Significance
We report a novel linear mixed-linkage (1→3)(1→4)-β-glucan produced by the bacterium Sinorhizobium meliloti upon raising cyclic diguanylate (c-di-GMP) intracellular levels. This unique bacterial polysaccharide resembles (1→3)(1→4)-β-glucans found in cereals and certain lichens but has a distinctive primary structure. Genes, proteins, and regulatory pathways for producing this new polymer are described. Our findings open the possibility of using bacteria to produce (1→3)(1→4)-β-glucans, which are receiving increasing interest as bioactive compounds, and provide new elements to disclose molecular mechanisms of c-di-GMP regulation as well as for investigating the evolution, activity, and specificity of glycosyl transferases.
Keywords: exopolysaccharides, cyclic diguanylate, plant–microbe interactions
Abstract
An artificial increase of cyclic diguanylate (c-di-GMP) levels in Sinorhizobium meliloti 8530, a bacterium that does not carry known cellulose synthesis genes, leads to overproduction of a substance that binds the dyes Congo red and calcofluor. Sugar composition and methylation analyses and NMR studies identified this compound as a linear mixed-linkage (1→3)(1→4)-β-d-glucan (ML β-glucan), not previously described in bacteria but resembling ML β-glucans found in plants and lichens. This unique polymer is hydrolyzed by the specific endoglucanase lichenase, but, unlike lichenan and barley glucan, it generates a disaccharidic →4)-β-d-Glcp-(1→3)-β-d-Glcp-(1→ repeating unit. A two-gene operon bgsBA required for production of this ML β-glucan is conserved among several genera within the order Rhizobiales, where bgsA encodes a glycosyl transferase with domain resemblance and phylogenetic relationship to curdlan synthases and to bacterial cellulose synthases. ML β-glucan synthesis is subjected to both transcriptional and posttranslational regulation. bgsBA transcription is dependent on the exopolysaccharide/quorum sensing ExpR/SinI regulatory system, and posttranslational regulation seems to involve allosteric activation of the ML β-glucan synthase BgsA by c-di-GMP binding to its C-terminal domain. To our knowledge, this is the first report on a linear mixed-linkage (1→3)(1→4)-β-glucan produced by a bacterium. The S. meliloti ML β-glucan participates in bacterial aggregation and biofilm formation and is required for efficient attachment to the roots of a host plant, resembling the biological role of cellulose in other bacteria.
In bacteria the second messenger cyclic diguanylate (c-di-GMP) regulates the transition from a motile to a sedentary lifestyle, so that high concentrations enhance the production of extracellular matrix components, exopolysaccharides (EPSs), and proteins (1–5). The c-di-GMP is synthesized from two molecules of GTP by the action of diguanylate cyclases (DGCs) and is hydrolyzed to 5′-phosphoguanylyl-(3′-5′)-guanosine by specific phosphodiesterases. It has been determined that artificial increments of the intracellular levels of c-di-GMP by the overexpression of DGCs often trigger phenotypes related to the synthesis of EPSs and biofilm formation (5, 6).
Rhizobia produce a complex array of glucidic molecules including LPSs, capsular polysaccharides (CPSs), cyclic glucans, high-molecular-weight neutral polysaccharide (glucomannan), gel-forming polysaccharide, and EPSs (7–9). In addition, these symbiotic bacteria secrete other compounds, such as lipo-chitin oligosaccharides, which specifically mediate the interaction with their legume hosts (10).
In contrast to structural polysaccharides (i.e., CPSs or LPSs), EPSs are weakly associated with the cell outer surface and often are released into the environment (8). EPSs are either branched or linear homo- or heteropolymers of a wide variety of d-monosaccharides and form a highly hydrated layer on the cell surface, providing protection against environmental biotic and abiotic stresses. EPSs also play a major role in cell aggregation, flocculation, and biofilm formation, critical processes in the initial stages of bacterial interaction with eukaryotic hosts (11, 12). Finally, many rhizobia also use specific EPSs as signaling molecules that are indispensable for the invasion of host plants (13, 14).
Sinorhizobium meliloti, an endosymbiont of alfalfa (Medicago sativa), produces two structurally distinct EPSs: EPS-I, or succinoglycan, and EPS-II, or galactoglucan; both are regulated by the ExpR/Sin quorum-sensing system and are important for nodulation of legume hosts (13, 15, 16). However, in contrast to many members of the Rhizobiaceae family, the model strain S. meliloti 8530 (Sme) lacks the gene clusters required for the biosynthesis of other common d-glucose homopolysaccharides such as cellulose or curdlan. Two types of cellulose synthase (CS) systems have been described in bacteria. Group I is found in the α, β, and γ subdivisions and is encoded by a three-gene operon bcsABC/acsABC, plus a glycosylhydrolase gene which can be linked to the main operon or located elsewhere in the genome (17, 18). Group II typically is found in α-Proteobacteria, such as Agrobacterium tumefaciens, and is encoded by two adjacent operons: celABC, homologs to group I bcsA, bcsB and the glycosylhydrolase, respectively, and celDE(FG) with no significant sequence similarity to group I genes (19). A. tumefaciens and other Rhizobiales also carry genes for the biosynthesis of curdlan (crdASC, crdR, and pssAG), another high-molecular-weight linear β-glucan composed of d-glucose residues linked by β(1→3) bonds instead of the β(1→4) bond found in cellulose (20). Production of both curdlan and cellulose seem to be activated by the second messenger c-di-GMP, although only the allosteric activation of CSs through c-di-GMP binding to a PilZ domain, located in the C-terminal region of BcsA/CelA proteins, has been demonstrated (21–23). A PilZ domain apparently is absent in curdlan synthases (24); nonetheless c-di-GMP activation could occur by a different mechanism, because this second messenger is able to regulate the production of other EPSs by interacting with proteins involved in EPS extrusion (25, 26). Here we have exploited c-di-GMP activation of bacterial EPSs to unveil an undisclosed linear mixed-linkage (ML) (1→3)(1→4)-β-glucan in a cellulose-nonproducing bacterium, S. meliloti 8530.
Results
Effects of Increased c-di-GMP Levels on the Production of EPSs by S. meliloti 8530.
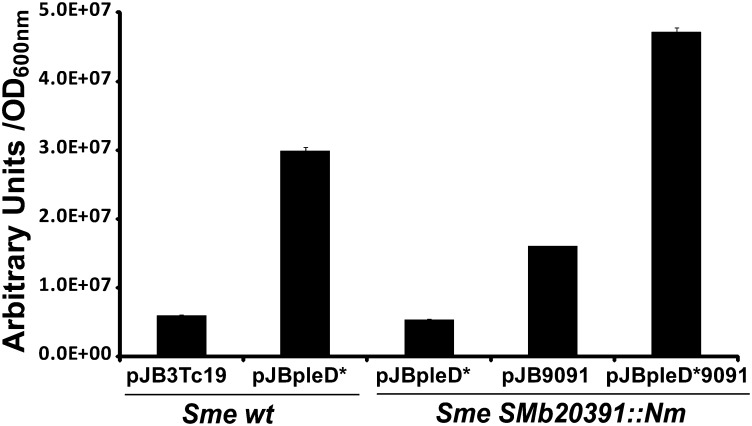
In cellulose-producing (Cel+) bacteria, raising c-di-GMP intracellular levels (i.e., by overexpression of a DGC) triggers production of cellulose and other extracellular molecules (5, 6, 27). S. meliloti 8530 does not bear any of the known CS operons in its genome; however, overexpression of the DGC PleD* generated wrinkled and red-stained colonies in medium with Congo red (CR), in deep contrast to the wild-type colonies (SI Appendix, Fig. S1A). Furthermore, the overexpression of pleD* (pJBpleD*) also increased the calcofluor (CF)-derived fluorescence in both solid and liquid cultures (Fig. 1 and SI Appendix, Fig. S1B).
Fig. 1.
Quantification of CF-derived fluorescence of S. meliloti 8530 and its derivative Smb20391::Nm mutant harboring different plasmid constructions. Results are expressed in arbitrary units standardized by the OD600 ± SE from three independent cultures.
In liquid media and particularly in minimal medium (MM), Sme overexpressing pleD* (Sme pJBpleD*) displayed a strong aggregative behavior with most bacteria forming flocs (SI Appendix, Fig. S2A). Scanning electron microscopy of flocs showed that bacteria were encased in a dense matrix of filaments (SI Appendix, Fig. S2B). Adherence to and biofilm formation on test tube walls also were enhanced (SI Appendix, Fig. S3A); moreover, those biofilms fluoresced in the presence of CF under UV light (SI Appendix, Fig. S3B), indicating that the CF+ substance was part of the biofilm matrix. However, biofilm formation could not be quantified because it detached easily from the abiotic surfaces and was removed during the Cristal Violet staining process (Materials and Methods). The PleD*-dependent EPS matrix produced by Sme also enhanced its adhesion to alfalfa roots and the formation of a thick CF+ biofilm coating the root surface (SI Appendix, Fig. S3C). All these phenotypes correlated with a strong increase in the c-di-GMP intracellular levels provoked by PleD*. Cell extracts from Sme harboring the plasmid pJBpleD* contained 596.86 pmol of c-di-GMP/mg total protein, whereas the amounts of c-di-GMP from Sme carrying the pJB3Tc19 empty vector were below the HPLC-MS limit of detection (SI Appendix, Supplementary Methods).
Characterization of a Novel β-Glucan Produced by S. meliloti.
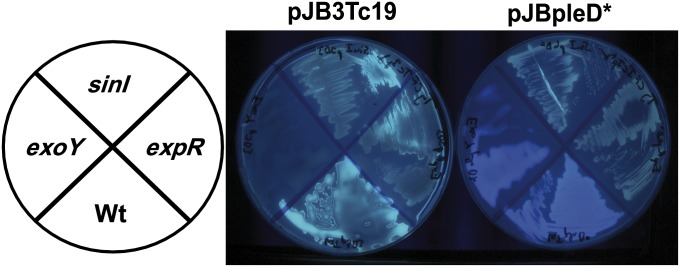
CR binds basic or neutral polysaccharides as well as some proteins, whereas CF is more specific for polymers containing β (1-3)- or β (1-4)-d-glucopyranosyl units (28, 29). Sme produces at least one EPS, known as “EPS-I” or “succinoglycan,” which binds CF but not CR (30, 31). Indeed, Sme shows a greenish CF-derived fluorescence; however pJBpleD* conferred a blue CF fluorescence (Fig. 2) which was retained in an exoY mutant lacking EPS-I (32). This finding suggested that high c-di-GMP levels generated by PleD* stimulated the production of a CF-binding polymer different from EPS-I.
Fig. 2.
CF-derived fluorescence of S. meliloti 8530 (Wt) and mutant derivatives carrying the plasmid pJBpleD* or the empty vector pJB3Tc19. (Right) Bacterial cultures were imaged under UV light after growth at 28 °C in solid TY medium supplemented with CF (200 µg/mL). (Left) The diagram depicts the positions of the strains.
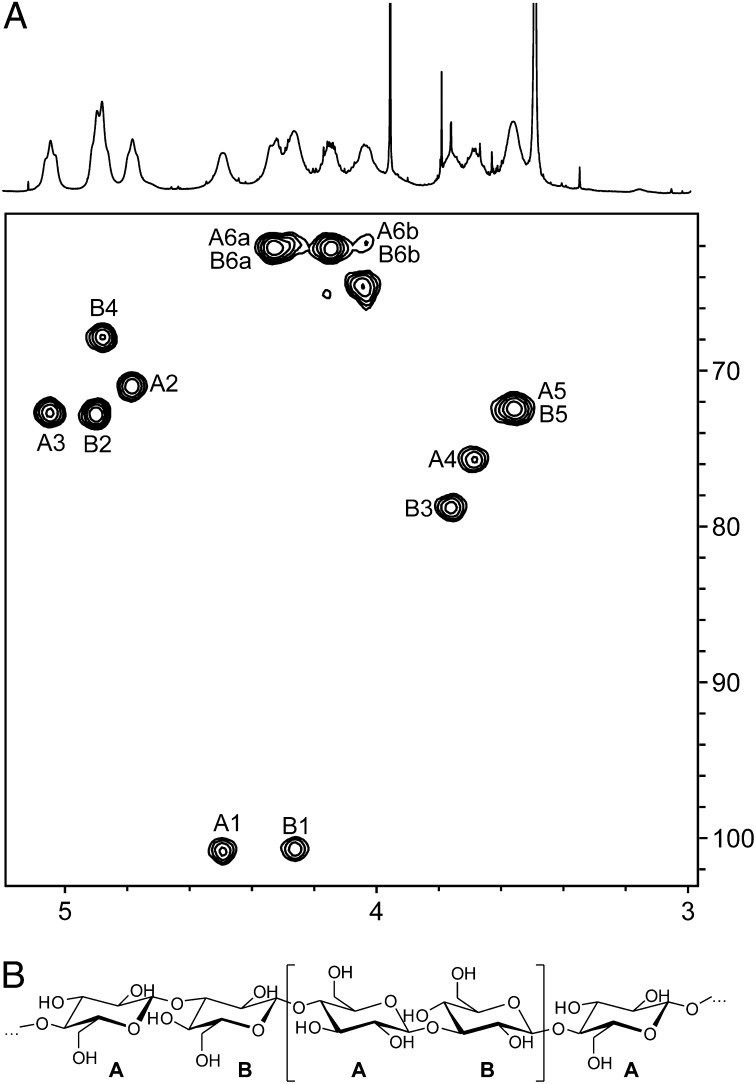
A purification approach was designed to isolate and identify this substance. The Sme pJBpleD* strain was grown in flasks with liquid MM, and the flocs generated were recovered after 3 d using a sieve (Materials and Methods). Like curdlan and cellulose, the Sme substance was water insoluble, but, unlike cellulose, it could be dissolved in diluted NaOH solutions (0.5 M). Curdlan is soluble in dilute alkali (0.25 M NaOH) and, after aqueous suspensions are heated above 80 °C and subsequently cooled, produces high-set thermo-irreversible gels (33). This gelling behavior was not observed with the Sme substance, so several rounds of boiling/cooling/centrifugation cycles were followed by a lyophilization step to obtain a substance with a cottony appearance. Monosaccharide analysis showed that d-glucose was the only component, and the methylation analysis indicated that these glucopyranoses were substituted at positions →3 and →4) in a 1:1 ratio. The polysaccharide was insoluble in water or in DMSO; therefore a few milligrams were treated with a mixture of acetic anhydride and pyridine to acetylate its hydroxyl groups and improve its solubility in organic solvents. After purification by size-exclusion chromatography (SEC), a fraction of the polysaccharide was solubilized in CDCl3 and was studied by NMR. Assignments were made using bidimensional experiments [DQF-COSY, total correlated spectroscopy (TOCSY), rotating-frame Overhauser spectroscopy (ROESY), and heteronuclear single-quantum coherence (HSQC)] and are listed in Table 1. NMR spectra showed two different units of glucose, labeled as “unit A” and “unit B,” with a ratio close to 1:1. Chemical shifts (1H and 13C) indicated their β configuration and that they were present in the pyranose form (Fig. 3A). Moreover, unit A was substituted at position →4), whereas unit B was substituted at position →3). The results were in good agreement with the chemical shifts of acetylated cellulose (34) and (1→3)-β-d-glucopentaose peracetate (35). Finally, the ROESY spectrum (SI Appendix, Fig. S4) allowed the sequence of residues in the polysaccharide to be deduced, because intense cross-peaks were found between signals H-1 in unit A and H-3 in unit B and between H-1 in unit B and H-4 in unit A. Considering all these data, we propose that the acetylated polysaccharide has the repeating unit →3)-β-d-Glcp-(1→4)-β-d-Glcp-(1→ (Fig. 3B).
Table 1.
Chemical shifts and assignments of acetylated ML β-glucan purified from S. meliloti 8530 overexpressing pleD* (Sme pJBpleD*)
| Position | ||||||||
| Unit | Nucleus | 1 | 2 | 3 | 4 | 5 | 6a | 6b |
| A: →4)-β-d-Glcp-(1→ | 1H | 4.49 | 4.78 | 5.05 | 3.69 | 3.56 | 4.3 | 4.1 |
| 13C | 100.9 | 71.0 | 72.7 | 75.7 | 72.4 | 62.2 | ||
| B: →3)-β-d-Glcp-(1→ | 1H | 4.26 | 4.9 | 3.76 | 4.88 | 3.56 | 4.3 | 4.1 |
| 13C | 100.7 | 72.8 | 78.8 | 67.8 | 72.4 | 62.2 | ||
Fig. 3.
Structure of the ML β-glucan. (A) 1H-NMR (500 MHz) and 1H-13C (125 MHz) HSQC spectra of the acetylated ML β-glucan purified from S. meliloti 8530 overexpressing pleD* (Sme pJBpleD*). (B) Proposed structure of the ML β-glucan. The repeating unit is indicated by brackets.
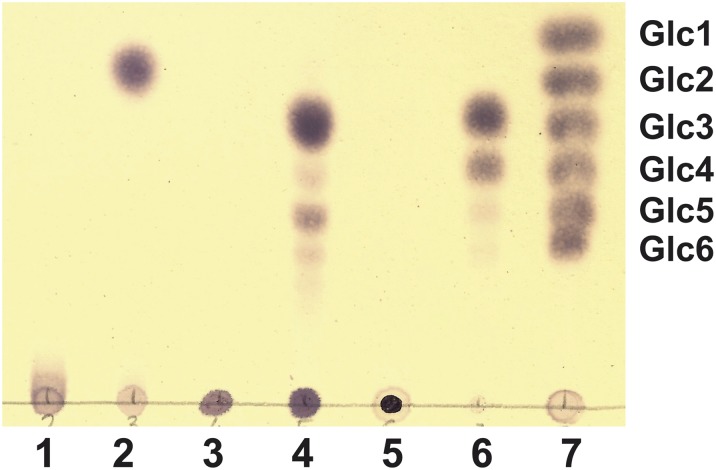
This β-glucan structure resembles ML β-glucans such as lichenan and barley β-glucan, which can be hydrolyzed specifically by lichenase, an endohydrolase whose target site is widely assumed to be all (1→4) bonds immediately following (1→3) bonds (36). It does not hydrolyze pure (1→3)-β-d-glucans (i.e., curdlan) nor (1→4)-β-d-glucans (cellulose). Incubation of lichenan and barley β-glucan with lichenase produced distinctive mixtures of tri-, tetra-, and larger cello-oligosaccharides which could be separated by TLC (Fig. 4), in agreement with previous reports (37). Lichenase also was able to hydrolyze the S. meliloti glucan, although in this case the only product of hydrolysis was a disaccharide (Fig. 4). This result agreed with the NMR data and further supported the notion that the repeating unit of the Sme β-glucan contains alternating β(1→3) and β(1→4) bonds (Fig. 3B).
Fig. 4.
TLC of β-glucans hydrolyzed with lichenase. Lane 1: untreated S. meliloti ML β-glucan; lane 2: S. meliloti ML β-glucan incubated with lichenase; lane 3: untreated lichenan; lane 4: lichenan + lichenase; lane 5: untreated barley β-glucan; lane 6: barley β-glucan + lichenase; lane 7: mixture of glucose, maltose, maltotriose, maltotetraose, maltopentaose, and maltohexaose.
The bgsBA Operon Is Required for ML β-Glucan Production.
The S. meliloti 8530 genome encodes at least two glycosyl transferases (GTs) of unknown function, SMb20391 and SMb202460, annotated as hypothetical CSs (38). The PleD*-dependent phenotypes were tested with transposon-induced mutants in both genes (SI Appendix, Table S1). The SMb20460 mutant behaved like the wild type; however, the SMb20391 mutant had lost the PleD*-dependent CR+ and CF+ phenotypes (SI Appendix, Fig. S5) and the aggregative behavior observed in the wild type.
The SMb20391 ORF overlaps by 19 bp the upstream ORF SMb20390, which encodes a putative membrane fusion protein (MFP) belonging to the AcrA/EmrA/HlyD family of transporters. RT-PCR experiments indicated that SMb20390 and SMb20391 are cotranscribed (SI Appendix, Fig. S6A).
An in-frame deletion of SMb20390 (ΔSMb20390) was constructed, and, like the SMb20391 mutant, the ΔSMb20390 pJBpleD* mutant showed a CR− and CF− phenotype and did not aggregate or form biofilm at high c-di-GMP levels. This result indicated that both the SMb20390 and SMb20391 genes are responsible for the c-di-GMP–induced phenotypes observed in Sme. This notion was confirmed further after genetic complementation. To avoid possible functional problems resulting from an imbalance between SMb20390 and SMb20391 protein levels, two new plasmids were constructed, one harboring the complete operon SMb20390-91 cloned in pJB3Tc19 (pJB9091) and a second one carrying SMb20390-91 and pleD* (pJBpleD*9091) (SI Appendix, Fig. S7). Both plasmid constructions were able to restore the CR+ and CF+ phenotypes in the SMb20391::Nm mutant (Fig. 1). Although the CF-derived fluorescence was much higher when pleD* was present, the two genes in multicopy were able to provide CR+ and CF+ phenotypes even under physiological c-di-GMP conditions. We also measured the relative yield of ML β-glucan in cultures, which represented 47.8% of the dry weight of the wild-type Sme (pJBpleD*) cultures. As expected, no ML β-glucan was produced by the SMb20391::Nm mutant overexpressing PleD*. The SMb20391::Nm mutant complemented with pJB9091 displayed a low (2.1% of culture dry weight) but nevertheless measurable yield of ML β-glucan. The ML β-glucan yield was highest in cultures of the mutant carrying both pleD* and SMb20390-20391 genes in multicopy (pJBpleD*9091), accounting for 77.5% of the culture dry weight. Therefore, two conditions seem to be required for high production of the ML β-glucan by Sme: (i) the presence of a functional copy of the SMb20390-91 operon and (ii) high intracellular levels of c-di-GMP. Given their requirement for ML β-glucan production, we propose that SMb20391 be renamed bgsA (with “bgs” standing for “β-glucan synthesis”) and that SMb20390 be renamed “bgsB.”
c-di-GMP Binding to BgsA.
The bgsB gene product contains an HlyD-like domain found in diverse proteins involved in the export of a broad variety of compounds (39) and shows homology to MFPs, accessory proteins that cooperate with a variety of transporters. MFPs span the periplasm, linking the inner and outer membranes and facilitating the formation of a channel to expel the substrate from the cell (40).
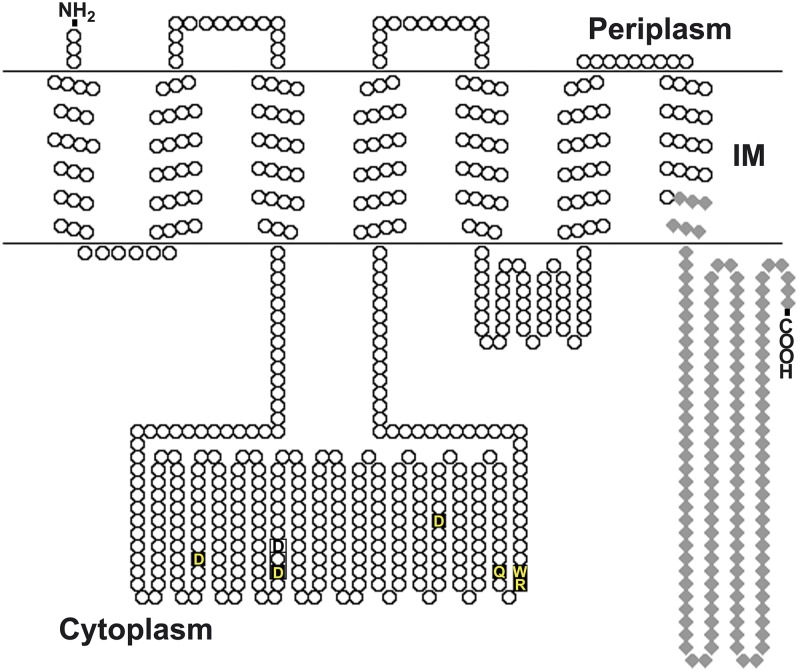
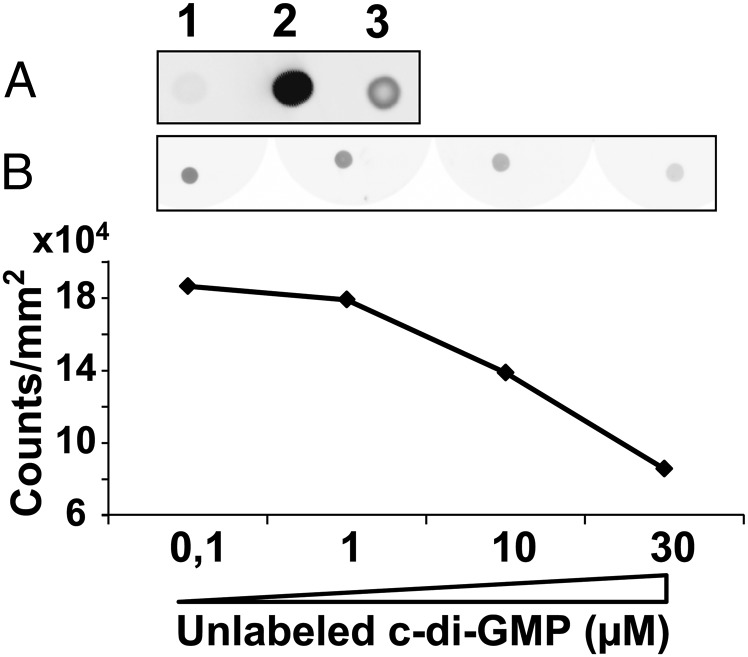
Hydropathy analysis of the 664-aa BgsA protein sequence using the TMHMM 2.0 prediction program (41) revealed seven potential transmembrane (TM) helices (Fig. 5), in contrast to the eight TMs present in CSs (42, 43). The large hydrophilic region between TM III and IV (amino acid residues 90–375) contains residues and motifs conserved in the GT-2 glycosyl transferases family (D,D,D35QxxRW), including the DXD signature involved in the addition of the UDP-sugar to the nascent polysaccharide (44) and the QXXRW sequence motif likely required for holding the growing glycan chain in the active site (Fig. 5) (45). After TM VII a C-terminal cytoplasmic segment was predicted (residues 532–664) which contains no known functional domains. This protein organization strongly resembles the structure predicted for the A. tumefaciens curdlan synthase (CrdS) (24) and CS; the latter contains a PilZ signature in its C-terminal cytoplasmic domain involved in c-di-GMP binding (22). However, neither CrdS nor BgsA seems to bear a PilZ domain nor the recently described c-di-GMP–binding GIL domain that is present in BcsE-like proteins (46). We nevertheless tested the ability of BgsA to bind c-di-GMP, using a previously reported fluorescence-based dot blot assay with 2′-Fluo-AHC-c-di-GMP (47). Crude extracts of Escherichia coli cells expressing full-length BgsA gave weak positive results, although BgsA expression probably was poor, because these cells had strongly delayed growth, and the protein could not be detected in SDS/PAGE gels. However, extracts of E. coli cells expressing the cytoplasmic C-terminal segment of BgsA (BgsA-C; the last 139 residues beginning with Ala526) (Fig. 5) showed significant binding to 2′-Fluo-AHC-c-di-GMP compared with cell extracts expressing the empty vector (Fig. 6A). The signal from the BgsA-C extract was lower than with the PilZ domain protein PA3353 (46), but this difference could be caused by different protein-expression levels and/or affinities for the ligand. Moreover, the binding of the BgsA-C extract to 2′-Fluo-AHC-c-di-GMP could be outcompeted by growing concentrations of pure, unlabeled c-di-GMP (Fig. 6B). These results provide strong evidence that BgsA can bind c-di-GMP through its C-terminal cytoplasmic domain, suggesting allosteric activation of this GT.
Fig. 5.
Topology model of BgsA (SMb20391) in the inner cell membrane (IM) predicted by TMHMM 2.0 (41) and presented by TOPO2 (www.sacs.ucsf.edu/cgi-bin/open-topo2.py). The putative catalytic residues of the D,D,D35QxxRW motif are shown in yellow, and the predicted DXD motif is enclosed in a box. The 139-aa C-BgsA peptide used in c-di-GMP–binding experiments is shown as gray diamonds.
Fig. 6.
Dot blot c-di-GMP–binding assays. (A) Dot fluorescence images of extracts of E. coli cells expressing the C-terminal fragment of BgsA (pQE80L::C-BgsA) (lane 3), the PilZ domain protein PA3353 (pET21::PA3353) (lane 2), or the empty vector (pQE80L) (lane 1) immobilized onto nitrocellulose membranes and incubated with 2′-Fluo-AHC-c-di-GMP. (B) Dot fluorescence of C-BgsA lysates incubated with a mix of 0.1 µM 2′-Fluo-AHC-c-di-GMP and increasing amounts of unlabeled c-di-GMP.
ExpR Activates Transcription of the bgsBA Operon.
The impact of c-di-GMP on the transcription of the bgsBA operon was assayed by quantitative RT-PCR (RT-qPCR) in the absence and presence of PleD*. Although bgsA transcription was slightly higher with PleD*, this variation was not statistically significant and suggested that c-di-GMP does not affect the transcription of bgsBA (SI Appendix, Fig. S6B). This suggestion was verified in a strain carrying a transcriptional bgsA:gus fusion. β-Glucuronidase activity in strain SMb20391::Nm-gus carrying pJBpleD* was 158.4 ± 13.85, similar to 171.7 ± 22.04 in the strain with the empty vector pJB3Tc19.
bgsA (SMb20391) was found previously among Sme genes whose expression was dependent on expR and sinI genes (48) and recently has been confirmed to be a member of the Sin/ExpR regulon (49). We confirmed the ExpR dependence of bgsA transcription (SI Appendix, Fig. S6B) and extended it to bgsB, which showed a 50-fold transcript reduction in the expR mutant. Moreover, expR and sinI mutants overexpressing PleD* were impaired in the emission of the c-di-GMP–induced blue CF fluorescence (Fig. 2), showed a CR− phenotype, and did not exhibit the PleD*-dependent flocculation in liquid media. Furthermore, the ExpR dependence of ML β-glucan production was confirmed in the expR mutant Rm1021 (38), which did not display any of the c-di-GMP–associated phenotypes described for Sme.
ML β-Glucan Is Involved in the Attachment of S. meliloti to Alfalfa Roots.
We evaluated the impact of the new ML β-glucan on bacterial colonization and attachment to alfalfa roots. For microscopy studies, variants of wild-type Sme and bgsA (SMb20391::Nm) mutant strains expressing two different fluorescent proteins, GFP and DsRed, were used. As a control for plasmid stability and to rule out any possible interference of the fluorescent proteins with bacterial physiology or ML β-glucan production, a flocculation assay was carried out first with mixtures of the Sme (pJBpleD*) strain tagged with either of the fluorescent proteins, and flocs formed after 3 d were observed under the epifluorescence microscope. These flocs were constituted by similar numbers of each tagged variant (SI Appendix, Fig. S8A). In contrast, when bacterial mixtures contained the wild-type (pJBpleD*, pBBR-gfp) and the bgsA mutant (pJBpleD*, pBBR-dsRed) cells, the wild-type cells were found almost exclusively in the flocs. Most mutant cells were in the surrounding medium, and few were located within the flocs (SI Appendix, Fig. S8B), suggesting that cells not producing the ML β-glucan were excluded from the cell network.
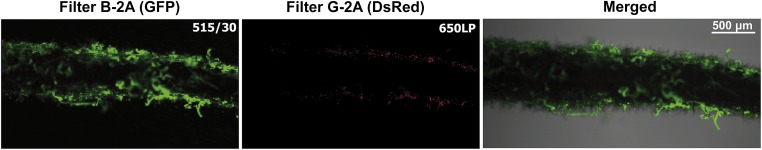
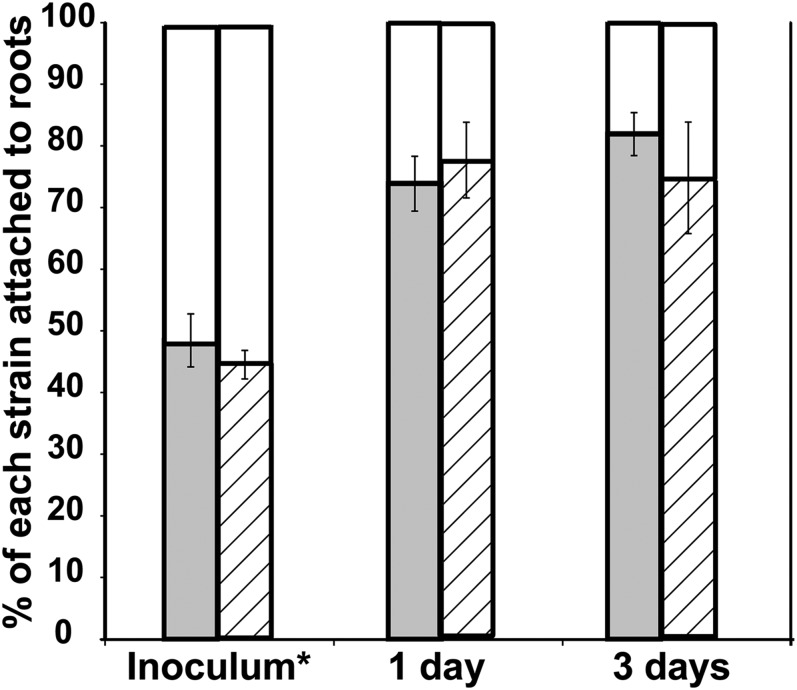
Confocal laser scanning microscopy (CLSM) was used to study competitive root attachment and colonization under high c-di-GMP conditions. Twenty-four hours after inoculation Bgs+ cells firmly attached to roots exceeded the bgsA mutant cells by more than 200-fold and formed a strong biofilm with large cell aggregates covering most of the root surface, whereas Bgs− cells were dispersed (Fig. 7). Bacterial attachment to roots also was studied at physiological levels of c-di-GMP. Plant roots were inoculated with 1:1 mixtures of the wild-type 8530 and the bgsA SMb20391::Nm strain at two different inoculum densities, 105 or 103 total cells applied per root. The BgsA− mutant cells firmly attached to roots were clearly outnumbered by the wild-type cells (Fig. 8), suggesting that Bgs+ bacteria attach to roots more effectively than those lacking the ML β-glucan. Taken together, the results support the notion that production of the ML β-glucan is required for efficient attachment of S. meliloti to the alfalfa root surface and consequently for root colonization.
Fig. 7.
Competitive root attachment and colonization with fluorescence-tagged strains. CLSM images of M. sativa roots inoculated with 1:1 mixtures of S. meliloti 8530-GFP and mutant SMb20391-dsRed, both overexpressing PleD*. (Magnification, 4×.) The wild type (green) forms dense biofilms coating the root, whereas SMb20391 cells (red) are found in low numbers and are dispersed.
Fig. 8.
Attachment of S. meliloti to alfalfa roots. Alfalfa roots were inoculated with 1:1 mixtures of wild-type strain 8530 and the bgsA mutant SMb20391::Nm (inoculum*) at two cell densities, 105 (Left Bars) or 103 (Right Bars). Cells firmly attached to roots were counted 1 and 3 d after inoculation. Bars show the proportion (%) of wild-type (gray and hatched portions of bars) and mutant (white portions of bars) cells attached to roots. Error bars indicate SDs.
Influence of ML β-Glucan on Nodulation.
Alfalfa nodulation with bacteria at high c-di-GMP levels could not be tested properly because the PleD* plasmid was lost rapidly under nonselective conditions. At physiological c-di-GMP levels the BgsA− mutant formed pink wild-type–like nodules indicative of nitrogen fixation. Nodulation kinetics by individual strains were determined at two different inoculum cell densities, 105 or 103 total cells applied per root; no significant differences between the wild-type and the bgsA SMb20391::Nm strain could be observed (SI Appendix, Fig. S9). Competitive nodulation was tested by inoculating roots with 1:1 mixtures of the wild type and the bgsA mutant, also applied at cell densities of 105 or 103 total cells per root. In both instances each strain occupied nearly 50% of the nodules, suggesting that under our experimental conditions the bgsA mutant is as competitive for nodulation as the wild type.
The bgsBA Operon Is Widespread Among Rhizobiales.
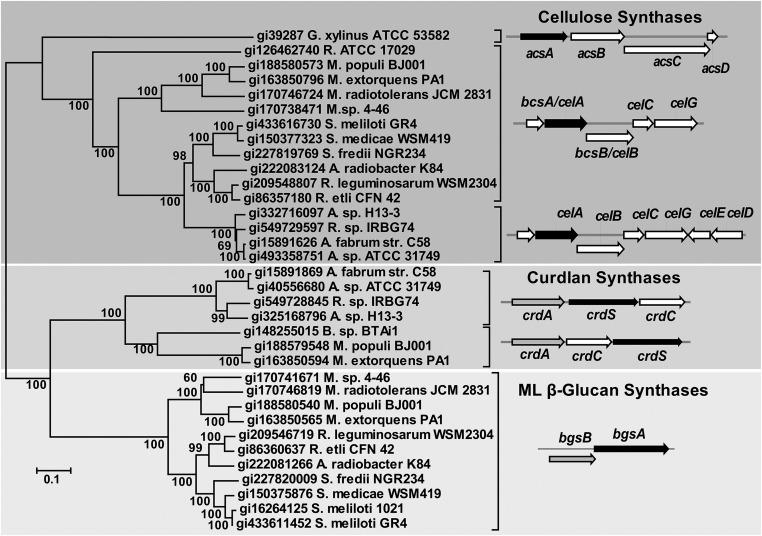
The bgsBA operon is required for the synthesis and/or secretion of the novel ML β-glucan uncovered in Sme. Given the features predicted for the encoded proteins, it is most likely that BgsA is the synthetase and that BgsB may participate in the export of the glucan chain. Similar to what occurs in cellulose or curdlan production (18, 20), it is likely that additional yet unknown components are necessary for full activity of the ML β-glucan synthesis system. We could identify bgsBA-like operons in genomes of the genera Sinorhizobium, Rhizobium, and Agrobacterium in the Rhizobiaceae and Methylobacterium in the Methylobacteriaceae, all within order Rhizobiales (Fig. 9). For instance, all available sinorhizobial (S. meliloti and S. fredii) and most Methylobacterium genomes bear a bgsBA-like operon, although only some strains of Rhizobium carry these genes. Among agrobacteria, only A. radiobacter K84 (a nonpathogenic biocontrol strain) carries a bgsBA-like operon. A phylogenetic analysis of BgsA (β-glucan), CrdS (curdlan), and CelA/BcsA/AcsA (cellulose) synthases showed two monophyletic clades separating Cel synthases from Crd and Bgs synthases (Fig. 9), with the two latter forming clearly diverging clusters. Unlike S. meliloti 8530, which carries no CS genes, a Cel system coexists with either a Bgs or a Crd system in most available Rhizobiales genomes. Coexistence of Crd and Bgs systems is far less frequent, although distinctive Cel, Crd, and Bgs synthases are borne by some strains of methylobacteria (Fig. 9). Taken together, the data suggest that CrdS and BgsA share a common ancestor and that divergent evolution may have shaped their specific activities toward synthesis of either (1,3)-β-glucan or (1,3) (1,4)-β-glucan, respectively.
Fig. 9.
Neighbor-joining phylogenetic analysis of BgsA proteins and cellulose and curdlan synthases. Bootstrap values (1,000 replicates) are indicated at branching points. Schematic diagrams of ORFs in representative operons are shown by black arrows for glycosyltransferases, gray arrows for membrane fusion proteins, and white arrows for all others.
Discussion
c-di-GMP is a common activator of several bacterial EPSs, such as cellulose in many bacteria, poly-β-1,6-N-acetilglucosamine in enterobacteria, or alginate, Psl, and Pel in Pseudomonads (50, 51). In this work, we have exploited this feature to uncover an otherwise cryptic EPS produced by the α-proteobacterium S. meliloti 8530 when its intracellular c-di-GMP levels are artificially increased. This new bacterial polymer is a linear ML (1→3)(1→4)-β-d-glucan. It resembles the ML β-glucans found in cereals and lichens, but the bacterial ML β-glucan presents a unique primary structure. Whereas the plant ML β-glucans contain distinctive ratios of cello-oligosaccharides of 3, 4, or higher degrees of polymerization, linked by (1→3) bonds (37), the bacterial ML β-glucan contains a →3)-β-d-Glcp-(1→4)-β-d-Glcp-(1→ repeating unit. ML β-glucans are attracting increasing industrial interest as bioactive ingredients and food additives (52). The structural features of ML β-glucans, including the ratio of β-(1→4)/(1→3) linkages, the abundance of long cellulosic fragments, and molecular size, determine their physical and physiological properties (53). The S. meliloti ML β-glucan shares some properties with other bacterial β-glucans such as its insolubility in water (i.e., cellulose and curdlan) and solubility in dilute alkali (curdlan); however, the precise description of its physical features and rheological properties shall be the subject of future specific studies.
At least two genes, bgsA and bgsB, likely encoding a GT-2 and a membrane fusion protein, respectively, are required for the production of this extracellular ML β-glucan. BgsA is a strong candidate to be the ML β-glucan synthase, because it shares sequence, domain conservation, and predicted secondary structure with other bacterial GT-2 proteins, particularly curdlan (CrdS) and Cel synthases. In Cel synthases the C-terminal region encloses the PilZ domain involved in c-di-GMP binding and required for enzyme activation (22). Although BgsA does not seem to contain a PilZ signature, we provide strong evidence that it binds c-di-GMP through its C-terminal region, which could represent a new c-di-GMP–binding domain that awaits characterization. Therefore, it is likely that induction of ML β-glucan by c-di-GMP takes place through allosteric activation of the BgsA GT. It also will be of interest to identify the DGC(s) involved in the provision of c-di-GMP to BgsA and the physiological conditions that promote its activation. Synthesis of the ML β-glucan also is dependent on the ExpR/SinI system which regulates bgsBA transcription, thereby linking ML β-glucan to the production of other S. meliloti EPSs (7) Thus, ML β-glucan biosynthesis seems to be regulated at multiple levels, a situation that also has been proposed for cellulose biosynthesis in A. tumefaciens (54).
We observed that high production of ML β-glucan facilitates aggregation and biofilm formation by S. meliloti 8530 under free-living conditions and also on the alfalfa root surface. ML β-glucan overproduction greatly enhances bacterial attachment to the host plant root and the formation of a dense biofilm on the root surface, from which mutants unable to produce ML β-glucan seem to be excluded. Moreover, under physiological c-di-GMP levels bgsA mutants show reduced attachment to alfalfa roots. The involvement of ML β-glucan in S. meliloti’s firm attachment to roots and its aggregation and biofilm formation on abiotic and biotic surfaces suggest that it can be of particular importance for bacterial persistence and multiplication in the soil and rhizosphere under field conditions. This evokes the biological role of related glucans such as cellulose, an important adherence factor in very diverse bacteria (55), including rhizobia. Like the S. meliloti ML β-glucan, cellulose also provides Rhizobium leguminosarum with a root attachment advantage; however, both cellulose-deficient and -overproducing strains show wild-type nodulation properties (56, 57) similar to those of the S. meliloti bgsA mutants. With resemblance, A. tumefaciens cellulose-deficient mutants also show reduced attachment to plant cells but remain fully virulent (54) except under very particular experimental conditions [e.g., when the infection sites are washed shortly after inoculation], suggesting that cellulose may be important for bacterial attachment to plant wound sites under adverse field conditions (58). Phylogenetic analyses suggest that, like S. meliloti, other Rhizobiales, including nonnodulating plant-interacting bacteria such as Agrobacterium and Methylobacterium, also are able to produce ML (1→3)(1→4)-β-d-glucan. This novel polymer adds to the plethora of surface polysaccharides that help rhizobia and other plant-associated bacteria thrive in the changing soil and rhizosphere environments.
Materials and Methods
Bacteria and Culture Conditions.
Bacterial strains and plasmids used in this work are listed in SI Appendix, Table S1. Starting cultures of S. meliloti strains were grown overnight at 28 °C on tryptone-yeast extract-CaCl2 (TY) broth (59) or MM medium (60). When required, antibiotics and other compounds were added at the following final concentrations: tetracycline, 10 μg/mL; CR, 50 μg/mL; CF, 200 μg/mL (in solid medium) or 100 μg/mL (in liquid medium). The plasmids pJBpleD*, pJB3Tc19, pJB9091, and pJBpleD9091 were introduced in rhizobia by conjugation using the E. coli β2163 donor strain (61) in matings as previously described (62).
Construction of S. meliloti Mutants and Complementing Plasmids.
Molecular biology techniques were performed according to standard protocols and manufacturers’ instructions. For the construction of Sme SMb20391::Nm and SMb20460::Nm mutants, the phage ΦM12 (63) was used to transduce mutations from the donors S. meliloti 2011mTn5STM.1.08.B12, 2011mTn5STM.4.13.E03, and 2011mTn5STM.4.03.E12 to Sme, as previously described (64). Mutants were verified by PCR and Southern hybridization with specific probes. For the construction of Sme ΔSMb20390 with an unmarked in-frame deletion, two fragments flanking the ORF were amplified separately, using primer pairs op90-F/20390L-R (496 bp) and 20390R-F/SM_b20391-R (408 bp) (SI Appendix, Table S2). These two fragments were purified and used as template DNA for an overlapping PCR with primers op90-F and SM_b20391-R. The final fragment was cloned into pCR-XL-TOPO (Invitrogen) and sequenced. A correct SphI/BamHI DNA insert was isolated and subcloned into the suicide plasmid pK18mobSacB (65), generating the pK18Δ20390. The pK18Δ20390 plasmid was introduced into Sme by biparental conjugation with an E. coli S17.1 donor, and the SMb20390 deletion of 1,035 bp (402490–403524) was generated by homologous recombination following the procedures described in ref. 65. The ΔSMb20390 mutant was corroborated by PCR and Southern hybridization with specific probes. Diagrams of the plasmid constructions used in the complementation experiments are depicted in SI Appendix, Fig. S7. For pJB9091 and pJBpleD*9091 constructions, two primers flanking the SMb2039-91 operon were used (op90-F and op91-R) in a PCR with Sme genomic DNA as a template. The 3,720-bp fragment containing both genes was cloned in pCR-XL-TOPO (Invitrogen) and sequenced. A correct EcoRI DNA insert was isolated and subcloned into pJB3Tc19 and pJBpleD* to generate pJB9091 and pJBpleD*9091, respectively. The correct orientation of both constructions was verified with appropriate restrictions enzymes.
Gene-Expression Assays.
For RT-PCR experiments, RNA extractions were carried out using the Qiagen RNeasy RNA purification kit (Qiagen). Total RNA (1 µg) treated with RNase-free DNase I Set (Roche) was reverse-transcribed using SuperScript III reverse transcriptase (Invitrogen) and random hexamers (Roche) as primers. RT-qPCR was performed on an iCycler iQ5 (Bio-Rad). Each 25-µL reaction contained 1 µL of cDNA, 200 nM of each primer, and iQ SyBrGreen Supermix (Bio-Rad). Control PCRs of the RNA samples also were performed to confirm the absence of contaminating DNA. Samples were denatured initially by heating at 95 °C for 3 min, followed by a 35-cycle amplification and quantification program (95 °C for 30 s, 55 °C for 45 s and 72 °C for 45 s). Melting-curve analysis was conducted to ensure amplification of a single product. The efficiency of each primer pair (E) was determined by running 10-fold serial dilutions (four dilution series) of Sme genomic DNA as template and generating a standard curve by plotting the log of the dilution factor against the cycle threshold (CT) value during amplification of each dilution. Amplification efficiency was calculated using the formula [E = (10(1/a) − 1) × 100], where a is the slope of the standard curve. The relative expression of the SMb20390 and SMb20391 genes was normalized to that of 16S rRNA gene.
A transcriptional reporter gene fusion also was used to study the effect PleD* on the expression bgsBA operon. A strain with an oriented miniTn5-gus inserted in bgsA was used (SMb20391::Nm-gus) (SI Appendix, Table S1), and β-glucuronidase was determined as previously described (62).
CF-Binding Assays.
Starting cultures of rhizobial strains were prepared as detailed above. After washing with MM, cultures were diluted 1:100 into 10-mL flasks containing MM supplemented with CF (100 μM final concentration) at 28 °C under agitation for 48 h. Then cultures of the different strains were centrifuged for 10 min at 3,220 × g. Supernatant containing broth with unbound CF of each biological replicate was removed. The pellet was suspended in 2 mL of distilled water and placed in the wells of 24-well plates. Similar growth of all strains was confirmed, and CF-binding measurements for three biological replicates of each strain were performed in a PTI fluorimeter (Photon Technology International); the results were expressed in arbitrary units ± SE.
Bacterial Attachment to Plant Roots.
Alfalfa (Medicago sativa L.) seeds were surface-sterilized and germinated as described by Olivares et al. (66). Seedlings were placed on 12 × 12 cm plates (12 seedlings per plate) containing nitrogen-free medium (66) solidified with 1.15% (wt/vol) purified agar and were incubated for 3 d in a plant-growth chamber (16-h light/8-h dark photoperiod, 24 °C/16 °C day/night temperature, 75% relative humidity) before inoculation. For root attachment of bacteria at high c-di-GMP levels, seedlings were transferred into a flask containing 25 mL of nitrogen-free plant nutrient solution with a mixture of the wild type and the SMb20391::Nm mutant (109 cells each), both expressing PleD* and tagged with GFP or dsRed, and were agitated gently at 28 °C for 24 h. After this time, roots were separated from shoots, and four groups of three roots each were prepared and washed vigorously four times with 1 mL of sterile deionized water in Eppendorf tubes to remove bacteria unbound or weakly bound to roots. After these washings, one group of three roots was used for CLSM with a Nikon Eclipse TE2000-U microscope at the technical service of Estación Experimental del Zaidín Consejo Superior de Investigaciones Cientificas. One milliliter of MM with 2 mM EDTA was added to each of the remaining three groups of roots, and bacteria firmly bound to roots were released by two cycles of vortexing for 1 min and sonication for 1 min. Serial dilutions were spread onto TY with and without Nm and were incubated at 28 °C for 3–4 d; then cfus of each strain were counted. For root attachment of bacteria at physiological c-di-GMP levels, a modification of the protocol described by Anollés and Favelukes was followed (67). Seedlings prepared as above were kept in plates, and each root was inoculated with 30 µL of nitrogen-free plant nutrient solution containing mixtures of Sme and SMb20391::Nm mutant in a 1:1 ratio. Two different inoculum densities were tested, so 105 or 103 cells were applied to each root. Plants were kept in the plant-growth chamber, and bacteria firmly attached to roots were recovered and counted as above, at 1 and 3 d after inoculation.
Nodulation Experiments.
Alfalfa seedlings were prepared as above, except that nine plants per plate were used. For nodulation kinetics each strain was inoculated onto the roots of 45 plants by spreading 30 µL of a bacterial suspension (prepared in a nitrogen-free plant nutrient solution) along each root surface. Two inoculum densities were tested, so 105 or 103 cells were loaded on each root. Plants were maintained in the growth chamber, and the number of visible nodules was recorded daily. For competitive nodulation, plants were prepared as above, but bacterial suspensions contained mixtures of the wild type and the SMb20391::Nm mutant in a 1:1 ratio. Two inoculum densities were tested, so 105 or 103 total cells were loaded on each root. Nodule occupancy was determined 15 d after inoculation. At least 100 nodules were collected, surface sterilized, and individually crushed. The contents of each nodule were spread on TY-agar plates with and without Nm and were incubated at 28 °C for 3–4 d.
ML β-Glucan Isolation and Chemical Characterization.
Starting cultures of Sme pJBpleD* were diluted 1:100 in 5-L flasks containing 0.5 L of liquid MM supplemented with tetracycline and were grown with slow shaking (80 rpm) for 3 d at 28 °C. The flocs formed were recovered using a sieve with a 500-µm cutoff and then were placed in a 50-mL Falcon tube (Fisher Scientific) and washed with 30 mL of boiling Milli-Q water (Millipore Corporation). After boiling for 5 min, the material was cooled at room temperature and centrifuged for 20 min at 3,220 × g. The supernatant was discarded, and the washing process was repeated four times. The β-glucan obtained was frozen and lyophilized overnight. For quantification of β-glucan yield, the different strains were grown in 35 mL of MM in 250-mL flasks and were incubated and processed as described above, except that the sieve runoffs were recovered and centrifuged, and bacterial pellets were lyophilized and weighed to calculate the amount of the β-glucan dry weight relative to the total dry weight of the culture. Monosaccharides of the β-glucan were identified upon gas liquid chromatography (GLC)-MS separation of their per-O-trimethylsilylated methyl glycosides (68). The absolute configuration of monosaccharides was assigned following GLC-MS analysis of their per-O-trimethylsilylated (S)- and (R,S)-2-butyl glycosides as described by Gerwig et al. (69). For methylation analysis, a further purification step on the β-glucan purification was applied to increase its final purity. The β-glucan was obtained as described above, and the flocs trapped on the sieve were dissolved in 15 mL of 1% NaOH for 1 h with shaking and were dialyzed in water using molecular porous-membrane tubing (Spectrum Labs) until neutral pH was obtain in the sample, which then was frozen and lyophilized overnight. Lyophilized samples were methylated using the method of Ciucanu and Costello (70). The methylated polysaccharide was purified by SEC on Sephadex LH-20, using CH2Cl2:MeOH (1:1) as the eluent. Hydrolysis was carried out first with 88% HCOOH (100 °C, 1 h) and then, after evaporation of the acid, with 2 M trifluoroacetic acid (120 °C, 4 h). Reduction with NaBD4 and acetylation were performed according to the methods of Kim and coworkers (71), yielding the corresponding partially methylated and acetylated alditols, which were solubilized in CH2Cl2 and analyzed by GLC-MS. For acetylation of polysaccharide, 6 mg of polysaccharide were resuspended in 1 mL of pyridine:acetic anhydride (1:1 ratio) for 24 h at room temperature The solution was poured into 5 mL of ice water, and the precipitate was recovered by centrifugation, rinsed with cold water, and freeze dried. This procedure was repeated once. The sample was resuspended in CH2Cl2:MeOH (1:1 ratio, vol/vol) and purified by SEC on Sephadex LH-20 with the same eluent. Fractions containing carbohydrates were vacuum-evaporated, dissolved in 300 μL of CDCl3, and studied by NMR. Spectra were recorded at 300 K on a Bruker Avance III 500 MHz spectrometer operating at 500.40 MHz (1H) and 125.8 MHz (13C). Chemical shifts are given in parts per million, using the residual CHCl3 signal (δH 7.26 ppm, δC 77.16 ppm) as reference. Standard Bruker sequences were used for 2D experiments (72). DQF-COSY, TOCSY, and ROESY were performed by using a data matrix of 256 × 2,048 points to digitize a spectral width of 5,515 × 5,515 Hz. TOCSY and ROESY experiments were acquired with mixing times of 80 and 100 ms, respectively. The 2D heteronuclear one-bond proton-carbon correlation experiment was registered in the 1H-detection mode via HSQC. A data matrix of 128 × 1,024 points was used to digitize a spectral width of 20,843 (F1) × 5,515 (F2) Hz.
Five milligrams of polysaccharides were digested with 10 U of a commercial lichenase (endo-1,3 (4)-β-glucanase; Megazyme) in 1 mL of water overnight at room temperature, including respective controls without the enzyme. Samples were centrifuged at 16,000 × g for 10 min; then supernatants were frozen and lyophilized, dissolved in water, and studied by TLC on silica gel 60 Alugram Sil G/UV (Macherey-Nagel) using butanol:acetic acid:water (2:1:1) as eluent. Carbohydrates were detected with orcinol:sulfuric acid (73). A mixture of glucose, maltose, maltotriose, maltotetraose, maltopentaose, and maltohexaose (Sigma-Aldrich) oligosaccharides were used as size markers.
Phylogenetic Analyses.
Protein sequence similarity searches were carried out with the BLASTP program from the National Center for Biotechnology Information (74). Alignment was performed with CLUSTALW (75). Phylogenetic and molecular evolutionary analyses were conducted using MEGA, version 5.2 (76), using the following phylogenetic parameters: analysis, phylogeny reconstruction; statistical method, neighbor-joining; number of bootstrap replications, 1,000; substitutions type, amino acid; model/method, Poisson model; rates among sites, uniform rates; pattern among lineages, same (homogeneous); gaps/missing data treatment; and pairwise deletion.
Dot Blot c-di-GMP–Binding Assays.
Plasmids pET28b::BgsA and pQE80L::C-BgsA were constructed to express the entire or the last 139 amino acids of BgsA, respectively. A 2,068-bp DNA fragment containing full-length bgsA was amplified with primers op91-F and op91-R, cloned into pCR-XL-TOPO, and subsequently subcloned as an EcoRI fragment into pET28b+. The correct orientation of the construction was checked using XhoI. Primers C-BgsA-F and C-BgsA-R were used to amplify a 488-bp DNA fragment encoding the last 139 amino acids of BgsA. The amplicon was cloned in pCR-XL-TOPO and subsequently was subcloned as a HindIII/BamHI fragment into pQE80L. E. coli strains carrying pET28b::BgsA, pQE80L::C-BgsA (SI Appendix, Table S1), respective empty vectors (as negative controls), and pET21:PA3353 overexpressing the Pseudomonas aeruginosa PilZ domain protein PA3353 [used as a positive control (47)] were grown to OD600 0.5–0.7 and induced with 1 mM of isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. After centrifugation, pellets from 8 mL of culture were resuspended in 500 µL of lysis buffer [50 mM Tris (pH 7.5), 10 mM NaCl, 1 mM DTT; and 5% glycerol] and were lysated by sonication (six cycles of 15 s on ice; Branson Sonifier 250). Protein expression was evaluated by SDS/PAGE, and all lysates were standardized with the total protein content determined by Bradford assay (77). For the c-di-GMP dot blot-binding assays, we modified a previously described protocol (47). Briefly, 3 µg of each lysate sample were spotted on 0.45-µm nitrocellulose membrane discs (Millipore) and left to dry. The membranes were blocked in Tris buffer saline with 0.2% Tween 20 (TBS-T) and 5% skim milk for 1 h at room temperature. Blocked membranes were incubated with 1 µM of 2′-O-(6-[Fluoresceinyl]aminohexylcarbamoyl)-c-di-GMP (2´-Fluo-AHC-c-di-GMP; Biolog) in TBS-T containing 5% skim milk for 1 h at room temperature in a dark room. After washing with TBS-T for 10 min, the dot blots were scanned with a Pharos FX Plus fluorescence imager (Bio-Rad) using the SYBR Green I settings and were quantified by densitometry with the Quantity One software (Bio-Rad). Competitive assays were performed as indicated above, but membranes were incubated with mixtures of 0.1 µM of 2´-Fluo-AHC-c-di-GMP and increasing quantities (0.1–30 µM) of unlabeled c-di-GMP.
Supplementary Material
Acknowledgments
We thank J. Nogales for Sme expR and sinI derivatives; M. J. Pérez-Mendoza, University of Granada, and I. Rodríguez-García, University of Almería, for help with initial chemical analyses; T. Felipe-Reyes and D. Rodríguez-Carbonell (Estación Experimental del Zaidín, EEZ) for excellent technical assistance; S. Muñoz for help with competitive nodulation; A. Olmedilla, EEZ, and G. Martín, University of Malaga, for help with scanning microscopy; J. Düvel, Helmholtz Centre for Infection Research, for assistance with dot blot assays and for providing cloned PA3353; and Centro de Investigación Tecnología e Innovación de la Universidad de Sevilla for support with NMR studies. This work was supported by Grant BIO2011-23032 from the Ministerio de Economía y Competitividad and Grant P10-CVI-5800 from the Junta de Andalucía, both cofinanced by the European Fund for Economic and Regional Development, and by Consejo Superior de Investigaciones Cientificas (CSIC) Grant 201440E026. D.P.-M. was supported by a Junta de Ampliación de Estudios (JAE)-doc CSIC contract and later by Grants P10-CVI-5800 and 201440E026; L.R.-J. was supported by a JAE-Pre fellowship; and G.d.A.F. was supported by a contract associated with Grant P10-CVI-5800.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421748112/-/DCSupplemental.
References
- 1.Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol. 2001;39(6):1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 2.Solano C, et al. Genetic analysis of Salmonella enteritidis biofilm formation: Critical role of cellulose. Mol Microbiol. 2002;43(3):793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- 3.Christen B, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281(42):32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 4.Recouvreux DO, et al. Cellulose biosynthesis by the beta-proteobacterium, Chromobacterium violaceum. Curr Microbiol. 2008;57(5):469–476. doi: 10.1007/s00284-008-9271-0. [DOI] [PubMed] [Google Scholar]
- 5.Pérez-Mendoza D, Coulthurst SJ, Sanjuán J, Salmond GPC. N-Acetylglucosamine-dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c-di-GMP levels. Microbiology. 2011;157(Pt 12):3340–3348. doi: 10.1099/mic.0.050450-0. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, et al. Genetic analysis of Agrobacterium tumefaciens unipolar polysaccharide production reveals complex integrated control of the motile-to-sessile switch. Mol Microbiol. 2013;89(5):929–948. doi: 10.1111/mmi.12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skorupska A, Janczarek M, Marczak M, Mazur A, Król J. Rhizobial exopolysaccharides: Genetic control and symbiotic functions. Microb Cell Fact. 2006;5:7. doi: 10.1186/1475-2859-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraysse N, Couderc F, Poinsot V. Surface polysaccharide involvement in establishing the rhizobium-legume symbiosis. Eur J Biochem. 2003;270(7):1365–1380. doi: 10.1046/j.1432-1033.2003.03492.x. [DOI] [PubMed] [Google Scholar]
- 9.Laus MC, et al. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol. 2006;59(6):1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- 10.Spaink HP, Lugtenberg BJ. Role of rhizobial lipo-chitin oligosaccharide signal molecules in root nodule organogenesis. Plant Mol Biol. 1994;26(5):1413–1422. doi: 10.1007/BF00016482. [DOI] [PubMed] [Google Scholar]
- 11.Danhorn T, Fuqua C. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol. 2007;61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 12.Rodríguez-Navarro DN, Dardanelli MS, Ruíz-Saínz JE. Attachment of bacteria to the roots of higher plants. FEMS Microbiol Lett. 2007;272(2):127–136. doi: 10.1111/j.1574-6968.2007.00761.x. [DOI] [PubMed] [Google Scholar]
- 13.Glazebrook J, Walker GC. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989;56(4):661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- 14.Janczarek M. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int J Mol Sci. 2011;12(11):7898–7933. doi: 10.3390/ijms12117898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurich N, González JE. Role of quorum sensing in Sinorhizobium meliloti-Alfalfa symbiosis. J Bacteriol. 2009;191(13):4372–4382. doi: 10.1128/JB.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glenn SA, Gurich N, Feeney MA, González JE. The ExpR/Sin quorum-sensing system controls succinoglycan production in Sinorhizobium meliloti. J Bacteriol. 2007;189(19):7077–7088. doi: 10.1128/JB.00906-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Römling U. Molecular biology of cellulose production in bacteria. Res Microbiol. 2002;153(4):205–212. doi: 10.1016/s0923-2508(02)01316-5. [DOI] [PubMed] [Google Scholar]
- 18.Whitney JC, Howell PL. Synthase-dependent exopolysaccharide secretion in Gram-negative bacteria. Trends Microbiol. 2013;21(2):63–72. doi: 10.1016/j.tim.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nobles DR, Jr, Brown RM., Jr . Many paths up the mountain: Tracking the evolution of cellulose biosynthesis. In: Brown RM Jr, Saxena I, editors. Cellulose Molecular and Structural Biology. Springer; Dordrecht, The Netherlands: 2007. pp. 1–15. [Google Scholar]
- 20.Stasinopoulos SJ, Fisher PR, Stone BA, Stanisich VA. Detection of two loci involved in (1—>3)-beta-glucan (curdlan) biosynthesis by Agrobacterium sp. ATCC31749, and comparative sequence analysis of the putative curdlan synthase gene. Glycobiology. 1999;9(1):31–41. doi: 10.1093/glycob/9.1.31. [DOI] [PubMed] [Google Scholar]
- 21.Ross P, et al. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987;325(6101):279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 22.Fujiwara T, et al. The c-di-GMP recognition mechanism of the PilZ domain of bacterial cellulose synthase subunit A. Biochem Biophys Res Commun. 2013;431(4):802–807. doi: 10.1016/j.bbrc.2012.12.103. [DOI] [PubMed] [Google Scholar]
- 23.Ruffing AM, Chen RR. Transcriptome profiling of a curdlan-producing Agrobacterium reveals conserved regulatory mechanisms of exopolysaccharide biosynthesis. Microb Cell Fact. 2012;11:17. doi: 10.1186/1475-2859-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karnezis T, Epa VC, Stone BA, Stanisich VA. Topological characterization of an inner membrane (1—>3)-beta-D-glucan (curdlan) synthase from Agrobacterium sp. strain ATCC31749. Glycobiology. 2003;13(10):693–706. doi: 10.1093/glycob/cwg093. [DOI] [PubMed] [Google Scholar]
- 25.Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol Microbiol. 2007;65(4):876–895. doi: 10.1111/j.1365-2958.2007.05817.x. [DOI] [PubMed] [Google Scholar]
- 26.Steiner S, Lori C, Boehm A, Jenal U. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 2013;32(3):354–368. doi: 10.1038/emboj.2012.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pérez-Mendoza D, et al. Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS ONE. 2014;9(3):e91645. doi: 10.1371/journal.pone.0091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wood PJ. Specificity in the interaction of direct dyes with polysaccharides. Carbohydr Res. 1980;85(2):271–287. [Google Scholar]
- 29.Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics. 2002;161(1):33–46. doi: 10.1093/genetics/161.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82(18):6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leigh JA, Lee CC. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J Bacteriol. 1988;170(8):3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glazebrook J, Walker GC. 1991. Genetic techniques in Rhizobium meliloti. Methods in Enzymology Bacterial Genetic Systems, ed Jeffrey HM (Academic, San Diego), Vol 204, pp 398–418.
- 33.Yotsuzuka F. Curdlan. In: Cho SS, Dreher ML, editors. Handbook of Dietary Fiber. Dekker; New York: 2001. pp. 737–757. [Google Scholar]
- 34.Kowsaka K, Okajima K, Kamide K. Two-dimensional nuclear magnetic resonance spectra of cellulose and cellulose triacetate. Polym J. 1988;20(12):1091–1099. [Google Scholar]
- 35.Sano J, Ikushima N, Takada K, Shoji T. NMR studies of (1—>3)-beta-D-glucooligosaccharide derivatives. Carbohydr Res. 1994;261(1):133–148. doi: 10.1016/0008-6215(94)80012-x. [DOI] [PubMed] [Google Scholar]
- 36.Planas A. Bacterial 1,3-1,4-beta-glucanases: Structure, function and protein engineering. Biochim Biophys Acta. 2000;1543(2):361–382. doi: 10.1016/s0167-4838(00)00231-4. [DOI] [PubMed] [Google Scholar]
- 37.Simmons TJ, et al. An unexpectedly lichenase-stable hexasaccharide from cereal, horsetail and lichen mixed-linkage β-glucans (MLGs): Implications for MLG subunit distribution. Phytochemistry. 2013;95:322–332. doi: 10.1016/j.phytochem.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 38.Galibert F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293(5530):668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 39.Zgurskaya HI, Yamada Y, Tikhonova EB, Ge Q, Krishnamoorthy G. Structural and functional diversity of bacterial membrane fusion proteins. Biochim Biophys Acta. 2009;1794(5):794–807. doi: 10.1016/j.bbapap.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Pimenta AL, Racher K, Jamieson L, Blight MA, Holland IB. Mutations in HlyD, part of the type 1 translocator for hemolysin secretion, affect the folding of the secreted toxin. J Bacteriol. 2005;187(21):7471–7480. doi: 10.1128/JB.187.21.7471-7480.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Morgan JL, Strumillo J, Zimmer J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature. 2013;493(7431):181–186. doi: 10.1038/nature11744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethaphong L, et al. Tertiary model of a plant cellulose synthase. Proc Natl Acad Sci USA. 2013;110(18):7512–7517. doi: 10.1073/pnas.1301027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gastinel LN, Cambillau C, Bourne Y. Crystal structures of the bovine beta4galactosyltransferase catalytic domain and its complex with uridine diphosphogalactose. EMBO J. 1999;18(13):3546–3557. doi: 10.1093/emboj/18.13.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saxena IM, Brown RM, Jr, Dandekar T. Structure—function characterization of cellulose synthase: Relationship to other glycosyltransferases. Phytochemistry. 2001;57(7):1135–1148. doi: 10.1016/s0031-9422(01)00048-6. [DOI] [PubMed] [Google Scholar]
- 46.Fang X, et al. GIL, a new c-di-GMP-binding protein domain involved in regulation of cellulose synthesis in enterobacteria. Mol Microbiol. 2014;93(3):439–452. doi: 10.1111/mmi.12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Düvel J, et al. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J Microbiol Methods. 2012;88(2):229–236. doi: 10.1016/j.mimet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 48.Hoang HH, Becker A, González JE. The LuxR homolog ExpR, in combination with the Sin quorum sensing system, plays a central role in Sinorhizobium meliloti gene expression. J Bacteriol. 2004;186(16):5460–5472. doi: 10.1128/JB.186.16.5460-5472.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Charoenpanich P, Meyer S, Becker A, McIntosh M. Temporal expression program of quorum sensing-based transcription regulation in Sinorhizobium meliloti. J Bacteriol. 2013;195(14):3224–3236. doi: 10.1128/JB.00234-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Franklin MJ, Nivens DE, Weadge JT, Howell PL. Biosynthesis of the Pseudomonas aeruginosa Extracellular Polysaccharides, Alginate, Pel, and Psl. Front Microbiol. 2011;2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: The first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev. 2013;77(1):1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brennan CS, Cleary LJ. The potential use of cereal (1→3,1→4)-β-d-glucans as functional food ingredients. J Cereal Sci. 2005;42(1):1–13. [Google Scholar]
- 53.Lazaridou A, Biliaderis CG, Micha-Screttas M, Steele BR. A comparative study on structure–function relations of mixed-linkage (1→3), (1→4) linear β-d-glucans. Food Hydrocoll. 2004;18(5):837–855. [Google Scholar]
- 54.Barnhart DM, Su S, Baccaro BE, Banta LM, Farrand SK. CelR, an ortholog of the diguanylate cyclase PleD of Caulobacter, regulates cellulose synthesis in Agrobacterium tumefaciens. Appl Environ Microbiol. 2013;79(23):7188–7202. doi: 10.1128/AEM.02148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaron S, Römling U. Biofilm formation by enteric pathogens and its role in plant colonization and persistence. Microb Biotechnol. 2014;7(6):496–516. doi: 10.1111/1751-7915.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Smit G, Kijne JW, Lugtenberg BJ. Involvement of both cellulose fibrils and a Ca2+-dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J Bacteriol. 1987;169(9):4294–4301. doi: 10.1128/jb.169.9.4294-4301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ausmees N, Jonsson H, Höglund S, Ljunggren H, Lindberg M. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology. 1999;145(Pt 5):1253–1262. doi: 10.1099/13500872-145-5-1253. [DOI] [PubMed] [Google Scholar]
- 58.Matthysse AG. Role of bacterial cellulose fibrils in Agrobacterium tumefaciens infection. J Bacteriol. 1983;154(2):906–915. doi: 10.1128/jb.154.2.906-915.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84(1):188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 60.Robertsen BK, Aman P, Darvill AG, Mcneil M, Albersheim P. The Structure of Acidic Extracellular Polysaccharides Secreted by Rhizobium-Leguminosarum and Rhizobium-Trifolii. Plant Physiol. 1981;67(3):389–400. doi: 10.1104/pp.67.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demarre G, et al. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol. 2005;156(2):245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Pérez-Mendoza D, et al. Identification of the rctA gene, which is required for repression of conjugative transfer of rhizobial symbiotic megaplasmids. J Bacteriol. 2005;187(21):7341–7350. doi: 10.1128/JB.187.21.7341-7350.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brewer TE, Stroupe ME, Jones KM. The genome, proteome and phylogenetic analysis of Sinorhizobium meliloti phage ΦM12, the founder of a new group of T4-superfamily phages. Virology. 2014;450-451:84–97. doi: 10.1016/j.virol.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 64.Finan TM, et al. General transduction in Rhizobium meliloti. J Bacteriol. 1984;159(1):120–124. doi: 10.1128/jb.159.1.120-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schäfer A, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145(1):69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 66.Olivares J, Casadesús J, Bedmar EJ. Method for Testing Degree of Infectivity of Rhizobium meliloti Strains. Appl Environ Microbiol. 1980;39(5):967–970. doi: 10.1128/aem.39.5.967-970.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Anollés GC, Favelukes G. Quantitation of adsorption of rhizobia in low numbers to small legume roots. Appl Environ Microbiol. 1986;52(2):371–376. doi: 10.1128/aem.52.2.371-376.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaplin MF. A rapid and sensitive method for the analysis of carbohydrate components in glycoproteins using gas-liquid chromatography. Anal Biochem. 1982;123(2):336–341. doi: 10.1016/0003-2697(82)90455-9. [DOI] [PubMed] [Google Scholar]
- 69.Gerwig GJ, Kamerling JP, Vliegenthart JFG. Determination of the d and l configuration of neutral monosaccharides by high-resolution capillary g.l.c. Carbohydr Res. 1978;62(2):349–357. [Google Scholar]
- 70.Ciucanu I, Costello CE. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J Am Chem Soc. 2003;125(52):16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- 71.Kim JS, Reuhs BL, Michon F, Kaiser RE, Arumugham RG. Addition of glycerol for improved methylation linkage analysis of polysaccharides. Carbohydr Res. 2006;341(8):1061–1064. doi: 10.1016/j.carres.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 72.Parella T. A complete set of novel 2D correlation NMR experiments based on heteronuclear J-cross polarization. J Biomol NMR. 2004;29(1):37–55. doi: 10.1023/B:JNMR.0000019514.04047.8b. [DOI] [PubMed] [Google Scholar]
- 73.Blom W, Luteyn JC, Kelholt-Dijkman HH, Huijmans JG, Loonen MC. Thin-layer chromatography of oligosaccharides in urine as a rapid indication for the diagnosis of lysosomal acid maltase deficiency (Pompe’s disease) Clin Chim Acta. 1983;134(1-2):221–227. doi: 10.1016/0009-8981(83)90200-0. [DOI] [PubMed] [Google Scholar]
- 74.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.