Abstract
DNA CpG methylation is a widespread epigenetic mark in high eukaryotes including mammals. DNA methylation plays key roles in diverse biological processes such as X chromosome inactivation, transposable element repression, genomic imprinting, and control of gene expression. Recent advancements in sequencing-based DNA methylation profiling methods provide an unprecedented opportunity to measure DNA methylation in a genome-wide fashion, making it possible to comprehensively investigate the role of DNA methylation. Several methods have been developed, such as Whole Genome Bisulfite Sequencing (WGBS), Reduced Representation Bisulfite Sequencing (RRBS), and enrichment-based methods including Methylation Dependent ImmunoPrecipitation followed by sequencing (MeDIP-seq), methyl-CpG binding domain (MBD) protein-enriched genome sequencing (MBD-seq), methyltransferase-directed Transfer of Activated Groups followed by sequencing (mTAG), and Methylation-sensitive Restriction Enzyme digestion followed by sequencing (MRE-seq). These methods differ by their genomic CpG coverage, resolution, quantitative accuracy, cost, and software for analyzing the data. Among these, WGBS is considered the gold standard. However, it is still a cost-prohibitive technology for a typical laboratory due to the required sequencing depth. We found that by integrating two enrichment-based methods that are complementary in nature (i.e., MeDIP-seq and MRE-seq), we can significantly increase the efficiency of whole DNA methylome profiling. By using two recently developed computational algorithms (i.e., M&M and methylCRF), the combination of MeDIP-seq and MRE-seq produces genome-wide CpG methylation measurement at high coverage and high resolution, and robust predictions of differentially methylated regions. Thus, the combination of the two enrichment-based methods provides a cost-effective alternative to WGBS. In this article we describe both the experimental protocols for performing MeDIP-seq and MRE, and the computational protocols for running M&M and methylCRF.
Keywords: DNA methylation, MeDIP-seq, MRE-seq, M&M, methylCRF
1. Introduction
DNA methylation typically refers to the methylation of the 5 position of cytosine (mC) by DNA methyltransferases (DNMT). It is a major epigenetic modification in human and many other species [1]. In somatic cells, 5-methylcytosine (5mC) is largely restricted to CpG sites [2], although some non-CpG (i.e., CHG, CHH) methylation is also observed in embryonic stem cell [3, 4]. DNA methylation profiles are highly variable across different genetic loci, cells and organisms, and are dependent on tissue, age, sex, diet and disease [5, 6]. DNA methylation plays a crucial role in normal development [1, 7, 8]. Aberrant methylation can lead to many diseases including cancers [9, 10].
Current genome-scale approaches for the determination of the modification status of CpG sites are largely based on detection of 5mC and can be divided into bisulfite conversion-based methods, affinity capture-based techniques, and restriction endonuclease-based methods [11–13]. The gold standard is bisulfite conversion coupled with sequencing due to its ability to map 5-modified cytosines at a single-base resolution (Whole Genome Bisulfite Sequencing, or WGBS) [2, 14]. However, bisulfite conversion detects fC and caC as unmodified C, and discrimination between 5mC and hmC requires additional cumbersome pretreatment steps [15, 16]. Overall, it is a labor-intensive technique that is still prohibitively expensive for large-scale populational studies, because it typically requires sequencing to 30× coverage [8]. Reduced Representation Bisulfite Sequencing (RRBS) provides a less expensive alternative, at the cost of reduced coverage of genome-wide CpGs [17, 18]. In affinity enrichment methods, methylated DNA fragments are non-covalently bound to 5mC-antibodies (MeDIP) [19, 20] or to the methyl-CpG binding domain of MBD2 or MeCP2 [21, 22]. These methods suffer from a poor coverage of the medium to low CpG density regions of the genome, and a relatively low resolution which is limited by the size of the fragments from immunoprecipitation [23, 24]. To enrich for unmethylated DNA fragments, unmodified CpG sites can be chemically modified via DNA methyltransferase, and tagged by covalent biotin for enrichment and sequencing (mTAG-seq) [25]. Restriction enzyme-based approaches permit interrogation of either the unmodified or modified fraction of genomic DNA (HELP [25], MRE-seq [26], Methyl-MAPS [27]). However, their coverage and resolution are inherently limited by the sequence- and modification-type-specificity of available enzymes. Aside from WGBS, most other methods are considered either low coverage, or low resolution.
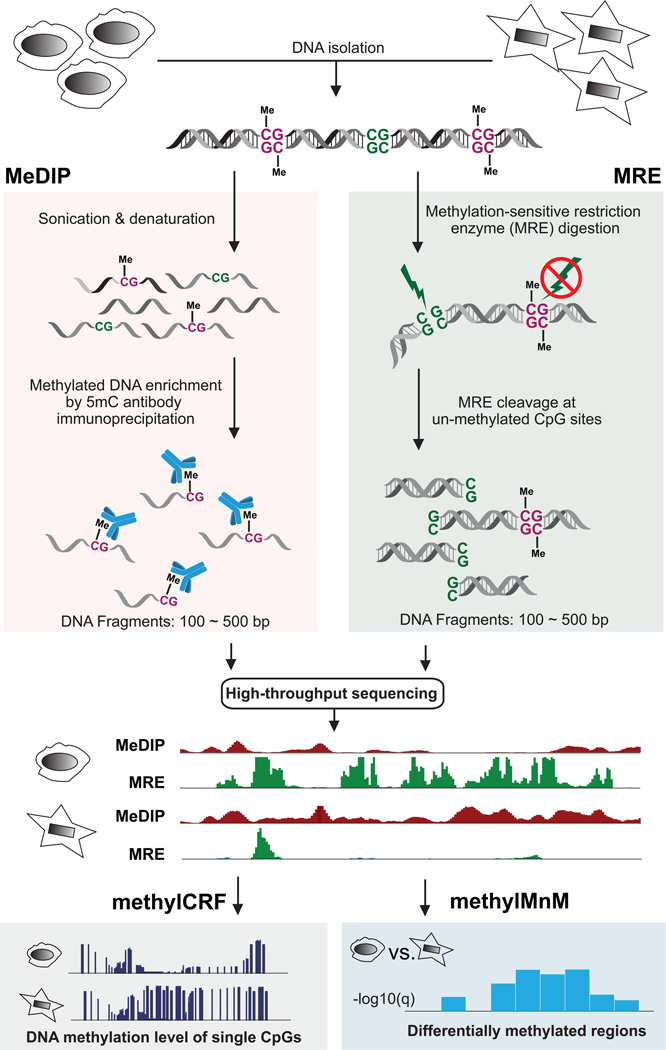
Our recent work suggests that by integrating two complementary technologies, MeDIP-seq and MRE-seq, we can effectively improve the coverage and resolution of DNA methylomes produced, and improve the accuracy of detection of differentially methylated regions (Figure 1). This experimental approach was chosen because, unlike the inefficiency of whole genome bisulfite sequencing (WGBS) in which 70–80% of sequence reads are uninformative [28] and 5-methylcytosine and 5-hydroxymethylcytosine are conflated, MeDIP-seq/MRE-seq detects 5-methylcytosine exclusively, and provides greater accuracy for several loci [29]. MeDIP-seq and MRE-seq each has its own advantages and disadvantages, and can be applied independently or jointly. MRE-seq provides DNA methylation estimates at single CpG resolution, but is considered low coverage due to the limit of CpG containing recognition sites. An important advantage of MeDIP over enzymatic digestion based methods is a lack of bias for a specific nucleotide sequence, other than CpGs. However, the relationship of enrichment to absolute methylation levels is confounded by variables such as CpG density [30]. Another inherent limitation of MeDIP-seq is its lower resolution (~150bp) compared to MRE-seq or bisulfite-based methods in that one or more of the CpGs in the immunoprecipitated DNA fragment could be responsible for the antibody binding.
Figure 1. Workflow of combining MeDIP-seq and MRE-seq.
Genomic DNA is isolated and purified. On the MeDIP-seq side, genomic DNA is sonicated to a specific size range, and a monoclonal anti-5’methylcytosine antibody is used to enrich for methylated DNA fragments. Immunoprecipitated DNA fragments are then sequenced and mapped back to the reference genome assembly to review methylated regions. On the MRE-seq side, several methylation sensitive restriction endonucleases are used to digest intact genomic DNA. The resulting DNA fragments are then size-selected and sequenced. When mapped back to the reference genome assembly, these sequencing reads can reveal locations of unmethylated CpG sites which are located within recognition sites of specific restriction enzymes. MeDIP-seq and MRE-seq data can then be integrated by applying methylCRF, which transform enrichment-based DNA methylation data to methylation level at single CpG resolution across the genome. To compare two samples and detect differentially methylated regions, M&M is applied in a region-specific fashion.
MeDIP-seq and MRE-seq represent complementary ways to enrich for either methylated portion of the genome, or unmethylated portion of the genome (Figure 1). Their protocols are simple and their data analyses do not require specialized sequence aligners to deal with bisulfite converted DNA [12, 26]. By using simple heuristics, the combination of these two methods gave promising results in identifying differentially methylated regions (DMRs) and intermediate or mono-allelic methylation [12]. Subsequently we developed two computational tools, M&M [31] and methylCRF [29], to integrate data from MeDIP-seq and MRE-seq and obtain additional, synergistic advantages. M&M is a new statistical framework that identifies differentially methylated regions (DMRs) by jointly modeling MeDIP-seq and MRE-seq data [31]. methylCRF is a Conditional Random Fields-based [32, 33] algorithm that integrates MeDIP-seq and MRE-seq data to predict genome-wide DNA methylation levels at single CpG resolution [29]. In this work, we present both experimental and computational protocols for combined application of MeDIP-seq and MRE-seq, and discuss how integrating the two complementary technologies significantly increase their values (Figure 1).
2. Materials
2.1. Buffers
| Buffer | Composition |
|---|---|
| Extraction buffer | 50 mM Tris-HCl pH 8.0 |
| 1 mM EDTA pH 8.0 | |
| 0.5% SDS | |
| 1 mg/ml proteinase K (add fresh) | |
| 0.1 M Na2HPO4/NaH2PO4 (pH 7.0) | 61 mM Na2HPO4 |
| 39 mM NaH2PO4, | |
| 10% Triton X-100 | 10 ml Triton X-100 |
| fill up with 90 ml ultrapure water | |
| MeDIP wash buffer | 10 mM Na2HPO4/NaH2PO4 (pH7.0) |
| (prepare fresh) | 140 mM NaCl |
| 0.05% Triton X-100 | |
| MeDIP elution buffer | 0.25 mg/ml proteinase K |
| (prepare fresh) | 0.25% SDS |
| fill up with TE buffer | |
| 0.1% Tween-20 | 100 µl Tween-20 |
| fill up with 99.9 ml ultrapure water |
2.2. Enzymes, antibodies and other reagents
| Material | Manufacturer |
|---|---|
| RNase A, DNase and protease-free | Thermo Scientific |
| DNA Polymerase I, Large (Klenow) Fragment | New England Biolabs |
| T4 DNA Polymerase | New England Biolabs |
| T4 Polynucleotide Kinase | New England Biolabs |
| Klenow Fragment (3´→5´ exo–) | New England Biolabs |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs |
| HpaII | New England Biolabs |
| HinP1I | New England Biolabs |
| AciI | New England Biolabs |
| HpyCH4IV | Thermo Scientific |
| Bsh1236I | New England Biolabs |
| Deoxynucleotide Solution Mix | New England Biolabs |
| Denosine 5´-Triphosphate (ATP) | New England Biolabs |
| dATP solution | New England Biolabs |
| T4 ligase reaction buffer (10×) | New England Biolabs |
| Quick Ligation Kit | |
| Agencourt AMPure XP beads | Beckman Coulter |
| Monoclonal Antibody against 5-Methylcytidine | Eurogentec |
| AffiniPure Rabbit Anti-Mouse IgG, Fcγ | Jackson ImmunoResearch |
| Fragment Specific | |
| Protein A/G agarose beads | Fisher Scientific |
| Phase lock gel light (2 ml) | 5 PRIM |
| Phenol/chloroform/isoamyl alcohol | Roche |
| Chloroform | Sigma-Aldrich |
| Sodium Acetate, pH 5.5 (3 M) | Life Technologies |
| Ethanol | Sigma-Aldrich |
| Qubit dsDNA HS Assay Kit | Life Technologies |
| Agarose, Molecular Biology Grade | VWR |
| Ethidium bromide | BioExpress |
| TAE 50× Reagent | 5 PRIME |
| Orange DNA loading dye (6×) | Thermo Scientific |
| 2-Log DNA ladder | New England Biolabs |
| MinElute PCR Purification Kit | Qiagen |
| Isopropanol | Sigma-Aldrich |
| iTaq universal SYBR Green supermix (2×) | Bio-Rad |
2.3. Oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| adapter: PE 1.0 | ACACTCTTTCCCTACACGACGCTCTTCCGATC*T |
| adapter: PE 2.0 | P-GATCGGAAGAGCACACGTCTGAACTCCAGTCAC |
| PCR primer: PE 1.0 | AATGATACGGCGACCACCGAGATCTACACTCTTTC CCTACACGACGCTCTTCCGATCT |
| PCR primer: PE 2.0 | CAAGCAGAAGACGGCATACGAGATNNNNNNNGTG ACTGGAGTTCAGACGTGTGCTCTTCCGA |
| SNRPN-F | CGCTCAACACCCCCTAAATA |
| SNRPN-R | GGTGGAGGTGGGTACATCAG |
| GABRB3-F | CCTGCAACTTTACTGAATTTAGC |
| GABRB3-R | GGAATCTCACTTTCACCACTGG |
| qPCR primer 1.0 | AATGATACGGCGACCACCGAGAT |
| qPCR primer 2.0 | CAAGCAGAAGACGGCATACGA |
Asterisk * indicates a phosphorothiate linkage, P indicates a phosphate group, and N indicates index code. The 5-terminal phosphate in adapter PE 2.0 facilitates the ligation of annealed adapters to DNA inserts. The phosphorothiate linkage in adapter PE 1.0 prevents nuclease cleavage of the T overhang, which is required for the adapter and A-tailed DNA fragment ligation.
2.4. Equipment
| Equipment | Manufacture |
|---|---|
| NanoVue spectrophotometer | GE Healthcare |
| Qubit fluorometer | Life Technologies |
| Qubit assay tubes | Life Technologies |
| Gel electrophoresis system | Thermo Scientific |
| Gel Doc Imaging System | Bio-Rad |
| Bioruptor Pico system | Diagenode |
| Thermal cycler CC007387 | Bio-Rad |
| Thermal cycler CC007277 CFX | Bio-Rad |
| Bioanalyzer | Agilent |
| Illumina HiSeq2500 | Illumina |
2.5. Software
| Name | Purpose | Website for download and documentation |
|---|---|---|
| Cutadapt | Adapter trimming and quality filtering of raw reads | https://code.google.com/p/cutadapt/ |
| BWA | Align raw or filtered reads to the corresponding genome assembly | http://bio-bwa.sourceforge.net/ |
| Samtools | Operating on read alignment files | http://samtools.sourceforge.net/ |
| R | Statistical computing environment | http://www.r-project.org/ |
| methylQA | Parsing aligned reads, generating bed files and bedGraph files. | http://methylqa.sourceforge.net/ |
| methylMnM | Calculating differentially methylated region (DMR) between two samples by integrating MeDIP-seq and MRE-seq data. | http://epigenome.wustl.edu/MnM/ |
| methylCRF | Predicting methylation level at single CpG resolution by combining MeDIP-seq and MRE-seq data. | http://methylcrf.wustl.edu/ |
2.6. Raw MeDIP-seq and MRE-seq data (for demonstration purpose)
| Sample | GEO number | Assay | URL |
|---|---|---|---|
| H1 ESC | GSM543016 | MeDIP-seq | http://wang.wustl.edu/MeDIP-MRE/H1Es_MeDIP.fq.gz |
| H1 ESC | GSM428286 | MRE-seq | http://wang.wustl.edu/MeDIP-MRE/H1Es_MRE.fq.gz |
| Brain | GSM669614 | MeDIP-seq | http://wang.wustl.edu/MeDIP-MRE/Brain_MeDIP.fq.gz |
| Brain | GSM669604 | MRE-seq | http://wang.wustl.edu/MeDIP-MRE/Brain_MRE.fq.gz |
2.7. Auxiliary genomic annotation files
| Used by | Description | URL |
|---|---|---|
| BWA | Human genome sequence | http://wang.wustl.edu/MeDIP-MRE/hg19.fa.gz |
| methylQA | Chromosome size file | http://wang.wustl.edu/MeDIP-MRE/ann/hg19.size |
| CpG sites file | http://wang.wustl.edu/MeDIP-MRE/ann/CpG.bed.gz | |
| 3 MRE enzyme fragment file | http://wang.wustl.edu/MeDIP-MRE/ann/TriMRE_frags.bed | |
| 5 MRE enzyme fragment file | http://wang.wustl.edu/MeDIP-MRE/ann/FiveMRE_frags.bed | |
| methylMnM | CpG sites in 500bp window | http://wang.wustl.edu/MeDIP-MRE/ann/num500_allcpg_hg19.bed |
| 3 MRE enzyme cut sites in 500bp window | http://wang.wustl.edu/MeDIP-MRE/ann/num500_Three_mre_cpg_hg19.bed | |
| 5 MRE enzyme cut sites in 500bp window | http://wang.wustl.edu/MeDIP-MRE/ann/num500_Five_mre_cpg_hg19.bed | |
| methylCRF | Model file | http://methylcrf.wustl.edu/h1es_mdl.tgz |
| Data files | http://methylcrf.wustl.edu/hg19/hg19_gdat.tgz | |
| 3 MRE enzyme fragment file | http://methylcrf.wustl.edu/hg19/MRE_frags/MRE_3enz_4_6000.bed | |
| 5 MRE enzyme fragment file | http://methylcrf.wustl.edu/hg19/MRE_frags/MRE_5enz_4_6000.bed |
3. Methods and applications
3.1 MeDIP-seq
The MeDIP-seq protocol was previously described in [26] with small modifications.
3.1.1. Genomic DNA extraction
High-quality, intact genomic DNA without degradation and denaturation, without protein and RNA contaminations is important for the success of library construction. Compared with commercial kits and the original DNA isolation protocol [26], our optimized protocol can generate higher quality DNA, and can be easily adapted for the isolation of genomic DNA from various cells and tissue types.
Cell pellets or finely sliced tissues are resuspended in 1.5 ml centrifuge tube containing 100–600 µl of extraction buffer. Allow frozen tissue to warm up slightly (5–10 sec), and chop it in a petri dish. The quantity of extraction buffer is dependent on cell numbers and tissue size. For instance, 600 µl of buffer is sufficient to lyse 1–5 million cells or up to 20 mg of tissue. Alternatively, small samples can be resuspended in 100 µl extraction buffer. Pipette up and down immediately to ensure that the samples suspend well. Please pipet gently, as genomic DNA can be easily sheared by vigorous mixing or pipetting. Incubate samples at 55 °C for 1 h or overnight if needed. The solution should look homogenous following digestion. Inadequate digestion will lead to lower yield and lower DNA quality. If additional digestion is required, add more extraction buffer and incubate until homogenous.
Centrifuge at maximum speed at 4 °C for 10 min. Then quickly transfer supernatant to a phase lock gel (pre-pellet phase lock gel (PLG) at 16000× g for 30 sec). Add an equal volume of phenol/chloroform/isoamyl alcohol (PCI) directly to the PLG tube. Thoroughly mix the organic and aqueous phases to form a transiently homogeneous suspension. Do not vortex. Centrifuge at 16000× g for 5 min to separate the phases. Carefully pipet upper phase to a fresh tube. Add DNase-free RNaseA (10 mg/ml) 1 µl and incubate for 1 h at 37 °C. Transfer the solution to a new pre-pelleted phase lock gel and repeat the process one more time.
Carefully transfer upper phase to a new pre-pelleted phase lock gel and repeat steps above one more time using chloroform instead of phenol/chloroform/isoamyl alcohol. Precipitate with 1/10 volume of 3 M sodium acetate (pH 5.5) and 2.5 volumes of 100% ethanol. Invert tubes several times to mix. At this point, a “ball” of DNA should appear in the tube. Incubate for 15 min at room temperature. If DNA “ball” cannot be observed, incubate at −20 °C for overnight.
Spin tubes at maximum speed for 30 min at 4 °C. Wash DNA pellet with 70% ethanol and spin 5 min. Resuspend DNA in a desired volume of elution buffer (Qiagen EB). Purified genomic DNA can be placed at −20 °C for long-term storage. Quantitate DNA by NanoVue Spectrophotometer. Qubit fluorometer and dsDNA HS assay kit can also be used to get accurate quantification. Validate DNA quality by running it on 1% TAE agarose gel. Intact DNA without random shearing should not appear as a smear.
3.1.2. DNA fragmentation
Resuspend 500–1,000 ng of intact genomic DNA in 30 µl of EB in a 1.5ml sterile DNase/RNase free tube and seal the tube with parafilm. Set 4 °C of the Diagenode Bioruptor Pico and pre-chill the sonicator water bath. Sonicate for 10–15 min with 30-sec on/off. Fragmented DNA can be stored at − 20 °C until needed. Run 3 µl of the sonicated DNA on 1% TAE agarose gel to validate DNA fragment size range. DNA fragment size range should be 100–500 bp. Continue sonication if the desired range is not achieved.
3.1.3. Library preparation I
DNA purification is necessary after each reaction. A lot of commercial kits can be used for DNA purification. It was reported that AMPure XP beads can reduce DNA loss during purification when compared to Qiagen kit [20]. The beads selectively bind DNA fragments of 100bp and bigger, thus excess adapter fragments and primer dimmers can be removed.
DNA end repair is performed to get blunt 5'-phosphorylated ends. The reaction is set up on ice in a 50 µl volume with 1× NEB T4 PNK buffer, fragmented DNA, 5 µl of 10 mM ATP, 2 µl of 10 mM dNTPs, 3 U of T4 DNA polymerase, 5 U of Klenow DNA polymerase and 10 U of T4 Polynucleotide Kinase. Incubate the mixture in a thermocycler at 20 °C for 30 min. Then spin briefly, and purify the DNA sample with AMPure XP beads. Elute in 34 µl of EB. Purified DNA can be stored at − 20 °C until needed.
dA-tailing reaction is set up in a 50 µl volume with 1× NEB klenow buffer 2, end-repaired DNA, 10 µl of 10 mM dATPs and 5 U of Klenow 3'–5' exo nimus. Incubate the mixture in a heat block for 30 min at 37 °C. Then spin briefly, and purify the DNA sample with AMPure XP beads. Elute in 23 µl of EB. Purified DNA can be stored at − 20 °C until needed.
Adapter ligation is set up in a 50 µl volume with 1× Quick ligation reaction buffer, end-repaired, dA-tailed DNA, 1 µl of 10 µM pre-annealed adapter oligo mix and 2,000 U of T4 DNA ligase. Incubate the mixture at room temperature for 15 min. Purify the DNA sample with AMPure XP purification beads. Elute in 30 µl of EB. Purified DNA can be stored at − 20 °C until needed.
3.1.4. MeDIP
DNA fragments need to be denatured to single strand for anti-methylcytosine immunoprecipitation. The denatured single-stranded DNA is not suitable for library construction. Therefore, the immunoprecipitation step is sandwiched between two library preparation steps.
Denature the adapter-ligated DNA at 95 °C for 10 min, then transfer immediately to ice to prevent re-annealing. Keep on ice for 10 min. A pre-mix is set up on ice in a 500 µl volume with 50 µl of 0.1 M Na2HPO4/NaH2PO4, 35 µl of 2 M NaCl, 2.5 µl of 10% triton X-100 and 1 µl of anti-methylcytidine antibody (1 mg/ml). Keep the mix on ice. Add the mix to the tube that contains the denatured adapter-ligated DNA. Incubate the reaction on a rotator at 4 °C overnight.
After overnight incubation, add 2 µl of secondary antibody of rabbit anti-mouse IgG (2.5 µg/µl) and 80 µl of protein A/G agarose beads. Incubate at 4 °C for 2 h on a rotator. Centrifuge the sample at 2000× g for 1 min in 4 °C microfuge. Carefully discard supernatant using a p1000 tip. Resuspend beads pellet in 1000 µl pre-cold MeDIP wash buffer (freshly prepared). Make sure that the pellet is fully resuspended. Repeat the wash 7~10 times. For final wash, centrifuge at 5000× g for 2 min, remove and discard supernatant. Spin briefly and remove all remaining liquid with a p10 tip. Add 200 µl of MeDIP elution buffer to the beads pellet. Resuspend thoroughly. Incubate at 55 °C for 2 h, mixing occasionally. Let the tube cool down to room temperature. Purify DNA with Qiagen MinElute PCR Purification Kit. Elute DNA in 30 µl EB. We choose not to use AMPure XP beads in this step because the elution buffer used in immunoprecipitation affects binding of DNA to the beads, especially when the DNA fragments are shorter than 300 bp. Purified DNA can be stored at −20 °C until needed.
3.1.5. Library preparation II
An adapter modified PCR is carried out on ice in a 50 µl volume with 1×Phusion HF buffer, 15 µl of MeDIP DNA, 1 µl of 10 µM dNTP, 2.5 µl of PCR primer PE 1.0, 2.5 µl of PCR primer PE 2.0 and 0.5 U of Phusion DNA polymerase.
Perform PCR in a preheated thermocycler with conditions of a denaturation of 30 sec at 98 °C, followed by 12 cycles of 10 sec at 98 °C, 30 sec at 64 °C and 30 sec at 72 °C, with a final extension of 5min at 72 °C. 12 cycles are enough in this step. Higher cycle numbers will result in higher PCR bias.
Purify the DNA sample with AMPure XP purification beads. Elute in 15 µl EB. Purified DNA can be stored at − 20 °C until needed.
3.1.6. Library size selection
A regular agarose gel extraction is performed for the final library size selection. Inserts of 100~400 bp are selected, which are around 220~520bp after the adapter ligation and PCR amplification. Gel selection excludes all the free adapters, primers, and dimerized oligonucleotides.
Prepare a 100 ml 2% TAE agarose gel containing ethidium bromide. Mix the PCR amplified DNA with 3 µl of 6× loading dye and load in the gel, leaving space between the ladder and sample wells. Carry out gel electrophoresis in 1× TAE buffer at 120 V until the orange dye runs to the two thirds of the gel. After gel electrophoresis, transfer the gel tray onto the gel doc imaging system under UV light, cut out 220–520 bp size range with a clean blade. Purify each gel slice with MinElute Gel DNA Extraction kit and elute in 15 µl EB buffer. We modified the Qiagen gel extraction protocol to melt agarose gel slices at 37 °C instead of at 50 °C to reduce G+C bias [34]. Use two Qiagen columns if a gel slice is over 400 mg and elute each column in 10 µl EB. Combine the two elutes.
Assess 2 µl of size-selected DNA on Qubit Fluorometer using Qubit dsDNA High-Sensitivity Assay kit and tubes according to the manufacturer’s instructions to determine the concentration of each library. Purified libraries can be stored at − 20 °C until needed.
3.1.7. Library QC
To assure libraries quality before sequencing, two independent qPCR reactions including one positive control (SNRPN promoter) and one negative control (a CpG-less sequence in the intron of GABRB3) are performed for MeDIP-seq libraries to confirm enrichment of methylated DNA and depletion of unmethylated DNA. Two sets of primers are designed to test for MeDIP enrichment for human samples (see 2.3.). qPCRs are set up in triplicates in 20 µl volume with 1× iTaq universal SYBR Green supermix, 2 µl of MeDIP library (prediluted to 10 nM in EB) and 0.8 µl of 10 µM primer mix (SNPRN or GABRB3). Perform qPCR in thermocycler CFX with conditions of a denaturation of 30 sec at 95 °C, followed by 50 cycles of 5 sec at 95 °C and 5 sec at 60 °C. A non-MeDIPed sonicated input control should be included if sufficient material is available. Non-MeDIPed sonicated input control should not exhibit any enrichment. It is recommended that the methylated fragments are at least 25-fold more enriched than the unmethylated fragments [29]. In practice our libraries show enrichment score higher than 2500.
Another qPCR quantification step is applied using the primers that match sequences within the linkers flanking library inserts. This step measures templates that have linker sequences on both ends. They will subsequently form clusters on a flowcell. Primers for the qPCR quantification are designed according to linker sequences which match both ends sequences of the adapter-modified PCR primers (see 2.3.). A control MeDIP-seq library that has been successfully sequenced should be selected. Add 2 µl of the control template (prediluted to 10 nM in buffer EB) to 198 µl of 0.1% Tween-20 solution to make a 100-fold dilution. Add 100 µl of the diluted template to 100 µl of 0.1% Tween-20 solution to make a titration curve of six 2× serial dilutions. This will give 7 control template dilutions in the range of 100-1.6 pM. Add 2 µl of each unknown library (prediluted to 10 nM in EB) to 998 µl of 0.1% Tween-20 solution to make a 500-fold dilution. This will give an approximate concentration of 20 pM. Three independent serial dilutions should be used if sufficient library material is available. qPCRs are set up in triplicates in 20 µl volume with 1× iTaq universal SYBR Green supermix, 2 µl of diluted control templates or unknown libraries, 0.4 µl of 10 µM qPCR primer 1.0 and 0.4 µl of 10 µM qPCR primer 2.0. Perform qPCR in thermocycler CFX with conditions of a denaturation of 30 sec at 95 °C, followed by 40 cycles of 5 sec at 95 °C and 5 sec at 60 °C. Generate a standard curve from the control template dilutions by plotting the Ct values against the log initial concentration. Ensure that the efficiency of the standard curve in is 90–110% and that the R2>0.9. Lock the threshold fluorescence based on the standard curve, calculate the initial concentration of the newly constructed libraries. The concentration of newly constructed libraries should be close to that of the control library. Determine samples’ loading concentration based on the control template’s loading and cluster density.
3.1.8. Next-generation sequencing
Indexed MeDIP-seq libraries quantified by a Qubit fluorometer are pooled after QC and are quantified again on an Agilent 2100 Bioanalyzer. 101-bp paired-end sequencing is run for MeDIP-seq libraries following Illumina’s standard protocol. On Illumina Hi-Seq 2500 platform, typically 150~200 million raw reads per lane can be obtained. Three MeDIP-seq libraries can be typically pooled per lane. Typically, 30~50 million mapped reads are sufficient for analysis included in this protocol, as suggested by saturation analysis [29].
3.2. MRE-seq
The MRE-seq protocol was previously described in [26] with small modifications. The table below contains MRE cut sites statistics for human, mouse, rat, and zebrafish genomes (supercontigs/scaffolds are excluded).
| Genome | Human hg19 | Mouse mm9 | Rat rn4 | Zebrafish danRer7 |
|---|---|---|---|---|
| CpG sites | 28217448 | 21342779 | 23932116 | 24222562 |
| 3 Enzymes CpG sites | 8062966 | 5198815 | 5609250 | 5709644 |
| 3 Enzymes fragments (100–500bp) | 2233827 | 1252814 | 1428220 | 1714471 |
| 5 Enzymes CpG sites | 10939789 | 7388695 | 8108261 | 8752292 |
| 5 Enzymes fragments (100–500bp) | 2908850 | 1744769 | 2040929 | 2725117 |
A summary of genomic coverage and distribution of MRE cut sites was also shown in the table below, specifically for human genome (hg19). The five MRE combinations can cover near 70% genome, and over 99% gene promoters.
| Human hg19# | Promoter# | CpG islands$ | |
|---|---|---|---|
| 3 Enzymes Coverage | 53.60% | 99.60% | 96.23% |
| 5 Enzymes Coverage | 69.02% | 99.97% | 96.41% |
: The coverage is calculated at 500bp window resolution;
: Promoter is defined 4KB region (−3KB to +1KB) around gene transcription start site (TSS), which is downloaded from UCSC genome browser and contains 24,944 RefSeq genes;
: CpG islands information is downloaded from UCSC genome browser, which define 28691 CpG islands.
3.2.1. Genomic DNA extraction
Same as 3.1.1.
3.2.2. DNA fragmentation
We recommend using five methylation-sensitive restriction endonucleases in DNA digestion for fragmentation. Three restriction endonucleases of HpaII (C↓CGG), HinP1I (G↓CGC) and AciI (C↓CGC) were used in the original protocol [26]. Five enzymes can be used to increase genome coverage. All five enzymes are sensitive to CpG methylation. Sequencing 5’ end of these digested fragments therefore provides information for unmethylated CpG.
Transfer 500 ng of genomic DNA to a 1.5 ml sterile DNase/RNase free tube for each reaction mix. A pre-mix is set up in a 20 µl volume with 1× restriction enzyme buffer and 2.5 U of restriction enzyme (HpaII, HinP1I, AciI, HpyCH4IV or Bsh1236I). Keep the mixture on ice. Incubate reaction in a heat block at 37 °C for 3 h. Five digests for each sample can be set up in parallel. Add additional 2.5 U of enzymes to each reaction after 3 h of incubation. Mix and incubate for another 3 h at 37 °C. After total 6 h digestion, incubate the reaction at 65 °C for 20 min to deactivate the all enzymes except for HpaII (NEB) which is deactivated at 80°C. Combine the five reactions. Each sample should have a total volume of 100 µl.
Purify the DNA sample with AMPure XP purification beads. AMPure XP beads are used in this step instead of phenol chloroform extraction which was used in the original protocol [26]. Elute in 15 µl EB. Purified DNA can be stored at − 20 °C until needed.
Prepare a 100 ml 1% TAE agarose gel containing ethidium bromide. Mix the digested DNA with 3 µl of 6× loading dye and load in the gel, leaving space between the ladder and sample wells. Carry out gel electrophoresis in 1× TAE buffer at 120 V until the orange dye runs to the two third of the gel. After gel electrophoresis, transfer the gel tray onto the gel doc imaging system under UV light, cut out 100–500 bp size range with a clean blade. Purify each gel slice with MinElute Gel DNA Extraction kit and elute in 30 µl EB buffer. We modified the Qiagen gel extraction protocol to melt agarose gel slices at 37 °C instead of at 50 °C to reduce G+C bias [34]. Use two Qiagen columns if a gel slice is over 400 mg and elute each column in 10 µl EB. Combine the two elutes.
3.2.3. Library preparation I
Same as 3.1.3, except that only Klenow fragment is used for end repair. Restriction endonuclease digested fragments contain 5'-phosphorylated and 3’-terminal recessive ends. Therefore, a Klenow DNA polymerase is used to finish filling-in.
3.2.4. Library preparation II
Same as 3.1.5.
3.2.5. Library size selection
Same as 3.1.6, except gel electrophoresis should run until the orange dye runs to the bottom of the gel. In MeDIP-seq, the unmethylated adapter cannot be precipitated, therefore there are no adapter dimers in its PCR product. In contrast, in MRE-seq, the dimerized oligonucleotides (~120bp in size) should be removed by running gel as long as possible.
3.2.6. Library QC
Same as 3.1.7, except to skip the MeDIP-specific primer sets. A control MRE-seq library that has been successfully sequenced should be used for qPCR quantification.
3.2.7. Next-generation sequencing
Indexed MRE-seq libraries quantified by a Qubit fluorometer are pooled after QC and are quantified again on an Agilent 2100 Bioanalyzer. 50-bp single-end sequencing is run for MRE-seq libraries following Illumina’s standard protocol. On the Illumina Hi-Seq 2500 platform, typically 150~200 million raw reads can be obtained per lane. Six MRE-seq libraries can be pooled per lane. As suggested by saturation analysis [29], 25~40 million mapped reads are typically sufficient for analysis included in this protocol.
3.3. Integrative analysis of MeDIP-seq and MRE-seq
To illustrate the computational integration of MeDIP-seq and MRE-seq data, we include data from two human samples – human embryonic stem cell H1 (H1 ESC) [12] and human brain [26]. Program executions are demonstrated in a Linux/Unix environment, in which Python and Perl are installed. We describe aligning raw reads to the reference genome, post processing MeDIP-seq and MRE-seq data, display the data on a Genome Browser, detect DMRs between H1 ESC and brain by using M&M, and transform MeDIP-seq and MRE-seq data into single CpG DNA methylation levels using methylCRF. Raw data are described in 2.6, and auxiliary data files are described in 2.7.
3.3.1. MeDIP-seq data processing
Investigators should use any standard next-gen sequencing read alignment tools. While we recommend BWA [35], many other alternatives exist. Typically the reference genome assembly sequences are indexed; sequencing data are first converted to be a specific data structure (e.g. suffix array), and aligned. The result can then be either in SAM format or BAM format. Investigators should consult User Instructions for any specific aligner to choose.
Once MeDIP-seq reads are aligned and stored in a BAM file, we recommend using methylQA for post-alignment processing. The methylQA package processes the MeDIP alignment result file (.bam) to generate alignment quality report and files formatted for various downstream needs. Resulting files include .bed file (aligned read location), bedGraph file (aligned read density), and .bigWig file (aligned read density, for display on a Genome Browser). Detailed documentation of methylQA can be obtained from its website (see 2.5).
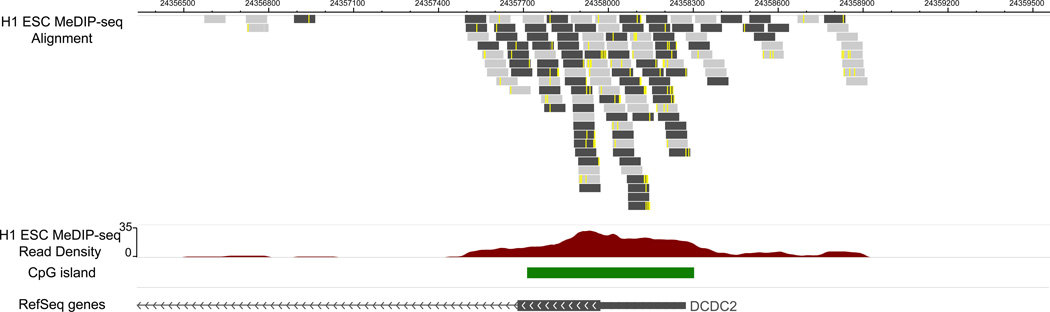
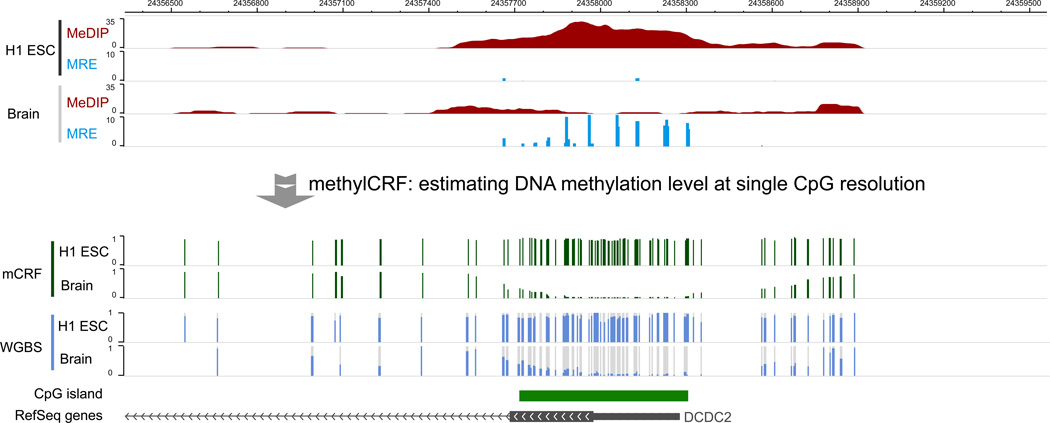
Figure 2 displays processed MeDIP-seq data on a Genome Browser. In this example, we displayed MeDIP-seq data of H1 ESC sample over a genomic region encompassing the DCDC2 gene promoter. Processed MeDIP-seq data are displayed on the WashU EpiGenome Browser [36]. DCDC2 stands for doublecortin domain containing 2 (DCDC2). This gene is specifically expressed in neuronal cells. It is thought to function in neuronal migration where it may affect the signaling of primary cilia. Mutations in this gene have been associated with reading disability (RD) type 2, also referred to as developmental dyslexia [37]. In Figure 2 the following tracks are included: The first track displays MeDIP-seq reads that are aligned to this region by BWA, with redundant reads removed. Note that reads that are mapped to the forward strand or reverse strand are colored differently (dark vs light). Mismatches between each read and the genome assembly are also visible as yellow ticks within the read. The second track shows MeDIP-seq read density (i.e., read peaks). The third track shows locations of CpG islands, and the last track displays RefSeq genes, in this case, DCDC2. MeDIP-seq data suggests that in embryonic stem cells, the promoter of DCDC2, which overlaps with a CpG island, strongly enriches for methylated CpGs. It is consistent with the expectation that this neuronal-specific gene is epigenetically silenced in ES cells.
Figure 2. Visualizing MeDIP-seq data on a Genome Browser.
H1 ESC MeDIP-seq data were visualized using the WashU EpiGenome Browser. The tracks from top to bottom were read alignment track, read density track, CpG island track, and refGene track. The genomic location was the promoter region of DCDC2 gene. DCDC2 is a neuron specifically expressed gene.
Note that the example we included here contains single-end reads. For paired-end sequencing data, methylQA has different parameters to optimize and process result.
3.3.2. MRE-seq data processing
Similar to processing of MeDIP-seq data, investigators should choose their favorite short-read alignment tools, and perform post-alignment processing using methylQA, using the “mre” option. The methylQA package processes the MRE alignment result file (.bam) to generate alignment quality report and files formatted for various downstream needs. Resulting files include .bed file (aligned read location), .bedGraph file (MRE score), and .bigWig file (MRE score, for display on a Genome Browser). Please note the MRE data used in this protocol were generated using three enzymes, therefore all data, MRE fragment file, MRE window file, were the 3-enzyme version. For experiments with different number or combination of enzymes, please use the corresponding annotation files.
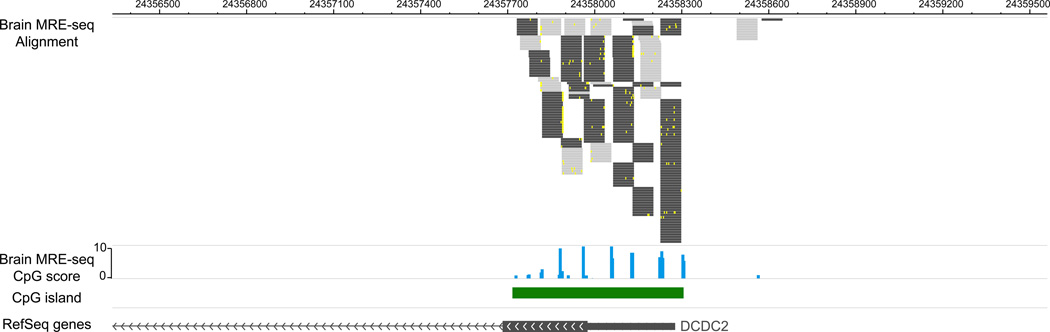
Figure 3 displays processed MRE-seq data on a Genome Browser. Processed MRE-seq data from a brain sample are displayed on the WashU EpiGenome Browser, across the same genomic region as in Figure 2, encompassing DCDC2 promoter. Four tracks are included. The first track displays MRE-seq reads that are uniquely aligned to this region, and filtered by restriction enzyme recognition sites. Because independent enzymatic cleavage of the same site will result in identical sequencing reads, all reads are kept for analysis. Note that reads that are mapped to the forward strand or reverse strand are colored differently (dark vs light). Mismatches between each read and the genome assembly are also visible as yellow ticks within the read. The second track shows MRE-seq score, which is a normalized read count per enzyme at a given CpG site. The third and fourth tracks are identical to those from Figure 2. MRE-seq data suggests that many CpG sites within this CpG island are unmethylated in the brain sample, which is consistent with the expectation that DCDC2, a neuronal-specific gene, is epigenetically active in the brain.
Figure 3. Visualizing MRE-seq data on a Genome Browser.
Brain MRE-seq data were visualized using the WashU EpiGenome Browser. The tracks from top to bottom are read alignment track, MRE CpG score track, CpG island track, and refGene track. The genomic region was the same as in Figure 2.
3.3.3. Run methylMnM to predict DMRs between H1 ESC and brain
M&M is a statistical framework that identifies differentially methylated regions (DMRs) by jointly modeling MeDIP-seq and MRE-seq data. Experimental measurements can be modeled as a function of the underlying methylation state and genomic context. Thus, independent measurements (i.e., MeDIP-seq and MRE-seq) of the same sample can be integrated on the same underlying methylation state. Detecting DMRs between two samples can then be modeled as a test of the null hypothesis, namely that the two samples have the same methylation state, given the observed measurements and genomic context. We formulated the statistical test and provided a numerical solution to compute a probability that a genomic region is differentially methylated given observed MeDIP-seq and MRE-seq measurements. This is implemented as a R-package called methylMnM [31].
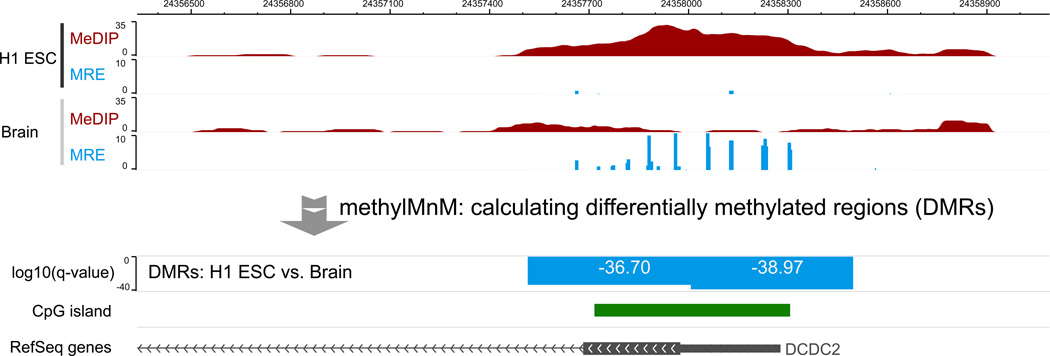
In this example, we partition the genome into genomic bins of 500bp in size. Then, we perform M&M test for each genomic bin to generate a statistical assessment of the probability that the methylation levels of the two samples within each bin are different, using the function “MnM.test()”. The function “MnM.qvalue()” estimates q-values based on all the p-values, and “MnM.selectDMR()” selects significant DMRs based on a cutoff provided by the user. The input files of methylMnM are methylQA processed alignment results in BED format. The output files contain genomic locations of statistically significant DMRs, their MeDIP-seq and MRE-seq values (in RPKM), as well as p-values and q-values. The software also generates a .bedGraph file that can be directly visualized on a Genome Browser. The absolute values of genomic regions are negative log10-transformed q-values. If the value is negative, it represents hypermethylation in sample 1 and hypomethylation in sample 2; if the value is positive, it represents hypomethylation in sample 1 and hypermethylation in sample 2.
Figure 4 displays a DMR between H1 ESC and brain, detected by using the methylMnM package. This differentially methylated region is located across a CpG island overlapping promoter of DCDC2, the same region featured in Figure 2 and 3. MeDIP-seq and MRE-seq data of H1 ESC and of the brain are displayed as the first four Browser tracks. The difference in DNA methylation between H1 ESC and brain can be visually appreciated, such that there is significantly higher MeDIP-seq signal in H1 ESC than in brain, and significantly higher MRE-seq signal in brain than in H1 ESC. This event is detected by M&M, which is represented by the fifth track that displays log10-transformed q-values. The q-values are resulted from M&M comparison between two samples at a 500bp resolution. Two genomic regions are detected as DMRs, each at a q-value of 1e-36.70, and 1e-38.97, respectively. The negative values indicate that between the two samples in comparison, the second sample (in this case brain) is hypomethylated. This DMR is biologically significant, and is directly linked to the regulation of DCDC2, which is expressed in neuronal cells but repressed in embryonic stem cells.
Figure 4. Visualizing M&M results.
DMR regions between H1 ESC and brain were visualized on the WashU EpiGenome Browser. The top four tracks displayed MeDIP and MRE signals from H1 ESC and brain. The y-axis of DMR track displayed –log10(q-value) of predicted DMRs.
3.3.4. Predict single CpG methylation level for H1 ESC and brain by methylCRF
methylCRF is a Conditional Random Fields-based [32, 33] algorithm that integrates MeDIP-seq and MRE-seq data to predict genome-wide DNA methylation levels at single CpG resolution. Because MeDIP-seq is enrichment-based, its resolution is limited by the size of the DNA fragments from immunoprecipitation. MRE-seq is a single CpG resolution method but is limited by the availability of restriction enzyme recognition sites in the genome. Based on the same principle that experimental measurements can be modeled as a function of the underlying methylation state and genomic context, Conditional Random Fields provide a machine learning framework to infer the absolute values of the underlying methylation state at single CpG level. We demonstrated that methylCRF transforms MeDIP-seq and MRE-seq data to the equivalent of a whole genome bisulfite sequencing (WGBS) experiment, which typically costs 20 times more to produce [29].
The input files of methylCRF are methylQA processed MeDIP-seq alignment file in BED format, and original MRE-seq alignment result in BAM format. Investigators need to download previously trained CRF models and species-specific genome annotation files, including genomic features (i.e., CpG island, repeats, etc) (see 2.7). The final output is a file of BED format, with genomic coordinates of each CpG site, and predicted methylation level ranging from 0 (completely unmethylated) to 1 (completely methylated).
Figure 5 displays methylCRF transformed DNA methylation data for the H1 ESC and brain samples. The same genomic region, DCDC2 gene promoter, is represented on a Browser view. MeDIP-seq and MRE-seq data from H1 ESC and brain are displayed as in Figure 4. In addition, two methylCRF tracks are included, displaying DNA methylation levels at single CpG resolution. CpGs in the DMR displayed in Figure 4 have high methylation levels in H1 ESC, and low methylation levels in brain. Two whole genome bisulfite sequencing tracks on matching samples are also included, in the format of methylC Track [38]. The two methylCRF tracks consistently recapitulate the methylation levels measured by WGBS.
Figure 5. Visualizing methylCRF results.
Single CpG methylation levels predicted by methylCRF were visualized on the WashU EpiGenome Browser. The top 4 tracks displayed MeDIP and MRE signals from H1 ESC and brain, and the bottom 4 tracks displayed methylCRF and WGBS methylation levels (0 for completely unmethylated, 1 for fully methylated) for H1 ESC and brain. The methylation values from methylCRF and WGBS were well correlated. Both data showed different methylation levels of this region between H1 ESC and brain, supporting the finding of DMRs using the M&M algorithm (Figure 4).
3.3.5. Genome-wide evaluation
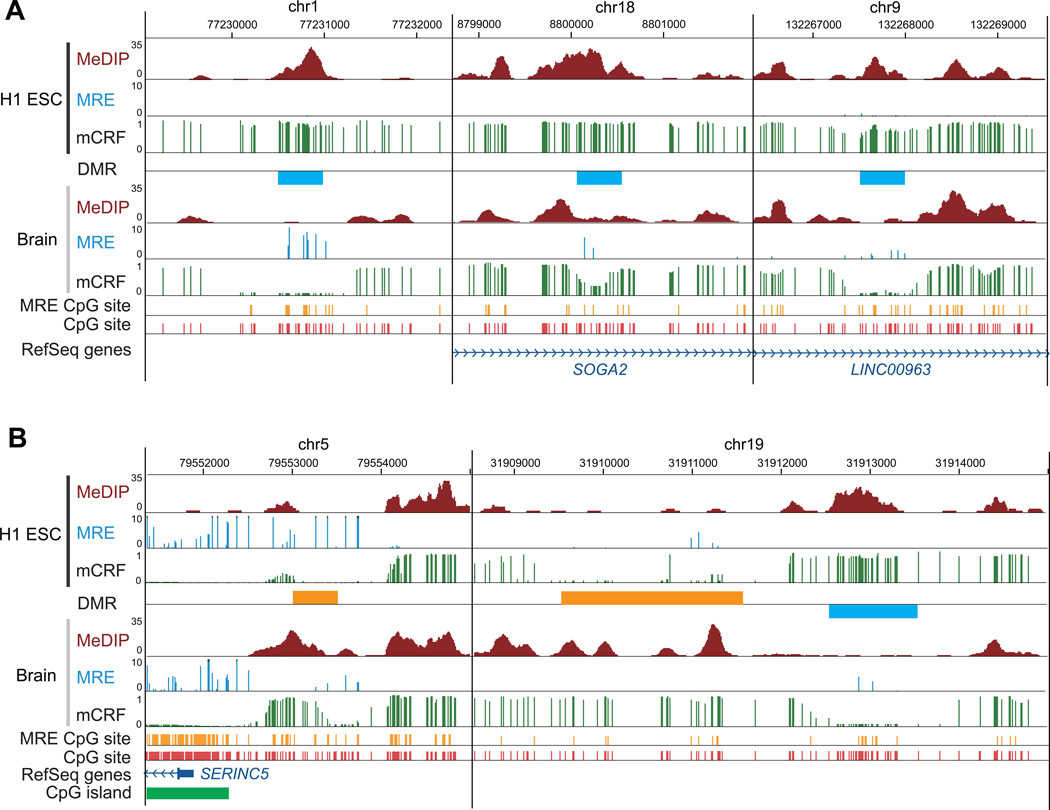
One key advantage of our method is that DNA methylation analysis can be performed at whole genome scale, but not be restricted to promoters or CpG islands. To illustrate this, we presented examples of six DMRs (Figure 6). These DMRs located in intergenic or intragenic regions, had moderate to low CpG density, and were not part of any CpG island.
Figure 6. DNA methylation changes in low CpG density regions.
Select non-CGI DMRs between H1 ESC and brain sample were visualized on the WashU EpiGenome Browser. (A) Brain-specific hypomethylated DMRs located in an intergenic region (left), in the intron of SOGA2 (middle), and in the intron of LINC00963 (right). (B) Left: a H1 ESC-specific hypomethylated DMR located in a CpG island (CGI) shore region. Right: a H1 ESC-specific hypomethylated DMR, and a neighboring H1 ESC-specific hypermethylated DMR, both located in an intergenic region.
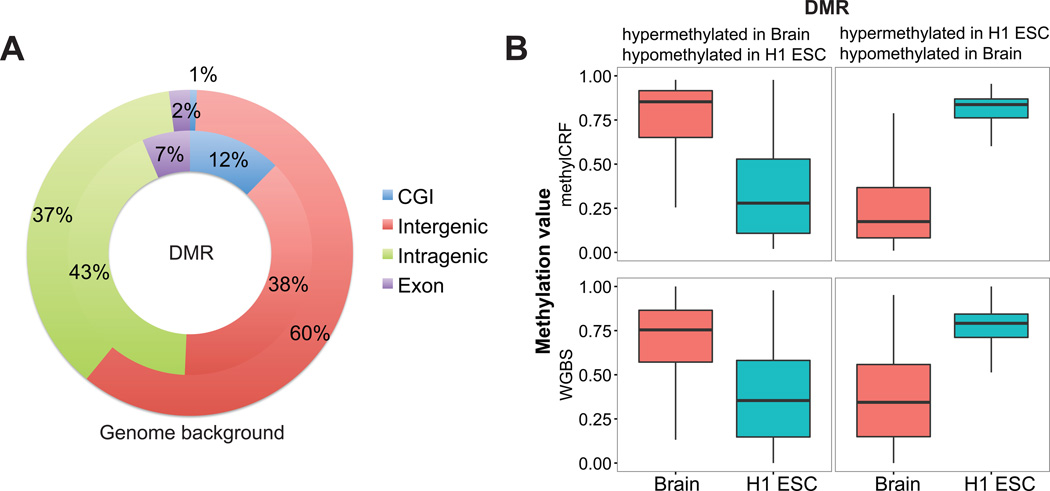
Indeed, when we examined the total of 14,866 DMRs we detected between H1 ESC and brain at a statistical cutoff q-value < 10−5, the large majority of them were outside CpG islands (Figure 7A). To further assess the accuracy of our method, we plotted DNA methylation levels of these DMRs in both H1 ESC and brain, and using both methylCRF predicted values and WGBS based values (Figure 7B). This comparison clearly demonstrate that the DMRs prediction was both sensitive and specific – in the tissue where the regions were called hypermethylated, the methylation levels were high; while in the tissue where they were called hypomethylated, the methylation levels were low. WGBS and methylCRF exhibited almost identical pattern, further supporting the accuracy of our method.
Figure 7. Genome-wide distribution and comparison of DMRs.
(A) A pie chart of locational analysis of DMRs between H1 ESC and brain. The outer pie represented genome-wide background. The inner pie represented distribution of DMRs. The majority of the DMRs were located in intergenic and intragenic regions. Only a small fraction overlapped with CpG islands, although they were enriched. (B) Distribution of DNA methylation levels of predicted DMRs. DMRs were grouped based on their hyper- or hypo-methylated status predicted by M&M. DNA methylation levels predicted by methylCRF (upper panel) and by WGBS (lower panel) were both plotted.
4. Concluding Remarks
Understanding the role of DNA methylation often requires accurate assessment and comparison of these modifications in a genome-wide fashion. Earlier genome-wide DNA methylation analyses have been focused on CpG dense regions including gene promoters and CpG islands, and more recently on CpG island shores [39]. However, only 7.4% of the 28M CpGs in human genome are within CpG islands, and an additional 4.9% within CpG island shores, leaving the majority of DNA methylome uncharacterized. The recent technological development, in particular next-gen sequencing technology, allowed much more comprehensive views of genome-wide DNA methylation patterns. Several groups have reported exciting findings on the dynamic nature of DNA methylation changes outside CpG islands and shores, but within or encompassing regulatory elements, especially distal enhancers [28, 31, 40, 41]. Among these, Stadler et al. discovered that DNA binding factors could lead to demethylation of distal regulatory regions in the mouse. By investigating intergenic hypomethylated regions in various human cell types, Schlesinger et al. suggested that de novo DNA demethylation defines distal regulatory elements [42]. Xie et al. discovered that thousands of transposable elements undergo DNA hypomethylation in a tissue-specific manner, and could serve as tissue-specific enhancers [43]. Hon et al. pointed out that identifying tissue-specific DMRs (tsDMRs) can be an alternative strategy for finding putative regulatory elements [40]. By profiling 30 different tissues and cell types, Ziller et al. estimated that 21.8% of the DNA methylome is dynamic [28]. These recent studies highlight the importance of measuring CpG methylation in a genome-wide, unbiased fashion.
We demonstrate that the integration of MeDIP-seq and MRE-seq is a method that provides the appropriate balance among genomic CpG coverage, resolution, quantitative accuracy, and cost, and comes with robust bioinformatics software for analyzing the data. MeDIP-seq, or methylation dependent immunoprecipitation followed by sequencing, uses an anti-methyl-cytosine antibody to enrich for methylated DNA fragments, and uses massively parallel sequencing to reveal identity of enriched DNA. MRE-seq, or methylation sensitive restriction enzyme digestion followed by sequences, relies on a collection of restriction enzymes that recognize CpG containing sequence motif but only cut when the CpG is unmethylated. Digested DNA fragments enrich for unmethylated CpGs at their ends, and these CpGs are revealed by massively parallel sequencing. The two computational methods, M&M and methylCRF, both implement advanced statistical algorithms that integrate MeDIP-seq and MREseq data. M&M is a statistical framework to detect differentially methylated regions between two samples. methylCRF is a machine learning framework that predicts CpG methylation levels at single CpG resolution, thus raising the resolution and coverage of MeDIP-seq and MRE-seq on CpGs to a comparable level of WGBS, but only incurring a cost of less than 5% of WGBS.
In the protocol we recommended running MeDIP-seq with 101bp paired-end reads, and running MRE-seq with 50bp single-end reads. However, this is only an empirical estimate and users should make decisions based on their specific application and available resource. As we know, there are many variables that could impact the optimal sequencing depth for an experiment. For example, the size of the genome, the read length, using single-ended reads or paired-ended reads, etc. Our experience suggested that for human samples, 50 million mapped MeDIP-seq reads and 30 million mapped MRE-seq reads would typically reach saturation [29]. We also note that MeDIP-seq data usually has less mapping efficiency, because relatively more MeDIP-seq reads are derived from repetitive regions of the genome which are often heavily methylated. Thus, we recommend using longer reads, and/or paired end reads to increase mapping efficiency. In contrast, relatively more MRE-seq reads are derived from unmethylated regions of the genome and are less repetitive. Thus, we recommend shorter reads for pure economical purpose.
Both methylMnM and methylCRF work with either MeDIP-seq data or MRE-seq data alone. However this will result in increased false positives. Thus, we do not recommend using methylMnM and methylCRF with only MeDIP-seq data or only MRE-seq data. Depending on total read counts from MeDIP-seq and MRE-seq data, methylMnM and methylCRF will need up to 20 CPU hours to finish calculation for a typical human experiment.
The main advantage of these two algorithms is the integration of independent, heterogeneous experiments on the same biological state that they measure – in this case, DNA methylation. All current genome-wide technologies for measuring DNA methylation have their inherent biases and limitations, but our confidence in inferring methylation state increases when results from two independent methods are integrated. For example, a decrease of MeDIP-seq signal could reflect a biological event (we infer that this region is demethylated) or could be a methodological artifact; but if it is corroborated by an increase of MRE-seq signal, then the inference of demethylation can be much more accurate. Taken together, the integrated methods we describe in this chapter provide a streamlined platform for investigators to explore DNA methylation with high coverage, high resolution, and low cost.
Supplementary Material
Acknowledgements
We thank Joseph F. Costello, Ravi Nagarajan, Chibo Hong for developing experimental protocols described in this chapter. We thank Michael Stevens for developing methylCRF. We thank Nan Lin, Yan Zhou, Boxue Zhang for developing M&M. We thank members of the Wang laboratory for testing and improving various parts of the methods. This work was supported by NIH grant U01ES017154, R01HG007354, R01HG007175, and R01ES024992 (TW), NIDA’s R25 program DA027995 (BZ), and American Cancer Society grant RSG-14-049-01-DMC (TW).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 2.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan J, Zierath JR, Barres R. Evidence for non-CpG methylation in mammals. Exp Cell Res. 2011;317:2555–2561. doi: 10.1016/j.yexcr.2011.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107:8689–8694. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- 7.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 8.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 9.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 10.Bergman Y, Cedar H. DNA methylation dynamics in health and disease. Nat Struct Mol Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 11.Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28:1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher A, Kapranov P, Kaminsky Z, Flanagan J, Assadzadeh A, Yau P, Virtanen C, Winegarden N, Cheng J, Gingeras T, Petronis A. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 2006;34:528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, Jacobsen SE. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 16.Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 20.Taiwo O, Wilson GA, Morris T, Seisenberger S, Reik W, Pearce D, Beck S, Butcher LM. Methylome analysis using MeDIP-seq with low DNA concentrations. Nat Protoc. 2012;7:617–636. doi: 10.1038/nprot.2012.012. [DOI] [PubMed] [Google Scholar]
- 21.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38:391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest. 2005;85:1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 23.Nair SS, Coolen MW, Stirzaker C, Song JZ, Statham AL, Strbenac D, Robinson MD, Clark SJ. Comparison of methyl-DNA immunoprecipitation (MeDIP) and methyl-CpG binding domain (MBD) protein capture for genome-wide DNA methylation analysis reveal CpG sequence coverage bias. Epigenetics. 2011;6:34–44. doi: 10.4161/epi.6.1.13313. [DOI] [PubMed] [Google Scholar]
- 24.Robinson MD, Stirzaker C, Statham AL, Coolen MW, Song JZ, Nair SS, Strbenac D, Speed TP, Clark SJ. Evaluation of affinity-based genome-wide DNA methylation data: effects of CpG density, amplification bias, and copy number variation. Genome Res. 2010;20:1719–1729. doi: 10.1101/gr.110601.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kriukiene E, Labrie V, Khare T, Urbanaviciute G, Lapinaite A, Koncevicius K, Li D, Wang T, Pai S, Ptak C. DNA unmethylome profiling by covalent capture of CpG sites. Nat Commun. 2013;4:2190. doi: 10.1038/ncomms3190. [DOI] [PubMed] [Google Scholar]
- 26.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziller MJ, Gu H, Muller F, Donaghey J, Tsai LT, Kohlbacher O, De Jager PL, Rosen ED, Bennett DA, Bernstein BE. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens M, Cheng JB, Li D, Xie M, Hong C, Maire CL, Ligon KL, Hirst M, Marra MA, Costello JF, Wang T. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res. 2013;23:1541–1553. doi: 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelizzola M, Koga Y, Urban AE, Krauthammer M, Weissman S, Halaban R, Molinaro AM. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Zhou Y, Lin N, Lowdon RF, Hong C, Nagarajan RP, Cheng JB, Li D, Stevens M, Lee HJ. Functional DNA methylation differences between tissues, cell types, and across individuals discovered using the M&M algorithm. Genome Res. 2013;23:1522–1540. doi: 10.1101/gr.156539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lafferty J, McCallum A, Pereira F. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. Departmental Papers. 2001 CIS-159. [Google Scholar]
- 33.Wallach H. Conditional Random Fields: An Introduction. Technical Report MS-CIS-04-21 Department of Computer and Information Science, University of Pennsylvania. 2004 [Google Scholar]
- 34.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P. The Human Epigenome Browser at Washington University. Nat Methods. 2011;8:989–990. doi: 10.1038/nmeth.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilcke A, Weissfuss J, Kirsten H, Wolfram G, Boltze J, Ahnert P. The role of gene DCDC2 in German dyslexics. Ann Dyslexia. 2009;59:1–11. doi: 10.1007/s11881-008-0020-7. [DOI] [PubMed] [Google Scholar]
- 38.Zhou X, Li D, Lowdon RF, Costello JF, Wang T. methylC Track: visual integration of single-base resolution DNA methylation data on the WashU EpiGenome Browser. Bioinformatics. 2014;30:2206–2207. doi: 10.1093/bioinformatics/btu191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, Ren B. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45:1198–1206. doi: 10.1038/ng.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 42.Schlesinger F, Smith AD, Gingeras TR, Hannon GJ, Hodges E. De novo DNA demethylation and noncoding transcription define active intergenic regulatory elements. Genome Res. 2013;23:1601–1614. doi: 10.1101/gr.157271.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie M, Hong C, Zhang B, Lowdon RF, Xing X, Li D, Zhou X, Lee HJ, Maire CL, Ligon KL. DNA hypomethylation within specific transposable element families associates with tissue-specific enhancer landscape. Nat Genet. 2013;45:836–841. doi: 10.1038/ng.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.