Abstract
Purpose
The genetic differences between Human papilloma Virus (HPV)-positive and negative head and neck squamous cell carcinomas (HNSCC) remain largely unknown. In order to identify differential biology and novel therapeutic targets for both entities we determined mutations and copy number aberrations in a large cohort of locoregionally-advanced HNSCC.
Experimental Design
We performed massively parallel sequencing of 617 cancer-associated genes in 120 matched tumor/normal samples (42.5% HPV-positive). Mutations and copy number aberrations were determined and results validated with a secondary method.
Results
The overall mutational burden in HPV-negative and HPV-positive HNSCC was similar with an average of 15.2 versus 14.4 somatic exonic mutations in the targeted cancer-associated genes. HPV-negative tumors showed a mutational spectrum concordant with published lung squamous cell carcinoma analyses with enrichment for mutations in TP53, CDKN2A, MLL2, CUL3, NSD1, PIK3CA and NOTCH genes. HPV-positive tumors showed unique mutations in DDX3X, FGFR2/3 and aberrations in PIK3CA, KRAS, MLL2/3 and NOTCH1 were enriched in HPV-positive tumors. Currently targetable genomic alterations were identified in FGFR1, DDR2, EGFR, FGFR2/3, EPHA2 and PIK3CA. EGFR, CCND1, and FGFR1 amplifications occurred in HPV-negative tumors, while 17.6% of HPV-positive tumors harbored mutations in Fibroblast Growth Factor Receptor genes (FGFR2/3) including six recurrent FGFR3 S249C mutations. HPV-positive tumors showed a 5.8% incidence of KRAS mutations, and DNA repair gene aberrations including 7.8% BRCA1/2 mutations were identified.
Conclusions
The mutational makeup of HPV-positive and HPV-negative HNSCC differs significantly, including targetable genes. HNSCC harbors multiple therapeutically important genetic aberrations, including frequent aberrations in the FGFR and PI3K pathway genes.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the fifth most common non-skin cancer worldwide with an annual incidence of 600,000 cases and a mortality rate of 40–50% despite aggressive treatment (1,2). The major known risk factors are environmental exposure to tobacco products, alcohol, and infection with high-risk Human Papilloma Viruses (HPV). The incidence of HPV-positive tumors is rising rapidly in Western countries and HPV-status is the strongest clinically-applicable prognostic marker, portending a favorable prognosis(3, 4).
While HNSCC is widely viewed as comprised of two distinct clinical entities, HPV-positive and HPV-negative tumors, a comprehensive list of differential molecular abnormalities, in particular therapeutically-relevant genetic aberrations has not been reported. A particular problem is the lack of study of HPV-positive HNSCC: Currently no large series of HPV-positive tumors exist and the upcoming cancer genome atlas (TCGA) cohort is compromised of 85% HPV-negative tumors(5). This bias is likely related to selection of surgically resected, earlier stage oral cavity and laryngeal tumors. This may not be representative for clinically more complex Stage IV tumors requiring multimodality or palliative treatments(6–8).
Unlike lung or breast adenocarcinomas, there are currently no defined targetable genetic aberrations for HNSCC, and no approved therapies are tied to genetic alterations as predictive biomarkers. All HNSCC patients are treated with a largely uniform approach based on stage and anatomic location, typically using surgery, radiation, and chemotherapy alone or in combination (9). Cetuximab, an anti-EGFR antibody, is the only approved targeted therapy for HNSCC with a single agent response rate of 10–13%. Despite the modest response rate there are no validated predictive biomarkers for benefit from cetuximab (10,11).
Previous studies have demonstrated frequent mutations of several genes in cohorts of largely HPV-negative HNSCC, most notably TP53, PIK3CA, CDKN2A, the TERT promoter, and NOTCH pathway gene alterations(12–16). However, the genetic makeup of HPV-positive HNSCC remains unclear (15).
In the current study, we investigated a fully annotated patient cohort of 120 locoregionally advanced HNSCC (including 42.5% HPV-positive tumors) treated uniformly with organ-preserving chemoradiotherapy using massively parallel sequencing, copy number profiling, and validation.
We discover distinct mutational and copy number profiles in HPV-positive and HPV-negative tumors and identify for the first time potentially targetable mutations and copy number aberrations that are of high translational relevance.
Materials and Methods
Chicago Head and Neck Cancer Genomics Cohort (CHGC)
Pre-treatment tumor tissues (n=120) and matched normal DNA for patients with locoregionally advanced HNSCC treated at the University of Chicago were obtained from the HNSCC tissue bank (UCCCC#8980).
Sample Preparation
An overview of the tissue-processing is provided in Supplementary Figure S1 and described in detail in the Supplemental Methods.
HPV consensus testing
HPV16/18 status was determined by E6/E7-specific qRT-PCR. Results were corroborated by additional tests to increase accuracy including an E6/E7 DNA based multiplex PCR for five high-risk HPV types (17) as well as p16/CDKN2A expression, and TP53 mutations (18).
Sequencing data generation and analysis
DNA sequencing libraries were prepared following published protocols (19) and enriched using custom capture reagents (Agilent, Nimblegen(validation)). 2×100bp paired-end sequencing occurred using Illumina HiSeq 2000/2500 sequencers.617 cancer-associated genes (Supplementary Table S1A) were targeted and sequenced at high-depth in tumor/normal pairs. Median coverage at targeted bases was 231X in the tumors and 254X in the matched normal. Mutation and indel calling was done using bioinformatic pipelines as described previously employing MuTect (20–22). Furthermore we used VarWalker to determine potentially relevant genetic aberrations (mutations and copy number aberrations) using prioritization via a protein-protein interaction network focusing on frequently mutated and cancer gene census genes (23)
We used an established machine learning based approach – Cancer-Specific High-Throughput Annotation of Somatic Mutations (CHASM) – to predict and prioritize missense mutations leading to functional changes and thus likely driving tumorigenesis (24). This approach was previously validated to show high specificity, and to a lesser degree sensitivity to identify “drivers” of oncogenesis(24).
Copy number (CN) analysis was performed using sequencing data and the CONTRA algorithm (25). Results were validated on the Nanostring nCounter using predefined/custom cancer gene panels (Nanostring, Seattle, WA).
Detailed methods are available in the Supplementary Methods.
Results
Patient Cohort Description and Sequencing Metrics
The median age of patients was 56 years with a median follow-up of 48 months (Table 1). 51 patients (42.5%) were HPV-positive. 45% of patients had no or light smoking histories while 55% had significant smoking-histories including 35% of HPV-positive tumors (N=18). The cohort consisted of locoregionally-advanced tumors with 67 (55.8%) oropharynx and 115(95.8%) stage IV tumors (Table 1).
Table 1.
Overview of clinical and histopathologic characteristics of samples
| Total | HPV Positive | HPV Negative | |
|---|---|---|---|
| Pt. (No.) | 120 (100%) | 51 (42.5%) | 69 (57.5%) |
| Age (Year) | 56.72 | 56.69 | 56.75 |
| *Gender (No.) | |||
| Female | 24 (20.0%) | 4 (3.3%) | 20 (16.7%) |
| Male | 96 (80.0%) | 47 (39.2%) | 49 (40.8%) |
| ***Anatomic Site (No.) | |||
| OROPHARYNX | 67 (55.8%) | 47 (39.2%) | 20 (16.6%) |
| ORAL CAVITY | 23 (19.2%) | 2 (1.7%) | 21 (17.5%) |
| LARYNX | 19 (15.8%) | 1 (0.8%) | 18 (15.0%) |
| HYPOPHARYNX | 8 (6.7%) | 0 (0%) | 8 (6.7%) |
| OTHERS | 3 (2.5%) | 1 (0.8%) | 2 (1.7%) |
| **Tobacco Use (No.) | |||
| Never/Light Smoker (<10 pack years) | 54 (45.0%) | 33 (27.5%) | 21 (17.5%) |
| Heavy Smoker (≥10 pack years) | 66 (55.0%) | 18 (15.0%) | 48 (40.0%) |
| Alcohol Use (No.) | |||
| Never/Light Drinker | 49 (40.8%) | 26 (21.7%) | 23 (19.1%) |
| Heavy Drinker | 71 (59.2%) | 25 (20.8%) | 46 (38.4%) |
| *T Stage (No.) | |||
| Small primary (T1–2) | 39 (32.5%) | 23 (19.2%) | 16 (13.3%) |
| Large Primary (T3–4) | 81 (67.5%) | 28 (23.3%) | 53 (44.2%) |
| N Stage (No.) | |||
| 0–1 | 20 (16.7%)**** | 5 (4.2%) | 15 (12.5%) |
| 2–3 | 100 (83.3%) | 46 (38.3%) | 54 (45.0%) |
| Clinical Stage (No.) | |||
| I–III | 5 (4.2%) | 3 (2.5%) | 2 (1.7%) |
| IV | 115 (95.8%) | 48 (40.0%) | 67 (55.8%) |
Detection of somatic aberrations
Analysis of targeted hybrid capture sequencing data identified a total of 5476 point mutations and 4562 insertion/deletion (indel) events across 120 tumor/normal pairs in targeted 617 genes (Supplementary Table S1A). 1536 of the point mutations and 200 of the indel events were in coding regions. Of the 1536 point mutations in coding regions, 1209 resulted in an amino acid changes in the corresponding proteins. Individuals displayed a mean point mutation rate of 2.33 mutations per Megabase (Mb) in the targeted regions and 0.22/Mb in the coding territory alone with a range of 0–96 mutations in the coding regions of the targeted genes. Among non-synonymous substitutions, transitions and transversions at CpG sites, which are typically associated with tobacco exposure (26), were the most commonly observed mutational context with a rate of 21/Mb. Mutations at Tp*Cp(A/C/T) sites, a substitution type commonly associated with virally induced cancers (26), were the second most commonly observed context at 13/Mb and enriched in the HPV-positive patients. We did not detect a significant difference in overall mutation rates by HPV status at all targeted bases (HPV-negative 2.22/Mb, HPV-positive 2.46/Mb, p = 0.64) or in the coding territory (HPV-negative 0.23/Mb, HPV-positive 0.21/Mb, p=0.79). Coding mutations were validated at a rate of 87% (338/390) in a second independent hybrid capture experiment when evaluating statistically enriched mutations (Tables S1B, C).
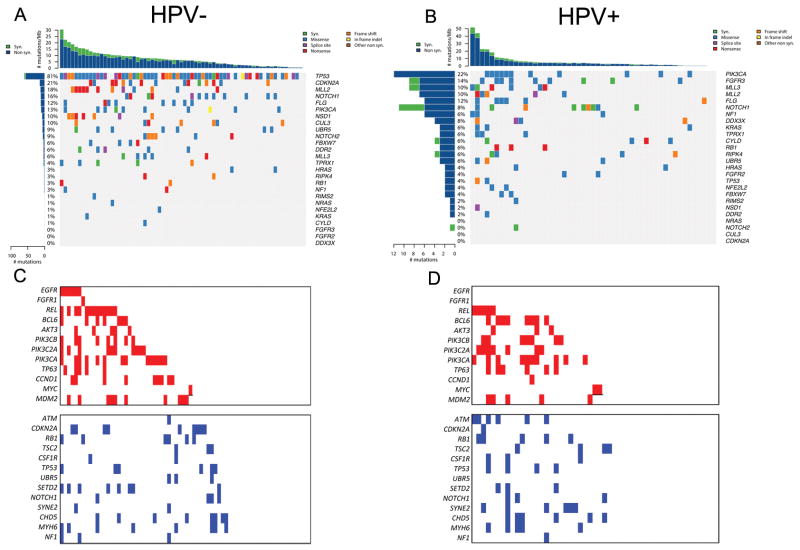
We applied the MutSig method to identify cancer-relevant genes demonstrating evidence of statistical selection for mutations in our cohort(20–22, 27). MutSig identified11 genes displaying significant enrichment for mutations as defined by a q-value of <0.1 in the overall cohort (Supplementary Figure S2): TP53, CDKN2A, PIK3CA, MLL2, TPRX1, CUL3, FLG, NSD1, DDX3X, RPIK4 and HRAS. FGFR3 and FBXW7 demonstrated q-values between 0.1 and 1. A second MutSig analysis, in which only genes annotated in the COSMIC database were considered, re-discovered TP53, CDKN2A, PIK3CA, HRAS, FGFR3 and FBXW7 and additionally nominated KRAS, MLL3, FGFR2, ZNF217 and RIMS2 as statistically significant with q<0.1. We performed additional significance analyses on the separate HPV-negative and HPV-positive cohorts using the same Mutsig algorithm. Analysis of mutated genes displaying enrichment in the HPV-negative cohorts demonstrated statistical enrichment for mutations of TP53, CDKN2A, MLL2/3, NOTCH1, PIK3CA, NSD1, FBXW7, DDR2 and CUL3, in the HPV-negative samples (Figure 1A). Genes displaying statistical enrichment in HPV-positive tumors were PIK3CA, MLL3, DDX3X, FGFR2/3, NOTCH1, NF1, KRAS, and FBXW7 (Figure 1B).
Figure 1. Mutation events sorted by HPV status.
(A) Left and right panel respectively: Left histogram: percentage of samples affected by an alteration in the corresponding gene on the right side, top histogram: number of mutations per megabase and sample, heat map:type of alteration events.(B) Copy number gains (red) and losses (blue) sorted by HPV status, samples are ordered by copy number events.
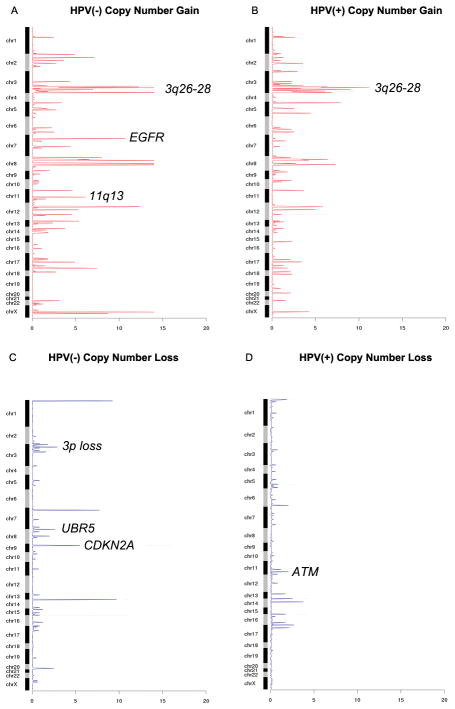
Somatic copy number alterations (SCNAs) were inferred from sequencing data by read-depth analysis using the CONTRA methodology (Supplementary Methods). We applied a modification of GISTIC 2.0 analysis (28) compatible with the focal sequencing data to identify recurrent peaks of amplification and deletion and identified 192 genes with false discovery rate of less than 0.25 (Supplementary Table S1D–G). Copy number alterations were validated at a rate of 83%(45/54) using the Nanostring nCounter, focusing on the genes found to be significantly altered in this dataset (Supplementary Table S1H). Copy number analysis demonstrated many previously demonstrated regions of amplification and deletion including focal gains of EGFR, REL, BCL6, PIK3CA, TP63, CCDN1 and MDM2 and losses of ATM, CDKN2A, RB1, NOTCH1 and NF1 (Figures 1C, 1D)(12–15, 29). Significantly amplified regions that occurred primarily in HPV-negative tumors were amplifications of 11q13 likely targeting CCND1, 7p11 (EGFR) (Figure 2A). Amplification of 3q26–28 a region containing SOX2/TP63/PIK3CA (Figures 2A, 2B) occurred in both HPV-positive and HPV-negative tumors. 3p loss and CDKN2A deletions (Figure 2C) occurred primarily in HPV-negative tumors while ATM deletions occurred primarily in HPV-positive tumors (Figure 2D).
Figure 2. GISTIC-like copy number analysis derived from next generation sequencing data.
Shown are the aberrant regions of the chromosome and the respective negative log2 of the q-value of the alteration in each panel, respectively. (A) Copy number gains in HPV(−) samples. (B) Copy number gains in HPV(+) samples. (C) Copy number losses in HPV(−) samples. (D) Copy number losses in HPV(+) samples.
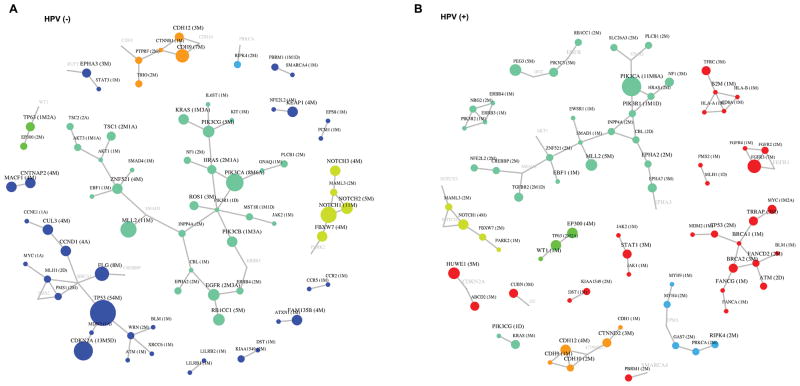
Comparison of altered networks for HPV-positive tumors versus HPV-negative tumors
To identify additional biologic differences between HPV-positive and HPV-negative tumors we employed an unbiased objective protein-protein interaction network based analysis of genetic aberrations (VarWalker) (Figure 3). The altered networks for HPV-negative tumors contained 84 proteins connected by 76 interactions (Figure 3A). For HPV-positive tumors, the altered networks contained 88 proteins connected by 83 interactions (Figure 3B). The two altered networks were substantially different: Alteration of p53 signaling and cell cycle pathway genes occurred almost exclusively in HPV-negative tumors. The alteration of oxidative stress pathway genes (CUL3, NFE2L2 and KEAP1) occurred more often in HPV-negative tumors (Figure 3A). The alteration of DNA damage pathway (BRCA1, BRCA2, FANCG, FANCA, FANCD2, and ATM), FGF signaling (FGFR2, FGFR3 and FGFR4), JAK/STAT signaling (STAT1, JAK1 and JAK2), and immunology related genes (HLA-A, HLA-B) favored HPV-positive tumors (Figure 3B). HPV-positive and HPV-negative tumors shared network alterations such as PI3K signaling, Notch aberrations, and SMAD signaling (Figures 3A, 3B).
Figure 3. Network based comparison of genetic aberrations in HPV-negative and HPV-positive tumors using protein-protein interaction (PPI) network prioritization (VarWalker) in order to identify significantly altered pathways/networks in each entity.
The size of the symbols correlates with frequency of aberrations. M=non-synonymous mutation/s, A=amplification/s, D=deletion/s. Color choices are random, but are intended to highlight similarities and differences between HPV-positive and HPV-negative tumors. PPI connections are shown with grey lines, and connecting genes without genetic aberrations are shown with grey font.
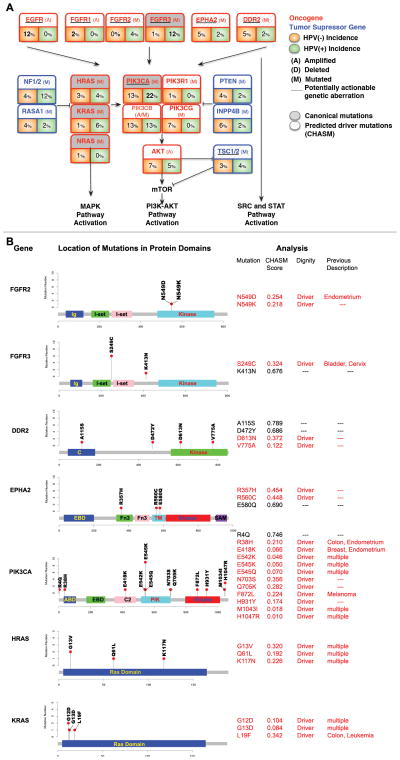
Targetable copy number aberrations and mutations
Somatic mutations in potentially targetable kinase genes occurred in FGFR2 and FGFR3 uniquely in HPV-positive tumors with an incidence of 17.6% among HPV-positive tumors. FGFR2 mutations included N569D and N569K (both in HPV-positive), and FGFR3 mutations included six S249C mutations, which were validated by Sanger Sequencing (all in HPV-positive), as well as a K413N somatic mutation (Figures 4A, 4B). We used a previously validated machine learning-based approach named CHASM to predict and prioritize missense mutations with a high likelihood of being drivers of oncogenesis(24). Both FGFR2 N569K and FGFR3 S249C have been described in several cancer types(30, 31) and showed low CHASM scores suggestive of oncogenic driver character (Figures 4A/B). Importantly the FGFR3 S249C mutation was identified recurrently in six HPV-positive tumors. S249C has been reported in one HPV-positive tumor in the TCGA HNSCC cohort and in the TCGA lung squamous cell and bladder carcinoma cohorts. A fraction of DDR2 and EPHA2 mutations also showed low CHASM scores, but were not recurrent, and did not cluster in any domain (Figures 4A, 4B).
Figure 4. Genomic alterations with translational or clinical relevance by HPV status.
(A) Shown is the incidence of selected potentially targetable and their association with key pathways in HPV(−) and HPV(+) samples.(B) Gene diagrams for a selection of key mutations in potentially targetable genes, or presumed driver genes. Shown are the genes domains that are affected by somatic mutations, the number of cases that harbor the respective mutation is indicated on the ordinate axis, the CHASM scores and p-values for each mutation (red = <0.5 CHASM score, which indicates predicted oncogenic driver character), as well prior reports of mutations based on occurrence in COSMIC, or prior publications.
PIK3CA was the most commonly altered oncogene including a number of established canonical mutations (E542K, E545K, H1047R) (Figures 4A, 4B). Other PI3K pathway mutations included somatic events in PIK3R1, PTEN, TSC1 and TSC2, all of which have been reported to harbor mutations in other tumor types (Figure 4A). PIK3CA mutations were more commonly observed in HPV-positive individuals though the difference was not statistically significant in our cohort (p=0.20).
Mutations in the MAPK pathway genes included seven canonical HRAS (G13V, Q61L, K117N), KRAS (G12D, G13D, L19F) and NRAS (Q61R) events (incidence 5.8%), as well as multiple mutations of unclear significance in the tumor suppressor NF1 (Figure 4A, 4B).
Clinical correlation
We first examined the mutation difference in smokers vs. non-smokers (Supplementary Table S2). In the overall cohort tumors from smokers displayed a higher overall mutation burden as compared to non-smokers 11.5 mut/pt (≥10 pack years) versus 5.6 mut/pt (non-smokers)(p = 0.0007, t test). We noted a significant association among smoking and mutations in TP53 (p = 0.0009), CSMD3 (p = 0.0062), RB1CC1 (p = 0.0161), THSD7A (p = 0.0319). Mutations in ZFHX4 (p = 0.0167, Fisher’s exact test) and TRRAP (p = 0.0392, Fisher’s exact test) were significantly enriched in smokers compared to non-smokers. Additional subgroup analyses including drinkers vs. non-drinkers, large primary versus small primary and node negative vs. node positive are provided in Supplementary Table 2, including subgroup analyses for oropharyngeal primary tumors.
The association among HPV status and prognosis was evaluated. As expected HPV positive patients had significantly better prognosis that HPV negative patients for both OS (p = 0.015) and PFS (p = 0.013), and this holds true for the overall cohort as well as oropharyngeal cancers (OS: p=4e-05, PFS: p = 0.0001) (Supplementary Figure S3A).
In an exploratory, hypothesis-forming analysis we correlated gene aberrations including mutations and copy number alterations with overall survival and progression-free survival. In the smaller sub-cohorts of HPV-positive and HPV-negative tumors most associations were not significant, though we did identify in HPV-negative tumors a possible correlation of PIK3CA mutations (OS: p = 0.017, PFS: p = 0.004)(Supplementary Figure S3B), as well as TP53 wildtype status with poor prognosis (OS: p = 0.262, PFS: p = 0.017) (Supplementary Figure S3C). These findings require validation in future studies.
Discussion
HPV-positive and HPV-negative head and neck cancers are distinct clinical entities. We report for the first time on the underlying differential mutational profiles, generated from a unique patient cohort of poor-prognosis, locoregionally-advanced tumors. Unlike prior exome-focused studies, we employed a focused 617-cancer gene-candidate approach, allowing deep coverage of cancer-relevant genes. This approach is similar, albeit significantly broader than current clinical grade next generation sequencing assays(32).
This is the first cohort to include a large number of HPV-positive tumors, including patients with a significant tobacco history (35% of HPV-positive tumors). While HPV-positive status confers a favorable prognosis, patients with >10 pack-year smoking history have a poorer prognosis (3) and we report a higher mutational burden including presence of KRAS mutationsin such HPV-positive tumors. We identify targetable genetic aberrations including both HPV-positive and HPV-negative tumors including frequent FGFR2/3 aberrations in later group.
Genetic aberrations correlate closely with HPV status, and less with anatomic site (Supplementary Table S2), suggesting that HPV status (not anatomic site) is the most important factor determining tumor biology. Furthermore it was recently reported that anatomic site cannot always be accurately determined and misclassification occurs(18).
The mutational spectrum in HPV-negative HNSCC is very similar to lung and esophageal squamous cell carcinomas with enrichment for mutations in TP53, CDKN2A, MLL2, CUL3, NSD1, PIK3CA and NOTCH genes, and copy number increases in EGFR, CCND1and FGFR1. In the future these genetic similarities may potentially allow biologically-driven cross-tumor drug development and biomarker discovery.
In contrast HPV-positive tumors show a distinct genetic profile with unique mutations in DDX3X, CYLD and FGFR and enrichment for PI3K pathway alterations and rarer KRAS mutations. Somatic aberrations in DNA-repair genes (BRCA1/2, Fanconi anemia genes, and ATM) may contribute to chemo- and/or radiosensitivity of HPV-positive tumors, and interestingly occurred in HPV-positive tumors in non-/light smokers. Both tobacco smoke and defects in DNA repair are known to induce a large number of genetic aberrations, and may be distinct ways to accumulate genetic aberrations required for the emergence of cancer. Of note a recent RAD51B was reported as an integration target for HPV16 leading to loss of its DNA repair function (5).
FGFR and PI3K pathway aberrations are potential therapeutic targets in this population. FGFR2/3 mutations are of particular interest as they occurred in 17.6% of HPV-positive tumors, most commonly the S249C mutation, which has been shown to be an oncogenic driven in bladder cancer.
One HPV-negative tumor harbored amplification of FGFR1, which is being explored clinically for lung squamous cell carcinomas (33, 34). Furthermore oncogenic FGFR3-TACC3 fusions were recently reported in HPV-positive HNC supporting a prominent role for oncogenic FGFR signaling for HPV-positive HNC while other kinases are genetically unaltered (33–37).
In contrast to prior smaller reports the overall mutational burden in HPV-negative and HPV-positive HNSCC was comparable (15.2 versus 14.4 somatic exonic mutations in the targeted cancer-associated genes), a finding corroborated when analyzing publically available data from the cancer genome atlas (TCGA) HNSCC cohort(5).
HPV-positive tumors in patients with significant tobacco use had a significantly higher mutational burden. While the number of KRAS (N=3) and TP53 (N=2) mutations in HPV-positive oropharyngeal tumors was small, they occurred in patients with significant tobacco history, and showed 4–6-fold higher average mutational burden (NS). Interestingly the two TP53 mutations (H179R, G361fs) co-occurred with KRAS (G12D, L19F) and one PIK3CA mutation (F872L) suggesting multiple oncogenic drivers in highly mutational altered HPV-positive tumors. The KRAS L19F mutation is thought to be modestly transforming (38).
Low-frequency canonical HRAS, KRAS, and NRAS mutations were identified in both HPV-positive and negative tumors. RAS mutations have been associated with poor outcome in other cancer types (e.g. lung adenocarcinomas), as well as resistance to cetuximab in colon cancer (39). Further study in HNC will be important, especially regarding the implications of KRAS mutations in otherwise good-prognosis HPV-positive tumors, which were present in 5.8% of HPV-positive tumors. One additional case of a canonical KRAS mutation in an HPV-positive tumor was previously reported (29).
The specific genetic events in HPV-positive or negative HNSCC likely contribute to the distinct biologic behavior and may impact therapy in the future. The lack of targeted therapies for squamous cell tumors remains challenging: In addition to FGFRs we identify multiple potentially targetable genomic alterations including mutations in the DDR2, EPHA2 kinases, PIK3CA and PI3K pathway genes (e.g. TSC1/2, NF2) that are predicted to be oncogenic drivers. Therapeutic implications for these particular changes are being investigated clinically and pre-clinically (33, 40, 41).
The lack of EGFR aberrations in HPV-positive tumors is consistent with prior reports (5, 42), and clinical data (10, 11). Other potential therapeutic targets include alterations of genes, which regulate the cell cycle (e.g. CCND1 amplification as a target for cyclin dependent kinase 4/6 (CDK4/6) inhibitors).
We identify a number of additional novel mutated pathways, providing further insight into the molecular pathogenesis of HNSCC: 1) Similar to lung squamous cell carcinoma, MLL2 and MLL3mutation were observed in both HPV-positive and negative tumors. MLL genes encode histone lysine methyltransferases that are involved in chromatin remodeling and recurrently mutated in several cancer types (43–46). Other studies have demonstrated an association with poor clinical outcomes (47). 2) CUL3, NFE2L2 and KEAP1mediate cellular responses to oxidative stress and are a frequent target for mutations in lung squamous cell carcinomas as well as the HPV-negative HNSCC cohort. KEAP1 binds to NFE2L2 and targets NFE2L2 for ubiquitination and subsequent degradation by the CUL3/RBX1 complex (48). Mutations are typically associated with heavy tobacco exposure, and distributed across the gene, which is suggestive of loss of function mutations. 3) The DDX3Xgene was exclusively mutated in HPV-positive tumors. DDX3X encodes an ATP-dependent RNA helicase and is involved in RNA processing (Supplementary Figure S4). It has been described to be significantly mutated in medulloblastomas, and can potentiate β-catenin activity by promoter transactivation (43, 44).4) In addition, HPV-positive tumors harbor mutations in CYLD, a gene that has not been implicated as a hotspot for mutations, but is well known for its’ role in HPV-positive cervical cancer: CYLD is involved in differentiation and NFκB signaling and in response to hypoxia activates pro-oncogenic processes (Supplementary Figure S4)(49).5) Lastly, UBR5 mutations are involved in DNA damage and apoptosis signaling, and in addition to our discovery in HPV-positive and negative tumors, UBR5 is recurrently mutated in mantle cell lymphomas (Supplementary Figure S4). Investigations into its role in radio- or chemosensitivity are promising(50).
In conclusion, the mutational landscape of HPV-positive HNSCC differs from HPV-negative HNSCC and may help explain the distinct clinical behavior and prognosis. Further study of clinical implications of the observed mutations will be vital with respect to targeted therapies, radiation, and chemotherapy. For the first time we identify targetable genetic aberrations in both HPV-positive and HPV-negative HNSCC. In particular, frequent aberrations in the FGFR family are promising as well asPI3K pathway and cell cycle abnormalities. The significant number of potential targets including rare aberrations (DDR2, TSC1/2, EPHA2), or candidate biomarkers such as HRAS/KRAS indicates a need for tumor profiling of HNSCC in the future- including on clinical trials, as well as functional validation in pre-clinical studies.
Supplementary Material
Statement of Translational Relevance.
Head and Neck Squamous Cell Carcinoma (HNSCC) is comprised of two distinct clinical entities, HPV-positive and HPV-negative HNSCC. A comprehensive list of differential molecular abnormalities, in particular therapeutically-relevant genetic aberrations has not been reported. Furthermore, while the incidence of HPV-positive tumors is rising rapidly, there is a lack of HPV-positive cohorts. In our current study, we investigate 120 locoregionally advanced HNSCC (including 42.5% HPV-positive tumors). Tumors show a differential genetic profile based on HPV-status. We discover multiple, novel therapeutic targets with predicted driver character that are of high translational relevance: EGFR, CCND1, and FGFR1 amplifications occurred in HPV-negative tumors, while 17.6% of HPV-positive tumors harbored mutations in Fibroblast Growth Factor Receptor genes (FGFR2/3). HPV-positive tumors showed a 5.8% incidence of KRAS mutations, and 7.8% incidence of BRCA1/2 mutations. Our study provides strong support for further clinical investigation of novel therapeutic targets and candidate biomarkers in patients with HNSCC.
Acknowledgments
Acknowledgements of research support for the study:
This work was supported by an ASCO Translational Professor ship award (EEV), the Grant Achatz and Nick Kokonas/Alinea Head and Neck Cancer Research Fund (EV, TYS), a Flight Attending Medical Research Institute Young Clinical Scientist Award (FAMRI, YCSA)(TYS), and a NCI K08 Award 1K08CA163677 (PSH).
Footnotes
The authors report no conflict of interest.
References
- 1.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 2.Vokes EE, Weichselbaum RR, Lippman SM, Hong WK. Head and neck cancer. N Engl J Med. 1993;328:184–194. doi: 10.1056/NEJM199301213280306. [DOI] [PubMed] [Google Scholar]
- 3.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. doi: 10.1056/NEJMoa0912217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Engels EA, Pfeiffer RM, Hernandez BY, Xiao W, Kim E, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29:4294–4301. doi: 10.1200/JCO.2011.36.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes David N, Grandis Jennifer R, El-Naggar Adel K. The Cancer Genome Atlas: Integrated analysis of genome alterations in squamous cell carcinoma of the head and neck. J Clin Oncol. 2013;31(suppl):abstr 6009. [Google Scholar]
- 6.Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5:489–500. doi: 10.1016/s1535-6108(04)00112-6. [DOI] [PubMed] [Google Scholar]
- 7.Leemans CR, Braakhuis BJM, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2010;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- 8.Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, et al. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12:701–709. doi: 10.1158/1078-0432.CCR-05-2017. [DOI] [PubMed] [Google Scholar]
- 9.Posner MR, Hershock DM, Blajman CR, Mickiewicz E, Winquist E, Gorbounova V, et al. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. N Engl J Med. 2007;357:1705–1715. doi: 10.1056/NEJMoa070956. [DOI] [PubMed] [Google Scholar]
- 10.Vermorken JB, Stöhlmacher-Williams J, Davidenko I, Licitra L, Winquist E, Villanueva C, et al. Cisplatin and fluorouracil with or without panitumumab in patients with recurrent or metastatic squamous-cell carcinoma of the head and neck (SPECTRUM): an open-label phase 3 randomised trial. Lancet Oncology. 2013;14:697–710. doi: 10.1016/S1470-2045(13)70181-5. [DOI] [PubMed] [Google Scholar]
- 11.Vokes EE, Seiwert TY. EGFR-directed treatments in SCCHN. Lancet Oncology. 2013;14:672–673. doi: 10.1016/S1470-2045(13)70215-8. [DOI] [PubMed] [Google Scholar]
- 12.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative Genomic Characterization of Oral Squamous Cell Carcinoma Identifies Frequent Somatic Drivers. Cancer Discov. 2013;3:770–781. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, Jr, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci US A. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sotlar K, Diemer D, Dethleffs A, Hack Y, Stubner A, Vollmer N, et al. Detection and typing of human papillomavirus by e6 nested multiplex PCR. J Clin Microbiol. 2004;42:3176–3184. doi: 10.1128/JCM.42.7.3176-3184.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo Z, Keck MK, Khattri A, Patel RD, Walter KZ, Lingen MW, et al. Multimodality determination of HPV status in head and neck cancers (HNC) and development of an HPV signature. J Clin Oncol. 31(suppl):abstr 6008. [Google Scholar]
- 19.Meyer M, Kircher M. Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5448. pdb.prot5448. [DOI] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cibulskis K, McKenna A, Fennell T, Banks E, DePristo M, Getz G. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2602. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia P, Zhao Z. VarWalker: Personalized Mutation Network Analysis of Putative Cancer Genes from Next-Generation Sequencing Data. PLoS Comput Biol. 2014;10:e1003460–e1003460. doi: 10.1371/journal.pcbi.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Research. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Lupat R, Amarasinghe KC, Thompson ER, Doyle MA, Ryland GL, et al. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–1313. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lechner M, Frampton GM, Fenton T, Feber A, Palmer G, Jay A, et al. Targeted next-generation sequencing of head and neck squamous cell carcinoma identifies novel genetic alterations in HPV+ and HPV− tumors. Genome Med. 2013;5:49. doi: 10.1186/gm453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cappellen D, De Oliveira C, Ricol D, de Medina S, Bourdin J, Sastre-Garau X, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 31.Liao RG, Jung J, Tchaicha J, Wilkerson MD, Sivachenko A, Beauchamp EM, et al. Inhibitor-Sensitive FGFR2 and FGFR3 Mutations in Lung Squamous Cell Carcinoma. Cancer Res. 2013;73:5195–5205. doi: 10.1158/0008-5472.CAN-12-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andre F, Ranson M, Dean E, Varga A, Noll R, Stockman PK, et al. Results of a phase I study of AZD4547, an inhibitor of fibroblast growth factor receptor (FGFR), in patients with advanced solid tumors[abstract]. Proceedings of the 104th Annual Meeting of the American Association for Cancer Research; 2013 Apr 6–10; Washington, DC. Philadelphia (PA): AACR; [Google Scholar]; Cancer Res. 2013;73(8 Suppl):Abstract nr LB-145. doi: 10.1158/1538-7445. AM2013-LB-145. [DOI] [Google Scholar]
- 34.Wolf J, LoRusso PM, Camidge RD, Perez JM, Tabernero J, Hidalgo M, et al. A phase I dose escalation study of NVP-BGJ398, a selective pan FGFR inhibitor in genetically preselected advanced solid tumors [abstract]. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research; 2012 Mar 31–Apr 4; Chicago, IL. Philadelphia (PA): AACR; [Google Scholar]; Cancer Res. 2012;72(8 Suppl):Abstract nr LB-122. 1538–7445.AM2012-LB-122. [Google Scholar]
- 35.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parker BC, Annala MJ, Cogdell DE, Granberg KJ, Sun Y, Ji P, et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J Clin Invest. 2013;123:855–865. doi: 10.1172/JCI67144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams SV, Hurst CD, Knowles MA. Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet. 2013;22:795–803. doi: 10.1093/hmg/dds486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith G, Bounds R, Wolf H, Steele RJC, Carey FA, Wolf CR. Activating K-Ras mutations outwith “hotspot” codons in sporadic colorectal tumours - implications for personalised cancer medicine. Br J Cancer. 2010;102:693–703. doi: 10.1038/sj.bjc.6605534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, et al. KRAS Mutation Is an Important Predictor of Resistance to Therapy with Epidermal Growth Factor Receptor Tyrosine Kinase Inhibitors in Non-Small-Cell Lung Cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 40.Licitra L, Mesia R, Rivera F, Remenár E, Hitt R, Erfán J, et al. Evaluation of EGFR gene copy number as a predictive biomarker for the efficacy of cetuximab in combination with chemotherapy in the first-line treatment of recurrent and/or metastatic squamous cell carcinoma of the head and neck: EXTREME study. Ann Onc. 2011;22:1078–1087. doi: 10.1093/annonc/mdq588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Young RJ, Rischin D, Fisher R, McArthur GA, Fox SB, Peters LJ, et al. Relationship between epidermal growth factor receptor status, p16(INK4A), and outcome in head and neck squamous cell carcinoma. J Clin Oncol. 2011;20:1230–1237. doi: 10.1158/1055-9965.EPI-10-1262. [DOI] [PubMed] [Google Scholar]
- 43.Jones DT, Jäger N, Kool M, Zichner T, Hutter B, Sultan M, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugh TJ, Weeraratne SD, Archer TC, Pomeranz Krummel DA, Auclair D, Bochicchio J, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y, Hammerman PS, Kim J, Yoon J-A. Integrative and Comparative Genomic Analysis of Lung Squamous Cell Carcinomas in East Asians. J Clin Oncol. 2014;32:121–128. doi: 10.1200/JCO.2013.50.8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.An J, Mo D, Liu H, Veena MS, Srivatsan ES, Massoumi R, et al. Inactivation of the CYLD Deubiquitinase by HPV E6 Mediates Hypoxia-Induced NF-κB Activation. Cancer Cell. 2008;14:394–407. doi: 10.1016/j.ccr.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meissner B, Kridel R, Lim RS, Rogic S, Tse K, Scott DW, et al. The E3 ubiquitin ligase UBR5 is recurrently mutated in mantle cell lymphoma. Blood. 2013;121:3161–3164. doi: 10.1182/blood-2013-01-478834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.