Abstract
There is a current interest in reducing the in vivo toxicity testing of nanomaterials in animals by increasing toxicity testing using in vitro cellular assays; however, toxicological results are seldom concordant between in vivo and in vitro models. This study compared global multi-walled carbon nanotube (MWCNT)-induced gene expression from human lung epithelial and microvascular endothelial cells in monoculture and coculture with gene expression from mouse lungs exposed to MWCNT. Using a cutoff of 10% false discovery rate and 1.5 fold change, we determined that there were more concordant genes (gene expression both up- or downregulated in vivo and in vitro) expressed in both cell types in coculture than in monoculture. When reduced to only those genes involved in inflammation and fibrosis, known outcomes of in vivo MWCNT exposure, there were more disease-related concordant genes expressed in coculture than monoculture. Additionally, different cellular signaling pathways are activated in response to MWCNT dependent upon culturing conditions. As coculture gene expression better correlated with in vivo gene expression, we suggest that cellular cocultures may offer enhanced in vitro models for nanoparticle risk assessment and the reduction of in vivo toxicological testing.
Keywords: Coculture, gene expression, in vivo, in vitro, correlation
1. Introduction
The current interest in reducing the in vivo toxicity testing of nanomaterials in animals by increasing toxicity testing using in vitro cellular assays has led to the observation that in vivo and in vitro nanomaterial toxicity results are seldom concordant (Rothen-Rutishauser et al. 2008; Sayes et al. 2007a; Sayes et al. 2007b; Seagrave et al. 2005). Several thorough reviews have recently highlighted the current state of knowledge concerning in vivo and in vitro nanoparticle toxicological testing and suggest that more advanced in vitro procedures are necessary to accurately representin vivo results in vitro(Klein et al. 2011; Snyder-Talkington et al. 2012).
Multi-walled carbon nanotubes (MWCNT) are nanomaterials consisting of concentric layers of cylindrical carbon tubes with a diameter less than 100 nm, light weight, extreme strength, electronic conductivity, and strong capillary forces that are being explored for numerous commercial applications, such as plastics, microelectronics, sporting goods, and resins, among many others (De Volder et al. 2013). Because of their light weight, MWCNT are easily aerosolized, and pulmonary exposure during production, use, and disposal is a major concern for the growing industry (Castranova 2011). A recent study concerning the potential toxicity of MWCNT showed that, although pathway-level cellular functions were similar between in vivo and in vitro experiments, microarray analysis determined different gene expression profiles between in vivo and in vitro models (Sos Poulsen et al. 2013). MWCNT induce numerous deleterious effects in mouse lungs after both aspiration and inhalation exposures (Mercer et al. 2011; Mercer et al. 2013; Porter et al. 2013; Porter et al. 2010; Sargent et al. 2014; Siegrist et al. 2014), and the ability to reflect these effects in vitro with an accurate transcriptomic profile remains difficult. Although in vitro global gene expression and microarray analysis may be useful for providing a generic overview of responses, cellular signaling from chronic exposure in vivo and acute signaling in vitro may produce different expression results (Klaper et al. 2014). As the majority of MWCNT in vitro toxicity testing is conducted in monoculture of a single cell type, the cellular communication and interactions inherent to the in vivo environment are lost. Several in vitro assay systems, such as coculture models and air-liquid interface, attempt to recapitulate the cellular communication and characteristics of the in vivo environment (Clift et al. 2014; Snyder-Talkington et al. 2013a). However, to the best of our knowledge, there are no transcriptomic comparisons of these advanced in vitro cellular models with in vivo gene expression.
This study was conducted to determine the overall gene expression correlation and concordance between in vitro monoculture or coculture of human small airway epithelial cells (SAEC) and human microvascular endothelial cells (HMVEC) and in vivo mouse lung gene expression following MWCNT exposure. SAEC and HMVEC were grown separately in monoculture or together in coculture, and total RNA was collected for microarray analysis and comparison with previously identified global gene expression from mouse lungs exposed to MWCNT (Guo et al. 2012). Through this global mRNA profiling and genome-wide correlation study, we determined that in vitro SAEC and HMVEC coculture mRNA expression after MWCNT exposure better correlated with in vivo mRNA expression than did monoculture mRNA expression. Therefore, in vitro cocultures may better mimic the in vivo signaling environment and provide enhanced in vitro methods that would reduce the need for in vivo toxicological testing. Those genes determined in vitro to be concordant with those expressed in vivo may additionally serve as potential biomarkers that can then be studied in vitro to determine potential biomarkers and mechanisms of MWCNT-induced human lung diseases.
2. Materials and Methods
2.1 MWCNT
Bulk MWCNT: The MWCNT used in this study were obtained from Mitsui & Company (MWCNT-7, lot #05072001K28) and have been previously characterized (Porter et al. 2010). Briefly, the bulk MWCNT exhibited a distinctive crystalline structure, with the number of walls ranging from 20 to 50 as determined by high resolution transmission electron microscopy. Overall, MWCNT trace metal contamination was 0.78%, including sodium (0.41%) and iron (0.32%), with no other trace metal contamination over 0.02%. Endotoxin contamination was below the level of detection. A survey-scan spectrum obtained by X-ray photoelectron spectroscopy measurement had a dominant C 1s peak (284.6 eV), with a small amount of oxygen and no other elements detected.
MWCNT in DM: Transmission electron microscopy (TEM) micrographs of MWCNT dispersed in DM demonstrated significant dispersion of MWCNT in DM. Quantitative analysis of the TEM micrographs determined the median length of the MWCNT sample to be 3.86 µm (GSD 1.94) and the count mean width to be 49 ± 13.4 (SD) nm. The zeta potential of the MWCNT in DM was −11 mV. The MWCNT used in this study were analyzed by electron spin resonance (ESR) and were found to decrease the ESR signal when added to the ESR reaction mixture, suggesting that MWCNT could scavenge •OH produced by the Fenton reaction. Additionally, when the MWCNT were substituted for the Fe in the Fenton reaction, no •OH was detected, suggesting that the iron present in the MWCNT was not capable of inducing reactive oxygen species (Porter et al. 2010).
2.2 In vitro MWCNT preparation
For cell culture exposures, MWCNT were prepared in DM. Transmission electron micrographs of MWCNT dispersed in DM determined that DM promoted significant dispersion of MWCNT. MWCNT were prepared in DM, followed by indirect sonication at 4°C for 5 min (Hielscher ultrasonic processor, UIS259L) at amplitude 100% and cycle 1. Following indirect sonication, the suspension was directly sonicated for 5 min at 5W output and 10% duty cycle (Branson Sonifier 450). The stock solution (0.5 mg/mL) of MWCNT was kept at 4°C and used within 2–3 weeks. Prior to cell culture experiments, the MWCNT stock solution was directly sonicated for 1 min at 5W output and 10% duty cycle.
2.3 Cell culture
SAEC were a kind gift from Dr. Tom K. Hei (Columbia University, New York, NY)(Piao et al. 2005). SAEC were cultured in serum free complete SAGM medium supplemented with various growth factors supplied by the manufacturer (Lonza Walkersville, Inc., Walkersville, MD). HMVEC were a kind gift from Dr. Rong Shao (Biomedical Research Institute, Baystate Medical Center/University of Massachusetts, Amherst, Springfield, MA)(Shao and Guo 2004). HMVEC were cultured in endothelial basal medium-2 (EBM-2) (Lonza) and supplemented with 10% fetal bovine serum (Atlanta Biological, Lawrenceville, GA), 100 U/mL penicillin and 10 µg/mL streptomycin (Lonza), 0.01 µg/mL epidermal growth factor (Sigma), and 1 µg/mL hydrocortisone (Sigma). All cells were maintained in an incubator at 37°C with 5% CO2.
To prepare cocultures, inserts were removed from 100 mm polycarbonate Transwell (Corning, Tewksbury, MA) dishes and hydrated in SAEC complete media in a companion 100 mm dish for at least 1 hour. HMVEC were plated at 1,000,000 cells in the bottom of each Transwell dish (growth area: 55 cm2) and allowed to adhere for at least 1 h without the apical chamber insert. Inserts were returned to the Transwell after hydration, and 1,000,000 SAEC were plated onto the Transwell insert (growth area: 44 cm2). Cells were maintained in 15 mL of complete EBM-2 media in the basolateral chamber and 10 mL of complete SAEC media in the apical chamber (Supplemental Figure 1).
For monoculture exposures, SAEC and HMVEC were plated directly into 100 mm cell culture dishes (Corning, Tewksbury, MA; growth area: 55 cm2), allowed to form intact barriers for 72 h, serum starved overnight, and exposed directly to MWCNT at a concentration of 1.2 µg/mL in 10 mL of their respective media for either 6 or 24 h. DM for 24 h was used as a negative control. Six biological replicates of each SAEC and HMVEC monoculture exposure condition were collected for microarray analysis. Both SAEC and HMVEC in monoculture have been shown to interact with MWCNT (Pacurari et al. 2012; Snyder-Talkington et al. 2013b).
For coculture exposures, SAEC and HMVEC were allowed to form intact epithelial and endothelial barriers for 72 h, serum starved overnight, and SAEC were exposed to MWCNT at a concentration of 1.2 µg/mL in 10 mL of SAEC media for either 6 or 24 h. DM for 24 h was used as a negative control. HMVEC in the coculture system were not directly exposed to MWCNT, and MWCNT are not apparent in HMVEC transmission electron microscopy preparations following SAEC exposure in coculture (Snyder-Talkington et al. 2013a).Six biological replicates of each SAEC and HMVEC coculture at each exposure condition were collected for microarray analysis. The total number of monoculture and coculture samples for microarray analysis was 72.
Images of cell morphology and confluency of SAEC and HMVEC grown in monoculture and coculture are shown in Supplemental Figure 2. SAEC and HMVEC were grown in monoculture in their respective mediums for 72 hours, serum starved, and imaged at 5 days after plating. SAEC and HMVEC were also grown in coculture in their respective mediums and serum starved as described. Light microscopy was imaged on an Olympus IX70 with a Retiga 2000R camera (QImaging, Surrey, British Columbia, Canada) at 10× magnification (Supplemental Figure 2A). Additionally, after imaging, cells were scraped from their dishes/membranes, counted, and 50,000 cells were used for cytospin preparations using a centrifuge (Shandon Elliot Cytocentrifuge, London). Cytospin preparations were imaged on an Olympus IX70 with a Retiga 2000R camera at 10× magnification (Supplemental Figure 2B). There were no significant differences in cell number (data not shown) between the monoculture and coculture conditions, and cells did not show any overt changes in confluency or morphology.
To accurately compare in vitro results with in vivo data, MWCNT mass concentrations/surface area of cells which mimic MWCNT mass burdens/alveolar surface area in animal studies must be used. The dose of 1.2 µg/mL MWCNT was chosen based upon in vivo alveolar surface area and occupationally observed peak airborne MWCNT concentrations. Peak MWCNT levels in an occupational setting were determined to be approximately 400 µg/m3 (Han et al. 2008). With regard to the average amount of human airway epithelial surface area, the amount of human exposure at 400 µg/m3 is 226 µg MWCNT per m2 of human alveolar epithelium (Porter et al. 2010). Taking into consideration MWCNT mass median aerodynamic diameter, minute ventilation, and human alveolar epithelial surface area, Porter, et al. (2010) determined that doses of 10, 20, 40 and 80 µg of MWCNT during in vivo mouse studies could accurately reflect increasing durations of human exposure to MWCNT. When extrapolated for use during in vitro studies, 1.2 µg/ml MWCNT was suggested to compare to the 40 and 80 µg in vivo doses of MWCNT which were shown to produce chronic inflammation and persistent fibrosis in a mouse model (Mercer et al. 2011; Porter et al. 2010). Total MWCNT exposure of 1.2 µg/mL was approximately equal to 0.25 µg/cm2.
2.4 Cellular RNA isolation
Total RNA was isolated from SAEC and HMVEC using RNAprotect Cell Reagent and an RNeasy Mini Kit from Qiagen according to the manufacturer’s protocol (Qiagen, Valencia, CA). RNA concentrations were determined using a NanoDrop 1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA).
2.5 Animals
The animal study described here has been previously published (Porter et al. 2010). Male C57BL/6J mice (7 weeks old) were obtained from Jackson Laboratories (Bar Harbor, ME). Individual mice were housed 1 per cage in polycarbonate isolator ventilated cages and provided HEPA-filtered air with fluorescent lighting from 0700 to 1900 hours. Autoclaved Alpha-Dri virgin cellulose chips and hardwood Beta-chips were used as bedding. Mice were monitored to be free of endogenous viral pathogens, parasites, mycoplasms, Helicobacter, and CAR Bacillus. Mice were maintained on Harlan Teklad Rodent Diet 7913 (Indianapolis, IN), and tap water was provided ad libitum. Animals were allowed to acclimate for at least 5 days before use. All animals in this study were housed in an AAALAC-accredited, specific pathogen-free, and environmentally controlled facility. All animal studies and procedures were approved by the National Institute for Occupational Safety and Health ACUC.
2.6 MWCNT pharyngeal aspiration exposure
Animals were exposed to MWCNT in DM as previously reported (Porter et al. 2010).Mice (8 per group) were exposed to DM, 10, 20, 40, or 80 µg MWCNT for each 1, 7, 28 or 56 days postexposure period (total n = 160). Mice were anesthetized with isoflurane (Abbott Laboratories, North Chicago, IL), positioned with their back against a slant board, and suspended by their incisor teeth using a rubber band. The mouth was opened and the tongue gently pulled aside from the oral cavity. A 50 µl aliquot of sample was pipetted at the base of the tongue, and the tongue was restrained until at least 2 deep breaths were completed (not for longer than 15 sec). Following release of the tongue, the mouse was gently lifted off the board, placed on its left side, and monitored for recovery from anesthesia.
At 1, 7, 28, and 56 days post-exposure, mice were euthanized by an intraperitoneal injection of sodium pentobarbital (>100 mg/kg body weight). Deep anesthesia was confirmed when the mouse no longer responded to a toe pinch. Transection of the abdominal aorta was completed to provide exsanguination. Lungs were rapidly removed, placed into RNAlater, and frozen at −80°C for future use.
2.7 Tissue RNA Extraction
Total RNA was extracted from 160 frozen mouse whole lung tissue samples stored at −80°C in RNAlater using a RNeasy Fibrous Tissue Mini Kit according to manufacturer’s protocol (Qiagen, USA) as previously reported (Pacurari et al. 2011). Total RNA was eluted in RNase-free water and stored at −80°C until further analysis. The quality and concentration of each RNA sample were determined using a NanoDrop-1000 Spectrophotometer (NanoDrop Tech, Germany).
2.8 Microarray expression profiling
In vivoexpression profiling of the 160 mouse lung samples was analyzed using Agilent Mouse Whole Genome Arrays as previously reported (Guo et al. 2012).In vitro extracted RNA from the 72 cell samples was analyzed for expression profiling using Agilent G3 Human Gene Expression 8 × 60k Arrays (Agilent, Santa Clara, CA). Total RNA quality for both microarray analyses was determined on an Agilent 2100 Bioanalyzer, with all samples having RNA integrity numbers greater than 8. Total RNA (250ng) was used for labeling using a QuickAmp labeling kit (Agilent). Extracted RNA was labeled with cyanine (Cy)-3-CTP (PerkinElmer, Waltham, MA) and reference RNA with (Cy)-5-CTP. Following purification of labeled cRNAs, 825 ng of Cy3-and Cy5-labeled cRNAs were combined and hybridized for 17 h at 65°C in an Agilent hybridization oven. Microarrays were then washed and scanned using an Agilent DNA Microarray Scanner.
2.9 Sample size justification
The study of Dobbin and Simon (2005) was followed to determine the number of microarrays to be performed for each experiment, and a reference design with a 2-color array system with no technical replicates was used. In each case, the design was a simple, case-control design with equal numbers of arrays for each sample. We used equation 4.2 from Dobbin and Simon (2005) to calculate the number of arrays required. Based on previous data, the variance for the in vitro study was estimated as 0.029, and the variance for the in vivo study was estimated as 0.25 (log base 2 expression ratio in both cases). In addition to the statistical power requirements, the analytical process described in Significance Analysis of Microarray (SAM)(Tusher et al. 2001)involves creating random permutations of the class labels of the samples. In order to generate at least 500 distinct permutations, a minimum of 6 samples in each class was needed, regardless of the considerations of statistical power. For thein vitroexperiment, a total of 18 arrays (6 control, 6 6 h, and 6 24h) for each treatment condition (SAEC monoculture, SAEC coculture, HMVEC monoculture, HMVEC coculture, 72 total) would achieve the required statistical power. Since this is the minimum required in order to perform SAM, we choose to use this number of arrays for the in vitro experiment. For the in vivo experiment, a total of 40 arrays (8 per each dose condition) per each time point (1, 7 28, or 56 days, 160 total) sufficed for all but the smallest numbers of true differentially expressed genes on the array.
2.10 Microarray data processing and statistical analysis to determine significant genes
In vitro microarray data were exported using Feature Extraction v10 as tab-delimited text files after background subtraction, log transformation, and lowess normalization and reported as log or relative expression of sample compared to universal reference. Data were read from each file into R using a custom script (Guo et al. 2012). For each array, values for control spots, spots which were saturated on either channel, spots which were reported by Feature Extraction as non-uniform outliers on either channel, and spots which were not well above background on at least one channel were considered unreliable and/or uninformative and replaced by “NA”. Values were collated into a single table, and probes for which fewer than 10 present values were available were removed. For probes spotted multiple times on the array, values were averaged across replicate probes. The in vivo microarray data is available in the NCBI Gene Expression Omnibus (GEO) repository with accession number GSE29042. The in vitro microarray data will be available in NCBI GEO upon publication of this study.
Missing data were imputed using the K-means nearest neighbor algorithm as implemented by the impute.knn function in the impute R package from Bioconductor(Bioconductor).For each dose and time point, a set of differentially expressed genes was identified by performing a two-class unpaired Significance Analysis of Microarrays (SAM) between the treated samples and the dose zero samples from the corresponding time point, using the Bioconductor package. A threshold delta value was chosen to produce a false discovery rate of 10% using the find Delta function from the same package. The list of probes called as significant were subsequently filtered by restricting to those probes which were at least 1.5 fold up- or downregulated (fold changes were computed from the data before imputation of missing values).
2.11 Gene pairing between human and mouse and correlation of gene expression analysis
The mouse and human genomes were matched by gene name using the data mining tool Biomart based on Ensembl Genes version 68 (http://www.ensembl.org/biomart/martview/) (Kasprzyk 2011). There were a total of 15,473 matched genes (orthologs) between human and mouse. For ortholog genes defined as a “one-to-many” or “many-to-many” relationship between the mouse and human genome, a randomly selected matched gene pair was chosen to compute the genome-wide Pearson’s correlation coefficient. This process was repeated 100 times, and the average correlation was reported as the final result. The whole genome-scale mRNA profile of each in vitro cell condition was correlated with the global mRNA profile in each in vivo dose/time condition of the animal study, respectively. The correlation coefficients were tested with statistical hypothesis testing and adjusted with multiple hypothesis testing, with a P< 0.05 defined as being statistically significant.
2.12 Ingenuity Pathway Analysis (IPA)
Data were analyzed through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Significant concordant genes identified at both 6 and 24 h were combined, and duplicates were only counted once. SAEC and HMVEC monoculture and coculture Core Analyses were compared using the Comparison Analyses tool. The p-value determined by IPA is a measure of significance based upon the number of genes/molecules that map to a biological function, pathway, or network.
3. Results
3.1 Correlation of in vivo and in vitro mRNA expression
Six biological replicates each of SAEC in monoculture, HMVEC in monoculture, and SAEC and HMVEC grown together in coculture were exposed to either DM for 24 h or 1.2 µg/mL MWCNT for 6 or 24 h (total n = 72). Global mRNA profiling was conducted by microarray and compared to global mRNA profiling in 8 biological replicates of mouse lungs each exposed to DM, 10, 20, 40, or 80 µg MWCNT by aspiration for 1, 7, 28, or 56 days (Porter et al. 2010)(total n = 160) using a genome-scale correlation study. Based upon in vivo alveolar surface area, occupationally observed MWCNT airborne concentrations, MWCNT mass median aerodynamic diameter, and minute ventilation, the 1.2 µg/mL concentration of MWCNT for in vitro exposure was considered a “high dose” to reflect the transient inflammation and chronic fibrosis seen following in vivo MWCNT exposure, although this is a relatively low dose when compared to other doses used throughout the literature (Mercer et al. 2011; Porter et al. 2010; Snyder-Talkington et al. 2012; Snyder-Talkington et al. 2013a).The correlation study was conducted by first matching the mouse and human genome by gene name using the data mining tool BioMart (Kasprzyk 2011). The whole genome expression profiles of each in vitro cell condition were next correlated with those of each in vivo treatment condition using Pearson’s correlation.
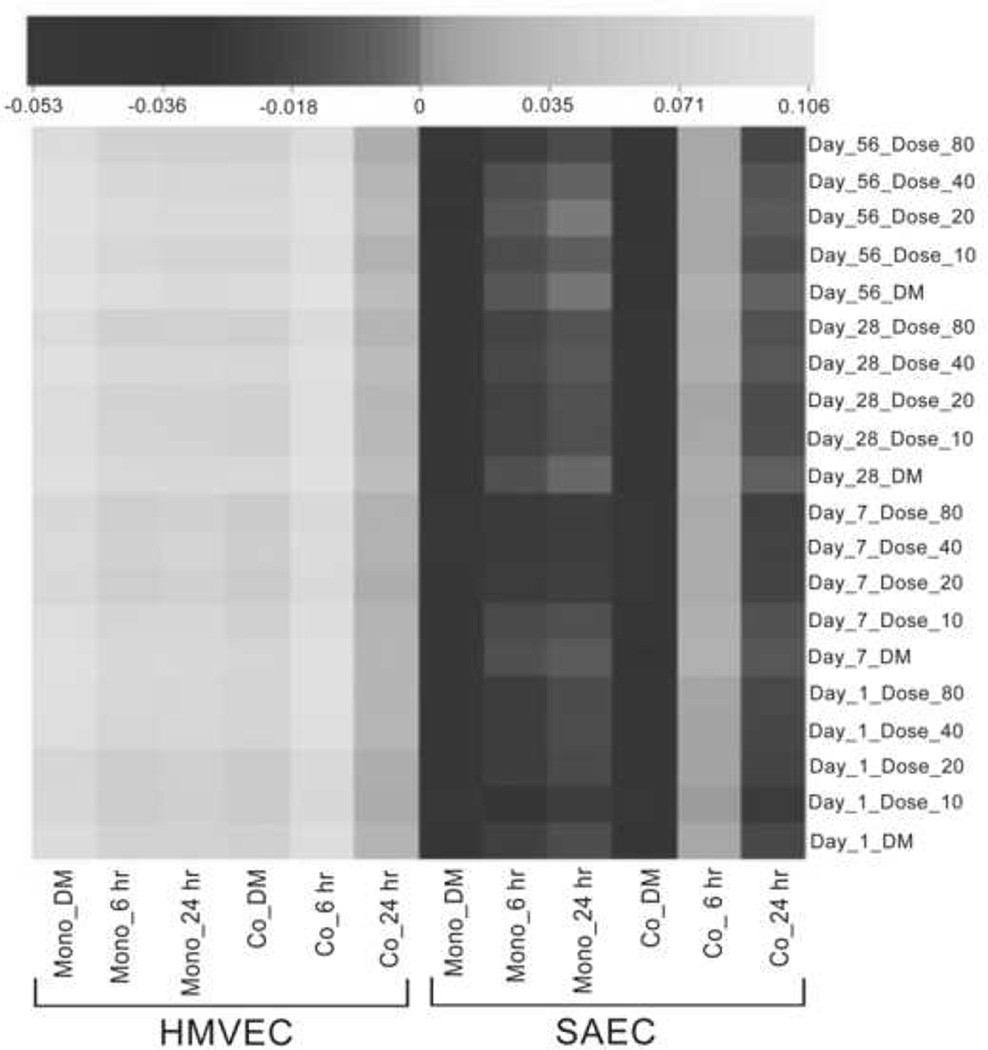
Gene expression in all HMVEC in vitro conditions, both monoculture and coculture, had a positive correlation with gene expression in vivo (Figure 1). With regard to SAEC, only the 6 h coculture condition showed a positive correlation with in vivo expression (Figure 1). The correlation coefficients and significance by single and multiple hypothesis testing are given in Supplemental Table 1. These results indicate that, at a genome-wide scale, the in vitro coculture model correlated better than the monoculture models with the in vivo transcriptomic profiles.
Figure 1.
Correlation coefficients of SAEC and HMVEC monoculture and coculture global mRNA expression with global mRNA expression from mouse lungs exposed to MWCNT. Total in vitro mRNA expression from 6 biological replicates of cell cultures exposed to DM or 1.2 µg/mL MWCNT for 6 and 24 h was compared to total mRNA expression from 8 biological replicates ofin vivo mouse lungs exposed to dispersion media, 10, 20, 40, or 80 µg MWCNT for 1, 7, 28, or 56 days. Human and mouse genomes were matched using the bioinformatics tool BioMart, and gene expression values were correlate ed using Pearson’s correlation.
3.2 Concordant genes between in vivo and in vitro conditions
Using a false discovery rate (FDR) ≤10% in SAM and fold change (FC) ≥1.5 as a cut off, the number of concordant genes (genes having the same over- or under-expression direction both in vivo and in vitro) was determined for each cell culture condition. With the exception of coculture SAEC 24 h, all coculture conditions had more significantly changed, concordant genes (FDR ≤10%, FC ≥1.5) than their monoculture counterparts (Table 1, Supplemental Table 2). These results confirmed that more concordant genes that reflected in vivo MWCNT-induced expression changes could be identified by the coculture system, potentially revealing MWCNT-induced pathogenesis and toxicity biomarkers for risk assessment.
Table 1.
Number of in vitro and in vivo concordant genes at FDR ≤10% and FC ≥1.5.
| Monoculture | Coculture | |
|---|---|---|
| SAEC 6hr | 106 | 290 |
| SAEC 24hr | 163 | 146 |
| HMVEC 6hr | 3 | 119 |
| HMVEC 24hr | 93 | 128 |
3.3 Pathway analysis of in vivo and in vitro concordant genes
To assess the different pathways that may be affected by culturing cells in monoculture or coculture, SAEC and HMVEC monoculture and coculture concordant genes were uploaded to IPA and analyzed using Core Analysis and Comparison Analyses tools. The p-value determined by IPA is a measure of significance based upon the number of genes/molecules that map to a biological function, pathway, or network.
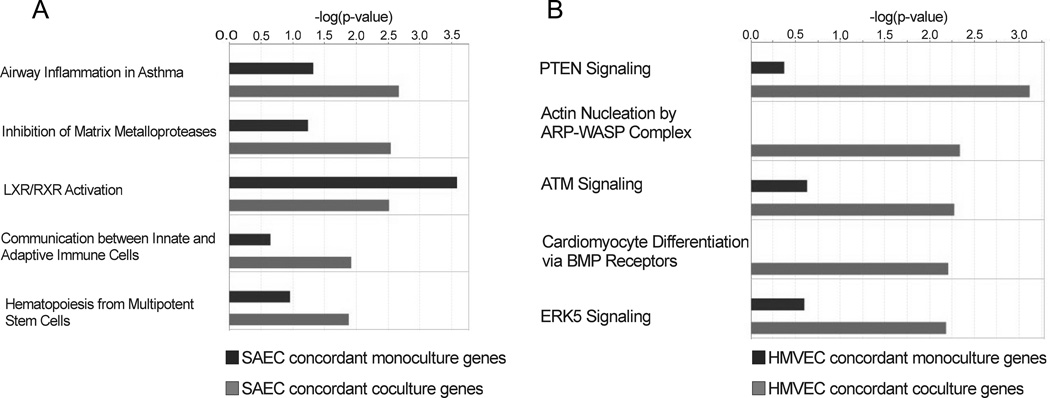
The SAEC monoculture dataset at both 6 and 24 h consisted of 203 unique genes; concordant genes that were significant at both time points were counted only once. The SAEC coculture dataset consisted of 319 unique genes. Each dataset was analyzed using a Core Analysis. The concordant SAEC monoculture and coculture core analyses were then compared using the Comparison Analyses tool. The top 5 networks related to those concordant genes significantly changed in SAEC monoculture and coculture were Airway Inflammation in Asthma, Inhibition of Matrix Metalloproteases, LXR/RXR Activation, Communication between Innate and Adaptive Immune Cells, and Hematopoiesis from Multipotent Stem Cells (Figure 2A). More SAEC coculture genes were involved in the Airway Inflammation in Asthma, Inhibition of Matrix Metalloproteases, Communication between Innate and Adaptive Immune Cells, and Hematopoiesis from Multipotent Stem Cells pathways than were SAEC monoculture genes. More SAEC monoculture genes were involved in the LXR/RXR Activation pathway than SAEC coculture genes.
Figure 2.
IPA analysis of monoculture and coculture concordant genes. All SAEC and HMVEC monoculture and coculture concordant genes were uploaded to IPA and analyzed using the Core Analysis tool. The core analyses were then compared using the Comparison Analyses tool. The top 5 pathways and p-values for SAEC (A) and HMVEC (B), based upon the number of genes represented in a given pathway, are shown.
The HMVEC monoculture dataset consisted of 93 unique genes at both 6 and 24 h, and the HMVEC coculture dataset consisted of 130 unique genes. Each dataset was analyzed using a Core Analysis, followed by comparison with the Comparison Analyses tool. The top 5 networks found in the HMVEC monoculture and coculture comparison were PTEN Signaling, Actin Nucleation by ARP-WASP Complex, ATM Signaling, Cardiomyocyte Differentiation via BMP Receptors, and ERK5 Signaling (Figure 2B).More HMVEC coculture genes were involved in each pathway than were HMVEC monoculture genes.
3.4 Disease-related concordant genes in vivo and in vitro
MWCNT induce lung inflammation and fibrosis in mice exposed by inhalation and aspiration (Porter et al. 2013; Porter et al. 2010). Using the Function and Disease Overlay function of IPA, significant concordant genes at 10% FDR in SAM that achieved a ≥1.5 FC (Table 1) and were involved in the Inflammatory Response and Fibrosis were identified. Aside from SAEC coculture 24 h, there were more concordant genes involved in both Inflammatory Response and Fibrosis disease overlays expressed in coculture than in monoculture (Table 2).
Table 2.
Concordant genes involved in fibrosis and inflammation by Ingenuity Pathway Analysis. Downregulated genes are shown in plain text. Upregulated genes are shown in italicized, bold text.
| Inflammation | Monoculture | Coculture |
|---|---|---|
| SAEC 6 h | SPHK1, IL33, SFRP1, CD9, MMP9, CFTR, INHBA, IL23A, EDNRA, EGF1, ATF3, CD47, FOXO3, HYAL1, PDE4D, RORA | SPHK1, CD38, MARCO, PNOC, CCL3, TIMD4, PRG2, ISG15, PTAFR, MMP14, INHBA, IL5, IL4, CCL24, ATF3, CD47, PDE4D, FOXO3, MSH2, RAG1, DEK, AOC3, HYAL1, PTX3, RORA, CAV1 |
| SAEC 24 h | CD9, ISG15, SFRP1, FOS, INHBA, IL1R1, IL5, EGR1, CFTR, ATF3, EDNRA,CD47, BLNK, PDE4D, HYAL1, FOXO3, RORA, IL16 | MARCO, ISG15, PTAFR, CCL3, SPHK1, MMP14, MMP9, INHBA, CD47, HYAL1, AOC3, PDE4D, DEK, CAV1 |
| HMVEC 6 h | XBP1 | SPHK1, RAG1, MSH2, DEK |
| HMVEC 24 h | CD69, XBP1, S11B, GRP, PRG2, BLNK, IL6, NR1D1, EFNB1, CXCR3 | SPHK1, RAG1, MSH2, DEK, CD47 |
| Fibrosis | Monoculture | Coculture |
| SAEC 6 h | INHBA, IL23A, EDNRA, EGR1, ATF3, CFTR, MMP9, BMP6, PLAC8, UBD, SNA1, HEY2, RORA | MMP14, INHBA, IL5, IL4, CCL24, ATF3, KCCN4, RGS16, KLK1, PTX3, RORA, CAV1, CBL, SMAD4, DAG1, PKD2 |
| SAEC 24 h | EGR1, CFTR, ATF3, EDNRA, IL5, IL1R1, INHBA, HEY2, CDK4, UBD, BMP6, HBEFG, SNAI1, IL16, RORA, MYLK3, P2RY1 | MMP14, MMP9, INHBA, KLK1, RGS16, KCCN4, CAV1 |
| HMVEC 6 h | CBL, CREB1, BMPR2, POSTN, SMAD4, PKD2 | |
| HMVEC 24 h | IL6, P2RY1, CXCR3 | CBL, CREB1, BMPR2, POSTN, SMAD4, PKD2, EGFR |
4. Discussion
This comparative mouse and human genome study following in vivo and in vitro MWCNT exposure, respectively, demonstrated that SAEC and HMVEC coculture gene expression better correlated with in vivo lung gene expression than either SAEC or HMVEC monoculture alone. This conclusion is supported by the higher overall genome-scale correlation of MWCNT -induced gene expression changes in in vitro coculture with in vivo lung exposure, as well as an increased number of concordant, significantly changed genes in the coculture model. The identified concordant genes between the coculture system and in vivo animal study could be used as potential biomarkers for risk assessment of MWCNT-induced human diseasesin vitro, particularly lung inflammation and fibrosis.
Molecular mechanisms and biomarkers of MWCNT-induced pulmonary diseases remain elusive, andin vitro information has been mainly generated from monoculture models, which do not account for cellular crosstalk between adjacent cells (Alfaro-Moreno et al. 2008; Aschberger et al. 2010; Johnston et al. 2010; Pacurari et al. 2010). The lung is composed of many different cell types, which undergo pivotal cellular communication in response to pulmonary exposures (Fehrenbach 2001; Kelly et al. 1998; Planus et al. 1999), including that following MWCNT exposure (Pacurari et al. 2010; Shvedova et al. 2009). Cytotoxicity profiling using conventional monoculturesis often markedly different from that of relevant in vivo models (Kasper et al. 2011; Rao et al. 2004). The coculture model used in this study measured MWCNT-induced gene expression changes in a representation of the alveolar-capillary unit of the lower respiratory tract, where MWCNT pose a serious point of attack (Castranova 2011; Hermanns et al. 2009; Hermanns et al. 2004; Muller et al. 2010), and represents a reasonable model for pulmonary exposure in view of the close proximity of alveolar epithelial cells and capillary endothelial cells in vivo(Balda and Matter 1998).
In the current study, human cells were surveyed in vitro, and their gene expression profiles were compared with those from mouse lungs in vivo. We showed in a previous study that genome-wide mRNA expression profiles from mouse lungs exposed to DM, 10, 20, 40, or 80 µg MWCNT were able to stratify human lung cancer patients into high and low cancer initiation and/or progression risk (Guo et al. 2012). Additionally, numerous studies have suggested that animal model-based gene expression profiling can successfully predict human toxicity for various diseases and substantiate the importance of using microarray-based gene signatures for toxicity prediction, risk assessment, and screening (Afshari et al. 2011; Aubrecht and Caba 2005; Bushel et al. 2007; Newton et al. 2004; Nuwaysir et al. 1999; Shi et al. 2010a). As these studies have shown the ability to translate animal model-based gene expression into predictive human toxicity data, we believe that this study can also relay potential human toxicity profiles through data from non-human exposures.
Traditional toxicological studies focus on observations of disease-specific models in vivo; however, there are limitations in the use of animals to reflect human responses given their differences in weight, life expectancy, and genotype (Hartung and Leist 2008). Using human cell lines during in vitro studies could bridge the gap between animal-based in vivo toxicological studies and human population risk assessments because human cell lines have similar phenotypic and genotypic characteristics to human beings (Suemori 2006). Several databases are available for comparative genomic analysis of human and mouse genomes. The Ensembl database analyzes protein-coding genes (Sanna et al. 2008), while Genome Pair View (Schneider et al. 2007) and MSOAR 2.0 (Shi et al. 2010b) can predict orthologs between multiple species by assigning orthologs based on sequence similarity/positions of genes in a genome and evolutionary history, including genome rearrangements after speciation events. These available databases and methods for classifying homologous relationships provide qualitatively similar results for comparative genomic studies (Shi et al. 2010b). A study of 13 species’ genomes combined with Gene Ontology (GO) annotation showed that orthologs tended to be more significantly similar in function than paralogs after controlling for confounding factors, such as authorship bias, variation of GO term frequency among species, variation of background similarity among specifies pairs, and propagated annotation bias (Altenhoff et al. 2012). In this study, the mouse and human genomes were matched by gene name using the data mining tool Biomart, which is based on the Ensembl Genes version 68 (Kasprzyk 2011) that focuses on orthologs. There were a total of 15,473 matched genes (one-to-one orthologs) between the human and mouse genomes. For ortholog genes defined as a “one-to-many” or “many-to-many” relationship between the mouse and human genome, a randomly selected matched gene pair was chosen to compute the genome-wide Pearson’s correlation coefficient and was averaged after 100 runs. The correlation results were then tested with statistical hypothesis testing and adjusted with multiple hypothesis testing. These rigorous analyses identified that the human coculture genomic profiles correlated with the in vivo animal profiles better than monoculture. Thus, a combined analysis of human in vitro coculture transcriptional profiles and in vivo animal profiles renders a better understanding of MWCNT-inducedin vivo human toxicity effects.
This study focused on broad, gene expression-based biological observations in vitro with the support of in vivo studies. In this case, the SAEC and HMVEC coculture model provided a better prediction of in vivo MWCNT-induced gene expression than either SAEC or HMVEC monoculture system. Through the use of IPA, it is possible to determine differences in the specific cellular signaling pathways between cells cultured in monoculture and those cultured in coculture. The top significant pathways involving concordant SAEC genes included those involved in chronic lung inflammation, the function and migration of inflammatory cells, regulators of inflammation, crosstalk between innate and adaptive immune systems, and the generation of blood cells. MWCNT and other nanomaterials are well-known for their ability to induce inflammation after in vivo pulmonary exposure. Through IPA analysis, this study showed that there are more genes involved in both overall inflammatory processes and specific inflammatory pathways expressed in SAEC coculture than in monoculture. With regard to HMVEC grown in monoculture or coculture, the top significant pathways involving concordant HMVEC genes included those involved in cellular growth and survival, activation of the actin cytoskeleton, repair of DNA damage, cardiovascular signaling, and cellular proliferation and differentiation. We have previously shown that HMVEC in coculture respond to SAEC exposure to MWCNT with increases in cell migration, cytokine signaling, and angiogenesis (Snyder-Talkington et al. 2012). In this study, we have additionally shown that more HMVEC coculture genes are related to pathways involved in cellular activation and repair than HMVEC monoculture. The increased cellular signaling between SAEC and HMVEC in coculture may allow for a better representation of signaling that would be encountered in vivo; therefore, coculture of cells may provide a better representation of the in vivosignaling environment.
In the in vitro study, SAEC and HMVEC were used to assess the effect of MWCNT exposure on global gene expression in a coculture system that attempted to mimic the small airways of the lung, which is the primary site of respiratory exposure and the route of exposure for our comparative in vivo study. As epithelial cells make up the inner lining of the lung, they represent a plausible first point of contact with MWCNT during inhalation exposure and a potential source of cellular signaling. As we have shown previously that the MWCNT in the coculture model do not appear to pass through the Transwell membrane into the endothelial layer (Snyder-Talkington et al. 2013a), we believe that changes in gene expression in the underlying HMVECs are the result of cellular signaling from the SAEC interaction with the MWCNT. It is very plausible that direct exposure of HMVEC to MWCNT in the monoculture models may result in differential gene expression than that seen in HMVEC in coculture due to a difference between direct and indirect MWCNT exposure and cellular signaling between the SAEC and HMVEC that is inherent to the coculture. In addition to coculture of SAEC and HMVEC, numerous cell types, such as macrophages and fibroblasts, could be incorporated into coculture models of the lung or other organs. Additionally, various epithelial and endothelial cell lines will produce their own exclusive gene expression. As each cell type provides its own unique signaling signature, combinations of different cell lines and types may provide different gene expression patterns (Tilton et al. 2014) that may more closely match the in vivo signaling environment. In addition to different cell types, different culture environments, such as 3D culture that allows cells to form more in vivo-like structures or air-liquid interface that allows for epithelial cells to be exposed to particles by air, as they would in the lung, may result in different gene expression profiles, each one an attempt at moving towards a more in vivo-like state (Alepee et al. 2014; Aufderheide et al. 2013; Brandenberger et al. 2010; Diabate et al. 2008; Rach et al. 2014; Rothen-Rutishauser et al. 2008).The inherent crosstalk between cells grown in coculture that cannot be achieved when cells are grown singly in monoculture allows for cellular signaling and response that may better mimic an in vivo environment and, thus, a more in vivo- like response.
While in vivo exposure of animals to MWCNT and other nanomaterials provides insight into the gross effects of nanoparticle exposure, we suggest that in vitro coculture of relevant cell types can provide an improved system for the development of high-throughput in vitro modeling that better mimics the in vivo environment. The use of relevant coculture models would allow for the performance of rapid mechanistic studies that are not feasible in vivo while maintaining similar cellular signaling cascades. The use of coculture models could both reduce the need for in vivo toxicological testing of nanomaterials and increase the ability for rapid hazard assessment of MWCNT and other nanomaterials for the development of strategies for improved occupational and environmental health protection and surveillance.
Supplementary Material
Acknowledgements
Nancy L. Guo is supported by NIH R01ES021764; LM009500. Julian Dymacek is supported by a NSF training grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the views of the National Institute for Occupational Safety and Health.
References
- Rothen-Rutishauser B, Blank F, Muhlfeld C, Gehr P. In vitro models of the human epithelial airway barrier to study the toxic potential of particulate matter. Expert opinion on drug metabolism & toxicology. 2008;4:1075–1089. doi: 10.1517/17425255.4.8.1075. [DOI] [PubMed] [Google Scholar]
- Sayes CM, Marchione AA, Reed KL, Warheit DB. Comparative pulmonary toxicity assessments of C60 water suspensions in rats: few differences in fullerene toxicity in vivo in contrast to in vitro profiles. Nano letters. 2007a;7:2399–2406. doi: 10.1021/nl0710710. [DOI] [PubMed] [Google Scholar]
- Sayes CM, Reed KL, Warheit DB. Assessing toxicity of fine and nanoparticles: comparing in vitro measurements to in vivo pulmonary toxicity profiles. Toxicological sciences : an official journal of the Society of Toxicology. 2007b;97:163–180. doi: 10.1093/toxsci/kfm018. [DOI] [PubMed] [Google Scholar]
- Seagrave J, McDonald JD, Mauderly JL. In vitro versus in vivo exposure to combustion emissions. Experimental and toxicologic pathology : official journal of the Gesellschaft fur Toxikologische Pathologie. 2005;1(57 Suppl):233–238. doi: 10.1016/j.etp.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Klein SG, Hennen J, Serchi T, Blomeke B, Gutleb AC. Potential of coculture in vitro models to study inflammatory and sensitizing effects of particles on the lung. Toxicology in vitro : an international journal published in association with BIBRA. 2011;25:1516–1534. doi: 10.1016/j.tiv.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Snyder-Talkington BN, Qian Y, Castranova V, Guo NL. New perspectives for in vitro risk assessment of multi walled carbon nanotubes: application of coculture and bioinformatics. Journal of toxicology and environmental health. Part B, Critical reviews. 2012;15:468–492. doi: 10.1080/10937404.2012.736856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Volder MF, Tawfick SH, Baughman RH, Hart AJ. Carbon nanotubes: present and future commercial applications. Science. 2013;339:535–539. doi: 10.1126/science.1222453. [DOI] [PubMed] [Google Scholar]
- Castranova V. Overview of current toxicological knowledge of engineered nanoparticles. Journal of occupational and environmental medicine / American College of Occupational and Environmental Medicine. 2011;53:S14–S17. doi: 10.1097/JOM.0b013e31821b1e5a. [DOI] [PubMed] [Google Scholar]
- Sos Poulsen S, Jacobsen NR, Labib S, Wu D, Husain M, Williams A, Bogelund JP, Andersen O, Kobler C, Molhave K, Kyjovska ZO, Saber AT, Wallin H, Yauk CL, Vogel U, Halappanavar S. Transcriptomic analysis reveals novel mechanistic insight into murine biological responses to multi-walled carbon nanotubes in lungs and cultured lung epithelial cells. PloS one. 2013;8:e80452. doi: 10.1371/journal.pone.0080452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RR, Hubbs AF, Scabilloni JF, Wang L, Battelli LA, Friend S, Castranova V, Porter DW. Pulmonary fibrotic response to aspiration of multi-walled carbon nanotubes. Particle and fibre toxicology. 2011;8:21. doi: 10.1186/1743-8977-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer RR, Scabilloni JF, Hubbs AF, Battelli LA, McKinney W, Friend S, Wolfarth MG, Andrew M, Castranova V, Porter DW. Distribution and fibrotic response following inhalation exposure to multi-walled carbon nanotubes. Particle and fibre toxicology. 2013;10:33. doi: 10.1186/1743-8977-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DW, Hubbs AF, Chen BT, McKinney W, Mercer RR, Wolfarth MG, Battelli L, Wu N, Sriram K, Leonard S, Andrew M, Willard P, Tsuruoka S, Endo M, Tsukada T, Munekane F, Frazer DG, Castranova V. Acute pulmonary dose-responses to inhaled multi-walled carbon nanotubes. Nanotoxicology. 2013;7:1179–1194. doi: 10.3109/17435390.2012.719649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter DW, Hubbs AF, Mercer RR, Wu N, Wolfarth MG, Sriram K, Leonard S, Battelli L, Schwegler-Berry D, Friend S, Andrew M, Chen BT, Tsuruoka S, Endo M, Castranova V. Mouse pulmonary dose- and time course-responses induced by exposure to multi-walled carbon nanotubes. Toxicology. 2010;269:136–147. doi: 10.1016/j.tox.2009.10.017. [DOI] [PubMed] [Google Scholar]
- Sargent LM, Porter DW, Staska LM, Hubbs AF, Lowry DT, Battelli L, Siegrist KJ, Kashon ML, Mercer RR, Bauer AK, Chen BT, Salisbury JL, Frazer D, McKinney W, Andrew M, Tsuruoka S, Endo M, Fluharty KL, Castranova V, Reynolds SH. Promotion of lung adenocarcinoma following inhalation exposure to multi-walled carbon nanotubes. Particle and fibre toxicology. 2014;11:3. doi: 10.1186/1743-8977-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist KJ, Reynolds SH, Kashon ML, Lowry DT, Dong C, Hubbs AF, Young SH, Salisbury JL, Porter DW, Benkovic SA, McCawley M, Keane MJ, Mastovich JT, Bunker KL, Cena LG, Sparrow MC, Sturgeon JL, Dinu CZ, Sargent LM. Genotoxicity of multi-walled carbon nanotubes at occupationally relevant doses. Particle and fibre toxicology. 2014;11:6. doi: 10.1186/1743-8977-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaper R, Arndt D, Bozich J, Dominguez G. Molecular interactions of nanomaterials and organisms: defining biomarkers for toxicity and high-throughput screening using traditional and next-generation sequencing approaches. The Analyst. 2014;139:882–895. doi: 10.1039/c3an01644g. [DOI] [PubMed] [Google Scholar]
- Clift MJ, Endes C, Vanhecke D, Wick P, Gehr P, Schins RP, Petri-Fink A, Rothen-Rutishauser B. A comparative study of different in vitro lung cell culture systems to assess the most beneficial tool for screening the potential adverse effects of carbon nanotubes. Toxicological sciences : an official journal of the Society of Toxicology. 2014;137:55–64. doi: 10.1093/toxsci/kft216. [DOI] [PubMed] [Google Scholar]
- Snyder-Talkington BN, Schwegler-Berry D, Castranova V, Qian Y, Guo NL. Multi-walled carbon nanotubes induce human microvascular endothelial cellular effects in an alveolar-capillary co-culture with small airway epithelial cells. Particle and fibre toxicology. 2013a;10:35. doi: 10.1186/1743-8977-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo NL, Wan YW, Denvir J, Porter DW, Pacurari M, Wolfarth MG, Castranova V, Qian Y. Multi walled carbon nanotube-induced gene signatures in the mouse lung: potential predictive value for human lung cancer risk and prognosis. Journal of toxicology and environmental health. Part A. 2012;75:1129–1153. doi: 10.1080/15287394.2012.699852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piao CQ, Liu L, Zhao YL, Balajee AS, Suzuki M, Hei TK. Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis. 2005;26:725–731. doi: 10.1093/carcin/bgi016. [DOI] [PubMed] [Google Scholar]
- Shao R, Guo X. Human microvascular endothelial cells immortalized with human telomerase catalytic protein: a model for the study of in vitro angiogenesis. Biochemical and biophysical research communications. 2004;321:788–794. doi: 10.1016/j.bbrc.2004.07.033. [DOI] [PubMed] [Google Scholar]
- Pacurari M, Qian Y, Fu W, Schwegler-Berry D, Ding M, Castranova V, Guo NL. Cell permeability, migration, and reactive oxygen species induced by multi walled carbon nanotubes in human microvascular endothelial cells. Journal of toxicology and environmental health. Part A. 2012;75:112–128. doi: 10.1080/15287394.2011.615110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder-Talkington BN, Pacurari M, Dong C, Leonard SS, Schwegler-Berry D, Castranova V, Qian Y, Guo NL. Systematic analysis of multiwalled carbon nanotube-induced cellular signaling and gene expression in human small airway epithelial cells. Toxicological sciences : an official journal of the Society of Toxicology. 2013b;133:79–89. doi: 10.1093/toxsci/kft019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JH, Lee EJ, Lee JH, So KP, Lee YH, Bae GN, Lee SB, Ji JH, Cho MH, Yu IJ. Monitoring multiwalled carbon nanotube exposure in carbon nanotube research facility. Inhalation toxicology. 2008;20:741–749. doi: 10.1080/08958370801942238. [DOI] [PubMed] [Google Scholar]
- Pacurari M, Qian Y, Porter DW, Wolfarth M, Wan Y, Luo D, Ding M, Castranova V, Guo NL. Multi-walled carbon nanotube-induced gene expression in the mouse lung: association with lung pathology. Toxicology and applied pharmacology. 2011;255:18–31. doi: 10.1016/j.taap.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbin K, Simon R. Sample size determination in microarray experiments for class comparison and prognostic classification. Biostatistics. 2005;6:27–38. doi: 10.1093/biostatistics/kxh015. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bioconductor. http://www.bioconductor.org/
- Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database : the journal of biological databases and curation. 2011;2011 doi: 10.1093/database/bar049. bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfaro-Moreno E, Nawrot TS, Vanaudenaerde BM, Hoylaerts MF, Vanoirbeek JA, Nemery B, Hoet PH. Co-cultures of multiple cell types mimic pulmonary cell communication in response to urban PM10. The European respiratory journal. 2008;32:1184–1194. doi: 10.1183/09031936.00044008. [DOI] [PubMed] [Google Scholar]
- Aschberger K, Johnston HJ, Stone V, Aitken RJ, Hankin SM, Peters SA, Tran CL, Christensen FM. Review of carbon nanotubes toxicity and exposure--appraisal of human health risk assessment based on open literature. Critical reviews in toxicology. 2010;40:759–790. doi: 10.3109/10408444.2010.506638. [DOI] [PubMed] [Google Scholar]
- Johnston HJ, Hutchison GR, Christensen FM, Peters S, Hankin S, Aschberger K, Stone V. A critical review of the biological mechanisms underlying the in vivo and in vitro toxicity of carbon nanotubes: The contribution of physico-chemical characteristics. Nanotoxicology. 2010;4:207–246. doi: 10.3109/17435390903569639. [DOI] [PubMed] [Google Scholar]
- Pacurari M, Castranova V, Vallyathan V. Single- and multi-wall carbon nanotubes versus asbestos: are the carbon nanotubes a new health risk to humans? Journal of toxicology and environmental health. Part A. 2010;73:378–395. doi: 10.1080/15287390903486527. [DOI] [PubMed] [Google Scholar]
- Fehrenbach H. Alveolar epithelial type II cell: defender of the alveolus revisited. Respiratory research. 2001;2:33–46. doi: 10.1186/rr36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JJ, Moore TM, Babal P, Diwan AH, Stevens T, Thompson WJ. Pulmonary microvascular and macrovascular endothelial cells: differential regulation of Ca2+ and permeability. The American journal of physiology. 1998;274:L810–L819. doi: 10.1152/ajplung.1998.274.5.L810. [DOI] [PubMed] [Google Scholar]
- Planus E, Galiacy S, Matthay M, Laurent V, Gavrilovic J, Murphy G, Clerici C, Isabey D, Lafuma C, d’Ortho MP. Role of collagenase in mediating in vitro alveolar epithelial wound repair. Journal of cell science. 1999;112(Pt 2):243–252. doi: 10.1242/jcs.112.2.243. [DOI] [PubMed] [Google Scholar]
- Shvedova AA, Kisin ER, Porter D, Schulte P, Kagan VE, Fadeel B, Castranova V. Mechanisms of pulmonary toxicity and medical applications of carbon nanotubes: Two faces of Janus? Pharmacology & therapeutics. 2009;121:192–204. doi: 10.1016/j.pharmthera.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Kasper J, Hermanns MI, Bantz C, Maskos M, Stauber R, Pohl C, Unger RE, Kirkpatrick JC. Inflammatory and cytotoxic responses of an alveolar-capillary coculture model to silica nanoparticles: comparison with conventional monocultures. Particle and fibre toxicology. 2011;8:6. doi: 10.1186/1743-8977-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao KM, Porter DW, Meighan T, Castranova V. The sources of inflammatory mediators in the lung after silica exposure. Environmental health perspectives. 2004;112:1679–1686. doi: 10.1289/ehp.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns MI, Fuchs S, Bock M, Wenzel K, Mayer E, Kehe K, Bittinger F, Kirkpatrick CJ. Primary human coculture model of alveolo-capillary unit to study mechanisms of injury to peripheral lung. Cell and tissue research. 2009;336:91–105. doi: 10.1007/s00441-008-0750-1. [DOI] [PubMed] [Google Scholar]
- Hermanns MI, Unger RE, Kehe K, Peters K, Kirkpatrick CJ. Lung epithelial cell lines in coculture with human pulmonary microvascular endothelial cells: development of an alveolo-capillary barrier in vitro. Laboratory investigation; a journal of technical methods and pathology. 2004;84:736–752. doi: 10.1038/labinvest.3700081. [DOI] [PubMed] [Google Scholar]
- Muller L, Riediker M, Wick P, Mohr M, Gehr P, Rothen-Rutishauser B. Oxidative stress and inflammation response after nanoparticle exposure: differences between human lung cell monocultures and an advanced three-dimensional model of the human epithelial airways. Journal of the Royal Society, Interface / the Royal Society. 2010;1(7 Suppl):S27–S40. doi: 10.1098/rsif.2009.0161.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balda MS, Matter K. Tight junctions. Journal of cell science. 1998;111(Pt 5):541–547. doi: 10.1242/jcs.111.5.541. [DOI] [PubMed] [Google Scholar]
- Afshari CA, Hamadeh HK, Bushel PR. The evolution of bioinformatics in toxicology: advancing toxicogenomics. Toxicological sciences : an official journal of the Society of Toxicology. 2011;1(120 Suppl):S225–S237. doi: 10.1093/toxsci/kfq373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrecht J, Caba E. Gene expression profile analysis: an emerging approach to investigate mechanisms of genotoxicity. Pharmacogenomics. 2005;6:419–428. doi: 10.1517/14622416.6.4.419. [DOI] [PubMed] [Google Scholar]
- Bushel PR, Heinloth AN, Li J, Huang L, Chou JW, Boorman GA, Malarkey DE, Houle CD, Ward SM, Wilson RE, Fannin RD, Russo MW, Watkins PB, Tennant RW, Paules RS. Blood gene expression signatures predict exposure levels. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:18211–18216. doi: 10.1073/pnas.0706987104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton RK, Aardema M, Aubrecht J. The utility of DNA microarrays for characterizing genotoxicity. Environmental health perspectives. 2004;112:420–422. doi: 10.1289/ehp.6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir EF, Bittner M, Trent J, Barrett JC, Afshari CA. Microarrays and toxicology: the advent of toxicogenomics. Molecular carcinogenesis. 1999;24:153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, Shaughnessy JD, Jr, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M, Chen M, Cheng J, Cheng J, Chou J, Davison TS, Delorenzi M, Deng Y, Devanarayan V, Dix DJ, Dopazo J, Dorff KC, Elloumi F, Fan J, Fan S, Fan X, Fang H, Gonzaludo N, Hess KR, Hong H, Huan J, Irizarry RA, Judson R, Juraeva D, Lababidi S, Lambert CG, Li L, Li Y, Li Z, Lin SM, Liu G, Lobenhofer EK, Luo J, Luo W, McCall MN, Nikolsky Y, Pennello GA, Perkins RG, Philip R, Popovici V, Price ND, Qian F, Scherer A, Shi T, Shi W, Sung J, Thierry-Mieg D, Thierry-Mieg J, Thodima V, Trygg J, Vishnuvajjala L, Wang SJ, Wu J, Wu Y, Xie Q, Yousef WA, Zhang L, Zhang X, Zhong S, Zhou Y, Zhu S, Arasappan D, Bao W, Lucas AB, Berthold F, Brennan RJ, Buness A, Catalano JG, Chang C, Chen R, Cheng Y, Cui J, Czika W, Demichelis F, Deng X, Dosymbekov D, Eils R, Feng Y, Fostel J, Fulmer-Smentek S, Fuscoe JC, Gatto L, Ge W, Goldstein DR, Guo L, Halbert DN, Han J, Harris SC, Hatzis C, Herman D, Huang J, Jensen RV, Jiang R, Johnson CD, Jurman G, Kahlert Y, Khuder SA, Kohl M, Li J, Li L, Li M, Li QZ, Li S, Li Z, Liu J, Liu Y, Liu Z, Meng L, Madera M, Martinez-Murillo F, Medina I, Meehan J, Miclaus K, Moffitt RA, Montaner D, Mukherjee P, Mulligan GJ, Neville P, Nikolskaya T, Ning B, Page GP, Parker J, Parry RM, Peng X, Peterson RL, Phan JH, Quanz B, Ren Y, Riccadonna S, Roter AH, Samuelson FW, Schumacher MM, Shambaugh JD, Shi Q, Shippy R, Si S, Smalter A, Sotiriou C, Soukup M, Staedtler F, Steiner G, Stokes TH, Sun Q, Tan PY, Tang R, Tezak Z, Thorn B, Tsyganova M, Turpaz Y, Vega SC, Visintainer R, von Frese J, Wang C, Wang E, Wang J, Wang W, Westermann F, Willey JC, Woods M, Wu S, Xiao N, Xu J, Xu L, Yang L, Zeng X, Zhang J, Zhang L, Zhang M, Zhao C, Puri RK, Scherf U, Tong W, Wolfinger RD, Consortium M. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nature biotechnology. 2010a;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T, Leist M. Food for thought … on the evolution of toxicology and the phasing out of animal testing. Altex. 2008;25:91–102. doi: 10.14573/altex.2008.2.91. [DOI] [PubMed] [Google Scholar]
- Suemori H. Establishment and therapeutic use of human embryonic stem cell lines. Human cell. 2006;19:65–70. doi: 10.1111/j.1749-0774.2006.00011.x. [DOI] [PubMed] [Google Scholar]
- Sanna CR, Li WH, Zhang L. Overlapping genes in the human and mouse genomes. BMC genomics. 2008;9:169. doi: 10.1186/1471-2164-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Dessimoz C, Gonnet GH. OMA Browser--exploring orthologous relations across 352 complete genomes. Bioinformatics. 2007;23:2180–2182. doi: 10.1093/bioinformatics/btm295. [DOI] [PubMed] [Google Scholar]
- Shi G, Zhang L, Jiang T. MSOAR 2.0: Incorporating tandem duplications into ortholog assignment based on genome rearrangement. BMC bioinformatics. 2010b;11:10. doi: 10.1186/1471-2105-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altenhoff AM, Studer RA, Robinson-Rechavi M, Dessimoz C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS computational biology. 2012;8:e1002514. doi: 10.1371/journal.pcbi.1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilton SC, Karin NJ, Tolic A, Xie Y, Lai X, Hamilton RF, Jr, Waters KM, Holian A, Witzmann FA, Orr G. Three human cell types respond to multi-walled carbon nanotubes and titanium dioxide nanobelts with cell-specific transcriptomic and proteomic expression patterns. Nanotoxicology. 2014;8:533–548. doi: 10.3109/17435390.2013.803624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepee N, Bahinski T, Daneshian M, De Wever B, Fritsche E, Goldberg A, Hansmann J, Hartung T, Haycock J, Hogberg H, Hoelting L, Kelm JM, Kadereit S, McVey E, Landsiedel R, Leist M, Lubberstedt M, Noor F, Pellevoisin C, Petersohn D, Pfannenbecker U, Reisinger K, Ramirez T, Rothen-Rutishauser B, Schafer-Korting M, Zeilinger K, Zurich MG. State-of-the-art of 3D cultures (organs-on-a-chip) in safety testing and pathophysiology. Altex. 2014 doi: 10.14573/altex1406111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aufderheide M, Halter B, Mohle N, Hochrainer D. The CULTEX RFS: a comprehensive technical approach for the in vitro exposure of airway epithelial cells to the particulate matter at the air-liquid interface. BioMed research international. 2013;2013:734137. doi: 10.1155/2013/734137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger C, Rothen-Rutishauser B, Muhlfeld C, Schmid O, Ferron GA, Maier KL, Gehr P, Lenz AG. Effects and uptake of gold nanoparticles deposited at the air-liquid interface of a human epithelial airway model. Toxicology and applied pharmacology. 2010;242:56–65. doi: 10.1016/j.taap.2009.09.014. [DOI] [PubMed] [Google Scholar]
- Diabate S, Mulhopt S, Paur HR, Krug HF. The response of a co-culture lung model to fine and ultrafine particles of incinerator fly ash at the air-liquid interface. Alternatives to laboratory animals : ATLA. 2008;36:285–298. doi: 10.1177/026119290803600306. [DOI] [PubMed] [Google Scholar]
- Rach J, Budde J, Mohle N, Aufderheide M. Direct exposure at the air-liquid interface: evaluation of an in vitro approach for simulating inhalation of airborne substances. Journal of applied toxicology : JAT. 2014;34:506–515. doi: 10.1002/jat.2899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.