Abstract
Motivation: Inferring how humans respond to external cues such as drugs, chemicals, viruses or hormones is an essential question in biomedicine. Very often, however, this question cannot be addressed because it is not possible to perform experiments in humans. A reasonable alternative consists of generating responses in animal models and ‘translating’ those results to humans. The limitations of such translation, however, are far from clear, and systematic assessments of its actual potential are urgently needed. sbv IMPROVER (systems biology verification for Industrial Methodology for PROcess VErification in Research) was designed as a series of challenges to address translatability between humans and rodents. This collaborative crowd-sourcing initiative invited scientists from around the world to apply their own computational methodologies on a multilayer systems biology dataset composed of phosphoproteomics, transcriptomics and cytokine data derived from normal human and rat bronchial epithelial cells exposed in parallel to 52 different stimuli under identical conditions. Our aim was to understand the limits of species-to-species translatability at different levels of biological organization: signaling, transcriptional and release of secreted factors (such as cytokines). Participating teams submitted 49 different solutions across the sub-challenges, two-thirds of which were statistically significantly better than random. Additionally, similar computational methods were found to range widely in their performance within the same challenge, and no single method emerged as a clear winner across all sub-challenges. Finally, computational methods were able to effectively translate some specific stimuli and biological processes in the lung epithelial system, such as DNA synthesis, cytoskeleton and extracellular matrix, translation, immune/inflammation and growth factor/proliferation pathways, better than the expected response similarity between species.
Contact: pmeyerr@us.ibm.com or Julia.Hoeng@pmi.com
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
From basic biology to translational medicine and clinical trials, animal models have been an invaluable tool for inferring human biological responses. Yet, in spite of the advances these models have facilitated, numerous findings have also been unsuccessfully translated to humans, as evidenced by the failure of many clinical trials. These failures could derive from species-specific differences in response to perturbations or stimuli that would preclude naively translating information learned in one animal model directly to another. Systems biology offers the means for understanding the limits of translatability of animal models in different settings, from clinical trials to toxicological assessments to basic cell biology. This approach can provide a more comprehensive predictive model because it considers changes at different levels of the entire system.
This is achieved through the development of systematic studies and integration of data over multiple experiments and data-generation platforms (Barabasi and Oltvai, 2004; Consortium, 2004, 2010; Gerstein et al., 2010; Goh et al., 2007; Meyer et al., 2012; Papin et al., 2005; Tarca et al., 2013). These more complete models will aid our understanding of at what regulatory levels and to what degree responses to different perturbations are translatable between species.
When developing models for species translation, orthologous genes are commonly thought to share the same or similar function. This assumption does not always hold, as several reports show that even among closely related species this is not necessarily the case (Gharib and Robinson-Rechavi, 2011). Such divergence goes beyond differences in function and can be seen in changes in essentiality; among 120 mouse orthologs of human essential genes, 27 (22.5%) were found to be non-essential (Liao and Zhang, 2008). In contrast, while paralogs may be expected to diverge more often than orthologs, it has been observed that changes in paralog function are observed with the same frequency as in orthologs (Studer and Robinson-Rechavi, 2009). Certainly, changes in the essentiality and functional role of a gene product are not solely driven by differences in gene sequence but also other factors (i.e. spatiotemporal expression of genes) must be considered when investigating species translation.
Gene expression, being at the core of biological function, is commonly used to evaluate changes between species and their response to perturbations. The conservation of promoters and transcription factor (TF) binding sites are important predictors of gene expression similarity, and there is a correlation between conservation of TF binding events and conservation of the target gene expression (Hemberg and Kreiman, 2011). TF promoter binding sites are conserved in liver cells for ∼ 30% of the cases when comparing human and mice (Odom et al., 2007), and most conserved non-coding DNA regions in vertebrates correspond to regulatory elements (Hemberg et al., 2012).
Existing species translation methods rely heavily on the concept of pathways for organization and prediction (Alleyne et al., 2009). Indeed, it seems that pathways may be better conserved than its individual components (i.e. genes and proteins; McGary et al., 2010; Subramanian et al., 2005), as groups of orthologous genes may continue to operate together between species. In such cases, pathway analysis provides important organizational information on the potential action of sets of genes. The two main approaches for deriving pathways or sets of functionally coherent genes are topology-driven and data-driven (Melas et al., 2011).
The sbv IMPROVER Species Translation Challenge (STC), using a systems biology approach, provided participants with both training and test datasets designed to assess the ability of methods to predict responses in normal human bronchial epithelial (NHBE) primary cells coming from two different donors from the responses observed in normal rat bronchial epithelial (NRBE) primary cells coming from an inbred laboratory strain. These cells were exposed to 52 different stimuli. Stimuli were chosen to ensure a broad spectrum of perturbations in the cellular system, and for each stimulus, samples were collected at different time points to generate phosphoproteomics, gene expression and secreted cytokines data. These data were used by 29 teams to make 49 predictions across four different sub-challenges that were each evaluated using multiple scoring metrics. The STC was centrally focused on two questions: (i) can the phosphoproteomic responses in human cells be predicted given responses generated by the same stimuli in rat cells? If so, does the accuracy of this prediction depend on the nature of the applied perturbation? (ii) Which gene expression regulatory processes (biological pathways/functions) are predictable across species?
2 METHODS
2.1 Data preparation
A complete description of the experimental design, data set generation and processing can be found in (Poussin et al., 2014). In brief, 19 phosphoproteins, 22 cytokines and genome-wide mRNA levels were measured under 52 different stimuli or Dulbecco’s Modified Eagle’s Medium (DME) control treatments (in triplicate), Table 1. The experiment was performed in two parts: 40 stimuli in the first experiment and 12 in the second. In each part, primary NHBE and NRBE cells were grown and exposed to the indicated number of stimuli. Cells were collected and lysed at different time points: 5 and 25 min. For phosphoprotein measurements, 6 h for gene expression measurements and 24 h for cytokine measurements. All cells were exposed to stimuli in triplicate, and DME controls were performed in 4-, 5- or 6-plicate.
Table 1.
STC datasets
| Dataset | Condition | Number of replicates | Number of measurements | Time point(s) | Total size |
|---|---|---|---|---|---|
| Phospho-proteomics | 52 stimuli | 3 biological replicates | 18 phosphoproteins | 5 min | 10 000+ data points |
| 25 min | |||||
| mRNA expression | 20 000 human genes | 6 h | 330+ CEL files | ||
| 19 000 rat genes | |||||
| Cytokine level | 22 cytokines | 24 h | 7000+ data points |
mRNA samples from the first experiment were processed in three batches. Each batch included human and rat mRNA for a subset of stimuli. DME control mRNA samples (four replicates) were measured for each batch. For the second experiment, all mRNA samples were processed together, including DME control mRNA samples (five replicates). Low-quality chips were excluded following quality control (QC) analysis. All remaining expression data including two to three replicates per stimulus were normalized using GC robust multiarray averaging within species. Probesets were mapped to gene symbols using Affymetrix annotations: HG-U133 Plus 2 (na33) and Rodent 230 2.0 (na32), for human and rat, respectively. Probesets mapping to multiple genes were excluded. In cases of multiple probesets mapping to the same gene, the probeset with the highest average expression over all experimental conditions was selected as representative. These high-quality normalized gene expression data in the gene symbol namespace were provided to the participants.
Protein phosphorylation status was measured independently for each experiment part in cell lysates collected at 5 and 25 min (in triplicates) using Luminex xMap (Dunbar, 2006). Experiment parts 1 and 2 have 6 and 5 DME controls, respectively. After QC, 16 phosphoproteins were kept for the challenge. Data were normalized using a robust regression, and normalized values were provided as the ratio of residuals to the root mean squared error of the fit. Cytokine data were similarly processed, though normalization was carried out by taking the Z-score of each cytokine across all stimuli within an experimental batch, including DME controls.
All data were divided into two equal groups, subsets A and B, by stimulus treatment to be used for training and testing of methods. To ensure similar distributions of signals in both data subsets, stimuli were separated through a data-driven approach that clustered stimuli according to phosphorylation level, gene set activation, gene expression (GEx) batch and differential gene expression. For each cluster, stimuli were randomly assigned to subset A or B.
Orthologs were identified using the HGNC Comparison of Orthology Predictions (downloaded December 19, 2012). Only gene symbol mappings between human and rat were used. A total of 12 458 orthologs were common between human and rat Affymetrix arrays after mapping of probesets to gene symbols.
Gene sets were based on the C2CP (Canonical Pathways) collection from MSigDB v3.1 of the Broad Institute (Subramanian et al., 2005). This collection was filtered to remove highly redundant gene sets, i.e. overlapping gene sets with many shared members, ensuring that remaining gene sets cover as many pathways/biological functions as possible. The resulting 246 gene sets were used for the STC. Gene set enrichment analysis (GSEA) was performed to assess co-regulation of genes representative of pathways/biological functions. For the analysis, genes were ranked based on calculated LIMMA t-values comparing respective DME control versus stimulus conditions (Smyth, 2004). LIMMA was performed using the lmFit and eBayes functions from the limma R package for the R Statistical Language with default parameters. The design matrix was constructed to compare the batch-specific DME control with each stimulus individually. Computed NES and associated significance values for each gene set were indicative of the activation/perturbation (increase or decrease) of pathways/biological functions by each stimulus in NHBE and NRBE cells (Subramanian et al., 2005). GSEA size parameters were min = 15 and max = 500. GSEA NES and FDR q-values were provided to participants.
2.2 Scoring
Sub-challenges 1 (SC1), 2 (SC2) and 3 (SC3) were scored as binary classification problems. Starting with the postulate that no single metric will capture all the attributes of a prediction, we used an aggregate of three metrics for evaluation. The metrics were proposed by IBM team members, and an independent panel of experts comprising the External Scoring Panel (ESP) decided on the final scoring approach. Participant identities were kept anonymous to the IBM team scoring the submissions. Five other metrics were considered but rejected as being redundant to the chosen three. The details of these metrics were not disclosed to the participants until the end of the challenge to avoid influencing method development toward optimizing for the scoring function rather than solving the scientific question posed. This practice is in keeping of other prediction evaluation challenges, like CASP, DREAM and a previous iteration of sbv IMPROVER.
We used non-redundant metrics that highlight three different qualities of a prediction: threshold versus non-threshold, order-based versus confidence-based and different ways of rewarding correct versus incorrect predictions. The chosen metrics were also selected to avoid rewarding pathological predictions, e.g. predicting all items to be of one class. Further complicating metric selection, the quantities of both classes (active and inactive) were imbalanced in the STC with active cases accounting for only ∼ 10% of all cases.
Participants were required to give confidence values for their predictions of either protein phosphorylation status or gene set activation (increase or decrease) to a given stimulus, depending on the sub-challenge. Confidence values could range between 0 and 1, where 1 represents the full confidence of an element being activated (either up- or downregulated) and 0 for full confidence of inactivation. A binarized gold standard (GS) was developed for protein phosphorylation status and gene set activation. For the phosphoprotein GS, normalized expression levels, which is akin to the standard deviation of a normal distribution, with an absolute value ≥3 were considered active, as agreed on by the ESP. Similarly for gene set activation, GSEA FDR q-values ≤0.25 were designated active, as recommended by GSEA.
The submitted matrix of predictions (stimuli versus protein or gene set response) could have been scored column-by-column or row-by-row and then aggregated together. However, given the sparseness of the GS for both protein phosphorylation status and gene set activation, we decided (in agreement with the ESP) to transform the matrix into a vector for scoring, i.e. columns of the matrix were joined to obtain single vector.
2.2.1 Metric descriptions
Area Under the Precision–Recall Curve (AUPR) is a well-known measure of classifier power. A list of items is ordered by descending confidence value (used only for ranking and not directly in the metric). The list is traversed corresponding to increasingly permissive confidence thresholds, and precision (fraction of ‘active’ predictions that are correct) is plotted versus recall (fraction of true ‘active’ class members correctly predicted). The area under this precision–recall curve is the AUPR score and is represented by a single number that summarizes the tradeoff between both measures.
Balanced Accuracy (BAC) avoids magnifying performance estimates of imbalanced datasets. It is computed as the average accuracy of either class.
| (1) |
where TP is the number of true positives, P is total number of positives, TN is the number of true negatives and N is the total number of negatives. For the STC, we used a confidence threshold of 0.5 to binarize the predictions as either positive (≥0.5) or negative (<0.5).
Pearson Correlation Coefficient (PCC) describes the linear dependence between two variables. In the STC, it was computed as the correlation between the predictions and the binarized GS, where 1 indicates an item is active and 0 inactive. PCC normally ranges from −1 to 1, but to be consistent with the AUPR and BAC measures, which range from 0 to 1, we used a normalized PCC:
| (2) |
For simplicity, we will refer to PCCnormalized as PCC when in reference to the challenge scoring metric.
2.2.2 Metrics aggregation
A rank-sum scheme to aggregate scoring metrics was proposed by the IBM team, along with one alternative, and was selected by the ESP because it equally weights each metric to produce an overall ranking. This rank-sum scheme was composed of ranking all teams within each respective metric. A team’s aggregate rank was then calculated by summing their rank across these three metrics. This rank sum was used for the final ordering of participants, with best performers achieving the lowest rank sum. To determine the robustness of these rankings, bootstrapping was performed to ensure that best performers were not highly sensitive to the exact configuration of GS. GS was sampled without replacement 1000 times, and the rankings recomputed each time. Given the imbalanced nature of GS, the bootstrapping was constrained to maintain the same proportion of active versus inactive items as observed in the entire GS.
2.3 Statistical significance of metrics
The null distribution for each metric in SC1-3 was generated by scoring 106 random predictions. To generate the confidences of a random submission, a uniform random number r (0 ≤ r ≤ 1) was generated for each ‘item’.
For SC1-3, the null hypothesis simulation was used to compute Z-scores. The mean (μ) and standard deviation (σ) of the scores obtained by the simulated prediction was computed and combined with an individual team’s score (x) to calculate the Z-score.
| (3) |
FDRs were computed for each metric using the R (Computing, 2013) function p.adjust with the method = ‘fdr’, which computes the Benjamini and Hochberg (1995) correction.
To compute a score’s P-value for each of the metrics, we counted the number of random predictions that were better than or equal to the observed score and divided it by the number of simulated predictions. FDR correction (Benjamini and Hochberg, 1995) was applied to the P-value, and a value of ≤0.05 was considered to be statistically significant.
The measure S represents the overall response similarity between human H and rat R GS, and is a Matthews correlation coefficient (MCC). The MCC represents a Pearson correlation between two binary vectors. A high S value would indicate a putatively conserved response and a signal that is expected to be translatable. Similarity measures can also be calculated per stimulus Ss = MCC(Rs, Hs), where Rs and Hs are binary vectors of phosphoprotein or gene set responses to stimulus s; per phosphoprotein Sp = MCC(Rp, Hp), where Rp and Hp are binary vectors of responses to stimuli for phosphoprotein p; and per gene set Sg = MCC(Rg, Hg), where Rg and Hg are binary vectors of responses to stimuli for gene set g.
Predictability Pr represents the overall similarity or agreement between the GS and a team’s or aggregate of teams’ predictions T, and is a MCC. A high Pr value would indicate good prediction performance and that the response was predictable. Like S, Pr can be calculated per stimulus Prs = MCC(GSs, Ts), where GSs and Ts are binary vectors of predicted phosphoprotein or gene set responses to stimulus s; per phosphoprotein Prp = MCC(GSp, Tp), where GSp and Tp are binary vectors of predicted responses to stimuli for phosphoprotein p; and per gene set Prg = MCC(GSg, Tg), where GSg and Tg are binary vectors of predicted responses to stimuli for gene set g.
The empirical P-values for the presence of genes in overlapping gene sets were calculated by sampling 105 times choosing a group of 25 gene sets of 246 gene sets. The frequency a gene is a member of the 25 randomly selected gene sets is recorded. The P-value is obtained by dividing the frequency a gene was found in at least x gene sets by 105.
3 RESULTS
The STC consisted of four sub-challenges, each addressing a different aspect of translatability: the intra-species protein phosphorylation prediction (SC1), the inter-species protein phosphorylation prediction (SC2), the inter-species pathway perturbation prediction (SC3) and the species-specific network inference (SC4). We explored the translatability of signals between different layers of transduction pathways by asking whether gene expression measurements are sufficient to predict upstream changes in protein phosphorylation. Furthermore, we examined across-species similarity in pathway activation by testing whether it was possible to predict the gene set activation and phosphorylation status of different pathways and important signaling proteins, respectively, in human lung epithelial cells given expression data in rat. These questions could reveal to what extent mathematical models are capable of recapitulating perturbed cellular functions from different data types within the same cell type and its across-species cell counterpart.
While the primary aim of SC2-4 was species translation, SC1 focused on assessing the informative power of transcriptional changes in response to different stimuli to infer phosphorylation responses. Transcriptional changes are typically the result of upstream signaling events driven by phosphorylation cascades. SC1 sought to address whether changes in gene expression are sufficiently informative to infer the molecular modifications observed upstream, in particular, the phosphorylation status of effector proteins. Furthermore, insights derived from this challenge could be informative for teams in the remaining sub-challenges. When making across-species predictions, it may be important to understand to what extent transcriptional data should be weighted when inferring phosphoproteomic responses.
Hence for SC1, participants were provided with GEx, protein phosphorylation (P) and secreted cytokine (Cy) data from stimuli subset A as training data (Fig. 1A). For testing, participants were asked to predict which proteins showed changes in their phosphorylation status (up- or downregulation is hereafter considered as an activation also stated as a response) for each stimulus in subset B. These predictions were to be reported as confidence values between 0 and 1, where 1 indicated the highest confidence of activation and 0 the lowest confidence. Phosphorylation levels were measured by the Luminex xMAP technology—a bead-based assay where microspheres are coated with antibodies designed to bind specifically to phosphorylated proteins—in primary NRBE cells under growing conditions (see methods).
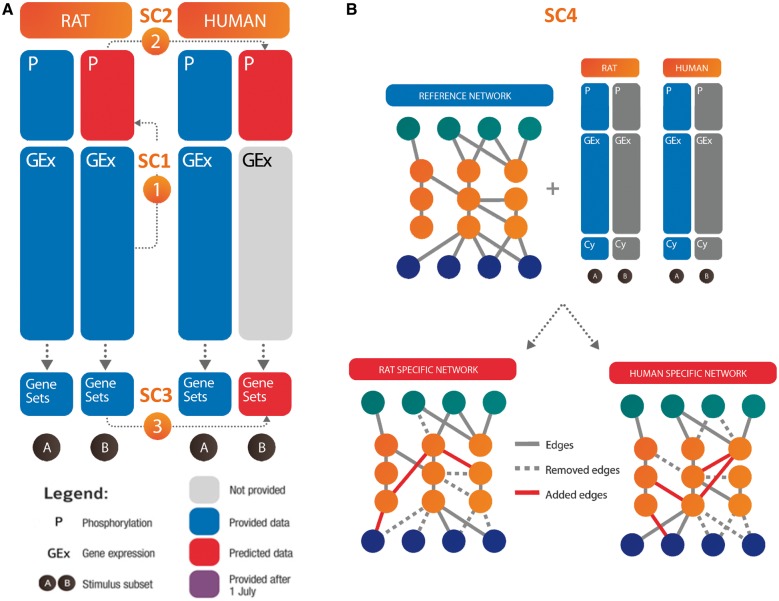
Fig. 1.
Overview of the STC: (A) Schematic of predictions to be made for each sub-challenge. Each sub-challenge required the prediction of the different sets of responses, indicated in red. (B) Schematic of SC4 to indicate utilization of a provided reference network with species-specific information from the training dataset to generate species-specific networks through the addition and removal of edges. Though cytokine measurements were made available to participants, they were not used in scoring, and for simplicity, were not included in this overview figure
As SC1 dealt with inferring protein phosphorylation status from downstream gene expression response, SC2 extended that aim to assess the across-species translatability of that phosphorylation status over the same set of proteins and stimuli. This sub-challenge required the prediction of human phosphoprotein activation in subset B based on equivalent data from homologous phosphoproteins in rat. The participants were provided with P, GEx and Cy data from subsets A and B in rat and subset A in human (Fig. 1A). Predictions could be based on translating signals directly from rat phosphoproteins to human phosphoproteins. They could also be made using GEx data to generate across-species inferences of gene expression that would then be used to predict human phosphoprotein status leveraging computational approaches developed for SC1. Similar to SC2, SC3 sought to explore the across-species translatability of molecular changes in the signaling response pathway, here focused on transcriptomic responses. Though orthologous genes by sequence conservation do not necessarily share the same pattern of expression changes, functionally coherent sets of genes representing biological pathways may often have a more conserved response between species or continue to operate as a group. It may also be the case that similar pathways are activated between species, but each uses different sets of genes from the same gene families. As such, SC3 asked for a prediction of the activation status of a broad range of gene sets in subset B of human cells (Fig. 1A), provided similar data as in SC2, along with gene set enrichment scores with associated significance values for subsets A and B in rat and subset A in human. From this sub-challenge, we hoped to identify which biological processes/pathways are similarly or differently perturbed between species, enabling the identification of conserved or divergent responses between biological systems.
The goal of SC4 was to infer human and rat networks given P, GEx and Cy data, as well as an a priori reference network (Fig. 1B). Participants were asked to use network inference methodologies to add or remove edges from the reference network to produce rat-specific and human-specific networks (see Bilal et al. in this issue). This sub-challenge differed from the others in that there is no obvious GS, but instead looked to leverage the wisdom of crowds to develop a consensus network that describes the conservation and divergence of biological pathways and interactions in response to the stimuli in subset A.
3.1 Challenge results
SC1-3 was scored in three different ways using different criteria and measured by the PCC, AUPR and BAC between the submitted confidence values and binarized GS. The ranks of the participants for each of these metrics were combined to obtain a final ranking (see methods). The robustness of these ranks was evaluated by subsampling 10% of the GS 1000×, while preserving the proportion of active/inactive calls, and calculating P-values (Supplementary Fig. S2).
As shown in Supplementary Figures S1A and S2A and Supplementary Table S1, from among 21 participating teams in SC1, the top three teams—teams 49, 50 and 75—could not be distinguished robustly between one another, and all were declared best performers (see Dayarian et al. in this issue). We compared these results to a series of aggregated ‘teams’ formed by averaging the prediction confidences of the best N teams to ascertain whether information could be gained by leveraging the wisdom of crowds. We found that the score for the aggregate of all teams ranks fourth overall and is better than the best performers in two out of three metrics (AUPR and PCC, Supplementary Fig. S3A). From among 13 participating teams in SC2, team 50 was clearly the best performer, followed by Team 111 [see (Biehl et al., 2014) in this issue]. In this sub-challenge, averaging the predictions of all teams did not fare better than the best performer, but ranked fifth overall and was better than the second best performer in two of three metrics (AUPR and PCC, Supplementary Fig. S3B). Finally in SC3, of 7 participating teams, team 50 was again the best performer, followed by Teams 49 and 111, which tied for second (see Hormoz et al. and Hafemeister et al. in this issue). As in SC2, averaging the prediction confidences of all teams did not fare better than the best performer, but the aggregate of all teams ranked fourth overall and was better than the second best performer in two of three metrics (AUPR and PCC, Supplementary Fig. S3C).
A known risk in classification problems is that some algorithms correctly separate the classes but label them incorrectly. Having seen such mislabeling in previous challenges, we attempted to identify similar occurrences in this challenge. Though reversing class labels may be less likely when datasets are highly imbalanced, as the STC was with only an ∼ 10% GS activation level in SC1-3, several teams from across different sub-challenges would have improved their rank if their class labels were reversed. It is important to note that if a prediction is close to random, then evaluating the reversed labels can give a small increase of performance. However, our aim was to look for predictions with large differences in their scores and where the prediction with reversed labels scores much higher. In SC1 four teams received a slightly better score when their prediction labels were reversed, and two teams achieved slightly better scores in SC2. SC3 stood out with one team, Team 111, having clearly reversed its labels, and its revised score would have positioned them as the best performer (see Hafemeister et al. in this issue).
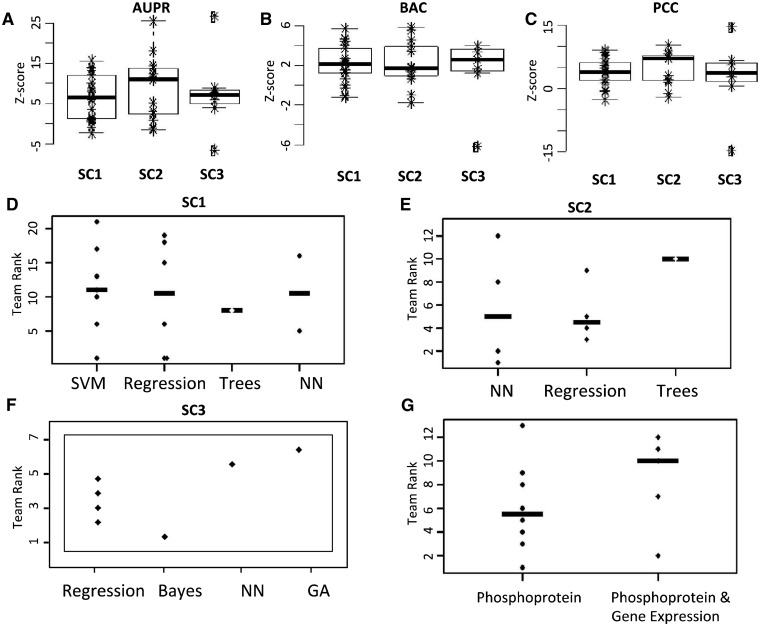
The overall success of participants in a sub-challenge can be measured by the median Z-score of the scoring metrics, which may be used to quantify the amount of predictive signal available in the provided data for a given classification problem. Z-scores offer a useful cross-challenge measure, as it takes into account size differences in the universe of predictions; important, since participants had to predict the activity of 16 × 26 phosphoprotein–stimulus pairs (SC1-2) and 246 × 26 gene set–stimulus pairs (SC3). Comparisons of the Z-scores for the three different metrics in Figures 2A–C suggest that protein phosphorylation was easier to translate across species (SC2) than solely within species from GEx (SC1), as reflected by higher Z-scores for AUPR and PCC. Inter-species protein phosphorylation also appeared easier to translate than inter-species pathway activation (SC3), as supported by the lower AUPR and PCC Z-scores for SC3 compared with SC2. The Z-scores for all three sub-challenges were tied for BAC (Fig. 2A–C).
Fig. 2.
Scores and computational methods used for solving the STC. The null hypothesis simulation was used to compute and plot team Z-scores of AUPR curve, balance accuracy (BAC) and PCC for SC1 (A), SC2 (B) and SC3 (C). Z-scores are used to compare the apparent difficulty of each of the sub-challenges. Panels (C–G) reflect actual performance differences—as measured by overall rank of three metrics—for different methodological approaches. Teams’ rank distributions are plotted separately by the type of approach for SC1 (D), SC2 (E) and SC3 (F). (G) In SC2, teams’ rank distribution is separated by usage of solely protein phosphorylation data or in combination with gene expression data. SVM: support vector machines, Trees: random forest and other tree-based methods, NN: neural networks, GA: genetic algorithm
The diversity of algorithms that participants deployed when solving the STC and broad rank distribution of similar approaches indicates these results were independent of the method used. Indeed, 7 teams used support vector machines (SVM), 14 teams used regression-based methods, 8 teams used decision trees or random forest, 4 teams used neural networks and 3 teams used a Bayesian approach. When the teams’ rank distribution was separated by the type of approach used for each sub-challenge, no clear tendency arose as the rankings of similar methods varied widely (Fig. 2D–F). Teams tried different combinations of feature selection approaches and classification algorithms. Although the sub-challenges shared similarities and a single team was best performer, no single combination of methods was universally advantageous across all sub-challenges. Consider that for SC2, 8 of 13 participants did not use GEx to infer phosphorylation activation in human and restricted their analysis to rat protein phosphorylation data. This seemed to be advantageous, as 5 of the 6 top-ranked submissions did not use gene expression, but no statistically significant difference was found between those who did and did not use GEx (P-value = 0.35, Fig. 2G). Nevertheless, there were some promising approaches arising from the STC. Neural network approaches ranked 1 and 2 for SC2, and it would have ranked 1 in SC3 had the class labels been reversed. The analysis of methods also suggested that a promising combination for the task of feature selection and classification is to select a subset of genes and use Linear Discriminant Analysis, an approach taken by half of the top 3 performing methods used for SC1 and SC2.
3.2 Analysis of stimulus prediction through gene sets and phosphoproteins
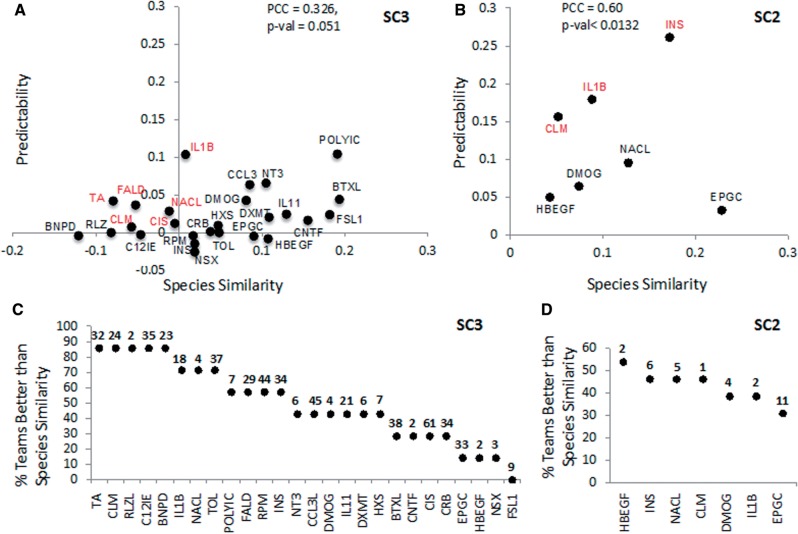
To assess how the accuracy of the participants’ predictions depended on the nature of the stimulus applied, we defined the species similarity S and the predictability or teams’ prediction performance Pr. Briefly, S is the MCC between rat and human GS, and Pr is the MCC between a team’s submission and the human GS. A high S value would indicate a putatively conserved response between rat and human; a high Pr suggests the signal is well translated by participants. S and Pr could be defined for stimuli based on gene set or protein phosphorylation activation. S and Pr could also be defined for gene set and phosphorylation activation based on response to stimuli (see methods for details). Figure 3 shows the mean Prs for all participants plotted against Ss based on the activation of gene sets (Fig. 3A) and protein phosphorylation (Fig. 3B).
Fig. 3.
Predictability versus species similarity for stimuli. (A) The y-axis indicates for each stimulus the mean predictability Prs of all team predictions when considering gene set activation in SC3. The x-axis is species similarity Ss of gene set activation. In red are stimuli where Prs > Ss > 0. (B) The y-axis indicates for each stimulus the mean predictability Prs of all team predictions when considering protein phosphorylation activation in SC2. The x-axis is Sp of phosphoprotein activation. In red are stimuli where Prs > Ss > 0. (C, D) Plots showing the percentage of teams where Prs > Ss for each stimulus when predicting gene set activation (C) or phosphoprotein activation (D). Stimuli are ordered by percentage of teams and the number of activated gene sets or phosphorylated proteins is indicated on top of each stimulus. The number of active calls per gene set is shown on the top of the graph. Nineteen stimuli are not shown in (B) and (D) because no proteins were measured as phosphorylated
Based on both gene set and phosphoprotein activation, clomipramine and IL1B were better predicted than expected by Ss (Prs > Ss > 0). In addition, formaldehyde, taurocholic acid, cisapride and activation, and insulin were better predicted based on protein phosphorylation. The correlation between Prs and Ss was higher for protein phosphorylation activation (PCC = 0.6, P-value < 0.013) than for gene set activation (PCC = 0.326, P-value < 0.051), perhaps reflecting not only a higher predictability for the protein phosphorylation data but also its smaller prediction space. Overall a higher percentage of teams performed better than Ss when predicting gene set activation in response to stimuli versus predicting phosphorylation status. Figure 3 shows that in 12 stimuli at least 50% of teams achieved a Prs > Ss when predicting gene set activation (Fig. 3C), but only in one stimulus, HBEGF, when predicting phosphorylation status (Fig. 3D). The individual team values Prs for protein phosphorylation and gene set activation are displayed in Supplementary Figure S4, and it shows that the translation of epigallocatechin and dimethyloxalyglycine was particularly difficult for both data types. Finally although aggregating the results of all teams did not yield a better overall prediction of stimuli effects when predicting protein phosphorylation, the aggregate of the five best teams performed better than individual predictions for insulin, clomipramine, IL1B, dimethyloxalyglycine, NaCl and epigallocatechin (Supplementary Fig. S4B).
3.3 Analysis of pathway predictions through gene sets and phosphoproteins
We also set out to assess the accuracy of the participants’ predictions regarding different biological pathways and to test which gene expression regulatory processes (biological pathways/functions) were translatable and therefore predictable across species. To do so, we defined the species similarity for protein and similar measures of predictability, or teams’ prediction performance, Prp and Prg (see methods).
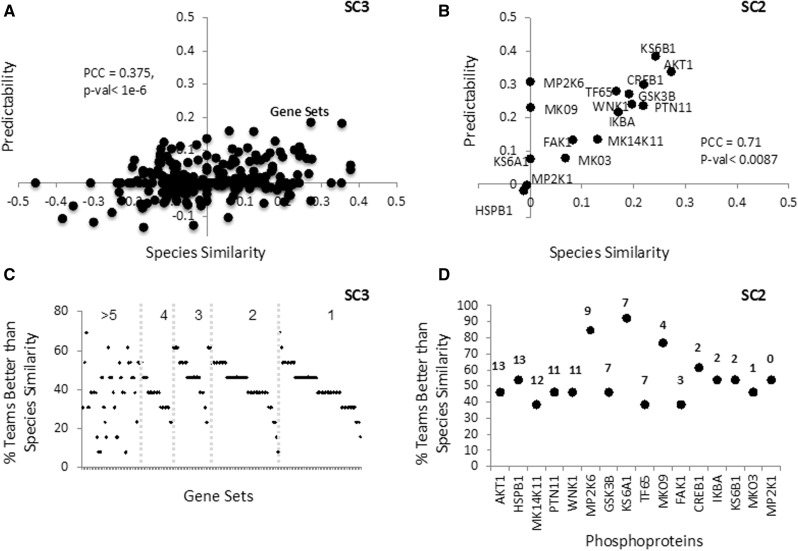
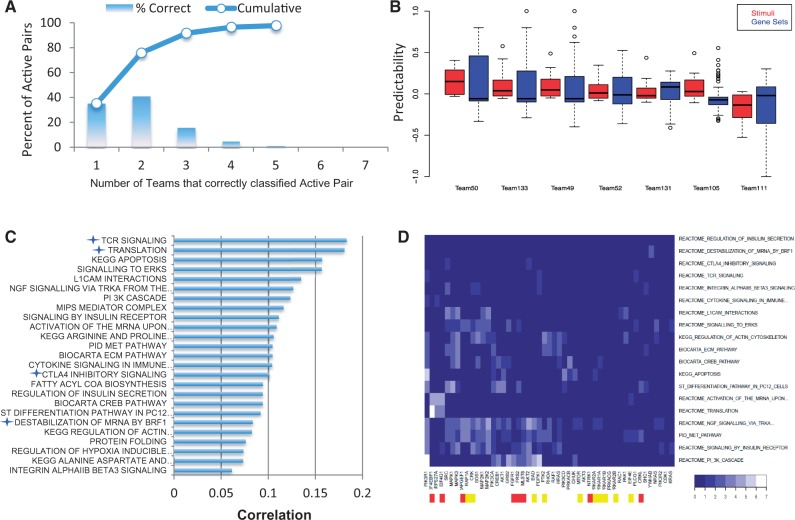
Figure 4A and B show the mean Prp and Prg for all participants plotted against Sp and Sg, respectively, based on activation in stimuli. A total of 49 of 246 gene sets were predicted better than expected by Sg (Prg > Sg > 0, Fig. 4A). Prediction performance per phosphoprotein Prp showed a ribosomal protein S6 kinase (KS6A1) and mitogen-activated protein kinases (MK09 and MP2K6) were predicted better than expected by Sp (Fig. 4B). Although aggregating all teams’ results did not yield a better overall prediction for protein phosphorylation activity, the aggregate of the five best teams performed better than individual predictions (Supplementary Fig. S5B). The high correlation between Prp and Sp (PCC = 0.71, P-value < 0.0087) reveals that most of the pathways defined by the protein phosphorylation activation were predicted with an accuracy expected by species similarity. We observed a similar situation for gene set activation prediction, with a lower but still significant correlation (PCC = 0.38, P-value < 1e-6). These results again suggested a slightly higher predictability in the protein phosphorylation data, though the prediction space was smaller. The individual team values for Prp and found that participants’ predictions were well translated for 71 of 176 active gene sets and for 8 of 16 phosphorylated proteins (Fig. 4A and B). Overall a higher percentage of teams performed better than species similarity when predicting protein phosphorylation activation (55%) versus predicting gene set activation (41%; see Fig. 4C and D). Nevertheless, when looking specifically at the set of active gene set and stimulus pairs (n = 560), 30% were correctly predicted by at least three teams (Fig. 5A), and in contrast to phosphorylation activation, six of seven teams in SC3 were better at globally translating the effects of stimuli than gene set activity (Fig. 5B).
Fig. 4.
Predictability versus species similarity for gene sets and phosphoproteins. (A) The y-axis indicates for each gene set the mean Prg of all team predictions when considering response to 26 stimuli in SC3. The x-axis is Sg of gene set activation. In red are stimuli where Prg > Sg > 0. (B) The y-axis indicates for each protein the mean Prp of all team predictions when considering response to 26 stimuli in SC2. The x-axis is Sp for phosphoprotein activation. (C and D) Plots showing the percentage of teams where Prg > Sg (C) and Prp > Sp gene sets and phosphoproteins are ordered by number of active calls, indicated on top of each black dot
Fig. 5.
Best translated gene sets representative of different pathways. (A) Histogram of the percentage of active gene set/stimulus pairs [560 pairs from 6396 (246 gene sets × 26 stimuli)] correctly predicted by N teams. Blue line represents the cumulative of the histogram values. (B) Distribution of teams’ Prg (blue) and Prs (red) values. (C and D) Best predicted gene sets as measured by Prg. (C) Barplot of 25 gene sets having a Prg Z-score ≥ 1.9. Blue star indicates a Sg Z-score ≥ 1.5. All gene sets are originally derived from Reactome unless otherwise indicated, according to MSigDB. (D) Hierarchical clustering of gene sets and genes that are present in at least 4 of the top 25 best predicted gene sets. Each cell is valued according to gene set membership and frequency the gene is found as part of that gene set’s GSEA CORE enrichment set. Gene/gene set pairs are assigned a 0 if the gene is not a member, 1 if only a member or 1 + C, where C is the number of stimuli under which the gene is found to be part of the CORE enrichment. Cells have a theoretical maximum value of 27. Cells are represented by a blue scale ranging from dark blue for 0 to white for the maximum value reached, here 7. Significantly overrepresented genes among these gene sets are labeled red (P-value < 0.01) or yellow (P-value < 0.05)
The 25 best-predicted gene sets showed some concordance in the biological processes they represent; in particular, translation and protein folding, apoptosis, metabolism, immune response (TCR, cytokine), growth signaling pathways (insulin, NGF, MET, TGFB), kinase signaling (ERK, PI3K), cell cytoskeleton and adhesion [extracellular matrix (ECM), integrin, actin, L1CAM] were well predicted (Fig. 5C). It was possible that specific genes were especially important for reaching high levels of predictability. To identify such biological drivers, the gene membership of the top 25 best-predicted gene sets (Z-score ≥ 1.9) was reviewed to identify genes that were consistently present. Moreover, from GSEA, genes identified as part of the CORE enrichment may be considered as the most biologically relevant as they contributed significantly to the enrichment score and were part of the ‘leading edge’ subset (Subramanian et al., 2005). Figure 5D reflects a hierarchical clustering of genes that were present in at least 4 of the top 25 best-predicted gene sets (49 genes among 19 gene sets), as well as the frequency they were found as part of the CORE enrichment for that gene set. The TF CREB1, the elongation factor eIF4EBP1 and kinases like AKT1, PIK3, PDPK1 and MAPK3 were in many of these gene sets and were also part of the CORE enrichment set for those gene sets, though their presence was not statistically significant. Notably, CREB1 and AKT1 phosphorylation activity was also well predicted by participants in SC2 (Fig. 4B). Yet, MAPK3 activity was not, showing some but not total coherence between the drivers of predictability in the two different data types, gene set and protein phosphorylation activation. Finally we performed a similar analysis looking for the most biologically relevant genes when considering the gene sets that were better predicted than expected by species similarity (Supplementary Fig. S6). Unexpectedly, we found that nuclear pore genes and replication factors were significantly enriched, as was a paxilin gene related to the FAK1 kinase, whose phosphorylation status was not very well translated by the participants (Fig. 4B).
4 DISCUSSION
We organized the STC as part of the sbv IMPROVER initiative and provided participants with experimental data describing multiple layers of different signaling pathways. The goal was to assess the ability of computational methods to predict biological responses in primary NHBE cells based on responses observed in primary NRBE cells. Several of our observations support the conclusion that changes in phosphorylation status and gene set activation induced by cellular response to 52 different perturbations in human cells can be predicted to some extent given responses generated in rat cells. Overall, stimuli caused more activation in rat than in human cells for most gene sets and for all phosphoproteins, except for phosphoproteins KS6A1 and HSPB1 (Supplementary Fig. S7). The differences in stimulus-induced activity could be due to a more homogenous biological sample in rats than in humans, or simply to higher sensitivity and faster signaling of NRBE cells compared with NHBE cells. Interestingly, differences between the kinetics of activation of homolog phosphoproteins at the 5 and 25 min time points were minimal [14 pairs for the whole dataset; see Figure 3 in (Biehl et al., 2014) in this issue].
Although not statistically significant, the average performance over all participants tended to be higher in SC2 than in SC3. This was seen in the higher Pr values when predicting stimuli activity across all phosphoproteins or gene sets and also when predicting the challenges’ respective signaling layer, gene set activation or phosphorylation responses across all stimuli. This observation holds when considering the performance of the best performer in both sub-challenges, where Team AMG’s prediction Pr values were higher for SC2 versus SC3.
When we considered cases where the majority of participants performed better than species similarity, i.e. a naïve, direct translation, 12 stimuli and 71 (of 176) active gene sets were well predicted in SC3, and 8 phosphoproteins and 1 stimulus in SC2 (Figs 3C and D and 4C and D). The greater number of well-predicted stimuli by gene set activation may have to do with the relatively lower levels of similarity Sg as compared with Sp, although, importantly teams were able to find informative biological signal in spite of these lower levels of response conservation. Finally, 10 teams in SC2 and 5 teams in SC3 submitted predictions that were statistically significantly different from random in two of the three metrics used (Supplementary Tables S1–S3). Overall for SC1-3, about two-thirds of the submissions were statistically significant (26 of 41), this indicates that in SC1, GEx data were sufficiently informative to infer upstream phosphorylation responses and that overall across-species predictions were achievable for specific stimuli, phosphoproteins and gene sets.
While many teams achieved a statistically significant result when considering random submissions as a null hypothesis, most teams in SC2 found their methods were unable to outperform a completely naïve approach to the challenge. If a team had submitted the rat’s subset B protein phosphorylation status in SC2 and the gene set activations in SC3 as their predictions, they would have ranked second and fifth, respectively. However, best-performer teams used approaches that did significantly better than the naïve approach, suggesting that their computational methods could capture additional informative biological signal in rat data [see in this issue (Biehl et al., 2014), Hafemeister et al. and Hormoz et al.]. Interestingly, there was also not a statistically significant difference between the five teams that used both P and GEx data and all others. The SC2 second place team, Team IGB, went further to test multiple variations of their Neural Network method to include GEx data and found it fared worse than methods using phosphorylation data alone [see (Biehl et al., 2014) in this issue]. This may be owed to the smaller difference in relative standard deviations (RSD) between human and rat phosphorylation response data versus GEx data (Supplementary Fig. S8). The similarity in phosphorylation response across species may have been difficult to detect due to the low number of replicate samples—only 3. These observations suggesting that it was relatively easier to predict a response of the phosphoproteomic layer are also reflected in the median Z-scores of the challenge metrics, where SC2 Z-scores were higher than SC3 for AUPR and PCC (Fig. 2A–C). These results likely reflect both the larger universe of predictions for SC3, which were an order of magnitude greater than for SC2 (246 × 26 = 6396 gene sets/stimuli pairs versus 16 × 26 = 416 phosphoproteins/stimuli pairs) and also a higher conservation between rat and human for protein phosphorylation activation (Sp = 0.71) compared with the gene set activation (Sg = 0.38). It is also possible that the higher similarity in phosphorylation response and smaller difference in the RSD between human and rat in the protein phosphorylation data with respect to the GEx data enabled more accurate predictions (Supplementary Fig. S8). This is likely due to the greater heterogeneity of human samples coming from two different donors compared with rat samples coming from an inbred laboratory strain but also given the increased complexity of human signaling. Transcriptional responses are a relatively downstream event from phosphorylation in signaling cascades. Hence, it is possible that when the signal finally propagates to the transcriptional layer, many other species-specific factors may amplify the differences of response between both species. An alternative explanation could be related to platform-specific biases due to inherent differences in mRNAs. However, we paid special attention to experimental design and execution as well as sample and data processing to minimize, as much as possible, experimental biases and avoid confounding effects within and between species (Poussin et al., 2014).
Additionally, phosphorylation response predictions benefited from more targeted experiments, which looked at 16 phosphoproteins enriched for active signals (Poussin et al., 2014) while GEx data was genome-wide and would only be expected to have a small percentage of genes differentially expressed. An interesting follow-up experiment would be to look at targeted gene expression for the pathway components using more specific measurements, like qRT-PCR or deep-sequencing. More sampled time points for GEx could provide greater granularity and may reveal patterns difficult to observe with a single 6 h exposure time point, and also confirm whether this choice left transcriptional responses to particular stimuli undetected.
The STC’s results indicate that GEx data were potentially noisier and the numbers of active gene set–stimulus pairs were low, however participants in SC3 made better predictions per stimulus than per gene sets (Fig. 5B). This shows that given sufficiently large datasets, it was still possible to extract a biologically relevant signal. Stimuli such as the chemical formaldehyde, cholesterol-derived taurocholic acid and the serotonin 5-HT4 receptor agonist cisapride were predicted better than the conservation of the response between both species. Similarly, the hydroxylase inhibitor dimethyloxalyglycine, the cytokine CCL3, the TLR activator PolyIC (Fortier et al., 2004) and the nerve growth factor NT3 were effectively translated and predictable at a level comparable with the observed conservation of response S (Fig. 3A) when based only on activation of protein phosphorylation. When we considered the overall prediction performance for phosphorylation response and gene set activation by all teams in SC2 and SC3, the antidepressant clomipramine and cytokine IL1B were better predicted than the levels of response conservation. This observation indicates that teams were able to identify human-specific signals that were not significantly present in the rat data. We also found that predictions based on the two data-generating platforms were not always in agreement, as in the case of insulin, which was well predicted based on protein phosphorylation but not based on gene set activation, and vice versa for NaCl (Fig. 3A and B).
Regarding the inference of biological processes, the gene sets predicted better than species similarity and/or best-predicted were related to DNA synthesis, cytoskeleton and ECM, translation, immune/inflammation and growth factor/proliferation (Fig. 5 and Supplementary Fig. S6). The gene sets associated with growth factor/proliferation processes shared a subset of CORE driver genes belonging to the following signaling pathways PI3K, RAS, RAF, MAPK, ERK and CREB. When considering only gene sets that were among the top 25 best predicted and had at least 1 member shared with at least 3 other best predicted gene sets, both CREB1 and AKT1 genes were present in, respectively, 4 and 6 of 19 gene sets (Fig. 5D). To note, the empirical P-values calculated for their presence across gene sets was not significant, indicating that these genes are frequently present in gene sets (see Fig. 5D). This may be due to a central role of these genes in many biological functions leading to a broad representation of those genes through C2CP gene sets. At the protein level, the activity of CREB1, AKT1 and MAP kinases such as MAPK9 and MP2K6 was well translated based on protein phosphorylation activation, showing a consistency in similar pathway perturbation prediction at different layers of the cellular system. It is interesting to note that the species similarity of MAPK9 and MP2K6 activation profile across stimuli was low, whereas the activation of both proteins was well predicted. The best predicted protein activity was that of KS6B (p70S6K). This protein is activated upstream by PI3K/AKT/mTOR protein kinases and regulates downstream phosphorylation of p70S6 protein, which is directly involved in the translation process found to be among the top best-predicted functions at the gene expression level. Interestingly, EIF4EBP1 and EIF4G1 factors involved in rate-limiting steps during the initiation phase of protein synthesis (Franke, 2008) were identified as CORE driver genes of translation-related gene sets, and the P-value associated to their presence across best-predicted gene sets was significant, indicating that those genes were rarely shared with other genes sets and therefore were specific to this biological function. Importantly, EIF4G1 gene also belonged to CORE genes contributing to the enrichment of translation-related gene sets in rat cells, suggesting a conserved role for this gene between human and rat species. Other proteins, such as TF65 and IKBA, associated to the NFkB signaling pathway showed good predictability in their activity in human cells. This result was coherent with the observation of good predictability for immune/inflammatory-related gene sets.
Except for some proteins such as MAPK9 and MP2K6 as mentioned above, many proteins of our measured panel have a level of species similarity that positively correlated with the level of predictability. This suggests that conserved responses at the protein levels possibly drive the translatability between both species in this cellular context.
Agreement in the biological processes that are similar between rat and human extends to the results of SC4, for the insulin, IL1R, MAPK, CREB1 and NFKB pathways (see Figs 2 and 5 in Bilal et al. in this issue). Participants of SC4 also found that regulation of RPS6KA1, active in human, and WNK1, in rat, differed between the two species (see Bilal et al. in this issue).
Finally, we observed that gene sets related to metabolism, which were generally expected to be conserved between species—such as insulin secretion, the CREB pathway, amino acid and fatty acid metabolism—were indeed well translated, although oxidative phosphorylation and gluconeogenesis were less well translated than expected by conservation (see Supplementary Table S6). The 49 different submissions to the STC used a diverse set of approaches including SVM, regression-based methods, tree-based methods like random forest, neural networks, Bayesian analyses and a genetic algorithm. The diversity of approaches might explain why the aggregate of all participant results performed better for two of three metrics in SC1 and SC2 (Supplementary Fig. S3). A lower participation level in SC3 may also explain why aggregation of all seven teams’ predictions did not perform better than the best performers. Interestingly, similar computational methods could have a wide range of performance within the same challenge, and no single method emerged as the clear winner (Fig. 2). Yet, methods that selected a subset of genes and used Linear Discriminant Analysis ranked among the top 3 performing approaches for SC1 and SC2. For SC2, one would have naively expected that using more data would benefit a prediction, and while not statistically significant, participants that used only protein phosphorylation data tended to rank higher than participants using protein phosphorylation data in conjunction with gene expression data (Fig. 2G).
A notable challenge in the STC was the imbalance in data, as only ∼ 10% of stimuli/phosphoprotein and stimuli/gene set pairs were active. Such a strong bias toward the inactive class complicates the training of models, although usage of ensemble methods that repeatedly sample the data, either over- or under-sampling, to converge on stable predictions could help overcome this imbalance. Generally, teams did not explicitly compensate for these imbalances, though methods like random forest inherently addressed such concerns. Yet as seen in the challenge results, random forests were not uniformly superior to all other methods and so there is potential for improved approaches that more explicitly account for class imbalance.
For their predictions, participants exclusively used data-driven approaches, and no computational method included a priori biological knowledge, as would be the case for topological approaches (Anvar et al., 2011; Melas et al., 2011). This rendered the interpretation of results with respect to biology more difficult. The construction and usage of a priori knowledge is characteristic of topology-based approaches. For example, Melas et al. used such approaches with data similar to the STCs to reconstruct pathways from input stimuli to the output cytokine release with phosphoprotein levels as intermediate signals. The initial construction of canonical pathways was based on gathering information from different databases (e.g. KEGG, Biocarta, etc.) combined with manual curation from the literature (e.g. reviews). Later, the Boolean networks were refined with a data-driven method using multi-linear regression on the phosphoprotein and cytokine data.
Alternative methods, beyond traditional machine learning approaches that assume training and test sets from the same dataset/domain, would be necessary to generalize predictions and enhance biological conclusions. Transfer learning and domain adaptation would be worth examining specially in problems that aim to integrate multiple layers of information—such as distances between stimuli based on the similarity of their chemical structure or their distance similarity in a protein interaction/transcriptional response network (Blitzer, 2006; Iorio et al., 2010; Napolitano et al., 2013; Pan et al., 2011).
The current work constitutes a proof of principle that predictability of responses in an in vitro system from one species is feasible to some extent given responses from another species. The results of this challenge provide insights on the predictability/accuracy in the context of diverse data types generated at various layers of the biological system studied; on the importance of time resolution to gain in prediction accuracy for species-specific sequential molecular events; on the different performance of similar computational methods due to variations in data preprocessing, feature selection and the classification algorithm (Tarca et al.); and on the different degrees of predictability of pathways/processes depending on the stimulus-induced perturbation in human and rat bronchial epithelial cells. Processes such as DNA synthesis, cytoskeleton and ECM, translation, immune/inflammation and growth factor/proliferation were better translated.
It will be important to test whether results and methods discussed here can be extended to more complex systems such as tissue, organ and whole organisms, the ultimate objective of translation between species. A better understanding of the range of applicability of the translation concept will impact the predictability of signaling responses, mode of action and efficacy of drugs in the field of systems pharmacology as well as increase the confidence in the estimation of human risk from rodent data for toxicological risk assessment.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the members of the Scoring Panel, Leonidas Alexopoulos, Jim Costello, Rudiyanto Gunawan, Torsten Schwede and Alfonso Valencia. We also thank Claudia Frei, Joanna Taylor and Immanuel Luhn for the organization of communications with the STC participants and of the IMPROVER symposium 2013; Jean Binder, Elise Blaese, Marianne Charaf, Alf Scotland, Peter Curle, Lionel Schilli and all the team members, who are not among the authors, for their contributions during discussion sessions and for the management of the project.
Funding: IBM and PMI authors performed this work under a joint research collaboration funded by PMI.
Conflict of interest: none declared.
REFERENCES
- Alleyne TM, et al. Predicting the binding preference of transcription factors to individual DNA k-mers. Bioinformatics. 2009;25:1012–1018. doi: 10.1093/bioinformatics/btn645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anvar SY, et al. Interspecies translation of disease networks increases robustness and predictive accuracy. PLoS Computat. Biol. 2011;7:e1002258. doi: 10.1371/journal.pcbi.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- Biehl M, et al. Inter-species prediction of protein phosphorylation in the sbv IMPROVER species translation challenge. Bioinformatics. 2015;31:453–461. doi: 10.1093/bioinformatics/btu407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzer JP, et al. EMNLP ‘06 Proceedings of the 2006 Conference on Empirical Methods in Natural Language Processing. 2006. Domain adaptation with structural correspondence learning; pp. 120–126. [Google Scholar]
- Computing,R.F.f.S. R:A Language and Environment for Statistical Computing. Austria: Vienna; 2013. [Google Scholar]
- Consortium,E.P. The ENCODE (ENCyclopedia Of DNA elements) project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- Consortium M, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin. Chim. Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortier ME, et al. The viral mimic, polyinosinic:polycytidylic acid, induces fever in rats via an interleukin-1-dependent mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2004;287:R759–R766. doi: 10.1152/ajpregu.00293.2004. [DOI] [PubMed] [Google Scholar]
- Franke TF. PI3K/Akt: getting it right matters. Oncogene. 2008;27:6473–6488. doi: 10.1038/onc.2008.313. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M. When orthologs diverge between human and mouse. Brief. Bioinformatics. 2011;12:436–441. doi: 10.1093/bib/bbr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh KI, et al. The human disease network. Proc. Natl Acad. Sci. USA. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberg M, Kreiman G. Conservation of transcription factor bindingevents predicts gene expression across species. Nucleic Acids Res. 2011;39:7092–7102. doi: 10.1093/nar/gkr404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemberg M, et al. Integrated genome analysis suggests that most conserved non-coding sequences are regulatory factor binding sites. Nucleic Acids Res. 2012;40:7858–7869. doi: 10.1093/nar/gks477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio F, et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl Acad. Sci. USA. 2010;107:14621–14626. doi: 10.1073/pnas.1000138107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao BY, Zhang J. Null mutations in human and mouse orthologs frequently result in different phenotypes. Proc. Natl Acad. Sci. USA. 2008;105:6987–6992. doi: 10.1073/pnas.0800387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGary KL, et al. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl Acad. Sci USA. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melas IN, et al. Combined logical and data-driven models for linking signalling pathways to cellular response. BMC Syst. Biol. 2011;5:107. doi: 10.1186/1752-0509-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, et al. Industrial methodology for process verification in research (IMPROVER): toward systems biology verification. Bioinformatics. 2012;28:1193–1201. doi: 10.1093/bioinformatics/bts116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano F, et al. Drug repositioning: a machine-learning approach through data integration. J. Cheminformatics. 2013;5:30. doi: 10.1186/1758-2946-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom DT, et al. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nature Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SJ, et al. Domain adaptation via transfer component analysis. IEEE Trans. Neural. Netw. 2011;22:199–210. doi: 10.1109/TNN.2010.2091281. [DOI] [PubMed] [Google Scholar]
- Papin JA, et al. Reconstruction of cellular signalling networks and analysis of their properties. Nat. Rev. 2005;6:99–111. doi: 10.1038/nrm1570. [DOI] [PubMed] [Google Scholar]
- Poussin C, et al. The species translation challenge—a systems biology perspective on human and rat bronchial epithelial cells. Scientific Data. 2014;1 doi: 10.1038/sdata.2014.9. , Article number: 140009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Studer RA, Robinson-Rechavi M. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet:TIG. 2009;25:210–216. doi: 10.1016/j.tig.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarca AL, et al. Strengths and limitations of microarray-based phenotype prediction: lessons learned from the IMPROVER Diagnostic Signature Challenge. Bioinformatics. 2013;29:2892–2899. doi: 10.1093/bioinformatics/btt492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.