Significance
The draft genome of Tibetan hulless barley provides a robust framework to better understand Poaceae evolution and a substantial basis for functional genomics of crop species with a large genome. The expansion of stress-related gene families in Tibetan hulless barley implies that it could be considered as an invaluable gene resource aiding stress tolerance improvement in Triticeae crops. Genome resequencing revealed extensive genetic diversity in Tibetan barley germplasm and divergence to sequenced barley genomes from other geographical regions. Investigation of genome-wide selection footprints demonstrated an adaptive correlation of genes under selection with extensive stressful environmental variables. These results reveal insights into the adaptation of Tibetan hulless barley to harsh environments on the highland and will facilitate future genetic improvement of crops.
Keywords: Tibetan hulless barley, Triticeae evolution, genetic diversity, adaptation, selective sweep
Abstract
The Tibetan hulless barley (Hordeum vulgare L. var. nudum), also called “Qingke” in Chinese and “Ne” in Tibetan, is the staple food for Tibetans and an important livestock feed in the Tibetan Plateau. The diploid nature and adaptation to diverse environments of the highland give it unique resources for genetic research and crop improvement. Here we produced a 3.89-Gb draft assembly of Tibetan hulless barley with 36,151 predicted protein-coding genes. Comparative analyses revealed the divergence times and synteny between barley and other representative Poaceae genomes. The expansion of the gene family related to stress responses was found in Tibetan hulless barley. Resequencing of 10 barley accessions uncovered high levels of genetic variation in Tibetan wild barley and genetic divergence between Tibetan and non-Tibetan barley genomes. Selective sweep analyses demonstrate adaptive correlations of genes under selection with extensive environmental variables. Our results not only construct a genomic framework for crop improvement but also provide evolutionary insights of highland adaptation of Tibetan hulless barley.
Genome sequences provide a substantial basis for understanding the biological essences of crops associated with biologically and economically essential traits. Therefore, decoding a genome will benefit crop development to feed the increasing demand for food brought by climate change and population growth. The Triticeae species, such as wheat (Triticum aestivum, 2n = 6x = 42) and barley (Hordeum vulgare, 2n = 2x = 14), which account for >30% of cereal production worldwide, are essential food and forage resources (faostat.fao.org). International efforts have been launched to decipher their genomes, and dramatic breakthroughs have been achieved on the reference genome of chromosome 3B (1), whole-genome sequencing (2, 3), and in-depth phylogenetic and transcriptome analyses (4, 5) of hexaploid wheat, as well as generations of draft genome sequences (6, 7) and construction of a physical map of its diploid A-genome (Triticum urartu, 2n = 2x = 14) and D-genome (Aegilops tauschii, 2n = 2x = 14) progenitors (6–8).
Barley, as one of the earliest domesticated crops and the world’s fourth most abundant cereal, is widely used in the brewing industry as a stock feed and in potential healthy food products (9, 10). As a diploid inbreeder, barley has long been considered as a genetic model for cereal crops in Triticeae. Recently, the International Barley Sequencing Consortium (IBSC) built a barley physical map of 4.98 giga base pairs (Gb), with more than 3.90 Gb anchored to a high-resolution genetic map (11). Supported by DNA and deep RNA sequence data, 26,159 “high-confidence” genes were identified. Furthermore, extensive single-nucleotide variation was found by survey sequencing of diverse barley accessions. These achievements provide unprecedented insight into the barley genome.
Compared with common cultivated barley with covered caryopsis, hulless barley (H. vulgare L. var. nudum), with naked caryopsis, is mainly cultivated in Tibet and its vicinity, which is one of the domestication and diversity centers for cultivated barley (12–16). The adaptation to extreme environmental conditions in high altitudes made it the staple food for Tibetans beginning at least 3,500–4,000 years ago (17, 18). It continues to be the predominant crop in Tibet, occupying ∼70% of crop lands (SI Appendix, Fig. S1). To facilitate the genetic development and gene identification of Tibetan hulless barley, we built the draft genome sequences of Lasa Goumang, a landrace of Tibetan hulless barley. Genome comparative analyses make deeper insights into the evolution among Poaceae species. Comparison analyses between Tibetan barleys and non-Tibetan barleys reveal genes associated with natural selections, which will help in the understanding of the adaptation biology of the barleys in the Tibetan Plateau.
Results
Genome Assembly and Annotation.
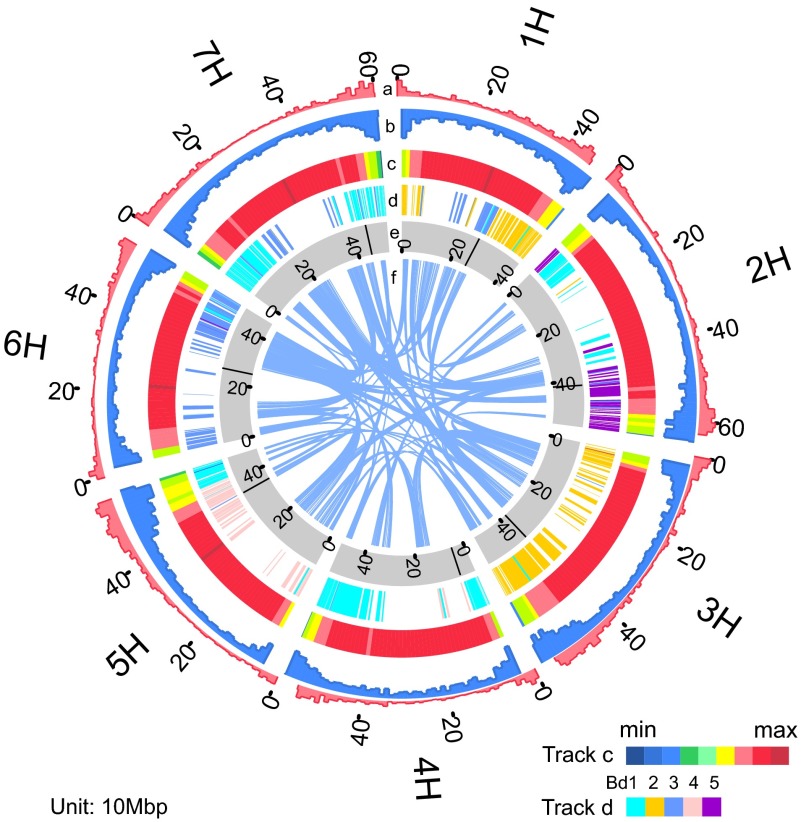
We used a whole-genome shotgun strategy to sequence a series of libraries from 250 base pairs (bp) to 40 kilobase pairs (kb) (SI Appendix, Table S1). A total of 797 Gb of high-quality sequence data were generated, which has an approximate 178-fold depth of the estimated genome size of 4.48 Gb (Table 1 and SI Appendix, Table S2 and Fig. S2). We built a de novo assembly of 3.89 Gb, with contig and scaffold N50 lengths of 18.07 kb and 242 kb, respectively (SI Appendix, Figs. S2–S4 and Tables S3 and S4). The assembly showed high genome coverage and single-base accuracy evaluated by bacterial artificial chromosome (BAC) sequences and RNA-seq data (SI Appendix, Fig. S5 and Tables S5–S7). We also anchored 28,374 scaffolds, representing 89.4% of the genome, onto seven chromosomes using the integrated and ordered physical and genetic map of cultivated barley (11) (Fig. 1 and SI Appendix, Tables S8 and S9).
Table 1.
Statistics of the draft genome of Tibetan hulless barley
| Genomic statistics | H. vulgare L. var. nudum |
| Estimated genome size | 4.48 Gb |
| Total size of assembled scaffolds, >200 bp | 3.89 Gb |
| Total sequence length anchored to chromosomes | 3.48 Gb |
| Percent of chromosomal sequences | 89.41% |
| N50 length, scaffolds | 242 kb |
| Longest scaffold | 3.07 Mb |
| Total size of assembled contigs | 3.64 Gb |
| Longest contig | 276.95 kb |
| N50 length, contig | 18.07 kb |
| GC content | 44.00% |
| Repeat content | 81.39% |
| Number of gene models | 36,151 |
Fig. 1.
Overview of Tibetan hulless barley genome. (Track a) Gene regions of cultivated barley cv. Morex (%) per 10 Mb—min., 0; max., 10. (Track b) Gene regions of Tibetan hulless barley (%) per 10 Mb—min., 0; max., 10. (Track c) LTR retro-transposon (%) per 10 Mb—min., 0; max., 100. (Track d) Synteny with the B. distachyon genome. (Track e) Tibetan hulless barley chromosomes with centromeres marked as black bands. (Track f) Syntenic blocks within and between chromosomes.
Using a combination of evidence-based and de novo approaches, ∼81.4% of our assembly were identified as repetitive elements (Table 1 and Fig. 1), which is similar to those of Morex (11) and maize (19) (SI Appendix, Tables S10–S12). In contrast to T. urartu and Ae. tauschii, the long terminal retrotransposons (LTRs) of Tibetan hulless barley contribute 68.3% to the whole genome, which is consistent with those obtained from gene-bearing BACs (11). The extent of divergence shows that the transposable element (TE) repeats were recently produced or anciently produced by transposition (SI Appendix, Fig. S6). We annotated 36,151 protein-coding genes combining various strategies of ab initio, homology, and transcriptome predictions (SI Appendix, Fig. S7 and Tables S13–S16) as well as the full-length cDNA of barley (11), which is comparable with those in T. urartu (34,879) (6) and Ae. tauschii (34,498) (7). As observed, the gene density is relatively lower surrounding the centromeres, where the repetitive contents are inversely higher, indicating the widely scattered gene deserts in the Tibetan hulless barley genome. Of the genes, 93.9% (33,928) were located on chromosomes, and 82.2% (29,730) were functionally annotated by multiple databases (SI Appendix, Tables S17 and S18).
We mapped the genome data of barley accessions sequenced by IBSC to the Tibetan hulless barley genome including Morex, Bowman, Barke, Hordeum spontaneum, Haruna Nijo, and Igri. The coverage rates range from 82.35% to 94.96% (SI Appendix, Table S19). There was no obvious difference in coverage rates across the seven chromosomes, which implies no preference of homogeny on different chromosomes. Moreover, we identified 113 megabase pairs (Mb) and 288 Mb of specific sequences for Morex (5.35%) and Tibetan hulless barley (7.90%), respectively (SI Appendix, Table S20). More than 4,500 genes are involved in Tibetan hulless barley-specific sequences. The overrepresenting KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways are flavonoid biosynthesis, stilbenoid, diarylheptanoid and gingerol biosynthesis, and plant hormone signal transduction (SI Appendix, Table S21). By comparison to 26,159 high-confidence gene sets of the Morex genome (11), we identified 22,673 reciprocal best hits, in which 7,224 (31.86%) gene pairs had identical sequences and 17,840 (78.68%) showed protein similarity higher than 95% (SI Appendix, Fig. S8A). Most of these gene pairs have similar lengths of coding DNA sequence (CDS) and gene body (CDS+intron), except that about 10.97% of genes in Tibetan hulless barley exhibited much longer gene body lengths than those in Morex (ratio, >3.0) (SI Appendix, Fig. S8B). These results indicate the level of genomic divergence between the two species.
Evolutionary Characteristics.
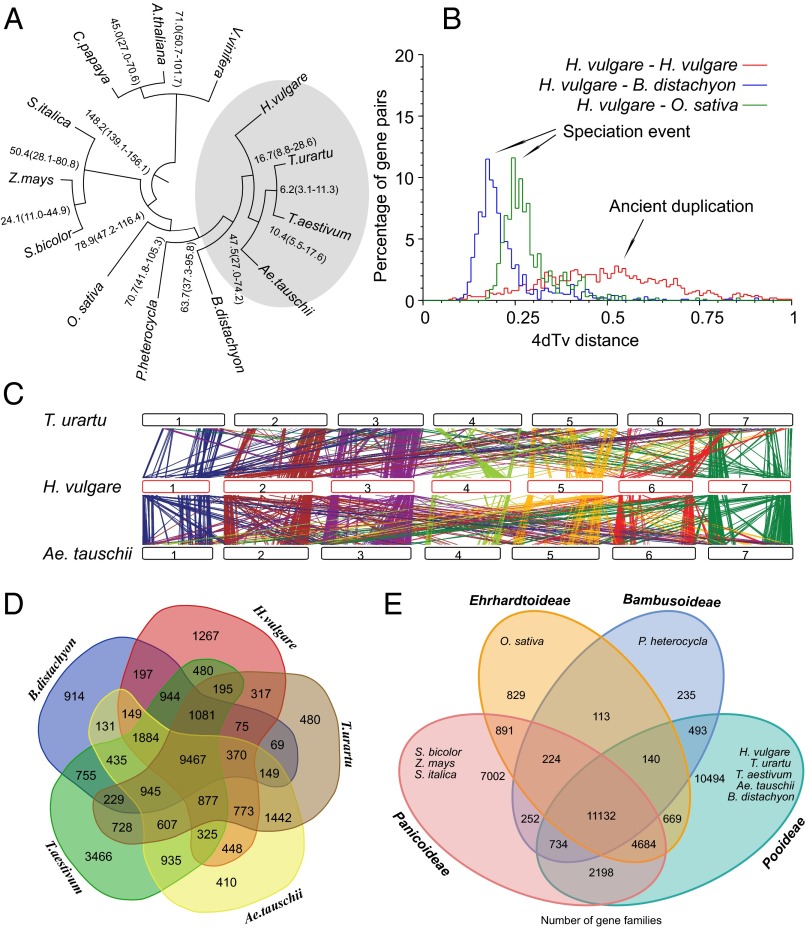
We estimated the divergence times of Tibetan hulless barley from other nine Poaceae species and three dicotyledons with 153 single-copy genes identified among these species (Fig. 2A and SI Appendix, Figs. S9 and S10 and Tables S22–S24). It showed that barley was separated from Ae. tauschii, T. urartu, and T. aestivum ∼17 million years ago (Mya), using the 95% credibility interval, and this divergence could have occurred ∼28.6–8.8 Mya (the same below) and speciated from Brachypodium distachyon, Phyllostachys heterocycla, and Oryza sativa ∼48 (74.2–27.0) Mya, ∼64(95.8–37.3) Mya, and ∼71(105.3–41.8) Mya, respectively. It was estimated that Ae. tauschii diverged from T. urartu ∼10 (17.6–5.5) Mya. To better understand Triticeae evolution at the genome level, we also analyzed the whole-genome duplication (WGD) event and synteny of Tibetan hulless barley and sequenced Triticeae species. A total of 1,320 syntenic blocks, comprising 10,502 paralogous gene pairs, in Tibetan hulless barley uncovered an ancient WGD, which is common to other Poaceae genomes (7) (Fig. 2B). In addition, 637 and 486 large syntenic blocks, comprising 6,209 and 4,346 orthologous gene pairs, were identified between barley and Ae. tauschii, and barley and T. urartu, respectively (Fig. 2C and SI Appendix, Fig. S11–S14 and Table S25). The orthologous relationship between barley and O. sativa chromosomes provides evidence that at least four major nested chromosome fusions occurred in barley from its intermediate ancestor (SI Appendix, Fig. S15). The regions outside these synteny blocks were rich in LTR repeats (Figs. 1 and 2C), which may be caused by retrotransposition after speciation.
Fig. 2.
Phylogeny, WGD, and gene families of Tibetan hulless barley compared with other plant genomes. (A) The divergence time tree estimated on fourfold degenerate sites of single-copy orthologous genes. (B) WGD and divergence of Tibetan hulless barley indicated by 4DTv (deviation at the fourfold degenerate third codon position) distribution. (C) Chromosomal syntenic relationship between Tibetan hulless barley and wheat A- and D-genome progenitors. (D) Gene families of H. vulgare, T. urartu, T. aestivum, Ae.tauschii, and B. distachyon. (E) Gene families of Ehrhardtoideae, Bambusoideae, Pooideae, and Panicoideae.
We compared gene families among barley and other Poaceae genomes (SI Appendix, Fig. S16 and Table S26). A total of 18,849 gene families are found in the Tibetan hulless barley genome, which is similar to those of Ae. tauschii, T. urartu, and B. distachyon. Within the Pooideae subfamily of Poaceae, 9,467 gene families are shared by five species, which is 40∼53% of the total number, respectively (Fig. 2D). For pairwise comparison, more than 60% of families are shared between any two species. These results reflect common origin and genome divergency among Poaceae genomes. For intersubfamilies comparison, 11,132 are shared by four subfamilies—Pooideae, Panicoideae, Ehrhardtoideae, and Bambusoideae—contributing to ∼84% of Bambusoideae and ∼36% of Pooideae, respectively (Fig. 2E).
We also analyzed the expansion and contraction of gene families excluding the hexaploid wheat. Compared with the common ancestor of the Triticeae tribe, we found 2,185 expanded families in barley, but only 510 in the Ae. tauschii–T. urartu branch (SI Appendix, Fig. S16). Among the 104 families significantly expanded in the barley lineage (P < 0.05), the overrepresented gene ontology (GO) terms are mainly related to gene regulation and stress resistance, such as regulation of transcription, transcription factor activity, defense response, response to stress, etc. (SI Appendix, Table S27). For example, we found more ethylene responsive factor (ERF) and dehydration-responsive element binding (DREB) transcription factors than those of T. urartu (2.0-fold), Ae. tauschii (1.6-fold), and Morex genomes (SI Appendix, Table S28). Extensive study revealed sound functions of both families in the regulation of responses to a wide range of abiotic and biotic stresses such as cold, drought, high salinity, heat shock, submergence, and multiple plant diseases, which made them ideal candidates for crop tolerance engineering (20, 21). Furthermore, using functionally known genes as queries (7), we found 230 cold-acclimated related genes in Tibetan hulless barley, which is also more abundant than those in Ae. tauschii, T. urartu, or other sequenced plant genomes analyzed in this study (SI Appendix, Table S28).
Analysis of the nonsynonymous to synonymous substitution ratio (Ka/Ks) among related plants found that the evolutionary process of Tibetan hulless barley diverged from Morex with more positively selected genes (Ka/Ks > 1) than those from other species (SI Appendix, Fig. S17). KEGG pathway analysis indicated that many of the positively selected genes are involved in pathways related to environmental responses and adaptation, such as plant hormone signal transduction, replication and repair, plant–pathogen interaction, circadian rhythm–plant, calcium signaling pathway, etc. (SI Appendix, Table S29).
Genetic Diversity Among Wild and Cultivated Barleys.
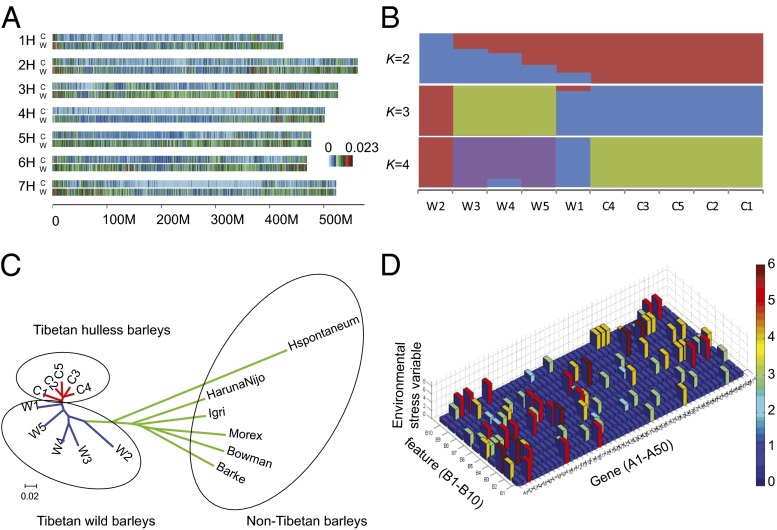
To understand the genetic diversity in Tibetan barley germplasms, we carried out whole-genome resequencing of 10 strains representing wild and cultivated accessions (SI Appendix, Table S30). Genome alignment averaged 96.5% sequencing coverage and a 16.4-fold depth for each individual. A total of 36,469,491 single nucleotide polymorphisms (SNPs) and 2,281,198 small insertions and deletions were identified in individuals. For the wild group, 34,064,490 SNPs were detected, nearly twice of that found in cultivated hulless barleys (SI Appendix, Tables S31 and S32 and Fig. 3A). The genome-wide pairwise nucleotide diversity within population θπ and Watterson’s estimator of segregating sites θw were 8.27 × 10−4 and 7.17 × 10−4 for wild and 3.31 × 10−4 and 2.78 × 10−4 for cultivated accessions, respectively. The principal component analysis (PCA) (22) and STRUCTURE (23) also indicated that the cultivated accessions cluster closely and wild groups are more divergent (Fig. 3B and SI Appendix, Fig. S18).
Fig. 3.
Population genetics of barleys. (A) SNP rate distribution for Tibetan wild barleys (W) and Tibetan hulless barley (C) across the seven chromosomes. There are 10 individuals including five Tibetan wild barleys and Tibetan hulless barleys. (B) STRUCTURE analysis of the 10 barley individuals. W1–W5 refer to five wild samples, and C1–C5 refer to five cultivated samples. (C) The neighbor-joining tree of barleys. The green lineages refer to six non-Tibetan barleys. The blue lineages refer to five Tibetan wild barleys. The red lineages refer to five Tibetan hulless barleys. (D) The adaptive correlation of randomly selected genes from SI Appendix, Table S34 (A1–A50) with 10 stressful environmental variables (B1–B10). The ecologically stressful variables include salinity, oxygen (low and high), solar radiation (especially high), CO2, drought, temperature (high and low), day length (short or long), and dormancy. The A1–A50 genes demonstrating adaptive correlation with environmental stress are directly or indirectly affecting other genes or gene networks related to metabolite cycles and other biological vital functions. Direct effects result in high correlations; indirect effects result in low correlations.
We further investigated the genomic divergence between barley accessions of the Tibetan Plateau (Tibetan group) and six additional barley accessions previously sequenced (non-Tibetan group) (11). From the PCA (SI Appendix, Fig. S19) and neighbor-joining tree (Fig. 3C), the Tibetan group and non-Tibetan group could be distinctly divided. The analysis of the population structure showed that a clear evolutionary divergence was evident between two groups (K = 2). When K > 2, considerable genetic composition was observed to be shared by wild and cultivated accessions within the Tibetan group but is distinct from the non-Tibetan group (SI Appendix, Fig. S20).
The genome regions under selective sweeps due to the plateau environment were also investigated. As the measures of genetic differentiation of different populations, Tajima’s D (24) and Fst (25) were introduced in this study. By comparison of two barley groups, the genome regions with Fst > 0.5 (or 5% top Fst windows) between the Tibetan group and non-Tibetan groups and Tajima’s D < –2 within the Tibetan group were determined as under selective sweeps. As a result, 1.07% of the genome and 1.23% (418) of the annotated genes were identified to be involved in selection (SI Appendix, Tables S33 and S34). KEGG analyses showed that some genes are enriched in pathways of plant hormone signal transduction, plant–pathogen interaction, phenylpropanoid biosynthesis, flavonoid biosynthesis, etc. (SI Appendix, Table S35). Adaptive correlations of 50 randomly chosen genes with 10 stressful environmental variables, including salinity, oxygen (low and high), solar radiation (especially high), CO2, drought, temperature (high and low), day length (short or long), and dormancy, were analyzed (SI Appendix, Table S36). A considerable number of these genes are found to be directly or indirectly associated with environmental stress adaptation (Fig. 3D).
Discussion
Increasing evidence supports the Tibetan Plateau as one of the centers for cultivated barley domestication. In this study, we provide a draft genome of Tibetan hulless barley. This genome assembly is 3.89 Gb, accounting for about 87% of the whole genome, with high gene space coverage and high single-base accuracy. A genomic frame has been built by anchoring more than 89.4% of the assembly, comprising 33,928 out of 36,151 predicted genes, onto seven chromosomes. These data allow us to reinspect the divergent time between barley and other important Poaceae crops, especially species of the Triticeae tribe, by whole genome-wide in-depth analyses, as well as the WGD, synteny relationship, chromosome rearrangements, and gene families. These results not only present an exhaustive delineation of the Poaceae families’ evolution but also build substantial genomic groundwork and a good reference for marker development and gene identification in Triticeae crops with huge and repetitive genomes.
The high genome coverage resequencing of 10 Tibetan wild and cultivated barley generated robust numbers of SNPs, among which the wild group contains nearly twice the SNP number than that of the cultivated group. θπ, θw, and Tajima’s D values also indicate the significantly lower genetic diversity in cultivated accessions, reflecting the genetic bottlenecks introduced during domestication. Although the divergences between the Tibetan wild and cultivated barley are evident, they are even closer in comparison with barley accessions from Europe, East Asia, and Israel, which are sequenced by IBSC. This result came from analyses of PCA, STRUCTURE, Fst, and cluster and is consistent with that obtained by transcriptome sequencing (12). Therefore, introducing the gene pool of Tibetan wild barley and those from non-Tibetan groups into Tibetan hulless barley may be helpful in widening the genetic background of Tibetan hulless barley.
To adapt to the harsh environments of the plateau, Tibetan hulless barley may have been domesticated under distinct processes of natural selection compared with the cultivated barley from other parts. Therefore, it will be interesting to uncover evolutionary evidence for its adaptation by comparative analyses. From a gene family comparison with Ae. tauschii–T. urartu, we found significant gene family expansion in the barley lineage (P < 0.05) with overrepresenting GO terms of regulation of transcription, transcription factor activity, defense response, etc. (SI Appendix, Table S27). This may enable Tibetan hulless barley more flexibility to regulate its adaptation to extreme environmental challenges at high altitudes. For example, the ERF and DREB transcription factors that related to biotic and biotic stresses (20, 21) and cold-acclimated related genes (7) were expanded in the Tibetan hulless barley lineage (SI Appendix, Table S28).
Moreover, Ka/Ks analysis showed more positively selected genes between Tibetan hulless barley and Morex (Ka/Ks > 1) than between other species (SI Appendix, Fig. S17). A considerable number of these genes are involved in pathways related to environmental responses and adaptation, such as plant hormone signal transduction, replication and repair, plant–pathogen interaction, etc. (SI Appendix, Table S29). Similar results come from the selective sweep analyses between Tibetan and non-Tibetan barley. The adaptive correlation of individual genes with extensive stressful environmental variables was demonstrated. They are enriched in pathways related to environmental adaptation or stress response. For example, phenylpropanoid biosynthesis and flavonoid biosynthesis are essential for accumulation of chemical sunscreens in protecting against ultraviolet (UV) radiation in plants (26). Plant hormone signal transduction pathways are not only important for germination, growth, and development, but also play key roles in responses to biotic and abiotic stress. Intriguingly, almost 45% of selected regions were linked to chromosome 7H, which indicated that more adaptation-related genes may lie on the chromosome that undergoes more severe selection. The selected regions provide targets for identification of adaptation-related genes from Tibetan barley. For example, a previously known drought stress-related quantitative trait locus (QTL), QRwc.TaEr-7H.2, which is associated with leaf relative water content, was found in selective sweep regions (SI Appendix, Fig. S21) (27). These results will facilitate crop improvement toward sustainable supplies of food, feed, and industry resources for people living on the highland and around the world.
Methods
Sequencing and Assembly.
Genomic DNA was isolated from a landrace of Tibetan hulless barley L. goumang. Paired-end sequencing (PE151, PE101, PE91, and PE50) was performed on the Illumina HiSeq2000 platform for short insert size libraries including 250, 500, and 800 base pairs (bps) and mate-pair libraries of 2 kb, 5 kb, 10 kb, 20 kb, and 40 kb. A stringent filtering process on raw reads was done to remove low quality, adapter contamination, small insert, and PCR duplicated reads. Short insert size data were further corrected by the ErrorCorrection module in SOAPdenovo2 (28). When conducting the assembly using SOAPdenovo2, we constructed contigs using corrected short insert size data with the K-mer parameter set to 67 and built a scaffold using all paired-end clean data. We used SSPACE-V1.1 (29) to construct a superscaffold with all mate-pair clean data. GapCloser (28) was used to fill gaps within the scaffold to improve the contig N50 length.
Genome Assembly Evaluation.
We mapped the assembled Tibetan hulless barley genome sequences—10 selected BAC sequences from a paper published by IBSC (BlastN, –e 1e-5; nucleotide identity, >0.97)—to check the quality and coverage of our assembly. We aligned 24.5-fold of clean data to BAC sequences by SOAPaligner (28) to observe the sequencing coverage depth across BAC sequences. The gene region coverage rate was calculated using BLAT (30), aligning Tibetan hulless barley scaffolds to Trinity (31)-assembled unigenes to determine the percentage of unigene bases covered by our assembly.
Repeat Annotation and Gene Annotation.
Genome sequences were scanned for known repeats by RepeatMasker and ProteinRepeatMask against Repbase (32). RepeatModeler (33) and LTR-FINDER (34), de novo prediction programs, were used to build the de novo repeat library based on the genome; then, contamination and multicopy genes in the library were removed. LTR_FINDER was used to search the whole genome for characteristic structures of the full-length LTRs (its ∼18-bp sequence was complementary to the 3′ tail of some tRNAs). Then, our in-house pipeline was used to filter the low-quality and falsely predicted LTRs. Using this library as a database, RepeatMasker was run to find and classify the repeats. Gene models were predicted using de novo software such as AUGUSTUS (35) and GENSCAN (36); homolog-based methods to map Arabidopsis thaliana, B. distachyon, O. sativa, Sorghum bicolor, and Zea mays protein sequences to Tibetan hulless barley genome sequences using TBlastN (37) and GENEWISE (38) software to infer gene structure; an RNA-seq–based method to map transcriptome data to the reference genome using TopHat (39); and assembling transcripts with Cufflinks (39) to obtain the gene structure. Finally, GLEAN (40) was used to make the high-confidence gene model by combining all of the evidence. Predicted protein-coding genes were further assigned functions by aligning them to the best matches in the SwissProt and TrEMBL databases (41), determining the motifs and domains using InterProScan (42), and assigning Gene Ontology (43) and KEGG pathway (44) annotations.
Chromosome Reconstruction Based on Barley Physical and Genetic Frameworks.
The Tibetan hulless barley scaffold sequences were aligned to the IBSC barley physical map sequences anchored to the high-resolution genetic map using BlastN (length, ≥200 bp; e-value, ≤1e−5; identity, ≥99%). Data for the IBSC barley physical map were downloaded from ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/anchoring/. The Tibetan hulless barley genome sequences were mapped to the IBSC barley sequences to determine their location in the genetic map. The best scoring matches were selected in the case of multiple matches.
Collinearity Among Poaceae Species.
We performed all-versus-all BlastP (e-value, <1e-5) and then detected syntenic blocks using MCscan. The 4DTv values (45) (deviation at the fourfold degenerate third codon position) among paralogous gene pairs of Tibetan hulless barley were calculated and revised in the Hasegawa, Kishino and Yano (HKY) model to analyze WGD events. The chromosomal collinearity of Poaceae species based on orthologous gene pairs was drawn among the species. The nested chromosomal fusion events were deduced by genome synteny between Tibetan hulless barley chromosomes and rice chromosomes.
Phylogenetic Analysis.
We constructed gene families for thirteen species including Hordeum vulgare, Triticum urartu, Triticum aestivum, Aegilops tauschii, Brachypodium distachyon, Phyllostachys heterocycla, Oryza sativa, Sorghum bicolor, Zea mays, Senna italic, Carica papaya, Arabidopsis thaliana, and Vitis vinifera with OrthoMCL (46) methods on the all-versus-all BlastP alignment (e-value, <1e-5). Single-copy ortholog protein sequences for each species were aligned by multiple sequence comparison by log-expectation (MUSCLE) (47), and the corresponding CDS sequences were concatenated to supergene sequences. To build a species phylogenetic tree by PhyML (48) under the GTR+gamma model with approximate likelihood ratio test (aLRT) assessment of branch reliability, 4D-sites were extracted. Divergence times were estimated using the Phylogenetic Analysis by Maximum Likelihood (PAML) mcmctree (49) program with the approximate likelihood calculation method, and the convergence was checked by Tracer (50) and confirmed by two independent runs.
Genome Diversity of Cultivars and Wild Barleys.
Ten barley accessions, including five cultivars and five wild barleys, were each sequenced for more than 15-fold in PE91. The clean reads were mapped to reference Tibetan hulless barley genomes by Burrows–Wheeler alignment tool (BWA) (51). We then used the Genome Analysis Toolkit (52) to identify SNP and insertions and deletions. PCA and population structure analysis were performed by EIGENSOFT (22) and FRAPPE (23), respectively. Genetic diversity of cultivars and wild barleys were compared by calculating the SNP rate in 500-kb windows across the genome.
Supplementary Material
Acknowledgments
We thank M. C. Luo (Department of Plant Sciences, University of California) for his valuable suggestions on data analyses. This work was supported by the following funding sources: the Tibet Autonomous Region Financial Special Fund (2011XZCZZX001), the National Science and Technology Support Program (2012BAD03B01), and the National Program on Key Basic Research Project (2011CB111512, 2012CB723006).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Sequence Read Archive, www.ncbi.nlm.nih.gov/sra (accession no. SRA201388).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423628112/-/DCSupplemental.
References
- 1.Choulet F, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345(6194):1249721. doi: 10.1126/science.1249721. [DOI] [PubMed] [Google Scholar]
- 2.Brenchley R, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491(7426):705–710. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Wheat Genome Sequencing Consortium (IWGSC) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345(6194):1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 4.Marcussen T, et al. International Wheat Genome Sequencing Consortium Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345(6194):1250092. doi: 10.1126/science.1250092. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer M, et al. International Wheat Genome Sequencing Consortium Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014;345(6194):1250091. doi: 10.1126/science.1250091. [DOI] [PubMed] [Google Scholar]
- 6.Ling HQ, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496(7443):87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- 7.Jia J, et al. International Wheat Genome Sequencing Consortium Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013;496(7443):91–95. doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- 8.Luo MC, et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc Natl Acad Sci USA. 2013;110(19):7940–7945. doi: 10.1073/pnas.1219082110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blake T, Blake V, Bowman J, Abdel-Haleem H. In: Barley: Production, Improvement and Uses. Ullrich SE, editor. Wiley-Blackwell, Ames, IA; 2011. pp. 522–531. [Google Scholar]
- 10.Collins HMea Variability in fine structures of noncellulosic cell wall polysaccharides from cereal grains: Potential importance in human health and nutrition. Cereal Chem. 2010;87(4):272–282. [Google Scholar]
- 11.Mayer KF, et al. International Barley Genome Sequencing Consortium A physical, genetic and functional sequence assembly of the barley genome. Nature. 2012;491(7426):711–716. doi: 10.1038/nature11543. [DOI] [PubMed] [Google Scholar]
- 12.Dai F, et al. Transcriptome profiling reveals mosaic genomic origins of modern cultivated barley. Proc Natl Acad Sci USA. 2014;111(37):13403–13408. doi: 10.1073/pnas.1414335111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao C, Li C, Baschan C. Origin and evolution of the cultivated barley. Acta Genet Sin. 1975;2(2):123–128. [Google Scholar]
- 14.Xu TW. On the origin and phylogeny of cultivated barley with reference to the discovery of ganze wild two-rowed barley. Hordeum spontaneum c. Koch. Acta Genet Sin. 1975;2(2):129–137. [Google Scholar]
- 15.Ma D, Xu T. The research on classification and origin of cultivated barley in Tibet autonomous region. Scientia Agricultura Sinica. 1988;21(5):7–14. [Google Scholar]
- 16.Dai F, et al. Tibet is one of the centers of domestication of cultivated barley. Proc Natl Acad Sci USA. 2012;109(42):16969–16973. doi: 10.1073/pnas.1215265109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang YS, et al. Discussion on the origin of the crops: Agriculture of Tibet Highland. Agri Sci and Tech in Tibet. 1999;21(4):4–11. [Google Scholar]
- 18.Fu DX, et al. A study on ancient barley, wheat and millet discovered at Changguo of Tibet. Acta Agron Sin. 2000;26(4):392–398. [Google Scholar]
- 19.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 20.Xu ZS, Chen M, Li LC, Ma YZ. Functions and application of the AP2/ERF transcription factor family in crop improvement. J Integr Plant Biol. 2011;53(7):570–585. doi: 10.1111/j.1744-7909.2011.01062.x. [DOI] [PubMed] [Google Scholar]
- 21.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta. 2012;1819(2):86–96. doi: 10.1016/j.bbagrm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genet Epidemiol. 2005;28(4):289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 24.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akey JM, Zhang G, Zhang K, Jin L, Shriver MD. Interrogating a high-density SNP map for signatures of natural selection. Genome Res. 2002;12(12):1805–1814. doi: 10.1101/gr.631202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130(1):234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diab AA, et al. Identification of drought-inducible genes and differentially expressed sequence tags in barley. Theor Appl Genet. 2004;109(7):1417–1425. doi: 10.1007/s00122-004-1755-0. [DOI] [PubMed] [Google Scholar]
- 28.Li R, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 30.Kent WJ. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110(1–4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 33. Smit AFA, Hubley R (2004) RepeatModeler. www.repeatmasker.org.
- 34.Xu Z, Wang H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(Web Server issue):W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanke M, et al. AUGUSTUS: Ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34(Web Server issue):W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268(1):78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birney E, Clamp M, Durbin R. GeneWise and Genomewise. Genome Res. 2004;14(5):988–995. doi: 10.1101/gr.1865504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elsik CG, et al. Creating a honey bee consensus gene set. Genome Biol. 2007;8(1):R13. doi: 10.1186/gb-2007-8-1-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28(1):45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zdobnov EM, Apweiler R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 43.Ashburner M, et al. The Gene Ontology Consortium Gene ontology: Tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang S, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41(12):1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- 46.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 49.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 50.Rambaut A, Drummond AJ. 2007 Tracer v1.4. beast.bio.ed.ac.uk/Tracer.
- 51.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.