Abstract
Scavenger receptor class B type I (SR-BI), the high density lipoprotein (HDL) receptor, is important for the delivery of HDL-cholesteryl esters to the liver for excretion via bile formation. The focus on therapeutic strategies aimed at reducing cholesterol levels highlights the critical need to understand the structural features of SR-BI that drive cholesterol removal. Yet, in the absence of a high-resolution structure of SR-BI, our understanding of how SR-BI interacts with HDL is limited. In this study, we have optimized the NMR solution conditions for the structural analysis of the C-terminal transmembrane domain of SR-BI that harbors putative domains required for receptor oligomerization. An isotopically-labeled SR-BI peptide encompassing residues 405–475 was bacterially-expressed and purified. [U-15N]-SR-BI(405–475) was incorporated into different detergent micelles and assessed by 1H-15N-HSQC in order to determine which detergent micelle best maintained SR-BI(405–475) in a folded, native conformation for subsequent NMR analyses. We also determined the optimal detergent concentration used in micelles, as well as temperature, solution buffer and pH conditions. Based on 1H-15N-HSQC peak dispersion, intensity, and uniformity, we determined that [U-15N]-SR-BI(405–475) should be incorporated into 5% detergent micelles consisting of 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-[1’-rac-glycerol] (LPPG) and data collected at 40°C in a non-buffered solution at pH 6.8. Furthermore, we demonstrate the ability of SR-BI(405–475) to form dimers upon chemical crosslinking. These studies represent the first steps in obtaining high-resolution structural information by NMR for the HDL receptor that plays a critical role in regulating whole body cholesterol removal.
Keywords: HDL, SR-BI, transmembrane, NMR, detergent, micelles, expression, purification
INTRODUCTION
Decades of epidemiological data strongly suggest that high density lipoproteins (HDL) protect against cardiovascular disease (reviewed in [1]). While HDL exerts numerous beneficial effects via inhibition of oxidative, apoptotic and inflammatory pathways (reviewed in [1]), its primary cardio-protective effect is attributed to its role in reverse cholesterol transport [2]. In this pathway, HDL removes cholesterol from peripheral tissues and transports it to the liver for excretion via bile formation [2]. Scavenger receptor class B type I (SR-BI), the HDL receptor, facilitates this transfer of cholesterol from HDL into cells. The physiological importance of SR-BI in mediating cholesterol flux in humans was obtained from the recent identification of three loss-of-function mutations in SCARB1 (the human SR-BI gene) in heterozygote carriers with high HDLcholesterol (HDL-C) levels [3,4]. These mutant SR-BI receptors were unable to mediate selective uptake of HDL-cholesteryl esters (CE) in cultured cells [4,5]. While the risk for cardiovascular disease has not been assessed in this small cohort of patients, the antiatherogenic properties of SR-BI and its ability to promote reverse cholesterol transport are firmly established in genetically-modified mouse models. Hepatic overexpression of SR-BI in mice [6–8] markedly lowered HDL-C, increased cholesterol catabolism and excretion, and reduced atherosclerosis [9–11]. On the other hand, a 50% reduction in SR-BI expression [12] or full disruption of the SR-BI gene [13,14] in mice significantly increased plasma HDL-C levels, yet dramatically accelerated atherosclerosis [14–16]. Since the delivery of HDL-CE to hepatic tissues only occurs upon binding of HDL to SR-BI, understanding the HDL/SR-BI interaction is critical as we strive to develop novel strategies to reduce whole body cholesterol levels.
There is currently no high-resolution structure available for SR-BI, an 82-kDa cell surface glycoprotein [17]. Based on hydropathy analyses [18], SR-BI (509 amino acids) consists of a short N-terminal cytoplasmic tail (~8 residues), followed by an N-terminal transmembrane domain (~28 residues), a large extracellular domain (~403 residues), a C-terminal transmembrane domain (~25 residues) and finally, a C-terminal cytoplasmic tail (~45 residues) (Figure 1A) [19]. Binding of HDL to the extracellular domain of SR-BI is necessary [20–24], but not sufficient for selective uptake of HDL-CE. It has been speculated that formation of a productive HDL/SR-BI complex [25] depends on the correct alignment of specific lipoprotein and receptor domains and/or the capacity of the receptor to undergo appropriate conformational changes that permit efficient lipid transport. Several studies [26–29], including our own [5,30–32], have demonstrated the important contributions of specific extracellular regions of SR-BI in mediating the selective uptake of HDL-CE. Indeed, the recent availability of the X-ray crystal structure of the extracellular domain of LIMP-2 [33], a scavenger receptor that shares 30% sequence identity with SR-BI, provides the opportunity to better recognize key structural features of this domain that contribute to its cholesterol transport functions.
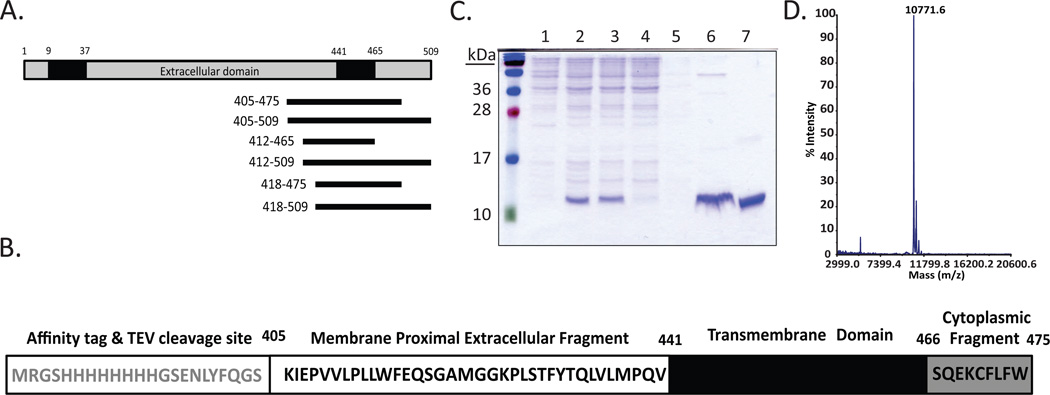
Figure 1. Purification of SR-BI(405–475).
(A) Schematic representation of linear murine SR-BI, where cytoplasmic domains (grey), transmembrane domains (TD; black) and the extracellular domain (white) are shown. SR-BI peptides containing the C-terminal transmembrane domain were cloned into a modified pQE30 vector and ranged from 53–104 residues in length. (B) The sequence of SR-BI(405–475), which includes the affinity tag and TEV cleavage site, membrane proximal extracellular fragment, transmembrane domain, and cytoplasmic fragment. (C) SR-BI(405–475) was purified as described in Materials and Methods. Lane 1, whole cell lysate of transformed SR-BI(405–475) in E. coli; lane 2, whole cell lysate following protein expression induced by IPTG; lane 3, supernatant fraction following solubilization of cell pellet in empigen detergent; lane 4, Ni-column flow through; lane 5, Ni-column wash; lane 6, Ni-column elution of protein with 500mM imidazole; lane 7, HPLC-purified peptide. (D) MALDI mass spectrometry analysis of SR-BI(405–475) peptide (expected mass is 10,778 Da).
The efficiency of HDL-CE selective uptake is also dependent on SR-BI oligomerization [34]. We [35,36] and others [37–39] have demonstrated the presence of SR-BI oligomers, and it has been postulated that HDL-CE uptake occurs via a non-aqueous pathway, possibly involving the formation of a “hydrophobic channel”[40]. Importantly, live cell fluorescence resonance energy transfer studies indicate that self-association of SR-BI is mediated by interaction between the C-terminal transmembrane domains [35], while another study identified a glycine dimerization motif in the N-terminal transmembrane domain that mediates SR-BI oligomerization [41]. In order to understand the mechanisms that regulate SR-BI oligomerization and the selective uptake of HDL-CE, it is critical that we understand the structure of the transmembrane domains of SR-BI.
In this study, we report the expression and purification of SR-BI(405–475), a peptide that encompasses residues 405–475 of SR-BI that contains the C-terminal transmembrane domain of SR-BI (residues 441–465), as well as portions of the extracellular domain (residues 405–440) and the C-terminal cytoplasmic domain (residues 466–475). We also describe optimization of conditions that will enable us to obtain high-resolution structural information of this peptide using nuclear magnetic resonance (NMR)-based strategies. These studies represent the first steps in obtaining structural information for the HDL receptor that plays a critical role in regulating whole body cholesterol removal.
METHODS
Materials
The following detergents were used: 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) (LPPG; Avanti Polar Lipids, Inc.); octyl β-D-glucopyranoside (OG; Sigma-Aldrich); n-dodecylphosphocholine (DPC; Affymetrix); empigen (Sigma-Aldrich); n-dodecyl β-D-maltopyranoside (DDM; Sigma-Aldrich); 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) (LMPG; Affymetrix); 1-oleoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol) (LOPG; Avanti Polar Lipids, Inc.). His60 Ni Superflow resin was purchased from Clontech. Protease inhibitor cocktail was purchased from Fermentas and RNAse was from Thermo Scientific. DNAse, glutaraldehyde, and 1-anilinonaphthalene-8-sulfonic acid (ANS) were purchased from Sigma-Aldrich. All other reagents were of analytical grade.
Plasmids
Sequences encompassing the C-terminal transmembrane domain of murine SR-BI cDNA were PCR-amplified and cloned into the BamHI and HindIII sites of a modified pQE30 vector (QIAGEN). The resulting plasmids encoded for peptides with an N-terminal eight-residue histidine tag and tobacco etch virus cleavage site, followed by SR-BI sequences of varying length. Protein purification, circular dichroism, and NMR experiments were performed using a peptide corresponding to SR-BI residues 405–475 [herein referred to as SR-BI(405–475)] following expression screening as described below.
Cell Culture and Protein Purification
Plasmids encoding SR-BI peptides were transformed in Escherichia coli (E. coli) strain BL21-pREP4 and plated onto ampicillin- and kanamycin-containing LB plates and cultured at 37°C overnight. Following transformation, colonies were used to inoculate 5mL LB broth and grown at 37°C overnight. Cells were then grown in M9 medium supplemented with either 15NH4Cl or 15NH4Cl and [U-13C]-glucose to produce [U-15N]- or [U-15N,13C]-labeled peptides, respectively, and incubated at 37°C overnight. The following day, the culture was used to inoculate 1L of M9 media and upon reaching O.D.600 0.6–0.8, protein expression was induced using 1mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 7–8 hours at 37°C. Cells were then centrifuged for 10 min at 5,000 × g at 4°C.
Protein was purified as previously described [42], using immobilized metal per manufacturer’s protocol with several modifications. Cell pellets were resuspended in Buffer A (50 mM sodium phosphate, 300 mM sodium chloride, 10 mM imidazole, pH 7.4) containing lysozyme, 2-mercaptoethanol, protease inhibitor cocktail, DNAse, and RNase. Lysate was gently rocked for 30 min at room temperature. Cells were disrupted by French pressure cell press. Following cell lysis, 10% empigen detergent was added to aid in solubilization of the membranes and lysate was rocked for an additional hour. Lysate was centrifuged for 30 min at 15,000 × g and supernatant was applied to a gravity flow column containing nickel (His60 Ni Superflow) resin. Resin was washed 5× with Buffer A containing 1.5% empigen and protein was eluted using Buffer B (50 mM sodium phosphate, 300 mM sodium chloride, 500 mM imidazole, pH 7.4) containing 1.5% empigen.
Protein was precipitated by excess addition of ammonium sulfate and the solution was centrifuged at 15,000 × g for 30 min. The resulting pellet was resuspended in Buffer A containing 6M guanidine HCl and 50 mM dithiothreitol. After 30 min, the solution pH was dropped below pH 3 and immediately separated by reversed phase HPLC using a C18 column (column buffer of 70% acetonitrile, 30% 2-propanol, and 0.1% trifluoroacetic acid). Purified protein (expected mass of 10,778 Da) was confirmed by MALDI mass spectrometry, lyophilized, and stored at −80°C until used for NMR and/or circular dichroism analysis. Prior to experiments, lyophilized protein was solubilized in water containing 5% (w/v) detergent, 10% (v/v) D2O, and 0.02% (v/v) sodium azide at pH 6.8, unless otherwise specified. Protein solubilization was aided by incubating the solution at 37–42°C. Based on visual inspection, lyophilized protein typically went into solution within 15 minutes after reconstitution into detergent micelles.
Critical Micelle Concentration (CMC) Measurement
The CMC of LOPG was determined using a fluorometric method as previously described [43]. Briefly, 30 µM ANS was added to series dilutions of LOPG in water. The CMC of DDM and DPC were determined at the same time to verify that our calculations were similar to previously reported values (reviewed in [44]). Non-Detergents Sulfobetaines (NDSB) served as a negative control for CMC calculations. The excitation (388 nm) and emission (480 nm) wavelengths were acquired using a FlexStation 3 MicroPlate Reader (Molecular Devices).
Circular Dichroism (CD)
CD data were acquired using 5–10 µM protein in 5% LPPG/Buffer A from 190 nm to 260 nm using a 1 mm path length cuvette at 25 °C in a Jasco J-710 circular dichroism spectropolarimeter. The digital response time was 2 seconds, the bandwidth was 1.0 nm, and the data pitch was set at 0.1 nm.
NMR Analyses
NMR samples (1 mM protein) were prepared in water containing 5% (98.7 mM) LPPG, 10% D2O, and 0.02% sodium azide at pH 6.8, unless otherwise specified. 1H-15N-Heteronuclear single quantum coherence (HSQC) and 1H-13C-15N-HNCACB NMR spectra [45,46] were obtained at 40 °C on either a Bruker Avance III 500 MHz or Bruker 600 MHz spectrometer. NMRPipe [47] was used for processing and data was analyzed using Xeasy [48].
Chemical Crosslinking of Peptide
LPPG detergent micelles (5%) containing 1mM SR-BI(405–475) at pH 6.8 was crosslinked for 30 minutes at room temperature with increasing concentrations of glutaraldehyde. Immediately following incubation, the sample was separated by 12% SDS-PAGE and visualized by immunoblot using an anti-histidine antibody, as previously described [32].
RESULTS
Expression and purification of SR-BI constructs
Based on hydropathy analyses [18], residues 441–465 of murine SR-BI are predicted to encompass the C-terminal transmembrane domain [18]. Therefore, we designed a series of SR-BI peptides that would span the C-terminal transmembrane domain and screened these peptides for expression and purity for subsequent NMR analyses. Six SR-BI constructs, ranging from 53 to 104 residues in length (Figure 1A), were screened. Based on yield of purified protein, we chose to pursue our studies with the SR-BI peptide that consisted of amino acids 405–475 [herein referred to as SR-BI(405–475)] (Figure 1B). Protein purification was optimized (Figure 1C) using the zwitterionic detergent, empigen, to aid in protein solubilization during cell lysis and Ni-column purification. After elution of the peptide from the column using 500 mM imidazole, [U-15N]- SR-BI(405–475) was further purified by reversed phase HPLC. The resulting purified protein was near the expected mass of 10,778 Da as determined by mass spectrometry (Figure 1D). The difference in actual mass can be attributed to approximately 95% 15N incorporation during protein expression.
Detergent screen for preparation of micelles containing SR-BI(405–475)
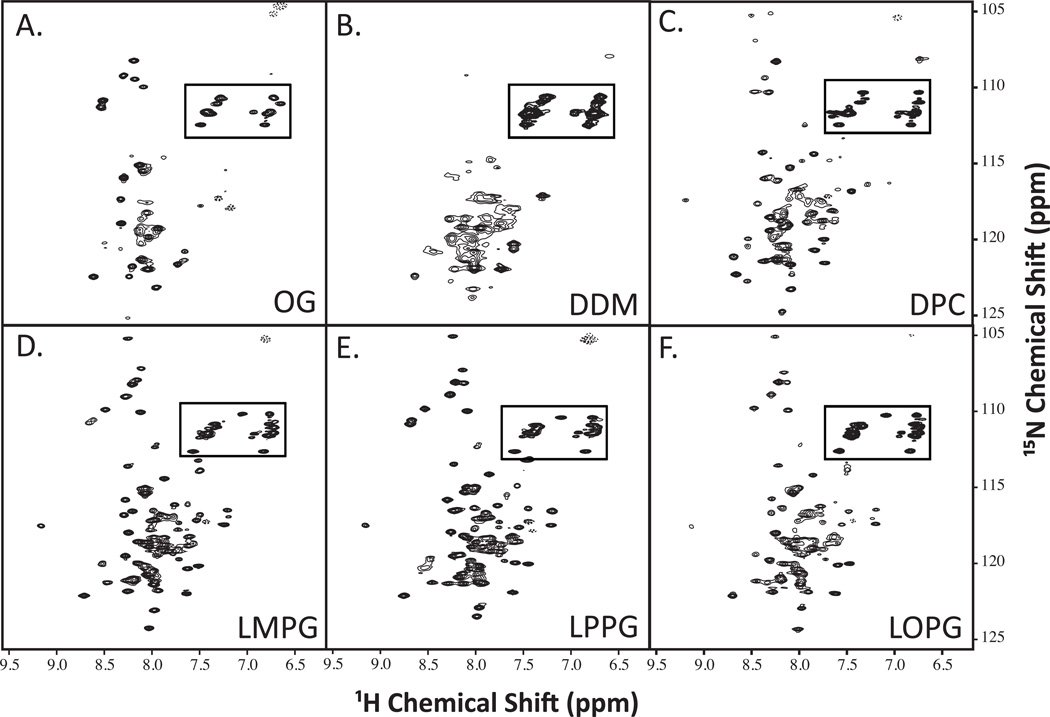
Detergent micelles are routinely used to study structures of insoluble proteins [44,49,50]. Multiple detergents were tested to determine which would best maintain SR-BI(405–475) in a folded, native conformation and produce the expected 105 peaks in 1H-15N-HSQC spectra. Using [U-15N]-SR-BI(405–475), we screened 6 different detergents by 1H-15N-HSQC NMR that differed in classification, as well as length of the carbon chain (Table 1). The detergent micelles that offered the best NMR peak dispersion were the lysophos-detergents ranging from 12:0–18:1 in carbon chain lengths (Figure 2), with LMPG (14:0 carbon chain) and LPPG (16:0 carbon chain) producing close to the correct number of 105 expected peaks (95 and 99 peaks, respectively; Table 1) and most uniform peak intensity.
Table 1. Detergents used for NMR analysis of SR-BI(405–475).
Chemical structures of detergents screened in Figure 2 are shown. The number of peaks observed on 1H-15N HSQC spectra is listed for each detergent tested. CMC values for all detergents were previously reported (reviewed in [56]), with the exception of the CMC for LOPG which we calculated as described in the Methods section.
| Detergent | CMC (mM) |
Detergent Structure |
1H/15N HSQC Peak Count |
|---|---|---|---|
| OG | 19–25 | 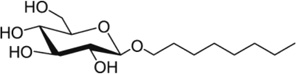 |
57 |
| DDM | 0.2 | 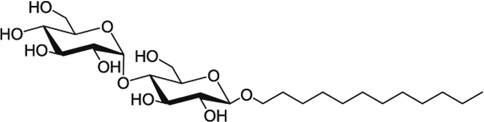 |
66 |
| DPC | 0.9–1.5 | 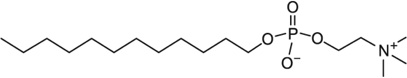 |
93 |
| LMPG (14:0 Lyso PG) | 0.2–3 | 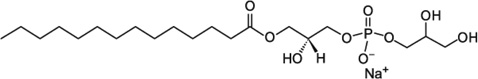 |
95 |
| LPPG (16:0 Lyso PG) | 0.02–0.6 | 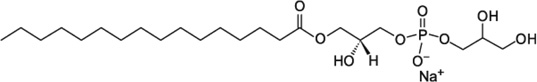 |
99 |
| LOPG (18:1 Lyso PG) | 8 | 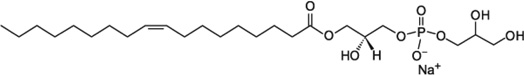 |
94 |
Figure 2. 1H-15N-HSQC spectra of SR-BI(405–475) in different detergent micelles.
Purified [U-15N]-SR-BI(405–475) was solubilized in water containing 5% detergent, 10% D2O, and 0.02% sodium azide at pH 6.8 and screened by NMR analyses. The following detergents were tested: (A) OG; (B) DDM; (C) DPC; (D) LMPG; (E) LPPG; and (F) LOPG. Anticipated side chain cross peaks are boxed.
LPPG, not LMPG, produces more stable detergent micelles
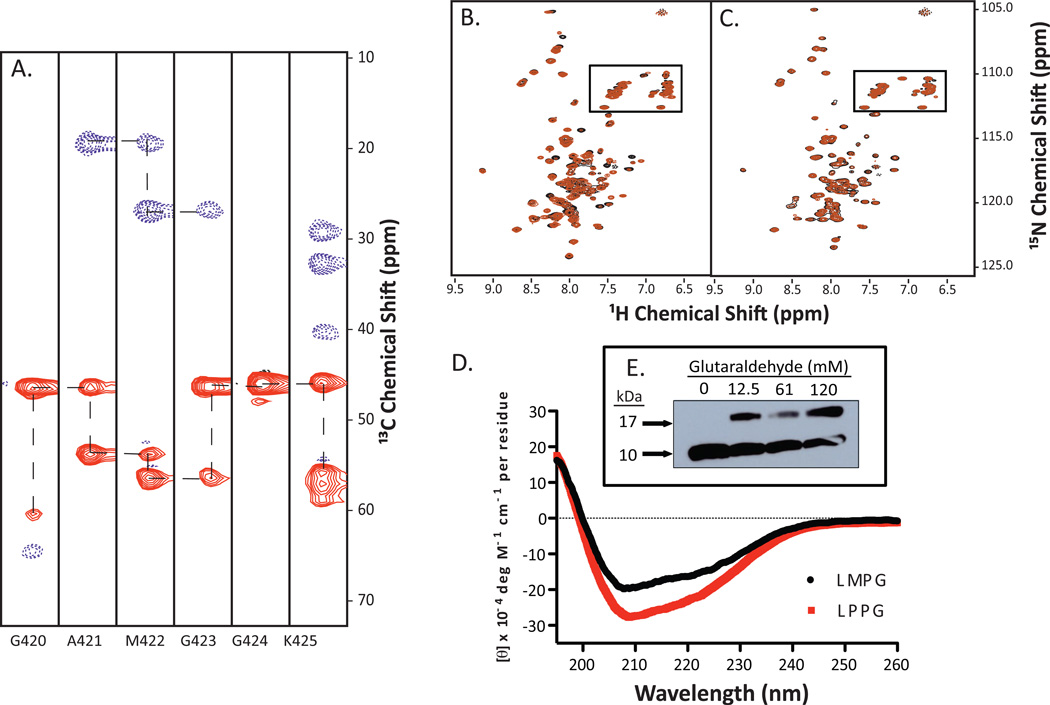
We collected a series of 3D NMR data using LMPG in order to begin structural studies using [U-13C,15N]- SR-BI(405–475) peptide. A series of 1H-13C strips from the HNCACB spectrum shows the sequential assignment of residues 420–425 spanning a putative GXXXG dimerization motif (Figure 3A). Unfortunately, over the period of several days required for data collection, numerous peak intensities significantly increased or decreased when the peptide was incorporated into LMPG-containing micelles (Figure 3B). With this change in stability, we decided to optimize the NMR solution conditions using a different detergent, LPPG, which also produced close to the expected number of peaks for this peptide (see Table 1). Unlike LMPG micelles, when SR-BI(405–475) was inserted into LPPG micelles, the mixture remained stable for at least one week with minimal variation in peak intensity or position (Figure 3C). Therefore, we chose to use LPPG as our detergent for subsequent NMR analyses.
Figure 3. Optimization of stability of SR-BI(405–475) in detergent micelles.
(A) 3D 1H-13C-15N-HNCACB NMR data was acquired for [U-13C,15N]-SR-BI(405–475) in LMPG-containing micelles. 1H-13C strips for residues G420-K425 are shown. (B) 1H-15N HSQC spectra of [U-15N]- SR-BI(405–475) in LMPG detergent micelles show changes in several peak intensities after 6 days, while (C) LPPG remains relatively similar. Overlay of peaks from Day 0 (black) and Day 6 (orange) are shown. Anticipated side chain cross peaks are boxed. (D) Circular dichroism analysis of SR-BI(405–475) reveals alpha helical content for peptide inserted into detergent micelles containing either LMPG (black spectrum) or LPPG (red spectrum). CD data represents the average from at least three independent experiments. (E) Immunoblot showing SR-BI(405–475) in 5% LPPG micelles crosslinked with increasing concentrations of glutaraldehyde (expected monomer size of ~11kDa).
Secondary structure analysis of SR-BI(405–475) by circular dichroism
The secondary structure of transmembrane domains often consists of α-helices, as this satisfies peptide hydrogen bonding in a hydrophobic environment [51]. Circular dichroism analysis of SR-BI(405–475) in LMPG- and LPPG-containing micelles revealed that, as expected, the peptide had α-helical content as demonstrated by the characteristic negative troughs at 208 nm and 222 nm (Figure 3D), with LPPG-containing micelles resulting in an apparent increase in overall helical content.
SR-BI(405–475) forms dimers in LPPG detergent micelles
Since SR-BI(405–475) harbors putative dimerization domains, we tested whether the peptide could form dimers in LPPG detergent micelles. As expected, dimeric species of SR-BI(405–475) were visualized by immunoblot analyses upon chemical crosslinking with increasing concentrations of glutaraldehyde (Figure 3E).
Optimization of NMR solution conditions for SR-BI(405–475) in LPPG micelles
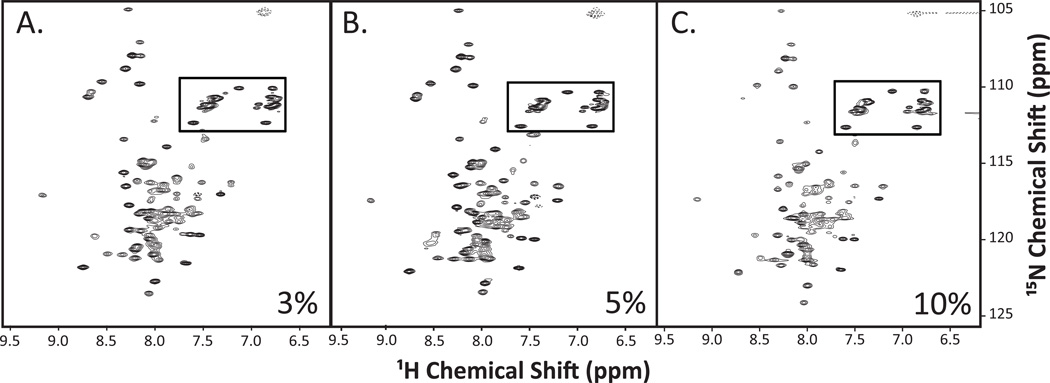
Using LPPG detergent micelles, we screened multiple NMR solution conditions to determine optimal parameters for data collection based on peak number, intensity, and uniformity. First, we collected spectra at temperatures ranging from 25–45°C, and found that 40°C was optimal for data collection as this temperature resulted in the best peak resolution (data not shown). Next, we tested the incorporation of SR-BI(405–475) into LPPG-containing micelles at concentrations of 3%, 5% and 10%, well within the range routinely used for structure determination [44,52]. Detergent concentrations lower than 3% were not tested due to insolubility of the peptide. Although there were differences between spectra (Figure 4A–C), samples containing 3% LPPG and 5% LPPG showed a more uniform peak intensity across the whole spectrum as compared to the 10% detergent solution. Therefore, we chose to collect data for subsequent analyses using micelles consisting of 5% LPPG.
Figure 4. 1H-15N-HSQC spectra for SR-BI(405–475) reconstituted into varying concentrations of LPPG detergent.
Purified [U-15N]-SR-BI(405–475) was solubilized in water containing either (A) 3%, (B) 5%, or (C) 10% LPPG detergent, as well as 10% D2O, and 0.02% sodium azide at pH 6.8. 1H-15N-HSQC spectra were acquired and analyzed as described in Materials and Methods. Anticipated side chain cross peaks are boxed.
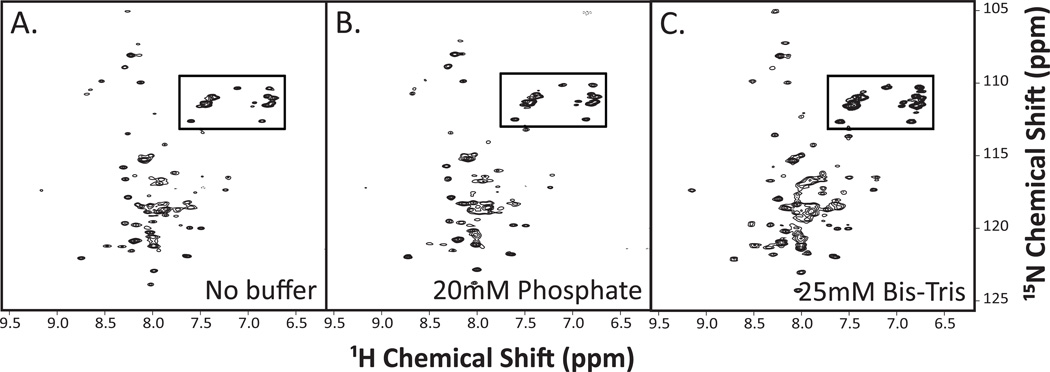
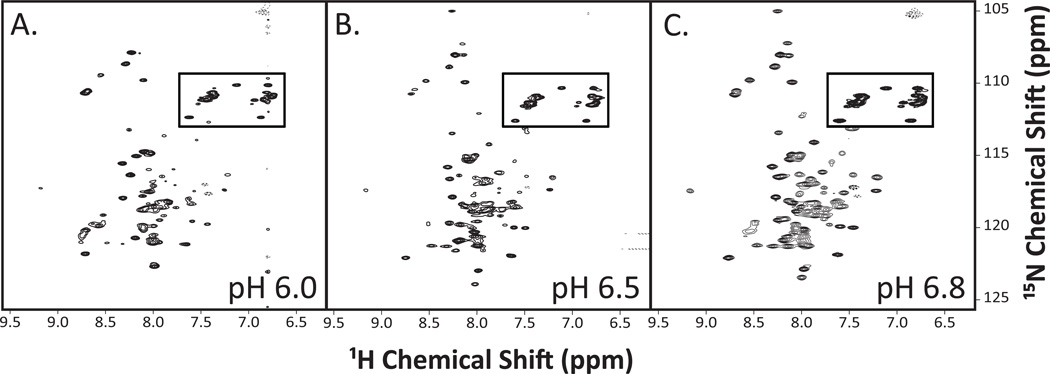
We also screened a series of commonly-used solution conditions which included peptide only, or peptide in buffers containing either 20 mM potassium phosphate or 25 mM bis-Tris buffer (Figure 5A–C). Between the three spectra, the water sample produced the spectrum with the greatest peak intensities and least background signal-to-noise ratio. Lastly, the optimal pH was determined. Since protein/detergent micelles precipitated over time at lower pHs (data not shown), we chose to test more neutral pH’s ranging from 6.0–6.8 (Figure 6A–C). The 1H-15N-HSQC spectra at higher pH values of 6.5 and 6.8 had more uniform peak shape and intensity.
Figure 5. 1H-15N-HSQC spectra for SR-BI(405–475) reconstituted into LPPG detergent micelles using different buffer systems.
Purified [U-15N]-SR-BI(405–475) was solubilized in either (A) water, (B) buffer containing 20 mM potassium phosphate, 100mM NaCl, or (C) buffer containing 25 mM D-Bis-Tris, 25 mM sodium acetate. All solutions also included 5% LPPG detergent, 10% D2O, and 0.02% sodium azide at pH 6.5. 1H-15N-HSQC spectra were acquired and analyzed as described in Materials and Methods. Anticipated side chain cross peaks are boxed.
Figure 6. 1H-15N-HSQC spectra of SR-BI(405–475) reconstituted into LPPG detergent micelles at various pH values.
Purified [U-15N]-SR-BI(405–475) was solubilized in water containing 5% LPPG detergent, 10% D2O, and 0.02% sodium azide at either (A) pH 6.0; (B) pH 6.5, or (C) pH 6.8. 1H-15N-HSQC spectra were acquired and analyzed as described in Materials and Methods. Anticipated side chain cross peaks are boxed.
DISCUSSION
In the current report, we report the expression, purification and reconstitution of an SR-BI peptide into detergent micelles for NMR analyses and structure determination. This peptide that encompasses the C-terminal transmembrane domain of SR-BI possesses α-helical secondary structure and is stable in LPPG micelles at 40 °C at pH 6.8 for the multiple days required to collect the required NMR spectra for structure determination.
To facilitate solubility in aqueous solutions, detergent micelles have routinely been used to determine structures of transmembrane proteins [53,54]. When used above the critical micelle concentration, detergent micelles allow the protein to insert into a membrane-mimicking environment, which proves advantageous for NMR studies since the micelle is relatively small compared to other membrane mimetics and, in favorable cases, permits the protein to fold into its native conformation. We screened six different detergents (Figure 2) in order to determine the most stable micellar environment for our construct that resulted in NMR spectra with the most uniform peak intensity, shape, and dispersion. 1H-15N-HSQC spectra were poor when SR-BI(405–475) was reconstituted into micelles containing either OG or DDM. However, the lyso-phospholipid detergents yielded much better quality spectra. Indeed, lyso-phosphatidylglycerols have recently been shown as favorable detergents for structural studies as they discourage aggregation without denaturing, and therefore improve protein stability [52]. The detergents with carbon chains of 14:0 (LMPG) and 16:0 (LPPG) were optimal, closely followed by the lyso-phospholipid detergents 12:0 (DPC) and 18:1 (LOPG). Since the 12:0 and 18:1 spectra exhibited reduced chemical peak dispersion and peak intensity compared to the 14:0 and 16:0 spectra, we chose not to explore detergents with fatty acyl chain lengths shorter than 12 carbons or longer than 18 carbons. Recent evidence has shown that micelle geometry has a marked influence on protein conformation [55]. Columbus et al reported that detergent complexes yielded well-resolved NMR spectra when the protein hydrophobic dimensions matched the dimensions of the micelle. We therefore anticipate that the LMPG and LPPG, with 14- and 16- carbon lengths, respectively, most closely mimic the SR-BI(405–475) hydrophobic surfaces as compared to detergent micelles of smaller and larger sizes, thereby yielding higher quality spectra.
Although the 1H-13C-15N-HNCACB NMR spectra looked promising, unexpected peak intensity shifting occurred in the LMPG-containing sample after several days. Perhaps this variation occurred because of conformational heterogeneity or changes in oligomeric state [52]. This apparent change in the NMR spectrum lead us to pursue subsequent NMR analyses using LPPG, as SR-BI(405–475) was more stable in this detergent with only slight changes in peak intensity occurring.
Further solution conditions were screened, which included a solution containing only water, potassium phosphate buffer, or deuterated bis-Tris buffer at varying pH values. Although the spectra were very similar, the water solution was ideal as the peaks were more intense and uniform compared to the buffered solutions. Only pH values ranging from 6.0–6.8 were tested by NMR, since the protein/micelles precipitated after only a few days in more acidic conditions, and the spectra collected at pH 6.8 were optimal.
We [35,36] and others [37–39] have shown that SR-BI oligomerization is essential for protein function, as SR-BI must form an oligomer for HDL-CE uptake into the cell and this is most likely mediated, in part, by homodimerization between the C-terminal transmembrane domains of SR-BI monomers [35]. Along with a GXXXG motif commonly found in dimerizing membrane-bound proteins [41], the C-terminal transmembrane domain of SR-BI also possesses a putative leucine zipper motif [36]. Ongoing efforts to determine the high-resolution NMR structure of SR-BI(405–475) will unambiguously define the oligomeric status of this peptide in micelles. Based on preliminary analyses, the calculated molar ratio of peptide:micelle for LPPG micelles is approximately 1.6:1, and as such, we anticipate that 1 mM SR-BI(405–475) exists as a dimer or a mix of monomers and dimers in 5% LPPG detergent micelles. Indeed, our immunoblot analyses reveal that glutaraldehyde crosslinking of the peptide yielded monomeric and dimeric species. A close examination of spectra in Figure 4C revealed that, upon increasing micellar detergent concentration, we observe several peak shifts and a slight, overall decrease in spectral quality. We believe this is most likely explained by an increase in peptide shifting from dimers to monomers, and that these dynamics increase conformational heterogeneity and cause signal degradation. Based on these findings, the oligomeric status of SR-BI(405–475) deserves further investigation. We acknowledge that the decrease in spectral quality may also be explained by an increase in the viscosity of the solution.
With optimized conditions in hand, we are now well-poised to determine the high-resolution NMR structure of SR-BI(405–475), a peptide that contains a transmembrane domain that may be critical for mediating receptor oligomerization and, ultimately, the delivery of HDL-CE into cells.
CONCLUSION
Although the structure of SR-BI has yet to be determined, we have made great strides towards optimizing the conditions required to collect NMR spectra for determination of the high-resolution structure of the C-terminal transmembrane domain of this receptor that is critical for whole body cholesterol disposal.
HIGHLIGHTS.
SR-BI is an HDL receptor that regulates whole body cholesterol removal.
SR-BI’s C-terminal transmembrane domain was recombinantly expressed and purified.
Detergent micelles and solution conditions were optimized for NMR analyses.
The SR-BI peptide forms dimers upon chemical crosslinking.
We can now collect high-resolution structural information by NMR for this peptide.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health grants R01HL58012 (D.S.), R01A1058072 (B.F.V.), R01GM097381 (B.F.V.), and AHA14PRE185800221 (to A.C.C.).
ABBREVIATIONS
- ANS
1-anilinonaphthalene-8-sulfonic acid
- CD
circular dichroism
- CE
cholesteryl ester
- CMC
critical micelle concentration
- DDM
n-dodecyl β-D-maltopyranoside
- DPC
n-dodecylphophocholine
- HDL
high density lipoprotein
- HDL-C
HDL-cholesterol
- HSQC
heteronuclear single quantum coherence
- IPTG
isoropyl β-D-1-thiogalactopyranoside
- LMPG
1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol)
- LOPG
1-oleoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol)
- LPPG
1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1’-rac-glycerol)
- NMR
nuclear magnetic resonance
- OG
octyl β-D-glucopyranoside
- SR-BI
scavenger receptor class B type I
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Navab M, Reddy ST, Van Lenten BJ, Fogelman AM. HDL and cardiovascular disease: atherogenic and atheroprotective mechanisms. Nat Rev Cardiol. 2011;8:222–232. doi: 10.1038/nrcardio.2010.222. [DOI] [PubMed] [Google Scholar]
- 2.Glomset JA. The plasma lecithins:cholesterol acyltransferase reaction. J Lipid Res. 1968;9:155–167. [PubMed] [Google Scholar]
- 3.Brunham LR, Tietjen I, Bochem AE, Singaraja RR, Franchini PL, et al. Novel mutations in scavenger receptor BI associated with high HDL cholesterol in humans. Clin Genet. 2011;79:575–581. doi: 10.1111/j.1399-0004.2011.01682.x. [DOI] [PubMed] [Google Scholar]
- 4.Vergeer M, Korporaal SJ, Franssen R, Meurs I, Out R, et al. Genetic variant of the scavenger receptor BI in humans. N Engl J Med. 2011;364:136–145. doi: 10.1056/NEJMoa0907687. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick AC, Sahoo D. Functional characterization of newly-discovered mutations in human SR-BI. PLoS One. 2012;7:e45660. doi: 10.1371/journal.pone.0045660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji Y, Wang N, Ramakrishnan R, Sehayek E, Huszar D, et al. Hepatic scavenger receptor BI promotes rapid clearance of high density lipoprotein free cholesterol and its transport into bile. J Biol Chem. 1999;274:33398–33402. doi: 10.1074/jbc.274.47.33398. [DOI] [PubMed] [Google Scholar]
- 7.Ueda Y, Royer L, Gong E, Zhang J, Cooper PN, et al. Lower plasma levels and accelerated clearance of high density lipoprotein (HDL) and non-HDL cholesterol in scavenger receptor class B type I transgenic mice. J Biol Chem. 1999;274:7165–7171. doi: 10.1074/jbc.274.11.7165. [DOI] [PubMed] [Google Scholar]
- 8.Wang N, Arai T, Ji Y, Rinninger F, Tall AR. Liver-specific overexpression of scavenger receptor BI decreases levels of very low density lipoprotein apoB, low density lipoprotein apoB, and high density lipoprotein in transgenic mice. J Biol Chem. 1998;273:32920–32926. doi: 10.1074/jbc.273.49.32920. [DOI] [PubMed] [Google Scholar]
- 9.Arai T, Rinninger F, Varban L, Fairchild-Huntress V, Liang CP, et al. Decreased selective uptake of high density lipoprotein cholesteryl esters in apolipoprotein E knock-out mice. Proc Natl Acad Sci USA. 1999;96:12050–12055. doi: 10.1073/pnas.96.21.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozarsky KF, Donahee MH, Glick JM, Krieger M, Rader DJ. Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler Thromb Vasc Biol. 2000;20:721–727. doi: 10.1161/01.atv.20.3.721. [DOI] [PubMed] [Google Scholar]
- 11.Ueda Y, Gong E, Royer L, Cooper PN, Francone OL, et al. Relationship between expression levels and atherogenesis in scavenger receptor class B, type I transgenics. J Biol Chem. 2000;275:20368–20373. doi: 10.1074/jbc.M000730200. [DOI] [PubMed] [Google Scholar]
- 12.Varban ML, Rinninger F, Wang N, Fairchild-Huntress V, Dunmore JH, et al. Targeted mutation reveals a central role of SR-BI in hepatic selective uptake of high density lipoprotein cholesterol. Proc Natl Acad Sci USA. 1998;95:4619–4624. doi: 10.1073/pnas.95.8.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rigotti A, Trigatti BL, Penman M, Rayburn H, Herz J, et al. A targeted mutation in the murine gene encoding the high density lipoprotein (HDL) receptor scavenger receptor class B type I reveals its key role in HDL metabolism. Proc Natl Acad Sci USA. 1997;94:12610–12615. doi: 10.1073/pnas.94.23.12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trigatti B, Rayburn H, Vinals M, Braun A, Miettinen H. Influence of the high density lipoprotein receptor SR-BI on reproductive and cardiovascular pathophysiology. Proc Natl Acad Sci USA. 1999;96:9322–9327. doi: 10.1073/pnas.96.16.9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun A, Trigatti BL, Post MJ, Sato K, Simons M, et al. Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ Res. 2002;90:270–276. doi: 10.1161/hh0302.104462. [DOI] [PubMed] [Google Scholar]
- 16.Covey SD, Krieger M, Wang W, Penman M, Trigatti BL. Scavenger receptor class B type I-mediated protection against atherosclerosis in LDL receptor negative mice involves its expression in bone marrow-derived cells.[see comment] Arterioscler Thromb Vasc Biol. 2003;23:1589–1594. doi: 10.1161/01.ATV.0000083343.19940.A0. [DOI] [PubMed] [Google Scholar]
- 17.Calvo D, Gomez-Coronado D, Lasuncion MA, Vega MA. CLA-1 is an 85-kD plasma membrane glycoprotein that acts as a high-affinity receptor for both native (HDL-LDL, and VLDL) and modified (OxLDL and AcLDL) lipoproteins. Arterioscler Thromb Vasc Biol. 1997;17:2341–2349. doi: 10.1161/01.atv.17.11.2341. [DOI] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Krieger M. Charting the fate of the "good cholesterol": identification and characterization of the high-density lipoprotein receptor SR-BI. Annu Rev Biochem. 1999;68:523–558. doi: 10.1146/annurev.biochem.68.1.523. [DOI] [PubMed] [Google Scholar]
- 20.Temel RE, Trigatti B, DeMattos RB, Azhar S, Krieger M, et al. Scavenger receptor B, type I (SR-BI) is the major route for the delivery of high density lipoprotein cholesterol to the steroidogenic pathway in cultured mouse adrenocortical cells. Proc Natl Acad Sci USA. 1997;94:13600–13605. doi: 10.1073/pnas.94.25.13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Connelly MA, de la Llera-Moya M, Monzo P, Yancey P, Drazul D, et al. Analysis of chimeric receptors shows that multiple distinct functional activities of scavenger receptor, class B, type I (SR-BI) are localized to the extracellular receptor domain. Biochemistry. 2001;40:5249–5259. doi: 10.1021/bi002825r. [DOI] [PubMed] [Google Scholar]
- 22.Connelly MA, Kellner-Weiber G, Rothblat GH, Williams DL. Scavenger receptor, class B, type I (SR-BI)-directed HDL-cholesteryl ester hydrolysis. J Lipid Res. 2003;44:331–341. doi: 10.1194/jlr.M200186-JLR200. [DOI] [PubMed] [Google Scholar]
- 23.Connelly MA, Klein SM, Azhar S, Abumrad NA, Williams DL. Comparison of class B scavenger receptors, CD36 and SR-BI, shows that both receptors mediate HDL-cholesteryl ester selective uptake but SR-BI exhibits a unique enhancement of cholesteryl ester uptake. J Biol Chem. 1999;274:41–47. doi: 10.1074/jbc.274.1.41. [DOI] [PubMed] [Google Scholar]
- 24.Gu X, Trigatti B, Xu S, Acton S, Babitt J, et al. The efficient cellular uptake of high density lipoprotein lipids via scavenger receptor class B type I requires not only receptor-mediated surface binding but also receptor-specific lipid transfer mediated by its extracellular domain. J Biol Chem. 1998;273:26338–26348. doi: 10.1074/jbc.273.41.26338. [DOI] [PubMed] [Google Scholar]
- 25.Liu T, Krieger M, Kan HY, Zannis VI. The effects of mutations in helices 4 and 6 of apoA-I on SR-BI-mediated cholesterol efflux suggest that formation of a productive complex between reconstituted HDL and SR-BI is required for efficient lipid transport. J Biol Chem. 2002;277:21578–21584. doi: 10.1074/jbc.M112103200. [DOI] [PubMed] [Google Scholar]
- 26.Guo L, Chen M, Song Z, Daugherty A, Li XA. C323 of SR-BI is required for SR-BI-mediated HDL binding and cholesteryl ester uptake. J Lipid Res. 2011;52:2272–2278. doi: 10.1194/jlr.M019091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vinals M, Xu S, Vasile E, Krieger M. Identification of the N-Linked Glycosylation Sites on the High Density Lipoprotein (HDL) Receptor SR-BI and Assessment of Their Effects on HDL Binding and Selective Lipid Uptake. J Biol Chem. 2003;278:5325–5332. doi: 10.1074/jbc.M211073200. [DOI] [PubMed] [Google Scholar]
- 28.Yu M, Lau TY, Carr SA, Krieger M. Contributions of a disulfide bond and a reduced cysteine side chain to the intrinsic activity of the high-density lipoprotein receptor SR-BI. Biochemistry. 2012;51:10044–10055. doi: 10.1021/bi301203x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connelly MA, De La Llera-Moya M, Peng Y, Drazul-Schrader D, Rothblat GH, et al. Separation of lipid transport functions by mutations in the extracellular domain of scavenger receptor class B, type I. J Biol Chem. 2003;278:25773–25782. doi: 10.1074/jbc.M302820200. [DOI] [PubMed] [Google Scholar]
- 30.Parathath S, Sahoo D, Darlington YF, Peng Y, Collins HL, et al. Glycine 420 near the C-terminal transmembrane domain of SR-BI Is critical for proper delivery and metabolism of high density lipoprotein cholesteryl ester. J Biol Chem. 2004;279:24976–24985. doi: 10.1074/jbc.M402435200. [DOI] [PubMed] [Google Scholar]
- 31.Papale GA, Hanson PJ, Sahoo D. Extracellular disulfide bonds support scavenger receptor class B type I-mediated cholesterol transport. Biochemistry. 2011;50:6245–6254. doi: 10.1021/bi2005625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papale GA, Nicholson K, Hanson PJ, Pavlovic M, Drover VA, et al. Extracellular hydrophobic regions in scavenger receptor BI play a key role in mediating HDL-cholesterol transport. Arch Biochem Biophys. 2010;496:132–139. doi: 10.1016/j.abb.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neculai D, Schwake M, Ravichandran M, Zunke F, Collins RF, et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature. 2013;504:172–176. doi: 10.1038/nature12684. [DOI] [PubMed] [Google Scholar]
- 34.Reaven E, Cortez Y, Leers-Sucheta S, Nomoto A, Azhar S. Dimerization of the scavenger receptor class B type I: formation, function, and localization in diverse cells and tissues. J Lipid Res. 2004;45:513–528. doi: 10.1194/jlr.M300370-JLR200. [DOI] [PubMed] [Google Scholar]
- 35.Sahoo D, Peng Y, Smith JR, Darlington YF, Connelly MA. Scavenger receptor class B, type I (SR-BI) homo-dimerizes via its C-terminal region: fluorescence resonance energy transfer analysis. Biochim Biophys Acta. 2007;1771:818–829. doi: 10.1016/j.bbalip.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sahoo D, Darlington YF, Pop D, Williams DL, Connelly MA. Scavenger receptor class B Type I (SR-BI) assembles into detergent-sensitive dimers and tetramers. Biochim Biophys Acta. 2007;1771:807–817. doi: 10.1016/j.bbalip.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Azhar S, Nomoto A, Reaven E. Hormonal regulation of adrenal microvillar channel formation. J Lipid Res. 2002;43:861–871. [PubMed] [Google Scholar]
- 38.Landschulz KT, Pathak RK, Rigotti A, Krieger M, Hobbs HH. Regulation of scavenger receptor, class B, type I, a high density lipoprotein receptor, in liver and steroidogenic tissues of the rat. J Clin Invest. 1996;98:984–995. doi: 10.1172/JCI118883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reaven E, Cortez Y, Leers-Sucheta S, Nomoto A, Azhar S. Dimerization of the scavenger receptor class B type I (SR-BI): formation, function and localization in diverse cells and tissues. J Lipid Res. 2004;45:513–528. doi: 10.1194/jlr.M300370-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Rodrigueza WV, Thuahnai ST, Temel RE, Lund-Katz S, Phillips MC, et al. Mechanism of scavenger receptor class B type I-mediated selective uptake of cholesteryl esters from high density lipoprotein to adrenal cells. J Biol Chem. 1999;274:20344–20350. doi: 10.1074/jbc.274.29.20344. [DOI] [PubMed] [Google Scholar]
- 41.Gaidukov L, Nager AR, Xu S, Penman M, Krieger M. Glycine dimerization motif in the N-terminal transmembrane domain of the high density lipoprotein receptor SR-BI required for normal receptor oligomerization and lipid transport. J Biol Chem. 2011;286:18452–18464. doi: 10.1074/jbc.M111.229872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian C, Karra MD, Ellis CD, Jacob J, Oxenoid K, et al. Membrane protein preparation for TROSY NMR screening. Methods Enzymol. 2005;394:321–334. doi: 10.1016/S0076-6879(05)94012-3. [DOI] [PubMed] [Google Scholar]
- 43.De Vendittis E, Palumbo G, Parlato G, Bocchini V. A fluorimetric method for the estimation of the critical micelle concentration of surfactants. Anal Biochem. 1981;115:278–286. doi: 10.1016/0003-2697(81)90006-3. [DOI] [PubMed] [Google Scholar]
- 44.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44(Spec No):S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]
- 45.Muhandiram DR, Kay LE. Gradient-Enhanced Triple-Resonance Three-Dimensional NMR Experiments with Improved Sensitivity. Journal of Magnetic Resonance, Series B. 1994;103:203–216. [Google Scholar]
- 46.Mori S, Abeygunawardana C, Johnson MO, van Zijl PC. Improved sensitivity of HSQC spectra of exchanging protons at short interscan delays using a new fast HSQC (FHSQC) detection scheme that avoids water saturation. J Magn Reson B. 1995;108:94–98. doi: 10.1006/jmrb.1995.1109. [DOI] [PubMed] [Google Scholar]
- 47.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 48.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 49.Arora A, Tamm LK. Biophysical approaches to membrane protein structure determination. Curr Opin Struct Biol. 2001;11:540–547. doi: 10.1016/s0959-440x(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 50.Etzkorn M, Raschle T, Hagn F, Gelev V, Rice AJ, et al. Cell-free expressed bacteriorhodopsin in different soluble membrane mimetics: biophysical properties and NMR accessibility. Structure. 2013;21:394–401. doi: 10.1016/j.str.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donnelly D, Overington JP, Ruffule SV, Nugent JHA, Blundell TL. Modeling a-helical transmembrane domains: The calculation and use of substitution tables for lipid-facing residues. Prot Sci. 1993;2:55–70. doi: 10.1002/pro.5560020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, et al. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;28:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 53.Gayen S, Li Q, Kang C. Solution NMR study of the transmembrane domain of single-span membrane proteins: opportunities and strategies. Curr Protein Pept Sci. 2012;13:585–600. doi: 10.2174/138920312803582979. [DOI] [PubMed] [Google Scholar]
- 54.Kang C, Li Q. Solution NMR study of integral membrane proteins. Curr Opin Chem Biol. 2011;15:560–569. doi: 10.1016/j.cbpa.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 55.Columbus L, Lipfert J, Jambunathan K, Fox DA, Sim AY, et al. Mixing and matching detergents for membrane protein NMR structure determination. J Am Chem Soc. 2009;131:7320–7326. doi: 10.1021/ja808776j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders CR, Sonnichsen F. Solution NMR of membrane proteins: practice and challenges. Magn Reson Chem. 2006;44(Spec No):S24–S40. doi: 10.1002/mrc.1816. [DOI] [PubMed] [Google Scholar]