Abstract
Microbial communities that underpin global biogeochemical cycles will likely be influenced by elevated temperature associated with environmental change. Here, we test an approach to measure how elevated temperature impacts the physiology of individual microbial groups in a community context, using a model microbial-based ecosystem. The study is the first application of tandem mass tag (TMT)-based proteomics to a microbial community. We accurately, precisely and reproducibly quantified thousands of proteins in biofilms growing at 40, 43 and 46 °C. Elevated temperature led to upregulation of proteins involved in amino-acid metabolism at the level of individual organisms and the entire community. Proteins from related organisms differed in their relative abundance and functional responses to temperature. Elevated temperature repressed carbon fixation proteins from two Leptospirillum genotypes, whereas carbon fixation proteins were significantly upregulated at higher temperature by a third member of this genus. Leptospirillum group III bacteria may have been subject to viral stress at elevated temperature, which could lead to greater carbon turnover in the microbial food web through the release of viral lysate. Overall, these findings highlight the utility of proteomics-enabled community-based physiology studies, and provide a methodological framework for possible extension to additional mixed culture and environmental sample analyses.
Introduction
The impacts of elevated temperature on microbial communities will have direct implications for ecosystem and global scale processes. Many microbial community studies have evaluated the effect of warming on overall community structure and on specific metabolic processes such as respiration (for example, Zogg et al., 1997; Finke and Jørgensen, 2008; Rose et al., 2009; Yergeau et al., 2012; Lindh et al., 2013; Wu et al., 2013; von Scheibner et al., 2014). Far fewer studies have comprehensively assessed functional responses across the entire community (for example, using ‘omic' approaches (Luo et al., 2013; Toseland et al., 2013) or functional gene arrays (Yergeau et al., 2012; Tu et al., 2014)).
Individual microbial groups (for example, genotypes, species or functional groups) will likely have different functional responses to elevated temperature, and yet an organism's response and adaptation to changing conditions in part relates to its behavior within a community. Thus, understanding the physiology and activity of individual microbial groups within a community context is essential for predicting the impact, resilience and response of ecological systems to changing conditions. This topic is relatively little studied, in part because it can be challenging to tease apart contributions of individual organisms from overall metabolic processes. Further, such investigations require a high level of taxonomic and functional resolution because closely related strains and species may respond very differently to temperature regime.
Quantitative proteomics can elucidate function of individual microbial groups within a community context by measuring protein abundance in a high-throughput manner. Both taxonomic and functional annotations are simultaneously assigned to unique proteins in the community proteome. Protein abundance can more accurately represent cellular activities than messenger RNA quantification, because messenger RNA abundance changes do not necessarily correlate with protein abundance change (for example, Pan et al., 2008). For example, some proteins may have long lifetimes, so that new production from messenger RNA is required infrequently. Conversely, cells may also have a low level of protein with abundant corresponding messenger RNA expression because of protein degradation and post-transcription regulation.
Recent advances in protein quantification using tandem mass tags (TMTs) and isobaric tags for relative and absolute quantification (iTRAQ) have improved measurement precision, accuracy and reproducibility (Thompson et al., 2003; Ross et al., 2004), surpassing label-free quantification methods such as spectral counting (Li et al., 2012). TMT/iTRAQ-based quantitative proteomics can be used with complex samples, including biological systems that are not amenable to efficient metabolic labeling with stable isotopes. In isobaric chemical labeling, peptides from different samples are labeled separately with different isotopic variants of the labeling reagent and then combined for analysis using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Each isotopic variant has the same overall mass but contains a reporter ion with a unique molecular mass, thus enabling accurate overall quantification alongside precise measurement of the relative protein abundance between samples. Currently, TMT/iTRAQ-based quantitative proteomics enables multiplexing of up to 8–10 samples with deep proteome coverage.
The objective of this study was to determine the impact of elevated temperature on the physiology of individual microbial groups in a community. The experiments were conducted at temperatures between the average in situ temperature and the maximum growth temperature, which was established in this study. We compared the protein expression levels using a new approach that combined shotgun community proteomics analysis with TMT quantification. The analyses targeted laboratory-grown acid mine drainage (AMD) biofilms that represent natural AMD populations (Belnap et al., 2010) and have served as a model microbial community system in many prior studies (Denef et al., 2010). The current research shows the utility of quantitative proteomics for understanding ecological processes by highlighting differential expression of closely related organisms.
Materials and methods
Sample collection and bioreactor growth
AMD biofilms were collected from the AB-muck site at the Richmond Mine on 7/15/11 (Iron Mountain near Redding, California), where pH is typically 0.85. For cultivation, biofilms were stored on ice for return to the laboratory. For fluorescence in situ hybridization (FISH) abundance analyses of the inoculum biofilm, biofilms were flash frozen on site in a dry ice/ethanol bath and then transferred to −80 °C upon return to the laboratory.
Biofilms were cultured in bioreactors using 9K-BR growth media as previously described (Belnap et al., 2010). The flow rate of the bioreactors was ∼200 μl per minute. Incubator temperature was monitored using HOBO Pendant Temperature Data Loggers (Onset Computer Corporation, Bourne, MA, USA). After 4 weeks of biofilm development at 40 °C, biofilms were regrown at 40, 43, 46 and 49 °C in separate reactors. Biofilms were harvested after 3 weeks and then reestablished (from residual planktonic cells) before a second harvest 5 weeks later. The two periods of biomass accumulation were treated as response replicates and their values analyzed together. This strategy was chosen to highlight proteins that responded similarly to temperature, regardless of growth period.
FISH
FISH was carried out on fixed (4% paraformaldehyde) AMD biofilm samples as described previously (Amann et al., 1995; Bond and Banfield, 2001). Oligonucleotide probes used in this study for identification of the dominant individual species and groups were as follows: EUBMIX (all bacteria); ARC915 (all archaea); EUKMIX (all eukaryotes); LF655 (all Leptospirillum bacteria); LF1252 (Leptospirillum group III bacteria); L2UBA353 (Leptospirillum group II UBA genotype); L2CG353 (Leptospirillum group II 5-way genotype); and SUL230 (Sulfobacillus spp.). For estimation of abundance, cell counts from both periods of biomass accumulation were averaged. For each temperature, a total of 2651–2828 cells were counted from 6–12 fields of view per probe (with an average of 468 cells counted per probe per sample). Counts were converted to a percentage of the total cell count found using the general nucleic acid stain 4',6-diamidino-2-phenylindole (DAPI).
Protein extraction
Proteins were extracted from the biofilms using an SDS protein extraction protocol based on previously reported methodology (Chourey et al., 2010). Biofilm from each sample was split into two, representing extraction replicates. Frozen biomass (between 500 and 750 mg) was resuspended in 1 ml SDS cell lysis buffer (5% SDS; 50 mM Tris–HCl, pH 8; 150 mM NaCl; 0.1 mM EDTA; 1 mM MgCl2) and 10 μl of 5 M dithiothreitol. The biofilm was dispersed in the buffer by vigorous vortexing for 2–3 min. Samples were heated at 99 °C for 15 min, followed by vigorous vortexing for 3 minutes. Cellular debris was pelleted by centrifugation at 10 000 r.p.m. for 10 min at 4 °C. The supernatant was transferred to a fresh tube, 300 μl cold 100% trichloroacetic acid was added and the proteins precipitated overnight at 4 °C. Precipitated proteins were centrifuged at 14 000 r.p.m. for 20 min at 4 °C and the concentrated protein pellet washed three times with cold acetone. The pellet was resuspended in a guanidinium chloride buffer (6 M guanidinium chloride, 10 mM CaCl2, 50 mM Tris pH 7.6) and reduced with 10 mM DTT.
Total protein concentrations were estimated with the bicinchoninic acid assay (Pierce BCA Protein Assay Kit; Thermo Fisher Scientific Inc. #23227, Waltham, MA, USA). A quantity of 50 μg of protein from each sample was further processed with the filter-aided sample preparation method (Wiśniewski et al., 2009) following the manufacturer's protocol (Expedeon, San Diego, CA, USA) with a minor modification by substituting urea with triethylammonium bicarbonate (TEAB) buffer for sample washes to avoid the primary amine group containing chemical that would interfere with TMT labeling. Each sample was digested with sequencing-grade trypsin (Promega, Fitchburg, WI, USA) in 500 mM TEAB buffer overnight in an enzyme:substrate ratio of 1:100 (wt:wt) at room temperature with gentle shaking, followed by a second digestion for 4 h with the same amount of trypsin (that is, 0.5 μg). The digested peptide samples were then eluted from the filter by centrifugation for TMT labeling.
Labeling of peptides with TMTs for quantification
For both sets of extraction replicates, a total of six samples (two response replicates for each of the three temperatures) were labeled with TMT 6-plex reagents (Thermo Fisher Scientific Inc.). For each extraction replicate set, each sample (50 μg) was individually labeled with one of the six TMT variants numbered by the distinct masses of their reporter ions: TMT126, TMT127, TMT128, TMT129, TMT130 and TMT131. After the labeling was finished, the six samples in the same replicate were combined into one aliquot for the two-dimensional liquid chromatography-tandem mass spectrometry (2D-LC-MS/MS) analysis in technical duplicates.
2D-LC-MS/MS proteomic measurements
The multidimensional protein identification technology (MudPIT) (Washburn et al., 2001) was used in our analytical workflow. In each MudPIT run, 50 μg of peptides were loaded offline into a 150 μm-I.D. Two-dimensional back column (Polymicro Technologies, Phoenix, AZ, USA) packed with 3 cm of C18 reverse phase (RP) resin (Luna, Phenomenex, Torrance, CA, USA) and 3 cm of strong cation exchange (SCX) resin (Luna, Phenomenex). The back column was connected to a 100-μm-I.D. front column (New Objective, Woburn, MA, USA) packed in-house with 15 cm of C18 RP resin. The back column and front column were placed in-line with a U3000 quaternary HPLC pump (Dionex, Sunnyvale, CA, USA). Prior to the measurement, the back column loaded with peptides was de-salted offline with 100% Solvent A (95% H2O, 5% CH3CN and 0.1% formic acid), and washed with a 1-h gradient from 100% Solvent A to 100% Solvent B (30% H2O, 70% CH3CN and 0.1% formic acid) to move peptides from RP resin to SCX resin. Each MudPIT run was configured with the 11 SCX-RP separations in 22 h. A quantity of 5%, 7%, 10%, 12%, 15%, 17%, 20%, 25%, 35%, 50% and 100% of Solvent D (500 mM ammonium acetate dissolved in Solvent A) was used in the 11 SCX fractionations. Each SCX fraction was separated by a 110-min RP gradient from 100% Solvent A to 60% Solvent B. The MS/MS measurements were performed on an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific Inc.) using the dual MS/MS scan method (Köcher et al., 2009): each selected precursor peptide ion was first fragmented by collision-induced dissociation (CID) for identification and then by higher CID (HCD) for quantification. The data were acquired with the following parameters: four CID–HCD dual MS/MS scans per full scan; CID MS/MS scans were acquired in LTQ; MS scans and HCD MS/MS scans were acquired in Orbitrap with the resolution 30 000 and 15 000, respectively; two-microscan averaging for all scan types; 35% normalized collision energy in CID and 55% normalized collision energy in HCD; dynamic exclusion enabled with ±1.5 m/z exclusion window.
Protein identification and quantification
All MS/MS spectra were searched with Sequest (Eng et al., 1994) against a database containing 79 633 proteins derived from ∼80 Gb of genomic information obtained from previous genomic characterizations of biofilms sampled from the Richmond Mine AMD system. Static modification of cysteine by iodoacetamide and static modification of N terminus, and dynamic modification of lysine by the TMT labeling reagent were specified for peptide identification. The output data files were then filtered using the DTASelect v1.9 algorithm (Tabb et al., 2002) with the following parameters: minimum XCorr score of 1.8, 2.5 and 3.5 for charge states (z)=+1, z=+2 and z=+3 precursor peptide ions, respectively; a minimum DeltCN value of 0.08; a minimum requirement of two peptides for each identified protein. These filtering criteria resulted in protein false discovery rate of less than 3% in each run estimated by the target decoy approach. Relative protein abundance changes were quantified using custom scripts, as described previously (Wang et al., 2013). Briefly, raw reporter ion intensities were extracted from HCD spectra and appended to the peptides. Protein intensities were summed from the intensities of their identified peptides. Only unique peptides were considered for protein quantification. Protein intensities from extraction replicates and technical runs were summed for each sample (for a total of four runs per sample). Raw protein intensities were normalized by making the total intensity of each sample (community-level analysis) or each organism (organism-level analysis) identical.
Proteome-based community structure and clustering analyses
For proteome-based community structure and clustering analyses, protein abundance values were normalized at the community level and then summed from both response replicates. Abundance estimates for each organismal group (for example, Leptospirillum group III) were calculated by summing the total intensities of the proteins for the organismal group and then dividing by the total sum of all proteins in a sample.
Hierarchical clustering was performed on protein abundance values with absolute intensities converted to percentages for each protein (the sum of the percentages for each protein is equal to 100%). The clustering method used an uncentered Pearson correlation distance matrix and average linkage clustering (using Multi-experiment Viewer; MeV_4_8; http://www.tm4.org/mev/) (Saeed et al., 2003). For clustering analyses only, 0.000001 was added to each number to avoid software adjustments of zero values.
Proteome-based functional analyses
Differentially expressed proteins were identified by statistically comparing the response replicates between two temperatures (for example, protein abundance of the two 40 °C response replicates versus protein abundance of the two 46 °C response replicates). Differentially expressed proteins were identified as those with normalized total intensity ratios (40 °C:46 °C) >1.2 or <0.8 combined with a Rank Product P-value ⩽0.05 (except where noted), similar to methods used in other studies (Williamson et al., 2008; Dobbin et al., 2010; Soares et al., 2010; Zhao et al., 2010; Han et al., 2011; Jain et al., 2011; Muthukrishnan et al., 2011). Rank Product, commonly used in microarray experiments, is a non-parametric statistical method based on ranks of fold changes (Breitling et al., 2004). Gene set enrichment analysis (Subramanian et al., 2005) was used to evaluate enrichment of proteins in the reductive tricarboxylic acid (rTCA) pathway with the following settings: gene set permutation; classic enrichment analysis; and log2 ratio of classes metric for ranking genes.
Functional categories of significant proteins were assigned upon manual review of annotations in ggKbase (http://ggkbase.berkeley.edu/) (including Clusters of Orthologous Groups (Tatusov et al., 1997) assignment), as well as reciprocal blast searches against the KEGG database (conducted using the KAAS server (Moriya et al., 2007)). Carbohydrate active enzymes (CAZymes) were predicted with the CAZymes Analysis Toolkit (http://mothra.ornl.gov/cgi-bin/cat.cgi) (Park et al., 2010) with an e-value threshold of 0.0001 for Pfam searches and 0.000001 for orthology searches with ‘domain consistent' and ‘length consistent' rules.
Validation of the TMT-based quantitative community proteomics method
The TMT-based quantitative community proteomics method was validated using standard mixtures of peptides extracted from an AMD biofilm sample (from the AB-muck site). Briefly, a large peptide sample was prepared from the AMD test sample with an established protocol using in-solution trypsin digestion and C18 solid-phase extraction clean-up as described (Ram et al., 2005). Two aliquots of the peptide sample (50 μg of each) were separately labeled with TMT126 and TMT127 and then mixed in three different ratios (∼2:1, 3:1 and 8:1 by pipetting volume). Each standard mixture was measured with two-dimensional liquid chromatography-tandem mass spectrometry and proteins were identified and quantified as described above.
Results
Validation of the TMT-based quantitative community proteomics method
To validate the application of the previously established (Li et al., 2012) TMT proteomics approach to microbial communities, we used peptides extracted from an AMD biofilm sample as a standard. The standard was labeled with two different TMTs in three mixing ratios (1.6:1, 2.8:1 and 8.3:1, based on the total intensities of the reporter ions). Histograms of the log2 scale abundance ratios of quantified proteins showed tight distribution of the measured protein abundance ratios in each standard mixture, indicating precise quantification across a large range of fold changes (Supplementary Figure S1). The 1.6:1 distribution (red) was clearly separated from the 1:1 ratio (log2 ratio=0), which validated the method's ability to distinguish small fold changes from no change. The 2.8:1 distribution (blue) had no overlap with the 1.6:1 distribution, which indicated that the method can resolve a 1.2-fold difference in protein abundance change. The 8.3:1 distribution (green) shows good quantification performance of this method for proteins with large fold changes.
Growth of AMD biofilms at different temperatures
In order to evaluate the effect of elevated temperature on community composition and function, AMD biofilms were grown in laboratory bioreactors from 40–49 °C. Mature biofilms were developed at 40, 43 and 46 °C. This temperature range corresponds with the normal range of temperatures associated with biofilm growth in the field (Supplementary Figure S2). There was no visible biofilm growth at 49 °C after 4 weeks. Biofilm growth rates may have differed between the temperature treatments; however, we expect that any differences that might have occurred were likely a result of temperature since all other growth conditions were identical.
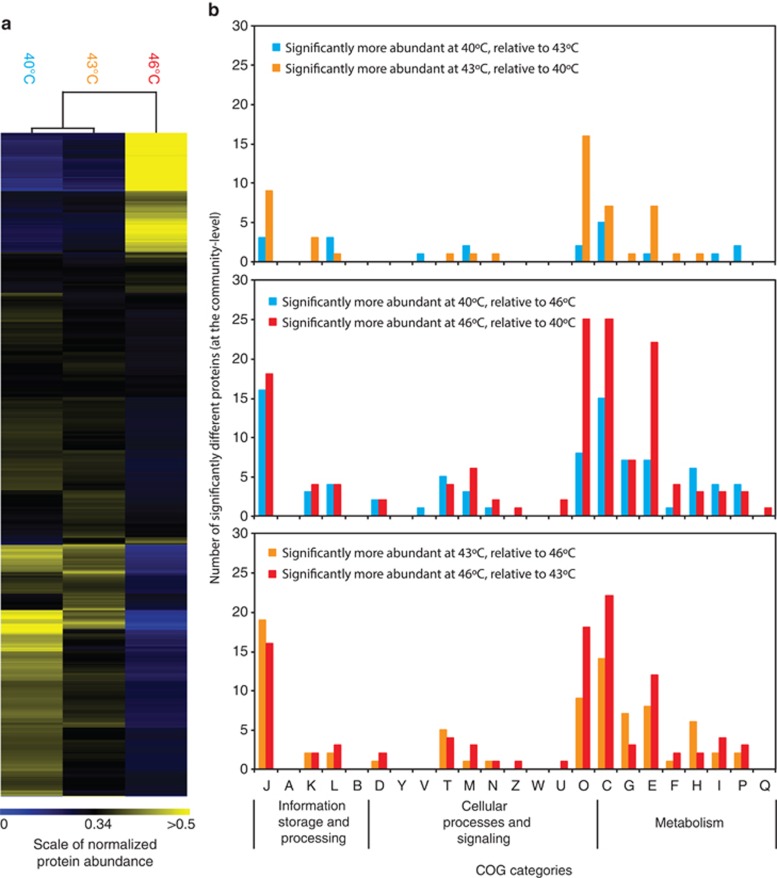
TMT proteomics was used to determine protein abundance and inferred function in bioreactor-grown biofilms grown at 40, 43, 46 °C. TMT proteomics identified 1724–1916 proteins from the biofilm communities (across all samples, extraction replicates and technical runs), 1596 of which could be uniquely assigned to one organism (Supplementary Table S1). Hierarchical clustering showed that the samples clustered into two groups based on their protein abundance levels (based on both community-level and organism-level normalizations): biofilms grown at 40 and 43 °C clustered together, whereas those grown at 46 °C clustered independently (Figure 1).
Figure 1.
Community-level changes in protein abundance. (a) Hierarchical clustering of protein abundance values normalized at the community level. (b) Number of proteins assigned to Clusters of Orthologous Groups that are significantly different between temperatures at the community level. COG categories J: Translation, ribosomal structure and biogenesis; A: RNA processing and modification; K: Transcription; L: Replication, recombination and repair; B: Chromatin structure and dynamics; D: Cell cycle control, cell division, chromosome partitioning; Y: Nuclear structure; V: Defense mechanisms; T: Signal transduction mechanisms; M: Cell wall/membrane/envelope biogenesis; N: Cell motility; Z: Cytoskeleton; W: Extracellular structures; U: Intracellular trafficking, secretion and vesicular transport; O: Posttranslational modification, protein turnover, chaperones; C: Energy production and conversion; G: Carbohydrate transport and metabolism; E: Amino-acid transport and metabolism; F: Nucleotide transport and metabolism; H: Coenzyme transport and metabolism; I: Lipid transport and metabolism; P: Inorganic ion transport and metabolism; and Q: Secondary metabolites biosynthesis, transport and catabolism.
Community composition of AMD biofilms grown at different temperatures based on FISH and protein abundance measurements
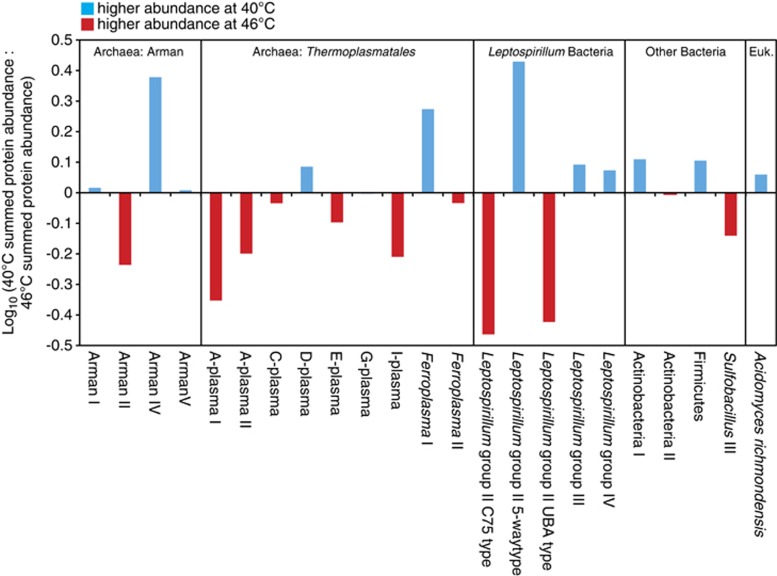
Proteins were quantified from 23 different bacterial, archaeal and eukaryal organisms (Supplementary Table S2). We evaluated community composition based on FISH and TMT proteomics measurements (Table 1, Figure 2, Supplementary Figure S3). FISH estimates indicated that archaea were very abundant in the bioreactor biofilms, making up 37% to 51% of the communities. Proteins were identified from many different archaea: ARMAN I, ARMAN II, ARMAN IV, ARMAN V, Ferroplasma I, Ferroplasma II, A-plasma I, A-plasma II, C-plasma, D-plasma, E-plasma, G-plasma and I-plasma. Although the overall abundance of archaea did not change significantly with increasing temperature (based on FISH and protein abundance estimates; Supplementary Figure S3), protein abundance of closely related organisms responded differently to temperature (Figure 2, Table 1). For instance, ARMAN II abundance increased with temperature, but ARMAN IV decreased with temperature. Two other ARMAN types had similar abundance levels at 40 and 46 °C, but lower abundance at 43 °C.
Table 1. Relative abundance of each organism at 40, 43 and 46 °C (based on summing the total intensities of the proteins for each organism and then dividing by the total sum of all proteins in a sample).
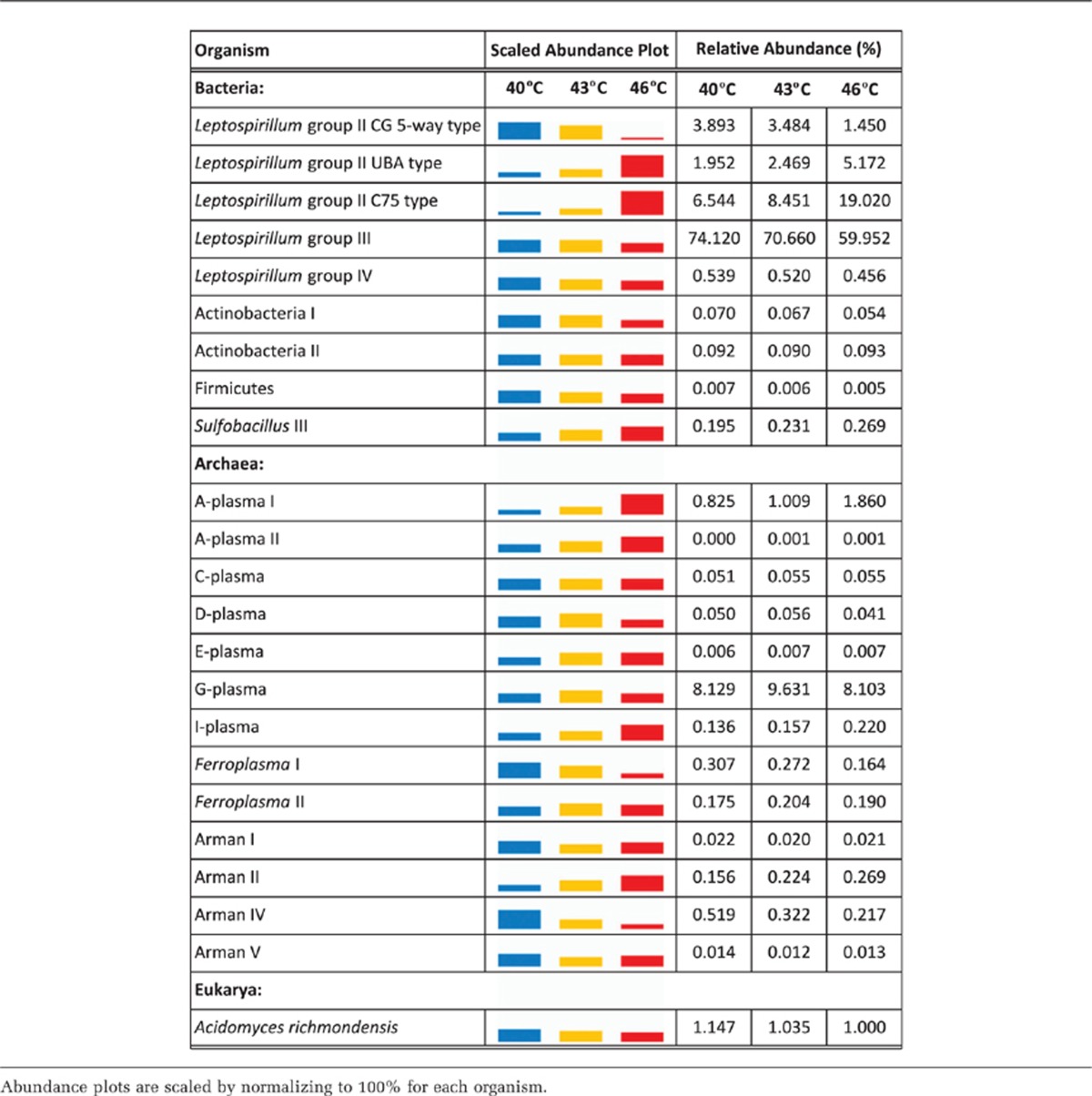
Figure 2.
Relative abundance of archaea, bacteria and eukarya in AMD biofilms grown at 40 and 46 °C (based on protein abundance as measured by TMT proteomics).
The chemolithoautotrophic, iron-oxidizing Leptospirillum group II bacteria were present in all of the biofilms. Elevated temperature differentially impacted the abundance of three distinct Leptospirillum group II organisms referred to as the Type I (5-way), Type III (C75) and Type VI (UBA) genotypic groups (Lo et al., 2007; Simmons et al., 2008; Denef et al., 2009; Denef and Banfield, 2012) (based on FISH and protein abundance estimates; Table 1, Figure 2, Supplementary Figure S3). The UBA and C75 genotypes increased in abundance from 40 to 46 °C, whereas the 5-way genotype abundance decreased. Leptospirillum group III bacteria were very abundant at all temperatures in the bioreactors.
The bioreactor-grown biofilms contained the same dominant organisms found in the in situ mine biofilm, including the Leptospirillum group II 5-way genotype, the Leptospirillum group II UBA genotype, Leptospirillum group III, Sulfobacillus and archaea (based on FISH estimates; Supplementary Figure S3). These organisms have been consistently documented in in situ biofilms from this mine ((Denef et al., 2010) and references therein]. The abundance estimates of archaea, the Leptospirillum group II 5-way genotype, Sulfobacillus and other bacteria (including Actinobacteria and Firmicutes, based on proteomic measurements) were similar between the mine biofilm and the 40 °C bioreactor biofilm. As seen previously (Mosier et al., 2013), the bioreactor biofilms had a much higher proportion of Leptospirillum group III bacteria than that seen in the in situ mine biofilm (31 to 41% compared with only 2% in the mine; Supplementary Figure S3). Leptospirillum Group III has been shown to dominate AMD biofilms in solutions with low Fe(II)/Fe(III) ratios (Spaulding et al., unpublished data).
Community function at low and high temperatures
Protein expression was further evaluated to determine if elevated temperature impacted metabolic function. Protein abundance was first normalized at the community level to determine each protein's abundance compared with all proteins in the sample (normalizing to account for biomass differences between samples, but not accounting for differences in each organism's abundance).
In a COG-based functional analysis (Figure 1), the greatest number of proteins with significantly different abundances occurred when comparing biofilms grown at 40 and 46 °C. There were fewer proteins with significantly different abundances in the 40:43 and 43:46 °C comparisons (78 significantly different protein abundances between 40 and 43 °C; 191 significantly different protein abundances between 43 and 46 °C; and 239 significantly different protein abundances between 40 and 46 °C).
Overall, increasing temperature led to an increasing number of significantly different proteins in the COG functional categories (E) amino-acid transport and metabolism, (C) energy production and conversion and (O) posttranslational modification, protein turnover, chaperones. In particular, more than three times as many proteins involved in the metabolism and transport of amino acids (COG E) were significantly more abundant at 46 °C than at 40 °C. Nearly twice as many proteins involved in energy production and conversion (COG C) were significantly more abundant at 46 °C compared with 40 °C. In addition, there were 3.1 times as many proteins in the functional category of posttranslational modification, protein turnover and chaperones (COG O) that were significantly more abundant at 46 °C than at 40 °C.
Function of individual organisms in biofilms growing at 40 and 46 °C
Protein abundance was evaluated at the organism level by normalizing individual proteins to the total protein abundance from each specific organism, allowing for evaluation of protein abundance for individual organisms. Organisms representing ⩾10% of the total proteins were analyzed, including three closely related Leptospirillum bacteria, as well as G-plasma archaea. In the significance analysis (based on fold change and Rank Product P-value), proteins that are considered as upregulated in one condition are concomitantly considered as downregulated under the other condition. Protein expression at the organism level was only analyzed between the 40 and 46 °C conditions because very few proteins were significantly different between 40 and 43 °C (ranging from 2 to 18 proteins per organism level comparison). In addition, the 40 and 43 °C community-level proteomes were similar (based on hierarchical clustering and community COG analysis).
Function of Leptospirillum bacteria in biofilms growing at 40 and 46 °C
Protein abundance was evaluated at the organism level for three closely related Leptospirillum bacteria: Leptospirillum group II UBA genotype, Leptospirillum group II 5-way genotype and Leptospirillum group III. Overall, 144 proteins were significantly different between 40 and 46 °C for the three organisms, spanning a broad range of functions.
Protein folding, sortingand degradation
Several proteins involved in protein degradation were significantly upregulated at 46 °C for Leptospirillum group III and the group II UBA genotype. Of the proteins with the highest total intensities in the entire data set (top 5% for each of the three organisms at 40 and 46 °C), 13% were chaperones. DnaK and ClpB chaperones were significantly upregulated at 40 °C for the group II 5-way genotype and at 46 °C for the UBA genotype. Leptospirillum group III bacteria had GroEL and HscA (P=0.06) chaperones and a chaperonin that were significantly upregulated at 46 °C. Two trigger factors (ribosome-associated chaperones) were upregulated at 40 °C (P=0.01, P=0.09).
Carbon transformation
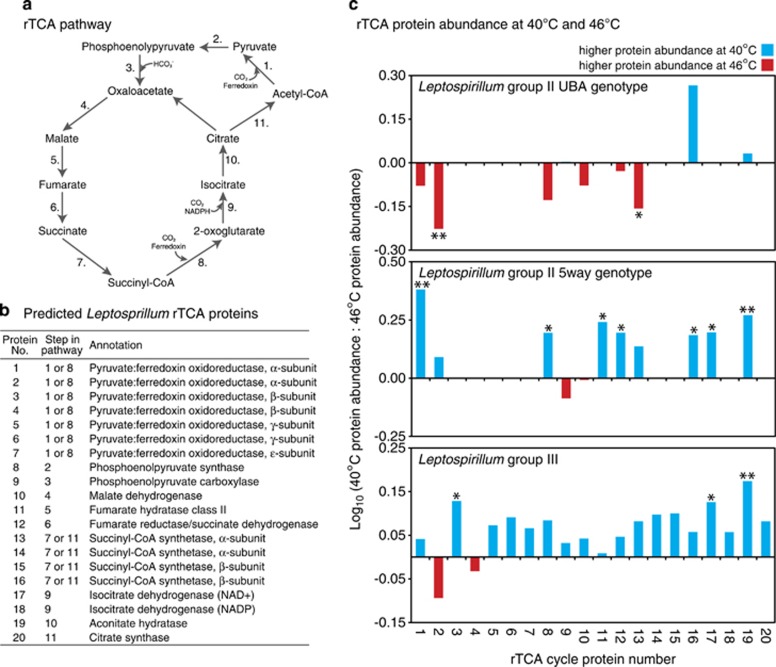
Proteins in the carbon fixation pathway of the Leptospirillum group II (UBA and 5-way genotypes) and III bacteria responded strongly to temperature (Figure 3). These organisms are believed to fix carbon via the rTCA cycle (Aliaga Goltsman et al., 2009). Of the 60 different Leptospirillum proteins predicted to be involved in rTCA, 41 were detected and quantified. Many of these proteins had very high total intensities: 11 were ranked in the top 5% highest total intensities. The majority of rTCA proteins from Leptospirillum group III and the group II 5-way genotype were more abundant at 40 °C than at 46 °C. Many of these proteins were significantly upregulated at 40 °C relative to 46 °C (seven proteins for 5-way and three for group III). Conversely, rTCA proteins for the Leptospirillum group II UBA genotype had the opposite abundance pattern. The majority of the rTCA proteins had higher intensities at 46 °C, two of which were significantly upregulated relative to 40 °C. Gene set enrichment analysis (a statistical method that evaluates enrichment of complete pathways, as opposed to individual genes) confirmed that the rTCA pathway was enriched at 40 °C for Leptospirillum group III and the group II 5-way genotype and at 46 °C for the Leptospirillum group II UBA genotype (P-value <0.005).
Figure 3.
Abundance of proteins involved in CO2 fixation for Leptospirillum group II (UBA and 5-way genotypes) and group III. (a) rTCA pathway, as proposed by Aliaga Goltsman et al., 2009. (b) Leptospirillum proteins predicted to be involved in rTCA (Aliaga Goltsman et al., 2009). (c) Abundance of rTCA proteins at 40 and 46 °C. Stars indicate proteins that are significantly different at a given temperature (fold change >1.2 or <0.8; one star P⩽0.1; two stars P⩽0.05).
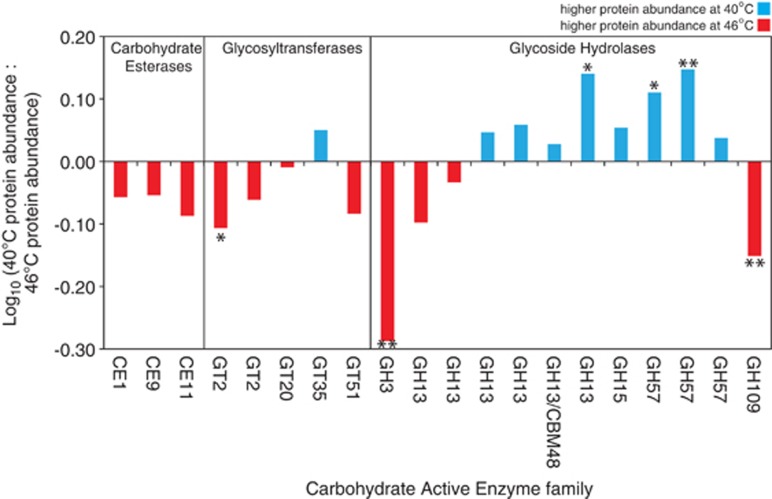
Twenty-five CAZymes were predicted among the quantified Leptospirillum proteins (additional CAZymes are found within the complete Leptospirillum genomes but were not measured here). CAZymes are classified as families of structurally related enzymes that degrade, modify or create glycosidic bonds. Only one CAZyme was predicted for the Leptospirillum group II 5-way genotype (GH57) and four for the Leptospirillum group II UBA genotype (CBM13, GH109 and two GH57s). Leptospirillum group III had 20 predicted CAZymes (Figure 4). Most (7 out of 8) of the carbohydrate esterases (CEs; hydrolysis of carbohydrate esters) and glycosyltransferases (GTs; biosynthesis of saccharides) had higher total intensities at 46 °C and one was significantly upregulated (GT2). Two glycoside hydrolases (GH3 with ß-N-acetylhexosaminidase activity and GH109 with an oxidoreductase domain) were also significantly upregulated at 46 °C. Half of the Leptospirillum group III CAZymes were related to GH families (GH13 and GH57) acting on substrates containing α-glucoside linkages including starch, glycogen and α-maltose: 8/10 of these proteins had higher total intensities at 40 °C, three of which were significantly upregulated relative to 46 °C. The Leptospirillum group II UBA genotype also had one GH57 protein that was also significantly upregulated at 40 °C. The Leptospirillum group II 5-way genotype had one predicted GH57 protein that had a higher total intensity at 46 °C but was not significantly different than 40 °C.
Figure 4.
Abundance of predicted CAZymes at 40 and 46 °C for Leptospirillum group III. Stars indicate proteins that are significantly different at a given temperature (fold change >1.2 or <0.8; *P⩽0.1; **P⩽0.05).
Energy production
Leptospirillum group III proteins involved in the various steps of the iron oxidation electron transport chain (Jeans et al., 2008; Singer et al., 2008; Blake and Griff, 2012; Bonnefoy and Holmes, 2012) differed in their response to temperature. Three cytochromes involved in the initial steps of iron oxidation were more abundant at 46 °C than at 40 °C (two of which were significantly upregulated: Cyt579 P=0.05; cytochrome C P=0.03). Conversely, 15 out of 20 downstream proteins involved in electron transfer, converting oxygen to water and generating ATP were more abundant at 40 °C (including three significant proteins: a cytochrome C oxidase and two ATP synthase subunits P=0.003, 0.03, 0.02). It is unclear how different temperature responses of these proteins affects energy yield within the cells.
Amino-acid metabolism
At higher temperatures, the Leptospirillum bacteria (the group II 5-way and UBA genotypes and group III) increased expression of proteins involved in amino-acid metabolism. Twelve amino-acid biosynthesis and degradation proteins were significantly upregulated at 46 °C, whereas only one was upregulated at 40 °C. Among those proteins upregulated at 46 °C were those involved in the biosynthesis of alanine, lysine, glutamate, cysteine, isoleucine and tryptophan. In addition, four separate proteins in the histidine biosynthesis pathway were upregulated at 46 °C for the Leptospirillum group II 5-way genotype (P=0.01–0.07).
Genetic information processing
Of proteins with the highest total intensities (top 5% for each of the three Leptospirillum bacteria), 31% were involved in genetic information processing functions. More than 3.4 times as many proteins involved in genetic information processing were significantly upregulated at 40 °C relative to 46 °C (24 at 40 °C versus 7 at 46 °C) including functions of replication, recombination and repair; transcription; translation; and nucleotide transport and metabolism. More ribosomal proteins were significantly upregulated at 40 °C relative to 46 °C for all three Leptospirillum bacteria, but most striking was Leptospirillum group III that had 13 ribosomal proteins significantly upregulated at 40 °C and only one at 46 °C.
Chemotaxis and Stress
Methyl-accepting chemotaxis sensory transducer proteins were significantly upregulated at 40 °C for each of the three Leptospirillum bacteria (P⩽0.05 except for the UBA genotype with P-values of 0.09 and 0.08). The Leptospirillum bacteria exhibited various stress responses: an oxidative stress protein was significantly upregulated at 46 °C for the UBA genotype; an osmotic stress protein was significantly upregulated at 40 °C for the 5-way genotype; and a metal stress protein was significantly upregulated at 40 °C for Leptospirillum group III. One phage integrase protein was upregulated at 40 °C for Leptospirillum group III. The Leptospirillum group III genome contains a cluster of Cas genes (Aliaga Goltsman et al., 2009), which are CRISPR-associated genes involved in viral defense. Five of these Cas proteins were upregulated at 46 °C (P=0.013–0.095; one with an abundance ratio of 0.84). Two phage proteins were also upregulated at 46 °C for the 5-way genotype (phage shock protein A P=0.01; phage integrase P=0.08). One viral protein was detected and quantified in the data set (AMDVIR_10150G0005).
Function of G-plasma archaea in biofilms growing at 40 and 46 °C
G-plasma (of the Thermoplasmatales order of Euryarchaeota) protein abundance was also evaluated at the organism level. Overall, only 24 proteins were significantly different between 40 and 46 °C. Functions of these significant proteins included chaperones, amino-acid metabolism, genetic information processing and transport.
A total of 34 proteins were quantified with predicted function in carbon transformation pathways (based on genomic analyses from (Yelton et al., 2013)), including the Entner–Doudoroff pathway, glycolysis, pyruvate dehydrogenase complex, TCA cycle and beta oxidation (Supplementary Figure S4). Of these, 29 proteins (85%) were more abundant at 40 than at 46 °C, although only two were significantly different.
Discussion
Experimentation on AMD biofilms
Here, AMD biofilms were used to test an approach to determine how elevated temperature regulates physiology of individual microbial groups in a community context. These communities have a level of complexity suitable for ecological experiments and are tractable for testing new proteomic methods. The biofilms contain organisms that represent all three domains of life (bacteria, archaea and eukaryotes; as well as viruses); span multiple trophic levels; and carry out many steps of the carbon cycle, including autotrophic carbon fixation, heterotrophic carbon consumption and turnover of fixed carbon during degradation ((Denef et al., 2010; Justice et al., 2012) and references therein).
Use of TMT proteomics to study microbial communities
Prior proteomics studies using TMT- or isobaric tags for relative and absolute quantification-based isobaric chemical labeling have been applied exclusively to human tissues, plants and cultured isolates (for example, Dayon et al., 2010; Lee et al., 2011; Li et al., 2012; Westman et al., 2012; Chen et al., 2013; Paulo et al., 2013; Li et al., 2014). Here, we validate the TMT-based quantitative proteomics approach as applied to microbial communities, showing precise quantification of thousands of proteins across a large range of fold changes. We show that TMT proteomics can provide mechanistic insights into enzymes and pathways of individual microbial groups in microbial communities and define their functional response to temperature change. By multiplexing our samples, we were able to obtain accurate, precise and reproducible quantification of proteins from three treatments with two response replicates and two technical replicates per treatment in just four LC-MS/MS runs. Across our samples, we identified an average of 1799 proteins from 25 different organisms including bacteria, archaea, eukaryotes and viruses. This technique is particularly useful in systems of intermediate complexity, where the most relevant proteins would be captured in a data set of 1500–2000 proteins. The total number of proteins quantified by TMT proteomics will increase as mass spectrometry instrumentation improves, making this approach more applicable to complex ecosystems.
Effect of warming on community structure
We found that a thermal shift from 40 to 46 °C caused a dramatic change in community composition (as reflected within the community proteome; Figure 2, Supplementary Figure S2), as has been reported in other warming studies in soils, oceans and freshwater (for example, Deslippe et al., 2012; Yergeau et al., 2012; Lindh et al., 2013; Luo et al., 2013; von Scheibner et al., 2014). Nearly a quarter of the organisms had a greater than twofold change in abundance between temperatures. Leptospirillum group III likely favors environmental conditions with lower stress, including lower temperature (Mueller et al., 2010). Here, we found that the overall abundance of Leptospirillum group III, the dominant organism in the cultivated biofilms, decreased by 14% from 40 to 46 °C (based on protein abundance) and ribosomal proteins were significantly downregulated at 46 °C, suggestive of reduced cell growth at elevated temperature.
The lack of visible biofilm growth at 49 °C suggests that persistent temperatures above 46 °C alter community structure and/or function in such a way that biofilm formation and development are hindered. Previous culture studies have shown that while some Leptospirillum isolates are capable of growth up to 45 °C, many others are unable to grow at that temperature or higher (Coram and Rawlings, 2002; Lo, 2006; Zhang et al., 2010). Thus, it may be the case here that the colonizing Leptospirillum bacteria have a maximum growth temperature around 46 °C, thereby preventing the initial stages of biofilm formation at higher temperatures. Studies such as these enable observations of growth in a community context where organisms experience competition for resources and interactions with other organisms, compared with static culture conditions.
Communities made up of both specialists and generalists are likely more productive and more stable over time under environmental fluctuations. Among groups of closely related organisms in the AMD biofilms, there appear to be a subset that are specialists in terms of their temperature optima, as well as generalists able to grow over a wider range of temperature. For instance, for the ARMAN archaea, ARMAN IV was more abundant at 40 °C, ARMAN II was more abundant at 46 °C and two other ARMAN types had similar abundance levels at both temperatures (Figure 2).
Effect of warming on Leptospirillum function
Elevated temperature differentially affected protein abundance in the carbon fixation pathway of closely related bacterial genotypes: high temperature repressed carbon fixation protein abundance by Leptospirillum group III and the group II 5-way genotype, whereas carbon fixation protein abundance was significantly upregulated at higher temperature by the Leptospirillum group II UBA genotype (Figure 3). Functional overlap of three Leptospirillum genotypes (iron oxidation coupled with carbon fixation) with different temperature responses may provide ecological insurance for community function under heterogeneous environments. Niche differentiation of Leptospirillum allows for asynchronous responses to fluctuating conditions and assists in preserving function within the community across changing environments.
Increasing temperatures have been shown to enhance the decomposition of organic matter and the extracellular release of carbohydrates in seawater (Wohlers et al., 2009; Engel et al., 2011). Here, 20 different CAZymes were quantified for Leptospirillum group III bacteria (Figure 4). CAZymes with different functionality were upregulated under different conditions. For instance, GT2 involved in biosynthesis of carbohydrates was upregulated at 46 °C, whereas GH57 involved in hydrolysis of carbohydrates was upregulated at 40 °C. The extracellular polymeric substance in AMD biofilms from the Richmond mine has been shown to contain abundant carbohydrates, including galactose, glucose, heptose, rhamnose and mannose (Jiao et al., 2010). Thus, Leptospirillum group III might act both as a source and sink to the carbohydrate pool in the biofilm matrix.
Several CRISPR-associated proteins were upregulated at 46 °C for the Leptospirillum group III bacteria. Most CRISPR-Cas systems confer resistance to foreign genetic elements, although some have been implicated in non-viral related functions (for example, Sampson and Weiss 2014). Among the upregulated Leptospirillum CRISPR-associated proteins was a Cas3 protein predicted to be responsible for cleavage of invading DNA (Brouns et al., 2008). Thus, the Leptospirillum group III bacteria may have been subjected to increased viral stress at elevated temperature. Viral-induced mortality impacts not only the abundance and composition of microbial communities but also system-level nutrient cycling. Viral lysis releases the contents of the host cell (including cytoplasmic and structural material) into the environment, thereby liberating a fraction of the organic matter pool and shifting nutrients from the particulate to dissolved states. Dissolved organic carbon (and other nutrients including phosphorus and nitrogen) released by viral lysis can stimulate the growth of non-infected populations, increase community respiration and decrease the efficiency of carbon transfer to higher trophic levels ((Gobler et al., 1997; Middelboe and Lyck 2002; Suttle, 2005) and references therein). Thus, increased susceptibility to viral stress at elevated temperature, as shown here, will likely lead to greater carbon turnover and altered community structure.
The expression of proteins involved in amino-acid metabolism was upregulated at higher temperatures both at the community level and for each Leptospirillum genotype (group II 5-way and UBA genotypes and group III). Several amino acids are thermolabile, and thus can have a reduced frequency in thermophilic proteomes (Russell et al., 1997; Hickey and Singer, 2004). AMD organisms may be increasing expression of amino-acid biosynthesis proteins at 46 °C to increase the size of the amino-acid pool available for making other cellular proteins that may be inactivated at higher temperature.
Temperature has been shown to affect bacterial movement via impacts on both chemotaxis and flagellar assembly (for example, Schneider and Doetsch 1977; Turner et al., 1999; Aygan and Arikan, 2007). Here, methyl-accepting chemotaxis sensory transducer proteins were significantly upregulated at 40 °C for each of the three Leptospirillum bacteria. Previous reports also showed that chemotaxis was strongly inhibited by high temperature in Escherichia coli (Morrison and McCapra, 1961; Adler and Templeton, 1967; Li et al., 1993). Structural studies of AMD biofilms show Leptospirillum group II at the base of mature biofilms and Leptospirillum group III as dispersed cells and microcolonies within the interior regions (Wilmes et al., 2009). Chemotaxis may be critical for positioning the bacteria within areas of the biofilm that are best suited for optimal growth. Decreased activity of chemotaxis proteins at elevated temperature may subject these bacteria to unfavorable geochemical conditions such as lower oxygen concentrations at the base of the biofilm or less nutrient availability in the interior. Nutrient limitation resulting from decreased chemotaxis at elevated temperature may be compensated for by enhanced nutrient scavenging, as indicated by upregulation of two nutrient assimilation proteins at 46 °C for the group II 5-way genotype (NifA and a periplasmic phosphate binding protein).
Conclusion
The current research shows the utility of quantitative proteomics for studies of ecological of phenomena such as niche differentiation. The approach provided information about differential expression of thousands of proteins involved in diverse functions including metabolism, growth, signaling and stress response. It enabled protein analysis at the level of individual microbial groups within a community context and across the whole community.
Acknowledgments
We thank the late T. W. Arman (President, Iron Mountain Mines) for providing access to the Richmond Mine. We also thank R. Sugarek (Environmental Protection Agency) for site access and R. Carver and M. Jones for on-site assistance. We thank Susan Spaulding, Nicholas Justice and Kyle Frischkorn for laboratory assistance. Funding was provided by the U.S. Department of Energy, through the Carbon-Cycling (DE-FG02-10ER64996) and Knowledgebase (DE-SC0004918) programs.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adler J, Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J Gen Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- Aliaga Goltsman DS, Denef VJ, Singer SW, VerBerkmoes NC, Lefsrud M, Mueller RS, et al. Community genomic and proteomic analyses of chemoautotrophic iron-oxidizing ‘Leptospirillum rubarum' (Group II) and ‘Leptospirillum ferrodiazotrophum' (Group III) bacteria in acid mine drainage biofilms. Appl Environ Microbiol. 2009;75:4599–4615. doi: 10.1128/AEM.02943-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygan A, Arikan B. An overview on bacterial motility detection. Int J Agr Biol. 2007;9:193–196. [Google Scholar]
- Belnap CP, Pan C, VerBerkmoes NC, Power ME, Samatova NF, Carver RL, et al. Cultivation and quantitative proteomic analyses of acidophilic microbial communities. ISME J. 2010;4:520–530. doi: 10.1038/ismej.2009.139. [DOI] [PubMed] [Google Scholar]
- Blake RC, Griff MN. In situ spectroscopy on intact Leptospirillum ferrooxidans reveals that reduced cytochrome 579 is an obligatory intermediate in the aerobic iron respiratory chain. Front Microbiol. 2012;3:136. doi: 10.3389/fmicb.2012.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond P, Banfield J. Design and performance of rRNA targeted oligonucleotide probes for in situ detection and phylogenetic identification of microorganisms inhabiting acid mine drainage environments. Microb Ecol. 2001;41:149–161. doi: 10.1007/s002480000063. [DOI] [PubMed] [Google Scholar]
- Bonnefoy V, Holmes DS. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ Microbiol. 2012;14:1597–1611. doi: 10.1111/j.1462-2920.2011.02626.x. [DOI] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-W, Scaria J, Mao C, Sobral B, Zhang S, Lawley T, et al. Proteomic comparison of historic and recently emerged hypervirulent Clostridium difficile strains. Environ Sci Technol. 2013;12:1151–1161. doi: 10.1021/pr3007528. [DOI] [PubMed] [Google Scholar]
- Chourey K, Jansson J, VerBerkmoes N, Shah M, Chavarria KL, Tom LM, et al. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J Proteome Res. 2010;9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- Coram N, Rawlings D. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptosphillum ferriphilum sp nov dominates South African commercial biooxidation tanks that operate at 40 degrees C. Appl Environ Microbiol. 2002;68:838–845. doi: 10.1128/AEM.68.2.838-845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayon L, Turck N, Scherl A, Hochstrasser DF, Burkhard PR, Sanchez J-C. From relative to absolute quantification of tryptic peptides with tandem mass tags: application to cerebrospinal fluid. Chimia (Aarau) 2010;64:132–135. doi: 10.2533/chimia.2010.132. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Banfield JF. In situ evolutionary rate measurements show ecological success of recently emerged bacterial hybrids. Science. 2012;336:462–466. doi: 10.1126/science.1218389. [DOI] [PubMed] [Google Scholar]
- Denef VJ, Mueller RS, Banfield JF. AMD biofilms: using model communities to study microbial evolution and ecological complexity in nature. ISME J. 2010;4:599–610. doi: 10.1038/ismej.2009.158. [DOI] [PubMed] [Google Scholar]
- Denef VJ, VerBerkmoes NC, Shah MB, Abraham P, Lefsrud M, Hettich RL, et al. Proteomics-inferred genome typing (PIGT) demonstrates inter-population recombination as a strategy for environmental adaptation. Environ Microbiol. 2009;11:313–325. doi: 10.1111/j.1462-2920.2008.01769.x. [DOI] [PubMed] [Google Scholar]
- Deslippe JR, Hartmann M, Simard SW, Mohn WW. Long-term warming alters the composition of Arctic soil microbial communities. FEMS Microbiol Ecol. 2012;82:303–315. doi: 10.1111/j.1574-6941.2012.01350.x. [DOI] [PubMed] [Google Scholar]
- Dobbin E, Graham C, Freeburn RW, Unwin RD, Griffiths JR, Pierce A, et al. Proteomic analysis reveals a novel mechanism induced by the leukemic oncogene Tel/PDGFRβ in stem cells: activation of the interferon response pathways. Stem Cell Res. 2010;5:226–243. doi: 10.1016/j.scr.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Engel A, Handel N, Wohlers J, Lunau M, Grossart HP, Sommer U, et al. Effects of sea surface warming on the production and composition of dissolved organic matter during phytoplankton blooms: results from a mesocosm study. J Plankton Res. 2011;33:357–372. [Google Scholar]
- Finke N, Jørgensen BB. Response of fermentation and sulfate reduction to experimental temperature changes in temperate and Arctic marine sediments. ISME J. 2008;2:815–829. doi: 10.1038/ISMEJ.2008.20. [DOI] [PubMed] [Google Scholar]
- Gobler CJ, Hutchins DA, Fisher NS, Cosper EM, Sanudo-Wilhelmy SA. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol Oceanogr. 1997;42:1492–1504. [Google Scholar]
- Han D, Moon S, Kim H, Choi S-E, Lee S-J, Park KS, et al. Detection of differential proteomes associated with the development of type 2 diabetes in the Zucker rat model using the iTRAQ technique. J Proteome Res. 2011;10:564–577. doi: 10.1021/pr100759a. [DOI] [PubMed] [Google Scholar]
- Hickey DA, Singer GAC. Genomic and proteomic adaptations to growth at high temperature. Genome Biol. 2004;5:117–117. doi: 10.1186/gb-2004-5-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S, Graham C, Graham RLJ, McMullan G, Ternan NG. Quantitative proteomic analysis of the heat stress response in Clostridium difficile strain 630. J Proteome Res. 2011;10:3880–3890. doi: 10.1021/pr200327t. [DOI] [PubMed] [Google Scholar]
- Jeans C, Singer SW, Chan CS, VerBerkmoes NC, Shah M, Hettich RL, et al. Cytochrome 572 is a conspicuous membrane protein with iron oxidation activity purified directly from a natural acidophilic microbial community. ISME J. 2008;2:542–550. doi: 10.1038/ismej.2008.17. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Cody GD, Harding AK, Wilmes P, Schrenk M, Wheeler KE, et al. Characterization of extracellular polymeric substances from acidophilic microbial biofilms. Appl Environ Microbiol. 2010;76:2916–2922. doi: 10.1128/AEM.02289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice NB, Pan C, Mueller R, Spaulding SE, Shah V, Sun CL, et al. Heterotrophic archaea contribute to carbon cycling in low-pH, suboxic biofilm communities. Appl Environ Microbiol. 2012;78:8321–8330. doi: 10.1128/AEM.01938-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köcher T, Pichler P, Schutzbier M, Stingl C, Kaul A, Teucher N, et al. High precision quantitative proteomics using iTRAQ on an LTQ orbitrap: a new mass spectrometric method combining the benefits of all. Environ Sci Technol. 2009;8:4743–4752. doi: 10.1021/pr900451u. [DOI] [PubMed] [Google Scholar]
- Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, et al. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol Syst Biol. 2011;7:1–12. doi: 10.1038/msb.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Louise CJ, Shi W, Adler J. Adverse conditions which cause lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2229–2235. doi: 10.1128/jb.175.8.2229-2235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Adams RM, Chourey K, Hurst GB, Hettich RL, Pan C. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ orbitrap velos. J Proteome Res. 2012;11:1582–1590. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- Li Z, Czarnecki O, Chourey K, Yang J, Tuskan GA, Hurst GB, et al. Strigolactone-regulated proteins revealed by iTRAQ-based quantitative proteomics in Arabidopsis. J Proteome Res. 2014;13:1359–1372. doi: 10.1021/pr400925t. [DOI] [PubMed] [Google Scholar]
- Lindh MV, Riemann L, Baltar F, Romero-Oliva C, Salomon PS, Granéli E, et al. Consequences of increased temperature and acidification on bacterioplankton community composition during a mesocosm spring bloom in the Baltic Sea. Environ Microbiol Rep. 2013;5:252–262. doi: 10.1111/1758-2229.12009. [DOI] [PubMed] [Google Scholar]
- Lo I, Denef V, Verberkmoes N, Shah M, Goltsman D, DiBartolo G, et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- Lo I. Diversification and recombination in Leptospirillum Group II. Thesis, University of California: Berkeley; 2006. [Google Scholar]
- Luo C, Rodriguez-R LM, Johnston ER, Wu L, Cheng L, Xue K, et al. Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl Environ Microbiol. 2014;80:1777–1786. doi: 10.1128/AEM.03712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M, Lyck PG. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat Microb Ecol. 2002;27:187–194. [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison RB, McCapra J. Flagellar changes in Escherichia coli induced by temperature of the environment. Nature. 1961;192:774–776. [Google Scholar]
- Mosier AC, Justice NB, Bowen BP, Baran R, Thomas BC, Northen TR, et al. Metabolites associated with adaptation of microorganisms to an acidophilic, metal-rich environment identified by stable-isotope-enabled metabolomics. mBio. 2013;4:00484–00412. doi: 10.1128/mBio.00484-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller RS, Denef VJ, Kalnejais LH, Suttle KB, Thomas BC, Wilmes P, et al. Ecological distribution and population physiology defined by proteomics in a natural microbial community. Mol Syst Biol. 2010;6:374. doi: 10.1038/msb.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukrishnan G, Quinn GA, Lamers RP, Diaz C, Cole AL, Chen S, et al. Exoproteome of Staphylococcus aureus reveals putative determinants of nasal carriage. J Proteome Res. 2011;10:2064–2078. doi: 10.1021/pr200029r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan C, Oda Y, Lankford PK, Zhang B, Samatova NF, Pelletier DA, et al. Characterization of anaerobic catabolism of p-coumarate in Rhodopseudomonas palustris by integrating transcriptomics and quantitative proteomics. Mol Cell Proteomics. 2008;7:938–948. doi: 10.1074/mcp.M700147-MCP200. [DOI] [PubMed] [Google Scholar]
- Park BH, Karpinets TV, Syed MH, Leuze MR, Uberbacher EC. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010;20:1574–1584. doi: 10.1093/glycob/cwq106. [DOI] [PubMed] [Google Scholar]
- Paulo JA, Kadiyala V, Banks PA, Conwell DL, Steen H. Mass spectrometry-based quantitative proteomic profiling of human pancreatic and hepatic stellate cell lines. Genomics Proteomics Bioinformatics. 2013;11:105–113. doi: 10.1016/j.gpb.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, et al. Community proteomics of a natural microbial biofilm. Science. 2005;308:1915–1920. [PubMed] [Google Scholar]
- Rose JM, Vora NM, Countway PD, Gast RJ, Caron DA. Effects of temperature on growth rate and gross growth efficiency of an Antarctic bacterivorous protist. ISME J. 2009;3:252–260. doi: 10.1038/ismej.2008.96. [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Russell RJ, Ferguson JM, Hough DW, Danson MJ, Taylor GL. The crystal structure of citrate synthase from the hyperthermophilic archaeon Pyrococcus furiosus at 1.9 Å resolution. Biochemistry. 1997;36:9983–9994. doi: 10.1021/bi9705321. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS. CRISPR-Cas systems: new players in gene regulation and bacterial physiology. Front Cell Infect Microbiol. 2014;4:1–8. doi: 10.3389/fcimb.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WR, Doetsch RN. Temperature effects on bacterial movement. Appl Environ Microbiol. 1977;34:695–700. doi: 10.1128/aem.34.6.695-700.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons SL, Dibartolo G, Denef VJ, Goltsman DSA, Thelen MP, Banfield JF. Population genomic analysis of strain variation in Leptospirillum group II bacteria involved in acid mine drainage formation. PLoS Biol. 2008;6:e177. doi: 10.1371/journal.pbio.0060177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SW, Chan CS, Zemla A, VerBerkmoes NC, Hwang M, Hettich RL, et al. Characterization of cytochrome 579, an unusual cytochrome isolated from an iron-oxidizing microbial community. Appl Environ Microbiol. 2008;74:4454–4462. doi: 10.1128/AEM.02799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares NC, Cabral MP, Gayoso C, Mallo S, Rodriguez-Velo P, Fernández-Moreira E, et al. Associating growth-phase-related changes in the proteome of Acinetobacter baumannii with increased resistance to oxidative stress. J Proteome Res. 2010;9:1951–1964. doi: 10.1021/pr901116r. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle C. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR. DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. Environ Sci Technol. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Toseland A, Daines SJ, Clark JR, Kirkham A, Strauss J, Uhlig C, et al. The impact of temperature on marine phytoplankton resource allocation and metabolism. Nat Clim Change. 2013;3:1–6. [Google Scholar]
- Tu Q, Yu H, He Z, Deng Y, Wu L, Van Nostrand JD, et al. 2014GeoChip 4: a functional gene-array-based high-throughput environmental technology for microbial community analysis Mol Ecol Resoure-pub ahead of print 12 February 2014doi: 10.1111/1755-0998.12239 [DOI] [PubMed]
- Turner L, Samuel AD, Stern AS, Berg HC. Temperature dependence of switching of the bacterial flagellar motor by the protein CheY(13DK106YW) Biophys J. 1999;77:597–603. doi: 10.1016/S0006-3495(99)76916-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Scheibner M, Dörge P, Biermann A, Sommer U, Hoppe H-G, Jürgens K. Impact of warming on phyto-bacterioplankton coupling and bacterial community composition in experimental mesocosms. Environ Microbiol. 2014;16:718–733. doi: 10.1111/1462-2920.12195. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ahn T-H, Li Z, Pan C. Sipros/ProRata: a versatile informatics system for quantitative community proteomics. Bioinformatics. 2013;29:2064–2065. doi: 10.1093/bioinformatics/btt329. [DOI] [PubMed] [Google Scholar]
- Washburn MP, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- Westman JO, Taherzadeh MJ, Franzén CJ. Proteomic analysis of the increased stress tolerance of Saccharomyces cerevisiae encapsulated in liquid core alginate-chitosan capsules. PLoS One. 2012;7:e49335. doi: 10.1371/journal.pone.0049335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson AJK, Smith DL, Blinco D, Unwin RD, Pearson S, Wilson C, et al. Quantitative proteomics analysis demonstrates post-transcriptional regulation of embryonic stem cell differentiation to hematopoiesis. Mol Cell Proteomics. 2008;7:459–472. doi: 10.1074/mcp.M700370-MCP200. [DOI] [PubMed] [Google Scholar]
- Wilmes P, Remis JP, Hwang M, Auer M, Thelen MP, Banfield JF. Natural acidophilic biofilm communities reflect distinct organismal and functional organization. ISME J. 2009;3:266–270. doi: 10.1038/ismej.2008.90. [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Wohlers J, Engel A, Zöllner E, Breithaupt P, Jürgens K, Hoppe H-G, et al. Changes in biogenic carbon flow in response to sea surface warming. Proc Natl Acad Sci USA. 2009;106:7067–7072. doi: 10.1073/pnas.0812743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Ke X, Hernández M, Wang B, Dumont MG, Jia Z, et al. Autotrophic growth of bacterial and archaeal ammonia oxidizers in freshwater sediment microcosms incubated at different temperatures. Appl Environ Microbiol. 2013;79:3076–3084. doi: 10.1128/AEM.00061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton AP, Comolli LR, Justice NB, Castelle C, Denef VJ, Thomas BC, et al. Comparative genomics in acid mine drainage biofilm communities reveals metabolic and structural differentiation of co-occurring archaea. BMC Genomics. 2013;14:485–485. doi: 10.1186/1471-2164-14-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yergeau E, Bokhorst S, Kang S, Zhou J, Greer CW, Aerts R, et al. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J. 2012;6:692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R-Y, Xia J-L, Peng J-H, Zhang Q, Zhang C-G, Nie Z-Y, et al. A new strain Leptospirillum ferriphilum YTW315 for bioleaching of metal sulfides ores. Trans Nonferrous Met Soc China. 2010;20:135–141. [Google Scholar]
- Zhao Z, Stanley BA, Zhang W, Assmann SM. ABA-regulated G protein signaling in Arabidopsis guard cells: a proteomic perspective. J Proteome Res. 2010;9:1637–1647. doi: 10.1021/pr901011h. [DOI] [PubMed] [Google Scholar]
- Zogg GP, Zak DR, Ringelberg DB, White DC, MacDonald NW, Pregitzer KS. Compositional and functional shifts in microbial communities due to soil warming. Soil Sci Soc Am J. 1997;61:475–481. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.