Significance
The tumor suppressor p53 is inactivated by aggregation in a substantial number of tumors, and those oncogenic mutants coaggregate with WT protein and other tumor suppressors. Inhibition of aggregation by small molecules is a possible drug therapy. p53 aggregation appears to have much simpler kinetics than commonly encountered in fibrillation, with two rate-determining sequential, apparently first-order, steps. We showed by combining mutagenesis and kinetics that the rate determining steps involve two molecules of p53 extensively unfolding and reacting in a bimolecular process that can appear first order. The mechanism provides a basis for understanding the progress of aggregation and coaggregation and points to the most effective drug targeting sites.
Keywords: protein, amyloid, folding, misfolding, cancer
Abstract
Many oncogenic mutations inactivate the tumor suppressor p53 by destabilizing it, leading to its rapid aggregation. Small molecule drugs are being developed to stabilize such mutants. The kinetics of aggregation of p53 is deceptively simple. The initial steps in the micromolar concentration range follow apparent sigmoidal sequential first-order kinetics, with rate constants k1 and k2. However, the aggregation kinetics of a panel of mutants prepared for Φ-value analysis has now revealed a bimolecular reaction hidden beneath the observed first-order kinetics. Φu measures the degree of local unfolding on a scale of 0–1. A number of sequential Φu-values of ∼1 for k1 and k2 over the molecule implied more than one protein molecule must be reacting, which was confirmed by finding a clear concentration dependence at submicromolar protein. Numerical simulations showed that the kinetics of the more complex mechanism is difficult, if not impossible, to distinguish experimentally from simple first order under many reaction conditions. Stabilization of mutants by small molecules will be enhanced because they decrease both k1 and k2. The regions with high Φu-values point to the areas where stabilization of mutant proteins would have the greatest effect.
The p53 tumor suppressor plays an essential role in genome surveillance and protects against cancer (1–5). p53 is inactivated in more than 70% of cancer types by missense point mutations, mainly located in the DNA-binding core domain (6–8). WT p53, with a melting temperature of 45 °C (9), is intrinsically thermodynamically unstable (10), as well as kinetically (11). Many of the oncogenic mutations further destabilize p53, leading to its rapid aggregation at body temperature (10–13). The increased aggregation propensity of the destabilizing mutants, such as R175H, Y220C, and R249S, not only causes loss of function but is also suggested to exert a dominant negative function by coaggregation with WT p53 (12, 14), p63, and p73 (15–19). p53 mutants with higher aggregation propensity are reported to impair p73- and p63-mediated transcription and apoptosis (15). These p53 mutants accumulate at high levels in most of tumors due to resistance to proteasome-dependent degradation. The high expression of mutant p53 in tumors makes stabilizing the mutants and restoring their WT functions an attractive strategy to eliminate selectively tumor cells.
Small molecules have been developed to stabilize such destabilizing mutants (20). Many of them are from cell or in vitro screening (21, 22), whereas others are rationally designed from the structures of WT p53 and mutants (21, 23). These small molecules can effectively restore WT activity of mutant p53 and specifically kill cancer cells.
The best-studied examples of protein aggregation are those that form regular repeating fibrillar structures (24–27). p53 aggregates have some of the diagnostic features of such fibrils, such as the binding of the dye thioflavine T (ThT), and will form regular fibrils under some conditions (28, 29). Generally, however, especially at physiological temperature and pH, p53 forms amorphous aggregates, which still contain the β-structural elements responsible for ThT binding and other properties (28, 29). Amorphous aggregates are very commonly found, but there are few systematic studies of their mechanism of formation (30). We are analyzing the mechanism of amorphous aggregation of p53 in its own right, as well as to aid in the design of inhibitors (31). The basic kinetics consists of two apparent sequential first-order steps to generate an aggregation competent intermediate that then rapidly forms amyloid structures that bind ThT and then undergo further slower rearrangements to generate large amorphous structures (31, 32).
Φ-value analysis is a powerful systematic experimental tool to map transition and intermediate states at a resolution of individual residues in ligand binding (33) and folding (34–36). For an unfolding process, the ratio of the change in free energy of activation of unfolding, ∆∆G‡-N, on mutation of a specific residue to the change in free energy change of denaturation, ∆∆GN-D, is defined as Φu. A value of Φu = 1 implies that the transition state for unfolding is as energetically weakened at the site of mutation as is the denatured state. Conversely, Φu = 0 implies that it has the same energy as the native state.
Here, we used Φ-value analysis to characterize the early transition states for the aggregation of p53. The panel of mutants synthesized exhibited a variety of kinetics and unexpected values of Φu, which uncovered a bimolecular reaction mechanism hidden within the observed sequential first-order kinetics in the usual experimental concentration range (31).
Results
Kinetics of Aggregation of Mutants.
Some mutants aggregated so fast that we could not use a standard parent protein for all mutations and make measurements at the same temperature. We used a series of different pseudo-WT reference constructs of different stability (Table 1) and made measurements at 30, 33.6, or 37 °C, the unstable mutants reacting too rapidly to measure at 33.6 or 37 °C and the more stable ones too slowly at 30 or 33.6 °C. We fitted the binding of ThT to the equation used previously for sequential first-order reactions, see Scheme 1 (31, 32)
| [1] |
where Ft is the intensity of ThT fluorescence at time t, and m, the amplitude, = [A]0f, where f is the specific fluorescence of ThT bound to the aggregate. The small linear term of k3t was included to allow for machine drift and the settling of particles over the time course, and the term a is included to allow for a nonzero intercept at t = 0. For short time courses, which we used for the Φu-value analysis, we omitted the k3t term. The yield of ThT emission is lower than that from the small proteins more usually studied because they form amyloid β-structures along most of their sequences, whereas only a small fraction of the p53 chain forms the fibrillar aggregate, and the remaining structure decorates it.
Table 1.
Pseudo-WT constructs and properties
| Construct | Residues | Mutations | T, °C | k1, min−1 | k2, min−1 | ∆GD-N, kcal/mol | Tm, °C |
| QCYC | 94–312 | M133L/V203A/N239Y/N268D/Y220C | 37 | 0.069 ± 0.004 | 0.26 ± 0.02 | 8.31 ± 0.35 | 44.3 ± 0.3 |
| 33.6 | 0.015 ± 0.007 | 0.14 ± 0.02 | |||||
| 30 | 0.0028 ± 0.0001 | 0.018 ± 0.001 | |||||
| WTC | 94–312 | 37 | 0.033 ± 0.007 | 1.2 ± 0.2 | 9.71 ± 0.03 | 45.8 ± 0.1 | |
| WTLC | 89–312 | 10.7 ± 0.1 | 47.2 ± 0.1 | ||||
| QMC | 94–312 | M133L/V203A/N239Y/N268D | 42.7 | 0.018 ± 0.002 | 0.07 ± 0.01 | 12.5 ± 0.03 | 52.4 ± 0.2 |
| 43.6 | 0.016 ± 0.003 | 0.10 ± 0.02 | |||||
| QMFLYC | 1–393 | M133L/V203A/N239Y/N268D/Y220C | 37 | 0.037 ± 0.002 | 0.21 ± 0.01 | — | 47.7 ± 0.2 |
| FL | 1–393 | 37 | 0.010 ± 0.001 | 0.40 ± 0.02 | — | 45.1 ± 0.2 (9) |
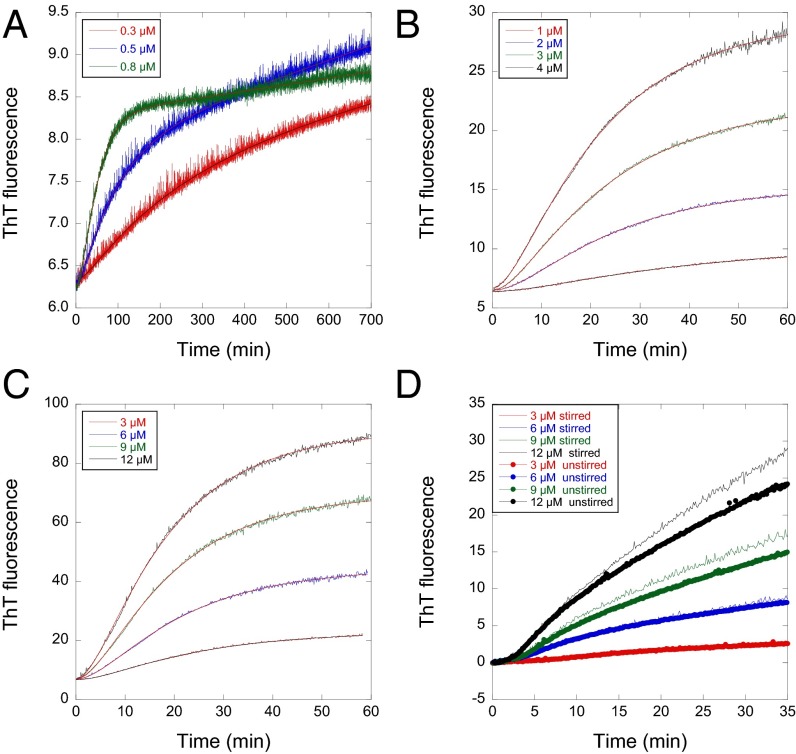
The progress curves for the core domain of Y220C in stabilized quadruple mutant construct QCYC fitted Eq. 1 very well from 0.3 to 12 µM (Fig. 1 A–C). Unlike aggregation of amyloid fibrils, which will be accelerated by stirring, the rate constants were relatively independent of whether the reaction mixtures were stirred or unstirred before dense aggregate settled. WT p53 core domain (WTC), unstirred, (Fig. 1D and Fig. S1), for example, had slightly lower amplitude terms but fitted similar rate constants as stirred.
Fig. 1.
Aggregation traces of p53 monitored by ThT can be fitted well with Eq. 1 at wide protein concentration range and different stirring conditions. (A–C) Time courses at 37 °C for QCYC from 0.3 to 12 µM. The linear drift term in Eq. 1 was used only when protein concentrations were lower than 0.8 µM. (D) WTC stirred and unstirred. They are fitted to Eq. 1 without the linear drift term. Unstirred had lower amplitudes but fitted to similar rate constants with Eq. 1.
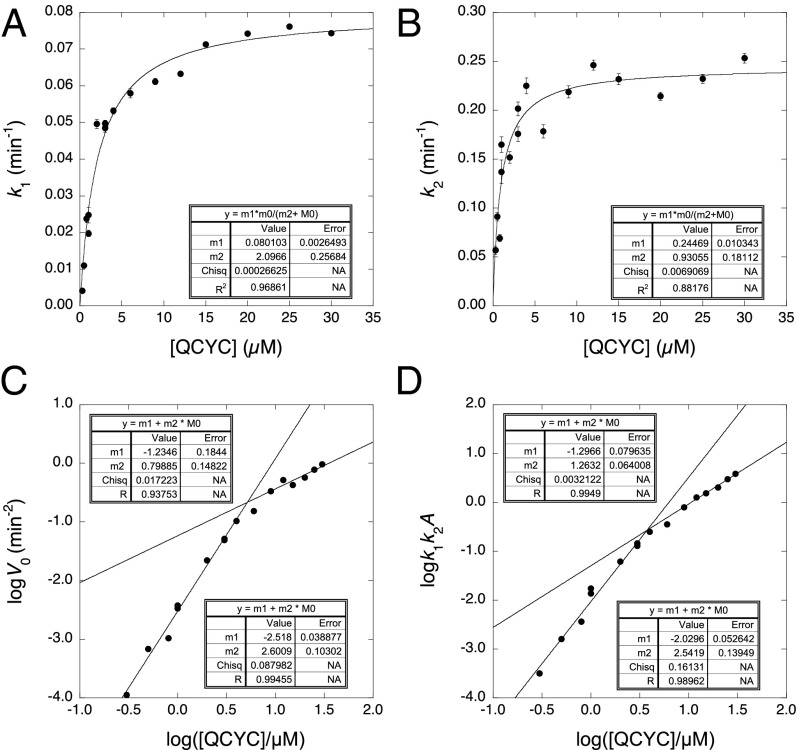
Plots of k1 and k2 vs. protein concentration (Fig. 2 A and B) were hyperbolic in nature, with apparent dissociation constants of ∼1.5 µM. The initial rate V0 (= dFt/dt2)t=0 of the increase in fluorescence vs. t2 when plotted as logV0 vs. log[protein] gives as slope the number of molecules in the nucleus − 2 for a homogeneous nucleation mechanism (37, 38). An equivalent plot in terms of the rate constants for sequential first-order reactions is log(k1k2A) vs. log[protein], where A is the amplitude of the curve (31). Such plots, from measuring V0 directly or indirectly from k1k2A, gave slopes that decreased with increasing concentration. The slopes tail off at 1 at high concentrations and tend to 2.6 at low (Fig. 2 C and D). Eq. 1 also held for the binding of ThT during the aggregation of some 30 mutants of the core. The derived plots of k1 and k2 vs. [protein] exhibited a variety of behavior (Fig. S1). Some showed peaks in k2 vs. [protein]. Mutants studied over a wide range of concentrations showed similar behavior to that of QCYC (Fig. S2), with slopes of log(k1k2A) vs. log[protein] decreasing from ∼2.6 to ∼1. Some mutants studied over short concentration ranges had slopes rolled over to 1 and others were close to 2 (Fig. S1).
Fig. 2.
Concentration dependence of the aggregation of p53 core domain Y220C in stabilized quadruple mutant construct (QCYC). (A and B) Hyperbolic concentration dependence of QCYC at low protein concentrations. (C) Concentration dependence of initial aggregation rate measured directly from initial rates. (D) Concentration dependence of initial aggregation rate calculated from k1 and k2 for first-order consecutive reactions.
Urea also significantly accelerated k1 and k2 (Fig. S3) according to the commonly found equation lnk = lnkH2O+mk[urea] (Table S1). The averaged fractional increases in solvent exposure of p53 variants in the transition state relative to that on complete unfolding were 0.35 ± 0.02 and 0.48 ± 0.04 for k1 and k2, respectively, which indicated a large degree of unfolding in each step (39).
The slopes are inconsistent with a homogeneous nucleation mechanism but are consistent with a reaction that is undergoing a transition from second or third order at low concentrations to first order at high concentrations. Further, a preliminary Φ-value analysis (see below) showed that the change in activation energy k1 and k2 [calculated from RTln(kmutant/kwild-type)] for some mutants approached the full change in their free energy of denaturation (Φu ∼1 for both k1 and k2), indicating that the mutated region fully unfolds at each step, implying that two or more protein molecules were unfolding during the process because one molecule cannot unfold twice in one process.
Kinetic Mechanism for Aggregation.
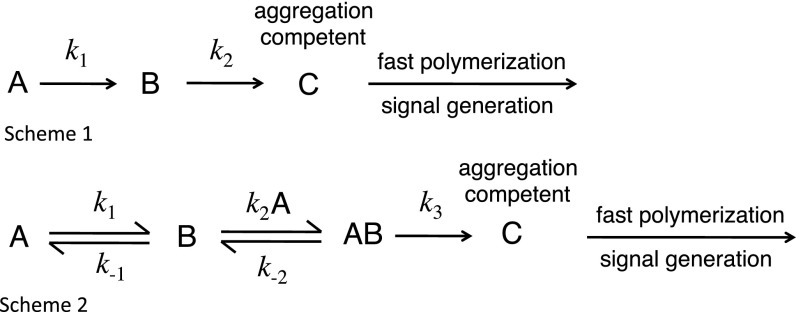
The previous kinetic scheme involved two sequential first-order steps to give an aggregation competent intermediate, C, which undergoes rapid polymerization (Scheme 1 in Fig. 3). That scheme must be expanded to include the progress curves fitting well to apparent sequential two-step first-order kinetics; the decrease in rate constants at low concentrations of protein; and the effects of urea and a preliminary Φ-value analysis indicating two molecules unfolding. The simplest mechanism that could lead to saturation kinetics is Scheme 2 (Fig. 3).
Fig. 3.
Schemes for aggregation of p53 variants.
In the first step, WT structure A forms B, which reversibly associates with A and then forms the aggregation competent state C (k2 > k1 for QCYC) (31). We must emphasize that many different mechanisms can have the same qualitative behavior, including schemes with off-pathway intermediates (30) or initial homodimerization. Scheme 2 is the simplest that can recapitulate the observed behavior. Its properties explain the basic features of the observed kinetics.
The kinetic equations are very difficult to solve analytically because they lead to second-order differential equations as the concentration of A in the scheme changes and are also involved in a second-order step. Qualitatively, we expect biphasic kinetics: a fast exponential initial lag phase as B and AB are formed, followed by a slow accumulation of C in a progress curve that changes from first-order to second-order kinetics as the concentration of A decreases from saturating to subsaturating for binding to B. We can, however, provide approximate solutions at extreme concentrations of A.
High concentrations of A.
At saturating concentrations of A ([A] ≫ KD, where KD = k-2/k+2), B and AB transiently reach a steady-state concentration, the approach to which should be exponential as the concentration of A is approximately constant over this time range. We assume the binding steps k-2 and k+2 are fast (and only their ratio KD is important). Assuming [A]k+1 is the slow step and [AB]k+3 is the fast step, we calculate the first-order approach to the steady state by dividing initial rate of formation of B + AB, (= k+1[A]), by the amplitude of the change, ([B] + [AB])ss
Therefore,
As [A] tends to infinity, k2 tends to k+3.
The initial part of the slow phase will also approximate to exponential when A remains high and is above KD. As [A] tends to infinity, k+2[A] and k+3 are fast, and for every molecule of A that reacts, a second molecule is also immediately lost via k+2[A] and k+3.
Low concentrations of A.
At the other extreme for [A] ≪ KD, the kinetics becomes simple. Here, B accumulates rapidly and reacts very slowly. The fast phase is for the accumulation of B, which should be exponential with rate constant
The slow phase for [A] ≪ KD will follow simple second-order kinetics. Solving the steady-state equations and approximating [A]/([A] + KD) to [A]/KD gives
Numerical Simulation.
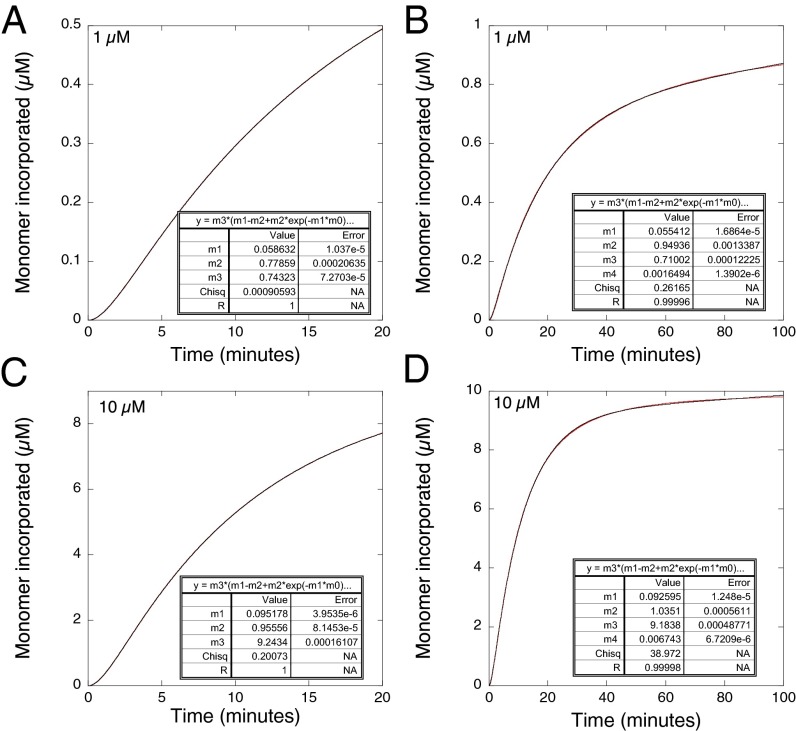
We simulated the progresses curves numerically, using the program Tenua (bililite.com/tenua). Typical simulations in Fig. 4 used Scheme 2, where k+1= 0.05 min−1, k-1 = 0.5 min−1, k-2/k+2 = KD = 2 µM, and k+3 = 1 min−1. The simulated output at higher concentrations of protein fitted exceptionally well to the analytical solution of a simple two-step first-order scheme A → B → C, using the equations used to analyze experimental data (note, reversible two-step first-order kinetics fit to the same curves as irreversible reactions, but where the observed values of k1 and k2 are functions of all of the rate constants, and we are not assuming that k1 and k2 are each unimolecular irreversible rate constants). Deviations from the fits to first-order kinetics should be seen as the concentration of protein decreases below KD so the kinetics becomes second order. That situation occurs toward the end of progress curves at initial concentrations ≫ KD or earlier on in the progress curves where initial concentration are <KD. The earlier the part of the time course that is analyzed, the closer should be the fit to first-order kinetics as the concentration of protein remains relatively constant. At 10 µM protein, 5 × KD, (Fig. 4D), the loss of monomers was 98% complete after 100 min and the curve fit had a correlation coefficient of R = 0.99998. At 20 min (Fig. 4C), the reaction was 78% completed, and the correlation coefficient was 1.0. Even at lower concentrations, the fits were good: at 1 µM protein, half the value of KD, the loss of monomers was 84% complete after 100 min (Fig. 4B), and the curve fit had a correlation coefficient of R = 0.99996. At 20 min (Fig. 4A), the reaction was 50% completed and the correlation coefficient was 1.0. These fits were so good that Schemes 1 and 2 could not be distinguished experimentally by the quality of the fits as the noise in the experimental data would be far greater than the theoretical deviations.
Fig. 4.
Simulated plots for Scheme 2 where k+1 = 0.05 min−1, k-1 = 0.5 min−1, k+2/k-2 = KD = 2 µM, k+3 = 1 min−1. Short and long time courses of protein aggregation at (A and B) low and (C and D) high concentrations. Data (red) were fitted (black) to Eq. 1, without the term a so the plots start from the origin. The 20-min time courses omitted the linear drift term from Eq. 1.
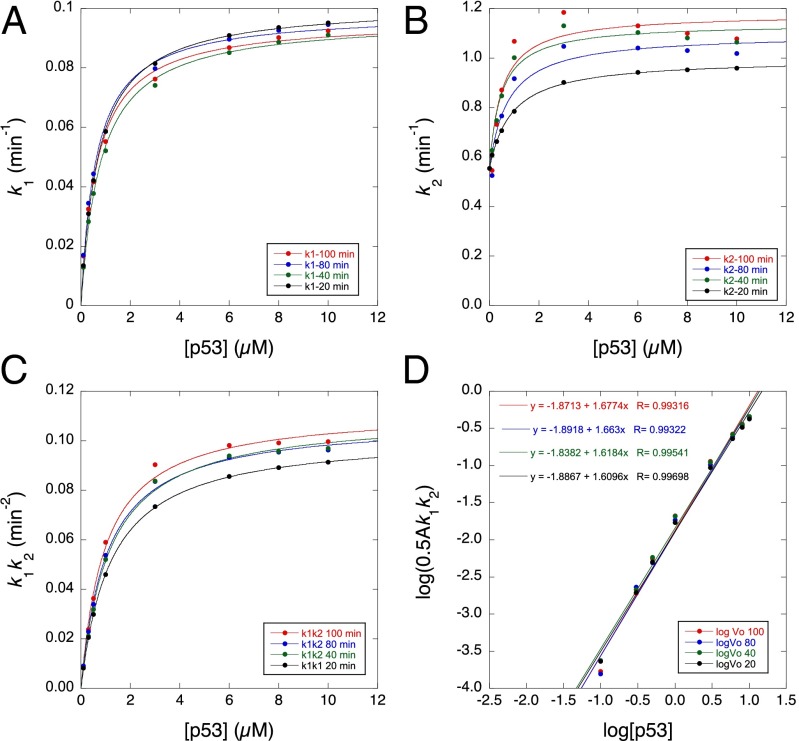
We plotted rate constants obtained from the first-order fits to the simulated data for Scheme 2 obtained from 20, 40, 80, and 100 min of the time courses from 0.03 or 0.1–10 µM protein (Fig. 5). The derived rate constants varied with degree of time course analyzed. k1 fitted hyperbolic curves with values of KD between 0.6 and 0.8 µM and maximum values between 0.1 min−1 (20-min fit) and 0.096 (100-min fit) (Fig. 5A); the theoretical value expected is 0.1 min−1, i.e., 2 × k+1. The curves for k2 (Fig. 5B) recapitulated the experimental curves, with the longer time courses peaking in the middle and the shorter time courses being smooth. Fitting hyperbolic curves to the data (Fig. 5B) gave values of KD between 0.5 and 0.9 µM, and the maximum values converge on the predicted value of 1.0 min−1 at [protein] = 5 × KD i.e., k+3. The values of k2 were approximately constant between 1 and 10 µM, despite KD being 2 µM. The product k1k2 was more robust with degree of time course (Fig. 5C), varying between 0.1 and 0.11 min−2, with apparent KD between 1.2 and 0.97 µM. A plot of logk1k2A vs. log[p53] gives a slope of 1.61–1.68 in the middle of the experimentally observed values. The value expected for purely first order is 1 and for purely second order is 2, so the data were consistent with a gradual changeover in the 0.1- to 10-µM range. Using numerical simulation, we also found that changing k+1, k+2, k-1, and k-2 greatly changed the concentration dependence of the observed rate constants k1 and k2. Hence, different concentration-dependent behaviors (Fig. S1) of the mutants may arise because mutation affects the different microscopic rate constant values of k+1, k+2, k-1, and k-2.
Fig. 5.
Derived rate constants from simulated plots using the same inputs as in Fig. 4 vs. protein concentration for time courses of 20, 40, 80, and 100 min (0.1–10 µM protein for k1 and 0.03–10 µM for k2). Concentration dependence of rate constants (A) k1, (B) k2, (C) k1k2, and (D) initial aggregation rate. Time courses over 20 and 40 min were fitted to Eq. 1 without the linear drift term, which was included for 80 and 100 min. The term a was included to mimic the fitting of data in experiments. The values of k1 and k2 plotted are the observed values from fitting aggregation curves given by Scheme 2 with Eq. 1 as shown in Fig. 4 and not the microscopic values in Scheme 2.
We saw experimentally the divergence from consecutive first-order reaction kinetics from individual curve fits at low protein concentrations, where the kinetics became second order as the reaction proceeds. If we fitted just the first part of the curve for the low concentrations, we got a good measurement of the fast rate constant, which is still first order, but the slower one changes according to the length of time course analyzed. At high concentrations of protein, we got very good fits to very long time courses, which gave very similar results to those for short time courses.
The important conclusions from the simulations are that the complex kinetics of Scheme 2 fit the solution of the simpler Scheme 1 to a good approximation over a reasonable range of concentrations and give a very good approximation of the values of k+1 and k+3 at high concentrations of protein. We now use that information to illuminate the stabilization of structure at various positions and seeding.
Φ-Value Analysis of k1 and k2.
The two empirical rate constants k1 and k2 were sensitive to mutation. We conducted a Φu-value analysis on their observed values at high concentrations of protein, without assuming any mechanism (Φu = Φ for unfolding; Fig. 6). We simply measured the ratios of the changes in their individual activation energies on mutation, ∆∆G‡-N, to that of the change in free energy of denaturation, ∆∆GD-N, measured from urea denaturation curves. [As for many other proteins, unfolding of p53 happens before aggregation (11).] The chosen mutation sites were mainly within the β sandwich region. Generally, one should use conservative mutations that delete parts of side chains (e.g., Ile to Val, Thr to Ser, etc.), which was done, but more radical mutations can be interpreted if the Φu-values are close to 0 or 1 (34–36). For accuracy, only mutants with ∆∆GD-N >0.6 kcal/mol were used (35). Although some of the rate constants were not highly accurate, the possible errors would make only small errors in the calculation of Φu as the changes in free energy of denaturation were large and calculations are on a ratio of logarithms. Several of the mutations were in the same region of structure, or even of the same residue, and the results were internally consistent. Most Φu-values were similar for both k1 and k2 and generally greater than 0.5, consistent with the high fraction of overall solvent exposure in the transition states as indicated by the effect of urea on unfolding rate constants.
Fig. 6.
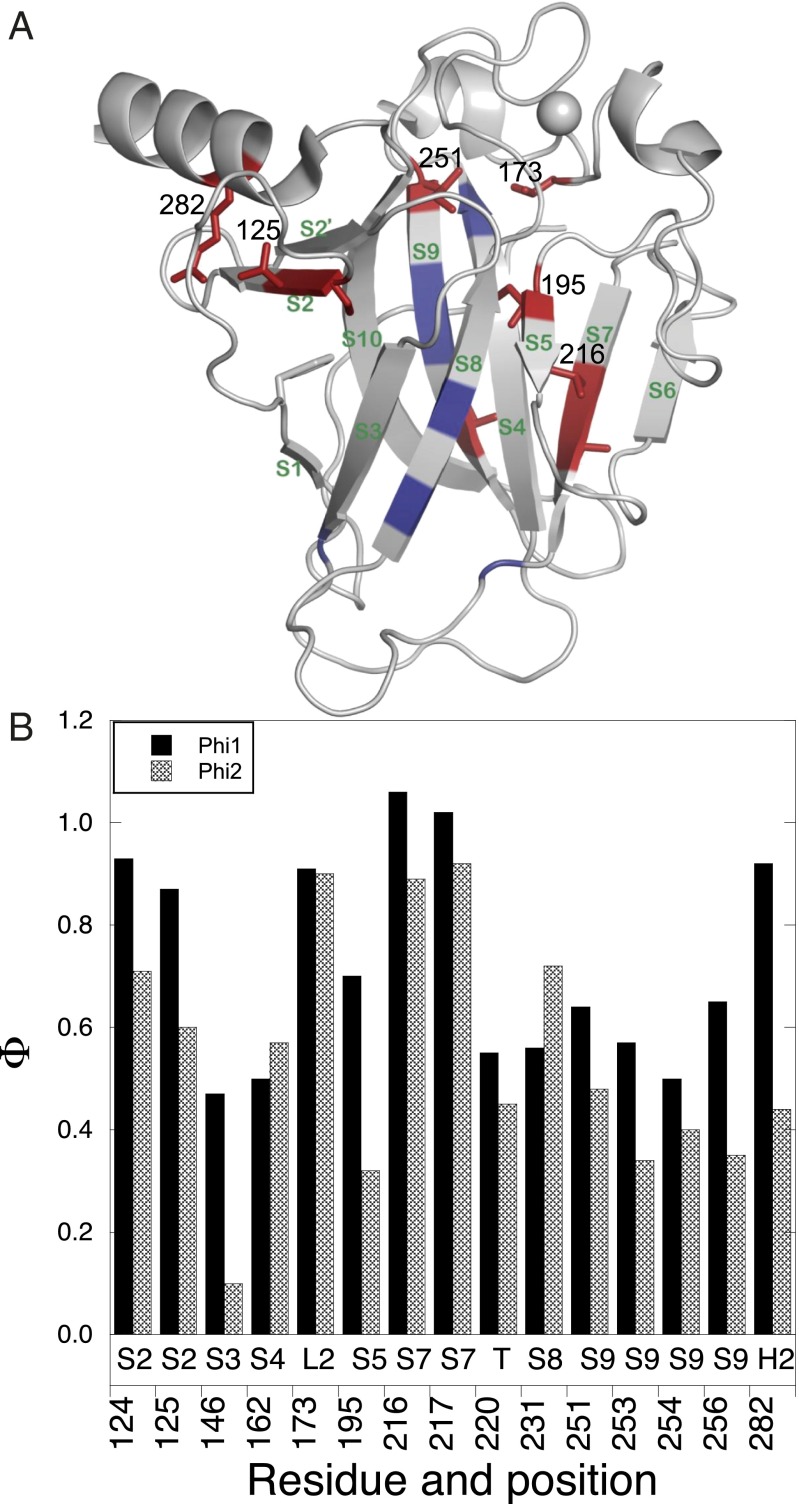
Φu values of p53 core domain during aggregation. (A) Φu values are mapped on the native structure and are color coded: residues with Φu values between 1.0 and 0.6, red; lower than 0.6, deep blue. The side chains of residues with Φu >0.6 are shown explicitly. (B) Φu values for different positions.
Low Φu-Value Regions.
The interactions around residues 146, 162, 220, 231/233, and 254 were significantly, but not fully, disrupted in the transition state.
High Φu-Value Regions.
The environment around loop regions 173 (L2), 256 (loop between S9 and S10), β-strand regions 124, 125 (S2), 195 (S4), 216, 217 (S7), and H2 helix 282 were largely disrupted in the transition state (Fig. 6; Φu values > 0.6–0.7). Most of these sites are in flexible regions of the p53 structure, including L1, L2, the loop between S9 and S10 [256–270, which is recognized by the antibody DO-12 (40)], and S7 [213–217, recognized by PAb240 (41)].
The major groove of the DNA binding region has helix H2 (278–287) and loop L1 (113–123) of the loop-sheet-helix motif. Arg282, a mutational oncogenic hotspot (8), plays a structural role in packing of the helix H2 onto the S2-S2′ β-hairpin. High Φu-values for R282A and T125A mutations, which disrupt the hydrogen bond between 125 and 282, showed the interactions were largely disrupted in the transition states (Fig. 6).
The side chain of V173 in loop L2, covering the β sandwich, interacts with residue I251. The Φu-value of V173A is ∼1 and Φu for I251V is also high, showing this part of structure was largely exposed in the transition state. V216 interacts with I195 in the hydrophobic core. The Φu-value for V216A was ∼1 and that of I195V was ∼0.6, implying the core was also largely disrupted in the transition state.
Discussion
Simple Kinetics Hides a More Complex Mechanism.
The initial rate-determining stages for the formation of amyloid structure from p53 before the subsequently slower rearrangement into large particles fit well within normal experimental error to consecutive first-order reaction kinetics, Scheme 1, with two observed first-order rate constants, k1 and k2 (31, 32) (Fig. 1). We now analyzed more than 30 mutants of p53 over a wide range of concentrations to conduct a Φu-value analysis. Such studies, in addition to the quantitative data obtained, can provide a series of in-built controls to discover new information about reactions. Different mutants had rate constants that depended in different ways on the concentration of protein, indicating clearly that a bimolecular step was also involved (Fig. 2 and Fig. S1). The Φu-value analysis indicated that each of k1 and k2 involved the unfolding of a molecule of p53, with full unfolding in several regions (Fig. 6). However, there would appear to be a contradiction between the bimolecular processes indicated by the Φu-value analysis and the change of rate constants over a wide range of concentrations on the one hand and, on the other hand, individual progress curves fitting well to sequential first-order kinetics. We could not solve the kinetics of Scheme 2 analytically and resorted to numerical simulation to understand the dilemma. Remarkably, simulation of Scheme 2 generated progress curves that are indistinguishable within reasonable experimental error from those of sequential first-order kinetics of Scheme 1 (Fig. 4). The numerical simulations recapitulated discontinuities that appeared in the experimental plots of observed rate constants vs. protein concentration (Fig. 5). Therefore, a more complex mechanism is not inconsistent with the simple observed kinetics.
Scheme 2 is the minimal reaction scheme that can account for those facts. Other schemes can be devised that would also generate the same kinetic curves, such as those involving off-pathway intermediates (30). Because of that, we will not try to assign at this stage k1 and k2 to microscopic rate constants as there will be other interpretations. We just draw conclusions from two observations that are independent of precise mechanism: there are two rate determining steps in the initial stages of aggregation of p53 that are sensitive to mutation; and we know the effects of mutation on those steps. The fundamentals of the kinetic mechanism provide a basis for understanding the kinetics of aggregation of full-length p53 and its possible seeding by oncogenic mutants.
Conclusions from Φ-Value Analysis.
Mutation of residues in L2 and S7 gave Φu-values of close to unity for both steps, and for other regions, the sum of both Φu-values was significantly greater than 1 (Fig. 6). Each step, therefore, involved the considerable unfolding of a molecule of p53, consistent with the concentration dependence of k1 and k2. The Φu-values point to regions that would be the most sensitive targets to compounds that inhibit the aggregation of p53 by binding to the native structure; tying together segments that fully unfold in the transition state and have large changes in energy would be the most effective. The high Φu-values in the DNA binding region, for example, explain why binding of cognate DNA stabilizes so well against aggregation (42). Conversely, the relatively low Φu-value of residue 220 explains why small molecules binding in the cavity caused by Y220C inhibit each of k1 and k2 by only 60–70% (31, 32). The region around Tyr220 is only partly disrupted in the transition state, so drugs targeting this site can only partly inhibit aggregation, and parallel aggregation pathways are observed as both the ligand-free protein and its complex with a small molecule can unfold within an order of magnitude of each other (31).
Methods
Protein Expression and Purification.
Mutants were constructed using QuikChange site-directed mutagenesis (Stratagene) (43). Recombinant p53 proteins including core domain and full-length variants were expressed and purified as previously described (32, 44). For full-length p53 variants, DsbA within the lysate was removed by washing with denatured Escherichia coli proteins after adsorption of target protein to a Ni column.
Equilibrium Denaturation.
Equilibrium denaturation of p53 core domain (94–312) variants was measured at 10 °C (10) in a buffer of 25 mM potassium phosphate, 150 mM NaCl, 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and various concentration of urea, pH 7.2. Denaturation was monitored by fluorescence using a Horiba FluoroMax-4 Spectrofluorometer with excitation at 280 nm and emission scanned from 300 to 370 nm. Slits for excitation and emission were 5 nm. Each sample was incubated at 10 °C for 12 h before fluorescence was measured. For p53 long core (89–312) and its mutants, protein was excited at 280 nm, and the emission spectrum was scanned from 300 to 500 nm. The fluorescence intensity change at 356 nm was used for WTC and integrated from 356 to 500 nm for WT long core (WTLC).
Measurement of Melting Temperatures.
The melting temperature of p53 variants were measured using SYPRO Orange as described previously with 10 µM protein in 25 mM potassium phosphate, 150 mM NaCl, and 1 mM TCEP, pH 7.2 (44).
Kinetics of Aggregation.
Aggregation kinetics of native p53 variants was monitored by light scattering and ThT fluorescence using a Horiba FluoroMax-3 spectrophotometer (31, 32) in buffer of 25 mM potassium phosphate, 150 mM NaCl, 1 mM TCEP, 5% (vol/vol) DMSO, pH 7.2 and, where required, 20 µM ThT.
Calculation of Φu.
Φu was calculated from the rate constants k1 and k2 using Φu = −RTln(kwt/kmut)/∆∆GD-N, where ∆∆GD-N is the difference in free energy of denaturation between WT and mutant (10). The values of k1 and k2 were mainly taken from the plateau regions in the curves of observed rate constant k vs. [p53] for both mutant and parent. Where curves of k vs. [p53] could be fitted well to a simple saturation isotherm, we used the numerical fit to the curve. For others, we used the highest measured value. Three core domain constructs were used (Table 1). For most of the mutants, the pseudo-WT was QCYC. For WTCI254D aggregating at 37 °C, the pseudo-WT was WTC.
The data from WTLC were not used because the rate constants did not sufficiently approach the plateau region.
Supplementary Material
Acknowledgments
We thank Dr. Andreas Joerger for advice on mutation sites and Dr. Miriana Petrovich for help with protein purification. This work was funded by European Research Council (ERC) Advanced Grant 268506 (to A.R.F.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1500243112/-/DCSupplemental.
References
- 1.Lane D, Levine A. p53 Research: The past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vazquez A, Bond EE, Levine AJ, Bond GL. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov. 2008;7(12):979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 3.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meek DW. Tumour suppression by p53: A role for the DNA damage response? Nat Rev Cancer. 2009;9(10):714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 5.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 6.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9(12):862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 7.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223(2):116–126. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 8.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science. 1994;265(5170):346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 9.Ang HC, Joerger AC, Mayer S, Fersht AR. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J Biol Chem. 2006;281(31):21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 10.Bullock AN, et al. Thermodynamic stability of wild-type and mutant p53 core domain. Proc Natl Acad Sci USA. 1997;94(26):14338–14342. doi: 10.1073/pnas.94.26.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedler A, Veprintsev DB, Hansson LO, Fersht AR. Kinetic instability of p53 core domain mutants: Implications for rescue by small molecules. J Biol Chem. 2003;278(26):24108–24112. doi: 10.1074/jbc.M302458200. [DOI] [PubMed] [Google Scholar]
- 12.Bullock AN, Fersht AR. Rescuing the function of mutant p53. Nat Rev Cancer. 2001;1(1):68–76. doi: 10.1038/35094077. [DOI] [PubMed] [Google Scholar]
- 13.Bullock AN, Henckel J, Fersht AR. Quantitative analysis of residual folding and DNA binding in mutant p53 core domain: Definition of mutant states for rescue in cancer therapy. Oncogene. 2000;19(10):1245–1256. doi: 10.1038/sj.onc.1203434. [DOI] [PubMed] [Google Scholar]
- 14.Butler JS, Loh SN. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry. 2003;42(8):2396–2403. doi: 10.1021/bi026635n. [DOI] [PubMed] [Google Scholar]
- 15.Gaiddon C, Lokshin M, Ahn J, Zhang T, Prives C. A subset of tumor-derived mutant forms of p53 down-regulate p63 and p73 through a direct interaction with the p53 core domain. Mol Cell Biol. 2001;21(5):1874–1887. doi: 10.1128/MCB.21.5.1874-1887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strano S, et al. Physical interaction with human tumor-derived p53 mutants inhibits p63 activities. J Biol Chem. 2002;277(21):18817–18826. doi: 10.1074/jbc.M201405200. [DOI] [PubMed] [Google Scholar]
- 17.Strano S, et al. Physical and functional interaction between p53 mutants and different isoforms of p73. J Biol Chem. 2000;275(38):29503–29512. doi: 10.1074/jbc.M003360200. [DOI] [PubMed] [Google Scholar]
- 18.Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol. 1999;19(2):1438–1449. doi: 10.1128/mcb.19.2.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milner J, Medcalf EA. Cotranslation of activated mutant p53 with wild type drives the wild-type p53 protein into the mutant conformation. Cell. 1991;65(5):765–774. doi: 10.1016/0092-8674(91)90384-b. [DOI] [PubMed] [Google Scholar]
- 20.Selivanova G, Wiman KG. Reactivation of mutant p53: Molecular mechanisms and therapeutic potential. Oncogene. 2007;26(15):2243–2254. doi: 10.1038/sj.onc.1210295. [DOI] [PubMed] [Google Scholar]
- 21.Wiman KG. Pharmacological reactivation of mutant p53: From protein structure to the cancer patient. Oncogene. 2010;29(30):4245–4252. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Vazquez A, Levine AJ, Carpizo DR. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21(5):614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilcken R, et al. Halogen-enriched fragment libraries as leads for drug rescue of mutant p53. J Am Chem Soc. 2012;134(15):6810–6818. doi: 10.1021/ja301056a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sipe JD, Cohen AS. Review: History of the amyloid fibril. J Struct Biol. 2000;130(2-3):88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 25.Chiti F, Dobson CM. Amyloid formation by globular proteins under native conditions. Nat Chem Biol. 2009;5(1):15–22. doi: 10.1038/nchembio.131. [DOI] [PubMed] [Google Scholar]
- 26.Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol. 2005;6(11):891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 27.Lansbury PT, Lashuel HA. A century-old debate on protein aggregation and neurodegeneration enters the clinic. Nature. 2006;443(7113):774–779. doi: 10.1038/nature05290. [DOI] [PubMed] [Google Scholar]
- 28.Ano Bom AP, et al. Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J Biol Chem. 2012;287(33):28152–28162. doi: 10.1074/jbc.M112.340638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishimaru D, et al. Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry. 2003;42(30):9022–9027. doi: 10.1021/bi034218k. [DOI] [PubMed] [Google Scholar]
- 30.Borgia MB, Nickson AA, Clarke J, Hounslow MJ. A mechanistic model for amorphous protein aggregation of immunoglobulin-like domains. J Am Chem Soc. 2013;135(17):6456–6464. doi: 10.1021/ja308852b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilcken R, Wang G, Boeckler FM, Fersht AR. Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc Natl Acad Sci USA. 2012;109(34):13584–13589. doi: 10.1073/pnas.1211550109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, Fersht AR. First-order rate-determining aggregation mechanism of p53 and its implications. Proc Natl Acad Sci USA. 2012;109(34):13590–13595. doi: 10.1073/pnas.1211557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fersht AR, Leatherbarrow RJ, Wells TN. Structure-activity relationships in engineered proteins: Analysis of use of binding energy by linear free energy relationships. Biochemistry. 1987;26(19):6030–6038. doi: 10.1021/bi00393a013. [DOI] [PubMed] [Google Scholar]
- 34.Matouschek A, Kellis JT, Jr, Serrano L, Fersht AR. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989;340(6229):122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- 35.Fersht AR, Sato S. Phi-value analysis and the nature of protein-folding transition states. Proc Natl Acad Sci USA. 2004;101(21):7976–7981. doi: 10.1073/pnas.0402684101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224(3):771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 37.Vitalis A, Pappu RV. Assessing the contribution of heterogeneous distributions of oligomers to aggregation mechanisms of polyglutamine peptides. Biophys Chem. 2011;159(1):14–23. doi: 10.1016/j.bpc.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thakur AK, Wetzel R. Mutational analysis of the structural organization of polyglutamine aggregates. Proc Natl Acad Sci USA. 2002;99(26):17014–17019. doi: 10.1073/pnas.252523899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4(10):2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vojtesek B, et al. Conformational changes in p53 analysed using new antibodies to the core DNA binding domain of the protein. Oncogene. 1995;10(2):389–393. [PubMed] [Google Scholar]
- 41.Stephen CW, Lane DP. Mutant conformation of p53. Precise epitope mapping using a filamentous phage epitope library. J Mol Biol. 1992;225(3):577–583. doi: 10.1016/0022-2836(92)90386-x. [DOI] [PubMed] [Google Scholar]
- 42.Ishimaru D, et al. Cognate DNA stabilizes the tumor suppressor p53 and prevents misfolding and aggregation. Biochemistry. 2009;48(26):6126–6135. doi: 10.1021/bi9003028. [DOI] [PubMed] [Google Scholar]
- 43.Petrovich M, Jonsson AL, Ferguson N, Daggett V, Fersht AR. Phi-analysis at the experimental limits: Mechanism of beta-hairpin formation. J Mol Biol. 2006;360(4):865–881. doi: 10.1016/j.jmb.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 44.Boeckler FM, et al. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci USA. 2008;105(30):10360–10365. doi: 10.1073/pnas.0805326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.