Abstract
Novel human immunodeficiency virus (HIV) protease inhibitors are urgently needed for combating the drug-resistance problem in the fight against AIDS. To facilitate lead discovery of HIV protease inhibitors, we have developed a safe, convenient, and cost-effective Escherichia coli-based assay system. This E. coli-based system involves coexpression of an engineered β-galactosidase as an HIV protease substrate and the HIV protease precursor comprising the transframe region and the protease domain. Autoprocessing of the HIV protease precursor releases the mature HIV protease. Subsequently, the HIV protease cleaves β-galactosidase, resulting in a loss of the β-galactosidase activity, which can be detected in high-throughput screens. Using Food and Drug Administration-approved HIV protease inhibitors, this E. coli-based system is validated as a surrogate screening system for identifying inhibitors that not only possess inhibitory activity against HIV protease but also have solubility and permeability for in vivo activity. The usefulness of the E. coli-based system was demonstrated with the identification of a novel HIV protease inhibitor from a library of compounds that were prepared by an amide-forming reaction with transition-state analog cores. A novel inhibitor with a sulfonamide core of amprenavir, E2, has shown good correlation with the in vitro enzymatic assay and in vivo E. coli-based system. This system can also be used to generate drug resistance profiles that could be used to suggest therapeutic uses of HIV protease inhibitors to treat the drug-resistant HIV strains. This simple yet efficient E. coli system not only represents a screening platform for high-throughput identification of leads targeting the HIV proteases but also can be adapted to all other classes of proteases.
Human immunodeficiency virus (HIV) protease, essential for processing HIV viral polyproteins into individual structural proteins and replication enzymes during virus maturation, has been an attractive target for antiviral therapy development (9). Several HIV protease inhibitors designed to bind the active site of HIV protease have been shown to have in vivo efficacy and are currently in clinical use. This line of therapy, by itself or in combination with reverse transcriptase inhibitors, has revolutionized antiviral treatments and dramatically lowered the number of deaths due to AIDS (32). However, widespread use of HIV protease inhibitors has caused the rapid emergence of drug-resistant HIV proteases (8), rendering AIDS with no definitive cure. In these HIV proteases, mutations have been found in 49 of the 99 amino acids of the coding sequence. Substitutions at 18 or more positions are directly correlated with loss of responsiveness to HIV protease inhibitor treatment. Since the existing HIV protease inhibitors target the active site of HIV protease and have similar structures, most of the drug-resistant HIV proteases confer cross-resistance to multiple HIV protease inhibitors. There is a great need for second-generation HIV protease inhibitors with different chemical structures and/or with an alternative mode of inhibition, such as targeting activation, i.e., dimerization and folding of HIV protease.
HIV protease is an aspartyl protease. The active HIV protease is an obligatory dimer, consisting of two identical and noncovalently associated monomers. The active site of the enzyme is formed at the dimer interface and contains two conserved catalytic aspartic acid residues, one from each monomer (5). Instead of being translated as an active enzyme, HIV protease is synthesized as an integral part of the viral Gag-Pol polyprotein and needs to be activated to gain catalytic activity. Activation of the HIV protease requires appropriate folding and dimerization of the protease domain in the viral Gag-Pol polypeptide to form an active site, which then can catalyze the hydrolysis of peptide bonds at cleavage sites to release the mature HIV protease (34).
The molecular mechanism of HIV protease activation is not completely clear yet. It has been suggested that the transframe region (TF), flanking the N terminus of the protease domain in the Pol gene, is involved in the regulation of HIV protease activity (26). The native TF comprises an 8-amino-acid transframe peptide and a 48-amino-acid p6pol protein. Purified recombinant p6pol protein was shown to have inhibitory effects on HIV protease activity (25). The intact HIV protease precursor, including TF and the protease domain, has a very low dimer stability relative to that of the mature enzyme and exhibits negligible levels of stable tertiary structure (33). Louis et al. (19) proposed that TF interacts with the dimer interface and thus destabilizes the dimeric structure to regulate HIV protease activity. The solution structure of the HIV protease precursor confirmed that TF hinders dimerization of the HIV protease domain and influences the catalytic activity of HIV protease (11).
Standard screening methods of HIV protease inhibitors include both in vitro and in vivo assays. While the in vitro enzyme kinetic assays use purified recombinant HIV proteases and specific substrates, the in vivo assays use mammalian cells and the HIV viruses to evaluate the permeability as well as the in vivo antiviral activities of the potential drug leads. The complexity and time-consuming nature of the current protocols prompted us to explore the use of an Escherichia coli-based system as a simple and safe alternative approach for HIV protease inhibitor screening.
Several E. coli-based systems have been developed for in vivo HIV protease activity assays. Some systems have used reporter enzymes, like thymidylate synthase (16) or β-galactosidase (2), while other systems used the proteins involved in regulatory pathways, such as the lytic/lysogenic cycle of λ phage (22, 29), transcriptional control (6, 12, 30), or the cyclic AMP signaling pathway (7). These systems have limited success, however, in adaptation for high-throughput screening or in the detection of in vivo activity of HIV protease inhibitors.
We report here the development of an E. coli-based system in a high-throughput format and with the sensitivity to detect in vivo activity of HIV protease inhibitors. This system involves an engineered β-galactosidase to report in vivo HIV protease activity, and the HIV protease precursor (TF-PR) that comprises TF and the protease domain from the Gag-Pol polyprotein. The autoproteolytic processing of the HIV protease precursor occurs in the E. coli cells and results in the release of the mature HIV protease. Subsequently, the mature HIV protease cleaves the β-galactosidase, causing a loss of β-galactosidase activity. β-Galactosidase activity, which can be further quantitated by the enzymatic assay with specific chromogenic substrates, is thus inversely related to HIV protease activity. The sensitive and robust enzymatic assay of β-galactosidase enables the detection of β-galactosidase activity by using either the purified enzyme, crude cell lysates (27), or whole cells (23).
The validation of this system was performed with Food and Drug Administration (FDA)-approved HIV protease inhibitors and a library of new inhibitors against both the wild-type HIV protease and the drug-resistant HIV proteases. The ranking of the inhibitory potency of FDA-approved HIV protease inhibitors against the wild-type HIV protease in this E. coli-based system was shown to correspond well with what was determined in standard antiviral assays. A novel HIV protease inhibitor, E2, was identified as a potential drug lead from a library of compounds generated through diversity-oriented synthesis in a microtiter plate for in situ screening. Also, the in vivo inhibitory activity of E2 obtained from the E. coli system correlates well with the in vitro activity. This E. coli-based system represents an efficient and cost-effective method for discovery of new drug leads targeting the HIV protease. Furthermore, it can also be applied to drug discovery for many other diseases, as long as a protease, regardless of the protease class to which it belongs, is involved in these disease processes.
MATERIALS AND METHODS
Materials.
The bacterial expression vector pBAD-TOPO-LacZ was obtained from Invitrogen Corp. (Carlsbad, Calif.). HIV-1 NL4.3 DNA was a gift from Jiing-Kuan Yee (The City of Hope, Duarte, Calif.). All DNA-modifying enzymes were from New England Biolabs (Beverly, Mass.), except that Taq polymerase was from Panvera (Madison, Wis.). Oligonucleotides for PCRs were synthesized by MWG Biotech (High Point, N.C.), and the amplified DNA fragments were then purified with ZymoClean (Zymo Research, Orange, Calif.). Pepstatin A was obtained from ICN Biochemicals (San Diego, Calif.). FDA-approved HIV protease inhibitors including amprenavir (APV), indinavir (IDV), nelfinavir (NFV), ritonavir (RTV), and saquinavir (SQV) were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program, Divisions of AIDS, National Institute of Allergy and Infectious Diseases, NIH. Analytical thin-layer chromatography was performed on precoated plates (silica gel 60F-254; Merck). Silica gel used for flash column chromatography was Mallinckrodt type 60 (230 to 400 mesh). Novex Bis-Tris polyacrylamide gels for protein separation were purchased from Invitrogen Corp., and the polyvinylidene fluoride membranes were from Millipore (Bedford, Mass.). Anti-β-galactosidase monoclonal antibodies were purchased from Roche Applied Science (Indianapolis, Ind.), and HIV type 1 (HIV-1) protease antiserum was provided by D. Bailey and Mark Page (Pfizer Inc.) through the NIH AIDS Research and Reference Reagent Program, Divisions of AIDS, National Institute of Allergy and Infectious Diseases, NIH. The horseradish peroxidase-conjugated secondary antibodies as well as the Lumi-Glo chemiluminescence reagent were purchased from KPL Inc. (Gaithersburg, Md.). Reagents for the highest purity were purchased from Aldrich, Sigma, Acros, Novabiochem, or Bachem, Inc.
Construction of expression plasmids.
The pBAD system, which uses the BAD promoter for recombinant protein expression, was used to construct all expression plasmids in this study. To engineer β-galactosidase as a substrate for the HIV protease, a pair of cleavage cassettes, encoding the decapeptide corresponding to the p6/PR cleavage site (Val-Ser-Phe-Asn-Phe-Pro-Gln-Ile-Thr-Leu) of the HIV Gag-Pol polyprotein, were synthesized and ligated with SauI-digested pBAD-TOPO LacZ (2). The construct was designated pβ-GalPR.
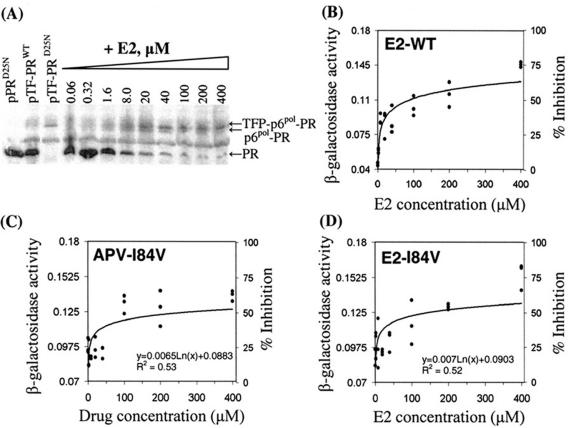
Coexpression of the HIV protease and β-GalPR was carried out by using a two-cistron approach (Fig. 1A). The first cistron encodes the HIV protease or TF-PR. Right before the stop codon of the first cistron was a Shine-Dalgarno sequence of AGGAGGAA for ribosome binding, which was followed with a start codon for translation of the second cistron, β-GalPR. The coding sequence for the HIV protease gene was amplified by PCR from HIV-1 NL4.3 DNA with oligonucleotide PR-F (ATACCATGGCCCCTCAGATCACTCTTTGGCAGCGACC) as the forward primer and oligonucleotide PR-R (AGCCCATGGGTTATTCCTCCTTAAAATTTA) as the reverse primer. The italicized nucleotides represent the NcoI restriction sites. Likewise, the coding sequence for the HIV protease precursor consisting of the TF and the protease domain was amplified with oligonucleotide TF-F (CATACCATGGGCTTTTTTAGGGAAGATCTGGCCTTC) as the forward primer and oligonucleotide PR-R as the reverse primer. After digestion with NcoI, the amplified fragments were then cloned into the unique NcoI site of pβ-GalPR. The resulting plasmid was designated pPRWT or pTF-PRWT, respectively. E. coli DH5α was used as the host for plasmid preparations and for recombinant protein expression (27).
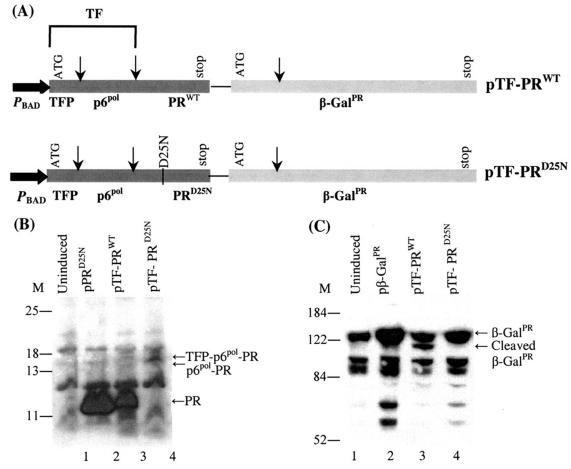
FIG. 1.
Coexpression of HIV protease and β-galactosidase in E. coli. (A) The two-cistron constructs used in this study. The first cistron is the HIV protease precursor, which consists of the TF, including the transframe peptide (TFP) and p6pol, and the protease domain (PRWT). The second cistron is the engineered β-galactosidase, containing an insertion of the protease cleavage site, Val-Ser-Phe-Asn-Phe-Pro-Gln-Ile-Thr-Leu, at amino acid 131 (β-GalPR). In the pTF-PRD25N construct, the inactive HIV protease with the catalytic Asp25-to-Asn mutation was used. The arrows represent the cleavage sites of HIV protease. (B) Autoprocessing of the HIV protease precursor detected with HIV-1 protease antiserum. Cells bearing individual plasmids (indicated at the top of the blots) were induced with 0.2% arabinose for 3 h at 30°C. The cells were then collected and processed for separation on 4 to 12% Bis-Tris gels followed by immunoblot analysis. M denotes the molecular weight standards. Each lane contained the lysate from 0.3 OD630 equivalents of cells, except the lane for PRD25N, which contained 0.1 OD630 equivalents of cells expressing PRD25N. (C) Cleavage of β-GalPR detected with antibodies against β-galactosidase. Lanes 1, 3, and 4 were the same as mentioned earlier, except that lane 2 contained the lysates from the cells expressing only β-GalPR. Each lane contained the lysate from 0.3 OD630 equivalents of cells.
In vitro PCR-mediated mutagenesis.
The HIV protease mutant with a mutation at residue 25 was constructed by PCR-mediated mutagenesis (1). The PCR-mediated mutagenesis includes three PCRs. The first PCR used pTF-PRWT as the template to generate the upstream fragment of the mutation site with TF-F as the forward primer and PRD25N-R (ATTTAATAGAGCTTC CTTTAATTTGC) as the reverse primer. The changed nucleotides are shown in boldface type, and the codons corresponding to the mutated residues are underlined. The second PCR was to generate the downstream fragment of the mutation site with PRD25N-F (GCAATTAAAGGAAGCTCTATTAAAT) as the forward primer and PR-R as the reverse primer. The upstream fragment from the first PCR and the downstream fragment from the second PCR were mixed and then used as templates for the final PCR. The final PCRs yielded either PRD25N if PR-F and PR-R were used as primers or TF-PRD25N if TF-F and PR-R were used. The amplified fragments were then digested with NcoI and subcloned into pβ-GalPR. The resulting construct was designated pPRD25Nor pTF-PRD25N, respectively.
The HIV protease precursor variant containing the D30N mutation or the I84V mutation was similarly constructed by PCR-mediated mutagenesis as described previously (1). The oligonucleotides TF-F and PRD30N-R (CCTGGCAAATTCATTTCTTCTAATACTGTGTT) or PRI84V-R (TA[C/T]GTTGACAGGTCTAGGTCCTACTAATACTGTACC) were used as primers for the first-round PCR to generate the upstream fragment of the mutation site. The oligonucleotides PRD30N-F (AACACAGTATTAGAAGAAATGAATTTGCCAGG) or PRI84V-F (GGTACAGTATTAGTAGGACCTACACCTGTCAAC[A/GTA]) and PR-R were used as primers for the second-round PCR to generate the downstream fragment containing the mutation site. The corresponding fragment pairs were then combined, and TF-F and PR-R were used as primers to amplify the final product, i.e., TF-PRD30N or TF-PRI84V. Again, these fragments were digested with NcoI and subcloned into pβ-GalPR. The resulting plasmid was designated pTF-PRD30N or pTF-PRI84V. The sequences of all constructs were confirmed by automatic DNA sequencing (DAVIS Sequencing, Davis, Calif.)
Diversity-oriented chemical synthesis of HIV protease inhibitors.
Fifty microliters of each different carboxylic acid (3 μmol) was added to each well of a 96-well microplate that contained 50 μl of N-[(1-H-benzotriazole-1-y) (dimethylamino) methylene]-N-methylmethanaminium hexafluorophosphate N-oxide (HBTU, 3 μmol), and N,N-disopropylethyl amine (DIEA, 6 μmol). To each reaction mixture, 50 μl of 2 μmol of amine core 1 (Fig. 2) dissolved in anhydrous dimethyl formamide (DMF) was added. All of the reaction mixtures were mixed and kept at room temperature. The reactions went to completion in 1 h, based on the disappearance of the free amine monitored by thin-layer chromatography (10:1 CHCl3/MeOH ratio; Rf = 0.28) and analysis of the crude reaction mixture by electrospray ionization mass spectrometry (3).
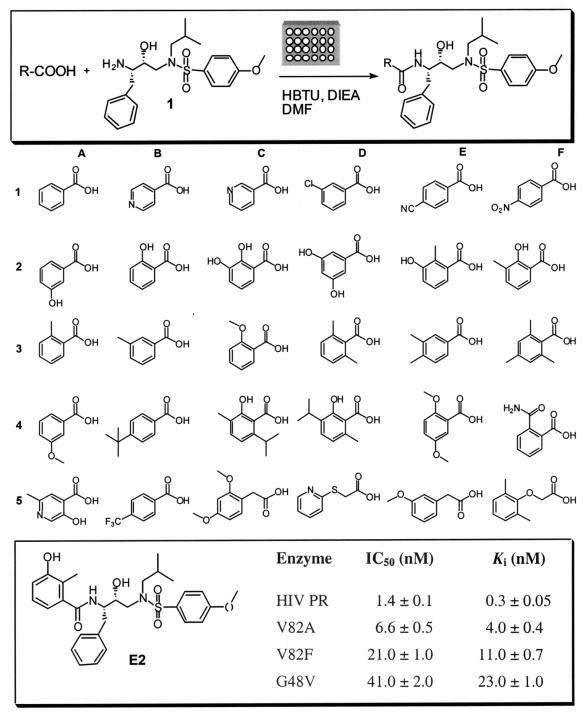
FIG. 2.
Preparation and in situ screening of different inhibitors with various P2 residues. The upper panel depicts the conditions used for the amide-forming reaction. Structures of the 30 carboxylic acids used for library compound synthesis are shown in the middle panel. In the lower panel, the structure of E2 and its Ki and in vitro IC50s are given.
Synthesis and purification of inhibitor E2.
Core 1 was prepared as previously reported (31). To a solution of a free amine 1 (200 mg, 0.45 mmol), 2-methyl-3-hydroxy benzoic acid (102 mg, 0.67 mmol) in 6 ml of dry DMF was added to HBTU (253 mg, 0.67 mmol) followed by DIEA (230 μl, 1.3 mmol) at 20°C in an argon atmosphere. The reaction mixture was stirred for 3 h, then quenched by the addition of brine, and extracted with ethyl acetate. The organic layer was washed with 1 N HCl, saturated aqueous NaHCO3, and brine, dried over MgSO4, filtered, and concentrated in vacuo. The crude product was purified by flash chromatography (CHCl3-MeOH), and the desired product was obtained in a 90% yield. The chemical structure of E2 was determined by nuclear magnetic resonance and electrospray ionization mass spectrometry (3).
Determination of in vitro IC50s of inhibitors against HIV protease.
Recombinant HIV protease for enzymatic assays was prepared as described previously (3). The determination of kinetic parameters of HIV protease was performed at 37°C at pH 5.6 by using an F-2000 fluorescence spectrophotometer (Hitachi) and a Packard fluorescence spectrophotometer (Fusion-Universal microplate analyzer) for the microplate assay. For HIV protease, the Km and Vmax values for the fluorogenic peptide substrate 2-aminobenzoyl (Abz)-Thr-Ile-Nle∼Phe-(p-NO2)-Gln-Arg-NH2 were determined by measuring the initial rate of hydrolysis at different substrate concentrations (2.5, 5.0, 10, 25, 50, and 100 μM) by monitoring the changes in fluorescence at a 420-nm emission and fitting the obtained data to the Michaelis-Menten equation with the GRAFIT program (version 3.0; Erithacus Software, Surrey, United Kingdom). Assays were run in 0.1 M morpholineethanesulfonic acid (MES) buffer containing 0.2 M NaCl and 1 mM dithiothreitol in a final volume of 200 μl in the wells of microplates. The enzyme concentration (30 μg/ml) that gave an ideal progress curve was used for the assays. The in vitro IC50, the concentration required for 50% inhibition of in vitro HIV protease activity, was determined from the percentage of inhibition rendered by E2 at various concentrations: 0.1, 0.5, 2.0, 4.0, 8.0, 12, and 16 nM. The Ki of the compound E2 was derived from the in vitro IC50 by using the formula for competitive inhibition: Ki = IC50/(1+[S]Km).
Treatment of E. coli cells with HIV protease inhibitors.
Pepstatin A was dissolved in water at 100 mM. Stock solutions of the FDA-approved HIV protease inhibitors were prepared by solubilizing the drugs in 100% dimethyl sulfoxide (DMSO) at 100 mM. A single colony of the cells bearing the respective plasmid, namely pTF-PRWT or pTF-PRmutant, was inoculated to 2 ml of Luria-Bertani medium with 100 μg of ampicillin/ml and grown overnight at 37°C. The culture was diluted 100-fold with fresh Luria-Bertani medium containing 100 μg of ampicillin/ml and incubated at 37°C for 2 h until the optical density at 630 nm (OD630) of the cells reached 0.6. The cells were then induced for protein expression with 0.2% arabinose and simultaneously treated with either HIV protease inhibitors at the indicated concentrations dissolved in 2% DMSO or 2% DMSO alone as the negative control. After incubation for an additional 3 h at 30°C, the cells were collected for Western blot analysis of proteins.
Western blot analysis of proteins.
After the cells were induced and treated as described above, they were collected by centrifugation and solubilized in Laemmli sample buffer (17) at a concentration of 0.01 OD630 cells per μl of sample buffer. The cellular proteins were separated on 4 to 12% Novex Bis-Tris polyacrylamide gels and electrotransferred onto polyvinylidene fluoride membranes for analysis with specific antibodies. The membranes were blocked with 5% nonfat milk in TTBS (10 mM Tris · HCl [pH 7.4], 500 mM NaCl, 0.1% Tween 20) for 1 h and then incubated with 1:5,000 anti-β-galactosidase monoclonal antibodies or 1:5,000 HIV-1 protease antiserum in 1% nonfat milk in TTBS for another hour. The bound antibodies were detected by chemiluminescence by horseradish peroxidase and Lumi-Glo chemiluminescence reagent.
Cell-based β-galactosidase activity assay.
E. coli cells bearing each of the expression plasmids were grown as described, except that after incubation at 37°C for 2 h to an OD630 of 0.6, the cells were transferred in triplicate sets to the wells of 96-well microplates. Similarly, the cells were subjected to simultaneous induction for recombinant protein expression as well as drug treatment with HIV protease inhibitors at the indicated concentrations in 2% DMSO. For the screening of the compound library generated by the amide-forming reaction, DMF was used in place of DMSO. After incubation for an additional 3 h at 30°C, the cells were made permeable for the entry of the β-galactosidase substrate, i.e., ortho-nitrophenyl-β-d-galactopyranoside (ONPG), by diluting the cells in assay buffer (10 mM NaP [pH 7.3], 10 mM NaCl, 1 mM MgSO4, 5 mM β-mercaptoethanol) that contained 50 μg of polymyxin B sulfate/ml and 2% Triton X-100 (28). The cells were then incubated for 5 min at room temperature in 96-well microplates. After the OD630 of the cells were determined by using an Ultramark microplate imaging system (Bio-Rad, Hercules, Calif.), ONPG was added at a final concentration of 200 μM to start the enzymatic reactions at 37°C. Changes in the OD415, indicating the production of ortho-nitrophenol, were continuously monitored with the Ultramark microplate imaging system. The linear portion of the progression curve was used to determine the initial velocity with MATLAB (The MathWorks Inc., Natick, Mass.). The difference in OD415 (ΔA) was converted to the increase in product concentration (ΔC) by using the equation ΔA = ɛ · ΔC · L, where L is the light path of 0.4 cm and the extinction coefficient (ɛ) of ortho-nitrophenol was 3,500 M−1 cm−1 under the assay conditions. The β-galactosidase activity was determined as the number of micromoles of ortho-nitrophenol produced per minute at 37°C. The β-galactosidase activity was then normalized against the amount of total cellular proteins, which was estimated by assuming that an OD630 unit corresponds to 1.4 × 109 cells and that every 109 cells yield approximately 150 μg of proteins (23).
RESULTS
Validation of detection of HIV protease activity in vivo in a high-throughput format.
It was reported that the engineered β-galactosidase that contains an insertion of HIV protease cleavage site at amino acid 131 still retains its activity, however, loses its activity when cleaved (2). Therefore, β-GalPR, i.e., β-galactosidase with an insertion of Val-Ser-Phe-Asn-Phe-Pro-Gln-Ile-Thr-Leu, corresponding to the sequence of the p6/PR junction in HIV Gag-Pol polyprotein, was used as the reporter of HIV protease activity in this E. coli-based model system.
Our model system uses the HIV protease precursor that comprises the TF, including the transframe peptide and p6pol protein, and the protease domain for coexpression together with β-GalPR (pTF-PRWT) (Fig. 1A). Coexpression under the same promoter control was carried out by using a two-cistron expression plasmid, where the HIV protease precursor was used as the first cistron and β-GalPR was used as the second cistron. A modified form of the HIV protease precursor that contained a D25N mutation at the catalytic residue Asp25 to abolish the proteolytic activity (14) was constructed by PCR-mediated mutagenesis and was used as a control (pTF-PRD25N) (Fig. 1A).
The expression of recombinant proteins in the E. coli cells bearing the expression plasmid, pTF-PRWT or pTF-PRD25N, was analyzed by Western blotting with HIV-1 protease antiserum (Fig. 1B) and with β-galactosidase antibodies (Fig. 1C). The immunoblots confirmed that both HIV protease and β-galactosidase could be translated from a single mRNA in this two-cistron system. Most of the wild-type HIV protease precursor was autoprocessed, and the mature HIV protease was released in the E. coli cells (Fig. 1B, lane 3). The mature HIV protease subsequently cleaves β-GalPR, resulting in the appearance of an extra band with a lower molecular weight in the lysates from the cells expressing TF-PRWT and β-GalPR (Fig. 1C).
The cleavage of the engineered β-galactosidase by HIV protease would cause a loss of β-galactosidase activity. In an effort to develop a high-throughput screening platform, we optimized the experimental procedures to quantitatively determine the β-galactosidase activity that requires minimal sample preparations, i.e., with no need for cell lysis and further protein purification steps. We used Triton X-100 and polymyxin B sulfate to increase the permeability of the β-galactosidase substrate (28) and then started the β-galactosidase reaction with the addition of ONPG. Using this protocol, we determined that uninduced E. coli contained no detectable β-galactosidase activity and the E. coli cells expressing only β-GalPR gave a β-galactosidase activity of 0.191 ± 0.005 μmol/min/mg of total E. coli proteins. When β-galactosidase was coexpressed with HIV protease, β-galactosidase activity changed to 0.041 ± 0.003 and 0.178 ± 0.007 μmol/min/mg for E. coli producing active and inactive HIV protease, respectively. These data confirmed E. coli β-galactosidase as a valid reporter for in vivo HIV protease activity.
Dose-dependent effects of the FDA-approved HIV protease inhibitors on the autoproteolytic processing of the HIV protease precursor.
The in vivo inhibitory effects of the HIV protease inhibitors were evaluated by a peptide-based protease inhibitor, pepstatin A, and the FDA-approved small-molecule HIV protease inhibitors APV, IDV, NFV, RTV, and SQV.
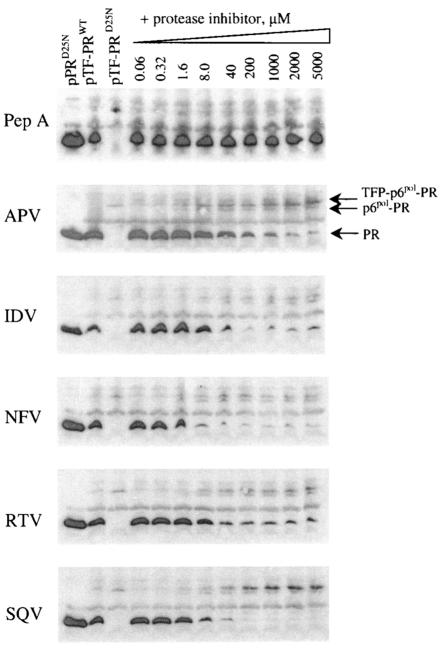
Drug permeability was evaluated by Western blot analysis in which E. coli cells were treated with HIV protease inhibitors without the use of permeabilization agents. As shown in Fig. 3, all small-molecule HIV protease inhibitors, except pepstatin A, inhibited the autoprocessing of the HIV protease precursor in a dose-dependent fashion. The dose-dependent inhibition of autoprocessing could be detected with 1.6 μM HIV protease inhibitors and reached a limit with 200 μM HIV protease inhibitors. The cleavage at the TFP-p6 pol site, resulting in the release of p6pol-PR, still occurred in the presence of 5 mM HIV protease inhibitors (Fig. 3). The concentration of IDV and RTV needed for 50% inhibition of the release of mature HIV protease was shown to be between 8 and 40 μM, and the concentrations of APV, NFV, and SQV needed for 50% inhibition of the release of mature HIV protease was between 1.6 and 8 μM.
FIG. 3.
In vivo dose-dependent effects of protease inhibitors on autoprocessing of the protease precursor, TF-PRWT, in E. coli cells. Protease inhibitors were pepstatin A (Pep A), APV, IDV, NFV, RTV, and SQV. The E. coli cells containing the expression construct pPRD25N, pTF-PRWT, or pTF-PRD25N were induced with 0.2% arabinose for 3 h at 30°C in the presence of 2% DMSO. Simultaneously, the E. coli cells expressing TF-PRWT were treated with the HIV protease inhibitors at the indicated concentrations in 2% DMSO. The cells were then collected, and the proteins were analyzed by Western blotting with HIV-1 antiserum. Each lane contained 0.3 OD630 equivalents of cells, except the lane for PRD25N, which contained 0.1 OD630 equivalents of cells expressing PRD25N.
Dose-dependent effects of the HIV protease inhibitor on β-galactosidase activity.
Since in vivo inhibitory effects of the FDA-approved HIV protease inhibitors in E. coli were confirmed by the inhibition of autoprocessing of the HIV protease precursor, we further investigated if we could detect the in vivo potency of the HIV protease inhibitors by an increase in the cleavage-induced loss of β-galactosidase activity. After drug treatment for 3 h, the cells were processed for detection of β-galactosidase activity in 96-well microplates as described. As shown in Fig. 4, β-galactosidase activity increased exponentially as the concentration of HIV protease inhibitors increased. Consistent with the inhibition of autoprocessing of the HIV protease precursor, the increase in β-galactosidase activity could be detected with as little as 1.6 μM HIV protease inhibitors and showed little differences between 200 μM and 400 μM HIV protease inhibitors, except with IDV. Additionally, with 400 μM HIV protease inhibitors, β-galactosidase activity was around 0.145 μmol/min/mg of total proteins, which was a little lower than the maximal β-galactosidase activity detected in the presence of inactive HIV protease. It is important that permeabilization reagents were used to allow the entry of ONPG for the in vivo β-galactosidase assay only after cells were harvested from drug treatment. Thus, it could not have had any effect on the permeability of HIV protease inhibitors. Indeed, pepstatin A with poor cell permeability did not show any in vivo inhibitory activity (data not shown).
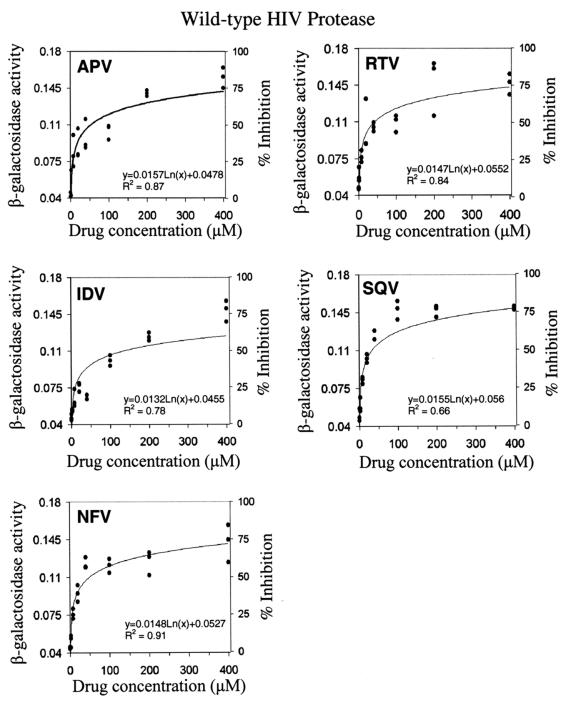
FIG. 4.
In vivo dose-dependent effects of HIV protease inhibitors against the wild-type HIV protease. The in vivo activity of the HIV protease inhibitors was determined by the inhibition of the cleavage-induced loss of β-galactosidase activity. The cells coexpressing TF-PRWT and β-GalPR were treated with HIV protease inhibitors at the indicated concentrations. Subsequently, β-galactosidase activities (micromoles per minute per milligram of total protein) in the treated E. coli cells were determined by using whole cells in a 96-well microplate format as described in Materials and Methods. Representative results from three separate experiments are shown, and each experiment had a triplicate set of each sample.
The IC50 of a given HIV protease inhibitor against the wild-type HIV protease in the E. coli system was determined by curve-fitting with MATLAB (Fig. 4 and Table 1). IC50 refers to the concentration of the HIV protease inhibitor required to reach a 50% inhibition of the cleavage-induced loss of β-galactosidase activity. The β-galactosidase activity detected in E. coli coexpressing TF-PRWT and β-GalPR in the absence of HIV protease inhibitors was referred to as 0% inhibition. The maximal β-galactosidase activity observed in E. coli containing pTF-PRD25N was regarded as 100% inhibition, which could theoretically be achieved by the most potent HIV protease inhibitor. As shown in Table 1, the IC50 for each HIV protease inhibitor was in the micromolar range, many times higher than what was determined in standard antiviral assays. Nevertheless, this E. coli-based system successfully detected the in vivo inhibitory activities of HIV protease inhibitors.
TABLE 1.
IC50s of protease inhibitors determined in our E. coli-based screening system and comparison of efficacies of HIV protease inhibitors determined in different systems
| HIV protease inhibitor | IC50 from our E. coli-based assay systema (μM) with:
|
EC90 from standard antiviral assayb (μM) with:
|
IC50 from another E. coli-based assay system with wild-type HIV proteasec (μM) | ||
|---|---|---|---|---|---|
| Wild-type HIV protease | D30N HIV protease | Wild-type HIV | D30N HIV strain | ||
| APV | 53.4 ± 17.4 | 35.9 ± 17.7 | —e | — | — |
| IDV | 132.5 ± 17.5 | 137.3 ± 13.7 | 0.060 | 0.010 | 100 |
| NFV | 48.4 ± 4.8 | >400.0 | 0.030 | 0.180 | — |
| RTV | 42.6 ± 9.1 | 26.9 ± 7.5 | 0.050 | 0.010 | — |
| SQV | 23.7 ± 3.4 | 9.3 ± 3.5 | 0.030 | 0.010 | 20 |
| Pepstatin A | NDd | ND | — | — | — |
IC50 refers to the inhibitor concentration required to reach a 50% inhibition of the loss of β-galactosidase activity caused by HIV protease induced cleavage. IC50s and standard errors were determined from three separate experiments as shown in Fig. 4 and 5.
Results are from reference 24. EC90 refers to the drug concentration that inhibited 90% of mammalian cell death induced by HIV infection.
Results are from reference 7. IC50 refers to the inhibitor concentration required to reach a 50% inhibition of the cyclic AMP production induced by HIV protease in the system.
ND, not detectable.
—, not reported in literature.
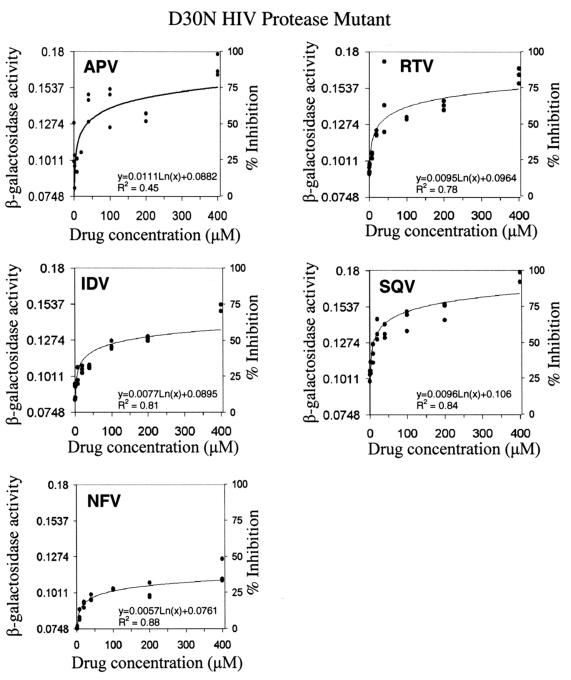
Dose-dependent effects of HIV protease inhibitor on a drug-resistant D30N HIV protease mutant.
The usefulness of this model system for drug-resistant HIV proteases was further validated with a D30N HIV protease mutant, a variant observed in HIV clinical isolates and specifically conferring drug resistance to NFV.
The HIV protease D30N mutant was created by PCR-mediated mutagenesis and was used to replace the wild-type HIV protease domain in the two-cistron expression construct for coexpression together with β-galactosidase to create the construct pTF-PRD30N. After the E. coli cells containing the expression plasmid pTF-PRD30N were induced for recombinant protein expression, β-galactosidase activity was determined by using whole cells in 96-well microplates. The β-galactosidase activity in the E. coli cells coexpressing TF-PRD30N and β-GalPR was 0.074 ± 0.011 μmol/min/mg of total proteins, which was higher than the β-galactosidase activity (0.041 ± 0.003 μmol/min/mg of total proteins) detected in the presence of the wild-type HIV protease. This indicated that the D30N HIV protease mutant has a lower catalytic efficiency upon cleaving the substrate, i.e., β-galactosidase, causing an increase in β-galactosidase activity compared with that determined in the presence of the wild-type HIV protease. Therefore, this system is suitable for detecting the HIV protease variants whose activities were different from the wild-type HIV protease activity.
The drug response of the D30N HIV protease to HIV protease inhibitors was investigated by using an experimental protocol similar to that used in the study of the wild-type HIV protease (Fig. 5). It is important to stress that, in this study, 0% inhibition was defined as the β-galactosidase activity detected in the E. coli cells coexpressing the D30N HIV protease and β-GalPR in the absence of HIV protease inhibitors. The reference to 100% inhibition was still the same as mentioned earlier. As shown in Fig. 5, the sensitivities toward APV, IDV, RTV, and SQV for the D30N HIV protease were similar to those for the wild-type HIV protease. On the other hand, compared to 75% inhibition against the wild-type HIV protease, 400 μM NFV only showed 30% inhibition against the D30N HIV protease. The IC50 of NFV for the D30N HIV protease mutant was thus determined to be greater than 400 μM, at least ninefold higher than the IC50 determined for the wild-type HIV protease (Table 1).
FIG. 5.
In vivo dose-dependent effects of HIV protease inhibitors against the D30N drug-resistant HIV protease. The in vivo activities of the HIV protease inhibitors were determined as described in the legend to Fig. 4 with the cells coexpressing TF-PRD30N and β-GalPR. β-Galactosidase activities (micromoles per minute per milligram of total protein) in the treated E. coli cells were determined, and the data were used to generate the dose-response curves by curve-fitting with MATLAB. Representative results from three separate experiments are shown, and each experiment had a triplicate set of each sample.
Preparation of a library of compounds by amide-forming reaction and in situ screening of in vitro HIV protease activity.
Starting from the sulfonamide isostere core 1, a compound library was prepared in microplates and was screened in situ against the HIV protease. The library was initially screened at 100 nM, and those reaching 50% or greater inhibition were selected for a second screening at 10 nM. The two rounds of screening revealed the preference of aromatic residues at the P2 position, such as benzoic acid. More than thirty acids of ortho-, meta-, para-substituted benzoic acids (Fig. 2) were coupled to the core and screened at 5 nM, in which E2 emerged as the best inhibitor from this library (Table 2). Following these results, E2 was synthesized and purified. The in vitro IC50s and Ki values of the purified E2 against the wild-type HIV protease and three other mutants were subsequently determined (Fig. 2).
TABLE 2.
Percentage of in vitro HIV protease activity detected in the presence of 5 nM inhibitors from the compound librarya
| R-COOH | HIV protease activity (%) with 5 nM inhibitor |
|---|---|
| A1 | 75 |
| B1 | 70 |
| C1 | 75 |
| D1 | 70 |
| E1 | 68 |
| F1 | 65 |
| A2 | 35 |
| B2 | 74 |
| C2 | 80 |
| D2 | 50 |
| E2 | 0 |
| F2 | 90 |
| A3 | 5 |
| B3 | 80 |
| C3 | 96 |
| D3 | 30 |
| E3 | 60 |
| F3 | 60 |
| A4 | 90 |
| B4 | 94 |
| C4 | 99 |
| D4 | 98 |
| E4 | 93 |
| F4 | 95 |
| A5 | 90 |
| B5 | 70 |
| C5 | 97 |
| D5 | 86 |
| E5 | 79 |
| F5 | 40 |
All measurements were run in triplicate, and the reported values are the means of these measurements.
Detection of in vivo inhibitory activity of the compound library against the wild-type HIV protease.
The library of compounds was also tested for inhibitory potency of HIV protease activity in the E. coli-based model system. The initial screening was conducted at 200 μM. Consistent with in vitro enzymatic studies, E2 was identified as the most potent compound in the E. coli-based model system. As shown in Fig. 6A, E2 inhibited autoprocessing of the HIV protease precursor at 1.6 μM and the inhibitory effects reached the maximum at 100 μM. After determining β-galactosidase activity in the cells treated with different concentrations of E2, the in vivo IC50 of E2 in the E. coli-based system was determined as 87.3 ± 25.4 μM, which is comparable to the potency of current drugs against HIV protease (Fig. 6B and Table 1).
FIG. 6.
In vivo dose-dependent effects of E2 against HIV proteases in the E. coli-based screening system. (A) Inhibition of autoprocessing of the wild-type HIV protease precursor. The drug treatment of E. coli cells containing the expression construct, pTF-PRWT, and Western blot analysis with HIV protease antiserum was performed as described. Each lane contained 0.3 OD630 equivalents of cells, except the lane for PRD25N, which contained 0.1 OD630 equivalents of cells expressing PRD25N. (B) Inhibition of cleavage-induced loss of β-galactosidase activity in the presence of the wild-type HIV protease. The E2-treated cells were processed for β-galactosidase activity detection by using whole cells in the 96-well microplates as described. (C and D) In vivo dose-dependent effects of APV (C) and E2 (D) against the I84V HIV protease mutant. The in vivo activity of the HIV protease inhibitors was determined by the inhibition of the cleavage-induced loss of β-galactosidase activity. The cells coexpressing TF-PRI84V and β-GalPR were treated with HIV protease inhibitors at the indicated concentrations. β-Galactosidase activities (micromoles per minute per milligram of total protein) in the treated E. coli cells were determined, and the data were used to generate the dose-response curves by curve-fitting with MATLAB. Representative results from three separate experiments are shown, and each experiment had a triplicate set of each sample.
Detection of in vivo inhibitory activity of E2 against the I84V HIV protease.
Since the core of inhibitor E2 is similar to that of APV, we were interested in investigating the drug response of E2 against the APV-resistant HIV protease mutants. One of the key signature substitutions for APV resistance resides at the amino acid residue I84 (10). The in vivo EC90, the inhibitor concentration that inhibits 90% of virus-induced cytotoxicity, of APV against I84V HIV protease mutant is twofold higher than the EC90 against the wild-type HIV protease (13). The in vitro enzymatic assay also showed a threefold increase of the Ki of APV against the I84V HIV protease mutant compared to that of the wild-type HIV protease (13).
Again, the I84V HIV protease mutant was created by PCR-mediated mutagenesis and used to replace the wild-type HIV protease domain in the two-cistron system to construct the plasmid pTF-PRI84V. After inducing E. coli cells bearing the expression plasmid pTF-PRI84V, β-galactosidase activity was determined as described. The β-galactosidase activity in the cells expressing TF-PRI84V and β-GalPR was 0.081 ± 0.001 μmol/min/mg of total proteins. Again, this was higher than the β-galactosidase activity (0.041 ± 0.003 μmol/min/mg of total proteins) detected in the presence of the wild-type HIV protease. This confirmed that the I84V HIV protease mutant has a lower catalytic efficiency than the wild-type HIV protease in this E. coli-based system.
The drug response of the I84V HIV protease mutant toward E2 was investigated by using similar experimental protocols, as described, and with APV as a control (Fig. 6C and D). In this study, 0% inhibition was defined as the β-galactosidase activity detected in the E. coli coexpressing the I84V HIV protease and β-GalPR without the treatment of HIV protease inhibitors. The IC50 of E2 against I84V HIV protease was determined as 141.3 ± 50 μM while the IC50 of APV was 280 ± 39 μM. Data obtained with E2 demonstrated that subtle changes of the structure at the P2 position could generate new HIV protease inhibitors with improved binding affinities toward drug-resistant HIV proteases.
The results indicated that this E. coli-based model system is useful in serving multiple purposes, including quick identification of drug leads against the protease target and establishment of differential inhibition profiles for drug-resistant variants against various drugs.
DISCUSSION
HIV protease inhibitors have been developed into effective antiviral therapeutics for treating AIDS in the past decade. However, the effectiveness of the existing HIV protease inhibitors has been hampered by the emergence of the drug-resistant HIV proteases. HIV protease variants can escape inhibition by current HIV protease inhibitors and thus pose a great medical need for novel HIV protease inhibitors. We report here a simple yet efficient E. coli-based system for screening the HIV protease inhibitors that are not only potent but also permeable for crossing the cell membrane. This system uses β-galactosidase as the reporter to adapt high-throughput screening and was validated with the in vivo inhibitory effects of existing HIV protease inhibitors against both the wild-type and drug-resistant HIV proteases. The ranking of IC50s of HIV protease inhibitors against the HIV proteases, the wild type and the drug-resistant variants, determined in our E. coli-based system corresponded well with published data (24). Furthermore, we demonstrated rapid identification of a novel HIV protease inhibitor from a library of compounds prepared by diversity-oriented synthesis in microtiter plates.
The usefulness of this E. coli-based system was demonstrated with the FDA-approved HIV protease inhibitors and pepstatin A, a peptide-based aspartic protease inhibitor, as a control for cell permeability. Concentrations of each HIV protease inhibitor in this E. coli-based system ranged from 1.6 μM to as high as 5 mM. The small-molecule HIV protease inhibitors exerted in vivo inhibitory effects on the autoprocessing of the HIV protease precursor at as low as 1.6 μM (NFV and SQV) or 8 μM (IDV and RTV) (Fig. 3), and these were three magnitudes greater than the in vitro IC50s in the nanomolar range against the purified wild-type HIV protease. Pepstatin A has an in vitro IC50 of 0.7 μM against the purified HIV protease (15). Therefore, if it were permeable, then one would expect to detect an in vivo IC50 for pepstatin A of around 700 μM. In fact, pepstatin A, up to 5 mM, failed to exert any inhibitory effects in the E. coli-based system. These data indicated that pepstatin A has low cell permeability; thus, it fails to diffuse passively across the cell membrane to inhibit autoprocessing of the HIV protease precursor intracellularly. This result suggested that the E. coli-based system could be used as an alternative method for rapidly selecting compounds with in vivo activities which require both potent inhibitory activity and desirable permeability.
The in vivo inhibition of HIV protease precursor autoprocessing by HIV protease inhibitors reached a limit at 200 μM. Interestingly, the cleavage at the TFP-p6pol site, resulting in the release of p6 pol-PR, still occurred in the presence of 5 mM HIV protease inhibitors (Fig. 3). Consistent with our results, in vivo studies with HIV-infected human T lymphocytes found an accumulation of the Pol intermediates, p6 pol-PR-RT-IN and p6-PR, in the presence of 1 μM SQV (18), which was many times higher than the nanomolar concentrations used in standard antiviral assays. Therefore, it was reasonable that we observed residual activity of autoproteolytic processing in the presence of 5 mM HIV protease inhibitor in the E. coli-based system.
The in vivo inhibitory effects of HIV protease inhibitors were quantitated by determining the IC50 of a given protease inhibitor by using an in vivo β-galactosidase activity assay. We compared the IC50s determined in our study with other reported data sets: (i) EC90s determined from antiviral assays with mammalian cells and (ii) IC50s obtained from another E. coli-based screening system (Table 1). Obviously, in E. coli-based systems, the drug concentrations needed to detect the in vivo inhibitory activity of HIV protease inhibitors are three magnitudes higher than what were needed for antiviral testing with mammalian cells. However, the ranking of IC50s determined in our system was similar to that determined with standard antiviral tests (24). The higher IC50s obtained from the E. coli system might be mainly due to an additional barrier posed by the bacterial cell wall.
We demonstrated the feasibility of this system for drug resistance enzymes by using an HIV protease variant that has a D30N mutation in the HIV protease coding sequence. The D30N mutation is an active-site mutation and represents the major mutation that confers drug resistance specifically against NFV. The structural analysis of binary complex D30N HIV protease with NFV indicated altered interactions with hydrophobic P2 side chains, resulting in variable substrate specificity and lower catalytic efficiency (20). The kcat/Km of D30N HIV protease was determined to be 0.089 min−1 μM−1 compared to 0.41 min−1 μM−1 for the wild-type HIV protease (21). This indicated that the D30N HIV protease has a catalytic efficiency fivefold lower than the wild-type HIV protease does. While using the β-galactosidase activity as the reporter, we detected a roughly twofold-lower activity for the D30N HIV protease compared to that for the wild-type enzyme. This system thus could be used to identify HIV protease variants that have different catalytic efficiencies than the wild-type HIV protease.
The drug response of the D30N HIV protease determined in our studies was similar to the drug resistance profile determined in standard antiviral assays (Table 1). In our HIV protease inhibitor studies with the E. coli-based model system, the drug-resistant D30N HIV protease showed low responsiveness specifically to NFV but not to other HIV protease inhibitors. The IC50s of APV, IDV, RTV, and SQV are similar or a little lower against the D30N HIV protease versus the wild-type HIV protease. However, the IC50 of NFV for the D30N HIV protease increased at least ninefold compared with that for the wild-type HIV protease. This result suggested that the most effective agent among the five tested in this study for treating patients infected with the HIV strain carrying a D30N mutation in its protease coding sequence may be SQV, followed by RTV. This E. coli-based system could potentially provide a simple alternative method for generating drug resistance profiles that could be used in turn to suggest therapeutic use of HIV protease inhibitors to treat drug-resistant HIV strains.
The amide-forming reaction has been successfully demonstrated as a rapid synthesis method for preparing diversity-oriented libraries against HIV protease for in situ screening (4). The method was used to prepare a library of compounds containing an APV-like core attached to various P2 residues, in which E2 emerged as a novel HIV protease inhibitor with potency against both the wild type and the I84V HIV proteases. The amino residue I84 resides in the S2 binding pocket, and its mutation occurs during most of the HIV protease inhibitor treatments. The inhibitor E2, containing the 2-methyl-3-hydroxyl-benzamide portion at the P2 residue rather than the tetrahydrofuran ring at the P2 residue of APV, showed higher potency against the I84V HIV protease mutant.
The amide-forming reaction could be used to combine different transition-state analog cores with various P2 or P3 residues to produce new inhibitors. Together with the E. coli-based screening platform, we should be able to perform rapid lead discovery of novel HIV protease inhibitors targeting the active site of HIV proteases. It is plausible that this E. coli-based screening method developed for the HIV protease inhibitor screening can be easily modified for inhibitor screening for other proteases, especially proteases from infectious agents that can cause serious diseases with unmet medical needs. Such proteases include the lethal factor from Bacillus anthracis and viral proteases from poliovirus and other viruses. The standard drug discovery process involves laborious in vitro kinetic assays followed by dangerous in vivo virus-induced cytotoxicity assays in biosafety level 3 and 4 labs. This E. coli-based assay system could be used as an efficient, convenient, and safe surrogate system for rapid identification of promising protease inhibitors with minimal exposure of human lives to infectious agents.
Acknowledgments
We thank David Galas for manuscript review and critical discussions.
This work was supported in part by the Keck Graduate Institute of Applied Life Sciences and the NIH (grant CA 99898 to C.-C.K.).
REFERENCES
- 1.Ansaldi, M., M. Lepelletier, and V. Mejean. 1996. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal. Biochem. 234:110-111. [DOI] [PubMed] [Google Scholar]
- 2.Baum, E. Z., G. A. Bebernitz, and Y. Gluzman. 1990. β-Galactosidase containing a human immunodeficiency virus protease cleavage site is cleaved and inactivated by human immunodeficiency virus protease. Proc. Natl. Acad. Sci. USA 87:10023-10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brik, A., Y.-C. Lin, J. Elder, and C.-H. Wong. 2002. A quick diversity-oriented amide-forming reaction to optimize P-subsite residues of HIV protease inhibitors. Chem. Biol. 9:891-896. [DOI] [PubMed] [Google Scholar]
- 4.Brik, A., and C.-H. Wong. 2003. HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 1:5-14. [DOI] [PubMed] [Google Scholar]
- 5.Cheng, Y. S. E., F. H. Yin, S. Foundling, D. Blomstrom, and C. A. Kettner. 1990. Stability and activity of human immunodeficiency virus protease: comparison of the natural dimer with a homologous, single-chain tethered dimer. Proc. Natl. Acad. Sci. USA 87:9660-9664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dasmahapatra, B., B. DiDomenico, S. Dwyer, J. Ma, I. Sadowski, and J. Schwartz. 1992. A genetic system for studying the activity of a proteolytic enzyme. Proc. Natl. Acad. Sci. USA 89:4159-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dautin, N., G. Karimova, A. Ullmann, and D. Ladant. 2000. A sensitive genetic screen for protease activity based on a cyclic AMP signaling cascade in Escherichia coli. J. Bacteriol. 182:7060-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson, J. W. 2001. HIV-1 protease as a target for AIDS therapy, p. 1-25. In R. C. Ogden and C. W. Flexner (ed.), Protease inhibitors in AIDS therapy. Marcel Dekker, Inc., New York, N.Y.
- 9.Frankel, A. D., and J. A. Yong. 1998. HIV-1: fifteen proteins and a RNA. Annu. Rev. Biochem. 67:1-25. [DOI] [PubMed] [Google Scholar]
- 10.Gong, Y.-F., B. S. Robinson, R. E. Rose, C. Deminie, T. P. Spicer, D. Stock, R. J. Colonno, and P.-F. Lin. 2000. In vitro resistance profile of the human immunodeficiency virus type 1 protease inhibitor BMS-232632. Antimicrob. Agents Chemother. 44:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishima, R., D. A. Torchia, S. M. Lynch, A. M. Gronenborn, and J. M. Louis. 2003. Solution structure of the mature HIV-1 protease monomer: insight into the tertiary fold and stability of a precursor. J. Biol. Chem. 278:43311-43319. [DOI] [PubMed] [Google Scholar]
- 12.Kim, S. Y., K. W. Park, Y. J. Lee, S. H. Back, J. H. Goo, O. K. Park, S. K. Jang, and W. J. Park. 2000. In vivo determination of substrate specificity of hepatitis C virus NS3 protease: genetic assay for site-specific proteolysis. Anal. Biochem. 284:42-48. [DOI] [PubMed] [Google Scholar]
- 13.Klabe, R. M., L. T. Bacheler, P. J. Ala, S. Erickson-Viitanen, and J. L. Meek. 1998. Resistance to HIV protease inhibitors: a comparison of enzyme inhibition and antiviral potency. Biochemistry 37:8735-8742. [DOI] [PubMed] [Google Scholar]
- 14.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krausslich, H. G., R. H. Ingraham, M. T. Skoog, E. Wimmer, P. V. Pallai, and C. A. Carter. 1989. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc. Natl. Acad. Sci. USA 86:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kupiec, J.-J., S. Hazebrouck, T. Leste-Lasserre, and P. Sonigo. 1996. Conversion of thymidylate synthase into an HIV protease substrate. J. Biol. Chem. 271:18465-18470. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Lindhofer, H., K. von der Helm, and H. Nitschko. 1995. In vivo processing of Pr160gag-pol from human immunodeficiency virus type 1 (HIV) in acutely infected, cultured human T-lymphocytes. Virology 214:624-627. [DOI] [PubMed] [Google Scholar]
- 19.Louis, J. M., C. M. Clore, and A. M. Gronenborn. 1999. Autoprocessing of HIV-1 protease is tightly coupled to protein folding. Nat. Struct. Biol. 6:868-875. [DOI] [PubMed] [Google Scholar]
- 20.Mahalingam, B., J. M. Louis, J. Hung, R. W. Harrison, and I. T. Weber. 2001. Structural implications of drug-resistant mutants of HIV-1 protease: high-resolution crystal structures of the mutant protease/substrate analogue complexes. Proteins 43:455-464. [DOI] [PubMed] [Google Scholar]
- 21.Mahalingam, B., J. M. Louis, C. C. Reed, J. M. Adomat, J. Krouse, Y.-F. Wang, R. W. Harrison, and I. T. Weber. 1999. Structural and kinetic analysis of drug resistant mutants of HIV-1 protease. Eur. J. Biochem. 263:238-245. [DOI] [PubMed] [Google Scholar]
- 22.Martínez, M.-A., M. Cabana, M. Parera, A. Gutierrez, J. A. Esté, and B. Clotet. 2000. A bacteriophage lambda genetic screen for characterization of the activity and phenotype of the human immunodeficiency virus type 1 protease. Antimicrob. Agents Chemother. 44:1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardee, A. B., F. Jacob, and J. Monod. 1959. The genetic control and cytoplasmic expression of inducibility in the synthesis of β-galactosidase of Escherichia coli. J. Mol. Biol. 1:165-168. [Google Scholar]
- 24.Patick, A. K., M. Duran, Y. Cao, D. Shugarts, M. R. Keller, E. Mazabel, M. Knowles, S. Chapman, D. R. Kuritzkes, and M. Markowitz. 1998. Genotypic and phenotypic characterization of human immunodeficiency virus type 1 variants isolated from patients treated with the protease inhibitor nelfinavir. Antimicrob. Agents Chemother. 42:2637-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paulus, C., S. Hellebrand, U. Tessmer, H. Wolf, H. G. Krauslich, and R. Wagner. 1999. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J. Biol. Chem. 274:21539-21543. [DOI] [PubMed] [Google Scholar]
- 26.Pettit, S. C., S. Gulink, L. Everitt, and A. H. Kaplan. 2003. The dimer interfaces of protease and extra-protease domains influence the activation of protease and the specificity of GagPol cleavage. J. Virol. 77:366-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Schupp, J. M., S. E. Travis, L B. Price, R. F. Shand, and P. Keim. 1995. Rapid bacterial permeabilization reagent useful for enzyme assays. BioTechniques 19:18-20. [PubMed] [Google Scholar]
- 29.Sices, H. J., and T. M. Kristie. 1998. A genetic screen for the isolation and characterization of site-specific proteases. Proc. Natl. Acad. Sci. USA 95:2828-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, T. A., and B. D. Kohorn. 1991. Direct selection for sequences encoding proteases of known specificity. Proc. Natl. Acad. Sci. USA 88:5159-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tung, R. D., D. J. Livingston, B. G. Rao, E. E. Kim, C. T. Baker, J. S. Boger, S. P. Chambers, D. D. Deininger, M. Dwyer, L. Elsayed, J. Fulghum, B. Li, M. A. Murcko, M. A. Navia, P. Novak, S. Pazhanisamy, C. Stuver, and J. A. Thomson. 2002. Design and synthesis of amprenavir, a novel HIV protease inhibitor. Infect. Dis. Ther. 25:101-118. [Google Scholar]
- 32.Wlodawer, A., and J. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543-5858. [DOI] [PubMed] [Google Scholar]
- 33.Wondrak, E. M., and J. M. Louis. 1996. Influence of flanking sequences on the dimer stability of human immunodeficiency virus type 1 protease. Biochemistry 35:12957-12962. [DOI] [PubMed] [Google Scholar]
- 34.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J. Virol. 69:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]