Abstract
This review examines oxidative protein folding within the mammalian endoplasmic reticulum (ER) from an enzymological perspective. In protein disulfide isomerase-first (PDI-first) pathways of oxidative protein folding, PDI is the immediate oxidant of reduced client proteins and then addresses disulfide mispairings in a second isomerization phase. In PDI-second pathways the initial oxidation is PDI-independent. Evidence for the rapid reduction of PDI by reduced glutathione is presented in the context of PDI-first pathways. Strategies and challenges are discussed for determination of the concentrations of reduced and oxidized glutathione and of the ratios of PDIred:PDIox. The preponderance of evidence suggests that the mammalian ER is more reducing than first envisaged. The average redox state of major PDI-family members is found to be largely to almost totally reduced. These observations are consistent with model studies showing that oxidative protein folding proceeds most efficiently at a reducing redox poise consistent with a stoichiometric insertion of disulfides into client proteins. Following a discussion of the use of natively-encoded fluorescent probes to report the glutathione redox poise of the ER, this review concludes with elaboration of a complementary strategy to discontinuously survey the redox state of as many redox-active disulfides that can be identified by ratiometric LC-MS-MS methods. Consortia of oxidoreductases which are in redox equilibrium can then be identified and compared to the glutathione redox poise of the ER to gain a more detailed understanding of the factors that influence oxidative protein folding within the secretory compartment.
Keywords: Disulfide exchange, Endoplasmic Reticulum, Ero1, Glutathione, Hydrogen peroxide, Oxidative Protein Folding, Peroxiredoxin, Protein Disulfide Isomerase, Ratiometric Mass Spectrometry, Redox Potential, Quiescin sulfhydryl oxidase
Introduction
Understanding the mechanisms by which disulfide bonds are both inserted and isomerized during oxidative protein folding remains a significant intellectual challenge some 50 years after the classic studies of Anfinsen and colleagues on the refolding of reduced ribonuclease A (RNase) [1]. Indeed, while the disulfide bond is one of the most recognized forms of post-translational modification, fundamental aspects of oxidative folding remain unclear. For example, why is there such a proliferation of cellular strategies for the formation of disulfides in higher eukaryotes? How can these various pathways operate with the prevailing concentrations of reduced glutathione (GSH) in mammalian cells, and how can futile cycles between them be avoided? Finally, how is the choreography between folding and disulfide generation managed from one protein to the next?
There have been a number of recent comprehensive reviews dealing with oxidative protein folding in a variety of organisms and cellular locales (e.g. see [2-9]). Here we principally address disulfide bond generation within the secretory apparatus of animal cells, while acknowledging the major contributions from complementary studies on yeast [10-12]. Rather than presenting a detailed account of the discovery and characteristics of the various catalysts of disulfide bond generation, we will focus on conceptual issues and future challenges that seem relevant to a deeper understanding of oxidative protein folding within the mammalian secretory apparatus. We will first briefly address key biochemical aspects of thiol-disulfide exchange reactions. Recognized actors in various oxidative folding pathways in the mammalian endoplasmic reticulum (ER) will then be introduced. We then discuss the role of redox poise in the ER from the perspective of two major participants: the small molecule glutathione, and oxidoreductases of the protein disulfide isomerase (PDI) family. We recount methodological advances in the estimation of glutathione and PDI redox states in the ER and discuss issues with their implementation. The likelihood of redox equilibration between glutathione and PDI-family members will be discussed and whether equilibration could be reconciled with the efficient generation of disulfides in the ER. Finally we propose an expanded toolbox of methods to complement the fluorescent probes that have so transformed the field of redox biology.
Chemical aspects of thiol-disulfide exchange reactions
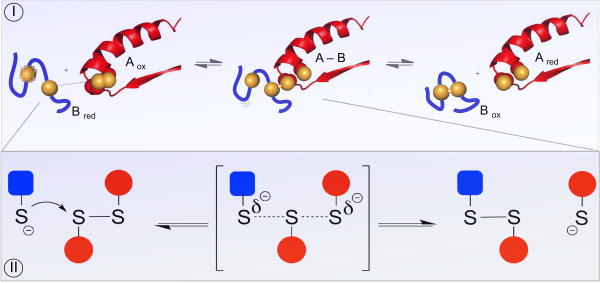
Thiol-disulfide exchange reactions are central to oxidative protein folding and key to the mechanism of almost all enzymes that generate and isomerize disulfide bonds [3, 13-15]. During the successful folding of a protein with multiple disulfide bonds an intensive iteration of disulfide exchanges precedes the emergence of the native fold (see later). Thiol-disulfide exchange reactions are initiated by the nucleophilic attack of a thiolate on a disulfide (Figure 1, Panel I). The thiolate is some 1010 – fold more reactive than its corresponding thiol form [14, 15]. The relationship between thiol pK values and the intrinsic nucleophilicity of their thiolates is described elsewhere [14, 15]. An important and still under-appreciated aspect of thiol-disulfide reactions is illustrated in Panel II. The attacking thiolate approaches along the disulfide axis and this requirement for colinearity establishes the orientation necessary for interactions between well-structured redox partners [15-19]. Thus disulfide exchange reactions have significant steric requirements that must be accommodated by the enzymes catalyzing oxidative protein folding. Additionally, these stringent requirements impact the rates by which disulfides can be reduced and rearranged non-enzymatically.
Figure 1. Mechanistic aspects of thiol-disulfide exchange reactions.
Panel I illustrates reduction of a CxxC disulfide motif in Aox by an unstructured reduced peptide dithiol (Bred; blue). Generation of the mixed-disulfide intermediate A-B involves an in-line transition state as depicted in the detail shown in Panel II.
A second nucleophilic attack in Figure 1, Panel I, discharges the mixed disulfide (providing that the resolving thiolate can approach in line) yielding a net redox reaction whose equilibrium constant is give by:
The magnitude of this equilibrium constant depends on which of the blue or red couples in Panel I is the stronger thermodynamic reductant. Several comprehensive reviews detail the application of the Nernst equation to thiol-disulfide equilibria (e.g. [20, 21]); the redox potential of a couple, E, is related to E′°, the corresponding value under standard conditions via:
The collected terms in parenthesis correspond to about a -30 mV change in the redox potential for every 10-fold increase in the ratio of [red]/[ox] for a particular 2-electron redox couple at 25 °C.
It is important to note that while the difference in standard redox potentials between the couples in Figure 1 dictate the overall equilibrium constant between reactants and products, such information provides no guidance concerning the rate of this equilibration. Thus the steric requirements for disulfide-exchange reactions may impose effective kinetic barriers preventing facile reactions between species that might be expected to react on thermodynamic grounds. Conversely, even if the reaction is thermodynamically far uphill - when products (Ared and Box) are strongly disfavored with respect to reactants (Aox and Bred) the A-B mixed-disulfide intermediate that forms between the reactants might be very stable thermodynamically [22]. Obviously arguments based on thermodynamics and redox potentials need to be applied with an appreciation of their limitations. A significant issue in this review is whether equilibrium conditions prevail between any of the thiol-disulfide couples in the secretory apparatus.
Oxidative protein folding – enzymes and oxidants
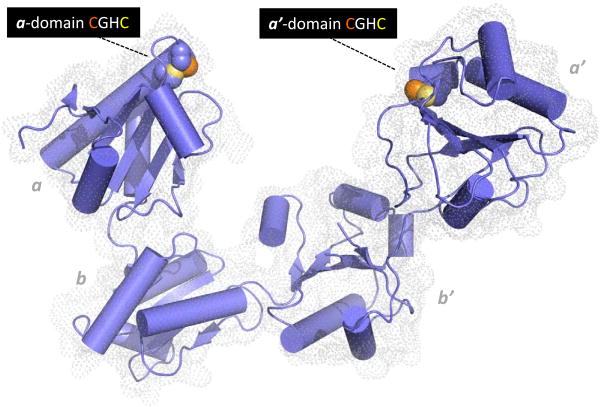
Oxidative protein folding comprises two conceptually separate stages. The first represents the net generation of disulfide bonds; each disulfide bond generated from a pair of thiols involves the removal of a pair of electrons. Since disulfides are frequently major determinants in the stability of secreted proteins [23], the insertion of disulfides linkages, particularly in early phases of oxidative folding where conformations resembling the native fold are likely to be poorly populated, is generally error-prone. Hence the second aspect of oxidative protein folding is the rearrangement of disulfide bonds via PDI enzymes. A strong case can be made that the isomerization of mispaired disulfide bonds is the linchpin of oxidative folding. In comparison, the net generation of disulfides is chemically much less demanding - from the inorganic oxidants that can be used in vitro (e.g. molecular iodine, ferricyanide, tetrathionate and hydrogen peroxide; [24, 25]) to the small molecule and enzymatic oxidants that contribute to disulfide formation in vivo (see later). The multiplicity of enzymes with PDI activity in various cell types perhaps speaks to the specialization of these catalysts for particular client types. There are about 20 proteins in the human PDI-family containing up to 5 thioredoxin domains and from 0-4 redox-active CxxC motifs [3, 26, 27]. A crystal structure of P4HB (hereinafter abbreviated PDI) is shown in Figure 2 [28]. PDI comprises 4 thioredoxin domain with the outer a and a′ domains carrying CxxC motifs. The first sulfur of each motif is comparatively solvent-accessible whereas its redox partner is buried behind it towards the core of the domain (Figure 2). The redox inactive b′ domain serves an important role in binding unstructured protein substrates [3, 29].
Figure 2.
Structure of human PDI. The coordinates for the oxidized protein are from Wang et al. [28]. Redox-active CxxC motifs are found in both a and a′ domains. The N-terminal cysteine sulfur atom of each motif (orange) is solvent accessible and engages in mixed disulfides with redox partners and proteins undergoing disulfide editing. The C-terminal cysteine by contrast is largely buried from solvent (yellow). The b and b′ domains are redox-inactive. Protein clients of PDI can occupy the central cavity with significant hydrophobic interactions with the b′ domain.
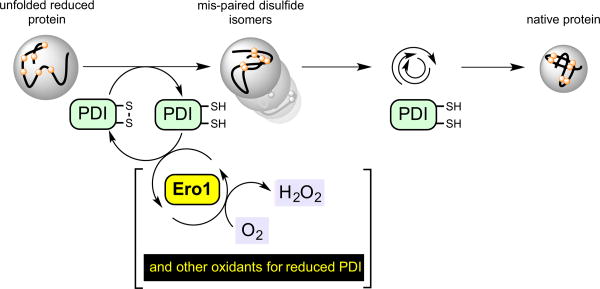
In many pathways for oxidative folding in the ER PDI is intimately involved in both the initial oxidation of client proteins and in the isomerization steps that are critical to correct mispairings. We will describe these as PDI-first pathways to distinguish them from PDI-second pathways (in which PDI's role is confined to the isomerization phase of oxidative folding). One PDI-first pathway is depicted in Figure 3. It should be noted at the outset that while oxidation and isomerization steps are depicted sequentially, the isomerase activity is likely engaged early in the oxidation phase to avoid the kinetic penalty associated with the accumulation of a concatenation of mispaired disulfides within a client protein.
Figure 3.
An example of a PDI-first pathway in oxidative protein folding. In the first phase the unfolded reduced protein is oxidized by PDIox. Here, reduced PDI is regenerated by the FAD-linked sulfhydryl oxidase, Ero1, with the formation of hydrogen peroxide. PDIred is then involved in correction of mispaired disulfides prior to the iterative emergence of the native protein fold.
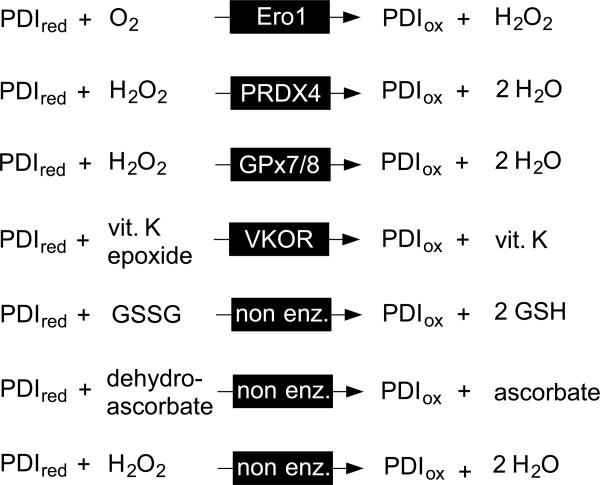
The PDIred generated upon oxidation of any given substrate dithiol in Figure 3 is recycled by Ero1-family proteins [10, 11, 30-34] with flavin-mediated transfer of a pair of electrons to molecular oxygen to yield hydrogen peroxide [35]. (Note that PDIred has negligible direct reactivity towards molecular oxygen and hence should never be described as an “oxidase”, or even a protein with “oxidase activity”. PDI is an oxidoreductase; the term oxidase is reserved for enzymes that oxidize substrates with the direct reduction of dioxygen to hydrogen peroxide [36]). Ero1p was first discovered as an essential protein in yeast [10, 11] and later two mammalian isoforms Ero1α and Ero1β were uncovered and subjected to detailed cell biological and biochemical scrutiny [30-34]. The general assumption that the mammalian Ero1 oxidases would prove essential in mammals was found to require reexamination as the result of an important study from the Ron laboratory [37]. The mild phenotype associated with the simultaneous ablation of both α– and β– isoforms [37] invigorated the search for other pathways for disulfide bond generation. Zito et al. [38] and Bulleid and coworkers [39] delineated pathways for the hydrogen peroxide-driven oxidation of PDIred catalyzed by peroxiredoxin 4 (PRDX4; Figure 4) [7, 8, 40]. Ruddock and colleagues described glutathione peroxidase 7 and 8 (PDI peroxidases) as a further means to the reoxidation of PDIred [41]. Rapoport and coworkers showed that vitamin K epoxide reductase provides yet another pathway for oxidative protein folding in the mammalian ER [42]. In addition to these enzymatic routes for the regeneration of PDIox, a number of small molecule oxidants for unfolded reduced proteins have been described (Figure 4). Oxidized glutathione (GSSG) was regarded for decades as an immediate oxidant for protein thiols, including PDIred, in the ER (reviewed in [21, 43]). More recently dehydroascorbate [44] and hydrogen peroxide [45] have also been described as candidate oxidants in the ER ([43, 46]).
Figure 4.
Some enzymatic and non-enzymatic oxidants for reduced PDI.
PDI – second pathways
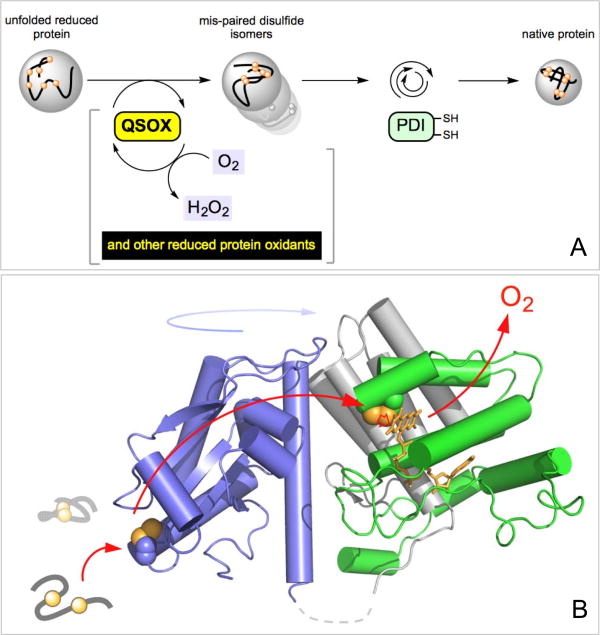
The small molecules listed in Figure 4 represent relatively non-specific oxidants of both protein and non-protein thiols. Hence, to the extent that GSSG, dehydroascorbate and hydrogen peroxide oxidize unfolded proteins directly, they can contribute to PDI-second routes to oxidative folding. However, there is one class of enzymes that is capable of directly oxidizing unfolded reduced peptides and proteins without significant collateral oxidation of PDIred [4, 18, 47-49]. Figure 5A shows that members of the Quiescin-sulfhydryl oxidase (QSOX) family of proteins transfer reducing equivalents from client protein dithiols directly to molecular oxygen. QSOX enzymes represent an intriguing amalgamation of PDI-like thioredoxin domains with helix-rich domains of the ERV family (Figure 5B) [4, 48, 50-52]. Later we show that efficient oxidative folding can occur when QSOX is the sole oxidant of client proteins and reduced PDI is confined to a role as an isomerization catalyst. These observations provide important perspective regarding the versatility and flexibility of pathways for oxidative folding (see later).
Figure 5.
PDI-second pathways of oxidative folding. The initial oxidation of reduced proteins is PDI-independent so that PDI is only engaged in the second phase of oxidative protein folding (Panel A). One facile direct oxidant of reduced conformationally-mobile proteins is Quiescin-sulfhydryl oxidase. Panel B shows the structure of the open form of QSOX from Trypanosoma brucei (3QCP; [52]).
PDI and Glutathione – interaction kinetics
Three redox systems that likely exert a strong influence on the redox poise within the ER are mentioned here. Glutathione is widely reported to be present at an aggregate concentration (reduced plus oxidized) of ~ 10 mM [53] with even higher concentrations proposed recently [54]. Mammalian PDI is a very abundant largely ER-resident protein with concentrations of perhaps up to 1 mM in certain cell types [3, 55]. The multiplicity of PDI family members will markedly inflate the total concentration of PDI-like thioredoxin domains in the ER. For example in HEK293T and HeLa cells the concentrations of PDI, ERp57, ERp72, and P5 were approximately comparable [56]. Finally, the ER of professional secretory cells may contain high concentrations of reduced and partially oxidized proteins in transit. While all these species contribute to the overall thiol-disulfide redox poise of the ER, we focus here on the interplay between the glutathione and PDI systems. How fast does glutathione react with PDI, and do measurements of the concentrations of GSH and GSSG when compared with the ratio of PDIred:PDIox suggest that these couples are in equilibrium in the ER? This discussion is largely focused on PDI since it is the best characterized representative of the mammalian PDI-family from both enzymological and cell biological aspects.
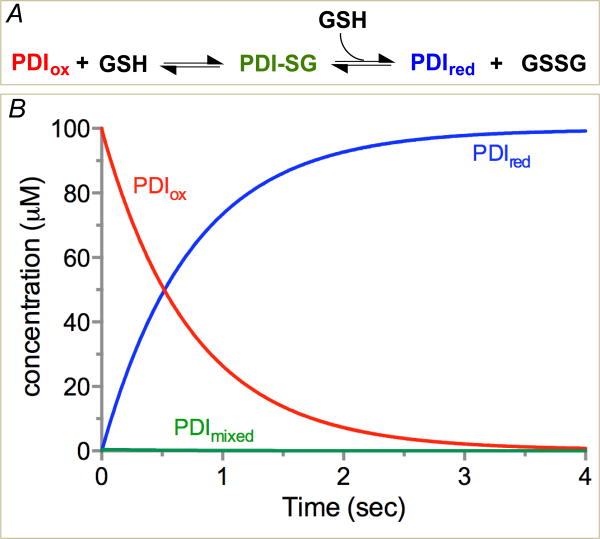
The classic studies of Darby and Creighton measured the rate constants for the interaction between the isolated a and a′ domains of human PDI in both reductive and oxidative directions using rapid mixing experiments [57]. Their studies suggested that PDI and glutathione could equilibrate with those concentrations of reduced and oxidized glutathione likely to prevail in the ER (see later) with sub-second half-times at pH 7.4 and 25 °C. A reinvestigation and extension of these studies by Lappi et al. reported comparable rate constants for the reduction of the PDIox a domain in the context of a full-length protein whose a′ domain was disabled by mutations to the CxxC motif [58]. This is to be expected because the a and a′ domains in human PDI seem to have very similar chemical reactivities and redox potentials (see later). To provide a graphic depiction of the reduction of PDI by 5 mM glutathione, the rate constants for the isolated a domain [59] were used to simulate the reaction in Figure 6A. Reduction shows a half-time of 0.52 seconds under these conditions with minimal glutathione mixed disulfide accumulation during this simulation (Figure 6B).
Figure 6.
Reduction of PDI by GSH. The model and rate constants of Darby and Creighton [57] were used for the a domain of human PDI reduced with glutathione (Panel A). Panel B shows the time course for oxidized, reduced and mixed disulfide forms of the single CxxC motif using 100 μM PDI and 5 mM GSH [59]. Under these conditions the half-time for equilibration is 0.52 sec; t1/2 values for 2.5 mM and 10 mM GSH are 1.12 and 0.25 sec respectively.
This rather rapid equilibration poses interesting issues for PDI-first pathways of oxidative protein folding in the ER. Thus the generally abundant GSH could compete with unfolded reduced proteins for PDIox [36]. To the extent that glutathione prevails, Ero1 would become a de facto glutathione oxidase. Possibly this is exactly what the cell intended such that GSH not only drives hydrogen peroxide generation but leverages a second disulfide bond formation via peroxiredoxin 4 and related pathways [7, 8, 38-41]. A series of comprehensive experiment describe the down-regulation of Ero1 activity when a regulatory disulfide is generated by PDI proteins so that overoxidation of the ER is avoided [32, 33, 60, 61]. These studies prompt an interesting question concerning the response to a decreased demand for disulfide-containing secreted proteins. Under this circumstance an increasing proportion of PDI might be reduced by GSH leading to the activation of Ero1 and an elevation of hydrogen peroxide production. The combined action of Ero1 and Prx4/peroxidases would then be expected to lead to a continuous generation of GSSG with the stoichiometry shown in Figure 7. The decreased luminal concentration of GSH might then stimulate ingress of cytosolic GSH via the bi-directional transporter [62]. These observation prompt a series of questions: is the ER a constitutive generator of GSSG in the absence of a heavy load of secreted proteins; what role would such a generalized loss of reducing equivalents play in the cellular energy economy; what is the fate of the accumulating impermeant GSSG under these circumstances; and do reservoirs of GSSG exist within the secretory apparatus that fulfill an analogous role to the vacuolar deposits uncovered in yeast by Dick and coworkers [63]?
Figure 7.
A potential pathway for redox cycling in the ER.
Preamble to redox poise measurements in the mammalian ER
As mentioned above, glutathione and PDI can rapidly equilibrate in vitro in a reaction that would be mostly complete in 1 sec under the conditions expected to prevail in the mammalian ER. In the following sections we first review experimental evidence for the concentration and redox poise of intraluminal glutathione. We then describe approaches and challenges to the determination of the redox status of PDI within the secretory apparatus. Given experimental values for glutathione and PDI, the extent to which these couples approach equilibrium can be assessed. Despite some skepticism concerning the applicability of redox potential measurements for intracellular thiol disulfide reactions involving glutathione [64] we note that the measurement of mass action ratios in cells has proved crucial in identifying the subset of steps in metabolic pathways that are far from equilibrium and thus candidates for control [65]. Hence it seems both relevant and informative to consider whether the PDI/glutathione reactions are close to equilibrium in the ER and if not, why not?
ER glutathione redox poise
Hwang et al. in 1992 provided the first quantitative estimation of the redox poise within the ER [66]. Using a membrane-permeant tetrapeptide that was subsequently glycosylated within the ER, they showed that the GSH:GSSG ratio in murine hybridoma cells ranged from 1:1 to 3:1 (Table 1). While this study selected the subset of peptides resident in the ER for analysis, later work (entries 2 and 3) determined the glutathione status of isolated microsomes by an initial alkylation with monobromobimane [67] or by acid treatment followed by conjugation with dansyl chloride [68]. Isolation of microsomes is associated with two potential complications that might lead to an overestimation of the oxidized status of the ER [54]. The first is the loss of GSH, but not GSSG, from microsomes via a bi-directional transport system [62]; a second is the tendency of the luminal contents to undergo enzyme-mediated oxidation prior to analysis [68]. As detailed earlier [3, 20, 21] the conversion of the ratio of GSH:GSSG deduced from the measurements in entries 1-3 to redox potentials requires knowledge of the net concentration of glutathione within the ER lumen. This is because the monothiol GSH appears as a squared term ([GSH]2/[GSSG]) in the Nernst equation. Here we have assumed a total luminal glutathione concentration ([GSH] + 2[GSSG]) of 10 mM to allow a comparison between the range of entries in Table 1. In another approach (entry 4) Montero et al. used a single cysteine glutaredoxin to deduce a GSH:GSSG ratio of <7:1 in HeLa cells [54]. In summary, entries 1-4 predict a redox potential for the ER luminal pool of between about -160 to -200 mV (assuming an aggregate 10 mM glutathione concentration).
Table 1.
Measurement of glutathione redox state in the mammalian ER. Since the redox potential of glutathione buffers depends on the [GSH]2/GSSG term, they are dependent on absolute concentration (see the Text). For the purposes of comparison, these values assume a total glutathione concentration (GSH + 2 GSSG) of 10 mM. At a ratio of GSH:GSSG of 3:1 the redox potential would be -186 mV; lower aggregate concentrations of 7.5 and 5 mM glutathione would yield somewhat more positive redox potentials of -182 mV and -177 mV respectively. We have arbitrarily used 10 mM total glutathione to present a comparison with the redox potentials reported for the fluorescent probes entries 5-7.
| Entry | Biological Sample | Method | GSH:GSSG | Reported redox potential | Adjusted Value | Year/Reference |
|---|---|---|---|---|---|---|
| 1 | Murine hybridoma CRL1606 cells | ER targeted peptide | 1:1 to 3:1 | -170 to 185 mV (8 mM) | -164 to -186 mV | 1992/[66] |
| 2 | Rat liver microsomes | Microsomal Isolation | 3:1 to 4:1 | NR | -186 to -192 mV | 2004/[67] |
| 3 | Rat liver microsomes | Microsomal Isolation | 4.7:1 to 5.5:1 | NR | -194 to -197 mV | 2008/[68] |
| 4 | HeLa cells | sCGrx1pER probe NEM soaking | <7:1 | NR | >-201 | 2013/[54] |
| 5 | Human fibroblast cells (HT1080) | roGFP1iL | NR | -231 mV | NA | 2011/[72] |
| 6 | CHO cells | CY_RL7 fluorescent probe | NR | -118 mV | NA | 2012/[75] |
| 7 | pancreatic acinar cells (AR42j) | roGFP1iE | NR | -236 mV | NA | 2013/[74] |
| 8 | HeLa cells | Grx1_roGFP1iE | NR | -208 mV | NA | 2013/[73] |
The landmark studies of Winther and colleagues introduced a redox-active disulfide bond within a yellow fluorescent variant of GFP (rxYFP) such that the fluorescence was responsive to thiol redox state [69]. Subsequently they fused rxYFP to glutaredoxin to make the probe communicate more specifically and rapidly with glutathione [70]. Remington, Tsien and their coworkers developed a ratiometric GFP probe (roGFP) [71] and variants of this construct have proved useful in interrogating the relatively oxidizing environment of the secretory system. Use of these probes in a range of cultured cell types yield the values shown in entries 5, 7 and 8 (Table 1). A value of -231 mV (entry 5) was obtained by Bulleid and colleagues using roGFP1iL expressed in the ER of human fibroblast cells [72]. A similar probe fused to glutaredoxin used by Appenzeller-Herzog, Dick and their colleagues (entry 8) gave a redox potential of – 208 mV in the ER of HeLa cells [73]. Ron and colleagues monitored the change in fluorescent lifetime (rather than ratiometric fluorescence intensity) between oxidized and reduced forms of roGFP1iE [74]. They obtained a redox poise for the ER lumen in a pancreatic acinar cell line of -236 mV (Table 1, entry 7). While entries 5, 7 and 8 reflect the use of different cell types and methodologies, they yield rather comparable redox potentials averaging – 225 mV. In contrast, a probe designed by Kolossov and colleagues exploits changes in FRET between enhanced versions of CYP and YFP fused via a cysteine-containing linker region. The construct records an extremely oxidizing potential of -118 mV in CHO cells (entry 6; [75]). It appears that this probe is responding to redox components within the ER that are not in effective communication with the glutathione redox pool [73] and so we will not consider it further here.
Assuming that the roGFP-derived probes are in equilibrium with glutathione pools within the ER, an average potential of – 225 mV would predict a GSH:GSSG ratio of 35:1 (using the standard redox potential for glutathione of -240 mV [13], and an aggregate glutathione concentration of 10 mM). In contrast, the experimentally reported values in entries 1-3 in Table 1 are considerably more oxidizing (with GSH:GSSG ratios between 1:1 to 5.5:1). In view of the significant experimental difficulties in accurately assessing intraluminal GSH and GSSG concentrations, the fluorescent probes in entries 5, 7 and 8 (Table 1) appear to provide the best current measures of redox poise. These probes respond rapidly to glutathione and they report directly from the ER lumen of live cells.
Redox state of PDI in the mammalian ER
A redox poise of -225 mV for glutathione in the ER corresponds to a predicted ratio of ~90:1 for PDIred:PDIox (see later). Two main methods have been used to measure the redox state of PDI family members within the mammalian ER. The experiments described in entries 1,2, 4 and 6 (Table 2) incubated intact cells with NEM to quench thiols. Subsequent quantitation of oxidized and reduced components employed gel-shift methods detected using Western blots [76]. These studies found various PDI family members to be largely reduced. Studies in HEK-293 cells (entry 4) applied a quantitative analysis procedure that allowed the percentage of each redox state of PDI to be quantified (here 67% of the CxxC motifs are reduced). A recent study (entry 7) by Inaba and colleagues, using HEK293 cells rapidly quenched in TCA showed that PDI was ~80 % reduced [77]. ERp46, ERp57, Erp72 and P5 showed levels of reduction from about 95 – 100% [77]. In contrast a protocol involving microsome isolation prior to trapping (entry 3) found that both PDI and ERp57 were predominantly oxidized [78]. As mentioned earlier, this divergent outcome might reflect selective loss of glutathione and/or oxidation of luminal contents of the ER.
Table 2.
Determination of the redox state of PDIs within the ER. In a number of references the redox state of PDI family members was not quantified densitometrically. In most cases the overall redox state of PDI family members that carry multiple CxxC motifs is reported in aggregate, without identification of the population of redox isoforms (but see entry 4; here, the percentage of CxxC domains that are reduced totals 67%).
| Entry | Biological Sample | Method | Protein | Redox State | Year/Reference |
|---|---|---|---|---|---|
| 1 | Hela, COS, U937 cells | TCA/intact cells | PDI and ERp57 | Predominantly Reduced | 2001/ [93] |
| 2 | HT1080 cells | NEM soaking/intact cells | PDI, ERp57, ERp72, P5, PDIr | Predominantly Reduced | 2004/ [53] |
| 3 | Rat Liver Microsomes | Microsome Isolation /TCA | PDI and ERp57 | Predominantly Oxidized | 2005/[78] |
| 4 | HEK-293 cells | NEM soaking/ intact cells | PDI TMX3 | 50% fully reduced, 18% a-oxidized, 15% a′-oxidized, and 16% fully oxidized Predominantly Reduced | 2008/[94] |
| 5 | Rat Heart Tissue | Tissue frozen in liquid nitrogen | PDI | Predominantly Reduced | 2011/ [95] |
| 6 | HeLa cells | NEM soaking/intact cells | ERp57 | Predominantly Reduced | 2013/[73] |
| 7 | HEK293 | TCA treatment intact cells | PDI, ERp46, ERp57, P5, ERp72 | Predominately Reduced 0%-20% | 2013/[77] |
Chemical trapping of PDI redox state
Several pioneering early studies of the redox poise of PDI soaked live cells using NEM before application of gel-shift analyses to measure redox state. The use of NEM was a logical choice because it reacts with thiols some thousand-fold faster than the iodoacetamide [13]. Further, NEM is membrane-permeant [3, 53, 79] - although this aspect does not seem to have been quantified. Cells were typically exposed to ice-cold NEM (in a concentration range of 1- 40 mM) prior to a disruption step and the determination of redox status by gel-shift. A relevant question is how fast does trapping have to be? If PDI and glutathione were at equilibrium with a t1/2 of about 500 msec (Figure 6), then NEM quenching would need to be completed in 50 – 100 msec to accurately reflect the intraluminal redox state of this oxidoreductase.
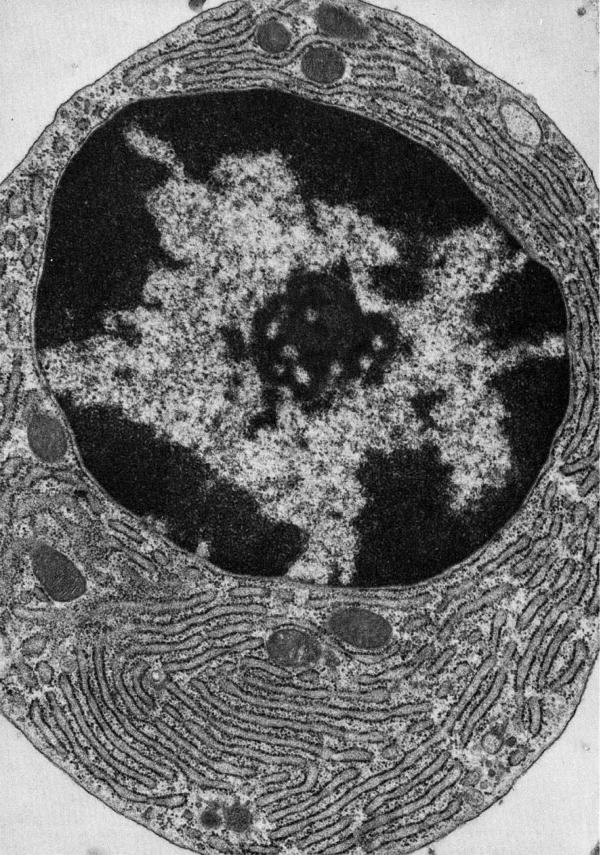
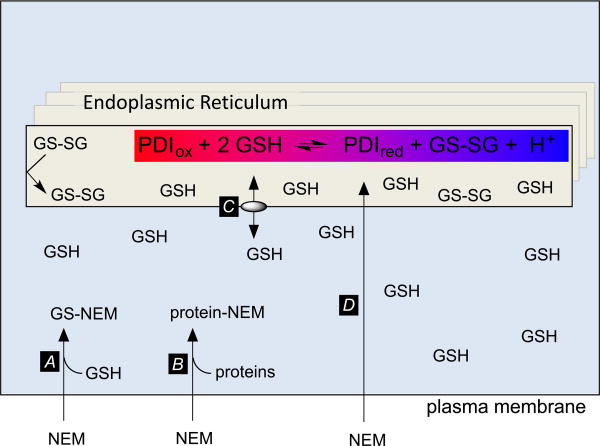
Figure 8 shows a micrograph to illustrate some of the cellular features that may delay quenching in an NEM soaking procedure. This professional secretory cell contains a packed reticular network of rough ER with associated post-ER compartments including the ER-Golgi intermediate complex, cis- and trans-Golgi networks and, finally, the vesicular/granular traffic destined for the plasma membrane. During NEM-soaking protocols the quenching agent must transit not just the plasma membrane but a multiplicity of internal membranes and/or navigate a convoluted path of cytosol separating adjacent luminal sections of ER (Figure 8). At the outset NEM diffusing across the plasma membrane will encounter a typical cytosolic GSH concentration of 5-10 mM (Figure 9, arrow A) [54, 80, 81]. Since the second-order rate constant for reaction with GSH at neutral pH values is about 104 M-1s-1 [13] the half-time for NEM capture under these conditions will be of the order of 10 msec. The high protein thiol content within the cytosol [81] will also delay accumulation of unmodified NEM (step B). As the depletion of cytosolic GSH continues release of ER-resident GSH (via step C) could bias the PDI/glutathione equilibrium in favor of PDIox because GSSG is reported to be impermeant [62]. Finally, as NEM gains entry to the ER by diffusion (step D) it will further deplete the level of thiols in that compartment. Which thiols are preferentially alkylated? If NEM reacts with luminal GSH faster than with PDIred, this would bias the equilibrium in favor of more PDIox. However, if PDIred is alkylated preferentially, then this will have the opposite effect, leading to an overestimation of PDIred. These issues were recognized as potential artifacts of in vitro trapping protocols by Darby and Creighton decades ago [57] In the present case we are unaware of data that allows for a comparison of the bimolecular rate constants for the reaction of NEM with GSH and PDIred at pH 7.4. Finally, while cooling cells on ice is often employed as part of alkylation quenching reactions, it is not immediately apparent that this will help: while a 35 °C drop in temperature will slow typical enzymatic reactions by a factor of about 5-10, it will also decrease the rate of diffusion of NEM across membranes, and slow the alkylation chemistry itself. These complexities make it uncertain whether the current methodologies used for trapping PDI redox state in live cells using NEM are demonstrably adequate. Trapping could be accomplished more rapidly with isolated microsomes, but the potential for leakage of GSH and for further enzymatic and non-enzymatic oxidation of PDI remain issues of concern (see earlier).
Figure 8.
Electron micrograph of a plasma cell from guinea pig bone marrow showing an extensive reticular network of ER. Reproduced from [103].
Figure 9.
The use of NEM to trap intraluminal redox state within the ER. The highlighted equilibrium depicts the reduction of one PDI CxxC motif by GSH. Approximately one proton is released at neutral pH values (see the text).
Other trapping methods have been utilized in which pelleted cells, cells in culture, or tissues samples are treated with TCA. These methods are likely to be much more robust providing that denaturing acidification is achieved by rapid mixing methods. An aggressive denaturation is important because some PDIs retain activity at low pH values [3]. In a related issue, it should be noted that the equilibrium between PDIox and GSH is itself pH dependent (Figure 9). Since the pK of the GSH thiol group is ~9.7 and that of the surface exposed N-terminal cysteine of reduced CxxC motifs in PDI is 4.4 - 6.7 [3], reduction of each redox-active CxxC disulfide in the isomerase would lead to the release of approximately one proton at pH 7.4. If the pH of the ER is not lowered rapidly enough, acidification will push the equilibrium towards PDIox before the isomerase is inactivated. The time resolution of acid trapping could be further improved by adopting those freeze-quenching procedures already employed for the isolation of enzyme intermediates and for the measurement of metabolite mass action ratios (see later).
The influence of PDI redox state on in vitro oxidative protein folding
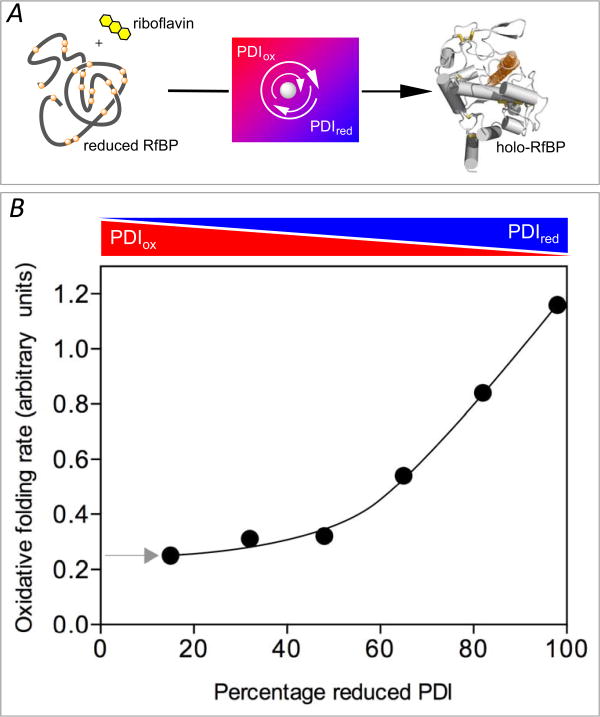
Table 3 lists the redox potentials of some other PDI family members and the predicted redox state if they, too, were to equilibrate at a redox poise of -225 mV. Although these predictions initially seem at variance with PDI-first pathways, earlier in vitro studies showed that oxidative protein folding occurs efficiently at very low levels of PDIox and that an accumulation of oxidized isomerase actually slowed the emergence of the native fold. The pioneering work of Gilbert and colleagues demonstrated that the oxidative refolding of RNase by PDI is facilitated when the isomerase is largely reduced [82]. RNase has proved a staple of oxidative folding studies and has 4 native disulfide bonds and consequently 105 disulfide isomers for the fully oxidized enzyme [83]. We wished to explore the influence of the redox state of PDI on the refolding of a protein with a much more complicated disulfide connectivity. Using avian riboflavin binding protein (RfBP; 9 disulfides and >34 million disulfide isomers) enabled us to continuously follow the recovery of riboflavin binding ability by the complete fluorescence quenching that accompanies the association of free riboflavin to apo-RfBP (Figure 10A) [49]. Mixtures of PDIox and PDIred with an aggregate concentration of 30 μM PDI served as redox buffers to drive the oxidative refolding of 1 μM reduced RfBP in the complete absence of glutathione [49]. The use of an enzyme in considerable molar excess over its substrate might initially appear unusual, however it is likely to be a frequent occurrence in cellular metabolism [84]. In particular, aggregate PDI concentrations probably approach several mM in the ER [56, 85] and are likely to be comparable to, or higher than, any one particular client protein. Figure 10 shows that under the most oxidizing conditions possible (far left; starting with 30 μM PDIox and no PDIred), the stoichiometric introduction of disulfides into RfBP will generate a redox buffer containing 25.5 μM PDIox and 4.5 μM PDIred (comprising 15% of the total PDI concentration; see arrow at left). Folding under these conditions is relatively slow, but accelerates as the percentage of PDIred in the redox buffer increases (rightwards in Figure 10). Under the most reducing environment, the redox buffer starts with 85% PDIred and 15% PDIox and ends with essentially 100% PDIred [49]. Oxidative folding is even faster when fully reduced PDI is used from the outset and disulfides are generated by nanomolar levels of QSOX [49]. Since QSOX cannot oxidize PDIred directly [49] the role of the isomerase in this PDI-second model is confined to correcting mispairings introduced by the oxidase.
Table 3. Redox potentials and projected redox state of selected human PDI-family members.
| Protein | Gene Name | Redox Potential (mV) | Reference | Red:Ox at -225 mV (assuming equilibration) |
|---|---|---|---|---|
| PDI | P4HB | -165 (avg) a | [96] | 90:1 |
| -176 (avg) | [97] | 40:1 | ||
| -160 (avg) | [98] | 130:1 | ||
| -162 (avg) | [56] | 110:1 | ||
| -163 (a) | [90] | 100:1 | ||
| -169 (a′) | [90] | 65:1 | ||
| ERp57 | PDIA3 | -167 (a) | [99] | 80:1 |
| -156 (a′) | 175:1 | |||
| -155 (avg) | [56] | 190:1 | ||
| ERp72 | PDIA4 | -155 (avg) | [56] | 195:1 |
| P5 | PDIA6 | -146 (avg) | [56] | 360:1 |
| ERdj5 | DNAJC10 | -190 (avg) | [100] | 14:1 |
| ERp18 | TXNDC12 | -165 | [101] | 90:1 |
| ERp46 | TXNDC5 | -161 (avg) | [56] | 120:1 |
| TMX3 | TMX3 | -157 | [102] | 160:1 |
Redox potentials for proteins with multiple CxxC motifs reflect the average redox behavior (denoted by avg) unless explicitly noted. ERp18 and TMX3 contain a single CxxC motif
Figure 10.
The rate of oxidative refolding of riboflavin binding protein in the presence of PDI redox buffers of defined concentration. Refolding was followed continuously by the quenching of riboflavin binding on association with folded oxidized RfBP in the absence of small molecule redox species and without other enzymatic catalysts of disulfide bond generation (panel A). Rates of refolding are plotted as a function of the percentage of PDIred in the PDI redox buffer (panel B). The concentration of reduced RfBP was 1 μM and the aggregate concentration of reduced and oxidized forms comprising the PDI redox buffer was 30-fold higher (see the Text). Data redrawn from Rancy and Thorpe [49].
These oxidative refolding model studies, using two widely different protein clients, show that efficient oxidative folding can occur under relatively reducing conditions and that increasing levels of PDIox actually slow acquisition of the native disulfide patterns in RfBP [49, 82]. Thus it seems likely that the accumulation of PDIox may be a general impediment to the efficient folding of a range of client proteins within the ER. PDI-first pathways may thus only need to maintain enough PDIox to stoichiometrically oxidize protein clients. In this way PDI-mediated oxidation does not run ahead of the isomerization of mispaired disulfides that is typically rate-limiting in oxidative protein folding [82]. This modest oxidation requirement may partly rationalize the seeming degeneracy of pathways for the oxidation of PDIred in mammalian cells (see above); protein folding will only be seriously compromised when the ER can no longer maintain sufficient PDIox to sustain the secretory load. Hence phenotypes associated with enzyme knockouts/knockdowns may emerge more clearly when secretion systems are stretched to capacity.
PDI within the ER – equilibrium or non-equilibrium states
Table 3 contains a prediction of the redox state of PDI-family members if they were in equilibrium with a redox potential of – 225 mV (corresponding to a GSH:GSSG ratio of 35:1 at 10 mM total glutathione). This potential is strongly reducing from the perspective of PDI-family members. Even if a potential of -200 mV is used (corresponding to a more oxidizing 5:1 ratio of GSH:GSSG; see earlier) most of the PDIs listed in Table 3 would still show reduced:oxidized ratios of >10. The experimental evidence for the redox state of PDI (Tables 2 and 3) seems generally consistent with both scenarios given the marked difficulties of the measurements.
While it appears that the redox potentials of the PDI and glutathione redox pools, when averaged over the ER system, are not widely different, a state of disequilibrium could be maintained if the oxidation of PDIred were to outstrip the rate at which PDI can equilibrate with glutathione. Another possibility is that the secretory system may provide structured microenvironments with clusters of enzymes, or sequestered luminal sections, that protect pools of PDI from equilibration with the bulk glutathione redox pool of the ER. The existence of a multiprotein complex containing molecular chaperones, the isomerases PDI and ERp72, and other component has already been reported in the ER of lymphoma cells [86]. Concentration gradients formed along stretches of the ER lumen might create “hot spots” of oxidation that may not be in equilibrium with the GSH levels that are averaged over the entire contents of the ER.
Future directions
Current procedures use gel-shift methods [76] combined with Western blots to estimate the redox state of one or other of the PDI family proteins resident in the ER. However what of all the other thiol-disulfide oxidoreductases that are directly or indirectly involved in oxidative folding, or otherwise responsive to changing redox states of the ER? While modern fluorescent probes offer the unparalleled advantage of combining a real-time appraisal of redox poise with the spatial resolution of current fluorescence microscopic methods, they cannot address multiple thiol-disulfide couples simultaneously. In the ER, for example, there are likely to be proteins with CxxC or other redox-active disulfides that are in equilibrium with the glutathione pool, and other protein consortia in redox equilibrium with themselves but not with the glutathione pool (Figure 11). There may be other proteins that are far from equilibrium with any of the thiol-disulfide couples that play major roles in modulating redox events within the ER. If measurement of a diverse array of oxidoreductases with known standard redox potentials (e.g. Table 3) predicts a common redox potential, and this consortium of proteins show coordinate redox changes with differing cellular conditions, then these proteins are likely equilibrium partners in a common redox environment [20]. Such equilibration could result from a direct communication between protein networks, as has recently been suggested by Nagata and colleagues [56], or via the intermediacy of a small molecule pool such as that provided by glutathione (Figure 11). Such analyses will likely identify components that are far from equilibrium with known redox pools/environments. We suggest that global approaches are likely to provide a useful complement to the real-time fluorescent probes mentioned earlier. All of the logistical steps for the implementation of such method have precedent – from freeze quenching and alkylation procedures, through procedures for the ratiometric mass spectrometric determination of multiple peptides carrying redox active dithiol/disulfide motifs [76, 87]. A flow diagram of this approach is presented in Figure 12.
Figure 11.
Schematic diagram of equilibrating and non-equilibrating redox species in the ER. Redoxactive proteins are distinguished by their shapes; the state of their redox active disulfides is shown by the blue and red shading. Some species are envisaged as directly in equilibrium with the glutathione redox pool and would have a range of redox states depending on their standard redox potentials. Other proteins are represented as interacting indirectly with the glutathione pool or with consortia of other proteins insulated from small molecular weight redox buffers.
Figure 12.
A suggested protocol for a more global analysis of the redox state of thiol-disulfide oxidoreductases within the Er (see the Text).
Well-established freeze quenching methodologies (step A) provide an alternative to passively soaking cells with NEM. Applied to tissues, they involve very rapid compression of small volumes of material on heavy metal plates cooled in liquid nitrogen [88]. Cell suspensions can be sprayed against rapidly rotating copper wheels that are cooled in liquid nitrogen and the accumulating frozen suspension scraped continuously into liquid nitrogen prior to TCA treatment [89]. Alternatively, a suspension of cells could be sprayed in fine droplets into rapidly stirred acetone at -78 °C followed by acidification with TCA and recovery of the dehydrated acidified powder by centrifugation at sub-zero temperatures. Cells on surfaces can be rapidly cooled by immersion in a slush of liquid/solid nitrogen (-210 °C) to avoid the generation of an insulating blanket of nitrogen gas [88]. In terms of quenching reagents (step B), while NEM is a facile alkylating agent (see above) it is generally not well suited for general mass spectrometric applications because NEM-adducts are prone to ring opening and are subject to dissociation by reverse Michael reactions [15]. Since the cell samples would be already quenched, a slower and more robust alkylating agent such as iodoacetate is more suitable. Iodoacetate is commercially available in 13C forms facilitating the application of ratiometric mass spectrometric approaches [76, 87]. In brief, forcing alkylation with normal iodoacetate under denaturing conditions will label all free thiols. Following rigorous removal of alkylating agent, disulfides are reduced with DTT or tris(2-carboxyethyl)phosphine and the liberated thiols labeled with excess 13C iodoacetate. The ratio of 13C /12C labeling peptides then signals their redox status in the original sample. In an important study providing precedent for the method, Bulleid and coworkers used ratiometric mass spectrometry to simultaneously determine the redox potentials of both CxxC motifs of full length PDI using in gel tryptic digestion of carboxymethylated PDI [90] (Table 3). The labeled 12C carboxymethylated CxxC peptides from both a and a′ domains of PDI show distinct masses (of 1944.866 and 1915.814; Table S1) and so can be readily distinguished from each other and from their corresponding mass-shifted forms reflecting the proportion of oxidized CxxC motifs in the sample. Importantly, inspection of the sequence databases for human PDI-family members that carry one or more CxxC motifs shows that of the 26 CxxC tryptic peptides from 13 PDIs, only two peptides had identical mass (the a′ domain of ERp57 and the third CxxC motif in ERp72; Table S1). The remaining peptides have distinguishable masses (from 819.287 to 5102.252 for their 12C carboxymethylated derivatives) and thus, in principle, can be discerned by mass spectrometry alone. Of course digestion of a whole cell will generate a very large number of peptides and, while a number of mammalian PDI family members are relatively abundant [3, 56, 85], pre-culling of proteins (e.g. by application of KDEL-specific antibodies; step E) may prove advantageous. In summary the scheme outlined in Figure 12 will allow searching for alkylated peptides whose mass is already known from the sequences of multiple cellular oxidoreductases both in the ER and beyond. None of the CxxC motifs in human PDI-family members contain inter-cysteine tryptic cleavage sites (Table S1). More generally, this method is amenable to any redox-active thiol in the proteome providing that it can be reliably identified mass spectrometrically.
Envoi
Understanding oxidative protein folding in the ER presents formidable challenges for the enzymologist. In a conventional metabolic pathway, individual steps are typically catalyzed by single enzymes that almost always show high specificity for their substrates and exhibit rate enhancements that are typically >1010-fold greater than the corresponding uncatalyzed reaction [91]. Non-enzymatic transformations very rarely contribute to metabolic flux and the role of an enzyme within a pathway can be identified with confidence. In contrast, many of the catalysts of oxidative folding, particularly PDI-family members, show pedestrian rate enhancements [13, 82, 92]. Further, the rather promiscuous chemical reactivity of thiols leads to the possibility that small molecule oxidants will contribute to the overall flux of disulfide bond generation [43, 44, 46]. Many of the enzymes of oxidative protein folding have broad and overlapping substrate preferences that compound difficulties in the interpretation of knock-down experiments. As an additional manifestation of the apparent plasticity of oxidative protein folding, the in vitro studies described here [49, 82] suggests that oxidative folding per se might tolerate all but the most oxidizing conditions within the ER. Nevertheless those model studies show that oxidative folding is more efficient under comparatively reducing conditions and this aligns with most reports of the prevailing averaged redox state for PDI family members within the ER (Table 2). It remains to be seen whether the ER shows a heterogeneity of redox environments and whether the redox poise of post ER compartments is significantly different from the bulk ER lumen. We further need to explore the redox protein consortia that are believed to populate the ER and the extent to which network communication is effected directly [56] or via the mediation of glutathione. Finally we suggest that a discontinuous interrogation of as broad a sampling of redox-active disulfides that can be identified by modern biological mass spectrometry approaches will prove a useful complement to the fluorescent probes that have transformed our understanding of redox homeostasis within cells.
Supplementary Material
Acknowledgments
This work was supported in part by NIH GM26643 (CT.) and USPHS Training Grant 1-T32-GM008550 (DH).
Abbreviations
- NEM
N-ethylmaleimide
- PDI
protein disulfide isomerase
- QSOX
Quiescin sulfhydryl oxidase
- RNase
ribonuclease A
- TCA
trichloroacetic acid
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Anfinsen CB. Principles that Govern the Folding of Protein Chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 2.Riemer J, Bulleid N, Herrmann JM. Disulfide formation in the ER and mitochondria: two solutions to a common process. Science. 2009;324:1284–1287. doi: 10.1126/science.1170653. [DOI] [PubMed] [Google Scholar]
- 3.Hatahet F, Ruddock LW. Protein Disulfide Isomerase: A Critical Evaluation of Its Function in Disulfide Bond Formation. Antioxid Redox Signalling. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 4.Kodali VK, Thorpe C. Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins. Antioxid Redox Signalling. 2010;13:1217–1230. doi: 10.1089/ars.2010.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Ann Rev Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- 6.Sevier C. Erv2 and Quiescin Sulfhydryl Oxidases: Erv-Domain Enzymes Associated with the Secretory Pathway. Antioxid Redox Signalling. 2012;16:800–808. doi: 10.1089/ars.2011.4450. [DOI] [PubMed] [Google Scholar]
- 7.Oka OB, Bulleid NJ. Forming disulfides in the endoplasmic reticulum. Biochim Biophys Acta. 2013;1833:2425–2429. doi: 10.1016/j.bbamcr.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Zito E. PRDX4, an endoplasmic reticulum-localized peroxiredoxin at the crossroads between enzymatic oxidative protein folding and nonenzymatic protein oxidation. Antioxid Redox Signalling. 2013;18:1666–1674. doi: 10.1089/ars.2012.4966. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Holmgren A. The Thioredoxin Superfamily in Oxidative Protein Folding. Antioxid Redox Signalling. 2014;21:457–470. doi: 10.1089/ars.2014.5849. [DOI] [PubMed] [Google Scholar]
- 10.Frand AR, Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 11.Pollard MG, Travers KJ, Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 12.Sevier CS, Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 14.Nagy P. Kinetics and Mechanisms of Thiol-Disulfide Exchange Covering Direct Substitution and Thiol Oxidation-Mediated Pathways. Antioxid Redox Signalling. 2013 doi: 10.1089/ars.2012.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winther JR, Thorpe C. Quantification of thiols and disulfides. Biochim Biophys Acta. 2014;1840:838–846. doi: 10.1016/j.bbagen.2013.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosenfield RE, Parthasarathy R, Dunitz JD. Directional Preferences of Nonbonded Atomic Contacts with Divalent Sulfur .1. Electrophiles and Nucleophiles. J Amer Chem Soc. 1977;99:4860–4862. [Google Scholar]
- 17.Bach RD, Dmitrenko O, Thorpe C. Mechanism of thiolate-disulfide interchange reactions in biochemistry. J Org Chem. 2008;73:12–21. doi: 10.1021/jo702051f. [DOI] [PubMed] [Google Scholar]
- 18.Codding JA, Israel BA, Thorpe C. Protein Substrate Discrimination in the Quiescin Sulfhydryl Oxidase (QSOX) Family. Biochemistry. 2012;51:4226–4235. doi: 10.1021/bi300394w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandes PA, Ramos MJ. Theoretical insights into the mechanism for thiol/disulfide exchange. Chemistry. 2004;10:257–266. doi: 10.1002/chem.200305343. [DOI] [PubMed] [Google Scholar]
- 20.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 21.Appenzeller-Herzog C. Glutathione- and non-glutathione-based oxidant control in the endoplasmic reticulum. J Cell Sci. 2011;124:847–855. doi: 10.1242/jcs.080895. [DOI] [PubMed] [Google Scholar]
- 22.Israel BA, Kodali VK, Thorpe C. Going through the Barrier: coupled disulfide exchange reactions promote efficient catalysis in Quiescin sulfhydryl oxidase. J Biol Chem. 2014;289:5274–5284. doi: 10.1074/jbc.M113.536219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fass D. Disulfide Bonding in Protein Biophysics. Annu Rev Biophys. 2012;41:43–59. doi: 10.1146/annurev-biophys-050511-102321. [DOI] [PubMed] [Google Scholar]
- 24.Jocelyn PC. Biochemistry of the SH Group. London: Academic Press; 1972. [Google Scholar]
- 25.Torchinsky YM. Sulfur in Proteins. New York: Pergamon Press; 1981. [Google Scholar]
- 26.Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics. 2012;6:6. doi: 10.1186/1479-7364-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Li W, Ren J, Fang J, Ke H, Gong W, Feng W, Wang CC. Structural insights into the redox-regulated dynamic conformations of human protein disulfide isomerase. Antioxid Redox Signaling. 2013;19:36–45. doi: 10.1089/ars.2012.4630. [DOI] [PubMed] [Google Scholar]
- 29.Klappa P, Ruddock LW, Darby NJ, Freedman RB. The b′ domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998;17:927–935. doi: 10.1093/emboj/17.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabibbo A, Pagani M, Fabbri M, Rocchi M, Farmery MR, Bulleid NJ, Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 31.Pagani M, Fabbri M, Benedetti C, Fassio A, Pilati S, Bulleid NJ, Cabibbo A, Sitia R. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 32.Tavender TJ, Bulleid NJ. Molecular mechanisms regulating oxidative activity of the Ero1 family in the endoplasmic reticulum. Antioxid Redox Signaling. 2010;13:1177–1187. doi: 10.1089/ars.2010.3230. [DOI] [PubMed] [Google Scholar]
- 33.Araki K, Inaba K. Structure, mechanism, and evolution of Ero1 family enzymes. Antioxid Redox Signaling. 2012;16:790–799. doi: 10.1089/ars.2011.4418. [DOI] [PubMed] [Google Scholar]
- 34.Ramming T, Appenzeller-Herzog C. The physiological functions of mammalian endoplasmic oxidoreductin 1: on disulfides and more. Antioxid Redox Signaling. 2012;16:1109–1118. doi: 10.1089/ars.2011.4475. [DOI] [PubMed] [Google Scholar]
- 35.Gross E, Sevier CS, Heldman N, Vitu E, Bentzur M, Kaiser CA, Thorpe C, Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci U S A. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: Emerging roles for a new flavoprotein family. J Biol Chem. 2007;282:13929–13933. doi: 10.1074/jbc.R600037200. [DOI] [PubMed] [Google Scholar]
- 37.Zito E, Chin KT, Blais J, Harding HP, Ron D. ERO1-beta, a pancreas-specific disulfide oxidase, promotes insulin biogenesis and glucose homeostasis. J Cell Biol. 2010;188:821–832. doi: 10.1083/jcb.200911086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zito E, Melo EP, Yang Y, Wahlander A, Neubert TA, Ron D. Oxidative protein folding by an endoplasmic reticulum-localized peroxiredoxin. Molecular Cell. 2010;40:787–797. doi: 10.1016/j.molcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tavender TJ, Springate JJ, Bulleid NJ. Recycling of peroxiredoxin IV provides a novel pathway for disulphide formation in the endoplasmic reticulum. EMBO J. 2010;15:4185–4197. doi: 10.1038/emboj.2010.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kakihana T, Nagata K, Sitia R. Peroxides and peroxidases in the endoplasmic reticulum: integrating redox homeostasis and oxidative folding. Antioxid Redox Signaling. 2012;16:763–771. doi: 10.1089/ars.2011.4238. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen VD, Saaranen MJ, Karala AR, Lappi AK, Wang L, Raykhel IB, Alanen HI, Salo KE, Wang CC, Ruddock LW. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J Mol Biol. 2011;406:503–515. doi: 10.1016/j.jmb.2010.12.039. [DOI] [PubMed] [Google Scholar]
- 42.Schulman S, Wang B, Li W, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc Natl Acad Sci U S A. 2010;107:15027–15032. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruddock LW. Low molecular weight oxidants involved in disulfide bond formation. Antioxid Redox Signaling. 2011 doi: 10.1089/ars.2011.4481. [DOI] [PubMed] [Google Scholar]
- 44.Banhegyi G, Csala M, Szarka A, Varsanyi M, Benedetti A, Mandl J. Role of ascorbate in oxidative protein folding. Biofactors. 2003;17:37–46. doi: 10.1002/biof.5520170105. [DOI] [PubMed] [Google Scholar]
- 45.Karala AR, Lappi AK, Saaranen M, Ruddock LW. Efficient Peroxide Mediated Oxidative Refolding of a Protein at Physiological pH and Implications for Oxidative Folding in the Endoplasmic Reticulum. Antioxid Redox Signaling. 2008;11:963–970. doi: 10.1089/ars.2008.2326. [DOI] [PubMed] [Google Scholar]
- 46.Margittai E, Banhegyi G. Oxidative folding in the endoplasmic reticulum: Towards a multiple oxidant hypothesis? FEBS Let. 2010;584:2995–2998. doi: 10.1016/j.febslet.2010.05.055. [DOI] [PubMed] [Google Scholar]
- 47.Hoober KL, Sheasley SS, Gilbert HF, Thorpe C. Sulfhydryl oxidase from egg white: a facile catalyst for disulfide bond formation in proteins and peptides. J Biol Chem. 1999;274:22147–22150. doi: 10.1074/jbc.274.32.22147. [DOI] [PubMed] [Google Scholar]
- 48.Heckler EJ, Rancy PC, Kodali VK, Thorpe C. Generating disulfides with the Quiescin-sulfhydryl oxidases. Biochim Biophys Acta. 2008;1783:567–577. doi: 10.1016/j.bbamcr.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rancy PC, Thorpe C. Oxidative Protein Folding in vitro: a Study of the Cooperation between Quiescin-sulfhydryl Oxidase and Protein Disulfide Isomerase. Biochemistry. 2008;47:12047–12056. doi: 10.1021/bi801604x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, Turi G, Coppock D. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys. 2002;405:1–12. doi: 10.1016/s0003-9861(02)00337-5. [DOI] [PubMed] [Google Scholar]
- 51.Heckler EJ, Alon A, Fass D, Thorpe C. Human quiescin-sulfhydryl oxidase, QSOX1: probing internal redox steps by mutagenesis. Biochemistry. 2008;47:4955–4963. doi: 10.1021/bi702522q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alon A, Grossman I, Gat Y, Kodali VK, DiMaio F, Mehlman T, Haran G, Baker D, Thorpe C, Fass D. The dynamic disulphide relay of quiescin sulphydryl oxidase. Nature. 2012;488:414–418. doi: 10.1038/nature11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jessop CE, Bulleid NJ. Glutathione directly reduces an oxidoreductase in the endoplasmic reticulum of mammalian cells. J Biol Chem. 2004;279:55341–55347. doi: 10.1074/jbc.M411409200. [DOI] [PubMed] [Google Scholar]
- 54.Montero D, Tachibana C, Rahr Winther J, Appenzeller-Herzog C. Intracellular glutathione pools are heterogeneously concentrated. Redox Biol. 2013;1:508–513. doi: 10.1016/j.redox.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Freedman RB, Hirst TR, Tuite MF. Protein disulfide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 56.Araki K, Iemura S, Kamiya Y, Ron D, Kato K, Natsume T, Nagata K. Ero1-alpha and PDIs constitute a hierarchical electron transfer network of endoplasmic reticulum oxidoreductases. J Cell Biol. 2013;202:861–874. doi: 10.1083/jcb.201303027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 58.Lappi AK, Ruddock LW. Reexamination of the role of interplay between glutathione and protein disulfide isomerase. J Mol Biol. 2011;409:238–249. doi: 10.1016/j.jmb.2011.03.024. [DOI] [PubMed] [Google Scholar]
- 59.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI--a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 60.Appenzeller-Herzog C, Riemer J, Christensen B, Sorensen ES, Ellgaard L. A novel disulphide switch mechanism in Ero1alpha balances ER oxidation in human cells. EMBO J. 2008;27:2977–2987. doi: 10.1038/emboj.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker KM, Chakravarthi S, Langton KP, Sheppard AM, Lu H, Bulleid NJ. Low reduction potential of Ero1alpha regulatory disulphides ensures tight control of substrate oxidation. EMBO J. 2008;27:2988–2997. doi: 10.1038/emboj.2008.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Banhegyi G, Lusini L, Puskas F, Rossi R, Fulceri R, Braun L, Mile V, di Simplicio P, Mandl J, Benedetti A. Preferential transport of glutathione versus glutathione disulfide in rat liver microsomal vesicles. J Biol Chem. 1999;274:12213–12216. doi: 10.1074/jbc.274.18.12213. [DOI] [PubMed] [Google Scholar]
- 63.Morgan B, Ezerina D, Amoako TNE, Riemer J, Seedorf M, Dick TP. Multiple glutathione disulfide removal pathways mediate cytosolic redox homeostasis. Nat Chem Biol. 2013;9:119–125. doi: 10.1038/nchembio.1142. [DOI] [PubMed] [Google Scholar]
- 64.Flohe L. The fairytale of the GSSG/GSH redox potential. Biochim Biophys Acta. 2013;1830:3139–3142. doi: 10.1016/j.bbagen.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 65.Heinrich R, Rapoport SM, Rapoport TA. Metabolic regulation and mathematical models. Prog Biophys Mol Biol. 1977;32:1–82. [PubMed] [Google Scholar]
- 66.Hwang C, Sinskey AJ, Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 67.Bass R, Ruddock LW, Klappa P, Freedman RB. A Major Fraction of Endoplasmic Reticulum-located Glutathione Is Present as Mixed Disulfides with Protein. J Biol Chem. 2004;279:5257–5262. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 68.Dixon BM, Heath SH, Kim R, Suh JH, Hagen TM. Assessment of Endoplasmic Reticulum Glutathione Redox Status Is Confounded by Extensive Ex Vivo Oxidation. Antioxid Redox Signaling. 2008;10:963–972. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bjornberg O, Ostergaard H, Winther JR. Mechanistic insight provided by glutaredoxin within a fusion to redox-sensitive yellow fluorescent protein. Biochemistry. 2006;45:2362–2371. doi: 10.1021/bi0522495. [DOI] [PubMed] [Google Scholar]
- 71.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 72.van Lith M, Tiwari S, Pediani J, Milligan G, Bulleid NJ. Real-time monitoring of redox changes in the mammalian endoplasmic reticulum. J Cell Sci. 2011 doi: 10.1242/jcs.085530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Birk J, Meyer M, Aller I, Hansen HG, Odermatt A, Dick TP, Meyer AJ, Appenzeller-Herzog C. Endoplasmic reticulum: reduced and oxidized glutathione revisited. J Cell Sci. 2013;126:1604–1617. doi: 10.1242/jcs.117218. [DOI] [PubMed] [Google Scholar]
- 74.Avezov E, Cross BC, Kaminski Schierle GS, Winters M, Harding HP, Melo EP, Kaminski CF, Ron D. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J Cell Biol. 2013;201:337–349. doi: 10.1083/jcb.201211155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kolossov VL, Leslie MT, Chatterjee A, Sheehan BM, Kenis PJA, Gaskins HR. Forster resonance energy transfer-based sensor targeting endoplasmic reticulum reveals highly oxidative environment. Exp Biol Med. 2012;237:652–662. doi: 10.1258/ebm.2012.011436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eaton P. Protein thiol oxidation in health and disease: techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radical Biol Med. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 77.Sato Y, Kojima R, Okumura M, Hagiwara M, Masui S, Maegawa K, Saiki M, Horibe T, Suzuki M, Inaba K. Synergistic cooperation of PDI family members in peroxiredoxin 4-driven oxidative protein folding. Sci Rep. 2013;3:2456. doi: 10.1038/srep02456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nardai G, Stadler K, Papp E, Korcsmaros T, Jakus J, Csermely P. Diabetic changes in the redox status of the microsomal protein folding machinery. Biochem Biophys Res Commun. 2005;334:787–795. doi: 10.1016/j.bbrc.2005.06.172. [DOI] [PubMed] [Google Scholar]
- 79.Hansen RE, Winther JR. An introduction to methods for analyzing thiols and disulfides: Reactions, reagents, and practical considerations. Anal Biochem. 2009;394:147–158. doi: 10.1016/j.ab.2009.07.051. [DOI] [PubMed] [Google Scholar]
- 80.Chakravarthi S, Jessop CE, Bulleid NJ. The role of glutathione in disulphide bond formation and endoplasmic-reticulum-generated oxidative stress. EMBO Rep. 2006;7:271–275. doi: 10.1038/sj.embor.7400645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hansen RE, Roth D, Winther JR. Quantifying the global cellular thiol-disulfide status. Proc Natl Acad Sci U S A. 2009;106:422–427. doi: 10.1073/pnas.0812149106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lyles MM, Gilbert HF. Catalysis of the oxidative folding of ribonuclease A by protein disulfide isomerase: dependence of the rate on the composition of the redox buffer. Biochemistry. 1991;30:613–619. doi: 10.1021/bi00217a004. [DOI] [PubMed] [Google Scholar]
- 83.Narayan M, Welker E, Wedemeyer WJ, Scheraga HA. Oxidative folding of proteins. Acc Chem Res. 2000;33:805–812. doi: 10.1021/ar000063m. [DOI] [PubMed] [Google Scholar]
- 84.Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J Theoret Biol. 1990;143:163–195. doi: 10.1016/s0022-5193(05)80266-8. [DOI] [PubMed] [Google Scholar]
- 85.Hillson DA, Lambert N, Freedman RB. Formation and isomerization of disulfide bonds in proteins: protein disulfide-isomerase. Methods Enzymol. 1984;107:281–294. doi: 10.1016/0076-6879(84)07018-x. [DOI] [PubMed] [Google Scholar]
- 86.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chouchani ET, James AM, Fearnley IM, Lilley KS, Murphy MP. Proteomic approaches to the characterization of protein thiol modification. Curr Op Chem Biol. 2011;15:120–128. doi: 10.1016/j.cbpa.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Severs N, Shotton D. Rapid Freezing of Biological Specimenst for Freeze Fracture and Deep Etching. In: Celis J, editor. Cell Biol. Elsevier; 2006. pp. 249–255. [Google Scholar]
- 89.Tanaka M, Matsuura K, Yoshioka S, Takahashi S, Ishimori K, Hori H, Morishima I. Activation of hydrogen peroxide in horseradish peroxidase occurs within approximately 200 micro s observed by a new freeze-quench device. Biophys J. 2003;84:1998–2004. doi: 10.1016/s0006-3495(03)75008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chambers JE, Tavender TJ, Oka OB, Warwood S, Knight D, Bulleid NJ. The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1alpha. J Biol Chem. 2010;285:29200–29207. doi: 10.1074/jbc.M110.156596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wolfenden R. Benchmark reaction rates, the stability of biological molecules in water, and the evolution of catalytic power in enzymes. Ann Rev Biochem. 2011;80:645–667. doi: 10.1146/annurev-biochem-060409-093051. [DOI] [PubMed] [Google Scholar]
- 92.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochim Biophys Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 93.Mezghrani A, Fassio A, Benham A, Simmen T, Braakman I, Sitia R. Manipulation of oxidative protein folding and PDI redox state in mammalian cells. EMBO J. 2001;20:6288–6296. doi: 10.1093/emboj/20.22.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Appenzeller-Herzog C, Ellgaard L. In vivo reduction-oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signaling. 2008;10:55–64. doi: 10.1089/ars.2007.1837. [DOI] [PubMed] [Google Scholar]
- 95.Toldo S, Boccellino M, Rinaldi B, Seropian IM, Mezzaroma E, Severino A, Quagliuolo L, Van Tassell BW, Marfella R, Paolisso G, Rossi F, Natarajan R, Voelkel N, Abbate A, Crea F, Baldi A. Altered oxido-reductive state in the diabetic heart: loss of cardioprotection due to protein disulfide isomerase. Mol Med. 2011;17:1012–1021. doi: 10.2119/molmed.2011.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lundstrom J, Holmgren A. Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry. 1993;32:6649–6655. doi: 10.1021/bi00077a018. [DOI] [PubMed] [Google Scholar]
- 97.Schwaller M, Wilkinson B, Gilbert HF. Reduction-reoxidation cycles contribute to catalysis of disulfide isomerization by protein-disulfide isomerase. J Biol Chem. 2003;278:7154–7159. doi: 10.1074/jbc.M211036200. [DOI] [PubMed] [Google Scholar]
- 98.Raturi A, Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic Biol Med. 2007;43:62–70. doi: 10.1016/j.freeradbiomed.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 99.Frickel EM, Frei P, Bouvier M, Stafford WF, Helenius A, Glockshuber R, Ellgaard L. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J Biol Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
- 100.Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 101.Jeong W, Lee DY, Park S, Rhee SG. ERp16, an endoplasmic reticulum-resident thiol-disulfide oxidoreductase: biochemical properties and role in apoptosis induced by endoplasmic reticulum stress. J Biol Chem. 2008;283:25557–25566. doi: 10.1074/jbc.M803804200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Haugstetter J, Blicher T, Ellgaard L. Identification and characterization of a novel thioredoxin-related transmembrane protein of the endoplasmic reticulum. J Biol Chem. 2005;280:8371–8380. doi: 10.1074/jbc.M413924200. [DOI] [PubMed] [Google Scholar]
- 103.Brandner D, Withers G. Image of Plasma Cell from Guinea Pig Bone Marrow CIL:10733. The Cell: An Image Library. 2010 http://www.cellimagelibrary.org;
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.