Abstract
Flavonol 3-O-diglucosides with a 1→2 inter-glycosidic linkage are representative pollen-specific flavonols that are widely distributed in plants, but their biosynthetic genes and physiological roles are not well understood. Flavonoid analysis of four Arabidopsis floral organs (pistils, stamens, petals and calyxes) and flowers of wild-type and male sterility 1 (ms1) mutants, which are defective in normal development of pollen and tapetum, showed that kaempferol/quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides accumulated in Arabidopsis pollen. Microarray data using wild-type and ms1 mutants, gene expression patterns in various organs, and phylogenetic analysis of UDP-glycosyltransferases (UGTs) suggest that UGT79B6 (At5g54010) is a key modification enzyme for determining pollen-specific flavonol structure. Kaempferol and quercetin 3-O-glucosyl-(1→2)-glucosides were absent from two independent ugt79b6 knockout mutants. Transgenic ugt79b6 mutant lines transformed with the genomic UGT79B6 gene had the same flavonoid profile as wild-type plants. Recombinant UGT79B6 protein converted kaempferol 3-O-glucoside to kaempferol 3-O-glucosyl-(1→2)-glucoside. UGT79B6 recognized 3-O-glucosylated/galactosylated anthocyanins/flavonols but not 3,5- or 3,7-diglycosylated flavonoids, and prefers UDP-glucose, indicating that UGT79B6 encodes flavonoid 3-O-glucoside:2″-O-glucosyltransferase. A UGT79B6-GUS fusion showed that UGT79B6 was localized in tapetum cells and microspores of developing anthers.
Keywords: glucosyltransferase, At5g54010, NM_124780, tapetum, pollen, glycosyltransferase, flavonol, flavonoid, Arabidopsis thaliana
Introduction
Flavonoids are a large group of plant secondary metabolites, and include the flavonols, flavones, anthocyanins and proanthocyanidins. They are one of the best-studied plant natural products in the genetics, biochemistry and molecular biology fields (Anderson and Markham, 2006; Grotewold, 2006). Over 9000 known compounds are widely distributed throughout the plant kingdom, including bryophytes (mosses and liverworts), pteridophytes (ferns), gymnosperms and angiosperms (Markham, 1988; Richardson, 1989; Williams and Grayer, 2004; Anderson and Markham, 2006).
Flavonoids play important roles as pigments, UV protectants, attractants of pollinators, phytoalexins, signaling molecules, and regulators of fertility and auxin transport (Gould and Lister, 2006). The precise relationships between flavonoid structures and their physiological functions are still largely unknown due to the huge diversity of flavonoid structures, their multiple roles in plant differentiation and function, and their complex distribution in various plant tissues and species. Based on the distribution of flavonoids and flavonoid biosynthetic genes among land plants, the time of appearance in evolutionary history and the primary advantages of producing each flavonoid class have been discussed (Rausher, 2006). Bryophytes are thought to be the oldest plant group to produce chalcones, flavanones, flavonols and flavones (Stafford, 1991; Rausher, 2006). Protection against UV irradiation and regulation of plant hormone action are two functions that have been proposed as initial advantages of flavonoid production (Stafford, 1991; Shirley, 1996; Rausher, 2006). Ferns and flowering plants (gymnosperms and angiosperms) are considered the oldest producers of proanthocyanidins and anthocyanins/isoflavonoids/aurones, respectively, although there are some exceptions (Stafford, 1991; Rausher, 2006). One of the primary functions of proanthocyanidins is thought to be defense against bacterial and fungal pathogens and herbivores. During evolution, these functions have greatly diversified, making it difficult to definitively address their current physiological roles, let alone their primordial activities.
Flavonols are believed to have appeared in the early period of land plant history, and their broad distribution at the tissue level and throughout the plant kingdom suggests that they play general roles as flavonoids. Anthocyanins, isoflavonoids and proanthocyanidins appear to be confined in the roles specific to seed plants.” to “In contrast, anthocyanins, isoflavonoids and proanthocyanidins may play roles specific to seed plants. Among a variety of flavonols with relatively similar structures, specific flavonol 3-O-diglucosides with a 1→2 inter-glycosidic linkage often accumulate as major flavonols in pollen, a tissue specific to seed plants (Pratviel and Perchero, 1972; Zerback et al., 1989; Price et al., 1998; Ross et al., 2005). Pollen of petunia (Petunia hybrida) predominantly accumulates kaempferol/quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-galactopyranosides (Zerback et al., 1989). Kaempferol and quercetin 3-O-glucosyl-(1→2)-glucoside accumulate in pollen of Cannabis sativa L. and florets of broccoli (Brassica oleracea) (Price et al., 1998; Ross et al., 2005). In addition, quercetin 3-O-glucosyl-(1→2)-glucoside is also found as a major constituent in the pollen of trees in the families Juglandaceae, Betulaceae, Fagaceae and Oleaceae (Pratviel and Perchero, 1972). These observations suggest that flavonols play a specific role in pollen.
In Arabidopsis, one of the best-studied model plants for the molecular biology of flavonoid metabolism (Debeaujon et al., 2003; Pourcel et al., 2005; Yonekura-Sakakibara et al., 2008; Nakabayashi et al., 2009; Stracke et al., 2010; Saito et al., 2013), flavonol 3-O-hexosylglucosides corresponding to flavonol 3-O-diglucosides with a 1→2 inter-glycosidic linkage were also detected in the pollen (Stracke et al., 2010). The genes involved in flavonoid skeleton biosynthesis have been completely identified in Arabidopsis. Several genes for flavonoid modification (eight glycosyltransferases, four acyltransferases and one methyltransferase) have also been isolated (Saito et al., 2013). In addition, MYB transcription factors for flavonols (PFG1/MYB11, PFG2/MYB12 and PFG3/MYB111), anthocyanins (PAP1/MYB75, PAP2/MYB90, MYB113 and MYB114) and proanthocyanidins (TT2/MYB123), and other transcription factors such as bHLH and WD, have been characterized (Koes et al., 2005; Dubos et al., 2010). However, the structures of some flavonoids in flowers and roots, and their corresponding flavonoid modification enzymes, including flavonol 3-O-hexosylglucosides (f21 and f26) and UDP-glycosyltransferases (UGTs), remain to be determined. Furthermore, flavonol 3-O-hexosylglucosides were the sole flavonols in pollen of triple knockout mutants deficient in MYB11/MYB12/MYB111, suggesting the existence of an unknown regulatory system for pollen flavonol biosynthesis (Stracke et al., 2010).
To solve the mystery of flavonoid biosynthesis and regulation in pollen and to achieve a comprehensive understanding of flavonoid metabolism, we analyzed flavonoid accumulation patterns in pistils, stamens, petals and calyxes. Among these four floral organs, stamens showed a distinctive flavonol distribution. Flavonol analysis of a mutant deficient in the MALE STERILITY 1 (MS1) gene, which is required for normal development of the pollen and tapetum, showed that two flavonol derivatives (f21 and f26) are absent from ms1 mutants. We identified flavonol f21 as kaempferol 3-O-glucosyl-(1→2)-glucoside and f26 as quercetin 3-O-glucosyl-(1→2)-glucoside, and identified UGT79B6 as a flavonol 3-O-glucoside:2″-O-glucosyltransferase for formation of pollen-specific flavonoids. Here, we discuss the pollen-specific regulation and function of flavonol metabolism in its evolutionary context.
Results
Structural identification of pollen-specific flavonols absent from ms1 mutants
To determine the structure of pollen-specific flavonoids, we used male sterility 1 (ms1) mutants. MS1 encodes a transcription factor with Leu zipper-like and PHD-finger motifs, and is required for normal tapetal development and pollen wall synthesis (Ito et al., 2007; Yang et al., 2007). The ms1 mutant lacks pollen walls, and microspores and tapetum cells degenerate (Wilson et al., 2001; Ito and Shinozaki, 2002; Ariizumi et al., 2005). Flowers of the ms1 mutant contained significantly lower levels of quercetin 3-O-hexosylglucoside (f26, m/z 627) than wild-type (Landsberg erecta, Ler) (Figures1a and S1). Isorhamnetin 3-O-glucoside-7-O-rhamnoside (f14, m/z 625) and/or kaempferol 3-O-hexosylglucoside (f21, m/z 611) were also slightly decreased in ms1 mutants. Extracted ion chromatograms of extracts from flowers of the ms1 mutant and the wild-type indicated that f26 and f21 were absent from the ms1 mutant, but f14 accumulated to a comparable level (Figure1b). Untargeted metabolite profiling indicated that at least seven compounds (C1-C7) were missing from ms1 plants, and C1, C2 and C4 were detected as major peaks in the wild-type (Figure S2A and Table S1). The mass spectra and retention times of C1 and C2 indicated that these compounds correspond to flavonol derivatives f26 and f21, respectively, and that the other compounds (C3-C7) are not flavonoid derivatives (Figure S2A and Table S1). It has been reported that pollen-specific kaempferol or quercetin 3-O-diglucosides correspond to flavonols f21 and f26, although the exact structures remain to be determined (Stracke et al., 2010; Fellenberg et al., 2012). These data suggest that flavonols f21 and f26 are major components of pollens.
Figure 1.
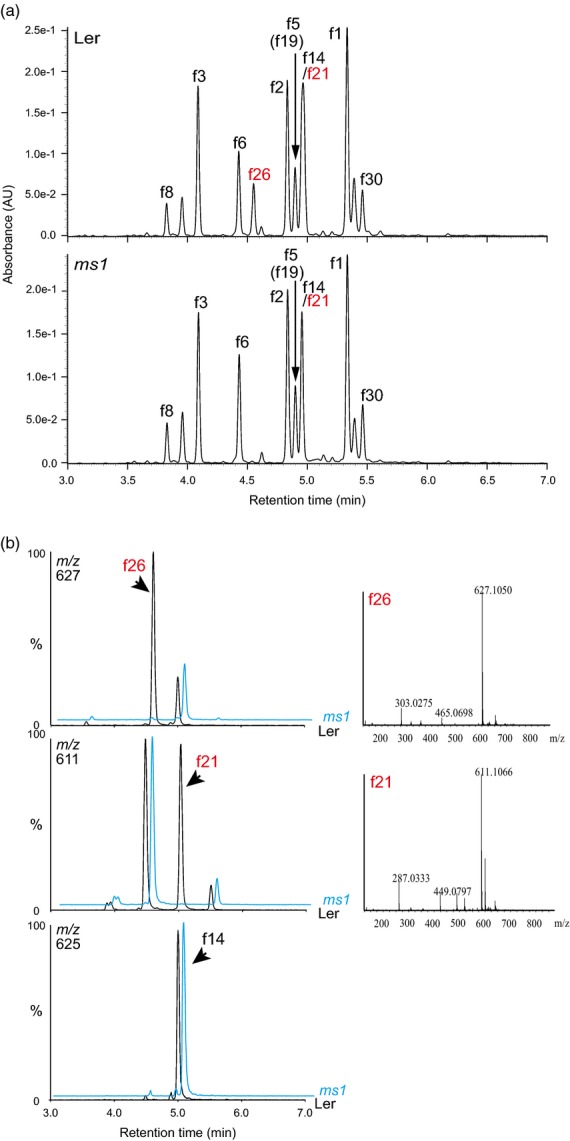
UPLC-PDA-MS analyses of extracts from flowers of wild-type and ms1 mutants.(a) UPLC-PDA chromatograms of aqueous methanol extracts from flowers of the Arabidopsis wild-type (Ler) and ms1 mutants. Absorbance at 320 nm was used for detection of flavonols.(b) Extracted fragment mass chromatograms (m/z 627, 611 and 625) of aqueous methanol extracts from flowers of the wild-type (Ler) and ms1 mutants. Mass spectra of f26 and f21 are shown on the right.
We used the Columbia-0 (Col-0) accession for further analyses because flavonol profiles in flowers of Arabidopsis accessions Col–0 and Ler showed that the qualitative compositions of flavonols in both accessions are very similar, although the quantitative profiles are slightly different (Figure S3). It has also been reported that the flavonol compositions in mature seeds in Col-0 and Ler are similar (Routaboul et al., 2006). Flavonol profiles in pistils, stamens, petals and calyxes of the Col-0 accession obtained using ultraperformance liquid chromatography (UPLC)/photodiode array (PDA)/electrospray ionization (ESI)/quadrupole time-of-flight (Q-TOF)/MS showed that quercetin 3-O-glucoside-7-O-rhamnoside (f6), quercetin 3-O-hexosylglucoside (f26) and isorhamnetin 3-O-glucoside-7-O-rhamnoside (f14) or kaempferol 3-O-hexosylglucoside (f21) are predominant in stamen tissue (Figure2).
Figure 2.
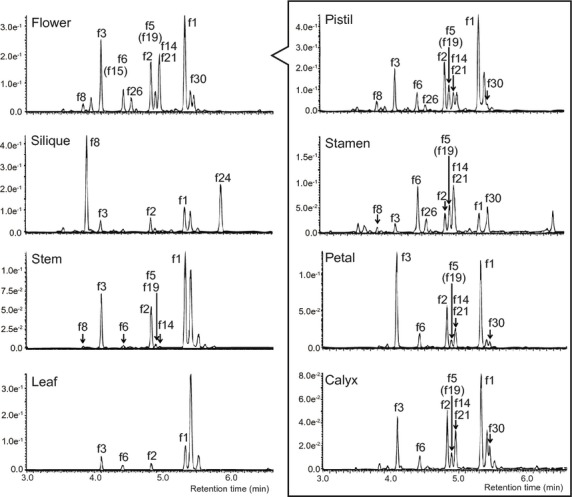
UPLC-PDA-MS analyses of extracts from various organs of Arabidopsis wild-type (Col-0).UPLC-PDA and mass chromatograms of aqueous methanol extracts from Arabidopsis wild-type. Absorbance at 320 nm was used for detection of flavonols. Labels correspond to compounds shown in Figure S1.
Flavonols f26 and f21 were isolated from floral buds of the wild-type (Col-0) by chromatographic techniques, and identified as quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranoside and kaempferol 3-O-β-d- glucopyranosyl-(1→2)-β–d-glucopyranoside, respectively (Figure S1) based on the data of appearance and specific optical rotation, NMR and MS.
A pollen-specific flavonol glycosyltransferase gene deduced by transcriptome analysis
The structures of f26 and f21 indicate that a previously unidentified flavonol 3-O-glucoside:2″-O-glucosyltransferase is present in floral tissues. f26 and f21 were missing from ms1 mutants, suggesting that expression of a previously unknown flavonol glycosyltransferase gene is suppressed, or that the gene is absent from ms1 mutants, and that expression of this gene is specific to pollen and/or tapetum.
Comparison of microarray data from ms1 and wild-type (Ler) using consecutive floral developmental stages from inflorescence meristems to mature flowers suggests that UGT76E2 (At3g46660), UGT78D2 (At5g17050), UGT79B6 (At5g54010), UGT85A1 (At1g22400), UGT85A3 (At1g22380) and UGT92A1 (At5g12890) are down-regulated in ms1 mutants (Alves-Ferreira et al., 2007). In other microarray experiments, UGT79B6 was the only UGT gene among 228 down-regulated genes in young buds of ms1 mutants (Yang et al., 2007). The Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) indicates that UGT79B6 is expressed predominantly at an early pollen developmental stage (Figure S4), but the other five UGT candidates are not. Based on these data, we selected UGT79B6 as the most likely candidate for flavonol 3-O-glucoside:2″-O-glucosyltransferase.
Phylogenetic analysis of flavonoid UGTs
Generally, flavonoid UGTs form a unique cluster based on their regio-specificity for sugar acceptors (i.e. the glycosylation position of sugar acceptors) (Yonekura-Sakakibara et al., 2007). UGT79B6 belongs to the cluster of UGTs that catalyze glycosylation at the sugar moiety attached to flavonoid aglycones (GGTs) (Figure3 and Appendix S1), strongly suggesting that UGT79B6 encodes a flavonoid GGT. UGT707B1 from saffron (Crocus sativa) is involved in formation of kaempferol and quercetin 3-O-glucosyl-(1→2)-glucoside (Trapero et al., 2012). However, UGT79B6 and other GGTs belong to a cluster distinct from UGT707B1. UGT79B6 has some amino acid sequence identity with known flavonoid GGTs, namely UGT79B1 (57%), AcA3Ga2″XylT (49%), IpA3G2″GlcT (46%), CsF7G6″RhaT (42%), PhA3G6″RhaT (39%), BpA3G2″GlcAT (26%) and CmF7G2″RhaT (26%) (Bar-Peled et al., 1991; Brugliera et al., 1994; Kroon et al., 1994; , Morita et al., 2005; Sawada et al., 2005; Montefiori et al., 2011; Yonekura-Sakakibara et al., 2012; Frydman et al., 2013). UGT79B6 has lower sequence identity with UGT707B1 (24%) (this work).
Figure 3.
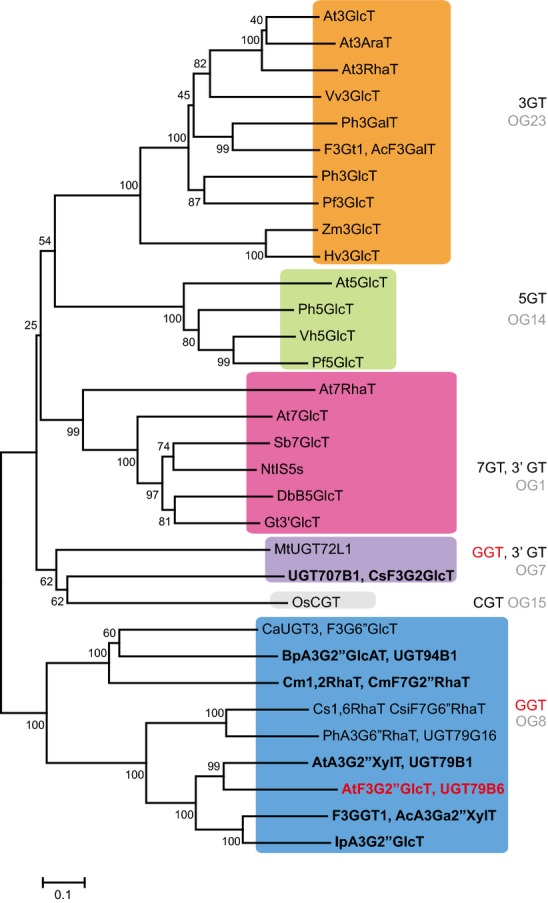
Non-rooted molecular phylogenetic tree of flavonoid glycosyltransferases.The phylogenetic tree was constructed as described in Experimental Procedures. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The alignment used for this analysis is provided in Appendix S1. The scale bar = 0.1 amino acid substitutions per site. Flavonoid 2″-O-glycosyltransferases are shown in bold and UGT79B6 is shown in red. The Genbank accession numbers for the sequences are shown in parentheses: At3RhaT (NM_102790, At1g30530); At3GlcT (NM_121711, At5g17050); At3AraT (NM_121709, At5g17030); Vv3GlcT (AF000371); Ph3GalT (AF316552); AcF3GalT (GU079683); Ph3GlcT (AB027454); Pf3GlcT (AB002818); Hv3GlcT (X15694); Zm3GlcT (X13501); At5GlcT (NM_117485, At4g14090); Ph5GlcT (AB027455); Pf5GlcT (AB013596); Vh5GlcT (AB013598); At7GlcT (NM_129234, At2g36790); At7RhaT (NM_100480, At1g06000); DbB5GlcT (Y18871); NtIS5a (AF346431); Gt3′GlcT (AB076697); Sb7GlcT (AB031274); MtUGT72L1 (EU434684); UGT707B1, CsaF3G2″GlcT (HE793682); OsCGT (FM179712); Cm1,2RhaT, CmF7G2″RhaT (AY048882); BpA3G2″GlcAT (AB190262); CaUGT3, F3G6″GlcT (AB443870); Cs1,6RhaT, CsiF7G6″RhaT (DQ119035); F3GGT1, AcA3Ga2″XylT (FG404013); AtA3G2″XylT, UGT79B1 (NM_124785, At5g54060); AtF3G2″GlcT, UGT79B6 (NM_124780, At5g54010); IpA3G2″GlcT (AB192315); PhA3G6″RhaT, UGT79G16 (Z25802). A3G, anthocyanin 3-O-glucoside; A3Ga, anthocyanin 3-O-galactoide; F3G, flavonol 3-O-glucoside; F7G, flavonoid 7-O-glucoside; 3AraT, 3-O-arabinosyltransferase; 3GlcT, 3-O-glucosyltransferase; 3′GlcT, 3′-O-glucosyltransferase; 3GalT, 3-O-galactosyltransferase; 3RhaT, 3-O-rhamnosyltransferase; 5GlcT, 5-O-glucosyltransferase; 7GlcT, 7-O-glucosyltransferase; 7RhaT, 7-O-rhamnosyltransferase; 2″GlcT, 2″-O-glucosyltransferase; 2″RhaT, 2″-O-rhamnosyltransferase; 2″XylT, 2″-O-xylosyltransferase; 6″RhaT, 6″-O-rhamnosyltransferase; CGT, C-glucosyltransferase; NtIS5a, salicylate-induced glucosyltransferase. Abbreviations for species: Ac, Actinidia chinensis; At, Arabidopsis thaliana; Bp, Bellis perennis; Cm, Citrus maxima; Csa, Crocus sativus; Csi, Citrus sinensis; Db, Dorotheanthus bellidiformis; Gt, Gentiana triflora; Hv, Hordeum vulgare; Ip, Ipomoea purpurea; Nt, Nicotiana tabacum; Os, Oryza sativa; Pf, Perilla frutescens; Ph, Petunia hybrida; Sb, Scutellaria baicalensis; Vh, Verbena hybrida; Vv, Vitis vinifera; Zm, Zea mays.
UGT79B6 encodes flavonoid 3-O-glucoside:2″-O-glucosyltransferase
To identify the function of UGT79B6, TILLING lines with point mutations in the coding region of UGT79B6 were screened using the services of the Seattle Tilling Project (http://tilling.fhcrc.org/) (Till et al., 2003). No sequence-indexed insertion mutants in which tag sequences were inserted into exon or intron regions were available from public resource centers. Of 49 TILLING alleles with point mutations in the coding region of UGT79B6, three alleles harbored nonsense mutations, resulting in a truncated UGT79B6 protein (W324stop, W118stop and W321stop in ugt79b6-1, ugt79b6-2 and ugt79b6-3, respectively) (Figure4). Thirteen of the mutations were silent, and 33 alleles contained missense mutations. Three mutant lines, ugt79b6-1, ugt79b6-2 and ugt79b6-3, were self-crossed and genotyped to produce homozygous plants. We excluded ugt79b6-2 from further analysis because it has a relatively low germination rate (41.6%) and a morphological floral aberration that results in sterility unlinked to UGT79B6 mutations. Morphological aberrations were also observed in flowers of some ugt79b6-1 progeny, but the phenotype was not linked with UGT79B6 mutations. The morphological aberrations in the flowers of ugt79b6-1 and ugt79b6-2 may be due to additional EMS-induced mutations in other loci. We therefore used ugt79b6-1 and ugt79b6-3 for further analyses. No apparent differences in the phenotype of mature pollen grains of wild-type and ugt79b6 mutants were observed by scanning electron microscopy (Figure4).
Figure 4.
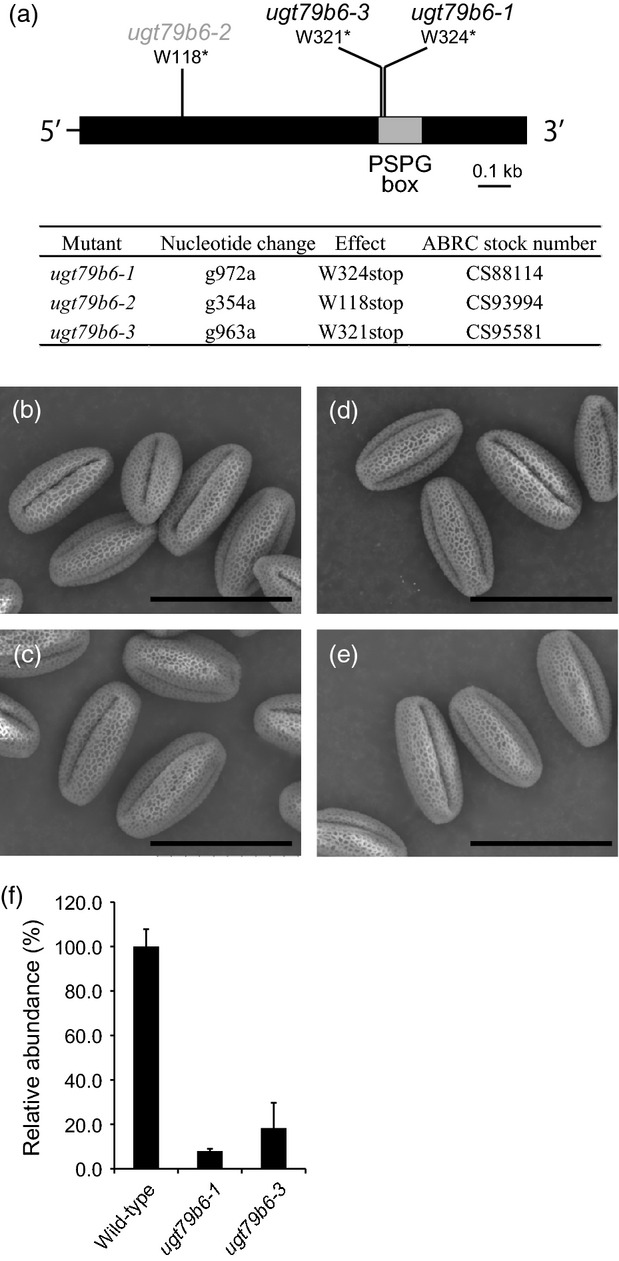
UGT79B6 mutants.(a) Schematic representation of UGT79B6 with three EMS-induced mutations. The box indicates the coding region, and the thinner line indicates the 5′ and 3′ untranslated regions. UGT79B6 has no introns. The gray box indicates the plant secondary product glycosyltransferase (PSPG) box.(b–e) Scanning electron micrographs of mature pollen grains of wild-type (Col-0, b), tt4 (c), and the ugt79b6 mutants ugt79b6-1 (d) and ugt79b6-3 (e). Scale bars = 30 μm.(f) Real-time PCR analysis of UGT79B6 transcripts in flowers of wild-type and ugt79b6 mutants. Values are means. Error bars represent the SD of three repetitions per sample.
The flavonoid profiles in flowers of wild-type, ugt79b6-1 and ugt79b6-3 plants were determined by UPLC/PDA/ESI/Q-TOF/MS (Figure5). Flavonols f21 and f26 were absent from the flowers of both homozygous lines. ugt79b6-3 plants were transformed with genomic clones of UGT79B6 (pKYS388 and pKYS389, 2.1 kb/3.4 kb UGT79B6 genomic fragments containing 683 or 2000 bp of the promoter region, respectively) to complement the ugt79b6-3 mutation. Independent transgenic lines of both genomic clones had essentially the same flavonoid profiles as wild-type (Figure5). The flavonol profile of ugt79b6-1 plants and transgenic lines expressing pKYS388/pKYS389 were essentially identical to those of ugt79b6-3 (Figure S5). These data indicate that UGT79B6 encodes a flavonoid 3-O-glucoside:2″-O-glucosyltransferase in vivo. They also suggest that the 2.1 kb UGT79B6 genomic fragment containing 683 bp of the promoter region, which starts just behind the stop codon of the adjacent At5g54020, is sufficient for functional complementation. Using the New PLACE database (https://sogo.dna.affrc.go.jp/cgi-bin/sogo.cgi?page=analysis&lang=en), predicted MYB binding sites (MYB1AT, MYBCORE, MYBCOREATCYCB1, MYBGAHV and MYBST) were found in the UGT79B6 promoter region (Table S2).
Figure 5.
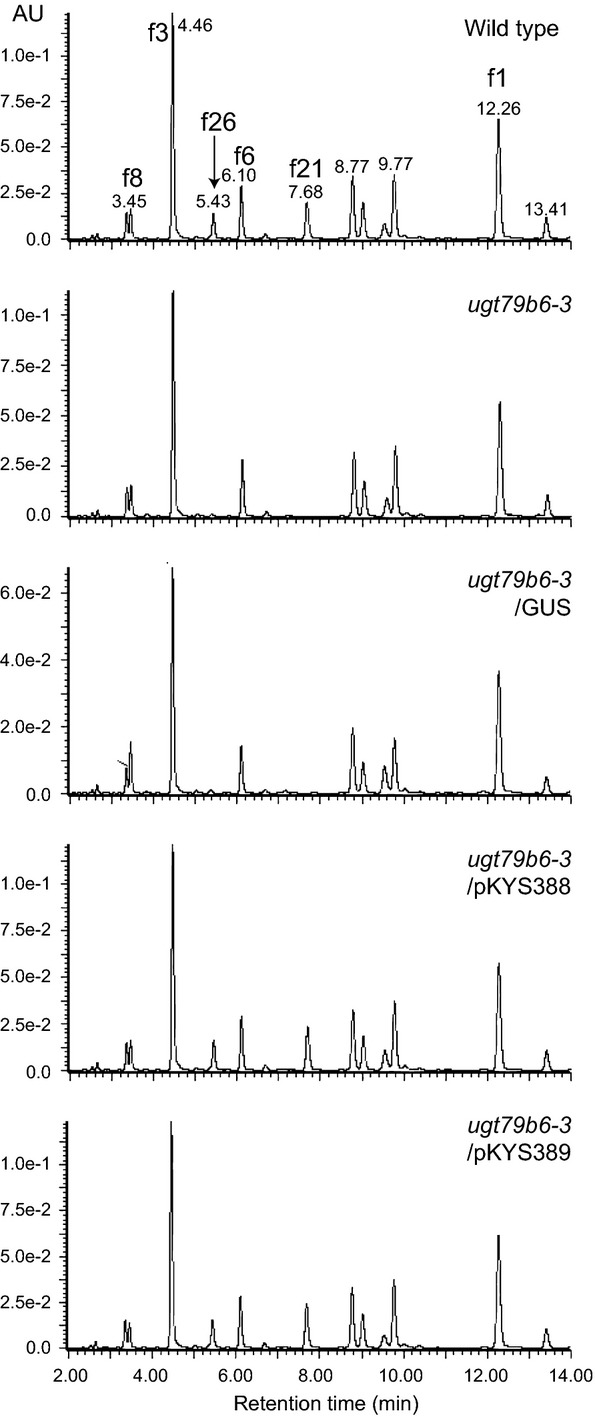
UPLC-PDA-MS analyses of the ugt79b6 mutant lines.Flavonol composition of flowers of wild-type (Col-0), a ugt79b6-deficient mutant (ugt79b6-3), a ugt79b6-deficient mutant complemented with GUS (ugt79b6–3/GUS) and ugt79b6-deficient mutants complemented with 2.1 kb/3.4 kb genomic UGT79B6 clones (ugt79b6-3/pKYS388 and ugt79b6-3/pKYS389, respectively). Labels correspond to the compounds shown in Figure S1.
Real-time PCR showed that accumulation of UGT79B6 transcripts in flowers of ugt79b6-1 and ugt79b6-3 mutants decreased to 8 and 18% of that in wild-type, respectively (Figure4). Stop codon-inserted transcripts were detected to a lesser extent in both ugt79b6 mutants compared to wild-type.
In vitro characterization of recombinant UGT79B6
Recombinant UT79B6 protein was expressed in Escherichia coli as a His/ProS2 tag-fused protein. After cleavage of the His/ProS2 tag, recombinant UT79B6 protein was used for enzymatic assays. The UGT79B6 protein catalyzed conversion of kaempferol 3-O-glucoside to a single product, kaempferol 3-O-glucosyl-(1→2)-glucoside, as confirmed by comparison of retention time and MS spectra with the standard compound (Figures6 and S6). The His/ProS2 tag alone as a negative control did not catalyze conversion to the 2″-O-glucoside. Thus, UGT79B6 may be defined as a flavonol 3-O-glucoside:2″-O-glucosyltransferase.
Figure 6.
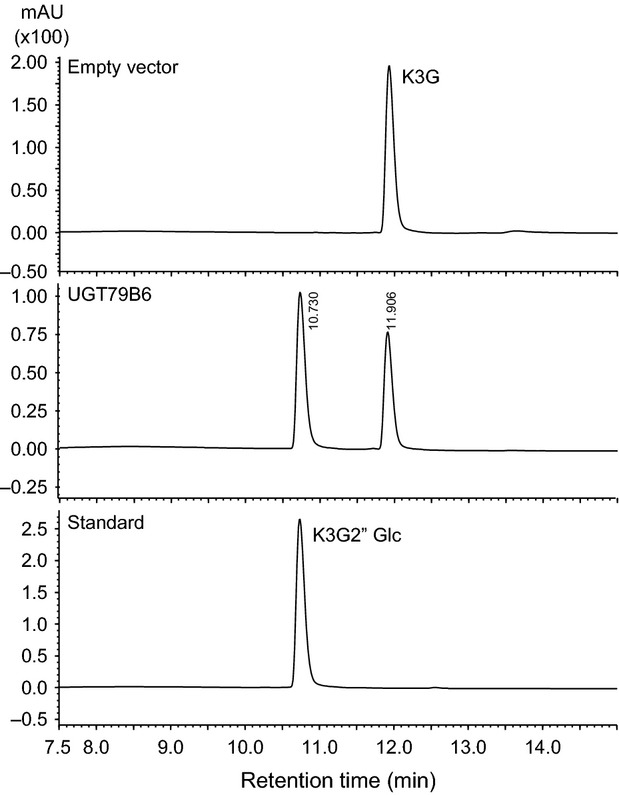
HPLC analyses of the reaction products of the UGT79B6 recombinant protein.Elution profile of reaction products of the His/ProS2 tag protein (empty vector) and the UGT79B6 protein (UGT79B6), and for standard (kaempferol 3-O-glucosyl-(1→2)-glucoside). K3G, kaempferol 3-O-glucoside; K3G2″Glc, kaempferol 3-O-glucosyl-(1→2)-glucoside.
The specificity of UGT79B6 as a sugar acceptor was also examined. UGT79B6 showed significant activity for 3-O-glucoside derivatives of flavonols and anthocyanins (Table1). To a lesser extent, UGT79B6 utilizes quercetin 3-O-galactoside as a sugar acceptor. Interestingly, UGT79B6 glucosylates kaempferol, quercetin and cyanidin 3-O-rhamnosyl-(1→6)-glucosides but not kaempferol 3-O-glucoside-7-O-rhamnoside or cyanidin 3-O-glucoside-5-O-glucoside, suggesting that glycosylation at C-5 or C-7 may occur after full modification at C-3.
Table 1.
Substrate specificity of UGT79B6 from Arabidopsis thaliana
| Relative activity (%) | |
|---|---|
| Sugar acceptora | |
| Kampferol (Kae) | ND |
| Kae 3-O-glucosideb | 100.0 ± 3.6 |
| Kae 3-O-rhamnosyl(1→6)glucoside | 104.9 ± 1.6 |
| Kae 3-O-glucoside-7-O-rhamnoside | 0.2 ± 0.0 |
| Kae 3-O-rhamnoside | ND |
| Quercetin (Que) | ND |
| Que 3-O-glucoside | 410.0 ± 41.5 |
| Que 3-O-rhamnosyl(1→6)glucoside | 289.9 ± 23.1 |
| Quercetin 3-O-galactoside | 20.3 ± 1.6 |
| Isorhamnetin 3-O-galactoside | 67.5 ± 7.7 |
| Cyanidin (Cya) 3-O-glucoside | 44.4 ± 0.7 |
| Cya 3-O-rhamnosyl(1→6)glucoside | 43.0 ± 2.9 |
| Cya 3-O-glucoside-5-O-glucoside | ND |
| Sugar donor | |
| UDP-glucose | 100.0 ± 0.7 |
| UDP-galactose | 5.1 ± 0.9 |
| UDP-rhamnose | ND |
| UDP-xylose | ND |
| UDP-arabinose | ND |
| UDP-glucuronic acid | ND |
ND, not detected.
The reactions were performed with UDP-glucose as the sugar donor.
The enzymatic products were identified based on comparisons with the standards.
The reactions were performed with kaempferol 3-O-glucoside as the sugar acceptor.
The sugar donor specificity of UGT79B6 was examined using UDP-glucose, UDP-galactose, UDP-rhamnose, UDP-xylose, UDP-arabinose and UDP-glucuronic acid as donors, and kaempferol 3-O-glucoside as the acceptor (Table1). No UGT activity was detected for UDP-sugars other than UDP-glucose and UDP-galactose. UGT79B6 has a much higher preference for UDP-glucose than for UDP-galactose, with only 5% activity relative to that for UDP-glucose.
Expression of UGT79B6 is specific to the tapetum and microspores
The accumulation of UGT79B6 transcripts in Arabidopsis organs was measured by real-time PCR. UGT79B6 transcripts were particularly abundant in floral buds but were nearly undetectable in stems and siliques (Figure7). This accumulation pattern was consistent with that described in the Arabidopsis eFP browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), in which UGT79B6 is expressed almost exclusively in early developmental stages of pollens (Figure S4).
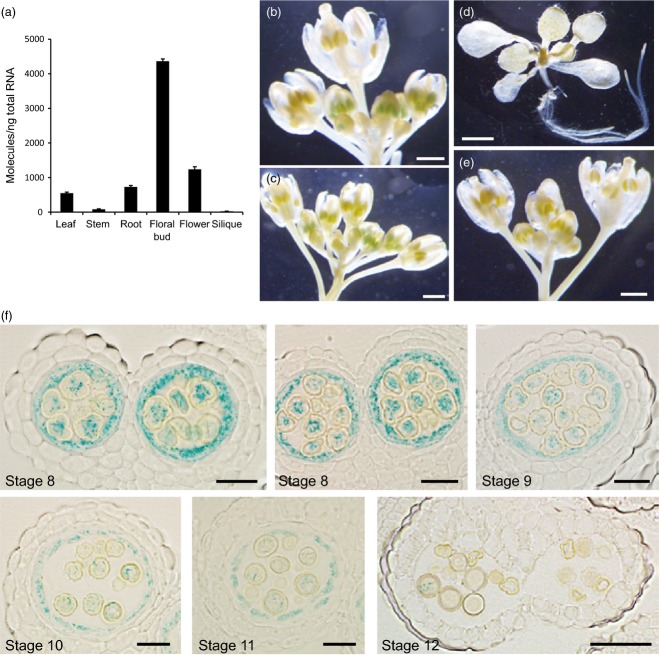
Figure 7.
Expression pattern of UGT79B6.(a) Real-time PCR analysis of UGT79B6 transcripts in organs of the Arabidopsis wild-type (Col-0).(b–d) GUS staining of plants expressing the GUS reporter gene driven by the UGT79B6 promoter (ProUGT79B6:GUS). Scale bars = 500 μm (b,c) and 2 mm (d).(e) GUS staining of inflorescences of wild-type (Col-0). Scale bars = 500 μm.(f) Sections (5 μm) of ProUGT79B6: GUS anthers stained with X-glc. Light microscopy of anthers at stages 8–11 shows tapetum- and microspore-specific expression. Stages of anther development are according to Sanders et al. (1999). Scale bars = 20 μm (stages 8–11) and 50 μm (stage 12).
To identify the tissue/cellular localization of UGT79B6, we generated transgenic lines harboring a 2 kb promoter of UGT79B6 fused to a β-glucuronidase (GUS) reporter gene. GUS expression was observed in young floral buds of transgenic lines, where it was confined to the developing anthers, but it was not evident in non-transgenic plants. No GUS staining was observed in older floral buds, open flowers or vegetative organs of seedlings (Figure7). At the cellular level, blue spotty coloration was observed in the tapetum and microspores of the anthers at developmental stages 8–11 as defined by Sanders et al. (1999) (Figure7). Blue colorization by GUS staining decreased with degeneration of the tapetum, and was not apparent in anthers by developmental stage 12 when the anthers become bilocular. GUS staining in the tapetum is consistent with the finding that flavonoids are present in tapetosomes of tapetum cells, and later on the mature pollen surface of Arabidopsis (Hsieh and Huang, 2007). UGT79B6 expression in microspores is consistent with the transcriptome data on the Arabidopsis eFP browser (Figure S4).
Discussion
Flavonoid biosynthesis and regulation in pollen
We identified the structure of pollen-specific flavonols f26 and f21 as quercetin 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranoside and kaempferol 3-O-β–d-glucopyranosyl-(1→2)-β-d-glucopyranoside, respectively, and revealed that UGT79B6 encodes a flavonol 3-O-glucoside:2″-O-glucosyltransferase, a key enzyme for determining pollen-specific flavonoid structures.
Pollens accumulate flavonols accounting for 2–4% of pollen dry weight in a variety of plant species (Wiermann and Vieth, 1983), and flavonol 3-O-diglucosides with a 1→2 inter-glycosidic linkage are frequently present as major flavonols in pollen (Pratviel and Perchero, 1972; Zerback et al., 1989; Price et al., 1998; Ross et al., 2005), suggesting that flavonol 3-O-glycoside:2″-O-glycosyltransferase is a characteristic UGT enzyme that is widely used for the biosynthesis of pollen-specific flavonols.
In Arabidopsis, flavonoid biosynthetic genes are regulated directly by several R2R3-MYB genes (MYB11/PFG2, MYB12/PFG1 and MYB111/PFG3 for flavonols, MYB75/PAP1, MYB90/PAP2, MYB113 and MYB114 for anthocyanins, MYB123/TT2 for proanthocyanidins, and MYB4 for phenylpropanoids) (Dubos et al., 2010). Interestingly, kaempferol/quercetin 3-O-glucosyl-(1→2)-glucosides (f21 and f26) accumulated as the sole flavonols in pollen of triple myb11/myb12/myb111 knockout mutants (Stracke et al., 2010). These pollen-specific flavonols also accumulated in loss-of-function mutants of other R2R3-MYBs, namely PAP1, PAP2, MYB113, MYB114, TT2 and MYB5 (Stracke et al., 2010), indicating that biosynthesis of pollen-specific flavonols is controlled by an unknown regulatory system, independent of those previously reported in other organs. Functional identification of UGT79B6 paves the way for a deeper understanding of pollen-specific flavonoid biosynthesis, including regulation.
In Arabidopsis, the evolutionary relationships of UGT79 genes are reported (Figure S7) (Wang et al., 2013). UGT79B9 (At5g53990) and UGT79B2 (At4g27560) may be parental loci duplicated by the most recent whole-genome duplication. UGT79B6 shows local proximal duplication relationships with UGT79B9 and UGT79B1 (At5g54060). UGT79B1, an anthocyanin 3-O-glucoside:2″-O-xylosyltransferase, is regulated by MYB genes for anthocyanin biosynthesis (PAP1, PAP2, MYB113 and/or MYB114), suggesting that additional acquisition of the UGT function and the regulatory system is required for the biosynthesis of pollen-specific flavonols.
The PHD-finger class transcription factor MS1 is located upstream in the regulatory cascade, because quercetin/kaempferol 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranosides are absent from ms1 mutants. However, it remains to be determined whether or not MS1 regulates flavonol biosynthesis directly or indirectly, or whether it only regulates the development of tapetum cells for flavonol biosynthesis and/or accumulation.
Diversity of UGTs that glycosylate the sugar moiety attached to aglycones of plant secondary metabolites
UGTs may be classified into 24 orthologous groups (OGs) based on phylogenetic analyses of six land plants (Yonekura-Sakakibara and Hanada, 2011). Flavonoid GGTs belong to either the UGT79 or UGT94 families in orthologous group 8 (OG8) (Figure6), and the function of flavonoid GGTs was most likely established before the divergence of UGT79 and UGT94 in OG8. Flavonoid GGTs in family UGT79 may be divided into two sub-clusters based on the position they glycosylate (i.e. the 2″- or 6″-hydroxy groups of sugar moieties attached to a flavonoid aglycone) (Figure3), suggesting that regio-selectivity in flavonoid GGTs was established before the divergence of plant species in the UGT79 family. Phylogenetic comparisons of flavonoid GGTs suggest that possible conserved amino acid residues are involved in the further regio-selectivity. Four amino acid residues (Ile35, Thr76, Pro81 and Phe339 in UGT79B6) are generally conserved throughout all known flavonoid 3-O-glycoside:2″-O-glycosyltransferases (Figure S8). The close relationship between UGT79B6 and UGT79B1 in the phylogenetic tree also indicates that the UDP-sugar specificity of flavonoid GGTs was established after species differentiation, at least in Arabidopsis, as is the case with flavonoid 3-O-glycosyltransferases.
Relationship of flavonoids to pollen fertility and evolutionary considerations
Flavonoids are known to be involved in pollen fertility. In petunia and maize (Zea mays), mutants deficient in flavonoids are unable to germinate pollen tubes, resulting in male sterility (Coe et al., 1981; Taylor and Jorgensen, 1992). However, Arabidopsis tt4 mutants, which lack chalcone synthase, the first committed enzyme in flavonoid biosynthesis, are fertile, with only a slight reduction in the number of seeds per silique (Burbulis et al., 1996; Ylstra et al., 1996). The present results also indicate that ugt79b6 mutants lacking pollen-specific flavonols are fertile, suggesting that the effect of flavonoids on fertility is not universal among plants, and that other compounds may fill the role of flavonoids in Arabidopsis. As pollens accumulate specific flavonol diglycosides, it is conceivable that these flavonols had characteristic functions during the process of evolution, despite an apparent lack of function in Arabidopsis.
In Brassica napus, flavonoids are detected exclusively in tapetum cells, first in the endoplasmic reticulum network with flavonoid 3′-hydroxylase and then in endoplasmic reticulum-derived tapetosomes. During the degradation of tapetum cells, flavonoids are transferred to the pollen surface (Hsieh and Huang, 2007). The presence of flavonoids in tapetosomes and subsequently on pollen surfaces was also observed in Arabidopsis (Hsieh and Huang, 2007). These reports support our observation that UGT79B6 is localized in tapetum cells and microspores of developing anthers. In petunia, it has been proposed that flavonol aglycones are synthesized in the tapetum, released into the locule, and taken up to be glycosylated in developing pollen grains, and that flavonol 3-O-galactoside:2″-O-glucosyltransferase associates with membranes, because detergents are required for enzyme solubilization (Vogt and Taylor, 1995; Taylor and Hepler, 1997; Xu et al., 1997). It remains to be seen whether or not the temporal and spatial distribution of flavonoid biosynthetic and modification enzymes for pollen-specific flavonoids also affects fertility.
Experimental Procedures
Plant materials
Arabidopsis thaliana accession Columbia-0 (Col-0; Lehle Seeds, http://www.arabidopsis.com/) was used as the wild-type in this study unless otherwise specified. The Arabidopsis TILLING lines CS88114, CS93994 and CS95581 for UGT79B6 (ugt79b6-1, ugt79b6-2 and ugt79b6-3, respectively), were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/). The TILLING mutants are in the Col er105 (Big Mama) background (Torii et al., 1996). The services of the Seattle Tilling Project (http://tilling.fhcrc.org/) were used to screen for lines with point mutations in the coding region of UGT79B6. Specific primers for UGT79B6 (UGT79B6 left and UGT79B6 right, Table S3), yielding a 1274 bp fragment starting 16 bp upstream from the first ATG, were used for screening. Homozygous knockout lines were screened by PCR using primers At5g54010(1f) and At5g54010(1362r) (Table S3). PCR products were sequenced to identify homozygous knockout lines using primer At5g54010(1258r) (Table S3). For phenotypic analyses, we used mutant lines back-crossed with Col–0 for three generations.
Chemicals
Chemicals of the highest grade commercially available were used unless specifically noted. Flavonoid standards were purchased from Extrasynthese (http://www.extrasynthese.com/http://www.extrasynthese.com/), AnalytiCon Discovery (http://www.ac-discovery.com/) and YouChemicals (http://www.youchemicals.com/). UDP-β-l-arabinose and UDP-α-d-xylose were purchased from CarboSource Services (http://www.ccrc.uga.edu/~carbosource/CSS_home.html, supported in part by National Science Foundation/Plant Cell Wall Biosynthesis Research Network grant 0090281).
Phylogenetic analysis
UGT protein sequences were aligned using clustal w implemented in mega5 (version 5.2.2, http://www.megasoftware.net/) (Tamura et al., 2011). A phylogenetic tree was constructed from aligned UGT protein sequences by mega5 using the neighbor-joining method (Saitou and Nei, 1987) with the following parameters: bootstrap method (1000 replicates), Poisson model, uniform rates, and complete deletion.
Flavonoid profiling and untargeted analyses by UPLC/PDA/ESI/Q-TOF/MS
Frozen tissues were homogenized in 5 μl extraction solvent (1:1 methanol/H2O) per mg fresh weight of tissue in a mixer mill (MM300; Retsch, http://www.retsch.com/retsch-international/) for 5 min at 30 Hz. Supernatants were immediately used for analysis after centrifugation at 12 000 g. Flavonol analyses were performed essentially as described previously (Yonekura-Sakakibara et al., 2008). Untargeted metabolome analyses of wild-type (Ler) and ms1 mutants were also performed as described previously (Yonekura-Sakakibara et al., 2008).
Isolation of flavonols from Arabidopsis flower buds
Flower buds (25.5 g fresh weight) were harvested at stages 6.00–6.50 (Boyes et al., 2001), flash-frozen in liquid nitrogen, and immediately extracted with methanol. After concentration in a rotary evaporator, the sample was sequentially extracted using n-hexane and then CHCl3 to remove non-polar components. After liquid–liquid partitioning and concentration, the MeOH phase (approximately 5 ml) was fractionated by open column chromatography using a Cosmosil 75C18-OPN column (column diameter 3.5 cm, length 5 cm; Nacalai Tesque, http://www.nacalai.co.jp/english/index.html), and separated by elution with a gradient of H2O and MeOH with the following elution profile (fraction A, 0% MeOH; fraction B, 10% MeOH; fraction C, 20% MeOH; fraction D, 30% MeOH; fraction E, 40% MeOH; fraction F, 50% MeOH; fraction G, 60% MeOH; fraction H, 100% MeOH; elution solvent, 30 ml per fraction) to give eight fractions. To trace target flavonols, HPLC/PDA/ESI/MS was performed on an Agilent HPLC 1100 series (Agilent Technologies, http://www.home.agilent.com/agilent/home.jspx) using an Atlantis® ODS column (column diameter 4.6 mm, length 250 mm; Waters, http://www.waters.com/waters/home.html) and a Finnigan LCQ-DECA mass spectrometer (ThermoQuest, http://www.thermoscientific.com/en/home.html) (Tohge et al., 2005). Next, fraction D (24.6 mg) was purified by preparative HPLC using an LC 10A system (Shimadzu,http://www.shimadzu.com/) with an Inertsil prep-ODS column (column diameter 10 mm, length 150 mm) at 30°C, with a linear gradient over 60 min from 0% solvent B (90% CH3CN and 10% H2O with 0.1% trifluoroacetic acid) in solvent A (10% CH3CN and 90% H2O with 0.1% trifluoroacetic acid) to 100% solvent B, at a flow rate of 2 ml min−1, to yield f21 (2.5 mg) and f26 (2.5 mg).
Structural identification of f21 and f26
Kaempferol 3-O-β-d-glucopyranosyl-(1→2)-β-d-glucopyranoside (f21) was identified on the basis of the following data: yellow amorphous solid; [α]D20 = 63.4° (c. 0.1, MeOH); UV λmax (MeOH) = 204.5, 266.5, 343; Orbitrap MS m/z 609.1464 ([M+H]+, calculated for C27H29O16, 609.1461); 1H NMR (CD3OD) δ = 6.19 (1H, d, J = 2.0 Hz, H-6), 6.39 (1H, d, J = 2.0 Hz, H-8), 8.04 (2H, d, J = 8.8 Hz, H-2′ and H-6′), 6.90 (2H, d, J = 8.8 Hz, H-3′ and H-5′) (kaempferol: H-6 and H-8, H-2′ to H-6′), 5.41 (1H, d, J = 7.5 Hz, Glc H-1″), 3.72 (1H, m, Glc H-2″), 3.58 (1H, m, Glc H-3″), 3.35 (1H, m, Glc H-4″), 3.18 (1H, m, Glc H-5″), 3.51 (1H, m, Glc H-6a″), 3.67 (1H, m, Glc H-6b″), 4.75 (1H, d, J = 7.2 Hz, Glc H-1‴), 3.35 (1H, m, Glc H-2‴), 3.36 (1H, m, Glc H-3‴), 3.35 (1H, m, Glc H-4‴), 3.38 (1H, m, Glc H-5‴), 3.68 (1H, m, Glc H-6a‴), 3.78 (1H, m, Glc H-6b‴); 13C NMR (CD3OD) δ = 158.5, 135.0, 179.0, 163.1, 99.9, 166.2, 94.8, 158.9, 105.7, 122.8, 132.4, 116.3, 161.6, 116.3, 132.4 (kaempferol: C-2 to C-10 and C-1′ to C-6′); 101.1, 82.6, 78.3, 71.3, 78.2, 62.5 (glucose: C-1″ to C-6″); 104.8, 75.6, 77.9, 71.1, 77.9, 62.4 (glucose: C-1‴ to C-6‴). Rotating-frame Overhauser Enhancement (ROE) correlation from the anomeric proton (glucose H-1‴, δ 4.75) to the proton of C-2″) position of glucose (H-2″, δ 3.72) was observed.
Quercetin 3-O-β-d-glucopryanosyl-(1→2)-β-d-glucopyranoside (f26) was identified on the basis of the following data: yellow amorphous solid; [α]D20 = 60.8° (c. 0.03, MeOH); UV λmax (MeOH) = 203.5, 257, 355; Orbitrap MS m/z 625.1416 ([M+H]+, calculated for C27H29O17, 625.1410); 1H NMR (CD3OD) δ = 6.20 (1H, br. s, H-6), 6.39 (1H, br. s, H-8), 7.67 (1H, d, J = 2.2 Hz, H-2′), 7.53 (1H, dd, J = 8.5, 2.2 Hz, H-5′), 6.88 (1H, d, J = 8.3 Hz, H-6′) (quercetin: H-6, H-8, H-2′, H-5′ and H-6′), 5.34 (1H, d, J = 7.7 Hz, Glc H-1″), 3.77 (1H, m, Glc H-2″), 3.58 (1H, m, Glc H-3″), 3.39 (1H, m, Glc H-4″), 3.19 (1H, m, Glc H-5″), 3.52 (1H, m, Glc H-6a″), 3.69 (1H, m, Glc H-6b″), 4.76 (1H, d, J = 7.2 Hz, Glc H-1‴), 3.38 (1H, m, Glc H-2‴), 3.40 (1H, m, Glc H-3‴), 3.39 (1H, m, Glc H-4‴), 3.32 (1H, m, Glc H-5‴), 3.69 (1H, m, Glc H-6a‴), 3.82 (1H, m, Glc H-6b‴); 13C NMR (CD3OD) δ = 158.5, 135.1, 179.8, 163.1, 99.8, 165.9, 94.6, 158.9, 105.8, 123.0, 117.7, 146.0, 149.8, 116.1, 121.7 (quercetin: C-2 to C-10 and C-1′ to C-6′); 101.2, 83.0, 77.9, 71.0, 78.3, 62.4 (glucose: C-1″ to C-6″); 105.0, 75.6, 77.9, 70.9, 78.0, 62.3 (glucose: C-1‴ to C-6‴). ROE correlation from the anomeric proton (glucose H-1″), δ 4.76) to the proton of C-2″ position of glucose (H-2‴), δ 3.77) was observed.
Evaluation of TILLING lines
For complementation tests, 3.4 and 2.1 kb genomic fragments covering 2000 and 683 bp of the promoter region, the entire UGT79B6 coding region, and 70 bp of the 3′ non-coding region were amplified by PCR using primers 5g54010-21936975r and 5g54010GW21939089f or 5g54010GW21940406f (Table S3). Amplified fragments were cloned into the pENTR/D-TOPO vector (Invitrogen, http://www.lifetechnologies.com/jp/en/home/brands/invitrogen.html) as entry vector, and sequenced to confirm the absence of PCR errors. pGWB1 was used as the destination vector, and the LR reactions for the binary vectors pKYS388 and pKYS389 were catalyzed using Gateway LR Clonase™ II enzyme mix (Invitrogen). pKYS388 (pGWB1/2.1 kb UGT79B6 genomic fragment) and pKYS389 (pGWB1/3.4 kb UGT79B6 genomic fragment) were transformed into Agrobacterium tumefaciens GV3101(pMP90), and Arabidopsis plants were transformed by the floral-dip method (Clough and Bent, 1998). Transgenic T2 plants were selected on half-strength Murashige & Skoog medium containing 50 mg L−1 kanamycin sulfate.
Quantitative real-time PCR
RNA extraction and cDNA synthesis were performed as described previously (Yonekura-Sakakibara et al., 2004). The developmental stage of each organ used for analysis was as described previously (Yonekura-Sakakibara et al., 2007). Accumulation of UGT79B6 transcripts was measured by real-time PCR with a StepOnePlus Real-Time PCR system using SYBR Green Master Mix (Applied Biosystems, http://www.lifetechnologies.com/jp/en/home/brands/applied-biosystems.html). The primers UGT79B6-1134F and UGT79B6-1195R (Table S3) were designed using Primer Express software (Applied Biosystems) and checked for specific product formation using a dissociation program. Plasmid DNA containing the corresponding gene was used as a template to generate a calibration curve. Real-time PCR was performed in triplicate on a single biological sample.
Production of recombinant UGT79B6 protein and glycosyltransferase assays
Full-length UGT79B6 was amplified by PCR using primers At5g54010_pColdProS2-5 and At5g54010_pColdProS2-3 to construct a protein expression vector (Table S3). The PCR product was cloned into pColdProS2 vector using an In-Fusion Advantage PCR cloning kit (Clontech, http://www.clontech.com). The nucleotide sequence of the resulting plasmid, pKYS448, was sequenced to confirm the absence of PCR errors.
Escherichia coli strain BL21star™ (DE3) was used as a host for expression. Transformed cells were cultivated at 37°C until an absorbance at 600 nm of 0.5. After addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mm, cells were cultured at 15°C for 24 h. The cells were collected by centrifugation, and the protein was purified as a His fusion using TALON® metal affinity resin (Clontech) according to the manufacturer's instructions. The ProS2 tag was removed using HRV3C protease (Novagen, www.novagen.com) according to the manufacturer's instructions. After exchanging the buffer for 50 mm HEPES–KOH, pH 7.5, proteins were concentrated using an Amicon Ultra filter (10 000 molecular weight cut-off; Millipore, http://www.merckmillipore.com/US/en).
The standard enzyme assay reaction mixture was described previously (Yonekura-Sakakibara et al., 2012). The mixture was pre-incubated at 30 °C for 2 min, and the reaction was started by addition of enzyme. Reactions were stopped after 0, 2, 4, 6, 60 or 80 min of incubation at 30°C by addition of 50 μl ice-cold 0.5% v/v trifluoroacetic acid/methanol for flavonols or 50 μl ice-cold 0.5% v/v HCl/methanol for anthocyanins. Supernatants were recovered by centrifugation at 12 000 g for 3 min. Flavonols in the resulting solution were analyzed using a Shimadzu HPLC system with a Unison UK–C18 column (column diameter 2.0 mm, length 150 mm, particle size 3 μm, Imtakt, http://www.imtaktusa.com/) at a flow rate of 0.2 ml min−1 at 35°C. The compounds were separated using a linear eluting gradient comprising solvent A (0.1% trifluoroacetic acid in water) and solvent B (0.1% trifluoroacetic acid in acetonitrile) according to the following profile: 0 min, 10% B; 3 min, 10% B; 18 min, 75% B; 18.01 min, 95% B; 20 min, 95% B; 30 min, 95% B. Anthocyanins in the resulting solution were analyzed using a Shimadzu HPLC system with a Unison UK–C18 column as above at a flow rate of 0.2 ml min−1 at 35°C. Compounds were separated using a linear eluting gradient comprising solvent A (0.5% trifluoroacetic acid in water) and solvent B (0.5% trifluoroacetic acid in acetonitrile) according to the following profile: 0.0 min, 16% B; 15.0 min, 16% B; 15.5 min, 100% B; 20.0 min, 100% B; 20.5 min, 16% B; 32.0 min, 16% B. PDA was used for detection of UV-visible absorption in the range 200–600 nm.
Scanning electron microscopy
Pollen grains of Arabidopsis were directly observed using a scanning electron microscope (TM–1000; Hitachi, http://www.hitachi-hitec.com/global/index.html).
Generation and analysis of GUS reporter lines
The 2000 and 683 bp fragments of the UGT79B6 promoter region were amplified by PCR using primers At5g54010promoter-R and At5g54010promoter-2000 or At5g54010promoter-683 (Table S3). Amplified fragments were cloned into the pENTR/D–TOPO vector (Invitrogen) as entry vector, and sequenced to confirm the absence of PCR errors. pBGGUS (Funakoshi, http://www.funakoshi.co.jp/export/index.php) was used as the destination vector, and the LR reactions for the binary vectors pKYS452 and pKYS451 were catalyzed using Gateway LR Clonase™ II enzyme mix (Invitrogen). pKYS452 (pBGGUS/2000 bp of the UGT79B6 promoter region) and pKYS451 (pBGGUS/683 bp of the UGT79B6 promoter region) were transformed into Agrobacterium and subsequently into Arabidopsis plants as described above. Transgenic T2 plants were selected on half-strength Murashige & Skoog medium containing 50 mg L−1 carbenicillin and 50 μm glufosinate ammonium.
For histochemical GUS assays, tissues were treated with ice-cold 90% acetone for 15 min on ice, and stained in 100 mm sodium phosphate buffer, pH 7.0, 10 mm EDTA, 0.1% Triton X-100, 0.5 mg ml−1 5-bromo-4-chloro-3-indolyl glucuronide, 5 mm potassium ferricyanide. After 15 min vacuum infiltration, samples were incubated overnight at 37°C and subsequently de-stained by a series of washes in 70% ethanol. Digital images were captured using a stereoscopic microscope (Leica MZ10F, http://www.leica-microsystems.com/), and processed using Adobe Photoshop (https://www.adobe.com/). For sectioning after GUS staining, tissue samples were fixed in 5% v/v acetic acid/45% v/v ethanol/5% v/v formaldehyde, dehydrated for 20 min using 50, 60, 70, 80, 90, 99.5 and 100% v/v ethanol sequentially, and then incubated in Technovit 7100 resin (Heraeus Kulzer, http://heraeus-kulzer.com/en/int/home_4/home_5.aspx) containing a 1:1 v/v ratio of Hardener I (Heraeus Kulzer) to ethanol at room temperature, 100% Technovit 7100 resin, and Technovit 7100 resin containing a 15:1 v/v ratio of Hardener I:Hardener II (Heraeus Kulzer). Tissue sections were sliced into 5 μm transverse sections using a rotary microtome (Leica RM2135) and tungsten carbide disposable blades (Leica TC–65). Sections were observed using an Olympus (http://www.olympus-global.com/en/) BX53 microscope.
Acknowledgments
We greatly appreciate T. Nakagawa (Shimane Universtiy) for kindly providing pGWB1 vector and T. Furuhashi, K. Kawade, K. Toyooka and M. Sato (RIKEN Center for Sustainable Resource Science) for their kind advice on microscopy analysis. We also thank M. Suzuki and Z. Yang (RIKEN Center for Sustainable Resource Science) for technical assistance with MS and NMR, respectively. This study was supported, in part, by JSPS KAKENHI program (grant numbers 22108008 to K.S. and 25440148 to K.Y.–S.), Japan Advanced Plant Science Network, and JST Strategic International Collaborative Research Program (SICORP).
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure S1. Flavonol glycosides in Arabidopsis.
Figure S2. UPLC/PDA/MS analyses.
Figure S3. UPLC/PDA/MS analyses of extracts from flowers of wild-type (Col-0 and Ler) and ms1 mutants.
Figure S4. Output image from the Arabidopsis eFP browser showing the expression pattern of At5g54010 in the developmental series.
Figure S5. UPLC/PDA/MS analyses of ugt79b6-1 mutant lines.
Figure S6. The MS/MS spectrum of the product catalyzed by UGT79B6 is identical to that of the standard kaempferol 3-O-glucosyl-(1→2)-glucoside (K3G2″Glc).
Figure S7. Modes of gene duplication in the Arabidopsis UGT79 sub-family.
Figure S8. Multiple alignment of flavonoid UGTs catalyzing glycosyl transfer to a sugar moiety of flavonoid glycosides.
Table S1. List of compounds missing from the ms1 mutants.
Table S2. Predicted MYB binding sites in 683 bp of the UGT79B6 promoter region.
Table S3. Primers used in this study.
Appendix S1. The alignment used for construction of the phylogenetic tree shown in Figure 3.
References
- Alves-Ferreira M, Wellmer F, Banhara A, Kumar V, Riechmann JL, Meyerowitz EM. Global expression profiling applied to the analysis of Arabidopsis stamen development. Plant Physiol. 2007;145:747–762. doi: 10.1104/pp.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson ØM, Markham KR. Flavonoids: Chemistry, Biochemistry and Applications. Boca Raton, FL: CRC Press; 2006. [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sex. Plant Reprod. 2005;18:1–7. [Google Scholar]
- Bar-Peled M, Lewinsohn E, Fluhr R, Gressel J. UDP-rhamnose:flavanone-7–O-glucoside-2″–O-rhamnosyltransferase. Purification and characterization of an enzyme catalyzing the production of bitter compounds in citrus. J. Biol. Chem. 1991;266:20953–20959. [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Gorlach J. Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugliera F, Holton TA, Stevenson TW, Farcy E, Lu CY, Cornish EC. Isolation and characterization of a cDNA clone corresponding to the Rt locus of Petunia hybrida. Plant J. 1994;5:81–92. doi: 10.1046/j.1365-313x.1994.5010081.x. [DOI] [PubMed] [Google Scholar]
- Burbulis IE, Iacobucci M, Shirley BW. A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell. 1996;8:1013–1025. doi: 10.1105/tpc.8.6.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coe EH, Mccormick SM, Modena SA. White pollen in maize. J. Hered. 1981;72:318–320. [Google Scholar]
- Debeaujon I, Nesi N, Perez P, Devic M, Grandjean O, Caboche M, Lepiniec L. Proanthocyanidin-accumulating cells in Arabidopsis testa: regulation of differentiation and role in seed development. Plant Cell. 2003;15:2514–2531. doi: 10.1105/tpc.014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Fellenberg C, van Ohlen M, Handrick V, Vogt T. The role of CCoAOMT1 and COMT1 in Arabidopsis anthers. Planta. 2012;236:51–61. doi: 10.1007/s00425-011-1586-6. [DOI] [PubMed] [Google Scholar]
- Frydman A, Liberman R, Huhman DV, Carmeli-Weissberg M, Sapir-Mir M, Ophir R, Sumner LW, Eyal Y. The molecular and enzymatic basis of bitter/non-bitter flavor of citrus fruit; evolution of branch-forming rhamnosyltransferases under domestication. Plant J. 2013;73:166–178. doi: 10.1111/tpj.12030. [DOI] [PubMed] [Google Scholar]
- Gould KS, Lister C. Flavonoid functions in plants. In: Anderson ØM, Markham KR, editors. Flavonoids: Chemistry, Biochemistry and Applications. Boca Raton, FL: CRC Press; 2006. pp. 397–442. [Google Scholar]
- Grotewold E. The Science of Flavonoids. New York, NY: Springer; 2006. [Google Scholar]
- Hsieh K, Huang AH. Tapetosomes in Brassica tapetum accumulate endoplasmic reticulum-derived flavonoids and alkanes for delivery to the pollen surface. Plant Cell. 2007;19:582–596. doi: 10.1105/tpc.106.049049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Shinozaki K. The MALE STERILITY1 gene of Arabidopsis, encoding a nuclear protein with a PHD-finger motif, is expressed in tapetal cells and is required for pollen maturation. Plant Cell Physiol. 2002;43:1285–1292. doi: 10.1093/pcp/pcf154. [DOI] [PubMed] [Google Scholar]
- Ito T, Nagata N, Yoshiba Y, Ohme-Takagi M, Ma H, Shinozaki K. Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell. 2007;19:3549–3562. doi: 10.1105/tpc.107.054536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Kroon J, Souer E, de Graaff A, Xue Y, Mol J, Koes R. Cloning and structural analysis of the anthocyanin pigmentation locus Rt of Petunia hybrida: characterization of insertion sequences in two mutant alleles. Plant J. 1994;5:69–80. doi: 10.1046/j.1365-313x.1994.5010069.x. [DOI] [PubMed] [Google Scholar]
- Markham KR. Distribution of flavonoids in the lower plants and its evolutionary significance. In: Harborne JB, editor. The Flavonoids. Advances in Research Since 1980. London: Chapman and Hall; 1988. pp. 427–468. [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis. Plant J. 2011;65:106–118. doi: 10.1111/j.1365-313X.2010.04409.x. [DOI] [PubMed] [Google Scholar]
- Morita Y, Hoshino A, Kikuchi Y, et al. Japanese morning glory dusky mutants displaying reddish-brown or purplish-gray flowers are deficient in a novel glycosylation enzyme for anthocyanin biosynthesis, UDP-glucose:anthocyanidin 3–O-glucoside-2″–O-glucosyltransferase, due to 4–bp insertions in the gene. Plant J. 2005;42:353–363. doi: 10.1111/j.1365-313X.2005.02383.x. [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Kusano M, Kobayashi M, Tohge T, Yonekura-Sakakibara K, Kogure N, Yamazaki M, Kitajima M, Saito K, Takayama H. Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana. Phytochemistry. 2009;70:1017–1029. doi: 10.1016/j.phytochem.2009.03.021. [DOI] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I. TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell. 2005;17:2966–2980. doi: 10.1105/tpc.105.035154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratviel F, Perchero F. Sophorosides of flavonols of some pollens. Phytochemistry. 1972;11:1809–1813. [in French] [Google Scholar]
- Price KR, Casuscelli F, Colquhoun IJ, Rhodes MJC. Composition and content of flavonol glycosides in broccoli florets (Brassica olearacea) and their fate during cooking. J. Sci. Food Agric. 1998;77:468–472. [Google Scholar]
- Rausher MD. The evolution of flavonoids and their genes. In: Grotewold E, editor. The Science of Flavonoids. New York, NY: Springer; 2006. pp. 175–211. [Google Scholar]
- Richardson PM. Flavonoids of the fern allies. Biochem. Syst. Ecol. 1989;17:155–160. [Google Scholar]
- Ross SA, ElSohly MA, Sultana GNN, Mehmedic Z, Hossain CF, Chandra S. Flavonoid glycosides and cannabinoids from the pollen of Cannabis sativa L. Phytochem. Anal. 2005;16:45–48. doi: 10.1002/pca.809. [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Kerhoas L, Debeaujon I, Pourcel L, Caboche M, Einhorn J, Lepiniec L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta. 2006;224:96–107. doi: 10.1007/s00425-005-0197-5. [DOI] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR. The flavonoid biosynthetic pathway in Arabidopsis: structural and genetic diversity. Plant Physiol. Biochem. 2013;72:21–34. doi: 10.1016/j.plaphy.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999;11:297–322. [Google Scholar]
- Sawada S, Suzuki H, Ichimaida F, Yamaguchi MA, Iwashita T, Fukui Y, Hemmi H, Nishino T, Nakayama T. UDP-glucuronic acid:anthocyanin glucuronosyltransferase from red daisy (Bellis perennis) flowers. Enzymology and phylogenetics of a novel glucuronosyltransferase involved in flower pigment biosynthesis. J. Biol. Chem. 2005;280:899–906. doi: 10.1074/jbc.M410537200. [DOI] [PubMed] [Google Scholar]
- Shirley BW. Flavonoid biosynthesis: ‘New’ functions for an ‘old’ pathway. Trends Plant Sci. 1996;1:377–382. [Google Scholar]
- Stafford HA. Flavonoid evolution: an enzymic approach. Plant Physiol. 1991;96:680–685. doi: 10.1104/pp.96.3.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B. Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol. 2010;188:985–1000. doi: 10.1111/j.1469-8137.2010.03421.x. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK. Pollen germination and tube growth. Annu. Rev. Plant Physiol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- Taylor LP, Jorgensen R. Conditional male-fertility in chalcone synthase-deficient petunia. J. Hered. 1992;83:11–17. [Google Scholar]
- Till BJ, Reynolds SH, Greene EA, et al. Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 2003;13:524–530. doi: 10.1101/gr.977903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, et al. Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 2005;42:218–235. doi: 10.1111/j.1365-313X.2005.02371.x. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapero A, Ahrazem O, Rubio-Moraga A, Jimeno ML, Gomez MD, Gomez-Gomez L. Characterization of a glucosyltransferase enzyme involved in the formation of kaempferol and quercetin sophorosides in Crocus sativus. Plant Physiol. 2012;159:1335–1354. doi: 10.1104/pp.112.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T, Taylor LP. Flavonol 3–O-glycosyltransferases associated with petunia pollen produce gametophyte-specific flavonol diglycosides. Plant Physiol. 1995;108:903–911. doi: 10.1104/pp.108.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YP, Tan X, Paterson AH. Different patterns of gene structure divergence following gene duplication in Arabidopsis. BMC Genomics. 2013;14:652. doi: 10.1186/1471-2164-14-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiermann R, Vieth K. Outer pollen wall, an important accumulation site for flavonoids. Protoplasma. 1983;118:230–233. [Google Scholar]
- Williams CA, Grayer RJ. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004;21:539–573. doi: 10.1039/b311404j. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- Xu P, Vogt T, Taylor LP. Uptake and metabolism of flavonols during in vitro germination of Petunia hybrida (L) pollen. Planta. 1997;202:257–265. [Google Scholar]
- Yang C, Vizcay-Barrena G, Conner K, Wilson ZA. MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell. 2007;19:3530–3548. doi: 10.1105/tpc.107.054981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylstra B, Muskens M, VanTunen AJ. Flavonols are not essential for fertilization in Arabidopsis thaliana. Plant Mol. Biol. 1996;32:1155–1158. doi: 10.1007/BF00041399. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Hanada K. An evolutionary view of functional diversity in family 1 glycosyltransferases. Plant J. 2011;66:182–193. doi: 10.1111/j.1365-313X.2011.04493.x. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Kojima M, Yamaya T, Sakakibara H. Molecular characterization of cytokinin-responsive histidine kinases in maize. Differential ligand preferences and response to cis-zeatin. Plant Physiol. 2004;134:1654–1661. doi: 10.1104/pp.103.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Niida R, Saito K. Identification of a flavonol 7–O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 2007;282:14932–14941. doi: 10.1074/jbc.M611498200. [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell. 2008;20:2160–2176. doi: 10.1105/tpc.108.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Fukushima A, Nakabayashi R, Hanada K, Matsuda F, Sugawara S, Inoue E, Kuromiri T, Ito T, Shinozaki K, Wangwattana B, Yamazaki M, Saito K. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012;69:154–167. doi: 10.1111/j.1365-313X.2011.04779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerback R, Bokel M, Geiger H, Hess D. A kaempferol 3–glucosylgalactoside and further flavonoids from pollen of Petunia hybrida. Phytochemistry. 1989;28:897–899. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Flavonol glycosides in Arabidopsis.
Figure S2. UPLC/PDA/MS analyses.
Figure S3. UPLC/PDA/MS analyses of extracts from flowers of wild-type (Col-0 and Ler) and ms1 mutants.
Figure S4. Output image from the Arabidopsis eFP browser showing the expression pattern of At5g54010 in the developmental series.
Figure S5. UPLC/PDA/MS analyses of ugt79b6-1 mutant lines.
Figure S6. The MS/MS spectrum of the product catalyzed by UGT79B6 is identical to that of the standard kaempferol 3-O-glucosyl-(1→2)-glucoside (K3G2″Glc).
Figure S7. Modes of gene duplication in the Arabidopsis UGT79 sub-family.
Figure S8. Multiple alignment of flavonoid UGTs catalyzing glycosyl transfer to a sugar moiety of flavonoid glycosides.
Table S1. List of compounds missing from the ms1 mutants.
Table S2. Predicted MYB binding sites in 683 bp of the UGT79B6 promoter region.
Table S3. Primers used in this study.
Appendix S1. The alignment used for construction of the phylogenetic tree shown in Figure 3.