Abstract
Four Xanthomonas species are known to cause bacterial spot of tomato and pepper, but the global distribution and genetic diversity of these species are not well understood. A collection of bacterial spot-causing strains from the Americas, Africa, Southeast Asia, and New Zealand were characterized for genetic diversity and phylogenetic relationships using multilocus sequence analysis of six housekeeping genes. By examining strains from different continents, we found unexpected phylogeographic patterns, including the global distribution of a single multilocus haplotype of X. gardneri, possible regional differentiation in X. vesicatoria, and high species diversity on tomato in Africa. In addition, we found evidence of multiple recombination events between X. euvesicatoria and X. perforans. Our results indicate that there have been shifts in the species composition of bacterial spot pathogen populations due to the global spread of dominant genotypes and that recombination between species has generated genetic diversity in these populations.
INTRODUCTION
Understanding the evolution and host specificity of plant-pathogenic bacteria is an ongoing challenge. Strains of phytopathogenic bacteria commonly exhibit high host specificity, with host ranges restricted to one or a few plant species (1, 2). Bacterial plant pathogens also exhibit biogeography, such that species can be limited in their geographic distributions (3). Globalization of agriculture has contributed to the dispersal of phytopathogenic bacteria, but the geographic ranges of species are not well characterized, in part because of the difficulty in differentiating phylogenetically distinct strains that have similar host specificities (4). Phenotypic characters can sometimes distinguish species with similar host specificities, but classification by molecular markers is often required due to variation in phenotypic traits within species (5). Phenotypes can also dramatically differ among strains within a species due to acquisition and loss of genes related to pathogenicity and fitness (4). Bacterial evolution is driven by point mutations, variation in gene content, recombination, and selection on the resulting phenotypes (6). Phylogenetic relationships among species are defined by point mutations in the genome that accumulate over time; however, these relationships can be obscured by polymorphisms that have been distributed to other closely related species via homologous recombination and horizontal gene transfer (7). These events can introduce conflicting phylogenetic signals between genes that have been vertically inherited versus horizontally acquired (8). The possibility of infection of a single host plant by multiple species may increase the probability of genetic exchange (9). Coinfection by multiple species may be more common as pathogens are moved out of their native geographic ranges.
Multilocus nucleotide-sequence-based approaches help in resolving phylogenetic relationships of bacteria within and between species (10). Multilocus sequence typing (MLST) and analysis (MLSA) are two approaches used to analyze multiple housekeeping genes that are conserved in sequence and present in strains of closely related species. MLST is useful in grouping strains from different groups of species but is limited by the sequence diversity based on allelic mismatches observed within the same species (10). In contrast, MLSA makes use of concatenated nucleotide sequences of the housekeeping genes for characterization of more diverse strains representing multiple species within a genus by constructing phylogenetic trees (10, 11). Since the method is based on the nucleotide sequence, it provides unambiguous results that are directly comparable, unlike randomly amplified polymorphic DNA (RAPD) or other anonymous marker systems. Since Gevers et al. (10) described the MLSA method; it has been applied to numerous pathogenic and nonpathogenic bacteria. Housekeeping genes also are subject to homologous recombination and help to approximate the extent and impact of recombination in bacterial evolution (12). As a result, MLSA has been used to estimate rates of recombination, which vary widely among bacterial species (13). Because recombination and horizontal gene transfer can result in qualitative differences in phenotype between phylogenetically closely related strains, pathogenicity tests and phenotypic assays remain critical to characterizing strains and interpreting MLSA results.
The genus Xanthomonas comprises numerous pathogenic species infecting approximately 400 different host plants (14). Phenotypic and phylogenetic analyses have shown a wide range of variation among Xanthomonas strains that cause bacterial spot of tomato and pepper (15, 16). Bacterial spot is caused by four different species: Xanthomonas euvesicatoria, X. vesicatoria, X. perforans, and X. gardneri (3, 17). Among the four species, X. euvesicatoria and X. gardneri strains are reported as pathogens of both tomato and pepper, X. perforans strains are reported only from tomato, and X. vesicatoria strains primarily infect tomato. Strains belonging to X. euvesicatoria and X. vesicatoria have a worldwide distribution (18). X. perforans and/or X. gardneri strains increasingly have been isolated in Canada (19), the United States and South America, and regions bordering the Indian Ocean (20–22). These bacterial populations can also change over time. For example, the bacterial spot pathogen population on tomato in Florida shifted from X. euvesicatoria to X. perforans. Prior to 1991, only X. euvesicatoria strains were found in Florida. In a survey in 2006 and 2007, only X. perforans strains were isolated (23), corresponding to a shift in tomato races. The origin of the X. perforans strains now responsible for bacterial spot in Florida tomatoes is unknown, in part because the global distribution of this species is not well characterized.
MLSA of Xanthomonas species has been used for phylogenetics of the genus and to examine evolution via recombination. An MLSA database of Xanthomonas strains has been created using six housekeeping genes (fusA, gapA, gltA, gyrB, lacF, and lepA) (15). MLSA has revealed recombination as a primary factor underlying the evolution of X. axonopodis (24). Some xanthomonad populations have been reported as highly clonal, with little variation among strains collected from geographically distant locations (25, 26). MLSA also has been applied to bacterial spot-causing xanthomonads. Strains causing bacterial spot of tomato and pepper in the southwest Indian Ocean region were examined; all four species were found (22). A recent study found three different species responsible for bacterial spot of tomato in Ethiopia (27), while another study found atypical strains in Grenada and India (28). Findings of species diversity and dynamic shifts in species reported from previous regional studies prompted us to apply MLSA and MLST to a collection of bacterial spot-causing xanthomonads from diverse geographic origins. Bacterial strains representing four Xanthomonas species associated with bacterial spot of tomato and pepper collected from the Americas, Africa, Southeast Asia, and New Zealand were examined using MLSA to understand the phylogeographic diversity of bacterial spot pathogens. Our objectives were to determine the geographic distribution of the four Xanthomonas species, the extent of diversity within species, and the role of homologous recombination in generating diversity in the bacterial spot pathogens.
MATERIALS AND METHODS
Bacterial strains.
Strains from multiple collections of xanthomonads isolated from tomato and pepper exhibiting bacterial spot were used (Table 1). The collections were mainly from the United States and Africa, including the southwest Indian Ocean (SWIO) islands previously reported by Hamza et al. (22), with smaller representative samples from elsewhere in the Americas, India, and New Zealand. These strains were subjected to MLSA using the six housekeeping genes fusA, gapA, gltA, gyrB, lacF, and lepA (16). Sequences were either obtained via Sanger sequencing or extracted from whole genome sequences. The sequenced strains were compared with type strains of X. vesicatoria and X. gardneri. Reference strains from X. euvesicatoria (strain 85-10) and X. perforans (strain 91-118) were used as both these strains have been extensively characterized in previous studies (16, 29) and have sequences identical to that of the type strain from their respective species for the six housekeeping genes (16).
TABLE 1.
Xanthomonas strains used for the MLSA study
| Species and groupa | Strain designation(s)b | Hostc | Location | Yr(s) |
|---|---|---|---|---|
| X. euvesicatoria | ||||
| Group 1 | 85-10R | T | Florida | 1985 |
| E3 | T | Florida | NAd | |
| 1085 | T | Mexico | 1992 | |
| 153, 155 | T | Florida | 1975, 1985 | |
| 157 | T | Australia | 1989 | |
| LB230-1 | T | Reunion | 2005 | |
| LB102-1 | T | Seychelles | 2005 | |
| LE84, LH5 | P | Mauritius | 2008, 2010 | |
| JW6 | T | Reunion | 2000 | |
| LB216 | P | Reunion | 2005 | |
| Xe072, Xe073 | P | North Carolina | 1993, 1994 | |
| Xe074, Xe075, Xe081, Xe077, Xe078, Xe079, Xe082, Xe083, Xe085, Xe086, Xe091 | P | Florida | 1994–2003 | |
| Xe076 | P | Kentucky | 1995 | |
| Xe101, Xe103, Xe104, Xe105, Xe106, Xe107, Xe108 | P | North Carolina | 2008–2012 | |
| Xe102 | P | Florida | 2008 | |
| Xe109, Xe110, Xe111, Xe112 | P | Georgia, USA | 2004 | |
| NI14, NI15, NI17 | P | Nigeria | 2012 | |
| LA88-3, LA88-5, LA84-1, LA85-1, LA88-1, LB223-1 | P | Comoros | 2004, 2005 | |
| LB226-1, LB226-4, LB215-1 | T | Comoros | 2005 | |
| LA127-1, LA127-4 | P | Reunion | 2004 | |
| LE82-2, LE83-2, LH4-1, LH4-2 | P | Mauritius | 2008, 2010 | |
| Group 2 | LMG907, LMG908 | NA | India | NA |
| LMG918 | P | India | 1957 | |
| 330, 338 | T | Barbados | 1990 | |
| LD50, LD53 | P | Grenada | 2007 | |
| ICMP3381 | P | India | 1971 | |
| Other | 1605 | T | Ohio | 1994 |
| X. gardneri | ATCC 19865T | T | Yugoslavia | 1953 |
| ETH8, ETH9, ETH15, ETH30 | T | Ethiopia | 2011 | |
| Furman-3e | T | Pennsylvania | NA | |
| 1782, 1783f | T | Brazil | 1991 | |
| 444, 451 | T | Costa Rica | 1991 | |
| JQ711, JQ725, JS749-1, JS749-3, JS750-1 | T | Reunion | 1995, 1997 | |
| JS750-3 | P | Reunion | 1997 | |
| O4T5g | T | Canada | 2004 | |
| OOT12Bg | T | Canada | NA | |
| Xg153, Xg164, Xg165, Xg173, Xg174, Xg177 | T | Ohio | 2010–2012 | |
| Xg156, Xg157, Xg159, Xg160 | T | Michigan | 2010 | |
| Other | ICMP7383 | T | New Zealand | 1980 |
| X. perforans | ||||
| Group 1 | 91-118R | T | Florida | 1991 |
| 1220 | T | Thailand | 1993 | |
| 1484 | T | Mexico | 1993 | |
| 938 | T | Florida | 1991 | |
| ETH5, ETH13, ETH21, ETH26 | T | Ethiopia | 2011 | |
| GEV872, GEV893, GEV904, GEV909, GEV915, GEV917, GEV936, GEV940, GEV968, GEV993, GEV1026 | T | Florida | 2012 | |
| LB101-1, LB101-2, LB102-2 | T | Seychelles | 2005 | |
| LB273-2, LB273-3 | T | Mayotte | 2005 | |
| LH3 | T | Mauritius | 2010 | |
| Xp1-7, Xp2-12, Xp5-6, Xp11-2, Xp15-11, Xp17-12, Xp18-15, Xp19-10 | T | Florida | 2006 | |
| Group 2 | Xp3-15, Xp4B, Xp7-12, Xp8-16, Xp9-5, Xp10-13 | T | Florida | 2006 |
| GEV839, GEV1001, GEV1044, GEV1054, GEV1063 | T | Florida | 2012 | |
| TB6, TB9, TB15 | T | Florida | 2013 | |
| Other | Xp4-20 | T | Florida | 2006 |
| X. vesicatoria | ||||
| Group 1 | ATCC 35937T | T | New Zealand | 1955 |
| ETH1 | T | Ethiopia | 2011 | |
| 141 | T | New Zealand | 1971 | |
| Group 2 | ETH17 | T | Ethiopia | 2011 |
| 144 | T | Argentina | NA | |
| 56 | T | Brazil | 1987 | |
| Group 3 | JS683-2 | T | Reunion | 1997 |
| LC161, LC162 | T | Madagascar | 2006 | |
| Atypical Nigerian strains | NI1, NI4, NI5, NI7 | T | Nigeria | 2012 |
| Xanthomonas sp. strain | ETH12h | T | Ethiopia | 2011 |
Strains are grouped based on allele type.
Superscript R, reference strain; superscript T, type strain.
T, tomato; P, pepper.
NA, not applicable.
Omnilytics, Inc., Sandy, UT.
Phytobacterial culture collection of Instituto Biológico, CEIB, Campinas, SP, Brazil.
D. Cuppels, Agriculture and Agri-Food Canada, London, Ontario, Canada.
ETH12 was first isolated from tomato but was identified as being nonpathogenic to tomato and pepper.
Phylogenetic analysis.
Sequences for six housekeeping genes from the worldwide strains along with reference Xanthomonas strains were aligned using MUSCLE within MEGA 5.2.1 (30). The alignments were further confirmed via BioEdit software (31). Nucleotide substitution models that best fit the aligned sequences were selected using the Akaike Information Criterion (AIC) within jModeltest 1.1 (32). The general time reversible model with gamma-distributed invariant sites (GTR+G+I) model was selected and used for construction of phylogenetic trees based on maximum likelihood (ML) and Bayesian inference. Maximum likelihood phylogenetic trees based on the six housekeeping genes were constructed individually and using concatenated sequences. The maximum likelihood tree, with 1,000 bootstrap samples, inferred using RaxML was compared to ML, maximum parsimony (MP), and neighbor-joining (NJ) trees constructed using the GTR+G+I model in MEGA 5.2.1 (29). MrBayes v.3.2 (33) was used for the Bayesian phylogeny, using the same substitution model with 1,000,000 Markov chain Monte Carlo (MCMC) steps, sampled every 500 steps. A burn-in period of 88,500 steps was used for the concatenated data set and 56,000 steps for the individual genes (33, 34). Consensus trees obtained from MrBayes were visualized using FigTree version 1.4 (Institute of Evolutionary Biology, University of Edinburgh [http://tree.bio.ed.ac.uk/software/figtree/]). Results from the phylogenetic analyses, along with phenotypic assays from previous studies, were used to delineate groups of strains into species.
Analysis of diversity and recombination.
Nucleotide diversity, the number of haplotypes, and the minimum number of recombination events were determined using DnaSP 5.0 (35). DnaSP also was used for calculating class I neutrality tests (Tajima's D and Fu and Li's D* and F*) for detecting departure from the mutation/drift equilibrium (36, 37). For these calculations, all strains were considered together and by species.
Multiple methods were used to detect recombination. Splits-decomposition trees were constructed (38), and the pairwise homoplasy index (PHI) was calculated using SplitsTree version 4.13.1 (39). These calculations used the concatenated genes for both the entire data set and a subset of the data that included only X. perforans and X. euvesicatoria strains. The Recombination Detection Program (RDP) version 4 combines seven nonparametric detection programs (3Seq, Chimaera, RDP, GENECONV, MaxChi, BootScan, and SiScan) to detect recombination and estimate breakpoints (40). The default settings and a Bonferroni step-down correction method with a P value cutoff of 0.05 were applied in the analysis of the concatenated data set. Recombination breakpoints also were identified using GARD (Genetic Algorithm for Recombination Detection) (41).
After detection of recombination, the concatenated data were used to reconstruct a nonrecombinant coalescent-based genealogy using ClonalFrame 1.1 with the default settings (42). The MCMC used a burn-in period of 50,000 steps sampled every 100th step. Mutational rate (θ), intragenic recombination rate (R), average length of recombination event (δ), and the rate of new polymorphism generated due to recombination were estimated along with time to most recent common ancestor (TMRCA) for all strains and for each species group. Outputs were used to calculate the impact of recombination to mutation (r/m) and to measure the rate of recombination per site relative to mutation rate (ρ/θ) for each species and the full collection.
Nucleotide sequence accession numbers.
The six housekeeping gene sequences for four reference Xanthomonas strains were obtained from the Plant-Associated and Environmental Microbes Database (PAMDB) online database (www.pamdb.org). The sequenced genes have been deposited into the National Center for Biotechnology Institute (NCBI) database under the following accession numbers: fusA, KF994809 to KF994819, KJ938581 to KJ938587, and KM491929 to KM492062; gapA, KF994820 to KF994830, KJ938588 to KJ938594, and KM492063 to KM492196; gltA, KF994831 to KF994841, KJ938595 to KJ938601, and KM492197 to KM492330; gyrB, KF994896 to KF994906, KJ938602 to KJ938608, and KM492331 to KM492464; lacF, KF994874 to KF994884, KJ938629 to KJ938635, and KM492465 to KM492598; and lepA, KF994885 to KF994895, KJ938636 to KJ938642, and KM492599 to KM492732.
RESULTS
Phylogenetic characterization.
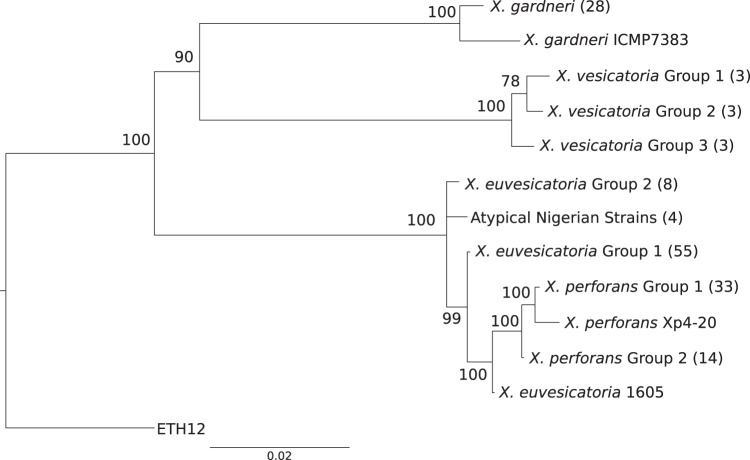
Phylogenetic analysis of the worldwide strain collection showed different patterns of variation within species as well as various species compositions across geographic locations (Fig. 1 and Table 1). Table 2 lists the haplotypes, based on nucleotide sequence, found for each housekeeping gene. In our sample of 29 X. gardneri strains, 28 had identical sequences across six housekeeping genes (Table 2; see Fig. S1 in the supplemental material). The exception was strain ICMP7383, isolated in New Zealand in 1980. This strain differed from the X. gardneri type strain in four of six housekeeping genes by 2, 4, 10, and 12 nucleotides in genes lacF, fusA, gyrB, and gapA, respectively.
FIG 1.
Bayesian phylogeny of clone-corrected Xanthomonas strains from tomato and pepper. The number of strains in each group is included in parentheses. Values on the branch indicate Bayesian posterior probabilities expressed as a percentage of the trees. The scale bar indicates the number of substitutions per site. The strains in each group are listed in Table 1. Group 1 of X. vesicatoria, X. perforans, and X. euvesicatoria includes type strains in addition to the strains listed in the table.
TABLE 2.
Sequence types of the Xanthomonas strains used in this study
| Strain, species, or groupa | Allele or sequence typeb |
|||||
|---|---|---|---|---|---|---|
| lacF | lepA | gyrB | fusA | gapA | gltA | |
| Xanthomonas sp. strain ETH12 | 1 | 1 | 1 | 1 | 1 | 1 |
| X. perforans | ||||||
| Group 1 (33) | 2 | 2 | 2 | 2 | 2 | 2 |
| Group 2 (14) | 2 | 2 | 3 | 2 | 3 | 2 |
| Xp4-20 | 2 | 3 | 3 | 2 | 2 | 2 |
| X. euvesicatoria | ||||||
| Group 1 (55) | 3 | 4 | 3 | 3 | 3 | 3 |
| Group 2 (8) | 3 | 4 | 3 | 4 | 4 | 3 |
| 1605 | 3 | 5 | 3 | 3 | 3 | 3 |
| Atypical Nigerian strains (4) | 3 | 6 | 2 | 2 | 5 | 3 |
| X. vesicatoria | ||||||
| Group 1 (3) | 4 | 7 | 4 | 5 | 6 | 4 |
| Group 2 (3) | 4 | 7 | 5 | 5 | 7 | 4 |
| Group 3 (3) | 4 | 7 | 5 | 6 | 8 | 5 |
| X. gardneri | ||||||
| Miscellaneous strains (28) | 5 | 8 | 6 | 7 | 9 | 6 |
| ICMP7383 | 6 | 8 | 7 | 8 | 10 | 6 |
Numbers in parentheses indicate the numbers of strains in each group. The strains are listed in Table 1. Type strains are included in group 1 in each species.
The same numbers in each column represent the same allele/type sequences.
Among the nine X. vesicatoria strains in our collection, three multilocus haplotypes were observed (Fig. 1 and Table 2; see Fig. S1 in the supplemental material). The multilocus haplotype that included the type strain of X. vesicatoria was found in strains from New Zealand and Ethiopia, and strains with this haplotype will be referred to as X. vesicatoria group 1. A second haplotype was identified from two strains isolated in South America and one strain from Ethiopia (strains 56, 144, and ETH17); these strains are collectively referred to as X. vesicatoria group 2. This haplotype varied in genes gyrB and gapA by 1 and 12 nucleotides, respectively. A third multilocus haplotype was identified only from the SWIO region, specifically the African islands of Reunion and Madagascar, and strains with this haplotype will be referred as X. vesicatoria group 3. This haplotype varied in gyrB and fusA genes by 1 and 2 nucleotides, respectively, and by 8 nucleotides in both gapA and gltA, respectively. The gyrB gene was identical in sequence in groups 2 and 3.
X. euvesicatoria had the greatest representation in our collection, and we detected at least three multilocus haplotypes. The first haplotype, represented as X. euvesicatoria group 1, with 55 strains, was identical to X. euvesicatoria reference strain 85-10 across the six genes (Fig. 1, Table 2, and Table 3). Eight strains (ICMP3381, LD50, LD53, LMG907, LMG908, LMG918, 330, and 338) shared a multilocus haplotype that was distinct from that of strain 85-10. These strains will be referred to as X. euvesicatoria group 2. The X. euvesicatoria group 2 strains had distinct sequences for genes fusA and gapA (Table 2) that varied by 2 and 8 nucleotides, respectively (see Fig. S2A and B in the supplemental material). Genealogies showed the fusA and gapA variant sequences form a sister clade to the X. perforans-X. euvesicatoria clade. Strain 1605, an amylolytic strain isolated from Ohio, contained a lepA haplotype that shared 100% sequence similarity with the X. perforans type strain, whereas the other genes produced the group 1 X. euvesicatoria haplotype (see Fig. S2F).
TABLE 3.
Sequence variation statistics for the collection of Xanthomonas strainsa
| Sequence set | Diversity parameterb |
Neutrality test |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | H | S | ND | θw | NM | NSM | Tajima's D | Fu and Li's D* | Fu and Li's F* | Rc | |
| All strains | 156 | 13 | 283 | 0.02554 | 50.379 | 307 | 61 | 0.545 (NS)d | −0.32 (NS) | 0.107 (NS) | 40 |
| X. euvesicatoria | 65 | 3 | 18 | 0.00099 | 3.087 | 18 | 8 | −1.06 (NS) | −1.78 (NS) | −1.81 (NS) | 0 |
| X. perforans | 52 | 4 | 27 | 0.00194 | 5.975 | 27 | 3 | −0.61 (NS) | 0.91 (NS) | 0.44 (NS) | 2 |
| X. vesicatoria | 9 | 3 | 21 | 0.00419 | 7.727 | 21 | 0 | 1.77 (NS) | 1.57 (P < 0.02) | 1.81541 (P < 0.02) | 0 |
| X. gardneri | 29 | 2 | 21 | 0.00058 | 5.347 | 21 | 21 | −2.57 (P < 0.001) | −4.57 (P < 0.02) | −4.63 (P < 0.02) | 0 |
| X. euvesicatoria and X. perforans group | 117 | 7 | 32 | 0.00483 | 12.067 | 32 | 0 | 3.04 | 1.99 (P < 0.02) | 2.89 (P < 0.02) | 4 |
All calculations were made using DNAsp v.5 software. While calculating different parameters for the whole set, strain ETH12 was also used, but since it was not possible to define a species group for that strain, it was not included in other parameter calculations.
n, number of strains; H, number of haplotypes; S, total number of segregating sites; ND, nucleotide diversity; θw, Watterson's theta; NM, number of mutations; NSM, number of singleton mutations.
R, minimum number of recombination events.
NS, not significant.
Many of the X. perforans strains had identical haplotypes to reference strain 91-118 (Fig. 1 and Table 2; see Fig. S1 in the supplemental material). Phylogenetic analysis based on individual genes showed that some of the X. perforans strains isolated from Florida in the years 2006 and 2012 had gapA and gyrB sequences identical to those of the X. euvesicatoria strains (see Fig. S2B and D in the supplemental material). These strains are collectively designated X. perforans group 2, and the collection of strains identical to the type strain have been designated X. perforans group 1. Surprisingly, strains NI1, NI4, NI5, and NI7 from Nigeria, which were identified as X. perforans based on phenotypic characterization and hrpB sequences (42), had a unique combination of housekeeping gene sequences (see Fig. S2A to F). These atypical Nigerian strains had fusA and gyrB genes identical to those of X. perforans group 1, but the gltA and lacF genes were identical to those in X. euvesicatoria group 1 (see Fig. S2A and D versus Fig. S2C and E). These strains also contained a gapA sequence that differed from the atypical gapA sequence of X. euvesicatoria group 2 by only one nucleotide, and the lepA gene sequence was distinct from those of all other X. euvesicatoria and X. perforans strains (see Fig. S2B and F).
Genealogy reconstruction and recombination analysis.
Nucleotide diversity and Watterson's theta (θw) showed greater sequence variation in our sample of X. vesicatoria than the other bacterial spot-causing Xanthomonas species (Table 3). Tajima's D and Fu and Li's D* and F* statistics showed that there was significant departure from the mutation drift equilibrium within X. vesicatoria and X. gardneri species, which may be explained by the equal distribution of polymorphisms in X. vesicatoria (positive values of the statistics) and low frequency of variants in X. gardneri (negative values). Mutation drift statistics were nonsignificant for X. perforans and X. euvesicatoria, but unique recombination events were observed within X. perforans species and when data for these two species were combined (Table 3).
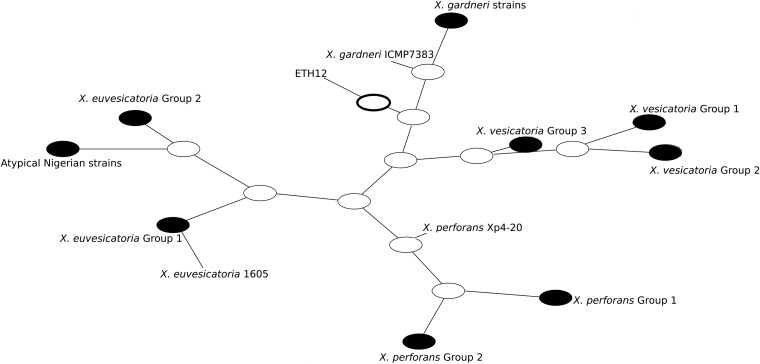
Phylogenetic networks were generated because individual maximum likelihood phylogenies showed incompatible topologies, suggesting recombination. The splits-decomposition phylogenetic tree inferred from concatenated housekeeping gene sequences confirmed incompatibilities and recombination within X. euvesicatoria and X. perforans (Fig. 2). The pairwise homoplasy index (PHI) rejected the hypothesis of no recombination in the whole set of strains, and the same result was obtained when only X. euvesicatoria and X. perforans were considered (Table 4). Genetic Algorithm for Recombination Detection (GARD) found evidence for 3 recombination breakpoints in the concatenated genes of the whole data set. The Kishino-Hasegawa test of tree congruency indicated one significant breakpoint in the lacF gene. The Recombination Detection Program (RDP) detected recombination between X. euvesicatoria and X. perforans in 6 out of 7 methods (Table 5). The genes lepA, fusA, and gapA were identified as potential recombinants, but the different algorithms varied in the strains identified as probable recombinants, which included X. euvesicatoria 1605, the X. perforans group 2 strains, and the atypical Nigerian strains. The X. euvesicatoria group 2 strains were identified as potential recombinants by only 2 algorithms (data not shown).
FIG 2.
Splits decomposition tree of the subset of strains from the X. euvesicatoria and X. perforans species groups. Parallel lines indicate conflicting phylogenetic relationships.
TABLE 4.
Test of recombination based on pairwise homoplasy index (ϕw)
| Sequence set | No. of polymorphic sites | Mean ϕw | P value |
|---|---|---|---|
| Xanthomonas collection | 229 | 0.0944 | <0.0001 |
| X. euvesicatoria and X. perforans | 32 | 0.32056 | <0.0005 |
TABLE 5.
Test of recombination between X. euvesicatoria and X. perforans strains using RDP4 with a step-down test at a probability of 0.05
| Programa | No. of: |
|
|---|---|---|
| Unique events | Recombination signals | |
| RDP | 2 | 30 |
| GENECONV | 4 | 47 |
| BootScan | 0 | 0 |
| MaxChi | 13 | 268 |
| Chimaera | 8 | 68 |
| SiScan | 15 | 121 |
| 3Seq | 19 | 181 |
| Total | 32 | 7,182 |
Recombination Detection Program (RDP) v.4 combines the seven nonparametric detection programs shown.
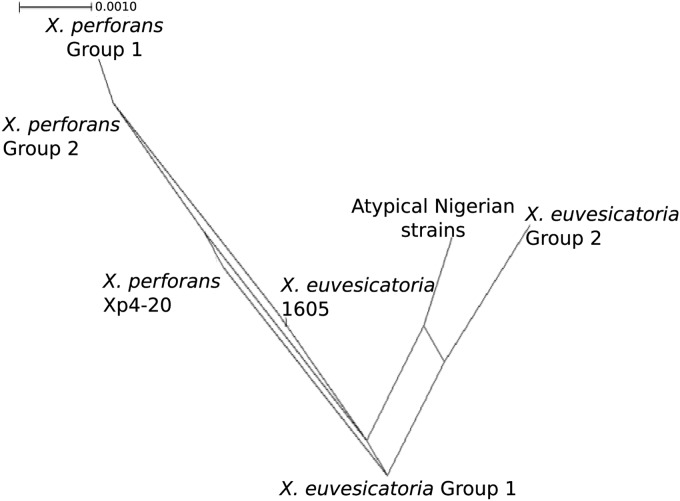
The program ClonalFrame was used to calculate the mean relative impact of recombination to mutation (r/m) on sequence variation. Recombination had 18 times higher impact than point mutation (r/m = 18.516) for the whole data set, but the rate of occurrence of recombination was only slightly higher than the rate of mutation (ρ/θ) at 1.97, The mean tract length of recombinant sequence (δ) was 843 bp (95% confidence interval [CI], 602 to 1,099 bp), which is approximately the length of two arbitrarily concatenated genes. When species groups were considered separately, recombination was more frequent than mutation within species (Table 6). A dot plot of the 50% consensus tree shows inferred ancestral relationships under clonal descent, meaning that it attempts to remove the effects of recombination (Fig. 3). The X. perforans group 2 strains share an ancestor with the X. perforans group 1 strains, as would be expected if the variant genes were introduced via homologous recombination. In contrast, the atypical Nigerian strains (NI1, NI4, NI5, and NI7) and X. euvesicatoria group 2 share an ancestor that is distinct from the ancestry of the X. euvesicatoria reference strain.
TABLE 6.
Parameter estimates for different sources of variation, including mutation and recombination, from ClonalFrame analysis
| Group or species | Result (95% CI) for indicated parametera |
|||||
|---|---|---|---|---|---|---|
| n | θ | V | R | TMRCA | ρ/θ | |
| All strains | 156 | 1.78 (0.430–4.180) | 0.023 (0.019–0.027) | 2.63 (1.410–4.030) | 2.81 (1.34–5.09) | 1.97 (0.48–5.69) |
| X. euvesicatoria and X. perforans group | 117 | 0.07 (0.010–0.260) | 0.011 (0.007–0.016) | 1.07 (0.440–1.900) | 1.84 (1.11–3.53) | 48.49 (3.33–204.40) |
| X. euvesicatoria | 65 | 0.03 (0.001–0.180) | 0.013 (0.007–0.021) | 0.28 (0.030–0.720) | 2.58 (0.99–5.32) | 98.26 (1.67–554.34) |
| X. vesicatoria | 9 | 0.04 (0.001–0.203) | 0.018 (0.006–0.035) | 0.57 (0.190–1.150) | 3.59 (1.66–6.79) | 161.12 (2.05–995.40) |
| X. perforans | 52 | 0.10 (0.002–0.600) | 0.013 (0.007–0.019) | 0.77 (0.340–1.560) | 2.49 (1.23–4.76) | 65.52 (1.012–371.48) |
| X. gardneri | 29 | 0.09 (0.001–0.380) | 0.010 (0.006–0.015) | 0.10 (0.003–0.360) | 1.67 (0.60–3.04) | 17.93 (0.02–145.78) |
95% CI, 95% confidence interval; n, number of strains; θ, number of mutation events; V, rate of substitution via recombination; R, number of recombination events; TMRCA, estimate of time to the most recent common ancestor; ρ/θ, rate of occurrence of recombination to mutation. Values in parentheses indicate 95% confidence intervals for each parameter. Calculations were made using ClonalFrame version 1.1.
FIG 3.
Dot plot diagram showing ancestral relationships among Xanthomonas strains generated using ClonalFrame (v.1.1). The diagram shows distinct lineages for each species group and their ancestries using a model of clonal descent. Each node represents an ancestor of sampled strains, with the inferred most recent common ancestor of all strains indicated by the boldface oval. The distance between nodes is arbitrary and does not indicate genetic distance. The larger groups of strains with identical haplotypes were collapsed to their shared ancestral node, shown in solid black, for presentation purposes.
DISCUSSION
Bacterial spot of pepper and tomato is caused by four different Xanthomonas species with dynamic global distributions. We characterized strains collected from different geographical locations by MLSA and found that recombination between species is shaping the diversity of some bacterial spot pathogen populations. Homologous recombination should be more likely between closely related strains due to sequence similarity (43). However, recombination is difficult to detect when it occurs between highly similar sequences; thus, some sequence divergence is required to identify recombinant sequences (44). In Xanthomonas, recombination in housekeeping genes has been observed when there is sequence variation within a species (45) and among closely related pathovars in a species complex (23). We found statistically supported recombination events in the phylogenetic clade, including both X. euvesicatoria and X. perforans. Phylogenetic analyses consistently show these species to be closely related (16, 29). At the same time, these species can be easily differentiated using nucleotide sequence variation in the hrpB gene (46) or the housekeeping genes used in this study, for which reference strains show about 1% divergence. Our sample included an X. euvesicatoria strain (strain 1605 from Ohio) that had apparently acquired DNA from X. perforans, as well as multiple X. perforans strains that contained sequences from X. euvesicatoria. Identification of such events in bacterial spot Xanthomonas points to potential sources of variation and diversity within the pathogen population.
One group of recombinant X. perforans strains was collected from tomato in Florida. Prior to 1991, only X. euvesicatoria was responsible for bacterial spot disease of tomato in Florida (47). X. perforans tomato race 3 was identified in Florida in 1991, followed by identification of tomato race 4 in 1998. A subsequent survey of 377 bacterial spot strains recovered only X. perforans from tomato lesions throughout Florida, with a nearly 2:1 frequency of race 4 to race 3 strains (23), and a recent survey of 175 strains in 2012 recovered only race 4 strains (S. Timilsina, G. E. Vallad, and J. B. Jones, unpublished data). In this study, we found that X. perforans race 4 strains collected in 2006 and 2012 contained two housekeeping genes from X. euvesicatoria. Other race 4 strains had the same multilocus haplotype as the X. perforans reference strain. We also found a single strain, Xp4-20, collected in 2006, that had X. euvesicatoria-derived sequence at a different gene, suggesting multiple independent events in which X. perforans acquired DNA from X. euvesicatoria. Frequent exchange of plasmid material has been reported between the strains of Xanthomonas (48). Horizontal gene transfer among the strains of X. axonopodis pv. vesicatoria also has been identified in planta (49). Although the frequency of exchange of genetic material is higher for plasmids than for the chromosome, these results suggest there is a potential for genetic exchange when species share the same host. Displacement of X. euvesicatoria by X. perforans on tomato may be attributed to production of bacteriocins by X. perforans, resulting in a competitive advantage over X. euvesicatoria (50). However, the reasons for the recent race shift in X. perforans in Florida, as well as the impact of recombination in race 4, remain unknown. In contrast to the dynamic changes in Xanthomonas populations on tomato, our results revealed that X. euvesicatoria strains collected from pepper in the United States have had the same multilocus haplotype over a 20-year period.
Our finding of an apparent mix of housekeeping genes from different Xanthomonas species in four of the sequenced Nigerian strains is perplexing. Of the six sequenced housekeeping genes, two genes were alleles common to X. perforans, two genes were alleles common to X. euvesicatoria, and another two genes are unique and may be from two other closely related but unknown Xanthomonas species. These strains were studied previously for pathogenicity and phenotypic characters (51). Based on reactions on tomato differentials and hrpB sequence, the atypical Nigerian strains were identified as X. perforans tomato race 3. Pectolytic and amylolytic activity also was observed in these atypical Nigerian strains. However, based on similarity to X. euvesicatoria strains at three of the MLSA genes, phylogenetic and ClonalFrame analysis of the concatenated genes grouped these atypical Nigerian strains with X. euvesicatoria, rather than X. perforans. Additional data will be required to understand the evolution of the atypical Nigerian strains.
Recombination was previously detected in X. euvesicatoria in the atpD gene, but this locus was not used in our MLSA study (28, 52). Using our MLSA genes, the X. euvesicatoria group 2 strains similarly contained a potentially recombinant sequence from an unknown donor. Two X. euvesicatoria group 2 strains were collected in India in 1957 and 1971 (28); therefore, X. euvesicatoriagroup 2 strain sequences are not a new sequence type. Together with results from the atypical Nigerian strains, these findings indicate that recombination between species is not limited to X. euvesicatoria and X. perforans but also may occur with other closely related Xanthomonas species. Identification of atypical and variant strains will be a key in understanding future population diversity and the evolution of those Xanthomonas spp. that cause bacterial spot.
Some of the strains analyzed here were genetically characterized in previous studies focused only on specific geographic locations (22, 27, 51). Examination of all strains together revealed unexpected patterns of genetic variation, including the global distribution of dominant multilocus haplotypes of X. euvesicatoria, X. gardneri, and X. perforans, possible regional differentiation of X. vesicatoria, and the presence of X. euvesicatoria group 2 in both India and the Americas. In a small sample of only nine strains of X. vesicatoria, three multilocus haplotypes were found: X. vesicatoria group 1 included the type strain along with strains from New Zealand and Ethiopia, X. vesicatoria group 2 included strains from South America and Ethiopia, and X. vesicatoria group 3 was identified only from the islands in southwest Indian Ocean (SWIO) region. These results are consistent with a previous analysis of strains from the SWIO region using a different MLSA scheme (22). In contrast to the other species, X. vesicatoria may exhibit regional differentiation in MLSA genes. A previous analysis, based on a different MLSA scheme supported by amplified fragment length polymorphism (AFLP) data, identified five clades in a worldwide strain collection of X. vesicatoria (28). The strains from South America and Ethiopia differed from the type strain by 12 nucleotides in the gapA gene. Compared to available sequences in the NCBI database, this gapA sequence was most closely related to X. arboricola, which is in the same MLSA clade as X. gardneri (16). A similar result was obtained in a study of bacterial spot strains from Tanzania, which found strains with an fyuA gene sequence similar to that of Xanthomonas arboricola (53).
We found only two haplotypes of X. gardneri, one represented by a single strain that was isolated in 1980 from New Zealand. The genetic divergence of this strain was previously reported (22). Although quite rare after its first report in 1957 as Pseudomonas gardneri, the global distribution of X. gardneri has increased dramatically over the past 2 decades (18). It is striking that there was no genetic variation in the six genes among strains from Canada, the United States, Costa Rica, Brazil, Ethiopia, and Reunion and in the four genes analyzed previously (28). Interestingly, no sequence variation was observed between the type strain of X. gardneri isolated in 1953 (reported in 1957) from the former Yugoslavia and those strains recently collected. The lack of diversity and the sudden geographic expansion of X. gardneri are likely associated with the global movement of seed (26).
International trade in seeds is likely affecting distribution of all four bacterial spot species. It is notable that in Ethiopia, Nigeria, Tanzania, and the SWIO islands, three or more different species are found within tomato-growing regions, whereas in the United States, there appears to be a single dominant species in each region. The presence of multiple species may be due to the import of seeds or plant material from multiple sources. Although we have good representation of strains from some geographic locations, more extensive sampling of different growing regions would be necessary to test this hypothesis. MLSA using six genes also may not capture enough of the variation within species and populations to make conclusions regarding the global movement of strains. A thorough understanding of the evolution of xanthomonads causing bacterial spot of tomato and pepper throughout the world would require a collaborative next-generation sequencing approach.
In conclusion, using a wide geographic representation of strains from Xanthomonas species responsible for bacterial spot of tomato and pepper, we were able to detect recombination among species and begin to characterize their global distribution. The recombinant sequences are conserved housekeeping genes that are not expected to confer a fitness advantage to the strain upon acquisition of a new allele. These results indicate that homologous recombination among xanthomonads could be occurring throughout the genome. Given the potential importance of interspecific recombination in shaping diversity of bacterial spot pathogen populations, it should be determined if genetic exchange among species has introduced variation in pathogenicity and other fitness-associated genes.
Supplementary Material
ACKNOWLEDGMENTS
We thanks Sally Miller and David Ritchie for the X. gardneri and X. euvesicatoria strains provided, respectively.
This research was supported in part with funds from a Specialty Crop Block Grant, award 18015, to G. E. Vallad and J. B. Jones from the Florida Department of Agriculture and Consumer Services and administered by the Florida Specialty Crop Foundation. This work was also financially supported in part by the European Union (ERDF), Conseil Régional de La Réunion, and CIRAD.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03000-14.
REFERENCES
- 1.Jones JB, Lacy GH, Bouzar H, Stall RE, Schaad NW. 2004. Reclassification of the xanthomonads associated with bacterial spot disease of tomato and pepper. Syst Appl Microbiol 27:755–762. doi: 10.1078/0723202042369884. [DOI] [PubMed] [Google Scholar]
- 2.Schaad NW, Postnikova E, Lacy G, Sechler A, Agarkova I, Stromberg PE, Stromberg VK, Vidaver AK. 2006. Emended classification of xanthomonad pathogens on citrus. Syst Appl Microbiol 29:690–695. doi: 10.1016/j.syapm.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR, Morin PJ, Naeem S, ℘vreås L, Reysenbach AL, Smith VH, Staley JT. 2006. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol 4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- 4.Martinez JL, Baquero F. 2002. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev 15:647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosselló-Mora R, Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. [DOI] [PubMed] [Google Scholar]
- 6.Bryant J, Chewapreecha C, Bentley SD. 2012. Developing insights into the mechanisms of evolution of bacterial pathogens from whole-genome sequences. Future Microbiol 7:1283–1296. doi: 10.2217/fmb.12.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicker E, Lefeuvre P, de Cambiaire J-C, Lemaire C, Poussier S, Prior P. 2012. Contrasting recombination patterns and demographic histories of the plant pathogen Ralstonia solanacearum inferred from MLSA. ISME J 6:961–974. doi: 10.1038/ismej.2011.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Philippe H, Douady CJ. 2003. Horizontal gene transfer and phylogenetics. Curr Opin Microbiol 6:498–505. doi: 10.1016/j.mib.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Wijayawardena KB, Minchella DJ, DeWoody JA. 2013. Hosts, parasites, and horizontal gene transfer. Trends Parasitol 29:329–338. doi: 10.1016/j.pt.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL, Swings J. 2005. Re-evaluating prokaryotic species. Nat Rev Microbiol 3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 11.Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A. 2008. Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214. doi: 10.1099/ijs.0.65392-0. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Valero L, Rusniok C, Jarraud S, Vacherie B, Rouy Z, Barbe V, Medigue C, Etienne J, Buchrieser C. 2011. Extensive recombination events and horizontal gene transfer shaped the Legionella pneumophila genomes. BMC Genomics 12:536. doi: 10.1186/1471-2164-12-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vos M, Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J 3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 14.Hayward AC. 1993. The host of Xanthomonas, p 51–54. In Swings J, and Civerolo EL (ed), Xanthomonas. Springer, Dordrecht, Netherlands. [Google Scholar]
- 15.Stall RE, Beaulieu C, Egel DS, Hodge NC, Leite RP, Minsavage GV, Bouzar H, Jones JB, Alvarez AM, Benedict AA. 1994. Two genetically diverse groups of strains are included in Xanthomonas campestris pv. vesicatoria. Int J Syst Bacteriol 44:47–53. doi: 10.1099/00207713-44-1-47. [DOI] [Google Scholar]
- 16.Almeida NF, Yan S, Cai R, Clarke CR, Morris CE, Schaad NW, Schuenzel EL, Lacy GH, Sun X, Jones JB. 2010. PAMDB, a multilocus sequence typing and analysis database and website for plant-associated microbes. Phytopathology 100:208–215. doi: 10.1094/PHYTO-100-3-0208. [DOI] [PubMed] [Google Scholar]
- 17.Jones JB, Bouzar H, Stall RE, Almira EC, Roberts PD, Bowen BW, Sudberry J, Strickler PM, Chun J. 2000. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int J Syst Evol Microbiol 50:1211–1219. doi: 10.1099/00207713-50-3-1211. [DOI] [PubMed] [Google Scholar]
- 18.Jones JB, Lacy GH, Bouzar H, Minsavage GV, Stall R, Schaad N. 2004. Bacterial spot—worldwide distribution, importance and review. 1st International Symposium on Tomato Diseases. Acta Hortic 695:27–33. [Google Scholar]
- 19.Cuppels DA, Louws FJ, Ainsworth T. 2006. Development and evaluation of PCR-based diagnostic assays for the bacterial speck and bacterial spot pathogens of tomato. Plant Dis 90:451–458. doi: 10.1094/PD-90-0451. [DOI] [PubMed] [Google Scholar]
- 20.Bouzar H, Jones J, Stall R, Louws F, Schneider M, Rademaker J, De Bruijn F, Jackson L. 1999. Multiphasic analysis of xanthomonads causing bacterial spot disease on tomato and pepper in the Caribbean and Central America: evidence for common lineages within and between countries. Phytopathology 89:328–335. doi: 10.1094/PHYTO.1999.89.4.328. [DOI] [PubMed] [Google Scholar]
- 21.Bouzar H, Jones JB, Somodi GC, Stall RE, Daouzli N, Lambe RC, Felix Gastelum R, Trinidad Correa R. 1996. Diversity of Xanthomonas campestris pv. vesicatoria in tomato and pepper fields of Mexico. Can J Plant Pathol 18:75–77. doi: 10.1080/07060669609500659. [DOI] [Google Scholar]
- 22.Hamza AA, Robene-Soustrade I, Jouen E, Gagnevin L, Lefeuvre P, Chiroleu F, Pruvost O. 2010. Genetic and pathological diversity among Xanthomonas strains responsible for bacterial spot on tomato and pepper in the southwest Indian Ocean region. Plant Dis 94:993–999. doi: 10.1094/PDIS-94-8-0993. [DOI] [PubMed] [Google Scholar]
- 23.Horvath DM, Stall RE, Jones JB, Pauly MH, Vallad GE, Dahlbeck D, Staskawicz BJ, Scott JW. 2012. Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS One 7:e42036. doi: 10.1371/journal.pone.0042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mhedbi-Hajri N, Hajri A, Boureau T, Darrasse A, Durand K, Brin C, Fischer-Le Saux M, Manceau C, Poussier S, Pruvost O. 2013. Evolutionary history of the plant pathogenic bacterium Xanthomonas axonopodis. PLoS One 8:e58474. doi: 10.1371/journal.pone.0058474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wichmann G, Ritchie D, Kousik CS, Bergelson J. 2005. Reduced genetic variation occurs among genes of the highly clonal plant pathogen Xanthomonas axonopodis pv. vesicatoria, including the effector gene avrBs2. Appl Environ Microbiol 71:2418–2432. doi: 10.1128/AEM.71.5.2418-2432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkinson N, Cowie C, Henney J, Stead D. 2009. Phylogenetic structure of Xanthomonas determined by comparison of gyrB seqeuences. Int J Syst Evol Microbiol 59:264–274. doi: 10.1099/ijs.0.65825-0. [DOI] [PubMed] [Google Scholar]
- 27.Kebede MG, Timilsina S, Ayalew A, Admassu B, Potnis N, Minsavage GV, Goss EM, Hong JC, Strayer A, Paret M, Jones JB, Vallad GE. 2014. Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur J Plant Pathol 140:677–688. doi: 10.1007/s10658-014-0497-3. [DOI] [Google Scholar]
- 28.Hamza AA, Robene-Soustrade I, Jouen E, Lefeuvre P, Chiroleu F, Fisher-Le SM, Gagnevin L, Pruvost O. 2012. Multilocus sequence analysis- and amplified fragment length polymorphism-based characterization of xanthomonads associated with bacterial spot of tomato and pepper and their relatedness to Xanthomonas species. Syst Appl Microbiol 35:183–190. doi: 10.1016/j.syapm.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Potnis N, Krasileva K, Chow V, Almeida NF, Patil PB, Ryan RP, Sharlach M, Behlau F, Dow JM, Momol MT, White FF, Preston JF, Vinatzer BA, Koebni R, Setubal JC, Norman DJ, Staskawicz BJ, Jones JB. 2011. Comparative genomics reveals diversity among xanthomonads infecting tomato and pepper. BMC Genomics 12:146. doi: 10.1186/1471-2164-12-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98. [Google Scholar]
- 32.Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 59:793–808. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- 33.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 34.Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 36.Tajima F. 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu Y-X, Li W-H. 1993. Statistical tests of neutrality of mutations. Genetics 133:693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph S, Forsythe SJ. 2012. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front Microbiol 3:397. doi: 10.3389/fmicb.2012.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 40.Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. 2010. RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26:2462–2463. doi: 10.1093/bioinformatics/btq467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pond SLK, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 42.Didelot X, Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts MS, Cohan FM. 1993. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics 134:401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posada D, Crandall KA, Holmes EC. 2002. Recombination in evolutionary genomics. Annu Rev Genet 36:75–97. doi: 10.1146/annurev.genet.36.040202.111115. [DOI] [PubMed] [Google Scholar]
- 45.Marcelletti S, Ferrante P, Scortichini M. 2010. Multilocus sequence typing reveals relevant genetic variation and different evolutionary dynamics among strains of Xanthomonas arboricola pv. juglandis. Diversity 2:1205–1222. doi: 10.3390/d2111205. [DOI] [Google Scholar]
- 46.Obradovic A, Mavridis A, Rudolph K, Janse JD, Arsemijevic M, Jones JB, Minsavage GV, Wang JF. 2004. Characterization of PCR-based typing of Xanthomonas campestris pv. vesicatoria from peppers and tomatoes in Serbia. Eur J Plant Pathol 110:285–292. doi: 10.1023/B:EJPP.0000019797.27952.1d. [DOI] [Google Scholar]
- 47.Astua-Monge G, Minsavage GV, Stall RE, Vallejos CE, Davis MJ, Jones JB. 2000. Xv4-Avrxv4: a new gene-for-gene interaction identified between Xanthomonas campestris pv. vesicatoria race T3 and the wild tomato relative Lycopersicon pennellii. Mol Plant Microbe Interact 13:1346–1355. doi: 10.1094/MPMI.2000.13.12.1346. [DOI] [PubMed] [Google Scholar]
- 48.Canteros BI, Minsavage GV, Jones JB, Stall RE. 1995. Diversity of plasmids in Xanthomonas campestris pv. vesicatoria. Phytopathology 85:1482–1486. doi: 10.1094/Phyto-85-1482. [DOI] [Google Scholar]
- 49.Basim H, Stall RE, Minsavage GV, Jones JB. 1999. Chromosomal gene transfer by conjugation in the plant pathogen Xanthomonas axonopodis pv. vesicatoria. Phytopathology 89:1044–1049. doi: 10.1094/PHYTO.1999.89.11.1044. [DOI] [PubMed] [Google Scholar]
- 50.Hert AP, Roberts PD, Momol MT, Minsavage GV, Tudor-Nelson SM, Jones JB. 2005. Relative importance of bacteriocin-like genes in antagonism of Xanthomonas perforans tomato race 3 to Xanthomonas euvesicatoria tomato race 1 strains. Appl Environ Microbiol 71:3581–3588. doi: 10.1128/AEM.71.7.3581-3588.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jibrin MO, Timilsina S, Potnis N, Minsavage GV, Shenge KC, Akpa AD, Alegbejo MD, Vallad GE, Jones JB. 2014. First report of Xanthomonas euvesicatoria causing bacterial spot disease in pepper in northwestern Nigeria. Plant Dis doi: 10.1094/PDIS-06-14-0586-PDN. [DOI] [PubMed] [Google Scholar]
- 52.Bui Thi Ngoc L, Vernière C, Jouen E, Ah-You N, Lefeuvre P, Chiroleu F, Gagnevin L, Pruvost O. 2010. Amplified fragment length polymorphism and multilocus sequence analysis-based genotypic relatedness among pathogenic variants of Xanthomonas citri pv. citri and Xanthomonas campestris pv. bilvae. Int J Syst Evol Microbiol 60:515–525. doi: 10.1099/ijs.0.009514-0. [DOI] [PubMed] [Google Scholar]
- 53.Mbega ER, Wulff EG, Mabagala RB, Adriko J, Lund OS, Mortensen CN. 2012. Xanthomonads and other yellow-pigmented Xanthomonas-like bacteria associated with tomato seeds in Tanzania. Afr J Biotechnol 11:14303–14312. doi: 10.5897/AJB12.1305. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.