Abstract
microRNAs (miRNAs) are short noncoding RNAs that regulate gene expression through posttranscriptional repression of target genes. miRNAs exert a fundamental level of control over many developmental processes, but their role in the differentiation and development of skeletal muscle from myogenic progenitor cells in humans remains incompletely understood. Using primary cultures established from human skeletal muscle satellite cells, we performed microarray profiling of miRNA expression during differentiation of myoblasts (day 0) into myotubes at 48 h intervals (day 2, 4, 6, 8, and 10). Based on a time-course analysis, we identified 44 miRNAs with altered expression [false discovery rate (FDR) < 5%, fold change > ±1.2] during differentiation, including the marked upregulation of the canonical myogenic miRNAs miR-1, miR-133a, miR-133b, and miR-206. Microarray profiling of mRNA expression at day 0, 4, and 10 identified 842 and 949 genes differentially expressed (FDR < 10%) at day 4 and 10, respectively. At day 10, 42% of altered transcripts demonstrated reciprocal expression patterns in relation to the directional change of their in silico predicted regulatory miRNAs based on analysis using Ingenuity Pathway Analysis microRNA Target Filter. Bioinformatic analysis predicted networks of regulation during differentiation including myomiRs miR-1/206 and miR-133a/b, miRNAs previously established in differentiation including miR-26 and miR-30, and novel miRNAs regulated during differentiation of human skeletal muscle cells such as miR-138-5p and miR-20a. These reciprocal expression patterns may represent new regulatory nodes in human skeletal muscle cell differentiation. This analysis serves as a reference point for future studies of human skeletal muscle differentiation and development in healthy and disease states.
Keywords: miRNA, transcriptome, skeletal muscle, differentiation, satellite cells
skeletal muscle displays remarkable plasticity in response to ageing, contractile activity, disease, or injury. Critical to the regulation of muscle growth and repair are myogenic progenitor cells, satellite cells, which are quiescent cells located adjacent to muscle fibers between the sarcolemma and beneath the fiber basal lamina (56). Satellite cells can be induced to divide during muscle damage or contractile activity. Subsequent fusion with an existing myofiber results in the addition of a myonucleus to the fiber syncytium. In human skeletal muscle, the large interindividual variability in the magnitude of the hypertrophic response to resistance exercise training is explained by the relative ability to mobilize satellite cells and add myonuclei to existing muscle fibers (32, 33).
Myotube differentiation is under the control of several basic helix-loop-helix transcription factors known as myogenic regulatory factors (MRFs), including myogenic factor 5 (MYF5), myoblast determination protein (MYOD1), and myogenin (MYOG), which promote terminal differentiation by regulating myogenic gene expression (7). Other transcription factors, including the paired box proteins PAX3 and PAX7, are involved in determination and proliferation of cells belonging to the myogenic lineage (7). However, detailed understanding of the transcriptional networks and genes controlling skeletal muscle differentiation is warranted, particularly for human skeletal muscle cells.
MicroRNAs (miRNAs) are a class of short, noncoding RNAs that show high sequence conservation throughout evolution (3). miRNAs regulate gene expression through posttranscriptional mechanisms, either by reducing mRNA abundance through complementary binding and direction of the RNA-induced silencing complex to targeted mRNAs or by attenuating protein translation (3, 20). Several miRNAs are enriched in heart and skeletal muscle (42) and play key roles in muscle differentiation and development (53). For example, miRNA biogenesis is indispensable for skeletal muscle fiber formation during embryogenesis (31). Myogenic miRNAs (termed “myomiRs”), including miR-1, miR-133, and miR-206, have dramatic effects on satellite cell proliferation and differentiation (8, 15, 24). Expression of the myomiRs is markedly induced during skeletal muscle differentiation through transcriptional regulation by several of the MRFs (37). miR-1 and miR-206 induce skeletal muscle differentiation by targeting repressors of differentiation, for example histone deacetylase 4 and PAX7, respectively (8, 15). Numerous non-myomiRs are also involved in the regulation of proliferation and differentiation of satellite cells (9, 30). Examples include the differentiation-induced miR-26a, which negatively regulates SMAD1 and SMAD4 protein abundance in transforming growth factor (TGF)-β signaling, and miR-351, which is induced after injury and during regeneration and affects myogenic progenitor cell proliferation and apoptosis (10, 16).
The role of miRNA in skeletal muscle differentiation has largely been elucidated in murine skeletal muscle cell lines (8, 9, 15, 24). Given the predicted species specificity of miRNA in regulating target genes (5, 35) and the debated differences in the phenotype and function of human and mouse satellite cells (6), studies on human skeletal muscle satellite cells are warranted. While this knowledge gap has been partially addressed (17, 26, 27), the understanding of miRNAs and their regulatory networks during human skeletal muscle differentiation remains underdeveloped. Thus, we performed simultaneous microarray profiling of miRNA and mRNA expression of human skeletal muscle cells during differentiation. We used a time course approach to achieve sufficient temporal resolution to examine miRNA-centered networks of biological regulation of skeletal muscle differentiation. We hypothesized that differential expression patterns of miRNA and mRNA would be present in “early” compared with “late” differentiation and that analysis of reciprocal miRNA-mRNA expression patterns would identify novel regulatory nodes involved in primary human skeletal muscle cell differentiation.
MATERIAL AND METHODS
Cell culture.
Using procedures as described (1), we harvested satellite cells from human vastus lateralis biopsies from six male volunteers with no known metabolic or muscle dysfunction. Informed consent was obtained from all participants in accordance with the Declaration of Helsinki, and the local ethics committee at the Karolinska Institute approved all study protocols. Muscle biopsies were obtained under local anesthesia (5 mg/ml lidocaine hydrochloride) from overnight fasted volunteers and placed in cold PBS supplemented with 1% PeSt (100 U/ml penicillin and 100 μg/ml streptomycin). Biopsies were dissected free from visible connective and fat tissues, minced finely, transferred to a digestion solution [0.015 g collagenase IV, 8% 10× trypsin, 0.015 g bovine serum albumin (BSA), and 1% PeSt in Ham's F-10 medium], and incubated with gentle agitation at 37°C for 20 min. Undigested tissue was allowed to settle, and the supernatant containing liberated satellite cells was collected and mixed 1:1 with growth medium (GM; DMEM/F-12 with 20% FBS, 1% PeSt, 1% Fungizone). This step was repeated with the undigested tissue, and the resultant supernatant was pooled with the previous cells and centrifuged for 10 min at 350 g. The cell pellet was resuspended in GM and incubated in a noncoated petri dish for 1 h to selectively promote adherence of nonmyogenic cells. The supernatant was transferred, and cells were seeded in 150 cm culture flasks. At confluence (>80%), cells were trypsinized and subcultured. The passage after the first trypsinization was designated passage 1, and differentiation experiments took place at passage 5.
Differentiation of myoblasts into myotubes was induced by switching to differentiation medium [DM; DMEM (low glucose) with 2% FBS, 1% PeSt, and 1% Fungizone] when cells were ∼80% confluent. Cells were differentiated for up to 10 days. Cultures were incubated at 37°C in 7.5% CO2 humidified chambers, and medium was changed every second day during growth and differentiation. All cell culture medium and medium components were from Life Technologies (Carlsbad, CA).
Human embryonic kidney cells (HEK293) were obtained from ATCC and cultured in high-glucose (4.5 g/l) DMEM supplemented with 10% (vol/vol) FBS. Cells were seeded in 96-well plates 24 h before experimentation.
Experimental design.
RNA and protein were isolated from cells before differentiation (i.e., as proliferating myoblasts; day 0) and at five time points during differentiation at 48 h intervals (day 2, 4, 6, 8, and 10). RNA was applied to miRNA microarrays for expression profiling (n = 6). For n = 3 subjects, the same RNA was applied to microarrays for profiling of gene expression at day 0, 4, and 10. The entire cohort could not be studied because of sample limitations. Based on the datasets of differentially expressed miRNA and mRNA, we built a model of predicted miRNA-regulated gene networks during differentiation.
RNA isolation.
Total RNA was isolated with TRIzol according to manufacturer's instructions (Life Technologies) with minor modifications. Separation of the aqueous and organic phases was performed with Phase-Lock Heavy Gel tubes (2302830; 5PRIME, Gaithersburg, MD). RNA was precipitated at −20°C for 20 min after addition of 15 μg of glycogen (AM9510, Life Technologies) per 1 ml of TRIzol initially used. Isolated RNA was stored at −80°C until further analysis. RNA concentration and purity was assessed by UV spectrophotometry with a Nanodrop ND-1000 (Thermo Fisher Scientific, Waltham, MA). For microarray analysis, RNA integrity was determined by gel electrophoresis using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). An RNA integrity number > 7 and spectrophotometric A260/A280 and A260/A230 ratios > than 1.8 and 1.5, respectively, were considered acceptable.
Microarray profiling of miRNA expression.
Total RNA (300 ng) from the samples and a pooled reference RNA was labeled with Hy3 and Hy5 fluorescent label, respectively, using the miRCURY LNA Array Power Labeling kit (Exiqon, Vedbæk, Denmark). The Hy3-labeled samples and the Hy5-labeled reference RNA sample were mixed pair-wise and hybridized to the miRCURY LNA array version 11.0 (Exiqon), which contains capture probes targeting all miRNAs for human, mouse, or rat sequences registered in the miRBase version 13.0 at the Sanger Institute. miRBase version 13.0 accounts for 1,276 of 2,588 miRNAs annotated in miRBase version 21 (released in June 2014). The hybridization was performed according to the miRCURY LNA array manual using a Tecan HS4800 hybridization station (Tecan, Austria). After hybridization, microarray slides were scanned and stored in an ozone-free environment (ozone level <2.0 ppb) to prevent potential bleaching of the fluorescent dyes. miRCURY LNA array microarray slides were scanned with the Agilent G2565BA Microarray Scanner System (Agilent Technologies), and the image analysis was carried out with the ImaGene 8.0 software (BioDiscovery, El Segundo, CA). The quantified signals were background corrected (Normexp with offset value 10) and normalized with the global Lowess (LOcally WEighted Scatterplot Smoothing) regression algorithm (38).
For expression analysis, only miRNAs with logarithmic Hy3 values >6 at one time point during differentiation were included (manufacturer's recommendation for abundance cut-off). Differences in miRNA expression during skeletal muscle differentiation was determined by significant analysis of microarrays (SAM) time course analysis with a false discovery rate (FDR) <5% (46). This analysis identifies miRNAs with a consistent change in expression over several time points. Changes in miRNA expression at individual time points compared with proliferating cells were also determined with SAM (two class paired analysis) with an FDR < 5%. The microarray data were submitted to the National Center for Biotechnology Information's (NCBI's) Gene Expression Omnibus (GEO). The data can be found under the GEO accession number GSE53383. A heat map of differentially expressed miRNAs with a time course approach analysis was constructed with the R software (version 3.0.1) using clustering performed with a hierarchical method with average linking based on Euclidean distance measures (34).
Relative miRNA expression determined by miRNA quantitative reverse transcriptase-polymerase chain reaction.
Validation of the microarray-derived expression profile of selected miRNAs was performed using custom miRNA reverse transcription primer pools according to the manufacturer's protocol (Life Technologies). Equal amounts of total RNA (1,000 ng) were used as input for each sample. miRNA TaqMan primers were used for measurement of relative expression of individual miRNAs, and quantitative reverse transcriptase-polymerase chain reaction (qPCR) was performed using a StepOne Plus Real-Time PCR system (Life Technologies). The optimal reference for normalization of expression was determined to be the geometrical mean of four reference genes (U18, RNU24, RNU44, and RNU48) according to the NormFinder algorithm in GenEx software (MultiD Analyses, Gothenburg, Sweden) (4). All reagents, including reverse transcription and TaqMan primers [U18 (001204), RNU24 (001001), RNU44 (001094), RNU48 (001006), miR-1 (002222), miR-20a-5p (000580), miR-26a-5p (000405), miR-30b-5p (000602), miR-30c-5p (000419), miR-133a-3p (002246), miR-133b (002247), miR-138-5p (002284), miR-199a-5p (000498), and miR-206 (000510)], were from Life Technologies.
Microarray profiling of gene expression.
Gene expression changes during differentiation were profiled by hybridizing biotinylated sense strand cDNA to Gene Chip Human Gene 1.0 ST arrays (Affymetrix, Santa Clara, CA). Sense strand cDNA was synthesized from total RNA with Ambion WT Expression Kit (Life Technologies) and biotin-labeled with the Affymetrix GeneChip WT Terminal Labeling Kit (Affymetrix) before being hybridized to arrays. Gene arrays were washed, stained, and scanned as instructed by Affymetrix. Preprocessing of data was performed using robust multiarray average with sketch quantile normalization using Expression Console software (Affymetrix).
Following exclusion of nonannotated genes, differential expression of transcripts was determined with a paired class comparison with univariate test using a random variance model comparing gene expression at day 0 vs. day 4 or day 10 using a software package developed by Dr. Richard Simon and the BRB-ArrayTools Development Team (BRB ArrayTools version 4.3.0, http://linus.nci.nih.gov/BRB-ArrayTools). Genes with an FDR of < 10% were considered to be differentially regulated. The microarray data were submitted to NCBI's GEO and can be found under the GEO accession number GSE53384. Significantly up- or downregulated genes were used as input in the Database for Annotation, Visualization, and Integrated Discovery (DAVID) to explore enrichment of biological processes in the Gene Ontology (GO) database (22, 23).
Relative mRNA expression determined qPCR.
Gene expression was evaluated following synthesis of cDNA using random primers with the High Capacity cDNA Reverse Transcription kit (Life Technologies). Equal amounts of total RNA (1,000 ng) were used as input for each sample. qPCR was performed using a StepOne Plus Real-Time PCR system (Life Technologies) with TaqMan Fast Universal PCR Master Mix and TaqMan primers [ADAM19 (Hs00224690_m1), ID1 (Hs03676575_s1), ITGA5 (Hs01547673_m1), MYF5 (Hs00929416_g1), MYOG (Hs01072232_m1), NAPG (Hs00909795_m1), PRDM1 (Hs00153357_m1), RASAL2 (Hs00183129_m1), SH2B3 (Hs00193878_m1), SNAI1 (Hs00195591_m1), TBP (4326322E), TNNT1 (Hs00162848_m1), and WNT5A (Hs00998537_m1)]. Threshold cycle (Ct) values were determined with the StepOne Software (version 2.3) and relative gene expression was calculated by the comparative Ct method relative to a reference gene. The most stable reference gene was determined to be TBP (out of three possible candidates) using the NormFinder algorithm.
Identifying regulatory networks and predicted target genes of miRNAs.
Significantly altered transcripts from analysis with BRB ArrayTools and miRNAs from SAM analysis between day 0 and day 10 of the differentiation process were used as input for the miRNA Target Filter function in Ingenuity Pathway Analysis (IPA, Ingenuity Systems, http://www.ingenuity.com) to find predicted miRNA-regulated target genes associated with the biological process of human skeletal muscle differentiation. Using the premise that reciprocal expression patterns exist between miRNA and their predicted gene target within the defined list of differentially expressed genes, networks of predicted miRNA-regulated genes were constructed to visualize the potential effects of individual miRNAs on networks and biological functions during differentiation. We used the IPA miRNA Target Filter function, which incorporates experimentally demonstrated and in silico predicted miRNA-mRNA interactions from the databases TargetScan, TarBase, and miRecords. IPA was used to perform functional gene enrichment analysis using predicted target genes from miRNA centered networks. Correlation of expression patterns of miRNAs and differentially expressed transcripts were performed with logarithmic array data representing day 0, day 4, and day 10 for both miRNA and transcript arrays. Pearson product moment correlation analysis, reported as the coefficient r, was performed using R software (version 3.0.1) with Benjamini-Hochberg (BH) correction of P values for multiple testing.
Western blot analysis.
For quantification of protein content in the proliferative state (day 0) and at each subsequent time point (day 2, day 4, day 6, day 8, and day 10) following induction of differentiation, cells were lysed at time points in ice-cold homogenization buffer [137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.5 mM Na3VO4, 1% (vol/vol) Triton X-100, 10% (vol/vol) glycerol, 20 mM Tris, 10 mM NaF, 1 mM EDTA, 1 mM PMSF, and 1% (vol/vol) protease inhibitor cocktail set 1 (Merck, Darmstadt, Germany)]. Homogenates were rotated for 30 min at 4°C and centrifuged at 12 000 g for 15 min at 4°C. Protein content of the supernatants was determined by BCA protein assay kit (Pierce Biotechnology, Rockford, IL). Samples were prepared for SDS-PAGE with Laemmli buffer to equal final protein concentrations, separated on Criterion XT Bis-Tris Gels (Bio-Rad, Hercules, CA) and transferred to PVDF membranes (Merck). After transfer, membranes were stained with Ponceau S to confirm equal loading of samples and quality control for the transfer procedure. After being blocked in 7.5% nonfat milk in Tris-buffered saline with Tween (TBST; 10 mM Tris·HCl, 100 mM NaCl, 0.02% Tween 20) for 2 h at room temperature, membranes were incubated overnight at 4°C with primary antibodies directed to β-actin (A5441; Sigma Aldrich, St. Louis, MO), desmin (ab15200, Abcam), MYOD1 (ab64159, Abcam), and myosin heavy chain (MHC) (Myh1/2, sc-53088; Santa Cruz Biotechnology, Dallas, TX). Membranes were washed with TBST and incubated with species-appropriate horseradish peroxidase-conjugated secondary antibody before proteins were visualized by enhanced chemiluminescence (Amersham ECL Western Blotting Detection Reagent, Little Chalfont, UK). When appropriate, protein content was quantified by densitometry (QuantityOne, Bio-Rad).
Immunocytochemistry.
Cells were fixed with 4% paraformaldehyde before the induction of differentiation (day 0) or following 10 days of differentiation. By indirect immunofluorescence, presence of the myogenic marker desmin and nuclear localization of the proliferative marker Ki67 was determined with primary antibodies targeting desmin and Ki67 (8D5) (#9449; Cell Signaling, Danvers, MA), respectively. Secondary antibodies Alexa Flour 488 IgG (rabbit anti-mouse) and Alexa Flour 594 IgG (goat anti-rabbit) were used for visualization of markers. Nuclei were stained with DAPI according to manufacturer's instructions (Life Technologies).
Overexpression of miR-30 using transient transfection of miRNA mimics.
Cells were grown to ∼80% confluence in six-well format. Using a double transfection protocol, we transfected cells first immediately prior to the induction of differentiation and second 48 h later with 50 nM of Ambion Pre miR miRNA precursors (Life Technologies) for miR-30b or miR-30c. Control cells were transfected with appropriate scrambled miRNA mimic (50 nM). Each transfection was performed for 5 h with transfection complexes formed in reduced serum media (OptiMEM, Life Technologies) with Lipofectamine RNAiMAX transfection reagent according manufacturer's protocol. RNA was isolated as described above 48 h after the second transfection (day 4 of differentiation) for the measurement of predicted target genes performed by qPCR.
Luciferase activity measurements.
The RASAL2 3′-untranslated region (UTR) (HmiT022785-MT05) and WNT5A 3′-UTR (HmiT018531a-MT05 and HmiT018531b-MT05; 3′-UTR was divided into two clones) luciferase reporter clones were purchased from Genecopoeia (Rockville, MD). Target Search in microRNA.org (http://www.microrna.org) was used for prediction of miRNA target sites. Predicted miR-30b/c-binding sites were mutated using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene, Agilent Technologies, Santa Clara, CA). Oligoprimers used for mutagenesis of the RASAL2 3′-UTR were 5′-CCTTTACCTAGCCCCTCCAGCGAACCAGAATGTTGCTACTTCAC-3′ and 5′-GTGAAGTAGCAACATTCTGGTTCGCTGGAGGGGCTAGGTAAAGG-3′, and oligoprimers used for mutagenesis of the WNT5A 3′-UTR were 5′-GTCCAAAAAAATGTATTTTTTTCTGTCGGGCGCACTGCAACTATTGCACCTCTCTATTTG-3′ and 5′-CAAATAGAGAGGTGCAATAGTTGCAGTGCGCCCGACAGAAAAAAATACATTTTTTTGGAC-3′.
3′-UTR promoter plasmids (100 ng per well) was transfected into HEK293 cells in 96-well plates using the transfection reagent Lipofectamine 2000 (Life Technologies) with Ambion Pre miR miRNA precursors (Life Technologies) for miR-30b or miR-30c. We used 50 nM of miRNA precursor for transfection of RASAL2 3′-UTR plasmid and 100 nM of miRNA precursor for WNT5A 3′-UTR. Control cells were transfected with appropriate scrambled miRNA mimic. After 24 h, the cell culture medium was collected and processed for luciferase assay using Secrete-PairTM Luminescence Assay Kit (GeneCopoeia). Assays were read in the GloMax 96 Microplate Luminometer (Promega, Madison, WI) and normalized with secreted alkaline phosphatase signals.
Statistical analysis.
Analysis of the array data has been described above. Other data are reported as means ± SE. Analysis of changes in miRNA or mRNA expression measured by qPCR was performed by a repeated-measures ANOVA followed by Dunnett's test for post hoc pair-wise comparisons or paired student's t-test when appropriate (GraphPad Prism; GraphPad Software, La Jolla, CA). Statistical significance was accepted at P < 0.05.
RESULTS
Differentiation of human skeletal muscle cells.
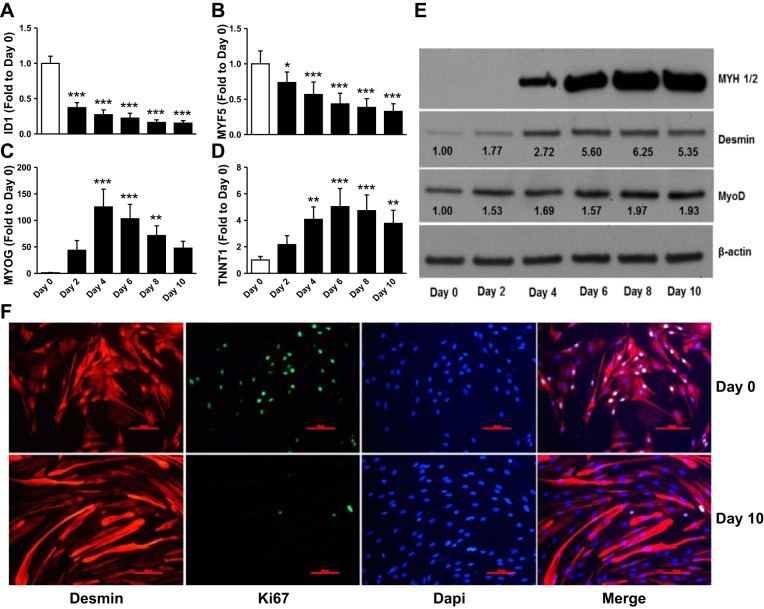
Satellite cells isolated from muscle biopsies from six male volunteers were cultured in GM and harvested as myoblasts (day 0) and at 48 h intervals up to 10 days after induction of differentiation by switching to low serum DM. As expected, differentiation was accompanied by a reduction in mRNA expression of MYF5 (P < 0.0001) and inhibitor of DNA binding 1, dominant negative helix-loop-helix protein (ID1) (P < 0.0001), and a concomitant increase in troponin T type 1 (TNNT1) (P < 0.0001) and MYOG (P < 0.0001) (Fig. 1, A–D). Protein content of the muscle differentiation markers desmin, MYOD1, and MHC also increased with the induction of the myogenic program (Fig. 1E). Fusion of myotubes was evident by the appearance of multinucleated, elongated desmin-positive cells after 10 days of differentiation (Fig. 1F). The number of proliferative cells decreased, as evident by the disappearance of Ki67-positive stained nuclei after differentiation compared with myoblast at day 0 (Fig. 1F).
Fig. 1.
Validation of myogenic marker expression during human skeletal muscle cell differentiation. Skeletal muscle cells cultured in vitro as myoblasts and induced to differentiate were harvested at various time points during differentiation for analysis of gene expression (A–D), protein abundance (E), or immunocytochemistry (F). A–D: quantitative reverse transcriptase-polymerase chain reaction (qPCR) analysis of proliferative and myogenic markers. White bars, proliferative cells (day 0); black bars, different days following differentiation induction. E: Western blot analysis of myogenic markers. When appropriate numbers inserted on the immunoblot indicate average fold change over day 0. Representative blots. F: immunocytochemistry the myogenic marker desmin, proliferative marker Ki67, and DAPI of skeletal muscle cells before and after 10 days of differentiation. Scale bars are 100 μm. Representative images (n = 3). Data are means ± SE across biological replicates (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001 by repeated-measures ANOVA.
Time course analysis of miRNA expression during differentiation.
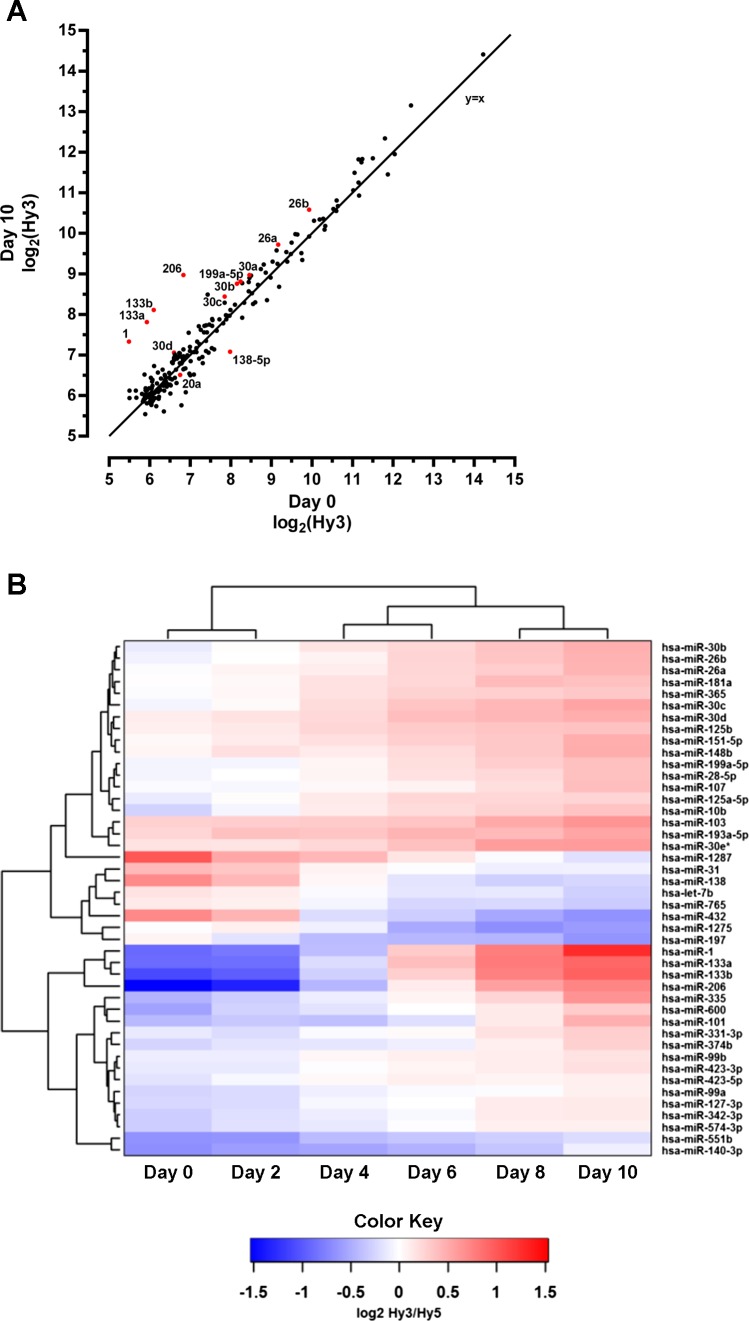
To identify miRNAs demonstrating altered expression during muscle differentiation, we utilized miRNA microarrays for expression profiling of 1,276 putative miRNAs at the six different time points selected. After removal of miRNAs with low expression, 206 miRNA with measureable expression in myoblasts or differentiating cells remained for further analysis. These 206 miRNAs show a wide range of abundance in skeletal muscle cells, with the canonical myomiRs (miR-1, miR-133a, miR-133b, and miR-206) showing the largest fold change in response to differentiation (Fig. 2A). Forty-four miRNAs, with a fold change of at least 20% compared with day 0, were differentially expressed according to a SAM time course analysis (FDR < 5%), indicating increased or decreased expression throughout the study (Fig. 2B). Based on a comparison of differential expression at any time point during differentiation compared with the myoblast stage (FDR < 5%), 103 miRNAs exhibited altered expression (Supplemental Table S1).1 A majority (58 of 103, 56%) of these miRNAs show changed expression at more than one time point. For the 45 miRNAs that showed altered expression at one time point only, 36 of 45 (80%) showed changes at later stages of myogenic differentiation (i.e., at day 8 or day 10). We also observed miRNAs exhibiting altered expression only early in differentiation (i.e., miR-665, -1246, and -1290 at day 2 and miR-125b-1*, -197, and -301 at day 4) (Supplemental Table S1).
Fig. 2.
Analysis of microRNA (miRNA) expression during human skeletal muscle cell differentiation. A: intensity (log2Hy3) scatter plot showing comparison of miRNA expression between undifferentiated (day 0) and differentiated (day 10) human skeletal muscle cells. The miRNAs that are the focus of our network analyses are highlighted in red and identified by the respective labels. Line (x = y). B: heat map of miRNAs with altered expression during skeletal muscle cell differentiation by SAM time-course analysis [false discovery rate (FDR) < 5%, fold change 1.2 cut-off; n = 5–6].
Several miRNAs not previously known to be regulated during skeletal muscle cell differentiation in humans or rodents were identified (Supplemental Table S2). Out of the 103 miRNAs identified as being regulated at any time point during differentiation, 31 have not previously been linked to this process in human or mouse, and 36 have not been identified as being regulated during human skeletal muscle cell differentiation. However, 26 out of these 36 show directional changes that are consistent with previous results from murine skeletal muscle cells (Supplemental Table S2).
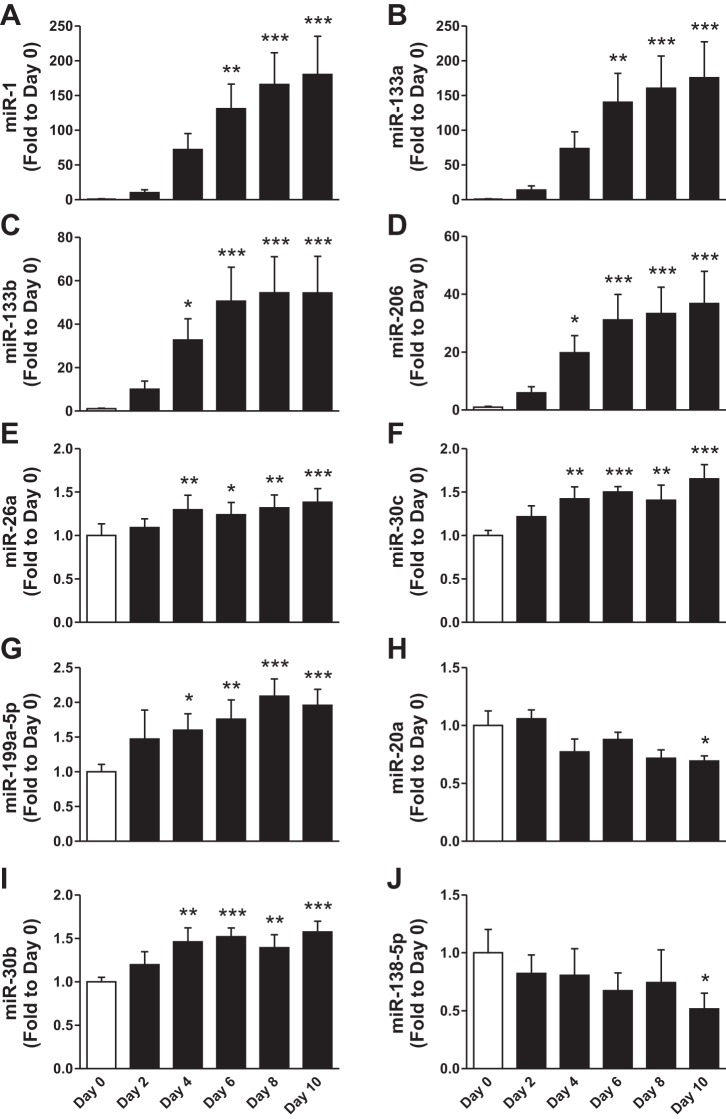
The expression of canonical myomiRs, miR-1, -133a, -133b, and -206, was dramatically altered during differentiation (Fig. 3, A–D). This finding was validated by qPCR analysis. The ∼50- to 200-fold change observed for these myomiRs was contrasted by modest changes in expression of other selected miRNAs for qPCR validation. Expression changes of miR-26a, miR-30c, and miR-199a-5p were validated to increase during differentiation (Fig. 3, E–G). We also validated several miRNAs that have not been previously shown to be regulated during skeletal muscle differentiation in human cells including miR-20a, miR-30b, and miR-138-5p (Fig. 3, H–J). On the basis of the detection cut-off threshold employed, we considered other “muscle-enriched” miRNAs including miR-208a, -208b, -486, and -499 to have low expression levels.
Fig. 3.
qPCR validation of differentially regulated miRNAs during human skeletal muscle cell differentiation. A–J: qPCR measurement of miRNA expression at indicated days of differentiation. White bar, proliferative cells (day 0); black bar, different days following differentiation induction. Data are means ± SE across biological replicates (n = 6). *P < 0.05, **P < 0.01, ***P < 0.001 by repeated-measures ANOVA.
Gene expression changes during differentiation.
Relative mRNA expression was determined by microarray analysis in a subset of samples at day 0, day 4, and day 10. There were 949 differentially expressed genes (FDR < 10%) in proliferating myoblasts vs. myotubes after 10 days of differentiation, with 437 increased and 512 decreased at day 10 (Supplemental Table S3). At day 4, 842 genes were differentially expressed (FDR < 10%), with 392 genes increased and 450 decreased (FDR < 10%) (Supplemental Table S4).
GO analysis (DAVID) of differentially expressed transcripts was used to explore biological processes altered during differentiation. This analysis revealed that the most enriched biological processes of upregulated genes participate in (striated) muscle contraction and development (Table 1). The most enriched biological processes of downregulated genes participate in regulation of the cell cycle processes (Table 1). Although the number of genes with differential expression at day 4 was marginally less than at day 10 (843 vs. 949, respectively), similarly enriched GO biological processes were noted at day 4 (Table 2).
Table 1.
Top five GO categories of biological process enriched by genes either up- or downregulated in human skeletal muscle cells after 10 days of differentiation
| GO Identity | GO Term | Genes, n | P Value | FDR |
|---|---|---|---|---|
| Upregulated genes | ||||
| GO:0006941 | striated muscle contraction | 15 | 1.9 × 10−12 | 3.7 × 10−9 |
| GO:0006936 | muscle contraction | 23 | 1.3 × 10−11 | 1.4 × 10−8 |
| GO:0003012 | muscle system process | 23 | 8.8 × 10−11 | 5.9 × 10−8 |
| GO:0007517 | muscle organ development | 20 | 7.6 × 10−7 | 3.8 × 10−4 |
| GO:0014706 | striated muscle tissue development | 14 | 5.4 × 10−6 | 2.2 × 10−3 |
| Downregulated genes | ||||
| GO:0000279 | M phase | 56 | 1.8 × 10−27 | 4.3 × 10−24 |
| GO:0022403 | cell cycle phase | 60 | 1.2 × 10−25 | 1.4 × 10−22 |
| GO:0000087 | M phase of mitotic cell cycle | 43 | 4.0 × 10−23 | 3.2 × 10−20 |
| GO:0000280 | nuclear division | 42 | 1.7 × 10−22 | 1.0 × 10−19 |
| GO:0007067 | mitosis | 42 | 1.7 × 10−22 | 1.0 × 10−19 |
Enrichment analysis of differentiation-related genes annotated with Gene Ontology (GO) terms demonstrates involvement of multiple molecular pathways and biological processes. Number (n) of regulated genes (D10/D0) is reported. P values were derived using Fisher's exact test. False discovery rate (FDR) determined by Benjamini-Hochberg adjusted P value.
Table 2.
Top five GO categories of biological process enriched by genes either up- or downregulated in human skeletal muscle cells after 4 days of differentiation
| GO Identity | GO Term | Genes, n | P Value | FDR |
|---|---|---|---|---|
| Upregulated genes | ||||
| GO:0006936 | muscle contraction | 29 | 1.4 × 10−18 | 2.4 × 10−15 |
| GO:0003012 | muscle system process | 29 | 1.8 × 10−17 | 1.6 × 10−14 |
| GO:0006941 | striated muscle contraction | 14 | 8.8 × 10−12 | 4.8 × 10−9 |
| GO:0007517 | muscle organ development | 24 | 1.4 × 10−10 | 6.2 × 10−8 |
| GO:0014706 | striated muscle tissue development | 15 | 2.4 × 10−7 | 8.5 × 10−5 |
| Downregulated genes | ||||
| GO:0007049 | cell cycle | 108 | 3.8 × 10−52 | 8.4 × 10−49 |
| GO:0000279 | M phase | 76 | 7.4 × 10−52 | 8.2 × 10−49 |
| GO:0022403 | cell cycle phase | 81 | 1.5 × 10−49 | 1.1 × 10−46 |
| GO:0000087 | M phase of mitotic cell cycle | 61 | 9.9 × 10−46 | 5.4 × 10−43 |
| GO:0000280 | nuclear division | 60 | 5.4 × 10−45 | 2.4 × 10−42 |
Enrichment analysis of differentiation-related genes annotated with GO terms demonstrates involvement of multiple molecular pathways and biological processes. Number of regulated genes (D4/D0) is reported. P values were derived using Fisher's exact test. FDR determined by Benjamini-Hochberg adjusted P value.
Prediction of miRNA-regulated networks during human skeletal muscle cell differentiation.
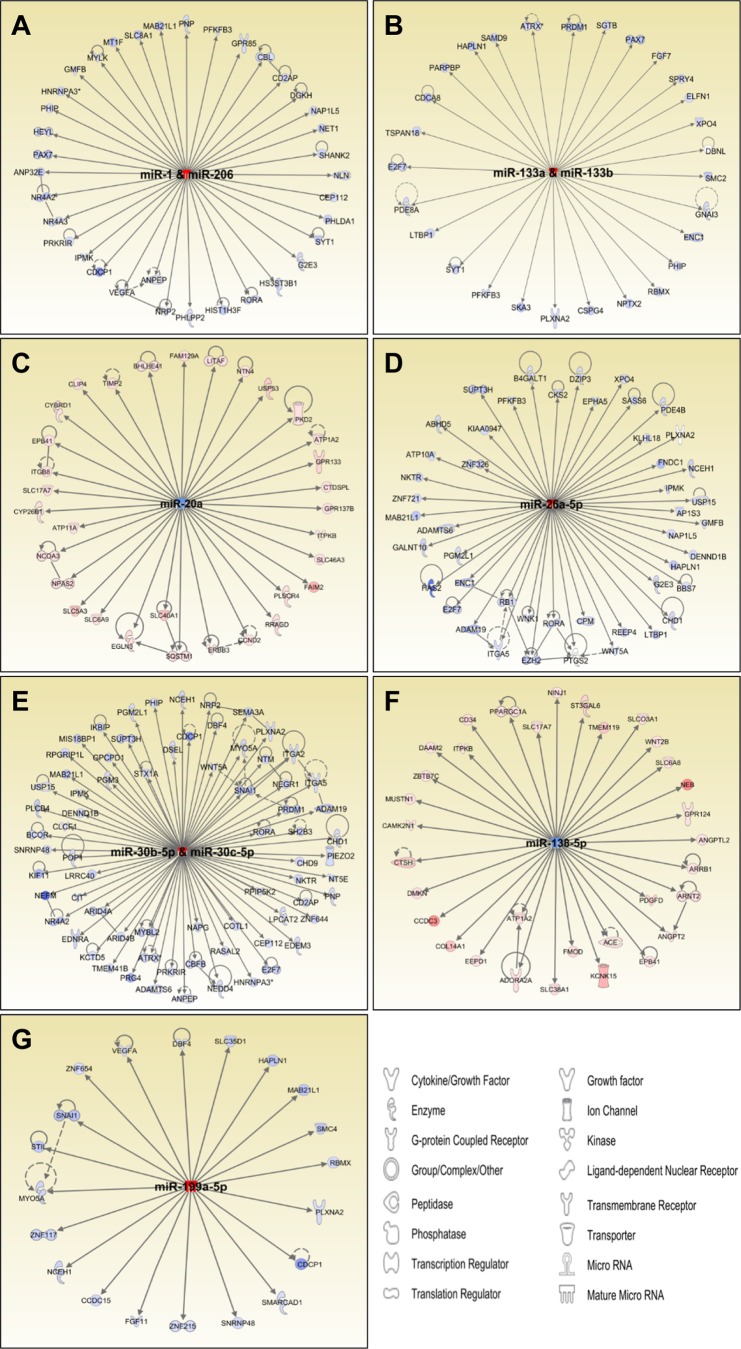
To identify potential miRNA-regulated target genes during skeletal muscle differentiation, we used IPA's microRNA Target Filter to integrate the datasets of all miRNAs and transcripts differentially expressed between day 0 and day 10. Of the 949 altered transcripts, 399 (42%) demonstrated reciprocal expression patterns in relation to the directional change of their predicted regulatory miRNAs, suggesting they could be under miRNA-dependent regulation during muscle differentiation. IPA also infers biological functionality from lists of differentially regulated genes and networks. Based on the IPA analysis, for all miRNAs validated by qPCR, miRNA-mRNA networks were identified as potential regulatory nodes in skeletal muscle differentiation (Fig. 4, A–G). For example, networks regulated by the myomiRs miR-1/206 (Fig. 4A) and miR-133 (Fig. 4B) contain genes, such as HEYL, NR4A2, NR4A3, PAX7, and PHIP, that are annotated as regulators of muscle development and differentiation (Supplementary Table S5). Several genes included in these networks are also annotated as being involved in cellular proliferation, indicating that many transcripts are involved in functions related to skeletal muscle differentiation.
Fig. 4.
miRNA-mRNA networks of predicted regulation during human skeletal muscle cell differentiation. A–G: miRNAs (central node) are connected to reciprocally regulated predicted target genes (peripheral nodes) by edges. Data represent differential expression comparing day 0 with day 10 of differentiation of skeletal muscle cells. The color blue indicates downregulated, and red indicates upregulated expression.
miR-30 as a potential regulatory node.
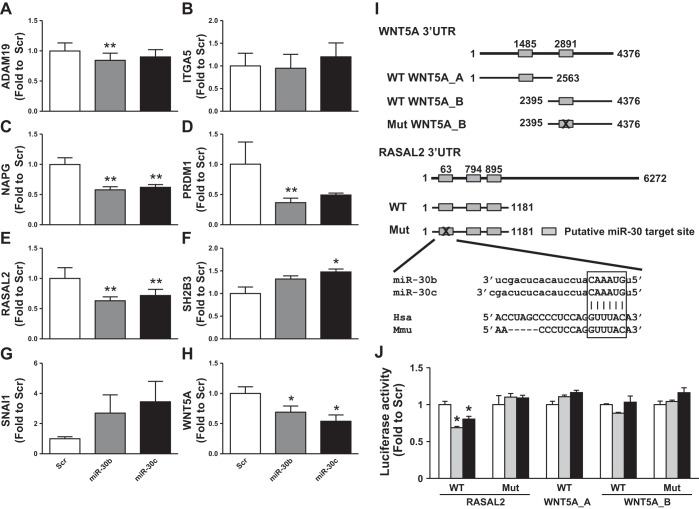
miR-30b and miR-30c likely have a regulatory role in the differentiation of human skeletal muscle cells by targeting several genes important in cell cycle, proliferation, and differentiation processes (Fig. 4E, Supplemental Table S5). Correlation analysis based on the expression data from day 0, day 4, and day 10 of miR-30 in silico-predicted targets identified several genes with strong inverse correlation to the expression of miR-30b and miR-30c (BH corrected P value < 0.05), including ADAM metallopeptidase domain 19 (ADAM19) (r = −0.944), integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (ITGA5) (r = −0.951), N-ethylmaleimide-sensitive factor attachment protein, gamma (NAPG) (r = −0.888), PR domain containing 1, with ZNF domain (PRDM1) (r = −0.910), RAS protein activator like 2 (RASAL2) (r = −0.870), SH2B adaptor protein 3 (SH2B3) (r = −0.892), snail family zinc finger 1 (SNAI1) (r = −0.878), and wingless-type MMTV integration site family, member 5A (WNT5A) (r = −0.944). We next transfected human skeletal muscle cells with mimics of miR-30b and miR-30c to validate these predictions and explore the expression of genes predicted to be regulated by these networks. Four days after transfection, five (ADAM19, NAPG, PRDM1, RASAL2, WNT5A) out of the aforementioned eight (63%) in silico-predicted gene targets display miR-30-responsive decreases (P < 0.05) in relative mRNA expression (Fig. 5, A–H). These five genes are all novel qPCR-validated targets of miR-30 in skeletal muscle cells, but expression of the remaining genes (ITGA5, SH2B3, and SNAI1) was unchanged or increased. The number of predicted miR-30 binding sites or the predicted total context score did not explain why the five qPCR-validated targets respond to miR-30 transfection relative to the other three predicted targets (Table 3).
Fig. 5.
qRT-PCR validation of miR-30b and miR-30c regulated targets in human skeletal muscle cells. A–H: negative control miRNA mimic (Scr, white bar), miR-30b (gray bar), or miR-30c (black bar) were overexpressed in skeletal muscle cells and expression of predicted targets were determined by qPCR. Expression of predicted target genes of cells transfected with miR30b or miR30c are expressed as fold change to Scr. Data are means ± SE (n = 3). *P < 0.05, **P < 0.01 by Student's paired t-test compared with Scr-transfected cells. I: overview of luciferase experiment setup with 3′-untranslated region (UTR) plasmids highlighting the putative binding sites of miR-30 isoforms. J: negative control miRNA mimic (Scr, white bar), miR-30b (gray bar), or miR-30c (black bar) were overexpressed in HEK293 cells together with indicated luciferase 3′-UTR plasmid. Luciferase activity was measured 24 h after transfection, and activity was expressed as fold change to Scr-transfected cells. Data are means ± SE (n = 3–6). *P < 0.001 by Student's t-test compared with Scr transfected cells.
Table 3.
Number of miR-30 target sites in 3′-UTR of qPCR-validated miR-30 predicted genes downregulated during human skeletal muscle cell differentiation
| Conserved Sites |
Poorly Conserved Sites |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Target Gene | total | 8mer | 7mer-m8 | 7mer-1A | total | 8mer | 7mer-m8 | 7mer-1A | Total Context+ Score |
| ADAM19 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.32 |
| ITGA5 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.14 |
| NAPG | 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | −0.26 |
| PRDM1 | 3 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | −0.46 |
| RASAL2 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | −0.04 |
| SH2B3 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | −0.23 |
| SNAI1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | −0.25 |
| WNT5A* | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
Number of conserved and poorly conserved target sites and total context score of miR-30 in the 3′-untranslated region (UTR) of miR-30 predicted target genes. Information obtained from the TargetScan database.
WNT5A is a pSILAC validated target of miR-30 (TarBase database) and has no predicted targets using TargetScan, while microRNA.org indicates 2 potential binding sites of miR-30 isoforms in the WNT5A 3′-UTR. N/A, not available.
To further validate the functional regulation of miR-30 isoforms on in silico-predicted target genes, we performed luciferase reporter assays for two of the novel qPCR-validated targets of miR-30, namely RASAL2 and WNT5A (Fig. 5I). Following overexpression of either miR-30b or miR-30c, luciferase activity for RASAL2 was decreased, whereas luciferase activity for WNT5A 3′-UTRs was unchanged following miR-30b overexpression (Fig. 5J). Mutagenesis of one of the predicted target sites of miR-30 in the RASAL2 3′-UTR, a conserved site between the human and mouse RASAL2 gene 3′-UTR, abolished all effects of miR-30 isoform overexpression on luciferase activity.
DISCUSSION
miRNAs are necessary for skeletal muscle development (31) and have regulatory roles throughout differentiation (8, 24, 30, 43, 48, 55). Nevertheless, only a few studies have examined miRNA expression profiles during differentiation in human skeletal muscle (17, 26, 27). Thus the primary objective of this study was to identify miRNA-centered networks of biological regulation in the context of human skeletal muscle cell differentiation by employing a time course analysis coupled to microarray profiling of changes in miRNA and mRNA expression. This integrated approach, which anticipates the reciprocal relationship between a miRNA and the expression of target genes, adds biological relevance to miRNA-mRNA predictions throughout skeletal muscle differentiation (17, 47). We report dynamic changes in miRNA expression during the differentiation of human skeletal muscle indicating that altered expression of specific miRNAs should be considered in terms of temporal periods during differentiation. Finally, we constructed networks of predicted regulation using IPA, integrating changes in expression of miRNAs and their predicted gene targets, which may serve as a reference point for future studies of human skeletal muscle differentiation and development.
In an unbiased approach, we utilized miRNA microarrays to profile expression of all annotated miRNAs (miRBase version 13) at 48 h intervals during the first 10 days of differentiation in primary human skeletal muscle cells. Results demonstrate that as many as 103 miRNAs exhibit altered expression during human skeletal muscle differentiation, with the majority (72%) being increased. These results are broadly in line with previous miRNA microarray profiling studies (9, 17, 47) and provide further evidence that numerous miRNAs have altered expression and likely exert regulatory control during skeletal muscle differentiation. Because skeletal muscle differentiation is a dynamic process, a key methodological element employed here was the multiple time point analysis to elucidate miRNA responses with greater temporal resolution than previously reported by single or endpoint studies. This type of analysis has been recently reported in the C2C12 immortalized murine muscle cell line and primary mouse skeletal muscle cells (9, 47), but this current work applies the approach to primary human skeletal muscle cells from multiple donors. In primary mouse cells, a hierarchal cluster analysis of differentially expressed miRNAs resulted in the identification of “clusters” of miRNAs exhibiting similar altered expression patterns (9). For example, the expression of the myomiRs miR-1, −133a, -133b, and -206 was increased early in differentiation and progressively increased thereafter. A similar increase was observed in the present study at day 4 of differentiation and continued to increase up to day 10. These miRNAs play a critical regulatory role in skeletal muscle satellite cell proliferation and differentiation (8, 15, 24). After induction by day 4, the largest fold changes in expression for any miRNA were observed for myomiRs miR-1, -133a, -133b, and -206. These miRNAs show low expression in proliferating human skeletal muscle cells but increase by ∼50- to 200 fold (validated by qPCR) at later time points during differentiation.
We also observed a small number of miRNAs with altered expression levels only at early time points during the differentiation process. Because the processes of proliferation and differentiation of muscle cells are mutually exclusive and regulated by the balance between activities of positive and negative cell cycle regulators, these early-responding miRNAs may trigger events essential for differentiation. Although these miRNAs (e.g., miR-10b, -665, -1246, and -1290) have yet to be studied in muscle cells, each is associated with regulation of cell cycle and cell transitions in other cell models (13, 18, 21). Finally, we studied muscle differentiation at 48 h intervals but cannot exclude the possibility that other miRNAs are altered during the first 48 h. To our knowledge this has not been studied at the level of miRNA expression, but changes in gene expression are extremely dynamic within the first 24 h of muscle differentiation (14, 36).
Results are broadly in agreement with data reported from primary human and mouse skeletal muscle cells and immortalized C2C12 mouse cells, with many miRNAs exhibiting same-directional changes including miR-1, -23b, -26b, -30a/c/d, -103/107, -133a/b, and -206 (8, 9, 11, 15, 17, 29, 45, 55). Additionally, we provide evidence that miR-26a and miR-30b, isoforms from the miR-26 and miR-30 families, as well as miR-20a, miR-138-5p, and members of the miR-199 family, are altered during differentiation. While miR-199a-5p increases during human skeletal muscle cell differentiation and regulates myoblast fusion into myotubes (2), the functional significance of other miRNAs, including miR-20a and miR-138-5p, remains to be determined. Furthermore, we observed 103 miRNAs with altered expression during differentiation compared with previous profiling (17), which identified 60 miRNAs with altered expression in differentiating human skeletal muscle cells. We identified 31 miRNAs that have not been previously shown to be altered during skeletal muscle differentiation in either human or mouse cells. Furthermore, we identified an additional 36 miRNAs that have previously been identified in mouse cells during skeletal muscle differentiation, but not in human cells. Methodological differences are likely to explain these discrepancies and novel findings. We used a multiple time point analysis rather than a single end-point design and profiled over 1,200 known and putative miRNAs, as opposed to the 365 miRNAs available on the TaqMan Low Density Array platform used previously (17). Furthermore, differences in establishment of muscle cultures could influence the results. We utilized primary human skeletal muscles cultures without further purification, whereas earlier workers (17) utilized a selection purification based on CD56-positive cells specifically targeting myogenic precursors. Despite some differences in specific miRNAs identified, the major GOs regulated during skeletal muscle differentiation are similar between the present and previous studies (17).
A further analysis revealed some species differences between mouse and human studies. Interestingly, out of the 36 miRNAs previously not shown to be regulated during human skeletal muscle cell differentiation, 10 miRNAs change in the opposite direction compared with mouse cells (9, 15, 29, 45, 52). For example, in microarray studies of murine cells, miR-138-5p expression is elevated during differentiation (9, 29), whereas in the present study using microarray analysis and qPCR-validation, we demonstrate that miR-138-5p expression is decreased in human skeletal muscle cells during differentiation. This indicates that species-specific differences in miRNA regulation and expression likely exist during differentiation.
Even when differentially expressed miRNAs are identified, confirming the target genes of these miRNAs is challenging. Numerous algorithms exist to predict miRNA-targeted genes, but these in silico prediction tools each use a variety of different assumptions for identifying target genes and often give different results (39). In addition, these prediction algorithms can result in large numbers of false positives and thereby have low precision (54). Since miRNAs mainly act to reduce target gene expression by mRNA degradation in mammals (20), a frequently utilized tool to identify context-dependent miRNA targets is microarray profiling of gene expression (54). By combining microarray analysis with in silico prediction tools, investigators can identify reciprocal regulation of miRNAs and their targets. Also, correlating data from such combinations may increase the sensitivity of predictions (40, 51) and identify novel miRNA-dependent targets in skeletal muscle differentiation (17, 47). In the present study, we identified 842 and 949 altered transcripts after 4 and 10 days of differentiation, respectively. Comparison of these gene sets based on GO at early and late differentiation identified functions related to proliferation and cell cycle progression being downregulated during differentiation, with functions related to muscle differentiation, development, and contraction being associated with genes upregulated during differentiation. Compared with previously published mRNA transcriptome analyses of skeletal muscle cell differentiation (17, 44), we have identified similar GOs. Comparison of gene expression changes with other studies (17, 44) shows positive correlations. Use of a set of 6,153 significantly regulated genes from Dmitriev et al. (17) for comparison of fold changes shows that 62% of the genes have fold change regulation that is similar to the current study; 62% when the current study is compared with Soleimani et al. (44); and 70% when Dmitriev et al. is compared with Soleimani et al. Altogether, this indicates similar regulation of genes during skeletal muscle cell differentiation is revealed in the present study compared with previous studies (17, 44).
The functions associated with altered genes in the miR-1/206 and miR-133a/b networks include cellular growth and proliferation and skeletal and muscular system development. Because the roles of canonical myomiRs are well established in skeletal muscle differentiation, we focused on potentially novel networks of regulation in human skeletal muscle cells. For example, analysis of miR-26- and miR-30-centered networks suggests a role for these miRNAs in differentiation through regulation of genes involved in cell cycle control, intracellular transport, and cytoskeleton organization. In murine muscle cells, miR-26a is markedly induced during differentiation and regulates myogenesis through regulation of Enhancer of Zeste homolog 2 (55) and coordinated repressive effects on the SMAD proteins within the TGF-β signaling pathway (16).
miR-30 is elevated in human skeletal muscle cells during differentiation (17). Little is known about role of miR-30 in the regulation of muscle differentiation, although by virtue of its repression of SNAI1/2 in Cos7 and C2C12 cells, it may be part of a network involving miR-206, SNAI1/2, and MYOD1 (44). To investigate miR-30-mediated regulation, we overexpressed miR-30b and miR-30c and examined the effects on in silico predicted gene targets that were also decreased during differentiation. Expression of five (ADAM19, NAPG, PRDM1, RASAL2, WNT5A) out of eight targets was decreased after miR-30b or miR-30c overexpression, but the remaining genes were unchanged (ITGA5, SNAI1) or increased (SH2B3). By cloning the 3′-UTR of RASAL2 into luciferase reporter plasmids, we provide evidence that RASAL2 is a direct target of miR-30b and -30c isoforms. Additionally, the conserved miR-30 target site at position 63 in the 3′-UTR of RASAL2 is sufficient to regulate gene expression, as shown by mutagenesis experiments. RASAL2, which encodes a RasGAP, is downregulated during differentiation in the present study and during mouse skeletal muscle differentiation (44), suggesting conserved regulation. Oncogenic activation of the Ras-MAPK pathway inhibits muscle differentiation (12). Moreover, downstream pathways of Ras, for example PI3K-AKT or PKC pathways, are important for muscle differentiation (25). Though the function of RASAL2 in skeletal muscle differentiation remains to be established, the inhibition of RASAL2 gene expression by miRNA-30 may be part of the regulatory process. WNT5A was another predicted target of miR-30, and miR-30 overexpression decreased WNT5A mRNA in skeletal muscle cells. Although WNT5A expression is responsive to miR-30 (41), here we provide evidence that the decrease in target abundance is not due to direct binding of miR-30 to the WNT5A 3′-UTR. While we cannot exclude the presence of a target site that we did not identify using the microRNA.org prediction, our results suggest that miR-30-mediated regulation of WNT5A is indirect.
PRDM1 expression was markedly decreased during differentiation and in response to miR-30 overexpression and is validated by luciferase assay as a target of miR-30 (57). PRDM1 (also known as BLIMP1) is a zinc finger transcriptional repressor that promotes oxidative muscle fiber differentiation by acting as a global repressor of glycolytic and fast-twitch-specific genes, as well as by abrogating the repression of oxidative and slow-twitch-specific genes, and is essential for muscle development in zebrafish (50). This function is not conserved in murine myotome where PRDM1 does not influence fiber type or development (49). However, PRDM1 is a predicted target of five miRNAs showing increased expression in this study, namely miR-30b, -125a-5p, -129*, -133b, and -365. Regulation by multiple miRNAs is likely to increase the specificity of target gene regulation and enhance the robustness of gene expression against fluctuations in levels of individual miRNAs (19). These repressed miR-30 targets can be considered qPCR-validated targets of miR-30, but the direction of SNAI1 expression was unexpected given the observation of direct regulation by miR-30b (28, 44). This highlights the importance of considering the specific context when investigating miRNA-mRNA relationships and associated physiological effects. These miRNA-mRNA expression associations are based solely on mRNA that are degraded after targeting by a respective miRNA, so a limitation is that relationships that affect translation and, therefore, protein abundance only would be missed. Whether this miR-30 regulatory network described in murine muscle cells is conserved in human skeletal muscle cells remains to be confirmed.
In conclusion, we have integrated simultaneous miRNA and mRNA expression data with target prediction algorithms and network analysis to identify regulatory nodes in a specific biological context. We report >100 miRNAs exhibiting differential expression during the time course of muscle differentiation, concomitant with reciprocal changes in target gene expression. We demonstrate several predicted networks of regulation of biological processes, but clearly bioinformatics-based deductions cannot substitute for experimental validation of miRNA functions. Therefore, these results provide the basis to support hypothesis-driven functional studies of these miRNAs and their gene targets in human skeletal muscle development.
GRANTS
This work was supported by grant funding from the Swedish Diabetes Association, the Novo Nordisk Foundation, the Swedish Research Council, the Swedish Foundation for Strategic Research, and the European Research Council Ideas program (ICEBERG, ERC-2008-AdG23285).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: R.J.S., B.E., J.R.Z., and A.K. conception and design of research; R.J.S., B.E., and M.K. performed experiments; R.J.S., B.E., and M.K. analyzed data; R.J.S., B.E., M.K., J.R.Z., and A.K. interpreted results of experiments; R.J.S. and B.E. prepared figures; R.J.S. and B.E. drafted manuscript; R.J.S., B.E., M.K., J.R.Z., and A.K. edited and revised manuscript; R.J.S., B.E., J.R.Z., and A.K. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bioinformatics and Expression Analysis core facility at Novum, which is supported by the board of research at the Karolinska Institute and the research committee at Karolinska University Hospital.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Al-Khalili L, Kramer D, Wretenberg P, Krook A. Human skeletal muscle cell differentiation is associated with changes in myogenic markers and enhanced insulin-mediated MAPK and PKB phosphorylation. Acta Physiol Scand 180: 395–403, 2004. [DOI] [PubMed] [Google Scholar]
- 2.Alexander MS, Kawahara G, Motohashi N, Casar JC, Eisenberg I, Myers JA, Gasperini MJ, Estrella EA, Kho AT, Mitsuhashi S, Shapiro F, Kang PB, Kunkel LM. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death Differ 20: 1194–1208, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature 431: 350–355, 2004. [DOI] [PubMed] [Google Scholar]
- 4.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boldrin L, Muntoni F, Morgan JE. Are human and mouse satellite cells really the same? J Histochem Cytochem 58: 941–955, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 12: 349–361, 2011. [DOI] [PubMed] [Google Scholar]
- 8.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38: 228–233, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Gelfond J, McManus LM, Shireman PK. Temporal microRNA expression during in vitro myogenic progenitor cell proliferation and differentiation: regulation of proliferation by miR-682. Physiol Genomics 43: 621–630, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Melton DW, Gelfond JA, McManus LM, Shireman PK. MiR-351 transiently increases during muscle regeneration and promotes progenitor cell proliferation and survival upon differentiation. Physiol Genomics 44: 1042–1051, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciarapica R, Russo G, Verginelli F, Raimondi L, Donfrancesco A, Rota R, Giordano A. Deregulated expression of miR-26a and Ezh2 in rhabdomyosarcoma. Cell Cycle 8: 172–175, 2009. [DOI] [PubMed] [Google Scholar]
- 12.Ciuffini L, Castellani L, Salvati E, Galletti S, Falcone G, Alema S. Delineating v-Src downstream effector pathways in transformed myoblasts. Oncogene 27: 528–539, 2008. [DOI] [PubMed] [Google Scholar]
- 13.Cui L, Shi Y, Zhou X, Wang X, Wang J, Lan Y, Wang M, Zheng L, Li H, Wu Q, Zhang J, Fan D, Han Y. A set of microRNAs mediate direct conversion of human umbilical cord lining-derived mesenchymal stem cells into hepatocytes. Cell Death Dis 4: e918, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delgado I, Huang X, Jones S, Zhang L, Hatcher R, Gao B, Zhang P. Dynamic gene expression during the onset of myoblast differentiation in vitro. Genomics 82: 109–121, 2003. [DOI] [PubMed] [Google Scholar]
- 15.Dey BK, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 31: 203–214, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dey BK, Gagan J, Yan Z, Dutta A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev 26: 2180–2191, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dmitriev P, Barat A, Polesskaya A, O'Connell MJ, Robert T, Dessen P, Walsh TA, Lazar V, Turki A, Carnac G, Laoudj-Chenivesse D, Lipinski M, Vassetzky YS. Simultaneous miRNA and mRNA transcriptome profiling of human myoblasts reveals a novel set of myogenic differentiation-associated miRNAs and their target genes. BMC Genomics 14: 265, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gabriely G, Teplyuk NM, Krichevsky AM. Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy 7: 1384–1386, 2011. [DOI] [PubMed] [Google Scholar]
- 19.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466: 835–840, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harazono Y, Muramatsu T, Endo H, Uzawa N, Kawano T, Harada K, Inazawa J, Kozaki K. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS One 8: e62757, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57, 2009. [DOI] [PubMed] [Google Scholar]
- 24.Kim HK, Lee YS, Sivaprasad U, Malhotra A, Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J Cell Biol 174: 677–687, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight JD, Kothary R. The myogenic kinome: protein kinases critical to mammalian skeletal myogenesis. Skelet Muscle 1: 29, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koning M, Werker PM, van Luyn MJ, Krenning G, Harmsen MC. A global downregulation of microRNAs occurs in human quiescent satellite cells during myogenesis. Differentiation 84: 314–321, 2012. [DOI] [PubMed] [Google Scholar]
- 27.Koutsoulidou A, Mastroyiannopoulos NP, Furling D, Uney JB, Phylactou LA. Expression of miR-1, miR-133a, miR-133b and miR-206 increases during development of human skeletal muscle. BMC Dev Biol 11: 34, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer 130: 2044–2053, 2012. [DOI] [PubMed] [Google Scholar]
- 29.Marzi MJ, Puggioni EM, Dall'Olio V, Bucci G, Bernard L, Bianchi F, Crescenzi M, Di Fiore PP, Nicassio F. Differentiation-associated microRNAs antagonize the Rb-E2F pathway to restrict proliferation. J Cell Biol 199: 77–95, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 8: 278–284, 2006. [DOI] [PubMed] [Google Scholar]
- 31.O'Rourke JR, Georges SA, Seay HR, Tapscott SJ, McManus MT, Goldhamer DJ, Swanson MS, Harfe BD. Essential role for Dicer during skeletal muscle development. Dev Biol 311: 359–368, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrella JK, Kim JS, Cross JM, Kosek DJ, Bamman MM. Efficacy of myonuclear addition may explain differential myofiber growth among resistance-trained young and older men and women. Am J Physiol Endocrinol Metab 291: E937–E946, 2006. [DOI] [PubMed] [Google Scholar]
- 33.Petrella JK, Kim JS, Mayhew DL, Cross JM, Bamman MM. Potent myofiber hypertrophy during resistance training in humans is associated with satellite cell-mediated myonuclear addition: a cluster analysis. J Appl Physiol (1985) 104: 1736–1742, 2008. [DOI] [PubMed] [Google Scholar]
- 34.R Core Team. R: A Language and Environment for Statistical Computing. 2013. [Google Scholar]
- 35.Ragan C, Cloonan N, Grimmond SM, Zuker M, Ragan MA. Transcriptome-wide prediction of miRNA targets in human and mouse using FASTH. PLoS One 4: e5745, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajan S, Chu Pham Dang H, Djambazian H, Zuzan H, Fedyshyn Y, Ketela T, Moffat J, Hudson TJ, Sladek R. Analysis of early C2C12 myogenesis identifies stably and differentially expressed transcriptional regulators whose knock-down inhibits myoblast differentiation. Physiol Genomics 44: 183–197, 2012. [DOI] [PubMed] [Google Scholar]
- 37.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci USA 103: 8721–8726, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics 23: 2700–2707, 2007. [DOI] [PubMed] [Google Scholar]
- 39.Ritchie W, Flamant S, Rasko JE. Predicting microRNA targets and functions: traps for the unwary. Nat Methods 6: 397–398, 2009. [DOI] [PubMed] [Google Scholar]
- 40.Ruike Y, Ichimura A, Tsuchiya S, Shimizu K, Kunimoto R, Okuno Y, Tsujimoto G. Global correlation analysis for micro-RNA and mRNA expression profiles in human cell lines. J Hum Genet 53: 515–523, 2008. [DOI] [PubMed] [Google Scholar]
- 41.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63, 2008. [DOI] [PubMed] [Google Scholar]
- 42.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol 5: R13, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Small EM, O'Rourke JR, Moresi V, Sutherland LB, McAnally J, Gerard RD, Richardson JA, Olson EN. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc Natl Acad Sci USA 107: 4218–4223, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soleimani VD, Yin H, Jahani-Asl A, Ming H, Kockx CE, van Ijcken WF, Grosveld F, Rudnicki MA. Snail regulates MyoD binding-site occupancy to direct enhancer switching and differentiation-specific transcription in myogenesis. Mol Cell 47: 457–468, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y, Ge Y, Drnevich J, Zhao Y, Band M, Chen J. Mammalian target of rapamycin regulates miRNA-1 and follistatin in skeletal myogenesis. J Cell Biol 189: 1157–1169, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98: 5116–5121, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Iterson M, Bervoets S, de Meijer EJ, Buermans HP, t Hoen PA, Menezes RX, Boer JM. Integrated analysis of microRNA and mRNA expression: adding biological significance to microRNA target predictions. Nucleic Acids Res 41: e146, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ Jr, Olson EN. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17: 662–673, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vincent SD, Mayeuf A, Niro C, Saitou M, Buckingham M. Non conservation of function for the evolutionarily conserved prdm1 protein in the control of the slow twitch myogenic program in the mouse embryo. Mol Biol Evol 29: 3181–3191, 2012. [DOI] [PubMed] [Google Scholar]
- 50.von Hofsten J, Elworthy S, Gilchrist MJ, Smith JC, Wardle FC, Ingham PW. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep 9: 683–689, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang YP, Li KB. Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data. BMC Genomics 10: 218, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei H, Li Z, Wang X, Wang J, Pang W, Yang G, Shen QW. microRNA-151–3p regulates slow muscle gene expression by targeting ATP2a2 in skeletal muscle cells. J Cell Physiol (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 53.Williams AH, Liu N, van Rooij E, Olson EN. MicroRNA control of muscle development and disease. Curr Opin Cell Biol 21: 461–469, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med 11: 93–109, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem 283: 9836–9843, 2008. [DOI] [PubMed] [Google Scholar]
- 56.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev 93: 23–67, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 113: 4586–4594, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.