Abstract
PPARα is well known as a master regulator of lipid metabolism. PPARα activation enhances fatty acid oxidation and decreases the levels of circulating and cellular lipids in obese diabetic patients. Although PPARα target genes are widely known, little is known about the alteration of plasma and liver metabolites during PPARα activation. Here, we report that metabolome analysis-implicated upregulation of many plasma lysoGP species during bezafibrate (PPARα agonist) treatment. In particular, 1-palmitoyl lysophosphatidylcholine [LPC(16:0)] is increased by bezafibrate treatment in both plasma and liver. In mouse primary hepatocytes, the secretion of LPC(16:0) increased on PPARα activation, and this effect was attenuated by PPARα antagonist treatment. We demonstrated that Pla2g7 gene expression levels in the murine hepatocytes were increased by PPARα activation, and the secretion of LPC(16:0) was suppressed by Pla2g7 siRNA treatment. Interestingly, LPC(16:0) activates PPARα and induces the expression of PPARα target genes in hepatocytes. Furthermore, we showed that LPC(16:0) has the ability to recover glucose uptake in adipocytes induced insulin resistance. These results reveal that LPC(16:0) is induced by PPARα activation in hepatocytes; LPC(16:0) contributes to the upregulation of PPARα target genes in hepatocytes and the recovery of glucose uptake in insulin-resistant adipocytes.
Keywords: lysoglycerophospholipid, mass spectrometry, liver, phospholipases/A2, omics
Obesity is recognized to have major adverse health effects and is a risk factor for serious chronic disorders (1–4), including diabetes and cardiovascular diseases. Dyslipidemia and hyperglycemia, in particular, result from obesity. Therefore, it is important to prevent dyslipidemia and hyperglycemia to avoid serious diseases.
PPARs are ligand-activated transcription factors and members of the nuclear hormone receptor superfamily (5–9). The family of PPARs comprises three isoforms: PPARα, PPARβ/δ, and PPARγ. PPARα is a very important factor in the regulation of lipid metabolism (5–7) and is expressed at high levels in the liver where it promotes β-oxidation, ketogenesis, and lipid transport (10, 11). During PPARα activation, transcription of PPARα-regulated genes [e.g., carnitine-O-palmitoyltransferase 1 (CPT1) and acyl-CoA oxidase (ACO)] is stimulated and β-oxidation is activated (12–14). This activation and increased PPARα sensing in the liver result in increased energy burning and reduced fat storage (15). It has been reported that PPARα activation enhances fatty acid oxidation and decreases the levels of circulating and cellular lipids in obese diabetic patients (9, 16). Regulating PPARα activity is therefore one of the most important means of managing chronic diseases related to the dysfunction of lipid metabolism.
PPARα has the capability to accommodate and bind to a variety of natural and synthetic lipophilic acids (15). It is well known that fibrates are widely prescribed hypolipidemic drugs that act in a PPARα-dependent mechanism. Bezafibrate is one of the fibrate drugs and improves dyslipidemia, slows the progression of focal coronary atherosclerosis, and reduces the number of coronary events in young survivors of myocardial infarction (17). It is interesting to note that fibrates, including bezafibrate, contribute to the improvement of both dyslipidemia and glucose metabolism disorder (18–20). Although PPARα target genes are widely known, little is known about the variation of metabolites in plasma and liver during the activation of these genes.
Over the past decade, the use of metabolomic techniques, including liquid chromatography coupled with ultraprecise mass spectrometry, to investigate animal or plant metabolism has increased dramatically (21, 22). These techniques provide molecular weight data with the precision of a few ppm, permitting the determination of chemical formulae. Metabolite investigation based on precise molecular weight data enables the large-scale analysis of animal metabolite dynamics during treatment with the PPARα agonist. The application of metabolomics is useful for monitoring the metabolism and helpful for identifying novel PPARα agonist-responsive pathways.
The aim of the present study is to identify the metabolite that is regulated by PPARα and to clarify its function. In the present study, metabolomics approach revealed that 1-palmitoyl lysophosphatidylcholine [LPC(16:0)] is induced by PPARα activation in the liver. Furthermore, LPC(16:0) activates PPARα in the hepatocytes and stimulates glucose uptake in insulin-resistant 3T3-L1 adipocytes. To the best of our knowledge, this is the first report stating that LPC(16:0) induces PPARα activation and is capable of improving dyslipidemia and hyperglycemia.
MATERIALS AND METHODS
Materials
Sigma (St. Louis, MO) supplied us with bezafibrate, LPC(16:0), GW7647, and GW6471. Fenofibrate were purchased from Tokyo Chemical Industry (Tokyo, Japan). Leucine enkephalin and insulin were purchased from Wako (Osaka, Japan). Dexamethasone, troglitazone, and 1-methyl-3-isobutylxanthine (IBMX) were purchased from Nacalai Tesque (Kyoto, Japan). Extrasynthese (Lyon, France) supplied us with 7-hydroxy-5-methylflavone (HMF). All the LC/MS solvents used in this study were purchased from Wako (Osaka, Japan). The LC/MS buffer (acetonitrile, ultrapure water, formic acid, and methanol) was LC/MS grade.
Animal experiments
All the animal experiments were approved by the Kyoto University Animal Care Committee. The mice were kept in individual cages in a temperature-controlled room at 23 ± 1°C and maintained under a constant 12 h light/dark cycle. Male KK-Ay mice were purchased from CLEA Japan (Tokyo, Japan). The 4-week-old mice were maintained for 7 days on a standard diet (SD) and then divided into two groups of similar average body weights. Each group was maintained on a 60%kcal high-fat diet (HFD) and HFD containing 0.2% (w/w) bezafibrate for 4 weeks. The energy intake of all the mice was adjusted by pair feeding. Male C57BL/6J mice were purchased from CLEA Japan. Mice were randomly divided into four groups (n = 4/group) and fed SD (16%kcal fat) or HFD for 8 weeks. The mice were then fed SD, HFD, SD plus fenofibrate, and HFD plus fenofibrate (200 mg/kg body weight/day, oral administration) for 2 weeks. At the end of the treatment period, anesthetized mice were euthanized by cervical dislocation after overnight fasting, and blood and organs samples were collected. Plasma TG and glucose levels were determined by the TG E-test and glucose CII-test (Wako), respectively.
Extraction of mouse plasma, tissues, and cell culture sample
Mouse heparin-blood samples were centrifuged at 10,000 rpm for 10 min at 4°C. After centrifugation, plasma samples (5 µl) were dissolved in 95 µl of extraction solvent (80% methanol containing HMF as an internal standard). Mouse liver samples (10 mg) were homogenized in 1 ml of extraction solvent. The partial cell culture medium (300 µl/well) was evaporated to dryness and redissolved in 100 µl of extraction solvent. The hepatocyte extraction was washed with PBS and dissolved in extraction solvent (200 µl/well). The hepatocyte extraction was ultrasonically fragmented.
After centrifugation (15,000 rpm, 10 min, 4°C), the supernatant was collected as extract. The extracts were filtered through a 0.2 µm pore polyvinylidene difluoride membrane (Whatman, Brentford, UK), and the filtrate was used in LC/MS.
Metabolomic analysis by HPLC-Orbitrap MS
LC/MS for metabolomics was performed using an HPLC system (Agilent) coupled to an LTQ Orbitrap XL-MS system (Thermo Fisher Scientific Inc., San Jose, CA), equipped with an electrospray source operating in the positive- and negative-ion modes. The spray voltage and capillary temperature were 4 kV and 250°C, respectively. This analysis consists of two scan events. Scan Event 1 is full mass type (Analyzer; FTMS, Resolution; 60,000). Scan Event 2 is MS/MS type (Analyzer; Ion Trap MS, Act Type; collision-induced dissociation, Normalized Collision Energy; 35.0). An aliquot of the extracted sample (10 µl) was injected into an Inertsil ODS-4 reversed-phase column (column size, 3.0 × 250 mm; particle size, 3.0 µm; GL Sciences Inc., Tokyo, Japan). The column temperature was set at 30°C. Mobile phases A (0.1% formic acid) and B (acetonitrile including 0.1% formic acid) were used. The buffer gradient consisted of 30.0% to 90.0% B for 0 to 30 min, 95.0% B for 30.0 to 60.0 min, 95.0% to 30.0% B for 60.0 to 60.1 min, and 30.0% B for 24.9 min before the next injection, at a flow rate of 250 µl/min. These data were acquired with X-Calibur software (Thermo Fisher Scientific Inc.) and PowerGet software (Kazusa DNA Research Institute, Japan) using a previously described methods (22; http://www.kazusa.or.jp/komics/software/PowerGet). Briefly, PowerGet is a Java software package for detection, alignment, and annotation of metabolite features from data obtained using LC/high-resolution MS. The peak area was divided by the area of the internal standard. This value was used to calculate the rate of change for the control group. Differences between groups were compared with the Student’s t-test.
Quantification of lysoGPs by ultraperformance liquid chromatography quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS)
LC/MS for target metabolites analysis was performed using a Acquity UPLC system coupled to a Xevo QTOF-MS system (Waters, Milford, MA), equipped with an electrospray source in the positive-ion mode operating with a lock-spray interface for accurate mass measurement. Leucine enkephalin was used as the lock-mass compound. It was infused straight into the MS system at a flow rate of 20 µl/min at 200 pg/µl (in 50% acetonitrile, 0.1% formic acid). During lysoGP analysis, the capillary, sampling cone, and extraction cone voltages were set at 3,500, 35, and 3.0 V, respectively. The source and desolvation temperatures were 120°C and 450°C, respectively. The cone and desolvation gas flow rates were set at 50 l/h and 800 l/h, respectively. Scan event is MS mode (scan range: 100–1,000 Da; scan time: 0.2 s). An aliquot of the extracted sample (3 µl) was injected into an Acquity UPLC BEH-C18 reversed-phase column (column size, 2.1 × 100 mm; particle size, 1.7 µm). Mobile phases A and B were used. The column temperature was set at 40°C. The buffer gradient of lysoGP analysis consisted of 5.0% to 99.0% B for 0 to 10 min, 99.0% B for 10 to 15 min, 99.0% to 5.0% B for 15 to 15.5 min, and 5.0% B for 4.5 min before the next injection, at a flow rate of 300 µl/min. These data were acquired with the MassLynx software (Waters). The amount of LPC(16:0) was estimated from calibration curves obtained using analytical-grade standard compound. The peak area of m/z [M-H] ± 0.05 Da was divided by the area of the internal standard. This value was used to generate the calibration curves.
Preparation of mouse primary hepatocytes
Mouse hepatocytes were prepared as previously described (23). Briefly, C57BL/6J male mice (wild type and PPARα−/−) were anesthetized with intraperitoneal administration of pentobarbital, and the liver was perfused with liver perfusion medium (Life Technologies Japan Ltd., Tokyo, Japan), followed by liver digestion medium (Life Technologies Japan Ltd.). After filtration through a 100 mm nylon mesh filter, hepatocytes were isolated by repeated centrifugation at 50 g for 3 min (three times). The isolated hepatocytes were cultured in type-1 collagen-coated 12-well plates at a cell density of 2.0 × 105 cells/well. After 5 h incubation at 37°C in 5% CO2 atmosphere, hepatocytes were cultured in serum-free DMEM with or without bezafibrate, fenofibrate, GW7647, or GW6471 for 24 h. The hepatocytes were used for mRNA quantification and LC/MS assay.
siRNA experiments
Mouse siRNA was chemically synthesized by Qiagen (Tokyo, Japan). Negative control siRNA (Block-it NC siRNA) and lipofectamine 2000 were purchased from Life Technologies Japan Ltd. The primary hepatocytes were seeded in 12-well plates and transfected with 40 nmol/well synthesized siRNA targeting mouse Pla2g7. The primary hepatocytes were transfected with siRNA-lipofectamine complexes and incubated for 12 h at 37°C in 5% CO2 atmosphere and then used in the experiments.
Cell culture
FAO cell lines from rat liver were cultured in growth medium, DMEM with 5% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. FAO cells were seeded on 12-well plates at a cell density of 2.0 × 105 cells/well for 24 h in growth medium and then serum-free DMEM with or without bezafibrate for 24 h.
The 3T3-L1 cells were cultured, maintained, and differentiated using a previously described method (24). Briefly, 3T3-L1 murine preadipocytes were cultured in a growth medium, DMEM supplemented with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. At 2 days after reaching confluence, the cells were incubated in a differentiation medium containing 0.25 μM dexamethasone, 1 µM troglitazone, 10 μg/ml insulin, and 0.5 mM IBMX for 48 h, and then in a medium containing 1 µM troglitazone and 5 μg/ml insulin for another 2 days. On the fourth day, the medium was replaced with the growth medium containing insulin (5 μg/ml). The medium was replaced with serum-free DMEM for 18 h before glucose-uptake experiments.
RAW264.7 macrophage (RAW) cell lines were cultured in a growth medium, DMEM with 10% (v/v) fetal bovine serum, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C in 5% CO2. RAW cells were seeded on 12-well plates at a cell density of 3.0 × 105 cells/well for 24 h in growth medium and then serum-free DMEM containing 0.5% BSA for 24 h. The medium was used as RAW-conditioned medium (RAW-CM).
Quantification of mRNA expression levels
Total RNA was prepared from primary hepatocytes, as well as liver and skeletal muscle, using Sepasol (Nacalai Tesque Inc., Kyoto, Japan), according to the manufacturer’s protocols. Using M-MLV reverse transcriptase (Life Technologies Japan Ltd.), total RNA was reverse-transcribed using a thermal cycler (Takara PCR Thermal Cycler SP; Takara Bio Inc., Shiga, Japan). To determine mRNA expression levels, real-time quantitative RT-PCR analysis was performed with a Light Cycler System (Roche Diagnostics) using SYBR green fluorescence signals, as described previously (25, 26). The oligonucleotide primer sets of mouse 36B4 and PPARa target genes were designed using a PCR primer selection program at the website of the Virtual Genomic Center from the GenBank database as follows: mouse Cpt1a (Fwd: 5′-CTCAGTGGGAGCGACTCTTCA-3′ Rev: 5′-GGCCTCTGTGGTACACGACAA-3′), mouse Cpt1b (Fwd: 5′-CTGTTAGGCCTCAACACCGAAC-3′ Rev: 5′-CTGTCATGGCTAGGCGGTACAT-3′), mouse Aco (Fwd: 5′-GCACCATTGCCATTCGATACA-3′ Rev: 5′-ACGGCTATTCTCACAGCAGTGG-3′), mouse Pla2g6 (Fwd: 5′-GTGCCTGTAACCTGTGTAGATGTC-3′ Rev: 5′-CTCTGAGTCCTAGTGTCAGATGGA-3′), mouse Pla2g7 (Fwd: 5′-ATTTCTTGGAACACCCAGTATTGT-3′ Rev: 5′-GAACATTCTATTGCTCTTTGCTGA-3′), mouse Pla2g12b (Fwd: 5′-ATCAAGGTACCAGGAAGTATGGAC-3′ Rev: 5′-TCATAGCTCTTCTTTCTCCTCCTC-3′), and mouse 36B4 as an internal control (Fwd: 5′-TCCTTCTTCCAGGCTTTGGG-3′ Rev: 5′-GACACCCTCCAGAAAGCGAG-3′). All data indicating mRNA expression levels are presented as a ratio relative to a control in each experiment.
Induction of insulin resistance
Two different insulin-resistant adipocyte models were established as previously described with modifications (27, 28). One of these used TNF-α. On the sixth day after induction, the fully differentiated 3T3-L1 adipocytes were treated with serum-free DMEM containing 10 ng/ml recombinant mouse TNF-α (Peprotech, Rocky Hill, NJ) for 18 h. In another method used, RAW-CM 3T3-L1 adipocytes were incubated with control medium (basal medium of serum-free DMEM containing 0.5% BSA) or basal medium conditioned by RAW-CM for 18 h. The 3T3-L1 adipocytes exposed to TNF-α or RAW-CM became insulin resistant, as assessed by the ability of insulin to stimulate glucose uptake. The level of uptake of 2-deoxy-d-[1,2-3H]glucose ([1,2-3H]-2DG) was measured, as previously described (29).
Luciferase assay
Luciferase assays were performed as previously described, using a GAL4/PPAR chimera system (25, 30). We transfected p4xUASg-tk-luc (a reporter plasmid), pM-hPPARa (an expression plasmid for a chimera protein for the GAL4 DNA-binding domain and each human PPAR-ligand-binding domain), and pRL-CMV (an internal control for normalizing transfection efficiency) into monkey CV1 kidney cells by using Lipofectamine (Life Technologies Japan Ltd.), according to the manufacturer’s protocol. Luciferase activity was assayed using the dual luciferase system (Promega, MO) according to the manufacturer’s protocol.
Statistical analyses
Data are presented as the mean ± SEM. Differences between groups were compared with the Student’s t-test (for two groups), Pearson correlation coefficient, and one-way ANOVA, followed by least-significant multiple comparison methods. Values of P < 0.05 were considered statistically significant.
RESULTS
LPC in plasma was increased by PPARα activation
We confirmed that plasma TG was decreased (to ∼70% vs. control; Table 1), and the expression of PPARα target genes in the liver was increased by bezafibrate treatment for 4 weeks (Table 1). These data suggest that bezafibrate treatment for 4 weeks was sufficient to activate hepatic PPARα in KK-Ay mice.
TABLE 1.
Effect of bezafibrate on plasma TG, glucose, and PPARα target gene expression in liver
| Group | Control | Bezafibrate |
| Body weight (g) | 43.97 ± 0.93 | 44.2 ± 0.59 |
| Food intake (g/day) | 4.09 ± 0.007 | 4.10 ± 0.006 |
| Plasma TG (mg/dl) | 172.11 ± 11.54 | 123.16 ± 7.45a |
| Plasma glucose (mg/dl) | 568.49 ± 33.10 | 349.95 ± 18.73b |
| Cpt1a (% of control) | 100.00 ± 9.91 | 136.56 ± 10.20a |
| Cpt1b (% of control) | 100.00 ± 18.27 | 2,778.23 ± 316.81b |
| Aco (% of control) | 100.00 ± 24.35 | 679.17 ± 95.81b |
Data are mean ± SEM (n = 5–7). Control: HFD; bezafibrate: 0.2% bezafibrate + HFD.
P < 0.05, versus control.
P < 0.001 versus control.
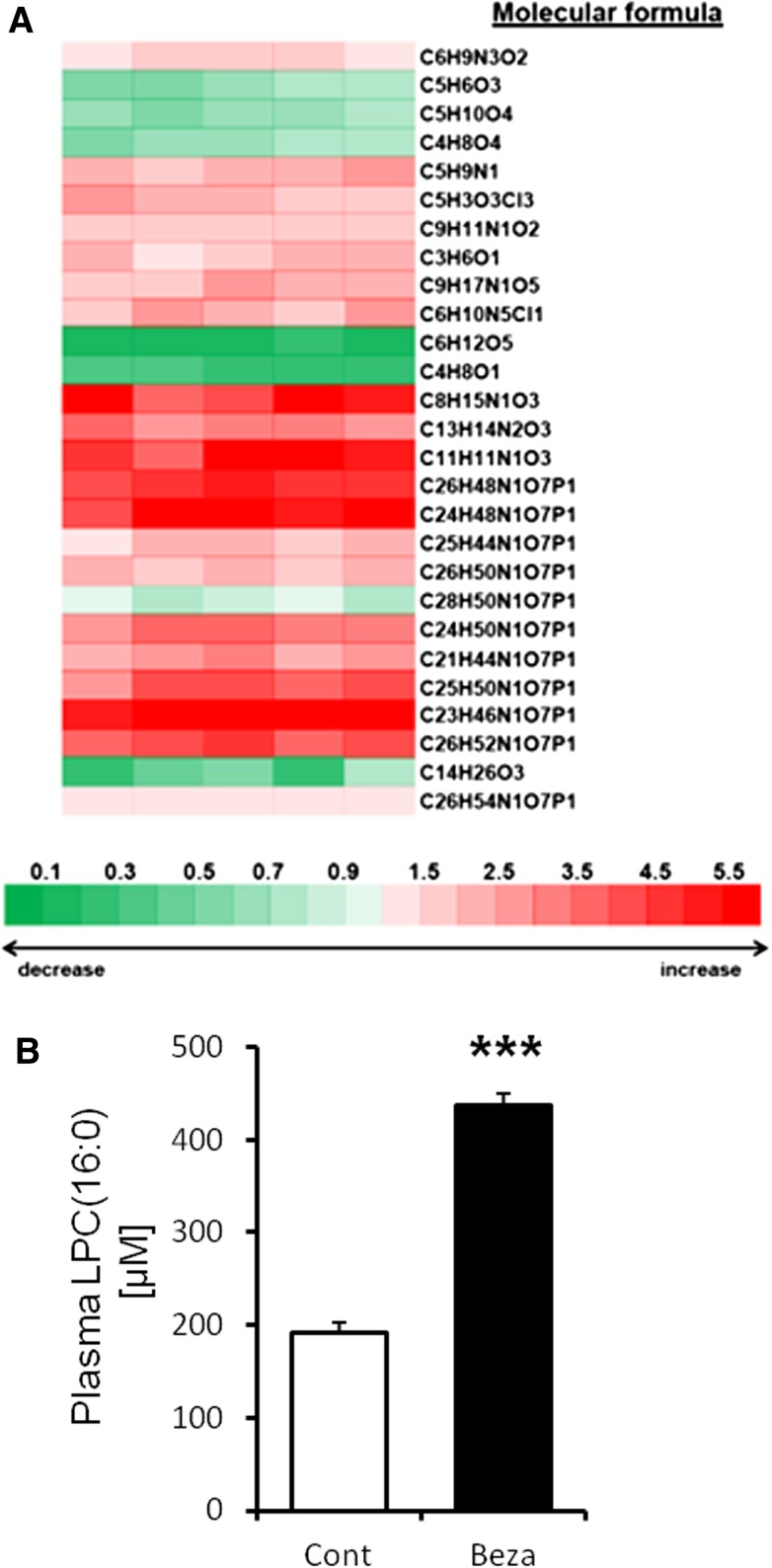
To identify the metabolites that were influenced by PPARα activation, we investigated the plasma metabolites profile in mice treated with bezafibrate for 4 weeks by metabolome analysis based on LC/MS and by using metabolite databases, including the Kyoto Encyclopedia of Genes and Genomes and Lipid Maps. We detected 887 peaks of metabolites in mice plasma (data not shown). Metabolome analysis was used to elucidate the patterns of plasma metabolic change in the control versus bezafibrate-fed mice. Differences between these groups were compared with the statistical analysis. This analysis revealed that 27 metabolites including lysoGP species influenced by PPARα activation (Fig. 1A; supplementary Table 1). Metabolome analysis showed that LPC(16:0) was one of the metabolites that was predicted to be upregulated by PPARα activation (C24H50N1O7P1; Fig. 1A; supplementary Table 1). A previous study reported that LPC(16:0) increased adipocyte glucose uptake (31). We confirmed that plasma glucose was decreased (to ∼60% vs. control) by bezafibrate treatment (Table 1).
Fig. 1.
High plasma LPC(16:0) levels in mice activated with PPARα. A: Metabolome analysis of mice plasma treated with bezafibrate for 4 weeks. The values are mean fold-change relative to control. B: Quantitative analysis of plasma LPC(16:0). Data are mean ± SEM (n = 5). *** P < 0.001 versus control. Beza, bezafibrate; Cont, control.
We identified LPC(16:0) using LC/MS (supplementary Fig. 1A–D). In the MS/MS data, we confirmed the fragment ion (m/z 184), which has been reported to be characteristic of LPC (31, 32). We also quantified plasma LPC(16:0) and showed that plasma LPC(16:0) was increased by bezafibrate treatment (control group: 192.45 ± 12.85 µM; bezafibrate: 437.16 ± 29.63 µM; P < 0.001; Fig. 1B). These data suggest that LPC(16:0) in plasma was increased by bezafibrate treatment.
Secretion of LPC(16:0) was increased by PPARα activation in liver
It is well known that PPARα is highly expressed in liver and skeletal muscle (10, 11). To elucidate the source of plasma LPC(16:0) in mice treated with bezafibrate, we quantified LPC(16:0) in these tissues.
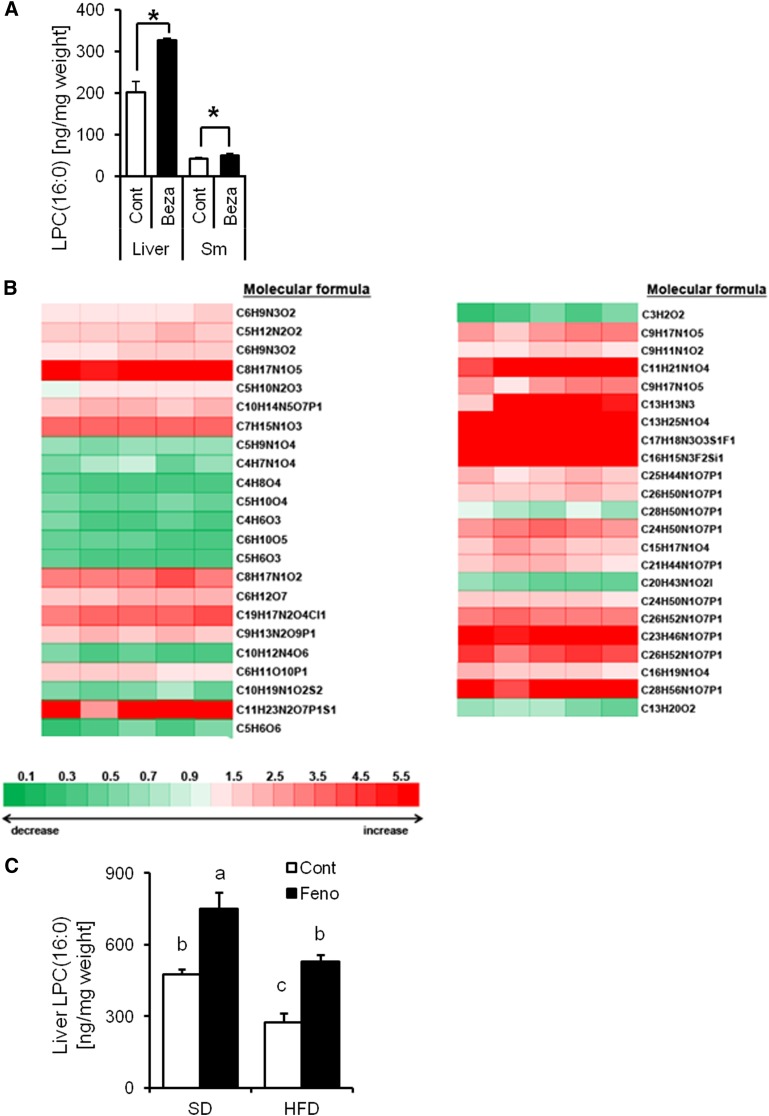
Quantitative analysis of LPC(16:0) in control mice tissue revealed that the high concentration of LPC(16:0) was in the liver where PPARα is highly expressed (202.61 ± 24.60 ng/mg liver weight, 41.66 ± 2.70 ng/mg skeletal muscle weight; Fig. 2A). We also demonstrated that the concentration of LPC(16:0) was markedly increased by PPARα activation in the liver, compared with skeletal muscle (control group: 202.61 ± 24.60 ng/mg liver weight; bezafibrate: 326.13 ± 5.57 ng/mg liver weight; P < 0.05; Fig. 2A). The metabolome analysis revealed that the concentrations of many lysoGPs in the liver, including LPC(16:0), were increased by bezafibrate treatment (Fig. 2B; supplementary Table 2). We also measured the LPC(16:0) level in mice liver receiving fenofibrate (specific PPARα activator) under SD or HFD eating conditions. These data showed that liver LPC(16:0) was increased by fenofibrate treatment under both SD (control group: 475.87 ± 20.11 ng/mg liver weight; fenofibrate group: 751.51 ± 68.40 ng/mg liver weight; P < 0.05; Fig. 2C) and HFD (control group: 273.37 ± 37.98 ng/mg liver weight; fenofibrate group: 528.73 ± 28.26 ng/mg liver weight; P < 0.05; Fig. 2C) conditions. We also showed that the amount of liver LPC(16:0) is lower under HFD condition than SD condition (SD group: 475.87 ± 20.11 ng/mg liver weight; HFD group: 273.37 ± 37.98 ng/mg liver weight P < 0.05; Fig. 2C).
Fig. 2.
Mouse liver induced by PPARα activation produces LPC(16:0). A: Comparative analysis of LPC(16:0) content in mouse liver and skeletal muscle (Sm) treated with or without bezafibrate for 4 weeks (n = 4–7). B: Metabolome analysis of mouse liver treated with bezafibrate for 4 weeks (n = 5). The values are mean fold-change relative to control. C: Comparative analysis of LPC(16:0) content in mouse liver treated with or without fenofibrate for 2 weeks (n = 4). Data are mean ± SEM. * P < 0.05 versus control. Feno; fenofibrate.
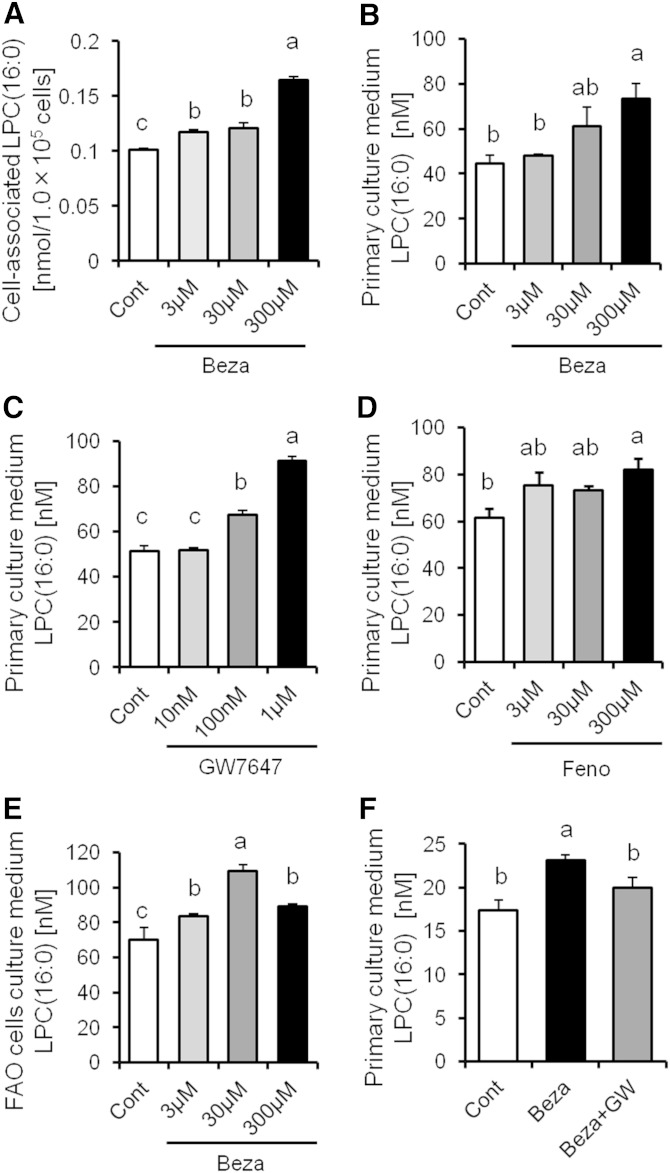
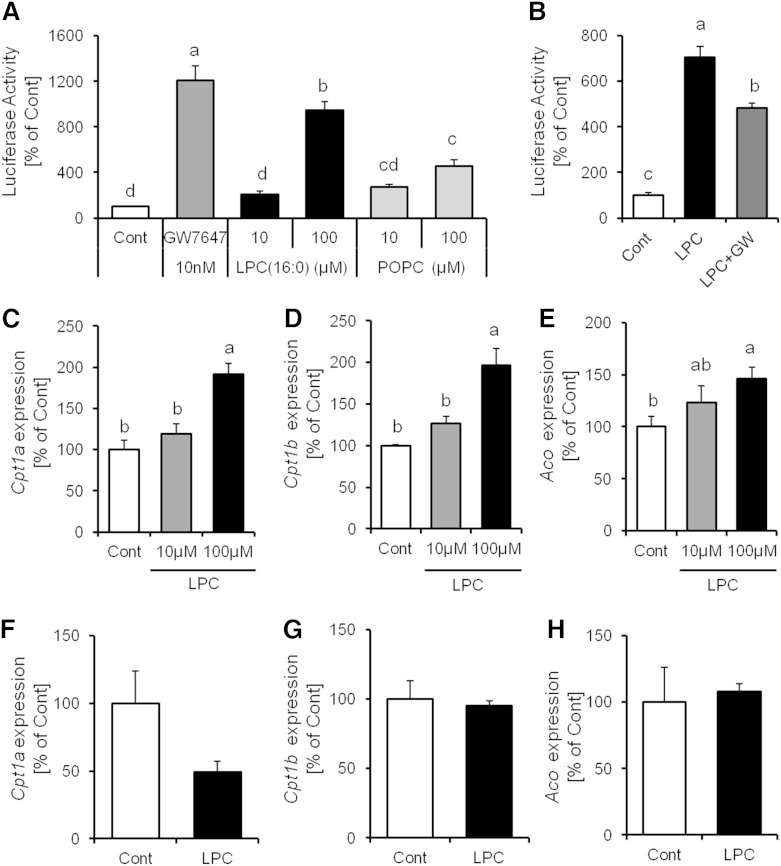
To further elucidate the mechanism of regulation of LPC(16:0) production, we analyzed the content of LPC(16:0) in primary hepatocytes treated with bezafibrate for 24 h. The content of LPC(16:0) in primary hepatocytes and culture medium increased in a dose-dependent manner (Fig. 3A, B). The concentration of LPC(16:0) in primary hepatocyte medium increased with GW7647 and fenofibrate (PPARα-specific agonist, respectively) in a dose-dependent manner (Fig. 3C, D). We also confirmed this effect in FAO rat hepatocytes treated with bezafibrate (Fig. 3E). Furthermore, the increase in LPC(16:0) secretion in primary hepatocytes treated with bezafibrate was diminished by GW6471 (PPARα antagonist) treatment (Fig. 3F). These findings show that production and secretion of LPC(16:0) were induced by PPARα activation in hepatocytes.
Fig. 3.
High LPC(16:0) levels in murine primary hepatocytes and culture medium treated with PPARα activator, and the secretion of LPC(16:0) suppressed by PPARα antagonist. Quantitative analysis of LPC(16:0) content in murine primary hepatocytes-associated (A) and culture medium treated with bezafibrate (B). Effect of GW7647 (C) or fenofibrate (D) on the secretion of LPC(16:0) in murine primary hepatocytes. E: Effect of bezafibrate on the secretion of LPC(16:0) in FAO cells. F: Quantitative analysis of LPC(16:0) content in murine primary hepatocytes culture medium treated with bezafibrate (300 µM) and/or GW6471 (100 nM). Data are mean ± SEM (n = 3–4). GW, GW6471.
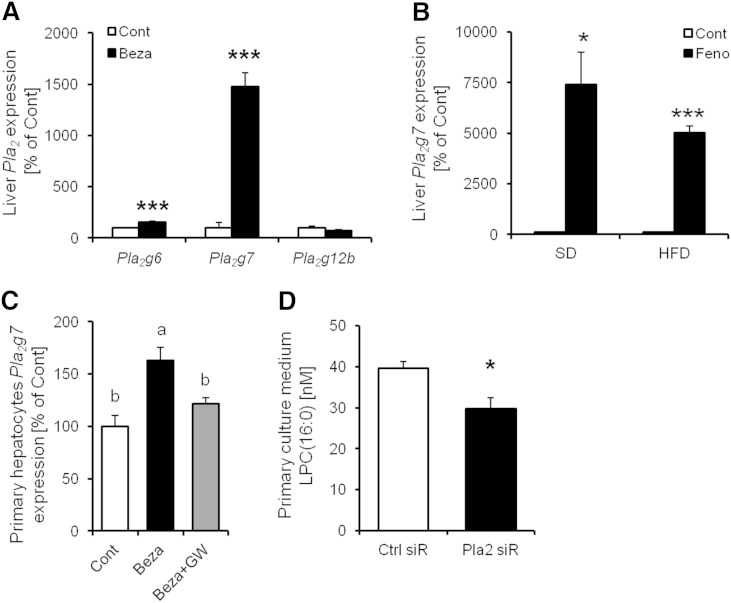
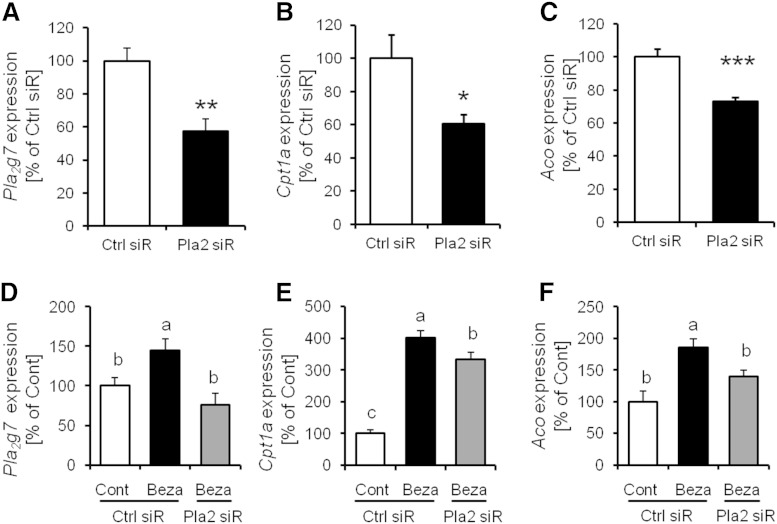
Pla2g7 gene expression levels in the hepatocytes were increased by PPARα activation, and the secretion of LPC(16:0) was suppressed by Pla2g7 siRNA treatment
LPC(16:0) is derived from phosphatidylcholine because of the action of phospholipase A2 (PLA2), which has many subtypes. To determine the main PLA2 subtype contributing to LPC(16:0) production in hepatocytes treated with bezafibrate, we analyzed the mRNA expression of Pla2 subtype in the liver. We determined that Pla2g6, Pla2g7, and Pla2g12b were mainly expressed in the liver and that the mRNA expression of Pla2g7 was markedly elevated (to ∼1,500% of the control) by bezafibrate treatment (Fig. 4A). The mRNA expression of Pla2g7 levels in mice liver under SD and HFD eating conditions were also increased (to ∼7,500% and 5,000% of the control group, respectively) by fenofibrate treatment (Fig. 4B). The mRNA expression of Pla2g7 was also increased by PPARα activation in murine primary hepatocytes, and the increase in mRNA expression of Pla2g7 was diminished by GW6471 treatment (Fig. 4C). These findings show that PPARα is involved in the expression level of Pla2g7.
Fig. 4.
The effect of Pla2g7 gene expression on LPC(16:0) production in mouse liver. A: Effect of bezafibrate on Pla2 genes expression in mouse liver (n = 3–4). B: Effect of fenofibrate on Pla2g7 gene expression in mouse liver (n = 4). C: Effect of bezafibrate on Pla2g7 gene expression in murine primary hepatocytes (n = 3–4). D: LPC(16:0) concentration in murine primary culture medium treated with Pla2g7 siRNA (n = 8–10). Data are mean ± SEM. * P < 0.05, *** P < 0.001 versus control. Ctrl siR, control siRNA treatment; Pla2 siR, Pla2g7 siRNA treatment.
To investigate the contribution of the expression of Pla2g7 to LPC(16:0) production, we analyzed LPC(16:0) production in murine primary hepatocytes treated with control siRNA or Pla2g7 siRNA. LPC(16:0) secretion in hepatocytes was attenuated by Pla2g7 siRNA treatment (control siRNA group: 39.57 ± 1.65 nM; Pla2g7 siRNA group: 29.71 ± 2.76 nM; P < 0.05; Fig. 4D). These data indicate that Pla2g7 contributes to the production and secretion of LPC(16:0) in hepatocytes.
LPC(16:0) activates PPARα and induces mRNA expressions of PPARα target genes in hepatocytes
To investigate the effect of Pla2g7 on PPARα target genes expression, we analyzed the expression of PPARα target genes in murine primary hepatocytes treated with control siRNA or Pla2g7 siRNA. We confirmed that control siRNA transfection has no effect on the expression of Pla2g7 (supplementary Fig. 2). The mRNA expressions of Aco and Cpt1a were attenuated by Pla2g7 siRNA treatment (to ∼60% and 70% vs. control siRNA group, respectively, P < 0.05; Fig. 5B, C). In murine primary hepatocytes treated with bezafibrate and Pla2g7 siRNA, we also observed that upregulated expression of PPARα target gene mRNA induced by bezafibrate treatment was suppressed on Pla2g7 siRNA treatment (Fig. 5E, F).
Fig. 5.
PPARα target gene expression levels in murine primary hepatocytes were suppressed by Pla2g7 siRNA treatment. A–C: The expression levels of Pla2g7, Cpt1a, and Aco in murine primary hepatocytes treated with Pla2g7 siRNA. D–F: The expression levels of Pla2g7, Cpt1a, and Aco in murine primary hepatocytes treated with bezafibrate (300 µM) and siRNA. Data are mean ± SEM (n = 5–7). * P < 0.05, ** P < 0.01, *** P < 0.001 versus Ctrl siR.
We hypothesized that PPARα activation is influenced by the LPC(16:0). To elucidate the function of LPC(16:0) on PPARα in hepatocytes, we investigated whether LPC(16:0) activated PPARα in a luciferase ligand assay. Compared with POPC, which is one of the precursors of LPC(16:0), LPC(16:0) increased luciferase activity remarkably (Fig. 6A). This activity was inhibited by GW6471 treatment (Fig. 6B). We also demonstrated that the mRNA expression levels in primary hepatocytes of PPARα target genes, such as Cpt1a, Cpt1b, and Aco, were increased by LPC(16:0) treatment in a dose-dependent manner (Fig. 6C–E). Furthermore, The effect of LPC(16:0) on the expression of PPARα target gene disappeared in PPARα KO primary hepatocytes (Fig. 6F–H). These findings indicate that LPC(16:0) has the ability to reinforce PPARα activation.
Fig. 6.
LPC(16:0) activated PPARα and PPARα target gene expression. A, B: Evaluation of PPARα activity in luciferase reporter assay (n = 5). Effect of LPC(16:0) on Cpt1a (C), Cpt1b (D), and Aco (E) PPARα target gene expression in wild-type primary hepatocytes (n = 3–4). F–H: Effect of LPC(16:0) on PPARα target gene expression in PPARα−/− primary hepatocytes (n = 6). Data are mean ± SEM. LPC, LPC(16:0).
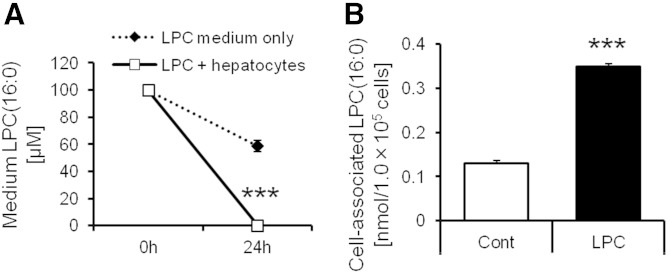
We added LPC(16:0) to the medium including or not including hepatocytes. The concentration of LPC(16:0) in the medium and cell-associated LPC(16:0) were measured by LC/MS. The concentration of LPC(16:0) in medium including only LPC(16:0) was decreased for 24 h (−40%; Fig. 7A, broken line). On the other hand, the concentration of LPC(16:0) in medium including LPC(16:0) plus hepatocytes was markedly decreased for 24 h (−99.8%; Fig. 7A, black line). Furthermore, cell-associated LPC(16:0) was increased by LPC(16:0) treatment (Fig. 7B). These data raise the possibility that part of LPC(16:0) entry into cells.
Fig. 7.
The quantification analysis of LPC(16:0) in the medium and cell-associated LPC(16:0) at 24 h after addition of LPC(16:0). A: The concentration of LPC(16:0) in the medium including (black line) or not including hepatocytes (broken line). B: The cell-associated LPC(16:0) treated with or without LPC(16:0). Data are mean ± SEM (n = 6). *** P < 0.001 versus LPC medium only or control.
LPC(16:0) stimulates glucose uptake in insulin-resistant 3T3-L1 adipocytes
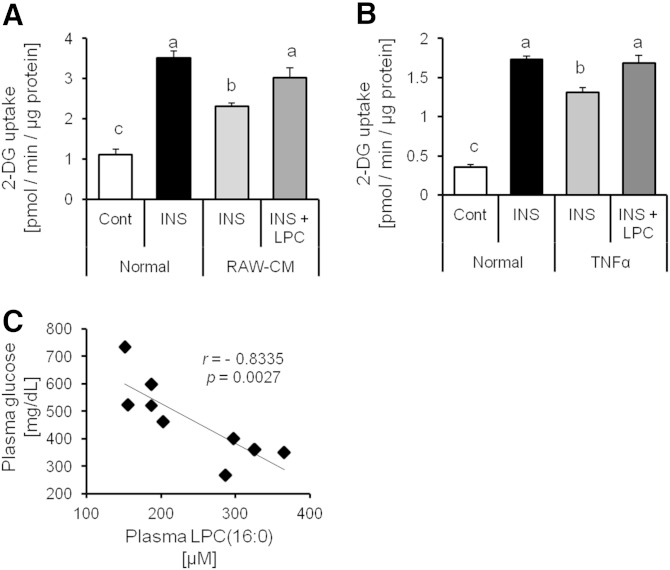
It is well recognized that LPC(16:0) increases adipocyte glucose uptake (31). We also confirmed the LPC(16:0) capacity to promote glucose uptake in 3T3-L1 adipocytes in a dose-dependent manner and LPC(16:0) had an additive effect increasing glucose uptake in 3T3-L1 adipocytes cotreated with insulin and LPC(16:0) (data not shown). We hypothesized that LPC(16:0) has the ability to promote glucose uptake under conditions of insulin resistance. To elucidate the function of LPC(16:0), we investigated whether it stimulates glucose uptake in insulin-resistant adipocytes. We revealed that LPC(16:0) has the ability to recover glucose uptake in insulin-resistant adipocytes, which were induced by TNF-α or RAW-CM (Fig. 8A, B). Furthermore, we analyzed a correlation between the concentration of plasma glucose and LPC(16:0) in mice treated with or without bezafibrate for 4 weeks. The results showed strong correlation between plasma glucose and LPC(16:0) (r = −0.8335, P = 0.0027; Fig. 8C). These findings suggest that LPC(16:0) is capable of recovering glucose uptake in insulin-resistant adipocytes, which raises the possibility that it contributes to the decrease in the plasma glucose level.
Fig. 8.
LPC(16:0) improves glucose uptake in insulin-resistant adipocytes. Effect of LPC(16:0) (300 µM) on glucose uptake in 3T3-L1 induced insulin-resistant by RAW-CM (A) and TNF-α (B) (10 ng/ml). Data are mean ± SEM (n = 3–5). INS, insulin (100 nM). C: Correlation between plasma glucose and LPC(16:0) concentration in KK-Ay mice treated with or without bezafibrate for 4 weeks. 2-DG, 2-deoxy-D-glucose.
DISCUSSION
In this study, metabolome analysis revealed that many types of plasma lysoGPs were increased by PPARα activation. LysoGPs have been found in a wide range of tissues and cell types (33). It has also been reported that lysoGPs play an important role in many physiological and pathophysiological processes (33–36). LysoGPs are well known to be lipid mediators (autacoid) that have topical effects on cells. However, our data showed that lysoGPs have not only topical but also whole-body effects throughout the bloodstream. LPC(16:0) was one of the lysoGPs that was increased by PPARα activation in the plasma. Previous studies have demonstrated that LPC(16:0) is a major component of lysoGPs in plasma and activates glucose uptake in adipose tissue (31, 37). Therefore, we focused here on LPC(16:0) and investigated the relationship between LPC(16:0) and PPARα activation.
Liver tissue is important in lipid metabolism. Although the lipidomics approach revealed in detail the hepatic lipid profiling including that of TGs, phospholipids, and cholesteryl esters (21, 38), little is known about the relationship between PPARα activation and LPC(16:0) production. In this study, we demonstrated that the activation of PPARα induced the upregulation of many types of lysoGP including LPC(16:0) production. Quantitative analysis of LPC(16:0) showed that LPC(16:0) was markedly increased in the liver, which expresses PPARα to a great extent. In the murine primary hepatocytes, secretion of LPC(16:0) was induced by PPARα agonist (bezafibrate, fenofibrate, and GW7647) treatment and suppressed by PPARα antagonist. These findings suggest that synthesis of LPC(16:0) in hepatocytes is induced by PPARα activation and that secretion of LPC(16:0) from hepatocytes contributes to elevated plasma LPC(16:0) concentration. Our metabolomic data also showed that not only LPC(16:0) but also many other lysoGPs were changed by bezafibrate treatment. In particular, lysophosphatidylethanolamine (LPE) (20:4), LPC(18:2), LPC(20:4), LPE(16:0), LPE(18:1), and LPC(18:1) were changed both plasma and liver. These findings suggest that the source of these lysoGPs in plasma is liver.
LPCs are mainly generated from PLA2-catalyzed hydrolysis of phosphatidylcholine (37, 39). The various subtypes of PLA2, which have different localizations, individually regulate synthesis of LPC (37). In the present study, we showed that the mRNA expression of Pla2g7 in murine liver or primary hepatocytes was elevated by PPARα activation. Furthermore, we also showed that LPC(16:0) secretion in hepatocytes was attenuated by Pla2g7 siRNA treatment. We demonstrated for the first time that LPC(16:0) is induced by PPARα activation via Pla2g7-dependent pathway in the liver. Interestingly, the present study revealed that the expression of PPARα target genes was suppressed by Pla2g7 siRNA treatment and that LPC(16:0) is not only induced by PPARα activation but also has the ability to activate PPARα and upregulates the mRNA expression of PPARα target genes. These findings raise the possibility of positive feedback regulation of PPARα and Pla2g7. The analysis of LPC(16:0) concentration in the medium and cell-associated LPC(16:0) at 24 h after addition of LPC(16:0) suggested that part of LPC(16:0) entry into cells. However, there is also the possibility that LPC(16:0) is metabolized in cells. These findings raise the possibility that LPC(16:0) activates PPARα both directly and indirectly.
Adipose tissue plays a key role in glucose homeostasis (40). A preceding study has indicated that LPC(16:0) stimulates glucose uptake in 3T3-L1 adipocytes (31). It is well recognized that obesity is characterized by chronic inflammation of adipose tissues and this inflammation contributes to the development of insulin resistance and type 2 diabetes (41, 42). Notably, macrophages contribute significantly to inflammation in adipose tissue (43). TNF-α is the major proinflammatory cytokine (44). Previous studies have reported that TNF-α, which is secreted by macrophages, induces additional infiltration of macrophages into adipose tissues (44–47). LPC(16:0) can increase glucose uptake in an insulin-independent manner (31). We hypothesize that LPC(16:0) has an ability to promote glucose uptake under conditions of insulin resistance. We revealed that LPC(16:0) has the ability to improve glucose uptake in insulin-resistant adipocytes induced by RAW-CM or TNF-α. We demonstrated for the first time that the ability of LPC(16:0) to induce glucose uptake is unaffected by insulin resistance. This effect raises the possibility that LPC(16:0) contributes to ameliorating hyperglycemia. Many previous studies indicate that the levels of LPCs and inflammation have crucial relevance. LPCs have been demonstrated to have both proinflammatory (48, 49) and anti-inflammatory (35, 50) effects. Therefore, additional work is required to completely determine the relationship between LPCs and inflammation.
It is interesting to note that fibrates, including bezafibrate, contribute to the improvement of not only dyslipidemia but also glucose metabolism disorder (18–20). However, the molecular basis of this effect remains unexplained. Excessive free fatty acids, particularly stearic acid (SA) and palmitic acid (PA), are also proinflammatory factors and induce insulin resistance (51, 52). Bezafibrate greatly facilitates lipid metabolism, thereby anticipating the reduction of SA or PA. However, our previous study showed that bezafibrate has no apparent effect on plasma SA and PA (53). The evidence suggested that SA or PA were not involved in the improvement of hyperglycemia in KK-Ay mice treated with bezafibrate. We hypothesize that the ability of LPC(16:0) to increase glucose uptake in adipocytes improves hyperglycemia during treatment with bezafibrate. To validate this hypothesis, we analyzed a correlation between plasma LPC(16:0) and plasma glucose in mice treated with or without bezafibrate for 4 weeks. The result showed strong correlation between plasma glucose and LPC(16:0). The previous study has provided evidence that plasma LPC levels are reduced in obesity and type 2 diabetes (54). Recently, administration of LPC(16:0) has been reported to reduce plasma glucose (31). We also confirmed that intraperitoneal injection of LPC(16:0) in mice induced reduction of plasma glucose (data not shown). Furthermore, the previous studies showed that plasma glucose level is reduced by fenofibrate (55) or Wy-14,643 [PPARα specific activator, (56)] treatment. The results of the previous studies support our findings. These data suggested that LPC(16:0) produced in liver by PPARα activation participates in the regulation of plasma glucose level.
In conclusion, metabolomics has revealed the upregulated LPC(16:0) in mice plasma and liver following bezafibrate treatment. PPARα activation induces the expression of Pla2g7 and secretion of LPC(16:0) from the hepatocytes. Furthermore, LPC(16:0) contributes to activate PPARα and induces the expression of PPARα target genes in hepatocytes. We also showed that LPC(16:0) has the ability to recover glucose uptake in insulin-resistant adipocytes. The data presented herein suggested that LPC(16:0) induced by PPARα activation improved dyslipidemia and hyperglycemia.
Note added in proof
The authors Rieko Nakata and Hiroyasu Inoue were inadvertently left out of the author list of the accepted version of this article. All other authors and the Journal’s Editors-in-Chief approved the addition after the article was in proof stage. Drs. Nakata and Inoue will appear as authors in all forms of the article except in the originally accepted Paper in Press.
Supplementary Material
Acknowledgments
The authors thank S. Shinoto and M. Sakai for secretarial and technical support, respectively.
Footnotes
Abbreviations:
- Aco
- acyl-CoA oxidase
- Cpt1
- carnitine-O-palmitoyltransferase 1
- HFD
- high-fat diet
- LPC
- lysophosphatidylcholine
- LPC(16:0)
- 1-palmitoyl lysophosphatidylcholine
- LPE
- lysophosphatidylethanolamine
- PA
- palmitic acid
- PLA2
- phospholipase A2
- SA
- stearic acid
- SD
- standard diet
This work was supported in part by Research and Development Projects for Application in Promoting New Policies Agriculture, Forestry, and Fisheries of Japan; by grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (22228001 and 24688015); and by Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists (24521).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and two figures.
REFERENCES
- 1.Reilly M. P., Rader D. J. 2003. The metabolic syndrome: more than the sum of its parts? Circulation. 108: 1546–1551. [DOI] [PubMed] [Google Scholar]
- 2.Miranda P. J., DeFronzo R. A., Califf R. M., Guyton J. R. 2005. Metabolic syndrome: definition, pathophysiology, and mechanism. Am. Heart J. 149: 33–45. [DOI] [PubMed] [Google Scholar]
- 3.Kahn B. B., Flier J. S. 2000. Obesity and insulin resistance. J. Clin. Invest. 106: 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ginsberg H. N. 2000. Insulin resistance and cardiovascular disease. J. Clin. Invest. 106: 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Escher P., Wahli W. 2000. Peroxisome proliferator-activated receptors: insight into multiple cellular functions. Mutat. Res. 448: 121–138. [DOI] [PubMed] [Google Scholar]
- 6.Desvergne B., Wahli W. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20: 649–688. [DOI] [PubMed] [Google Scholar]
- 7.Chinetti G., Fruchart J. C., Staeles B. 2000. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm. Res. 49: 497–505. [DOI] [PubMed] [Google Scholar]
- 8.Duval C., Chinetti G., Trottein F., Fruchart J. C., Staeles B. 2002. The role of PPARs in atherosclerosis. Trends Mol. Med. 8: 422–430. [DOI] [PubMed] [Google Scholar]
- 9.Goldenberg I., Benderly M., Goldbourt U. 2008. Update on the use of fibrates: focus on bezafibrate. Vasc. Health Risk Manag. 4: 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernal-Mizrachi C., Weng S., Feng C., Finck B. N., Knutsen R. H., Leone T. C., Coleman T., Mecham R. P., Kelly D. P., Semenkovich C. F. 2003. Dexamethasone induction of hypertension and diabetes is PPARα dependent in LDL receptor-null mice. Nat. Med. 9: 1069–1075. [DOI] [PubMed] [Google Scholar]
- 11.Reddy J. K., Hashimoto T. 2001. Peroxisomal β-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21: 193–230. [DOI] [PubMed] [Google Scholar]
- 12.Tugwood J. D., Issemann I., Anderson R. G., Bundell K. R., McPheat W. L., Green S. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11: 433–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roepstorff C., Halberg N., Hillig T., Saha A. K., Ruderman N. B., Wojtaszewski J. F., Richter E. A., Kiens B. 2005. Malonyl-CoA and carnitine in regulation of fat oxidation in human skeletal muscle during exercise. Am. J. Physiol. Endocrinol. Metab. 288: E133–E142. [DOI] [PubMed] [Google Scholar]
- 14.Miller C. W., Ntambi J. M. 1996. Peroxisome proliferators induce mouse liver stearoyl-CoA desaturase 1 gene expression. Proc. Natl. Acad. Sci. USA. 93: 9443–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grygiel-Gόrniak B. 2014. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications – a review. Nutr. J. 13: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim S., Shin H. J., Kim S. Y., Kim J. H., Lee Y. S., Kim D. H., Lee M. O. 2004. Genistein enhances expression of genes involved in fatty acid catabolism through activation of PPARalpha. Mol. Cell. Endocrinol. 220: 51–58. [DOI] [PubMed] [Google Scholar]
- 17.Ericsson C. G., Hamsten A., Nilsson J., Grip L., Svane B., de Faire U. 1996. Angiographic assessment of effects of bezafibrate on progression of coronary artery disease in young male postinfarction patients. Lancet. 347: 849–853. [DOI] [PubMed] [Google Scholar]
- 18.Tenenbaum A., Fisman E. Z., Boyko V., Benderly M., Tanne D., Haim M., Matas Z., Motro M., Behar S. 2006. Attenuation of progression of insulin resistance in patients with coronary artery disease by bezafibrate. Arch. Intern. Med. 166: 737–741. [DOI] [PubMed] [Google Scholar]
- 19.Tenenbaum A., Motro M., Fisman E. Z., Schwammenthal E., Adler Y., Goldenberg I., Leor J., Boyko V., Mandelzweig L., Behar S. 2004. Peroxisome proliferator-activated receptor ligand bezafibrate for prevention of type 2 diabetes mellitus in patients with coronary artery disease. Circulation. 109: 2197–2202. [DOI] [PubMed] [Google Scholar]
- 20.Tenenbaum A., Motro M., Fisman E. Z., Tanne D., Boyko V., Behar S. 2005. Bezafibrate for the secondary prevention of myocardial infarction in patients with metabolic syndrome. Arch. Intern. Med. 165: 1154–1160. [DOI] [PubMed] [Google Scholar]
- 21.Hu C., Hoene M., Zhao X., Häring H. U., Schleicher E., Lehmann R., Han X., Xu G., Weigert C. 2010. Lipidomics analysis reveals efficient storage of hepatic triacylglycerides enriched in unsaturated fatty acids after one bout of exercise in mice. PLoS ONE. 5: e13318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iijima Y., Nakamura Y., Ogata Y., Tanaka K., Sakurai N., Suda K., Suzuki T., Suzuki H., Okazaki K., Kitayama M., et al. 2008. Metabolite annotations based on the integration of mass spectral information. Plant J. 54: 949–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim Y. I., Hirai S., Takahashi H., Goto T., Ohyane C., Tsugane T., Konishi C., Fujii T., Inai S., Iijima Y., et al. 2011. 9-oxo-10(E),12(E)-octadecadienoic acid derived from tomato is a potent PPAR α agonist to decrease triglyceride accumulation in mouse primary hepatocytes. Mol. Nutr. Food Res. 55: 585–593. [DOI] [PubMed] [Google Scholar]
- 24.Goto T., Nagai H., Egawa K., Kim Y-I., Kato S., Taimatsu A., Sakamoto T., Ebisu S., Hohsaka T., Miyagawa H., et al. 2011. Farnesyl pyrophosphate regulates adipocyte functions as an endogenous PPARγ agonist. Biochem. J. 438: 111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi N., Kawada T., Goto T., Yamamoto T., Taimatsu A., Matsui N., Kimura K., Saito M., Hosokawa M., Miyashita K., et al. 2002. Dual action of isoprenols from herbal medicines on both PPARγ and PPARα in 3T3–L1 adipocytes and HepG2 hepatocytes. FEBS Lett. 514: 315–322. [DOI] [PubMed] [Google Scholar]
- 26.Goto T., Takahashi N., Kato S., Egawa K., Ebisu S., Moriyama T., Fushiki T., Kawada T. 2005. Phytol directly activates peroxisome proliferator-activated receptor α (PPARα) and regulates gene expression involved in lipid metabolism in PPARα-expressing HepG2 hepatocytes. Biochem. Biophys. Res. Commun. 337: 440–445. [DOI] [PubMed] [Google Scholar]
- 27.Chen X. H., Zhao Y. P., Xue M., Ji C. B., Gao C. L., Zhu J. G., Qin D. N., Kou C. Z., Qin X. H., Tong M. L., et al. 2010. TNF-alpha induces mitochondrial dysfunction in 3T3–L1 adipocytes. Mol. Cell. Endocrinol. 328: 63–69. [DOI] [PubMed] [Google Scholar]
- 28.Permana P. A., Menge C., Reaven P. D. 2006. Macrophage-secreted factors induce adipocyte inflammation and insulin resistance. Biochem. Biophys. Res. Commun. 341: 507–514. [DOI] [PubMed] [Google Scholar]
- 29.Goto T., Lee J. Y., Teraminami A., Kim Y-I., Hirai S., Uemura T., Inoue H., Takahashi N., Kawada T. 2011. Activation of peroxisome proliferator-activated receptor-alpha stimulates both differentiation and fatty acid oxidation in adipocytes. J. Lipid Res. 52: 873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi N., Kawada T., Goto T., Kim C. S., Taimatsu A., Egawa K., Yamamoto T., Jisaka M., Nishimura K., Yokota K., et al. 2003. Abietic acid activates peroxisome proliferator-activated receptor-γ (PPARγ) in RAW264.7 macrophages and 3T3–L1 adipocytes to regulate gene expression involved in inflammation and lipid metabolism. FEBS Lett. 550: 190–194. [DOI] [PubMed] [Google Scholar]
- 31.Yea K., Kim J., Yoon J. H., Kwon T., Kim J. H., Lee B. D., Lee H., Lee S. J., Kim J., Lee T. G., et al. 2009. Lysophosphatidylcholine activates adipocyte glucose uptake and lowers blood glucose levels in murine models of diabetes. J. Biol. Chem. 284: 33833–33840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X., Peter A., Fritsche J., Elcnerova M., Fritsche A., Häring H. U., Schleicher E. D., Xu G., Lehmann R. 2009. Changes of the plasma metabolome during an oral glucose tolerance test: is there more than glucose to look at? Am. J. Physiol. Endocrinol. Metab. 296: E384–E393. [DOI] [PubMed] [Google Scholar]
- 33.Birgbauer E., Chun J. 2006. New developments in the biological functions of lysophospholipids. Cell. Mol. Life Sci. 63: 2695–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y., Fang X. J., Casey G., Mills G. B. 1995. Lysophospholipids activate ovarian and breast cancer cells. Biochem. J. 309: 933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenblat M., Oren R., Aviram M. 2006. Lysophosphatidylcholine (LPC) attenuates macrophage-mediated oxidation of LDL. Biochem. Biophys. Res. Commun. 344: 1271–1277. [DOI] [PubMed] [Google Scholar]
- 36.Murugesan G., Sandhya Rani M. R., Gerber C. E., Mukhopadhyay C., Ransohoff R. M., Chisolm G. M., Kottke-Marchant K. 2003. Lysophosphatidylcholine regulates human microvascular endothelial cell expression of chemokines. J. Mol. Cell. Cardiol. 35: 1375–1384. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz G., Ruebsaamen K. 2010. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 208: 10–18. [DOI] [PubMed] [Google Scholar]
- 38.Balogun K. A., Albert C. J., Ford D. A., Brown R. J., Cheema S. K. 2013. Dietary omega-3 polyunsaturated fatty acids after the fatty acid composition of hepatic and plasma bioactive lipids in C57BL/6 mice: a lipidomic approach. PLoS ONE. 8: e82399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan W., Jenkins C. M., Han X., Mancuso D. J., Sims H. F., Yang K., Gross R. W. 2005. The highly selective production of 2-arachidonoyl lysophosphatidylcholine catalyzed by purified calcium-independent phospholipase A2γ. J. Biol. Chem. 280: 26669–26679. [DOI] [PubMed] [Google Scholar]
- 40.Hespel P., Vergauwen L., Vandenberghe K., Richter E. A. 1996. Significance of insulin for glucose metabolism in skeletal muscle during contractions. Diabetes. 45: S99–S104. [DOI] [PubMed] [Google Scholar]
- 41.Fernández-Real J. M., Ricart W. 2003. Insulin resistance and chronic cardiovascular inflammatory syndrome. Endocr. Rev. 24: 278–301. [DOI] [PubMed] [Google Scholar]
- 42.Dandona P., Aljada A., Bandyopadhyay A. 2004. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 25: 4–7. [DOI] [PubMed] [Google Scholar]
- 43.Wellen K. E., Hotamisligil G. S. 2003. Obesity-induced inflammatory changes in adipose tissue. J. Clin. Invest. 112: 1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hotamisligil G. S., Shargill N. S., Spiegelman B. M. 1993. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 259: 87–91. [DOI] [PubMed] [Google Scholar]
- 45.Hotamisligil G. S., Murray D. L., Choy L. N., Spiegelman B. M. 1994. Tumor necrosis factor alpha inhibits signaling from the insulin receptor. Proc. Natl. Acad. Sci. USA. 91: 4854–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hotamisligil G. S., Arner P., Caro J. F., Atkinson R. L., Spiegelman B. M. 1995. Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J. Clin. Invest. 95: 2409–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saghizadeh M., Ong J. M., Garvey W. T., Henry R. R., Kern P. A. 1996. The expression of TNF α by human muscle. Relationship to insulin resistance. J. Clin. Invest. 97: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parks B. W., Gambill G. P., Lusis A. J., Kabarowski J. H. 2005. Loss of G2A promotes macrophage accumulation in atherosclerotic lesions of low density lipoprotein receptor-deficient mice. J. Lipid Res. 46: 1405–1415. [DOI] [PubMed] [Google Scholar]
- 49.Huang F., Subbaiah P. V., Holian O., Zhang J., Johnson A., Gertzberg N., Lum H. 2005. Lysophosphatidylcholine increases endothelial permeability: role of PKCα and RhoA cross talk. Am. J. Physiol. Lung Cell. Mol. Physiol. 289: L176–L185. [DOI] [PubMed] [Google Scholar]
- 50.Yamamoto M., Hara H., Adachi T. 2002. The expression of extracellular-superoxide dismutase is increased by lysophosphatidylcholine in human monocytic U937 cells. Atherosclerosis. 163: 223–228. [DOI] [PubMed] [Google Scholar]
- 51.Suganami T., Tanimoto-Koyama K., Nishida J., Itoh M., Yuan X., Mizuarai S., Kotani H., Yamaoka S., Miyake K., Aoe S., et al. 2007. Role of the Toll-like receptor 4/NF-kappaB pathway in saturated fatty acid-induced inflammatory changes in the interaction between adipocytes and macrophages. Arterioscler. Thromb. Vasc. Biol. 27: 84–91. [DOI] [PubMed] [Google Scholar]
- 52.Ohto U., Fukase K., Miyake K., Satow Y. 2007. Crystal structures of human MD-2 and its complex with antiendotoxic lipid Iva. Science. 316: 1632–1634. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi H., Suzuki H., Suda K., Yamazaki Y., Takino A., Kim Y-I., Goto T., Iijima Y., Aoki K., Shibata D., et al. 2013. Long-chain free fatty acid profiling analysis by liquid chromatography-mass spectrometry in mouse treated with peroxisome proliferator-activated receptor α agonist. Biosci. Biotechnol. Biochem. 77: 2288–2293. [DOI] [PubMed] [Google Scholar]
- 54.Barber M. N., Risis S., Yang C., Meikle P. J., Staples M., Febbraio M. A., Bruce C. R. 2012. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE. 7: e41456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srivastava R. A. 2009. Fenofibrate ameliorates diabetic and dyslipidemic profiles in KKAy mice partly via down-regulation of 11beta-HSD1, PEPCK and DGAT2. Comparison of PPARalpha, PPARgamma, and liver x receptor agonists. Eur. J. Pharmacol. 607: 258–263. [DOI] [PubMed] [Google Scholar]
- 56.Tsuchida A., Yamauchi T., Takekawa S., Hada Y., Ito Y., Maki T., Kadowaki T. 2005. Peroxisome proliferator-activated receptor (PPAR)alpha activation increases adiponectin receptors and reduces obesity-related inflammation in adipose tissue: comparison of activation of PPARalpha, PPARgamma, and their combination. Diabetes. 54: 3358–3370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.