Abstract
Xenopus has become an important tool for dissecting the mechanisms governing craniofacial development and defects. A method to quantify orofacial development will allow for more rigorous analysis of orofacial phenotypes upon abrogation with substances that can genetically or molecularly manipulate gene expression or protein function. Using two dimensional images of the embryonic heads, traditional size dimensions-such as orofacial width, height and area- are measured. In addition, a roundness measure of the embryonic mouth opening is used to describe the shape of the mouth. Geometric morphometrics of these two dimensional images is also performed to provide a more sophisticated view of changes in the shape of the orofacial region. Landmarks are assigned to specific points in the orofacial region and coordinates are created. A principle component analysis is used to reduce landmark coordinates to principle components that then discriminate the treatment groups. These results are displayed as a scatter plot in which individuals with similar orofacial shapes cluster together. It is also useful to perform a discriminant function analysis, which statistically compares the positions of the landmarks between two treatment groups. This analysis is displayed on a transformation grid where changes in landmark position are viewed as vectors. A grid is superimposed on these vectors so that a warping pattern is displayed to show where significant landmark positions have changed. Shape changes in the discriminant function analysis are based on a statistical measure, and therefore can be evaluated by a p-value. This analysis is simple and accessible, requiring only a stereoscope and freeware software, and thus will be a valuable research and teaching resource.
Keywords: Developmental Biology, Issue 93, Orofacial quantification, geometric morphometrics, Xenopus, orofacial development, orofacial defects, shape changes, facial dimensions
Introduction
Among the most prevalent and devastating types of human birth defects are those affecting the mouth and face, such as orofacial clefts1. Children with malformed orofacial structures undergo multiple surgeries throughout their lifetime and struggle with facial disfigurements, speech, hearing and eating problems. Therefore, facilitating new research in cranio- and orofacial development is paramount to prevention and treatment of these types of birth defects in humans. Xenopus laevis has emerged as a new tool for dissecting the mechanisms governing craniofacial development (some examples include2,3,4-11). Therefore, a quantitative method to analyze size and shape changes during development of the head and face of this species could be very powerful 3.
Here, we present such a method; combining traditional size measurements with geometric morphometrics adapted from a Xenopus study12 and a wealth of studies analyzing human facial form13-15. The goal of this protocol is to allow researchers to quantify facial size and shapes to distinguish between different orofacial phenotypes during normal and abnormal development. This analysis will allow for better differentiation between subtle craniofacial defects such as those arising from synergistic effects of genes and/or environmental factors. Additionally, this quantification method could also reveal even slight improvement or rescue of an orofacial defect. This therefore makes it a useful guide in analyzing potential therapeutics.
The combination of facial measurements and geometric morphometrics that we present here allows for a more comprehensive statistical analysis of both size and shape of the orofacial region than current protocols which largely utilize only one or the other15-18. Further, we present a simple way to assess both the medial and lateral planes of the face without requiring sophisticated three-dimensional imaging equipment used in current studies13,19.
We demonstrate this protocol on Xenopus laevis embryos treated with a retinoic acid receptor inhibitor that induces abnormal orofacial development and a median cleft palate2,3. Quantification of the dimensions and shape of the orofacial region in these embryos has revealed changes in the midface that is analogous to humans with similar palatal clefts and mouse models 20,21. However, this protocol can be utilized to assess the effects of other compounds on orofacial development such as natural substances, herbicides, or proteins such as growth factors. Further, orofacial size and shape changes arising from perturbation of gene expression via loss or gain of function experiments (using antisense morpholinos or Crispers/Talens) can also be quantified using this protocol. Finally, we developed this method specifically to assess Xenopus morphology; however, it is easily modified for analysis of any vertebrate. Other applications could also include using this protocol for comparing closely related species for evolutionary or ecological studies. While the example we provide here utilizes this protocol to describe analysis of the orofacial region, it could easily be modified for analysis of other regions, organs, or structures.
This orofacial quantification protocol will become a valuable resource for the research community, as well as an excellent teaching tool for undergraduate students as a video demonstration.
Protocol
All experiments performed using Xenopus laevis have been approved by IACUC (protocol #AD20261) .
1. Preparing Reagents and Required Materials
- Reagents:
- Make 1 L of 10x MBS (Modified Barth’s Saline) solution22. Add NaCl (880 mM), KCl (10 mM), MgSO4 (10 mM), HEPES (50 mM, pH 7.8), and NaHCO3 (25 mM) to 1 L of distilled water. Adjust pH to 7.8 with NaOH.
- Make 1 L of 1x MBS by diluting 100 ml of 10x MBS solution in 900 ml of distilled water. Add 0.7 ml of 1 M CaCl2 solution.
- Make 1 L of 0.1x MBS by diluting 100 ml of 1x MBS solution in 900 ml of distilled water. To 1 L of 0.1x MBS solution, add 1 ml of 10 mg/ml gentamicin solution for use in embryo culture.
- Make high salt MBS by dissolving 4 ml of 5 M NaCl in 100 ml of 10x MBS. Then add distilled water until total volume is 1 L.
- Make 70% ethanol by diluting 100% ethanol to 70% ethanol using distilled water.
- Make BMS-543 by preparing a 10 mM stock solution in dimethyl sulfoxide (DMSO).
- Make 4% paraformaldehyde (PFA) by adding 4 g of paraformaldehyde powder to 50 ml distilled water. Heat on a hot plate to approximately 65 °C, at which point add 5 M NaOH drop-wise until solution clears.
- Once the solution clears, remove from heat and add 50 ml 2x PBS and 100 µl Tween-20. Allow to cool to RT on ice. Adjust pH to between 7.2 and 7.5 using HCl.
- Make a phosphate buffered saline solution with Tween (PBT) by dissolving 8 g NaCl, 0.2 g KCl, 1.44 g of Na2HPO4, and 0.24 g of KH2PO4 in 800 ml distilled water. Using HCl, adjust the pH to 7.4. Add 2 ml of Tween-20. Adjust final volume with distilled water to equal 1 L.
- Make a cysteine solution by dissolving 1 g of cysteine in 50 ml distilled water or 0.1x MBS. Add 1.5 ml of 5 M NaOH and adjust the pH to 7.8.
- Make a 10% Benzocaine stock solution by adding 10 g of Benzocaine powder to 100 ml 100% EtOH. For euthanasia of adult males and embryos ready for fixation, dilute to a 0.05% solution.
- Make a testes storage solution by adding 1 ml of calf serum to 9 ml of L15 buffer.
- Required Tools and Equipment:
- Obtain the equipment listed in the Table of Materials and Equipment, shown in Figure 1.
- Modify a pipette tool. Place the tip of a glass Pasteur pipette into a flame from a Bunsen burner. Rotate the pipette tip until it melts and forms a rounded sealed end.
- Flatten modeling clay into the bottom of a plastic Petri dish, smoothing out to cover the entire plate (any size can be used) (Figure 1Aiv).
- Lightly press a straight teasing needle into the clay-lined dish at a 45° angle. Hold the needle there, and slowly move the Petri dish horizontally to create a depressed line (Figure 1Bi). Repeat for the desired number of rows.
- Use the end of a glass pipette tool to make shallow, circular depressions in each row of the clay-lined dish to place embryo heads (Figure 1Bii) and aid in organizing the embryos while photographing.
- For photography, fill dish with PBT (Figure 1Biii). Between uses, thoroughly wash the dish and clay surface with sterile water, followed by 70% ethanol.
2. Xenopus laevis Embryo Culture and Inhibitor Treatments
- In vitro fertilization and culture of Xenopus eggs:
- Obtain and culture Xenopus laevis embryos using standard published methods22,23.
- Briefly, euthanize an adult male by submersion in a 0.05% Benzocaine solution. Dissect testes and store in testes storage buffer. Inject adult females with human chorionic gonadotropin (HCG), collect eggs in high salt MBS, and fertilize in vitro with dissected testes (Figure 2A,B).
- Culture embryos in 0.1x MBS at 15 °C until the desired stage is reached (stage embryos according to the Normal table of Xenopus laevis by Nieuwkoop and Faber24,25).
- Here, culture embryos until stage 22 (24 hr post fertilization (hpf)), and remove the vitelline membrane under a stereomicroscope using two pairs of sharpened forceps. Transfer embryos with a standard, disposable transfer pipette to a clean Petri dish (100 mm in diameter) containing 30-40 ml of 0.1x MBS.
- Inhibitor treatments of Xenopus embryos (illustrated in Figure 2):
- Fill the necessary wells of a 24-well culture dish with 1 ml of 0.1x MBS using a calibrated pipette-man. Demarcate the outside of the well with a fine point marker (Figure 2D, inset). Use the marker to also label the contents to be added to each well on the 24-well culture dish lid. Copy this information onto a 24-well set-up page in the laboratory notebook.
- Transfer 5-10 embryos into each well using a standard, disposable transfer pipette (Figure 2C). Place the same number of embryos in each well.
- Top up or remove the 0.1x MBS so that it is level with the 1 ml marked line (Figure 2D). Add the appropriate volume of DMSO (so that the final volume of DMSO is 1%) and gently swirl. Be careful not to add DMSO too close to the embryos, and avoid contact of the embryos with the surface of the buffer.
- Add the appropriate volume of chemical to the designated wells and gently swirl again. Here, add 1 μl of 10 mM BMS-453 (a retinoic acid receptor antagonist) and 9 µl of DMSO to 1 ml of 0.1x MBS.
- If the chemical is light sensitive, loosely wrap the culture dish in aluminum foil. Culture embryos in a 15 °C incubator until the desired stage. In this example, treat embryos for 16-18 hr.
- Washing embryos out of the treatment and rearing until tadpole stages:
- Remove one-half to three-quarters of the solution with a disposable transfer pipette. Be careful not to disturb the embryos. Using a new transfer pipette, refill the well with 0.1x MBS. Repeat this 3-6 times.
- Transfer embryos to a large Petri dish (150 mm in diameter) containing 100 ml of 0.1x MBS with gentamicin (1 ml/L). Incubate the embryos at 15 °C until they reach stage 42-45 (82-100 hpf).
- Change the 0.1x MBS solution containing gentamicin daily, and remove dead embryos promptly to prevent contamination or death of other embryos. Record the number of dead embryos in laboratory notebook.
- Fixing embryos in paraformaldehyde (PFA):
- Fix embryos between stages 42 and 45 (82 and 100 hpf, respectively) by transferring them into a new 24-well culture dish containing 1-2 ml of 0.1x MBS. Label the lid with the same information as the treatment set-up described above (section 2.2.1).
- Remove as much 0.1x MBS from each well as possible, while still leaving embryos covered and ensuring minimal contact to prevent injury. Add 1-2 ml of 0.05% Benzocaine solution in 0.1x MBS and incubate until embryos are no longer responsive to stimuli.
- Add 1-2 ml of 4% PFA. Alternatively, place embryos in labeled microcentrifuge tubes for fixation, if preferred.
- Incubate embryos in PFA for 24 hr at 4 °C on a rotating shaker. Remove PFA by performing 3-4 washes with PBT over a 3-5 hr period.
3. Photographing the Orofacial Region of Xenopus Tadpoles
- Pre-photography preparation (this is illustrated in Figure 3):
- Place fixed embryos in a large Petri dish (150 mm in diameter) containing approximately 100 ml of PBT.
- Make two incisions to sever the head from the body (Figure 3A, solid black lines). First, hold the embryo with a pair of forceps and make an incision on the posterior side of the gut with a sterile scalpel (Figure 3B). Make a second incision on the anterior side of the gut, near the heart, to completely remove the head (Figure 3C).
- Pipette severed embryo heads into a clay-lined dish (part 1.2.2) using a standard, disposable transfer pipette.
- For frontal views, use two pairs of forceps to position embryo heads posterior side down inside the circular depressions. Using the forceps to manipulate both the embryo head and the surrounding clay, position embryos such that they are facing the camera and are not tilted backward or to either side (Figure 3D). Gently push the surrounding clay around the head using forceps and/or the glass pipette tool to secure the head in place.
- For lateral views, use forceps to position embryo heads inside the circular depressions such that they are on their side, facing the same direction, and are flat against the clay. Manipulate the embryo head and the surrounding clay so that the cement glands of all the embryos are positioned downward at the same angle (Figure 3E). Use the rows drawn with the teasing needle as a guide to ensure accuracy when taking lateral measurements.
- Capturing images of the tadpole head:
- Photograph the head of the tadpole using any dissecting microscope with an attached digital camera and the corresponding camera software. Use the highest magnification that can be kept constant for all embryos.
- Center embryos and face them all in the same direction. Capture images using the best lighting conditions that allow for the most accurate image of the orofacial region. Position lights to avoid excessive shadows. Save images as .tiff files with the highest resolution possible.
4. Measuring and Analyzing Facial Size Dimensions in Xenopus Tadpoles
- Measure orofacial dimensions (summarized in Figure 4):
- Determine the face width. Identify the points at which the ventral part of each eye meets the peripheries of the face (Figure 4A, arrows), and measure the distance between these points (Figure 4A, red line).
- Determine the face height. First, determine the midline by measuring the distance between each eye and calculating the halfway mark. Next, draw horizontal lines that mark the dorsal most edges of the eyes and the dorsal most point of the cement gland (Figure 4B, white lines). At the midline, measure the distance between the horizontal lines (Figure 4B, red line).
- Determine the orofacial area. Use the same horizontal lines marking the dorsal edges of the eyes and cement gland to guide area measurement (Figure 4C, white lines). Start at the point where the horizontal line marking the top of the cement gland meets the periphery of the left side of the face (Figure 4Ca).
- Trace along the edge of the face encompassing the eye until the point where the horizontal line marking the top of the eyes meets the periphery of the face (Figure 4Cb). Continue to trace along this dorsal most horizontal line until the point where it meets the periphery of the right side of the face (Figure 4Cc).
- Trace downwards along the edge of the face encompassing the eye until the point where the ventral most horizontal line marking the top of the cement gland meets the periphery of the right side of the face (Figure 4Cd). Continue to trace along this bottom horizontal line until it meets the point where tracing began. Use photo editing software to calculate the area inside the tracing.
- Determine the snout length. On lateral images, make a vertical line that marks the anterior of the eye. Next, measure the horizontal distance from the anterior most point where the face meets the dorsal edge of the cement gland to the vertical line marking the anterior of the eye (Figure 4D).
- Determine the mouth width. Measure the horizontal distance between left and right points where the top and bottom “lips” of the mouth opening meet (Figure 4E). These could be considered the frog equivalent of the labial commissures.
- Determine the mouth roundness. Calculate mouth roundness as an inverse aspect ratio where (4 × [Area]) / (π × [Major axis]2).
- Use the Lasso tool in a photo-editor to trace the edges of the mouth opening as shown in Figure 4F. Select Analyze in the main toolbar, and choose Measure. Alternatively, calculate roundness from the measurement of area and diameter of the mouth opening.
- Data analysis of facial dimensions:
- Input the measurements of facial dimensions into a data analysis program to compare the treatment group with the control. Determine the mean of each measurement for both experimental and control groups to create bar graphs. Determine standard deviation in order to create error bars. Perform Student’s t-tests to statistically compare groups. NOTE: This data can be quite variable due to the slightly different rates of development among embryos.
5. Quantitative Analysis of Orofacial Shape and Morphometrics
- Place landmarks and create landmark coordinate file (illustrated in Figure 5A).
- Choose Landmarks such that they capture the shape of the target image, and can be repeatedly placed in the same relative location in all samples being analyzed. If a structure does not exist in all samples, do not place a landmark on this structure.
- Here, place 24 landmarks to represent the midface with distinguishing marks such as the eyes, nostrils, and mouth. For consistency, place landmarks halfway between previously placed landmarks (for example, mouth landmarks, Figure 5Ai).
- Capture landmark coordinates and create “landmark coordinate file”. Open the first image of a tadpole face (for example, the first control embryo).
- Select the Plugins tab on the main toolbar and choose Pointpicker from the dropdown menu. Select the Add Points tab and place landmarks in desired locations as landmarks will appear as multicolored crosses (Figure 5Ai).
- Move landmark locations after placement by selecting the Move crosses tool. When all landmarks have been placed on the image, select the Display Results Tab on the main toolbar and choose Show (Figure 5Aii) to display landmark coordinates and copy into a spreadsheet program (Figure 5Aiii).
- When pasting these coordinates from this first embryo into a spreadsheet program, leave an empty row above the data. In column B of this empty row (row 1), type “LM= ” with the number of landmarks in the sample. Here, enter “LM= 24” since 24 landmarks were created (Figure 5Aiii, red box).
- In the row below the data points, type “ID=” with a name identifying the sample. Here in Figure 5Aiii, enter “ID=CON1” since this is the landmark coordinate data of the first control embryo (Figure 5Aiii, red arrow).
- Repeat this process with faces of every embryo in the experiment (experimental and controls) until all landmark coordinates have been entered into the spreadsheet. NOTE: It is important that each embryo is given a unique name in the “ID=” row. Here for example, the next set of landmark coordinates is given the ID of “CON2” since it was the second embryo in the control group.
- Copy the entire second and third columns into a text document and save as a .txt file.
- Set up the morphometric software program for data analysis (illustrated in Figure 5B).
- Here, select Create New Data Set on the main menu in the morphometric software program. Name the file and the data set in the appropriate boxes (for example, “Retinoic Acid Inhibition” and “Facial landmarks”, respectively) in the pop up screen (Figure 5Bi). Import the file as a TPS, and select the text file of coordinates to create the dataset (Figure 5Bi, red arrow).
- Data alignment and procrustes fit:
- Perform a procrustes fit and alignment. Highlight the data set in the Project Tree tab, choose Preliminaries on the main toolbar, and New Procrustes Fit in the dropdown menu.
- Choose Align by Principal Axis and select Perform Procrustes Fit at the bottom of the box (Figure 5Bii, red arrow). Return to the Preliminaries drop down menu, select Generate Covariance Matrix (Figure 5Biii), and select Execute.
- View the correlations of the Covariance Matrix in the Results tab.
- Create a “classifier file” by assigning each sample in the data set to the appropriate classifier variable to distinguish whether it belongs in the experimental group or the control group.
- Label two columns in a spreadsheet. Label the header of the first column with “ID” and input the same ID’s given for each sample (for example, CON1, RARINHIB1, and so on; Figure 5Biv). Label the header of the second column with “TREATMENT” and input into each cell the treatment group that corresponds to the sample listed in the adjacent ID cell.
- Here, use “CONTROL and RAR INHIBITOR” labels as classifiers (Figure 5Biv). Copy into a text file and save as a .txt file. Import Classifier Variables into the morphometric software program. Choose Match by Identifier, and open the text file (Figure 5Bv).
- Create a principal component analysis (PCA) scatter plot by selecting the Variation dropdown menu and choosing Principal Component Analysis (Figure 6Ai). Next, select the Graphics tab to see three additional tabs: PC shape changes, eigenvalues, and PC scores (Figure 6Aii). The PC scores tab displays the bivariate principal component scatterplot of the Procrustes fit landmark data (Figure 6Aii).
- To change which principal components are plotted, bring up the pop-up menu in the plot space and select the desired axis to change (Figure 6Aiii, red arrow).
- Select the Color Data Points tool (Figure 6Aiii). Select classifiers previously imported (Section 5.2.2) in the second pop-up menu that appears to determine the color of each category (Figure 6Aiv).
- View the amount of variance captured by each principal component in the Results tab (Figure 6Av). Export graph as a .bmp file.
- Create a discriminant function analysis (DFA) transformation grid in the morphometric software program by choosing Discriminant Function Analysis under the Comparison tab (Figure 6Bi). In the pop-up menu, ensure that the appropriate dataset is selected and the type of data is the Procrustes-fit coordinates.
- Highlight the pairs of groups to be compared, and run 1,000 permutation tests (Figure 6Bii).
- Under Graphics, view the vector map of the coordinates in the Shape Difference tab (Figure 6Biii). If the image is inverted, bring up the pop-up menu in the plot space to flip the diagram in the correct orientation (Figure 6Biii).
- The direction of vectors reflects the shape difference from the first group compared to the second, which is displayed in the Discriminant Function Menu prior to running the analysis (Figure 6Bii). To reverse the direction of the vectors, bring up the pop-up menu in the plot space and change the sign of the scale factor so that it is negative (Figure 6Biii,iv). The value of the scale value is set to a factor of ten that best represents the statistical differences in landmark position between the two groups without distortion, and is usually left at the default (Figure 6Biv).
- Also in the pop-up menu, change the graph to a transformation grid to superimpose the vectors onto a grid (Figure 6Biii,v).
- Specify the number of horizontal and vertical lines of the grid in the pop-up menu (Figure 6Biii,v).
- Export the graph as a .bmp file.
- View the Procrustes and Mahalinobis distances and p-values under the Results tab (Figure 6Bvi).
- Additional morphometric analyses that are useful for assessment of more than one treatment group
- Create a principal component analysis transformation grid. The PC Shape Changes tab displays the vector map of shape changes accounting for variance within each sample group. Use the pop-up menu in the plot area to orient the image properly and superimpose vectors onto a transformation grid. Set the scale factor to the value on the x-axis of the PC scores graph that corresponds to the estimated center of the control group. Export the image as a .bmp file.
- Create a CVA scatterplot. Under the Comparison tab on the main toolbar, select Canonical Variate Analysis, and highlight the classifier variable set that was previously imported (see Section 5.2.3). Select 1,000 iterations for permutation tests, and select Execute. Select the CV scores tab to view the bivariate CVA scatterplot. This is color coded based on the uploaded classifiers. Change the color scheme by bringing up the pop-up menu in the plot space (see Section 5.3.3). Export as a .bmp file.
- Create a CVA transformation grid. A transformation grid of the CVA results can be generated when comparing more than two groups. Under the Graphics tab, choose CV Shape Changes to visualize the transformation grid of the landmark coordinates. Bring up the pop- up menu in the plot space to change the direction of the vector, superimpose results onto a transformation grid, and specify the number of gridlines. Export the graph as a .bmp file.
Representative Results
Here, a quantitative analysis of orofacial size and shape was demonstrated to compare embryos treated with a retinoic acid receptor inhibitor (RAR inhibitor) to untreated controls. Embryos were treated with a 1 μM concentration of this chemical inhibitor from stage 24 to 30 (26-35 hpf), washed out, and fixed at stage 42 (82 hpf). They were then processed and analyzed as described in the protocol. Results are original data, but consistent with observations in previous publications2,3. Control embryos were treated with the vehicle, DMSO, and developed normally (Figure 7Ai,ii). Embryos treated with a 1 μM concentration of the RAR inhibitor showed slight narrowing of the face, eye anomalies, and a malformed embryonic mouth opening that was more triangular shaped (Figure 7Aiii,iv).
First, traditional orofacial dimensions were measured and are summarized in Figure 7B. Statistical significance was determined by performing a Student’s t-test assuming unequal variance between inhibitor treated embryos and controls for each measurement. We found that both snout length and face width were significantly decreased in RAR inhibitor treated embryos compared to controls (p-values = 0.0062 and 0.0058, respectively; Figure 7Bi,ii). While face height and mouth roundness were significantly increased (p-values = 3.7772 x 10-6, 1.4812 x 10-7), mouth width was significantly decreased (p-value = 2.5175 x 10-10; Figure 7Biii-v).The results showed no significant difference in the overall orofacial area between the two groups (p-value = 0.3754; Figure 7Bvi). These data show that loss of retinoic acid signaling at a specific time in development results in a shorter snout, slight narrowing of the midface region, and malformation of the embryonic mouth opening.
To provide a sophisticated view of the shape changes of the embryonic orofacial region in response to reduced retinoic acid signals, we next utilized geometric morphometric analyses. After identifying and aligning orofacial landmarks using morphometric analysis software, we then examined the variance within each group via principal component analysis (PCA). When the first two principal components were plotted against each other, RAR inhibitor treated embryos were clearly distinguished from controls along the PC1 axis (Figure 7C). This test also showed the outliers in the sample set- illustrated by two inhibitor treated embryos that did not cluster with the rest of the group (Figure 7C, arrows).
Next, the statistical differences in the shape of the orofacial region between RAR inhibitor treated embryos and controls were assessed and visualized by performing a discriminant function analysis (DFA). The Procrustes distance between the two groups was significantly different (distance = 0.2665, p-value < 0.0001, Figure 7D), indicating a change in orofacial shape when retinoic acid signaling is disrupted. Indeed, dramatic shifts in the position of lateral landmarks in the orofacial region indicate a narrowing of the face shape respective to the height in inhibitor treated embryos (Figure 7D). In addition, the slight outward shift in position of the nasal landmarks (Figure 7D, arrows) reveals the abnormality in nostril position in these embryos that is consistent with decreased outgrowth of the snout. The shifts in landmarks that define the edges of the mouth opening show position changes that reflect the formation of a triangular shaped mouth opening that is consistent with the median cleft reported in our previous studies 2,3. In addition to vector shifts, the warping pattern of the transformation grid also illustrates shape changes in the orofacial region. Warping in the midface region is consistent with the midface hypoplasia and overall facial narrowing seen in embryos with decreased retinoic acid signals (Figure 7D).
The results of the discriminant function analysis (DFA) show shape changes that were consistent with our qualitative analysis, as well as revealing some changes that were not adequately captured by traditional size measurements alone. For instance, while the orofacial area was not significantly different between controls and inhibitor treated embryos (Figure 7Bvi), the DFA transformation grid revealed dramatic changes in this region consistent with the facial narrowing seen in inhibitor treated embryos (Figure 7A,D). Further, the warping pattern and landmark shifts of the mouth opening, coupled with the significant change we saw in mouth roundness, illustrate the malformation of the mouth opening shape in inhibitor treated embryos. In summary, a combination of traditional measurements of facial dimensions and geometric morphometric analysis illustrates the changes in shape and size of the orofacial region when retinoic acid signals are disrupted.
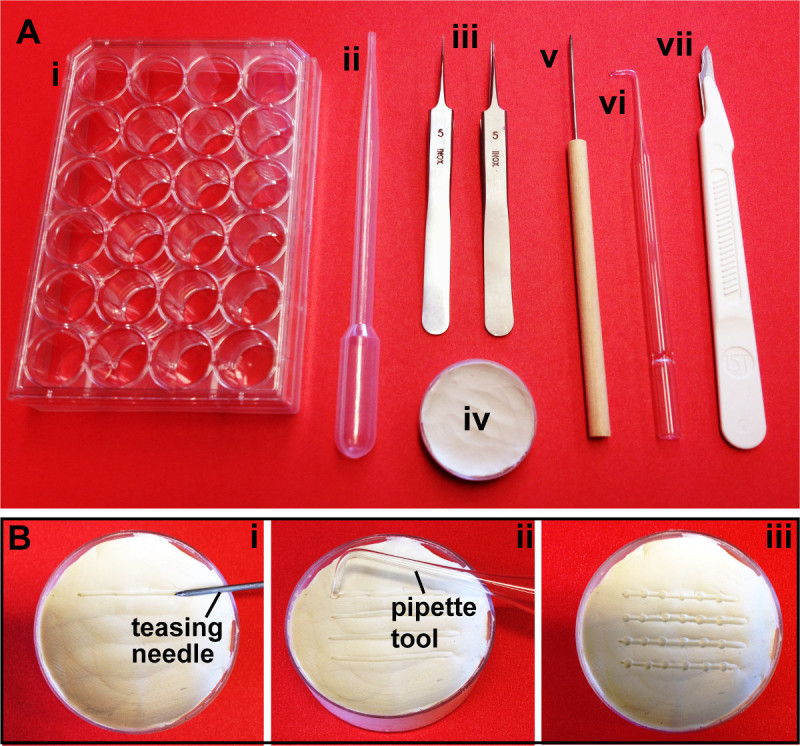 Figure 1. Required materials. (A) Tools for data analysis. (i) 24-well plate, (ii) standard disposable transfer pipette, (iii) Dumont #5 Inox forceps, (iv) clay-lined Petri dish, (v) straight teasing needle, (vi) glass pipette tool, (vii) sterile, disposable scalpel. (B) Preparation of the clay-lined dish. (i) A straight teasing needle is used to draw horizontal lines in the clay. (ii) A glass pipette tool is used to make circular depressions along each row. (iii) The dish is filled with PBT for imaging.
Figure 1. Required materials. (A) Tools for data analysis. (i) 24-well plate, (ii) standard disposable transfer pipette, (iii) Dumont #5 Inox forceps, (iv) clay-lined Petri dish, (v) straight teasing needle, (vi) glass pipette tool, (vii) sterile, disposable scalpel. (B) Preparation of the clay-lined dish. (i) A straight teasing needle is used to draw horizontal lines in the clay. (ii) A glass pipette tool is used to make circular depressions along each row. (iii) The dish is filled with PBT for imaging.
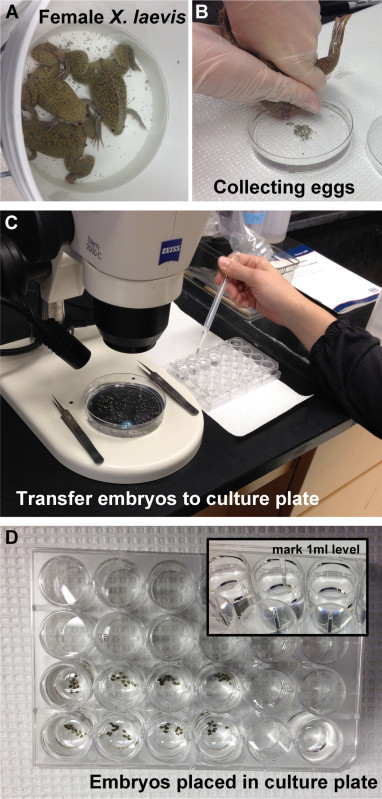 Figure 2. In vitro fertilization and culture of Xenopus eggs. (A) Following HCG injection, adult Xenopus laevis females are induced to lay eggs. (B) Eggs are collected in high salt MBS, fertilized with testes extracted from a male, and cultured using standard methods. (C) Embryos are transferred to a 24-well dish containing 0.1x MBS using a standard, disposable transfer pipette. (D) A calibrated pipette-man is used to measure 1 ml into an empty well, and a marker is used to demarcate this level on the outside of all wells containing embryos (inset). 0.1x MBS is then added or taken away so that it is level with this mark.
Figure 2. In vitro fertilization and culture of Xenopus eggs. (A) Following HCG injection, adult Xenopus laevis females are induced to lay eggs. (B) Eggs are collected in high salt MBS, fertilized with testes extracted from a male, and cultured using standard methods. (C) Embryos are transferred to a 24-well dish containing 0.1x MBS using a standard, disposable transfer pipette. (D) A calibrated pipette-man is used to measure 1 ml into an empty well, and a marker is used to demarcate this level on the outside of all wells containing embryos (inset). 0.1x MBS is then added or taken away so that it is level with this mark.
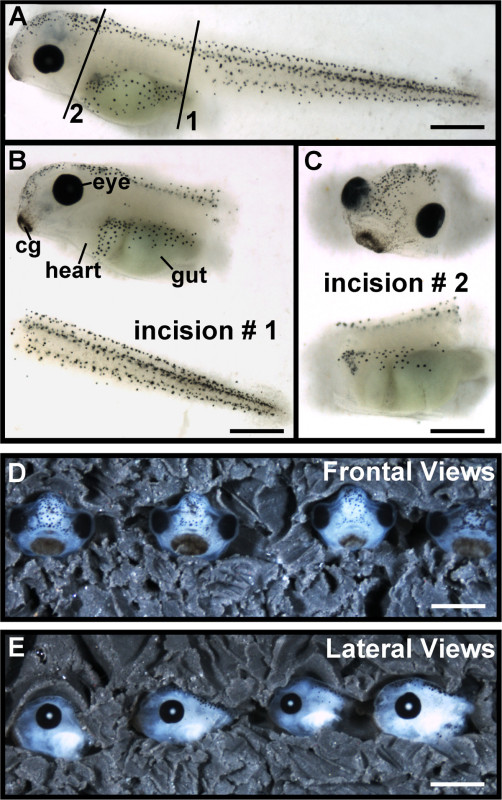 Figure 3. Preparation of embryo heads for imaging. (A) Diagram of the two incisions required to remove heads, solid black lines. Scale bar = 400 μm. (B) The first incision is made at the posterior end of the gut to remove the tail and release pressure from the scalpel. Scale bar = 400 μm. (C) The second incision is made at the anterior end of the gut, near the heart, to completely sever the head. Scale bar = 400 μm. (D) Frontal views of row of embryo heads positioned in clay. Scale bar = 650 µm. (E) Lateral views of row of embryo heads position in clay. Scale bar = 500 µm cg: cement gland.
Figure 3. Preparation of embryo heads for imaging. (A) Diagram of the two incisions required to remove heads, solid black lines. Scale bar = 400 μm. (B) The first incision is made at the posterior end of the gut to remove the tail and release pressure from the scalpel. Scale bar = 400 μm. (C) The second incision is made at the anterior end of the gut, near the heart, to completely sever the head. Scale bar = 400 μm. (D) Frontal views of row of embryo heads positioned in clay. Scale bar = 650 µm. (E) Lateral views of row of embryo heads position in clay. Scale bar = 500 µm cg: cement gland.
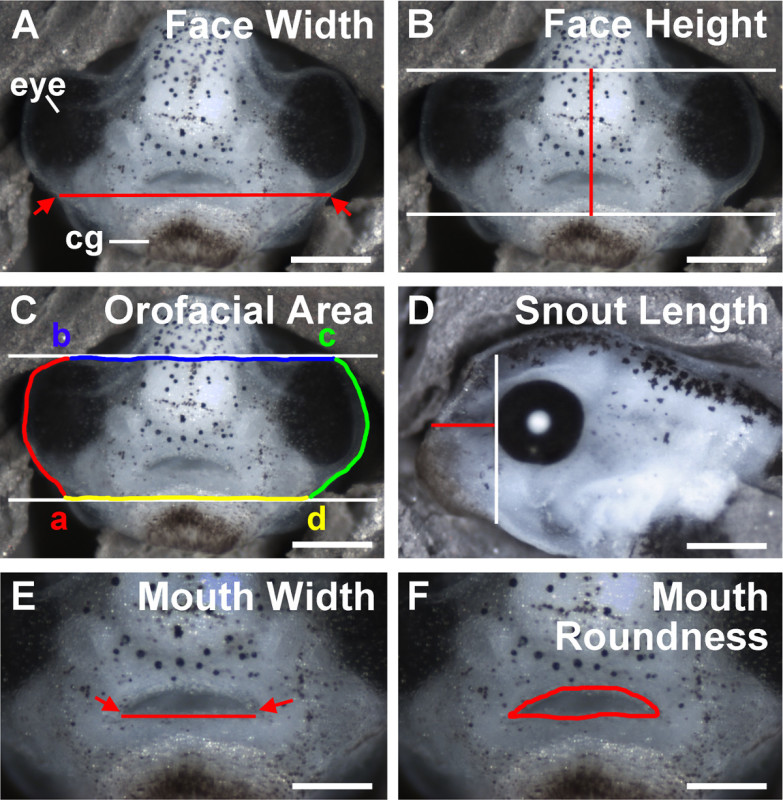 Figure 4. Traditional size measurements of orofacial dimensions. (A)Face width. Arrows indicate the points where the ventral portion of the eye meets the periphery of the face. Red line is the face width, measured as the distance between these points. Scale bar = 210 µM. (B)
Face Height. White lines are guides drawn prior to measurement at the dorsal edge of the eyes and the dorsal edge of the cement gland. Red line is the face height, measured as the distance between these two guides at the midline of the face. Scale bar = 210 µM. (C)
Orofacial area. White lines are guides drawn prior to measurement. (a) Point where the bottom guide meets the ventral edge of the left eye. Red line shows the tracing around the left eye. (b) Point where the dorsal edge of the eye meets the top guide. Blue line shows the dorsal boundary of orofacial area, traced along the top guide at the dorsal edge of the eyes. (c) Point where the top guide meets the right facial periphery. Green line shows the tracing around the right eye. (d) Point where the ventral edge of the right eye meets the bottom guide. Yellow line shows ventral boundary of orofacial area, traced along the bottom guide at the dorsal edge of the cement gland. Scale bar = 210 µM. (D)
Snout length. White line is the anterior edge of the eye and is drawn as a guide prior to measurement. Red line is snout length, measured from this line to the point where the dorsal edge of the cement gland meets the lateral periphery of the face. Scale bar = 300 µM. (E)
Mouth width. Arrows are the points where the dorsal and ventral lips meet. The red line is the mouth width, measured as the distance between these two points. Scale bar = 200 µM. (F)
Mouth roundness. The perimeter of the mouth opening is traced and shown in red. cg: cement gland. Scale bar = 200 µM.
Figure 4. Traditional size measurements of orofacial dimensions. (A)Face width. Arrows indicate the points where the ventral portion of the eye meets the periphery of the face. Red line is the face width, measured as the distance between these points. Scale bar = 210 µM. (B)
Face Height. White lines are guides drawn prior to measurement at the dorsal edge of the eyes and the dorsal edge of the cement gland. Red line is the face height, measured as the distance between these two guides at the midline of the face. Scale bar = 210 µM. (C)
Orofacial area. White lines are guides drawn prior to measurement. (a) Point where the bottom guide meets the ventral edge of the left eye. Red line shows the tracing around the left eye. (b) Point where the dorsal edge of the eye meets the top guide. Blue line shows the dorsal boundary of orofacial area, traced along the top guide at the dorsal edge of the eyes. (c) Point where the top guide meets the right facial periphery. Green line shows the tracing around the right eye. (d) Point where the ventral edge of the right eye meets the bottom guide. Yellow line shows ventral boundary of orofacial area, traced along the bottom guide at the dorsal edge of the cement gland. Scale bar = 210 µM. (D)
Snout length. White line is the anterior edge of the eye and is drawn as a guide prior to measurement. Red line is snout length, measured from this line to the point where the dorsal edge of the cement gland meets the lateral periphery of the face. Scale bar = 300 µM. (E)
Mouth width. Arrows are the points where the dorsal and ventral lips meet. The red line is the mouth width, measured as the distance between these two points. Scale bar = 200 µM. (F)
Mouth roundness. The perimeter of the mouth opening is traced and shown in red. cg: cement gland. Scale bar = 200 µM.
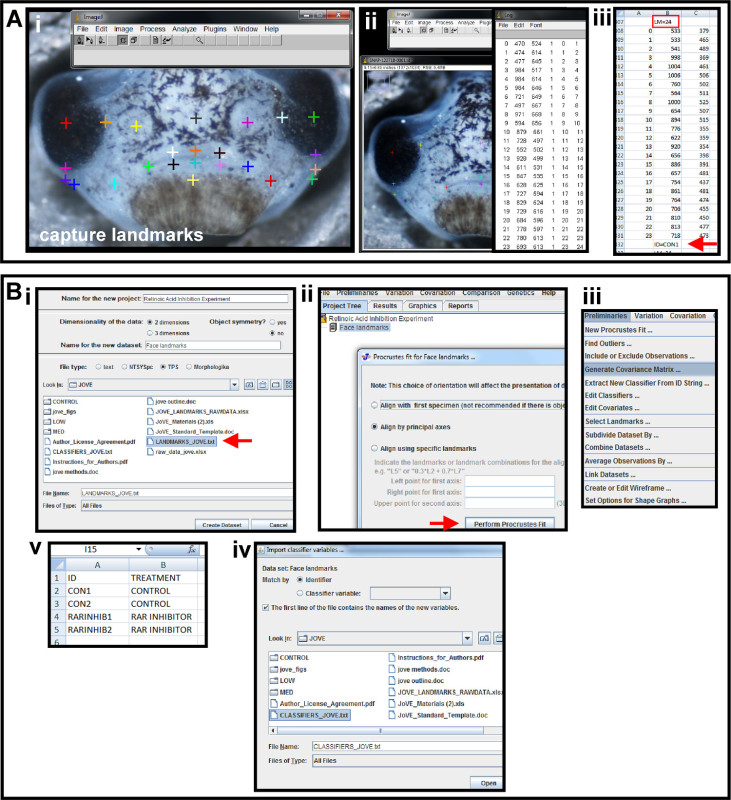 Figure 5. Capturing landmarks and preliminaries for geometric morphometric analysis. (A) Using photo-editing software and a spreadsheet program to place landmarks and capture coordinates. (i) Multicolored crosses are the landmarks placed on the image using the Add points tool in ImageJ to represent the shape of the orofacial region. (ii) Landmark data is displayed by using the Display Results Tool. (iii) Landmark data is copied and pasted into a spreadsheet. Above the second column is a header identifying the number of landmarks and denoted by "LM=24" (red box). Below the second column of data, the sample is given a unique name and denoted by "ID=CON1" (red arrow). This is repeated for all images in a sample set and the data is saved as a text file. (B) Preliminary data analysis in a geometric morphometric software program. (i) The text file created in from the photo-editing software is imported into the morphometric program, MorphoJ, as a TPS file. File is indicated by red arrow. (ii) Landmark coordinate data is aligned by Procrustes fit by principal axes. Red arrow indicates execution of alignment. (iii) A covariance matrix of Procrustes fit landmarks is generated in the Preliminaries menu. (iv) A classifier file is created in a spreadsheet. Column A and B are given headers “ID” and “TREATMENT”, respectively. The ID’s given to each sample in landmark data collection are input under column A, and the treatment group to which each sample belongs is input in column B. (v) The classifier file is imported into the morphometric program as a classifier variable set and Matched by Identifier for the chosen data set. Please click here to view a larger version of this figure.
Figure 5. Capturing landmarks and preliminaries for geometric morphometric analysis. (A) Using photo-editing software and a spreadsheet program to place landmarks and capture coordinates. (i) Multicolored crosses are the landmarks placed on the image using the Add points tool in ImageJ to represent the shape of the orofacial region. (ii) Landmark data is displayed by using the Display Results Tool. (iii) Landmark data is copied and pasted into a spreadsheet. Above the second column is a header identifying the number of landmarks and denoted by "LM=24" (red box). Below the second column of data, the sample is given a unique name and denoted by "ID=CON1" (red arrow). This is repeated for all images in a sample set and the data is saved as a text file. (B) Preliminary data analysis in a geometric morphometric software program. (i) The text file created in from the photo-editing software is imported into the morphometric program, MorphoJ, as a TPS file. File is indicated by red arrow. (ii) Landmark coordinate data is aligned by Procrustes fit by principal axes. Red arrow indicates execution of alignment. (iii) A covariance matrix of Procrustes fit landmarks is generated in the Preliminaries menu. (iv) A classifier file is created in a spreadsheet. Column A and B are given headers “ID” and “TREATMENT”, respectively. The ID’s given to each sample in landmark data collection are input under column A, and the treatment group to which each sample belongs is input in column B. (v) The classifier file is imported into the morphometric program as a classifier variable set and Matched by Identifier for the chosen data set. Please click here to view a larger version of this figure.
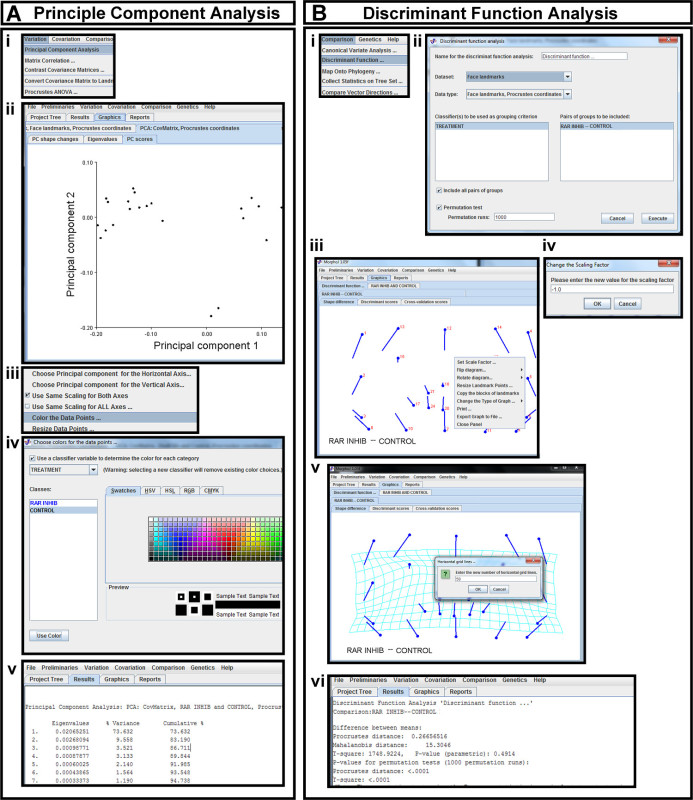 Figure 6. Statistical analysis in morphometric software. (A) Principal Component Analysis (PCA) (i) PCA is selected from the Variation tab. (ii) The first two principal components of Procrustes landmarks are displayed as a scatterplot in the PC scores tab. (iii) A pop-up menu is brought up in the plot space. This menu is used to change which principal components are plotted against each other (red arrow) and to color the data points (highlighted in blue). (iv) The data points are colored according to the classifier variables in the pop-up menu. (v) Percentage of variance captured by each principal component is viewed in the Results tab. (B) Discriminant Function Analysis (DFA) (i) DFA is selected from the Comparison tab. (ii) The data set of Procrustes coordinates is selected for DFA, and the previously uploaded classifiers are chosen for grouping. The desired groups to be compared are chosen and permutation tests are run. (iii) DFA results are displayed as a vector map in the Shape Difference tab. A pop-up menu in the plot space can be used to orient the image correctly. (iv) By selecting the Set Scale Factor tab in the pop-up menu, the sign of the scale factor can be changed. (v) The vector map is changed to a transformation grid with the desired number of grid lines by choosing Change the Type of Graph in the pop-up menu of the vector map. (vi) The Mahalanobis and Procrustes distances and corresponding p-values are viewed under the Results tab. Please click here to view a larger version of this figure.
Figure 6. Statistical analysis in morphometric software. (A) Principal Component Analysis (PCA) (i) PCA is selected from the Variation tab. (ii) The first two principal components of Procrustes landmarks are displayed as a scatterplot in the PC scores tab. (iii) A pop-up menu is brought up in the plot space. This menu is used to change which principal components are plotted against each other (red arrow) and to color the data points (highlighted in blue). (iv) The data points are colored according to the classifier variables in the pop-up menu. (v) Percentage of variance captured by each principal component is viewed in the Results tab. (B) Discriminant Function Analysis (DFA) (i) DFA is selected from the Comparison tab. (ii) The data set of Procrustes coordinates is selected for DFA, and the previously uploaded classifiers are chosen for grouping. The desired groups to be compared are chosen and permutation tests are run. (iii) DFA results are displayed as a vector map in the Shape Difference tab. A pop-up menu in the plot space can be used to orient the image correctly. (iv) By selecting the Set Scale Factor tab in the pop-up menu, the sign of the scale factor can be changed. (v) The vector map is changed to a transformation grid with the desired number of grid lines by choosing Change the Type of Graph in the pop-up menu of the vector map. (vi) The Mahalanobis and Procrustes distances and corresponding p-values are viewed under the Results tab. Please click here to view a larger version of this figure.
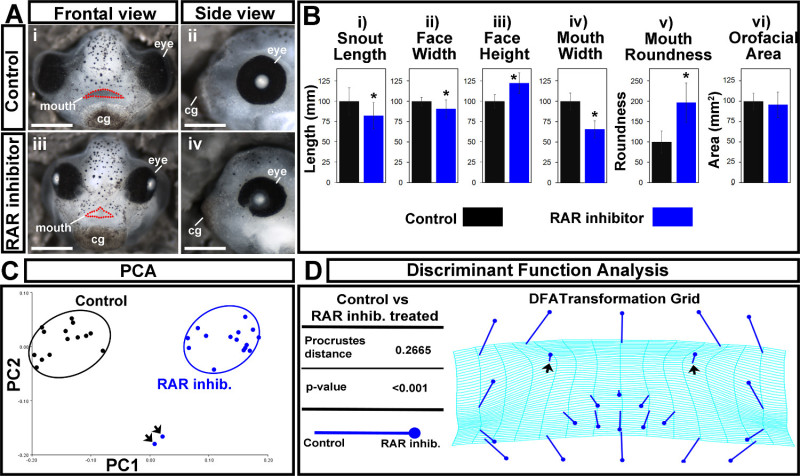 Figure 7. Orofacial analysis of control and RAR inhibited treated embryos. (A) (i,ii) Representative images of controls. Scale bars = 270 µm. (iii,iv) Embryos treated with a 1 μM concentration of the RAR inhibitor, BMS-453. Scale bars = 260 µm. (i,iii) Frontal views. Mouth opening is outlined in red dots. (ii,iv) Side views. cg: cement gland. (B) Traditional orofacial dimensions of control (black) and inhibitor treated (blue) embryos. (i) snout length, in mm (ii) face width, in mm (iii) face height, in mm (iv) mouth width, in mm (v) mouth roundness, a unit-less number determined in ImageJ using the equation: (4 × [Area]) / (π × [Major axis]2). (vi) Orofacial area, in mm2. Asterisks indicate significance as determined by Student’s T-test assuming unequal variance. α < 0.05. (C) Principal Component Analysis. Controls are in black and RAR inhibitor treated embryos are in blue. Black arrows indicate outliers. PC1 = 73.63%, PC2 = 9.56%. (D) Discriminant Function Analysis displaying the Procrustes distance and p-value, in addition to a transformation grid. Closed circle end of vector is landmark position in RAR inhibitor treated embryos. The end of the line of the vector is the landmark position in controls. Black arrows indicate shift in nasal landmarks. Please click here to view a larger version of this figure.
Figure 7. Orofacial analysis of control and RAR inhibited treated embryos. (A) (i,ii) Representative images of controls. Scale bars = 270 µm. (iii,iv) Embryos treated with a 1 μM concentration of the RAR inhibitor, BMS-453. Scale bars = 260 µm. (i,iii) Frontal views. Mouth opening is outlined in red dots. (ii,iv) Side views. cg: cement gland. (B) Traditional orofacial dimensions of control (black) and inhibitor treated (blue) embryos. (i) snout length, in mm (ii) face width, in mm (iii) face height, in mm (iv) mouth width, in mm (v) mouth roundness, a unit-less number determined in ImageJ using the equation: (4 × [Area]) / (π × [Major axis]2). (vi) Orofacial area, in mm2. Asterisks indicate significance as determined by Student’s T-test assuming unequal variance. α < 0.05. (C) Principal Component Analysis. Controls are in black and RAR inhibitor treated embryos are in blue. Black arrows indicate outliers. PC1 = 73.63%, PC2 = 9.56%. (D) Discriminant Function Analysis displaying the Procrustes distance and p-value, in addition to a transformation grid. Closed circle end of vector is landmark position in RAR inhibitor treated embryos. The end of the line of the vector is the landmark position in controls. Black arrows indicate shift in nasal landmarks. Please click here to view a larger version of this figure.
Discussion
Xenopus laevis has become a useful tool for dissecting the developmental mechanisms underlying orofacial development; however, there are currently no protocols describing size and shape changes of this region in frogs. The method described here will contribute significantly to the field of orofacial development by allowing for more rigorous quantification of orofacial phenotypes in Xenopus and other vertebrates.
The first, most critical aspect of properly executing this protocol is the ability to measure facial dimensions and determine landmark placement both accurately and reproducibly. To this end, it is crucial that embryonic faces be photographed at the same angle, direction and magnification. Particular care must be taken to obtain precise measurements of the snout length, as it is difficult to manipulate embryos laterally and achieve consistent placement. Having a single person perform all measurements on the same day minimizes this type of error, while maximizing the reproducibility of the results. The second most critical aspect of ensuring good results is the reduction of unnecessary variability, such as developmental differences or genetic background. This is especially important when assessing statistical significance between subtle defects. Embryos, therefore, need to be at the same stage when treated and photographed. Moreover, care should be taken to ensure developmental rates are equivalent between and among treatment groups. To decrease such problems with variability, make sure that all embryos are from the same parents and are stage matched at the beginning of the experiment. Also reduce external sources of variability such as different buffer sources and volumes, different numbers of embryos, and distributions in the culture dishes (for example, prevent crowding).
A key step in assessment of shape changes through geometric morphometrics is the alignment of landmark coordinates via Procrustes fit. By application of this mathematical algorithm, any information about size or differences in rotation of the image is removed. The subsequently generated covariance matrix determines the unstandardized correlations of landmark coordinates among all the embryos in the data set, on which multivariate statistical techniques- such as principal component or discriminant function analyses- can be performed.
A principal component analysis (PCA) reduces a complex sample to a smaller set of variables called principal components26. The first component accounts for the most variance within the sample set, with each subsequent component accounting for the rest. By plotting the first two components against each other using morphometric software, samples that are most similar cluster together. In this way, PCA discriminates groups within a sample set, while simultaneously determining the variation within them. In some cases, groups may not be clearly distinguished from each other and overlap along either or both axes. In this case, it is advantageous to plot other components (for example, PC3) in order to reveal subtle features by which groups are discriminated. This is especially relevant when the total variance is more evenly distributed among the first several variables.
The discriminant function analysis (DFA) of Procrustes fit data determines whether samples in a data set are effectively discriminated into groups by the continuous variables that define them. Samples are therefore classified into groups prior to analysis, and the variables are translated into components called discriminate functions to determine the statistical relationship between the groups27. Prior to running this analysis, it is important to indicate the number of permutation tests. These permutation tests randomize the data such that any assumptions about its distribution are eliminated. More permutation test iterations increase the accuracy of the p-value. When visualized as a transformation grid, significant changes in landmark position between the groups being compared are displayed. Further, the warping pattern of the grid reveals where shape changes occur. The drawback of this analysis is that it can only be utilized for comparison of two groups. If there are three or more groups in a sample set for comparison, it is better to perform a canonical variate analysis. This is similar to a DFA in that it generates a statistical p-value; however, it shows changes occurring over the entire sample set in addition to those occurring between individual groups within that set.
The major limitation of this orofacial quantification protocol is that it can only be applied to two-dimensional data. Our future goals include using CT scanning or confocal microscopy to develop similar methods for analysis of three-dimensional data. On the other hand, working with two-dimensional images is also one of the major strengths of this protocol. Only basic stereoscopes fitted with cameras are needed for capturing images of the embryonic face. The Openware utilized for this data analysis also increases the accessibility of this method, while decreasing the cost. Further, an advanced knowledge of sophistical imaging, statistical analyses, or computer programming is not required to extract meaningful and significant results from the data. In fact, this technique is currently being taught as part of an undergraduate laboratory course in the VCU department of biology. Thus, the orofacial quantification protocol presented here is easy to learn and apply in a short period of time. As a video representation, critical steps such as positioning embryos and navigating the software are highlighted to ensure the protocol can be successfully utilized by even untrained students and researchers. In summary, this protocol will provide a valuable resource for the research community and as a teaching tool.
Disclosures
The authors have nothing to disclose.
Acknowledgments
Start-up monies to A. Dickinson from VCU supported this work.
The authors wish to acknowledge Dan Nacu for his artistic talent in creating the schematic illustration.
References
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374:1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- Kennedy AE, Dickinson AJ. Median facial clefts in Xenopus laevis: roles of retinoic acid signaling and homeobox genes. Dev Biol. 2012;365:229–240. doi: 10.1016/j.ydbio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- Kennedy AE, Dickinson AJ. Quantitative Analysis of Orofacial Development and Median Clefts in Xenopus Laevis. Anat Rec (Hoboken) 2014. [DOI] [PubMed]
- Dickinson A, Sive H. Positioning the extreme anterior in Xenopus: cement gland, primary mouth and anterior pituitary. Semin Cell Dev Biol. 2007;18:525–533. doi: 10.1016/j.semcdb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Sive H. Development of the primary mouth in Xenopus laevis. Dev Biol. 2006;295:700–713. doi: 10.1016/j.ydbio.2006.03.054. [DOI] [PubMed] [Google Scholar]
- Dickinson AJ, Sive HL. The Wnt antagonists Frzb-1 and Crescent locally regulate basement membrane dissolution in the developing primary mouth. Development. 2009;136:1071–1081. doi: 10.1242/dev.032912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett C, et al. Syndrome Transcription Factor is critical for neural crest cell function in Xenopus laevis. Mech Dev. 2012;129:324–338. doi: 10.1016/j.mod.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales B, Yang H, Henning D, Valdez BC. Cloning and functional characterization of the Xenopus orthologue of the Treacher Collins syndrome (TCOF1) gene product. Gene. 2005;359:73–80. doi: 10.1016/j.gene.2005.04.042. [DOI] [PubMed] [Google Scholar]
- Reisoli E, De Lucchini S, Nardi I, Ori M. Serotonin 2B receptor signaling is required for craniofacial morphogenesis and jaw joint formation in Xenopus. Development. 2010;137:2927–2937. doi: 10.1242/dev.041079. [DOI] [PubMed] [Google Scholar]
- Schuff M, et al. FoxN3 is required for craniofacial and eye development of Xenopus laevis. Dev Dyn. 2007;236:226–239. doi: 10.1002/dvdy.21007. [DOI] [PubMed] [Google Scholar]
- Slater BJ, Liu KJ, Kwan MD, Quarto N, Longaker MT. Cranial osteogenesis and suture morphology in Xenopus laevis: a unique model system for studying craniofacial development. PLoS One. 2009;4 doi: 10.1371/journal.pone.0003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg LN, Adams DS, Levin M. Normalized shape and location of perturbed craniofacial structures in the Xenopus tadpole reveal an innate ability to achieve correct morphology. Dev Dyn. 2012;241:863–878. doi: 10.1002/dvdy.23770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaighis I, Mattick CR, Tiddeman B, Hobson R. 3D Facial Morphometry in Children with Oral Clefts. Cleft Palate Craniofac J. 2013. [DOI] [PubMed]
- Farkas LG, Katic MJ, Forrest CR. Surface anatomy of the face in Down's syndrome: anthropometric proportion indices in the craniofacial regions. J Craniofac Surg. 2001;12:519–524. doi: 10.1097/00001665-200111000-00003. [DOI] [PubMed] [Google Scholar]
- Scheuer HA, Holtje WJ, Hasund A, Pfeifer G. Prognosis of facial growth in patients with unilateral complete clefts of the lip, alveolus and palate. J Craniomaxillofac Surg. 2001;29:198–204. doi: 10.1054/jcms.2001.0227. [DOI] [PubMed] [Google Scholar]
- Parsons KJ, Andreeva V, James Cooper W, Yelick PC, Craig Albertson R. Morphogenesis of the zebrafish jaw: development beyond the embryo. Methods Cell Biol. 2011;101:225–248. doi: 10.1016/B978-0-12-387036-0.00011-6. [DOI] [PubMed] [Google Scholar]
- Farkas LG, Katic MJ, Forrest CR. Surface anatomy of the face in Down's syndrome: age-related changes of anthropometric proportion indices in the craniofacial regions. J Craniofac Surg. 2002;13:368–374. doi: 10.1097/00001665-200205000-00002. [DOI] [PubMed] [Google Scholar]
- Cooper WJ, et al. Bentho-pelagic divergence of cichlid feeding architecture was prodigious and consistent during multiple adaptive radiations within African rift-lakes. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP, et al. Prenatal alcohol exposure alters the patterns of facial asymmetry. Alcohol. 2010;44:649–657. doi: 10.1016/j.alcohol.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, et al. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc Natl Acad Sci U S A. 1999;96:15002–15006. doi: 10.1073/pnas.96.26.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allam KA, et al. The spectrum of median craniofacial dysplasia. Plast Reconstr Surg. 2011:812–821. doi: 10.1097/PRS.0b013e318200aa08. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger R, Harlard R. Early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Cross MK, Powers M. Obtaining eggs from Xenopus laevis females. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus Laevis (Daudin) Garland Publishing Inc; 1967. [Google Scholar]
- Nieuwkoop PDaFJ. Normal Table of Xenopus laevis (Daudin): A Systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis. Garland Publishing Inc; 1994. [Google Scholar]
- Abdi H, Williams LJ. Principal Component Analysis. WIREs Computational Statistics. 2010;2 [Google Scholar]
- Hill TL. P. STATISTICS: Methods and Applications. Tulsa, OK: StatSoft, Inc; 2013. [Google Scholar]


