Abstract
The immunoglobulin M heavy-chain locus contains two poly(A) sites which are alternatively expressed during B-cell differentiation. Despite its promoter proximal location, the secretory poly(A) site is not expressed in undifferentiated cells. Crucial to the activation of the secretory poly(A) site during B-cell differentiation are changes in the binding of cleavage stimulatory factor 64K to GU-rich elements downstream of the poly(A) site. What regulates this change is not understood. The secretory poly(A) site contains two downstream GU-rich regions separated by a 29-nucleotide sequence. Both GU-rich regions are necessary for binding of the specific cleavage-polyadenylation complex. We demonstrate here that U1A binds two (AUGCN1-3C) motifs within the 29-nucleotide sequence and inhibits the binding of cleavage stimulatory factor 64K and cleavage at the secretory poly(A) site.
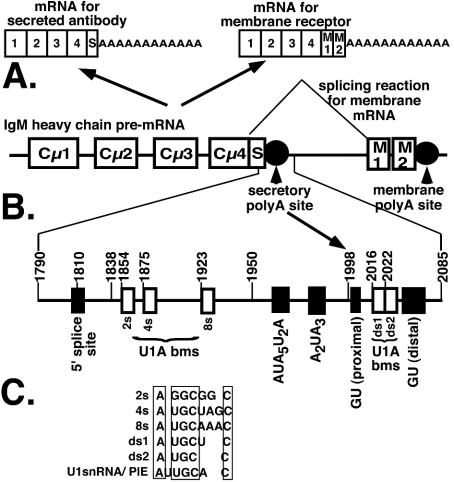
The immunoglobulin M (IgM) heavy-chain pre-mRNA (μ) is alternatively processed into mRNAs encoding a membrane receptor or secreted antibody during differentiation (4) (Fig. 1). It is the classic model for an important pattern of alternative processing which involves competition between splicing and cleavage-polyadenylation, and it includes a number of important receptors involved in growth and differentiation (for a review, see reference 6). In undifferentiated cells, exons encoding a membrane tail are spliced on and the mRNA is cleaved at a downstream, membrane poly(A) site, resulting in mRNA encoding the heavy chain of the membrane receptor. When cells differentiate into Ig-secreting cells, an upstream, secretory poly(A) site is activated within the intron involved in the splicing of the membrane exons. This results in the secretory form of mRNA which encodes the heavy chain of a secreted antibody. The secretory form of μ-mRNA is expressed in differentiated cells by a combination of increased cleavage at the secretory poly(A) site and increased stability of the secretory mRNA itself (1, 3, 10, 12).
FIG. 1.
Schematic model of the Ig secretory poly(A) site. (A) The genetic organization of the IgM heavy chain and its alternative processing to a secretory or a membrane form of mRNA. (B) The location of the secretory poly(A) site and relative location of the 5′ splice site, the U1A binding motifs, the hexanucleotide sequence, and the downstream GU-rich regions. Numbers indicate the positions referred to in the text. (C) A comparison of the AUGCN1-3C sequences for the 2s, 4s, 8s, ds1, and ds2 U1A binding sites.
3′ end cleavage in metazoans takes place on the recognition of a bipartite poly(A) signal consisting of a consensus AAUAAA and a less-defined GU-rich sequence, upstream and downstream of the cleavage site, respectively, by components of the cleavage-polyadenylation complex. These consist of the multimeric cleavage polyadenylation specificity factor (CPSF) and cleavage stimulatory factor (CstF), of which the 64-kDa component (CstF64K) recognizes the GU-rich region, as well as cleavage factors I and II and poly(A) polymerase (reviewed in reference 31). After cleavage, the RNA is specifically polyadenylated by poly(A) polymerase tethered to the RNA via the 160-kDa component of CPSF, bound by the AAUAAA sequence (16).
The secretory poly(A) site is unusual in that it contains dual elements for both CPSF and CstF binding (23). The hexanucleotide sequence is situated within an AU-rich region that retains residual activity even when the consensus sequence is mutated, suggesting a mechanism by which CPSF may be recruited away from its optimal binding site. In addition, there are two GU-rich regions, one is suboptimally located too close to the cleavage site and the other requires to be presented in the form of a stem-loop structure to be operational (21). Both GU-rich regions are necessary for full expression of the secretory poly(A) site (23). This bipartite structure suggests a mechanism by which this poly(A) site is weak. However, experiments involving progressive depletion of CstF64K from chicken B cells showed that this poly(A) site is particularly sensitive to CstF64K concentration (25), suggesting supplementary mechanisms to prevent its activation in undifferentiated B cells. Indeed there is an early report of an inhibitory factor whose binding site is coincident with the poly(A) site (30).
A comparison of ratios of usage of tandem splice sites and tandem poly(A) sites in cell lines representing different stages of B-cell differentiation indicated that it is changes in poly(A) site expression that regulate the switch from the membrane to secretory form of mRNA (18). Furthermore, a non-Ig gene with a similar balance and arrangement of competing cleavage-polyadenylation reactions is alternatively processed and regulated in murine splenic B cells at a twofold lower level than is a coexpressed IgM heavy-chain gene, suggesting that additional mechanisms unique to the IgM heavy-chain gene regulate its expression (24). Artificial introduction of CstF64K can activate the secretory poly(A) site in a chicken B-cell line which normally produces the membrane form of mRNA (27), suggesting that CstF64K binding strength plays a crucial role in the activation. However, this does not appear to be via an increase in overall CstF64K levels in physiologically relevant cells but rather due to a differentiation-specific transacting factor that alters CstF64K binding strength at this site (5, 11).
It has previously been shown that the change in stability of the secretory mRNA is regulated by the addition of a poly(A) tail to the secretory mRNA (20). The U1A protein binds upstream of the secretory poly(A) site and inhibits poly(A) addition to the secretory mRNA in a developmentally regulated manner. This inhibition depends on sequences that resembled the U1 snRNA consensus U1A binding site (AUUGCAC) but are nonconsensus [A(U/G)GCN1-3C] (20) (see Fig. 1B for location and C for sequences).
We now demonstrate the existence of two U1A binding sites downstream of the secretory poly(A) site between the two GU-rich regions which have the same AUGCN1-3C motif as those upstream (see Fig. 1B and C for location and sequences). However, these downstream U1A sites inhibit secretory poly(A) site expression by a completely different mechanism. They play a unique role by inhibiting cleavage at the secretory poly(A) site via impeding the binding of the cleavage complex and in particular CstF64K.
MATERIALS AND METHODS
Plasmid constructs.
For in vitro transcription, the PCR products from the secretory poly(A) site sequences, containing mutations and 5′ EcoRI and 3′ XbaI sites, introduced as part of the synthetic primers, were cloned into pGem 3Zf (containing a T7 promoter in the forward direction) between the EcoRI and XbaI sites. For transfection, a 5′ BglII site replaced the EcoRI site, and PCR products were cloned into pPKLT55 (22) containing the Firefly luciferase cDNA between the BglII and XbaI sites, replacing the poly(A) site. The forward primers (5′GAC TCT AGA [or AGATCT] GGA CCG TGG ACA AGT CC3′ [1790 wild type] and 5′GAC TCT AGA [or AGATCT] GGA CCG TCC ACA AGT CCA CTG CAA ACC CCA CAC TGT ACA ATG3′ [1790 5′ splice site mutation]) were combined with the reverse primer (5′GCG TCT AGA TAG GGT GGA GGC AAG TAT GC3′ [2085]) to amplify from position 1790 to 2085 (nucleotide positions are numbered according to the mouse IgM sequence with accession number [emb] V00818). The mutations in the downstream U1A sequences were incorporated using crossover PCR as previously described (21). The outside forward and reverse primers were combined with the internal reverse and forward primers, respectively, to produce two PCR products which when combined formed the template for a second PCR using the outside primers. The forward internal primers were as follows: for mutant (mut) ds1, 5′CGT CAC TGG TTT TGA TTA TAC AAA AAT CAT GCC TGC TGA GAC AG3′; for mut ds2, 5′CGT CAC TGG TTT TGA TTA TAC AAT GCT CAT CGG ACG TGA GAC AGT TGT GTT TTG CTT GC3′; for mut ds12, 5′CGT CAC TGG TTT TGA TTA TAC AAA AAT CAT CGG ACG TGA GAC AGT TGT GTT TTG CTTGC3′. The introduced mutations are highlighted in bold. The reverse internal primers were the exact reverse of the forward internal primers.
RNA substrates for footprinting, electrophoretic mobility shift assays (EMSAs), UV cross-linking, and cleavage assays.
Templates for synthesis of radiolabeled RNA were obtained by XbaI digestion of the pGem 3Zf constructs. Uniformly labeled RNA substrates were synthesized by in vitro transcription using T7 RNA polymerase and [α-32P]UTP as previously described (23). 5′ end-labeled transcripts were obtained by the same protocol, substituting [γ-32P]GTP as the radiolabeled nucleotide. 3′ end-labeled RNA substrates were synthesized by the same protocol omitting [α-32P]UTP. The cold RNA was subsequently labeled at the 3′ end with 5′-32pCp using T4 RNA ligase as previously described (21). All transcripts were purified by extraction after electrophoresis on 8% polyacrylamide gels containing 7 M urea.
Recombinant proteins.
Untagged recombinant wild-type U1A and U1A with the scrambled dimerization domain were purified from E. coli as described previously (2). Recombinant bovine poly(A) polymerase, tagged at the carboxy terminus with six histidines, was purified from E. coli on Ni2+ nitrilotriacetic acid. The His-tagged carboxy terminus ensured that all of the C-terminal residues were present after purification (8). The first 108 amino acids of CstF64K (spanning the RNA binding domain [RBD]) fused to glutathione S-transferase (GST) at the N-terminal end (26) were purified from E. coli on glutathione Sepharose (Amersham Pharmacia) and eluted in 20 mM reduced glutathione (Sigma) as described in reference 26.
Cell culture and transfection.
HeLa cells were plated at 104 per ml and grown overnight to ensure that they were in log phase. Plasmids were transfected into cells using Superfect (QIAGEN) at 20 μl/106 cells. Transfection efficiency was measured by cotransfection of Renilla SV40. Firefly and Renilla luciferase activities were measured using a dual luciferase kit from Promega.
RNA secondary structural probing and footprinting.
Probing of the secondary structure of RNA was carried out as previously described (21). Briefly, all RNA substrates were first refolded by heating in reaction buffer at 70°C for 2 min followed by cooling to 37°C over a 30-min period. For RNase T1 cleavage, 10,000 cpm of 3′ or 5′ end-labeled RNA substrate was incubated at 37°C in a total volume of 5 μl with 0.001 U of RNase T1 (Boehringer) in 50 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7.0, 100 mM NaCl, and 10 mM MgCl2 for 5 min. The reactions were stopped by the addition of 10 M urea and 25 mM EDTA. Samples were analyzed on 15% polyacrylamide gel electrophoresis (PAGE) containing 7 M urea. A reference nucleotide ladder was made by boiling of substrates with 50 mM NaOH and 10 mM EDTA for 10 s. Footprints were obtained by adding U1A before T1 digestion. These reactions were stopped by the addition of 180 μl of proteinase K buffer (50 mM Tris, pH 7.9, 10 mM EDTA, 230 mM NaCl, 0.2% sodium dodecyl sulfate [SDS]) and 50 μg of proteinase K and incubated for 10 min at 30°C. This was followed by phenol-chloroform extraction and ethanol precipitation before loading on PAGE as above.
EMSAs.
U1A binding assays were performed as previously described (20, 28) by using 50 fmol of 32P-labeled RNA substrate per lane and recombinant U1A as indicated.
UV cross-linking assays.
The UV cross-linking assays were performed as previously described (22) by using conditions described by Takagaki and Manley for gel shift assays (26). Briefly, recombinant GSTCstF64KRBD (2.5 μM) and increasing concentrations of recombinant U1A (as indicated) were incubated with 50 fmol of uniformly 32P-labeled RNA in 8 mM HEPES (pH 7.9), 40 mM NaCl, 2 mM EDTA, 0.2 mM dithiothreitol, 0.5 μg of tRNA, and 8% glycerol in a total volume of 12.5 μl. Products were cross-linked on ice under a handheld UV lamp at 245 nM for 10 min. These were incubated with 1 μg of RNase A at 30°C for 30 min and immediately run on SDS-12% PAGE. Cross-links were visualized by phosphorimagery.
Cleavage assays.
Cleavage assays were performed as described in references 14 and 15, by using 250 fmol of 32P-labeled RNA substrate and 7 μl of HeLa cell nuclear extract (10 μg of total protein/μl), 1.25 mM 3′ dATP, 0.7 mM MgCl2, and 3% polyvinyl alcohol in a total reaction volume of 20 μl. Increasing amounts of recombinant U1A were added immediately before the nuclear extracts. These were incubated for 2 h at 30°C and digested with proteinase K. RNA products were then extracted and precipitated with phenol-chloroform and ethanol, respectively, and run on 8% denaturing PAGE.
RESULTS
U1A protects AUGCN1-3C sequences downstream of the cleavage site between the two GU-rich regions, as well as the distal GU-rich region.
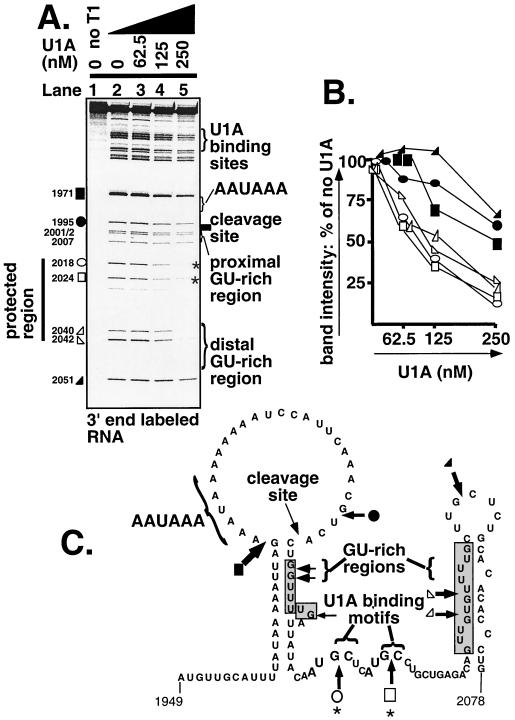
A possible protection downstream of the cleavage site had previously been noticed while investigating U1A's ability to protect the upstream U1A binding sites in the context of an RNA substrate spanning 295 nucleotides (nt), position 1790 to 2085 encompassing a possible cloverleaf structure (19). To investigate the downstream footprints more closely, we 3′ end-labeled the substrate (position 1790 to 2085) (for location sequence and motifs spanned, see Fig. 1B). We added increasing amounts of recombinant U1A and looked for protection from partial digestion with RNase T1 (which cuts 3′ of accessible guanosines) (7). As can be seen in Fig. 2A, a protection is seen downstream of the proximal GU-rich region which extends into the distal GU-rich region (Fig. 2A). The indicated bands were quantitated by phosphorimager analysis to measure the effect of increasing concentrations of U1A at each site of RNase T1 digestion. Bands both 5′ and 3′ of the protected region were quantitated as controls. As can be seen in Fig. 2B, the intensity of the bands inside the protected regions decreased more rapidly than those outside the region, demonstrating protection by U1A. Furthermore, within the protected cluster, it can be seen that the two 5′ bands are protected to a greater extent than those which are within the distal GU-rich region. Examination of the sequences of these two bands revealed two AUGCN1-3C motifs, which is the sequence we had previously determined to bind U1A upstream of the secretory poly(A) site (20) (Fig. 2A and C). Figure 2C shows the predicted secondary structure of this region. The structure of the stem-loop spanning the downstream GU-rich region (position 2024 to 2077) has been systematically mapped (21). The structure surrounding the hexanucleotide sequence 1960 to 2012 was predicted by MFOLD. As can be seen, the GU-rich regions flank the potential U1A binding sites, and the U1A protected region extends to cover the distal GU-rich region. We named the possible downstream U1A binding motifs ds1 and ds2, with ds standing for downstream.
FIG. 2.
U1A protects AUGCN1-3C sequences downstream of the cleavage site between the two GU-rich regions. (A) 3′ end-labeled RNA. The indicated amounts of U1A were allowed to bind the RNA before partial RNase T1 digestion. Samples were treated with proteinase K before PAGE. Sites of RNase T1 digestion are labeled with position numbers according to the mouse IgM sequence with accession number V00818. The downstream footprint is labeled with a line, the positions of the two AUGCN1-3C sequences are indicated with asterisks. The protected bands are indicated with open symbols, and those used as comparison are indicated with solid symbols. Symbols relate to quantitation in panel B. (B) Quantitation of the bands marked with symbols in panel A by phosphorimager analysis. Results were normalized to zero U1A. Symbols refer to bands marked with equivalent symbols in panel A. (C) Schematic diagram of predicted RNA structure of the poly(A) site and the downstream region (position 1949 to 2085). The position of the RNase T1 cuts are indicated with arrows of the size proportional to the intensity of the cut. Positions of RNase T1 cuts quantitated in panel B are indicated with the equivalent symbol. The AUGCN1-3C motifs are indicated with brackets and labeled with asterisks. The positions of the GU-rich sequences are indicated by arrows and shaded.
Mutations in either or both of the downstream AUGCN1-3C sequences reduced U1A binding to the secretory poly(A) site.
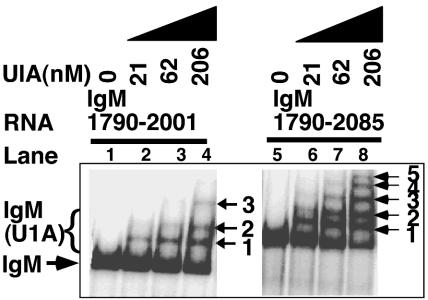
We obtained further evidence for U1A binding sites downstream of the secretory poly(A) site in binding studies. We have previously reported that three U1A molecules bind a substrate spanning the region upstream of the poly(A) site IgM 1790-2001 (20). When this substrate is expanded in a 3′ direction to include the poly(A) site and downstream GU-rich regions (IgM 1790-2085), we obtained five bands representing U1A complexed with RNA (Fig. 3, compare lane 4 [IgM 1790-2001] and lane 8 [IgM 1790-2085]). This suggests five binding sites on the longer substrate and that the extra two binding sites are in the region 2001 to 2085. We examined a series of truncation mutants to determine more precisely the sequences required for the binding of the two extra complexes and were able to discern bands in substrates spanning both regions 1951 to 2030 and 1924 to 2071, suggesting that U1A binds in the 1924 to 2030 region (data not shown). However, these bands were not very intense, suggesting low-affinity binding without the surrounding sequences. As long-range RNA-RNA interactions control the accessibility of the upstream sites (19), RNA outside the 1924 to 2030 region may contribute to the structure of these sites and their accessibility to U1A.
FIG. 3.
Inclusion of sequences downstream of the secretory poly(A) site results in the formation of two extra U1A(IgM) complexes. EMSAs using recombinant U1A and uniformly radiolabeled RNA spanning the indicated sequences. IgM(U1A) complexes are labeled with arrows and numbers representing the number of U1A molecules in each complex.
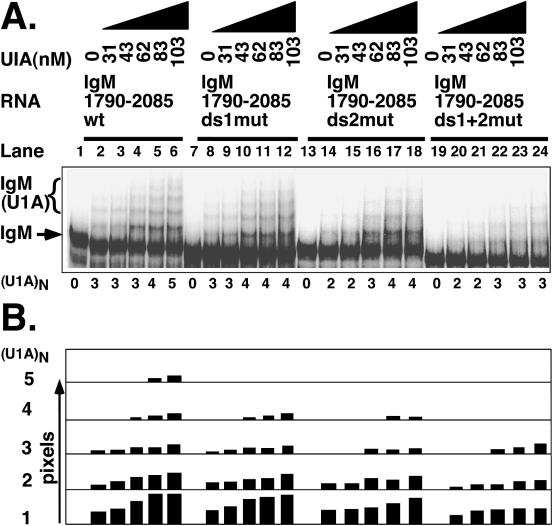
Therefore, to test whether the two AUGCN1-3C motifs within this region are responsible for these additional bands, we introduced mutations into both sequences in the context of the whole IgM 1790-2085 substrate. This region also contains the three upstream U1A binding sites that we described previously. We mutated ds1 from AUGCUC to AAAAUC but ds2 from AUGCCUGC to AUCGGACG, so as not to introduce too many A's in close proximity (see Materials and Methods for primers). We introduced the mutations into the two downstream U1A binding motifs independently or in combination with each other and examined the number of U1A-RNA complexes formed on each substrate. As can be seen in Fig. 4, five complexes are formed with the wild-type substrate (Fig. 4A, lanes 1 to 6; Fig. 4B, phosphorimager quantitation directly below lanes 1 to 6). Three of these are U1A binding the upstream sites and the other two are those found when the substrate was extended into the 2002 to 2085 region in Fig. 3. Mutation of either the ds1 or ds2 U1A binding sites reduced the number of complexes to four in each case (Fig. 4A, lanes 7 to 12 [ds1] and lanes 13 to 18 [ds2]; Fig. 4B, phosphorimager quantitation directly below the equivalent lanes), and both mutations in combination reduced this to three (Fig. 4A, lanes 19 to 24; Fig. 4B, phosphorimager quantitation directly below each lane). Mutations do not sizably reduce the intensity of the complexes which form on nonmutated sites but result in the disappearance of the fifth complex (in the case of the single mutations) or the fourth and fifth complex (in the case of the double mutant). Furthermore, mutation of the two downstream sites in the context of the mutated upstream site abolished the residual complexes (data not shown). We have previously shown that mutation of sequences 100 nt upstream can affect the accessibility and the binding of U1A to the upstream AUGCN1-3C motifs (19). Therefore, we cannot formally rule out the possibility that mutations in the downstream motifs cause conformational changes which allow U1A to bind to other sites. However, the fact that U1A protects these sites and that they match the motifs that we have shown to bind U1A upstream of the poly(A) site are strong arguments that the extra two U1A(RNA) complexes are formed on the two downstream AUGCN1-3C sites.
FIG. 4.
Mutations in either or both of the downstream AUGCN1-3C sequences reduced U1A binding to the secretory poly(A) site in the context of intact upstream U1A binding sites. EMSAs used recombinant U1A and uniformly radiolabeled RNA containing the indicated mutation. (A) Mutation of the downstream U1A binding sites results in fewer IgM(U1A) complexes. Bands representing the unbound substrate (IgM) and the IgM(U1A) complexes are indicated with an arrow or bracket, respectively. The number of complexes formed [(U1A)N] are indicated below each lane. wt, wild type. (B) A phosphorimager quantitation of the bands in panel A. Bars represent pixels (arbitrary units) for each of the five complexes. The number of U1A molecules in the complex is indicated on the left. Each set of bar graphs appears directly below the lane it represents.
U1A binding between the downstream GU-rich regions inhibits secretory poly(A) site expression in vivo.
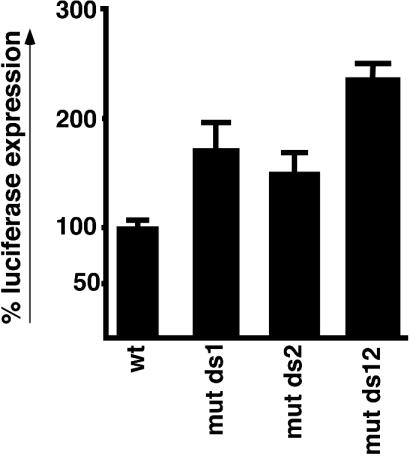
In order to discover a functional role for the downstream U1A, we next examined how mutation of the downstream U1A binding sites affects the expression of the secretory poly(A) site in vivo. For this we used a luciferase reporter system that has been described previously (20-23). We found that mutation of either of the downstream sites enhanced the luciferase activity (Fig. 5, compare wild type with mut ds1 and mut ds2), and in combination this was further enhanced to double that of the wild type (compare wild type with mut ds12). All constructs used in Fig. 5 contained intact upstream U1A binding sites which were previously shown to inhibit expression of the secretory poly(A) site (20). That we obtained such a significant increase in expression upon mutation of the downstream sites even in the context of intact upstream sites suggests that the downstream sites make an important contribution to the inhibition of secretory poly(A) site expression and raises the possibility that they may play a separate role in its regulation.
FIG. 5.
U1A binding between the downstream GU-rich regions inhibits secretory poly(A) site expression in vivo. Luciferase constructs containing the wild-type secretory poly(A) site from position 1790 to 2085 or that containing single (mut ds1 and mut ds2) or double mutations (mut ds12) in the downstream U1A binding sites were transfected into HeLa cells, and luciferase activity was measured 24 h later. Bars represent the mean of triplicates ± SE. Activity was expressed as a percentage of the wild type (wt).
U1A binding between the two GU-rich regions inhibits CstF64K binding.
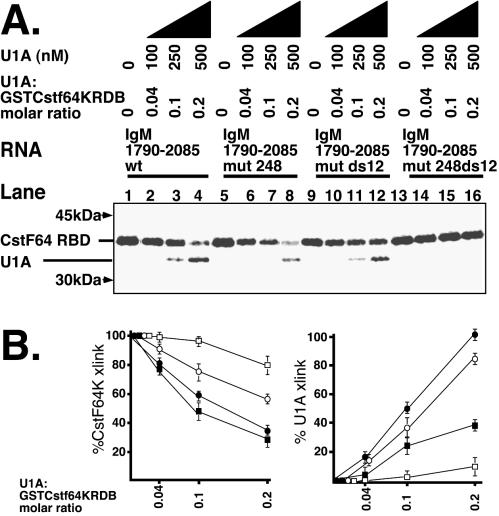
As the downstream U1As are located between the two GU-rich regions which bind CstF64K and the protected regions extended to cover the distal GU-rich region, we next directly assessed the effect of U1A on the binding of CstF64K in UV cross-linking assays. For this we used the N-terminal amino acids 1 to 108 of CstF64K, which spans the RBD, fused with GST (GSTCstF64KRBD), which has been previously shown to cross-link to RNA on its own (26). The full-length CstF64K does not bind to RNA alone but requires the other components of CstF as well as CPSF to stabilize its binding. This is because full-length CstF64K contains domains which partially occlude its binding to RNA, thereby increasing its dependence on the simultaneous binding of CPSF to an AAUAAA motif. In this way, the specificity of the full polyadenylation complex is maintained (26). Takagaki and Manley showed that the RBD of CstF64K binds RNA alone, as it does not include domains which occlude the binding site (26). The RBD alone is sufficient to define a functional downstream element and, in SELEX experiments, selected GU-rich sequences closely matching those present in natural poly(A) sites (26). Thus, the isolated CstF64KRBD provides a convenient tool to investigate the effect of U1A on CstF64 binding to downstream elements.
By titrating the amount of GSTCstF64KRBD, we determined that at 2.5 μM, a cross-link was clearly discernible by phosphorimagery but did not completely saturate the available RNA (4 nM). This is comparable to the 7.5 μM found by Takagaki and Manley to completely shift 1.5 nM 59-nt GU-rich SELEX-selected RNA (26). Furthermore, the assay is internally controlled, as the U1A cross-link is in the same lane as the GSTCstF64KRBD cross-link. We found that the introduction of a very low relative concentration of U1A (0.04 to 0.2 U1A-to-GSTCstF64KRBD molar ratio) inhibits GSTCstF64KRBD binding to the wild-type substrate (Fig. 6A, lanes 1 to 4). This is consistent with the very low binding affinity of GSTCstF64KRBD to RNA shown by Takagaki and Manley compared with the relatively stronger U1A binding to the nonconsensus U1A binding sites in the vicinity of the secretory poly(A) sites (19). Figure 6B shows the quantitation of the results.
FIG. 6.
U1A binding between the two GU-rich regions inhibits CstF64K binding. (A) CstF64K UV cross-linking assays. Uniformly radiolabeled RNA was incubated with recombinant GSTCstF64KRBD (2.5 μM) and increasing concentrations of U1A (as indicated), also expressed as a molar ratio of U1A to GSTCstF64KRBD. Products were cross-linked with UV light, subjected to RNase A digestion, and run on SDS-12% PAGE. wt, wild type. (B) Phosphorimager quantitation of panel A. CstF64K cross-link, results expressed as a percentage of CstF64K binding without added U1A; U1A cross-linking, results expressed a percentage of 500 nM U1A binding the wild-type substrate (lane 4, U1A cross-link). Solid circles, wild type; solid squares, IgM 1790-2085 mut 248; open circles, IgM 1790-2085 mut ds12; open squares, IgM 1790-2085 mut 248 ds12. Data are means of triplicates from three separate cross-linkings ± SE.
Mutating the upstream U1A binding sites (248 mutant) does not reduce the inhibition of the GSTCstF64KRBD cross-link as increasing amounts of U1A are added (Fig. 6A, compare lanes 5 to 8 with 1 to 4; Fig. 6B, compare wild type and IgM 1790-2085 mut 248 for the CstF64K cross-link). From this we conclude that the upstream sites do not contribute greatly to the inhibition of CstF64K binding. Note, however, that the amount of U1A which cross-links to the substrate is reduced, consistent with mutation of three major U1A binding sites (Fig. 6A, compare lanes 5 to 8 with 1 to 4; Fig. 6B, compare wild type and IgM 1790-2085 mut 248 for the U1A cross-link).
In contrast, mutation of the downstream sites (ds12) considerably reduces the inhibitory effect of U1A (Fig. 6A, compare lanes 9 to 12 with 1 to 4; Fig. 6B, compare wild-type with IgM 1790-2085 mut ds12). Note, however, that the total amount of U1A which cross-links to this substrate is intermediate between the wild type and the 248 mutant (Fig. 6A, compare lanes 9 to 12 with 1 to 4 and 5 to 8), consistent with the mutation of two rather than the three U1A binding sites in the case of the 248 mutant.
We note that by mutation of ds1 from AUGCUC to AAAAUC and ds2 from AUGCCUGC to AUCGGACG (mutated UG sequences are in boldface), we have eliminated three UG dinucleotides in the intervening sequence. As CstF64K is known to prefer to bind GU-rich sequences (26), this raises the possibility that these mutations affect the binding affinity of CstF64K to the downstream region. In addition, as the substrate is uniformly labeled by including [32P]UTP in the transcription reaction, elimination of three U's in the intervening sequence may decrease the labeling of the CstF64K cross-link. In fact, a slight diminution in CstF64K binding between the wild type and mut ds12 is seen (Fig. 6, compare lanes 1 and 9). To correct for this, we normalized each cross-linked product obtained after competition with U1A to its respective cross-link obtained with zero U1A (Fig. 6B). In this way, we measured the effect of U1A for each substrate individually. If CstF64K does binds more weakly to the mutant substrate due to the elimination of three UG's in the intervening sequence, a greater effect of U1A competition would be expected, all else being equal. In contrast, we obtained a reduction in the effect of U1A, compounding the result. Taken together, these results show that the downstream U1A has a unique role in inhibiting CstF64K binding: it is not the total amount of U1A that binds the substrate but the binding of U1A to the downstream sites that has the inhibitory effect.
We note, however, that there is still a residual inhibitory effect of U1A even when the downstream sites are mutated which is abolished when the upstream sites are mutated in addition (Fig. 6A, compare lanes 9 to 12 with 13 to 16; Fig. 6B, compare IgM 1790-2085 mut ds12 and IgM 1790-2085 mut 248 ds12 for the CstF64K cross-link). Thus, we do not exclude the possibility that U1A binding upstream of the poly(A) site may have a minor effect on CstF64K binding. Nevertheless, it is clear that the U1A binding between the two GU-rich regions downstream of the poly(A) site exerts the major inhibitory effect on GSTCstF64KRBD binding to this substrate.
U1A binding between the two GU-rich regions inhibits poly(A) cleavage at the secretory poly(A) site.
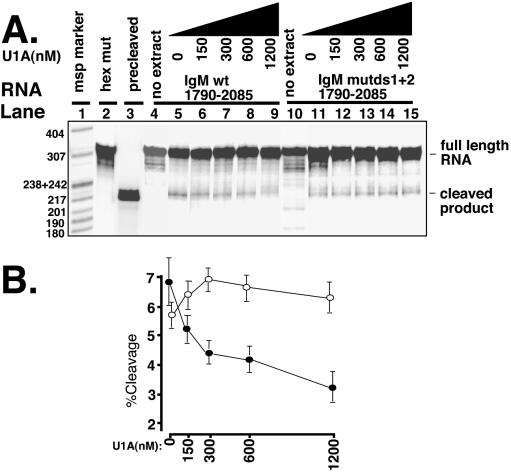
We next examined whether U1A binding between the two downstream GU-rich regions affected cleavage at the secretory poly(A) site. For this we performed in vitro cleavage assays in HeLa cell nuclear extracts (14, 15) (Fig. 7). By including 3′ dATP in the reaction mixture, the addition of a poly(A) tail is prevented and the cleavage product is seen as a discrete band. The unreacted substrate is 305 nt and the cleavage products are 218 and 87 nt. The latter runs off the gel. A substrate containing an extended hexanucleotide mutation (AGA5G2AGA2GA3) (Fig. 7A, lane 2) does not cleave, showing that the cleavage of the wild type and U1A mutants is hexanucleotide specific (lanes 5 and 11). Unreacted wild-type and mutated substrates are included as controls (lanes 4 and 10, respectively). An in vitro-transcribed precleaved substrate is included as a reference (lane 3). Under these conditions, we achieved 6.9% ± 0.8% standard error (SE) (lane 5) cleavage of the wild-type substrate and 5.7% ± 0.4% SE cleavage of the mutated substrate (lane 11), which compares to 32% cleavage of an adenovirus L3 substrate under the same conditions (data not shown). Addition of increasing concentrations of U1A significantly inhibits cleavage of the wild-type substrate (Fig. 7A, lanes 5 to 9; Fig. 7B, wild type). In contrast, when the U1A binding sites between the two GU-rich regions are mutated, addition of U1A did not inhibit cleavage and a slight enhancement was seen, although this was not a significant effect (Fig. 7A, lanes 11 to 15; Fig. 7B, mutated substrate). Similarly, U1A had no significant effect on the adenovirus L3 substrate (data not shown). Interestingly, increasing concentrations of U1A have a biphasic effect on cleavage (Fig. 7B, wild type). Lower concentrations of U1A produce an immediate effect on cleavage which then plateaus into a slower decline. This, taken together with the footprinting results in Fig. 2 (which showed that U1A protected the distal GU-rich region and not the proximal GU-rich region), is consistent with U1A blocking one GU-rich element more efficiently than the other.
FIG. 7.
U1A binding between the two GU-rich regions inhibits cleavage at the secretory poly(A) site. Uniformly radiolabeled RNA was incubated with increasing concentrations of recombinant U1A and HeLa cell extracts for 2 h at 30°C. After proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation, products were run on 8% denaturing PAGE. The full-length RNA and cleavage products are indicated on the right. The controls were an extended hexanucleotide mutant spanning position 1790 to 2085 which did not cleave in HeLa cell extracts (lane 2) and a precleaved RNA substrate (position 1790 to 1998) as a marker of the correct position for the cleaved product (lane 3). Lanes 4 and 10 are input RNA without nuclear extracts. wt, wild type. (B) Phosphorimager quantitation of panel A. Results are expressed as percent cleavage. Solid squares are the quantitation of the wild type (lanes 5 to 9) with increasing concentrations of recombinant U1A; open circles represent the same for the substrate containing mutations in the downstream U1A (lanes 11 to 15). Data are means of triplicates from three separate cleavage assays ± SE.
We note a second band that appears above the indicated cleavage product and is more highly represented in the mut ds12 than in the wild type in the image chosen for this figure. This band was present to a greater or lesser degree in the other images which were quantitated to form Fig. 7B and appeared in wild type and mutant alike. We cannot rule out the possibility that this is an alternative cleavage site 8 to 12 nt upstream of the mapped cleavage site but favor the interpretation that it is a nuclease site: it is close to a nuclease site which appears in the unreacted substrate which is obscured by the more intense site directly above it, and its appearance correlated with the age of the substrate. From the data discussed above, we conclude that U1A binding between the two GU-rich downstream elements inhibits cleavage at the secretory poly(A) site.
DISCUSSION
We have identified two U1A binding sites downstream of the secretory poly(A) site between the two GU-rich regions which have an inhibitory effect on the expression of the μ-secretory poly(A) site. These inhibit the binding of CstF64K, which binds the GU-rich sequences and subsequent cleavage at the secretory poly(A) site.
U1A may selectively block the distal GU-rich region.
We found that U1A's footprint extends into the distal GU-rich element which raises the possibility that U1A binds within this region also. However, mutation of the two AUGCN1-3C motifs completely abolished the inhibitory effect of U1A, demonstrating that these motifs play the major role in inhibiting cleavage. These motifs show a clustering in this vicinity of much greater than expected frequency, suggesting specific targeting to this region (20). We therefore favor the model that U1A binding the AUGCN1-3C motifs protects the adjacent distal GU-rich region in addition. We cannot rule out the possibility that U1A binds the GU-rich region and requires U1A to bind the ds12 motifs to stabilize its binding to the GU-rich region. This would be difficult to test directly, as mutating the GU-rich element would affect CstF64K binding, making interpretation of the results inconclusive. Nevertheless, the footprint extending into the distal GU-rich region rather than the proximal GU-rich region suggests that U1A blocks CstF64K's access to the distal rather than the proximal GU-rich region.
It has previously been shown that deletion of the distal downstream GU-rich element reduced but did not eliminate expression in vivo (23). Interestingly, increasing concentrations of U1A have a biphasic effect on cleavage (Fig. 7). Low concentrations of U1A produce an immediate effect on cleavage which then plateaus into a slower decline consistent with U1A blocking one GU-rich element more efficiently than the other. Thus, the effect of U1A on cleavage may be to eliminate the contribution of the distal GU-rich region to polyadenylation efficiency.
U1A as a negative regulator of CstF64K binding.
The downstream U1As are located between the two GU-rich regions which are both necessary for full polyadenylation activity at this site. We showed that a relatively small amount of U1A can inhibit CstF64K binding to this substrate and that the downstream U1As have the major effect on CstF64K binding. A negative regulator of the activation of the secretory poly(A) site has been detected previously (30). The binding of that factor was mapped to between the hexanucleotide sequence and the GU-rich regions, the location which we now discover for U1A. U1A as a negative regulator of CstF64K binding would explain why the activity of the secretory poly(A) site has been ranked as one of the weakest poly(A) sites despite its prominent GU-rich regions (25).
Regulation of U1A during B-cell differentiation.
U1A was first identified as a component of U1 snRNP involved in splicing. It was subsequently shown that it could autoregulate its production by preventing poly(A) tail addition and thus allowing degradation of its own mRNA (2). This could imply that U1A that is not U1 snRNP bound would quickly shut off its own production and that the amount of snRNP-free U1A would be severely limited. However, we showed that U1A also binds μ-secretory mRNA via novel U1A binding sites and similarly regulates poly(A) addition. We have found evidence that these novel U1A binding sites are used by other genes which follow the same pattern of regulation (20), thus demonstrating the existence of a pool of U1A-binding pre-mRNA in the nucleus. As U1A can be imported into the nucleus independent of U1 snRNP (reviewed in reference 9), this U1A would presumably be available to bind either U1 snRNP or pre-mRNA. U1A bound by heterologous mRNAs would be unavailable to U1A's own mRNA, and autoregulation would be avoided. Other mechanisms that prevent autoregulation may also exist. For instance, U1A has been found free of U1 snRNP in large complexes which can be immunoprecipitated by antibodies which recognize an epitope that is buried when U1A binds U1 RNA (17). This complex may target the free U1A to mRNA. However, once U1A binds mRNA, it will presumably no longer be recognized by this antibody.
By using two antibodies that recognize different epitopes on the U1A molecule, one hidden when U1A binds RNA (12E12) and one which is exposed whether or not U1A is bound to RNA (10E3), Milcarek et al. found that approximately 16% of U1A in human B cells is not bound by RNA, and this does not change upon differentiation (13). However, the total amount of U1A (recognized by 10E3) was significantly reduced upon differentiation relative to other snRNP proteins, suggesting a redistribution of U1A between the U1 snRNP and mRNA pools. They concluded that this changing level of U1A may be important for influencing Ig heavy-chain mRNA processing. Furthermore, we have evidence that both the total level of U1A and the proportion that is not snRNP bound is raised in undifferentiated cells (our unpublished data). Thus U1A may be an important regulator of the expression of the secretory poly(A) site both at the level of cleavage and poly(A) addition.
U1A may regulate a group of poly(A) sites coordinately during differentiation.
Other factors have been shown to be involved in regulating the activation of the secretory poly(A) site. It is likely that a number of mechanisms would be employed to control a poly(A) site, whose expression could unleash pathogenic amounts of antibodies of poor or dangerous specificity. It has been shown that B cells from transgenic mice containing non-Ig genes modified to have an Ig gene-like structure with the same balance and positioning of two poly(A) sites and an intron show an upregulation in the usage of the proximal poly(A) site upon stimulation with lipopolysaccharide (24). Thus, changes in the amounts or activities of general RNA processing components play a role in activating the secretory poly(A) site. However, those experiments consistently showed at least a twofold higher level of activation of the endogenous secretory poly(A) site above the introduced proximal non-Ig poly(A) site in eight different experiments with cells from three lines of transgenic mice (Fig. 2 and Table 1 of reference 24). This shows that additional factors bind sequences in the vicinity of the secretory poly(A) site to regulate the activation of the secretory poly(A) site over and above the introduced gene. A number of factors may coordinately regulate specific groups of poly(A) sites which contain binding sites for these factors. For instance, the ratio of hnRNP F to H or H′ changes between memory and plasma stage B cells and may affect the ability of CstF64K to bind the secretory poly(A) site (29). Here we have shown that U1A inhibits the binding of CstF64K to the secretory poly(A) site by binding between the two downstream GU-rich regions. U1A may regulate other poly(A) sites in a similar manner. Thus, changing amounts of U1A available to interact with mRNA during differentiation could regulate a specific U1A binding group of poly(A) sites coordinately.
Acknowledgments
We thank Yoshio Takagaki for the GSTCstF64KRBD plasmid, Chris Milcarek and Ravinder Singh for advice, and Steve Jung for technical assistance.
The work was supported by AHA 0430004N Scientist Development Grant to C. Phillips and NIH GM57286 to S. Gunderson.
REFERENCES
- 1.Berberich, I., and A. Schimpl. 1990. Regulation of Ig gene expression in normal lymphocytes. I. The half-life of secreted mu chain mRNA differs from that of membrane mu chain mRNA in resting and activated B cells. Eur. J. Immunol. 20:445-448. [DOI] [PubMed] [Google Scholar]
- 2.Boelens, W. C., E. J. R. Jansen, W. J. van Venrooij, R. Stripecke, I. W. Mattaj, and S. I. Gunderson. 1993. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell 72:881-892. [DOI] [PubMed] [Google Scholar]
- 3.Cox, A., and J. S. Emtage. 1989. A 6-fold difference in the half-life of immunoglobulin m heavy chain mRNA in cell lines representing two stages of B cell differentiation. Nucleic Acids Res. 17:10439-10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early, P., J. Rogers, M. Davis, K. Calame, M. Bond, R. Wall, and L. Hood. 1980. Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell 20:313-319. [DOI] [PubMed] [Google Scholar]
- 5.Edwalds-Gilbert, G., and C. Milcarek. 1995. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 15:6420-6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwalds-Gilbert, G., K. L. Veraldi, and C. Milcarek. 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25:2547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehresmann, C., F. Baudin, M. Mougel, P. Romby, J. P. Ebel, and B. Ehresmann. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15:9109-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gunderson, S. I., S. Vagner, M. Polycarpou-Schwarz, and I. W. Mattaj. 1997. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 11:761-773. [DOI] [PubMed] [Google Scholar]
- 9.Kuersten, S., M. Ohno, and I. W. Mattaj. 2001. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11:497-503. [DOI] [PubMed] [Google Scholar]
- 10.Lamson, G., and M. E. Koshland. 1984. Changes in J-chain and μ-chain RNA expression as a function of B-cell differentiation. J. Exp. Med. 160:877-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martincic, K., R. Campbell, G. Edwalds-Gilbert, L. Souan, M. T. Lotze, and C. Milcarek. 1998. Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc. Natl. Acad. Sci. USA 95:11095-11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mason, J. O., G. T. Williams, and M. S. Neuberger. 1988. The half-life of immunoglobulin mRNA increases during B-cell differentiation: a possible role for targeting to membrane-bound polysomes. Genes Dev. 2:1003-1010. [DOI] [PubMed] [Google Scholar]
- 13.Milcarek, C., K. Martincic, L.-H. Chung-Ganster, and C. S. Lutz. 2003. The snRNP-associated U1A levels change following IL-6 stimulation of human B-cells. Mol. Immunol. 39:809-814. [DOI] [PubMed] [Google Scholar]
- 14.Moore, C. L., and P. A. Sharp. 1985. Accurate cleavage and polyadenylation of exogenous RNA substrate. Cell 41:845-855. [DOI] [PubMed] [Google Scholar]
- 15.Moore, C. L., and P. A. Sharp. 1984. Site-specific polyadenylation in a cell-free reaction. Cell 36:581-591. [DOI] [PubMed] [Google Scholar]
- 16.Murthy, K. G., and J. L. Manley. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672-2683. [DOI] [PubMed] [Google Scholar]
- 17.O'Connor, J. P., J. C. Alwine, and C. S. Lutz. 1997. Identification of a novel, non-snRNP protein complex containing U1A protein. RNA 3:1444-1455. [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson, M. L., E. R. Gimmi, and R. P. Perry. 1991. The developmentally regulated shift from membrane to secreted m mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol. Cell. Biol. 11:2324-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips, C., and S. I. Gunderson. 2003. Sequences adjacent to the 5′ splice site control U1A binding upstream of the IgM heavy chain secretory poly(A) site. J. Biol. Chem. 278:22102-22111. [DOI] [PubMed] [Google Scholar]
- 20.Phillips, C., S. Jung, and S. I. Gunderson. 2001. Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J. 20:6443-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips, C., C. B. Kyriakopoulou, and A. Virtanen. 1999. Identification of a stem-loop structure important for polyadenylation at the murine IgM secretory poly(A) site. Nucleic Acids Res. 27:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phillips, C., A. Schimpl, W. Dietrich-Goetz, J. B. Clements, and A. Virtanen. 1996. Inducible nuclear factors binding the IgM heavy chain pre-mRNA secretory poly(A) site. Eur. J. Immunol. 26:3144-3152. [DOI] [PubMed] [Google Scholar]
- 23.Phillips, C., and A. Virtanen. 1997. The murine IgM secretory poly(A) site contains dual upstream and downstream elements which affect polyadenylation. Nucleic Acids Res. 25:2344-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seipelt, R. L., B. T. Spear, E. C. Snow, and M. L. Peterson. 1998. A nonimmunoglobulin transgene and the endogenous immunoglobulin mu gene are coordinately regulated by alternative RNA processing during B-cell maturation. Mol. Cell. Biol. 18:1042-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takagaki, Y., and J. L. Manley. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2:761-771. [DOI] [PubMed] [Google Scholar]
- 26.Takagaki, Y., and J. L. Manley. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17:3907-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takagaki, Y., R. L. Seipelt, M. L. Peterson, and J. L. Manley. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941-952. [DOI] [PubMed] [Google Scholar]
- 28.van Gelder, C. W. G., S. I. Gunderson, E. J. R. Jansen, W. C. Boelens, M. Polycarpou-Schwarz, I. W. Mattaj, and W. J. van Venrooij. 1993. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J. 12:5191-5200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veraldi, K. L., G. K. Arhin, K. Martincic, L. H. Chung-Ganster, J. Wilusz, and C. Milcarek. 2001. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 21:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan, D.-H., E. A. Weiss, and J. R. Nevins. 1995. Identification of an activity in B-cell extracts that selectively impairs the formation of an immunoglobulin μs poly(A) site processing complex. Mol. Cell. Biol. 15:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]