ABSTRACT
Pneumocystis species are fungal parasites of mammal lungs showing host specificity. Pneumocystis jirovecii colonizes humans and causes severe pneumonia in immunosuppressed individuals. In the absence of in vitro cultures, the life cycle of these fungi remains poorly known. Sexual reproduction probably occurs, but the system of this process and the mating type (MAT) genes involved are not characterized. In the present study, we used comparative genomics to investigate the issue in P. jirovecii and Pneumocystis carinii, the species infecting rats, as well as in their relative Taphrina deformans. We searched sex-related genes using 103 sequences from the relative Schizosaccharomyces pombe as queries. Genes homologous to several sex-related role categories were identified in all species investigated, further supporting sexuality in these organisms. Extensive in silico searches identified only three putative MAT genes in each species investigated (matMc, matMi, and matPi). In P. jirovecii, these genes clustered on the same contig, proving their contiguity in the genome. This organization seems compatible neither with heterothallism, because two different MAT loci on separate DNA molecules would have been detected, nor with secondary homothallism, because the latter involves generally more MAT genes. Consistently, we did not detect cis-acting sequences for mating type switching in secondary homothallism, and PCR revealed identical MAT genes in P. jirovecii isolates from six patients. A strong synteny of the genomic region surrounding the putative MAT genes exists between the two Pneumocystis species. Our results suggest the hypothesis that primary homothallism is the system of reproduction of Pneumocystis species and T. deformans.
Importance Sexual reproduction among fungi can involve a single partner (homothallism) or two compatible partners (heterothallism). We investigated the issue in three pathogenic fungal relatives: Pneumocystis jirovecii, which causes severe pneumonia in immunocompromised humans; Pneumocystis carinii, which infects rats; and the plant pathogen Taphrina deformans. The nature, the number, and the organization within the genome of the genes involved in sexual reproduction were determined. The three species appeared to harbor a single genomic region gathering only three genes involved in sexual differentiation, an organization which is compatible with sexual reproduction involving a single partner. These findings illuminate the strategy adopted by fungal pathogens to infect their hosts.
Importance
Sexual reproduction among fungi can involve a single partner (homothallism) or two compatible partners (heterothallism). We investigated the issue in three pathogenic fungal relatives: Pneumocystis jirovecii, which causes severe pneumonia in immunocompromised humans; Pneumocystis carinii, which infects rats; and the plant pathogen Taphrina deformans. The nature, the number, and the organization within the genome of the genes involved in sexual reproduction were determined. The three species appeared to harbor a single genomic region gathering only three genes involved in sexual differentiation, an organization which is compatible with sexual reproduction involving a single partner. These findings illuminate the strategy adopted by fungal pathogens to infect their hosts.
INTRODUCTION
Pneumocystis species constitute a group of fungi belonging to the Taphrinomycotina subphylum of the Ascomycota which colonize the lungs of mammals. Genetic and transmission analyses revealed that each Pneumocystis species infects specifically a single mammalian host species. Pneumocystis jirovecii, the species colonizing humans, can turn into an opportunistic pathogen in immunosuppressed individuals and cause severe, sometimes fatal pneumonia (Pneumocystis pneumonia [PCP]). PCP is nowadays the second most frequent life-threatening invasive fungal infection worldwide, with above 400,000 annual cases (1). Comparative genomics suggested the loss of synthesis and assimilation pathways in Pneumocystis species and thus that these fungi are obligate parasites without free-living forms (2–4). The characteristics of these fungi suggest that they are biotrophs of mammals, i.e., parasites retrieving energy and compounds from host cells without killing them (4–7).
The life cycle of Pneumocystis species remains poorly known, mostly because of the absence of a method for long-term culture in vitro. During the infection, two types of cellular structures are observed: the trophic cells and the asci (also called trophs and cysts, respectively). Microscopic and cytological studies suggested that the trophic cells may undergo binary fission but also diploidization upon conjugation, followed by meiosis and mitosis to produce asci containing eight haploid ascospores (8, 9). The observation of synaptonemal structures within Pneumocystis cells (10, 11) and the demonstration of the expression of sex-related genes (5, 12) further confirmed that the reproduction of these fungi probably includes a sexual phase. Quantitative experiments suggested that the main mode of reproduction of Pneumocystis species might be meiotic division, whereas the contribution of mitotic division of trophic cells is still unclear (8, 13, 14). Therefore, the sexual phase might be obligatory for the growth of these fungi. Obligate sexuality is also consistent with the fact that the asci might be the only particles aerially transported and responsible for transmission between hosts (14, 15). Obligate sexuality would imply that trophic cells of compatible mating types must always be present within host lungs in order to allow sexual reproduction.
There are two main systems of sexual reproduction among fungi: heterothallism, involving two compatible mating types, and homothallism, involving a single self-compatible mating type. In heterothallic ascomycetes, the MAT loci contain divergent genes (idiomorphs) in opposite mating types (16). In so-called primary homothallic species, the genes of the two idiomorphs are present in the same genome, closely located or not. Another form of homothallism, called secondary, is observed in ascomycetous yeasts such as Saccharomyces cerevisiae and Schizosaccharomyces pombe. In this case, the homothallic behavior results from mating type switching and the presence of three MAT loci in the same genome: one active and two silent. In S. pombe, the MAT loci are flanked by cis-acting sequences that are involved in a switching mechanism which exchanges the expressed cassette. Primary homothallism is observed in many filamentous ascomycetes (Pezizomycotina or Euascomycetes). As far as Pneumocystis species are concerned, their particular system of reproduction is still unknown. The genomic region surrounding the pheromone receptor ste3 gene has been postulated to constitute a MAT locus in these fungi. It would also include genes encoding the protein kinase Ste20 and the homeodomain transcriptional regulator Ste12 and thus, surprisingly, would resemble the MAT locus of the basidiomycete Cryptococcus neoformans (17).
In the present study, we investigated the system of reproduction of P. jirovecii and Pneumocystis carinii, the species infecting rats, by the analysis of their genome content and comparative genomics. We also investigated the issue in the Taphrinomycotina relative Taphrina deformans, a plant pathogen whose sexual reproduction is also poorly characterized. In order to identify the sex-related genes present in the genomes of these species, genes from the well-annotated Taphrinomycotina member S. pombe were used as homology references.
RESULTS
Identification of sex-related genes.
Due to the low conservation and great diversity of the sex-related genes among fungi, the identification of homologs of sex-related S. pombe proteins within the genomes of P. jirovecii, P. carinii, and T. deformans was done using the bioinformatics strategy shown in Fig. S1 in the supplemental material. The procedure relied on tBLASTn search and recognition by manual inspection of specific domain architecture (see details in Materials and Methods). The 103 S. pombe genes used in sequence queries were selected on the basis of their annotation in UniProt and Gene Ontology (GO) terms, as well as of their belonging to relevant protein complexes (see Table S1 in the supplemental material). Genes were classified in one or two of seven role categories: cell fusion, signal transmission, signal transduction, signal regulation, meiosis, mating type locus silencing (RNA interference pathway), and mating type locus switching. Using this strategy, we established the presence or absence of sex-related genes within the genomes of the three fungi (see Table S2 in the supplemental material; the locus reference or genomic location of the genes identified is given in Table S3 in the supplemental material).
Genes homologous to all seven role categories of sex-related S. pombe proteins were present in the three genomes investigated (see Table S2 in the supplemental material, green areas). In agreement with their notoriously difficult identification, the presence or absence of the pheromone precursor genes could not be assessed (map2, mfm1, mfm2, and mfm3). The genes encoding the two pheromone receptors (mam2 and ste3) and the elements of the pheromone-induced signaling cascade (e.g., gpa1, byr2, byr1, spk1, and ste11) were present in all three genomes investigated, except that T. deformans lacked mam2. There was a remarkable similarity of the presence and absence of the genes in the three species investigated, with 51 genes present (see Table S2 in the supplemental material, green areas), and 25 absent (brown areas). Twelve genes were absent only in the two Pneumocystis species, suggesting specific features of this genus (signal transmission, pmd1; signal transduction, ste4; signal regulation, rst2, sxa1, and sxa2; mating type silencing: ago1, arb1, arb2, chp1, dcr1, hst2, obr1, and rdr1). Thus, more genes were detected in T. deformans than in the two Pneumocystis species. The Argonaute small interfering RNA chaperone (ARC) complex was absent in the Pneumocystis species (mating type silencing: ago1, arb1, and arb2), a feature which we already reported (4). The presence or absence of the gene products of the three species within the reconstructed pathways of S. pombe is shown in the supplemental material (see Fig. S2 in the supplemental material).
MAT genes.
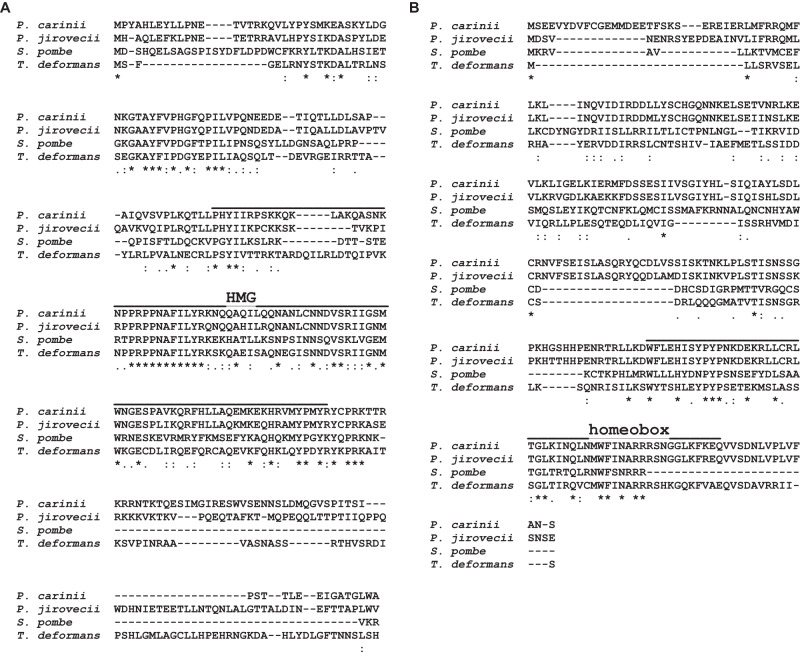
Genes homologous to the four S. pombe MAT genes were sought in the three species investigated (see Table S2 in the supplemental material, boxed genes). Extensive in silico searches using our bioinformatics strategy (see Fig. S1 in the supplemental material) revealed the presence of only three genes in each Pneumocystis species and in T. deformans, namely, the homologs of S. pombe matMc, matMi, and matPi. The gene matMc encodes a transcription factor with the high-mobility-group domain (PTHR10270:SF176 or PTHR10270:SF159), matMi encodes a mating type M-specific polypeptide, and matPi encodes a transcription factor with a homeobox domain (PTHR11850 or PTHR11850:SF31). The matMi-encoded proteins have no known domains and are poorly conserved between species, so that their identification did not rely on domain architecture as for the other genes but on their localization close to matMc and opposite orientation relative to matMc.
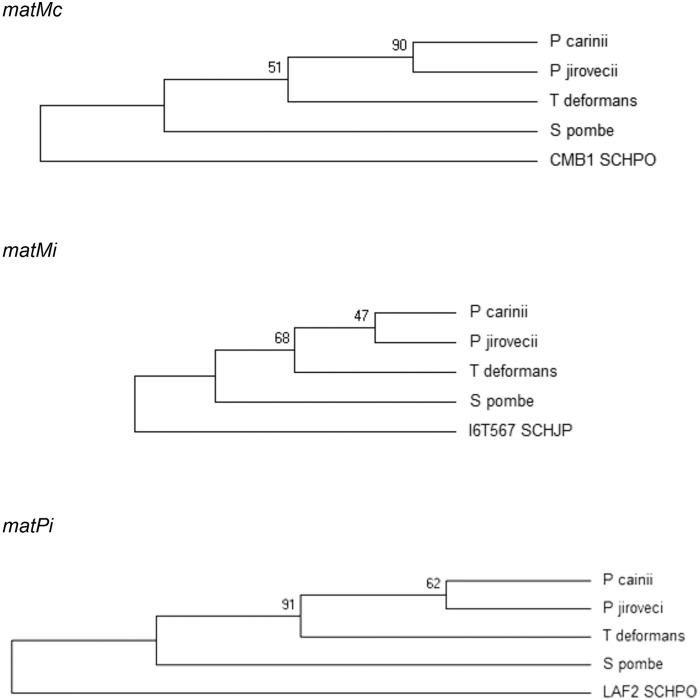
Multiple sequence alignment revealed a fair degree of similarity of the matMc-encoded proteins identified, in particular at the high-mobility-group domain, with 27% overall identity with the S. pombe protein for all three P. jirovecii, P. carinii, and T. deformans proteins (Fig. 1A). The degree of similarity was lower between the matPi proteins and concentrated at the homeobox domain present at the end of the proteins, with 19% overall identity with the S. pombe protein for P. jirovecii and 18% for both P. carinii and T. deformans (Fig. 1B). Reasonable alignment of the matMi proteins could not be generated because of their low similarity, the identity with the S. pombe protein being only 12, 15, and 20% for P. jirovecii, P. carinii, and T. deformans proteins, respectively. Despite this low similarity, the phylogeny of the three putative MAT genes was inferred using the maximum likelihood method (Fig. 2). Although the bootstrap values were low because of the diversity of these proteins, the trees were consistent with the phylogeny of the species that we observed previously by concatenation of 458 orthologs (3). Manual inspection failed to detect putative cis-acting sequence motifs homologous to those which flank the S. pombe MAT loci and which are involved in mating type switching (H1, H2, and H3 [Fig. 3]).
FIG 1 .
Multiple sequence alignment of S. pombe matMc (A) and matPi (B) proteins against homologs identified in P. carinii, P. jirovecii, and T. deformans. The identical, strongly, and weakly conserved residues identified using T-Coffee (47) are indicated by asterisks, double points, and single points, respectively. Dashes indicate gaps. The high-mobility group and homeobox domains are shown in panels A and B, respectively.
FIG 2 .
Phylogeny of the MAT genes of S. pombe, P. jirovecii, P. carinii, and T. deformans. The maximum likelihood method based on the JTT matrix-based model (48) was used to infer the evolutionary history of matMc, matMi, and matPi genes.
FIG 3 .
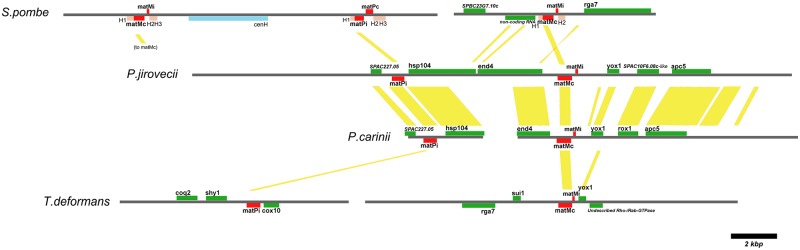
Schematic representation of the MAT loci of S. pombe, P. jirovecii, P. carinii, and T. deformans. Genes are shown in green, and their synteny with those of the other species is shown by the yellow rectangles. The sequence contigs carrying the genes are shown as gray lines. The centromere-homologous sequence (cenH) is shown in blue. The cis-acting sequence motifs H1, H2, and H3 involved in mating type switching in S. pombe are shown in light brown. The gene name or detected domain name is indicated for each identified open reading frame.
Synteny of MAT loci.
The genomic region surrounding the MAT genes appeared to be fully syntenic between the two Pneumocystis species with six genes conserved (Fig. 3). The three putative MAT genes were located on a single contig in P. jirovecii and thus on a single DNA molecule in the genome. These genes are on two contigs in P. carinii and T. deformans, but this may result from DNA fragmentation during genome sequencing and assembly, especially in P. carinii because of its overall high synteny with P. jirovecii. Limited synteny existed between the other species, with only one gene conserved in addition to the MAT genes (yox1 between Pneumocystis species and T. deformans; rga7 between S. pombe and T. deformans). The finding of rga7 next to the matMc/matMi gene pair in T. deformans is particularly relevant since this arrangement is conserved in all Schizosaccharomyces species (18).
Analysis of the putative MAT genes of several P. jirovecii isolates.
The putative MAT genes matMc, matMi, and matPi were amplified from the P. jirovecii isolates present in six different patients with PCP and sequenced. The sequences of all isolates were exactly the same as that present in the P. jirovecii genome sequence, except in the isolates of three patients where a deletion of 5 bp was located between the putative matMc and matMi genes (CCTTG at positions 57275 to 57279 in sequence CAKM01000262.1 of the P. jirovecii genome [contig 262]). One of the three patients harbored at least two coinfecting genotypes, one in the majority which presented the deletion and one in the minority which did not.
Genomic region surrounding the gene encoding the pheromone receptor Ste3.
We analyzed the genomic region surrounding the ste3 gene because it has been postulated to constitute a MAT locus in P. carinii (17). This genomic region was reported to include the protein kinase Ste20 and the homeodomain transcriptional regulator Ste12. In both P. carinii and P. jirovecii, BLAST analyses using S. pombe sequences as queries revealed that the so-called ste20 gene was in fact an shk2 homolog with a pleckstrin homology-like domain (IPR011993) and the so-called ste12 gene was a truncated version of ste11, a signal transduction kinase gene. We identified ste3 and shk2 on the same contig in P. carinii, whereas shk2 was on the same contig as the three putative MAT genes in P. jirovecii but fairly distant (ca. 80 kb). The complete ste11 gene was identified in distinct locations in the two Pneumocystis genomes (see Table S3 in the supplemental material).
DISCUSSION
In order to better characterize the life cycle of Pneumocystis species and T. deformans, we searched for sex-related genes in their genomes. We detected many genes involved in various sex-related processes, such as mating, pheromone signalizing, and meiosis. Some of these genes have been previously identified in P. carinii (17, 19–23) and T. deformans (24), but we report them in P. jirovecii for the first time. The expression of several putative mating or meiosis genes in P. carinii has been documented elsewhere (5, 12). Together, these observations strongly support the hypothesis that a sexual phase occurs in the life cycle of the Pneumocystis species and T. deformans.
Our analyses reveal a conserved syntenic genomic conformation of the MAT locus between the two Pneumocystis species, despite a high level of divergence between the MAT genes. This suggests that the conservation of the locus configuration might be more critical for sexual reproduction than that of the mating genes. The most striking feature of our findings was that only three genes homologous to the four S. pombe MAT genes were identified in each Pneumocystis species, as well as in T. deformans (matMc, matMi, and matPi). It must be stressed that the matMi genes identified remain hypothetical because of their low similarity with that of S. pombe. It is likely that no other MAT genes were present because (i) the results were similar in the two Pneumocystis species and (ii) the genomic and transcriptomic data analyzed are expected to cover most of the genomes. Moreover, we did observe the same synteny and three MAT genes also in the Pneumocystis murina genome, but our data are unpublishable because this genome sequence was released prior to publication under specific terms (see Pneumocystis murina Sequencing Project, Broad Institute of Harvard and MIT, http://www.broadinstitute.org/). One limitation of our study is that MAT genes too divergent from those of S. pombe would not have been detected. However, S. pombe harbors two different types of DNA binding domains (high-mobility group and homeobox), which increased the probability of detecting such genes. The proximity of three putative MAT genes identified in the Pneumocystis species and T. deformans is atypical and suggests a fusion of two MAT loci, one of type minus (M) with matMc and matMi and one incomplete of type plus (P) with only matPi. Consequently, the transcription factors encoded by matMc (high-mobility group) and matPi (homeobox) would be sufficient to trigger sexual development. Such fusion of two MAT loci has been previously observed in other fungi, for example, in the Stemphylium genus (25), and the loss of MAT transcription factors occurred in Candida species other than C. albicans (26). A similar fusion of the two MAT loci M and P is also present in three Schizosaccharomyces species other than S. pombe, with only matMc in their type M loci (18).
The number and the organization of the putative MAT genes in the Pneumocystis species and T. deformans allow formulation of a hypothesis concerning the system of sexual reproduction of these fungi. First, the presence of only three MAT genes seems incompatible with secondary homothallism. Indeed, this system requires generally more genes, i.e., six corresponding to three MAT loci in S. pombe and S. cerevisiae and four corresponding to two MAT loci in some methylotrophic yeasts, as recently described (27). This is consistent with the fact that we did not detect any cis-acting sequences for mating type switching flanking the putative MAT genes. Second, the three MAT genes in P. jirovecii are present on a single DNA molecule (Fig. 3), and this is also the case in P. murina (our unpublishable findings). The close proximity of these genes on a single DNA molecule is particularly important because it is not compatible with heterothallism. Indeed, if the genome sequences had been derived from a mixture of two compatible heterothallic mating types, two MAT loci would have been observed located on two distinct DNA molecules, not on a single one. The alternative possibility, i.e., that the genome sequences correspond to a single heterothallic mating type, is also unlikely. First, the MAT locus identified appears to include both types, M and P. Second, asci are present in most Pneumocystis infections, if not all, i.e., sexuality appears obligatory. In particular, this was the case in the clinical specimen used for P. jirovecii genome sequencing because the diagnosis was made by silver staining of the walls of asci (3).
The loss of the transcription factor gene matPc (high-mobility group) in the Pneumocystis species and T. deformans suggests that the P-specific genes may not be expressed and that P cells may be lacking. Thus, these species may use a single mating type for sexual reproduction, a system previously observed in C. neoformans (28) and Candida albicans (29). An alternative hypothesis is that expression of the P-specific genes, including that encoding the P-factor, is ensured thanks to a rewiring of the MAT pathways, a phenomenon frequently observed in fungi (30, 31). The latter hypothesis would be consistent with the presence of the receptor mam2 to the P-factor in the Pneumocystis species. The presence of matPi in all three genomes investigated might reflect that both this gene and the matMi gene seem necessary for the expression of mei3 and thus for entry into meiosis (32). Further experiments are required in order to decipher the system of sexual reproduction in the Pneumocystis species and T. deformans.
Our results do not allow us to ascertain that the genes identified are bona fide MAT genes of these species, because we cannot rule out the possibility that there are other transcription factors involved in mating type determination elsewhere in the genomes. Also, one cannot formally exclude secondary homothallism. However, the close vicinity and arrangement of the putative MAT genes are consistent with primary homothallism in the Pneumocystis species as well as in T. deformans. The latter system of reproduction implies the presence of the same MAT locus in all isolates of each species that we investigated. This is what we found in the P. jirovecii isolates of six patients, further supporting the hypothesis of primary homothallism. This hypothesis is also compatible with the T. deformans cell cycle. Indeed, single haploid yeast cells inoculated on peach leaves are able to produce, without cell-cell conjugation, dikaryotic hyphae that give rise to asci containing eight ascospores (33, 34).
Primary homothallism has been hypothesized to be advantageous for pathogens (35), including Pneumocystis species (28, 29). Our results are compatible with this hypothesis. Although it involves a single strain, primary homothallism has been shown in C. neoformans to avoid accumulation of deleterious mutations, as well as to increase genetic diversity and virulence (36). In the case of P. jirovecii, the fact that the majority of infections involve two or more distinct genotypes (37) suggests that the genetic diversity may be further increased by mating among these genetic variants.
The presence of many genes that we classified in the silencing and switching categories in the Pneumocystis species and T. deformans appears contradictory to the putative absence of secondary homothallism. Nevertheless, their presence could be simply due to their involvement in other cellular or metabolic processes. For example, some swi-encoded proteins are known to be necessary for DNA metabolism. On the other hand, the absence of two silencing (clr1 and swi6) and two switching (sap1 and swi2) genes in all three species investigated might be significant because these genes are apparently dedicated to silencing or switching of MAT genes (see Table S1 in the supplemental material).
As far as P. carinii is concerned, we confirmed the location of ste3, the so-called ste20 gene, and the so-called ste12 gene within the genomic region which was postulated to be a MAT locus resembling those of the basidiomycete C. neoformans (17). However, the so-called ste20 and ste12 genes turned out to be shk2 and a truncated version of ste11, respectively. These different findings may result from the constant improvement of the databases, the increase in knowledge about these genes since 2001, and the fact that in this study we dealt with more than one genome. The confusion between ste12 and ste11 probably results from the fact that S. cerevisiae ste12 is the ortholog of S. pombe ste11. Consequently, the genomic region surrounding ste3 in the Pneumocystis species may constitute a cluster of sex-related genes, rather than a MAT locus resembling that of C. neoformans.
The computational observation of putative MAT genes does not prove conclusively that a species is sexual, because these genes may have been conserved from a sexual ancestor. In conclusion, our analyses suggest the working hypothesis that primary homothallism is the system of reproduction of the Pneumocystis species as well as of T. deformans.
MATERIALS AND METHODS
Source data.
The annotated genome sequences of P. jirovecii and T. deformans were obtained from the European Nucleotide Archive (38) (CAKM00000000 and CAHR00000000, last accessed on 14 May 2013). The P. jirovecii transcriptome assembly (HAAA01000001 to HAAA01003261) was downloaded from EBI (http://www.ebi.ac.uk, last accessed 11 November 2013). These genome sequences and transcriptome correspond to the data that we published previously (3, 24). P. carinii assembled contigs were downloaded from the project site (http://pgp.cchmc.org/, last accessed 14 April 2013) and correspond to data published by Slaven et al. (39). The S. pombe protein sequences were obtained from the annotated proteome present in UniProt (http://www.uniprot.org, last accessed 30 July 2014) and correspond to the data published previously (40).
Search for candidate genes and manual curation.
The bioinformatics strategy used to identify homologs of S. pombe proteins within P. jirovecii, P. carinii, and T. deformans genomes is shown in Fig. S1 in the supplemental material. Each S. pombe sequence used as a query was characterized in order to identify specific domains or domain architecture using the InterProScan4 tool (41), with a particular consideration for the PANTHER subfamilies (42). tBLASTn analyses (Blast+ suite, NCBI, version 64bit) (43) with relaxed parameters (E value from 1E−4 down to its default value) were used in order to obtain a list of candidate genomic regions within the target genome, which presumably included the genes of interest. The already existing annotation of the genes was considered, and existing collinear coding sequences (CDSs) were retained after assessment using the InterProScan4 tool. When no CDSs were already annotated, a CDS was proposed based on the alignment of the S. pombe query protein against the genomic region using GeneWise (44). Only GeneWise global alignments matching most of the query were retained. The start and stop codons of the CDSs were assessed, and the respective exons were corrected manually to obtain the longest CDSs possible. Both existing and newly derived CDSs translated using the EMBOSS_transeq program (45) were submitted to the InterProScan4 tool. The predicted domains and their architecture were compared to those retained for the corresponding S. pombe protein. When strong domain evidence was detected, the CDS was considered the valid homolog. When no or ambiguous domain architecture was detected, the CDSs from all the investigated species were compared in multiple sequence alignments and associated phylogenetic trees. The latter analysis permitted us to validate some homologs whose domain architecture was ambiguous. The overall synteny between the genomic region of interest and that of S. pombe was also taken into account. If no CDS could be validated, it was assumed that no homolog existed in the genome investigated. Whenever possible, the original genome annotation was retained, but a complete reappraisal of the CDS definition and of its annotation was required in several cases (see Table S3 in the supplemental material).
Phylogeny analysis.
To address specific cases of domain architecture ambiguity, formal phylogeny analysis was carried out using MEGA6 (46) on alignments made by T-Coffee (47). The resulting topologies were chosen on consensus between the results of the different methods and the statistical support for branching. All positions containing gaps and missing data were eliminated. For the inference of the evolutionary history of matMc, matMi, and matPi genes, the maximum likelihood approach was used. Initial trees for the heuristic search were obtained by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model (48). The discrete gamma distribution was used to model the evolutionary rate differences among sites, and the distribution parameter value was determined through maximum likelihood based on the sequence alignment. For every analysis, appropriate outgroup sequences were chosen based on length, similar domain architecture, and distinct function.
Synteny analysis.
The assessment of the degree of synteny between the relevant genomic regions was carried out using the ACT tool (49). Whenever necessary, sequences were trimmed and/or concatenated. BLASTN comparisons were used for less divergent genomes (Pneumocystis), whereas tBLASTn was used for comparison of the other genomes.
PCR amplification and sequencing of the putative MAT genes from P. jirovecii isolates.
A genomic region of 1,240 bp encompassing the putative P. jirovecii matMc and matMi genes was amplified using primers 5′ ATC CGA TAA ATA CAT ACA CG 3′ (positions 56360 to 56379 in sequence CAKM01000262.1 of P. jirovecii genome [contig 262]) and 5′ GAG GCT GTA AAA AGC ATA AG 3′ (positions 57581 to 57600). The template was genomic DNA extracted as previously described (37) from the bronchoalveolar lavage samples of patients with PCP. The PCR began by an initial denaturation step of 3 min at 94°C, which was followed by 35 cycles consisting of 30 s at 94°C, 30 s at the annealing temperature of 56°C, and 90 s at 72°C. The last reaction cycle was terminated with 10 min of extension at 72°C. The final MgCl2 concentration in the PCR mixtures was 4.5 mM. A region of 889 bp including the putative P. jirovecii matPi gene was amplified using primers 5′ ATG TTA CTA TAT ATA AGA CC 3′ (positions 47271 to 47290) and 5′ ATG AAT TAG AAG GCA AAA AG 3′ (positions 48141 to 48160). The PCR conditions were exactly the same as for amplification of matMc and matMi, except that the annealing temperature was 54°C. Sequencing of both strands of the PCR products was performed with the two primers used for PCR amplification, as well as the BigDye Terminator DNA sequencing kit and the ABI Prism 3100 automated sequencer (both from PerkinElmer Biosystems, Rotkreuz, Switzerland).
Data accessibility.
Curated annotations and scripts are in the folder Pneumocystis_Mating available from GitHub (https://github.com/ocisse/Pneumocystis_comparative).
SUPPLEMENTAL MATERIAL
Function of sex-related genes used as sequence queries.
Sex-related genes identified in two Pneumocystis species and T. deformans using S. pombe sequences as queries.
Locus reference or genomic location of the sex-related genes identified in two Pneumocystis species and T. deformans
Bioinformatics strategy used for homolog identification. The strategy involved a tBLASTn search using S. pombe proteins as queries and domain architecture analysis of the candidate genes (see details in Materials and Methods). Analyses of proteins are in blue, those of DNA are in red, and those of domains are in green. Download
Circuitries of pheromone signal transmission and transduction (A) and mitosis/meiosis balance control (B). The interactions of the gene products involved in the transmission and transduction of the sexual pheromones in S. pombe were reconstructed. The presence or absence of the gene products of P. carinii, P. jirovecii, and T. deformans is shown. The diagrams were based on UniProt annotation and built with CellDesigner (celldesigner.org). Download
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation grant 310030-146135 to P.M.H. and M.P. O.H.C. is supported by the Swiss National Science Foundation fellowship grant 151780.
We thank Sophie Chevalley for excellent technical assistance.
Footnotes
Citation Almeida JMGCF, Cissé OH, Fonseca Á, Pagni M, Hauser PM. 2015. Comparative genomics suggests primary homothallism of Pneumocystis species. mBio 6(1):e02250-14. doi:10.1128/mBio.02250-14.
REFERENCES
- 1.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 2.Hauser PM, Burdet FX, Cissé OH, Keller L, Taffé P, Sanglard D, Pagni M. 2010. Comparative genomics suggests that the fungal pathogen Pneumocystis is an obligate parasite scavenging amino acids from its host’s lungs. PLoS One 5:e15152. doi: 10.1371/journal.pone.0015152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cissé OH, Pagni M, Hauser PM. 2012. De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. mBio 4(1):e00428-12. doi: 10.1128/mBio.00428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cissé OH, Pagni M, Hauser PM. 2014. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol Evol 6:1938–1948. doi: 10.1093/gbe/evu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cushion MT, Smulian AG, Slaven BE, Sesterhenn T, Arnold J, Staben C, Porollo A, Adamczak R, Meller J. 2007. Transcriptome of Pneumocystis carinii during fulminate infection: carbohydrate metabolism and the concept of a compatible parasite. PLoS One 2:e423. doi: 10.1371/journal.pone.0000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cushion MT, Stringer JR. 2010. Stealth and opportunism: alternative lifestyles of species in the fungal genus Pneumocystis. Annu Rev Microbiol 64:431–452. doi: 10.1146/annurev.micro.112408.134335. [DOI] [PubMed] [Google Scholar]
- 7.Hauser PM. 2014. Genomic insights into the fungal pathogens of the genus Pneumocystis: obligate biotrophs of humans and other mammals. PLoS Pathog 10:e1004425. doi: 10.1371/journal.ppat.1004425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aliouat-Denis CM, Martinez A, Aliouat EM, Pottier M, Gantois N, Dei-Cas E. 2009. The Pneumocystis life cycle. Mem Inst Oswaldo Cruz 104:419–426. doi: 10.1590/S0074-02762009000300004. [DOI] [PubMed] [Google Scholar]
- 9.Chabé M, Aliouat-Denis CM, Delhaes L, Aliouat EM, Viscogliosi E, Dei-Cas E. 2011. Pneumocystis: from a doubtful unique entity to a group of highly diversified fungal species. FEMS Yeast Res 11:2–17. doi: 10.1111/j.1567-1364.2010.00698.x. [DOI] [PubMed] [Google Scholar]
- 10.Matsumoto Y, Yoshida Y. 1984. Sporogony in Pneumocystis carinii: synaptonemal complexes and meiotic nuclear divisions observed in precysts. J Protozool 31:420–428. doi: 10.1111/j.1550-7408.1984.tb02989.x. [DOI] [PubMed] [Google Scholar]
- 11.Peters SE, English K, Rana A, Akter S, Malik S, Warburton NC, Duckett JG. 2001. Synaptonemal complexes in the pre-cyst of Pneumocystis carinii. J Eukaryot Microbiol 48:134s. doi: 10.1111/j.1550-7408.2001.tb00485.x. [DOI] [PubMed] [Google Scholar]
- 12.Vohra PK, Park JG, Sanyal B, Thomas CF Jr.. 2004. Expression analysis of PCSTE3, a putative pheromone receptor from the lung pathogenic fungus Pneumocystis carinii. Biochem Biophys Res Commun 319:193–199. doi: 10.1016/j.bbrc.2004.04.154. [DOI] [PubMed] [Google Scholar]
- 13.Martinez A, Aliouat EM, Standaert-Vitse A, Werkmeister E, Pottier M, Pinçon C, Dei-Cas E, Aliouat-Denis CM. 2011. Ploidy of cell-sorted trophic and cystic forms of Pneumocystis carinii. PLoS One 6:e20935. doi: 10.1371/journal.pone.0020935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez A, Halliez MC, Aliouat EM, Chabé M, Standaert-Vitse A, Fréalle E, Gantois N, Pottier M, Pinon A, Dei-Cas E, Aliouat-Denis CM. 2013. Growth and airborne transmission of cell-sorted life cycle stages of Pneumocystis carinii. PLoS One 8:e79958. doi: 10.1371/journal.pone.0079958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cushion MT, Linke MJ, Ashbaugh A, Sesterhenn T, Collins MS, Lynch K, Brubaker R, Walzer PD. 2010. Echinocandin treatment of Pneumocystis pneumonia in rodent models depletes cysts leaving trophic burdens that cannot transmit the infection. PLoS One 5:e8524. doi: 10.1371/journal.pone.0008524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni M, Feretzaki M, Sun S, Wang X, Heitman J. 2011. Sex in fungi. Annu Rev Genet 45:405–430. doi: 10.1146/annurev-genet-110410-132536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smulian AG, Sesterhenn T, Tanaka R, Cushion MT. 2001. The ste3 pheromone receptor gene of Pneumocystis carinii is surrounded by a cluster of signal transduction genes. Genetics 157:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, Young SK, Furuya K, Guo Y, Pidoux A, Chen HM, Robbertse B, Goldberg JM, Aoki K, Bayne EH, Berlin AM, Desjardins CA, Dobbs E, Dukaj L, Fan L, FitzGerald MG, French C, Gujja S, Hansen K, Keifenheim D, Levin JZ, Mosher RA, Müller CA, Pfiffner J, Priest M, Russ C, Smialowska A, Swoboda P, Sykes SM, Vaughn M, Vengrova S, Yoder R, Zeng Q, Allshire R, Baulcombe D, Birren BW, Brown W, Ekwall K, Kellis M, Leatherwood J, Levin H, Margalit H, Martienssen R, Nieduszynski CA, Spatafora JW, Friedman N, Dalgaard JZ, Baumann P, Niki H, Regev A, Nusbaum C. 2011. Comparative functional genomics of the fission yeasts. Science 332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vohra PK, Puri V, Kottom TJ, Limper AH, Thomas CF Jr. 2003. Pneumocystis carinii STE11, an HMG-box protein, is phosphorylated by the mitogen activated protein kinase PCM. Gene 312:173–179. doi: 10.1016/S0378-1119(03)00614-0. [DOI] [PubMed] [Google Scholar]
- 20.Vohra PK, Puri V, Thomas CF Jr. 2003. Complementation and characterization of the Pneumocystis carinii MAPK, PCM. FEBS Lett 551:139–146. doi: 10.1016/S0014-5793(03)00914-1. [DOI] [PubMed] [Google Scholar]
- 21.Burgess JW, Kottom TJ, Limper AH. 2008. Pneumocystis carinii exhibits a conserved meiotic control pathway. Infect Immun 76:417–425. doi: 10.1128/IAI.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krajicek BJ, Kottom TJ, Villegas L, Limper AH. 2010. Characterization of the PcCdc42 small G protein from Pneumocystis carinii, which interacts with the PcSte20 life cycle regulatory kinase. Am J Physiol Lung Cell Mol Physiol 298:L252–L260. doi: 10.1152/ajplung.00191.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutty G, Achaz G, Maldarelli F, Varma A, Shroff R, Becker S, Fantoni G, Kovacs JA. 2010. Characterization of the meiosis-specific recombinase Dmc1 of Pneumocystis. J Infect Dis 202:1920–1929. doi: 10.1086/657414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cissé OH, Almeida JM, Fonseca A, Kumar AA, Salojärvi J, Overmyer K, Hauser PM, Pagni M. 2013. Genome sequencing of the plant pathogen Taphrina deformans, the causal agent of peach leaf curl. mBio 4(3):e00055-13. doi: 10.1128/mBio.00055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inderbitzin P, Harkness J, Turgeon BG, Berbee ML. 2005. Lateral transfer of mating system in Stemphylium. Proc Natl Acad Sci U S A 102:11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, Cuomo CA. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson SJ, Byrne KP, Wolfe KH. 2014. Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system. Proc Natl Acad Sci U S A 111:E4851–E4858. doi: 10.1073/pnas.1416014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X, Hull CM, Heitman J. 2005. Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021. doi: 10.1038/nature03448. [DOI] [PubMed] [Google Scholar]
- 29.Alby K, Schaefer D, Bennett RJ. 2009. Homothallic and heterothallic mating in the opportunistic pathogen Candida albicans. Nature 460:890–893. doi: 10.1038/nature08252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reedy JL, Floyd AM, Heitman J. 2009. Mechanistic plasticity of sexual reproduction and meiosis in the Candida pathogenic species complex. Curr Biol 19:891–899. doi: 10.1016/j.cub.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherwood RK, Scaduto CM, Torres SE, Bennett RJ. 2014. Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature 506:387–390. doi: 10.1038/nature12891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly M, Burke J, Smith M, Klar A, Beach D. 1988. Four mating-type genes control sexual differentiation in the fission yeast. EMBO J 7:1537–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mix AJ. 1935. The life history of Taphrina deformans. Phytopathology 25:41–66. [Google Scholar]
- 34.Kramer CL. 1960. Morphological development and nuclear behavior in the genus Taphrina. Mycologia 52:295–320. doi: 10.2307/3756013. [DOI] [Google Scholar]
- 35.Heitman J. 2010. Evolution of eukaryotic microbial pathogens via covert sexual reproduction. Cell Host Microbe 8:86–99. doi: 10.1016/j.chom.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roach KC, Heitman J. 2014. Unisexual reproduction reverses Muller’s ratchet. Genetics 198:1059–1069. doi: 10.1534/genetics.114.170472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nahimana A, Blanc DS, Francioli P, Bille J, Hauser PM. 2000. Typing of Pneumocystis carinii f. sp. hominis by PCR-SSCP to indicate high frequency of co-infections. J Med Microbiol 49:753–758. [DOI] [PubMed] [Google Scholar]
- 38.Brooksbank C, Bergman MT, Apweiler R, Birney E, Thornton J. 2014. The European Bioinformatics Institute’s data resources 2014. Nucleic Acids Res 42:D18–D25. doi: 10.1093/nar/gkt1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slaven BE, Meller J, Porollo A, Sesterhenn T, Smulian AG, Cushion MT. 2006. Draft assembly and annotation of the Pneumocystis carinii genome. J Eukaryot Microbiol 53(Suppl 1):S89–S91. doi: 10.1111/j.1550-7408.2006.00184.x. [DOI] [PubMed] [Google Scholar]
- 40.Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schäfer M, Müller-Auer S, Gabel C, Fuchs M, Düsterhöft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dréano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sánchez M, del Rey F, Benito J, Domínguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. 2002. The genome sequence of Schizosaccharomyces pombe. Nature 415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 41.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mi H, Muruganujan A, Thomas PD. 2013. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Birney E, Durbin R. 2000. Using GeneWise in the Drosophila annotation experiment. Genome Res 10:547–548. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 48.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 49.Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Function of sex-related genes used as sequence queries.
Sex-related genes identified in two Pneumocystis species and T. deformans using S. pombe sequences as queries.
Locus reference or genomic location of the sex-related genes identified in two Pneumocystis species and T. deformans
Bioinformatics strategy used for homolog identification. The strategy involved a tBLASTn search using S. pombe proteins as queries and domain architecture analysis of the candidate genes (see details in Materials and Methods). Analyses of proteins are in blue, those of DNA are in red, and those of domains are in green. Download
Circuitries of pheromone signal transmission and transduction (A) and mitosis/meiosis balance control (B). The interactions of the gene products involved in the transmission and transduction of the sexual pheromones in S. pombe were reconstructed. The presence or absence of the gene products of P. carinii, P. jirovecii, and T. deformans is shown. The diagrams were based on UniProt annotation and built with CellDesigner (celldesigner.org). Download
Data Availability Statement
Curated annotations and scripts are in the folder Pneumocystis_Mating available from GitHub (https://github.com/ocisse/Pneumocystis_comparative).