Abstract
The development of high-throughput methods, such as the construction of 18S rRNA gene clone or pyrosequencing libraries, has allowed evaluation of ciliate community composition in hundreds of samples from the rumen and other intestinal habitats. However, several genera of mammalian intestinal ciliates have been described based only on morphological features and, to date, have not been identified using molecular methods. Here, we isolated single cells of one of the smallest but widely distributed intestinal ciliates, Charonina ventriculi, and sequenced its 18S rRNA gene. We verified the sequence in a full-cycle rRNA approach using fluorescence in situ hybridization and thereby assigned an 18S rRNA gene sequence to this species previously known only by its morphology. Based on its full-length 18S rRNA gene sequence, Charonina ventriculi was positioned within the phylogeny of intestinal ciliates in the subclass Trichostomatia. The taxonomic framework derived from this phylogeny was used for taxonomic assignment of trichostome ciliate 18S rRNA gene sequence data stemming from high-throughput amplicon pyrosequencing of rumen-derived DNA samples. The 18S rRNA gene-based ciliate community structure was compared to that obtained from microscopic counts using the same samples. Both methods allowed identification of dominant members of the ciliate communities and classification of the rumen ciliate community into one of the types first described by Eadie in 1962. Notably, each method is associated with advantages and disadvantages. Microscopy is a highly accurate method for evaluation of total numbers or relative abundances of different ciliate genera in a sample, while 18S rRNA gene pyrosequencing represents a valuable alternative for comparison of ciliate community structure in a large number of samples from different animals or treatment groups.
INTRODUCTION
Ciliate protozoa have been found to colonize the intestinal tracts of a wide range of ruminant and nonruminant herbivores (1). In the rumen ecosystem, ciliates can account for up to 50% of the total microbial nitrogen, reaching densities of 105 to 106 cells/ml rumen fluid (2, 3). Although ciliates are not essential for feed degradation and survival of the host (4), it is believed that they contribute to overall gut function, for example, by adding degradative complexity (5), by their ability to scavenge oxygen (6), or by their grazing behavior, which helps to shape and regulate prokaryotic populations (4, 7, 8). During fermentation of ingested plant material, some of the known rumen ciliates release large amounts of hydrogen produced in their hydrogenosomes (9–11), thereby providing ideal conditions for commensal hydrogenotrophic methanogens. As a consequence, the total number of rumen ciliate protozoa as well as ciliate community composition appears to influence the amount of methane emitted by the host (12–14). However, studies so far have used limited numbers of animals to analyze the impact of ciliate community structure or the roles of individual members of ciliate communities in ruminant methane emissions in vivo. This may be largely due to the great time and effort involved in microscopic evaluation of ciliate community composition and a lack of validated alternative methods.
It is widely accepted that microscopic identification and counting represent the gold standard for analyzing ciliate community structure in intestinal samples. This technique relies on a high level of expertise and experience of the researcher in recognizing the features that differentiate between ciliate genera or species. In addition, the natural variation between animals may render it essential to examine large numbers of animals in order to test for correlations between ciliate community structure and host phenotype (15). Molecular tools based on marker gene surveys are very useful for this scale of investigation. However, despite ongoing efforts, several genera of intestinal ciliates that have been observed under the microscope have so far eluded characterization at the molecular level. Therefore, along with the need to pass on and acquire laboratory skills in handling and identifying rumen and other intestinal ciliates, there is the need to expand the number of ciliate genera and species represented by 18S rRNA gene reference sequences through the isolation of single cells. These data can be used to build a robust phylogeny and taxonomy, which can serve as references for molecular-based methods of surveying ciliate communities in large numbers of samples. Finally, there is a need to compare both methods and evaluate the strengths and weaknesses of each for addressing different types of research questions.
Here, we isolated single cells of one of the smallest rumen ciliates, Charonina ventriculi, and verified the origin of the novel 18S rRNA gene sequence type by fluorescence in situ hybridization in a full-cycle rRNA approach (16). No 18S rRNA gene sequence data were previously associated with this species. Based on this information, we recalculated the 18S rRNA gene phylogeny of intestinal ciliates (subclass Trichostomatia). We also used the resulting taxonomy as a reference database for BLAST-based taxonomic assignment of sequencing reads stemming from high-throughput 454-based pyrosequencing of ciliate 18S rRNA genes amplified from rumen DNA samples. Community structure results obtained by pyrosequencing were compared to microscopic counts in the same samples. These results allowed us to make recommendations for the types of research questions that may be addressed using 18S rRNA gene sequence-based community analyses and those that should not.
MATERIALS AND METHODS
Rumen sampling and sample fractionation.
Rumen sampling was approved by the AgResearch Grasslands Animal Ethics Committee, Palmerston North, New Zealand, under approvals 12391 and 12174. Rumen contents were collected via the rumen fistula from a pasture-fed sheep (S4) and a pasture-fed cow (C1) that were likely to harbor different types of ciliate communities, as established earlier (17). Samples were immediately transferred to 39°C, and all further sample fractionation steps were carried out at this temperature. The total sample (fraction 1) was filtered through a double layer of cheesecloth, and approximately 150 ml of rumen filtrate (fraction 2) was directly transferred into a separation funnel containing 150 ml of anaerobic dilution solution (ADS) (18). ADS was prepared as follows: 75 ml of mineral solution I (0.3% K2HPO4) and 75 ml of mineral solution II [0.3% KH2PO4, 0.6% (NH4)2SO4, 0.6% NaCl, 0.06% MgSO4, 0.06% CaCl2] were diluted with 337 ml of distilled water, spiked with 5 drops of resazurin (0.1%), and boiled and gassed with CO2 until reduced. A total of 12.5 ml Na2CO3 solution (12%) and 0.25 g of cysteine were added, the bottle was sealed, and the solution was autoclaved. The sample in the funnel was left to settle for 50 min before glucose was added to a concentration of 1 g liter−1 of diluted rumen liquor. After a further 10 min, a sample was taken from the supernatant (fraction 3), and the settled ciliate fraction (fraction 4) was released through the tap of the funnel. Further fractionation of the ciliate fraction was achieved by consecutively using a series of filters with decreasing pore sizes (Saati, Appiano Gentile, Italy): 90 μm (62-64 Ultra Orange Plain Weave; retentate designated fraction 5), 55 μm (90-48 Ultra Yellow Plain Weave; retentate, fraction 6), and 23 μm (180-31 Ultra Yellow Twill Weave; retentate, fraction 7; filtrate, fraction 8). Retentates on filters were washed twice with 20 ml of ADS and carefully rinsed from the filter into a sterile flask with 20 ml of ADS delivered from a sterile plastic syringe. Ciliate community composition in each fraction was evaluated by microscopic counting of species as well as by amplicon pyrosequencing of rumen ciliate 18S rRNA genes. Subsamples for light microscopy were fixed and stained in a 10% (wt/vol) formaldehyde solution containing 0.6 g of methyl green and 8 g of sodium chloride per liter and incubated at room temperature in the dark for at least 3 days. Subsamples for molecular analysis were immediately frozen and stored at −20°C until further use.
Microscopic identification and counting of ciliate cells.
Fixed and stained subsamples for light microscopy were centrifuged at 1,000 × g, and the formaldehyde fixative was replaced with 30% glycerol in water (vol/vol) for unrestricted air transportation of samples to the Agricultural Research and Development Center, Wooster, OH, where samples were subjected to microscopic analysis and taxonomic identification according to Dehority (19). A total of at least 100 cells were counted per slide, and mean values of at least two slides per fraction are reported.
Isolation of single ciliate cells.
Single cells of the rumen ciliate Charonina ventriculi were isolated from fraction 3 of a rumen sample of cow C1 collected at a later date using the same sampling and fractionation method as described above, but without the addition of ADS. Subsamples (10 ml) obtained from fraction 3 were centrifuged at 500 × g for 5 min. The pellet was resuspended in 5 ml of fixing solution made up of 50% (vol/vol) ethanol, 15% (vol/vol) glycerol, and 35% (vol/vol) phosphate-buffered saline, and this was centrifuged again at 500 × g for 5 min. Phosphate-buffered saline (PBS) contained 10 mM Na2HPO4, 10 mM NaH2PO4, and 130 mM NaCl, and the pH was adjusted to 7.4 with HCl. The pellet was resuspended in 5 ml of fixing solution and stored at 4°C until further use. Subsequently, a 30-μl subsample from the settled pellet was washed twice by the addition of 500 μl of PBS and subjected to centrifugation at 1,000 × g for 2 min before the pellet was resuspended in 30 μl of PBS and 30 μl of 99.5% glycerol. A droplet of the solution was transferred onto a well of a 10-well diagnostic slide. The isolation of single cells of C. ventriculi was performed under an inverted IX71 microscope (Olympus, Melbourne, Victoria, Australia) with MMI CellCut Plus system software (Eching, Germany). Single cells were first identified according to Dehority (19) and then isolated using a bent glass micropipette at a magnification of ×10. Cells were washed by transfer into a free well on the microscopic slide containing a drop of PBS. This process was carried out sequentially at least three times. Single cells were transferred into a PCR tube containing 5 μl of 10× PCR buffer plus MgCl2 (Promega, Alexandria, NSW, Australia) and 20 μl of sterile water, and the tube was kept on ice until a total of 10 single cells were isolated and transferred into the same tube. This procedure yielded sufficient PCR product for further molecular analysis of the cells.
PCR amplification and sequencing of 18S rRNA genes from isolated cells.
Nested-PCR amplification on a pool of 10 isolated cells was performed as previously described but with certain modifications (20). Briefly, each 50-μl PCR mixture contained 1× PCR buffer plus MgCl2 (Promega), 50 μM each of the deoxynucleoside triphosphates (dNTPs; Promega), 1 μg of bovine serum albumin (BSA; Ambion, Carlsbad, CA, USA), 500 nM each of the primers Euk-F (5′-AYC TGG TTG ATY YTG CCA G-3) (21) and Euk-R (5′-TGA TCC ATC TGC AGG TTC ACC T-3′) (21), and 0.75 U of Taq DNA polymerase (Promega). Amplification was performed as follows on a ProS thermocycler (Eppendorf, Hamburg, Germany): 10 cycles of denaturation (94°C for 40 s), annealing (55°C for 40 s), and elongation (72°C for 2 min), followed by 20 cycles of denaturing (92°C for 30 s), annealing (55°C for 40 s), and elongation (72°C for 2.5 min), with a final 6-min extension at 72°C. PCR amplicons were purified using a QIAquick purification kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations and eluted in 40 μl of elution buffer (EB; 10 mM Tris, pH 8.5 with HCl). Twenty microliters of the purified PCR product was evaporated for 2 h at 60°C to concentrate the DNA before the addition of the following PCR reagents to a total of 50 μl for amplification of rumen ciliate 18S rRNA genes: 1× PCR buffer plus MgCl2, 50 μM each of the dNTPs, 1 μg of BSA, 500 nM each of the rumen ciliate-specific primers PSSU54F (5′-CAY GTC TAA GTA TAA ATA ACT AC-3′) (3) and PSSU1747R (5′-CTC TAG GTG ATW WGR TTT AC-3′) (3) and 0.75 U of Taq DNA polymerase. The correct sizes of PCR products were verified by agarose gel electrophoresis. The almost full-length ciliate 18S rRNA genes were cloned using a Topo TA cloning kit (Invitrogen, Carlsbad, CA, USA). DNA of 30 randomly selected clones was subjected to vector-targeting PCR with primers Gem2987F (5′-CCC AGT CAC GAC GTT GTA AAA CG-3′) and Top168R (5′-ATG TTG TGT GGA ATT GTG AGC GG-3′). The resulting PCR products were column purified as described above, quantified using a Nanodrop instrument (NanoDrop Technologies, Wilmington, DE, USA), and Sanger sequenced using sequencing primer RP841F (5′-GAC TAG GGA TTG GAG TGG-3′) (17) at the Alan Wilson Centre Genome Sequencing Service (Massey University, Palmerston North, New Zealand). Out of a total of 30 clones, one sequence was mixed and was therefore omitted from further analyses. The remaining 29 clones showed almost 100% sequence identity across the sequenced region (∼635 bp in length). Three of these clones were randomly selected to be sequenced with three different additional sequencing primers to cover the full sequence (primers PSSU54F, Syl316F [5′-GCT TTC GWT GGT AGT GTA TT-3′], and Syl539R [5′-CTT GCC CTC YAA TCG TWC T-3′]) (3). Sequence contigs were assembled from shorter fragments using ContigExpress, version 11.1, as part of the VectorNTI Advance 11 suite (Invitrogen).
Probe design and fluorescence in situ hybridization.
For fluorescence in situ hybridization, samples derived from fraction 3 of the rumen sample of cow C1 were immediately fixed by adding 3 volumes of a 4% (wt/vol) paraformaldehyde solution (dissolved in PBS) and stored at −20°C until further use. An oligonucleotide probe (CHA1350) specific for the putative 18S rRNA gene sequence of Charonina ventriculi was designed according to Hugenholtz et al. (22) (Table 1). Fluorescence in situ hybridization was performed as described previously with slight modifications (22), using different fluorescently labeled oligonucleotide probes (Table 1). A 250-μl aliquot from the fixed sample was centrifuged at 10 × g for 3 min. The pellet was then washed six times by the addition of 450 μl of PBS–96% ethanol (1:1 [vol/vol]); cells were harvested each time by centrifugation at 10 × g for 3 min, and the supernatant was discarded. Subsequently, the pellet was resuspended in 50 μl of PBS–96% ethanol (1:1 [vol/vol]). This sample was left to settle for 5 min, without further centrifugation. Then 3 μl of supernatant was applied to each well of a 10-well microscopic slide, and the slide was air dried overnight. Prior to hybridization, the air-dried slides were subjected to an ascending ethanol series of 50%, 80%, and 100% ethanol for 3 min each. Hybridization was generally performed with two probes simultaneously, the Charonina-specific probe CHA1350-Cy3 and the universal eukaryote probe EUKb1193-Cy5 (Table 1) (23). Throughout the experiment, one sample on each slide was hybridized with the nonsense probe NON1350-Cy5 to serve as a negative control (Table 1). The optimal formamide concentration in the hybridization buffer for the newly designed probe was 25 to 30%. Hybridization buffer and 50 ng of the fluorescently labeled probes were applied to each well, and hybridization was carried out at 46°C for 90 min. Slides were briefly rinsed with prewarmed washing buffer (48°C) and transferred into a 50-ml Falcon tube containing washing buffer to incubate at 48°C for 15 min. Finally, slides were dried at 39°C, spiked with Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA), and covered with a coverslip. Visualization of cells was carried out using a DM2500 fluorescence microscope (Leica Microsystems, Wetzlar, Germany) and a magnification of ×200. Microscopic images were obtained using a Leica DFC420C camera (Leica Microsystems) and the Leica Application suite software (Leica Microsystems), and colors were exchanged in Corel Paint Shop Pro Photo XI (Ottawa, Canada).
TABLE 1.
Oligonucleotide probes used for fluorescence in situ hybridization of rumen ciliates in this study
| Name of probe | Fluorescent dyea | 5′–3′ sequence | Reference or source |
|---|---|---|---|
| CHA1350 | Cy3 | TGA ATA TTC ACC GAG TGA ATA CCA | This study |
| EUKb1193 | Cy5 | GGG CAT MAC DGA CCT GTT | 23 |
| NON1350b | Cy5 | ACT TAT AAG TGG CTC ACT TAT GGT | This study |
Probes were fluorescently labeled using cyanine 3 (Cy3) and cyanine 5 (Cy5). The optimal formamide concentration for a hybridization temperature of 46°C was 25 to 30%.
The nonsense probe was designed as the reverse of the Charonina ventriculi-specific probe and served as a negative control.
Expansion of the intestinal ciliate 18S rRNA gene taxonomic framework.
A total of 123 reference sequences of ≥1,500 bp in length from all publicly available sequences obtained from isolated trichostome ciliates to date were downloaded from NCBI. Our database further included three almost full-length 18S rRNA gene sequences obtained from Charonina ventriculi (this study) and the 18S rRNA gene sequence of Didinium nasutum which served as an outgroup (order Haptorida; GenBank accession number U57771). All sequences were aligned using SINA (24). The alignment was imported into ARB and manually corrected where necessary (25). Phylogenetic trees were calculated based on aligned sequences from an extracted region between Saccharomyces cerevisiae 18S rRNA gene positions 83 to 1727. Phylogenetic tree reconstruction was performed using the randomized accelerated maximum likelihood (RAxML) method with 1,000 bootstrap replications (26). The sequence of Balantidium entozoon was added to the tree by using positions 87 to 1684 and the ARB parsimony tool. A total of 40 sequences that were ≥1,300 bp but <1,500 bp in length and obtained from isolated ciliate cells in a previous study (27) were added to the tree by using positions 317 to 1727 and the ARB parsimony tool to obtain genus-level taxonomic assignments for these sequences. This phylogeny, consisting of a total of 168 sequences, formed the basis of the newly established sequence database and taxonomic framework containing reference sequences of all (to date) molecularly described intestinal ciliate species within the subclass Trichostomatia.
Positional coverage and variability of trichostome 18S rRNA genes.
Sequence variability of bases in each alignment position was analyzed in R (28) using the seqinR library (29), and results were plotted using the ggplot2 library (30). Shannon diversity indices were calculated for each alignment position using the frequencies of bases A, T (U and T were used synonymously), G, and C (31). The Shannon diversity index is zero if all the bases at a certain position in the alignment are the same, and the index increases as the position becomes more variable.
Amplicon pyrosequencing of rumen ciliate 18S rRNA genes.
A 1.5-ml aliquot from each fraction of the sheep and cow rumen sample was centrifuged at 500 × g, and the pellet was resuspended in 282 μl of buffer A (NaCl 0.2 M, Tris 0.2 M, EDTA 0.02 M; pH 8). Nucleic acids were extracted using a combined bead-beating, phenol-chloroform, and column purification method (32). Briefly, the resuspended pellet was subjected to bead beating for 45 s at 6.5 m s−1 in a FastPrep FP120 instrument (Qbiogene, Carlsbad, CA, USA) in the presence of 0.7 g of zirconium beads (0.1-mm diameter), 200 μl of sodium dodecyl sulfate (20% [wt/vol]), 268 μl of buffer PM (QIAquick 96 PCR purification kit, Qiagen), and 550 μl of phenol-chloroform-isoamyl alcohol (25:24:1; pH 8). After centrifugation at 18,000 × g for 20 min at 4°C, the supernatant was mixed with 650 μl of buffer PM in a sterile deep 96-well plate, and the mixture was transferred onto a QIAquick 96 plate (QIAquick 96 PCR purification kit; Qiagen). DNA from each sample was individually column purified according to the manufacturer's recommendation and eluted in 80 μl of EB. Roche 454 GS FLX Titanium amplicon pyrosequencing was performed simultaneously with all samples using barcoded primers RP841F and Reg1302R (5′-AAT TGC AAA GAT CTA TCC C-3′), which flank variable regions V5 to V8 of the ciliate 18S rRNA gene, as described earlier (15, 33). The resultant sequence data were processed in QIIME v.1.8 (34). Quality control and mapping of sequence reads to the corresponding rumen sample based on inspection of the unique Golay barcodes were carried out as described previously (33). Samples for which fewer than 750 sequence reads were obtained after quality filtering were excluded from further analysis. Sequence reads were phylogenetically assigned using the novel sequence and taxonomy files derived from the expanded phylogeny of trichostome ciliates (see File S1 in the supplemental material) and the BLAST algorithm (35). Relative abundance tables were generated at the genus level (level 10 [-L 10]) using the summarize_taxa.py QIIME script.
Nucleotide sequence accession numbers.
Almost full-length sequences of Charonina ventriculi were deposited in the GenBank database under accession numbers KJ870172, KJ870173, and KJ870174 . Pyrosequencing data obtained in this study were deposited in the NCBI Sequence Read Archive (SRA) under BioProject number PRJNA247677.
RESULTS
Ciliate community composition as evaluated by microscopic counting.
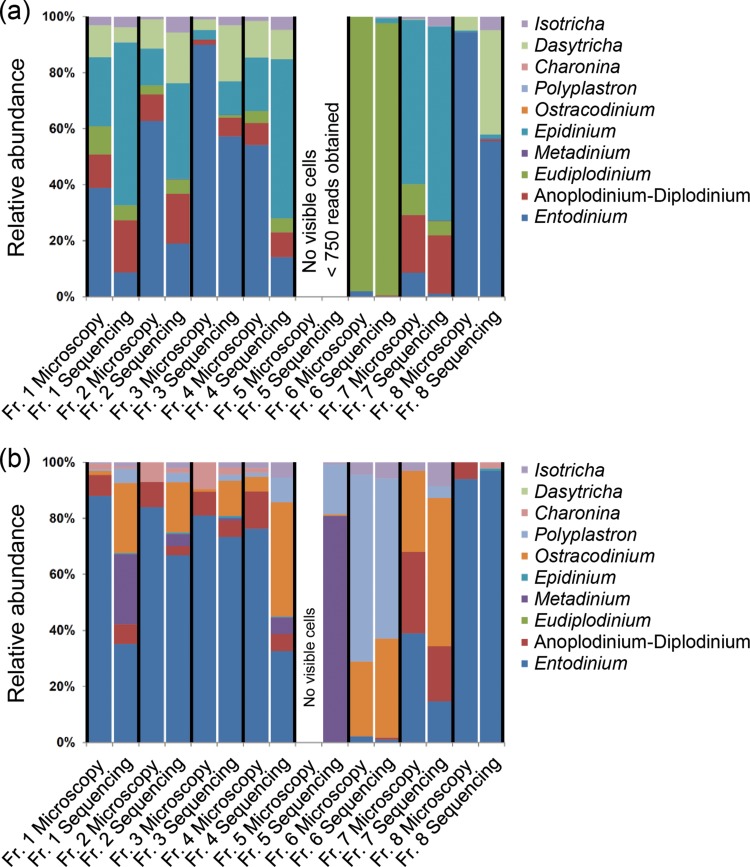
Rumen samples were collected from a cow and a sheep that had previously been shown to have different ciliate communities (17). As expected, microscopic counts revealed that the different fractions of the rumen samples collected from sheep S4 and cow C1 contained different relative abundances of rumen ciliates (Fig. 1; see also Table S1 in the supplemental material). The sample of sheep S4 harbored a B-type ciliate community. In the classification developed by Eadie (36), B-type ciliate communities are characterized by the presence of the key species Epidinium caudatum and/or Eudiplodinium maggii. Here, the following species were observed: Entodinium spp., Epidinium caudatum, Eudiplodinium maggii, Dasytricha ruminantium, Diplodinium spp., Isotricha spp., and very rarely Buetschlia parva (Fig. 1a). The untreated sample, fraction 1, contained a mixed community of ciliates: Entodinium spp. (38.9% of total observed ciliates), Epidinium caudatum (24.6%), Diplodinium spp. (11.9%), Dasytricha ruminantium (11.4%), Eudiplodinium maggii (10.1%), and Isotricha spp. (3.1%). Fractions 2, 3, and 4 as well as fraction 8 were dominated by Entodinium spp. (75.4% ± 19.9%, average ± standard deviation). In contrast, fractions 6 and 7 were dominated by Eudiplodinium maggii (98.0%) and Epidinium caudatum (58.7%), respectively, while fraction 5 did not contain any visible cells.
FIG 1.
Rumen ciliate community composition in fractions of a rumen sample, evaluated by microscopic counting of ciliate cells (microscopy) and next-generation amplicon sequencing of ciliate 18S rRNA genes (sequencing). (a) Rumen sample obtained from sheep S4. (b) Rumen sample obtained from cow C1. No visible cells were detected in fractions 5 of sheep S4 and cow C1. Fraction 5 of sheep S4 did also not provide ≥750 sequencing reads. Fr, fraction. Buetschlia parva was observed only rarely and is not shown.
The sample obtained from cow C1 harbored an A-type ciliate community. Eadie's A-type ciliate communities are characterized by the presence of the predatory ciliate Polyplastron multivesiculatum (36). In our sample, the following species were detected by microscopy: Charonina ventriculi, Diplodinium spp., Entodinium spp., Isotricha spp., Ostracodinium spp., and Polyplastron multivesiculatum (Fig. 1b). Fractions 1, 2, 3, 4, and 8 were dominated by Entodinium spp. (84.7% ± 6.7%). Fraction 6 was largely dominated by Polyplastron multivesiculatum (66.7%) and Ostracodinium spp. (26.7%), whereas fraction 7 was more diverse. Fraction 5 did not contain any visible cells. Fractions 1, 2, 3, and 4 harbored considerable proportions of cells identified as Charonina ventriculi (5.1% ± 3.8%).
Isolation of single cells of Charonina ventriculi and sequencing of cloned 18S rRNA genes.
Single cells of Charonina ventriculi were isolated from the fixed rumen content of cow C1. Ten cells were pooled, and amplicons of the 18S rRNA gene were generated using a nested PCR. A clone library was constructed from almost full-length 18S rRNA gene amplicons. Partial sequencing of clones and preliminary BLAST analysis revealed a novel sequence type only distantly related to sequences of other molecularly characterized intestinal ciliates. We hypothesized that this sequence type was derived from Charonina ventriculi. Almost full-length 18S rRNA gene sequence information was obtained from three clones. These sequences showed ≥99.8% sequence similarity among each other based on S. cerevisiae positions 83 to 1727.
Verification of 18S rRNA gene sequence types by fluorescence in situ hybridization.
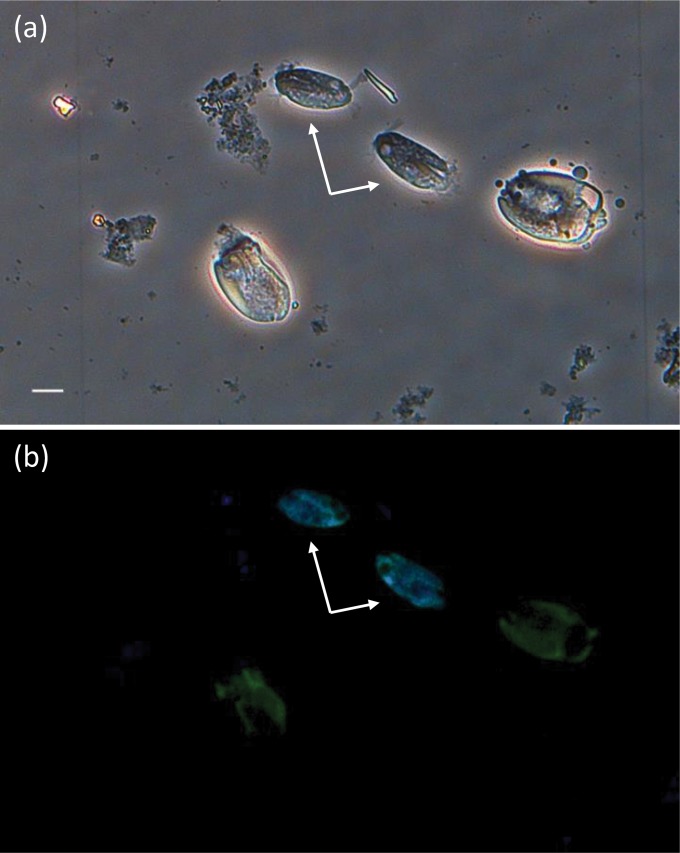
A specific 18S rRNA-targeting fluorescent probe was designed based on the almost full-length 18S rRNA gene sequence information of this novel sequence type likely derived from Charonina ventriculi. This probe, named CHA1350 (according to its 5′ binding position to the S. cerevisiae 18S rRNA gene), binds to the V7 region of the ciliate 18S rRNA gene. Fluorescence in situ hybridization was performed to determine the morphology of the cells with this particular 18S rRNA sequence type. Cells in the samples were subjected to double hybridization with the Charonina ventriculi-specific probe labeled with Cy3 and the universal eukaryotic probe EUKb1193 labeled with Cy5. A nonsense probe, NON1350, was applied to at least one sample on every slide and consistently gave no signal. The Charonina ventriculi probe gave a strong and specific signal for cells identified as Charonina ventriculi based on morphological features using bright-field microscopy on the same field (Fig. 2a and b; see also Fig. S1 and S2 in the supplemental material). Cells of Charonina ventriculi could be distinguished from other rumen ciliate cells based on their extremely small body size (less than 40 μm in length) with cilia on the anterior end and two characteristic tufts of cilia on the posterior end of the cell (Fig. 2a) (19). The eukaryotic probe hybridized to all ciliate cells present in the sample (Fig. 2b; see also Fig. S1 and S2).
FIG 2.
Bright-field and fluorescence in situ hybridization identification of Charonina ventriculi cells. The images were taken from the same field of fraction 3 of a rumen sample collected from cow C1. (a) Bright-field image. Scale bar, 10 μm. (b) Overlay fluorescence in situ hybridization image of the same field as shown in panel a. Ciliates were simultaneously hybridized with the universal eukaryotic probe EUKb1193 (Cy5) and the Charonina ventriculi-specific probe CHA1350 (Cy3). Color replacement was used to show cells stained with EUKb1193 in green and cells stained with CHA1350 in blue. In the overlay of both images, cyan indicates cells that were stained with both probes. Charonina ventriculi cells are indicated by arrows. Separate images are shown in Fig. S1 in the supplemental material.
Phylogenetic placement of Charonina ventriculi within the Trichostomatia.
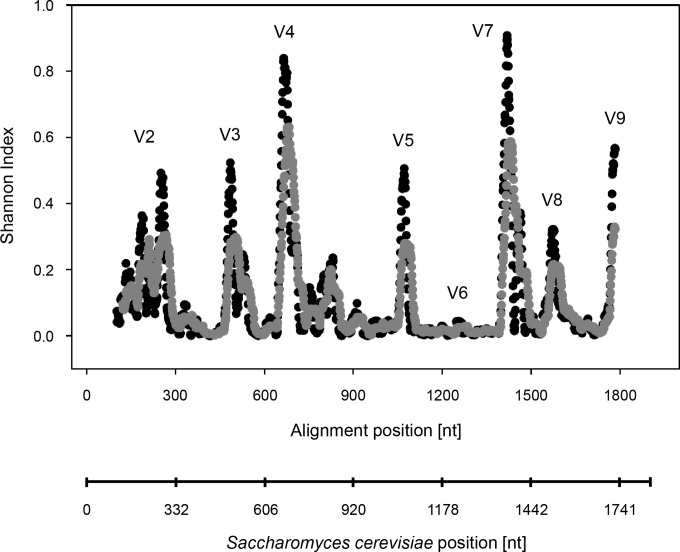
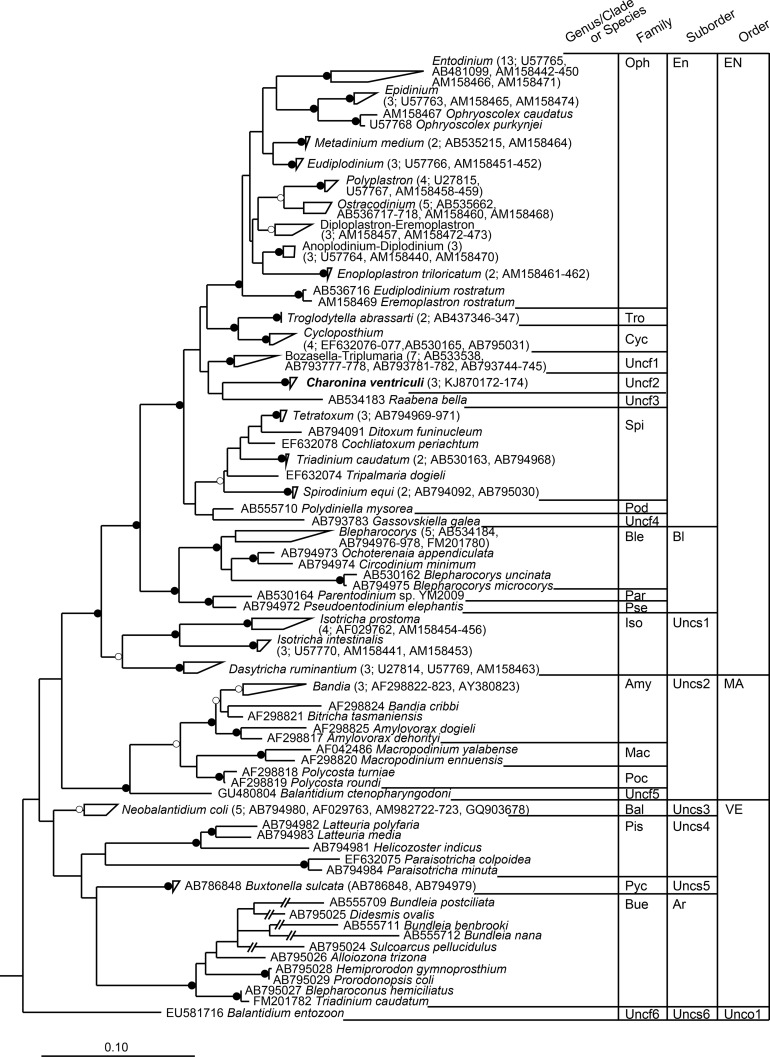
To assess the phylogenetic position of Charonina ventriculi within the intestinal ciliates (subclass Trichostomatia), three almost full-length 18S rRNA gene sequences, obtained from cloned Charonina ventriculi cells, were aligned with 123 reference sequences from isolated and described ciliate species inhabiting the intestinal tracts of herbivorous animals (subclass Trichostomatia). The alignment included variable regions V2 to V9. The variability of each alignment position was evaluated by calculation of Shannon's diversity index for each alignment position across the almost full-length trichostome 18S rRNA gene (Fig. 3). A RAxML tree was constructed with 1,000 bootstrap replications (Fig. 4; see also Fig. S3 in the supplemental material). The 18S rRNA gene of Didinium nasutum served as an outgroup sequence. In this tree, Charonina ventriculi formed a new clade within the Trichostomatia. It shared only 97.1% sequence similarity with its closest relative, Triplumaria selenica, which was isolated from a fecal sample from an Asian elephant (GenBank accession number AB533538) (Fig. 4) (37). Forty sequences generated in a previous study (27), covering S. cerevisiae positions 317 to 1710, were added to the tree using the ARB parsimony tool. From this information and based on the recommendations of Ito et al. (38), we derived a sequence database and taxonomic framework. This framework is compatible with the QIIME pipeline (see File S1 in the supplemental material) (34) and can be used for BLAST-based assignment of intestinal ciliate 18S rRNA gene amplicon pyrosequence data. The framework comprises a total of 168 sequences derived from isolated cells of intestinal ciliates, including our new data from Charonina ventriculi. In this framework, genera that clustered monophyletically were assigned the common genus name. Genera that clustered tightly but contained members of other genera were assigned a combined group name at genus level. Several genera, namely, Balantidium, Bandia, Blepharocorys, and Bundleia, formed two different clusters in the tree that were only distantly related. Therefore, these genera were divided into subgroups. In the future, these groups should undergo revision and potentially be reclassified into novel genera. The framework comprises a total of 11 taxonomic levels (kingdom, superphylum, phylum, subphylum, class, subclass, order, suborder, family, genus, and species) (see File S1 in the supplemental material). We recommend use at the genus level (pass option “-L 10” with QIIME script summarize_taxa.py) for high-resolution ciliate community structure analysis based on short-read high-throughput sequence data.
FIG 3.
Sequence variability of the trichostome 18S rRNA gene. Sequence variability is expressed as the Shannon index for each alignment position of the 126 aligned trichostome ciliate sequences (black circles). Shannon indices for a subset of 54 aligned sequences from genera known to occur in the rumen are shown in gray circles. The sequence of Balantidium entozoon was omitted because it was shorter than all other remaining reference sequences. A 20-bp moving average was used. Approximate positions of frequently targeted variable regions (V2 to V9) for community structure analysis are shown for orientation. Labeling of variable regions corresponds to eukaryotic 18S rRNA gene variable region designations (75). V6 is highly conserved among eukaryotes in general and therefore indistinct (76). Nucleotide numbering corresponds to base positions in the Saccharomyces cerevisiae strain NCYC 505 18S rRNA gene (GenBank accession number Z75578).
FIG 4.
Randomized accelerated maximum likelihood (RAxML) tree of almost full-length trichostome ciliate 18S rRNA marker gene sequences (S. cerevisiae positions 83 to 1727) calculated from 1,000 replications (open circles indicate nodes with bootstrap values of ≥75%; filled circles indicate bootstrap values of ≥95%; bootstrap values of <75% are not shown). The scale bar indicates 0.1 nucleotide substitution per nucleotide position. For clarity, some coherent groups of sequences are indicated only as triangles or trapezoids, with the number of sequences clustering into these groups and accession numbers of sequences obtained from isolates given in parentheses behind the cluster name. Genus/clade designations used in the taxonomic framework provided in File S1 in the supplemental material are shown in Fig. S3. Branches marked with double slashes (//) are drawn at 25% of the actual branch length. The 18S rRNA gene sequence of Didinium nasutum (order Haptorida; GenBank accession number U57771) served as the outgroup. Oph, family Ophryoscolecidae; Tro, family Troglodytellidae; Cyc, family Cycloposthiidae; Uncf1 to Uncf6, unclassified family 1 to 6; Spi, family Spirodiniidae; Pod, family Polydiniellidae; Ble, family Blepharocorythidae; Par, family Parentodiniidae; Pse, family Pseudoentodiniidae; Iso, family Isotrichidae; Amy, family Amylovoracidae; Mac, family Macropodiniidae; Poc, family Polycostidae; Pis, family Paraisotrichidae; Pyc, family Pycnotrichidae; Bal, family Balantidiidae; Bue, family Buetschliidae; En, suborder Entodiniomorphina; Bl, suborder Blepharocorythina; Uncs1 to Uncs6, unclassified suborder 1 to 6; Ar, suborder Archistomatina; EN, order Entodiniomorphida; MA, order Macropodiniida; VE, order Vestibuliferida; Unco1, unclassified order 1.
Ciliate community composition as evaluated from 18S rRNA gene pyrosequencing.
We used the trichostome ciliate 18S rRNA gene reference database and taxonomic framework described above with BLAST to assign the ciliate 18S rRNA gene pyrosequencing reads amplified from DNA extracted from the fractionated rumen samples used earlier in this study.
Based on the pyrosequencing results, ciliate communities in both the sheep and the cow samples could be categorized into one of the ciliate community types described by Eadie (36). The sheep sample harbored a B-type and the cow sample harbored an A-type ciliate community, in agreement with the microscopy-based assessment (Fig. 1; see also Table S1 in the supplemental material).
In the sheep sample, all species detected microscopically were also found using the molecular approach (Fig. 1a), with the exception of the rarely observed Buetschlia parva for which there is so far no 18S rRNA gene reference sequence available. In terms of relative abundances of the observed genera, we detected a quantitative discrepancy between both methods. In particular, the small-celled genus Entodinium was detected at significantly lower relative abundances by the molecular approach than were obtained from the microscopy data. The larger-celled genera (Epidinium, Anoplodinium-Diplodinium, and Eudiplodinium) were detected at higher relative abundances than with microscopy.
After inclusion of Charonina ventriculi in the BLAST database, all genera present in the cow sample detected using microscopy were also detected using the molecular approach (Fig. 1b). Similar to the sheep sample, discrepancies were observed in relative abundances of the different genera when the sample was assessed using the two different methods. Smaller-celled genera such as Entodinium, Charonina, and Diplodinium spp. made up smaller proportions of the communities according to the molecular approach, while larger genera such as Polyplastron and Ostracodinium appeared to be more abundant. Initially, the large-celled rumen ciliate Metadinium medium was not detected using microscopic analysis of the samples, while amplicon pyrosequencing revealed its presence in almost every fraction of the sample of cow C1. The presence of M. medium in an A-type community was surprising, given that earlier studies had reported antagonism between M. medium and P. multivesiculatum (39). However, repeated analysis of single fractions under the microscope eventually confirmed the presence of M. medium in the rumen sample of cow C1.
DISCUSSION
Ciliate protozoa make up a large proportion of the microbial biomass in the intestinal tracts of ruminant and nonruminant herbivores. However, knowledge of the involvement of the different ciliate genera in intestinal fermentation processes is limited, and their interactions with other members of the microbial community (in particular, methanogenic archaea) or their potential impact on production traits of the host are not well understood. This is due in part to the difficulty of culturing intestinal ciliate protozoa and the challenge of developing a sound high-throughput alternative to microscopic counting for ciliate community structure analysis. Here, we obtained the 18S rRNA gene sequence of the common rumen ciliate Charonina ventriculi and demonstrated that expanding and curating the 18S rRNA gene reference database will significantly improve ciliate community structure analysis based on next-generation amplicon sequencing. These advances are needed to make this technology a useful alternative to microscopic counting, especially when dealing with a large number of rumen samples.
Charonina ventriculi and phylogenetic placement of its 18S rRNA gene sequence.
Charonina ventriculi was first described by Jameson as Charon ventriculi, family Blepharocorythidae, in rumen contents from sheep and cattle (40). Almost at the same time, Dogiel reported this protozoon as Blepharocorys bovis (41) and later proposed the name Blepharocorys ventriculi (42). Strelkov (43) and Wolska (44), however, after thorough analysis of its infraciliature and morphogenesis, placed this species into the more primitive genus Charonina Strand (45). To date, the genus Charonina comprises nine different species, with C. ventriculi representing the type species. Both C. nuda and C. equi have been observed in cattle (46, 47), but C. equi was first reported to occur in the colon of the horse (48). C. dicerotis, C. odontophora, C. tenuis, C. tetragona, and C. tortuosa have been found in the colon of wild African rhinoceroses (49), while C. hippopotami was reported from the stomach of the hippopotamus (50). Charonina spp. are infrequently observed but can occur at high relative abundances, sometimes making up more than 30% (51) or even 50% (52) of the total ciliate community. Dehority and Mattos (51) reported that C. ventriculi concentrations were much lower when readily digested carbohydrates were included in the diet. Whether this was the result of an inability to utilize these substrates or of a lowered pH from rapid fermentation of these substrates by bacteria is not known. Interestingly, these authors also found that concentrations of C. ventriculi in the rumen followed a diurnal cycle more closely related to the entodiniomorphs than the holotrichs, namely, Dasytricha and Isotricha spp. (51). This observation was supported by Ito et al. (38), who found that Raabena bella, which is considered a close relative to C. ventriculi based on morphological features, is phylogenetically more closely related to the Entodiniomorphina than to the holotrich genera Dasytricha and Isotricha. The phylogenetic placement of the 18S rRNA gene sequence of Charonina ventriculi within the trichostome ciliates indicates that Charonina and Raabena do not belong to the family Blepharocorythidae (suborder Blepharocorythina) but to as yet unclassified families within the suborder Entodiniomorphina.
Intestinal ciliate 18S rRNA gene phylogeny and taxonomy.
Until the late 1990s, understanding of the taxonomy and evolution of the intestinal ciliates of herbivorous mammals was purely based on morphological characteristics (53–55). Despite the fact that intestinal ciliates occur in a great variety of shapes and sizes and harbor a great number of distinguishable features, classification of species into higher taxonomic ranks such as genera, families, and orders based on ultrastructural features has proved challenging (56–61). With the advances in molecular methods, the acquisition and analysis of molecular marker genes, e.g., 18S rRNA genes, became an important tool for unambiguous species identification and the development of a phylogeny-based classification scheme. Embley et al. (62) were the first to describe the monophyletic position of the anaerobic rumen ciliates Dasytricha ruminantium and Polyplastron multivesiculatum (originally named Entodinium simplex) among the free-living aerobic ciliates based on 18S rRNA genes. This work was closely followed by Wright et al. (63–65), who provided molecular data for Diplodinium dentatum, Entodinium caudatum, Epidinium caudatum, Eudiplodinium maggii, Isotricha intestinalis, Ophryoscolex purkynjei, and Polyplastron multivesiculatum. Since then, numerous 18S rRNA gene reference sequences have been deposited in public databases, which has allowed phylogenetic reconstruction of intestinal ciliate evolution. Despite these novel insights, relicts of a purely morphology-based phylogeny still remain apparent in the taxonomy to date, and several polyphyletic genera, families, suborders, and orders have recently been identified (38). To carry out high-resolution community structure analysis based on ciliate 18S rRNA genes, an exact taxonomic nomenclature is desirable, yet it is not absolutely essential. However, to detect differences between samples, it is essential that distinct sequence clusters are given unique taxonomic identifiers (Fig. 4; see also Fig. S3 in the supplemental material). The following reclassifications were made to develop a working 18S rRNA gene taxonomy of mammalian intestinal ciliates.
Genus-level reclassifications.
The genus Blepharocorys clustered into two distant groups in the phylogenetic tree based on RAxML. To distinguish between the two groups, we subdivided this genus into Blepharocorys_a and Blepharocorys_b. The group containing the type species, Blepharocorys uncinata (GenBank accession number AB530162), was called Blepharocorys_a. The same was done for the genus Bundleia with its type species Bundleia postciliata (Bundleia_a). Species with the genus name Balantidium occurred in two different clusters in the phylogenetic tree. The polyphyletic nature of this genus has been described previously (66). The type species, Balantidium entozoon, was given the group name Balantidium_a, to distinguish it from Balantidium ctenopharyngodoni (group name Balantidium_b). The new genus name Neobalantidium was adopted for the cluster of sequences stemming from Neobalantidium coli (formerly Balantidium coli), as suggested in a recent study on the genetic diversity of the genus Balantidium (66). The genus Bandia was also represented by two sequence clusters. Since no 18S rRNA gene sequence is currently available for the type species Bandia beveridgei, the cluster containing the sequences of Bandia smalesae, Bandia tammar, and Bandia deveneyi was named Bandia_a, while the sequence of Bandia cribbii was classified as Bandia_b. A total of three sequences are named Triadinium caudatum according to NCBI. Two of these clustered into the Spirodiniidae (GenBank accession numbers AB530163 and AB794968), while the remaining one clustered into the Buetschliidae (GenBank FM201782) and was closely related to Blepharoconus hemiciliatus (GenBank AB795027). For separation on genus and species level, the former group was designated Triadinium (containing Triadinium caudatum_a), while the latter was assigned to the “Blepharoconus-Triadinium” group (containing Triadinium caudatum_b).
One of the distinct sequence clusters in the tree was comprised of sequences of species identified as Diploplastron spp. and Eremoplastron spp., which clustered polyphyletically. Until this genus is formally reappraised, we classified all sequences within this cluster as belonging to the combined “Diploplastron-Eremoplastron” group. Similarly, sequences of species identified as Anoplodinium spp. and Diplodinium spp. clustered closely together, but polyphyletically, and were assigned to the “Anoplodinium-Diplodinium” group. Sequence clusters containing two sequences with different genus designations that shared ≥99.5% sequence similarity were given a combined group name. This was done for two sequences, one each from Eudiplodinium and Eremoplastron, that grouped together but distantly from the “true” members of these two genera. We assigned them to the “Eudiplodinium-Eremoplastron” group, but a new formal genus name should be assigned in future. This rule also applied to sequences within the “Hemiprorodon-Prorodonopsis” and “Blepharoconus-Triadinium” groups.
Family-level reclassifications.
Several families that had been described based on morphological characteristics appeared to cluster polyphyletically based on the molecular analysis. For example, the family Cycloposthiidae formally comprises the genus Cycloposthium as well as the genera Bozasella and Triplumaria, but these genera do not form a monophyletic cluster. Based on our data, which is in accordance with Ito et al. (38), we suggest the transfer of Bozasella and Triplumaria from the family Cycloposthiidae to an “unclassified family 1 (Uncf1).” The family Blepharocorythidae was found to be monophyletic with the exception of the genera Charonina and Raabena. These genera were therefore assigned to two novel families, the so far unclassified families 2 (Uncf2) and 3 (Uncf3), respectively. Based on morphological characteristics, especially the skeletal plates and the macronucleus, it seems justifiable to include Tripalmaria dogieli (formerly assigned to the family Cycloposthiidae) in the family Spirodiniidae (38). This reclassification achieved monophyly of both these families.
In contrast to the results of a study by Ito et al. (38), the family Paraisotrichidae was monophyletic in this study, albeit with low bootstrap support of 42%. The sequence of Gassovskiella galea (family Spirodiniidae) did not cluster monophyletically with the remaining members of the family Spirodiniidae but with Polydiniella mysorea (family Polydiniellidae). Therefore, Gassovskiella galea was classified into unclassified family 4 (Uncf4) until more information is available. The sequence of Balantidium ctenopharyngodoni was isolated from the grass carp hindgut (GenBank accession number GU480804). Clustering of this sequence into the trichostome ciliates is surprising since so far this subclass of ciliates was thought to be exclusively comprised of ciliates from mammalian hosts. Balantidium ctenopharyngodoni clustered separately from the members of the Balantidiidae. Instead, it clustered as a sister group to the order Macropodiniida. Since its sequence type was clearly different from the other three designated families in this order, it was assigned to as yet unclassified family 5 (Uncf5). The 18S rRNA gene sequence of Balantidium entozoon also clustered clearly separately from the members of the Balantidiidae and was thus assigned to as yet unclassified family 6 (Uncf6).
Suborder-level reclassifications.
Based on the phylogenetic clustering, the suborders Entodiniomorphina and Blepharocorythina are polyphyletic. We have therefore adopted the suggestion of Ito et al. (38) to assign the as yet unclassified families Uncf2 (represented by Charonina) and Uncf3 (represented by Raabena) to the suborder Entodiniomorphina and to assign the families Parentodiniidae and Pseudoentodiniidae to the suborder Blepharocorythina. The family Isotrichidae had so far not been assigned to a suborder. For consistency, and until more molecular and morphological evidence is available, we suggest assigning this family to the as yet unclassified suborder 1 (Uncs1). Similarly, the three described families within the order Macropodiniida and the so far unclassified family 5 are assigned to the as yet unclassified suborder 2 (Uncs2). Members of the families Balantidiidae, Paraisotrichidae, and Pycnotrichidae clustered as sister groups to the Buetschliidae and can be placed in the suborder Archistomatina. However, until this is revisited, and due to the low bootstrap support, the three families are individually categorized into the as yet unclassified suborders Uncs3 (Balantidiidae), Uncs4 (Paraisotrichidae), and Uncs5 (Pycnotrichidae). Balantidium entozoon was assigned to as yet unclassified suborder 6 (Uncs6).
Order-level reclassifications.
The order Entodiniomorphida was found to be polyphyletic, as reported previously (38), with the members of the suborder Archistomatina (family Buetschliidae) clustering distantly from the remaining members of the order Entodiniomorphida (suborders Entodiniomorphina, Blepharocorythina, and Uncs1). Thus, we adopted the recommendation to move the suborder Archistomatina into the order Vestibuliferida (38). The former vestibuliferid family Isotrichidae formed a monophyletic sister group to the order Entodiniomorphida with 96% bootstrap support and was thus reclassified into this order, as suggested by Ito et al. (38). The species Balantidium entozoon clustered outside the three described orders and was therefore placed into the unclassified order 1 (Unco1) until its relationship to the other intestinal ciliates can be fully resolved.
All reclassifications described above have been incorporated into a new database and taxonomic framework of trichostome ciliates (see File S1 in the supplemental material). This working taxonomic framework is compatible with software such as QIIME (34) as described previously (33), which allows taxonomic assignment of sequence data obtained from high-throughput sequencing technologies based on BLAST (35) and can be used for 18S rRNA gene-based community structure analysis of mammalian intestinal ciliate protozoa (Fig. 1).
To fully address and resolve the remaining discrepancies (e.g., clustering at family and suborder level), marker gene information of further representatives of mammalian intestinal ciliates, more comprehensive taxon sampling, and the calculation of more reliable phylogenies are needed (38).
Comparison of ciliate community structure as assessed by microscopic counts and amplicon pyrosequencing.
This study compared two methods of ciliate community structure analysis in order to determine if high-throughput amplicon pyrosequencing could serve as a valid alternative to microscopic counting when the experiment requires a large number of samples to be analyzed. To date, microscopic counting is still largely regarded as the gold standard for studying the diversity and the relative and absolute abundance of rumen ciliates in individual samples. Microscopy holds several advantages over PCR-based molecular methods for studying ciliate protozoa. First, while the vast majority of intestinal ciliates has been characterized morphologically, there is a lack of 18S rRNA gene reference sequences for many of the observed genera and species. Second, copy number variation of rRNA genes across the different genera may skew the observed proportions of these genera in a sample (67). Previous studies using 18S rRNA gene surveys, however, have revealed a high “hidden” diversity of intestinal ciliates that may only have been poorly identified by conventional morphological methods (17, 68), and recent interest in understanding the involvement of rumen ciliates in livestock methane emissions requires the analysis of large numbers of samples. Therefore, research into the comparability of microscopic and molecular methods seemed more than justified. In our study, both microscopic and molecular methods were successfully applied to identify the ciliate protozoa present in the analyzed rumen samples. Both methods may therefore be used to address the question of “who is there.” This could, for example, include studies aiming at describing geographic extension or host specificity of intestinal ciliates. While microscopic analysis will allow detection of species that are still elusive to molecular characterization, pyrosequencing may allow detection of rare taxa that may easily be missed by microscopy (such as Metadinium medium in this study).
The two different methods produced quantitatively different results in regard to relative abundances of different ciliate genera. While the smaller-celled genera such as Entodinium, Charonina, and Diplodinium tended to be underrepresented, larger-celled genera such as Metadinium, Epidinium, Eudiplodinium, Ostracodinium, and Polyplastron tended to be overrepresented by the pyrosequencing approach compared to microscopy. This result corroborates earlier findings from the molecular and microscopic assessment of alveolate diversity and abundance in a freshwater lake (67). Overrepresentation may be explained by primer bias toward certain genera or, more likely, by the presence of different numbers of 18S rRNA gene copies in the genomes of the different species. rRNA copy number was found to be correlated with genome size (69), and it was suggested by Cavalier-Smith (70) and later demonstrated for phytoplankton by Boucher et al. that genome size (DNA content) is also correlated with body size (71). A direct correlation between rRNA gene copy number and body size was presented by Zhu et al. (72). Similarly, more recent literature on rumen ciliates suggests that Epidinium caudatum, which is approximately five times larger by volume than Entodinium caudatum (19), also has approximately five times more 18S rRNA gene copies encoded in its genome (73). These results agree with the finding that PCR-based methods return lower estimates of abundance of small Entodinium spp. (27) and overestimates of the abundance of, e.g., Polyplastron spp. (74). For this reason, the pyrosequencing method cannot be used to estimate α-diversity or relative abundances of different genera and species in a sample. However, it may turn out that 18S rRNA gene-based analyses are a proxy for the biomass of individual genera. What we can measure reliably using molecular methods, despite copy number differences, are trends in relative abundances of genera and species between different samples (β-diversity). The higher sample throughput that can be achieved by using molecular tools may allow even subtle shifts or differences between treatment groups to be detectable.
In conclusion, both methods have advantages and disadvantages. As with every method, limitations need to be acknowledged, and results must be interpreted correspondingly. Here, the best method to be used strongly depends on the research question to be addressed and the number of samples to be analyzed. If exact determination of the proportions of certain ciliates in a given sample is crucial, we recommend the use of microscopic counting. However, if exact proportions are not essential and if the overall aim is to estimate β-diversity by comparing ciliate community structure in hundreds of samples derived from different animals, geographic locations, treatment groups, time points, etc., then high-throughput amplicon pyrosequencing will provide a valuable alternative. In the future, more detailed knowledge on rRNA gene copy number in the various species (e.g., by sequencing and assembly of whole genomes) may provide a means to correct molecular marker gene data for those differences. To further improve molecular community structure analysis of intestinal ciliates, the aim should be to obtain and deposit 18S rRNA gene reference sequences from microscopically described species so that they can be incorporated into existing taxonomic frameworks. Deposition of reference sequences of these species would help conserve centuries of knowledge by linking the species names described in early and current literature with unique molecular marker gene sequence information.
Supplementary Material
ACKNOWLEDGMENTS
This research was carried out under contract to the Pastoral Greenhouse Gas Research Consortium (PGgRc) and the New Zealand Agricultural Greenhouse Gas Research Centre (NZAGRC). The visit by S.K. to the lab of B.A.D. in Wooster, OH, was supported by funding from the PGgRc.
We thank Jeffrey I. Gordon for access to the 454 Life Sciences sequencer and Su Deng (Center for Genome Sciences and Systems Biology, Washington University in St. Louis, Missouri) and Jason Peters (AgResearch) for excellent technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03697-14.
REFERENCES
- 1.Dehority BA. 1986. Protozoa of the digestive tract of herbivorous mammals. Int J Trop Insect Sci 7:279–296. doi: 10.1017/S1742758400009346. [DOI] [Google Scholar]
- 2.Hobson PN, Stewart CS. 1997. The rumen microbial ecosystem, 2nd ed. Springer, New York, NY. [Google Scholar]
- 3.Sylvester JT, Karnati SKR, Yu Z, Morrison M, Firkins JL. 2004. Development of an assay to quantify rumen ciliate protozoal biomass in cows using real-time PCR. J Nutr 134:3378–3384. [DOI] [PubMed] [Google Scholar]
- 4.Williams AG, Coleman GS. 1997. The rumen protozoa, p 73–139. In Hobson PN, Stewart CS (ed), The rumen microbial ecosystem, 2nd ed. Blackie Academic and Professional, London, United Kingdom. [Google Scholar]
- 5.Coleman GS. 1986. The metabolism of rumen ciliate protozoa. FEMS Microbiol Lett 39:321–344. doi: 10.1111/j.1574-6968.1986.tb01864.x. [DOI] [Google Scholar]
- 6.Ellis JE, Williams AG, Lloyd D. 1989. Oxygen consumption by ruminal microorganisms: protozoal and bacterial contributions. Appl Environ Microbiol 55:2583–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonhomme A. 1990. Rumen ciliates: their metabolism and relationships with bacteria and their hosts. Anim Feed Sci Technol 30:203–266. doi: 10.1016/0377-8401(90)90016-2. [DOI] [Google Scholar]
- 8.Coleman GS. 1989. The role of protozoa and fungi in ruminant digestion. Penambul Books, Armidale, Australia. [Google Scholar]
- 9.Yarlett N, Hann AC, Lloyd D, Williams A. 1981. Hydrogenosomes in the rumen protozoon Dasytricha ruminantium Schuberg. Biochem J 200:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarlett N, Hann AC, Lloyd D, Williams AG. 1983. Hydrogenosomes in a mixed isolate of Isotricha prostoma and Isotricha intestinalis from ovine rumen contents. Comp Biochem Physiol B Biochem Mol Biol 74:357–364. doi: 10.1016/0305-0491(83)90025-1. [DOI] [PubMed] [Google Scholar]
- 11.Yarlett N, Coleman GS, Williams AG, Lloyd D. 1984. Hydrogenosomes in known species of rumen entodiniomorphid protozoa. FEMS Microbiol Lett 21:15–19. doi: 10.1111/j.1574-6968.1984.tb00178.x. [DOI] [Google Scholar]
- 12.Jouany J, Zainab B, Senaud J, Groliere C, Grain J, Thivend P. 1981. Role of the rumen ciliate protozoa Polyplastron multivesiculatum, Entodinium sp. and Isotricha prostoma in the digestion of a mixed diet in sheep. Reprod Nutr Dev 21:871–884. doi: 10.1051/rnd:19810701. [DOI] [PubMed] [Google Scholar]
- 13.Kreuzer M, Kirchgessner M, Müller H. 1986. Effect of defaunation on the loss of energy in wethers fed different quantities of cellulose and normal or steamflaked maize starch. Anim Feed Sci Technol 16:233–241. doi: 10.1016/0377-8401(86)90114-8. [DOI] [Google Scholar]
- 14.Whitelaw F, Eadie JM, Bruce L, Shand W. 1984. Methane formation in faunated and ciliate-free cattle and its relationship with rumen volatile fatty acid proportions. Br J Nutr 52:261–275. doi: 10.1079/BJN19840094. [DOI] [PubMed] [Google Scholar]
- 15.Kittelmann S, Pinares-Patiño CS, Seedorf H, Kirk MR, Ganesh S, McEwan JC, Janssen PH. 2014. Two different bacterial community types are linked with the low-methane emission trait in sheep. PLoS One 9:e103171. doi: 10.1371/journal.pone.0103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amann RI, Ludwig W, Schleifer K-H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kittelmann S, Janssen PH. 2011. Characterization of rumen ciliate community composition in domestic sheep, deer, and cattle, feeding on varying diets, by means of PCR-DGGE and clone libraries. FEMS Microbiol Ecol 75:468–481. doi: 10.1111/j.1574-6941.2010.01022.x. [DOI] [PubMed] [Google Scholar]
- 18.Bryant MP, Burkey LA. 1953. Cultural methods and some characteristics of some of the more numerous groups of bacteria in the bovine rumen. J Dairy Sci 36:205–217. doi: 10.3168/jds.S0022-0302(53)91482-9. [DOI] [Google Scholar]
- 19.Dehority BA. 1993. Laboratory manual for classification and morphology of rumen ciliate protozoa, CRC Press, Boca Raton, FL. [Google Scholar]
- 20.Dyal P, Hope S, Roberts D, Embley T. 1995. Use of the PCR and fluorescent probes to recover SSU rRNA gene sequences from single cells of the ciliate protozoon Spathidium. Mol Ecol 4:499–504. doi: 10.1111/j.1365-294X.1995.tb00244.x. [DOI] [PubMed] [Google Scholar]
- 21.Medlin L, Elwood HJ, Stickel S, Sogin ML. 1988. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 22.Hugenholtz P, Tyson GW, Blackall LL. 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol Biol 179:29–42. doi: 10.1385/1-59259-238-4:029. [DOI] [PubMed] [Google Scholar]
- 23.Baker BJ, Hugenholtz P, Dawson SC, Banfield JF. 2003. Extremely acidophilic protists from acid mine drainage host Rickettsiales-lineage endosymbionts that have intervening sequences in their 16S rRNA genes. Appl Environ Microbiol 69:5512–5518. doi: 10.1128/AEM.69.9.5512-5518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig W, Strunk O, Westram R, Richter L, Meier H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 27.Tymensen L, Barkley C, McAllister TA. 2012. Relative diversity and community structure analysis of rumen protozoa according to T-RFLP and microscopic methods. J Microbiol Methods 88:1–6. doi: 10.1016/j.mimet.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 28.R Development Core Team. 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 29.Charif D, Lobry JR. 2007. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis, p 207–232. In Bastolla U, Porto M, Roman HE, Vendruscolo M (ed), Structural approaches to sequence evolution. Springer, New York, NY. [Google Scholar]
- 30.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer, New York, NY. [Google Scholar]
- 31.Shannon CE. 1948. A mathematical theory of communication. Bell System Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 32.Rius AG, Kittelmann S, Macdonald KA, Waghorn GC, Janssen PH, Sikkema E. 2012. Nitrogen metabolism and rumen microbial enumeration in lactating cows with divergent residual feed intake fed high-digestibility pasture. J Dairy Sci 95:5024–5034. doi: 10.3168/jds.2012-5392. [DOI] [PubMed] [Google Scholar]
- 33.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Gonzales-Pena A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widman J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 36.Eadie JM. 1962. Inter-relationships between certain rumen ciliate protozoa. J Gen Microbiol 29:579–588. doi: 10.1099/00221287-29-4-579. [DOI] [Google Scholar]
- 37.Ito A, Honma H, Gürelli G, Göçmen B, Mishima T, Nakai Y, Imai S. 2010. Redescription of Triplumaria selenica (Ciliophora, Entodiniomorphida) and its phylogenetic position based on the infraciliary bands and 18SSU rRNA gene sequence. Eur J Protistol 46:180–188. doi: 10.1016/j.ejop.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 38.Ito A, Ishihara M, Imai S. 2014. Bozasella gracilis n. sp. (Ciliophora, Entodiniomorphida) from Asian elephant and phylogenetic analysis of entodiniomorphids and vestibuliferids. Eur J Protistol 50:134–152. doi: 10.1016/j.ejop.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 39.Dogiel VA, Fedorowa T. 1925. Über den Bau und die Funktion des inneren Skellets des Ophryoscoleciden. Zool Anz 62:97–107. [Google Scholar]
- 40.Jameson AP. 1925. A new ciliate, Charon ventriculi n. g., n. sp., from the stomach of ruminants. Parasitology 17:403–405. doi: 10.1017/S0031182000004807. [DOI] [Google Scholar]
- 41.Dogiel VA. 1926. Une nouvelle espèce du genre Blepharocorys, B. bovis n. sp. habitant l'estomac du boeuf. Ann Parasitol Hum Comp 4:61–64. [Google Scholar]
- 42.Dogiel VA. 1934. Angaben über die Ophryoscolecidae des Wildschafes aus Kamtschatka, des Elches und des Yaks, nebst deren zoogeographischen Verwertung. Arch Protistenkd 82:290–297. [Google Scholar]
- 43.Strelkov A. 1939. Parasitic infusoria from the intestine of Ungulata belonging to the family Equidae. Uchenye Zapiski Leningradskii Gosudarstvennyi Pedagogicheskii Institut Gercena 17:1–262. [Google Scholar]
- 44.Wolska M. 1967. Study on the family Blepharocorythidae Hsiung. II. Charonina ventriculi (Jameson). Acta Protozool 4:279–283. [Google Scholar]
- 45.Strand E. 1928. Miscelanea nomenclatorica et paleontologica I und II. Arch Naturgesch 92:30–75. [Google Scholar]
- 46.Clarke RTJ. 1964. Ciliates of the rumen of domestic cattle (Bos taurus L.). New Zeal J Agr Res 7:248–257. doi: 10.1080/00288233.1964.10416409. [DOI] [Google Scholar]
- 47.Hsiung TS. 1932. A general survey of the protozoan fauna of the rumen of the Chinese cattle. Bull Fan Mem Inst Biol 3:87–104. [Google Scholar]
- 48.Hsiung TS. 1930. Some new ciliates from the large intestine of the horse. Trans Am Microsc Soc 49:34–41. doi: 10.2307/3222293. [DOI] [Google Scholar]
- 49.Gilchrist F, Van Hoven W, Stenson M. 1994. Five new species of Trichostomatida (ciliated protozoa) from the colon of wild African rhinoceroses. Syst Parasitol 28:187–196. doi: 10.1007/BF00009517. [DOI] [Google Scholar]
- 50.Thurston JP, Grain J. 1971. Holotrich ciliates from the stomach of Hippopotamus amphibius, with descriptions of two new genera and four new species. J Protozool 18:133–141. doi: 10.1111/j.1550-7408.1971.tb03295.x. [DOI] [Google Scholar]
- 51.Dehority B, Mattos W. 1978. Diurnal changes and effect of ration on concentrations of the rumen ciliate Charon ventriculi. Appl Environ Microbiol 36:953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Syrjälä L, Saloniemi H, Laalahti L. 1976. Composition and volume of the rumen microbiota of sheep fed on grass silage with different sucrose, starch and cellulose supplements. J Sci Agric Soc Finl 48:138–153. [Google Scholar]
- 53.Dogiel VA. 1927. Monographie der Familie Ophryoscolecidae. Arch Protistenkd 59:1–288. [Google Scholar]
- 54.Dogiel VA. 1925. New parasitic infusoria from the stomach of reindeer (Rangifer tarandus). Arch Russ Protistol 4:43–65. [Google Scholar]
- 55.Dogiel VA. 1947. The phylogeny of the stomach-infusorians of ruminants in the light of palaeontological and parasitological data. Q J Microsc Sci 3:337–344. [Google Scholar]
- 56.de Puytorac P. 1994. Traité de Zoologie: anatomie, systématique, biologie. Infusoires ciliés biologie. Infusoires ciliés, tome II. Masson, Paris, France. [Google Scholar]
- 57.de Puytorac P, Grain J, Mignot J-P. 1987. Précis de protistologie. Société Nouvelle des Éditions Boubée, Paris, France. [Google Scholar]
- 58.de Puytorac P, Legendre P, Devaux J. 1984. Essai d'application de l'analyse phénétique à la classification du phylum des Ciliophora. J Protozool 31:496–507. doi: 10.1111/j.1550-7408.1984.tb05491.x. [DOI] [Google Scholar]
- 59.Lynn DH, Corliss JO. 1991. Ciliophora. In Harrison FW, Corliss JO (ed), Microscopic anatomy of invertebrates. Wiley-Liss, New York, NY. [Google Scholar]
- 60.Small EB, Lynn DH. 1981. A new macrosystem for the phylum Ciliophora Doflein, 1901. Biosystems 14:387–401. doi: 10.1016/0303-2647(81)90045-9. [DOI] [PubMed] [Google Scholar]
- 61.Small EB, Lynn DH. 1985. Phylum Ciliophora Doflein, 1901. In Lee JJ, Hutner SH, Bovee ED. (ed), An illustrated guide to the protozoa. Allen Press, Lawrence, KS. [Google Scholar]
- 62.Embley TM, Finlay BJ, Dyal PL, Hirt RP, Wilkinson M, Williams AG. 1995. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc R Soc Lond B Biol Sci 262:87–93. doi: 10.1098/rspb.1995.0180. [DOI] [PubMed] [Google Scholar]
- 63.Wright A-DG, Dehority BA, Lynn DH. 1997. Phylogeny of the rumen ciliates Entodinium, Epidinium and Polyplastron (Litostomatea: Entodiniomorphida) inferred from small subunit ribosomal RNA sequences. J Eukaryot Microbiol 44:61–67. doi: 10.1111/j.1550-7408.1997.tb05693.x. [DOI] [PubMed] [Google Scholar]
- 64.Wright A-DG, Lynn DH. 1997. Monophyly of the trichostome ciliates (phylum Ciliophora: class Litostomatea) tested using new 18S rRNA sequences from the Vestibuliferids, Isotricha intestinalis and Dasytricha ruminantium, and the Haptorian, Didinium nasutum. Eur J Protistol 33:305–315. doi: 10.1016/S0932-4739(97)80008-9. [DOI] [Google Scholar]
- 65.Wright A-DG, Lynn DH. 1997. Phylogenetic analysis of the rumen ciliate family Ophryoscolecidae based on 18S ribosomal RNA sequences, with new sequences from Diplodinium, Eudiplodinium, and Ophryoscolex. Can J Zool 75:963–970. doi: 10.1139/z97-117. [DOI] [Google Scholar]
- 66.Pomajbíková K, Oborník M, Horák A, Petrželková KJ, Grim JN, Levecke B, Todd A, Mulama M, Kiyang J, Modrý D. 2013. Novel insights into the genetic diversity of Balantidium and Balantidium-like cyst-forming ciliates. PLoS Negl Trop Dis 7:e2140. doi: 10.1371/journal.pntd.0002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medinger R, Nolte V, Pandey RV, Jost S, Ottenwaelder B, Schloetterer C, Boenigk J. 2010. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol 19:32–40. doi: 10.1111/j.1365-294X.2009.04478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moon-van der Staay SY, van der Staay GW, Michalowski T, Jouany JP, Pristas P, Javorský P, Kičidayová S, Varadyova Z, McEwan NR, Newbold CJ, van Alen T, de Graaf R, Schmid M, Huynen MA, Hackstein JHP. 2014. The symbiotic intestinal ciliates and the evolution of their hosts. Eur J Protistol 50:166–173. doi: 10.1016/j.ejop.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Prokopowich CD, Gregory TR, Crease TJ. 2003. The correlation between rDNA copy number and genome size in eukaryotes. Genome 46:48–50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- 70.Cavalier-Smith T. 1985. Eukaryote gene numbers, non-coding DNA and genome size. John Wiley & Sons, Ltd., New York, NY. [Google Scholar]
- 71.Boucher N, Vaulot D, Partensky F. 1991. Flow cytometric determination of phytoplankton DNA in cultures and oceanic populations. Mar Ecol Prog Ser 71:75–84. doi: 10.3354/meps071075. [DOI] [Google Scholar]
- 72.Zhu F, Massana R, Not F, Marie D, Vaulot D. 2005. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol Ecol 52:79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 73.Sylvester JT, Karnati SKR, Dehority BA, Morrison M, Smith GL, St-Pierre NR, Firkins JL. 2009. Rumen ciliated protozoa decrease generation time and adjust 18S ribosomal DNA copies to adapt to decreased transfer interval, starvation, and monensin. J Dairy Sci 92:256–269. doi: 10.3168/jds.2008-1417. [DOI] [PubMed] [Google Scholar]
- 74.Ishaq SL, Wright A-DG. 2014. Design and validation of four new primers for next-generation sequencing to target the 18S rRNA gene of gastrointestinal ciliate protozoa. Appl Environ Microbiol 80:5515–5521. doi: 10.1128/AEM.01644-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Neefs J-M, Van de Peer Y, De Rijk P, Chapelle S, De Wachter R. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res 21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neefs J-M, Van de Peer Y, Hendriks L, De Wachter R. 1990. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res 18(Suppl):2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.