Abstract
Many of the developmental responses and behaviors in plants that occur throughout the year are controlled by photoperiod; among these, seasonal flowering is the most characterized. Molecular genetic and biochemical analyses have revealed the mechanisms by which plants sense changes in day length to regulate seasonal flowering. In Arabidopsis thaliana, induction of the expression of a florigen, FLOWERING LOCUS T (FT) protein, is a major output of the photoperiodic flowering pathway. The circadian clock coordinates the expression profiles and activities of the components in this pathway. Light-dependent control of CONSTANS (CO) transcription factor activity is a crucial part of the induction of the photoperiodic expression of FT. CO protein is stabilized only in the long day afternoon, which is when FT is induced. In this review, we summarize recent progress in the determination of the molecular architecture of the circadian clock and mechanisms underlying photoperiodic flowering. In addition, we introduce the molecular mechanisms of other biological processes, such as hypocotyl growth and reactive oxygen species production, which are also controlled by alterations in photoperiod.
There are seasonal latitude-dependent fluctuations in photoperiod due to the rotation of the earth on its tilted axis and its revolution around the sun. Because the degree of day length change increases as one moves away from the equator toward the poles, photoperiod correlates with and can serve as an indicator of seasonal change.
Plants use photoperiodic information to predict environmental change and to regulate their developmental processes so that they occur under favorable conditions. To maximize reproductive success, molecular mechanisms that are regulated through the integration of environmental and endogenous cues to control the timing of flowering have evolved. Plants measure changes in day length using their circadian clocks and by sensing the surrounding light environment.1−3 Circadian clock-controlled transcription and light perception by the photoreceptor must coincide to allow photoperiodic responses.1,3 Here we summarize recent advances regarding the molecular mechanisms of the circadian clock and photoperiodic responses such as flowering time and hypocotyl elongation. Lastly, we introduce recent findings that reactive oxygen species (ROS) homeostasis is also modulated by photoperiod.
Molecular Mechanisms of the Circadian Clock in Arabidopsis
The circadian clock is an endogenous molecular oscillator with period of approximately 24 h. There are a wide range of processes in plants that show circadian rhythms, including the movement of leaves, stomatal opening, stem elongation, metabolic processes, such as photorespiration and photosynthesis, and the expression of a large set of genes.1,3−5 Thus, the circadian clock plays an important role as a pacemaker for various physiological events that occur throughout the day. Plants also use their circadian clocks to pace changes that occur throughout the year and to regulate developmental transitions, such as flowering and dormancy.1,6 Initially, the circadian clock was described as a single loop comprising CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), and TIMING OF CAB EXPRESSION 1 (TOC1).7−10 The current view of the Arabidopsis clock is more complex. The original loop is one part of interconnected multiple feedback loops that contain the morning- and evening-expressed clock proteins. Our view of the clock architecture is also constantly improving. For instance, recent identification of transcriptional activators/coactivators for the evening element (EE)-regulated genes, REVEILLE8 (RVE8), NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENES 1 (LNK1), and LNK2, has helped us to create a more accurate explanation of the induction of evening-expressed clock genes.11−14 Here, we describe our current understanding of the complex transcriptional, posttranscriptional, and posttranslational regulatory mechanisms that make up the circadian clock in Arabidopsis.
Transcriptional Regulation of the Circadian Clock
Similar to biological clocks in other organisms, the Arabidopsis circadian clock consists of multiple interlocking feedback loops.1,4,5 The first characterized feedback loop, which connects morning clock components with evening ones, consists of two related morning MYB transcription factors, CCA1 and LHY, and evening-expressed TOC1 proteins (Figure 1). During the daytime, CCA1 and LHY form a negative-feedback loop with PSEUDO-RESPONSE REGULATOR 7 (PRR7) and PRR9.15,16 CCA1 and LHY directly induce the expression of PRR7 and PRR9 genes. PRR7 and PRR9 encode proteins that directly repress CCA1 and LHY expression during the daytime, although the timing of the peak expression of PRR7 and PRR9 proteins differs (Figure 1A,B). CCA1 and LHY also act as transcriptional repressors. During the morning, they concomitantly suppress the expression of afternoon- and evening-phased circadian clock genes, such as PRR5, TOC1, GIGANTEA (GI), LUX ARRHYTHMO (LUX), and EARLY FLOWERING 4 (ELF4)10,17−19 (Figure 1B). To repress the expression of TOC1, GI, and most likely other evening genes, CCA1 and LHY bind to the CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10)–DE-ETIOLATED 1 (DET1)–DAMAGED DNA BINDING 1 (DDB1) complex (the CDD complex). DET1 serves as a transcriptional corepressor necessary for CCA1 and LHY to inhibit TOC1 and GI expression, and the recruitment of DET1 to the TOC1 promoter is dependent on the presence of CCA1 and LHY20 (Figure 1B). As DET1 is a part of the CCA1/LHY repressor complex, the det1 mutant demonstrates a short period phenotype similar to that of cca1 or lhy mutants. Although CCA1 and LHY likely confer repressive activity through interaction with the CDD complex, an understanding of how CCA1 and LHY act as transcriptional activators (for PRR9, PRR7, and other genes) at the molecular level remains elusive.
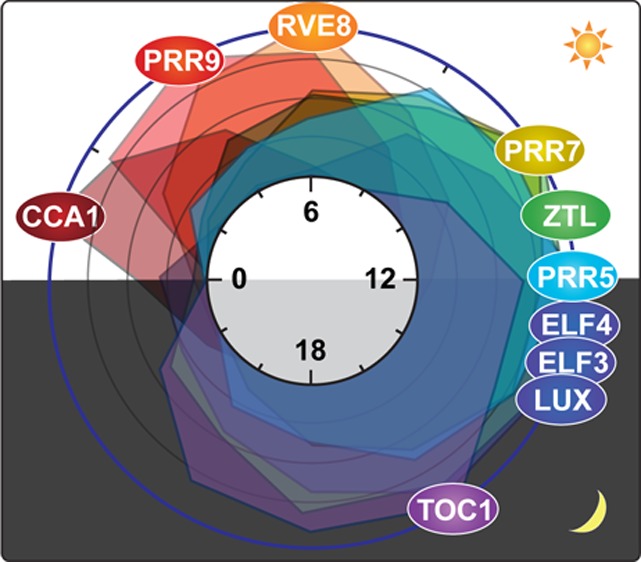
Figure 1.
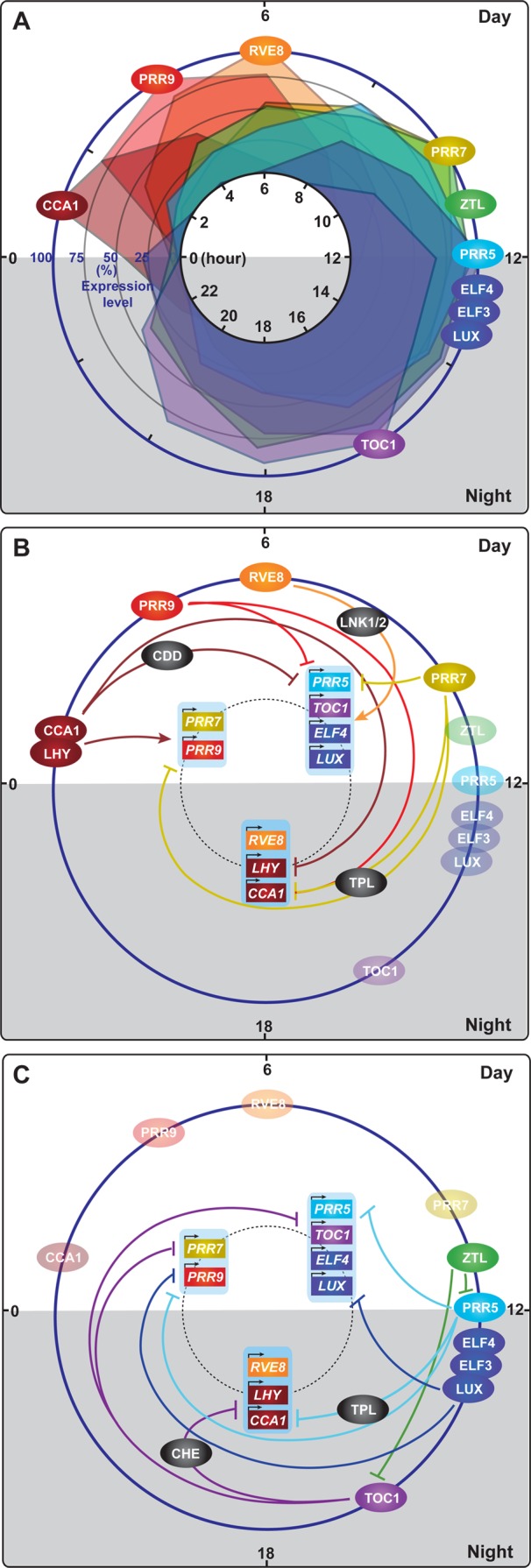
Expression of circadian clock proteins and the architecture of the clock in Arabidopsis thaliana. (A) Daily protein expression profiles of circadian clock components. The expression profiles of the clock proteins are based on the following: CCA1,8 RVE8,32 PRR9,21 PRR7,21ZTL,55 PRR5,21 Evening Complex (ELF4–ELF3–LUX),28 and TOC1.57 The peak expression levels of these proteins were set to 100%, and the rest of the expression levels were calculated against the peak levels. The levels of expression are plotted at 2 h intervals throughout the day. (B) Molecular events occurring from morning to afternoon in the circadian clock. The interactions of clock components and their transcriptional targets, which mainly happen from morning to afternoon, are depicted. In the morning, CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY) activate the expression of PSEUDO-RESPONSE REGULATOR 9 (PRR9) and PRR7 and suppress the expression of most evening phase genes such as PRR5, TIMING OF CAB EXPRESSION 1 (TOC1), LUX ARRHYTHMO (LUX), and EARLY FLOWERING 4 (ELF4). CCA1 and LHY form a repressor complex with the CONSTITUTIVE PHOTOMORPHOGENIC 10 (COP10)–DE-ETIOLATED 1 (DET1)–DAMAGED DNA BINDING 1 (DDB1) complex (CDD) to suppress the expression of the evening genes. CCA1 and LHY also suppress their own expression. In the early afternoon, PRR9 and PRR7 suppress the expression of CCA1 and LHY using TOPLESS (TPL) as a corepressor. Downregulation of CCA1 and LHY expression results in derepression of evening phase genes. RVE8, together with NIGHT LIGHT-INDUCIBLE AND CLOCK-REGULATED GENES 1 and 2 (LNK1/2) as coactivators, directly activates expression of evening phase genes, such as PRR5, TOC1, ELF4, and LUX. (C) Molecular events occurring from evening to night in the circadian clock. The interactions of clock components and their transcriptional targets, which mainly happen from evening to the end of the night, are depicted. PRR5 protein continuously suppresses the expression of CCA1 and LHY. PRR5 also represses its own expression. TOC1 suppresses the expression of CCA1 and LHY by itself or interacting with CCA1 HIKING EXPEDITION (CHE). TOC1 also suppresses the expression of other clock components, PRR9, PRR7, TOC1, GI, LUX, and ELF4. ZTL interacts with PRR5 and TOC1 to degrade them in the dark. The Evening Complex (ELF4–ELF3–LUX) suppresses the expression of PRR9 to complete the cycle by induction of CCA1 and LHY. The positions of the circles in panels B and C indicate the timing of peak protein accumulation (based on panel A) of each component.
Throughout the day, the sequential expression of PRR9, PRR7, and PRR5 reciprocally contributes to the regulation of CCA1 and LHY expression (Figure 1A,B). PRR9, PRR7, and PRR5 physically associate with the promoters of CCA1 and LHY to repress their expression.21 PRR9, PRR7, and PRR5 also repress their own expression and that of each other by directly binding to their promoters.21 This repression occurs through the direct interaction of these PRRs with TOPLESS/TOPLESS-RELATED (TPL/TPR) corepressors22 (Figure 1B,C). As a consequence, PRR-dependent suppression of CCA1 and LHY during the day results in the derepression of the evening phase genes. Once TOC1, a founding member of the PRRs, can be expressed during the night, TOC1 physically associates with the promoters of CCA1 and LHY to suppress their expression23 (Figure 1A,C). TOC1 binds to the TOC1 morning element (TIME) present in the CCA1 and LHY promoter.23 TOC1 also physically associates with the CCA1 promoter by interacting with CCA1 HIKING EXPEDITION (CHE), the TCP transcription factor that represses the expression of CCA1 and LHY (Figure 1C). Similar to TOC1, the expression of CHE is also directly repressed by CCA1 and LHY during the morning.24 Recently, a genomic-scale chromatin immunoprecipitation assay revealed that TOC1 binds to and represses not only CCA1 and LHY but also many other clock genes, such as PRR9, PRR7, TOC1, GI, LUX, and ELF4(25) (Figure 1C). This clearly shows that TOC1 forms an important hub structure in the clock from evening to night.
In addition to TOC1, the Evening Complex (EC, ELF4–ELF3–LUX) also indirectly contributes to the dawn expression of CCA1 and LHY during the night.26 The Evening Complex is formed using ELF3 as an adaptor protein between ELF4 and LUX whenever these three components are expressed. The LUX transcription factor contains the GARP domain that binds to the GATWCG site in the PRR9 promoter and recruits the Evening Complex to the promoter region. The Evening Complex suppresses the expression of PRR9, which in turn activates expression of CCA1 and LHY(26−29) (Figure 1B,C). Expression of LUX and ELF4 is also under the control of CCA1 and LHY, as CCA1 and LHY directly bind to the promoters of LUX and ELF4 to suppress their expression18,30 (Figure 1B). In addition, the self-negative-feedback regulation of LUX forms an additional loop of the Evening Complex29 (Figure 1C).
As discussed above, multiple intertwined negative-feedback loops exist in the architecture of the circadian clock. Several positive regulators of the clock have been identified, and the characterization of these factors has provided a better understanding of the molecular architecture of the circadian clock. LIGHT REGULATED WD1 (LWD1) and LWD2 genetically function as activators in the light input pathway as well as in the central oscillator.14,31 In the lwd1 lwd2 mutant, the amplitudes of the daily PRR9, PRR7, and PRR5 oscillations are reduced and the period length is shorter than that of the wild-type plants. LWD1, a nuclear localized WD repeat protein, directly binds to the promoters of PRR9, PRR5, and TOC1 and positively regulates their expression. In addition, PRR9 and PRR7 positively regulate the expression levels of LWD1 and LWD2.14 Therefore, LWDs and PRRs form a positive-feedback loop in the regulation of the circadian clock. Interestingly, in the lwd1 lwd2 double mutant, the expression of CCA1 and LHY also shows a reduced degree of oscillation under continuous light conditions and an arrhythmic expression pattern under constant dark conditions. It seems as though LWDs affect the expression of CCA1 and LHY through a PRR-independent mechanism.14RVE8 has been identified as an EE binding transcriptional activator that functions as a positive regulator13,32 (Figure 1A,B). The RVE genes belong to the MYB transcription factor gene family that includes CCA1 and LHY genes.32 Even though the EE is regarded as the CCA1 and LHY binding site for negative-feedback regulation of the circadian clock, it was suggested that an unknown transcriptional activator(s) could also bind to the EE to activate the afternoon- and evening-phased gene expression.33RVE8 mRNA peaks at the subjective dawn, similar to CCA1 and LHY, but RVE8 protein accumulates 3–6 h after the subjective dawn, which is later than the CCA1 and LHY peak32 (Figure 1A). RVE8 induces the expression of afternoon- and evening-phased genes, such as PRR5, TOC1, GI, ELF4, and LUX. The expression of RVE8 is reciprocally controlled by PRR5, as PRR5 suppresses the expression of RVE8(32) (Figure 1B). Similar to RVE8, LNK1 and LNK2 also contribute to the activation of afternoon- and evening-phased genes, such as PRR5, ELF4, and FKF1(12) (Figure 1B). Both rve4 rve6 rve8 and lnk1 lnk2 mutants shows similar long period phenotypes with a lower amplitude of PRR5 and TOC1 oscillations.12,13 In addition, recent work revealed that LNK1 and LNK2 form complexes with RVE8, RVE4, CCA1, and LHY in vivo.11 Together with RVE8, LNK1 and LNK2 directly contribute to the regulation of PRR5 and TOC1 expression as transcriptional coactivators (Figure 1B). Interestingly, LNK1 and LNK2 also interact with CCA1 and LHY. It will be of interest to know the function of the LNK1/2–CCA1/LHY complexes. PRR5 and TOC1 form negative-feedback loops with LNK1/2. PRR5 and TOC1 (as well as PRR9 and PRR7) bind to the LNK promoters to repress the expression of LNKs.12,34 Therefore, PRRs and TOC1 regulate the expression patterns of both positive and negative factors, both of which reciprocally control their expression. As the Arabidopsis circadian clock is operated by multiple negative and positive-feedback loops throughout the day, it is important to study the kinetic changes of their interactions as well as to integrate more precise spatial (tissue-specific) and temporal expression profiles of these factors.
Alternative Splicing Regulation of the Circadian Clock Transcripts
Although the transcriptional network of the Arabidopsis clock is complicated, posttranscriptional and posttranslational regulation in the system provides additional complexity and contributes to the precision and robustness of the circadian clock.35 One type of posttranscriptional regulation, alternative splicing, operates widely in eukaryotic organisms and greatly increases both transcriptional and translational diversity. In the transcription process, precursor mRNAs undergo splicing events to remove the introns and join exons to generate mature mRNAs. Through alternative splicing, different mRNA variants can be produced from the same gene. These mRNA variants can give rise to protein variants, which may have different functions by altering protein activity, cellular localization, and posttranslational modification.36
Alternative splicing modulates the function of the circadian clock components.35,37 The Arabidopsis PROTEIN ARGININE METHYL TRANSFERASE 5 (AtPRMT5) gene encodes a type II protein, arginine methyltransferase, that mediates the methylation of diverse substrates, including components of the spliceosome. Defects in the spliceosome caused by the atprmt5 mutation result in splicing defects in PRR7 and PRR9 transcripts and alter the output of the circadian clock.38−40 Two other splicing factors, SNW/SKI-INTERACTING PROTEIN (SKIP) and SPLICEOSOMAL TIMEKEEPER LOCUS 1 (STIPL1), also affect the regulation of the circadian clock in Arabidopsis.41,42 The level of alternative splicing of PRR7 and PRR9 transcripts is reduced in the skip mutant, and the expression of CCA1, LHY, PRR9, GI, and TOC1 is altered in this mutant. In the skip mutant, longer period length changes were observed when the surrounding temperature was reduced.35 This indicates that alternative splicing may contribute to the temperature compensation function of the clock. Other results also indicate the presence of this mechanism. The CCA1 locus generates two alternative splicing isoforms, CCA1α and CCA1β. CCA1β encodes a protein that has a dimerization domain with CCA1 and LHY but lacks the DNA binding domain. The abundance of the CCA1β variant increases with heat (37 °C) exposure but decreases with cold (4 °C) treatment under continuous light conditions.43 The protein translated from the CCA1β transcript competes with the one derived from CCA1α to form a CCA1α–CCA1β homodimer and CCA1α–LHY heterodimer. The formation of the CCA1β–CCA1α and CCA1β–LHY dimers weakens their DNA binding affinity, thus affecting the activity of the dimer, and subsequently changing the pace of the clock. The alternative splicing variants derived from the LHY, PRR7, PRR5, and TOC1 loci also affect the overall transcripts or protein levels.44 Some splice variants of these genes are targeted for the nonsense-mediated mRNA decay (NMD) pathway.44 These findings suggest that alternative splicing gives an additional regulatory mechanism in the circadian clock and is likely involved in attaining the temperature compensation characteristic of the clock.
Phosphorylation and Degradation of the Circadian Clock Proteins
In addition to the complex transcriptional and posttranscriptional regulation, posttranslational regulation also plays an important role in the clock mechanism by changing the stability, activity, and cellular localization of clock components.45−47 One typical protein modification is phosphorylation. Within the plant clock, CCA1 is phosphorylated by CASEIN KINASE 2 (CK2). CCA1 phosphorylation affects its DNA binding activity.45,48 The CK2-dependent phosphorylation of CCA1 interferes with the binding of CCA1 to its target gene promoter.49 Overexpression of CK2 β-subunit 3 (CKB3) or CKB4 leads to a short period phenotype similar to that of the cca1 mutant.50 Phosphorylation of clock components by CK seems to be a conserved mechanism in both the animal and plant clocks. In both cases, this regulation influences the period length of the clock.
Another important posttranslational regulation is protein stability regulation. The regulation of clock protein stability is an essential part of proper clock progress. For instance, the single-cell green alga Ostreococcus tauri possesses a clock simpler than that in Arabidopsis in which CCA1 and TOC1 homologues form a single negative-feedback loop,51 and thus, it became the model for analyzing the minimal requirements for circadian oscillation. Pharmacological inhibition of proteasomal function stopped circadian oscillation, suggesting that proteolysis of clock components is required to maintain the function of the biological clock.47
In Arabidopsis, ZEITLUPE (ZTL) protein plays an important role in the regulation of period length.52−54 ZTL is a blue light photoreceptor F-box protein that is unique to plants, and blue light regulates ZTL stability throughout the day.53 Even though ZTL mRNA is constitutively expressed, the amount of ZTL protein oscillates with the peak at the end of the day46 (Figure 1A). This daily oscillation of ZTL abundance is regulated by the blue light-dependent interaction with GI.55 ZTL absorbs blue light with its LOV (light, oxygen, or voltage) domain, and absorbing blue light triggers the interaction with GI though the LOV domain. The ZTL–GI complex stabilizes ZTL by protecting it from proteasome-dependent degradation. This interaction also helps to keep GI in the cytosol.56 The daily ZTL stability change controls the abundance of clock components that are degraded by ZTL.52−54,57 ZTL physically interacts with the pseudoreceiver domains of TOC1 and PRR5 through its LOV domain and degrades these proteins in the dark52−54 (Figure 1C). This light-dependent posttranslational regulation controls the period length and amplitude of the circadian clock gene expression.52−54
In addition to ZTL, its homologues, FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), and LOV KELCH RPOTEIN2 (LKP2), also participate in this posttranslational regulation by directly interacting with both TOC1 and PRR5.57 In the ztl fkf1 and ztl fkf1 lkp2 mutants, TOC1 and PRR5 proteins are more stable than in the ztl single mutant. Moreover, both the ztl fkf1 and ztl fkf1 lkp2 mutants show a period phenotype longer than that of the ztl single mutant, indicating that both FKF1 and LKP2 contribute to regulate the stability of TOC1 and PRR5.57 Thus, ZTL, FKF1, and LKP2 regulate TOC1 and PRR5 degradation to determine the period length of circadian oscillation. Phosphorylation of TOC1 and PRRs also affects their interactions with ZTL.58 Phosphorylation of TOC1 leads to its nuclear localization.58 As ZTL exists in the cytosol, this regulation prevents ZTL-mediated degradation of TOC1.58 The direct interaction of TOC1 with PRR3 and/or PRR5 also stabilizes TOC1.59,60 Interaction of TOC1 with PRR5 also enhances the nuclear localization of TOC1.60 Light and the circadian clock regulate ZTL protein abundance, which in turn regulates the stability of TOC1 and PRR5 and controls the periodicity and robustness of the clock.
In addition to the ZTL group proteins, several other E3 ubiquitin ligases also participate in the regulation of circadian clock protein stability. CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) physically interacts with ELF3 to degrade it.61 This degradation allows the recruitment of newly synthesized ELF3 into the COP1–ELF3 complex. Formation of the COP1–ELF3–GI complex at night leads to the destabilization of GI. Another E3 ligase, SINAT5, participates in the regulation of LHY protein stability.61 SINAT5, an Arabidopsis homologue of the Drosophila SINA ring-finger protein, physically interacts with LHY. Ubiquitination of LHY by SINAT5 is suppressed by nuclear protein DET1.62 In addition to ubiquitination, the deubiquitination enzymes UBP12 and UBP13 also affect clock progression. Expression of UBP12 and UBP13 is under the control of the circadian clock, and the ubp12 ubp13 double mutant shows a short period phenotype, indicating that the deubiquitination process of clock proteins may fine-tune the degradation kinetics.63 As described in this section, the proper dynamics of posttranslational regulation of the clock proteins determine the proper pace of the circadian oscillation.
Photoperiodic Flowering in Arabidopsis
The intricate networks of the circadian clock allow plants to regulate diverse diurnal and seasonal physiological and developmental events. Within the circadian clock-regulated responses, one of the most characterized is the photoperiodic flowering response. Arabidopsis is a facultative long-day (LD) plant, in which flowering is accelerated under LD conditions through the function of FLOWERING LOCUS T (FT) protein.64,65 FT is a small mobile protein regarded as a florigen (flowering hormone).66 It is synthesized from the leaf vasculature and transported to the shoot apical meristem through the phloem.67 The amount of FT transcript influences the floral induction of Arabidopsis thaliana.68 Induction of FT transcription occurs under LD conditions, and this induction is regulated by the CONSTANS (CO) transcriptional activator.65 The circadian clock and light signaling tightly regulate the timing of CO transcript expression and CO protein activity in LD.69,70 In this section, we summarize recent advances in the mechanisms by which photoperiodic information regulates expression of CO and FT. Many recent reviews cover the floral induction initiated by FT at the shoot apical meristem in detail;66,71,72 therefore, we will not discuss those mechanisms here.
Transcriptional Regulation of the CO Gene
Similar to circadian clock regulation, both transcriptional and posttranslational regulation of CO are crucial for plants to measure changes in day length for photoperiodic flowering. To regulate daily CO expression patterns, the circadian clock-regulated FKF1, GI, and CYCLING DOF FACTORs (CDFs) play major roles73−76 (Figure 2). CO expression profiles are at trough level in the morning and then show a daytime peak in the late afternoon, with the highest peak at night under LD conditions. This rhythmic expression pattern of CO is regulated by time-dependent expression and degradation of CDF (CDF1–CDF3 and CDF5) transcriptional repressors. CDF1 protein directly binds to the CO promoter to repress the expression of CO and contributes to reducing the level of expression in the early part of the day.74,77 Expression of the CDF1 transcript is under control of the circadian clock. CCA1 and LHY positively regulate the expression of CDF1 at dawn,16 and in the afternoon, PRR9, PRR7, and PRR5 negatively regulate expression of CDF1, CDF2, CDF3, and CDF5.16,34 Consequently, circadian clock-dependent transcriptional regulation of CDFs consists of a basal regulatory loop to determine the timing of CO expression.
Figure 2.
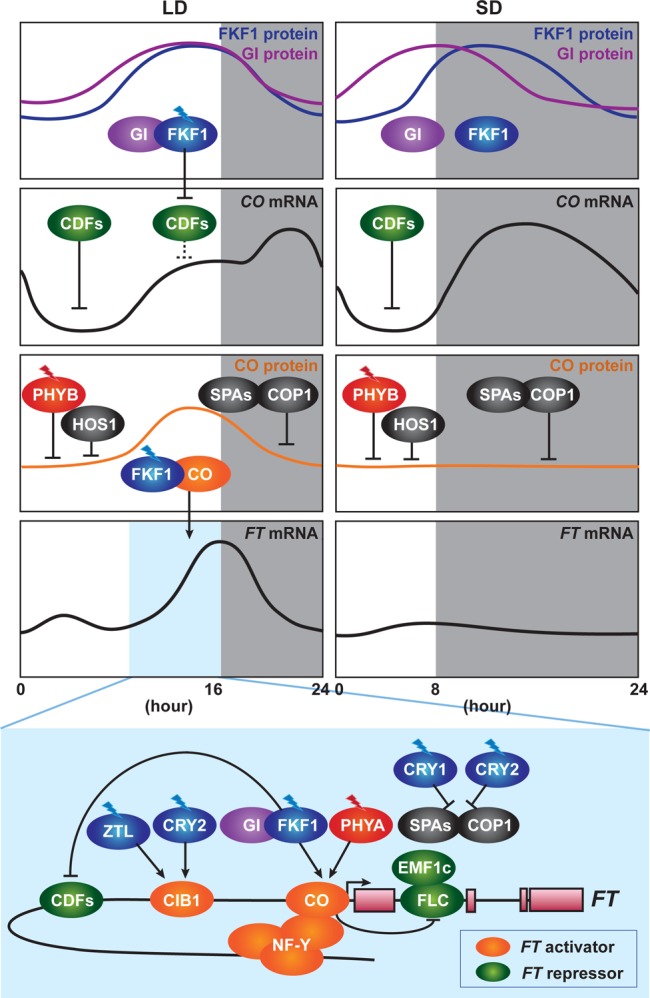
Photoperiodic regulation of FLOWERING LOCUS T (FT) expression. In LD, FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), and GIGANTEA (GI) form a complex, when their expression patterns coincide and FKF1 absorbs blue light. The FKF1–GI complex degrades CYCLING DOF FACTOR (CDF) proteins on the CONSTANS (CO) promoter in the afternoon. FKF1–GI-dependent degradation of CDFs results in derepression of CONSTANS (CO) expression. The same mechanism of degradation of CDFs by the FKF1–GI complex also exists on the FT promoter. FKF1 physically interacts with CO protein to stabilize it. Far-red light-absorbed PHYTOCHROME A (PHYA) also stabilizes CO protein. Stabilized CO protein binds to the FT promoter to activate FT expression. The NUCLEAR FACTOR-Y (NF-Y) complex enhances the binding of CO protein to the FT promoter. CO protein antagonizes the function of the EMBRYONIC FLOWERING 1 complex (EMF1c) and the FLOWERING LOCUS C (FLC) complex that suppresses the expression of FT. CRYPTOCHROME-INTERACTING BASIC HELIX–LOOP–HELIX 1 (CIB1) is activated by blue light absorbed by CRYPTOCHROME 2 (CRY2) and stabilized by blue light absorbed by ZEITLUPE (ZTL). CIB1 directly activates the expression of FT in the afternoon. In the morning, PHYB that has absorbed red light and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 (HOS1) degrade CO protein. At night, the CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1)–SUPPRESSOR OF PHYA-105s (SPAs) complex degrades CO protein. They prevent flowering under unfavorable conditions, such as SD. In SD, the expression peaks of FKF1 and GI do not coincide. Without the FKF–GI complex, CO expression is continuously suppressed by CDF proteins during the daytime.
Expression of CO in the afternoon is important for inducing FT expression and is induced by an interaction between FKF1 and GI.75 GI is a plant-specific large nuclear protein without any known functional domains.78 FKF1 interacts with GI in a blue light-dependent manner in the LD afternoon, and the FKF1–GI complex degrades CDF proteins in the afternoon, resulting in the derepression of the CO promoter under LD conditions (Figure 2). Under short day (SD) conditions, the expression peaks of FKF1 and GI do not coincide, and the FKF1 peak occurs at night. Without forming the light-dependent FKF1–GI complex during the daytime, CDF proteins continuously repress CO expression during the day in SD75 (Figure 2). ZTL and LKP2, which interact with FKF1 and GI, are involved in the destabilization of the CDF2 protein.77 Once CDF repressors are removed by the FKF1–GI complex, the bHLH (basic helix–loop–helix) transcription factors, FLOWERING BHLH1 (FBH1), FBH2, FBH3, and FBH4, activate the transcription of CO. These FBH proteins bind to the E-box elements located in the CO promoter. Chemically induced expression of FBH1 and FBH2 leads to an increased amount of CO transcript in the late afternoon and the dark under both LD and SD.79
Other components also participate in the regulation of CO expression. RED AND FAR-RED INSENSITIVE 2 (RFI2) is a nuclear protein that possesses a C3H2C3-type zinc finger and RING domains. RFI2 might work together with GI to negatively regulate the evening and night expression of CO.80 LONG VEGETATIVE PHASE1 (LOV1) is a NAC domain protein. Its mutation affects flowering time under LD. LOV1 represses the expression of CO at the end of day and during the night.81 FIONA1 also represses the expression of CO in both LD and SD.82 DAY NEUTRAL FLOWERING (DNF) is a membrane-bound E3 ligase that represses CO expression in morning and early afternoon in SD.83 The chromatin remodeling component MULTICOPY SUPPRESSOR OF IRA 1 (MSI1) also participates in the regulation of CO expression. MSI1 is a component of the Polycomb Group Repressive Complex 2 (PRC2) and also interacts with LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) and FVE, all of which participate in chromatin remodeling. MSI1 positively regulates expression of CO and GI in response to photoperiod.84CO-EXPRESSED WITH CLOCK GENES LHY AND CCA1 1 (CEC1) (also known as LNK2) and CEC2/LNK3 negatively regulate expression of CO and FT. This is interesting because LNK1 and LNK2 were originally characterized as activators of the evening clock genes as well as FT, as the level of FT expression is diminished in the lnk1-1 lnk2-1 allele.12 On the other hand, in the cec1 (=lnk2-2) and cec2 (lnk3) single mutants, the level of CO and FT expression in the afternoon and night is elevated.85 Some of the difference in CO and FT expression may be caused by a lnk2 allele difference. These results indicate that LNKs may function as activators (LNK1 and LNK2) and repressors (LNK3, and potentially LNK4). Taken together, CO expression throughout the day is tightly controlled by complex transcriptional regulation.
Posttranslational Regulation of CO Protein
Posttranslational regulation of CO protein is crucial for photoperiod-dependent activation of FT.2,70,76 Even though the CO transcript is strongly expressed at night in both LD and SD, the induction of FT expression occurs only at dusk in LD. At night, CO protein is actively degraded by COP1 and the SUPPRESSOR OF PHYA-105 1 (SPA1) complex86−88 (Figure 2). HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1 (HOS1) encodes a RING finger containing E3 ubiquitin ligase. Similar to COP1 and SPA1, HOS1 binds to the CO CCT motif to degrade CO protein during the early hours of the day89 (Figure 2). Two red/far-red light photoreceptors, phytochrome A (PHYA) and PHYB, antagonistically regulate the stability of CO protein.90 CO protein is degraded in the morning and stabilized in the late afternoon. PHYB mediates the degradation of CO in the morning, whereas PHYA stabilizes CO protein in the afternoon90 (Figure 2). Recently, PHYTOCHROME-DEPENDENT LATE FLOWERING (PHL) was characterized as an interacting partner of PHYB and CO. PHL accumulates in the nucleus in the afternoon to suppress PHYB-mediated degradation of CO protein.91 Two blue light photoreceptors, cryptochrome 1 (CRY1) and CRY2, also participate in the regulation of CO protein stability.92−94 CRY1 physically interacts with SPA1 to interrupt the formation of the COP1–SPA1 complex93,94 (Figure 2). CRY2 also interacts with SPA1 in response to blue light. Light-dependent CRY2–SPA1 interaction enhances the CRY2–COP1 interaction to suppress the function of COP1 in CO degradation92 (Figure 2). Several photoreceptors and E3 ubiquitin ligases control the stability of CO protein. However, the molecular mechanism of late afternoon-specific stabilization of CO protein is not clearly understood, because those regulators are constitutively expressed throughout the day.
The daytime-specific stabilization of CO protein is regulated by the blue light photoreceptor FKF1.73−76 FKF1 protein accumulates from the afternoon to the end of the day. FKF1 interacts with CO protein through its LOV domain to stabilize CO protein in a blue light-dependent manner. Both FKF1 and CO exist on the FT promoter region where the CORE sequences are located. Therefore, FKF1 and light determine the timing of accumulation of CO protein within a day, and this is crucial for the induction of FT(76) (Figure 2).
Transcriptional Regulation of the FT Gene
The circadian clock and light signaling coordinate to control CO protein activity and FT expression.2,70,76 Several classes of transcriptional repressors participate in the regulation of FT expression. MADS-box proteins, FLOWERING LOCUS C (FLC) and SHORT VEGETATIVE PHASE (SVP), form a heterodimer. The FLC–SVP complex directly suppresses the expression of FT and another flowering gene, SUPPRESSOR OF OVEREXPRESSION OF CO 1 (SOC1).95 FLC also forms a nuclear protein complex with its homologue FLOWERING LOCUS M (FLM) and MADS AFFECTING FLOWERINGs (MAFs) to suppress the expression of FT.96 The Polycomb group (PcG) complex also participates in FLC clade-mediated repression of FT expression. EMBRYONIC FLOWER1 (EMF1), LHP1, and histone H3 lysine-4 demethylase form a distinct Polycomb group complex (PRC1-like complex) and act as a transcriptional repressor complex. The EMF1 complex physically associates with FT chromatin and represses the expression of FT.97 EMF1 directly interacts with FLC and FLM.97 Thus, even though the EMF complex lacks a DNA binding protein, FLC and FLM may recruit the PcG complex to the FT promoter. FLC clade- and PcG-mediated repression prevents the induction of FT under noninductive SD conditions. The binding of EMF1 to the FT promoter is disrupted in the afternoon by CO overexpression. Thus, daytime-specific stabilization of CO may antagonize the binding of the EMF complex to the FT chromatin at dusk, leading to proper activation of FT(97) (Figure 2).
TEMPRANILLO1 (TEM1) and TEM2 belong to the RAV subfamily of transcription factors and bind to the FT promoter and negatively regulate its expression.98SCHLAFMÜTZE (SMZ) encodes the APETALA 2 (AP2)-related transcription factor. SMZ directly binds to the FT promoter and represses its expression.99 The microRNAs, miR156 and miR172, participate in the control of SMZ expression to determine the timing of FT expression.100 miR156 is expressed strongly in early developmental stages and weakly in late developmental stages. miR156 targets SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors, suppressing their expression by transcriptional cleavage. Thus, the age-dependent decrease in miR156 is accompanied by an increase in the level of SPL expression. SPL9 and SPL10 directly activate the expression of miR172 in an age-dependent manner. miR172 accumulates to high levels in the leaves and floral buds.101 The amount of miR172 is more abundant in LD than in SD. In addition, GI promotes the accumulation of miRNA172 under LD conditions.102 miR172 suppresses the expression of AP2-type floral repressor transcripts, such as AP2, TARGET OF EAT 1 (TOE1), TOE2, TOE3, SMZ, and SCHNARCHZAPFEN (SNZ).100,102 CDF1, a transcriptional repressor of CO, also associates with the promoter of FT and represses the expression of FT in the morning. Repression of FT by CDF1 is removed by the blue light-dependent FKF–GI complex in the afternoon under LD conditions.76 Thus, FKF1 determines the daytime-specific expression of FT by removing the CDF transcriptional repressors and stabilizing the CO transcriptional activator (Figure 2).
In the activation of FT expression, two transcriptional activators, CO and CRYPTOCHROME-INTERACTING BASIC HELIX–LOOP–HELIX 1 (CIB1), play major roles.90,103−105 CO directly binds to the CONSTANS responsive element (CORE) in the FT promoter but its binding activity to the FT promoter is weak in vitro.106 It is suggested that CO interacts with other proteins and this interaction increases the binding affinity of CO for the FT promoter. CO and CO-like protein physically associate with NUCLEAR FACTOR-Y (NF-Y)/HEME ACTIVATOR PROTEIN (HAP).107 The NF-Y/HAP complex consists of NF-YA, NF-YB, and NF-YC and binds to the CCAAT site located in the FT promoter. Chromatin looping of the FT promoter is accelerated in the late afternoon and keeps the NF-Y complex very close to the CORE. The NF-Y complex physically interacts with CO protein to stabilize the binding of CO protein to the CORE in the FT promoter108 (Figure 2).
CIB1, together with CIB2, CIB4, and CIB5, is also involved in FT induction.104 CIB1 directly binds to the promoter of FT and activates its expression (Figure 2). CIB proteins form heterodimer complexes, and the heterodimerization increases the DNA binding activity of CIBs with the FT promoter. The function of CIB1 is restricted from the afternoon to early night. CIB1 protein forms a complex with CRY2 in a blue light-dependent manner; CIB protein accumulates only in the presence of blue light, whereas it is actively degraded without blue light. Interestingly, blue light-dependent stabilization of CIB1 is mediated by ZTL and LKP2, not CRY2. ZTL and LKP2 stabilize CIB1 under blue light. Therefore, CIB1 is activated by CRY2 and stabilized by ZTL and LKP2 under blue light to promote FT expression103 (Figure 2). In summary, information about changing photoperiods in growing conditions is monitored using large numbers of components, many of which are regulated by the circadian clock. The information is utilized by plants to precisely determine the expression of FT protein, which consequently determines the timing of flowering.
Photoperiodic Regulation of Hypocotyl Growth
In addition to the timing of flowering, the growth rates of hypocotyls and petioles also differ depending on photoperiod. LD-grown seedlings show hypocotyls and petioles much shorter than those of SD-grown seedlings.109 To regulate day length-dependent differences in hypocotyl growth rates, plants also use the circadian clock to sense photoperiod changes. In Arabidopsis, phytochromes and their downstream components PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) and its homologue PIF5 play prominent roles in determining the timing of hypocotyl growth within a day.110−112 PIF4 and PIF5 are bHLH transcription factors that promote elongation of the hypocotyl. PIF4 and PIF5 proteins are stable in the dark and destabilized under light mainly by PHYB. SD-induced elongation of the hypocotyl is diminished in the pif4 pif5 double mutants, indicating that photoperiodic information is transmitted through PIF4 and PIF5 function to regulate hypocotyl length. PIF4 and PIF5 induce the expression of ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2 (ATHB2) and other genes that are involved in hypocotyl elongation.113 PHYB acts as a repressor of hypocotyl elongation by degrading PIF4 protein under light.114 Therefore, the hypocotyl length of the phyB mutant in LD and SD is always longer than that of wild-type plants, regardless of photoperiod.109
The circadian clock contributes to photoperiodic hypocotyl growth regulation in part by regulating the timing of the expression of PIF4 and PIF5. The expression patterns of both PIF4 and PIF5 transcripts show diurnal oscillation with peaks in the daytime.109 During the evening, the Evening Complex (ELF4–ELF3–LUX) directly represses the expression of PIF4 and PIF5 transcription28 (Figure 3). Following the changes in mRNA expression levels, PIF4 protein accumulates at a high level at the end of the night in SD, while the PIF4 protein level is low at night in LD.115 As elongation occurs around dusk,110 this may cause the difference in growth.
Figure 3.
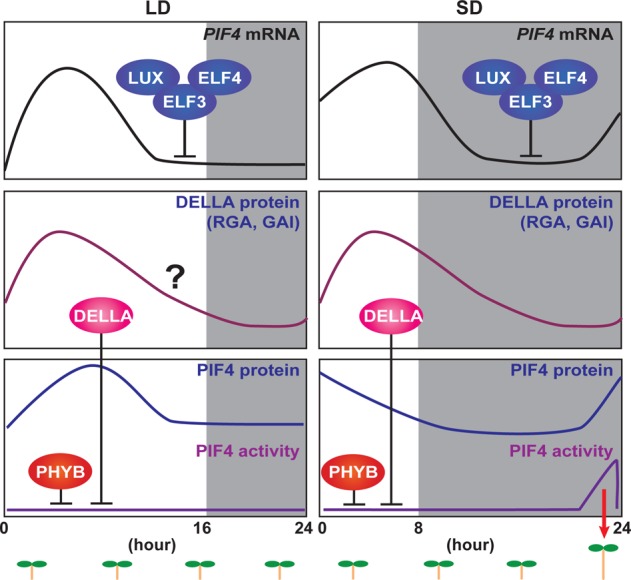
Photoperiodic regulation of hypocotyl elongation. The hypocotyl elongation rate of plants is determined by a combination of the internal circadian clock and external photoperiodic information. Expression of PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) is controlled by the circadian clock. The Evening Complex (ELF4–ELF3–LUX) directly suppresses the expression of PIF4. Under LD conditions, both transcription and translation of PIF4 occur during the day. PIF4 that accumulates during the daytime is inactivated by PHYB and DELLA protein. PHYB interacts with PIF4 to degrade it, and DELLAs interact with residual PIF4 to inactivate it by interrupting its DNA binding activity. Under SD conditions, PIF4 transcription and translation occur during day and night. PIF4 that accumulates during the nighttime activates its downstream target genes, which regulate hormonal responses that trigger hypocotyl elongation. We used the experimental data of DELLA protein expression under SD conditions117 to illustrate DELLA protein expression in both LD and SD.
Phytohormone gibberellin (GA) signaling regulates the activity of PIF proteins. DELLA proteins, repressors of GA signaling, bind to PIF4 to interrupt its DNA binding activity.116 The expression of DELLA proteins is also regulated by the circadian clock and peaks at the end of the day.117 This expression pattern contributes to the weakening of the DNA binding ability of PIF4 during the daytime116,117 (Figure 3). Therefore, the mechanism governing photoperiodic hypocotyl growth also can be explained by the coincidence between the interaction of internal clock components and external light cues.
Photoperiodic Regulation of ROS Homeostasis
Daily environmental changes regulate not only plant development but also cellular homeostasis, such as metabolism. In the final section, we introduce recent advances in circadian clock-regulated redox homeostasis and the effect of photoperiod changes on this regulation, as an example of photoperiodic regulation of cellular homeostasis. Reactive oxygen species (ROS), which include hydroxyl radical (HO–), superoxide (O2–), and hydrogen peroxide (H2O2), are oxygen-containing molecules that have chemical reactivity higher than that of molecular oxygen (O2). ROS is a byproduct of many cellular metabolic processes, such as photosynthesis and respiration. Recent evidence suggests that ROS are not only byproducts but also signaling molecules in various biological processes, including biotic and abiotic stress responses, stomatal movement, development, and cell expansion.118−120 Inside cells, ROS are generated in chloroplasts, peroxisomes, and mitochondria.119 Despite their importance as signaling molecules, the ways in which plants regulate the status of cellular ROS throughout the day remain elusive.
Circadian Regulation of ROS Homeostasis and Their Roles in Plant Development
Cellular ROS homeostasis is tightly regulated by several antioxidant systems for protecting cell function. Failure to regulate cellular ROS levels results in severe cellular damage, such as excessive oxidative stresses.121 There is emerging evidence that the circadian clock contributes to cellular processes that maintain ROS at physiological levels in diverse organisms. Peroxiredoxins (PRXs) are highly conserved antioxidant proteins that contain Cys residues, which can be oxidized by peroxide. PRX oxidation patterns show circadian oscillation from bacteria to mammals.122,123 Oxidation patterns of other antioxidants, such as glutathione and ascorbic acid, also show circadian oscillation and contribute to the determination of the excitability of neurons in the mouse suprachiasmatic nucleus (SCN).124 Transcriptional regulation of nuclear factor erythroid-derived 2-like 2 (NRF2) is another example of circadian clock-dependent regulation of cellular ROS homeostasis.121 Disruption of NRF2 expression by mutations in ClockΔ19 results in decreasing levels of cellular reduced glutathione as well as increasing levels of protein oxidative damage.121 In the case of zebrafish, cellular ROS act as input signals for the circadian clock.125 Light-dependent accumulation of hydrogen peroxide, which is controlled by a diurnal change in catalase activity, induces the expression of zebrafish Cryptochrome 1a (zCry1a) and Period (zPer2).125 This mechanism is part of the light-dependent induction of zCry1a and zPer2 that entrains the circadian clock.125 As the enzymatic activity of the catalase involved in this regulation diurnally oscillates, the circadian clock and ROS homeostasis regulate each other through complicated feedback mechanisms under daily light–dark cycles.
The plant circadian clock also regulates ROS homeostasis. In the case of Arabidopsis, hydrogen peroxide (H2O2) levels diurnally oscillate and this oscillation is sustained under constant light conditions,126 indicating the involvement of the circadian clock in this regulation. In constant light, H2O2 production peaks during the afternoon following the peak expression of photosynthesis genes and reaches trough level at midnight. The mutation or overexpression of the CCA1 gene abolishes the rhythmicity of endogenous H2O2 levels, catalase activity, and ROS-related gene expression. Mutations in other circadian clock components, such as ELF3, LUX, and TOC1, also disrupt the rhythmic expression patterns of CAT genes and ROS-related genes.126
The oscillation of cellular H2O2 levels is closely linked to the antioxidant enzyme catalase. Diurnal catalase activity patterns are similar to the patterns of cellular H2O2 levels126 (Figure 4A). Hydrogen peroxide is generated in peroxisomes via enzymatic reactions catalyzed by glycolate oxidase and/or superoxide dismutase. Catalase is the first peroxisomal antioxidant enzyme that detoxifies cellular hydrogen peroxide to be characterized.119 There are three catalase genes (CAT1, CAT2, and CAT3) in Arabidopsis.127 Expression of all three is controlled by the circadian clock.126 Circadian oscillation of catalase expression is diminished in the CCA1 overexpressor and the cca1 lhy double mutant.126 The daily production of H2O2 also becomes constant in these plants. In addition, the expression patterns of multiple ROS responsive genes are also regulated by CCA1, indicating that the circadian clock gates the sensitivity of ROS signaling through CCA1 function in Arabidopsis.
Figure 4.
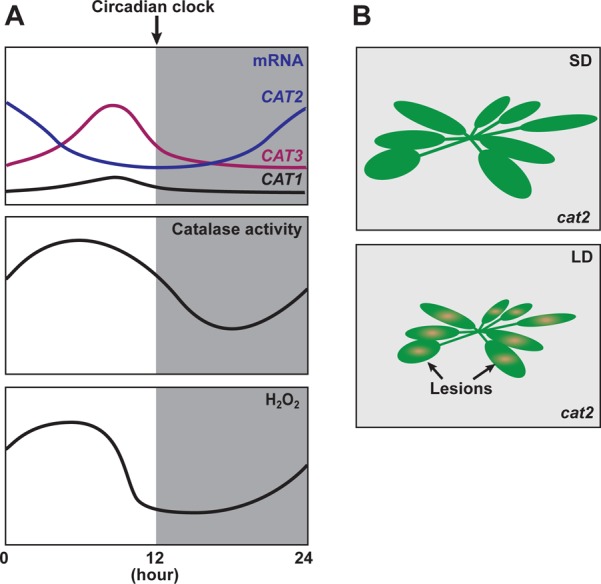
Circadian regulation of ROS homeostasis. (A) Circadian regulation of ROS homeostasis. The circadian clock regulates the daily expression patterns of three catalase genes (CAT1–CAT3) and their activities to maintain cellular hydrogen peroxide (H2O2) levels. (B) Day length-dependent phenotype of the cat2 mutant. The cat2 mutant shows the lesion (cell death) phenotype when it is grown only under LD conditions.
Circadian clock-dependent apoplastic ROS homeostasis also regulates leaf growth.128,129 Leaves show diurnal growth patterns with maximal expansion in the morning.130 One circadian clock-regulated mechanism of leaf growth is associated with the function of the MYB-like transcriptional regulator KUODA1 (KUA1).128,129 The diurnal expression pattern of the KUA1 gene shows a morning peak. The morning expression of KUA1 is directly controlled by CCA1 and LHY,128,129 both of which affect the leaf growth rate.130 Leaf size is reduced in the kua1 mutant, and this is due to the reduction of the cell expansion rate. The KUA1 protein directly binds to the promoters of seven apoplastic peroxidase genes to repress their expression. Consequently, the level of H2O2 in the kua1 mutant is higher than that in the wild-type plant. In addition, the small leaf size phenotype of the kua1 mutant is rescued by exogenous application of either the peroxidase inhibitor or the hydrogen peroxide scavenger, indicating that the basal cellular H2O2 levels that are controlled by the circadian clock influence the cell expansion rate during leaf development. In the roots, the regulation of apoplastic ROS homeostasis controlled by the UPBEAT1 (UPB1) bHLH transcription factor is the most important mechanism for modulating the balance between cell proliferation and differentiation.131 Although it is still underexplored, the findings introduced here imply that the circadian clock regulates various plant developmental processes in part by modulating H2O2 levels. As H2O2 mediates plant development, defense responses, and abiotic stress responses,129,132 the circadian regulation of H2O2 production described above could contribute to the appropriate time-of-day production of ROS signals in these biological responses.
Photoperiodic Regulation of Hydrogen Peroxide Levels
Although oscillation of hydrogen peroxide levels and catalase activity are regulated by the circadian clock,126 differences in photoperiod also affect these levels. In the presence of light, the inactivation of antioxidant enzyme (catalase) activity enhances hydrogen peroxide accumulation and increases the level of expression of ROS-regulated genes in green algae.133 In rye leaves, light accelerates the inactivation of catalase.134 Catalase is degraded under light, and the degradation is clearly observed from 16 h after the onset of light.134 These examples suggest that the duration of light within a day can be an important factor that affects the activity of catalase. In addition, prior to the exposure of ROS-inductive conditions, growing plants under different day length conditions (LD vs SD) changes how plants deal with new ROS-inductive conditions by changing both leaf catalase activity and antioxidant content.135 In plants that were previously grown in LD but not in SD, the activity of catalase is increased to cope with the increase in the level of ROS. This indicates that, between LD and SD conditions, different mechanisms are utilized to regulate daily ROS homeostasis. As expected, the catalase activity in LD is higher than that in SD.
Among the three Arabidopsis catalases, CAT2 is the major one that deals with LD-dependent oxidative stresses.136 The leaves of cat2 mutants show lesions (H2O2-induced cell death) when they are grown in LD, but the phenotype is not visible in SD (Figure 4B), indicating that the H2O2 level in cat2 increases in LD. In addition, this phenotype is also regulated by light intensity, as higher light levels during the first half of the day enhance the phenotype. Similar to the day length-specific lesion formation observed in the cat2 mutant, pathogenesis-related genes such as PR1 and camalexin, both of which are induced by increasing H2O2 levels, are induced only in LD.137 Another lesion formation mutant, lesion simulating disease 1 (lsd1), also shows a similar photoperiod-dependent phenotype. LSD1 encodes a novel zinc finger protein. Under LD conditions, low light intensity is enough to develop lesions in lsd1 mutant leaves.138 In contrast, under SD conditions, lesion formation is not observed even under high-light conditions. Interestingly, LSD1 interacts with catalases through its zinc finger domain. In the lsd1 mutant, catalase activity is lower than that in wild-type plants,139 indicating that LSD1 enhances the activity of catalases. These observations suggest that photoperiod information can affect cellular ROS signal generation in part through the day length-specific regulation of catalase activity.
Conclusion
In this review, we discuss recent advances in our understanding of the molecular mechanisms of the circadian clock and photoperiodic response in Arabidopsis. Within the last several years, identification of positive regulators (RVE8 and LNK1/2) for EE-regulated genes, as well as comprehensive genome-scale analyses of the DNA binding sites of PRRs and TOC1 proteins, has drastically changed our view of the molecular architecture of the circadian clock. Additionally, characterizing new posttranscriptional regulatory mechanisms, such as alternative splicing of clock gene transcripts, has allowed us to understand that the circadian clock is composed of many layers of regulation. Although our knowledge regarding the molecular clock is improving rapidly, unidentified mechanisms and regulators within the circadian clock still exist. For instance, we do not know the identities of the positive regulator(s) that induce the expression of morning components such as CCA1, LHY, and RVE8. In addition, PRR7, PRR5, and TOC1 repress the gene expression of many clock components simultaneously around the end of the day and the beginning of the night. We are not yet sure if they act additively, repressing the same targets, or if any synergistic functional interactions exist (in addition to the PRR5-dependent TOC1 activity regulation described here). It is of great interest to us to perform more quantitative analyses of spatial and temporal expression patterns of the clock components and the interactions among them. Currently, both phosphorylation of CCA1 protein and alternative splicing of CCA1 transcripts are implicated in the temperature compensation mechanism of the circadian clock.43,49 Are these mechanisms equally important for temperature compensation? A more comprehensive understanding of the timing of each modification would be helpful to help us develop an entire view of how the clock times its 24 h rhythms regardless of ambient temperature changes. Moreover, insights into molecular clock structures and mechanisms in plants other than Arabidopsis are still lacking. Studying the circadian clock in other plants at molecular levels will facilitate the determination of the similarities and differences in clocks among plant species. It will also help us to understand how plants regulate the timing of various species-specific responses and/or development, which do not exist in Arabidopsis.
Our current understanding of the molecular mechanisms of photoperiodic flowering and hypocotyl growth regulation has given us seminal examples of how environmental (photoperiodic) information is integrated into internal regulatory networks (which are regulated by the circadian clock). These coordinate the timing of crucial developmental processes that occur over longer time scales. In the regulatory pathway of photoperiodic flowering, the presence of circadian clock-controlled components and light at the same time are critical to induce the expression of CO and FT. Recent findings in photoperiod-dependent activation and stabilization of CO protein and in the alteration of the chromatin structure of the FT promoter suggest that multilayers of regulatory mechanisms concomitantly control the timing of flowering to make it occur under the most favorable conditions. One of the next challenges will be investigating the combinational effects of various environmental stimuli, such as light, temperature, nutrient availability, humidity, and pathogen attacks.
In addition to flowering and growth rate regulation, there is a growing body of evidence that metabolic pathways, such as carbon allocation, starch metabolism, and cellular ROS homeostasis, could also be under the control of photoperiod differences.137,140,141 Currently, we have little understanding of the molecular mechanisms underlying these responses. Unveiling the mechanisms of the photoperiodic regulation of metabolic pathways will provide new insights into the physiological strategies of plants as they adapt to seasonal environmental changes. Some of the known components in photoperiodic responses may also function in the metabolic pathways. For instance, LD-specific activation of CO protein contributes to photoperiodic starch accumulation through direct induction of GRANULE BOUND STARCH SYNTHASE gene expression.140 In addition, differences in photoperiod affect the generation of ROS signals that regulate several biological processes under control of the circadian clock. Therefore, we are likely to be able to apply our current knowledge of the circadian clock and photoperiodic flowering to further understand how other biological processes could be regulated under different photoperiods.
Adapting to seasonal environmental change must have been a challenge for land plants. Many plants, including major crops, possess similar molecular mechanisms that allow them to determine the timing of flowering using photoperiod as an indicator of seasonal change.1,2,142 In addition, similar mechanisms are utilized to control seasonal responses in tree species. In poplar, for example, FT homologues regulate not only the timing of flowering but also that of growth cessation.143
We do not know when the photoperiodic time measurement mechanism was generated during the history of land plant evolution. Interestingly, a recent report demonstrated that FKF1 and GI are conserved in not only angiosperm species but also nonvascular bryophyte species.144 The FKF1–GI complex controls the photoperiodic growth phase transition in the liverwort Marchantia polymorpha.144 This suggests that this photoperiod-sensing module developed in the early lineage of land plants. Together with comparative genomic analyses, this type of analysis will help us speculate how plants have obtained the ability to use photoperiodic differences as seasonal cues for adjusting their developmental programs to fit their surrounding environments.
In summary, we are in the middle of a very exciting time in which we are able to understand the complex but sophisticated molecular mechanisms by which plants adapt to dynamic changes that occur daily and seasonally, using their time-keeping mechanisms, the circadian clocks.
Acknowledgments
We thank Greg Golembeski and Hannah Kinmonth-Schultz for critical reading of the manuscript.
This work was supported by National Institutes of Health Grant GM079712 to T.I.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Song Y. H.; Ito S.; Imaizumi T. (2010) Similarities in the circadian clock and photoperiodism in plants. Curr. Opin. Plant Biol. 13, 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres F.; Coupland G. (2012) The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 13, 627–639. [DOI] [PubMed] [Google Scholar]
- Kinmonth-Schultz H. A.; Golembeski G. S.; Imaizumi T. (2013) Circadian clock-regulated physiological outputs: Dynamic responses in nature. Semin. Cell Dev. Biol. 24, 407–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel D. H.; Kay S. A. (2012) Complexity in the wiring and regulation of plant circadian networks. Curr. Biol. 22, R648–R657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R.; Mas P. (2013) Chromatin remodeling and alternative splicing: Pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24, 399–406. [DOI] [PubMed] [Google Scholar]
- Penfield S.; Hall A. (2009) A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 21, 1722–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer R.; Ramsay N.; Samach A.; Corden S.; Putterill J.; Carre I. A.; Coupland G. (1998) The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93, 1219–1229. [DOI] [PubMed] [Google Scholar]
- Wang Z. Y.; Tobin E. M. (1998) Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Strayer C.; Oyama T.; Schultz T. F.; Raman R.; Somers D. E.; Mas P.; Panda S.; Kreps J. A.; Kay S. A. (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. [DOI] [PubMed] [Google Scholar]
- Alabadi D.; Oyama T.; Yanovsky M. J.; Harmon F. G.; Mas P.; Kay S. A. (2001) Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. [DOI] [PubMed] [Google Scholar]
- Xie Q.; Wang P.; Liu X.; Yuan L.; Wang L.; Zhang C.; Li Y.; Xing H.; Zhi L.; Yue Z.; Zhao C.; McClung C. R.; Xu X. (2014) LNK1 and LNK2 are transcriptional coactivators in the Arabidopsis circadian oscillator. Plant Cell 26, 2843–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone M. L.; Faigon Soverna A.; Sanchez S. E.; Schlaen R. G.; Hernando C. E.; Seymour D. K.; Mancini E.; Chernomoretz A.; Weigel D.; Mas P.; Yanovsky M. J. (2013) LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. U.S.A. 110, 12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. Y.; Devisetty U. K.; Harmer S. L. (2013) Accurate timekeeping is controlled by a cycling activator in Arabidopsis. Elife 2, e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wu J. F.; Nakamichi N.; Sakakibara H.; Nam H. G.; Wu S. H. (2011) LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré E. M.; Harmer S. L.; Harmon F. G.; Yanovsky M. J.; Kay S. A. (2005) Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15, 47–54. [DOI] [PubMed] [Google Scholar]
- Nakamichi N.; Kita M.; Niinuma K.; Ito S.; Yamashino T.; Mizoguchi T.; Mizuno T. (2007) Arabidopsis clock-associated pseudo-response regulators PRR9, PRR7 and PRR5 coordinately and positively regulate flowering time through the canonical CONSTANS-dependent photoperiodic pathway. Plant Cell Physiol. 48, 822–832. [DOI] [PubMed] [Google Scholar]
- Doyle M. R.; Davis S. J.; Bastow R. M.; McWatters H. G.; Kozma-Bognar L.; Nagy F.; Millar A. J.; Amasino R. M. (2002) The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419, 74–77. [DOI] [PubMed] [Google Scholar]
- Hazen S. P.; Schultz T. F.; Pruneda-Paz J. L.; Borevitz J. O.; Ecker J. R.; Kay S. A. (2005) LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl. Acad. Sci. U.S.A. 102, 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T.; Wright L.; Fujiwara S.; Cremer F.; Lee K.; Onouchi H.; Mouradov A.; Fowler S.; Kamada H.; Putterill J.; Coupland G. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O. S.; Huang X.; Charron J. B.; Lee J. H.; Li G.; Deng X. W. (2011) Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43, 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N.; Kiba T.; Henriques R.; Mizuno T.; Chua N.-H.; Sakakibara H. (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Kim J.; Somers D. E. (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U.S.A. 110, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron J. M.; Pruneda-Paz J. L.; Doherty C. J.; Gross A. M.; Kang S. E.; Kay S. A. (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J. L.; Breton G.; Para A.; Kay S. A. (2009) A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323, 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.; Perez-Garcia P.; Pokhilko A.; Millar A. J.; Antoshechkin I.; Riechmann J. L.; Mas P. (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. [DOI] [PubMed] [Google Scholar]
- Herrero E.; Kolmos E.; Bujdoso N.; Yuan Y.; Wang M.; Berns M. C.; Uhlworm H.; Coupland G.; Saini R.; Jaskolski M.; Webb A.; Goncalves J.; Davis S. J. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24, 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer A.; Nusinow D. A.; Chow B. Y.; Gehrke A. R.; Bulyk M. L.; Kay S. A. (2011) LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21, 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D. A.; Helfer A.; Hamilton E. E.; King J. J.; Imaizumi T.; Schultz T. F.; Farre E. M.; Kay S. A. (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475, 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow B. Y.; Helfer A.; Nusinow D. A.; Kay S. A. (2012) ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signaling Behav. 7, 170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G.; Siddiqui H.; Teng Y.; Lin R.; Wan X. Y.; Li J.; Lau O. S.; Ouyang X.; Dai M.; Wan J.; Devlin P. F.; Deng X. W.; Wang H. (2011) Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13, 616–622. [DOI] [PubMed] [Google Scholar]
- Wu J. F.; Wang Y.; Wu S. H. (2008) Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 148, 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R.; Takahashi N.; Hsu P. Y.; Jones M. A.; Schwartz J.; Salemi M. R.; Phinney B. S.; Harmer S. L. (2011) REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7, e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S. L.; Kay S. A. (2005) Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17, 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N.; Kiba T.; Kamioka M.; Suzuki T.; Yamashino T.; Higashiyama T.; Sakakibara H.; Mizuno T. (2012) Transcriptional repressor PRR5 directly regulates clock-output pathways. Proc. Natl. Acad. Sci. U.S.A. 109, 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z.; Xu Q.; Wang X. (2014) Regulation of the circadian clock through pre-mRNA splicing in Arabidopsis. J. Exp. Bot. 65, 1973–1980. [DOI] [PubMed] [Google Scholar]
- Wahl M. C.; Will C. L.; Luhrmann R. (2009) The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718. [DOI] [PubMed] [Google Scholar]
- Petrillo E.; Sanchez S. E.; Kornblihtt A. R.; Yanovsky M. J. (2011) Alternative splicing adds a new loop to the circadian clock. Commun. Integr. Biol. 4, 284–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez S. E.; Petrillo E.; Beckwith E. J.; Zhang X.; Rugnone M. L.; Hernando C. E.; Cuevas J. C.; Godoy Herz M. A.; Depetris-Chauvin A.; Simpson C. G.; Brown J. W.; Cerdan P. D.; Borevitz J. O.; Mas P.; Ceriani M. F.; Kornblihtt A. R.; Yanovsky M. J. (2010) A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116. [DOI] [PubMed] [Google Scholar]
- Deng X.; Gu L.; Liu C.; Lu T.; Lu F.; Lu Z.; Cui P.; Pei Y.; Wang B.; Hu S.; Cao X. (2010) Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. U.S.A. 107, 19114–19119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.; Song H. R.; Lutz K.; Kerstetter R. A.; Michael T. P.; McClung C. R. (2010) Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 21211–21216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Wu F.; Xie Q.; Wang H.; Wang Y.; Yue Y.; Gahura O.; Ma S.; Liu L.; Cao Y.; Jiao Y.; Puta F.; McClung C. R.; Xu X.; Ma L. (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24, 3278–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. A.; Williams B. A.; McNicol J.; Simpson C. G.; Brown J. W.; Harmer S. L. (2012) Mutation of Arabidopsis spliceosomal timekeeper locus1 causes circadian clock defects. Plant Cell 24, 4066–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J.; Park M. J.; Lim M. H.; Kim S. G.; Lee M.; Baldwin I. T.; Park C. M. (2012) A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24, 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James A. B.; Syed N. H.; Bordage S.; Marshall J.; Nimmo G. A.; Jenkins G. I.; Herzyk P.; Brown J. W. S.; Nimmo H. G. (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24, 961–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J.; Mas P. (2014) Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y.; Geng R.; Somers D. E. (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome. Proc. Natl. Acad. Sci. U.S.A. 100, 4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen G.; Dixon L. E.; Troein C.; Millar A. J. (2011) Proteasome function is required for biological timing throughout the twenty-four hour cycle. Curr. Biol. 21, 869–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.; Andronis C.; Green R. M.; Wang Z. Y.; Tobin E. M. (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. U.S.A. 95, 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portoles S.; Mas P. (2010) The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6, e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M.; Portoles S.; Mas P. (2006) The proteasome-dependent degradation of CKB4 is regulated by the Arabidopsis biological clock. Plant J. 46, 849–860. [DOI] [PubMed] [Google Scholar]
- O’Neill J. S.; van Ooijen G.; Dixon L. E.; Troein C.; Corellou F.; Bouget F. Y.; Reddy A. B.; Millar A. J. (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P.; Kim W. Y.; Somers D. E.; Kay S. A. (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426, 567–570. [DOI] [PubMed] [Google Scholar]
- Ito S.; Song Y. H.; Imaizumi T. (2012) LOV domain-containing F-box proteins: Light-dependent protein degradation modules in Arabidopsis. Mol. Plant 5, 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiba T.; Henriques R.; Sakakibara H.; Chua N. H. (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by an SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19, 2516–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. Y.; Fujiwara S.; Suh S. S.; Kim J.; Kim Y.; Han L.; David K.; Putterill J.; Nam H. G.; Somers D. E. (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449, 356–360. [DOI] [PubMed] [Google Scholar]
- Kim J.; Geng R.; Gallenstein R. A.; Somers D. E. (2013) The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140, 4060–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A.; Ito S.; Song Y. H.; Strait A. A.; Kiba T.; Lu S.; Henriques R.; Pruneda-Paz J. L.; Chua N. H.; Tobin E. M.; Kay S. A.; Imaizumi T. (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22, 606–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S.; Wang L.; Han L.; Suh S. S.; Salome P. A.; McClung C. R.; Somers D. E. (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283, 23073–23083. [DOI] [PubMed] [Google Scholar]
- Para A.; Farre E. M.; Imaizumi T.; Pruneda-Paz J. L.; Harmon F. G.; Kay S. A. (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19, 3462–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Fujiwara S.; Somers D. E. (2010) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29, 1903–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J. W.; Rubio V.; Lee N. Y.; Bai S.; Lee S. Y.; Kim S. S.; Liu L.; Zhang Y.; Irigoyen M. L.; Sullivan J. A.; Zhang Y.; Lee I.; Xie Q.; Paek N. C.; Deng X. W. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32, 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. S.; Eo H. J.; Jang I.-C.; Kang H.-G.; Song J. T.; Seo H. S. (2010) Ubiquitination of LHY by SINAT5 regulates flowering time and is inhibited by DET1. Biochem. Biophys. Res. Commun. 398, 242–246. [DOI] [PubMed] [Google Scholar]
- Cui X.; Lu F.; Li Y.; Xue Y.; Kang Y.; Zhang S.; Qiu Q.; Cui X.; Zheng S.; Liu B.; Xu X.; Cao X. (2013) Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162, 897–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P. A. (2011) FT, a mobile developmental signal in plants. Curr. Biol. 21, R374–R378. [DOI] [PubMed] [Google Scholar]
- Song Y. H.; Ito S.; Imaizumi T. (2013) Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.; Zhu Y.; Shen L.; Yu H. (2013) Emerging insights into florigen transport. Curr. Opin. Plant Biol. 16, 607–613. [DOI] [PubMed] [Google Scholar]
- Corbesier L.; Vincent C.; Jang S.; Fornara F.; Fan Q.; Searle I.; Giakountis A.; Farrona S.; Gissot L.; Turnbull C.; Coupland G. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Pin P. A.; Nilsson O. (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant, Cell Environ. 35, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Suarez-Lopez P.; Wheatley K.; Robson F.; Onouchi H.; Valverde F.; Coupland G. (2001) CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120. [DOI] [PubMed] [Google Scholar]
- Song Y. H.; Ito S.; Imaizumi T. (2013) Flowering time regulation: Photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18, 575–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giakountis A.; Coupland G. (2008) Phloem transport of flowering signals. Curr. Opin. Plant Biol. 11, 687–694. [DOI] [PubMed] [Google Scholar]
- Pin P. A.; Nilsson O. (2012) The multifaceted roles of FLOWERING LOCUS T in plant development. Plant, Cell Environ. 35, 1742–1755. [DOI] [PubMed] [Google Scholar]
- Imaizumi T.; Tran H. G.; Swartz T. E.; Briggs W. R.; Kay S. A. (2003) FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature 426, 302–306. [DOI] [PubMed] [Google Scholar]
- Imaizumi T.; Schultz T. F.; Harmon F. G.; Ho L. A.; Kay S. A. (2005) FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309, 293–297. [DOI] [PubMed] [Google Scholar]
- Sawa M.; Nusinow D. A.; Kay S. A.; Imaizumi T. (2007) FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318, 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y. H.; Smith R. W.; To B. J.; Millar A. J.; Imaizumi T. (2012) FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science 336, 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F.; Panigrahi K. C. S.; Gissot L.; Sauerbrunn N.; Rühl M.; Jarillo J. A.; Coupland G. (2009) Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev. Cell 17, 75–86. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T.; Wright L.; Fujiwara S.; Cremer F.; Lee K.; Onouchi H.; Mouradov A.; Fowler S.; Kamada H.; Putterill J.; Coupland G. (2005) Distinct roles of GIGANTEA in promoting flowering and regulating circadian rhythms in Arabidopsis. Plant Cell 17, 2255–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S.; Song Y. H.; Josephson-Day A. R.; Miller R. J.; Breton G.; Olmstead R. G.; Imaizumi T. (2012) FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 3582–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.; Ni M. (2006) RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J. 46, 823–833. [DOI] [PubMed] [Google Scholar]
- Yoo S. Y.; Kim Y.; Kim S. Y.; Lee J. S.; Ahn J. H. (2007) Control of flowering time and cold response by a NAC-domain protein in Arabidopsis. PLoS One 2, e642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.; Kim Y.; Yeom M.; Kim J.-H.; Nam H. G. (2008) FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. Plant Cell 20, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris K.; Thornber S.; Codrai L.; Richardson C.; Craig A.; Sadanandom A.; Thomas B.; Jackson S. (2010) DAY NEUTRAL FLOWERING represses CONSTANS to prevent Arabidopsis flowering early in short days. Plant Cell 22, 1118–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach Y.; Hennig L. (2014) Arabidopsis MSI1 functions in photoperiodic flowering time control. Front. Plant Sci. 5, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara M.; Kamada H.; Mizoguchi T. (2014) CO-EXPRESSED WITH CLOCK GENES LHY AND CCA1 1 (CEC1) is regulated by LHY and CCA1 and plays a key role in phase setting of GI in Arabidopsis thaliana. Plant Biotechnol. 31, 35–41. [Google Scholar]
- Laubinger S.; Marchal V.; Le Gourrierec J.; Wenkel S.; Adrian J.; Jang S.; Kulajta C.; Braun H.; Coupland G.; Hoecker U. (2006) Arabidopsis SPA proteins regulate photoperiodic flowering and interact with the floral inducer CONSTANS to regulate its stability. Development 133, 3213–3222. [DOI] [PubMed] [Google Scholar]
- Jang S.; Marchal V.; Panigrahi K. C.; Wenkel S.; Soppe W.; Deng X. W.; Valverde F.; Coupland G. (2008) Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27, 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.-J.; Zhang Y.-C.; Li Q.-H.; Sang Y.; Mao J.; Lian H.-L.; Wang L.; Yang H.-Q. (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20, 292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A.; Valverde F.; Pineiro M.; Jarillo J. A. (2012) The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell 24, 982–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F.; Mouradov A.; Soppe W.; Ravenscroft D.; Samach A.; Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303, 1003–1006. [DOI] [PubMed] [Google Scholar]
- Endo M.; Tanigawa Y.; Murakami T.; Araki T.; Nagatani A. (2013) PHYTOCHROME-DEPENDENT LATE-FLOWERING accelerates flowering through physical interactions with phytochrome B and CONSTANS. Proc. Natl. Acad. Sci. U.S.A. 110, 18017–18022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z.; Liu H.; Liu B.; Liu X.; Lin C. (2011) Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21, 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H. L.; He S. B.; Zhang Y. C.; Zhu D. M.; Zhang J. Y.; Jia K. P.; Sun S. X.; Li L.; Yang H. Q. (2011) Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25, 1023–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Zuo Z.; Liu H.; Liu X.; Lin C. (2011) Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D.; Liu C.; Shen L.; Wu Y.; Chen H.; Robertson M.; Helliwell C. A.; Ito T.; Meyerowitz E.; Yu H. (2008) A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15, 110–120. [DOI] [PubMed] [Google Scholar]
- Gu X.; Le C.; Wang Y.; Li Z.; Jiang D.; Wang Y.; He Y. (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat. Commun. 4, 1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Gu X.; Yuan W.; Schmitz R. J.; He Y. (2014) Photoperiodic control of the floral transition through a distinct polycomb repressive complex. Dev. Cell 28, 727–736. [DOI] [PubMed] [Google Scholar]
- Castillejo C.; Pelaz S. (2008) The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr. Biol. 18, 1338–1343. [DOI] [PubMed] [Google Scholar]
- Mathieu J.; Yant L. J.; Murdter F.; Kuttner F.; Schmid M. (2009) Repression of flowering by the miR172 target SMZ. PLoS Biol. 7, e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanudakis E.; Jackson S. (2014) The role of microRNAs in the control of flowering time. J. Exp. Bot. 65, 365–380. [DOI] [PubMed] [Google Scholar]
- Wu G.; Park M. Y.; Conway S. R.; Wang J. W.; Weigel D.; Poethig R. S. (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. H.; Seo Y. H.; Seo P. J.; Reyes J. L.; Yun J.; Chua N. H.; Park C. M. (2007) The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Wang Q.; Liu Y.; Zhao X.; Imaizumi T.; Somers D. E.; Tobin E. M.; Lin C. (2013) Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 110, 17582–17587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Yu X.; Li K.; Klejnot J.; Yang H.; Lisiero D.; Lin C. (2008) Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322, 1535–1539. [DOI] [PubMed] [Google Scholar]
- Putterill J.; Robson F.; Lee K.; Simon R.; Coupland G. (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80, 847–857. [DOI] [PubMed] [Google Scholar]
- Tiwari S. B.; Shen Y.; Chang H. C.; Hou Y.; Harris A.; Ma S. F.; McPartland M.; Hymus G. J.; Adam L.; Marion C.; Belachew A.; Repetti P. P.; Reuber T. L.; Ratcliffe O. J. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 187, 57–66. [DOI] [PubMed] [Google Scholar]
- Wenkel S.; Turck F.; Singer K.; Gissot L.; Le Gourrierec J.; Samach A.; Coupland G. (2006) CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18, 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S.; Kumimoto R. W.; Gnesutta N.; Calogero A. M.; Mantovani R.; Holt B. F. III (2014) A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26, 1009–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y.; Yamashino T.; Mizuno T. (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 50, 838–854. [DOI] [PubMed] [Google Scholar]
- Nozue K.; Covington M. F.; Duek P. D.; Lorrain S.; Fankhauser C.; Harmer S. L.; Maloof J. N. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361. [DOI] [PubMed] [Google Scholar]
- Nomoto Y.; Kubozono S.; Yamashino T.; Nakamichi N.; Mizuno T. (2012) Circadian clock- and PIF4-controlled plant growth: A coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana. Plant Cell Physiol. 53, 1950–1964. [DOI] [PubMed] [Google Scholar]
- de Lucas M.; Prat S. (2014) PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 202, 1126–1141. [DOI] [PubMed] [Google Scholar]
- Kunihiro A.; Yamashino T.; Nakamichi N.; Niwa Y.; Nakanishi H.; Mizuno T. (2011) Phytochrome-interacting factor 4 and 5 (PIF4 and PIF5) activate the homeobox ATHB2 and auxin-inducible IAA29 genes in the coincidence mechanism underlying photoperiodic control of plant growth of Arabidopsis thaliana. Plant Cell Physiol. 52, 1315–1329. [DOI] [PubMed] [Google Scholar]
- Lorrain S.; Allen T.; Duek P. D.; Whitelam G. C.; Fankhauser C. (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53, 312–323. [DOI] [PubMed] [Google Scholar]
- Yamashino T.; Nomoto Y.; Lorrain S.; Miyachi M.; Ito S.; Nakamichi N.; Fankhauser C.; Mizuno T. (2013) Verification at the protein level of the PIF4-mediated external coincidence model for the temperature-adaptive photoperiodic control of plant growth in Arabidopsis thaliana. Plant Signaling Behav. 8, e23390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M.; Daviere J. M.; Rodriguez-Falcon M.; Pontin M.; Iglesias-Pedraz J. M.; Lorrain S.; Fankhauser C.; Blazquez M. A.; Titarenko E.; Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484. [DOI] [PubMed] [Google Scholar]
- Arana M. V.; Marín-de la Rosa N.; Maloof J. N.; Blázquez M. A.; Alabadí D. (2011) Circadian oscillation of gibberellin signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 108, 9292–9297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper C.; Dolan L. (2006) Control of plant development by reactive oxygen species. Plant Physiol. 141, 341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A.; Queval G.; Chaouch S.; Vanderauwera S.; Van Breusegem F.; Noctor G. (2010) Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 61, 4197–4220. [DOI] [PubMed] [Google Scholar]
- Baxter A.; Mittler R.; Suzuki N. (2014) ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Pekovic-Vaughan V.; Gibbs J.; Yoshitane H.; Yang N.; Pathiranage D.; Guo B.; Sagami A.; Taguchi K.; Bechtold D.; Loudon A.; Yamamoto M.; Chan J.; van der Horst G. T. J.; Fukada Y.; Meng Q.-J. (2014) The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev. 28, 548–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. S.; Reddy A. B. (2011) Circadian clocks in human red blood cells. Nature 469, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. S.; Green E. W.; Zhao Y.; van Ooijen G.; Olmedo M.; Qin X.; Xu Y.; Pan M.; Valekunja U. K.; Feeney K. A.; Maywood E. S.; Hastings M. H.; Baliga N. S.; Merrow M.; Millar A. J.; Johnson C. H.; Kyriacou C. P.; O’Neill J. S.; Reddy A. B. (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. A.; Yu Y. V.; Govindaiah G.; Ye X.; Artinian L.; Coleman T. P.; Sweedler J. V.; Cox C. L.; Gillette M. U. (2012) Circadian rhythm of redox state regulates excitability in suprachiasmatic nucleus neurons. Science 337, 839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J.; Cho S.; Sassone-Corsi P. (2007) Circadian control by the reduction/oxidation pathway: Catalase represses light-dependent clock gene expression in the zebrafish. Proc. Natl. Acad. Sci. U.S.A. 104, 15747–15752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai A. G.; Doherty C. J.; Mueller-Roeber B.; Kay S. A.; Schippers J. H.; Dijkwel P. P. (2012) CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. U.S.A. 109, 17129–17134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frugoli J. A.; Zhong H. H.; Nuccio M. L.; McCourt P.; McPeek M. A.; Thomas T. L.; McClung C. R. (1996) Catalase Is Encoded by a Multigene Family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 112, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O.; Michael T. P.; Burger B. T.; Le Metté C.; Mockler T. C.; Weigel D.; Chory J. (2008) A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105, 17193–17198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D.; Wang T.; Persson S.; Mueller-Roeber B.; Schippers J. H. (2014) Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat. Commun. 5, 3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruts T.; Matsubara S.; Wiese-Klinkenberg A.; Walter A. (2012) Diel patterns of leaf and root growth: Endogenous rhythmicity or environmental response?. J. Exp. Bot. 63, 3339–3351. [DOI] [PubMed] [Google Scholar]
- Tsukagoshi H.; Busch W.; Benfey P. N. (2010) Transcriptional regulation of ROS controls transition from proliferation to differentiation in the root. Cell 143, 606–616. [DOI] [PubMed] [Google Scholar]
- Spoel S. H.; van Ooijen G. (2014) Circadian redox signaling in plant immunity and abiotic stress. Antioxid. Redox Signaling 20, 3024–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N.; Beck C. F.; Lemaire S. D.; Krieger-Liszkay A. (2008) Photosynthetic electron flow affects H2O2 signaling by inactivation of catalase in Chlamydomonas reinhardtii. Planta 228, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertwig B.; Streb P.; Feierabend J. (1992) Light dependence of catalase synthesis and degradation in leaves and the influence of interfering stress conditions. Plant Physiol. 100, 1547–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B.; Holtgrefe S.; Jung S.; Wunrau C.; Kandlbinder A.; Baier M.; Dietz K. J.; Backhausen J. E.; Scheibe R. (2006) Influence of the photoperiod on redox regulation and stress responses in Arabidopsis thaliana L. (Heynh.) plants under long- and short-day conditions. Planta 224, 380–393. [DOI] [PubMed] [Google Scholar]
- Queval G.; Issakidis-Bourguet E.; Hoeberichts F. A.; Vandorpe M.; Gakière B.; Vanacker H.; Miginiac-Maslow M.; Van Breusegem F.; Noctor G. (2007) Conditional oxidative stress responses in the Arabidopsis photorespiratory mutant cat2 demonstrate that redox state is a key modulator of daylength-dependent gene expression, and define photoperiod as a crucial factor in the regulation of H2O2-induced cell death. Plant J. 52, 640–657. [DOI] [PubMed] [Google Scholar]
- Chaouch S.; Queval G.; Vanderauwera S.; Mhamdi A.; Vandorpe M.; Langlois-Meurinne M.; Van Breusegem F.; Saindrenan P.; Noctor G. (2010) Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 153, 1692–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateo A.; Muhlenbock P.; Rusterucci C.; Chang C. C.; Miszalski Z.; Karpinska B.; Parker J. E.; Mullineaux P. M.; Karpinski S. (2004) LESION SIMULATING DISEASE 1 is required for acclimation to conditions that promote excess excitation energy. Plant Physiol. 136, 2818–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Chen L.; Mu J.; Zuo J. (2013) LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 163, 1059–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Marchena M. I.; Albi T.; Lucas-Reina E.; Said F. E.; Romero-Campero F. J.; Cano B.; Ruiz M. T.; Romero J. M.; Valverde F. (2014) Photoperiodic control of carbon distribution during the floral transition in Arabidopsis. Plant Cell 26, 565–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; Gehan J. P.; Sharkey T. D. (2005) Daylength and circadian effects on starch degradation and maltose metabolism. Plant Physiol. 138, 2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha R.; Gómez-Ariza J.; Brambilla V.; Fornara F. (2014) Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 114, 1445–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlenius H.; Huang T.; Charbonnel-Campaa L.; Brunner A. M.; Jansson S.; Strauss S. H.; Nilsson O. (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. [DOI] [PubMed] [Google Scholar]
- Kubota A.; Kita S.; Ishizaki K.; Nishihama R.; Yamato K. T.; Kohchi T. (2014) Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 5, 3668. [DOI] [PubMed] [Google Scholar]


