Abstract
The ability of the site-frequency spectrum (SFS) to reflect the particularities of gene genealogies exhibiting multiple mergers of ancestral lines as opposed to those obtained in the presence of population growth is our focus. An excess of singletons is a well-known characteristic of both population growth and multiple mergers. Other aspects of the SFS, in particular, the weight of the right tail, are, however, affected in specific ways by the two model classes. Using an approximate likelihood method and minimum-distance statistics, our estimates of statistical power indicate that exponential and algebraic growth can indeed be distinguished from multiple-merger coalescents, even for moderate sample sizes, if the number of segregating sites is high enough. A normalized version of the SFS (nSFS) is also used as a summary statistic in an approximate Bayesian computation (ABC) approach. The results give further positive evidence as to the general eligibility of the SFS to distinguish between the different histories.
Keywords: coalescent, multiple mergers, population growth, approximate maximum likelihood test, approximate Bayesian computation, site-frequency spectrum
THE site-frequency spectrum (SFS) at a given locus is one of the most important and popular statistics based on genetic data sampled from a natural population. In combination with the postulation of the assumptions of the infinitely-many-sites mutation model (Watterson, 1975) and a suitable underlying coalescent framework, the SFS allows one to draw inferences about evolutionary parameters, such as coalescent parameters associated with multiple-merger coalescents or population-growth models.
The Kingman coalescent, developed by Kingman (1982a,b,c), Hudson (1983a,b), and Tajima (1983), describing the random ancestral relations among DNA sequences drawn from natural populations, is a prominent and widely used model from which one can make predictions about genetic diversity. Many quantities of interest, such as the expected values and covariances of the SFS associated with the Kingman coalescent, are easily computed thanks to results by Fu (1995). The robustness of the Kingman coalescent is quite remarkable; indeed, a large number of genealogy models can be shown to have the Kingman coalescent or a variant thereof as their limit process (cf., e.g., Möhle 1998). A large volume of work is thus devoted to inference methods based on the Kingman coalescent [see, e.g., Donnelly and Tavaré (1995), Hudson (1990), Nordborg (2001), Hein et al. (2005), and Wakeley (2007) for reviews].
However, many evolutionary histories can lead to significant deviations from the Kingman coalescent model. Such deviations can be detected using a variety of statistical tools, such as Tajima’s D (Tajima 1989a), Fu and Li’s D (Fu and Li 1993), and Fay and Wu’s H (Fay and Wu 2000), which are all functions of the SFS. However, they do not always allow one to identify the actual evolutionary mechanisms leading to such deviations. Developing statistical tools that allow one to distinguish between different evolutionary histories is therefore of fundamental importance.
This work focuses on properties of the (folded and unfolded) SFS in the infinitely-many-sites model for three population histories: (1) classical Kingman coalescent, (2) population growth, in particular, exponential population growth, and (3) high fecundity coupled with skewed offspring distributions (HFSODs), resulting in gene genealogies being described by so-called Lambda-coalescents (Sagitov 1999; Pitman 1999; Donnelly and Kurtz 1999). Briefly, multiple-merger coalescents may be more appropriate for organisms exhibiting HFSODs than the Kingman coalescent (cf., e.g., Beckenbach 1994; Árnason 2004; Eldon and Wakeley 2006; Sargsyan and Wakeley 2008; Hedgecock and Pudovkin 2011) [see also the review by Tellier and Lemaire (2014)].
Recent population growth as well as multiple-merger coalescents may lead to an excess of singletons in the SFS compared with the classical Kingman coalescent–based SFS, which e.g., contributes to shifting Tajima’s D values (Tajima 1989b) to the negative. Indeed, Durrett and Schweinsberg (2005) proved that Tajima’s D will be negative, at least for large sample size, under fairly general multiple-merger coalescents.
The associated genealogical trees are, however, qualitatively different. While moderate fluctuations in population size lead to a time change of the Kingman coalescent (Kaj and Krone 2003), multiple-merger coalescents by definition change the topology of the genealogical tree. There is thus hope that each demographic effect leaves specific signatures in the resulting SFS not only with respect to an excess of singletons but also, e.g., with respect to its right tail.
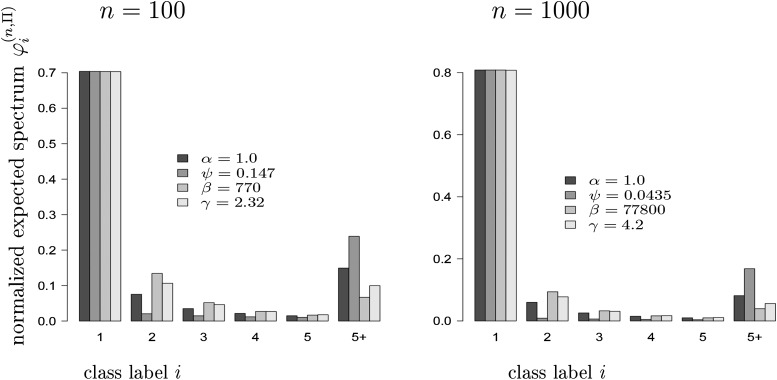
Indeed, one observes that the Kingman coalescent will not be a good match to genetic data containing a large fraction of singleton polymorphisms (relative to the total number of polymorphisms) because of a lack of free (coalescent) parameters as opposed to multiple-merger and population-growth models, both of which can predict an excess of singletons. Encouragingly, multiple-merger and population-growth models exhibit noticeable differences in the bulk of the site-frequency spectrum, in particular, in the lumped tail (Figure 1; see also Figures 4 and S2 in Neher and Hallatschek 2013. (Neher and Hallatschek 2013). In Figure 1, the normalized expected spectrum [see Equation (2)] for a given coalescent Π, i.e., the expected spectrum scaled by the expected total number of segregating sites, is compared for different multiple-merger coalescents [B = beta-coalescents (Schweinsberg, 2003) or D = Dirac coalescents (Eldon and Wakeley, 2006)] and exponential (E) and algebraic (A) growth models leading to time-changed Kingman coalescents for sample size (number of leaves) n as shown. Details for these coalescent models are given at the beginning of File S1. The first five classes (representing relative length of external branches, two-leaf branches, etc.) are shown, with classes from six onward collected together (labeled 5+). In Figure 1, the relative external branch lengths were matched between the different coalescent processes. Even though the relative external branch lengths and, by implication, the number of singletons relative to the total number of segregating sites can be matched between the different processes, the collapsed tail (group 5+ in Figure 1) differs noticeably between the multiple-merger coalescents and the growth models. One also observes that the parameters have been chosen to match for with when α = 1, where α is the coalescent parameter associated with B. Thus, is maximized for the given n (because ), but , , and all can increase by increasing the relevant parameters (ψ, β, or γ).
Figure 1.
Matching [see Equation 2] for the different coalescent processes Π ∈ {A, B, D, E} with number of leaves n as shown. Expected values were computed exactly. The processes and their associated parameters are algebraic growth (A, γ), beta(2 − α, α)-coalescent (B, α), Dirac coalescent (D, ψ), and exponential growth (E, β). The values with label 5+ represent the collapsed tail .
Figure 4.
The distance for X ∈ {B, D} of the normalized expected spectra [see Equation 2] and as a function of α (X = B) [resp. ψ (X = D)] and γ (A) for number of leaves n = 100. Expected values were computed exactly. The grid points are and α ∈ {1, 1.025, …, 2}; for γ as shown.
Matching the relative external branch lengths [see Equation 2] and observing how the rest of the normalized expected spectrum behaves, as illustrated in Figure 1, give hope that multiple-merger processes may be distinguished from (at least) particular population-growth models with adequate statistical power. In the limit of large n, for the Kingman coalescent, .
Inference methods for distinguishing population growth from the usual Kingman coalescent have been studied extensively (see, e.g., Tajima 1989a; Slatkin and Hudson 1991; Rogers and Harpending 1992; Kaj and Krone 2003; Sano and Tachida 2005). Simulation-based work includes Ramírez-Soriano et al. (2008), who considered the statistical power of several tests under population size increase and decrease and the impact of recombination. Ramos-Onsins and Rozas (2002) considered the statistical power of statistics based on the site-frequency spectrum to distinguish deterministic population growth from the Kingman coalescent. On the theoretical side, Myers et al. (2008), Bhaskar and Song (2014), and Kim et al. (2014) considered principal questions of identifiability of demographic histories. In particular, Bhaskar and Song (2014) showed theoretically that complete knowledge of the SFS for large sample sizes carries enough information to fully recover demographic history under mild assumptions on the possible fluctuations of the demography.
Detecting multiple-merger coalescents in populations deviating from the Kingman coalescent assumptions is a relatively new direction of research. Indeed, deriving inference methods based on multiple-merger coalescents has only just begun (Eldon and Wakeley 2006; Birkner and Blath 2008; Eldon 2011; Birkner et al. 2011, 2013a, b; Steinrücken et al. 2013; Rödelsperger et al. 2014; Koskela et al. 2015). In particular, Birkner et al. (2013b) obtained recursions for the expected site-frequency spectrum associated with Lambda-coalescents. In this work, we address the issue of distinguishing multiple-merger coalescents from exponential population growth by proposing statistical tests based on the (normalized) SFS, estimating statistical power for interval hypotheses via simulation. Because we can only work with approximate likelihood functions and our methods, in particular, the so-called fixed-s method, can be sensitive to an (unknown) true coalescent mutation rate θ/2, we complement our analysis by an approximate Bayesian computation approach (ABC) (Rubin 1984; Tavaré et al. 1997; Pritchard et al. 1999; Cucala and Marin 2013; Baragatti and Pudlo 2014).
Materials and Methods
Basic properties of the site-frequency spectrum
Consider a sample of n DNA sequences taken at a given genetic locus, and assume that we can distinguish between derived (new mutations) and ancestral states. For , let n := {1,…,n} {1, …, n}. We denote by the total number of sites at which the mutant base appears i ∈ [n − 1] times. Then
is referred to as the unfolded site-frequency spectrum based on the n DNA sequences. If mutant and wild type cannot be distinguished, one often considers the folded spectrum , where ancestral and derived states are not distinguished and hence
(Fu 1995), where δi,j = 1 if i = j and 0 otherwise. In this study, we will mostly be concerned with the unfolded site-frequency spectrum. Define , where denotes the total number of segregating sites. Thus, is the “normalized” unfolded SFS (nSFS), with the convention that in the trivial case of complete absence of segregating sites .
In order to compute expected values, variances, and covariances of the SFS, an explicit underlying probabilistic model is needed. In the following, we assume that the genealogy of a sample can be described by a coalescent process, more precisely by either (a time change of) the Kingman coalescent or a multiple-merger coalescent. In addition, the infinitely-many-sites mutation model (Watterson 1975) is assumed, and mutations are modeled by a Poisson process on the coalescent branches with rate θ/2. With this parameterization, the expected number of segregating sites in a sample of size 2, and hence the expected number of pairwise differences in a sample from the population, equals θ.
Closed-form expressions for the expected values and (co-)variances of have been determined by Fu (1995) when associated with the Kingman coalescent. One can represent the expected values of in a unified way using the results of Griffiths and Tavaré (1998), Kaj and Krone (2003), and Birkner et al. (2013b), which allow one to treat the expected values (and covariances) of the SFS for all coalescent models in question.
Let be a (partition-valued exchangeable) coalescent process started from n leaves (partition blocks) corresponding to the random genealogy of a sample of size n. By discussing leaves rather than DNA sequences, we are emphasizing our viewpoint of the genealogy as a random graph, where the leaves are a particular kind of vertex. Our interest is in the topology of the genealogy and how it is reflected in the associated site-frequency spectrum.
If the initial number of leaves is not specified, we simply speak of Π. One may think of Π as the Kingman coalescent, but the point is that the following result will also stay true for externally time-changed Kingman coalescents as well as multiple-merger coalescents (a.k.a. Lambda- or Xi-coalescents in the mathematical literature) and even externally time-changed multiple-merger coalescents.
Given n and a coalescent model Π, let be the block-counting process of the underlying coalescent Π(n) started from n lineages; i.e., gives the number of ancestral lines (blocks) present/active at (backward) time t. For , let be the random amount of time that spends in state k. Given a coalescent Π(n) started from n (unlabeled) lineages, denote by p(n),Π[k, i] as the probability that conditional on the event that for some time point t a given one of the k blocks subtends exactly i ∈ [n − 1] leaves. A general representation of is then
| (1) |
The normalized expected SFS for i ∈ [n − 1] is defined as
| (2) |
where the denominator in Equation 2 is the expected total tree length when starting from n leaves. One can interpret the quantity as the probability that a mutation, under the infinitely-many-sites assumption and the coalescent model Π, with known ancestral types, appears i times in a sample of size n. Importantly, is not a function of the mutation rate, unlike . One also can view as a first-order approximation of the expected value of the nSFS.
As examples for Π, we will consider the classical Kingman coalescent (K), exponential (E) and algebraic (A) growth models, and the beta(2 − α, α) (B) and Dirac (D) multiple-merger coalescents, as shown in File S1. Simulations suggest that is a good approximation of when α is not too close to 1 and n and θ are not too small (Birkner et al. 2013b). Similar conclusions hold in the case of exponential and algebraic growth (results not shown).
One can use the recursive formulas obtained by Birkner et al. (2013b) to compute associated with Lambda-coalescents. To compute associated with growth models, we use the results of Polanski and Kimmel (2003), whose recursions are given in File S1.
A comparison of the observed (instead of ) with an expected value —obtained under a particular coalescent model Π—enables one to do inference without having to jointly estimate the mutation rate θ using, e.g., a minimum-distance statistic. Indeed, it appears that under any coalescent model Π, is almost constant as a function of the mutation rate θ (unless θ is very small); we provide some evidence for this in Equation S20 in File S1. Unfortunately, there seems to be no explicit way of representing as a simple function of the associated coalescent parameters and sample size n. As mentioned earlier, one may instead work with .
Time scales, segregating sites, and mutation rates
The choice of a multiple-merger coalescent model (i.e., demographic history) Π and its underlying parameters strongly affects classical estimates of the coalescent mutation rate θ/2 (i.e., the Poisson rate at which mutations appear on coalescent branches). Assume without loss of generality for all multiple-merger coalescents in question that the underlying coalescent measure Λ is always a probability measure: this normalization fixes the coalescent time unit as the expected time to the most recent common ancestor of two individuals sampled uniformly from the population.
Given an observed number of segregating sites S in a sample of size n, a common estimate of the scaled mutation rate θ associated with coalescent model Π is the Watterson estimate, i.e.,
| (3) |
where is the expectation of the total tree length B(n) of an (n-)coalescent model Π. One can, of course, also estimate θ as a (different) linear combination of the site-frequency spectrum [cf. Achaz (2009) in the case of the Kingman coalescent]. Using the recursions for obtained by Birkner et al. (2013b), one also can estimate θ using either Equation 3 or a linear combination of the expected SFS in the case of a Lambda-coalescent.
Given an estimate and knowledge of the mutation/substitution rate per year at the locus under consideration, one can find a real-time embedding of the coalescent history via the approximate identity
| (4) |
(see Steinrücken et al. 2013, Section 4.2), which, of course, depends on Π.
If one has additional information on the specific reproductive mechanisms of an approximating population model, this can even enable one to estimate the model census population size. For example, given a Cannings population model (Cannings, 1974, 1975) of fixed size N, let cN be the probability that two gene copies, drawn uniformly at random and without replacement from a population of size N, derive from a common parental gene copy in the previous generation. While for the usual haploid Wright-Fisher model cN = 1/N, in a class of population models studied by Schweinsberg (2003) leading to the beta(2 − α, α)-coalescent, cN is proportional to 1/Nα−1 for α ∈ (1, 2] (but note that the proportionality constant depends on finer details of the particular model). By a limit theorem for Cannings models by Möhle and Sagitov (2001), one coalescent time unit corresponds to approximately 1/cN generations in the original model with population size N. Thus the mutation rate at the locus under consideration per individual per generation must be scaled with 1/cN [as noted e.g., in Eldon and Wakeley (2006)], and the relation between , the coalescent mutation rate θΠ/2, and cN is then given by the (approximate) identity
| (5) |
In particular, if the Cannings model class (and thus cN as a function of N) is known, N can be estimated via Equation 5.
In this context, it is important to note that different population models on very different time scales still can have the Kingman coalescent as their ancestral limit process; two examples are the Wright-Fisher and Moran models. This is certainly also the case for multiple-merger coalescents. In particular, cN is a priori not a function of the limiting coalescent model (this appears to be a rather frequent misperception).
Distinguishing real and coalescent time scales is important because nonlinear scaling otherwise may easily lead to confusion: for example, the expected total tree length (measured in coalescent time units) decreases as a function of α ∈ (1, 2], while the corresponding quantity (measured in real-time generations) increases in α ∈ (1, 2] (cf. Figure S1 in File S1).
The time scaling applied to a classical Wright-Fisher model with fluctuating population size [as in Kaj and Krone (2003)] in order to obtain a (time-changed) Kingman coalescent is shown in particular in Equation (S10) in File S1. Again, the estimate (Equation 3) of θ depends on the growth model and growth parameter.
Approximate likelihood-ratio tests for the SFS
Our aim is to construct a statistical test to distinguish among the model classes E, A, D, and B (which intersect exactly in the Kingman coalescent K). In order to distinguish, say, E from B based on an observed site-frequency spectrum with sample size n and segregating sites , a natural approach is to construct a likelihood-ratio test.
Recall that we think of our observed spectrum as a realization of a coalescent tree with n leaves obtained from a coalescent model Π, with mutations distributed on the tree according to an independent Poisson (θ/2) process. For each model Π from classes {E, A, D, B}, the coalescent will be uniquely determined by a single coalescent parameter β ∈ [0, ∞) (for E), γ ∈ [0, ∞) (for A), ψ ∈ [0, 1] (for D), and α ∈ [1, 2] (for B). [Note that the beta-coalescent is well defined for α ∈ (0, 2], but we restrict to a smaller parameter range corresponding to the population model in Schweinsberg (2003).]
Suppose that our null hypothesis H0 is the presence of recent exponential population growth (E) with (unknown) parameter β ∈ [0, ∞), and we wish to test it against the alternative H1 hypothesis of a multiple-merger coalescent, say, the beta(2 − α, α)-coalescent (B) for (unknown) α ∈ [1, 2], where β = 0 and α = 2 correspond to the Kingman coalescent. In this framework, the coalescent mutation rate θ is not directly observable but plays the role of a nuisance parameter. In particular, it is the interplay of the coalescent model Π and the mutation-rate parameter θ that governs the law of the observed number of mutations (see the discussion in the preceding section). To take θ explicitly into account, one could test
(exponential growth) against
if the beta-coalescent family is the alternative [by slight abuse of notation, we identify the coalescent model Π with the corresponding coalescent parameter β (resp. α) in each model class when appropriate]. The underlying parameter ranges are two-dimensional, and although an explicit likelihood-ratio test based on methods described in Simonsen et al. (1995) can be constructed, it will likely pose computational challenges.
Instead, given an observed number of segregating sites S = s, we simplify our framework by employing the fixed-s method discussed, e.g., in Depaulis and Veuille (1998) and Ramos-Onsins and Rozas (2002). Here we treat the observed number of segregating sites as a fixed parameter , not as (observation of a) random variable S. We will thus obtain the empirical distributional quantities of our test by Monte Carlo simulations, placing uniformly at random s mutations along the branches of the simulated tree.
The fixed-s method is different from generating samples for a given θ by conditioning on S = s, yet the fixed-s method usually leads to reasonable tests when the true θ is close to the Watterson estimate based on s (Wall and Hudson 2001). However, it can lead to substantial deviations from the conditional distribution if θ is extreme [see, e.g., Markovtsova et al. (2001), who show that for a test in a related framework, the probability of rejection can be substantially different from 5%]. Regarding this caveat, Depaulis et al. (2001) took a Bayesian viewpoint and showed that the values of θ that lead to unreliable tests are highly unlikely given s. We address this issue by using rejection sampling to check the robustness of the fixed-s method against varying θ (see File S1). Our analysis is complemented with an ABC approach using the normalized frequency spectrum , which should be insensitive to the actual value of θ as long as θ is not too small [cf. Equation (S20) in File S1].
By fixing S = s and treating it as a parameter of our test, we may consider the new pair of hypotheses
and
Define a likelihood function for the observed frequency spectrum with fixed under the coalescent model (resp. ) by
| (6) |
where are the random lengths of branches subtending i ∈ [n − 1] leaves, and B(n) is the total branch length of the coalescent under Π. The fixed-s paradigm thus leads to a mixture of multinomial distributions where the parameters are given by the respective relative branch lengths. The hope is that the location of the maximum of is typically not far from the location of the corresponding coordinate of the maximizer in the full two-dimensional explicit-θ model, in which one can additionally maximize over all θ ∈ [0, ∞).
Now we can construct a likelihood-ratio test based on via the likelihood-ratio function
| (7) |
Given a significance level a ∈ (0, 1) (say, a = 0.05), let be the a-quantile of under E, chosen as the largest values so that
| (8) |
The decision rule that constitutes the fixed-s likelihood-ratio test, given s and sample size n, is
This formulation is free of the nuisance parameter θ. To assess the justification for the fixed-s assumption, we investigate how close this is to the corresponding quantiles for different values of θ, including the Watterson estimator
| (9) |
[cf. (Equation 3)], for selected choices of Π. The agreement appears reasonably good and seems to increase with sample size (see File S1).
The corresponding power function of the test, i.e., the probability of rejecting a false null hypothesis, is given by
| (10) |
The likelihood (Equation 6) cannot be represented as a simple formula involving the coalescent parameters; one can approximate (Equation 6) via a Monte Carlo approach, but this is computationally expensive. An approximation is
| (11) |
where we replaced the random quantities in Equation 6 by (Equation 2).
Interestingly, an approximate maximum likelihood method based on Equation 11 is equivalent to the following approach: consider a family of (approximate) likelihood functions
| (12) |
where is the Watterson estimator for the mutation rate under a Π-coalescent with n leaves when S = s segregating sites are observed [recall Equation 3]. In Equation 12, is well defined even if .
The rationale behind Equation 12 is simple: it pretends that the classes are approximately independent and Poisson distributed (this is, of course, not literally true but encouraged by the fact that the off-diagonal entries of the covariance matrix of are small compared with the diagonal terms) (see Birkner et al. 2013b). Equation 12 is indeed equal to the one obtained from the Poisson random field (PRF) of Sawyer and Hartl (1992), which considers unlinked sites. Within the PRF framework, Equation 12 is an exact likelihood function. In our model of completely linked sites at a single locus, the assumption of independence is merely a convenient computational tool. An analogous approximation of likelihood functions is considered by Bhaskar et al. (2015) in the context of varying population sizes; these authors also provide a detailed discussion of the intermediate situation when there is some but not too much recombination between sites at a given locus but free recombination between loci.
For fixed s, we can view as parameterizing a one-dimensional curve in the full two-dimensional space H0 ∪ H1 defined by the requirement that . The two approximate maximum likelihood approaches based on Equations 11 and 12 are equivalent. Indeed,
| (13) |
because the sum to 1. Hence, both likelihood functions differ only by the fixed prefactor e−s, so they attain their maximum at the same position.
Thus now we consider the statistic
| (14) |
[where ΘE and ΘB refer to the projection of H0 (resp. H1) on the coalescent parameter]. For a given value of s, we can then (by simulations using the fixed-s approach) determine approximate quantiles associated with a significance level a as in Equation 6 and base our test on the criterion . Similarly, the (approximate) power function can be estimated using simulations.
An alternative approach to the (approximate) likelihood-based tests would be rejection rules based on minimal-distance statistics, i.e.,
| (15) |
for some suitable distance measure d (e.g., the distance with p = 2) with corresponding power function . We will not discuss the theoretical justification for this method. However, we will use the distance between normalized expected spectra under various coalescent models to produce three-dimensional heat maps that give some intuitive insight into how a pair of different models out of {E, B, A, D} relates to each other depending on the underlying pair of coalescent parameters (cf. Results).
We conclude this section with a remark on lumping. One often observes for most i greater than some (small) number m in observed data, in particular, for large n. It thus seems natural to consider (approximate) likelihood functions for lumped spectra (e.g., collapsing all entries in classes to the right of some number m into one class m+), as we have done, e.g., in Figure 1. Another natural type of lumping may be to collect together classes so that for some x ∈ (0, 1/2]. This may not always be feasible, though, if the individual quickly become quite small, and we will refrain from going into a more detailed theoretical discussion of optimal lumpings. However, we will see in our subsequent ABC analysis that adequate lumping can improve the reliability of our model-selection procedure.
Approximate Bayes factors and model selection
In view of the approach and notation of the preceding section, an analogous method of model selection could be based on a Bayes factor of the form
| (16) |
(and similar for all other combinations of classes A, D, E, and B) given a pair of priors πB, πE on . While the approach will also work in principle for the two-dimensional prior ranges ΘB and ΘE, we will present the (approximate) Bayesian methods with one-dimensional prior ranges (where S = s is treated as a fixed parameter, motivated from the fixed-s approach) so that they complement our previous methods.
Our simulations will be obtained using the rationale behind Equation 12; i.e., after simulating a tree according to a given coalescent parameter, say, α from πB, mutations are placed on the tree according to a Poisson process, with mutation rate estimated using Equation 3. However, in Equation 16, we use the normalized site-frequency spectrum as observed statistics because it should be more insensitive to the true coalescent mutation rate θ [potentially deviating from ], as argued in the corresponding section in File S1, and thus yield more robust results. in Equation 16 thus denotes the likelihood function of the observed nSFS under the chosen coalescent model, with the mutation parameter given by Watterson’s estimator based on s. Because we estimate the mutation rate based on s, the information loss of using the nSFS instead of the SFS should be only slight. We also experimented with ABC based on simulations using the fixed-s method, i.e., distributing a fixed number of mutations uniformly on the simulated tree, and generally found higher misclassification probabilities (results not shown).
To overcome the problem of exact computation of , which appears infeasible in practice, we employ approximate Bayesian methods (see, e.g., Beaumont 2010) based on the distance between observed and simulated nSFS. Bayes factors based on further (lumped) distances d and/or the folded nSFS may, of course, also be considered. In line with classical Bayes factor philosophy (cf., e.g., Kass and Raftery 1995), one interprets an observed value of as evidence in favor of over .
For the ABC analyses, we consider as before on exponential growth (E), algebraic growth (A), and beta- and Dirac coalescents (B and D). Given sample size n and number of segregating sites s, again the coalescent model classes can be parameterized by a single parameter each, which are the exponential growth rate β ∈ [0, ∞), algebraic growth rate γ ∈ [0, ∞), beta-coalescent parameter α ∈ [1, 2], and mass point location ψ ∈ (0, 1] for the Dirac coalescent.
For convenience, we employ a simple rejection-based ABC scheme to approximate the Bayes factor for the model (class) comparison given an observed nSFS (resp. folded and/or lumped versions, which can be treated analogously). First, select a number of models (out of {E, B, A, D}) that should be compared, say, E and B, and choose the corresponding prior distributions on the coalescent parameter ranges. To simulate, say, nr independent samples of the nSFS from each model, say, from E, independently generate nr coalescent parameters from the prior πE and a corresponding coalescent tree Π for each generated coalescent parameter. Distribute independently Poisson mutations with parameter on each such tree, and record the corresponding normalized site-frequency spectra. This should be done independently for all models.
Then fix a tolerance level x ∈ (0, 1) and count the number of simulations NE, NB from each model that are among the 100 ⋅ x% best fits with respect to the distance to the observed nSFS (the “accepted” simulations). Here we use an additional scaling by dividing each class (resp. lumped class) in the nSFS by the median (if nonzero) within this class observed in all simulations as implemented in the R package abc (Csilléry et al. 2012). The Bayes factor for model E vs. B then can be approximated by
To assess how well this ABC approach allows one to distinguish, say, E from B (or, more generally, simultaneously among {E, B, A, D}), we use two approaches from the R package abc. Both are based on leave-one-out cross-validation. More precisely, we pick ncv simulations at random from each model, treat them as the observed value of the nSFS, and then run the ABC approach with the same parameters and simulations as earlier. For each cross-validation sample, say, from model E, we record the counts of accepted simulations , and from the model classes A, B, D, and E (recall that the chosen cross-validation sample is left out). As measures for the distinction ability of this approach, we record for each model class, borrowing notation from Stoehr et al. (2014):
- The (estimated) mean posterior probabilities π for model B given the observed nSFS under the true model E, say,
where is the number of accepted simulations. - The (estimated) mean misclassification probabilities
for Y ∈ {A, E, D}. To ease the notation, we will from now on omit n in the formulas.
In practice, we need to efficiently generate samples of the nSFS under the different models that can be achieved by backward-in-time coalescent simulations. For the exponential growth models (E), we use Hudson’s ms (Hudson 2002), as implemented in the R package (R Core Team 2012) phyclust (Chen 2011). For algebraic growth models (A), the beta-coalescents (B), and the Dirac coalescents (D), we use custom R and C scripts to generate samples of the nSFS (available at: http://page.math.tu-berlin.de/~eldon/programs.html). To conduct the actual ABC analysis including cross-validation techniques, we employed the R package abc (Csilléry et al. 2012). Additionally, because we use Watterson’s estimator to set the mutation rate within each model, we compute the mean total length of each coalescent model as described in File S1.
Results
Power estimates of approximate likelihood-ratio tests
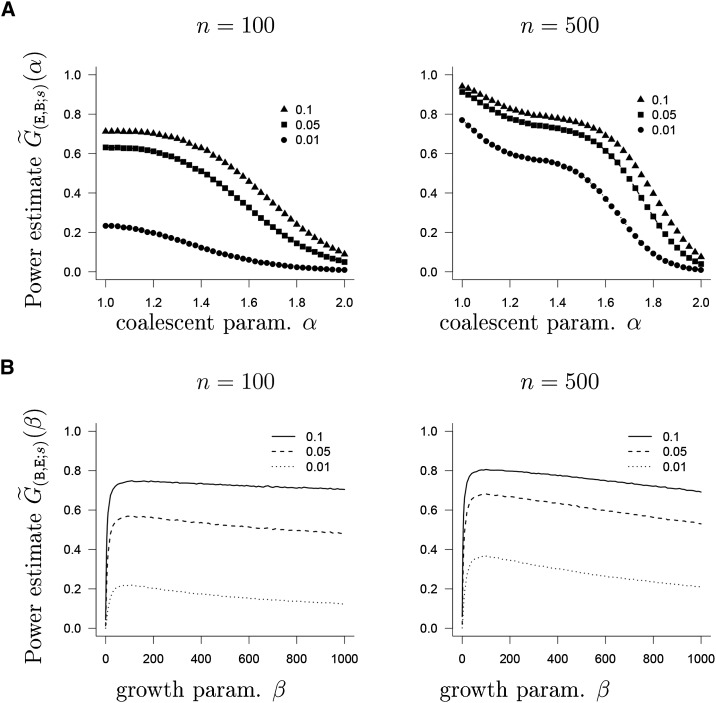
To assess the sensitivity of our approximate likelihood-ratio test associated with the likelihood-ratio function (Equation 7), we estimate its power from the analog of Equation 10 based on the approximate likelihood from Equation 12 as a function of α (Figure 2A) with and and estimate as a function of β with and (Figure 2B).
Figure 2.
(A) Estimate of from Equation 10 based on the approximate likelihood from Equation 12 as a function of α (no lumping) with number of leaves n as shown and s = 50. (B) Estimate of from Equation 10 based on the approximate likelihood from Equation 12 as a function of β (no lumping) with number of leaves n as shown and s = 50. The symbols denote the size of the test, as shown in the legend. The interval hypotheses are discretized to and. . In A, the beta(2 − α, α)-coalescent is the alternative; in B, exponential growth is the alternative.
As shown in Figure 2, reasonably high power is obtained to reject for n = 500 and even for a smaller sample size n = 100, but the power also depends, as one would expect, on the size of the test. As a side note, we remark that the power estimates , as a function of α, are right at the size of each corresponding test when α = 2 (the Kingman case) as required.
The mitochondrial DNA (mtDNA)–genome analysis of Carr and Marshall (2008), who scanned whole mitochondrial genomes (15,655 bp) of the highly fecund Atlantic cod (Gadus morhua), prompted us to briefly investigate the power (Figures S3 and S4 in File S1) with the number of segregating sites s = 300. This is nearly the total number of polymorphisms (298) observed among the 32 mtDNA genomes sampled by Carr and Marshall (2008). Our results show that while we may not quite have enough power when n = 30 and s = 300 (Figure S4A in File S1), we would be in good shape for n = 100 (Figures S3 and S4B in File S1). It would be very interesting to analyze such a sample, once available, because it appears to be an open debate whether beta-coalescents should be favored over classical models (including recent population growth) in HFSOD populations (cf., e.g., Steinrücken et al. 2013).
Another quite striking observation is that the power of our test is apparently nonmonotone as a function of β when , in particular, for a smaller type I error. We will present a possible heuristic explanation for this in the Discussion section. A rather high power in general is obtained when comparing and associated with the Dirac coalescent (Figure S2 in File S1) for (n, s) = (100, 50). For further combinations, we refer to File S1, where , associated with algebraic growth, is compared with in Figure S6 and with in Figure S5. The power functions (Figure S5 in File S1) are decidedly nonmonotone, as is (Figure S6A in File S1).
We conclude with a short remark on the sensitivity of our results on lumping of classes in the observed spectrum. Indeed, our power estimates suggest that keeping at least the first five classes of the SFS intact and collecting the rest into one other class have little effect on the power of the test (results not shown). Keeping only the singleton class intact and collecting all the rest into one class, however, significantly diminish power (results not shown). C code (cf. Kernighan and Ritchie 1988) written for estimating the power of our tests, where use was made of the GNU Scientific Library (Galassi et al. 2013), is available at http://page.math.tu-berlin.de/~eldon/programs.html.
Mean-squared distance landscapes for the normalized expected SFS under different growth and coalescent models
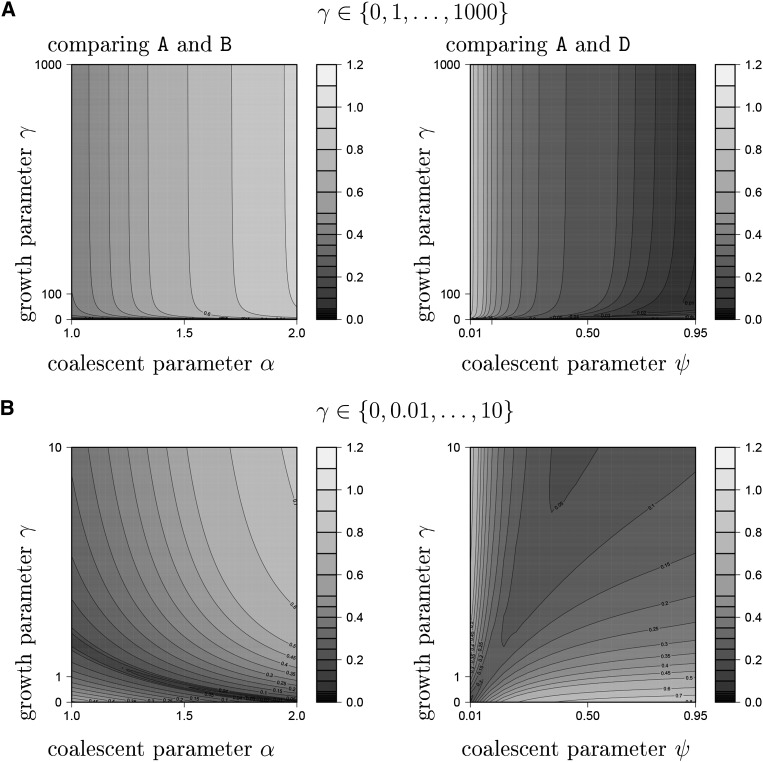
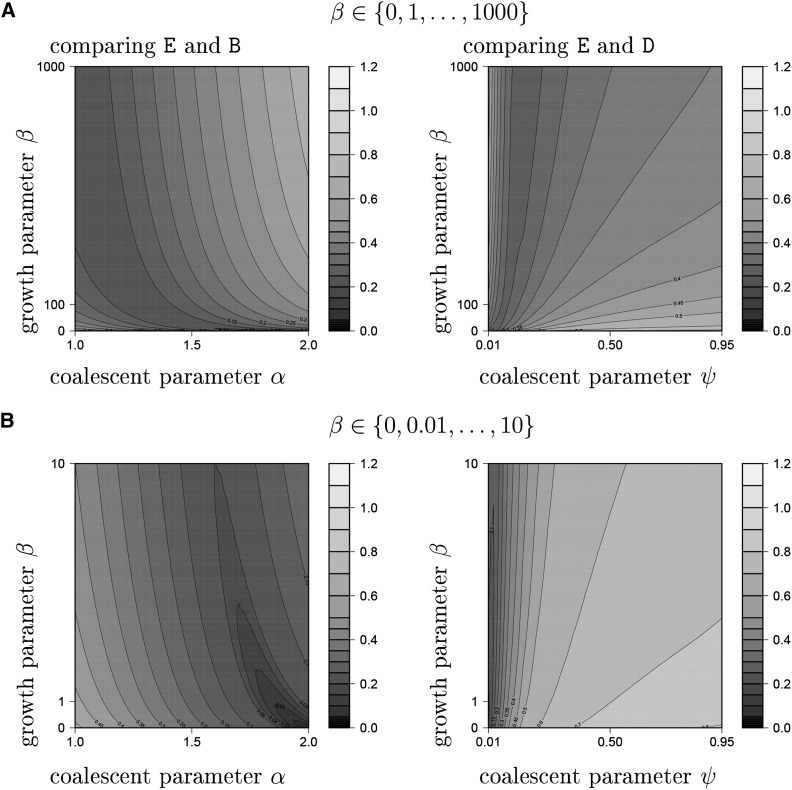
Given the potential ability to distinguish between growth and multiple-merger coalescent models, the following questions arise: how does the distance between and behave as a function of the underlying coalescent and growth parameters? Is it possible to visibly identify a one-dimensional curve given by coalescent parameter pairs corresponding to (Π1, Π2) along which minimal distance is achieved? Figure 3 and Figure 4 are a brief effort to understand the relation between the expected nSFS for the models in question by graphing the distance as a function of the coalescent and growth parameters associated with X and Y. In Figure 3, E is compared with B and D. In Figure 4, A is compared with B and D. In Figure 3 and Figure 4, the upper panels show the distance as the respective growth-parameter ranges from 0 to 1000, while the lower panels zoom in on the range from 0 to 10.
Figure 3.
The distance for of the normalized expected spectra [see Equation 2] and as a function of α (X = B) [resp. ψ (X = D)] and β (E) for number of leaves n = 100. Expected values were computed exactly. The grid points are α ∈ {1,1.025,…,2} and ; for β as shown.
Figure 3 indicates the presence of a region, essentially a curve in the two-dimensional (α, β) parameter space, along which the lowest distance is reached. However, one should be aware that this curve shifts in space when sample size n is increased (data not shown).
Figure 3 suggests that we should have good power to distinguish between algebraic growth and beta-coalescents. However, this seems not to be the case for distinguishing algebraic growth from Dirac coalescents: extreme growth (large γ) seems to produce an almost star-shaped genealogy—consequently, the distance to a Dirac coalescent with ψ close to 1 becomes very small (recall that ψ = 1 exactly corresponds to the star-shaped coalescent).
Mean misclassification and posterior probabilities for the ABC approach
In this section we analyze how far an ABC approach using the nSFS (resp. the folded nSFS, abbreviated as nfSFS) and the lumped variants as summary statistics supports our claim that one can distinguish between exponential growth E and the beta(2 − α, α)-coalescent B as well as between E, B, and the Dirac coalescent D or between B, D, and algebraic growth A. The distinction ability of the ABC model comparison is assessed based on the simulation procedure and notation described in Materials and Methods. Priors were uniform over the full range of coalescent parameters in models B and D and uniform until a maximal cutoff for models E (on [0, βmax]) and A (on [0, γmax]). We discretized the parameter range for the growth models by using increments of 1 or 10 for exponential growth (the first used in all multiple-model comparisons, the latter in the pairwise comparisons between E and B) and increments of 1 for algebraic growth. If not specified otherwise, we used βmax = γmax = 1000. We fixed a sample size n = 200. The number of replications was set to nr = 2 × 105. See Table 1, Table 2, and Table 3 and Tables S4–S8 in File S1 for the estimates of posterior probabilities and misclassification probabilities (some with one replication) with various degrees of lumping and various parameter settings for n = 200. For an example with higher sample size n = 1278, see Table S9 in File S1.
Table 1. Approximations of the mean posterior probabilities and misclassification probabilities for the ABC model comparison between models E and B for tolerance x = 0.01, s = 15 segregating sites, and using either the nSFS or the nfSFS as summary statistics.
| Fold | Lump | ncv | ||||
|---|---|---|---|---|---|---|
| No | 10+ | 24,000 | 0.30 | 0.25 | 0.26 | 0.13 |
| No | 50+ | 12,000 | 0.32 | 0.29 | 0.26 | 0.17 |
| No | 100+ | 1,200 | 0.33 | 0.29 | 0.28 | 0.17 |
| Yes | 10+ | 24,000 | 0.32 | 0.25 | 0.28 | 0.12 |
| Yes | 50+ | 12,000 | 0.34 | 0.29 | 0.28 | 0.16 |
| Yes | No | 12,000 | 0.34 | 0.29 | 0.29 | 0.16 |
ncv denotes the number of cross-validations; lump indicates which mutation classes are lumped into one class.
Table 2. Approximations of the mean posterior probabilities for the ABC model comparison among models E, B, and D for tolerance x = 0.005, sample size n = 200, and s = 15 or 75.
| s | Lump | ncv | ||||||
|---|---|---|---|---|---|---|---|---|
| 15 | 10+ | 24,000 | 0.29 | 0.12 | 0.24 | 0.02 | 0.55 | 0.03 |
| 15 | 50+ | 12,000 | 0.39 | 0.10 | 0.22 | 0.02 | 0.54 | 0.05 |
| 15 | no | 12,000 | 0.42 | 0.10 | 0.21 | 0.02 | 0.55 | 0.07 |
| 75 | 10+ | 24,000 | 0.22 | 0.09 | 0.12 | 0.00 | 0.11 | 0.01 |
| 75 | 50+ | 12,000 | 0.26 | 0.10 | 0.12 | 0.00 | 0.11 | 0.01 |
The nfSFS was used as summary statistics.
Table 3. Approximations of the mean posterior probabilities for the ABC model comparison among models A, B, and D for tolerance x = 0.005, sample size n = 200, and s = 15 or 75.
| s | Lump | ncv | ||||||
|---|---|---|---|---|---|---|---|---|
| 15 | 10+ | 24,000 | 0.02 | 0.12 | 0.01 | 0.03 | 0.16 | 0.55 |
| 15 | 50+ | 12,000 | 0.02 | 0.11 | 0.01 | 0.03 | 0.24 | 0.57 |
| 75 | 10+ | 24,000 | 0.01 | 0.09 | 0.01 | 0.05 | 0.10 | 0.29 |
| 75 | 50+ | 12,000 | 0.01 | 0.08 | 0.01 | 0.05 | 0.15 | 0.32 |
The nfSFS was used as summary statistics.
The estimated error probabilities range from moderate to low values. Mean posterior probabilities indicate a correct classification probability , which shows that our method has good distinguishing ability. As expected, lower tolerance generally leads to smaller errors, as do larger mutation rates, while using the folded nSFS increases them. Appropriate lumping seems to decrease the error probabilities on many occasions; see, e.g., Table 1, where a positive effect for strong lumping is observed for s = 15 segregating sites, whereas for s = 75 in Table S4 in File S1, moderate lumping seems to be more appropriate (both tables show the comparison of models B and E). Not surprisingly, exponential growth rates closer to zero are harder to distinguish from the beta(2 − α, α)-coalescent models than higher growth rates (see Tables S4 and S6 in File S1). The ABC model comparison distinguishes especially well, even for s = 15, between exponential growth from Dirac coalescents and algebraic growth from beta(2 − α, α)-coalescents (see Table 2, Table 3, and Tables S7 and S8 in File S1). For a relatively low number of segregating sites (s = 15), some comparisons (e.g., algebraic growth with Dirac coalescents and beta-coalescents with Dirac coalescents) can lead to common misclassification, but this effect vanishes for larger s. For s = 75, Dirac coalescents can be distinguished relatively well from beta(2 − α, α)-coalescents (Table 2, Table 3, and Tables S7 and S8 in File S1).
Discussion
The development of methods to distinguish between different (time-changed) coalescent scenarios for the underlying genealogy of a population on the basis of observed data is an important task, in particular, because the choice of an underlying coalescent model affects the estimated coalescent mutation rate via (3) and a potential real-time embedding of the genealogy based on (5). Identification of an appropriate coalescent model also may give hints about the underlying reproductive mechanisms present in a population. By way of example, multiple-merger coalescents may indicate the presence of HFSODs in the population.
While inference methods for distinguishing population growth from the usual Kingman coalescent have been studied extensively (see, e.g., Tajima 1989a; Slatkin and Hudson 1991; Rogers and Harpending 1992; Kaj and Krone 2003; Sano and Tachida 2005) and sophisticated theoretical results on the question of identifiability of demographic histories have been obtained (cf., e.g., Myers et al. 2008; Bhaskar and Song 2014;Kim et al. 2014), none of these studies has addressed multiple-merger coalescents. In fact, only a few results, e.g., on the statistical properties of the SFS in multiple-merger coalescents (see Birkner et al. 2013b) are available.
For the particular case of distinguishing multiple-merger coalescents from population-growth scenarios, this decision problem is complicated by the fact that the patterns of genetic variation produced by the two demographic effects and summarized in the SFS are expected to be similar: both lead to an excess of singletons compared with a classical Kingman coalescent–based genealogy. However, while it is usually possible to match the predicted number of singletons with the observed number in various special cases for both models, the bulk and tail of the spectrum typically will differ (cf. Figure 1 for some examples).
This paper thus is aimed at exploiting and quantifying these differences. However, for feasibility, we had to restrict both the scope and employed methods of our analysis. The first (restrictive) decision in the design of our analysis was the selection of certain subfamilies of Lambda-coalescents and demographic growth scenarios that we deemed suitable for investigation. The reason for restricting to subclasses of Lambda-coalescents is that the full class of multiple-merger coalescents is in one-to-one relation with the uncountable and nonparametric set of finite measures Λ on [0, 1], which drastically complicates statistical questions, while most of these coalescents do not appear to have a clear biological motivation in terms of a natural underlying population model. Considering the whole Lambda-coalescent class also would raise theoretical questions concerning the unique identifiability of multiple-merger coalescents on the basis of the SFS related to Myers et al. (2008) and Bhaskar and Song (2014), and this is mathematically challenging and outside the scope of this work.
Hence, in case of the multiple-merger coalescents, we restricted our attention to the class of beta-coalescents (B) and the Dirac coalescents (D). These classes appeared particularly interesting to us because they both interpolate in a parametric way between the boundary points of the Kingman coalescent (K) via the Bolthausen-Sznitman coalescent (B at α = 1) to the star-shaped coalescent (B, α = 0; D, ψ = 1)—where the whole genealogy collapses to a single line in a single large merger event—among the multiple-merger coalescents. Beta-coalescents have been studied frequently in the literature (see, e.g., Birkner et al. 2005; Bertoin and Le Gall 2003; Hallatschek and Neher 2013; Steinrücken et al. 2013; Birkner et al. 2013b) and are related to a population model with HFSODs (Schweinsberg 2003). Dirac coalescents have been chosen for their simplicity from a mathematical standpoint and also have been investigated by Eldon and Wakeley (2006). The parameter of the Dirac coalescent has a clear interpretation as the fraction of the population that is replaced in each single HFSOD reproductive event.
For similar reasons, we restricted demographic scenarios to two basic parametric growth models. Exponential growth (E) is certainly a natural model in the presence of a supercritical branching population model without geographic or resource restrictions. Our second choice, the algebraic growth model (A), appears perhaps less natural but can reflect situations in which there are spatial or resource limitations and has been analyzed in the mathematical literature (e.g., Schweinsberg 2010). We refrain from more complicated scenarios, such as models with different epochs of exponential growth and recent models including superexponential growth (Reppell et al. 2014), which indeed could be investigated with similar methods.
Regarding statistical methodology, one could construct likelihood-based tests on the full two-dimensional parameter spaces for Π and θ given by ΘE and ΘB, as outlined in Materials and Methods, but this likely would yield considerable computational challenges. Instead, we opted to employ approximate likelihood methods based on the fixed-s method as, e.g., done by Ramos-Onsins and Rozas (2002), reducing our test to a one-dimensional situation, where Equation 14 does not depend on θ at all.
Based on this method, we derive an approximate likelihood-ratio test based on a Poissonization of the SFS via Equation 12 for interval hypotheses, including large ranges of parameters such as the growth parameter β in model E and the coalescent parameter α in model B. By considering the power of our test, a key result in this setup is that even for moderate sample sizes, B and E can be distinguished reasonably well for substantial parts of the parameter space of α and β.
A well-known criticism of this method is its sensitivity on the true yet unknown coalescent mutation rate θ (cf. Markovtsova et al. 2001). We checked by rejection sampling (cf. File S1), conditioning on S = s, that for various fixed values of (Π, θ) the rejection probability in our proposed test would be reasonably close to the true rejection probability as long as the true θ is close enough to the Watterson estimate , in line with similar observations (for different test statistics) made by Wall and Hudson (2001).
Additional information about the exact coalescent and growth parameters could lead one to test the point hypotheses (e.g., B with fixed α vs. E with fixed β). Indeed, in this case, higher power can be achieved, even for relatively small numbers of segregating sites (s = 20), as expected (data not shown).
Distance plots (e.g., Figure 3) over two-dimensional parameter ranges indicate a one-dimensional curve along which the minimal distance is reached. Note that both approaches [maximum (approximate) likelihood and minimum distance] could be linked if asymptotic normality of our estimators could be established—this is a theoretical question for future work.
Finally, we consider decision rules for the normalized spectrum associated with models A, B, E, and D based on a simple rejection-based ABC analysis. More sophisticated techniques are available [see Beaumont (2010) for an overview] that may improve the prediction accuracy. Empirical misclassification probabilities show, for a reasonable sample size of n = 200, at least moderate success in distinguishing among the four model classes even for as few as s = 15 segregating sites. Note, though, that depending on the model class comparison to be performed, reasonable error probabilities may be achieved only at higher mutation rates (a higher number s). This indicates that the genealogies produced by the different model classes (at least for suitable sample sizes) are different enough to be distinguished but that mutation rates have to be high enough that these differences are mirrored in the SFS.
In practice, our results could be used to design studies that allow one to distinguish between different conjectured scenarios with suitable power. For example, in marine species, such as Atlantic cod (cf., e.g., Birkner et al. 2013b) and Pacific oysters (cf. Sargsyan and Wakeley 2008), it has been suggested that certain multiple-merger coalescents could be more appropriate to describe underlying genealogies, and a reproductive mechansism (HFSOD) for population models has been proposed. We have put this to a test by performing an ABC model comparison among our four model classes for the Atlantic cod data of Árnason (2004). The model comparison clearly rejected both Dirac coalescents and algebraic growth as potential models.
While our ABC analysis indicates that exponential growth is slightly favored over the beta(2 − α, α)-coalescent, evidence is not really strong enough to rule out the latter model class. This may indicate that the SFS information of the Árnason (2004) data does not have enough polymorphic sites to distinguish between E and B clearly (our posterior predictive checks revealed that likely neither model class explains the data completely). However, rejection of the D and A model classes suggests that models that predict star-shaped genealogies do not fit the data well. Árnason (2004) used a maximum likelihood estimation method (Kuhner et al. 1998) and standard tests of neutrality (Tajima, 1989b; Fu, 1997) to rule out exponential population growth.
At this point, we would like to point out that while our methods and results are exemplified in certain special coalescent and growth models, they could be modified to cover different frameworks.
Before ending the discussion, we wish to comment on a few interesting side issues that appeared during the analysis.
Nonmonotonicity of the power function:
At first glance, the observed nonmonotonicity of the power function in the exponential growth parameter β when compared with certain multiple-merger coalescents (cf., e.g., Figure 2) may appear strange. However, the following example may suggest a heuristic way to understand such behavior in a relatively simple special case. Suppose that one wants to distinguish between an exponential growth model and a multiple-merger coalescent with a substantial Kingman component and a small weight on large multiple mergers (e.g., as in the Dirac coalescent with ψ close to 1). This means that most of the time the multiple-merger coalescent will behave like a Kingman coalescent (producing frequent binary mergers), but with a small rate, comprehensive multiple mergers may occur. Certainly, when the growth parameter β is small, the exponential growth model will yield a pattern of variability close to a Kingman coalescent, and hence the power of a test to distinguish between both will be small if the Kingman component has a weight close to 1. As β increases, the power to distinguish from a Kingman coalescent will increase, in line with intuition. However, as β becomes very large, lineages will coalesce after a very short time in the exponential growth model. Such a scenario is certainly different from a Kingman coalescent but could produce patterns of variability closer to a multiple-merger coalescent with a drastic merger after a very short time. Seeing such a merger in the very recent past has some cost (according to the weight of the Dirac component near 1) but appears more likely than observing large amounts of Kingman-like mergers within a unnaturally short time interval, thus leading to a relative decrease in power of associated test. This last effect is nicely illustrated by the upper-right scenario in Figure 4 in the case of a large (algebraic) growth parameter.
Effect of lumping:
It is intriguing to see that using the complete nSFS as summary statistics in the ABC approach can yield higher errors than using intermediate (resp. strong) lumpings of the nSFS. A possible explanation is as follows: consider the approximate likelihood function (Equation 12). Assume that the distribution of the SFS is approximately composed of independent Poisson distributions with parameter for i ∈ [n − 1]. For a Poisson-distributed random variable X with parameter κ, we have , thus showing that smaller Poisson parameters yield a higher amount of variation relative to their expected value. Hence classes in the SFS with small underlying branch lengths (which tend to be in the right tail of the SFS) and/or a low mutation rate show relatively more variation compared with their contribution to the total number of mutations than those with longer branches or if the mutation rate is higher. Lumping such classes together, under Equation 12, yields again a Poisson-distributed lumped class but with the Poisson parameter being the sum of parameters from the classes lumped together. Thus, the variation within this class relative to its contribution to the total number of mutations is reduced by lumping. If different coalescent models show different mean behavior of (lumped) classes, lumping reduces noise and thus increases the chance to correctly identify the underlying model. Naturally, this effect is weakened by higher mutation rates and/or higher sample size n [e.g., consider the limit results for the SFS in Berestycki et al. (2014) and Kersting and Stanciu (2015)].
Thus, using an appropriate weighing of the variables in the nSFS (resp. SFS) should improve the power to distinguish between model classes. It also would be a worthwhile future study to see whether a one-dimensional summary of the SFS similar to Tajima’s D or Fay and Wu’s H, as described in Achaz (2009), could yield a similar or even higher power to distinguish between the model classes than the complete (possibly reweighted) nSFS.
Supplementary Material
Acknowledgments
F. Freund thanks Luca Ferretti and Guillaume Achaz (SMILE, Collège de France, Paris) for discussions about the site-frequency spectrum. The authors thank two anonymous referees whose insightful comments and constructive criticism helped to improve the presentation. J. Blath and B. Eldon were supported by Deutsche Forschungsgemeinschaft (DFG) grant BL 1105/3-1 and M. Birkner by DFG grant BI 1058/2-1 as part of the SPP Priority Programme 1590.
Footnotes
Supporting information is available online at http://www.genetics.org/lookup/suppl/doi:10.1534/genetics.114.173807/-/DC1
Communicating editor: Y. S. Song
Literature Cited
- Achaz G., 2009. Frequency spectrum neutrality tests: one for all and all for one. Genetics 183: 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Árnason E., 2004. Mitochondrial cytochrome b variation in the high-fecundity Atlantic cod: trans-Atlantic clines and shallow gene genealogy. Genetics 166: 1871–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baragatti M., Pudlo P., 2014. An overview on approximate Bayesian computation. ESAIM Proc. 44: 291–299. [Google Scholar]
- Beaumont M. A., 2010. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 41: 379–406. [Google Scholar]
- Beckenbach A. T., 1994. Mitochondrial haplotype frequencies in oysters: neutral alternatives to selection models, pp. 188–198 in Non-Neutral Evolution, edited by Golding B. Chapman & Hall, New York. [Google Scholar]
- Berestycki J., Berestycki N., Limic V., 2014. Asymptotic sampling formulae for lambda-coalescents. Ann. Inst. H. Poincaré Probab. Statist. 50: 715–731. [Google Scholar]
- Bertoin J., Le Gall J.-F., 2003. Stochastic flows associated to coalescent processes. Probab. Theory Relat. Fields 126: 261–288. [Google Scholar]
- Bhaskar A., Song Y., 2014. Descartes’ rule of signs and the identifiability of population demographic models from genomic variation data. Ann. Stat. 42: 2469–2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar A., Wang Y. X. R., Song Y. S., 2015. Efficient inference of population size histories and locus-specific mutation rates from large-sample genomic variation data. Genome Res. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkner M., Blath J., 2008. Computing likelihoods for coalescents with multiple collisions in the infinitely many sites model. J. Math. Biol. 57: 435–465. [DOI] [PubMed] [Google Scholar]
- Birkner M., Blath J., Capaldo M., Etheridge A., Möhle M., et al. , 2005. Alpha-stable branching and beta-coalescents. Electron. J. Probab. 10: 303–325. [Google Scholar]
- Birkner M., Blath J., Eldon B., 2013a An ancestral recombination graph for diploid populations with skewed offspring distribution. Genetics 193: 255–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkner M., Blath J., Eldon B., 2013b Statistical properties of the site-frequency spectrum associated with lambda-coalescents. Genetics 195: 1037–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkner M., Blath J., Steinrücken M., 2011. Importance sampling for lambda-coalescents in the infinitely many sites model. Theor. Popul. Biol. 79: 155–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannings C., 1974. The latent roots of certain Markov chains arising in genetics: a new approach. I. Haploid models. Adv. Appl. Probab. 6: 260–290. [Google Scholar]
- Cannings C., 1975. The latent roots of certain Markov chains arising in genetics: a new approach. II. Further haploid models. Adv. Appl. Probab. 7: 264–282. [Google Scholar]
- Carr S. M., Marshall H. D., 2008. Intraspecific phylogeographic genomics from multiple complete mtDNA genomics in Atlantic cod (Gadus morhua): origins of “codmother,” transatlantic vicariance, and midglacial population expansion. Genetics 180: 381–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W.-C., 2011 Overlapping codon model, phylogenetic clustering, and alternative partial expectation conditional maximization algorithm. Ph.D. Thesis, Iowa State University, Ames, IA. Available at: http://gradworks.umi.com/34/73/3473002.html. [Google Scholar]
- Csilléry K., François O., Blum M. G. B., 2012. ABC: an R package for approximate Bayesian computation (ABC). Methods Ecol. Evol. 3: 475–479. [DOI] [PubMed] [Google Scholar]
- Cucala L., Marin J., 2013. Bayesian inference on a mixture model with spatial dependence. J. Comput. Graph. Stat. 22: 584–597. [Google Scholar]
- Depaulis F., Mousset S., Veuille M., 2001. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol. Biol. Evol. 18: 1136–1138. [DOI] [PubMed] [Google Scholar]
- Depaulis F., Veuille M., 1998. Neutrality tests based on the distribution of haplotypes under an infinite-site model. Mol. Biol. Evol. 15: 1788–1790. [DOI] [PubMed] [Google Scholar]
- Donnelly P., Kurtz T. G., 1999. Particle representations for measure-valued population models. Ann. Probab. 27: 166–205. [Google Scholar]
- Donnelly P., Tavaré S., 1995. Coalescents and genealogical structure under neutrality. Annu. Rev. Genet. 29: 401–421. [DOI] [PubMed] [Google Scholar]
- Durrett R., Schweinsberg J., 2005. A coalescent model for the effect of advantageous mutations on the genealogy of a population. Stochastic Processes Appl. 115: 1628–1657. [Google Scholar]
- Eldon B., 2011. Estimation of parameters in large offspring number models and ratios of coalescence times. Theor. Popul. Biol. 80: 16–28. [DOI] [PubMed] [Google Scholar]
- Eldon B., Wakeley J., 2006. Coalescent processes when the distribution of offspring number among individuals is highly skewed. Genetics 172: 2621–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wu C.-I., 2000. Hitchhiking under positive darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X., 1995. Statistical properties of segregating sites. Theor. Popul. Biol. 48: 172–197. [DOI] [PubMed] [Google Scholar]
- Fu Y. X., 1997. Statistical tests of neutrality against population growth, hitchhiking, and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. X., Li W. H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galassi M., Davies J., Theiler J., Gough B., Jungman G., et al. , 2013. GNU Scientific Library Reference Manual, Ed. 3 Free Software Foundation, Boston. [Google Scholar]
- Griffiths R. C., Tavaré S., 1998. The age of a mutation in a general coalescent tree. Stoch. Models 14: 273–295. [Google Scholar]
- Hallatschek O., Neher R.-H., 2013. Genealogies of rapidly adapting populations. Proc. Natl. Acad. Sci. USA 110: 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock D., Pudovkin A. I., 2011. Sweepstakes reproductive success in highly fecund marine fish and shellfish: a review and commentary. Bull. Mar. Sci. 87: 971–1002. [Google Scholar]
- Hein J., Schierup M. H., Wiuf C., 2005. Gene Genealogies, Variation and Evolution. Oxford University Press, Oxford, UK. [Google Scholar]
- Hudson R. R., 1983a Properties of a neutral allele model with intragenic recombination. Theor. Popul. Biol. 23: 183–201. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 1983b Testing the constant-rate neutral allele model with protein sequence data. Evolution 37: 203–217. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., 1990. Gene genealogies and the coalescent process, pp. 1–44 in Oxford Surveys in Evolutionary Biology, Vol. 7, edited by Futuyma D. J., Antonovics J. Oxford University Press, Oxford, UK. [Google Scholar]
- Hudson R. R., 2002. Generating samples under a wright-fisher neutral model. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Kaj I., Krone S., 2003. The coalescent process in a population with stochastically varying size. J. Appl. Probab. 40: 33–48. [Google Scholar]
- Kass R. E., Raftery A. E., 1995. Bayes factors. J. Am. Stat. Assoc. 90: 773–795. [Google Scholar]
- Kernighan B. W., Ritchie D. M., 1988. The C Programming Language, Ed. 2 Prentice-Hall, Englewood Cliffs, NJ. [Google Scholar]
- Kersting, G., and I. Stanciu, 2015 The internal branch lengths of the Kingman coalescent. Ann. Appl. Probab. (in press).
- Kim J., Mossel E., Rácz M. Z., Ross N., 2015. Can one hear the shape of a population history? Theor. Popul. Biol. 100: 26–38. [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982a The coalescent. Stochic Processes Appl. 13: 235–248. [Google Scholar]
- Kingman J. F. C., 1982b Exchangeability and the evolution of large populations, pp. 97–112 in Exchangeability in Probability and Statistics, edited by Koch G., Spizzichino F. North-Holland, Amsterdam. [Google Scholar]
- Kingman J. F. C., 1982c On the genealogy of large populations. J. Appl. Probab. 19: 27–43. [Google Scholar]
- Koskela J., Jenkins P., Spanò D., 2015. Computational inference beyond Kingman’s coalescent. J. Appl. Probab. (in press).
- Kuhner M. K., Yamato J., Felsenstein J., 1998. Maximum likelihood estimation of population growth rates based on the coalescent. Genetics 149: 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsova L., Marjoram P., Tavaré S., 2001. On a test of Depaulis and Veuille. Mol. Biol. Evol. 18: 1132–1133. [DOI] [PubMed] [Google Scholar]
- Möhle M., 1998. Robustness results for the coalescent. J. Appl. Probab. 35: 438–447. [Google Scholar]
- Möhle M., Sagitov S., 2001. Classification of coalescent processes for haploid exchangeable coalescent processes. Ann. Probab. 29: 1547–1562. [Google Scholar]
- Myers S., Fefferman C., Patterson N., 2008. Can one learn history from the allelic spectrum? Theor. Popul. Biol. 73: 342–348. [DOI] [PubMed] [Google Scholar]
- Neher R. A., Hallatschek O., 2013. Genealogies of rapidly adapting populations. Proc. Natl. Acad. Sci. USA 110: 437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., 2001. Coalescent theory, pp. 179–212 in Handbook of Statistical Genetics, Ed. 2, edited by Balding D. J., Bishop M. J., Cannings C. John Wiley & Sons, Chichester, UK. [Google Scholar]
- Pitman J., 1999. Coalescents with multiple collisions. Ann. Probab. 27: 1870–1902. [Google Scholar]
- Polanski A., Kimmel M., 2003. New explicit expressions for relative frequencies of single-nucleotide polymorphisms with application to statistical inference on population growth. Genetics 165: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Seielstad M. T., Perez-Lezaun A., Feldman M. W., 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Ramírez-Soriano A., Ramos-Onsins S. E., Rozas J., Calafell F., Navarro A., 2008. Statistical power analysis of neutrality tests under demographic expansions, contractions and bottlenecks with recombination. Genetics 179: 555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Onsins S. E., Rozas J., 2002. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19: 2092–2100. [DOI] [PubMed] [Google Scholar]
- R Core Team , 2012. R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Reppell M., Boehnke M., Zöllner S., 2014. The impact of accelerating faster than exponential population growth on genetic variation. Genetics 196: 819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödelsperger C., Neher R. A., Weller A. M., Eberhardt G., Witte H., et al. , 2014. Characterization of genetic diversity in the nematode pristionchus pacificus from population-scale resequencing data. Genetics 196: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A. R., Harpending H. C., 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9: 552–569. [DOI] [PubMed] [Google Scholar]
- Rubin D. B., 1984. Bayesian justifiable and relevant frequency calculations for the applied statistician. Ann. Stat. 12: 1151–1172. [Google Scholar]
- Sagitov S., 1999. The general coalescent with asynchronous mergers of ancestral lines. J. Appl. Probab. 36: 1116–1125. [Google Scholar]
- Sano A., Tachida H., 2005. Gene genealogy and properties of test statistics of neutrality under population growth. Genetics 169: 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargsyan O., Wakeley J., 2008. A coalescent process with simultaneous multiple mergers for approximating the gene genealogies of many marine organisms. Theor. Popul. Biol. 74: 104–114. [DOI] [PubMed] [Google Scholar]
- Sawyer S. A., Hartl D. L., 1992. Population genetics of polymorphism and divergence. Genetics 132: 1161–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweinsberg J., 2003. Coalescent processes obtained from supercritical Galton-Watson processes. Stochastic Processes Appl. 106: 107–139. [Google Scholar]
- Schweinsberg J., 2010. The number of small blocks in exchangeable random partitions. ALEA Lat. Am. J. Probab. Math. Stat. 7: 217–242. [Google Scholar]
- Simonsen K. L., Churchill G. A., Aquadro C. F., 1995. Properties of statistical tests of neutrality for DNA polymorphism data. Genetics 141: 413–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M., Hudson R. R., 1991. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics 129: 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrücken M., Birkner M., Blath J., 2013. Analysis of DNA sequence variation within marine species using beta-coalescents. Theor. Popul. Biol. 87: 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehr, J., P. Pudlo, and L. Cucala, 2014 Geometric summary statistics for ABC model choice between hidden Gibbs random fields. arXiv:1402.1380 [Math. ST].
- Tajima F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989a The effect of change in population size on DNA polymorphism. Genetics 123: 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989b Statistical methods for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., Balding D. J., Griffiths R. C., Donnelly P., 1997. Inferring coalescence times from DNA sequence data. Genetics 145: 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellier A., Lemaire C., 2014. Coalescence 2.0: a multiple branching of recent theoretical developments and their applications. Mol. Ecol. 23: 2637–2652. [DOI] [PubMed] [Google Scholar]
- Wakeley J., 2007. Coalescent Theory, Roberts & Company, Greenwood Village, CO. [Google Scholar]
- Wall J. D., Hudson R. R., 2001. Coalescent simulations and statistical tests of neutrality. Mol. Biol. Evol. 18: 1134–1135. [DOI] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 1539–1546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.