Abstract
Human immunodeficiency virus type 1 (HIV-1) transmission and infection occur mainly via the mucosal surfaces. The commensal bacteria residing in these surfaces can potentially be employed as a vehicle for delivering inhibitors to prevent HIV-1 infection. In this study, we have employed a bacteria-based strategy to display a broadly neutralizing antibody VRC01, which could potentially be used to prevent HIV-1 infection. The VRC01 antibody mimics CD4-binding to gp120 and has broadly neutralization activities against HIV-1. We have designed a construct that can express the fusion peptide of the scFv-VRC01 antibody together with the autotransporter β-barrel domain of IgAP gene from Neisseria gonorrhoeae, which enabled surface display of the antibody molecule. Our results indicate that the scFv-VRC01 antibody molecule was displayed on the surface of the bacteria as demonstrated by flow cytometry and immunofluorescence microscopy. The engineered bacteria can capture HIV-1 particles via surface-binding and inhibit HIV-1 infection in cell culture.
Introduction
The Acquired Immunodeficiency Syndrome (AIDS) pandemic caused by the human immunodeficiency virus type 1 (HIV-1) has been ongoing for over three decades (Barre-Sinoussi et al., 1983; Schupbach et al., 1984). It is estimated that 39 million people have already died of AIDS-related diseases; 35 million people are living with HIV-1, and 2.1 million people are newly infected each year (UNAIDS, 2014). In spite of the recent optimism regarding a functional cure for HIV-1, there are still substantial obstacles for achieving a sterilizing cure with the current available treatment regimens. Therefore, preventing new infection is still the priority for curbing the AIDS epidemic. So far, much effort has been dedicated towards the development of an effective AIDS vaccine, but none has yet been successful (Burton et al., 2012; Johnston and Fauci, 2011; Lifson and Haigwood, 2012; O’Connell et al., 2012; Xiang, 2013). Therefore, other alternative prevention approaches are needed in order to curtail HIV-1 transmission and infection.
There are several reports on a new prevention approach using gram-positive bacteria, such as lactobacilli, for surface display of CD4 (2-domains) (Liu et al., 2008), HIV-1 entry inhibitor cyanovirin-N (CV-N) (Lagenaur et al., 2011; Liu et al., 2006; Yamamoto et al., 2013), or for secretion of a CCR5 antagonist RANTES (Vangelista et al., 2010) and HIV-1 fusion peptide inhibitors (Pusch et al., 2006) to prevent HIV-1 infection. These approaches involve the use of bacteria to synthesize anti-HIV-1 inhibitors that can be secreted or displayed on the cell surface so that the inhibitors can bind to the virus and prevent infection. Since these commensal bacteria inhabit the mucosal sites which are the port of entry for HIV-1, these protein-based inhibitors can be produced by the bacteria which can be maintained as part of the normal mucosal bacterial flora, this approach could thus be employed as a potential strategy to prevent HIV-1 infection.
Gram-negative bacteria have also been used for a number of therapeutic applications including gene delivery into the gut (Castagliuolo et al., 2005). Succesful attempts include surface display of lipase (Lee et al., 2013), antimicrobial peptides (Shin et al., 2013), p-glucuronidase (Cheng et al., 2013) and also for the secretion of single-chain or Fab antibody fragments (Cheung et al., 1992; Fallecker et al., 2013; Fernandez et al., 2000). However, very few studies have been reported in the use of gram-negative bacteria as potential agents for the delivery (Abdel-Mohsen et al., 2013) of anti-HIV regimens. So far only one study has been reported in which Escherichia coli Nissle 1917 was used to secrete an anti-HIV-1 fusion peptide inhibitor to target gp41 glycoprotein of the virus. The fusion peptides from bacterial secretion can inhibit viral infection in cell culture experiments, and bacterial colonization in mice can last for weeks, or in some cases even months, however, the viral challenge experiment in vivo has not yet been carried out as the mice cannot directly be used for HIV-1 infection (Rao et al., 2005). Surface display of anti-HIV-1 inhibitors on gram-negative bacteria is another approach in this commensal bacterial strategy, but it has not yet been tested. For surface display, the bacterial transporter genes must be used to translocate the molecules of interest onto the cell surface (Castagliuolo et al., 2005; Fairman et al., 2011; Jose et al., 2012). Among the known transporters, the autotranspoter (AT) is one of the most studied, and its structure and translocating mechanisms have been reported recently (Benz and Schmidt, 2011; Ieva and Bernstein, 2009; Rutherford and Mourez, 2006; van den Berg, 2010). More importantly, these autotransporters are shown to be able to translocate single-chain antibody molecules onto the bacterial surface (Pyo et al., 2009; Veiga et al., 1999, 2003).
In this report, we have used the gram-negative bacteria for surface display of anti-HIV-1 antibody molecules. The autotransporter, an immunoglobulin A (IgA) protease gene (IgAP) of Neisseria gonorrhoeae, was used because its C-terminal domain has been shown to efficiently translocate the passenger domain at its N-terminal (Dautin and Bernstein, 2007; Pohlner et al., 1987). We therefore employed this gram-negative bacterial autotransporter for surface display of a potent broadly neutralizing antibody (VRC01) against HIV-1 infection. VRC01 is a potent neutralizing antibody isolated from an HIV-1 patient, which can neutralize about 90% of HIV-1 isolates tested (Wu et al., 2010). VRC01 binds to the CD4-binding site (CD4-BS) and can mimic CD4-binding to gp120 (Zhou et al., 2010). We have generated the single-chain variable fragment (scFv) of VRC01 antibody (scFv-VRC01) and displayed it on the surface of E. coli to test its ability to inhibit HIV-1 infection.
Results
Design of the scFv-VRC01 surface-display constructs
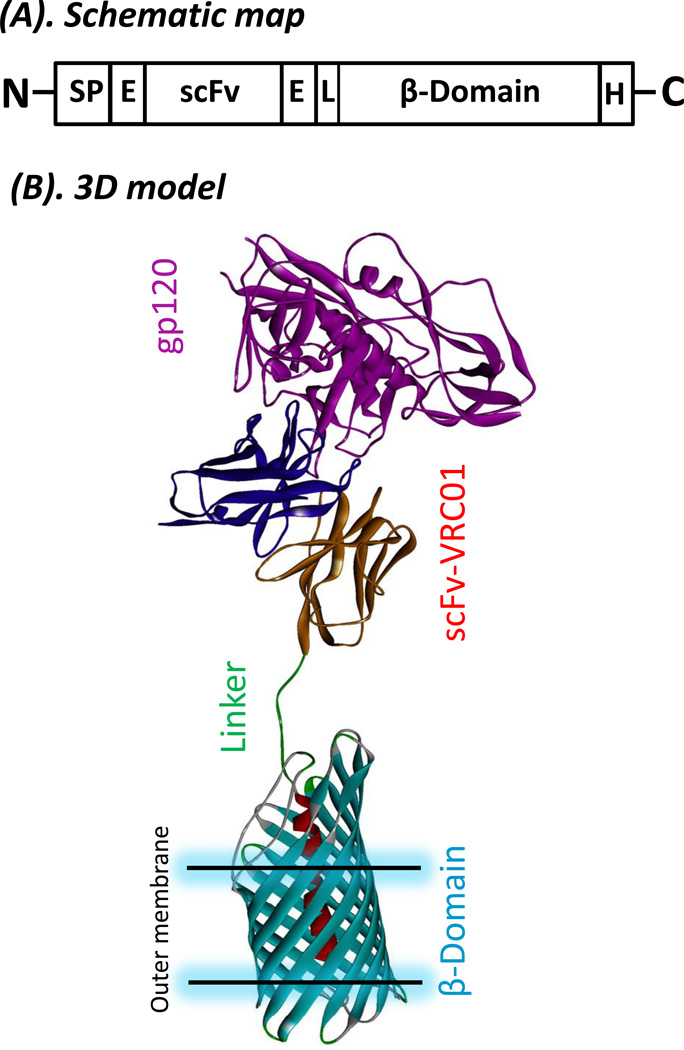
The scFv-VRC01 was designed using a two-step approach. The first was the designing of the single-chain (scFv) VRC01 antibody domain for expression. The VRC01 antibody gene was used to generate the single-chain antibody (scFv). A linker (-GGGGSGGGGSGGGGS-) was used to link the heavy chain (VH) and light chain (VL) gene fragments. Two E-tags were inserted into the recombinant gene; one was located between the β-barrel domain and the single-chain antibody, another was added to the N-terminus of the single-chain antibody (Fig. 1A). The resulting recombinant protein would display the His-tag at the C-terminus when expressed in the pET22b vector, and will be 257 amino acids in length with an expected molecular weight of about 27kDa. The designed peptide was codon-optimized and synthesized for the E. coli expression system. The second step was to link scFv-VRC01 fragment to the translocator β-barrel domain (C-IgAP) from bacterial (N. gonorrhoeae) autotransporter (434aa), which will then generate a fusion protein (scFv-VRC01-β-barrel domain (C-IgAP)) of about 75kDa. The proposed structural model of the fusion recombinant protein molecule is shown in Fig. 1B. The scFv-VRC01 fusion upon expression is then expected to be displayed on the surface of the bacterial cell and bind to gp120 on the surface of the HIV-1 virion to inhibit viral infection.
Fig. 1.
Construction of fusion protein of single-chain antibody VRC01 and autotransporter β-domain from N. gornohoeae(A). Schematic representation of the fusion protein construct for surface display. Sp, signal peptide; E, E-tag; L, linker (GSG); scFv, single-chain variable fragment; p-domain, the c-terminal part of autotransporter serine proteinase gene of N. gornohoeae. H, His-tag. (B). Three dimensional (3D) model of the fusion protein for surface display and its binding to gp120. The linker is GSG plus the E-tag, β-barrel domain (PDB 1UYN (van den Berg, 2010)), scFV-VRC01 and gp120 of HIV-1 (PDB 3NGB).
Expression of scFv-VRC01 antibody in E. coli
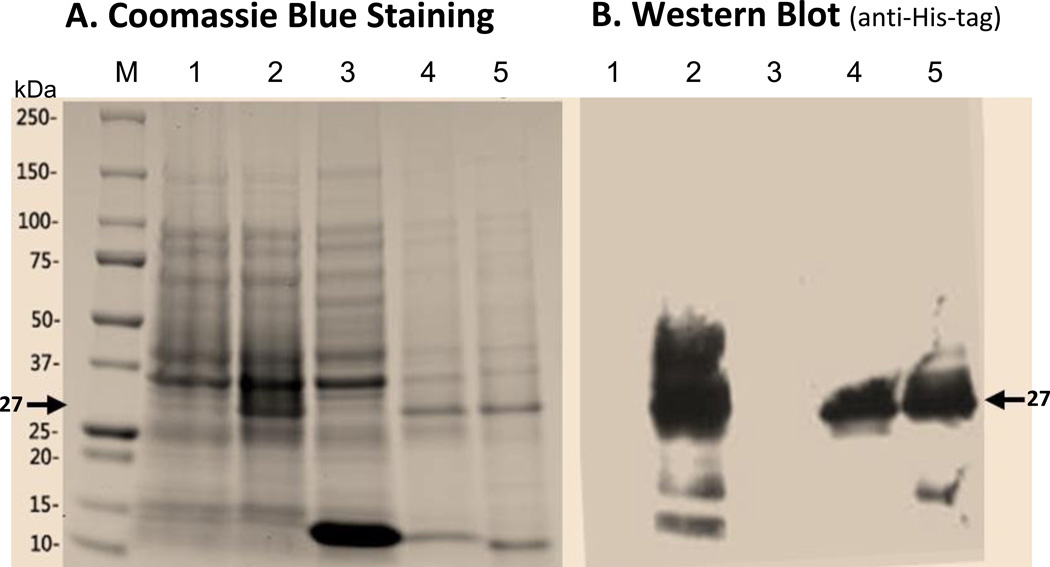
The synthesized single-chain VRC01 antibody (scFv-VRC01) gene was first cloned into the pET28b expression vector to test for expression of the scFv-VRC01 protein with the predicted molecular weight of about 27 kDa. As shown in Fig. 2A, a band of 27 kDa could be detected in total bacterial lysates upon induction by IPTG, but was absent in both un-induced lysates or in the supernatant of induced samples. This suggests that the single-chain VRC01 antibody protein molecule could be expressed in E. coli, and found mostly in the insoluble fractions of the bacterial lysates. The recombinant protein was then further purified using the Ni-NTA column and the presence of the protein was confirmed by western blot analysis using anti-His-tag antibody. An example of the blot is shown in Fig. 2B, confirming that most of the expressed protein could be found in the insoluble fraction of the bacterial lysate.
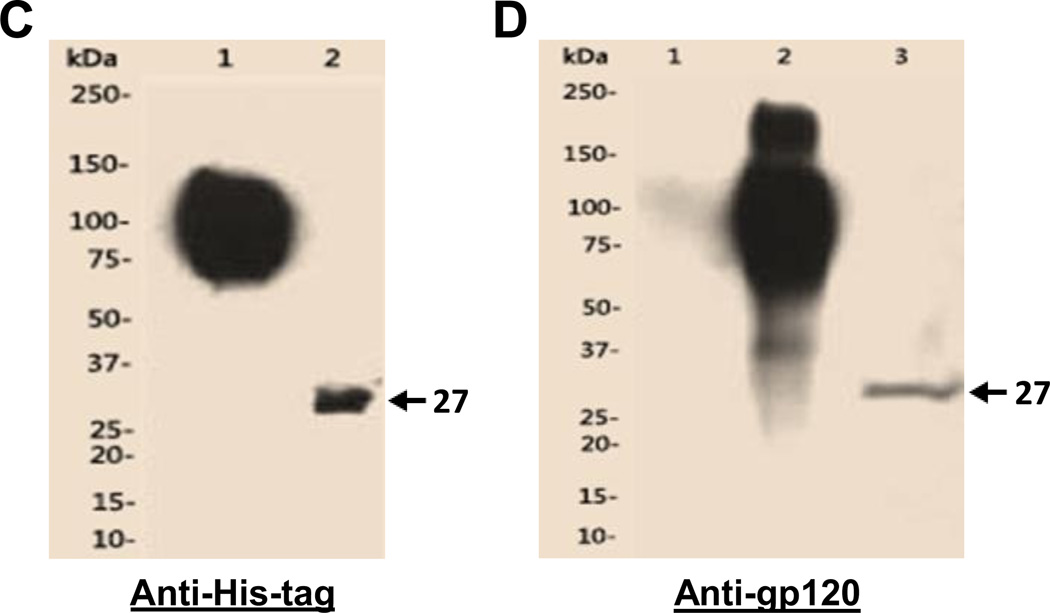
Fig. 2.
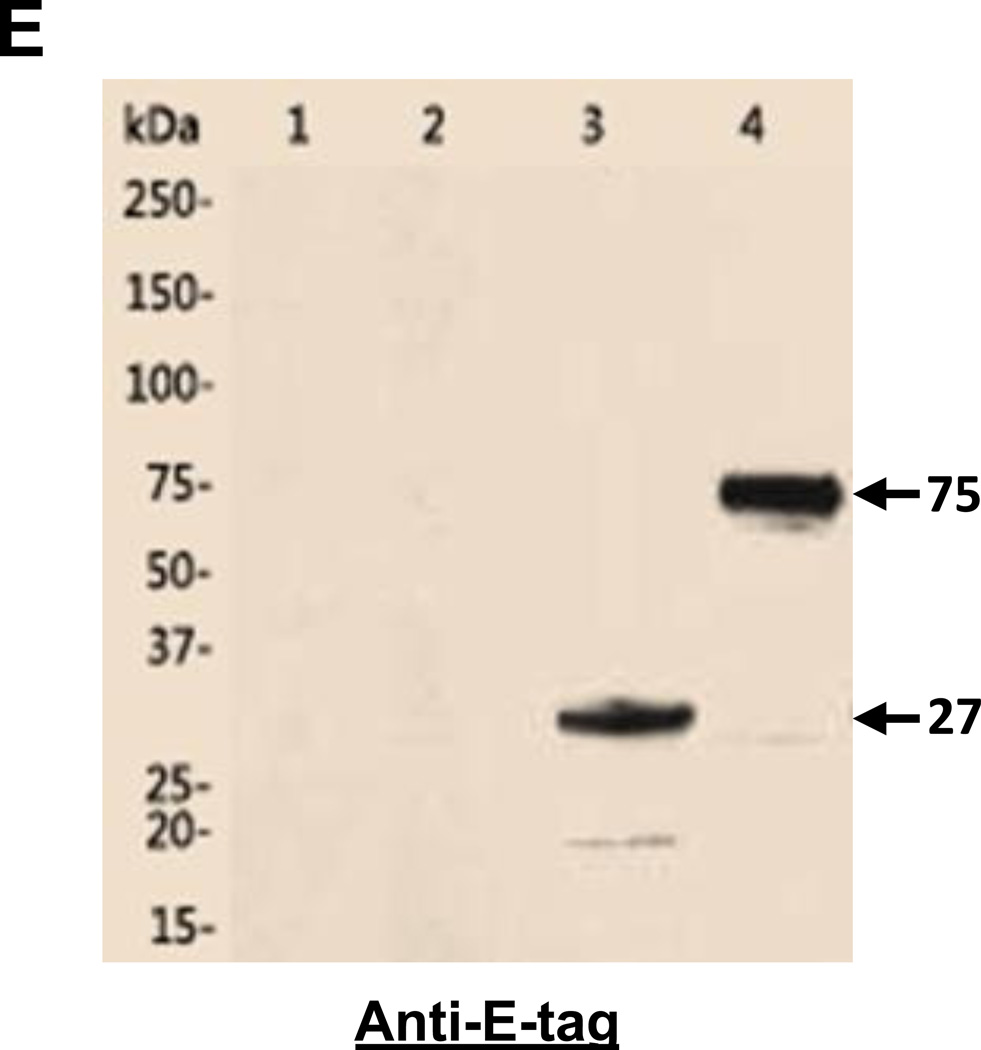
Expression and purification of the designed single-chain antibody VRC01 in E. coli(A). Polyacrylamide gel with Coomassie blue staining. Lane 1, cell pellet total proteins of un-induced; 2, cell pellet total proteins of induced; 3, supernatant of cell lysate; 4, pellet of cell lysate; 5, Urea-solubilized inclusion bodies. (B). Western blot of the same purified protein samples from Fig. 2A using anti-His-tag antibody. The specific band (~27kDa) of scFv antibody VRC01 is indicated by the arrow in Fig. 2A and 2B. Western blot to verify the purified single-chain antibody VRC01. (C). Western blot with anti-His-tag antibody. Lane1, gp120, 2, scFv_VRC01. (D). Far-Western blot first with the binding of bait protein gp120, then with the anti-gp120 antibody. Lane 1, BSA; 2, gp120, 3, scFv-VRC01. (E). Western blot of scFv-VRC01 and scFv-VRC01-β-domain fusion protein (~75kDa). Both of the samples are labeled with as follows: Lane 1, Supernatant of strain containing plasmid scFv-VRC01; 2, Supernatant of strain containing plasmid scFv-VRC01-β-domain; 3, Cell pellet of strain containing plasmid scFv-VRC01; 4, Cell pellet of strain containing plasmid scFv-VRC01-β-domain.
We then further characterized the expressed single-chain antibody VRC01 by determining whether it could be refolded into a native conformation, and Far-Western blotting was used to analyze whether it could bind gp120. This was carried out using anti-gp120 antibody to detect gp120 that was captured by the refolded scFv-VRC01, and the result is shown in Fig. 2D. The gp120 molecule was found to be bound by the refolded scFv-VRC01 on the membrane, resulting in the presence of a 27 kDa band which corresponds with expected size of the scFv-VRC01 antibody molecule and with the size of purified scFV-VRC01 shown by Western blot analysis using anti-His-tag antibody (Fig. 2C). Our results thus confirmed that the detected 27kDa recombinant protein was indeed scFv-VRC01 and when refolded, could bind gp120.
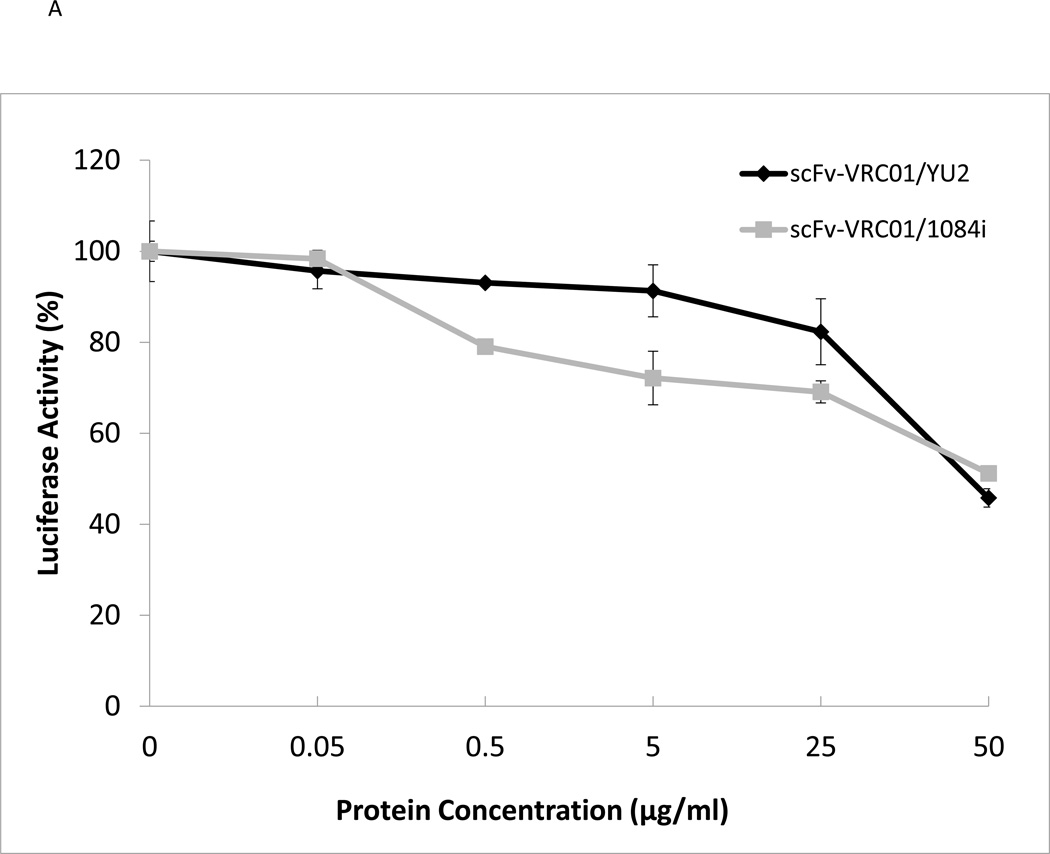
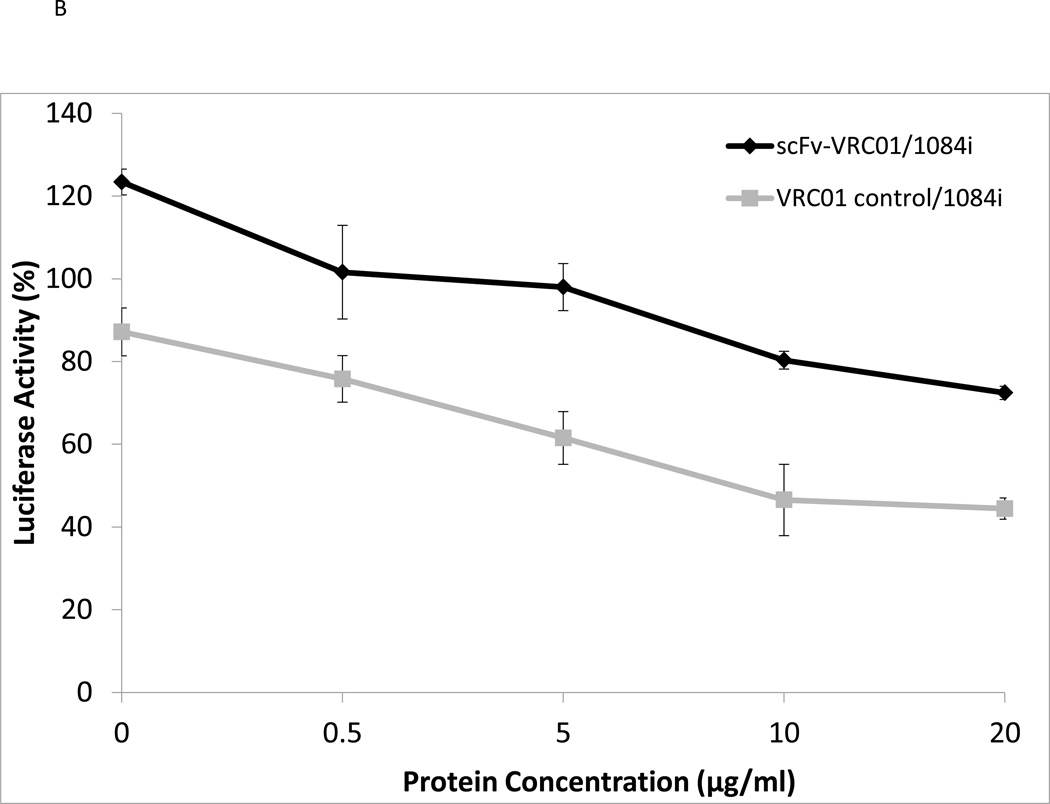
Inhibition of HIV-1 by the scFv-VRC01 antibody
Since the recombinant scFv-VRC01 can bind gp120 as determined by Far-Western blotting analysis, this suggested that the refolded SCFV-VRC01 can possibly bind and neutralize HIV-1. We then tested whether the expressed single-chain antibody VRC01 had neutralizing activity for different HIV-1 subtype strains. We tested the activities of SCFv-VRC01 against two different HIV-1 subtypes, subtype B (YU2) and subtype C (1084i), and the results are shown in Fig. 3A. The sc-Fv-VRC01 was found to have neutralizing activity against both HIV-1 strains. However, its activity was found to be several folds lower than the full length VRC01 IgG (Fig. 3B). Ten µg/ml of VRC01 was able to achieve 50% neutralization of 1084i but 50 ug/ml of sc-Fv-VRC01 was needed to achieve similar levels of neutralization (see Fig. 3A). This was not unexpected since a single-chain antibody is generally known to be weaker than the full-length antibody in binding activity.
Fig. 3.
Functional analysis of purified single-chain antibody VRC01. (A). Neutralization assay of the scFv-VRC-1 against different strains of HIV-1: YU2 (subtype B), 1084i (subtype C). (B). Comparison of the scFv VRC01 antibody and corresponding VRC01 IgG antibody.
Surface display of the scFv-VRC01 antibody on E. coli
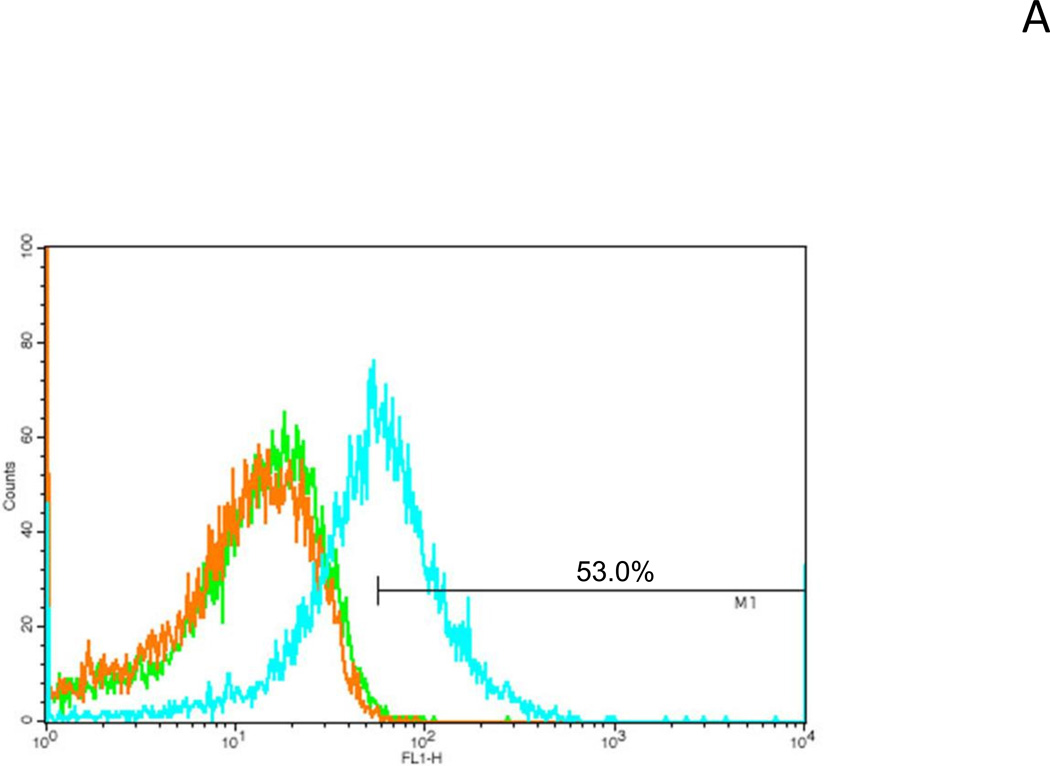
In order to display the scFv-VRC01 on the bacterial surface, an anchor protein domain (β-barrel domain) of IgAP from the autotransporter was added. The IgAP β-barrel domain can be integrated into the outer membrane of the bacterial cell wall and will translocate the N-terminal domain scFv-VRC01 from the cytoplasm to the cell surface. We then generated a construct (Fig. 1A and 1B) that included the signal peptide (sp), the single-chain antibody (scFv-VRC01), the anchor domain (C-IgAP) and the E-tag for the detection. The induced bacterial lysate was tested for the expression of the recombinant fusion protein by western blot analysis, and as expected a 75 kDa fusion protein was detected by anti-E-tag antibody (Fig. 2E). To verify that this fusion protein molecule could be displayed on the surface of the bacterium, flow cytometry was carried out to identify the presence of the recombinant scFv-VRC01 using FITC-conjugated rabbit anti-human IgG antibody. Over 50% of the bacteria were shown to express scFv-VRC01 on the cell surface (Fig. 4A).
Fig. 4.
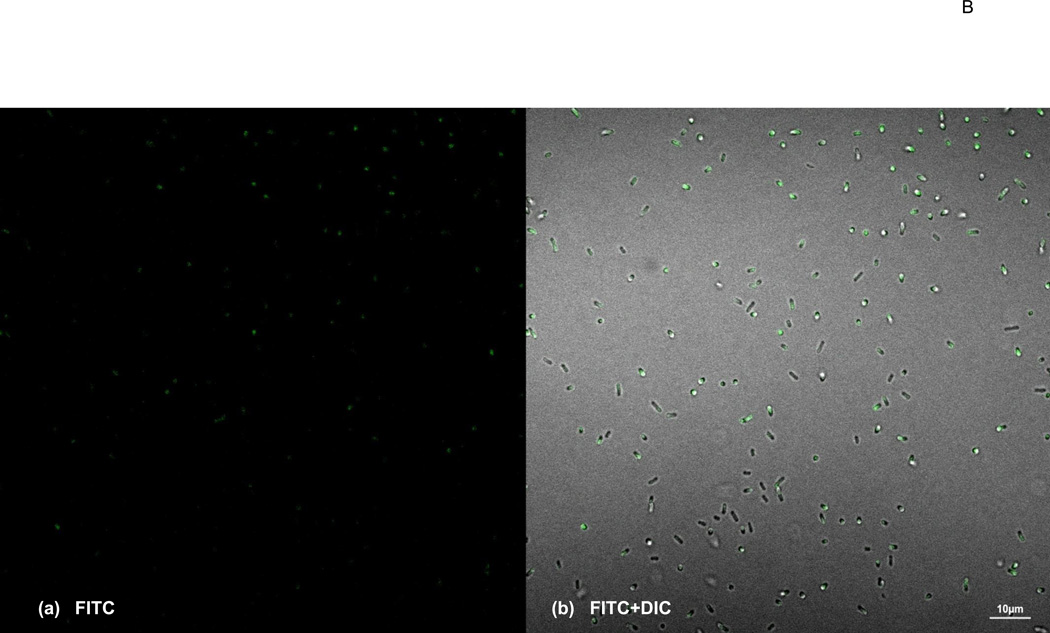
Identification of Surface display of fusion protein on E. coli(A). Bacterial sample cells (E. coli) were treated with FITC-conjugated anti-rabbit IgG antibody and analyzed by Flow cytometer. Orange, unstained bacterial cells as negative control; green, stained bacteria cells harboring the vector only; cyan, stained bacteria harboring the scFv-VRC01-β-domain plasmid. The total gated positive cell percentage (M1) is shown in the figure. (B). Bacterial Cells displaying scFv-VRC01 on the surface using confocal microscopy. The positive stained bacterial cells by FITC-conjugated anti-rabbit IgG antibody are shown in green. The same image is shown in (a), dark background window (FITC). (b), the bright background window (FITC) + (DIC) (differential interference contrast). The size bar showing in the picture is 5.0µ in (a) and 10.0µ in (b).
To further confirm the expression of the surface scFV-VRC01 confocal microscopy was carried out to directly visualize the presence of the molecule on the bacterial surface using FITC labelled antibodies. More than half of the cells were found to be positive (Fig. 4B). The results corresponded well with the number of positive cells determined by flow cytometry analysis (Fig. 4A), and suggest that the scFv-VRC01 molecules can be displayed on the bacterial surface.
Binding and inhibition of HIV-1 infection by the bacterial displayed scFv-VRC01
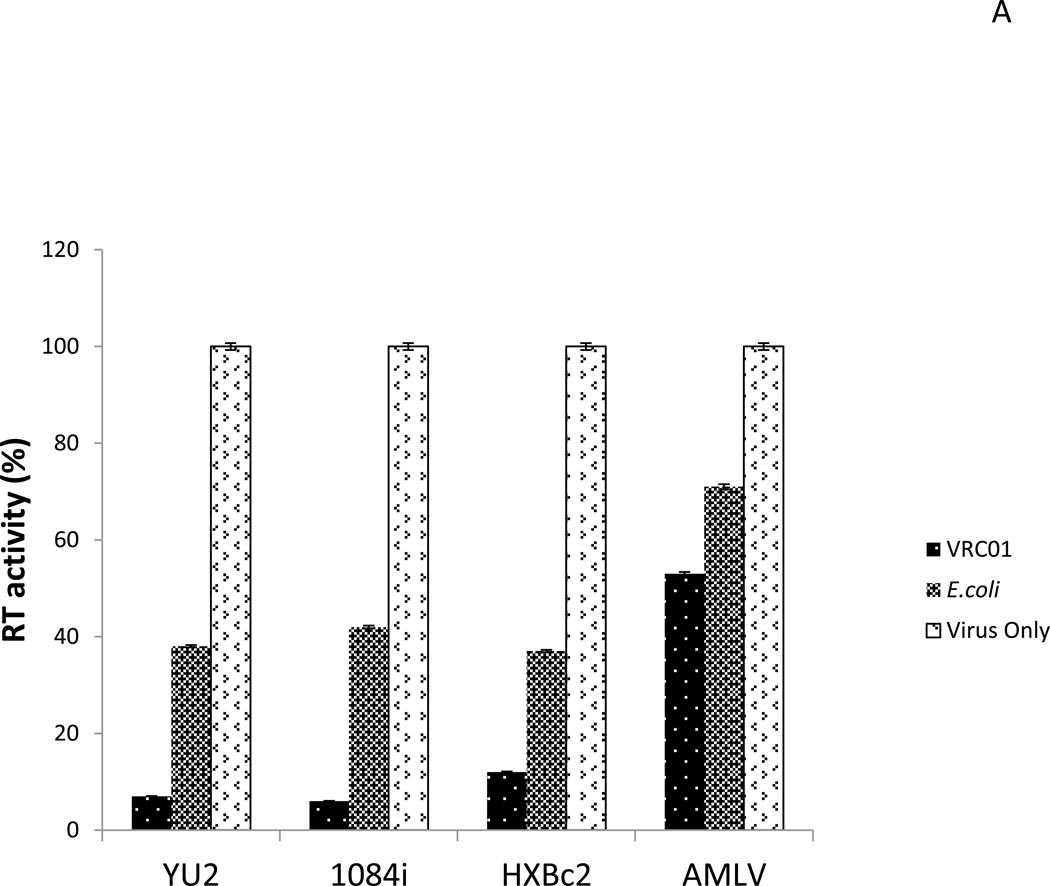
Since scFv-VRC01 could be expressed in a large number of bacteria, it was important to determine whether they could bind to HIV-1 and prevent its infection. To demonstrate that the bacteria which displayed surface scFv-VRC01 could adsorb viral particles, they were mixed with viral particles (100µl of 108/ml bacteria, and 12,500 RT units of HIV-1) and the amount of unbound viral particles were determined by measuring the residual RT (reverse transcriptase) activities in the supernatant after binding. As shown in Fig. 5A, the presence of bacteria expressing surface scFv-VRC01 could reduce the RT activity in the supernatant by over 90% as compared to 60% non-specific adsorption of the virion by control bacteria. The adsorption was specific for HIV-1 since similar levels of inhibition were not observed with MuLV control. As expected the scFv-VRC01 bacteria were found to be equally effective against multiple strains of HIV-1 (Fig. 5A).
Fig. 5.
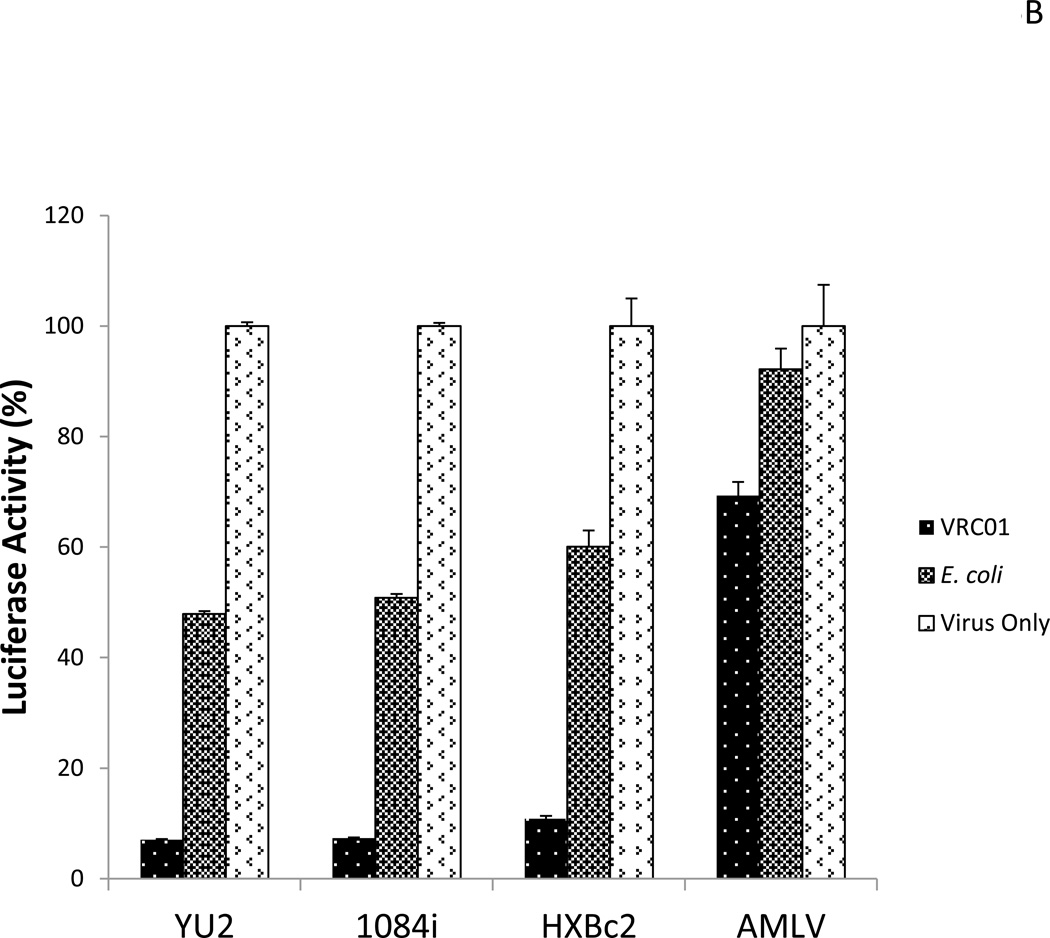
(A).Adsorption of HIV-1 particles by scFv-VRC01 surface displayed bacteria. The engineered bacteria with the concentration of 108/ml mixed with the viral stock can reduce the viral titer by surface binding. HIV-1 strains: YU2 (R5 virus, subtype B), 1084i (R5 virus, subtype C), HXBc2 (X4 virus, subtype B), AMLV, unrelated virus. RT, reverse transcriptase. (B). Inhibition of viral infection by scFv-VRC01 surface displayed bacteria in vitro. The target cells (Cf2Th) stably expressed receptor CD4 and co-receptor CCR5 were used for infection, and the luciferase was a reporter gene. HIV-1 strains: YU2 (R5 virus, subtype B), 1084i (R5 virus, subtype C), HXBc2 (X4 virus, subtype B) as negative control, AMLV, unrelated virus.
To further confirm the effectiveness of the displayed scFc-VRC01 bacteria in inhibiting HIV-1 infection, infectivity assays using CF2-Th cells were carried out. Following incubation of the virus with the bacteria, the residual virus in the supernatant was used to infect CF2-Th and the infectivity was determined by measuring luciferase activity in the target cells. These results support the adsorption data demonstrating that HIV-1 infectivity was reduced by over 90% after incubation with the scFc-VRC01 bacteria, while much lower levels of inhibition were observed with the control bacteria (Fig. 5B). Thus, the bacterial cells with surface displayed scFv-VRC01 have the ability to block HIV-1 infection in vitro.
Discussion
Using commensal bacteria as anti-HIV infection agents is a novel initiative, and investigation in this area is still lacking with only very few relevant published reports. In this report, we have demonstrated as a proof of principle that the use of the p-domain of an autotransporter is able to translocate a single-chain anti-HIV antibody molecule from the cytoplasm to efficiently display it on the cell surface. The displayed molecule retained its anti-HIV activities and was able to bind to different subtypes of HIV-1, however, our expression system still needs to be optimized. Flow cytometry analyses data showed that only about 53% of bacterial cells positively displayed the scVRC01 on the cell surface. This suggests that either some bacteria are either not expressing the recombinant protein or their expression levels were below the detection limit. Another possibility is that some of the bacteria might have lost the plasmid upon passaging and propagation, or over expression of the recombinant molecule could affect the bacterial viability, and only certain bacteria with lower levels of expression had survived. These issues will need to be further investigated since stable expression and appropriate display are important factors contributing to the success of this approach.
Another issue we encountered is the weaker neutralizing activities by the single chain antibody compared to a full length antibody, also indicated in a recent research paper that included evalutions of several single-chain antibodies against HIV infection including the scFv-VRC01 (West et al., 2012). This phenomenon is actually well documented, single-chain antibody has weaker binding affinity, and will result in weaker neutralizing activity when compared to the native antibody molecule. As we know that the Fab fragment antibody has a better affinity than the single-chain antibody for binding to its antigen. Therefore, it will be significant for us to display the Fab fragment of the VRC01 antibody to increase the neutralizing activity and improve this strategy for application.
Nevertheless, our in vitro study here has shown that the use of bacteria expressing anti-HIV inhibitors has the potential to be an alternate approach for the prevention of HIV-1 infection. Following this study and after the optimization of expression binding of the antibody molecule, it will be necessary to deliver these engineered bacteria into mice for colonization studies, examining how long the bacteria can stay viable in the animals and how well the bacteria are able to disseminate to different tissue sites. Further, it will be necessary to test our system in an animal model, such as a humanized mouse or the SHIV macaque model, for viral challenge experiments. This will demonstrate the role and the efficacy of the engineered bacteria in preventing HIV-1 infection in vivo before testing any human trials.
Conclusions
This is the first report of using gram-negative commensal bacteria for surface display of anti-HIV single-chain antibody to prevent HIV-1 infection. We have successfully displayed the anti-HIV-1 single-chain antibody VRC01 on the surface of E. coli bacteria and the engineered bacteria have shown neutralizing activity in vitro. It is likely that this approach could be expanded to be used in vivo as an alternative approach to prevent HIV-1 transmission and infection.
Materials and methods
Bacterial strains and vectors
Escherichia coli K-12 strain DH5α, Escherichia coli BL21 (λDE3) (Novagen) E. coli K12 strain UT5600 (ΔompT) were used for surface display (Lum and Morona, 2012; Maurer et al., 1997). pET22b, pET28b (Invitrogen) and PUC57 (GenScript) were used for cloning.
Design of the single-chain antibody and fusion protein
The single-chain antibody VRC01 gene was designed using a linker sequence (GGGGSGGGGSGGGGS) to connect the two domains of antibody light chain (VH), and an E-tag fragment was added to the N-terminus of the recombinant molecule to be used for detection of protein expression. A 789-bp fragment with BamH1 and Xho1 restriction sites was synthesized by GenScript (Piscataway, NJ).
The 410 amino acids of the β-barrel domain (C-IgAP) fragment were synthesized from the Neisseria gonorrhoeae IgA1 protease gene of C-terminal region from position 3364 to the end of the gene at position 4599. An E-tag was added to the N-terminus, and a His-tag was added to the end of the β-domain segment. The entire 1302 bp fragment was synthesized by GenScript.
The entire fusion protein consists of the signal peptide (sp), scFV-VRC01, the β-domain (C-IgAP) plus the tags and linkers, which total ~2100 bp and encodes a peptide of ~700 amino acids. The structure of the recombinant gene and the predicted 3D structure of the fusion protein are shown in Fig. 1A and 1B respectively.
Expression and purification of scFv-VRC01
The synthesized scFv-VRC01 gene fragment was removed from the original vector pUC57 using the restriction enzymes BamH1 and Xhol, and then ligated into an E. coli expression vector pET28b (Invitrogen). The vector containing scFv-VRC01 gene was transformed into E. coli BL21 (ADE3) bacterial cells for protein expression. After transformation, the selected positive colonies were cultured and induced by 0.5mM IPTG (isopropylthio-p-galactoside) for 3 hrs at 37°C. After induction, the cells were collected by centrifugation and then disrupted by sonication. To purify the protein from inclusion bodies, the precipitates were dissolved in 8M urea and purified by Ni-NTA affinity chromatography under denaturing conditions. The protein was dialyzed for 24 hrs at 4°C in refolding buffer containing 50mM Tris-HCL, 0.5M L-Arginine, 50mM NaCl and 10% glycerol. The protein concentration of purified scFv-VRC01 protein was measured by BCA protein assay (Pierce) and stored at −80°C.
Surface expression of scFv-VRC01
The E. coli K-12 strain UT5600 was used as host cells for surface display of the fusion protein. The designed β-domain(C-IgAP) gene (1302bp) was synthesized and cloned into the pUC57 vector (GenScript). To construct the recombinant expression vector pET22b–scFv-VRC01-β-domain(C-IgAP), the β-domain-E-tag encoding fragment was cloned into the pET22b–scFv-VRC01 vector at the Xhol site to generate the vector pET22b–scFv-VRC01-β-domain which harbors the main fusion structure of scFv-VRC01, the transporter C-terminal β-domain of IgAP and the E-tag connecting the two fragments. Transformation of E. coli K-12 strain UT5600 was performed as previously described (Lum and Morona, 2012; Maurer et al., 1997). For induction of the fusion protein, single colonies were picked and inoculated into liquid LB containing 2% (w/v) glucose and antibiotics, and grown at 30°C until OD600 was ~0.5. Bacterial cells were then harvested by centrifugation, resuspended into similar volume of fresh liquid medium with antibiotics and 0.25mM or 0.5mM IPTG, and incubated further at 30°C for 3 hrs.
To verify the expression of the scFv and β-domain of IgAP fusion protein, 0.25mM or 0.5mM IPTG-induced E. coli cells containing the vectors pET22b–scFv-VRC01-β-domain or control empty vector pET22b were centrifuged and resuspended in 20mM phosphate buffer. The bacterial suspensions were mixed with 2×SDS sample buffer. After 5 min of boiling, samples were briefly centrifuged and subjected to Western blot analysis. To verify that the fusion protein was expressed in the insoluble fraction including membranes and inclusion bodies, the bacterial cells, both before and after IPTG-induction, were resuspended in PBS and sonicated at 15W for 30 seconds 5 times to break open the cells. After sonication the samples were spun at 10,000×g for 20 minutes to pellet any unbroken cells or large cellular debris. The supernatant was then centrifuged at 100,000×g for 1 hr at 4°C to pellet the membrane, and then subjected to Western blot analysis.
SDS-PAGE, Western blotting and Far-Western blotting
Samples were run on a 10% SDS-polyacrylamide gel and stained with Coomassie blue to verify protein expression.
For Western blotting analysis, the SDS-PAGE gel was further transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, USA) and the membrane was blocked with blocking buffer (5% skim milk in TBS) for 1 hr at room temperature and reacted with primary antibody (5000-fold diluted anti-His-tag antibody HRP conjugated or 500-fold diluted rabbit anti-E-tag antibody) in blocking buffer for 16 hr at 4°C. The membrane was washed 3 times with wash buffer (0.05% tween-20 in TBS) and incubated with a secondary antibody (HRP conjugated anti-rabbit IgG antibody) for 1 hr at room temperature. After washing 3 times, the membrane was developed with TMB membrane peroxidase substrate (KPL, USA). The anti-His-tag antibody HRP conjugated (Abcam) or rabbit anti-E-tag antibody HRP conjugated (Bioss Inc) was used for detection.
To examine the single-chain VRC01 antibody (scFV-VRC01) in the fusion molecule, far-Western blotting was performed. BSA was used as a negative control and HIV-1 gp120 was used as a positive control. The purified scFv-VRC01 and the fusion protein containing scFV-VRC01 were first separated by SDS-PAGE and then transferred to PVDF membrane. In order to refold the proteins on the membrane to generate an intact binding site, a denaturation step was added. Briefly, immediately after transfer, the blot was incubated in 2% AC denaturing buffers (100 mM NaCl, 20 mMTris (pH 7.6), 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20, 2% skim milk powder and 1 mM DTT) for 16 hrs at 4°C, the membrane was then blocked in 5% skim milk and incubated with 1:250 diluted gp120 in 2% AC buffer for 16 hrs at 4°C. The membrane was washed 3 times with wash buffer (0.05% tween-20 in TBS) and incubated with HRP conjugated anti-gp120 for 1 hr at room temperature. After washing 3 times, the membrane was developed with TMB membrane peroxidase substrate (KPL, USA).
Flow cytometry analysis
Bacteria were grown in the presence of IPTG as described above until the log phase, 10µl of each E. coli culture (106/ml bacteria) containing the vectors pET22b–scFv-VRC01-β-domain(C-IgAP) or empty vector pET22b were washed three times in PBS by centrifugation (10,000g for 15 min) before they were resuspended in 50µl of PBS. A 20-fold diluted rabbit anti-E-tag rabbit antibody conjugated with Fluorescein isothiocyanate (FITC) (Bioss Inc) was added and the samples were incubated for 1 hr at 4°C. The washing procedure in PBS was repeated once and the samples were resuspended in 300µl of PBS. Bacterial flow cytometry analysis was carried out with FACSCalibur (Becton Dickinson) as described previously (Gunasekera et al., 2000).
Confocal Microscopy
After growth in the presence of IPTG as described above, 10µl of each E. coli culture (106/ml bacteria) containing the vectors pET22b–scFv-VRC01-β-domain or empty vector pET22b were washed 3 times in PBS, spunned at 10,000g for 15 min before resuspending in 50µl of PBS. A 20-fold diluted rabbit anti-E-tag antibody conjugated-FITC (Bioss, Inc) was added and the samples were incubated for 1 hr at 4°C. The washing procedure in PBS was repeated and the samples were resuspended in 50ul of 2% PFA in PBS. Images were collected using a Leica TCS-NT/SP confocal microscope (Leica Microsystems, Mannheim, Germany). Standard filters were used for FITC. The detector slits for the FITC channel were configured to collect between 500 and 547 nm to minimize overlapping of the green and yellow channels. All images were analyzed using the same gain, and organized using the Adobe Photoshop (Adobe Systems, San Jose, CA).
Single-chain antibody neutralization assay
Cf2Th cells with CD4 and CCR5 receptors were used to test the sensitivity of the viruses to neutralization by scFv-VRC01. 2,500 RT units of luciferase-expressing recombinant viruses were incubated with 50µg, 25µg, 5µg, and 0.5µg, and 0.05µg/ml of scFv-VRC01 in DMEM for 60 min at 37°C. The virus-antibody mixtures were then transferred to wells containing 105/well of target Cf2Th cells. After overnight incubation, 0.1ml of fresh medium was added to each well, and the cells were cultured for 2 more days. The cells were then lysed and assayed for luciferase activity (Tuner 20; Promega). In another independent experiment, we compared the neutralizing activity between scFv-VRC01 and the native IgG VRC01 antibody (20µg, 10µg, 5µg, and 0.5µg/ml) using a subtype C HIV-1 recombinant luciferase-expressing virus 1084i.
Viral particles absorption assay
To determine the ability of E. coli expressing scFv-VRC01 on the cell surface to bind HIV-1, the bacteria (100µl of 108/ml) were mixed with 12,500 RT units of viral stock (YU2, 1084i, HXBc2, AMLV) and incubated with rocking for 1 hr at 37°C, then spun down to remove the bacteria and RT was measured in the supernatant. RT assay performed as described previously (Rho et al., 1981).
Bacteria neutralization assay
To determine the HIV neutralizing activity of E. coli expressing scFv-VRC01 on the cell surface, the bacteria (100µl of 108/ml) were mixed with 2,500 RT units of viral stock (YU2, 1084i, HXBc2, AMLV) and incubated with rocking for 1h at 37°C, then spun down to remove the bacteria. The supernatant was then transferred to wells containing the target Cf2Th cells. The cells were then lysed and assayed for luciferase activity. The luciferase activity assay was performed as described previously (Xiang et al., 2002).
Highlights.
Designed single-chain VRC01 antibody was demonstrated to bind HIV-1 envelope gp120.
Single-chain VRC01 antibody was successfully displayed on the surface of E. coli.
Engineered bacteria can absorb HIV-1 particles and prevent HIV-1 infection in cell culture.
Acknowledgments
This work was supported in part by the Bill and Melinda Gates Foundation to SHX. GM103509, the Fogarty AIDS International Training and Research Program D43TW01492 and T32 AI060547 from the NIH to C.W. L.X.W. was a Fogarty Fellow. DB was an NIH Ruth Kirschstein Fellow. We would like to thank Dr. You Zhou of the Microscopy core facility for his help with the study. We would also like to thank the Nebraska Center for Virology Core facility for its support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Mohsen M, Raposo RA, Deng X, Li M, Liegler T, Sinclair E, Salama MS, Ghanem Hel D, Hoh R, Wong JK, David M, Nixon DF, Deeks SG, Pillai SK. Expression profile of host restriction factors in HIV-1 elite controllers. Retrovirology. 2013;10:106. doi: 10.1186/1742-4690-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- Benz I, Schmidt MA. Structures and functions of autotransporter proteins in microbial pathogens. Int J Med Microbiol. 2011;301:461–468. doi: 10.1016/j.ijmm.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, Scanlan CN, Schief WR, Silvestri G, Streeck H, Walker BD, Walker LM, Ward AB, Wilson IA, Wyatt R. A Blueprint for HIV Vaccine Discovery. Cell host & microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagliuolo I, Beggiao E, Brun P, Barzon L, Goussard S, Manganelli R, Grillot-Courvalin C, Palu G. Engineered E. coli delivers therapeutic genes to the colonic mucosa. Gene Ther. 2005;12:1070–1078. doi: 10.1038/sj.gt.3302493. [DOI] [PubMed] [Google Scholar]
- Cheng CM, Chen FM, Lu YL, Tzou SC, Wang JY, Kao CH, Liao KW, Cheng TC, Chuang CH, Chen BM, Roffler S, Cheng TL. Expression of beta-glucuronidase on the surface of bacteria enhances activation of glucuronide prodrugs. Cancer gene therapy. 2013;20:276–281. doi: 10.1038/cgt.2013.17. [DOI] [PubMed] [Google Scholar]
- Cheung SC, Dietzschold B, Koprowski H, Notkins AL, Rando RF. A recombinant human Fab expressed in Escherichia coli neutralizes rabies virus. J Virol. 1992;66:6714–6720. doi: 10.1128/jvi.66.11.6714-6720.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- Fairman JW, Noinaj N, Buchanan SK. The structural biology of beta-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol. 2011;21:523–531. doi: 10.1016/j.sbi.2011.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallecker C, Tarbouriech N, Habib M, Petit MA, Drouet E. Structural and functional characterization of the single-chain Fv fragment from a unique HCV E1E2-specific monoclonal antibody. FEBS letters. 2013 doi: 10.1016/j.febslet.2013.07.057. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Sola I, Enjuanes L, de Lorenzo V. Specific secretion of active single-chain Fv antibodies into the supernatants of Escherichia coli cultures by use of the hemolysin system. Appl Environ Microbiol. 2000;66:5024–5029. doi: 10.1128/aem.66.11.5024-5029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunasekera TS, Attfield PV, Veal DA. A flow cytometry method for rapid detection and enumeration of total bacteria in milk. Appl Environ Microbiol. 2000;66:1228–1232. doi: 10.1128/aem.66.3.1228-1232.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci U S A. 2009;106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston MI, Fauci AS. HIV vaccine development--improving on natural immunity. N Engl J Med. 2011;365:873–875. doi: 10.1056/NEJMp1107621. [DOI] [PubMed] [Google Scholar]
- Jose J, Maas RM, Teese MG. Autodisplay of enzymes--molecular basis and perspectives. J Biotechnol. 2012;161:92–103. doi: 10.1016/j.jbiotec.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Lagenaur LA, Sanders-Beer BE, Brichacek B, Pal R, Liu X, Liu Y, Yu R, Venzon D, Lee PP, Hamer DH. Prevention of vaginal SHIV transmission in macaques by a live recombinant Lactobacillus. Mucosal immunology. 2011;4:648–657. doi: 10.1038/mi.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Park SJ, Han MJ, Eom GT, Choi MJ, Kim SH, Oh YH, Song BK, Lee SH. Expression of a lipase on the cell-surface of Escherichia coli using the OmpW anchoring motif and its application to enantioselective reactions. Biotechnology letters. 2013 doi: 10.1007/s10529-013-1260-0. [DOI] [PubMed] [Google Scholar]
- Lifson JD, Haigwood NL. Lessons in nonhuman primate models for AIDS vaccine research: from minefields to milestones. Cold Spring Harb Perspect Med. 2012;2:a007310. doi: 10.1101/cshperspect.a007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lagenaur LA, Lee PP, Xu Q. Engineering of a human vaginal Lactobacillus strain for surface expression of two-domain CD4 molecules. Applied and environmental microbiology. 2008;74:4626–4635. doi: 10.1128/AEM.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Lagenaur LA, Simpson DA, Essenmacher KP, Frazier-Parker CL, Liu Y, Tsai D, Rao SS, Hamer DH, Parks TP, Lee PP, Xu Q. Engineered vaginal lactobacillus strain for mucosal delivery of the human immunodeficiency virus inhibitor cyanovirin-N. Antimicrobial agents and chemotherapy. 2006;50:3250–3259. doi: 10.1128/AAC.00493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum M, Morona R. IcsA autotransporter passenger promotes increased fusion protein expression on the cell surface. Microb Cell Fact. 2012;11:20. doi: 10.1186/1475-2859-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer J, Jose J, Meyer TF. Autodisplay: one-component system for efficient surface display and release of soluble recombinant proteins from Escherichia coli. J Bacteriol. 1997;179:794–804. doi: 10.1128/jb.179.3.794-804.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RJ, Kim JH, Corey L, Michael NL. Human immunodeficiency virus vaccine trials. Cold Spring Harb Perspect Med. 2012;2:a007351. doi: 10.1101/cshperspect.a007351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- Pusch O, Kalyanaraman R, Tucker LD, Wells JM, Ramratnam B, Boden D. An anti-HIV microbicide engineered in commensal bacteria: secretion of HIV-1 fusion inhibitors by lactobacilli. Aids. 2006;20:1917–1922. doi: 10.1097/01.aids.0000247112.36091.f8. [DOI] [PubMed] [Google Scholar]
- Pyo HM, Kim IJ, Kim SH, Kim HS, Cho SD, Cho IS, Hyun BH. Escherichia coli expressing single-chain Fv on the cell surface as a potential prophylactic of porcine epidemic diarrhea virus. Vaccine. 2009;27:2030–2036. doi: 10.1016/j.vaccine.2009.01.130. [DOI] [PubMed] [Google Scholar]
- Rao S, Hu S, McHugh L, Lueders K, Henry K, Zhao Q, Fekete RA, Kar S, Adhya S, Hamer DH. Toward a live microbial microbicide for HIV: commensal bacteria secreting an HIV fusion inhibitor peptide. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11993–11998. doi: 10.1073/pnas.0504881102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho HM, Poiesz B, Ruscetti FW, Gallo RC. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology. 1981;112:355–360. doi: 10.1016/0042-6822(81)90642-5. [DOI] [PubMed] [Google Scholar]
- Rutherford N, Mourez M. Surface display of proteins by gram-negative bacterial autotransporters. Microb Cell Fact. 2006;5:22. doi: 10.1186/1475-2859-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach J, Popovic M, Gilden RV, Gonda MA, Sarngadharan MG, Gallo RC. Serological analysis of a subgroup of human T-lymphotropic retroviruses (HTLV-III) associated with AIDS. Science. 1984;224:503–505. doi: 10.1126/science.6200937. [DOI] [PubMed] [Google Scholar]
- Shin JR, Lim KJ, Kim da J, Cho JH, Kim SC. Display of multimeric antimicrobial peptides on the Escherichia coli cell surface and its application as whole-cell antibiotics. PLoS One. 2013;8:e58997. doi: 10.1371/journal.pone.0058997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UNAIDS. Data and analysis. 2014 http://www.unaids.org/en/
- van den Berg B. Crystal structure of a full-length autotransporter. J Mol Biol. 2010;396:627–633. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- Vangelista L, Secchi M, Liu X, Bachi A, Jia L, Xu Q, Lusso P. Engineering of Lactobacillus jensenii to secrete RANTES and a CCR5 antagonist analogue as live HIV-1 blockers. Antimicrob Agents Chemother. 2010;54:2994–3001. doi: 10.1128/AAC.01492-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga E, de Lorenzo V, Fernandez LA. Probing secretion and translocation of a beta-autotransporter using a reporter single-chain Fv as a cognate passenger domain. Mol Microbiol. 1999;33:1232–1243. doi: 10.1046/j.1365-2958.1999.01571.x. [DOI] [PubMed] [Google Scholar]
- Veiga E, De Lorenzo V, Fernandez LA. Neutralization of enteric coronaviruses with Escherichia coli cells expressing single-chain Fv-autotransporter fusions. J Virol. 2003;77:13396–13398. doi: 10.1128/JVI.77.24.13396-13398.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Galimidi RP, Jr, Gnanapragasam PN, Bjorkman PJ. Single-chain Fv-based anti-HIV proteins: potential and limitations. Journal of virology. 2012;86:195–202. doi: 10.1128/JVI.05848-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, Longo NS, McKee K, O’Dell S, Louder MK, Wycuff DL, Feng Y, Nason M, Doria-Rose N, Connors M, Kwong PD, Roederer M, Wyatt RT, Nabel GJ, Mascola JR. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH. Recent advances on the use of structural biology for the design of novel envelope immunogens of HIV-1. Current HIV Research. 2013;11:1–6. doi: 10.2174/1570162x113116660053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang SH, Kwong PD, Gupta R, Rizzuto CD, Casper DJ, Wyatt R, Wang L, Hendrickson WA, Doyle ML, Sodroski J. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. Journal of virology. 2002;76:9888–9899. doi: 10.1128/JVI.76.19.9888-9899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto HS, Xu Q, Fichorova RN. Homeostatic properties of Lactobacillus jensenii engineered as a live vaginal anti-HIV microbicide. BMC Microbiol. 2013;13:4. doi: 10.1186/1471-2180-13-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]