substrate; however, these measurements were problematic because OxlT is not stable in the absence of substrate (see Figure S4).
Table 1. Room-Temperature CW EPR Distance Measurements in DDMa.
| mutant | CW distance (Å) | DEER (Å) | n |
|---|---|---|---|
| L141R1/S336R1 | 15 ± 0.3 | 2 | |
| L141R1/I338R1 | >20 | 29 ± 0.5 | 2 |
| A142R1/S336R1 | 15 ± 0.3 | 2 | |
| S143R1/S336R1 | 8 ± 0.1 | 3 | |
| S143R1/D337R1 | 15 ± 0.2 | 2 | |
| S143R1/I338R1 | 17 ± 0.2 | 2 | |
| G144R1/S336R1 | 15 ± 0.6 | 2 | |
| G144R1/D337R1 | 17 ± 0.0 | 2 |
The distance shown for each pair is the mean distance with the half-range for two independent experiments except for S143R1/S336R1, which shows the mean ± standard deviation for three independent experiments. Additional distance distributions obtained by pulse EPR are presented in Figure 7.
The Periplasmic and Cytoplasmic Regions of OxlT Sample Both Open and Closed States
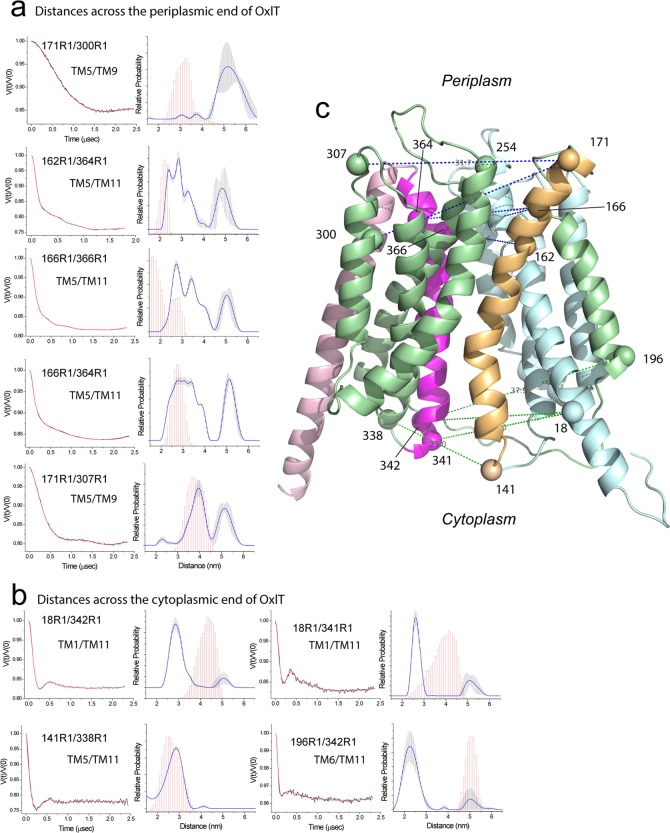
Pulse EPR measurements, such as double electron–electron resonance (DEER), yield both distances and distance distributions between spin labels beyond 20 Å. Shown in Figure 7 are DEER measurements and distance distributions obtained for OxlT in the presence of substrate in DDM micelles. Shown in Figure 7a are measurements made across the periplasmic surface of OxlT, and distances measured across the cytoplasmic region are shown in Figure 7b. Also shown are the predicted R1–R1 distance distributions obtained from the homology model.
Figure 7.
Distances and distance distributions measured by double electron–electron resonance (DEER) for several spin pairs in OxlT. (a) Measurements across the periplasmic end of OxlT. The background-corrected DEER data is shown on the left (black traces) along with the fits to the data using a model-free approach (red traces). The distributions obtained are shown on the right, and an error range is indicated by the vertical error bars (shaded in gray) in the distribution. This error range is based on uncertainty in the background subtraction and dimensionality in the background form factor that produces fits within 15% of the RMSD of the best fits. These errors were obtained using the validation routine in DeerAnalysis. Shown in magenta are predictions of the distances and distance distributions based on the OxlT homology model using the PyMOL plug-in mtsslWizard.52 (b) Measurements across the cytoplasmic end of OxlT. (c) Homology model of OxlT along with the labeled sites used for DEER. Raw DEER data were processed and analyzed using the Matlab software package DEER Analysis.35
As seen in Figure 7, there are multiple distances in most of these distributions. A portion of these distributions may be contributed by spin-labeled side chain (R1) rotamers; however, a comparison of the predicted distributions with the experimental distributions indicates that R1 rotamers cannot account for the broad distributions seen. These distributions indicate the presence of conformational heterogeneity in OxlT, perhaps due to conformational exchange between two or more states. On the periplasmic surface, two or more major distances are usually seen in the distribution, and, in many cases, the shorter distance corresponds closely with that expected from the homology model. On the cytoplasmic surface, there is less heterogeneity, but the predicted distances from the homology model are in 3 out of 4 cases examined significantly longer than distances measured by DEER. It should be noted that the spin pair L141R1/I338R1 yields a mean interspin distance of 28.5 Å, which is in agreement with the model and consistent with the lack of any significant dipolar broadening in the corresponding CW spectrum seen in Figure 6.
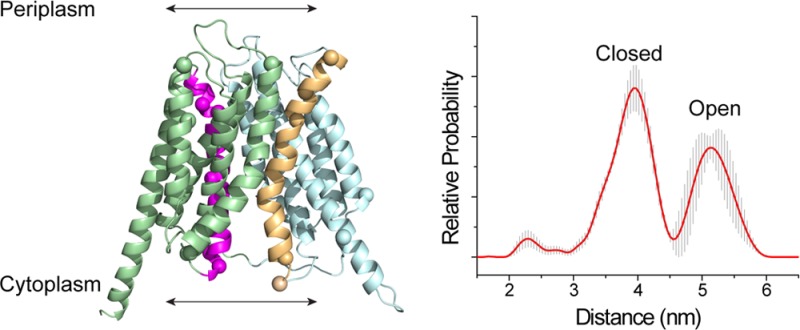
Several additional measurements made across OxlT are shown in Figure S6. These measurements include distances across the length of OxlT as well as one additional measurement between TM5 and TM8 at the periplasmic interface. The distances measured are in general agreement with predictions based on the homology model, but there is clear heterogeneity when the measurement is made from the cytoplasmic end of TM1 to the periplasmic end of TM8. Taken together, the data in Figures 7 and S6 indicate that OxlT samples multiple structural states, where one highly populated state found in the presence of substrate is more compact on the cytoplasmic surface and more expanded on the periplasmic surface than the homology model.
Several Labels at the Cytoplasmic Ends of TM5 and TM11 Inhibit Transport
A number of the spin-labeled and reconstituted OxlT mutants were assayed for substrate binding and function. These spin-labeled mutants were found to bind substrate (Figure S4) by criteria used previously;39,40 however, many of the spin-labeled mutants located near the cytoplasmic and periplasmic interfaces of TM5 and TM11 showed reduced or no transport. As seen in Figure S7, treatment of proteoliposomes with externally added DTT restored function in many cases, suggesting that the OxlT mutants must be correctly folded into the membrane and that chemical modification of the cysteines is primarily responsible for the loss of transport activity. Helices TM5 and TM11 line the oxalate permeation pathway,5,23−25 and the spin label may act to block access to the pathway or interfere with substrate movement along the pore. Consistent with this idea, OxlT mutants retain function when the R1 labels are placed at locations more distant from the permeation pathway.
Discussion
High-resolution crystal structures are now available for a number of membrane transport proteins, including the MFS family of transporters.5−20 However, in the case of the MFS transporters, as is the case with many transport proteins, the detailed steps in transport remain unknown. Recent work using spectroscopic methods indicates that membrane transporters can assume multiple states, which are often in equilibrium.41,42 Such conformational sampling is difficult to characterize but is thought to underlie protein–protein interactions, allostery, and protein function.43 Crystal structures do not reveal these conformational equilibria, and they often trap the protein in a specific substate, which may be populated as a result of the crystallization conditions.44
In the present study, cysteine mutagenesis,45,46 disulfide trapping,47 and EPR spectroscopy48 were used to examine conformational changes at the cytoplasmic ends of TM5 and TM11, which line the transport pore. Pulse EPR was also used to characterize the overall conformation of OxlT in the context of a previously generated homology model. One motivation for this study was the presence of GxxxG and GxxxG-like motifs at the cytoplasmic ends of helices 5 and 11 of OxlT and several well-studied MFS members. Since these motifs are known to mediate helix–helix interactions,26,27 they might mediate helix–helix contact to close off the substrate pore at the cytoplasmic interface during the transport cycle.
Previous work demonstrated that there were substrate-dependent conformational changes in S143C (a position in a GxxxG motif in the cytoplasmic end of TM5),24 and the observation that S336C resides in the symmetry-related SxxxG motif of TM11 (also at the cytoplasmic end of helix 11) suggested that there might be similar conformational changes at this site. The results of site-directed alkylation (Figure 2) and disulfide cross-linking (Figures 3–5) indicate that substrate-coupled conformational changes occur in OxlT and that these conformational changes place the cytoplasmic ends of helices 5 and 11 in close proximity. Moreover, SDSL demonstrates that S143R1/S336R1 are in close proximity, in agreement with the cross-linking of this pair. From this work, the cytoplasmic poles of helices 5 and 11 must move closer, relative to the inward-open GlpT-based homology model23 and the occluded electron crystallographic structure of OxlT.5
A partial closure of the cytoplasmic end of GlpT, mediated by TM5 and TM11, has been observed in molecular dynamics simulations.49 In that study, which used the crystal structure of GlpT in the inward-open conformation,6 substrate binding was observed to stabilize a partially occluded state where TM5 and TM11 approach each other on the cytoplasmic side at regions that constitute GxxxS/GxxxG motifs in GlpT. The findings presented here are consistent with this proposal, but they indicate that larger movements of these helices take place to bring the cytoplasmic regions of TM5 and TM11 together.
The data presented here indicate that OxlT samples an outward-open state that resembles the FucP crystal structure.9 A comparison of several sites between the FucP structure and the OxlT data is shown in Figure S8. Among the currently available MFS structures, the FucP structure places the cytoplasmic poles of helices 5 and 11 in the closest proximity. This is consistent with the short distances measured by CW EPR between the cytoplasmic ends of TM5 and TM11 in OxlT and the DEER measurements on the periplasmic ends of TM5 and TM11, Figure 7a, which are much longer than those predicted by the homology model. As seen in Figure 7b, the most populated distances measured by DEER for the cytoplasmic surface of OxlT are shorter than those predicted by the model, with longer distances being populated on the periplasmic surface. Although an outward-facing state is present and may even dominate the population under these conditions for OxlT, it should be noted that multiple distances are present, particularly at the periplasmic interface, and that some combination of both outward-open and inward-open states are likely to be present and in equilibrium. This finding is similar to that seen for the aspartate transporter, GltPh, where multiple distances are seen, indicating that both inward- and outward-facing states appear to be present in bilayers.42,50
The “rocker-switch” hypothesis22 proposes that transport is mediated by a relative scissor-like motion of the N- and C-terminal domains. This hypothesis implies that there must be contact at the cytoplasmic and periplasmic ends of OxlT that create effective seals during OxlT turnover. In support of this hypothesis, cross-linking has been observed between TM1 (N-terminal domain) and TM7 (C-terminal domain) at the periplasmic end of OxlT,23 as well as substrate-dependent proximity between TM1 and TM7 at the center of FucP.51 The present work demonstrates that there is an interaction between TM5 (N-terminal domain) and TM11 (C-terminal domain) at the cytoplasmic side and is also consistent with this hypothesis.
In summary, the results presented here demonstrate that interactions take place between the cytoplasmic ends of TM5 and TM11 and that OxlT samples an inward-closed state in the presence of substrate. Moreover, pulse EPR measurements show that the protein samples more than one state when substrate is present and may be in exchange between states that can be described as both open and closed. These data are consistent with the expectations drawn from molecular dynamics work on GlpT,49 and they suggest that OxlT may sample a structure that resembles the structure obtained for FucP.9 The cytoplasmic ends of TM5 and TM11 contain GxxxG and GxxxG-like motifs that are frequently found at the cytoplasmic ends of TM5 and TM11 in MFS members. Although not specifically tested in the present study, the results are consistent with the idea that these motifs can function to transiently close and limit cytoplasmic access to the transport pathway.
Acknowledgments
We are grateful to Dr. Albert S. Mildvan (Johns Hopkins Medical School) for technical advice in the early stages of this project, Drs. David R. Shortle, Rajini Rao, and Dax Fu (all of Johns Hopkins Medical School) for critical comments, and Dr. Jinming Cui for help with EPR figures. Support from the Turock Young Scientist Award, Johns Hopkins University (to O.I.), is acknowledged.
Glossary
Abbreviations
- CuPhe
copper(II) (1,10)-(phenanthroline)3
- DDM
n-dodecylmaltoside
- DEER
double electron–electron resonance
- DMSO
dimethyl sulfoxide
- DTDP
4,4′-dithiodipyridine
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetate
- EPR
electron paramagnetic resonance
- IPTG
isopropyl-β-d-thiogalactopyranoside
- MFS
major facilitator superfamily
- MTS
methanethiosulfonate
- MTS-1-MTS
1,1-methanediyl bismethanethiosulfonate
- MTS-2-MTS
1,2-ethanediyl bismethanethiosulfonate
- MTSCE
methanethiosulfonate ethyl carboxylate
- MTSL
methanethiosulfonate spin label or S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methylmethanesulfonothioate
- NEM
N-ethylmaleimide
- OD
optical density
- R1
thiol adduct after derivatization of cysteine with MTSL
- SDSL
site-directed spin labeling
- TM
transmembrane helix
- TCEP
tris(2-carboxyethyl)phosphine
Supporting Information Available
Figure S1: Control for cross-linking in a cysteine-less OxlT mutant (GA) in liposomes. Figure S2: X-band CW EPR of S143R1/I338R1 and L141R1/S336R1. Figure S3: X-band CW EPR spectra of other spin-labeled OxlT mutants. Figure S4: Substrate stabilizes spin-labeled OxlT. Figure S5: Fast protein liquid chromatography of OxlT and transport measurements indicate S143C/S336C is a monomer. Figure S6: Additional distances and distance distributions measured by double electron–electron resonance for spin pairs in OxlT. Figure S7: Transport profiles of MTSL-labeled OxlT mutants. Figure S8: Comparison of DEER-derived distance distributions obtained from OxlT with the predictions from similar sites on FucP. Table S1: Occurrence of GxxxG and GxxxG-like motifs in the cytoplasmic regions of TM5 and TM11 of transporters within the major facilitator superfamily. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by grants from the National Institutes of Health, NIGMS, R01 GM024195 (P.C.M.) and R01 GM035215 (D.S.C.).
The authors declare no competing financial interest.
Author Status
§ Peter C. Maloney: deceased (Dec. 12, 2013).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Allison M. J.; Dawson K. A.; Mayberry W. R.; Foss J. G. (1985) Oxalobacter formigenes gen. nov., sp. nov.: oxalate-degrading anaerobes that inhabit the gastrointestinal tract. Arch. Microbiol. 141, 1–7. [DOI] [PubMed] [Google Scholar]
- Stewart C. S.; Duncan S. H.; Cave D. R. (2004) Oxalobacter formigenes and its role in oxalate metabolism in the human gut. FEMS Microbiol. Lett. 230, 1–7. [DOI] [PubMed] [Google Scholar]
- Maloney P. C. (1994) Bacterial transporters. Curr. Opin. Cell Biol. 6, 571–582. [DOI] [PubMed] [Google Scholar]
- Pao S. S.; Paulsen I. T.; Saier M. H. Jr. (1998) Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai T.; Heymann J. A.; Shi D.; Sarker R.; Maloney P. C.; Subramaniam S. (2002) Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 9, 597–600. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Lemieux M. J.; Song J.; Auer M.; Wang D. N. (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301, 616–620. [DOI] [PubMed] [Google Scholar]
- Abramson J.; Smirnova I.; Kasho V.; Verner G.; Kaback H. R.; Iwata S. (2003) Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615. [DOI] [PubMed] [Google Scholar]
- Yin Y.; He X.; Szewczyk P.; Nguyen T.; Chang G. (2006) Structure of the multidrug transporter EmrD from Escherichia coli. Science 312, 741–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S.; Sun L.; Huang Y.; Lu F.; Liu Y.; Gong H.; Wang J.; Yan N. (2010) Structure of a fucose transporter in an outward-open conformation. Nature 467, 734–738. [DOI] [PubMed] [Google Scholar]
- Newstead S.; Drew D.; Cameron A. D.; Postis V. L.; Xia X.; Fowler P. W.; Ingram J. C.; Carpenter E. P.; Sansom M. S.; McPherson M. J.; Baldwin S. A.; Iwata S. (2011) Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 30, 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solcan N.; Kwok J.; Fowler P. W.; Cameron A. D.; Drew D.; Iwata S.; Newstead S. (2012) Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 31, 3411–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.; Zeng X.; Yan C.; Sun X.; Gong X.; Rao Y.; Yan N. (2012) Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490, 361–366. [DOI] [PubMed] [Google Scholar]
- Zheng H.; Wisedchaisri G.; Gonen T. (2013) Crystal structure of a nitrate/nitrite exchanger. Nature 497, 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H.; Huang W.; Yan C.; Gong X.; Jiang S.; Zhao Y.; Wang J.; Shi Y. (2013) Structure and mechanism of a nitrate transporter. Cell Rep. 3, 716–723. [DOI] [PubMed] [Google Scholar]
- Iancu C. V.; Zamoon J.; Woo S. B.; Aleshin A.; Choe J. Y. (2013) Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc. Natl. Acad. Sci. U.S.A. 110, 17862–17867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethayathulla A. S.; Yousef M. S.; Amin A.; Leblanc G.; Kaback H. R.; Guan L. (2014) Structure-based mechanism for Na+/melibiose symport by MelB. Nat. Commun. 5, 3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen B. P.; Kumar H.; Waight A. B.; Risenmay A. J.; Roe-Zurz Z.; Chau B. H.; Schlessinger A.; Bonomi M.; Harries W.; Sali A.; Johri A. K.; Stroud R. M. (2013) Crystal structure of a eukaryotic phosphate transporter. Nature 496, 533–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar H.; Kasho V.; Smirnova I.; Finer-Moore J. S.; Kaback H. R.; Stroud R. M. (2014) Structure of sugar-bound LacY. Proc. Natl. Acad. Sci. U.S.A. 111, 1784–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D.; Xu C.; Sun P.; Wu J.; Yan C.; Hu M.; Yan N. (2014) Crystal structure of the human glucose transporter GLUT1. Nature 510, 121–125. [DOI] [PubMed] [Google Scholar]
- Jiang D.; Zhao Y.; Wang X.; Fan J.; Heng J.; Liu X.; Feng W.; Kang X.; Huang B.; Liu J.; Zhang X. C. (2013) Structure of the YajR transporter suggests a transport mechanism based on the conserved motif A. Proc. Natl. Acad. Sci. U.S.A. 110, 14664–14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doki S.; Kato H. E.; Solcan N.; Iwaki M.; Koyama M.; Hattori M.; Iwase N.; Tsukazaki T.; Sugita Y.; Kandori H.; Newstead S.; Ishitani R.; Nureki O. (2013) Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc. Natl. Acad. Sci. U.S.A. 110, 11343–11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. J.; Maloney P. C.; Wang D. N. (2008) Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q.; Wang X.; Ye L.; Mentrikoski M.; Mohammadi E.; Kim Y. M.; Maloney P. C. (2005) Experimental tests of a homology model for OxlT, the oxalate transporter of Oxalobacter formigenes. Proc. Natl. Acad. Sci. U.S.A. 102, 8513–8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Ye L.; McKinney C. C.; Feng M.; Maloney P. C. (2008) Cysteine scanning mutagenesis of TM5 reveals conformational changes in OxlT, the oxalate transporter of Oxalobacter formigenes. Biochemistry 47, 5709–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D.; Sarker R. I.; Abe K.; Bolton E.; Maloney P. C. (2001) Structure/function relationships in OxlT, the oxalate-formate transporter of Oxalobacter formigenes. Assignment of transmembrane helix 11 to the translocation pathway. J. Biol. Chem. 276, 8753–8760. [DOI] [PubMed] [Google Scholar]
- Russ W. P.; Engelman D. M. (2000) The GxxxG motif: a framework for transmembrane helix-helix association. J. Mol. Biol. 296, 911–919. [DOI] [PubMed] [Google Scholar]
- Senes A.; Engel D. E.; DeGrado W. F. (2004) Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14, 465–479. [DOI] [PubMed] [Google Scholar]
- Fu D.; Maloney P. C. (1998) Structure–function relationships in OxlT, the oxalate/formate transporter of Oxalobacter formigenes. Topological features of transmembrane helix 11 as visualized by site-directed fluorescent labeling. J. Biol. Chem. 273, 17962–17967. [DOI] [PubMed] [Google Scholar]
- Fu D.; Maloney P. C. (1997) Evaluation of secondary structure of OxlT, the oxalate transporter of Oxalobacter formigenes, by circular dichroism spectroscopy. J. Biol. Chem. 272, 2129–2135. [DOI] [PubMed] [Google Scholar]
- Kim Y. M.; Ye L.; Maloney P. C. (2001) Helix proximity in OxlT, the oalate:formate antiporter of Oxalobacter formigenes. Cross-linking between TM2 and TM11. J. Biol. Chem. 276, 36681–36686. [DOI] [PubMed] [Google Scholar]
- Abe K.; Ruan Z. S.; Maloney P. C. (1996) Cloning, sequencing, and expression in Escherichia coli of OxlT, the oxalate:formate exchange protein of Oxalobacter formigenes. J. Biol. Chem. 271, 6789–6793. [DOI] [PubMed] [Google Scholar]
- Riener C. K.; Kada G.; Gruber H. J. (2002) Quick measurement of protein sulfhydryls with Ellman’s reagent and with 4,4′-dithiodipyridine. Anal. Bioanal. Chem. 373, 266–276. [DOI] [PubMed] [Google Scholar]
- Altenbach C.; Oh K. J.; Trabanino R. J.; Hideg K.; Hubbell W. L. (2001) Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry 40, 15471–15482. [DOI] [PubMed] [Google Scholar]
- Pannier M.; Veit S.; Godt A.; Jeschke G.; Spiess H. W. (2000) Dead-time free measurement of dipole–dipole interactions between electron spins. J. Magn. Reson. 142, 331–340. [DOI] [PubMed] [Google Scholar]
- Jeschke G.; Chechik V.; Ionita P.; Godt A.; Zimmermann H.; Banham J.; Timmel C. R.; Hilger D.; Jung H. (2006) DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498. [Google Scholar]
- Futai M. (1974) Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J. Membr. Biol. 15, 15–28. [DOI] [PubMed] [Google Scholar]
- Anantharam V.; Allison M. J.; Maloney P. C. (1989) Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J. Biol. Chem. 264, 7244–7250. [PubMed] [Google Scholar]
- Rabenstein M. D.; Shin Y. K. (1995) Determination of the distance between two spin labels attached to a macromolecule. Proc. Natl. Acad. Sci. U.S.A. 92, 8239–8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney P. C.; Anantharam V.; Allison M. J. (1992) Measurement of the substrate dissociation constant of a solubilized membrane carrier. Substrate stabilization of OxlT, the anion exchange protein of Oxalobacter formigenes. J. Biol. Chem. 267, 10531–10536. [PubMed] [Google Scholar]
- Wang X.; Sarker R. I.; Maloney P. C. (2006) Analysis of substrate-binding elements in OxlT, the oxalate:formate antiporter of Oxalobacter formigenes. Biochemistry 45, 10344–10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores Jimenez R. H.; Do Cao M. A.; Kim M.; Cafiso D. S. (2010) Osmolytes modulate conformational exchange in solvent-exposed regions of membrane proteins. Protein Sci. 19, 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva E. R.; Borbat P. P.; Ginter C.; Freed J. H.; Boudker O. (2013) Conformational ensemble of the sodium-coupled aspartate transporter. Nat. Struct. Mol. Biol. 20, 215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker B. A.; Portman J. J.; Wolynes P. G. (2000) Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. U.S.A. 97, 8868–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed D. M.; Horanyi P. S.; Wiener M. C.; Cafiso D. S. (2010) Conformational exchange in a membrane transport protein is altered in protein crystals. Biophys. J. 99, 1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R. T.; Maloney P. C. (1993) Identification of a residue in the translocation pathway of a membrane carrier. Cell 75, 37–44. [PubMed] [Google Scholar]
- Karlin A.; Akabas M. H. (1998) Substituted-cysteine accessibility method. Methods Enzymol. 293, 123–145. [DOI] [PubMed] [Google Scholar]
- Careaga C. L.; Falke J. J. (1992) Thermal motions of surface alpha-helices in the d-galactose chemosensory receptor. Detection by disulfide trapping. J. Mol. Biol. 226, 1219–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbell W. L.; Cafiso D. S.; Altenbach C. (2000) Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 7, 735–739. [DOI] [PubMed] [Google Scholar]
- Enkavi G.; Tajkhorshid E. (2010) Simulation of spontaneous substrate binding revealing the binding pathway and mechanism and initial conformational response of GlpT. Biochemistry 49, 1105–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanelt I.; Wunnicke D.; Bordignon E.; Steinhoff H. J.; Slotboom D. J. (2013) Conformational heterogeneity of the aspartate transporter Glt(Ph). Nat. Struct. Mol. Biol. 20, 210–214. [DOI] [PubMed] [Google Scholar]
- Sugihara J.; Sun L.; Yan N.; Kaback H. R. (2012) Dynamics of the l-fucose/H+ symporter revealed by fluorescence spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 109, 14847–14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagelueken G.; Ward R.; Naismith J. H.; Schiemann O. (2012) MtsslWizard: in silico spin-labeling and generation of distance distributions in PyMOL. Appl. Magn. Reson. 42, 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.