Abstract
Background and Aims The largest subfamily of orchids, Epidendroideae, represents one of the most significant diversifications among flowering plants in terms of pollination strategy, vegetative adaptation and number of species. Although many groups in the subfamily have been resolved, significant relationships in the tree remain unclear, limiting conclusions about diversification and creating uncertainty in the classification. This study brings together DNA sequences from nuclear, plastid and mitochrondrial genomes in order to clarify relationships, to test associations of key characters with diversification and to improve the classification.
Methods Sequences from seven loci were concatenated in a supermatrix analysis for 312 genera representing most of epidendroid diversity. Maximum-likelihood and parsimony analyses were performed on this matrix and on subsets of the data to generate trees and to investigate the effect of missing values. Statistical character-associated diversification analyses were performed.
Key Results Likelihood and parsimony analyses yielded highly resolved trees that are in strong agreement and show significant support for many key clades. Many previously proposed relationships among tribes and subtribes are supported, and some new relationships are revealed. Analyses of subsets of the data suggest that the relatively high number of missing data for the full analysis is not problematic. Diversification analyses show that epiphytism is most strongly associated with diversification among epidendroids, followed by expansion into the New World and anther characters that are involved with pollinator specificity, namely early anther inflexion, cellular pollinium stalks and the superposed pollinium arrangement.
Conclusions All tested characters show significant association with speciation in Epidendroideae, suggesting that no single character accounts for the success of this group. Rather, it appears that a succession of key features appeared that have contributed to diversification, sometimes in parallel.
Keywords: Epidendroideae, Orchidaceae, diversification, epiphytism, phylogenetics, missing data
INTRODUCTION
Ever since the first classification of Orchidaceae (Swartz, 1800), systematists have sought to reveal the relationships in this largest plant family. Numerous classifications have been proposed, each re-evaluating and adding character evidence and incorporating new taxa as they were discovered and material became available for study. The past 20 years have seen an especially striking amount of progress in understanding phylogenetic patterns in orchids. Prior to that time, 200 years of morphological study had delimited the more clearly demarcated groups – Apostasioideae, Cypripedioideae, Orchidoideae in a narrow sense, comprising most terrestrial orchids with sectile pollinia, and Epidendroideae, generally accepted to be those orchids possessing an incumbent or inflexed anther at maturity (Dressler, 1981). The essentially unplaced remaining orchids comprised an assemblage of soft-pollinium-bearing, primarily terrestrial species that seemed to have affinities to various of the major well-defined taxa, but did not share clear synapomorphies with any of them (Rasmussen, 1982). These ‘neottioid’ orchids were eventually shown by molecular analyses to comprise the early diverging elements of broadened Orchidoideae and Epidendroideae. The other group of uncertain affinities, Vanilla and its relatives, showed affinities with epidendroids (the incumbent anther), but shared many plesiomorphies with neottioids. Elucidation of their position as a distinct lineage (Vanilloideae) near the base of the family (Cameron et al., 1999) further clarified the circumscription of Epidendroideae and revealed the incumbent anther to have been gained in parallel in epidendroids and vanilloids. Subfamilial circumscriptions having been clarified, progress was then made on relationships in each of these units. Whereas much progress has been made resolving the composition of tribal and subtribal groups of epidendroids, understanding relationships among these has remained challenging.
Today we recognize Epidendroideae as the largest of the five subfamilies, comprising approximately 21 160 species or 76 % of the family (Govaerts et al., 2014). Given that they constitute the major part of the orchid radiation and have many of the signature characteristics of orchids, understanding diversification of epidendroids must rank among the primary goals of current orchid systematics. As with most phylogenetic studies undertaken today, reconstructing a well-supported hypothesis of epidendroid relationships is important not just for a stable classification for the subfamily, but also to provide a basis for understanding the pattern of diversification – specifically in this case to seek correlations of character change with species richness.
This study is an analysis of the major DNA sequence datasets that have been produced over the past 20 years. Its primary objectives are to: (1) assemble the broadest sampling of epidendroids thus far for as many loci as possible in order to further resolve relationships across the subfamily; (2) interpret patterns of morphological characters on the resulting trees and seek correlations between specific character transformations and species diversity; and (3) evaluate recent classifications of the subfamily and propose realignments where necessary in light of these results.
MATERIALS AND METHODS
Eight DNA sequence data sets were assembled, derived from seven loci. Some new sequences were produced for the analysis and were combined with pre-existing sequences from GenBank (Supplementary Data File S1). The loci/regions used are (1) a section of the nuclear ribosomal repeat comprising internal transcribed spacer (ITS)1–5.8S–ITS2, (2) nuclear xanthine dehydrogenase (XDH), (3) plastid matK, (4) plastid rbcL, (5) plastid trnL-F spacer + intron, (6) plastid ycf1a, (7) plastid ycf1b and (8) the mitochondrial nad1 b-c intron. ycf1a and ycf1b refer to the approx. 1780 bp 3′ region of the gene and the approx. 1200 bp 5′ region of the gene, respectively, that were used in the study of Neubig et al. (2009). This analysis focuses on discovering relationships among genera of Epidendroideae, so genera were used as terminals. Because many available sequences were produced using different DNA accessions and even different species, in many cases sequences from more than one species were concatenated to create synthetic generic terminals. As long as a sequence was from the target genus, it was eligible for inclusion. Although in some cases sequences from multiple species were used, in most cases the majority of sequences for a genus were derived from just one or two species; in all cases an attempt was made to minimize the number of species utilized to represent a genus (details given in File S1). Some genera that appear in the analysis have been subsumed into other genera in recent treatments; we maintain the names here to give a sense of the taxonomic scope sampled, but also provide the current generic names. Given the large amount of phylogenetic work done with orchid genera over the past 30 years, we are confident that our terminals in general represent monophyletic groups and therefore that error due to the combining of sequences from non-monophyletic units is minimal. Five outgroup genera were chosen from other orchid subfamilies: Neuwiedia (Apostasioideae), Spiranthes and Platanthera (Orchidoideae) and Paphiopedilum (Cypripedioideae).
Alignments were created for each locus individually by aligning in MUSCLE (Edgar, 2004) and adjusting manually. Only the ITS and trnL-F alignments had regions with questionable homology; some of these were excluded from the analyses (see matrices). A total of 41 indels were coded for ITS, matK, trnL-F, ycf1 and nad1 using the simple coding procedure of Simmons and Ochoterena (2000). The alignments were loaded into an application called GENERATOR (Matt Yoder, Illinois Natural History Survey, Champaign, IL, USA, unpubl.) that facilitated selection of individual sequences for each generic terminal and then output an interleaved data matrix along with a listing of the details for the sequences that were chosen for each genus. The general rule for inclusion of a genus was that sequences for at least three of the eight loci needed to be present. A few exceptions were made for unusual or particularly important genera for which only fewer sequences were available.
The full matrix, comprising 312 ingroup genera and five outgroups, contained 55 % missing values (including indels). The matrices are provided in Files S2 and S3 (Supplementary Information). To evaluate the effect of missing data, reduced taxon matrices were assembled for which missing values were also reduced. The first comprised 175 genera, for each of which at least five sequences were present, and the second 49 genera, which focused on representing each major group (i.e. subtribe) with one or two of the most completely represented genera. Missing data comprised 40 % for the 175-taxon matrix and 34 % for the 49-taxon matrix (the missing data were largely indels). Tree search and jackknife analysis were performed for all matrices using parsimony in TNT (Goloboff et al., 2008). For initial tree search and strict consensus tree reconstruction the following settings were used: hold 20 000 trees, collapse level = 6, xmult = level 8, bbreak = tbr fill. Parsimony jackknifing (Farris et al., 1996) by clade frequency was performed, holding up to 100 trees per replicate for 5000 replicates, utilizing collapse level = 6 and a search strategy of xmult = level 5, bbreak = tbr fill for each replicate and with a character deletion probability of 0·37.
Maximum-likelihood (ML) tree search and bootstrapping were simultaneously performed using the ‘-f a’ option in RaxML version 7.4.2 (Stamatakis, 2006) after excluding gap-only columns in the matrix. A run with 500 replicates was performed that contributed to the search for the best tree (as every fifth bootstrap replicate is also used as a starting tree for the ML tree analysis in RaxML), with an additional 500 replicates performed that were used only for the bootstrap analysis – giving 1000 replicates in total for the bootstrap. Each of the eight sequence subsets was coded as a separate partition, and the GTR + Γ model was used. Indels were included in a single additional partition, and the Mk model (Lewis, 2001) was specified. For the reduced taxon matrices, columns comprising only missing data were removed before the analyses were performed. Analyses of plastid loci only and nuclear loci only were also performed using 500 replicates for the simultaneous tree search and bootstrap procedure to determine the degree to which incongruence might be present among loci from different genomes.
Analysis of patterns of diversification was carried out in Mesquite (Maddison and Maddison, 2011; Midford and Maddison, 2011) using the BiSSE approach of Maddison et al. (2007). The ML tree was used and one parsimonious tree was arbitrarily selected and imported into RaxML where ML branch lengths were calculated; this tree was then exported and also used for the BiSSE calculations. Three morphological characters (superposed pollinia, early anther inflexion and cellular pollinium stalk) were analysed along with habit (terrestrial/epiphyte) and geography (Old World/New World). The log likelihood difference test was used to compare constrained versus unconstrained scenarios to test for significant association of individual characters with speciation on the tree. Specifically, pairs of runs compared the situation where the instantaneous speciation rates (λ0 and λ1) are allowed to vary with the constrained situation, λ0 = λ1, testing whether rates are symmetric or not. The log likelihood difference was used as the test statistic, and twice this difference is expected to follow a chi-square distribution with one degree of freedom (because of six free parameters in the unconstrained case and five in the constrained). The optimal ML tree and a single most parsimonious tree also were used for character mapping, with character optimizations produced in Mesquite using the parsimony option. Although >20 000 most parsimonious trees were found, a high level of structure is present in the strict consensus, arguing for the representativeness of this approach for the characters that we are using.
RESULTS
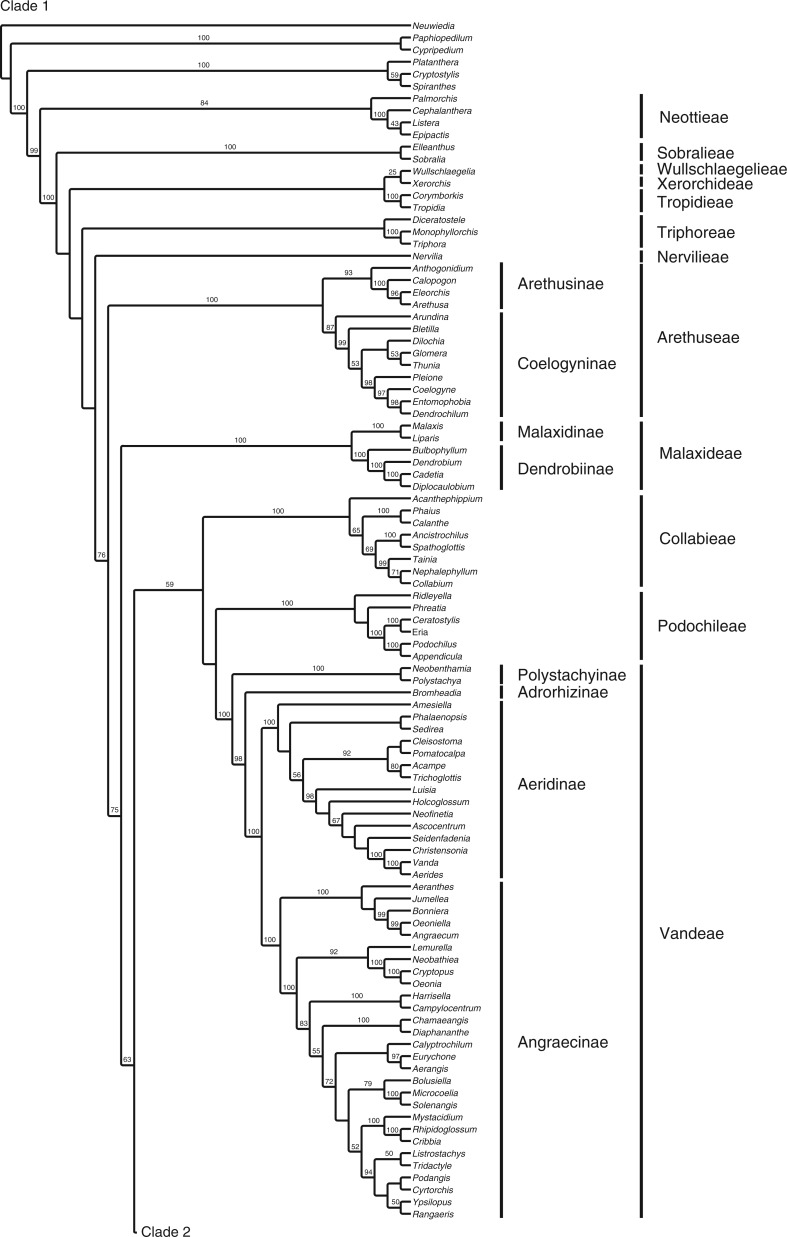
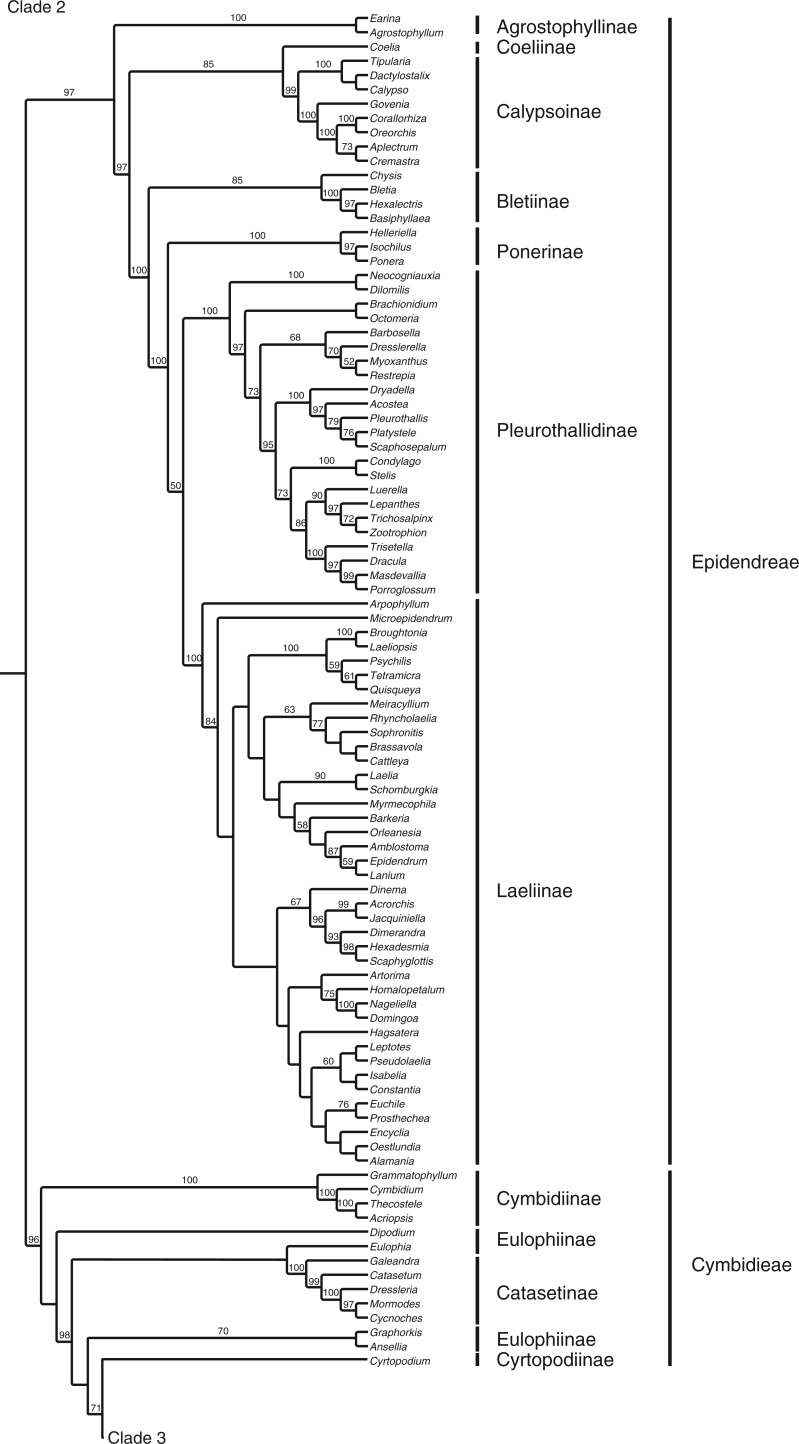
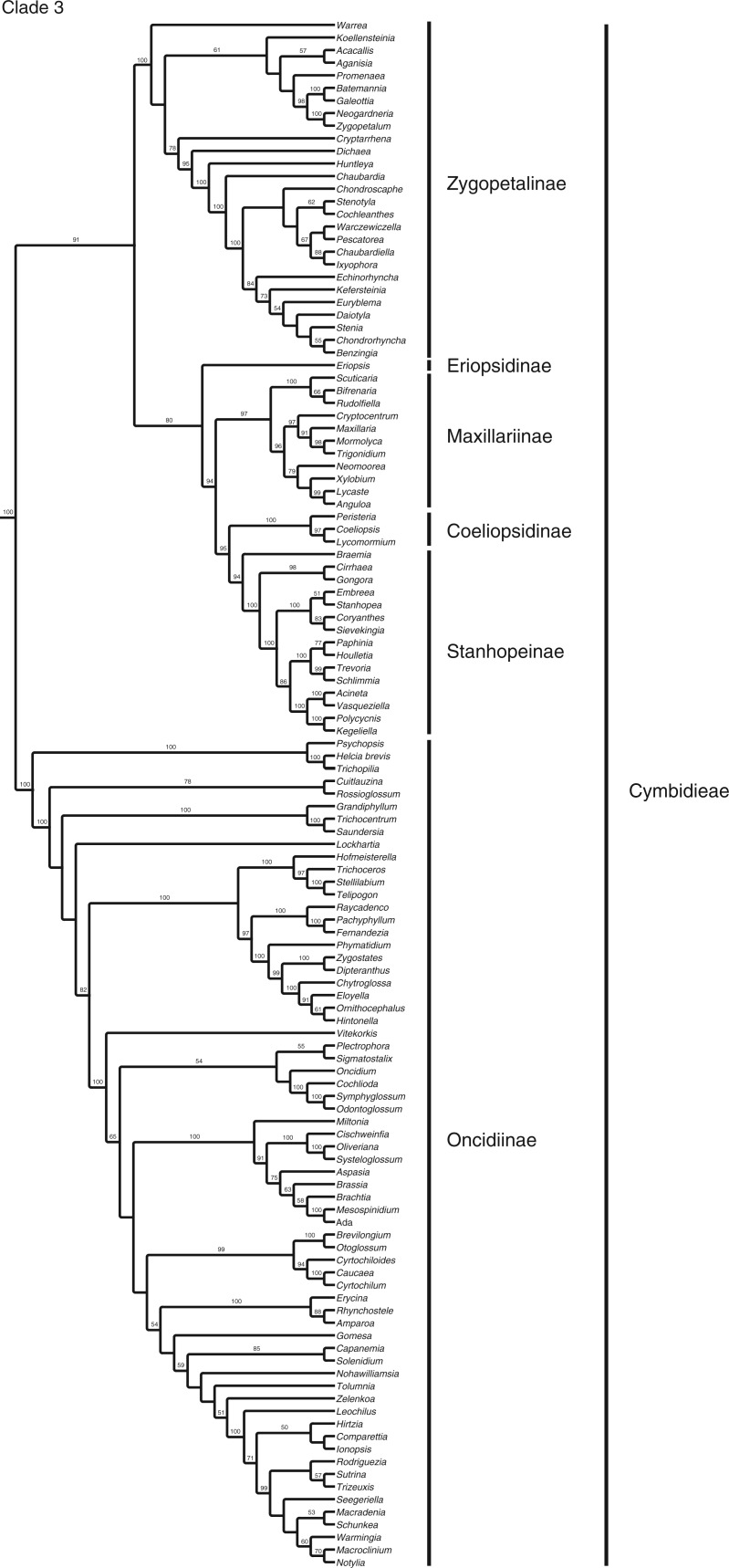
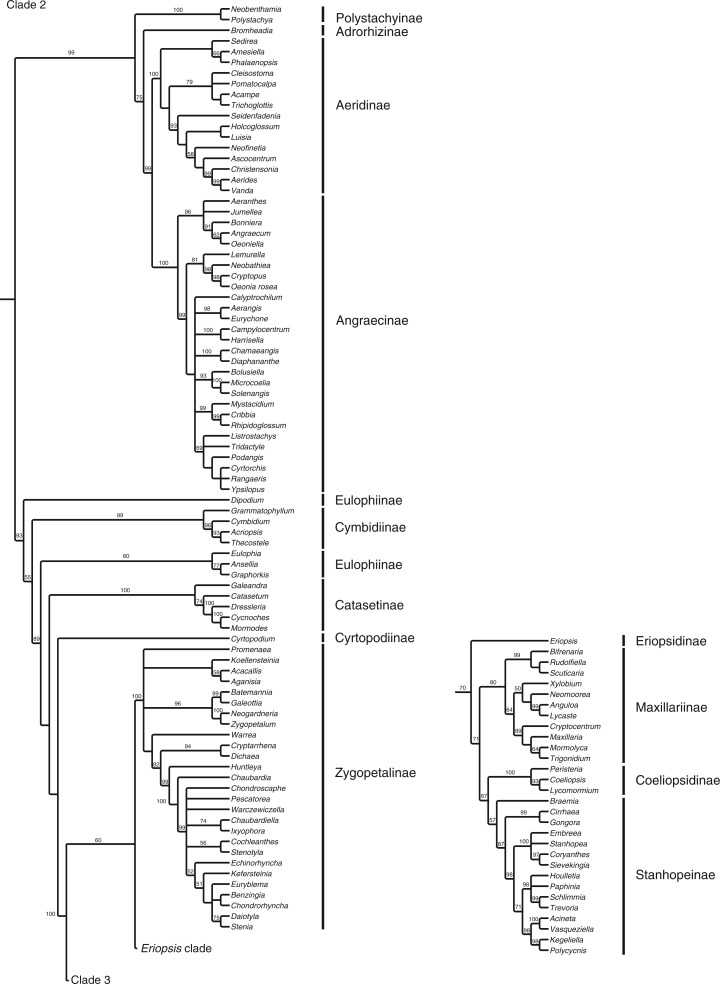
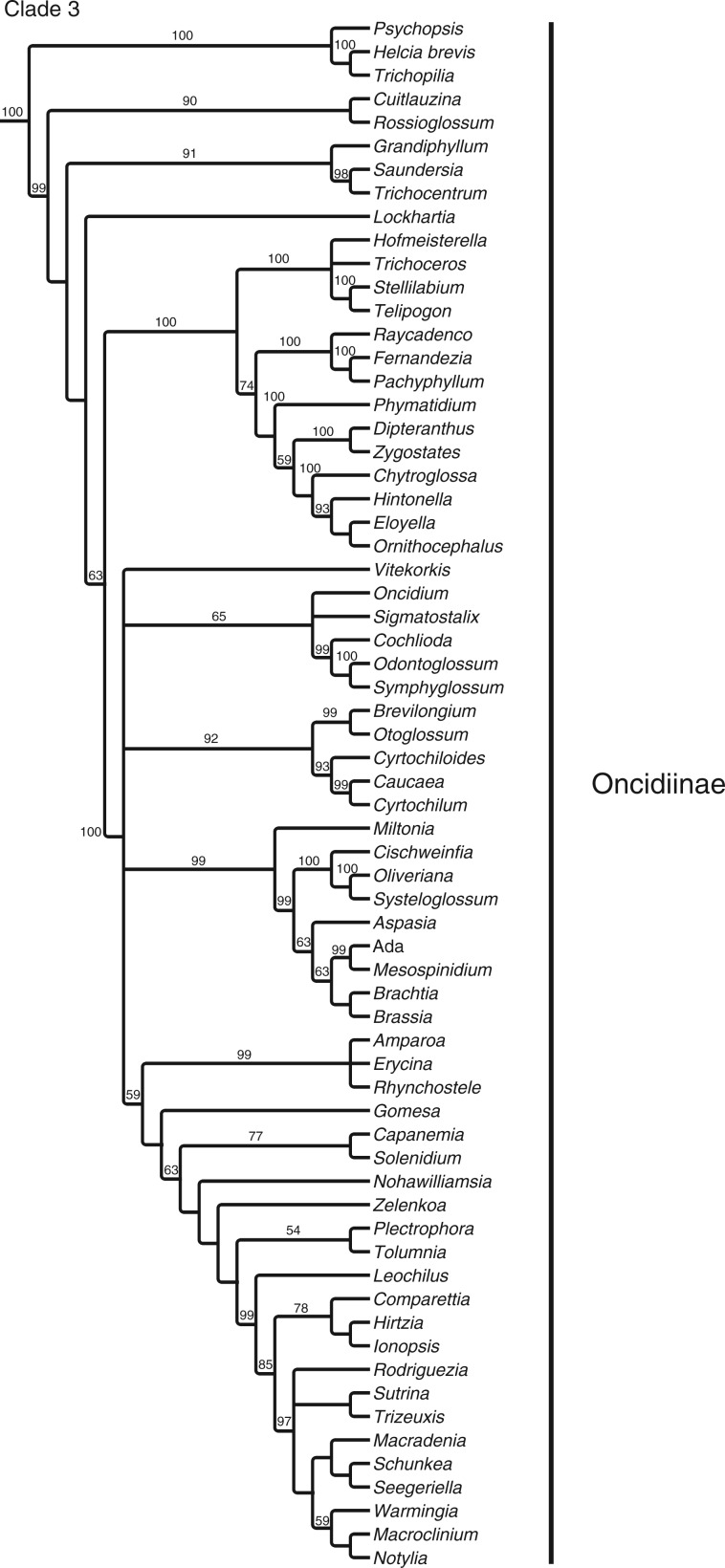
The full matrix comprised 12 462 aligned positions and 41 indels. ML analysis yielded a tree with significant support for many clades (Fig. 1; Supplementary Data Fig. S1 shown with branch lengths). The parsimony analysis yielded >20 000 most parsimonious trees [length = 36 420, consistency index (CI) = 0·183, retention index (RI) = 0·179]. The strict consensus is very similar to the ML tree (Fig. 2). Although there are some differences in topology, in almost all cases the differences between these two trees involve branches that have weak (<60) support. There are no examples of strongly supported major groups that conflict in the two trees. There are only three examples of individual genera for which well-supported placement differs in the two trees: Phreatia, Lepanthes and Dichaea. The last two involve local rearrangements within small clades, but the first is more striking. In the parsimony tree, Phreatia is united with Malaxis + Liparis [maximum parsiomony (MP): 87] and then Bulbophyllum + Dendrobium + Diplocaulobium + Cadetia, whereas in the likelihood tree it falls in a clade composed of Ridleyella + Ceratostylis + Eria + Appendicula + Podochilus (ML: 100).
Fig. 1.
ML tree with branch support values and the tribal/subtribal classification used here.
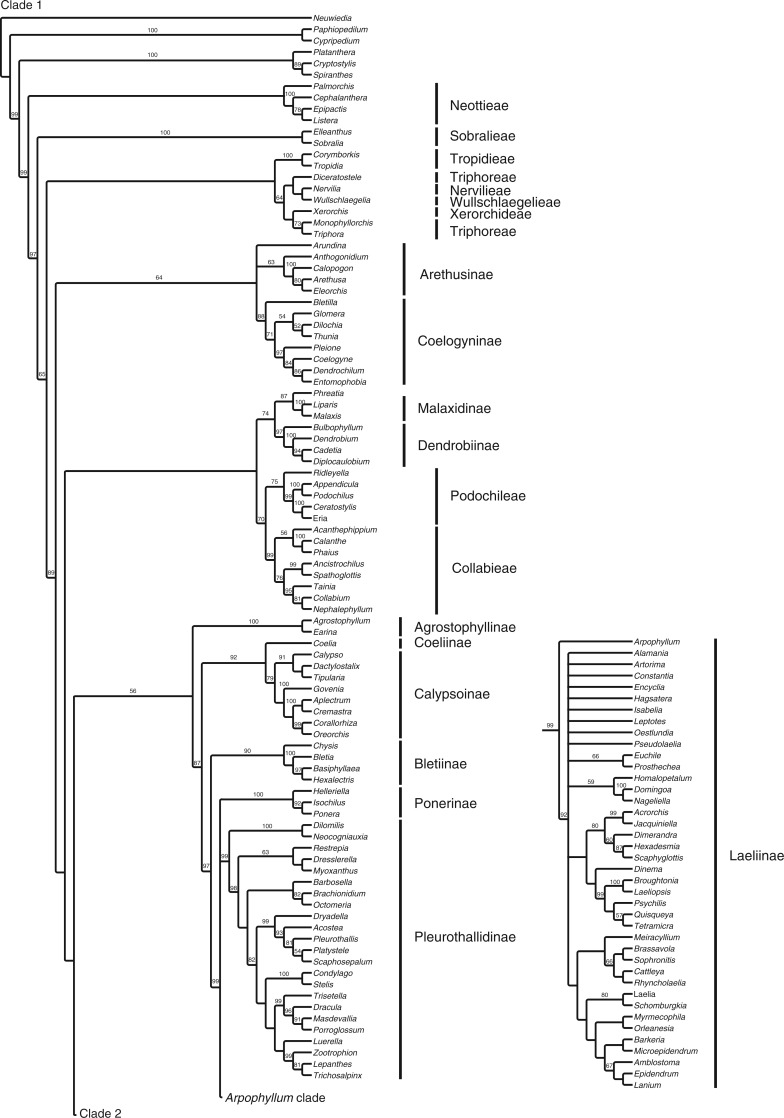
Fig. 2.
Parsimony consensus tree with jackknife values and the classification used here.
Comparing the plastid (matK + rbcL + trnL-F + ycf1a + ycf1b) ML analysis with the nuclear (ITS region + XDH) ML analysis, there were no supported (>50) incongruent relationships among groups at the subtribal level or above. All supported incongruences involved minor rearrangements of genera within subtribes (Supplementary Data Figs S2 and S3).
Comparison of full and reduced taxon data sets
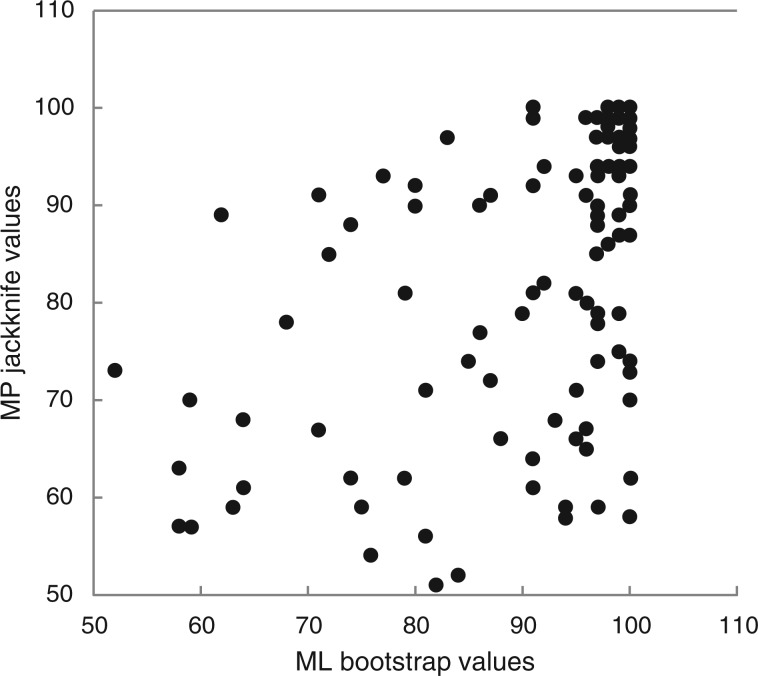
Parsimony jackknife percentages of the full matrix were in general somewhat lower than those for corresponding clades using ML bootstrap (Fig. 3). Seven times as many clades with ML support of 50 % or above showed a reduction in support on the parsimony tree of 10 points or more as showed an increase of the same magnitude. A correlation analysis of support values for individual clades (excluding numbers <50) gave r = 0·63, indicating fairly strong correspondence between the two approaches (the slope of the regression line is 0·48). Comparing the 317- and 175-taxon matrices between analyses, the parsimony support values changed somewhat more (a total of 518 points for clades with ≥10 point change) than the ML values (435 points for similar clades). For both ML and MP analyses, more clades showed an increase in support than a decrease (nine vs. five with ML and 13 vs. four with MP for clades that changed ≥10 points). In no case was there a disagreement in relationship for clades supported at the 60 % level or greater when comparing among the 317-, 175- and 49-taxon matrices in ML or MP analysis sets (Supplementary Data Figs S4–S7).
Fig. 3.
Graph of parsimony jackknife values against ML bootstrap support values.
One notable pattern of change in support values between matrix sizes that appeared for both ML and MP concerns three adjacent nodes in Oncidiinae (a: Rhynchostele + Erycina and its sister; b: Gomesa and its sister; c: Capanemia + Solenidium and its sister). In each case there was an increase in support from the 317- to 175-taxon matrices (ML: 54 to 97, 49 to 93 and 59 to 98; MP: 59 to 93, <50 to 86 and 63 to 91, respectively). There were decreases exhibited by both analyses as well: Zygopetalinae + Eriopsidinae + Maxillariinae + Coeliopsidinae + Stanhopeinae fell in support in ML from 91 to 83 to 68, whereas in MP it was reduced from 60 to 71 to 54. Additional notable changes in support and position were shown in the MP analysis. The clade comprising Malaxis + Liparis + Phreatia + Bulbophyllum + Dendrobium + Cadetia + Diplocaulobium showed 74 MP support with the 317-taxon matrix, <50 with the 175-taxon matrix and 73 with the 49-taxon matrix. At the same time, Phreatia shifted position from that described above with the 317-taxon matrix to sister to Podochilus + Eria with 82 MP support in the 49-taxon matrix. As another example, Earina + Agrostophyllum are sister to a clade comprising Coelia + Calypsoinae + Laeliinae + Pleurothallidinae with 56 MP support in the 317-taxon matrix, whereas support increased to 77 in the 175-taxon matrix (it is always >90 in the ML analyses). Finally, although a sister relationship between Vandeae (including Polystachyinae) and Cymbidieae is not supported in the 317- and 175-taxon matrices, it received 88 MP support in the 49-taxon analysis.
Diversification analyses
The results of the diversification analyses using BiSSE are shown in Table 1. For both ML and parsimony trees, all characters tested here are significant for their association with speciation on both the likelihood and parsimony trees. The most significant relationship is between diversification and the shift from terrestrial to epiphytic habit. The second most significant test was not with a morphological feature but with geography, the shift from Old World to New World. Following these, superposed pollinia, cellular pollinium stalk and early anther inflexion were less significant, but all still highly significant at P < 0·005. Tests for significant association with extinction were also run for both trees and all five characters, but none of them was significant at the P < 0·05 level (results not shown).
Table 1.
Results of BiSSE analyses.
| Tree | Character | Unconstrained | Constrained | Likelihood Δ | 2 × ln Δ | P |
|---|---|---|---|---|---|---|
| ML | Habit | −795·50164395 | −774·017225996 | 21·48441795 | 42·968834 | <0·0005 |
| ML | Distribution | −821·10233274 | −813·26639135 | 7·83594138 | 15·671882 | <0·0005 |
| ML | Superposed pollinia | −846·24835521 | −841·29297584 | 4·95537937 | 9·9107586 | <0·0025 |
| ML | Cellular pollinium stalk | −848·97211750 | −844·40586568 | 4·56625182 | 9·1325036 | <0·005 |
| ML | Early anther inflexion | −853·18216746 | −848·68035906 | 4·50180840 | 9·0036168 | <0·005 |
| MP | Habit | −789·72008491 | −771·53784604 | 18·18223887 | 36·364476 | <0·0005 |
| MP | Distribution | −816·60665361 | −809·08041921 | 7·52623440 | 15·052468 | <0·0005 |
| MP | Superposed pollinia | −842·54765661 | −837·51177948 | 5·03587713 | 10·071754 | <0·0025 |
| MP | Cellular pollinium stalk | −848·02432112 | −843·93252768 | 4·09179343 | 8·1835868 | <0·005 |
| MP | Early anther inflexion | −852·51204453 | −848·52491453 | 3·98712999 | 7·9745998 | <0·005 |
DISCUSSION
Missing data and supermatrix analysis
Because of the way in which molecular datasets have been assembled over time for smaller studies within the subfamily, to utilize the available data at the scale of the entire subfamily an approach that allows the creation of synthetic terminals at some level was chosen. The upper bound on this data combination used here is the genus, meaning that to the extent that genera chosen here are monophyletic, there should be no error due to loci for some terminals being more closely related to loci from others. We feel confident that this is the case for the great majority of these genera. However, beyond issues with synthesizing terminals, there is also the question of the effect of missing data. Supermatrix analyses often have large numbers of missing data, and some investigators have commented on the shortcomings of supermatrix analysis that are due to that issue (Thomson and Shaffer, 2010; Simmons and Norton, 2013). The effects of missing data on cladistic analysis have been studied in some detail (e.g. Nixon and Davis, 1991; Maddison, 1993; Wiens, 2006). Wiens (2003) concluded that it is not just the number of missing data but rather their distribution that is important, which makes it difficult to predict their effects on a particular analysis; most importantly, it is clear that successful analyses can be carried out even in the presence of large numbers of missing data. More recently, attention has been paid to the effects of missing data on support analyses, where it is clear that they can have an important effect on support values (Thomson and Shaffer, 2010; Simmons and Freudenstein, 2011; Simmons and Norton, 2013). Here we conducted three levels of analysis, with 317, 75 and 49 taxa, respectively, which had proportionally fewer missing data as the number of taxa decreased. We found little difference in the topologies of the support trees among these analyses, although support was seen to vary. In general, support increased with fewer taxa and fewer missing data, which might be expected in general because with characters spread over fewer branches they should yield higher support, although this was not always the case. This suggests that the effects of sampling sometimes interact with branch lengths in unpredictable ways. Perhaps most striking here is the effect that we observed with the only well-supported disagreement in placement between the MP and ML analyses – involving Phreatia. We usually value the effect of increasing sample size – the densest sampling is expected to give us our best estimate of the phylogenetic pattern. Here the position of Phreatia with Malaxidinae in the full MP analysis is distinct from its more expected (consistent with morphology) position with Podochileae seen with ML. However, when the MP dataset is reduced to 49 taxa, Phreatia assumes a position with Podochileae as well, suggesting that sparser sampling may be giving a better result.
Patterns of generic and tribal relationship: taxonomic implications
In the following analysis of relationships, names of tribes and subtribes are those used by Chase et al. (2015). ML and parsimony MP support values for clades are given as (ML/MP); support values ≤50 % are described as ‘no support’.
The ‘basal’ epidendroids have long been problematic in their composition and placement. Some of these lineages appear to be ‘relics’, in the sense that they comprise one or a few species on fairly long branches and do not have clear morphological synapomorphies linking them to other groups. Many of these species, also known as part of the ‘neottioid’ orchids, were allied with members of Orchidoideae/Spiranthoideae by early authors and even up to Dressler (1993) because of their soft pollinia, terrestrial habit and anthers that are often more erect than incumbent; the assemblage was studied by Rasmussen (1982), who agreed with Dressler (1974) that it was probably not a natural group. The morphological cladistic study of Freudenstein and Rasmussen (1999) and to a greater extent the rbcL study of Cameron et al. (1999) helped in clarifying the boundaries of Epidendroideae by sorting ‘neottioids’ into appropriate orchidoid, vanilloid and epidendroid groups. Although a relatively small assemblage, the truly ‘basal’ (early diverging) epidendroids have been fairly heavily split into tribes, reflecting their lack of unifying features (e.g. Dressler, 1981, recognized ten subtribes and Chase et al., 2015, recognized nine). In addition, several genera traditionally placed among these primitive epidendroids are leafless; Gastrodieae comprise only leafless species, many rarely seen, and Epipogiinae are also leafless. Most were not included in the present study due to lack of material or difficulty in amplifying plastid DNA loci (Wullschlaegelia is the only such genus included here).
Although some previous studies have suggested the basal placement of Neottieae in the subfamily (e.g. Goldman et al., 2001; Freudenstein et al., 2004; Górniak et al., 2010), here it is strongly supported as sister to the remainder of epidendroids based on multiple loci, as it was by Xiang et al. (2012). This is a wholly terrestrial group of wide distribution, with temperate and tropical members. Their terrestrial habit, in combination with the primarily terrestrial habit of the other early-branching epidendroids and of the other subfamilies, clearly fixes the plesiomorphic habit of the subfamily as terrestrial (Supplementary Data Figs S8 and S9).
The placement of the Neotropical Palmorchis as sister to Neottieae is supported in the ML analysis (84) and the genus also falls there in the parsimony analysis, but not with support >50 %. A close affinity between Palmorchis and Neottieae was indicated by Freudenstein et al. (2004) and Górniak et al. (2010); Xiang et al. (2012) recovered a Bayesian posterior probability of 0·98 for the Palmorchis + Neottieae clade based on three plastid loci. This relationship has not been suggested previously based on morphology, and we can suggest no morphological features that link them. The broad geographical distribution of such a clade would be consistent with its being an old group within the subfamily. Szlachetko (1995) placed Diceratostele with Neottieae in a reduced Neottioideae, but we found no evidence for that relationship here.
Although usually placed among the early-branching epidendroids based on structural features, Wullschlaegelia has been suggested to belong with the relatively advanced Calypsoinae (Calypso and related genera have often been treated as a tribe, but in Chase et al., 2015, they are a subtribe) based on 18S gene sequence data (Molvray et al., 2000) and a putative matK gene sequence (Freudenstein et al., 2004). Like Górniak et al. (2010), and in part because of their XDH data, this study places Wullschlaegelia outside the advanced epidendroids with relatively strong support (76/89), although it is not placed with any other early-branching genera with significant support. Morphological features also would not place Wullschlaegelia with advanced groups, especially Calypsoinae, given that the former has sectile, simple pollinia, which do not occur among advanced epidendroids including Calypsoinae. It is unclear why the 18S and matK sequences indicate a relationship with advanced groups; for matK it could be the presence of a paralogue, as was seen in the leafless Corallorhiza (Freudenstein and Senyo, 2008), whereas with the18S analysis it could have been the relatively meagre sampling. Studies that continue to include Wullschlaegelia among Calypsoinae (e.g. Zhai et al., 2013) without including as well many of the neottioid clades must be careful about interpretation.
Both ML and parsimony analyses place Elleanthus with Sobralia and Tropidia with Corymborkis with strong support. These groups have long been recognized as tribal or subtribal groups (e.g. Sobralieae, Tropidieae). Their specific affinities to other early-branching epidendroids remain uncertain. Szlachetko (1995) placed Elleanthus and Sobralia, which have long been recognized as closely related, in different subfamilies.
The enigmatic Neotropical Xerorchis is placed with Tropidia and Corymborkis in the ML analysis (<50) and with Triphora and Monophyllorchis in the parsimony tree (<50). We are unaware of morphological characters that link Xerorchis with either of these pairs of genera; Xerorchis is similar to Sobralieae in that it has eight pollinia, but a close relationship between the groups is not indicated here.
The African monospecific Diceratostele is placed in a clade with Sobralieae and Triphoreae in the ML tree, but with no support >50 %; the parsimony consensus places it with Triphoreae with weak support (64). Its Old World distribution contrasts with the New World distribution of Triphoreae sensu Dressler (1993), but such an association could be an old one and is similar to other putatively ancient South American–African disjunctions (e.g. among some genera of Caricaceae and Strelitziaceae). Nervilia is also African–Asian, but shares no clear characters with the other genera; it has an isolated position in the ML analysis, but with low support, and is a member of a weakly supported clade of early-branching genera in the parsimony analysis.
The remainder of the epidendroid genera comprise a clade (76/89) that corresponds to the ‘advanced’ Epidendroideae – those epidendroids that have more elaborate pollinium/pollinarium structures and are primarily epiphytic. The first branch of this clade contains a strongly supported group (100/64) that corresponds to Arethuseae of Chase et al. (2015). The only difference in clade membership here with respect to that classification is that Arundina is placed with some support (87/–) with Coelogyninae rather than with Arethusinae, which are themselves strongly supported (93/63) here.
The remainder of the epidendroids are supported with ML (76) and <50 % with MP. The next subclade is well supported (100/74) and corresponds to Malaxideae and Dendrobiinae of Chase et al. (2003), with each group also being well supported. Dendrobiinae was left as ‘unplaced’ in epidendroids by Chase et al. (2003), but in Chase et al. (2015) Dendrobiinae are included in Malaxideae. Dendrobiinae fit well here in that both Malaxidinae (recognized at the subtribal level in Chase et al., 2015) and Dendrobiinae are characterized by largely naked pollinia (but see Li and Yan, 2013). A clade of Dendrobiinae (including Bulbophyllum and allies) and Malaxideae was also suggested by Yukawa et al. (2000). As noted above, placement of Phreatia is one of very few instances in which ML and MP disagree among well-supported clades. In the full analyses, Phreatia is sister to Malaxidinae in MP, whereas in ML it is placed with Eriinae. Phreatia remains with Malaxidinae in the 175-taxon in the MP analysis, but with reduced support (66). However, in the 49-taxon analyses, both MP (82) and ML (100) place Phreatia with Podochilus + Appendicula, even though Liparis is still in the matrix as a representative of Malaxidinae. With respect to morphology, we view it as properly placed in Podochileae.
The next subclade, comprising (Collabieae + Podochileae + Vandeae), is only weakly supported by ML (59) and does not appear as a group with MP. Collabieae are strongly supported (100/99), as are Podochileae (100/75), although the specific relationship of those clades to one another is not; they are successive clades in ML and sister clades in MP. Collabieae were another group left as unplaced by Chase et al. (2003).
There is a potential structural synapomorphy to link Podochileae and Vandeae – spherical stegmata. Møller and Rasmussen (1984) found spherical stegmata in Vandeae and Podochileae, although no stegmata were observed in examples of Collabieae investigated. Spherical stegmata are known otherwise only from Dendrobiinae (Møller and Rasmussen, 1984), which according to the topology presented here would be an independent derivation, although the intervening clade is not well supported, leaving open the possibility that Malaxidinae, Podochileae, Collabieae and Vandeae could form a clade. Stegmata are not known from Malaxidinae or Bulbophyllum of Dendrobiinae (Møller and Rasmussen, 1984). The only other discordant observation in this scenario is that Dressler and Cook (1988) observed conical stegmata in Eria javanica, a member of Podochileae, although other species of Eria were shown by Møller and Rasmussen (1984) to have spherical stegmata.
Vandeae sensu Chase et al. (2003) comprise Polystachyinae (here, Neobenthamia and Polystachya; the former now considered a synonym of the latter; Russell et al., 2010), which is strongly supported in both ML and MP (100/100), and the more traditional circumscription (‘core’ Vandeae) of Aeridinae + Angraecinae, which is also strongly supported (100/99) as a group; Polystachyinae + ‘core’ Vandeae are also strongly supported (100/99) as a group. It was not until molecular analyses such as Freudenstein et al. (2004) that a sister relationship between the two primarily palaeotropical groups Polystachyinae and core Vandeae was suggested.
One genus not previously associated with Vandeae, but now coming between Polystachyinae and core Vandeae in both ML and MP analyses with strong support, is the Old World Bromheadia. Previously placed with Cymbidieae by Dressler (1981, 1993), Bromheadia fits well in a broad Vandeae due to its Asian distribution. Its affinities to Cymbidieae were questioned by Dressler (1993), being held there primarily by sharing the Cymbidium velamen type. Szlachetko (1995) placed Bromheadia in his Polystachyeae, but Collabiinae were also included there.
Within ‘core’ Vandeae, two principal clades are resolved, one equivalent to Aeridinae (100/100) and the other to Angraecinae (100/100). These correspond to clades resolved by Carlsward et al. (2006). They concluded that Angraecinae and the previously recognized Aerangidinae (cf. Dressler, 1993) were both polyphyletic based on their analysis and, at the generic level, at least Angraecum is grossly polyphyletic. Although monophyletic units that include Angraecum and Aerangis, respectively, could be recognized as subtribes, we find no compelling reason to do so based on morphological or chromosome data, and so recognize a broader Angraecinae as in Carlsward et al. (2006) and Chase et al. (2003, 2015).
Agrostophyllinae, here represented by Agrostophyllum and Earina and placed together strongly by both analyses (100/100), were an unplaced clade in the system of Chase et al. (2003). Here they are strongly supported by the ML analysis (97) and weakly so by MP (56) as sister to a large, primarily New World clade of subtribes in Epidendreae. Agrostophyllum had been included previously in Podochilinae (Dressler, 1981) and was allied to it by Szlachetko (1995), but it is anomalous there in its possession of conical stegmata (Møller and Rasmussen, 1984). Earina had previously been associated with Glomera (Dressler, 1993), but that genus now falls among Coelogyninae. Both Earina and Agrostophyllum were placed near Vandeae and apart from Podochilinae based on plastid sequence data by van den Berg et al. (2005). The plastid sequence sub-analysis performed here places them with Epidendreae as in the full analysis.
The first group in the next, primarily New World clade comprises genera belonging to Calypsoinae except for one – Coelia – which is sister to those subtribes in both ML (85) and MP (92) analyses. Coelia was included in Epidendreae but with no subtribal affiliation by Chase et al. (2003). It was shown to be sister to Calypsoinae by van den Berg et al. (2005), but with no support. It is a small Neotropical genus with eight pollinia (but no cellular pollinia stalks) and distinct pseudobulbs. Calypsoinae are largely temperate (Govenia extends in temperate habitats through the Neotropical zone; Garcia-Cruz and Sosa, 2005); as in Freudenstein (1994), Calypso + Tipularia + Dactylostalix are sister to the formerly recognized Corallorhizinae, with Govenia sister to core Corallorhizinae (Aplectrum, Corallorhiza, Cremastra and Oreorchis). Zhai et al. (2013) presented an analysis of Calypsoeae, but almost all Bayesian posterior probabilities for their clades relating genera were not significant (<90), so the relationships depicted there are unreliable.
Chysis, which has also been previously of uncertain placement (Chase et al., 2003), is here strongly united (85/90) with Bletia, Basiphyllaea and Hexalectris (comprising Bletiinae). These genera in turn are strongly supported as sister to Poneriinae + Laeliinae + Pleurothallidinae. Ponerinae, Laeliinae and Pleurothallidinae were all recovered as strongly supported groups (100/100, 100/99 and 100/99, respectively). Sister to the rest of this group are Ponerinae, but their relationships to the remaining subtribes in this group are less clear. The ML analysis shows Pleurothallidinae + Laeliinae to be monophyletic, although with only weak support (50), whereas the parsimony analysis resolves Ponerinae in a polytomy with Pleurothallidinae and Laeliinae. van den Berg et al. (2009) also found Laeliinae + Pleurothallidinae to be sisters based on plastid and nuclear sequences. Within Laeliinae, Arpophyllum is resolved as sister to the remainder of the subtribe in both of the present analyses. The relationships within these subtribes have been studied in detail by van den Berg et al. (2000, 2009) and Pridgeon et al. (2001).
The remainder of the genera comprise Cymbidieae, which are strongly supported in both analyses (96/93). The early-branching elements in both analyses are members of Cymbidiinae and Eulophiinae, although some details of relationships remain unclear because of poor support. Grammatophyllum, Cymbidium, Thecostele and Acriopsis are strongly supported as a group (Cymbidiinae, 100/99). The monophyly of Eulophiinae as a group of any size is questionable here – the subtribe comprises Ansellia + Eulophia + Graphorkis in MP, but with only weak support (60). The genera do not come together in a single group with ML; rather, Ansellia + Graphorkis is supported (70), but Eulophia is attached to a well-supported Catasetinae (100/100) with no support. Cymbidiinae are separated from most of the remainder of the tree by a strongly supported node (98/89). The positions of Cyrtopodium and Dipodium remain somewhat uncertain, moving among the more strongly supported groups in this part of the tree. Neither analysis supports the placement of these genera in Eulophiinae, as proposed by Chase et al. (2003). A recent analysis by Batista et al. (2014) that focused on placement of Cyanaeorchis also showed a lack of support for relationships among Eulophia, Geodorum, Ansellia, Cyrtopodium and Catasetinae, depending on which locus was used, but agrees with our strongly supported relationships in this part of the tree.
The rest of the tree comprises a strongly supported (100/100) New World clade corresponding to Maxillarieae of Dressler (1993) and Chase et al. (2003, 2015). One of two major subclades receives strong ML support (91), with less from MP (70). That subclade comprises several well-supported subtribal groups: Eriopsidinae, Maxillariinae, Stanhopeinae and Zygopetalinae. The other major subclade, Oncidiinae, is strongly supported in both analyses (100/100). Details of relationships within these subtribes have been analysed by Whitten et al. (2000, 2005, 2007, 2014) and Neubig et al. (2012). The relationships among some of these subtribes continue to be uncertain or poorly supported; however, our study provides some further resolution among them. Coeliopsidinae and Stanhopeinae were shown to be strongly supported as sisters by Whitten et al. (2000) and that relationship is confirmed here (95/67). The relationship between (Coeliopsidinae + Stanhopeinae) and Maxillariinae has been less clear, shown as unresolved by Whitten et al. (2014) and as a poorly supported group by Whitten et al. (2000). Here, the three subtribes are strongly supported as a clade (94/71). Eriopsis (Eriopsidinae) is then sister to that group with moderate support (80/70). Zygopetalinae are in turn sister to that whole assemblage with moderate to strong support (91/60), and finally Oncidiinae are strongly supported as sister to all of the other New World subtribes (100/100).
Major character patterns and diversification in epidendroids
Previous analyses of the subfamily had not yielded enough resolution and/or support to make an analysis of character patterns across the subfamily possible. Although the present analyses do not resolve all relationships with high support, there is enough resolution across the subfamily to make such an analysis meaningful. In this study, we analysed characters and their relationship to diversity on a tree using BiSSE. However, a number of assumptions of that method were violated here, as they probably are in many empirical studies. First, our trees are not ultrametric; real trees almost never are. Second, we do not have complete sampling of the subfamily; in fact we have only included the equivalent of 312 species out of approx. 19 785, or 1·6 %. However, we have covered much of the phylogenetic breadth of the subfamily (based on results of other broadly sampled studies of the family). One place in which our study has strength is in the numbers of terminals necessary for the statistical test to have sufficient power; Davis et al. (2013) suggested that studies with <300 terminals need to be interpreted with caution. Hence we interpret our BiSSE results with caution and in a more relative way with respect to the significant of association of characters with diversification rather than assigning meaning to the absolute likelihood values obtained.
Habit, vegetative structure and ecology
The epidendroids are a fundamentally tropical group; of approx. 19 785 species, about 99 % are tropical. No major radiations have occurred in temperate regions. The largest single temperate radiation of epidendroids is undoubtedly Neottieae, with approx. 85 temperate species, although it appears that some lineages in this tribe have crept back into the tropics (Aphyllorchis, some Neottia, Cephalanthera and Epipactis), based on the patterns shown in Pridgeon et al. (2005) and Xiang et al. (2012). The tropical/temperate distinction is intimately related to the epiphytic/terrestrial habit distinction. Although there are tropical terrestrial species, there are essentially no temperate epiphytic orchids. Although Epidendroideae are largely an epiphytic group (approx. 91 %; Atwood, 1986), it is clear that the plesiomorphic state is the terrestrial habit; all early-branching clades of the subfamily comprise terrestrial species (Supplementary Data Figs S8 and S9). The first branching clade that contains epiphytes is Sobralieae; Elleanthus and Sobralia each contain terrestrial and epiphytic species, as well as species in which plants may show either habit, suggesting that epiphytism has arisen independently in each. Arethuseae appear to be the only plesiomorphically terrestrial ‘advanced’ epidendroids, as the sister to Arethuseae, the remainder of the subfamily, is primarily epiphytic. Arethuseae are also relatively primitive in some floral characters (with soft or sectile pollinia; Dressler, 1993). Collabieae appear to represent a reversion to the terrestrial habit, as do Calypsoinae + Corallorhizinae, Bletia + Hexalectris + Basiphyllaea and most species of Eulophia. Pleurothallidinae appear to have shifted between epiphytism and the terrestrial habit a number of times. Dipodium also represents a terrestrial reversion, at least in the holomycotrophic species. Epiphytism appears to have arisen independently in Coelogyninae.
In this study, the ‘character’ that is most significantly associated with species diversity is epiphytism, as it was in the family-wide study of Gravendeel et al. (2004). However, epiphytism is not a simple character, but rather a syndrome associated with a suite of morphological features, including roots with velamen and thickened leaves and stems. The family could be viewed as being pre-adapted for epiphytism in that it has dust seeds that can easily be transported onto bark substrates. At the same time, its mycotrophic dependence could be a limitation for epiphytism, as the correct fungus needs to be present in a situation in which it can effectively aid in seed germination; any such limitation among epidendroids appears to have been overcome, given the high diversity of epiphytes in the subfamily.
Vegetative structure is also important for the ecology of epidendroids and is clearly related to habit. Development of a velamen of some type is necessary to prevent roots from drying between periods of moisture in epiphytes (cf. Porembski and Barthlott, 1988), but there is no key stem modification that appears to be necessary for success as an epiphyte, although thickened leaves are typical. A corm/pseudobulb type of thickened stem arises early in the tree, just past the ‘basal’ groups (i.e. with terrestrial Arethuseae and beyond; Supplementary Data Figs S10 and S11). However, this structure is lost in some species-rich groups (Aeridinae + Angraecinae; Pleurothallidinae). The former shifted from sympodial to monopodial growth and retained thickened leaves, whereas the latter often have wiry stems and thickened leaves but may be diminutive plants and occupy a less clearly epiphytic habit, which may also limit their water loss. Loss of distinct pseudobulbs also appears to have occurred multiple times in Laeliinae, but again thickened leaves are retained. Pseudobulbs have also been lost in some Oncidiinae; this includes the case of some ‘twig epiphytes’, in which a psygmoid (fan-shaped; e.g. Erycina) leaf arrangement occurs, as well as larger plants with elongate fan-shaped habits such as Lockhartia and Dichaea. Holomycotrophic groups among the advanced epidendroids, such as Corallorhiza, Hexalectris and Dipodium, have also lost corms/pseudobulbs, but these are always terrestrial and so are presumably less subject to the kind of moisture fluctuations that epiphytes encounter.
Corms are essentially the same as pseudobulbs except that they occur below ground; they are found among some terrestrial epidendroids and in fact probably have facilitated their entry into temperate regions in groups such as Arethuseae, Calypsoinae, Malaxidinae and Calanthe (see next section), as they serve to bring the plant through unfavourable seasons. Temperate orchid floras are in general dominated by members of Orchidoideae, most of which have root tubers that serve the same function. Many members of Neottieae [especially in Listera (=Neottia), Epipactis and Cephalanthera] are temperate but are plesiomorphically without stem thickenings.
Biogeography
The BiSSE analysis identified geographical shifts as the second most significant associations with speciation. Geography plotted on the trees reveals that major shifts between the Old World and the New World (those that resulted in highly species-rich groups) have happened only twice (Figs S12 and S13). The ML tree suggests that New World distribution is the plesiomorphic state, whereas on the MP tree it is ambiguous. This ambiguity is due to poor resolution among the small early-branching groups, suggesting that confidently resolving these will ultimately be essential for reconstructing this pattern on the tree. However, beyond the small early-branching groups (i.e. beginning with Arethuseae), the optimization for both topologies is Old World. There is a shift to the New World in Epidendreae where there is a major radiation that yielded, among others, Laeliinae and Pleurothallidinae (approx. 6300 species), and another in Cymbidieae that gave rise to a clade including major subtribes such as Maxillariinae and Oncidiinae, comprising approx. 2700 species. Both of these shifts correspond with groups that are optimized as epiphytic at their base (Supplementary Data Figs S8 and S9), suggesting that it may have been the introduction of these epiphytic groups into at least partially unoccupied epiphytic habitats in the New World that spurred much of their diversification there. Based on the dating of extant epidendroid lineages at about 50 Ma provided by Ramírez et al. (2007), these clades would be too young to have been the result of vicariance due to the Africa–South America split, suggesting long-distance dispersal. However, given that this dating was provided by a single relatively derived orchidoid pollinarium fossil, it must be viewed as extremely tentative. Another analysis using three fossil calibration points and Bayesian methods (Gustafsson et al., 2010, using the same fossil as Ramírez et al., 2007, plus two more recently discovered ones from Epidendroideae, Dendrobium and Earina, in Conran et al., 2009) reached similar general conclusions, although the dates were slightly more recent.
Several geographical shifts involving smaller groups of genera have also occurred; of these on the ML tree (Supplementary Data Fig. S12), 11 occur in fundamentally temperate groups or represent a shift from the tropics to temperate regions, and eight are within tropical groups. Among the temperate shifts, some are in groups that exhibit the classic temperate eastern Asia–eastern North America vicariance pattern (Calopogon/Arethusa/Eleorchis, Tipularia, Aplectrum/Cremastra, possibly Oreorchis/Corallorhiza), which probably represent Tertiary flora elements that have become disjunct. Others are circumboreal groups [Listera (=Neottia), Calypso]. Still others (Malaxis, Liparis, Calanthe) may emphasize connections between Asia and Mexico. Based on the analysis of Cameron (2005), and with the broader context presented here, Malaxidinae appear to be a fundamentally Old World group, with two probable incursions into the New World (possibly three, depending on the placement of Malaxis paludosa, which was not included there). The distribution of habit states shown here also supports the conclusion of Cameron (2005) that Malaxidinae are plesiomorphically epiphytic; he concluded that a single shift to the terrestrial habit occurred in the group. Because of the uncertainty that remains in relationships at the base of Cymbidieae (notably with respect to Eulophia and Cyrtopodium), the number and nature of geographical shifts in that part of the tree remain uncertain.
Among geographical shifts that occur in tropical genera, Tropidia, Polystachya and Eulophia are fundamentally Old World genera each with a small number of New World species. Corymborkis is found in Asia, Africa and Central–South America, with a slight majority (four of seven) of species occurring in the last. With respect to their early-branching position in the tree, Corymborkis and Tropidia have the possibility of being much older shifts than the others. Polystachya is clearly a recent arrival in the New World (and Asia) from Africa (Russell et al., 2010). Campylocentrum/Harrisella represents an unusual radiation of Vandeae into the New World, comprising >50 species, some of which are leafless (but with photosynthetic roots). The analysis of Carlsward et al. (2003) indicated that this assemblage represents a single dispersal to the New World.
Column structure – anther characters
The morphological feature that is the key synapomorphy for Epidendroideae is the incumbent anther, although not all epidendroids have this (and it is occasionally present in some non-epidendroid groups; Rasmussen, 1982). Even in groups that are considered to have an incumbent anther the degree of inflexion may be variable. Rasmussen (1982) investigated this feature in the ‘neottioid’ portion of the epidendroids and found it to be variable among, for example, members of Neottieae. This feature characterizes most of the more advanced epidendroids, but clearly has undergone a reversal in species that are bird-pollinated (e.g. Elleanthus spp., Dendrobium secundum) and in some taxa with small flowers and short columns (e.g. some Malaxidinae). In many epidendroids, inflexion clearly occurs late in ontogeny (Dressler, 1981; Kurzweil, 1987) whereas in others (the vandoids) there was a question as to whether the anther inflexed early (Hirmer, 1920) or not at all (Dressler, 1981). Kurzweil (1987), Freudenstein and Rasmussen (1996) and Freudenstein et al. (2002) illustrated the ontogenetic changes occurring in ‘basal’ and vandoid epidendroids, and it is clear that the latter exhibit not an early inflexion of a well-formed anther, but rather an initial reorientation of growth such that the anther develops ab initio in an incumbent orientation.
Two other features are almost always associated with the early-inflexing anther: superposed pollinia and a cellular pollinium stalk. Together these three features are the floral components of what has been termed the ‘vandoid’ morphology (Dressler, 1981, 1993). Freudenstein et al. (2002) suggested that the vandoid anther was the result of a paedomorphic shift, allowing optimal positioning of pollinia for attachment to a rostellar-derived stalk. Superposed pollinia result from a ‘flattening’ of the anther either by a lack of full reorientation of the thecae to yield a juxtaposed arrangement or by differential overgrowth of the thecae (Freudenstein and Rasmussen, 1996; Freudenstein et al., 2002); either mechanism results in pairs of pollinia that are dorsally stacked rather than laterally adjacent to each other. It is difficult to assess pollinium arrangement in anthers with only two pollinia, as it is defined by the relative position of pollinia in each theca. In many cases, each pollinium when there are two has a bilobed or cleft appearance, presumably the rudiments of a partition that has either been lost or is developing. Many of these are angled, suggesting the superposed arrangement. For the purposes of this study, those genera with two pollinia were coded as having the superposed state. Cellular pollinium stalks (stipes; Bentham, 1881; Hirmer, 1920; Rasmussen, 1982) are the most highly developed accessory structures present in orchid pollinaria. They are derived from the rostellum, utilizing varying amounts of this structure (Rasmussen, 1986). Rasmussen (1982) distinguished two types, the hamulus, being the entire distal portion of the rostellum where the apex attaches to the pollinia, and the tegula, comprising the adaxial cuticle of the rostellum with subterminal pollinium attachment. Many vandoids have a stalk with subterminal pollinium attachment that comprises one to several layers of the adaxial portion of the rostellum in addition to the cuticle (J. V. Freudenstein and F. N. Rasmussen, unpubl. res.) and that variant is included here.
Individual characters of the vandoid syndrome can occur independently, meaning that the development of a single component character does not necessarily result in the full syndrome. For example, some members of Coelogyneae (e.g. Entomophobia, Dendrochilum, Pholidota) have superposed pollinia (Supplementary Data Figs S14 and S15). Although they do not have a cellular pollinium stalk, they do have elongate pollinia and caudicles that function in a similar way. It may be that the superposed morphology facilitates attachment of the caudicles to the viscidium in these cases. Sobralia exhibits what may be considered a ‘half-superposed’ pollinium arrangement; it also has unusually elongate and partially twisted pollinia/caudicles. Cellular pollinium stalks are known in Tropidia and Corymborkis at the base of the subfamily (Figs S16 and S17; Rasmussen, 1982) and also occur rarely outside Epidendroideae (Zeuxine; Rasmussen, 1982), where the other vandoid characters are not found in association.
The one vandoid character that always appears to lead to the full vandoid syndrome is early anther inflexion (Supplementary Data Figs S18 and S19). Coelia is resolved here as sister to Calypsoinae and exhibits early anther inflexion, but it does not have superposed pollinia – rather it has eight pollinia that are elongate–clavate. Early anther inflexion may facilitate the coordination between the apex of the rostellum (viscidium) and the elongate pollinia in this genus. This ontogenetic shift may then have made possible the development of the cellular stalks and superposed pollinia present in all Calypsoinae (Freudenstein, 1994).
The vandoid floral morphology, along with lateral inflorescence position, has been used to delimit a unit at subfamily level (Vandoideae; Dressler, 1981), implying a single origin, but that conclusion was subsequently reconsidered (Dressler, 1990). The present analysis indicates that the vandoid syndrome of these three floral character states has arisen at most two or three times – once in Calypsoinae (Epidendreae) and then also in Vandeae and Cymbidieae (using presence of cellular pollinium stalk as a proxy for the vandoid condition; Supplementary Data Fig. S16 and S17). In the ML tree, Cymbidieae and Epidendreae are sisters, implying that the vandoid syndrome in Vandeae and Cymbidieae arose separately, whereas in the MP tree Vandeae and Cymbidieae are sisters and thus could share the vandoid syndrome as a synapomorphy.
Although all examined species of Cymbidieae and Vandeae have early anther inflexion (cf. Kurzweil, 1987) and at least a rudimentary stipe, Polystachya (Vandeae) appears to be variable in whether the pollinia are superposed or not. Most species have only two pollinia, but Dressler (1993) noted that, in those with four he examined, the pollinia appear to assume a superposed arrangement only after they are pulled from the anther. Here they were coded as being juxtaposed, but more study is needed. Other than this anomaly, and some uncertainty in the juxtaposed/superposed state for those anthers with only two pollinia, the vandoid syndrome characterizes both Cymbidieae and Vandeae consistently. If the syndrome is hypothesized to be homologous, and there is no empirical evidence to suggest that it is not, then the MP topology may be correct.
Diversification conclusions
With respect to the features that led to success in diversification among epidendroids, we suggest that the story is not a simple one. Clearly, epiphytism is an important factor – perhaps the most important factor – in diversification among epidendroids. Indeed, some epiphytic groups have become diverse without the structural specializations comprising the vandoid anther. A case in point is Bulbophyllum, the largest genus in Orchidaceae, with approx. 1867 species (species numbers all from Govaerts et al., 2014). They are epiphytic, but appear to have structural pollinarium specializations in only a minority of species (Rasmussen, 1985; Gravendeel and Vermeulen, 2014). The closely related Dendrobium is similarly depauperate in advanced anther features, but has produced approx. 1509 species. Pleurothallidinae, with about 4570 species in the New World, are variably epiphytic–terrestrial and also have no highly specialized pollinarium structures. This is not to say that they do not have specialized pollination strategies, however, perhaps based on other features such as the perianth that may have contributed to their diversity.
Nonetheless, having admitted the importance of epiphytism in epidendroid diversity, we should not discount the role of the vandoid anther specializations, which have evolved primarily within the epiphytic groups. Their association with diversity was also shown to be highly significant in the BiSSE analysis. Individual vandoid anther features examined here have not led to large radiations when they occur in terrestrial groups, whereas they have in epiphytes. Tropidia, for example, which has a cellular pollinium stalk, comprises only 31 terrestrial species, whereas the superposed pollinia of Coelogyninae occur in up to 744 primarily epiphytic species. The vandoid syndrome as a whole occurs outside of epiphytes only in Calypsoinae; our trees suggest that two of the three vandoid features in this group evolved only after the lineage became terrestrial and thus far have yielded a group of only about 78 species.
It may well be that in these epidendroid clades the successive evolution of these features has propelled their diversification. Epiphytism, followed by the vandoid anther features and pollination specificity that they enable, may have been one successful combination. Production of large numbers of dust seeds such as is found in most orchids facilitates epiphytism, as they may easily be carried into the canopy; however, the number of successful germinations is undoubtedly reduced by the requirement for a suitable fungal host that may be unevenly distributed. This in turn may lead to a patchiness in distribution that requires a specific pollinator interaction in order to be effective in pollinium transfer, making the advanced anther features advantageous because they lead to precise and efficient placement and retrieval of pollinaria. Add to this the opening of new areas suggested by the strong significance in association of shifts to the New World and the effect may be multiplied.
In the end, it is important to remember that we have analysed here only a small part of the structural diversity, in terms of characters, of the epidendroids, even among anther features, and have not considered fungal associations at all. Correlation does not prove causation, meaning that the hypotheses of significant innovations proposed here are just that – hypotheses that await further testing and comparison with other potentially important features of epidendroids.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. File S1: Accessions used in this study with GenBank numbers for each sequence. File S2: Data matrix for TNT with details of loci and excluded positions. File S3: Data matrix for RaxML (PHYLIP format). Figure S1: ML tree with proportional branch lengths. Figure S2: ML tree based on plastid DNA sequences with bootstrap support values. Figure S3: ML tree based on nuclear DNA sequences with bootstrap support values. Figure S4: ML tree for 175 taxa with bootstrap values. Figure S5: ML tree for 49 taxa with bootstrap values. Figure S6: MP tree for 175 taxa with jackknife values. Figure S7: MP tree for 49 taxa with jackknife values. Figure S8: ML tree with habit plotted. Figure S9: MP tree with habit plotted. Figure S10: ML tree with presence of corm/pseudobulb plotted. Figure S11: MP tree with presence of corm/pseudobulb plotted. Figure S12: ML tree with geography plotted. Figure S13: MP tree with geography plotted. Figure S14: ML tree with pollinium orientation plotted. Figure S15: MP tree with pollinium orientation plotted. Figure S16: ML tree with presence of pollinarium stipe plotted. Figure S17: MP tree with presence of pollinarium stipe plotted. Figure S18: ML tree with early anther inflexion plotted. Figure S19: MP tree with early anther inflexion plotted.
ACKNOWLEDGEMENTS
We thank Matthew Yoder for use of his program GENERATOR and Erik Rothacker and Ryan Kitko for use of unpublished sequences. This work was supported by the National Science Foundation of the USA (DEB-9615437 to J.V.F.).
LITERATURE CITED
- Atwood JT. 1986. The size of the Orchidaceae and the systematic distribution of epiphytic orchids. Selbyana 9: 171–186. [Google Scholar]
- Batista JAN, Mota ACM, Proite K, et al. 2014. Molecular phylogenetics of Neotropical Cyanaeorchis (Cymbidieae, Epidendroideae, Orchidaceae): geographical rather than morphological similarities plus a new species. Phytotaxa 156: 251–272. [Google Scholar]
- Bentham G. 1881. Notes on Orchideae. Journal of The Linnean Society of London, Botany 18: 281–360. [Google Scholar]
- Cameron KM. 2005. Leave it to the leaves: a molecular phylogenetic study of Malaxideae (Epidendroideae, Orchidaceae). American Journal of Botany 92: 1025–1032. [DOI] [PubMed] [Google Scholar]
- Cameron KM, Chase MW, Whitten WM, et al. 1999. A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. American Journal of Botany 86: 208–224. [PubMed] [Google Scholar]
- Carlsward BS, Whitten WM, Williams NH. 2003. Molecular phylogenetics of Neotropical leafless Angraecinae (Orchidaceae): reevaluation of generic concepts. International Journal of Plant Sciences 164: 43–51. [Google Scholar]
- Carlsward BS, Whitten WM, Williams NH, Bytebier B. 2006. Molecular phylogenetics of Vandeae (Orchidaceae) and the evolution of leaflessness. American Journal of Botany 93: 770–786. [DOI] [PubMed] [Google Scholar]
- Chase MW, Cameron KM, Barrett RL, Freudenstein JV. 2003. DNA data and Orchidaceae systematics: a new phylogenetic classification. In: Dixon KL, Kell SP, Barrett RL, Cribb PJ, eds. Orchid conservation. Kota Kinabalu: Natural History Publications, 69–89. [Google Scholar]
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, van den Berg C, Schuiteman A. 2015. An updated classification of Orchidaceae. Botanical Journal of the Linnean Society, in press. [Google Scholar]
- Conran JG, Bannister JM, Lee DE. 2009. Earliest orchid macrofossils: early Miocene Dendrobium and Earina (Orchidaceae: Epidendroideae) from New Zealand. American Journal of Botany 96: 466–474. [DOI] [PubMed] [Google Scholar]
- Davis MP, Midford PE, Maddison W. 2013. Exploring power and parameter estimation of the BiSSE method for analyzing species diversification. BMC Evolutionary Biology 13: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressler RL. 1974. Classification of the orchid family. Seventh world orchid conference. Medellin, Colombia. [Google Scholar]
- Dressler RL. 1981. The orchids: natural history and classification . Cambridge, MA: Harvard University Press. [Google Scholar]
- Dressler RL. 1990. The major clades of the Orchidaceae-Epidendroideae. Lindleyana 5: 117–125. [Google Scholar]
- Dressler RL. 1993. Phylogeny and classification of the orchid family . Portland, OR: Timber Press. [Google Scholar]
- Dressler RL, Cook SL. 1988. Conical silica bodies in Eria javanica . Lindleyana 3: 224–225. [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farris JS, Källersjö M, Lipscomb D, Kluge A. 1996. Parsimony jackknifing outperforms neighbor-joining. Cladistics 12: 99–124. [DOI] [PubMed] [Google Scholar]
- Freudenstein JV. 1994. Gynostemium structure and relationships of the Corallorhizinae (Orchidaceae: Epidendroideae). Plant Systematics and Evolution 193: 1–19. [Google Scholar]
- Freudenstein JV, Rasmussen FN. 1996. Pollinium development and number in the Orchidaceae. American Journal of Botany 83: 813–824. [Google Scholar]
- Freudenstein JV, Rasmussen FN. 1999. What does morphology tell us about orchid relationships? – A cladistic analysis. American Journal of Botany 86: 225–248. [PubMed] [Google Scholar]
- Freudenstein JV, Senyo DM. 2008. Relationships and evolution of matK in a group of leafless orchids (Corallorhiza and Corallorhizinae; Orchidaceae: Epidendroideae). American Journal of Botany 95: 498–505. [DOI] [PubMed] [Google Scholar]
- Freudenstein JV, Harris EM, Rasmussen FN. 2002. The evolution of anther morphology in orchids: incumbent anthers, superposed pollinia, and the vandoid complex. American Journal of Botany 89: 1747–1755. [DOI] [PubMed] [Google Scholar]
- Freudenstein JV, van den Berg C, Goldman DH, Kores PJ, Molvray M, Chase MW. 2004. An expanded plastid DNA phylogeny of Orchidaceae and analysis of jackknife branch support strategy. American Journal of Botany 91: 149–157. [DOI] [PubMed] [Google Scholar]
- Garcia-Cruz J, Sosa V. 2005. Phylogenetic relationships and character evolution in Govenia (Orchidaceae). Canadian Journal of Botany 83: 1329–1339. [Google Scholar]
- Goldman DH, Freudenstein JV, Kores PJ, et al. 2001. Phylogenetics of Arethuseae (Orchidaceae) based on plastid matK and rbcL sequences. Systematic Botany 26: 670–695. [Google Scholar]
- Goloboff PA, Farris JS, Nixon KC. 2008. TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- Górniak M, Paun O, Chase MW. 2010. Phylogenetic relationships within Orchidaceae based on a low-copy nuclear coding gene, xdh: congruence with organellar and nuclear ribosomal DNA results. Molecular Phylogenetics and Evolution 56: 784–795. [DOI] [PubMed] [Google Scholar]
- Govaerts R, et al. 2014. World Checklist of Orchidaceae. Facilitated by the Royal Botanic Gardens, Kew. http://apps.kew.org/wcsp/ (accessed 8 October 2014). [Google Scholar]
- Gravendeel B, Vermeulen JJ. 2014. Bulbophyllum. In: Pridgeon AP, Cribb PJ, Chase MW, Rasmussen FN, eds. Genera orchidacearum, vol. 6, part 3 Oxford: Oxford University Press, 4–51. [Google Scholar]
- Gravendeel B, Smithson A, Slik FJW, Schuiteman A. 2004. Epiphytism and pollinator specialization: drivers for orchid diversity? Philosophical Transactions of the Royal Society of London Series B-Biological Sciences 359: 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson Al, Verola CF, Antonelli A. 2010. Reassessing the temporal evolution of orchids with new fossils and a Bayesian relaxed clock, with implications for the diversification of the rare South American genus Hoffmannseggella (Orchidaceae: Epidendroideae). BMC Evolutionary Biology 10: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirmer M. 1920. Beiträge zur Organographie der Orchideenblüte. Flora 113: 213–310. [Google Scholar]
- Kurzweil H. 1987. Developmental studies in orchid flowers I: epidendroid and vandoid species. Nordic Journal of Botany 7: 427–442. [Google Scholar]
- Lewis PO. 2001. A likelihood approach to estimating phylogeny from discrete morphological character data. Systematic Biology 50: 913–925. [DOI] [PubMed] [Google Scholar]
- Li L, Yan H. 2013. A remarkable new species of Liparis (Orchidaceae) from China and its phylogenetic implications. PLoS One 8: e78112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP. 1993. Missing data versus missing characters in phylogenetic analysis. Systematic Biology 42: 576–581. [Google Scholar]
- Maddison WP, Maddison DR. 2011. Mesquite: a modular system for evolutionary analysis. Version 2.75. http://mesquiteproject.org/ [Google Scholar]
- Maddison WP, Midford PE, Otto SP. 2007. Estimating a binary character’s effect on speciation and extinction. Systematic Biology 56: 701–710. [DOI] [PubMed] [Google Scholar]
- Midford P, Maddison WP. 2011. Diverse package for Mesquite, Version 2.75. http://mesquiteproject.org/ [Google Scholar]
- Møller JD, Rasmussen H. 1984. Stegmata in the Orchidales: character state distribution and polarity. Botanical Journal of the Linnean Society 89: 53–76. [Google Scholar]
- Molvray M, Kores PJ, Chase MW. 2000. Polyphyly of mycoheterotrophic orchids and functional influences on floral and molecular characters. In: Wilson KL, Morrison DA, eds. Monocots: systematics and evolution. Collingwood: CSIRO, 441–448. [Google Scholar]
- Neubig KM, Whitten WM, Carlsward BS, et al. 2009. Phylogenetic utility of ycf1 in orchids: a plastid gene more variable than matK. Plant Systematics and Evolution 277: 75–84. [Google Scholar]
- Neubig KM, Whitten WM, Williams NH, et al. 2012. Generic recircumscriptions of Oncidiinae (Orchidaceae: Cymbidieae) based on maximum likelihood analysis of combined DNA datasets. Botanical Journal of the Linnean Society 168: 117–146. [Google Scholar]
- Nixon KC, Davis JI. 1991. Polymorphic taxa, missing values and cladistic analysis. Cladistics 7: 233–241. [DOI] [PubMed] [Google Scholar]
- Porembski S, Barthlott W. 1988. Velamen radicum micromorphology and classification of the Orchidaceae. Nordic Journal of Botany 8: 117–137. [Google Scholar]
- Pridgeon AM, Solano R, Chase MW. 2001. Phylogenetic relationships in Pleurothallidinae (Orchidaceae): combined evidence from nuclear and plastid DNA sequences. American Journal of Botany 88: 2286–2308. [PubMed] [Google Scholar]
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. (eds.) 2005. Epidendroideae (Part one). Genera orchidacearum. Oxford: Oxford University Press. [Google Scholar]
- Ramírez SR, Gravendeel B, Singer RB, Marshall CR, Pierce NE. 2007. Dating the origin of the Orchidaceae from a fossil orchid with its pollinator. Nature 448: 1042–1045. [DOI] [PubMed] [Google Scholar]
- Rasmussen FN. 1982. The gynostemium of the neottioid orchids. Opera Botanica 65: 1–96. [Google Scholar]
- Rasmussen FN. 1985. The gynostemium of Bulbophyllum ecornutum . Botanical Journal of the Linnean Society 91: 447–456. [Google Scholar]
- Rasmussen FN. 1986. On the various contrivances by which pollinia are attached to viscidia. Lindleyana 1: 21–32. [Google Scholar]
- Russell A, Samuel MR, Rupp B, Barfuss MHJ, Šafran M, Besendorfer V, Chase MW. 2010. Phylogenetics and cytology of a pantropical orchid genus Polystachya (Polystachyinae; Vandeae; Orchidaceae): evidence from plastid DNA sequence data. Taxon 59: 389–404. [Google Scholar]
- Simmons MP, Freudenstein JV. 2011. Spurious 99 % bootstrap and jackknife support for unsupported clades. Molecular Phylogenetics and Evolution 61: 177–191. [DOI] [PubMed] [Google Scholar]
- Simmons MP, Norton AP. 2013. Quantification and relative severity of inflated branch-support values generated by alternative methods: an empirical example. Molecular Phylogenetics and Evolution 67: 277–96. [DOI] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology 49: 369–381. [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Swartz O. 1800. Afhandling om Orchidernes slaegter och deras systematiska indelning. Kongliga Vetenskaps Academiens Nya Handlingar 21: 115–139. [Google Scholar]
- Szlachetko DL. 1995. Systema orchidalium. Fragmenta Floristica et Geobotanica Supplementum 3: 1–152. [Google Scholar]
- Thomson RC, Shaffer HB. 2010. Sparse supermatrices for phylogenetic inference: taxonomy, alignment, rogue taxa, and the phylogeny of living turtles. Systematic Biology 59: 42–58. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Higgins WE, Dressler RL, et al. 2000. A phylogenetic analysis of Laeliinae (Orchidaceae) based on sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA. Lindleyana 15: 96–114. [Google Scholar]
- van den Berg C, Goldman DH, Freudenstein JV, Pridgeon AM, Cameron KM, Chase MW. 2005. An overview of the phylogenetic relationships within Epidendroideae inferred from multiple DNA regions and recircumscription of Epidendreae and Arethuseae (Orchidaceae). American Journal of Botany 92: 613–624. [DOI] [PubMed] [Google Scholar]
- van den Berg C, Higgins WE, Dressler RL, Whitten WM, Soto-Arenas MA, Chase MW. 2009. A phylogenetic study of Laeliinae (Orchidaceae) based on combined nuclear and plastid DNA sequences. Annals of Botany 104: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitten WM, Williams NH, Chase MW. 2000. Subtribal and generic relationships of Maxillarieae (Orchidaceae) with emphasis on Stanhopeinae: combined molecular evidence. American Journal of Botany 87: 1842–1856. [PubMed] [Google Scholar]
- Whitten WM, Williams NH, Dressler RL, Gerlach G, Pupulin F. 2005. Generic relationships of Zygopetalinae (Orchidaceae: Cymbidieae): combined molecular evidence. Lankesteriana 5: 87–107. [Google Scholar]
- Whitten WM, Blanco MA, Williams NH, et al. 2007. Molecular phylogenetics of Maxillaria and related genera (Orchidaceae: Cymbidieae) based on combined molecular data sets. American Journal of Botany 94: 1860–1889. [DOI] [PubMed] [Google Scholar]
- Whitten WM, Neubig KM, Williams NH. 2014. Generic and subtribal relationships in Neotropical Cymbidieae (Orchidaceae) based on matK/ycf1 plastid data. Lankesteriana 13: 375–392. [Google Scholar]
- Wiens JJ. 2003. Missing data, incomplete taxa, and phylogenetic accuracy. Systematic Biology 52: 528–538. [DOI] [PubMed] [Google Scholar]
- Wiens JJ. 2006. Missing data and the design of phylogenetic analyses. Journal of Biomedical Informatics 39: 34–42. [DOI] [PubMed] [Google Scholar]
- Xiang X-G, Li D-Z, Jin W-T, Zhou H-L, Li J-W, Jin X-H. 2012. Phylogenetic placement of the enigmatic orchid genera Thaia and Tangtsinia: evidence from molecular and morphological characters. Taxon 61: 45–54. [Google Scholar]
- Yukawa T, Kita K, Handa T. 2000. DNA phylogeny and morphological diversification of Australian Dendrobium (Orchidaceae). In: Wilson KL, Morrison DA, eds. Monocots: systematics and evolution. Collingwood: CSIRO, 465–471. [Google Scholar]
- Zhai JW, Zhang GQ, Chen LJ, et al. 2013. A new orchid genus, Danxiaorchis, and phylogenetic analysis of the tribe Calypsoeae. PLoS One 8: e60371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.