Abstract
Recombination in meiosis is a fascinating case study for the coordination of chromosomal duplication, repair, and segregation with each other and with progression through a cell-division cycle. Meiotic recombination initiates with formation of developmentally programmed DNA double-strand breaks (DSBs) at many places across the genome. DSBs are important for successful meiosis but are also dangerous lesions that can mutate or kill, so cells ensure that DSBs are made only at the right times, places, and amounts. This review examines the complex web of pathways that accomplish this control. We explore how chromosome breakage is integrated with meiotic progression and how feedback mechanisms spatially pattern DSB formation and make it homeostatic, robust, and error-correcting. Common regulatory themes recur in different organisms or in different contexts in the same organism. We review this evolutionary and mechanistic conservation but also highlight where control modules have diverged. The framework that emerges helps explain how meiotic chromosomes behave as a self-organizing system.
Keywords: Spo11, DNA double-strand breaks, cell cycle, ATM, DNA replication
INTRODUCTION
Meiosis is the specialized cell division that generates gametes in sexually reproducing organisms. It appends two rounds of chromosome segregation to one round of DNA replication, thereby achieving the necessary genome reduction prior to gamete fusion, which restores proper ploidy (117) (Figure 1a). The second meiotic division is like mitosis in that it separates centromeres of sister chromatids, but the first meiotic division is different: It separates homologous maternal and paternal chromosomes instead. Meiosis I poses unique challenges because homologous chromosomes need not share any special spatial relationship before meiosis, unlike sister chromatids, which are born alongside one another when DNA is replicated. To segregate accurately, homologous chromosomes must find one another, pair up, and form temporary physical connections that stabilize them on the metaphase I spindle.
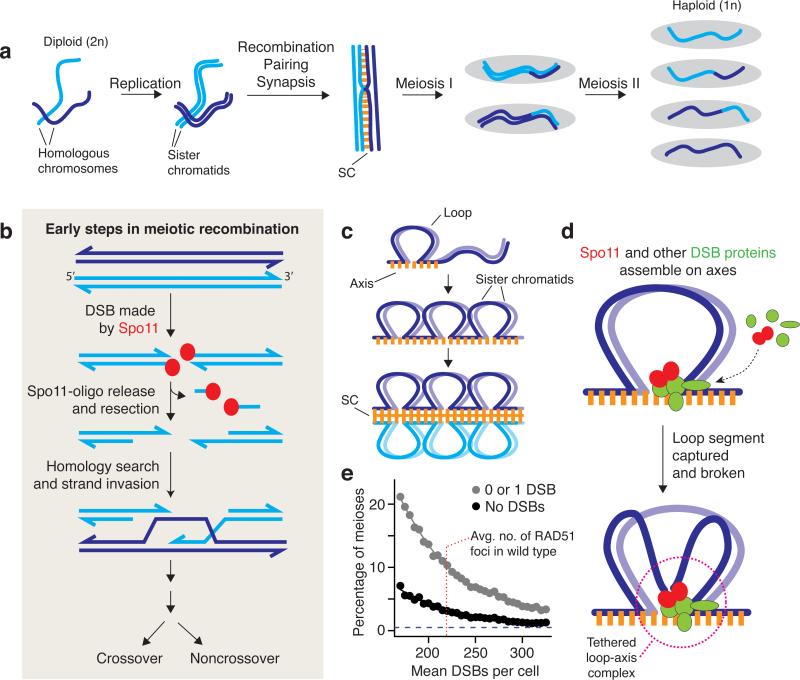
Figure 1.
Chromosome behaviors in meiosis. (a) Stages in the progression through meiosis. Homologous chromosomes undergo replication to form pairs of sister chromatids, which are held together by cohesin (not shown). Replicated chromosomes initiate recombination, then pair and become closely juxtaposed along their lengths. The aligned configuration is transiently stabilized by the synaptonemal complex (SC). A subset of recombination events are resolved by reciprocal exchange of chromosome arms (crossing over), which, in conjunction with sister chromatid cohesion, provide physical connections between homologous chromosomes that allow them to be aligned on the spindle at metaphase of the first meiotic division. Meiosis I separates homologous chromosomes, then Meiosis II separates sister centromeres. The two divisions yield progeny with half the genetic content of the parent and with new combinations of parental alleles in some of the meiotic products because of crossing over. (b) Overview of early steps in meiotic recombination. Recombination is initiated by a double-strand break catalyzed by the topoisomerase-like protein Spo11. The DNA cleavage reaction leaves a pair of Spo11 molecules attached to the 5′ DNA ends on either side of the DSB. Endonucleolytic cleavage releases Spo11 attached to a short oligonucleotide, and further exonucleolytic resection generates 3′ single-stranded tails that are bound by strand-exchange proteins (Dmc1 and/or Rad51; not shown). Once a homologous DNA duplex is located, the ssDNA tail invades the intact homologous duplex. Further DNA transactions (not pictured) give rise to mature recombination products in which homologs have exchanged arms (crossover) or not (noncrossover). (c) Loop-axis structure of meiotic chromosomes. Early in prophase of Meiosis I, each pair of sister chromatids develops a proteinaceous axis with chromatin extending out in loops. As prophase proceeds, the axes elongate and axes of homologous chromosomes are brought together and joined via the zipper-like SC. (d) Model for integration of DSB formation with loop-axis organization of chromosomes. Recombination occurs in spatial proximity to axes, but DSBs usually form in DNA thought to be in the chromatin loops. To reconcile this apparent paradox, it has been proposed that most of the DSB-forming machinery--- including Spo11 itself---assembles on chromosome axes and then captures and breaks a DNA segment from a nearby loop, forming a tethered loop-axis complex (15, 76, 120). (e) Monte Carlo simulations were used to evaluate whether randomly distributing DSBs among chromosomes could support the efficient pairing and synapsis seen in normal mouse spermatocytes [<0.5% of cells with an unsynapsed pair of autosomes (e.g., 68)]. We varied the mean number of DSBs per cell and mimicked natural cell-to-cell fluctuation estimated from variability in numbers of RAD51 foci (219.2 ± 69.8, mean ± SD, from 36). For each value of the mean, we simulated a population of 10,000 cells with that mean and with a coefficient of variation of 30%. For each simulated cell, DSBs were then randomly distributed among the 19 pairs of autosomes, with each chromosome weighted in proportion to its axis length (from 45). The figure plots the fraction of simulated cells in which at least one chromosome pair had no DSBs (black points) or had one or no DSBs (gray), as such chromosomes would probably fail to synapse (68). The results indicate that a random distribution could not provide the very low failure rate seen in normal cells (horizontal dashed line), even if DSB numbers were much higher than the wild-type average.
In most species, the physical connections are formed by reciprocal exchange of chromosome arms via homologous recombination in conjunction with sister chromatid cohesion (117). Recombination also fosters genetic diversification by breaking up linkage groups. In many taxa, including fungi, plants, and mammals, it promotes chromosome pairing by providing a mechanism for identifying DNA sequence homology (14). Recombination failure often leads to meiotic arrest or chromosome segregation failure, with dire consequences for fertility (109, 117).
Recombination initiates with DNA double-strand breaks (DSBs), which cells inflict on their own genomes (39, 72) (Figure 1b). Major steps in the recombination pathway are best defined in the budding yeast Saccharomyces cerevisiae, but conservation of key players indicates that many of the events proceed in similar fashion in different species (39). DSBs are formed by Spo11, a conserved topoisomerase relative that cleaves DNA via a covalent protein-DNA intermediate (12, 73). This intermediate is then endonucleolytically cleaved to release Spo11 attached to a short oligonucleotide (oligo) (73, 98, 110). Because Spo11-oligo complexes are a quantitative by-product of DSB formation---each DSB results in the generation of two SPO11-oligo complexes---they have proven useful in quantifying total DSB levels, even in organisms like mice, where direct molecular detection of DSBs is thus far not possible (e.g., 78).
Endonucleolytic release of Spo11-oligo complexes frees DSB ends so that the 5′ strand termini can be exonucleolytically resected to yield 3′ single-stranded tails. These tails invade intact homologous duplexes in reactions dependent on strand-exchange proteins related to bacterial RecA (Rad51 and, in some species, its meiosis-specific paralog Dmc1), ultimately giving rise to recombinant products (61). The repair of any given DSB can result in either the reciprocal exchange of chromosome arms flanking the break (a crossover) or no exchange of flanking arms (a noncrossover) (Figure 1b). The crossovers help link homologous chromosomes on the metaphase I spindle, but all interhomolog recombination events (including those leading to noncrossovers) promote pairing in those organisms that rely on recombination for this process.
Recombination is closely integrated with the development of meiosis-specific higher-order chromosome structures (76). Early in prophase I, sister chromatids develop a proteinaceous axis (the axial element), with chromatin extending out in loops (Figure 1c). As chromosomes pair, their axes align and are held together to form the zipper-like synaptonemal complex (SC). The tripartite SC comprises the juxtaposed chromosome axes plus central region components including transverse filaments (coiled-coil proteins spanning the gap between axes). The SC has as-yet poorly understood roles in promoting completion of recombination and may also foster exchange of chromosome axes at crossover sites, modulate sister chromatid cohesion, and/or sense and help resolve instances where nonhomologous chromosomes have become topologically intertwined (known as interlocks) (76). Direct cytological visualization shows that recombination protein complexes reside on chromosome axes, but molecular studies in yeast place the most frequently cleaved DNA sequences on chromatin loops; this paradox has led to the proposal that DSBs are formed within loop segments that become transiently tethered to chromosome axes (15, 76) (Figure 1d). Because Spo11 and its accessory factors are also enriched on chromosome axes, it is thought that DSB machinery assembled on axes captures and breaks loop segments (1, 76, 99, 120, 146). In fungi, plants, and mammals, DSBs form at approximately the same time as axes are forming, and recombination is completed within the context of the SC (39, 61, 76).
The positive roles DSBs play in promoting normal meiotic chromosome behavior come with risk because errors in DSB repair can lead to mutation, cell death, aneuploid gametes, and/or infertility (57, 109, 134). The potentially lethal nature of these lesions puts a premium on the cell's ability to control the timing, number, and location of DSBs to foster their essential functions and minimize deleterious effects. Here, we review recent discoveries that illuminate the molecular underpinnings of this control.
RECOMBINATION INITIATION IS A ROBUST, SELF-ORGANIZING PROCESS
DSBs are more likely to occur in some genomic regions than in others (10, 69, 82). Depictions of this nonrandom distribution often focus on hot spots, the small regions (typically ~150--250 base pairs wide in budding yeast) where DSBs occur most often. However, hot spots are only one aspect of the DSB distribution because essentially every base pair in the genome is a potential substrate for Spo11, with cleavage probability varying over orders of magnitude (119). The shape of this probability distribution, i.e., the DSB landscape, is molded by the combinatorial action of many factors (chromosomal proteins and the DNA they bind) that interact hierarchically over different size scales (69, 76, 82, 119, 157). The number of potential break sites is thus enormous, so each cell ends up with a different array of DSB positions that are chosen on the fly as meiosis proceeds. Likewise, the exact final number of DSBs is also not genetically predetermined and varies substantially from cell to cell (e.g., with a coefficient of variation of >30% in mouse) (33, 36).
Despite these stochastic aspects, DSB formation is robust, homeostatic, and error-correcting (27, 48, 68, 156). The geneticist's stock-in-trade is analysis of how things go wrong in mutants, but of course the implicit starting point for most studies is the fact that things generally go right in wild type. For example, the great majority of cells achieve a sufficient number of DSBs that are distributed appropriately so that each chromosome pair has enough for at least one crossover to form (33, 64, 97, 128). Moreover, in organisms with recombination-promoted pairing, sufficient DSBs nearly always form to support that process as well (55, 68, 155). Simple modeling suggests that randomly distributing DSBs among and along chromosomes would not achieve such a high success rate (Figure 1e), implying the existence of mechanisms that control DSB number and distribution to ensure proper chromosome behavior.
Although there is substantial cell-to-cell variation of DSB numbers within species, greater differences are seen when comparing between organisms. Relatively few DSBs occur in species that can pair chromosomes without recombination, such as Drosophila melanogaster and Caenorhabditis elegans (~20--30 per cell on average) (62, 96, 111, 131). In contrast, more DSBs tend to be generated in organisms that rely on recombination for efficient pairing, such as S. cerevisiae (average of ~150--200 per cell), plants (e.g., ~200--300 in Arabidopsis thaliana and >1,500 per cell in lily), or mammals (e.g., ~200--300 per cell in mouse) (25, 31, 36, 119, 123, 154, 162). These apparent species-specific set points, without genetic (or epigenetic) predetermination of precise numbers, imply that DSB regulation is self-organizing and homeostatic, which also provides potential for robustness and error correction (27, 68, 156).
DSB formation is a suicide reaction for Spo11 because the endonucleolytic release pathway leaves the protein's active-site tyrosine residue covalently linked to DNA (Figure 1b). In principle, this feature could be a means of controlling total DSB numbers, but in fact most Spo11 protein never makes a break (78, 110) and Spo11 and other proteins essential for DSB formation remain abundant on chromatin after most DSBs have formed (72). These features suggest that mechanisms that control DSB numbers work in part by restraining Spo11 activity.
Understanding of these DSB-regulating mechanisms has grown substantially in recent years. Studies in several species have uncovered elements that integrate DSB formation with progression through meiosis and that coordinate DSB formation with other chromosomal events (e.g., DNA replication). Recent studies have also revealed an intersecting network of negative feedback circuits that work locally along chromosomes to fine-tune the control of DSBs. In its broad framework, DSB regulation appears to be evolutionarily conserved, but many details differ strikingly between organisms. Figure 2 summarizes the known regulatory circuits, which affect DSB formation to different degrees depending on where the cell is within S phase or prophase. Each circuit is discussed in detail below.
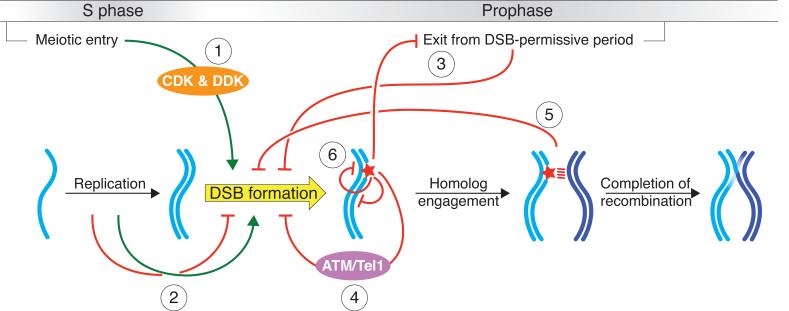
Figure 2.
Overview of the network of intersecting regulatory circuits controlling timing, number, and distribution of meiotic double-strand breaks (DSBs). Circuit 1: Cell cycle regulatory kinases tie DSB formation to meiotic progression. Circuit 2: DNA replication influences the spatial and temporal patterning of DSBs (green arrow), and replication stress inhibits DSB formation (red bar-headed arrow). Circuit 3: Progression through prophase closes a window of opportunity for DSB formation. Problems in recombination and/or certain other chromosome behaviors invoke signaling pathways that extend the DSB- permissive period. Circuit 4: DSBs activate the damage-responsive kinase ATM/Tel1, which then restrains SPO11 activity via a negative feedback loop. Circuit 5: Engagement of homologous chromosomes leads to changes in chromosome structure that inhibit further DSB formation. Circuit 6: Local DSB patterning is shaped by communication between potential DSB sites, both in cis along the same DNA molecule and in trans between sister chromatids or homologous chromosomes.
CELL CYCLE KINASES TIE DOUBLE-STRAND BREAKS TO MEIOTIC PROGRESSION
It is clear that DSB formation is usually restricted to a specific window of time during the first meiotic prophase, based on direct detection of DSBs in yeasts and on immunostaining to detect cytological DSB markers such as Rad51 foci in other organisms (e.g., 30, 35, 62, 100, 116, 162). This constrained window is important for recombination to serve its functions in connecting homologous chromosomes. For example, recombination must be integrated with sister chromatid cohesion to form the chiasmata that hold [**ED: Note, “chiasmata” is plural.**]chromosome pairs together at metaphase I (76). Proper timing is probably also important to minimize potential for genomic havoc: DSBs formed before DNA replication or after commitment to chromosome segregation may put cells at risk of mutation, aneuploidy, or meiotic arrest (57, 103).
One layer of temporal control involves developmentally regulated expression of Spo11 and other proteins required to make DSBs. Different species have evolved a range of strategies to restrict expression of these and other meiotic proteins to appropriate times, including control of transcription, splicing, mRNA stability, and translation (e.g., 23, 51, 89, 137). This type of control (gene regulation tied to differentiation itself), although important, is not considered further except for a few specific scenarios in budding and fission yeasts. Instead, we focus here and in subsequent sections on other layers of temporal control that involve more direct regulation of the activity or chromosomal association of Spo11 and its accessory factors.
The cell cycle regulatory kinases CDK (cyclin-dependent kinase) and DDK (Dbf4-dependent kinase) are key drivers of progression in meiosis, as in mitosis (90, 91). In S. cerevisiae, DSB formation is directly promoted by both kinases. Cdc28 (the principal cell cycle CDK), in association with the S-phase cyclins Clb5 or Clb6, phosphorylates the Spo11-accessory protein Mer2; this phosphorylation is essential for DSB formation (54, 145). Mer2 must also be phosphorylated by DDK, which comprises the kinase Cdc7 and its regulatory subunit Dbf4. DDK directly phosphorylates multiple sites on Mer2, some of which depend on prior phosphorylation of a neighboring residue by CDK (91, 135, 163, 164). Phosphorylation by both kinases apparently promotes the ability of Mer2 to interact with other proteins needed for DSB formation and thereby to recruit those proteins to chromatin (54, 120, 135).
Hsk1, the Schizosaccharomyces pombe ortholog of Cdc7, is essential for DSB formation and recruitment of the Spo11 ortholog Rec12 to chromatin (112, 135). Rec7 [a homolog of the Spo11-accessory protein Rec114 required for DSB formation in budding yeast (83, 99)] is phosphorylated by Hsk1 and phosphorylation-blocking rec7 mutations reduce recombination, making Rec7 a likely target of Hsk1 relevant to DSB control (H. Masai, personal communication). Whether other targets exist is not yet known.
In mice, normal CDC7 levels are required for meiosis (75) but whether the kinase functions in DSB formation or some other process is unknown, and meiotic analyses of DDK homologs have not been reported in other taxa. Furthermore, although CDKs or cyclins have been clearly implicated in recombination and/or other aspects of meiotic chromosome dynamics in many organisms (e.g., 6, 161), it has not yet been established whether DSB formation itself is controlled by CDK in species other than S. cerevisiae.
COORDINATING DOUBLE-STRAND BREAKS WITH DNA REPLICATION
To fulfill their functions in promoting pairing, generating connections between homologous chromosomes, and transmitting a haploid DNA content to gametes, DSBs need to form at a time when sister chromatids exist, i.e., after DNA replication has occurred locally. Cell-wide oscillation of CDK or DDK activity provides one means to control the timing of these meiotic events, but such global regulation does not by itself allow replication and DSB formation to be fully coordinated with one another. Studies in budding and fission yeast have uncovered paradigmatic mechanisms that provide this coordination and allow for error correction.
Temporospatial Coordination in Normal Meiosis
In S. cerevisiae, DSBs usually form approximately 90 minutes after replication (18). Pioneering studies by Lichten's group uncovered the remarkable finding that delaying replication of a chromosomal segment by deleting replication origins also delays DSB formation in that segment by the same margin (18). In strains heterozygous for the origin deletions, DSBs are delayed only on the mutated chromosome, so this temporal control of DSBs works in cis (102). Because DSB timing is dictated by local replication timing, replication and recombination initiation must be mechanistically coupled to one another (18, 102, 103).
A possibly related phenomenon occurs in S. pombe (167). If sporulation proceeds in the presence of a nitrogen source rather than in commonly used nitrogen-starvation conditions, more replication origins are utilized and relative replication times change for large swathes of the genome. How nitrogen levels effect this alteration is not known, but one consequence is clear: Regions that shift to earlier replication also display an increase in DSB formation.
One way coupling could occur is if replication is a strict prerequisite for DSBs (18, 145). However, Spo11 efficiently breaks chromosomes that remain unduplicated because the replication initiation factor Cdc6 has been depleted in S. cerevisiae (16, 58) or because of hydroxyurea treatment or replication factor depletion in checkpoint-defective S. pombe mutants (106, 112, 158). Thus, replication is dispensable for DSBs per se.
Instead, it has been proposed that temporospatial coupling of replication and DSB formation in S. cerevisiae operates at least in part by recruitment of DDK to the replication machinery, thereby preferentially targeting Mer2 in replicating regions for phosphorylation (103, 177) (Figure 3a). Mer2 binds chromatin independently of phosphorylation (54, 120) and DDK activity is limiting early in meiosis (91, 163). These features create a window of opportunity where selective targeting of DDK to replicating chromatin could confer a head start toward DSB formation. Supporting this model, replication-DSB coordination is eliminated by overexpressing DDK (177). Coordination is also eliminated by removing the replication fork protection complex (FPC) (177), a group of proteins that travels with replisomes and helps stabilize replication machinery during replicative stress (94). The FPC physically associates with DDK in budding and fission yeast and becomes dispensable for replication-DSB coordination if DDK is artificially tethered to replisomes (92, 140, 177).These and other findings indicate that DDK recruited by FPC to replisomes phosphorylates Mer2 in the wake of the replication fork, thus synchronizing replication with an early prerequisite for DSB formation. It remains to be seen whether S. pombe uses a similar mechanism.
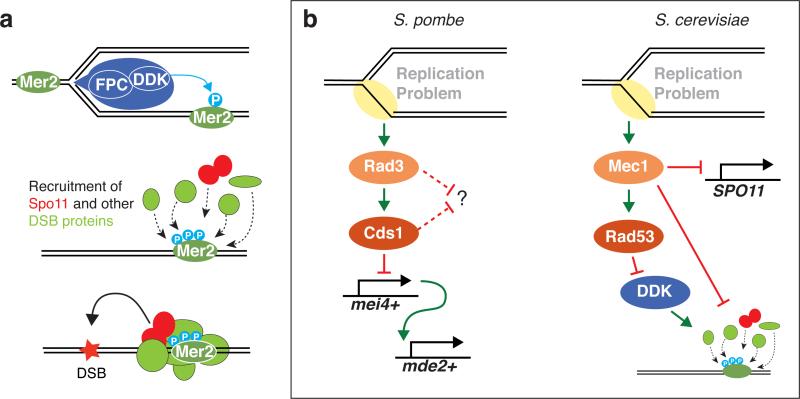
Figure 3.
Coordinating replication and double-strand break (DSB) formation. (a) Model for temporospatial coupling of DSBs with replication in Saccharomyces cerevisiae. The graphic at top depicts a replication fork with a replisome (blue) consisting of the replication machinery and accessory proteins, such as the fork protection complex (FPC). DDK (Dbf4- dependent kinase) is recruited to the replisome via interaction with the FPC, resulting in preferential phosphorylation of the Mer2 protein bound to replicating chromatin. Mer2 phosphorylation promotes recruitment of Rec114 and other DSB proteins, including Spo11, ultimately resulting in DSB formation. (b) Downregulation of DSB formation in the face of replication stress. In Schizosaccharomyces pombe, replication defects activate the kinase Rad3 (ortholog of mammalian ATR) and its effector kinase Cds1. By an unknown mechanism, these kinases inhibit the transcription of mei4+, which encodes a transcription factor that is needed for expression of mde2+, which encodes a protein essential for DSB formation. Whether Rad3 and Cds1 also suppress DSB formation via additional pathways is unknown. In S. cerevisiae, replication defects activate the Rad3 ortholog Mec1, which impinges on SPO11 transcription by an unknown mechanism. Mec1, via its effector kinase Rad53, also inhibits DDK, which likely inhibits recruitment of DSB proteins dependent on Mer2 phosphorylation (as depicted in panel a). Mec1 also appears to inhibit chromatin recruitment of the DSB-promoting proteins Rec114 and Mre11 by a separate, undefined pathway.
Responding to Replication Stress
Cells also coordinate replication and DSB formation by downregulating DSB machinery in the face of replication problems. In S. pombe meiosis, inhibiting replication with hydroxyurea invokes cellular responses via activation of the DNA damage response kinase Rad3 (the ortholog of mammalian ATR) and its downstream effector kinase Cds1 (104, 105). These responses include inhibition of DSB formation, attributed to Rad3- and Cds1-dependent inhibition of transcription of the mei4+ and mde2+ genes (99, 113) (Figure 3b, left). Mei4 is a Forkhead-like transcription factor required for meiosis-specific expression of a number of meiotic genes, including mde2+ (49, 60). Mde2 has essential functions in DSB formation: It bridges interactions between other DSB-promoting proteins and integrates DSB formation with higher-order chromosome structure (49, 99). It is not yet known how Rad3 and Cds1 activation impinges on mei4+ transcription or whether inhibition of Mei4 expression is the sole means by which the replication checkpoint inhibits DSB formation. Interestingly, artificial expression of Mde2 is not sufficient to rescue DSB formation in the presence of hydroxyurea, suggesting there are other critical Mei4-dependent targets or that Rad3 and Cds1 have additional means of inhibiting DSB formation (K. Ohta, personal communication).
Hydroxyurea treatment also blocks DSB formation in S. cerevisiae via Mec1 (ortholog of S. pombe Rad3 and mammalian ATR) (17) (Figure 3b, right). Analogous to fission yeast, replication checkpoint activation inhibits expression of a DSB-promoting protein, but in budding yeast it is SPO11 transcription that is targeted, and only partially. As in S. pombe, the mechanism of transcription inhibition is unknown. Importantly, however, Mec1 and its effector kinase Rad53 (ortholog of S. pombe Cds1) also inhibit DDK by phosphorylating Dbf4, thereby preventing Mer2 phosphorylation and reducing or altering chromatin association of several DSB-promoting factors. Replication stress thus downregulates Spo11 activity through multiple intersecting mechanisms; this DSB inhibition may promote genome stability by preventing formation of DSBs ahead of replication forks (17). In principle, this mode of DSB regulation could contribute to spatially patterned coordination with replication. For example, ongoing replication could set up a nucleus-wide block to DSB formation that is then removed locally in conjunction with replication fork passage (57). However, it has been argued that this model does not account for properties of the FPC in replication-DSB coordination discussed above (177). Further studies are needed to determine the extent to which Mec1- and Rad53-dependent processes contribute to DSB control when S phase is unperturbed.
CLOSING THE WINDOW OF OPPORTUNITY FOR DOUBLE-STRAND BREAK FORMATION
Restricting Spo11 activity to a narrow window of prophase I requires that cells control the end of the window, not just the beginning. Insight into the regulatory systems involved has come from studies in S. cerevisiae, D. melanogaster, and C. elegans.
Regulated Exit from Meiotic Prophase in Saccharomyces cerevisiae
In budding yeast, exit from the pachytene stage of prophase is controlled by the Ndt80 transcription factor, which activates expression of more than 200 genes, including those encoding the polo-like kinase Cdc5 and M-phase cyclins Clb1 and Clb3 (34, 147, 169). Recombination products and DSBs accumulate in mutants that lack Ndt80 or that have defects in pachytene exit because of attenuated CDK activity (2, 142, 169). These observations led to the proposal that pachytene exit ends a period permissive for DSB formation, i.e., that Ndt80 acts as an indirect negative regulator of Spo11 activity (2, 54, 71) (Figure 4a). Recent studies confirmed this hypothesis by providing multiple lines of evidence that ndt80 mutants make more DSBs (5, 27, 48, 128, 156).
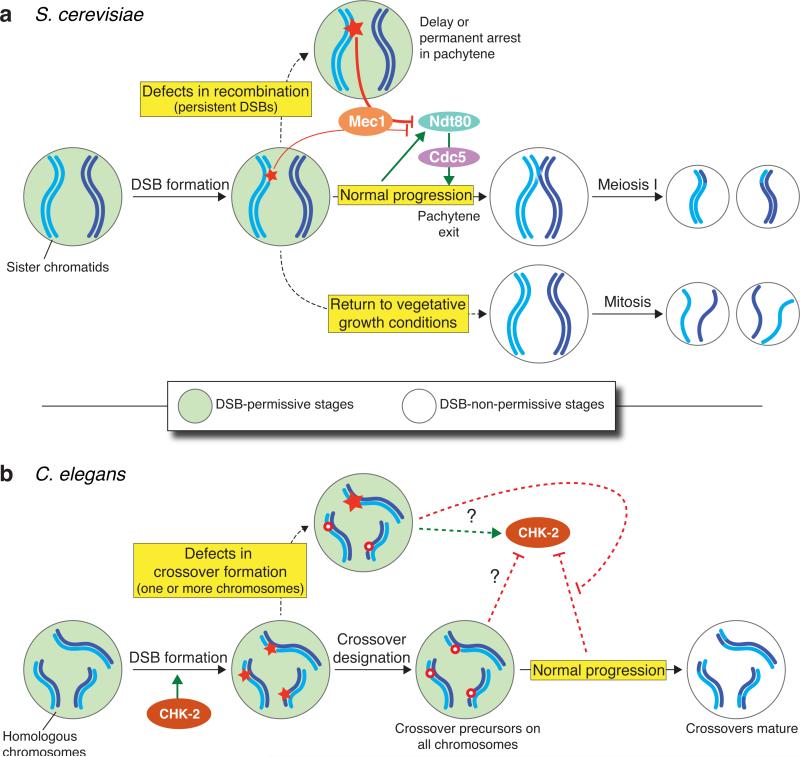
Figure 4.
Analogous but mechanistically distinct nucleus-wide systems in Saccharomyces cerevisiae and Caenorhabditis elegans link the potential to form double-strand breaks (DSBs) with meiotic progression. (a) In S. cerevisiae, normal progression through meiosis witnesses activation of Ndt80, a transcription factor that governs exit from the pachytene stage of prophase. Ndt80 activation leads to synaptonemal complex (SC) disassembly and removal of DSB-promoting proteins from chromosomes, ending a DSB-permissive period as cells commit to meiotic divisions. The gene encoding the polo-like kinase Cdc5 is an Ndt80 target that may be important for downregulating DSB potential. If recombination defects result in persistent DSBs, a response mediated by the kinase Mec1 (ATR) is provoked; this attenuates Ndt80 activity and delays or blocks pachytene exit. Cross talk between ongoing recombination and Ndt80 activation also occurs via Mec1 to control the length of prophase in normal cells. If cells are exposed to appropriate nutrients before Ndt80 becomes active, they exit meiosis and resume mitotic divisions. As in normal meiotic progression, this mode of exiting prophase also appears to include nucleus-wide loss of DSB potential. (b) In C. elegans, a DSB-permissive state is marked by active CHK-2 kinase and by chromosomal association of the DSB-1 and DSB-2 proteins (not shown). A coordinated transition during the pachytene stage results in shutdown of CHK-2 activity and removal of DSB-1 and DSB-2, ending the DSB-permissive period. However, this period can be extended if one or more chromosome pairs fail to acquire an appropriate number or distribution of crossover precursors. It is possible that proper acquisition of crossover precursors sends a signal that downregulates CHK-2 activity or that recombination failure sends a signal that keeps CHK-2 active (adapted from132, 148).
When recombination and/or chromosome synapsis are defective, checkpoint responses mediated by Mec1 (ATR) cause Ndt80 to be hypophosphorylated and less abundant. These alterations block or attenuate Ndt80 activity, leading to meiotic arrest or delay (115, 118, 159) (Figure 4a). But DSB-Ndt80 cross talk is not restricted to repair-defective cells, as it clearly also occurs in normal meiosis, where it extends the length of prophase I (115). Operation of the Ndt80 circuit tends to obscure the effects of mutations that impair the DSB-forming machinery, such as partial loss-of-function spo11 mutations, and it is thought to make DSB formation homeostatic in wild-type cells (5, 27, 48, 128). An interesting implication is that Mec1 has opposing effects depending on context: It inhibits DSB formation during S phase via control of Spo11 expression and DDK activity but promotes breakage during prophase via control of Ndt80 activity. S. cerevisiae will abort meiosis and resume mitotic divisions if transferred to vegetative growth medium before pachytene exit (143). Under these conditions, DSBs disappear rapidly even if they would have continued to accumulate while still in sporulation medium (4, 138, 173; M. Lichten, personal communication). This feature implies that DSBs stop forming (71), although this has not been directly demonstrated. If so, it provides another example in which DSB potential is tied to cell cycle status (Figure 4a).
The Ndt80 and return-to-growth systems for shutting down DSB formation make sense because new DSBs would not be useful in either case. It is too late to induce recombination once cells move on to segregating their chromosomes in meiosis I, and recombination is not needed at all if cells are returning to vegetative growth.
These two systems also share a plausible mechanism for downregulating Spo11. When Ndt80 is activated in meiosis or when cells return to vegetative growth, meiotic chromosome structures, such as SCs, are rapidly disassembled (38, 147, 173). When caused by Ndt80 activation, this disassembly includes turnover of Spo11 and Rec114 and removal of other proteins, such as Red1, that are important for DSB formation (27, 147, 156). (Return to growth has not been as systematically evaluated.) Artificial induction of Cdc5 is sufficient to trigger this disassembly even in the absence of Ndt80 (147). Thus, DSB formation is shut down upon exit from prophase by virtue of regulated disruption of the chromosome structures and proteins needed to support Spo11 activity.
CHK-2 and Control of a Double-Strand-Break-Permissive Stage in Caenorhaditis elegans
Altered crossover distributions and/or persistent RAD-51 foci are seen in numerous C. elegans mutants that reduce or eliminate proteins needed for SC formation, or in strains heterozygous for chromosome translocations that impair synapsis (3, 28, 35, 52, 53, 56, 108). To account for these findings, Villeneuve and coworkers proposed that meiotic cells in these mutants were experiencing greater numbers of DSBs and responding to incomplete synapsis and/or lack of crossover-designated recombination intermediates by extending the time during which DSB formation can occur (53, 56, 108) (Figure 4b).
Recent studies provide key insights into this DSB control pathway. DSB-1 and DSB-2, proteins required for DSB formation, both localize to chromosomes during early prophase at the time DSBs normally form. However, in mutants that cannot generate crossovers, DSB-1 and DSB-2 remain on chromosomes much longer than normal, consistent with a prolonged DSB-permissive period (132, 148). Continued DSB-1 and DSB-2 localization on chromosomes occurs in mutants with very different primary defects, including those that eliminate DSB formation entirely (spo-11), those that disrupt recombinational repair of DSBs (e.g., msh-5, rad-54), and those that affect SC formation (e.g., syp-1). However, mutations eliminating HORMA-domain proteins HTP-1 and HTP-3 (homologs of yeast Hop1 and mouse HORMAD-1 and -2) cause less (DSB-1) or none (DSB-2) of this response despite causing recombination defects, suggesting that these proteins are themselves needed for the response (132, 148).
Interestingly, a crossover defect on just one chromosome is sufficient to trigger retention of DSB-1 on all chromosomes, and animals in which crossover defects happen in only a subset of nuclei show DSB-1 retention only in that defective subset. Thus, the response to crossover defects is nucleus autonomous and genome wide (28, 148). When examined, no changes in crossing over were seen on normal chromosomes in translocation heterozygotes (e.g., 95, 174), but it is important to note that excess DSBs generally do not yield additional crossovers in C. elegans because of extremely robust homeostatic control of crossover formation (172). Assuming extra DSBs do form on other chromosomes in translocation heterozygotes, they are presumably repaired as noncrossovers or by genetically silent recombination between sister chromatids.
The mutations that prolong DSB-1 and DSB-2 presence on chromosomes also prolong the period when the nuclear envelope protein SUN-1 is phosphorylated and rapid chromosome movements occur, suggesting that several distinct aspects of early meiotic prophase are coordinately regulated (28, 132, 165). The CHK-2 kinase (homologous to S. pombe Cds1 and S. cerevisiae Rad53) is an attractive candidate to mediate this coordinated response because CHK-2 is required for DSB formation, normal chromatin association of DSB-1 and DSB-2 proteins, and SUN-1 phosphorylation by both CHK-2 and the polo-like kinase PLK-2 (86, 121, 132, 148, 165).
These findings suggest that nuclei monitor recombination progression and, if needed, maintain CHK-2 activity to continue to initiate recombination (132, 148) (Figure 4b). One possibility is that acquisition of sufficient crossover-competent recombination intermediates feeds back to shut down CHK-2 activity (132). An alternative possibility is that chromosomes lacking a crossover-competent intermediate generate a signal that upregulates CHK-2 or prevents CHK-2 shutoff (148). The latter scenario would account for the ability of a single misbehaving chromosome pair to prolong DSB-1 presence. How CHK-2 activity is controlled and how it in turn affects DSB-1 and DSB-2 localization remain to be determined.
The molecular details are very different between this system and the Ndt80 circuit in S. cerevisiae, not least in terms of the dissimilar roles played by kinases traditionally viewed as DNA-damage responsive (CHK-2 in C. elegans, Mec1 in S. cerevisiae). Nonetheless, there are intriguing parallels: Both systems operate nucleus-wide; both have checkpoint-like properties; both require HORMA-domain proteins; both work by controlling chromosomal association of DSB-promoting factors; and both involve a coordinated transition from a state in which interhomolog interactions are favored to a state in which completion of recombination and preparation for chromosome segregation are favored. Interestingly, the duration of meiotic prophase is also extended in A. thaliana mutants that have recombination and/or synapsis defects, suggesting that analogous regulation occurs in plants (178, 179, 180). These similarities highlight that the imperative to integrate recombination initiation with meiotic progression is evolutionarily conserved despite extensive variation in the specific regulatory modules involved.
The Interchromosomal Effect and Precondition Mutants in Drosophila melanogaster
Very early studies of recombination in flies showed that suppression of crossing over on one chromosome (e.g., because of translocation heterozygosity) caused increased crossover frequencies on other chromosomes (125, 150, 151). This phenomenon, dubbed the interchromosomal effect, implies that oocytes experiencing difficulty in some aspect of crossover formation mount a nucleus-wide response. There are obvious parallels with the response to single-chromosome defects in C. elegans (28) but with different quantitative effects on crossing over possibly due to the less robust crossover control in flies. Global alterations in crossover distribution are also seen in flies with any of a number of mutations that interfere specifically with crossover formation. These mutations were called precondition mutants on the hypothesis that the affected genes acted before recombination to establish the normal pattern of crossing over (29). However, it was later recognized that similar changes in crossover distributions can be caused by many different defects in the core recombination machinery, including hypomorphic mutations in the SPO11 homolog Mei-W68 or strong alleles of the RAD54 homolog okra (e.g., 13). Rather than envision that all of these recombination factors also have additional (precondition-like) roles before recombination begins, McKim and colleagues proposed that cells monitor whether chromosomes have acquired an appropriate number of crossover-designated recombination intermediates and, if not, respond by increasing the total number of DSBs (13). This idea is attractive viewed through the lens of what is known about S. cerevisiae and C. elegans. It was recognized that such a system would also explain the interchromosomal effect (13, 28), although it is also possible that these are separate phenomena (65). Direct evidence for altered DSB numbers is lacking, and indirect cytological measures show no evidence of DSB increases in translocation heterozygotes (65). Nevertheless, if changes in DSB number do occur in these pathological situations, important unanswered questions include what aspect of meiotic chromosome behavior is monitored and which signaling pathways mount the global response. Recent work suggests that the interchromosomal effect involves monitoring of chromosome axis organization by the AAA+ ATPase PCH2 [homologous to S. cerevisiae Pch2 (pachytene checkpoint) and mouse TRIP13 proteins, discussed further below] (65).
ATM-DEPENDENT NEGATIVE FEEDBACK CONTROL OF DOUBLE-STRAND BREAK FORMATION
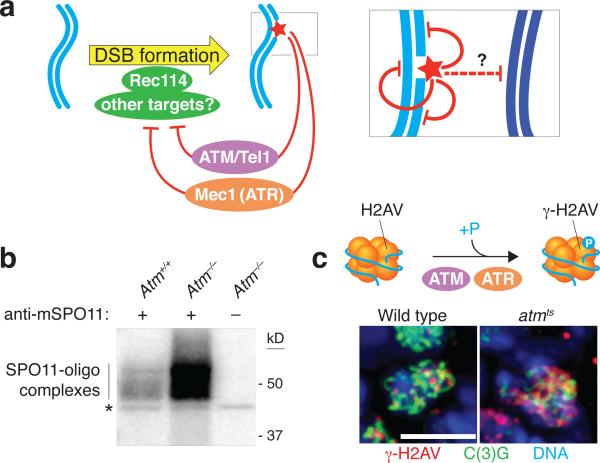
ATM (for ataxia telangiectasia mutated) is a serine/threonine kinase defective in the cancer-prone disease ataxia telagiectasia (A-T) (136). ATM activated by DSBs triggers cell cycle checkpoints and promotes DNA repair in somatic cells (40). A-T patients display gonadal dysgenesis (139) and Atm–/– mice are sterile, with chromosome synapsis defects and arrest during meiotic prophase (7, 8, 41, 42, 171). ATM is thus essential for normal, unperturbed meiosis, but until recently it was unclear what specific roles it plays. Independent studies in mouse, flies, and yeast suggest that ATM controls a negative feedback circuit that inhibits SPO11 activity (Figure 5a).
Figure 5.
The inhibitory feedback circuit between the ATM/Tel1 and ATR/Mec1 kinases and SPO11 operates in Saccharomyces cerevisiae, mice, and Drosophila melanogaster. a) In S. cerevisiae, Tel1 and Mec1 respond to double-strand breaks (DSBs) by negatively regulating local DSB formation on the same DNA molecule and on the sister chromatid, and possibly also around allelic positions on the homologous chromosome. In this pathway, phosphorylation targets of Tel1 and Mec1 may include Rec114, which is required for DSB formation. ATM acts in an analogous pathway in mice and D. melanogaster, but whether ATR contributes to feedback control in these organisms remains unknown. (b) More than 10-fold elevation in DSB levels in mice lacking ATM. The panel shows an autoradiograph of SPO11-oligo complexes isolated from wild-type and Atm-null mouse testes. Because two SPO11-oligo complexes are released from each DSB, they are quantitative by-products of meiotic recombination initiation that can serve as a measure of relative whole-testis DSB levels. SPO11-oligo complexes were immunopurified from testis lysates using an anti- SPO11 antibody and then radiolabeled and fractionated by electrophoresis. Asterisk indicates a SPO11-independent labeling artifact. Image adapted from Reference 78. (c) Increased phosphorylation of histone variant H2AV in ATM-deficient D. melanogaster oocytes. In response to DSBs, H2AV is phosphorylated by ATM or ATR to form γ-H2AV (equivalent of γ-H2AX in mammals). The micrographs show pachytene oocytes from a wild-type fly or an ATM-deficient fly. The cells were immunostained for γ-H2AV and C(3)G (a component of the synaptonemal complex that identifies oocytes within the ovary) and stained with DAPI to detect DNA. γ-H2AV levels are elevated in ATM-deficient oocytes and interpreted as being the result of an elevated DSB number yielding higher ATR activity. Images adapted from Reference 66 with permission of the authors.
In Atm–/– mice, testes have >10-fold higher steady-state levels of SPO11-oligo complexes (78) (Figure 5b). SPO11-oligo complexes have a long life span in wild-type spermatocytes, and the amount of the complexes is already elevated when they first appear in Atm−/− juvenile testes. Thus, the steady-state increase in SPO11-oligo complexes likely reflects increased DSB numbers rather than increased life span of the complexes. Interestingly, absence of ATM also renders DSB formation more sensitive to SPO11 expression level, suggesting that the normal robustness of DSB number control in wild-type spermatocytes depends on ATM (78).
D. melanogaster females homozygous for a temperature-sensitive allele of the fly Atm ortholog display 1.5- to 3-fold higher levels of γ-H2AV in oocytes and neighboring nurse cells at the restrictive temperature (66) (Figure 5c). γ-H2AV is the equivalent of mammalian γ-H2AX (62), a phosphorylated form of histone variant H2AX that arises in response to DSBs made in nonmeiotic contexts (129) or by SPO11 (87). In flies, meiotic γ-H2AV is generated redundantly by ATM and ATR (product of the mei-41 gene) (66). The elevated γ-H2AV levels thus suggest that more DSBs are made in ATM-deficient flies, resulting in higher ATR activity (66).
Similarly, several independent lines of evidence point to elevated meiotic DSB formation in S. cerevisiae cells lacking the ATM ortholog Tel1: a higher frequency of detectable recombinants (measured in an otherwise wild-type background) and DSBs (measured in a rad50S background) at an artificial recombination hot spot (175), elevated DSBs at a natural hot spot and modestly higher DSB frequency on at least one whole chromosome (also measured in a rad50S background) (27), and a greater number of DSBs genome-wide, assayed by immunoprecipitation of covalent Spo11-oligo complexes in a RAD50+ SAE2+ background (N. Mohibullah & S. Keeney, unpublished results). (Deletion of the SAE2 gene or rad50S (for separation of function) mutations block the removal of Spo11 from DSB ends and thereby prevent DSB resection; see Figure 1b.) Two studies reported instead that DSBs are reduced by tel1 mutation in rad50S or sae2 mutant backgrounds (5, 17). The reasons for the differences in results are unclear, but the preponderance of the data (and, in particular, data from otherwise wild-type backgrounds) indicates that DSBs are elevated in tel1 mutant yeast, similar to ATM-deficient mice and flies.
These findings suggest that an evolutionarily conserved pathway for DSB control entails activation of ATM by DSBs, which then feeds back to inhibit further DSB formation by other SPO11 molecules (Figure 5a). The mechanism is not yet well understood. Is ATM kinase activity required, and if so, what is the relevant phosphorylation target(s)? One candidate target in yeast is Rec114, a meiosis-specific protein required for DSB formation: Rec114 is phosphorylated in vivo in response to DSBs. This phosphorylation does not occur in mutants lacking both Tel1 and Mec1 activity; Rec114 can be phosphorylated in vitro by Mec1 (which has similar substrate preferences as Tel1); and mutant Rec114 protein lacking putative phosphorylation sites yields signs of increased or faster DSB formation, whereas potentially phosphomimetic mutations inhibit DSB formation (27, 135). Whether Rec114 is a direct Tel1 target has not yet been established, and it remains unclear whether the absence of Rec114 phosphorylation phenocopies the absence of Tel1, so other (possibly redundant) substrates may exist. Furthermore, the fact that Mec1 can phosphorylate Rec114 suggests this kinase (like Tel1) may also inhibit DSB formation directly, either in wild-type cells or perhaps only when Mec1 is hyperactivated (i.e., in recombination mutants). This possibility adds further complexity to the mix of positive and negative roles that Mec1 plays (see above).
In flies, γ-H2AV turns over quickly in the absence of ongoing ATM and/or ATR signaling (66). Assuming similar rapid turnover for ATM targets relevant to DSB inhibition, it seems likely that ATM-dependent feedback is transient, occurring only as long as ATM itself remains active. Moreover, it is likely that this DSB inhibition works at least partly at a local level because ATM is activated in direct spatial proximity to the sites of DSBs, as judged, for example, by the location of γ-H2AX formed in mouse meiosis (87). Thus, this pathway might serve to discourage the formation of multiple DSBs near one another on the same chromatid or on sister chromatids (27, 78) (Figure 5a, inset). In fact, S. cerevisiae and S. pombe cells rarely cut the same chromatid at adjacent hot spots, much less frequently than expected from the DSB frequencies of the individual hot spots (M. Lichten, M. Neale, G. Smith, personal communication). This behavior, i.e., interference between potential DSB sites on the same DNA molecule, requires Tel1 in both yeasts (M. Neale & G. Smith, personal communication). A further prediction, supported by DSB mapping via deep-sequencing of Spo11 oligos in S. cerevisiae (N. Mohibullah & S. Keeney, unpublished results) and mouse (J. Lange, M. Jasin, & S. Keeney, unpublished results), is that the DSB landscape in wild-type meiosis is shaped in part by the spatial patterning of ATM-dependent feedback. Additional implications of this form of DSB control are discussed in the final section.
NEGATIVE FEEDBACK TIED TO HOMOLOG ENGAGEMENT
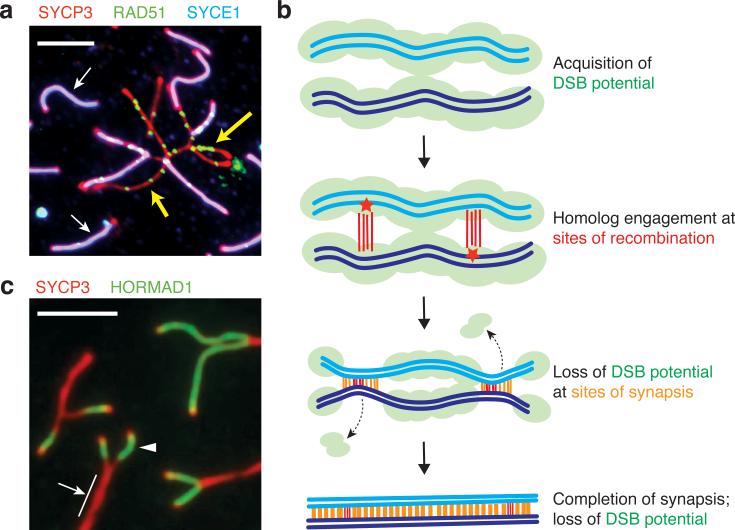
Another pathway for restraining Spo11 activity has been illuminated by independent studies in mice, nematodes, and budding yeast. A mouse transgenic construct expressing Spo11β (one of several Spo11 splicing isoforms) behaves as a hypomorphic Spo11 mutation when the transgene insertion is in a single copy and is the only source of SPO11 protein, i.e., in mice with the genotype Spo11–/– Tg(Spo11β)+/–. Spermatocytes from these mice generate approximately half the normal number of DSBs as assessed by quantification of SPO11-oligo complexes or RAD51 and DMC1 foci (68). This reduction is accompanied by defects in synapsis of homologous chromosomes: Many chromosomes successfully locate their partners and form normal SC, but in most cells several chromosomes fail to synapse properly. The unlucky chromosomes end up in topologically constrained tangles, where some axis segments remain unsynapsed and other segments show homologous or even nonhomologous synapsis.
Remarkably, even though the cell-wide DSB level is reduced in these mice, the unsynapsed chromosome axes in tangles continue to accumulate RAD51 foci, in some cases reaching a density comparable to wild type (Figure 6a). Moreover, in wild-type mice, RAD51 foci continue to accumulate on the portion of the X chromosome that is not homologous to the Y and that thus naturally remains unsynapsed. These results suggest that unsynapsed axes continue to accumulate DSBs, or put another way, that synapsis is normally accompanied by cessation of DSB formation (68). Importantly, this effect acts locally within the unsynapsed regions, which distinguishes it from the nucleus-wide responses discussed above.
Figure 6.
Feedback tied to engagement of homologous chromosomes. (a) Continued accumulation of double-strand breaks (DSBs) on segments of mouse chromosomes that fail to synapse. The panel shows a micrograph of some of the chromosomes from a mouse spermatocyte spread on a glass slide and immunostained for SYCP3 (a component of the axial elements), SYCE1 [a component of the central element of the synaptonemal complex (SC)] and RAD51 (a strand-exchange protein). Unsynapsed axes appear red, and mature SC appears magenta from the overlap of the red SYCP3 and blue SYCE1 signals. Each green focus is a site at which a DSB has been made and is in the process of being repaired by RAD51 and other factors. This spermatocyte is from a mouse that has reduced DSB formation overall because the only source of SPO11 protein is a single copy of a transgene expressing the Spo11β splicing isoform. Some chromosomes synapse normally (examples indicated with white arrows), but some display synaptic failure and are trapped in tangles. The unsynapsed axes in these tangles continue to accumulate RAD51 foci (examples indicated with yellow arrows), suggesting that DSBs continue to form. Image adapted from Reference 68. (b) Schematic illustrating how DSB potential (i.e., the ability of chromosomes to be a substrate for SPO11) is lost as homologous chromosomes engage each other during recombination and SC assembly. (c) Depletion of DSB-promoting protein HORMAD1 from chromosome axes after SC formation. A portion of a spread mouse spermatocyte nucleus is shown immunostained for SYCP3 and HORMAD1. Axial elements that have not yet synapsed with one another show strong staining for HORMAD1 (arrowhead), whereas regions that have synapsed do not (arrow; note that a single red signal is seen because conventional light microscopy cannot resolve the two SYCP3-staining axial elements of the synapsed homologous chromosomes). Image adapted from Reference 166 with permission of the authors.
C. elegans oocytes lacking X-chromosome pairing centers display X-chromosome-specific defects in homologous pairing and synapsis, plus numerous RAD-51 foci present on the X chromosomes much later than normal (85). Although it is possible these foci are simply persisting because of repair defects, it is attractive to think that they instead represent additional DSBs above the number that would have formed in wild type. If so, these extra DSBs appear to be more numerous than those on autosomes in the same cells, which would be consistent with a local effect on DSB formation specific to the misbehaving chromosomes, layered on top of the nucleus-wide prolongation of the DSB-permissive stage (28, 85, 132, 148).
These observations dovetail with independent studies of the S. cerevisiae ZMM proteins (Zip1--4, Msh4--5, Mer3, and others), a biochemically diverse suite of factors needed to ensure that crossover-designated recombination intermediates do indeed become crossovers (21, 84). ZMM proteins are needed for normal SC formation; in fact, one of them (Zip1) is a core structural component of the SC (152). Given these functions in recombination and synapsis, ZMM proteins were typically thought of as acting strictly downstream of DSB formation. However, surprisingly, zmm mutants accumulate more DSBs than wild type based on multiple lines of evidence, including elevated DSB levels by direct physical assays, increased frequency of detectable recombination products, and more Spo11-oligo complexes (156).
In principle, increased DSBs could have been an indirect consequence of the meiotic delay or arrest caused by the DSB repair defect in zmm mutants, which is known to trigger Mec1-dependent inhibition of Ndt80 (84, 159). However, epistasis tests show this not to be the case: zmm ndt80 double mutants make more DSBs than either zmm or ndt80 single mutants (156). These results indicate that a ZMM-dependent process that fosters engagement of homologous chromosomes is more directly responsible for inhibiting DSB formation in wild-type cells. A similar line of reasoning has been proposed to explain inferred increases in DSB formation in other yeast mutants with defects in homolog engagement (27, 79).
A plausible mechanism that accounts for results in both yeast and mouse is that SC formation leads to structural changes that render chromosomes unfit substrates for Spo11 (27, 68, 156, 166) (Figure 6b). This model was first proposed from studies of the mouse HORMA-domain proteins HORMAD1 and HORMAD2 (166). These proteins localize to unsynapsed chromosome axes on which RAD51 foci form during the leptotene and zygotene stages but are displaced soon after synapsis (46, 141, 166) (Figure 6c). Likewise in A. thaliana, the HORMA-domain protein ASY1 becomes similarly depleted from axes after chromosomes synapse (F.C. Franklin, personal communication). Tóth and colleagues proposed that HORMAD displacement might suppress further DSB formation because homologous proteins, such as S. cerevisiae Hop1, are needed for normal DSB levels (59, 166) [shown later to be true for mouse HORMAD1 as well (37, 141)]. Earlier work had demonstrated that numerous DSB-promoting factors, including Hop1, Mei4 (not related to the S. pombe Mei4 transcription factor discussed earlier), Red1, Rec102, Rec104, and Rec114 are displaced from chromosomes after synapsis in S. cerevisiae (20, 70, 80, 88, 144). These yeast results were not originally interpreted in terms of DSB regulation, but they fit nicely with this model in retrospect (27, 156).
The mechanism behind the chromosome structure changes remains to be determined. In mouse and A. thaliana, HORMA-domain protein displacement occurs with a delay after SC formation, so synapsis cannot be an instantaneous trigger (166; F.C. Franklin, personal communication). A temporal offset for removal of DSB-promoting proteins is also seen in yeast (e.g., 70). Moreover, mouse HORMAD displacement even occurs in chromosomal segments that synapse nonhomologously, e.g., in mice lacking SPO11 (166). Thus, recombination is not essential, and although SC formation between homologous chromosomes may be the normal conduit for DSB control in wild-type cells, homolog engagement per se is also dispensable.
The AAA+ ATPase TRIP13 is required for HORMAD proteins to be displaced from synapsed axes in mice (130, 166), as is the TRIP13 ortholog PCH2 in A. thaliana (F.C. Franklin, personal communication). Similarly, Pch2 governs the normal nonuniform localization of Hop1 along SCs in S. cerevisiae (20), probably by directly remodeling Hop1 protein structure (32) (N. Kleckner, personal communication). However, available evidence does not strongly support the view that TRIP13 or Pch2 are net-negative regulators of DSB formation. In yeast, DSB numbers are not increased in pch2 mutants (20, 44). In mouse, the fact that RAD51 foci and γH2AX are present on synapsed chromosomes during pachynema in Trip13-deficient spermatocytes may indicate that DSB formation continues longer than normal (68). However, global DSB levels do not appear to be increased based on SPO11-oligo quantification (S. Pacheco, M. Marcet-Ortega, J. Lange, M. Jasin, S. Keeney & I. Roig, unpublished results), so DSB persistence rather than increased DSB numbers may underlie these cytological patterns (81, 130).
It is interesting that this mode of DSB control is analogous to how Ndt80- and CHK-2-influenced circuits in S. cerevisiae and C. elegans may work, although the specific mechanisms regulating chromosomal association of DSB-promoting factors differ in each case. We also note that this feedback mechanism may provide a more permanent shutdown of DSB-forming potential compared with the proposed transience of the ATM/Tel1 circuit. Feedback tied to homolog engagement may operate over longer physical distances as well, given the potential for SC to polymerize further than ATM/Tel1 kinase signaling is likely to spread.
COMMUNICATION BETWEEN POTENTIAL DOUBLE-STRAND BREAK SITES
Four further layers of DSB regulation have been documented in S. cerevisiae, each involving apparent communication in trans between sister chromatids or homologous chromosomes, or in cis between regions on the same chromosome. First, DSB frequency at a hot spot can be affected by sequences at the same position on the homologous partner (26, 127, 170). In at least some cases, allelic sequences that match one another yield the most DSBs (127, 170), possibly because they also yield the most accessible (DNase I-hypersensitive) chromatin structure (74). These findings suggest that homology-dependent physical interactions between chromosomes influence chromatin structure and DSB formation, but the mechanism is not known.
Two more layers also work in trans between DNA molecules. Specifically, a strong hot spot can suppress DSB formation at the allelic position and at hot spots nearby (within a few kilobases) on the homologous chromosome (170, 175). Separately, numerical patterns of recombination in tetrads suggest that wild-type cells rarely break both sister chromatids at the same hot spot (175). Thus, it appears that any given cell usually experiences at most one DSB at the same place among four chromatids, even at an exceedingly strong hot spot (175). In mutants lacking Tel1 or Mec1, recombination patterns suggest loss of one of these control layers, i.e., DSBs can often occur at the same place on two DNA molecules in the same cell. This has been interpreted to mean that Tel1 and Mec1 convey an inhibitory signal in trans between homologous chromosomes (175) (Figure 5a, inset). However, the data are equally consistent with one or both kinases preventing breakage of both sister chromatids, as has been proposed for Tel1 in yeast (27; M. Neale, personal communication) and ATM in mouse (78). Further studies are needed to dissect the mechanisms and interplay between these pathways.
The fourth layer is revealed by the fact that creating new hot spots---whether by inserting an artificial hot spot-specifying DNA construct or by fusing Spo11 to a sequence-specific DNA binding domain to target cleavage to new positions---suppresses DSB formation in neighboring regions on the same chromosome (43, 47, 63, 114, 126, 168, 170). The magnitude of suppression decays with distance but has been detected 30--60 kilobases away and may extend further in some contexts (47, 63, 126, 168). S. pombe behaves similarly (149; G. Smith, personal communication). In principle, this phenomenon could involve competition between hot spots for a limiting pool of DSB-promoting factors and could occur before DSB formation. However, site-specific DSBs made by an endonuclease also suppress Spo11-generated DSBs nearby (47): Assuming that the suppression mechanism from endonuclease-directed DSBs is the same as for Spo11, this finding implies that hot-spot competition involves the spread of an inhibitory signal after DSB formation. Tel1 appears to be dispensable (N. Mohibullah & S. Keeney, unpublished results), distinguishing this form of DSB suppression from the Tel1-mediated DSB interference discussed above (i.e., less frequent double-cutting of the same chromatid than expected by chance). One possible mechanism derives from the proposal by Kleckner and colleagues that mechanical stress and stress relief drive DNA metabolic events and chromosome morphogenesis (77, 176). As a topoisomerase relative, Spo11 is predicted to be an inherently stress-sensitive enzyme (72). Thus, the same patterning forces that are proposed to shape crossover distributions (77, 176) might also shape DSB distributions (71, 72, 77).
CONCLUSIONS AND IMPLICATIONS
From the first proposals that DSBs might be the initiators of meiotic recombination (124, 153), it has been appreciated that this is a dangerous game for the cell to play. Work reviewed here brings into focus how cells accommodate this danger by controlling when, where, and how many DSBs are made by Spo11. The emerging view is that DSB control involves a drive toward DSB formation (promoted in part by cell cycle regulators and by development of meiosis-specific chromosome structures) that is subject to quantitative, spatial, and temporal restraints from distinct but intersecting negative influences (Figure 2). This view explains the basis of self-organization of recombination initiation and clarifies how DSB formation can be homeostatic and therefore robust against cell-to-cell variation and environmental perturbation.
We envision that this robustness helps cells cope with unbalanced karyotypes, such as might be encountered in outcrosses or with heterozygous de novo chromosome rearrangements. For example, the proclivity of SC to form between nonhomologous chromosome segments in maize, mice, and other organisms (e.g., 93, 101) may provide a means to eventually suppress DSB formation in regions that are unable to locate a homologous partner (156). In normal male meiosis in mammals, the X and Y chromosome share only a small segment of homology, the pseudoautosomal region (PAR) (133). The PAR must receive at least one DSB in order to support pairing and crossing over between the sex chromosomes (67), yet the cell must also cope with the presence of the large portions of the X and Y that cannot engage one another homologously. The network of DSB-regulating pathways, and in particular the ability to terminate DSB formation genome-wide at or before prophase exit, provides a means to accommodate this feature of male meiosis.
An important implication of the circuits that control Spo11 activity is that they can complicate genetic analyses, especially if a mutation impinges on multiple circuits at once. For example, S. cerevisiae zmm mutations simultaneously disrupt homolog engagement (removing a negative DSB regulator), inhibit Ndt80 activation via DSB repair defects (removing yet another negative DSB regulator), and hyperactivate Mec1 (possibly adding Tel1-like DSB inhibition) (156, 159). Thus, when homeostatic DSB control pathways respond to pathological defects in recombination or other processes, the phenotypic endpoints differ from the wild-type situation in many complex ways. By the same token, it is important to view cautiously the widespread use of recombination-defective mutant backgrounds (e.g., rad50S or dmc1 in S. cerevisiae) to determine what DSB numbers would have been in wild type or to query the effects of other mutations on DSB numbers.
Another important implication is that Spo11-regulating processes strongly influence DSB locations by virtue of being spatially patterned, e.g., via local activity of ATM/Tel1 near DSBs or by tying feedback to homolog engagement at sites of recombination (13, 156) (N. Mohibullah & S. Keeney, unpublished results; J. Lange, M. Jasin & S. Keeney, unpublished results). We can thus divide architects of the DSB landscape into intrinsic factors (chromosomal features that govern accessibility or activity of Spo11 toward specific locations, such as loop-axis structure, nucleosome positions, or histone modifications) and more extrinsic factors layered on top (feedback and other regulatory circuits). This division fits well with the view that the factors shaping the DSB landscape work in a hierarchical fashion (119).
A related point is that modes of Spo11 regulation explain otherwise puzzling features of set1 mutant yeast and Prdm9–/– mutant mice. In S. cerevisiae, trimethylation of histone H3 lysine 4 by the Set1 methyltransferase plays a key role in directing Spo11 to cleave preferentially in nucleosome-depleted regions in promoters (1, 146). In mouse and human, DSB hot-spot locations are defined by the DNA binding specificity of PRDM9, which sports a histone methyltransferase module and a zinc-finger DNA binding domain that evolves rapidly (9, 11, 24, 107). These methyltransferases have a near-absolute role in targeting Spo11 to particular sites in the genome, yet they are almost completely dispensable for DSB formation per se: In their absence, DSBs form in relatively normal numbers but different locations (19, 24). This behavior is explained by recognizing that the default is for Spo11 to make breaks until restrained. Even if the DSB-forming machinery is crippled by absence of targeting factors such as Set1 and PRDM9, the homeostatic responses of Spo11-regulating circuits will adjust, driving break formation at high frequency but in highly abnormal positions.
Similar reasoning affects understanding of the relationship between hot-spot evolutionary dynamics and the ability to execute recombination genome-wide. Because of bias in the direction of information transfer during gene conversion (the broken chromosome copies information from the intact donor), sequence polymorphisms that inactivate a hot spot tend to be overrepresented in offspring from heterozygous individuals (e.g., 50). Thus, hot spots are expected to become colder or disappear over evolutionary timescales and new hot-spot alleles should rarely if ever go to fixation in a population. The existence of hot spots despite these headwinds has been called the hot-spot paradox (22). Many attempts to model this phenomenon implicitly or explicitly assume that recombination cannot occur if hot spots are lost (122, 160). However, the logic of DSB control makes it impossible for inactivation of even large numbers of individual hot spots to render chromosomes immune to Spo11. Thus, the need to retain recombination proficiency is unlikely by itself to be a selective constraint in favor of hot-spot retention. Moreover, when a hot spot becomes cooler or disappears, homeostatic DSB regulation is predicted to elevate DSB frequency in neighboring regions: Like a genomic Whac-a-Mole® game, new hot spots pop up whenever a hot spot decays (hat tip: M. Lichten).
The broad outlines and basic concepts of DSB regulation appear to be evolutionarily conserved. This conservation seems fitting given the universality of the risk/reward tradeoff for DSB formation and recombination in meiosis. Nonetheless, detailed mechanisms differ widely between species. For example, even though involvement of DNA damage response kinases and control of the chromosomal association of DSB-promoting factors are common themes, they are often used in different contexts in different organisms and even within different regulatory pathways in the same organism. In many cases, this evolutionary plasticity clearly reflects differences between organisms’ cellular and developmental constraints on sexual reproduction, such as genome size and karyotype; repetitive DNA content; recombination (in)dependence of chromosome pairing; and the organism's lifestyle, e.g., reversibility of meiotic entry in yeast versus irreversibility in metazoans. Although much remains to be learned about detailed mechanisms of the regulatory modules involved, current knowledge provides molecular frameworks to guide future experiments.
ACKNOWLEDGMENTS
We thank Abby Dernberg, Chris Franklin, Maria Jasin, Liisa Kauppi, Nancy Kleckner, Michael Lichten, Hong Ma, Hisao Masai, Matthew Neale, Kunihiro Ohta, Gerry Smith, Drew Thacker, and Anne Villeneuve for discussions and/or sharing unpublished data. We thank Kim McKim and Attila Tóth for permission to reproduce published images. Our research is supported by grants from the NIH (GM058673 to S.K.; GM105421 to M. Jasin and S.K.; HD053855 to S.K. and M. Jasin) and the Howard Hughes Medical Institute. J.L. was supported in part by a fellowship from the American Cancer Society (grant # PF-12-157-01-DMC). S.K. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, et al. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–-18. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- 2.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–-57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 3.Alpi A, Pasierbek P, Gartner A, Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–-16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- 4.Arbel A, Zenvirth D, Simchen G. Sister chromatid--based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–-58. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argunhan B, Farmer S, Leung WK, Terentyev Y, Humphryes N, et al. Direct and indirect control of the initiation of meiotic recombination by DNA damage checkpoint mechanisms in budding yeast. PLOS ONE. 2013;8:e65875. doi: 10.1371/journal.pone.0065875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azumi Y, Liu D, Zhao D, Li W, Wang G, et al. Homolog interaction during meiotic prophase I in Arabidopsis requires the SOLO DANCERS gene encoding a novel cyclin-like protein. EMBO J. 2002;21:3081–-95. doi: 10.1093/emboj/cdf285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barchi M, Mahadevaiah S, Di Giacomo M, Baudat F, de Rooij DG, et al. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell. Biol. 2005;25:7203–-15. doi: 10.1128/MCB.25.16.7203-7215.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–-71. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 9.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–-40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794–-806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 11.Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, et al. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat. Genet. 2010;42:859–-63. doi: 10.1038/ng.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–-17. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 13.Bhagat R, Manheim EA, Sherizen DE, McKim KS. Studies on crossover-specific mutants and the distribution of crossing over in Drosophila females. Cytogenet. Genome Res. 2004;107:160–-71. doi: 10.1159/000080594. [DOI] [PubMed] [Google Scholar]
- 14.Bhalla N, Dernburg AF. Prelude to a division. Annu. Rev. Cell Dev. Biol. 2008;24:397–-424. doi: 10.1146/annurev.cellbio.23.090506.123245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111:791–-802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 16.Blitzblau HG, Chan CS, Hochwagen A, Bell SP. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLOS Genet. 2012;8:e1002643. doi: 10.1371/journal.pgen.1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blitzblau HG, Hochwagen A. ATR/Mec1 prevents lethal meiotic recombination initiation on partially replicated chromosomes in budding yeast. Elife. 2013;2:e00844. doi: 10.7554/eLife.00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borde V, Goldman ASH, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–-9. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 19.Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–-111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borner GV, Barot A, Kleckner N. Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proc. Natl. Acad. Sci. USA. 2008;105:3327–-32. doi: 10.1073/pnas.0711864105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–-45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 22.Boulton A, Myers RS, Redfield RJ. The hotspot conversion paradox and the evolution of meiotic recombination. Proc. Natl. Acad. Sci. USA. 1997;94:8058–-63. doi: 10.1073/pnas.94.15.8058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–-57. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–-45. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLOS Biol. 2007;5:e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bullard SA, Kim S, Galbraith AM, Malone RE. Double strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 1996;93:13054–-59. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, et al. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLOS Genet. 2013;9:e1003545. doi: 10.1371/journal.pgen.1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlton PM, Farruggio AP, Dernburg AF. A link between meiotic prophase progression and crossover control. PLOS Genet. 2006;2:e12. doi: 10.1371/journal.pgen.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter AT, Sandler L. On recombination-defective meiotic mutants in Drosophila melanogaster. Genetics. 1974;76:453–-75. doi: 10.1093/genetics/76.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol. Cell. 2000;5:883–-88. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- 31.Chelysheva L, Gendrot G, Vezon D, Doutriaux MP, Mercier R, Grelon M. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLOS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen C, Jomaa A, Ortega J, Alani EE. Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proc. Natl. Acad. Sci. USA. 2014;111:E44–-53. doi: 10.1073/pnas.1310755111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, et al. Global analysis of the meiotic crossover landscape. Dev. Cell. 2008;15:401–-15. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol. Cell. 1998;1:685–-96. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- 35.Colaiacovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–-74. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 36.Cole F, Kauppi L, Lange J, Roig I, Wang R, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 2012;14:424–-30. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniel K, Lange J, Hached K, Fu J, Anastassiadis K, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat. Cell Biol. 2011;13:599–-610. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dayani Y, Simchen G, Lichten M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLOS Genet. 2011;7:e1002083. doi: 10.1371/journal.pgen.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 2013;47:563–-99. doi: 10.1146/annurev-genet-110711-155423. [DOI] [PubMed] [Google Scholar]
- 40.Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584:3675–-81. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Giacomo M, Barchi M, Baudat F, Edelmann W, Keeney S, Jasin M. Distinct DNA damage--dependent and independent responses drive the loss of oocytes in recombination-defective mouse mutants. Proc. Natl. Acad. Sci. USA. 2005;102:737–-42. doi: 10.1073/pnas.0406212102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, et al. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA. 1996;93:13084–-89. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan QQ, Xu F, White MA, Petes TD. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics. 1997;145:661–-70. doi: 10.1093/genetics/145.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farmer S, Hong EJ, Leung WK, Argunhan B, Terentyev Y, et al. Budding yeast Pch2, a widely conserved meiotic protein, is involved in the initiation of meiotic recombination. PLOS ONE. 2012;7:e39724. doi: 10.1371/journal.pone.0039724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Froenicke L, Anderson LK, Wienberg J, Ashley T. Male mouse recombination maps for each autosome identified by chromosome painting. Am. J. Hum. Genet. 2002;71:1353–-68. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukuda T, Daniel K, Wojtasz L, Toth A, Hoog C. A novel mammalian HORMA domain-containing protein, HORMAD1, preferentially associates with unsynapsed meiotic chromosomes. Exp. Cell Res. 2010;316:158–-71. doi: 10.1016/j.yexcr.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 47.Fukuda T, Kugou K, Sasanuma H, Shibata T, Ohta K. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res. 2008;36:984–-97. doi: 10.1093/nar/gkm1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gray S, Allison RM, Garcia V, Goldman AS, Neale MJ. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol. 2013;3:130019. doi: 10.1098/rsob.130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, et al. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr. Biol. 2005;15:1663–-69. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 50.Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:317–-37. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature. 2006;442:45–-50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi M, Chin GM, Villeneuve AM. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLOS Genet. 2007;3:e191. doi: 10.1371/journal.pgen.0030191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayashi M, Mlynarczyk-Evans S, Villeneuve AM. The synaptonemal complex shapes the crossover landscape through cooperative assembly, crossover promotion and crossover inhibition during Caenorhabditis elegans meiosis. Genetics. 2010;186:45–-58. doi: 10.1534/genetics.110.115501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–-32. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc. Natl. Acad. Sci. USA. 2004;101:4519–-24. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henzel JV, Nabeshima K, Schvarzstein M, Turner BE, Villeneuve AM, Hillers KJ. An asymmetric chromosome pair undergoes synaptic adjustment and crossover redistribution during Caenorhabditis elegans meiosis: implications for sex chromosome evolution. Genetics. 2011;187:685–-99. doi: 10.1534/genetics.110.124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr. Biol. 2006;16:R217–-28. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Hochwagen A, Tham WH, Brar GA, Amon A. The FK506 binding protein Fpr3 counteracts protein phosphatase 1 to maintain meiotic recombination checkpoint activity. Cell. 2005;122:861–-73. doi: 10.1016/j.cell.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 59.Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–-62. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horie S, Watanabe Y, Tanaka K, Nishiwaki S, Fujioka H, et al. The Schizosaccharomyces pombe mei4+ gene encodes a meiosis-specific transcription factor containing a forkhead DNA-binding domain. Mol. Cell. Biol. 1998;18:2118–-29. doi: 10.1128/mcb.18.4.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hunter N. In: Meiotic recombination. In Molecular Genetics of Recombination. Aguilera A, Rothstein R, editors. Springer-Verlag; Berlin: 2007. pp. 381–-442. [Google Scholar]
- 62.Jang JK, Sherizen DE, Bhagat R, Manheim EA, McKim KS. Relationship of DNA double-strand breaks to synapsis in Drosophila. J. Cell Sci. 2003;116:3069–-77. doi: 10.1242/jcs.00614. [DOI] [PubMed] [Google Scholar]
- 63.Jessop L, Allers T, Lichten M. Infrequent co-conversion of markers flanking a meiotic recombination initiation site in Saccharomyces cerevisiae. Genetics. 2005;169:1353–-67. doi: 10.1534/genetics.104.036509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones GH, Franklin FC. Meiotic crossing-over: obligation and interference. Cell. 2006;126:246–-48. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 65.Joyce EF, McKim KS. Chromosome axis defects induce a checkpoint-mediated delay and interchromosomal effect on crossing over during Drosophila meiosis. PLOS Genet. 2010;6:e1001059. doi: 10.1371/journal.pgen.1001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joyce EF, Pedersen M, Tiong S, White-Brown SK, Paul A, et al. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J. Cell Biol. 2011;195:359–-67. doi: 10.1083/jcb.201104121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science. 2011;331:916–-20. doi: 10.1126/science.1195774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 2013;27:873–-86. doi: 10.1101/gad.213652.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 2004;5:413–-24. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- 70.Kee K, Protacio RU, Arora C, Keeney S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J. 2004;23:1815–-24. doi: 10.1038/sj.emboj.7600184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Keeney S. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 2001;52:1–-53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 72.Keeney S. Spo11 and the formation of DNA double-strand breaks in meiosis. In: Lankenau DH, editor. Recombination and Meiosis. Springer-Verlag; Heidelberg, Ger.: 2008. pp. 81–-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–-84. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 74.Keeney S, Kleckner N. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells. 1996;1:475–-89. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 75.Kim JM, Takemoto N, Arai K, Masai H. Hypomorphic mutation in an essential cell-cycle kinase causes growth retardation and impaired spermatogenesis. EMBO J. 2003;22:5260–-72. doi: 10.1093/emboj/cdg497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma. 2006;115:175–-94. doi: 10.1007/s00412-006-0055-7. [DOI] [PubMed] [Google Scholar]
- 77.Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, et al. A mechanical basis for chromosome function. Proc. Natl. Acad. Sci. USA. 2004;101:12592–-97. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–-40. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lao JP, Cloud V, Huang CC, Grubb J, Thacker D, et al. Meiotic crossover control by concerted action of Rad51-Dmc1 in homolog template bias and robust homeostatic regulation. PLOS Genet. 2013;9:e1003978. doi: 10.1371/journal.pgen.1003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, Hooker GW, Roeder GS. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics. 2006;173:1969–-81. doi: 10.1534/genetics.106.058768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li XC, Schimenti JC. Mouse pachytene checkpoint 2 (trip13) is required for completing meiotic recombination but not synapsis. PLOS Genet. 2007;3:e130. doi: 10.1371/journal.pgen.0030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lichten M. Meiotic chromatin: The substrate for recombination initiation. In: Egel R, Lankenau DH, editors. Recombination and Meiosis: Models, Means, and Evolution. Springer-Verlag; Berlin: 2008. pp. 165–-93. [Google Scholar]
- 83.Lin Y, Larson KL, Dorer R, Smith GR. Meiotically induced rec7 and rec8 genes of Schizosaccharomyces pombe. Genetics. 1992;132:75–-85. doi: 10.1093/genetics/132.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lynn A, Soucek R, Borner GV. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 2007;15:591–-605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 85.MacQueen AJ, Phillips CM, Bhalla N, Weiser P, Villeneuve AM, Dernburg AF. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell. 2005;123:1037–-50. doi: 10.1016/j.cell.2005.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.MacQueen AJ, Villeneuve AM. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–-87. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mahadevaiah SK, Turner JMA, Baudat F, Rogakou EP, de Boer P, et al. Recombinational DNA double strand breaks in mice precede synapsis. Nat. Genet. 2001;27:271–-76. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- 88.Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma. 2007;116:471–-86. doi: 10.1007/s00412-007-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Margolin G, Khil PP, Kim J, Bellani MA, Camerini-Otero RD. Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics. 2014;15:39. doi: 10.1186/1471-2164-15-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Marston AL, Amon A. Meiosis: cell-cycle controls shuffle and deal. Nat. Rev. Mol. Cell Biol. 2004;5:983–-97. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 91.Matos J, Lipp JJ, Bogdanova A, Guillot S, Okaz E, et al. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell. 2008;135:662–-78. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 92.Matsumoto S, Ogino K, Noguchi E, Russell P, Masai H. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J. Biol. Chem. 2005;280:42536–-42. doi: 10.1074/jbc.M510575200. [DOI] [PubMed] [Google Scholar]
- 93.McClintock B. The association of non-homologous parts of chromosomes in the midprophase of meiosis in Zea mays. Z. Zellforsch. Mikrosk. Anat. 1933;19:191–-237. [Google Scholar]
- 94.McFarlane RJ, Mian S, Dalgaard JZ. The many facets of the Tim-Tipin protein families’ roles in chromosome biology. Cell Cycle. 2010;9:700–-5. doi: 10.4161/cc.9.4.10676. [DOI] [PubMed] [Google Scholar]
- 95.McKim KS, Howell AM, Rose AM. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics. 1988;120:987–-1001. doi: 10.1093/genetics/120.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mehrotra S, McKim KS. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLOS Genet. 2006;2:e200. doi: 10.1371/journal.pgen.0020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–-86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol. Cell. Biol. 2009;29:5998–-6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, et al. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell. 2012;47:722–-33. doi: 10.1016/j.molcel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 100.Moens PB, Chen DJ, Shen Z, Kolas N, Tarsounas M, et al. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma. 1997;106:207–-15. doi: 10.1007/s004120050241. [DOI] [PubMed] [Google Scholar]
- 101.Moses MJ, Dresser ME, Poorman PA. Composition and role of the synaptonemal complex. Symp. Soc. Exp. Biol. 1984;38:245–-70. [PubMed] [Google Scholar]
- 102.Murakami H, Borde V, Shibata T, Lichten M, Ohta K. Correlation between premeiotic DNA replication and chromatin transition at yeast recombination initiation sites. Nucleic Acids Res. 2003;31:4085–-90. doi: 10.1093/nar/gkg441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murakami H, Keeney S. Regulating the formation of DNA double-strand breaks in meiosis. Genes Dev. 2008;22:286–-92. doi: 10.1101/gad.1642308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murakami H, Nurse P. Meiotic DNA replication checkpoint control in fission yeast. Genes Dev. 1999;13:2581–-93. doi: 10.1101/gad.13.19.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Murakami H, Nurse P. DNA replication and damage checkpoints and meiotic cell cycle controls in the fission and budding yeasts. Biochem. J. 2000;349:1–-12. doi: 10.1042/0264-6021:3490001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murakami H, Nurse P. Regulation of premeiotic S phase and recombination-related double-strand DNA breaks during meiosis in fission yeast. Nat. Genet. 2001;28:290–-93. doi: 10.1038/90142. [DOI] [PubMed] [Google Scholar]
- 107.Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, et al. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–-79. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nabeshima K, Villeneuve AM, Hillers KJ. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–-92. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nagaoka SI, Hassold TJ, Hunt PA. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012;13:493–-504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–-57. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nottke AC, Beese-Sims SE, Pantalena LF, Reinke V, Shi Y, Colaiacovo MP. SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc. Natl. Acad. Sci. USA. 2011;108:12805–-10. doi: 10.1073/pnas.1102298108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, et al. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc. Natl. Acad. Sci. USA. 2006;103:8131–-36. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ogino K, Masai H. Rad3-Cds1 mediates coupling of initiation of meiotic recombination with DNA replication. Mei4-dependent transcription as a potential target of meiotic checkpoint. J. Biol. Chem. 2006;281:1338–-44. doi: 10.1074/jbc.M505767200. [DOI] [PubMed] [Google Scholar]
- 114.Ohta K, Wu TC, Lichten M, Shibata T. Competitive inactivation of a double-strand DNA break site involves parallel suppression of meiosis-induced changes in chromatin configuration. Nucleic Acids Res. 1999;27:2175–-80. doi: 10.1093/nar/27.10.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, et al. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell. 2012;151:603–-18. doi: 10.1016/j.cell.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 116.Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–-56. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 117.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–-89. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 118.Pak J, Segall J. Role of Ndt80, Sum1, and Swe1 as targets of the meiotic recombination checkpoint that control exit from pachytene and spore formation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2002;22:6430–-40. doi: 10.1128/MCB.22.18.6430-6440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–-31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell. 2011;146:372–-83. doi: 10.1016/j.cell.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 121.Penkner AM, Fridkin A, Gloggnitzer J, Baudrimont A, Machacek T, et al. Meiotic chromosome homology search involves modifications of the nuclear envelope protein Matefin/SUN-1. Cell. 2009;139:920–-33. doi: 10.1016/j.cell.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 122.Pineda-Krch M, Redfield RJ. Persistence and loss of meiotic recombination hotspots. Genetics. 2005;169:2319–-33. doi: 10.1534/genetics.104.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Plug AW, Xu J, Reddy G, Golub EI, Ashley T. Presynaptic association of Rad51 protein with selected sites in meiotic chromatin. Proc. Natl. Acad. Sci. USA. 1996;93:5920–-24. doi: 10.1073/pnas.93.12.5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Resnick MA. The repair of double-strand breaks in DNA; a model involving recombination. J. Theor. Biol. 1976;59:97–-106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 125.Roberts P. Interchromosomal effects and the relation between crossing-over and nondisjunction. Genetics. 1962;47:1691–-709. doi: 10.1093/genetics/47.12.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, et al. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:1868–-80. doi: 10.1128/MCB.02063-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rocco V, Nicolas A. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells. 1996;1:645–-61. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- 128.Rockmill B, Lefrancois P, Voelkel-Meiman K, Oke A, Roeder GS, Fung JC. High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLOS Genet. 2013;9:e1003932. doi: 10.1371/journal.pgen.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–-68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 130.Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, Keeney S. Mouse TRIP13/PCH2 is required for recombination and normal higher-order chromosome structure during meiosis. PLOS Genet. 2010;6:e1001062. doi: 10.1371/journal.pgen.1001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rosu S, Libuda DE, Villeneuve AM. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number. Science. 2011;334:1286–-89. doi: 10.1126/science.1212424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rosu S, Zawadzki KA, Stamper EL, Libuda DE, Reese AL, et al. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLOS Genet. 2013;9:e1003674. doi: 10.1371/journal.pgen.1003674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rouyer F, Simmler MC, Johnsson C, Vergnaud G, Cooke HJ, Weissenbach J. A gradient of sex linkage in the pseudoautosomal region of the human sex chromosomes. Nature. 1986;319:291–-95. doi: 10.1038/319291a0. [DOI] [PubMed] [Google Scholar]
- 134.Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat. Rev. Mol. Cell Biol. 2010;11:182–-95. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sasanuma H, Hirota K, Fukuda T, Kakusho N, Kugou K, et al. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 2008;22:398–-410. doi: 10.1101/gad.1626608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–-53. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 137.Schmid R, Grellscheid SN, Ehrmann I, Dalgliesh C, Danilenko M, et al. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res. 2013;41:10170–-84. doi: 10.1093/nar/gkt811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–-35. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 139.Sedgwick RP, Boder E. Ataxia-Telangiectasia. In: de Jong JMBV, editor. Handbook of Clinical Neurology. Elsevier Sci. Publ.; Amsterdam: 1991. pp. 347–-423. [Google Scholar]
- 140.Shimmoto M, Matsumoto S, Odagiri Y, Noguchi E, Russell P, Masai H. Interactions between Swi1-Swi3, Mrc1 and S phase kinase, Hsk1 may regulate cellular responses to stalled replication forks in fission yeast. Genes Cells. 2009;14:669–-82. doi: 10.1111/j.1365-2443.2009.01300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shin YH, Choi Y, Erdin SU, Yatsenko SA, Kloc M, et al. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLOS Genet. 2010;6:e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shuster EO, Byers B. Pachytene arrest and other meiotic effects of the start mutations in Saccharomyces cerevisiae. Genetics. 1989;123:29–-43. doi: 10.1093/genetics/123.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Simchen G. Commitment to meiosis: what determines the mode of division in budding yeast? Bioessays. 2009;31:169–-77. doi: 10.1002/bies.200800124. [DOI] [PubMed] [Google Scholar]
- 144.Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 1997;136:957–-67. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Smith KN, Penkner A, Ohta K, Klein F, Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr. Biol. 2001;11:88–-97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 146.Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell. 2013;49:43–-54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 147.Sourirajan A, Lichten M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008;22:2627–-32. doi: 10.1101/gad.1711408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLOS Genet. 2013;9:e1003679. doi: 10.1371/journal.pgen.1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA breaks at the S. pombe recombination hot spot M26. Mol. Cell. 2002;9:847–-55. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- 150.Sturtevant AH. Contributions to the Genetics of Drosophila melanogaster. Carnegie Inst. Wash; Washington, DC: 1919. Inherited linkage variations in the second chromosome. pp. 305–-41. [Google Scholar]
- 151.Suzuki DT. Interchromosomal effects on crossing over in Drosophila melanogaster. II. A reexamination of X chromosome inversion effects. Genetics. 1963;48:1605–-17. doi: 10.1093/genetics/48.12.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Sym M, Engebrecht JA, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–-78. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 153.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–-35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 154.Terasawa M, Shinohara A, Hotta Y, Ogawa H, Ogawa T. Localization of RecA-like recombination proteins on chromosomes of the lily at various meiotic stages. Genes Dev. 1995;9:925–-34. doi: 10.1101/gad.9.8.925. [DOI] [PubMed] [Google Scholar]
- 155.Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. Localization and roles of Ski8p in Sordaria macrospora meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc. Natl. Acad. Sci. USA. 2003;100:12865–-70. doi: 10.1073/pnas.2034282100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510:241–-46. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tischfield SE, Keeney S. Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle. 2012;11:1496–-503. doi: 10.4161/cc.19733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tonami Y, Murakami H, Shirahige K, Nakanishi M. A checkpoint control linking meiotic S phase and recombination initiation in fission yeast. Proc. Natl. Acad. Sci. USA. 2005;102:5797–-801. doi: 10.1073/pnas.0407236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tung KS, Hong EJ, Roeder GS. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA. 2000;97:12187–-92. doi: 10.1073/pnas.220464597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ubeda F, Wilkins JF. The Red Queen theory of recombination hotspots. J. Evol. Biol. 2011;24:541–-53. doi: 10.1111/j.1420-9101.2010.02187.x. [DOI] [PubMed] [Google Scholar]
- 161.Viera A, Rufas JS, Martinez I, Barbero JL, Ortega S, Suja JA. CDK2 is required for proper homologous pairing, recombination and sex-body formation during male mouse meiosis. J. Cell Sci. 2009;122:2149–-59. doi: 10.1242/jcs.046706. [DOI] [PubMed] [Google Scholar]
- 162.Vignard J, Siwiec T, Chelysheva L, Vrielynck N, Gonord F, et al. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLOS Genet. 2007;3:1894–-906. doi: 10.1371/journal.pgen.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wan L, Niu H, Futcher B, Zhang C, Shokat KM, et al. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008;22:386–-97. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wan L, Zhang C, Shokat KM, Hollingsworth NM. Chemical inactivation of Cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–-74. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Woglar A, Daryabeigi A, Adamo A, Habacher C, Machacek T, et al. Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLOS Genet. 2013;9:e1003335. doi: 10.1371/journal.pgen.1003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu H, et al. Mouse HORMAD1 and HORMAD2, two conserved meiotic chromosomal proteins, are depleted from synapsed chromosome axes with the help of TRIP13 AAA-ATPase. PLOS Genet. 2009;5:e1000702. doi: 10.1371/journal.pgen.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wu PY, Nurse P. Replication origin selection regulates the distribution of meiotic recombination. Mol. Cell. 2014;53:655–-62. doi: 10.1016/j.molcel.2014.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wu T-C, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–-66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:6572–-81. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–-28. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–-22. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 172.Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149:75–-87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zenvirth D, Loidl J, Klein S, Arbel A, Shemesh R, Simchen G. Switching yeast from meiosis to mitosis: double-strand break repair, recombination and synaptonemal complex. Genes Cells. 1997;2:487–-98. doi: 10.1046/j.1365-2443.1997.1370335.x. [DOI] [PubMed] [Google Scholar]
- 174.Zetka MC, Rose AM. The meiotic behavior of an inversion in Caenorhabditis elegans. Genetics. 1992;131:321–-32. doi: 10.1093/genetics/131.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang L, Kleckner NE, Storlazzi A, Kim KP. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc. Natl. Acad. Sci. USA. 2011;108:20036–-41. doi: 10.1073/pnas.1117937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Zhang L, Liang Z, Hutchinson J, Kleckner N. Crossover patterning by the beam-film model: analysis and implications. PLOS Genet. 2014;10:e1004042. doi: 10.1371/journal.pgen.1004042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Murakami H, Keeney S. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014 doi: 10.1016/j.cell.2014.06.028. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, Franklin FCH. Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. EMBO J. 2006;25:1315–-1323. doi: 10.1038/sj.emboj.7600992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Higgins JD, Sanchez-Morán E, Armstrong SJ, Jones GH, Franklin FCH. The Arabidopsis synaptonemal complex protein ZYP1 is required for normal fidelity of crossing-over and chromosome synapsis. Genes Dev. 2005;19:2488–-2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18:2557–-2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]