Abstract
Carbonic anhydrases (CA) are zinc-containing metalloenzymes that catalyze the reversible hydration of CO2. The three evolutionarily unrelated families of CAs are designated α-, β-, and γ-CA. Aquatic photosynthetic organisms have evolved different forms of CO2 concentrating mechanisms (CCMs) to aid Rubisco in capturing CO2 from the surrounding environment. One aspect of all CCMs is the critical roles played by various specially localized extracellular and intracellular CAs. Five CAs have previously been identified in Chlamydomonas reinhardtii, a green alga with a well-studied CCM. Here we identify a sixth gene encoding a β-type CA. This new β-CA, designated Cah6, is distinct from the two mitochondrial β-CAs in C. reinhardtii. Nucleotide sequence data show that the Cah6 cDNA contains an open reading frame encoding a polypeptide of 264 amino acids with a leader sequence likely targeting the protein to the chloroplast stroma. We have fused the Cah6 open reading frame to the coding sequence of maltose-binding protein in a pMal expression vector. The purified recombinant fusion protein is active and was used to partially characterize the Cah6 protein. The purified recombinant fusion protein was cleaved with protease Factor Xa to separate Cah6 from the maltose-binding protein and the purified Cah6 protein was used to raise an antibody. Western blots, immunolocalization studies, and northern blots collectively indicated that Cah6 is constitutively expressed in the stroma of chloroplasts. A possible role for Cah6 in the CCM of C. reinhardtii is proposed.
Carbonic anhydrase (CA) is a zinc-containing metalloenzyme that catalyzes the reversible interconversion of CO2 and HCO3−. The enzyme was first discovered in human erythrocytes (Meldrum and Roughton, 1933) but has since been found in many organisms, including animals, plants, archaebacteria, and eubacteria (Hewett-Emmett and Tashian, 1996). CA plays an important role in many physiological functions that involve decarboxylation or carboxylation reactions, including both photosynthesis and respiration. It also participates in the transport of inorganic carbon (Ci) to actively photosynthesizing cells or away from actively respiring cells (Henry, 1996). The known CAs can be grouped broadly into three independent families (Hewett-Emmett and Tashian, 1996) called α-CA, β-CA, and γ-CA. These three families have no significant sequence identities and are an example of convergent evolution of catalytic function (Hewett-Emmett and Tashian, 1996). Most α-CAs are active as monomers of about 30 kD with three His coordinating the zinc atom and are generally highly susceptible to inhibition by sulfonamide compounds (Moroney et al., 1985, 2001). All active β-CAs are either monomers, or homo/heteromers with a His and two Cys residues that serve as the zinc coordinating residues (Bracey et al., 1994; Rowlett et al., 1994). The structure of the γ-CA is remarkably different from that of α-CA or β-CA (Alber and Ferry, 1994). The γ-CA functions as a trimer of identical subunits. As in α-CAs, three His coordinate the zinc atom but the His residues are provided by two subunits. Like the α-CAs, γ-CAs are highly sensitive to sulfonamide compounds (Moroney et al., 1985, 2001). Although the primary sequences of these CA families are different, the active sites of these three types of CAs contain Zn2+ and all of them employ a two-step catalytic mechanism (Lindskog, 1997).
Five CAs have previously been identified in the green alga Chlamydomonas reinhardtii (Moroney et al., 2001). These known CAs include three α-CAs and two β-CAs. Two of these α-CAs, Cah1 and Cah2, are localized to the periplasmic space (Fujiwara et al., 1990; Fukuzawa et al., 1990). Cah1 and Cah2 are very similar proteins although they are differentially regulated. Cah1 is expressed under low CO2 conditions (0.035% CO2 [v/v]) but not under high CO2 (5% CO2 [v/v] in air) conditions. The role of Cah1, the most abundant CA in C. reinhardtii, is to facilitate the movement of CO2 across the plasma membrane, particularly when the pH in the medium is alkaline (Moroney et al., 1985). Cah2 is present in much lower amounts. Cah2 is poorly expressed under low CO2 and only slightly up-regulated under high CO2 conditions. The third α-CA, Cah3, is localized to the thylakoid lumen (Karlsson et al., 1998). Mutants defective in Cah3 require elevated CO2 levels for growth compared to wild-type cells. The primary function of Cah3 is to provide CO2 to Rubisco (Hanson et al., 2003), which is localized to the pyrenoid of the chloroplast (Borkhsenious et al., 1998).
Two β-CAs have also been identified in C. reinhardtii (Eriksson et al., 1996). Both of these β-CAs are localized to the mitochondria. Expression of the β-CAs in C. reinhardtii is strongly influenced by the CO2 concentration during growth (Eriksson et al., 1998) as these CAs are induced strongly under low CO2 conditions but not under high CO2 conditions. This report describes the identification of a newly discovered β-CA gene (Cah6) in C. reinhardtii and the partial biochemical characterization of the recombinant Cah6 protein. Sequence and immunolocalization studies suggest that the Cah6 protein is targeted to the chloroplast stroma. The expression of Cah6 is constitutive and is slightly up-regulated under low CO2 (air) conditions.
RESULTS
Screening of the Cosmid and cDNA Library for Genomic and cDNA Clones of Cah6
To find new β-CA genes, the mitochondrial β-CA protein (Ca1) sequence of C. reinhardtii was used to BLAST the expressed sequence tag (EST) database of C. reinhardtii. The search yielded many ESTs that were the gene products of the mitochondrial β-CA genes. However, several other ESTs also were found that, while encoding a β-CA, did not belong to either of the two known mitochondrial β-CA genes. These ESTs were from the same gene and were analyzed by contig assembly program (CAP; http://www.infobiogen.fr/services/analyseq/cgi-bin/cap_in.pl) to form a consensus Cah6 sequence. In order to amplify Cah6 and screen for the Cah6 cDNA, several PCR primers were designed based on this Cah6 contig.
The PCR primers F4 and R5 were used on an indexed cosmid library to isolate a cosmid carrying the Cah6 gene (Colombo et al., 2002). After several rounds of screening by PCR, two cosmid clones, designated 72-E-6 and 29-D-12, were isolated. Positive cosmid clones were verified as Cah6 clones after each round of screening by PCR, followed by sequencing of the PCR product using different Cah6 primers. PCR using primers F4 and R5 yielded a product of 2.8 kb when used on the isolated cosmid clones. The same 2 primers used on the cDNA core library yielded a PCR product of 2.4 kb which is 6 bp short from being a full-length Cah6 cDNA clone (excluding the poly A tail).
Sequencing and Homology Search
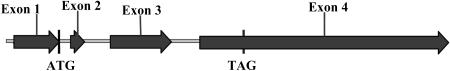
The cosmid clones 72-E-6 and 29-D-12, and the 2.4-kb cDNA PCR product mentioned above were sequenced in both directions. The sequencing results were confirmed by the EST and genomic Chlamydomonas databases (http://www.biology.duke.edu/chlamy_genome/). The Cah6 gene is 2,886-bp long with 4 exons and 3 introns (Fig. 1). The exons range in size from 93 bp to 1,652 bp while the introns range from 75 bp to 189 bp long. The genomic and cDNA sequence of Cah6 can be obtained from the GenBank (accession nos. AY463238 and AY463239).
Figure 1.
The genomic map of Cah6. Cah6 is 2,886 bp in length. The arrows represent the four exons while the horizontal line represents the three introns. The start and stop codons are labeled by black vertical lines.
The Cah6 cDNA is 2,452-bp long and encodes a putative protein of 264 amino acids. It contains a translation start site at nucleotide 299 and a stop site at nucleotide 1,091. It has an extremely long 3′ untranslated region (UTR) of more than 1,300 bp. Predictions based on various protein prediction programs (SORT P, CHLOR P, AND TARGET P) listed under ExPasy tools (http://ca.expasy.org/tools/#translate) indicate that the Cah6 protein is likely to be targeted to the chloroplast, as the protein contains a putative chloroplastic transit peptide of 39 amino acids. A protein database search using the Cah6 protein sequence showed that it is similar to β-CAs from Escherichia coli, green algae, and higher plants. A multiple sequence alignment of the Cah6 protein with that of β-CAs from other green algae and higher plants shows that it contains the characteristic one His and two Cys residues as zinc coordination residues, characteristic of enzymatically active β-CAs (Fig. 2).
Figure 2.
Alignment of Cah6 protein sequence with those of other well-characterized β-CAs. Ca1 represents the C. reinhardtii mitochondrial β-CA. Mitochondrial β-Ca1 and β-Ca2 (sequence not shown in the alignment) are almost identical in amino acid sequence and have only one amino acid difference in their sequences. Coccomyxa represents the cytosolic β-CA from the symbiotic alga Coccomyxa. Active site residues are in bold and highlighted. Asterisks represent a completely conserved amino acid; colons represent conserved amino acid substitutions; and periods represent semiconserved amino acid substitutions.
Cloning of the Cah6 in an Overexpression Vector
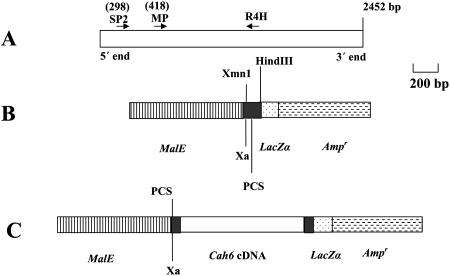
Cah6 was cloned into the expression vector pMal to study the properties of Cah6 protein and to raise an antibody against it. Two different MalE-Cah6 recombinant overexpression constructs were generated. One contained the cDNA sequence coding for the full-length open reading frame (ORF) of the Cah6 protein and the other contained the cDNA sequence that would code for the mature Cah6 protein predicted by the web protein prediction programs. The SP2 + R4H primers and the MP + R4H primers were used to amplify the cDNA coding for the full-length and putative mature Cah6 proteins, respectively (Fig. 3A).
Figure 3.
Generation of the recombinant pMal-Cah6 expression construct. A, A schematic figure showing the alignment of primers used to amplify Cah6 cDNA. PCR primers are denoted by black arrows. SP2 and R4H primers were used to amplify a cDNA PCR product that codes for the full-length protein while MP and R4H primers were used to amplify a cDNA product that codes for the putative mature protein. R4H primer has a HindIII site incorporated at the 5′ end. The numbers on the top of the primers denote the primer position in bp. B, A part of the pMal vector showing the polylinker cloning site (PCS),β-galactosidase (LacZα), and β-lactamase (Ampr) genes, XmnI and HindIII recognition sites. Xa denotes Factor Xa cleavage site. MalE codes for MBP. C, A schematic figure of the pMal-Cah6 recombinant construct.
Amplified cDNAs were purified from the gel and cloned into the overexpression vector pMal-c2x (Fig. 3B) to generate two different types of recombinant constructs. An in-frame insertion of Cah6 with the sequence of MBP in the recombinant clone was verified by restriction enzyme digestion analyses and DNA sequencing. Two clones were selected out of the 25 recombinant clones of each type. Clone B48 had a cDNA insert coding for the full-length Cah6 protein while clone B3 had one coding for the mature Cah6 protein.
Overexpression of MBP-Cah6
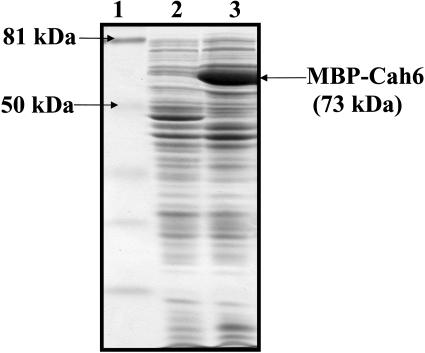
E. coli cells harboring the recombinant B48 and B3 recombinant constructs were induced with 1 mm isopropylthio-β-galactoside for 2 h at 37°C to overexpress the MBP-Cah6 fusion protein. Equal amounts of proteins from induced and uninduced cells were loaded on a 12% SDS-polyacrylamide gel and subjected to electrophoresis (Fig. 4). The overexpressed recombinant Cah6 fusion protein was 15% of the total E. coli cell protein.
Figure 4.
A 12% SDS-polyacrylamide gel showing overexpression of recombinant MBP-Cah6. Lane 1 represents prestained low molecular mass markers. Lanes 2 and 3 represent 30 μg of proteins from uninduced and induced E. coli cells, respectively.
Purification and Activity Assays of the Recombinant Cah6
Crude cell extracts from the B48 and B3 clones were used for CA activity assays. CA activity was detected in cell extracts of the induced B48 clone but not in that of the B3 clone. The B48 clone was selected for further study. This clone had the entire ORF of Cah6. The overexpressed recombinant Cah6 protein was purified by affinity chromatography using amylose resin following a protocol in the New England Biolabs technical catalog. Purified recombinant Cah6 was further concentrated by using a 100-kD cut-off centricon column.
The CA activity in the sample at each step of purification was assayed to check the purity of the Cah6 sample (Table I). The recombinant Cah6 protein was found to have a specific activity of 400 Wilbur-Anderson units (WAU)/mg. This calculation of specific activity was based on the total amount of recombinant fusion protein in the sample. CA activity assays were done using the method of Wilbur and Anderson (1948). CA activity was not detected in the extracts from uninduced cells containing the B48 clone or E. coli cells containing only the pMal vector.
Table I.
Purification of the chimeric MBP-Cah6
| Step | Total Activity | Protein | Specific Activity | Recovery 100% | Purification Fold |
|---|---|---|---|---|---|
| WAUa | mgb | WAU/mg | |||
| Sonicated cells | 1,200 | 8,000 | 0.15 | 100 | 1 |
| Amylose column | 800 | 3 | 300 | 67 | 2,000 |
| Centricon column | 400 | 1 | 400 | 33 | 2,567 |
One WAU = (t0−t)/t where t0 is the time for uncatalyzed reaction and t is the time for the enzyme catalyzed reaction.
Determined by Lowry's protein assay (Lowry et al., 1951).
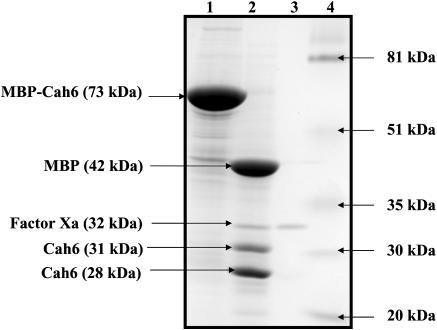
The fusion protein purified from the B48 clone was cleaved by the protease Factor Xa for 4 h at 23°C to separate Cah6 from the MBP. Factor Xa cleaved the fusion protein to yield 42 kD MBP and 31 kD Cah6 protein. It also nonspecifically cleaved the 31-kD Cah6 protein band to yield a 28-kD fragment. Factor Xa sometimes cleaves nonspecifically at other basic residues depending on the conformation of the protein substrate. The mature Cah6 protein starts with an Arg residue. This can explain the generation of the 28-kD fragment (similar in molecular mass to that of the mature Cah6 protein) by the nonspecific cleavage of 31 kD Cah6 protein. Use of common protease inhibitors like leupeptine did not prevent this nonspecific cleavage of Cah6. Purification and cleavage of the fusion protein was confirmed by performing SDS-PAGE (Fig. 5). There was no significant difference between the CA activities of the Factor Xa cleaved MBP-Cah6 fusion protein and undigested fusion protein. The 31-kD Cah6 protein band was excised from the gel to be used as an antigen for production of polyclonal Cah6 primary antibodies.
Figure 5.
A 12% SDS-polyacrylamide gel showing undigested and Factor Xa digested purified MBP-Cah6 protein. Lane 1, 55 μg of undigested purified recombinant protein; lane 2, 55 μg of purified recombinant protein digested by 1 μg of Factor Xa; lane 3, 1 μg of Factor Xa; lane 4, prestained low molecular mass markers.
Western-Blotting and Northern-Blotting Analyses of Cah6 Expression
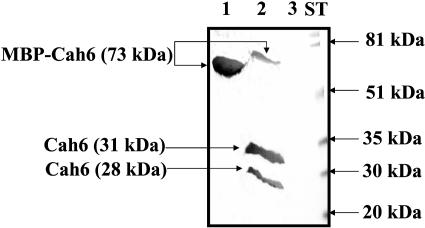
To test the specificity of the Cah6 antibody, Factor Xa (protease) cleaved MBP-Cah6 and purified MBP-Cah6 fusion proteins were separated by 12% SDS-PAGE and probed with the Cah6 antibody (Fig. 6). The antibody did not react with the MBP with induced E. coli cells containing only the pMal-c2x vector (data not shown). Proteins extracted from high CO2 and air acclimated D66 cells were separated by electrophoresis and probed with the Cah6 and mitochondrial β-CA primary antibodies (Fig. 7). The mitochondrial CAs are expressed only under low CO2 conditions but not under high CO2 conditions. Hence, the mitochondrial β-CA primary antibodies were used as a control to confirm that the cells used for the experiment were high and low CO2 adapted. The Cah6 antibody detected the Cah6 protein (28.5-kD band) in both the high CO2 and air acclimated cells. The air acclimated cells showed slight up-regulation of the Cah6 protein compared to that of the high CO2 acclimated cells (Fig. 7A). The mitochondrial β-CA antibody recognized the 22-kD mitochondrial β-CA protein in the air acclimated cells but not in the high CO2 grown cells (Fig. 7B), in agreement with earlier observations (Eriksson et al., 1998).
Figure 6.
A western blot probed by the Cah6 antibody using purified overexpressed MBP-Cah6 protein. Lane 1, 20 μg of purified undigested fusion protein; lane 2, 20 μg of fusion protein cleaved by 1 μg of Factor Xa; lane 3, 1 μg of Factor Xa. ST represents prestained low molecular mass markers.
Figure 7.
Western blots probed by the Cah6 and mitochondrial β-CA antibodies using wild-type Chlamydomonas cells. A, A western blot probed by Cah6 antibody using high (5% CO2 in air [v/v]) and air (0.035% CO2 [v/v]) acclimated wild-type Chlamydomonas cells grown in minimal medium. B, A western blot probed by mitochondrial β-CA antibody using high and air acclimated wild-type Chlamydomonas cells grown in minimal medium.
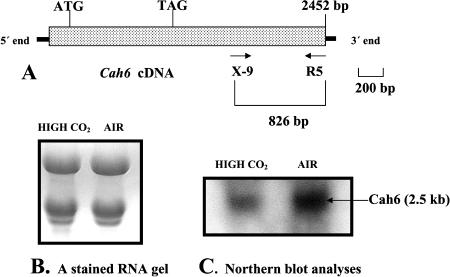
The primers X-9 and R5 were used to amplify an 826-bp PCR product from the cDNA core library (Fig. 8A). This 826-bp PCR product is contained within the 3′ UTR of Cah6 and was used as a probe for northern-blot analyses using mRNA extracted from high and low CO2 acclimated C. reinhardtii (strain D66) cells grown in minimal medium. Twenty micrograms of RNA from both cell types were loaded on the RNA gel (Fig. 8B). Northern blotting shows that Cah6 is expressed under both low and high CO2 conditions, but is slightly up-regulated in cells acclimated to air levels of CO2 (Fig. 8C).
Figure 8.
Northern-blot analyses of Cah6 expression. A, A schematic figure showing the alignment of primers used for making the probe for northern-blot analyses of Cah6 expression. B, A stained RNA gel showing RNA extracted from high and low CO2 acclimated wild-type cells grown in minimal medium. Each lane contains 20 μg of total RNA. C, Northern-blot result using 826-bp PCR product as a probe.
Immunolocalization of Cah6
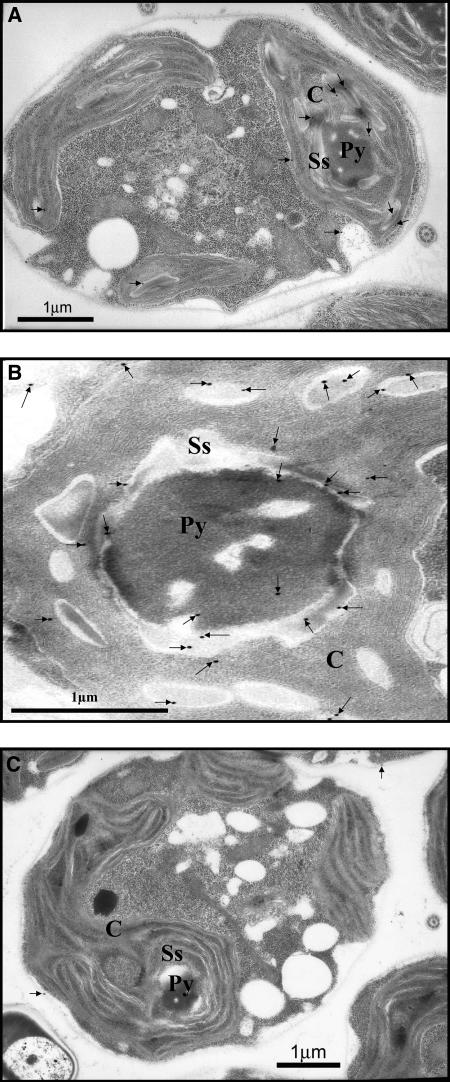
Air acclimated D66 and CC-124 cells grown in minimal medium were used for immunolocalization of Cah6. C. reinhardtii cell sections were probed with the Cah6 antibody or the preimmune serum and observed under a transmission electron microscope (Fig. 9). Immunogold densities in different cell compartments are given in Table II. The immunogold densities in different cell compartments in sections were calculated by dividing the number of immunogold particles in a particular cell organelle with the area of that cell organelle. Immunolocalization results demonstrated that Cah6 is located in the stroma of the chloroplast (Fig. 9A) and is 4-fold more abundant in the area around the pyrenoid (Fig. 9B) compared to the other areas of the stroma. C. reinhardtii cell sections probed with the preimmune serum served as negative controls for the immunolocalization study (Fig. 9C).
Figure 9.
A, Transmission electron micrograph showing the immunogold labeling of C. reinhardtii CC-124 cells probed with the Cah6 antibody. Cells grown under low CO2 (0.035%) conditions in minimal medium were probed with the Cah6 antibody. B, Transmission electron micrograph showing the immunogold density around the pyrenoid in C. reinhardtii cells probed with the Cah6 antibody. Cells grown under low CO2 (0.035%) conditions in minimal medium were probed with the Cah6 antibody. C, Transmission electron micrograph showing the immunogold labeling of C. reinhardtii cells probed with the preimmune serum. Low CO2 (0.035%) acclimated CC-124 cells grown in minimal medium were used. For all micrographs, Ss, Py, and C denote starch sheath, pyrenoid, and chloroplast, respectively. Immunogold labelings are shown by small black arrows.
Table II.
Intracellular localization of Cah6 using wild type C. reinhardtii cells
| Location | Area | Immunogold Density
|
||
|---|---|---|---|---|
| Immune | Preimmune | Difference | ||
| μm2 | No. of Immunogold Particles/μm2 | |||
| Outside | 7.80 ± 0.38 | 1.67 ± 0.10 | 1.54 ± 0.11 | 0.13 ± 0.19 |
| Cytoplasm | 6.84 ± 1.2 | 0.44 ± 0.02 | 0.30 ± 0.02 | 0.14 ± 0.03 |
| Nucleus | 0.36 ± 0.05 | 0.47 ± 0.02 | 0.47 ± 0.02 | 0.00 ± 0.01 |
| Stroma | 5.93 ± 1.07 | 3.0 ± 0.14 | 0.85 ± 0.04 | 2.15 ± 0.13 |
| Pyrenoid | 1.85 ± 0.30 | 0.99 ± 0.08 | 0.97 ± 0.7 | 0.02 ± 0.03 |
| Starch sheath | 0.86 ± 0.11 | 10.9 ± 1.9 | 2.3 ± 0.14 | 8.6 ± 1.95 |
Immunogold densities in different cell compartments in sections were calculated by dividing the number of immunogold particles in a particular cell organelle with the area of that cell organelle. The data presented in the table is the average ± sd of 15 cell sections. The stroma includes the thylakoid area. The cytoplasmic area was calculated by subtracting the total area of chloroplast and the nucleus from the cell area. The Cah6 antibody dilution used for immunolocalization is 1:10. The cells used for immunolocalization are air acclimated D66 cells grown in minimal medium.
Partial Characterization of the Cah6 Activity
The effects of sulfonamide and anion inhibitors on the CA activity of recombinant Cah6 were studied. Table III shows the inhibition of recombinant Cah6 by sulfonamides and anions. Cah6 was comparatively less inhibited by the sulfonamides and more inhibited by the anions azide and cyanide than bovine CAII, which is an α-CA. Generally, all β-CAs are less sensitive to sulfonamide inhibition than are α-CAs and have sulfonamide I50 ranging from 2 μm to 10 μm (Johansson and Forsman, 1993). The I50 of Cah6 falls within this range.
Table III.
The inhibition constants of the bovine CAII and heterologously produced Cah6
| Inhibitor | I50 of Bovine CA | I50 of Cah6 |
|---|---|---|
| M | M | |
| Acetazolamide | 1.4 × 10−8 | 2 × 10−6 |
| Ethoxyzolamide | 1.2 × 10−9 | 9 × 10−6 |
| Azide | 1.1 × 10−3 | 1.5 × 10−5 |
| Cyanide | 4.9 × 10−5 | 5 × 10−6 |
DISCUSSION
β-CAs were first recognized in photosynthetic organisms (Burnell et al., 1990; Fawcett et al., 1990) but later have been identified in eubacteria, cyanobacteria, yeast, micro-algae, and higher plants. They play important physiological roles in these organisms. For example, a carboxysomal β-CA in cyanobacteria, coded by icfA gene, converts HCO3− to CO2 (Badger and Price, 1992). This increases the concentration of CO2 at the site of Rubisco, ensuring efficient CO2 fixation. In Saccharomyces cerevisiae, deletion of the β-CA-like gene NCE103 causes an oxygen-sensitive growth defect (Götz et al., 1999). It has also been found that the tobacco salicylic acid-binding protein 3 (SABP3) is a chloroplast β-CA that exhibits antioxidant activity and plays a role in the hypersensitive defense response (Slaymaker et al., 2002). In E. coli, the cynT gene that codes for a β-CA is a part of the cyn operon. CO2 is produced in the reaction of cyanate with HCO3−, and this β-CA recycles the CO2 back to HCO3−, so that it does not diffuse out of the cell (Guilloton et al., 1993). Finally, a β-CA gene identified in Corynebacterium glutamicum has been shown to be essential for achieving normal growth under atmospheric conditions (Mitsuhashi et al., 2003). These researchers have shown that the effect of this β-CA is most likely due to its ability to maintain favorable intracellular HCO3− levels, particularly during exponential growth phases and during l-lysine overproduction, both of which are conditions of higher HCO3− demand (Mitsuhashi et al., 2003).
Two nearly identical β-CAs were identified in 1996 in C. reinhardtii (Eriksson et al., 1996). Both the β-CAs (Ca1 and Ca2) are located in the mitochondria. It has been suggested that the mitochondrial CAs of C. reinhardtii may play roles in recycling both respiratory and photorespiratory CO2 by converting it to HCO3− in the mitochondrial matrix (Raven, 2001). This HCO3− then would leak back into the cytosol where it would be available for transport into the chloroplast stroma. Recently, it has been shown that the expression of mitochondrial CAs (Ca1 and Ca2) decreases when the external NH4+ concentration decreases, to the point of being undetectable when the supply of NH4+ restricts the rate of photoautotrophic growth (Giordano et al., 2003). The expression of these CAs was induced at 0.2% CO2 condition by increasing the NH4+ concentration in the growth medium. These researchers have proposed that the mitochondrial CAs are involved in supplying HCO3− for anaplerotic assimilation catalyzed by phosphoenolpyruvate carboxylase, which in turn provides carbon skeletons for nitrogen assimilation.
Researchers in several laboratories have tried to assay CA activity in C. reinhardtii chloroplasts. Using mass-spectrometric measurements of 18O exchange, Sültemeyer et al. (1995) have characterized two chloroplastic CA activities in C. reinhardtii cells. One CA activity is an insoluble form associated with the thylakoid fraction and is less sensitive to ethoxyzolamide (EZ) while the other is a soluble form and sensitive to EZ (Amoroso et al., 1996). Villarejo et al. (2001) have found a new chloroplast envelope CA activity that is sensitive to EZ and is constitutively expressed. Support for the presence of the insoluble chloroplastic CA activity is provided by the identification of the 29-kD Cah3, which is located on the lumenal side of thylakoid membrane (Karlsson et al., 1998).
Here we report the identification of a nuclear gene encoding a chloroplastic β-CA, Cah6, in C. reinhardtii. Cah6, the sixth carbonic anhydrase gene and the third β-CA gene to be identified in C. reinhardtii, is located in the stroma of the chloroplast and likely represents the soluble chloroplastic CA activity detected earlier. The Cah6 protein has two Cys residues and one His residue that coordinate zinc, similar to all known enzymatically active β-CAs (Moroney et al., 2001). Furthermore, like all other β-CAs, Cah6 is 100- to 1,000-fold less sensitive to sulfonamide compounds like acetazolamide and EZ than the bovine CAII, an α-CA (Table III). Cah6 was 100-fold and 10-fold more sensitive to azide and cyanide, respectively, than the bovine alpha CAII. The full-length protein has a calculated molecular mass of 28 kD and a pI of 7.0. It has an apparent weight of 31 kD on a SDS-polyacrylamide gel. The mature Cah6 protein (beginning from amino acid residue 40) has a calculated molecular mass of 26 kD and a pI of 6.58. It has an apparent molecular mass of 28.5 kD on SDS-polyacrylamide gels (Fig. 7A). Cah6 is similar to β-CAs from E. coli, green algae, and higher plants, with an amino acid identity of 23% to 34%. Cah6 most closely resembles the green alga Coccomyxa β-CA with a 34% identity.
CA activity was detected in cell extracts of the induced B48 clone but not in that of the B3 clone of E. coli expressing the Cah6 gene. The B48 clone contains the entire ORF of Cah6, whereas the B3 clone had only the cDNA coding for the putative mature Cah6 protein. One explanation for the lack of activity is that Cah6 is cleaved at a point different than that predicted by the different target prediction programs. A second possible explanation for the lack of activity of the protein from B3 clone is that the first amino acid of the mature Cah6 was eliminated to facilitate cloning. During the cloning of the putative mature Cah6, the codon for Arg was substituted as Factor Xa does not cleave any protein that starts with an Arg after the Factor Xa recognition sequence (Ile-[Glu-Asp]-Gly-Arg). Thus the mature Cah6 protein in the B3 clone started with a Ser instead of an Arg residue. It is therefore possible that this change caused the recombinant fusion protein to become insoluble or misfold. It is clear however that Cah6 is an active CA as the full-length protein had a specific activity well within the normal range found for β-CAs.
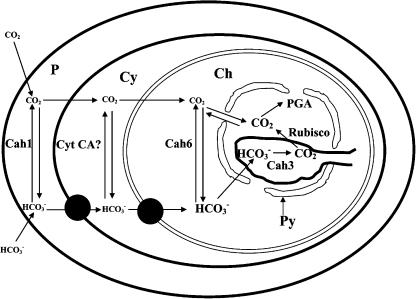
Western- and northern-blotting analyses show that Cah6 is constitutively expressed and slightly up-regulated under low CO2 conditions. Immunolocalization shows that Cah6 is localized in the stroma of the chloroplast and is not present in the pyrenoid. In Chlamydomonas, 90% of the Rubisco is localized inside the pyrenoid under low CO2 conditions (Borkhsenious et al., 1998). Western blots and the lack of immunogold labeling of the pyrenoid indicated that the Cah6 antibody is not cross reacting with Rubisco. Interestingly, in C. reinhardtii immunogold density is4-fold higher in the area around the pyrenoid, particularly around the starch sheath that surrounds the pyrenoid, compared to that in the other areas in the stroma of the chloroplast (Table III). Based on these observations, we hypothesize that Cah6 indirectly plays a role in the CCM by trapping CO2 diffusing out from the pyrenoid, the site of localization of Rubisco in C. reinhardtii, and converting it to HCO3− (Fig. 10). This conversion of CO2 to HCO3− would increase the HCO3− pool in the stroma, thereby retaining inorganic carbon within the chloroplast.
Figure 10.
A model showing the potential role of Cah6 and other known CAs in the operation of CCM in C. reinhardtii. The font sizes of CO2 and HCO3− indicate the relative concentrations of these Ci species. Cyt CA? represents a putative cytoplasmic carbonic anhydrase. P, Cy, Ch, and Py represent periplasm, cytoplasm, chloroplast, and pyrenoid, respectively. Cah1 and Cah3 represent the periplasmic and thylakoid CAs, respectively. Putative HCO3− transporters are denoted by small black circles.
A mutant of C. reinhardtii, cia5, which requires elevated levels of CO2 for growth, fails to induce CCM and does not synthesize any of the low CO2 inducible mitochondrial and periplasmic CAs along with other low CO2 inducible polypeptides (Manuel and Moroney, 1988). It is deficient in a putative transcription factor coded by the Cia5 gene (Fukuzawa et al., 2001; Xiang, et al., 2001). Katzman et al. (1994) used a 14C assay to measure the CA activity in the chloroplasts of cia5 and wild-type cells of Chlamydomonas and found that the CA activity of cia5 is almost identical to that in wild-type cells. Cah3 is an α-CA that is localized to the lumenal side of thylakoids in the chloroplast (Karlsson et al., 1998). The Cah3 and the Cah6 antibodies specifically detect the Cah3 and Cah6 protein in the cia5 cells (data not shown). So it appears that both Cah3 and Cah6 contributed to the total CA activity measured by Katzman et al. (1994) in the chloroplasts of cia5. This also suggests that Cah6 expression is probably not under the regulation of the Cia5 gene. Immunoblotting and northern-blotting analysis (using Cah3 and Cah6 specific probes) of cia5 and wild-type cells of C. reinhardtii could show the expression pattern of Cah6 gene in this mutant and could confirm if these genes are under the regulation of the Cia5 gene. Generation of mutants of Cah6 could help to confirm the physiological role of Cah6 in photosynthesis and CCM.
There has been a surge of interest in CAs from plants and algae over the past decade. This interest began with the discovery of the β-CA in plants in 1990 (Fawcett et al., 1990) and has continued with the finding of multiple α- and β-CAs in C. reinhardtii and Arabidopsis and the determination of the critical physiological roles CAs have in cyanobacteria and macro-algae. Analysis of the Arabidopsis genomic database reveals at least 14 genes potentially encoding CAs that have homologies with ESTs and are expressed in cells (Moroney et al., 2001). Clearly the number of CAs in plants is much greater than previously thought. C. reinhardtii, a unicellular green alga, is not far behind with six CAs already characterized. The availability of Arabidopsis and Chlamydomonas genome sequences and EST databases can be used to find out the exact number of expressed CA isoforms in these organisms. As there appears to be a large number of isoforms in plants and algae, the challenge for future researchers will be to determine the expression patterns, localization, and physiological roles for each of these isoforms.
MATERIALS AND METHODS
Strains and Media
Chlamydomonas reinhardtii strain D66 (nit2−, cw15, mt+) was obtained from Rogene Schnell, University of Arkansas (Little Rock, AR; Schnell and LeFebvre, 1993). The wild-type strain CC-124 was obtained from the Duke culture collection. To start cultures, cells from yeast acetate medium plates were inoculated into 100 mL of Tris-acetate phosphate (TAP) medium (Sueoka, 1960) and grown with continuous shaking and light (300 μmol photons m−2 s−1) for 2 d. An aliquot of the culture was then transferred to 1.5 L of minimal medium (Sueoka, 1960) and bubbled with high CO2 (5% CO2 [v/v] in air) until it reached a cell density of about 2 × 106 cells mL−1. The culture was diluted with an equal volume of fresh medium and split into two flasks. One was bubbled with high CO2 (5% CO2 [v/v] in air) and the other with air (0.035% [v/v] CO2). The cells were low CO2 acclimated for 12 h. The high and low CO2 acclimated D66 cells were used for RNA isolation, measurement of chlorophyll content, and western blotting. Only low CO2 acclimated cells were used for immunolocalization studies.
Total RNA Isolation and Northern-Blot Analysis
Extraction of total RNA from low CO2 and high CO2 acclimated D66 cells and RNA gel-blot analyses were performed by standard procedures (Sambrook et al., 1989). RNAs from high and low CO2 acclimated cells were transferred to a BA-S 85 nitrocellulose membrane (Schleicher and Schuell Bioscience, Keene, NH). When Cah6 5′end primer X-9 (5′AAACTCAACTCCTTCATAATAGGC-3′) and 3′end primer R5 (5′-TGCGGTACAGATTACAGTCA-3′) were used to perform PCR on the cDNA core library, an 826-bp PCR product was generated. This product was used to make a radioactive probe to study the expression pattern of Cah6 under low and high CO2 conditions. The PCR product corresponded to a unique 826 bp region in the 3′ UTR of Cah6. 32P-dCTP labeled probes were prepared using a random primer procedure (Sambrook et al., 1989).
DNA Preparation, Sequencing, and Homology Analysis
Plasmid and cosmid DNA was purified using a combination of the standard ethanol precipitation method (Sambrook et al., 1989) followed by the purification using the spin columns from a commercial kit (Qiagen, Chatsworth, CA). cDNA and genomic PCR products were purified from the 0.8% [w/v] agarose gels. Amplified bands were digested from the gel and were treated with 6 m NaI at 55°C to melt the gel piece. DNA was purified from the liquefied gel using the mini spin columns from the commercial kit mentioned above. DNA was sequenced using ABI dye terminators (Perkin Elmer Applied Biosystems, Foster City, CA; for some PCR fragments and cosmids enriched in GC content, the use of dGTP-BigDye generated better sequences than dITP-BigDye).
Homology searches (against Chlamydomonas EST and the full database) were performed using the BLAST server (http://www.ncbi.nlm.nih.gov/BLAST; Altschul et al., 1997) Exon/intron splice sites and ORFs were identified manually (Silflow, 1998) as well as by using GreenGenie (Kulp et al., 1996; http://www.cse.ucsc.edu/%7Edkulp/cgi-bin/greenGenie). Signal peptide analysis, molecular mass, and pI calculations were determined with different protein prediction programs like CHLOR P, TARGET P, and SORT P, which had hyperlinks in the ExPasy server (http://ca.expasy.org/tools/#translate).
cDNA Library Preparation and Amplification of Cah6 cDNA from the Prepared Library
A cDNA core library (in bacteriophage λ) was obtained from the Chlamydomonas Genetics Center at Duke University (Durham, NC). This core library was made from cDNAs prepared from CC-1690 cells grown to mid-log phase in TAP (acetate-containing) medium in the light, TAP medium in the dark, high salt (HS; minimal) medium in ambient levels of CO2, and HS medium bubbled with 5% CO2. cDNAs were cloned into the lambda Zap II (Stratagene, La Jolla, CA) in the EcoRI (5′) and XhoI (3′) sites. This cDNA library was amplified following the protocol given in the Stratagene manual. Cah6 primers F4 (5′-GCACGAGGCAACATTAAACA-3′) and R5 when used on the cDNA core library yielded a PCR product of 2446 bp. This product codes for the full-length Cah6 protein.
Screening of the Cosmid Library
To obtain wild-type genomic clones of the Cah6 gene, a PCR-based screen of an indexed cosmid library was used (Pollock et al., 2003). An indexed cosmid library was constructed using a cosmid library from Saul Purton, University of London (Purton and Rochaix, 1994). Briefly, 7,680 different Escherichia coli lines carrying single cosmids were grown in Luria-Bertani media on 80 different 96-well microtiter plates. Using this indexed library, 80 pools of cells, each containing 96 single cosmids, were generated. DNA from each pool, obtained by common alkaline lysis procedures (Sambrook et al., 1989), was used to create 10 superpools (each containing about 768 individual cosmids) that were suitable for PCR.
The mitochondrial β-CA protein (Ca1) sequence of C. reinhardtii was used to BLAST the EST database of C. reinhardtii. The search yielded many ESTs that were not the gene products of the mitochondrial β-CA genes although they encoded a β-CA. These ESTs were all from the same gene and were analyzed by contig assemblage program to form a consensus Cah6 sequence. Based on this Cah6 contig, primer sets were designed and used to screen the superpools. Cah6 primers F4 and R4 (5′-TTGCGCCATGAAGTCCCTAA-3′) were used for PCR. These primers amplified a Cah6 product of 1.8 kb. Once a plate carrying the correct cosmid was identified, a new set of pools was generated (12 pools, each containing 8 single cosmids). Finally, a new PCR reaction was performed with the single cosmid from the positive pool described above. Using this protocol, after four rounds of PCR, two cosmids, 72-E-6 and 29-D-12, containing the Cah6 gene were isolated from the cosmid library. PCR using primers F4 and R5 yielded a product of 2,880 bp when used on the isolated cosmid clones. This PCR product is just short of 6 bp from being the full-length genomic sequence of Cah6.
Production of Overexpression Constructs
Cah6 was cloned into the pMal-c2x overexpression vector (NEB, Beverly, MA) and fused to the MalE gene that encodes MBP. Two different pMal-Cah6 recombinant overexpression vectors were constructed. One contained the cDNA sequence coding for the ORF of Cah6 protein and the other contained the cDNA sequence that would code for the putative mature Cah6 protein. SP2 (5′ A TGGGATGCGGTGCCAGCGTG 3′) + R4H (5′-ATATAAGCTTTTGCGCCATGAAGTCCCT AA-3′) primers and MP (5′- AGCAACCGCAGCAGCCTT-3′) + R4H primers were used to amplify the cDNA coding for the full-length and mature Cah6 protein, respectively. SP2 + M1RH and MP + M1RH primers yielded a PCR product of 1.1 kb and 979 bp, respectively. A high fidelity DNA polymerase (Platinum Pfx DNA polymerase from Invitrogen, Carlsbad, California) was used for PCRs. The vector was double digested with the XmnI and HindIII. Amplified cDNAs were purified from the DNA gel using Qiagen spin columns and were digested with HindIII. Ligation of the insert to the overexpression vector pMal-c2x vector was performed following the protocol in the NEB technical manual. Transformations of DH5α cells were performed following the protocol in Sambrook et al. (1989). In-frame insertion of Cah6 with the sequence of MBP in the recombinant clone was verified by double restriction enzyme digestion analyses with SacI and HindIII and DNA sequencing.
Overexpression and Purification of MBP Fusion Proteins
Selected clones of Cah6 were grown at 37°C in 2 L Luria-Bertani medium + Glc (0.2%) + Amp (100 μg mL−1) cultures on a rotary shaker. Glc was added to the growth medium to repress the maltose genes on the chromosome of the E. coli host, one of which codes for amylase that can degrade the amylose on the affinity resin that is used for purification. The cells were induced for 2 h with 1 mm isopropylthio-β-galactoside at 37°C when the culture OD600 was between 0.6 and 0.7. Both induced and uninduced cells were harvested and resuspended in column buffer (20 mm Tris-HCl [pH 7.4], 200 mm NaCl, and 1 mm EDTA with or without 10 mm 2-mercaptoethanol) and ruptured in a prechilled French pressure cell. Equal amounts of protein samples of ruptured induced and uninduced cells were loaded on a 12% SDS-polyacrylamide gel and subjected to electrophoresis to verify the overexpression of the recombinant protein. The fusion protein was purified by one-step affinity chromatography using amylose resin. Amylose resin (1 mL of amylose resin binds 3 mg of the recombinant protein) was mixed with the crude ruptured cell extract on a shaker at room temperature for 2 h and poured into a 2.5-cm × 10-cm column to perform batch purification. The column was washed with 6 L of column buffer to remove other proteins. At the final step, fusion proteins were eluted from the column by column buffer containing 10 mm maltose. Purified recombinant fusion proteins were further concentrated by passing the extract through the 100-kD centricon columns (Amicon, Billerica, MA).
Recombinant proteins were cleaved from the MBP by digestion with Factor Xa. Fifty micrograms of the recombinant protein was digested by 1 μg of Factor Xa enzyme in the Factor Xa digestion buffer (20 mm Tris-HCl, 100 mm NaCl, and 2 mm CaCl2 [pH 8.0]) at 23°C for 4 to 6 h. Purification and Factor Xa digestion of the recombinant protein were verified using SDS-PAGE.
Generation of Polyclonal Cah6 Primary Antibodies
Factor Xa digested purified recombinant proteins were separated on a 12% gel by SDS-PAGE at 15 mA for 18 to 20 h. The 31-kD Cah6 protein band was excised carefully from the polyacrylamide gel. The gel pieces were shipped to Strategic BioSolutions (Ramona, CA) for production of antibody. Antibodies were raised against Cah6 proteins by a standard 70-d protocol using two pathogen-free rabbits. Approximately 1.6 mg of the Cah6 protein was used to raise the antibody.
Carbonic Anhydrase Assays and CA Inhibition Studies
CA activity was assayed electrometrically using a modification of the Wilbur-Anderson method (1948). The samples were assayed at 4°C by adding 50 to 200 μL of the test sample to 3.5 mL of 20 mm 4-(2-hydroxyethyl)-1-piperazine propane sulfonic acid (EPPS, pH 8.0). The reaction was initiated by addition of 1.5 mL of ice-cold CO2 saturated water. The time required for the pH drop from 7.7 to 6.3 was measured. The activity of the test sample was calculated using the equation: WAU = t0/t−1 where t is the time required for the pH change when the test sample is present and t0 is the time required for the pH change when the buffer is substituted for the test sample. Bovine CAII (Sigma, St. Louis) was used as a positive control. Both Factor Xa digested and undigested purified Cah6 fusion proteins were used for activity assays.
The effects of the sulfonamide inhibitors ethoxyzolamide and acetazolamide and the anions azide and cyanide on CA activities of recombinant Cah6 were studied. I50 was determined by plotting the percentage of inhibition versus the concentration of the inhibitor. Sodium salts of azide and potassium salts of cyanide were used. All the inhibitors were purchased from Sigma.
Electrophoresis and Immunoblotting
For protein analyses and western blots, cells were harvested, washed twice with fresh medium, and resuspended in 10 mm Tris-HCl, 10 mm EDTA, and 150 mm NaCl, pH 7.5. Proteins were separated on 12% and 15% polyacrylamide gels (0.8% bis-acrylamide) as described previously (Laemmli, 1970). SDS-PAGE was performed using prestained low molecular mass markers as protein standards (Bio-Rad, Hercules, CA).
Immunoblotting was performed as described in the protocol from Bio-Rad. Cah6 primary antibody was used to probe proteins from high and low CO2 acclimated D66 as well as the Factor Xa digested and undigested purified MBP-Cah6 fusion proteins. The Cah6 antibody was diluted with the antibody buffer (Tris-buffered saline + 0.005% Tween 20 + 1% bovine serum albumin, pH 7.4) in the ratio of 1:1,000, before being used as a probe. The secondary antibody used for western blotting was conjugated to the enzyme horseradish peroxidase (Bio-Rad) and was diluted at a ratio of 1:3,000 with the antibody buffer. Western blots were developed following the protocol from Bio-Rad using a mixture of the horse radish peroxidase color development reagent (Bio-Rad) in ice-cold 100% methanol (20 mL), Tris-buffered saline (80 mL, pH 7.4, and 30% H2O2, 60 μL).
Immunolocalization Studies Using Electron Microscopy
Air acclimated D66 or CC-124 cells were fixed in a mixture of 1% OsO4, 2% formaldehyde, and 0.5% glutaraldehyde in a 1:1 ratio for 15 min. The sample was then fixed for an additional 15 min in 1% OsO4, 2% formaldehyde, 0.5% glutaraldehyde, and 0.1 mm sodium cacodylate buffer. Materials were rinsed with distilled water and stained with 0.5% uranyl acetate for 30 min. After this, excess stain was rinsed and the samples were dehydrated in ethyl alcohol series. Samples were then infiltrated and embedded in LR White resin (EMS, Fort Washington, PA). Embedded tissues were sectioned with a DuPont Sorvall microtome (Wilmington, DE). The sections were 70-μm thick.
The immunocytochemical procedure was similar to the method of Borkhsenious et al. (1998) with some modifications. The sections were incubated for 90 min with diluted primary antibody (1:10 dilutions of the Cah6 primary antibody) or with the preimmune serum diluted similarly (used as a negative control). The grids were transferred to 1:50 dilution of Protein A (Sigma) conjugated to 20 nm colloidal gold particles for 1 h. Protein A was diluted with 1% bovine serum albumin and 0.1% Tween 20 in phosphate buffered saline (Sigma). Finally the sections were rinsed with distilled water and photographed under transmission electron microscopy.
Other Methods
The CO2 concentration in the growth chambers was measured using an infrared gas analyzer (Analytical Development, Hoddesdon, UK), which reads at an accuracy of ±2%. The CO2 concentration was checked at least once every day, while the alga was growing in the high and low CO2 growth chambers. Protein concentration was determined by the method of Lowry et al. (1951) with bovine serum albumin as standard. Chlorophyll concentration was determined spectrophotometrically (Arnon, 1949) using the equations of Holden (1976). The solvent used for extraction of chlorophyll was 100% methanol. Cell density values were determined by direct counting in a hemacytometer chamber.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AY463238 and AY463239.
Acknowledgments
The authors thank Steve Pollock for helping with the northern blot and Catherine Mason and Patricia Moroney for their critical reading of the manuscript. We also thank Göran Samuelsson, Umeå University, Sweden, for providing us with the mitochondrial β-CA primary antibody.
Supported by National Science Foundation (grants IBN–9904425 and IBN–0212093 to J.V.M.) and the Howard Hughes Memorial Institute undergraduate research program.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037283.
References
- Alber BE, Ferry JG (1994) A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc Natl Acad Sci USA 91: 6909–6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso G, Weber C, Sültemeyer DF, Fock HP (1996) Intracellular carbonic anhydrase activities in Dunaliella tertiolecta (Butcher) and Chlamydomonas reinhardtii (Dangeard) in relation to inorganic carbon concentration during growth: further evidence for the existence of two distinct carbonic anhydrases associated with the chloroplast. Planta 199: 177–184 [Google Scholar]
- Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Price GD (1992) The CO2 concentrating mechanism in cyanobacteria and microalgae. Physiol Plant 84: 606–615 [Google Scholar]
- Borkhsenious ON, Mason CB, Moroney JV (1998) The intracellular localization of ribulose-1, 5-bisphosphate carboxylase/oxygenase in Chlamydomonas reinhardtii. Plant Physiol 116: 1585–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracey MH, Christiansen J, Tovar P, Cramer SP, Bartlett SG (1994) Spinach carbonic anhydrase: investigation of the zinc-binding ligands by site-directed mutagenesis, elemental analysis and EXAFS. Biochemistry 33: 13126–13131 [DOI] [PubMed] [Google Scholar]
- Burnell JN, Gibbs MJ, Mason JG (1990) Spinach chloroplastic carbonic anhydrase-nucleotide sequence analysis of cDNA. Plant Physiol 92: 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo SL, Pollock SV, Eger KA, Godfrey AC, Adams JE, Mason CB, Moroney JV (2002) Use of the bleomycin resistance gene to generate tagged insertional mutants of Chlamydomonas reinhardtii that require elevated CO2 for optimal growth. Funct Plant Biol 29: 231–241 [DOI] [PubMed] [Google Scholar]
- Eriksson M, Karlsson J, Ramazanov Z, Gardeström P, Samuelsson G (1996) Discovery of an algal mitochondrial carbonic anhydrase: molecular cloning and characterization of a low-CO2-induced polypeptide in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 93: 12031–12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson M, Villand P, Gardeström P, Samuelsson G (1998) Induction and regulation of expression of a low-CO2-induced mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 116: 637–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett TW, Browse JA, Volokita M, Bartlett SG (1990) Spinach carbonic-anhydrase primary structure deduced from the sequence of a cDNA clone. J Biol Chem 265: 5414–5417 [PubMed] [Google Scholar]
- Fujiwara S, Fukuzawa H, Tachiki A, Miyachi S (1990) Structure and differential expression of two genes encoding carbonic anhydrase in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87: 9779–9783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S (1990) cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii- regulation by environmental CO2 concentration. Proc Natl Acad Sci USA 87: 4383–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho K, Saito T, Kohinata T, Ohyama K (2001) Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc Natl Acad Sci USA 98: 5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Norici A, Forssen M, Eriksson M, Raven J (2003) An anaplerotic role for mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Plant Physiol 132: 2126–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz R, Gnann A, Zimmermann FK (1999) Deletion of the carbonic anhydrase-like gene NCE103 of the yeast Saccharomyces cerevisiae causes an oxygen-sensitive growth defect. Yeast 15: 855–864 [DOI] [PubMed] [Google Scholar]
- Guilloton MB, Lamblin AF, Kozliak EI, Gerami-Nejad M, Tu C, Silverman D, Anderson PM, Fuchs JA (1993) A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J Bacteriol 175: 1443–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson DT, Franklin LA, Samuelsson G, Badger MR (2003) The Chlamydomonas reinhardtii cia3 mutant lacking a thylakoid lumen-localized carbonic anhydrase is limited by CO2 utilization by Rubisco and not PSII function in vivo. Plant Physiol 132: 2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RP (1996) Multiple roles of carbonic anhydrase in cellular transport and metabolism. Annu Rev Physiol 58: 523–538 [DOI] [PubMed] [Google Scholar]
- Hewett-Emmett D, Tashian RE (1996) Functional diversity, conservation, and convergence in the evolution of the α-, β-, and γ-carbonic anhydrase gene families. Mol Phylogenet Evol 5: 50–77 [DOI] [PubMed] [Google Scholar]
- Holden M (1976) Chlorophylls. In TW Goodwin, ed, Chemistry and Biochemistry of Plant Pigments. Academic Press, London, pp 2–37
- Johansson IM, Forsman C (1993) Kinetic-studies of pea carbonic-anhydrase. Eur J Biochem 218: 439–446 [DOI] [PubMed] [Google Scholar]
- Karlsson J, Clarke AK, Chen ZY, Hugghins SY, Park YI, Husic HD, Moroney JV, Samuelsson G (1998) A novel alpha-type carbonic anhydrase associated with the thylakoid membrane in Chlamydomonas reinhardtii is required for growth at ambient CO2. EMBO J 17: 1208–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman GL, Carlson SJ, Marcus Y, Moroney JV, Togasaki RK (1994) Carbonic anhydrase activity in isolated chloroplasts of wild-type and high-CO2-dependent mutants of Chlamydomonas reinhardtii as studied by a new assay. Plant Physiol 105: 1197–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp D, Haussler D, Reese MG, Eeckman FH (1996) A generalized hidden Markov model for the recognition of human genes in DNA. Proc Int Conf Intell Systems Mol Biol 4: 134–142 [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lindskog S (1997) Structure and mechanism of carbonic anhydrase. Pharmacol Ther 74: 1–20 [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Fart AL, Randall RJ (1951) Protein measurement with Folin phenol reagent. J Biol Chem 193: 265–275 [PubMed] [Google Scholar]
- Manuel LJ, Moroney JV (1988) Inorganic carbon accumulation by Chlamydomonas reinhardtii: new proteins are made during adaptation to low CO2. Plant Physiol 88: 491–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldrum NU, Roughton FJW (1933) Carbonic anhydrase. Its preparation and properties. J Physiol 80: 113–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhashi S, Ohnishi J, Hayashi M, Ikeda M (2003) A gene homologous to beta-type carbonic anhydrase is essential for the growth of Corynebacterium glutamicum under atmospheric conditions. Appl Microbiol Biotechnol (in press) [DOI] [PubMed]
- Moroney JV, Bartlett SG, Samuelsson G (2001) Carbonic anhydrases in plants and algae. Plant Cell Environ 24: 141–153 [Google Scholar]
- Moroney JV, Husic HD, Tolbert NE (1985) Effect of carbonic anhydrase inhibitors on inorganic carbon accumulation by Chlamydomonas reinhardtii. Plant Physiol 79: 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock SV, Colombo SL, Prout DL Jr, Godfrey AC, Moroney JV (2003) Rubisco activase is required for optimal photosynthesis in the green alga Chlamydomonas reinhardtii in a low CO2 atmosphere. Plant Physiol 133: 1854–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton S, Rochaix JP (1994) Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol Biol 24: 533–537 [DOI] [PubMed] [Google Scholar]
- Raven JA (2001) A role for mitochondrial carbonic anhydrase in limiting CO2 leakage from low CO2-grown cells of Chlamydomonas reinhardtii. Plant Cell Environ 24: 261–265 [Google Scholar]
- Rowlett RS, Chance MR, Wirt MD, Sidelinger DE, Royal JR, Woodroffe M, Wang YFA, Saha RP, Lam MG (1994) Kinetic and structural characterization of spinach carbonic-anhydrase. Biochemistry 33: 13967–13976 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schnell RA, Lefebvre PA (1993) Isolation of the Chlamydomonas reinhardtii regulatory gene NIT2 by transposon tagging. Genetics 134: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silflow CD (1998) Organization of the nuclear genome. In JD Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 25–40
- Slaymaker DH, Navarre DA, Clark D, del Pozo O, Martin GB, Klessig D (2002) The tobacco salicylic acid binding protein 3 (SABP3) is the chloroplast carbonic anhydrase which exhibits antioxidant activity and plays a role in the hypersensitive defense response. Proc Natl Acad Sci USA 99: 11640–11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueoka N (1960) Mitotic replication of deoxyribonucleic acids in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 46: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sültemeyer DF, Price GD, Yu JW, Badger MR (1995) Characterization of carbon dioxide and bicarbonate transport during steady state photosynthesis in the marine cyanobacterium Synechococcus strain PCC 7002. Planta 197: 597–607 [Google Scholar]
- Villarejo A, Rolland N, Martinez F, Sültemeyer D (2001) A new chloroplast envelope carbonic anhydrase activity is induced during acclimation to low inorganic carbon concentrations in Chlamydomonas reinhardtii. Planta 213: 286–295 [DOI] [PubMed] [Google Scholar]
- Wilbur KM, Anderson NG (1948) Electrometric and colorimetric determination of carbonic anhydrase. J Biol Chem 176: 147–154 [PubMed] [Google Scholar]
- Xiang YB, Zhang J, Weeks DP (2001) The Cia5 gene controls formation of the carbon concentrating mechanism in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 98: 5341–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]