Abstract
AIM: To establish the most common vacA alleles in Helicobacter pylori (H pylori) strains isolated from Chilean patients and its relationship with gastritis and gastroduodenal ulcers.
METHODS: Two hundred and forty five H pylori clinical isolates were obtained from 79 biopsies from Chilean infected patients suffering from gastrointestinal diseases. An average of 2-3 strains per patient was isolated and the vacA genotype was analyzed by PCR and 3% agarose electrophoresis. Some genotypes were checked by DNA sequencing.
RESULTS: The most prevalent vacA genotype in Chilean patients was s1b m1 (76%), followed by s1a m1 (21%). In contrast, the s2 m2 genotype was scarcely represented (3%). The s1b m1 genotype was found most frequently linked to gastropathies (P<0.05) rather than ulcers. Ulcers were found more commonly in male and older patients. Curiously, patients living in cities located North and far South of Santiago, the capital and largest Chilean city, carried almost exclusively strains with the s1b m1 genotype. In contrast, patients from Santiago and cities located South of Santiago carried strains with either one or both s1a m1 and s1b m1 genotypes. Regarding the s2 m2 genotype, comparison with GenBank sequences revealed that Chilean s2 sequence was identical to those of Australian, American, and Colombian strains but quite different from those of Alaska and India.
CONCLUSION: Differences in geographic distribution of the s and m vacA alleles in Chile and a relationship of s1b m1 genotype with gastritis were found. Sequence data in part support a hispanic origin for the vacA genotype. Asymmetric distribution of genotypes s1b m1 and s2 m2 recedes H Pylori strain distribution in Spain and Portugal.
Keywords: H pylori, vacA alleles, Chilean isolates, s1, s2, m1 and m2 sequences
INTRODUCTION
Helicobacter pylori (H pylori) is a curved Gram-negative bacterium frequently present in the human stomach. Once the host has been colonized, this microorganism persists for years or decades[1]. This microorganism has a worldwide distribution and its prevalence increases with age[2], and is a major etiological agent in the development of peptic ulcer and gastric cancer[3,4]. Several potential virulence factors have been suggested to play a role in the pathogenesis caused by this microorganism. The most studied H pylori virulence factors are CagA and VacA. The first one, called the cytotoxin associated antigen[5] is one of the proteins encoded by the pathogenicity island, a unique genomic fragment containing about thirty genes and some of them encode the machinery required for the secretion of the CagA protein. Strains carrying a functional cagA gene have been associated with a variety of clinical outcomes[5].
VacA is a vacuolating cytotoxin which induces cytoplasmic vacuolation in a variety of mammalian cell lines in vitro[6]. It is responsible for the gastric epithelial damage and causes mucosal ulceration when administered intragastrically to mice[7]. Recently, the VacA cytotoxin has been described as a permease that promotes urea diffusion across the epithelia[8] providing an additional source of nutrients to sustain H pylori growth in vivo. It has been recently reported that the cytotoxin is able to induce apoptosis in epithelial cells as well as a specific inhibition of the immune response[9,10]. Different allelic variants in the vacA sequence in two regions of the gene have been described: the s allele located in the region encoding the N-terminal signal region which may occurs as the alleles s1- (s1a or s1b) or s2 type, and the m allele located at the middle of the vacA gene, which is present as m1 or m2 type[11]. The variable structure resulting of different arrangements of these alleles in the gene has been related to the differences in the level of cytotoxin production and to distinct clinical outcomes of the H pylori infection[12-14].
In Chile, a country with one of the higher mortality rates of gastric cancer, gastroduodenal diseases are common causes of medical consultations[15]. Serological studies have indicated that over 70% of asymptomatic adults older than 35 years old are infected by H pylori[16]. Our previous studies[17] have revealed that 83% of Chilean infected patients from Santiago have antibodies against VacA or CagA virulence factors determined by Western blot using purified recombinant proteins and by ELISA tests based on the detection of CagA and VacA antibodies[17].
Few studies have been carried out on strains from the South Cone and in particular from Chilean patients[18]. Recent data from H pylori strains from Argentina have revealed that most of the vacA alleles are s1b m1[19]. Reports about the characterization of strains isolated from Brazilian children indicated that s1 allele is associated with the presence of peptic ulcer[20] and genotype cagA+vacA s1b m1 is the most frequent in Brazilian adults with non-mixed infections[21]. Recent studies done with strains from 63 patients from the central area in Santiago, Chile, found that s1m1 is also the most common allele associated to ulcer patients[22].
Since information in Latin American countries is rather scarce, the present study adds new pieces of information in this respect and it has been focused on the distribution of vacA alleles of H pylori strains obtained from infected patients along this country (more than 5 000 kilometers long) to identify, as a preliminary study, the most frequent vacA alleles in this population and their relationship with different pathologies.
MATERIALS AND METHODS
Strains, growth and collection of clinical specimens
E. coli DH5α was used as a host for cloning experiments. Campylobacter yeyuni VPI H840 (ATCC 29428), kindly provided by Dr. H. Fernández (Universidad Austral de Chile) was used as DNA source of a negative vacA genotype control. Strains 26695 (ATCC 700392), 60190 (ATCC 49503), T×30a (ATCC 51932), NCTC 11637 (ATCC 43504) and J99 (ATCC 700824) were used as well characterized vacA alleles sources.
H pylori clinical strains were isolated from human gastric biopsies obtained from each patient who underwent upper gastrointestinal endoscopy for medical indication. Patients were recruited from different hospitals and clinics from 10 Chilean cities. Biopsies were transported to Santiago as frozen samples in dry ice and immediately after arrival suspended in PBS (500 μL), manually grinded and plated on Brucella agar plates. This medium contained 5% fresh horse blood, 5 mg vancomycin, 2.5 mg cefsulodin, 2.5 mg trimethoprime lactate, 2.5 mg amphotericin B, and 1.5 g agar per 100 mL of medium. Colonies were grown for three to four days under microaerophilic conditions using the microaerophilic system envelopes with palladium catalyst. Two or three specimens per biopsy were obtained and a total of 245 isolates were characterized.
Patients
Seventy- nine consent patients who gave informed consent
including 35 males and 44 females aged between 3 and 79 years old with symptoms warranted upper gastrointestinal endoscopies. Twenty- two patients from Iquique (IQ), 1 from La Calera (LC), 3 from Quillota (QU), 6 from Valparaíso (VA), 12 from Santiago (SA), 5 from Linares (LI), 6 from Los Angeles (LA), 2 from Temuco (TE), 8 from Valdivia (VD) and 14 from Punta Arenas (PA) were analyzed in this study.
Criteria for enrollment included nocturnal or burning abdominal pain, chronic vomiting, hematemesis or history of recurrent abdominal pain plus a first degree relative with an endoscopically proven diagnosis of peptic ulcer disease. Exclusion criteria included hemodynamically unstable patients and recent antibiotic therapy, antisecretory drugs or bismuth compounds in the last 4 wk. Signed informed consent forms by the patients or their parents in the case of minors were obtained. The institutional Review Board of the local participating institutions reviewed and approved the study.
Endoscopy, histology and criteria of H pylori infection
The endoscopic findings in the esophagus, stomach and duodenum were described in a standardized protocol. The macroscopic appearance was recorded according to the presence of nodularity, erosion or ulceration. Diagnosis of gastritis and duodenitis were done on histological basis. Gastropathy was diagnosed when abnormalities such as erosions or nodularity were observed. Ulcer was diagnosed when either a circumscribed break in the gastroduodenal mucosa that measured at least 5 mm in diameter or a scar was observed. Two biopsies were obtained from the distal antrum of each patient to determine H pylori status. One of the biopsies was used for rapid urease test (Hepy test, BiosChile).
Serial sections of formalin-fixed and embedded in paraffin gastric tissue from the second antrum biopsy of each patient were stained either with Warthin-Starry silver or H&E stain and examined for the presence of H pylori and associated pathology for pathologists who were unaware of the results of other tests. Bacteria were defined as H pylori on the basis of size, spiral morphology, and tissue location. A subject was considered colonized by H pylori when both invasive diagnostic techniques (rapid urease test and H pylori staining) were positive. A patient was considered negative for H pylori when both invasive diagnostic tests were negative. We confirmed this assignment in most of the cases by isolation of colonies which gave positive for urease, catalase, and peroxidase enzyme tests.
Isolation of H pylori chromosomal DNA
DNA was obtained by using the method described by Owen and Bickley[28], with CTAB (Cetyltrimethylammonium bromide) as the detergent for cell lysis. Briefly, single colonies were grown in plates as described above. Cells coming from half a plate were collected and transferred to an Eppendorf tube using a sterile loop and processed as described[23].
PCR reactions, primers, cloning and sequencing
Reactions were done according to the conditions established by Atherton et al[11,12]. Cloning of s1a, s1b, s2, m1, m2 PCR fragments from some isolates were done by ligation into the pGEM-T vector (Promega) and electroporation into E. coli DH5α cells which were plated on Luria agar plates containing ampicillin 50 μg/mL. Some cloned PCR fragments were sequenced by using ABI Prism-3 100 genetic analyzer (Applied Biosystems).
Study groups and statistical analysis
H pylori infected patients were assigned to one of three groups according to their endoscopical or histological findings as follows: Chronic Active Gastritis group (CG) included patients with normal or non-specific endoscopic findings, plus chronic gastritis by histology; Gastropathy Group (GG) included patients with significant endoscopic abnormalities (nodularity, polyps or erosions but not ulcers) and chronic gastritis by histology; and Peptic Ulcer Disease group (PUD) included patients with gastric or duodenal ulcer at endoscopy and chronic gastritis by histology.
To asses whether the presence or absence of ulcers in infected patients could be related with the gender, age, or allele type, cross tabulation and χ2 test were performed using the Minitab release 12 program (State College, PA, USA). A P value < 0.05 was considered significant. The s2 m2 strains were not included in this analysis due to its low frequency (3%).
Accession numbers
The GenBank accession numbers of sequences used in this study were: Sant4, # AY167579; Sant50, #AY167580; Sant51, #AY167581; Sant52, #AY167582; Oroan10, #AY168871; Oroan15, #AY168872; Sant53, #AY167583; 95-54, #U95971; 90, #AF2201191; 87-203, #U05677; V-296, #ABO57273; CHCTX-1, #AF479032; LA-12, #AY840127; CHN5027ass, #AF050379.1; IQ-61, #AY858851; QU-61, #AY858850; QU-31 #AY858849; AR-312, #AY185131; AR-710, #AY185128; SA-64, #AY839241; USA2781, # ABO572201; SS-1, #AY049006; NA2010, #AB057202.1; NA1986, #AB077199.1; India 3, #AF217727.1; Alaska5, #AB057171.1; Alaska20, #AB057186.1; Alaska08, #AB057174.1; Alaska03, #AB057169.1; Alaska04, #AB057170.1; India27, #AF217733.1.
RESULTS
Detection of the m1 and m2 alleles in Chilean H pylori strains
Seventy nine biopsies from H pylori infected patients living at 10 Chilean cities were used as a source to isolate 245 H pylori colonies. Amplification of the m2 vacA allele was obtained for two patients from Iquique and one from Santiago. Among the 245 studied strains, 97% of them harbored the m1 allele and 3% the m2 allele. Hybrids corresponding to m1/m2 alleles or non-typable alleles were not detected.
Detection of the s1 and s2 alleles in Chilean H pylori strains
Most of the strains carried the s1 allele (97%). The 72.1% of these was s1b and 24.1% was s1a. A summary of these results according to the strain origin (city) and the s and m alleles is displayed in Figure 1.
Figure 1.
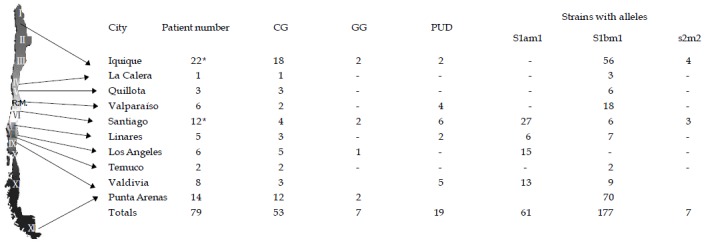
Chilean map and strain distribution according to the vacA alleles. Map of Chile including the location of the cities where biopsies were taken. The corresponding vacA alleles, as well as the number of strains and patients are displayed (*). Two patients, one from Iquique and the other from Santiago, had mixed infections. Chronic Active Gastritis (CG), Gastropathy Group (GG), Peptic Ulcer Disease (PUD). Total amount of strains = 245.
Identification of the m1 and m2 alleles of Chilean strains by DNA sequencing
Strain CHCTX-1 and LA-12 were utilized to amplify the m1 vacA allele. The PCR fragments were cloned in the pGEM-T vector and then sequenced. The results are shown in Figure 2A. After the alignment of the amino acid sequences with those available at in the GenBank, we found that the Chilean m1 sequences were similar to the Indian m1 deposited sequences. Seven strains out of 245 isolates presented the genotype s2m2. A further characterization of the m2 vacA allele was done on strain SA-64 by DNA sequencing the amplified cloned fragment. Comparison of this m2 allele sequence (Figure 2B) with other m2 sequences taken from the GenBank revealed almost identical amino acid sequences (one amino acid difference) with the m2 sequence of the Italian strain 95-54. Conversely, we found 45% amino acid identity for the SA-64 m2 sequence when compared to the Taiwanese strain V 296 as displayed in Figure 2B. These findings put the Chilean strain SA-64 closer to the strains that share a common ancestor from hispanic origin and far from those strains isolated from Asiatic patients.
Figure 2.

Alignment of s and m alleles as vacA amino acid partial sequences. A: Alignment of amino acid sequences corresponding to the amplified m1 vacA allele of Chilean strains CH-CTX1 and LA-12 with other representative m1 Indian sequences obtained from the GenBank; B: Alignment of amino acid sequences corresponding to the amplified m2 vacA allele and other representative m2 sequences obtained from the GenBank. The m2 allele from strain SA-64 was amplified using primers VA4F and VA4R and both strands were sequenced. The oligonucleotide sequence was translated (126 amino acids) and compared with other representative m2 allele sequences. These m2 sequences were obtained from GenBanK and corresponded to strains 95-54 from Italy, India 90, 97-203 from USA and V-296 from Taiwan; C: Alignment of amino acid sequences corresponding to Chilean strains having the s1a allele (CH-CTX1 and LA-12) and comparison with a s1a sequences from China. Sequences of Chilean s1b strains (IQ-61, QU-31 and QU-61) and comparison with other s1b sequences obtained from the GenBank from Argentinean strains AR-312 and AR-710; D: Alignment and comparison of amino acid sequences corresponding to the s2 sequence of the Chilean strain SA-64 and other sequences obtained from the GenBank of strains isolated in different locations. Sequences of the s2 vacA allele from different strains were aligned starting from amino acid position 30 in the vacA precursor sequence and the segment was fit with the ends defined by the Atherton primers for amplification of the s2 allele. ◆ Includes identical s2 sequences for strains: SA-64 from Chile, USA2781 from Arizona, USA, SS-1 from Australia, NA2010 and NA1986 from Colombia. ●Represents identical s2 sequences from strains NA1995, NA1992 and NA1685 from Colombia, India 3 and Alaska 5 from USA. Accession numbers are displayed in Materials and methods.
Sequences of the s2 and s1 alleles of Chilean strains
Strains CHCTX-1 and LA-12 were used as DNA source for amplification and sequencing of the s1a alleles, strains IQ-61, QU-31 and QU-61 for sequencing s1b allele (Figure. 2C), and strain SA-64 for sequencing the s2 allele (Figure. 2D). Comparison with other s1a, s1b, and s2 sequences available at the GenBank are displayed in Figures 2C and D. It was found that the s2 allele sequence from Chilean strain SA-64 was identical to those reported for American (USA2781), Australian (SS-1) and Colombian (NA2010) strains but far away from those of Alaskian (Alaska 4) and Indian (India 27) strains (in a segment of 66 amino acids there were 8 and 6 amino acid differences, respectively).
Correlation of vacA alleles and patient gastric diseases
The s1bm1 allele combination was more common in patients with chronic gastritis without ulcers and other endoscopic findings (P<0.05). The s1am1 has a homogeneous distribution among patients with chronic gastritis, gastrophaties or peptic ulcer diseases (Figure 3A). Figure 3B shows patient gender and disease occurrence. There was not significant difference between males and females regarding different clinical outcomes (Figure 3B). There was an age dependent increase in the frequency of peptic ulcer disease and an age dependent decrease in the frequency of chronic gastritis, without reaching statistical significance (Figure 3C).
Figure 3.
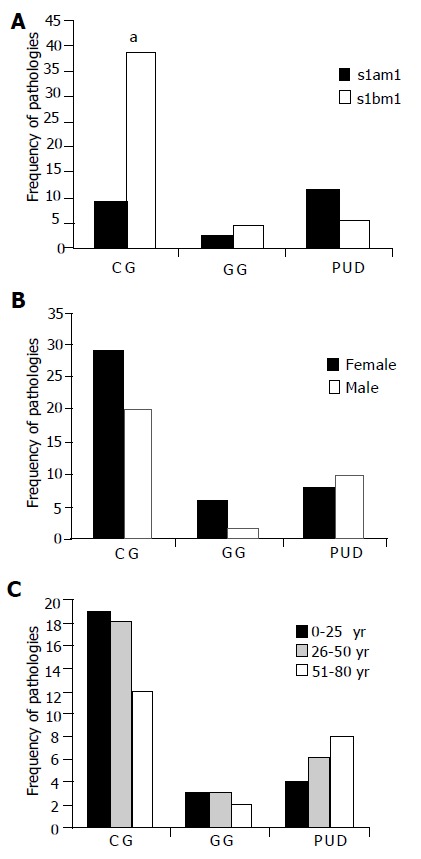
Analysis of gastropathies outcome frequencies respect to VacA. genotype, gender and patient age; A: Relationship between pathologies and alleles. Minitab Release 12 PA.USA and χ2 test for categorical variables was used (P<0.05). P <0.05 was considered significant. The s2m2 allele was excluded of the statistics test due a low frequency (3%); B: Relationship between gastropathies and gender. F: female, M: male. A total of 43 females and 32 males were studied (P >0.5), four patients carrying strains with s2 m2 allele were excluded from this study; C: Relationship between gastropathies and patient age (P>0.5).
DISCUSSION
A variety of structural and functional characteristic of the H pylori virulence factors have been used for strain genotyping, since they define a particular genotype which can be associated with an increased risk of peptic ulcer in human beings. Among these, genes of virulence factors such as cagA, vacA, hpaA, iceA and babA have been used before for classification purposes[24]. However one of the most extensively studied has been the vacA cytotoxin genotypes. This gene has a mosaic-like structure which is very uncommon in bacterial cytotoxins. Moreover, it is also unique for H pylori because the VacA polypeptide precursor promotes its own secretion. A similar auto-promoted mechanism has been observed for IgA proteases secreted by Neisseria gonorrhoeae and Haemophilus influenzae[25,26]. Some vacA gene regions have been used for genotyping and to evaluate the relationship between some vacA allele and a particular disease caused by H pylori[22].
We have studied 245 H pylori Chilean strains collected from patients belonging to different cities along the country. Most of the samples were collected from patients living at the central valley and some from the North and South regions as displayed in Figure 1. It should be noted that to the North of Chile is located the Atacama desert, one of the driest places in the world and this region do not support the development of cities. On the other hand, Southern Chile is under-populated because vast areas are preserved as ecological native resources and inhabited zones are rather minimal. Northern Chile also includes indigenous population. We included in the group of patients living in IQ, some strains isolated from native descendants from ancestral Aimara population. Curiously, all strains isolated from these patients had s1bm1 genotype. Also, a “Mapuche” native from Valdivia city (Southern Chile) presented the same genotype. A Mapuche autochthonous is considered a descendant of native Chileans before Hispanic colonization. Further analyses will be required to establish if the vacA genotype and other virulence markers within the autochthonous population remain homogeneous.
The analysis of the signal sequence s region showed that barely 3% of the isolated strains exhibited the s2 allele (just 7 out of 245 strains) and 97.1% was classified as s1 type. Therefore, most of the strains were classified either as s1a or s1b alleles. The analysis of the medium region (“m” alleles) revealed that only 3% was m2 type, which was always associated to the s2 allele. Considering simultaneously both “s” and “m” alleles, 72% of all characterized strains was s1b m1 and 25.1% was s1a m1. These results are quite different from those reported by Martínez et al[27], in particular to the frequency of the s2 and m2 alleles for one Chilean city Concepción, not included in our study. They found a higher proportion (44%) of this genotype in 34 infected patients and, considering each allele independently, 32% was s2 and 58% was m2 in an extended study for this city with 50 patients. Compared to our data, this could be an overestimation of the s2m2 genotype, since the screened patients were all recruited from a single city. In addition, they used in some cases a different set of primers to detect the m2 allele, based on a 6 bp difference in fragment length (m1 = 116 bp; m2 = 122 bp) which is difficult to distinguish even in a 3% agarose gel. Moreover, their results were not confirmed by DNA sequencing of the amplified alleles. In any case, our data support the idea of a putative hispanic origin for the Latin American strains, considering that the preferred s1bm1 allele distribution was also detected in the strains from patients from Spain and Portugal, as reported by van Doorn et al[28]. In addition, it is noticeable that the allele s2 m2 that we described for 7 Chilean strains has not frequently been described in other South American countries yet. Also, the s2 and m2 alleles occur in the lowest proportion in Spain and Portugal[28]. The s2 m2 Chilean allele seemed to be interesting to sequence since, as mentioned, it represents a local uncommon vacA genotype. The m2 allele cloned from strain SA-64 was sequenced. The amino acid sequence revealed a closer similarity to the m2 allele of strain 95-54 isolated from an Italian patient (one aminoacid difference). A feasible explanation for the low occurrence of this genotype could be that this allele may confer a disadvantage for natural in vivo selection of the strain such as a lower VacA activity, a feature proposed by some authors[11,29]. Alternatively, it may have resulted from a recombination event that actually occurs at very low frequency. The latter argument is sustained in part by the finding that some patients infected with multiple H pylori strains allow the exchange of bacterial DNA along the course of the disease[30,31].
Results of H pylori strain typing based on vacA allele done in other countries indicate that differences between m1 and m2 strains raise diversity in the level of cytotoxin activity. However, at present it is not clear if differences between s1 and s2 phenotypes also affect the level of cytotoxin expression by a reduction in the amount of exported cytotoxin. One hypothesis strongly proposes that S2 type allele containing a different signal sequence exports a cytotoxin precursor through the inner membrane in a less efficient way[13]. On the other hand, it has been demonstrated that when a short oligonucleotide extension encoding 12 amino acids (as part of the s2 allele) is transferred to a vacA gene displaying the s1 genotype. This completely abolishes the vacuolating activity but has no effect on VacA cytotoxin production[11,29].
A strong correlation between the severity of the clinical diagnosis and vacA genotype has been proposed by several authors. For instance, in South African patients, the allele s1m1 is associated to ulcer disease[32] and in other cases to gastric cancer, as described for German patients[33]. The s2 allele has also been associated with the lack of peptic ulcer[11]. Indeed, in the present analysis those strains whose s2 and m2 alleles were sequenced and reported here were isolated from patients suffering from mild gastritis and corresponded to supposedly less aggressive genotypes. However, one patient had a mixed infection carrying, in addition to the genotype s2m2, other strains with the s1am1 and s1bm1 genotypes. It remains to be established if the strains with genotype s2m2 are actually producing a VacA protein with attenuated cytotoxic properties.
We have reported here that most of the patients carrying the s1b m1 VacA allele suffered chronic gastritis (females mainly) and this genotype was preponderant in patients living in cities located to the north of Santiago. The s1am1 genotype was most frequently associated to isolates from patients suffering from ulcers (mild male predominance). Patients from Santiago and cities located south of Santiago (Central Chile) carried strains with either one or both s1am1 and s1bm1 genotypes. It should be noted that Santiago, as the Chilean capital city, attracts immigrants not only from the neighboring countries but also from Asian countries, and in a lesser extent, natives from Northern and Southern cities searching for jobs, a fact that may explain a mixed allele distribution found in strains isolated from patients living in Santiago.
Finally, this study supports the fact that H pylori is actually one of the microorganisms with the highest genetic variability and this feature may complicate genotyping[34]. The genomic variability is so high that some researchers consider H pylori as a quasi strain[34]. Also, in some cases, mutations in other genetic loci may be responsible for the generation of unexpected and more virulent strains.
ACKNOWLEDGMENTS
We gratefully acknowledge Emily Miller, from New York University, USA, for reading the manuscript and helpful suggestions.
Footnotes
Supported by FONDECYT, Comisión Nacional Científica y Tecnológica, Chile No.1000730 No.1030894 and No. 1000734 from and NIH No.DK54495
Science Editor Guo SY Language Editor Elsevier HK
References
- 1.Blaser MJ. Helicobacter pylori and the pathogenesis of gastroduodenal inflammation. J Infect Dis. 1990;161:626–633. doi: 10.1093/infdis/161.4.626. [DOI] [PubMed] [Google Scholar]
- 2.Graham DY, Klein PD, Opekun AR, Boutton TW. Effect of age on the frequency of active Campylobacter pylori infection diagnosed by the [13C]-urea breath test in normal subjects and patients with peptic ulcer disease. J Infect Dis. 1988;157:777–780. doi: 10.1093/infdis/157.4.777. [DOI] [PubMed] [Google Scholar]
- 3.Blaser MJ. Helicobacter pylori: its role in disease. Clin Infect Dis. 1992;15:386–391. doi: 10.1093/clind/15.3.386. [DOI] [PubMed] [Google Scholar]
- 4.Cover TL, Blaser MJ. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996;41:85–117. [PubMed] [Google Scholar]
- 5.Censini S, Lange C, Xiang Z, Crabtree JE, Ghiara P, Borodovsky M, Rappuoli R, Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leunk RD, Johnson PT, David BC, Kraft WG, Morgan DR. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 7.Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proc Natl Acad Sci USA. 2004;101:7727–7732. doi: 10.1073/pnas.0401528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tombola F, Morbiato L, Del Giudice G, Rappuoli R, Zoratti M, Papini E. The Helicobacter pylori VacA toxin is a urea permease that promotes urea diffusion across epithelia. J Clin Invest. 2001;108:929–937. doi: 10.1172/JCI13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer W, Gebert B, Haas R. Novel activities of the Helicobacter pylori vacuolating cytotoxin: from epithelial cells towards the immune system. Int J Med Microbiol. 2004;293:539–547. doi: 10.1078/1438-4221-00300. [DOI] [PubMed] [Google Scholar]
- 10.Willhite DC, Blanke SR. Helicobacter pylori vacuolating cytotoxin enters cells, localizes to the mitochondria, and induces mitochondrial membrane permeability changes correlated to toxin channel activity. Cell Microbiol. 2004;6:143–154. doi: 10.1046/j.1462-5822.2003.00347.x. [DOI] [PubMed] [Google Scholar]
- 11.Atherton JC, Cao P, Peek RM Jr, Tummuru MKR, Blaser MJ, Cover LT. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 12.Atherton JC, Peek RM, Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 13.McClain MS, Cao P, Iwamoto H, Vinion-Dubiel AD, Szabo G, Shao Z, Cover TL. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J Bacteriol. 2001;183:6499–6508. doi: 10.1128/JB.183.22.6499-6508.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Telford JL, Ghiara P, Dell'Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce MF, Censini S, Covacci A, Xiang Z. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duarte I, Ohmke J, Ciani S, Villaroel L. Patrones de carcinoma en gastrectomías de adultos chilenos: estudio multivariado en un país de alto riesgo. Gastr Latinoam. 2001;12:12–18. [Google Scholar]
- 16.Medina E. [Digestive diseases in Chile: epidemiologic outlook] Rev Med Chil. 1988;116:282–288. [PubMed] [Google Scholar]
- 17.Opazo P, Müller I, Rollan A, Valenzuela P, Yudelevich A, García-de la Guarda R, Urra S, Venegas A. Serological response to Helicobacter pylori recombinant antigens in Chilean infected patients with duodenal ulcer, non-ulcer dispepsia and gastric cancer. APMIS. 1999;107:1069–1078. [PubMed] [Google Scholar]
- 18.Müller I, Medina-Selby A, Palacios JL, Martínez P, Opazo P, Bruce E, Mancilla M, Valenzuela P, Yudelevich A, Venegas A. Cloning and comparison of ten gene sequences of a Chilean H pylori strain with other H pylori strains revealed higher variability for VacA and CagA virulence factors. Biol Res. 2002;35:67–84. doi: 10.4067/s0716-97602002000100010. [DOI] [PubMed] [Google Scholar]
- 19.Catalano M, Matteo M, Barbolla RE, Jiménez Vega DE, Crespo O, Leanza AG, Toppor J, Antelo P. Helicobacter pylori vacA genotypes, cagA status and ure A-B polymorphism in isolates recovered from an Argentinean population. Diagn Microbiol Infect Dis. 2001;41:205–210. doi: 10.1016/s0732-8893(01)00307-8. [DOI] [PubMed] [Google Scholar]
- 20.De Gusmão VR, Nogueira Mendes E, De Magalhães Queiroz DM, Aguiar Rocha G, Camargos Rocha AM, Ramadan Ashour AA, Teles Carvalho AS. vacA genotypes in Helicobacter pylori strains isolated from children with and without duodenal ulcer in Brazil. J Clin Microbiol. 2000;38:2853–2857. doi: 10.1128/jcm.38.8.2853-2857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashour AA, Magalhães PP, Mendes EN, Collares GB, de Gusmão VR, Queiroz DM, Nogueira AM, Rocha GA, de Oliveira CA. Distribution of vacA genotypes in Helicobacter pylori strains isolated from Brazilian adult patients with gastritis, duodenal ulcer or gastric carcinoma. FEMS Immunol Med Microbiol. 2002;33:173–178. doi: 10.1111/j.1574-695X.2002.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 22.Faundez G, Troncoso M, Figueroa G. cagA and vacA in strains of Helicobacter pylori from ulcer and non-ulcerative dyspepsia patients. BMC Gastroenterol. 2002;2:20. doi: 10.1186/1471-230X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owen RJ, Bickley J. Insolation of H pylori genomic DNA and restriction analysis. In: Clayton CL, Mobley HLT, editors. Methods in Molecular Medicine: Helicobacter pylori protocols, Tolowa, N J, Humana Press; 1997. pp. 81–88. [DOI] [PubMed] [Google Scholar]
- 24.Höcker M, Hohenberger P. Helicobacter pylori virulence factors--one part of a big picture. Lancet. 2003;362:1231–1233. doi: 10.1016/S0140-6736(03)14547-3. [DOI] [PubMed] [Google Scholar]
- 25.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 26.Veiga E, Sugawara E, Nikaido H, de Lorenzo V, Fernández LA. Export of autotransported proteins proceeds through an oligomeric ring shaped by C-terminal domains. EMBO J. 2002;21:2122–2131. doi: 10.1093/emboj/21.9.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez A, González C, Kawaguchi F, Montoya R, Corvalán A, Madariaga J, Roa J, García A, Salgado F, Solar H, et al. [Helicobacter pylori: cagA analysis and vacA genotyping in Chile. Detection of a s2/m1 strain] Rev Med Chil. 2001;129:1147–1153. [PubMed] [Google Scholar]
- 28.Van Doorn LJ, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz DM, Carneiro F, Vanderborght B, Pegado MD, Sanna R, et al. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 29.Letley DP, Atherton JC. Natural diversity in the N terminus of the mature vacuolating cytotoxin of Helicobacter pylori determines cytotoxin activity. J Bacteriol. 2000;182:3278–3280. doi: 10.1128/jb.182.11.3278-3280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31:31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 31.Marshall DG, Dundon WG, Beesley SM, Smyth CJ. Helicobacter pylori--a conundrum of genetic diversity. Microbiology. 1998;144(Pt11):2925–2939. doi: 10.1099/00221287-144-11-2925. [DOI] [PubMed] [Google Scholar]
- 32.Kidd M, Lastovica AJ, Atherton JC, Louw JA. Heterogeneity in the Helicobacter pylori vacA and cagA genes: association with gastroduodenal disease in South Africa? Gut. 1999;45:499–502. doi: 10.1136/gut.45.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miehlke S, Kirsch C, Agha-Amiri K, Günther T, Lehn N, Malfertheiner P, Stolte M, Ehninger G, Bayerdörffer E. The Helicobacter pylori vacA s1, m1 genotype and cagA is associated with gastric carcinoma in Germany. Int J Cancer. 2000;87:322–327. [PubMed] [Google Scholar]
- 34.Covacci A, Rappuoli R. Helicobacter pylori: molecular evolution of a bacterial quasi-species. Curr Opin Microbiol. 1998;1:96–102. doi: 10.1016/s1369-5274(98)80148-3. [DOI] [PubMed] [Google Scholar]


