ABSTRACT
Hepatitis C virus (HCV) infection is characterized by persistent replication of a complex mixture of viruses termed a “quasispecies.” Transmission is generally associated with a stringent population bottleneck characterized by infection by limited numbers of “transmitted/founder” (T/F) viruses. Characterization of T/F genomes of human immunodeficiency virus type 1 (HIV-1) has been integral to studies of transmission, immunopathogenesis, and vaccine development. Here, we describe the identification of complete T/F genomes of HCV by single-genome sequencing of plasma viral RNA from acutely infected subjects. A total of 2,739 single-genome-derived amplicons comprising 10,966,507 bp from 18 acute-phase and 11 chronically infected subjects were analyzed. Acute-phase sequences diversified essentially randomly, except for the poly(U/UC) tract, which was subject to polymerase slippage. Fourteen acute-phase subjects were productively infected by more than one genetically distinct virus, permitting assessment of recombination between replicating genomes. No evidence of recombination was found among 1,589 sequences analyzed. Envelope sequences of T/F genomes lacked transmission signatures that could distinguish them from chronic infection viruses. Among chronically infected subjects, higher nucleotide substitution rates were observed in the poly(U/UC) tract than in envelope hypervariable region 1. Fourteen full-length molecular clones with variable poly(U/UC) sequences corresponding to seven genotype 1a, 1b, 3a, and 4a T/F viruses were generated. Like most unadapted HCV clones, T/F genomes did not replicate efficiently in Huh 7.5 cells, indicating that additional cellular factors or viral adaptations are necessary for in vitro replication. Full-length T/F HCV genomes and their progeny provide unique insights into virus transmission, virus evolution, and virus-host interactions associated with immunopathogenesis.
IMPORTANCE
Hepatitis C virus (HCV) infects 2% to 3% of the world’s population and exhibits extraordinary genetic diversity. This diversity is mirrored by HIV-1, where characterization of transmitted/founder (T/F) genomes has been instrumental in studies of virus transmission, immunopathogenesis, and vaccine development. Here, we show that despite major differences in genome organization, replication strategy, and natural history, HCV (like HIV-1) diversifies essentially randomly early in infection, and as a consequence, sequences of actual T/F viruses can be identified. This allowed us to capture by molecular cloning the full-length HCV genomes that are responsible for infecting the first hepatocytes and eliciting the initial immune responses, weeks before these events could be directly analyzed in human subjects. These findings represent an enabling experimental strategy, not only for HCV and HIV-1 research, but also for other RNA viruses of medical importance, including West Nile, chikungunya, dengue, Venezuelan encephalitis, and Ebola viruses.
INTRODUCTION
Hepatitis C virus (HCV) is a positive-strand RNA virus in the Hepacivirus genus of the Flaviviridiae family and an important blood-borne pathogen (1, 2). HCV infects an estimated 2% to 3% of the world population, and it is a major cause of hepatitis, cirrhosis, and hepatocellular carcinoma (3). HCV replicates with an error-prone RNA-dependent RNA polymerase (RdRp) and persists as a diverse quasispecies in infected individuals (4). The virus also demonstrates remarkable diversity at a global level, with at least seven major genotypes that differ by approximately 30% at the nucleotide level (5, 6).
In contrast to the generally high intrahost diversity of HCV in chronic infection, virus transmission is associated with a significant population genetic bottleneck (7–9). Previous studies based on single-genome sequencing (SGS) of acute infection plasma viral RNA (vRNA) revealed that HCV generally exhibits early random diversification, thereby allowing for a precise phylogenetic inference and enumeration of transmitted/founder (T/F) virus sequence lineages based on partial genome sequences (9, 10). Extension of this strategy to span the complete genome represents a different strategy for characterization of complete HCV genomes compared with previous studies, which used population sequencing of plasma vRNA to generate a consensus of the circulating quasispecies (11–14). The T/F strategy is notable for identifying actual genomes that are responsible for transmission and productive clinical infection and that by inference encode all of the essential viral elements necessary and sufficient for productive infection of human liver cells in vivo. Moreover, T/F viruses and their progeny represent the initial targets of host immune defenses.
Consensus sequence based approaches to clone representative HCV genomes have been used to generate complete genomes for genotypes 1 to 4. Clones developed using this approach contain intact viral open reading frames (ORFs), and introduction of these clones into chimpanzees led to productive infection (11–14). There is a relative paucity of full-length genotype 3 and 4 genomes, which are of interest because of their distinct clinical profile (genotype 3) (15) and their extraordinarily high prevalence in Egypt (genotype 4) (16). The first complete genotype 3 sequences were reported for genotype 3g, 3h, 3i, and 3k isolates in 2011 (17, 18) and for genotypes 3a and 4a in 2013 (19). Other consensus-based genotype 3a and 4a genomes lack the terminal 3′ untranslated region (UTR) but have been complemented with 3′ UTR sequences from homologous or heterologous genotypes to successfully generate subgenomic replicons (20, 21) and molecular clones that are infectious in chimpanzees (14). Despite the success of consensus clones in supporting virus replication in vivo, only JFH-1 (and its derivatives) and other highly adapted virus strains (22–24) replicate efficiently in in vitro tissue culture systems.
The precise molecular identification of T/F genomes by SGS is a recently developed strategy for studying the transmission and early immunopathogenesis of RNA virus infections. It was first developed for human immunodeficiency virus type 1 (HIV-1) (25, 26) and validated with simian immunodeficiency virus (SIV) (27). The identification of T/F HIV-1 and SIV genomes has become a central feature of studies aimed at characterizing viral transmission, natural history, immunopathogenesis, and candidate vaccines (28–35). Such studies revealed that in HIV-1 infection, transmission generally resulted from acquisition of a single virus, that multiplicity of infection varied with clinicoepidemiological infection risk, and that T/F genomes replicated preferentially in CD4 T cells (30, 31). In addition, T/F HIV-1 viruses exhibited distinctive patterns of coreceptor utilization, neutralization sensitivity, potential N-linked glycosylation (PNG) distribution, dendritic cell interaction, envelope content, and sensitivity to type I interferon (28, 31, 36–40). HIV-1 and SIV vaccine studies in humans and animal models used T/F analyses to detect sieving of the virus quasispecies at transmission or shortly thereafter and may indicate vaccine-mediated antiviral activity (32, 34, 35, 41). These findings in HIV-1 infection provide a strong scientific rationale for analysis of T/F genomes of HCV as a means to probe transmission, virus biology, and virus-host interactions relevant to vaccine development.
HCV has distinctive features in its RNA sequence, genome organization, life cycle, replication strategy, and early evolution compared with HIV-1 and SIV that could pose challenges for inferring full-length T/F genomes. This includes a highly ordered secondary RNA structure (42), 5′ and 3′ termini lacking repeated elements, a 3′ terminus that consists of a poly(U/UC) tract of variable length preceding a highly conserved 98-nucleotide (nt) X-tail (43, 44), a prolonged infected-cell life span associated with the accumulation of as many as 40 replication complexes per cell (45), nonuniform evolution across the genome (8, 46–48), and reports of selective sweeps, population bottlenecks, shifts in viral lineage predominance, or compartmentalized infection early after infection (4, 47, 49). To account for these challenges, we developed two complementary mathematical models to explore early HCV evolution (9) and an experimental strategy based on SGS (9, 25, 26) of acute infection plasma vRNA sequences to amplify large overlapping internal segments of the viral genome, followed by adapter-primed 5′- and 3′-terminal amplifications and a “bridging” SGS method that incorporated a molecular cloning step to span the poly(U/UC) tract. This strategy allowed us to generate 14 full-length molecular clones containing variable-length poly(U/UC) tracts corresponding to 7 distinct T/F genomes representing genotypes 1a, 1b, 3a, and 4a and to perform a comparative analysis of acute-phase, chronic-phase, and T/F HCV genomes.
RESULTS
Single-genome sequencing of acute and chronic infection plasma vRNA.
Serially collected plasma specimens from 18 acutely infected plasma donors and 11 chronically infected control subjects were used as sample material for SGS (Table 1). The acute infection samples were seroconversion panels used in the development and validation of clinical diagnostic tests (Zeptometrix, Inc., and SeraCare Life Sciences, Inc.). The individuals who contributed these plasma specimens were qualified for plasma donation and had been screened extensively for findings of risk factors associated with the acquisition of HCV infection. Despite these efforts, they were found in the course of once- or twice-weekly plasma donations to become HCV RNA positive. Viral load, anti-HCV antibody kinetic data, and sample time point selection used for generation of full-length genome sequences and clones in the present study are shown in Fig. 1. Because plasma samples were deidentified and study subjects had previously denied risk factors for HCV infection, it was not possible to further assess risk behaviors that might have been associated with viral acquisition or monitor subsequent disease progression in these individuals. Chronically infected subjects were from clinical outpatient services at the University of Alabama at Birmingham. All subjects were treatment naive for anti-HCV therapeutics for the duration of the sampling period.
TABLE 1 .
Viral sequence characteristics in acute and chronic infection
| Infection status | Subject no. | Genotype | Region(s) covered | No. of amplicons | Mean intralineage nucleotide diversity (%)a | No. of T/F genomesb |
|---|---|---|---|---|---|---|
| Acute | 10002 | 1a | 5′ half | 31 | 0.104 | 13 |
| 10012 | 1a | 5′ half | 129 | 0.035 | 3 | |
| 10017 | 1a | 5′ half | 192 | 0.037 | 4 | |
| 10020 | 1a | 5′ half | 64 | 0.046 | 10 | |
| 10021 | 1a | 5′ UTR | 6 | 0.000 | 1 | |
| 5′ half | 96 | 0.027 | ||||
| 3′ half | 27 | 0.031 | ||||
| Poly(U/UC) | 46 | NAc | ||||
| X-tail | 4 | 0.000 | ||||
| 10024 | 1a | 5′ half | 112 | 0.025 | 6 | |
| 10025 | 1a | 5′ UTR | 5 | 0.000 | 1 | |
| 5′ half | 92 | 0.044 | ||||
| 3′ half | 16 | 0.042 | ||||
| Poly(U/UC) | 22 | NA | ||||
| X-tail | 5 | 0.460 | ||||
| 10029 | 1a | 5′ half | 201 | 0.037 | 9 | |
| 10062 | 1a | 5′ half | 140 | 0.025 | 3 | |
| 105686 | 1a | 5′ half | 23 | 0.055 | 2 | |
| 106889 | 1a | 5′ half | 87 | 0.024 | >30 | |
| 110069 | 1a | 5′ UTR | 8 | 0.000 | 4 | |
| 5′ half | 118 | 0.021 | ||||
| 3′ half | 22 | 0.035 | ||||
| Poly(U/UC) | 33 | NA | ||||
| X-tail | 12 | 0.000 | ||||
| 6213 | 1a | 5′ half | 41 | 0.000 | 3 | |
| 6222 | 1a | 5′ half | 17 | 0.082 | 4 | |
| 10051 | 1b | 5′ UTR | 10 | 0.250 | 1 | |
| 5′ half | 124 | 0.020 | ||||
| 3′ half | 28 | 0.042 | ||||
| Poly(U/UC) | 38 | NA | ||||
| X-tail | 11 | 0.235 | ||||
| 9055 | 3a | 5′ UTR | 8 | 0.031 | 1 | |
| 5′ half | 157 | 0.034 | ||||
| 3′ half | 9 | 0.020 | ||||
| Poly(U/UC) | 45 | NA | ||||
| X-tail | 4 | 0.000 | ||||
| 10003 | 3a | 5′ half | 133 | 0.031 | >30 | |
| 105431 | 4a | 5′ UTR | 40 | 0.026 | 2 | |
| 5′ half | 59 | 0.017 | ||||
| 3′ half | 43 | 0.029 | ||||
| Poly(U/UC) | 69 | NA | ||||
| X-tail | 15 | 0.000 | ||||
| Chronic | ARJA6267 | 1a | 5′ half | 43 | 1.249 | NA |
| Poly(U/UC) | 8 | NA | ||||
| BLMI6862 | 1a | 5′ half | 22 | 1.013 | NA | |
| Poly(U/UC) | 14 | NA | ||||
| JOTO6422 | 1a | 5′ half | 21 | 1.555 | NA | |
| Poly(U/UC) | 9 | NA | ||||
| KNPH3730 | 1a | 5′ half | 13 | 0.929 | NA | |
| Poly(U/UC) | 23 | NA | ||||
| LAST90001 | 1a | 5′ half | 19 | 0.730 | NA | |
| ROMI6847 | 1a | 5′ half | 18 | 0.879 | NA | |
| Poly(U/UC) | 9 | NA | ||||
| SLRO5563 | 1a | 5′ half | 29 | 1.087 | NA | |
| Poly(U/UC) | 13 | NA | ||||
| WEPA5774 | 1a | 5′ half | 44 | 1.708 | NA | |
| Poly(U/UC) | 10 | NA | ||||
| WHRO3882 | 1a | 5′ half | 22 | 0.504 | NA | |
| WIMI4025 | 1a | 5′ half | 36 | 1.323 | NA | |
| Poly(U/UC) | 16 | NA | ||||
| WIMI90003 | 1a | 5′ half | 28 | 1.013 | NA |
Percent divergence scores for all regions except the poly(U/UC) were determined by multiple pairwise alignment analysis using the DIVEIN software suite.
T/F estimates are based on combined quarter- and half-genome analysis.
NA, not applicable.
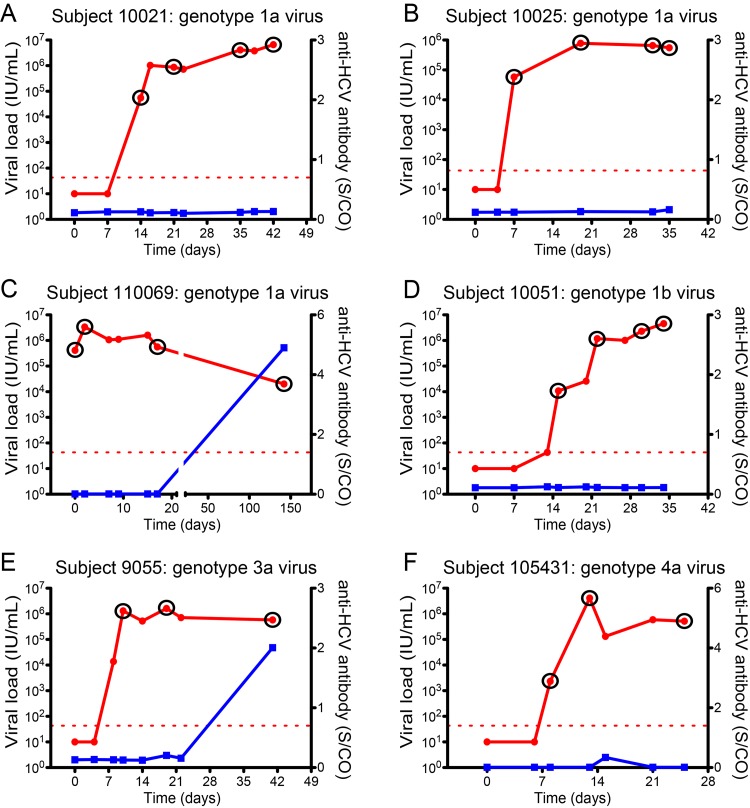
FIG 1 .
Plasma viral RNA and anti-HCV antibody kinetics in acutely infected subjects. Kinetics of viral RNA (red line) and anti-HCV antibody (blue line) are depicted. The red dashed line is the lower limit of sensitivity of the viral RNA assay. S/CO denotes the signal to cutoff ratio. Circled time points are samples that were used to generate full or partial genomes for genetic analysis and molecular clone construction.
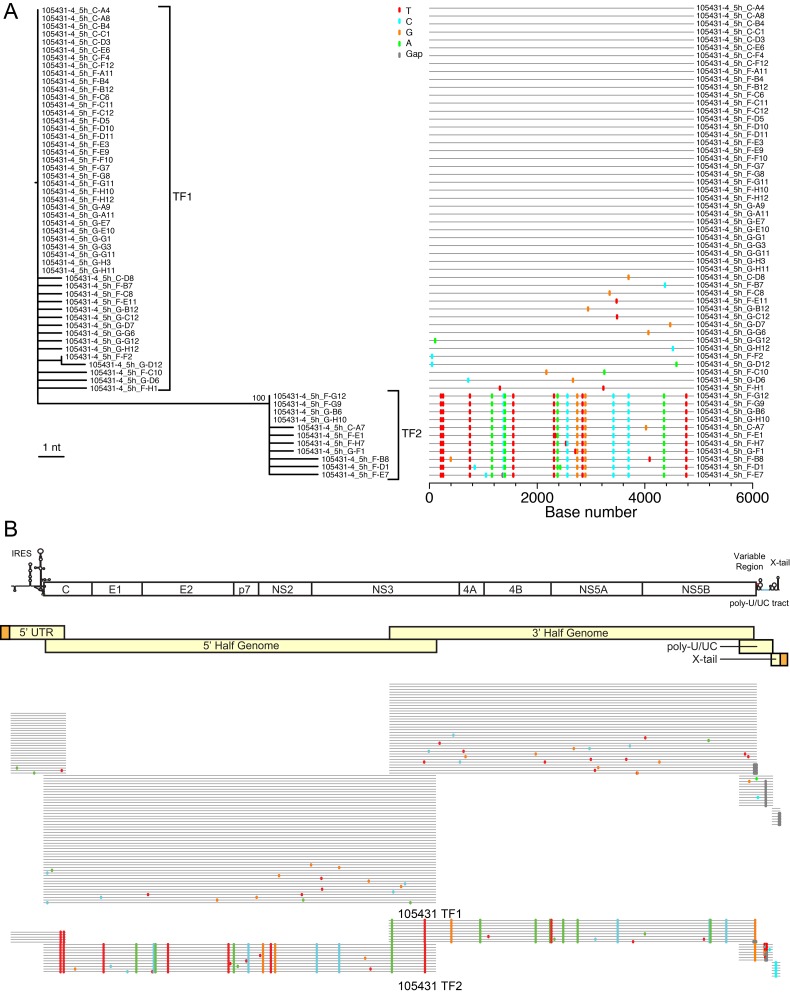
SGS was initially performed on 5′ half genomes from acute and chronic infection samples in order to identify and enumerate T/F HCV sequence lineages (Table 1). Figure 2A shows a representative maximum likelihood phylogenetic tree and Highlighter plot for 5′-half-genome sequences from subject 105431, revealing two discrete genotype 4a lineages. The inferred T/F genomes of these two lineages differed by 0.35% nucleotides (17/4,906 bases), whereas sequence diversity within each T/F lineage was exceedingly low, with a mean of 0.017% diversity (range, 0.000 to 0.082%). This pattern of extremely limited early viral sequence variation within discrete viral lineages was typical of all acutely infected subjects and adhered to a model of random early diversification that exhibited near star-like phylogeny. These low-diversity sequence lineages coalesced to distinct, unambiguous T/F genomes. In all, we studied 18 subjects with acute HCV infection, including 14 who were productively infected by more than one genetically distinguishable virus. The average interlineage nucleotide diversity among these subjects infected by more than one T/F virus ranged from 0.123 to 6.188%, which is typical of the spectrum of diversity found in chronically infected subjects. Figure 2B shows the strategy utilized to infer full-length T/F genomes from overlapping amplicons consisting of the 5′ UTR, 5′ and 3′ half genome, poly(U/UC) tract, and X-tail fragments. In all cases, diversification in regions of the HCV 5′ UTR and open reading frames (ORFs) conformed to a model of random diversification from discrete genomes. These results for complete viral genomes substantially extend our earlier findings based on quarter-genome analyses of some of these same study subjects (9). Importantly, additional T/F lineages were not identified as overlapping genome segments were amplified and sequenced using different sets of primers targeting different portions of the viral genome, indicating that primer-dependent selective amplification was not a confounder. In chronically infected subjects, HCV sequences were heterogeneous (range, 0.062% to 3.934% diversity) and did not show evidence of the low-diversity lineages typical of acute infection. An important feature of SGS is that it precludes artifactual in vitro recombination between genetically distinct target genomes due to Taq polymerase template switching (25, 26, 28). This enabled us to look across the viral genomes of subjects infected by more than one T/F virus for evidence of in vivo recombination. Among 1,589 sequences and 6,960,866 nucleotides analyzed by Highlighter plot inspection and the GARD (50) and Recco (51) recombination identification tools, we failed to identify a single sequence with evidence of recombination.
FIG 2 .
Identification of two full-length T/F viral genomes in acutely infected subject 105431. (A) The viral diversity found in subject 105431 is depicted for the 5'-half-genome fragment by a maximum likelihood phylogenetic tree and Highlighter plot. (B) The HCV genome is depicted with gene organization and major RNA secondary structures at the top followed by the amplification strategy for generating full-length HCV genomes in light yellow. Adaptors added to the 5' and 3' termini are depicted in orange. Highlighter plots for each region shown below demonstrate that a high degree of homology is maintained throughout the genomes within the T/F lineages, which are distinguished by large sets of shared polymorphisms.
Genomic organization and phylogenetic analysis of T/F sequences.
The genome organization of each T/F sequence revealed intact ORFs. Figure S1 in the supplemental material depicts a maximum likelihood tree of these sequences, together with representative globally circulating strains corresponding to the major HCV genotypes. The T/F genomes from subjects 10021, 10025, and 110069 were all genotype 1a viruses, whereas that from subject 10051 was a subtype 1b virus. Subject 9055 was infected with a genotype 3a T/F virus and subject 105431 with genotype 4a T/F viruses. The T/F virus from 10051 was most closely related to the consensus clone Con1 from Germany at 92.8% nucleotide identity (52). The virus from subject 10021 was most closely related to isolate AX663428 from a patient in France with 94.9% identity, while those from subjects 10025 and 110069 were most closely related to isolate HEC278830 sequenced from a British patient with 93.2% and 93.1% identity, respectively (53). The two genotype 4a T/F viruses from subject 105431 demonstrated close phylogenetic linkage, the next most closely related virus being isolate 01-09 from the United States, with 94.2% nucleotide identity (54). Genotype 3a T/F 9055 was most closely related to the Japanese isolate TYMM at 91.6% nucleotide identity (55). These results indicate that the T/F sequences generated were well distributed throughout the phylogeny of HCV and its globally circulating genotypes.
Potential N-linked glycosylation site analysis of T/F versus chronic envelope glycoproteins.
In HIV-1 infection, T/F viruses generally have fewer potential N-linked glycosylation (PNG) sites than viruses sampled in chronic infection (36, 38, 40), with N-linked glycans playing a protective role against antibody neutralization (56, 57). In HCV infection, a study in chimeric immunodeficient mice transplanted with human hepatocytes reported a distinctive amino acid signature pattern in envelope (198T, 448D, 474Y, and 570D/A), including one E2 PNG site that was associated with selective HCV transmission (58). Our data set included T/F HCV envelope sequences from 18 acutely infected humans and a corresponding set from 11 chronically infected patients. Figure S2 depicts the percentage of PNG motifs found throughout the E1 and E2 proteins for 274 chronic and 51 T/F genotype 1a envelope sequences. No significant differences in PNG sites between T/F and chronic HCV genomes were observed, nor were any differences found in the 4 amino acids reported for the chimeric mouse model of HCV transmission.
Hypervariation in the 3′ poly(U/UC) tract.
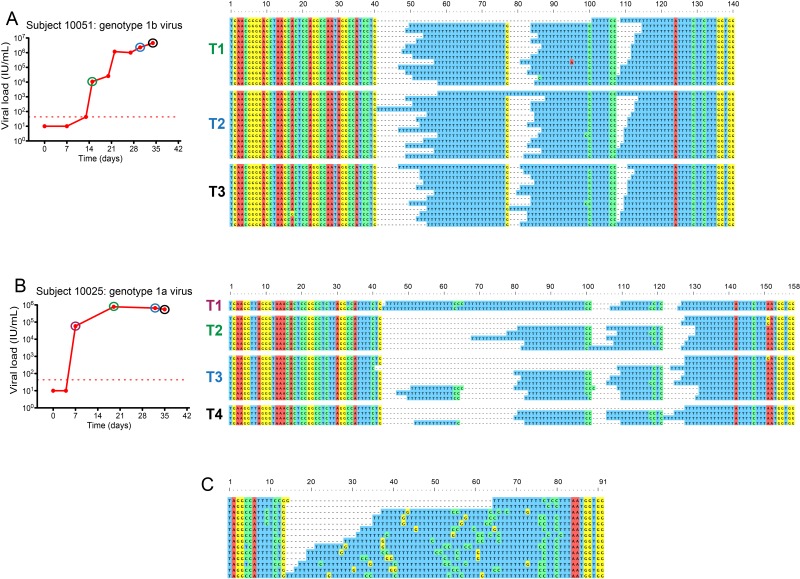
The HCV 3′ UTR consists of a variable region containing two stem-loops (59), a poly(U/UC) tract of various lengths, a transitional region with a higher concentration of non-U nucleotides, and the highly conserved 98-nucleotide X-tail (43, 44, 60). Reverse transcription, amplification, and sequencing of long homopolymer tracts are technically challenging because of the potential for polymerase slippage (61–64). In longitudinal acute-phase samples from subjects 10021, 10025, 110069, 10051, 9055, and 105431 and from single samples from chronically infected subjects ARJA6267, BLMI6862, JOTO6422, KNPH3730, ROMI6847, SLRO5563, WEPA5774, and WIMI4025, we observed striking variability in sequence lengths of poly(U/UC) sequences within and among all subjects (Fig. 3; see Fig. S3 in the supplemental material). In contrast, there was virtually no length variation in any of 3,652,088 nucleotides corresponding to the remaining ~98% of the genome exclusive of the poly(U/UC) tract. In acutely infected subjects, the viral sequences with the longest poly(U/UC) tracts exhibited the greatest variation in sequence length, and this was concentrated in the homopoly(U) stretches (Fig. 3B; see Fig. S3B and S3C). These sequence sets were characterized by a longer maximum poly(U/UC) tract length (112 to 162 nucleotides), longer maximum homopoly(U) tract lengths (36 to 115 nucleotides), and a smaller average proportion on non-U nucleotides as a percentage of the poly(U/UC) tract (10%). In subjects with shorter poly(U/UC) tracts, the length variability in homopoly(U) stretches was reduced (Fig. 3A; see Fig. S3A and S3D). In these latter sequence sets, the maximum poly(U/UC) tract length was 66 to 95 nucleotides, the maximum homopoly(U) tract length was 23 to 47 nucleotides, and the average proportion of non-U nucleotides interspersed in the poly(U/UC) tract was 13%. In subjects 10021, 10025, 10051, 9055, and 105431, we analyzed multiple early time points to look for progressive loss or gain of homopoly(U) sequences during early infection (Fig. 3A and B; see Fig. S3). Within each subject, we could identify no such pattern, and similar poly(U/UC) length variation was present at every time point.
FIG 3 .
Length variation in the HCV poly(U/UC) tract. Shown are various degrees of length and sequence variation in the poly(U/UC) tract in acute infection subjects 10051 (A) and 10025 (B) and in chronic infection subject BLMI6862 (C). See also Fig. S3 and S4 in the supplemental material.
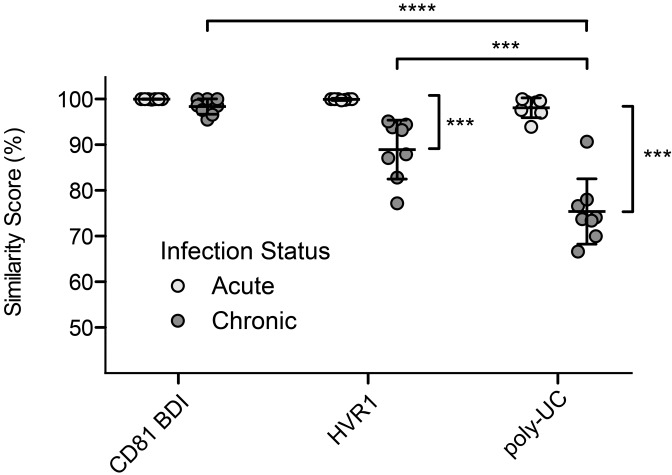
Variation in the length of the poly(U/UC) tract in viral genomes emanating from single T/F genomes could result from slippage of the HCV RdRp in vivo or slippage by any of the polymerases used in the amplification and sequencing steps in vitro. HCV RdRp is highly processive (65, 66) and tightly grips the template RNA (67, 68). In contrast, the Moloney murine leukemia virus (MMLV) reverse transcriptase (RT) and Thermus aquaticus (Taq) polymerase enzymes used to generate and amplify cDNA from vRNA are prone to slippage and template switching (69–72), likely due to inherent structure-function relationships of the enzymes (73–77). Additionally, both MMLV and Taq enzymes are particularly error prone in homopolymer tracts (61, 74, 78, 79). To evaluate potential sources of the variability that we observed in poly(U/UC) sequences, we analyzed each step in the analysis of vRNA sequences individually. We found that the Escherichia coli enzymes involved in plasmid replication during subcloning did not introduce frequent variation in the poly(U/UC) tract: the vast majority (96.6%) of bacterial colonies that had been transformed with a plasmid containing a known homopoly(U) tract nucleotides yielded the exact same plasmid after culture (see Fig. S5A in the supplemental material). Amplification by Taq polymerase potentially contributed more to the observed variation in poly(U/UC) sequences than did the E. coli DNA polymerase but had a minor overall effect on the median length of poly(U/UC) tracts: amplification of a molecularly cloned DNA construct with a known homopoly(U) sequence followed by plasmid subcloning and sequencing yielded a median length that was 99.4% of the expected length, but 83.3% of the poly(U/UC) tract sequences contained additions, deletions, or substitutions (see Fig. S5B). To evaluate the impact of the MMLV RT infidelity on the length and sequence variation observed in the poly(U/UC) tract, we chemically synthesized homologous 40-nucleotide RNA oligomers that differed only by the substitution of two cytidine residues within the 40-nucleotide homopoly(U) tract (see Fig. S5C to S5F). This was done to test directly MMLV polymerase slippage and the hypothesis that non-U nucleotides within a homopoly(U) tract would diminish its frequency. The two RNA templates were subjected to reverse transcription, subcloning, and SGS. Among colonies derived from the 40-nucleotide homopoly(U) RNA template, we observed wide variation in the length of the amplified homopoly(U) sequences that resulted in a trend toward truncation of the homopoly(U) tract. A total of 96.9% of the sequences contained differences from the input RNA template, and the median tract length was 80% of the expected length (see Fig. S5C). Conversely, there was less variation in the median length of the homopoly(U) tract in products sequenced from the RNA template that contained two cytidine residues (see Fig. S5D). The median length of the homopoly(U) tracts was 97.5% of the expected length. The difference in sequence lengths generated from the two otherwise identical homopoly(U) RNA templates was highly significant (P < 0.0001). Altogether, these findings indicate that the MMLV RT step is most error prone with regard to processivity and template slippage, slippage most often results in shortening of sequences, and RT slippage is potentially the primary contributor to the length variation that we observed in the poly(U/UC) region. However, these findings also highlighted other changes in the poly(U/UC) tract that could not be attributed to RT slippage and that were most notable in chronic HCV sequences (Fig. 3C; see Fig. S4 in the supplemental material). This included the interspersion of numerous C and G substitutions in the homopoly(U) region and variation in the transitional region. In order to quantify and compare this variation in sequences from acute- and chronic-phase subjects, we subjected a 45-nucleotide region of the poly(UC) tract region immediately 5′ of the X-tail to Needleman-Wunsch pairwise analysis. We compared variation in this region to that in the highly conserved CD81 binding domain I (BDI) (80) and hypervariable region 1 (HVR1) of the envelope gene E2 (81) (Fig. 4; see Table S2 in the supplemental material). Acute infection sequences did not differ significantly in diversity among the poly(U/UC), HVR1, and CD81 BDI regions in this analysis, consistent with the recent evolution of these sequences from discrete T/F genomes. However, among chronic infection sequences, the HVR1 (77.2 to 95.1% similarity score) and poly(UC) (66.6 to 90.7% similarity score) sequences exhibited significantly greater diversity than CD81 BDI sequences (95.5 to 100% similarity score; P < 0.001 for each comparison). Surprisingly, sequences of chronic infection poly(UC) exhibited even greater mean diversity (66.6 to 90.7% similarity score) than sequences of chronic infection HVR1 (77.2 to 95.1% similarity score, P < 0.001), a region that has been previously described as harboring the highest within-host variation (82).
FIG 4 .
Nucleotide sequence similarity scores of the poly(UC) tract, Env E2 hypervariable region and CD81 binding domain I in acute and chronic infection. The y axis denotes the mean percent similarity score from Needleman-Wunsch pairwise analysis of genomic regions corresponding to the CD81 binding domain I (AF009606 positions 1491 to 1530), the E2 hypervariable region (AF009606 positions 1575 to 1614), and a 45-nucleotide portion of the poly(U/UC) tract immediately 5' of the X-tail (AF009606 positions 9504 to 9548). Means and standard deviations are indicated (***, P < 0.001; ****, P < 0.0001).
Analysis of 3′ UTR sequence motifs.
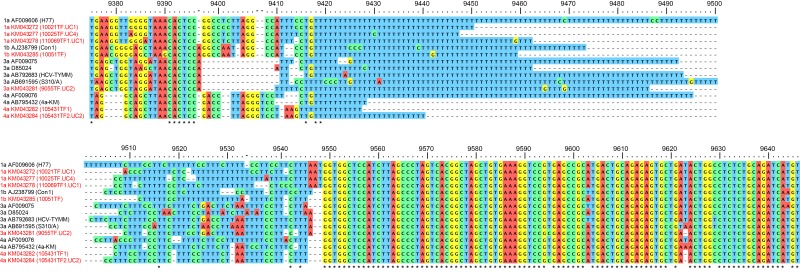
For detailed analysis of the 3′ UTR, we aligned this region from the 7 full-length T/F genomes we inferred along with representative HCV sequences from the Los Alamos HCV database (Fig. 5). The variable region immediately 5′ of the poly(U/UC) tract showed sequence patterns shared within genotypes but not between genotypes. This region is predicted to form two stem-loop structures that play a role in the viral life cycle (59, 83). Small insertions and deletions in this region are well tolerated in vitro and in vivo, but large deletions have a significant impact on optimal RNA replication (59, 83, 84). Interestingly, and consistent with previous reports, genotype 3a sequences had an abbreviated variable region approximately 50% shorter than that of genotypes 1a, 1b, 2a, and 4a. Nevertheless, a short island of conservation constituting the predicted 3′ miR-122 binding motif “ACACUCC” (85) is preserved immediately 5′ of the gap in the 3a genomes. The homopoly(U) tracts of the genotype 4a viruses sequenced in this study were shorter than those for the other genotypes studied. The length of the homopoly(U) and poly(U/UC) are known to have an important impact on viral replication in vitro. Minimal lengths of >16 nt (86) for the homopoly(U) and approximately >40 nt for the poly(U/UC) (59, 83) have been reported in vitro. The length of the poly(U/UC) tract is also known to play a critical role in permitting robust viral replication in vivo, with the longer poly(U/UC) structures generally favored over shorter structures (11). The X-tails of T/F viruses were highly conserved within and between genotypes, with the exception of a short polymorphic region spanning nt 9620 to 9623 that is predicted to be located at the tip of the X-tail stem-loop 1 (43) and would not be expected to alter the RNA folding structure of these regions (87). These data indicate that the variable region sequences 5′ of the poly(U/UC) tract of genotype 1, 3, and 4 genomes are conserved within, but not between, genotypes, while the X-tail is highly conserved and the poly(U/UC) tract is always highly variable.
FIG 5 .
T/F genotypes 1a, 1b, 3a, and 4a 3' UTR sequences compared with representative HCV reference sequences. The numbering on the top corresponds to the H77 AF009606 genome. Asterisks indicate positions with nucleotide identity. Dashes indicate deletions. The 5' codon represents the stop codon at the end of the NS5B gene. T/F genomes generated in this study are indicated in red.
Construction and analysis of T/F molecular clones.
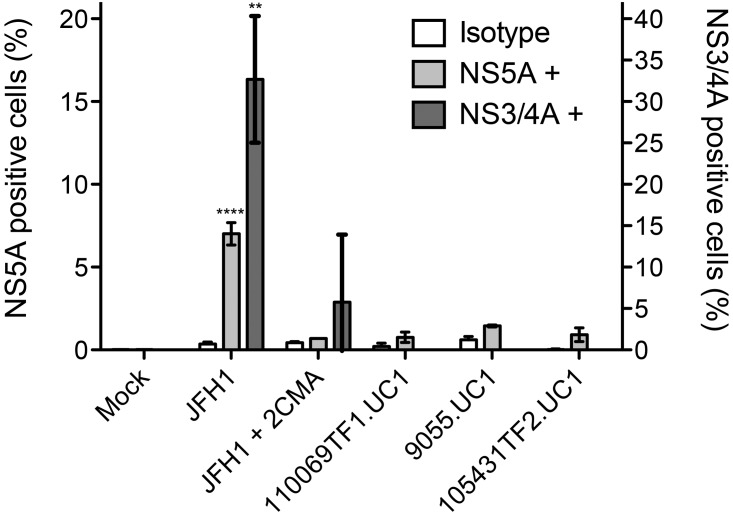
Excluding the poly(U/UC) region, the sequences of seven T/F genomes corresponding to genotypes 1a, 1b, 3a, and 4a from six subjects could be inferred unambiguously. These sequences were chemically synthesized and subcloned into plasmids together with poly(U/UC) sequences representing the spectrum of variation evident in the sequences from multiple early time points (see Fig. S6 in the supplemental material). Plasmids contained a 5′ T7 promoter and 3′ linearization restriction site to facilitate in vitro transcription. Plasmids containing HCV genomic clones were grown large scale, and sequence was confirmed. T/F molecular clones were used to generate vRNA for studies of innate immune signaling, with findings reported in the accompanying article (88) and elsewhere (89). vRNAs expressed from these clones were also tested for translation and replication competence by electroporation into Huh 7.5 cells stably expressing an HCV replication-dependent fluorescence relocalization reporter (90). Expression of functional HCV NS3/4A protein in these cells results in the nuclear localization of otherwise mitochondrially tethered red fluorescent protein (RFP). Viral protein expression was assessed 48 h after electroporation by monitoring the translocation of the RFP and by flow cytometry for NS5A protein expression. Figure 6 shows that electroporation of Huh 7.5 cells with vRNA from the replication-competent molecular clone JFH-1 yielded significant numbers of cells positive for NS5A (P < 0.0001) and NS3/4A (P < 0.01) expression compared with cells treated with the HCV polymerase inhibitor 2′CMA (91) or with mock-infected cells. In contrast, Huh 7.5 cells electroporated with T/F vRNA corresponding to HCV genotype 1a, 3a, or 4a showed no evidence of virus replication. Additional attempts at launching other T/F molecular clones (see Fig. S6) in Huh 7.5 cells under various conditions yielded similarly negative results. Moreover, efforts to launch these clones in primary fetal hepatoblasts (92) and stem-cell-derived differentiated hepatocyte-like cells (93) were largely unsuccessful; low levels of NS3/4A activity were observed particularly for T/F clone 10051TF.UC1, but successful passage of virus was not achieved (unpublished observations). These data suggest that similar to other unadapted HCV clones (14, 23), T/F molecular clones cannot replicate efficiently in the existing cell culture systems, and additional cellular factors or viral adaptations will be necessary to achieve robust in vitro replication.
FIG 6 .
Comparative launch of T/F and JFH-1 vRNA in Huh 7.5 cells. Huh 7.5 cells were electroporated with in vitro-transcribed RNA from pJFH-1 and T/F molecular clones 110069TF1.UC1, 9055.UC1, and 105431TF2.UC1. NS5A expression, determined by flow cytometry, is represented in white and light gray bars. NS3/4A activity, determined by translocation of the MAVS-NLS-RFP signal, is presented in the dark gray bars. Error bars represent the standard deviation (**, P < 0.01; ****, P < 0.0001).
DISCUSSION
Previously, we described a conceptual and mathematical model of early HCV sequence diversification that allowed for unambiguous identification of T/F genomes based on an analysis of quarter- or half-genome sequences (9). This work in HCV was adapted from earlier studies in HIV-1 that included both partial and full-length genome analyses as well as an empirical validation of this experimental strategy in the SIV-infected Indian rhesus macaque model (25–28, 94, 95). Because of differences in the primary and secondary RNA sequences, genome organizations, life cycles, kinetics of target cell turnover, sequence evolutions, and immunopathogeneses of HCV versus HIV-1 infection, it was not obvious a priori whether HCV diversification across the complete genome would exhibit essentially random diversification early in infection, which is a prerequisite for inferring T/F genomes. Indeed, some models suggested that violations to the Poisson distribution of near-random mutations might be more common in HCV than in HIV-1 (9) due to differences in viral replication strategies or that viral recombination (96–98), compartmentalized infection (4, 99, 100), early selective sweeps (8, 49), population bottlenecking or shifts in predominant virus populations (47, 49), and nonuniform evolution across the genome (8, 46–48) might all obscure virus lineages evolving from discrete T/F genomes. In this study, we found that none of these potential complexities obscured a precise and unambiguous inference of T/F genomes. Only polymerase slippage, presumably due primarily to reverse transcriptase infidelity in the cDNA synthesis step, complicated the identification of T/F genomes. We thus were able to infer full-length T/F genome sequences corresponding to all structural genes and 5′ and 3′ regulatory sequences exclusive of the poly(U/UC) tract from six subjects shortly after the first appearance of plasma viremia (Fig. 1 and 2). The rapid exponential rise in plasma virus load and comparable increase in numbers of productively infected hepatocytes and viral replication complexes likely explain the essentially random diversification that we observed throughout the viral genome. In a separate study where we characterized T/F HCV genomes in human-to-human and human-to-chimpanzee linked transmission pairs, we confirmed that T/F genomes inferred from early viral sequence coalesced to actual transmitted viral genomes found in the donor subjects (M. B. Stoddard, G. M. Shaw, and H. Li, unpublished data).
While it was not the original focus of our study, we had a unique opportunity to look with great sensitivity and specificity for viral recombination in subjects acutely infected by more than one genetically distinct virus. The SGS strategy for generating long amplicons and sequences (5 to 9 kb) is particularly useful in this regard because it is based on Taq polymerase amplification of endpoint-diluted, single cDNA molecules, which obviates the appearance of Taq polymerase-mediated recombination events in finished sequences (25, 26, 28). Based on this powerful technique and an examination of 1,589 partial or full-length sequences from 14 subjects infected with multiple genetically distinct T/F viruses, we failed to identify even one recombinant sequence. This stands in contrast to acute infection by genetically distinct genomes of HIV-1, where most of the circulating HIV-1 sequences are recombinants by 6 weeks of infection (26, 30, 31). This suggests that while HCV recombination is a theoretical concern (98, 101), it is exceedingly uncommon in the setting of acute and early infection. Altogether, the data reported here and previously (9, 10) reach the important conclusion that in most cases of acute HCV infection, discrete T/F viral genomes can be unambiguously identified and their early evolution mapped precisely. This permits the identification of viral genes and expressed proteins that are well suited for transmission and early replication. This in turn allows for comprehensive proteome-wide mapping of selective changes on the evolving viral quasispecies resulting from innate or adaptive immune pressures or drug therapy, and it enables a genetic analysis of virus sieving that could result from prior vaccination with candidate immunogens (102). It further allows for an analysis of viral superinfection in the setting of preexisting immunity to HCV.
Variation in T/F E1 and E2 envelope glycoproteins has been hypothesized to contribute to selective transmission of HCV viruses and the observed population bottleneck (58). Both E1 and E2 are heavily glycosylated, and these glycans are critically important for appropriate protein folding (103) and receptor engagement (104). This glycosylation is highly conserved among published HCV sequences (105) and plays a role in determining viral neutralization sensitivity (106). Brown and colleagues recently reported an envelope signature pattern associated with HCV transmission in human hepatocyte engrafted immunocompromised mice. This motif consisted of 4 amino acid changes (198T, 448 D, 474Y, and 570D/A), including an ablated PNG site at position 448 that distinguished transmitted viruses from the vast majority of viruses in the inoculum. This mutation further altered the entry phenotype of HCV pseudoparticles, consistent with the notion the altered PNG profile contributed to selective virus transmission (58). Our findings comparing 51 HCV T/F genome sequences and their evolved progeny with chronic HCV sequences of the same genotype (1a) failed to confirm this signature of HCV transmission fitness in acutely infected humans (see Fig. S2 in the supplemental material).
Another region of the HCV genome that represents a target of the immune response is the poly(U/UC) tract of the 3′ UTR. The 3′ UTR is of importance because it plays a critical role in the viral life cycle (59, 83, 86) and is the principal pathogen-associated molecular pattern (PAMP) that stimulates the retinoic acid-inducible gene 1 (RIG-I)-dependent innate response to HCV RNA (107, 108). The HCV poly(U/UC) tract and X-tail are notably difficult to amplify and sequence, as reported in the original descriptions of the HCV 3′ UTR (11, 43). This continues to be a challenge for generating full-length HCV sequences. In this study, we found notable length variation in the poly(U/UC) tract even in the setting of acute infection. We thus examined the likely causes of this length variation. HCV RdRp is highly processive (65, 66), and thus it seemed likely that one of the steps in the in vitro synthesis or amplification of HCV cDNA was a more likely cause of poly(U) length variability observed. Not unexpectedly, we found that the MMLV RT was the principal source of poly(U) length variation (see Fig. S5C and S5D in the supplemental material). Retroviral RTs exhibit an “open” configuration that loosely wraps around the RNA/DNA hybrid permitting homologous recombination, an essential property of the viral life cycle (62, 76, 109). Template switching and slippage due to the RT have been well documented in homopolymer tracts (61, 78, 79). Taq polymerase also exhibits a relatively “open” configuration (73), allowing for slippage on homopolymer tracks, which we showed contributes part of the length variation that we observed. Interestingly, we found that introduction of as few as two tandem cytosine residues in a homopoly(U) tract 40 nt in length could substantially anchor the RT and Taq polymerases and limit polymerase slippage. In turn, this likely accounts for the fidelity that we observed in poly(UC) sequences adjacent to homopoly(U) sequences. Given these findings (Fig. 3; see Fig. S3 to S5 in the supplemental material), we concluded that the longer poly(U/UC) tracts that we observed were more likely to be reflective of T/F sequences in vivo than were shorter poly(U/UC) tracts.
For HIV-1, in addition to anchoring viral immunopathogenesis and vaccine studies to unambiguous T/F proteomes, the identification of T/F HIV-1 genomes has been an enabling strategy for elucidating unique biological properties of those viruses responsible for transmission and productive clinical infection. For HCV, we hypothesized that the same could be true. We thus attempted to launch the replication of T/F HCV genomes in the Huh 7.5 cells, which support the replication of JFH-1 and its derivatives but not other primary virus strains. We reasoned that T/F genomes, which reached high viral titers in humans in vivo within weeks of infection (Fig. 1), might be better suited for replication in Huh 7.5 cells, which have defective RIG-I- and Toll-like receptor 3 (TLR3)-mediated cell-intrinsic innate signaling pathways (110, 111), than were consensus clones derived from plasma of chronic infection HCV patients. Unfortunately, this was not the case. While JFH-1 exhibited robust replication in Huh 7.5 cells (Fig. 6), T/F genomes did not. We obtained similar results with primary human fetal hepatoblasts and human embryo-derived differentiated liver cells: both supported replication of JFH-1 but not that of T/F genomes (unpublished data). This observation is consistent with previous reports for unadapted genomes from various genotypes (14, 23, 112) and suggests that the development of cell culture systems that more closely reflect the liver microenvironment or cell culture adaptation of T/F molecular clones may be required for their in vitro propagation (113, 114). In the meantime, the experimental strategy outlined here can be used to study the immunopathogenesis of HCV and structure-function properties of T/F HCV genes and their encoded proteins. Importantly, the approaches described in this study to characterize virus transmission can be extended to other RNA viruses of medical importance, including West Nile, chikungunya, dengue, Venezuelan encephalitis, and Ebola viruses.
MATERIALS AND METHODS
Samples.
All samples were obtained with informed consent and according to the Declaration of Helsinki. Acute-phase samples were obtained from regular source plasma donors via Zeptometrix, Inc., and SeraCare Life Sciences, Inc., while samples from chronically HCV-infected subjects were obtained from the University of Alabama at Birmingham Center for AIDS Research Network of Integrated Clinical Systems. vRNA load was determined by isolation of total nucleic acid using the COBAS AmpliPrep TNAI kit and COBAS TaqMan HCV test v2.0 (Roche Diagnostics). Samples obtained from Zeptometrix were also tested for anti-HCV antibody using the Murex Anti-HCV assay (Abbott Diagnostic Division). SeraCare samples were tested for anti-HCV antibodies with the HCV Ortho ELISA v3.0 assay (Ortho-Clinical Diagnsotics).
Single-genome sequencing, alignment, and phylogenetic analysis of complete HCV genomes.
Complete HCV genomes were amplified, sequenced, and analyzed as previously described (9) with additional modifications to permit study of the complete genomes as described in the Materials and Methods in Text S1 in the supplemental material.
Potential N-linked glycosylation site analysis.
The E1 and E2 regions of HCV from the full panel of chronic and T/F sequences were subjected to PNG analysis using the N-GlycoSite online tool (http://hcv.lanl.gov/content/sequence/GLYCOSITE/glycosite.html) with the AF009606 H77 sequence as a reference (5). The frequency of glycosylation for each site was calculated for each subject, and the difference between acute- and chronic-phase frequencies was assessed by multiple Mann-Whitney tests and the Bonferroni method to account for multiple comparisons. (P values of <0.0026 were considered significant.)
Poly(UC) tract similarity scoring analysis.
Similarity scores for the poly(UC) or transitional region immediately 5′ of the conserved X-tail “GGTGG” motif (corresponding to positions 9504 to 9548 for AF009606) were generated using the EMBOSS Needleman-Wunsch pairwise sequence alignment tool (http://variome.bic.nus.edu.sg/cgi-bin/emboss/needleall/) with the DNAfull cost matrix. Polymerase-associated truncations due to polymerase slippage were described in this study as heavily concentrated in HCV homopoly(U) sequences, and apparent homopoly(U) tract deletions were excluded from similarity score analyses. The HVR1 and CD81BD regions corresponding to AF009606 positions 1491 to 1530 and 1575 to 1614, respectively, were analyzed using the same process. Percent similarity scores were natural log transformed. Differences in variation in either acute or chronic data sets were investigated using one-way analysis of variance (ANOVA). Differences in variation within the CD81 BDI, HVR1, and poly(UC) region between acute and chronic infection data sets were analyzed using independent t test comparisons. The Bonferroni method was used to account for multiple comparisons. (P values of <0.0083 were considered significant.) In cases where homopoly(U) or poly(U/UC) tract lengths are compared based on percent scores, natural log transformation followed by two-tailed unpaired t tests was used for statistical analysis.
Molecular clone construction and RNA transcription.
Fourteen molecular clones corresponding to seven separate transmitted founder viruses were chemically synthesized (Blue Heron Biotech) in four or five sequential fragments with a 5′ T7 promoter, a 3′ in vitro transcription runoff site, and terminal 5′ and 3′ restriction sites to facilitate cloning into the pBR322 or pCR-XL-TOPO vectors (see Fig. S6 in the supplemental material). Plasmid DNA stock was grown in MAX Efficiency Stbl2 cells (Life Technologies), and DNA was purified with the Purelink Maxiprep (Life Technologies) according to the manufacturer’s specifications. Plasmids were linearized using the restriction enzymes described in Fig. S6 (New England Biolabs) and phenol-chloroform purified. vRNA was transcribed using the T7 RiboMAX Express large-scale RNA production system (Promega) and purified using the RNeasy minikit (Qiagen). RNA quality was assessed by agarose gel electrophoresis and quantified with the Qubit assay kit (Life Technologies).
Cell culture, electroporation, and analysis for HCV protein expression.
Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies) containing 10% fetal bovine serum, 10 mM HEPES buffer (Life Technologies), 200 mM GlutaMAX I (Life Technologies), and 100 U/ml penicillin-streptomycin solution (Life Technologies). Five micrograms of vRNA was electroporated into 3 million Huh 7.5 cells using an ECM 830 electroporator (Harvard Apparatus), BTX 0.2-cm cuvettes (Harvard Apparatus), and BTXpress Electro solution (Harvard Apparatus). The instrument settings were 820 V, s pulse length of 99 µs, 5 pulses, and an interval of 1.1 s. The mitochondrial antiviral signaling (MAVS)-based NS3/4A reporter system was visualized on a DMRE fluorescence microscope (Leica Microscopy). Cells were prepared for flow cytometry 48 h to 7 days after electroporation. Briefly, cells were trypsinized and fixed with 2% paraformaldehyde for 20 min at room temperature and then washed and permeabilized with 0.1% saponin–phosphate-buffered saline (PBS). Cells were incubated for 1 h with the anti-NS5A IgG2a antibody 9E10 at a 1:2,000 concentration in 3% FBS–0.1% saponin–PBS, washed, and stained with goat anti-mouse allophycocyanin (APC)-conjugated secondary antibody (Life Technologies) for 1 h. Flow cytometry was performed using a FACSAria II (BD Biosciences) instrument, and the results were analyzed using FlowJo 10 (Tree Star, Inc.). Results were statistically assessed using one-way ANOVA with Bonferroni multiple comparison tests.
Statistical analysis.
Statistics were performed using the SPSS software suite (IBM Corporation) and Prism 5.0d (GraphPad Software). For specific methods, see the respective methods described above.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the full nucleotide sequences of the transmitted/founder viruses are as follows: subject 10021, KM043272 to KM043273; subject 10025, KM043272 to KM043273; subject 10025, KM043274 to KM043277; subject 110069, KM043278 to KM043279; subject 9055, KM043280 to KM43281; subject 105431, KM043282 to KM043284; subject 10051, KM043285; and additional acute and chronic infection sequences, KP666311-KP668776.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
T/F genomes are representative of three of the major HCV genotypes found globally. The maximum likelihood nucleotide tree is based on the full ORFs with T/F genomes identified in this study highlighted in red. GenBank accession numbers are shown with the names of the strains or clones shown in parentheses. Bootstrap values are included for bootstraps >70% to the left of their respective nodes. The bar represents 0.1% variation at a nucleotide level. Download
Potential N-linked glycan sites are highly conserved between acute T/F lineage sequences and chronic infection sequences. The amino acid (AA) position is based on the H77 AF009606 reference sequence and is indicated on the x axis. The error bars represent the standard error of the mean. Download
Poly(U/UC) tract sequence variation in acute infection subjects. Shown are poly(U/UC) tract sequences for acute infection subjects 10021 (A), 110069 (B), 9055 (C), and 105431 (D). Serial samples are color coded with the corresponding viral kinetics presented on the left. Download
Poly(U/UC) tract sequence variation from chronic infection subjects. Shown are poly(U/UC) tract sequences for chronic infection subjects SLRO5563 (A), WEPA5774 (B), WIMI4025 (C), ARJA6267 (D), JOTO6422 (E), KNPH3730 (F), and ROMI6847 (G). Download
Contributions of E. coli, Taq polymerase, and MMLV reverse transcriptase to poly(U/UC) tract variation. (A) DNA from a molecular clone containing a 41-nucleotide homopoly(U) tract (input) was used to transform E. coli. A total of 60 colonies were sequenced revealing rare deletions and no nucleotide substitutions. (B) Plasmid DNA containing an 87-nucleotide homopoly(U) tract was endpoint diluted and subjected to 45 PCR cycles of amplification, cloned into a plasmid, and transfected into E. coli, and 36 colonies were analyzed, revealing contributions of Taq polymerase slippage to poly(U/UC) length variation. (C) Reverse transcription, single-genome amplification, and subcloning were performed on input synthetic RNA molecules containing a 40-nucleotide homopoly(U) tract, revealing substantial poly(U/UC) length variation—more so than in panel A, B, or D. (D) Reverse transcription, single-genome amplification, and subcloning were performed on input synthetic RNA molecules containing a 30-nucleotide homopoly(U) tract. (E) Electrospray ionization mass spectrometry (ESI-MS) demonstrates that the synthetic RNAs used as the input in panel C are homogeneous in length with the expected molecular weight for RNA oligonucleotide UC1 (predicted, 25,019.4; observed, 25,019.3). (F) The synthetic RNA used as the input in panel D is similarly homogeneous in length and with the expected molecular weight for UC2 (predicted, 25,017.4; observed, 25,017.5). Download
Full-length T/F HCV molecular clones. (A to G) All clones include a 5' T7 transcription site and 3' linearization site for use in generating viral RNA in vitro. Restriction sites used to concatenate the full genomes are labeled in green, and the restriction sites used to introduce the genomes into the vectors are labeled in red. In cases where the terminal restriction site and the RNA runoff sites are different, the runoff site is labeled in blue. (F) The distinctive poly(U/UC) tracts for each molecular clone are listed with the corresponding accession number and clone designation given in parentheses. Download
Sample origin and characteristics of single-genome amplification fragments used for analyses. (a) ZM-SP, Zeptometrix seroconversion panel. (b) SC-SP, SeraCare seroconversion panel. (c) UAB-C, University of Alabama at Birmingham, Center for AIDS Research Network of Integrated Clinical Systems. (d) T/F estimates are based on combined quarter- and half-genome analysis. (e) Percent divergence scores for all regions, except the poly(U/UC) tracts, were determined by DIVEIN multiple pairwise alignment analysis. Needleman-Wunsch pairwise dynamic programming was used for poly(U/UC) comparisons, and the results are presented in Fig. 4 and in Table S2. (f) NA, not applicable.
Poly(UC) tract hypervariation as revealed by Needleman-Wunsch pairwise analysis. Needleman-Wunsch similarity scores are presented as a percentage of the maximum possible score. (a) The short poly(U/UC) fragment denotes the first 45 nucleotides immediately 5’ of the X-tail. (b) The long poly(U/UC) fragment denotes the first 60 nucleotides immediately 5’ of the X-tail. (c) In the case of subjects 105431 and ARJA6267, the poly(U/UC) tract was not sufficiently long for a 60- or 65-nt analysis. NA, not applicable.
Oligonucleotide primers used to amplify and sequence viral genome fragments. Primers used to generate cDNA, amplify genome fragments, and sequence partial or complete genomes are listed with the corresponding AF009606 location where applicable.
ACKNOWLEDGMENTS
This work was supported by grants from the NIH AI106000 (G.M.S.), AI007324 (M.B.S.), AI107301 (A.P.), AI099284, CA057973, and DK085713 (C.M.R.) and by a Helmsley Postdoctoral Fellowship for Basic and Translation Research on Disorders of the Digestive System at the Rockefeller University (M.S.). M.v.S. is a recipient of a fellowship from the German Research Foundation.
Footnotes
Citation Stoddard MB, Li H, Wang S, Saeed M, Andrus L, Ding W, Jiang X, Learn GH, von Schaewen M, Wen J, Goepfert PA, Hahn BH, Ploss A, Rice CM, Shaw GM. 2015. Identification, molecular cloning, and analysis of full-length hepatitis C virus transmitted/founder genotypes 1, 3, and 4. mBio 6(2):e02518-14. doi:10.1128/mBio.02518-14.
REFERENCES
- 1.Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152. In Knipe DM, Howley PM (ed), Fields virology, vol 1, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Ray SC, Thomas DL. 2009. Hepatitis C, p 2157–2185. In Mandell GL, Bennett JE, Colin R (ed), Principles and practice of infectious diseases, 7th ed. Churchill Livingstone, Philadelphia, PA. [Google Scholar]
- 3.Averhoff FM, Glass N, Holtzman D. 2012. Global burden of hepatitis C: considerations for healthcare providers in the United States. Clin Infect Dis 55(Suppl 1):S10–S15. doi: 10.1093/cid/cis361. [DOI] [PubMed] [Google Scholar]
- 4.Farci P. 2011. New insights into the HCV quasispecies and compartmentalization. Semin Liver Dis 31:356–374. doi: 10.1055/s-0031-1297925. [DOI] [PubMed] [Google Scholar]
- 5.Simmonds P, Bukh J, Combet C, Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin-I T, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962–973. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- 6.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang GP, Sherrill-Mix SA, Chang K-M, Quince C, Bushman FD. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J Virol 84:6218–6228. doi: 10.1128/JVI.02271-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, Lloyd AR. 2011. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog 7:e1002243. doi: 10.1371/journal.ppat.1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Stoddard MB, Wang S, Blair LM, Giorgi EE, Parrish EH, Learn GH, Hraber P, Goepfert PA, Saag MS, Denny TN, Haynes BF, Hahn BH, Ribeiro RM, Perelson AS, Korber BT, Bhattacharya T, Shaw GM. 2012. Elucidation of hepatitis C virus transmission and early diversification by single genome sequencing. PLoS Pathog 8:e1002880. doi: 10.1371/journal.ppat.1002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribeiro RM, Li H, Wang S, Stoddard MB, Learn GH, Korber BT, Bhattacharya T, Guedj J, Parrish EH, Hahn BH, Shaw GM, Perelson AS. 2012. Quantifying the diversification of hepatitis C virus (HCV) during primary infection: estimates of the in vivo mutation rate. PLoS Pathog 8:e1002881. doi: 10.1371/journal.ppat.1002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 12.Yanagi M, Purcell RH, Emerson SU, Bukh J. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci U S A 94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanford RE, Lee H, Chavez D, Guerra B, Brasky KM. 2001. Infectious cDNA clone of the hepatitis C virus genotype 1 prototype sequence. J Gen Virol 82:1291–1297. [DOI] [PubMed] [Google Scholar]
- 14.Gottwein JM, Scheel TK, Callendret B, Li Y-P, Eccleston HB, Engle RE, Govindarajan S, Satterfield W, Purcell RH, Walker CM, Bukh J. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol 84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui JM, Kench J, Farrell GC, Lin R, Samarasinghe D, Liddle C, Byth K, George J. 2002. Genotype-specific mechanisms for hepatic steatosis in chronic hepatitis C infection. J Gastroenterol Hepatol 17:873–881. doi: 10.1046/j.1440-1746.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- 16.Talaat M, Kandeel A, Rasslan O, Hajjeh R, Hallaj Z, El-Sayed N, Mahoney FJ. 2006. Evolution of infection control in Egypt: achievements and challenges. Am J Infect Control 34:193–200. doi: 10.1016/j.ajic.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 17.Lu L, Li C, Yuan J, Lu T, Okamoto H, Murphy DG. 2013. Full-length genome sequences of five hepatitis C virus isolates representing subtypes 3g, 3h, 3i and 3k, and a unique genotype 3 variant. J Gen Virol 94:543–548. doi: 10.1099/vir.0.049668-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, Lu L, Murphy DG, Negro F, Okamoto H. 2014. Origin of hepatitis C virus genotype 3 in Africa as estimated through an evolutionary analysis of the full-length genomes of nine subtypes, including the newly sequenced 3d and 3e. J Gen Virol 95:1677–1688. doi: 10.1099/vir.0.065128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsume A, Tokunaga Y, Hirata Y, Munakata T, Saito M, Hayashi H, Okamoto K, Ohmori Y, Kusanagi I, Fujiwara S, Tsukuda T, Aoki Y, Klumpp K, Tsukiyama-Kohara K, El-Gohary A, Sudoh M, Kohara M. 2013. A serine palmitoyltransferase inhibitor blocks hepatitis C virus replication in human hepatocytes. Gastroenterology 145:865–873. doi: 10.1053/j.gastro.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Saeed M, Scheel TK, Gottwein JM, Marukian S, Dustin LB, Bukh J, Rice CM. 2012. Efficient replication of genotype 3a and 4a hepatitis C virus replicons in human hepatoma cells. Antimicrob Agents Chemother 56:5365–5373. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saeed M, Gondeau C, Hmwe S, Yokokawa H, Date T, Suzuki T, Kato T, Maurel P, Wakita T. 2013. Replication of hepatitis C virus genotype 3a in cultured cells. Gastroenterology 144:56–58.e7. doi: 10.1053/j.gastro.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich H-G, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yi M, Villanueva RA, Thomas DL, Wakita T, Lemon SM. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc Natl Acad Sci U S A 103:2310–2315. doi: 10.1073/pnas.0510727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi M, Hu F, Joyce M, Saxena V, Welsch C, Chavez D, Guerra B, Yamane D, Veselenak R, Pyles R, Walker CM, Tyrrell L, Bourne N, Lanford RE, Lemon SM. 2014. Evolution of a cell culture-derived genotype 1a hepatitis C virus (H77S.2) during persistent infection with chronic hepatitis in a chimpanzee. J Virol 88:3678–3694. doi: 10.1128/JVI.03540-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, Derdeyn CA, Farmer P, Hunter E, Allen S, Manigart O, Mulenga J, Anderson JA, Swanstrom R, Haynes BF, Athreya GS, Korber BT, Sharp PM, Shaw GM, Hahn BH. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol 82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J Exp Med 206:1117–1134. doi: 10.1084/jem.20082831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J Exp Med 206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goonetilleke N, Liu MK, Salazar-Gonzalez JF, Ferrari G, Giorgi E, Ganusov VV, Keele BF, Learn GH, Turnbull EL, Salazar MG, Weinhold KJ, Moore S, CHAVI Clinical Core B, Letvin N, Haynes BF, Cohen MS, Hraber P, Bhattacharya T, Borrow P, Perelson AS, Hahn BH, Shaw GM, Korber BT, McMichael AJ. 2009. The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J Exp Med 206:1253–1272. doi: 10.1084/jem.20090365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bar KJ, Li H, Chamberland A, Tremblay C, Routy JP, Grayson T, Sun C, Wang S, Learn GH, Morgan CJ, Schumacher JE, Haynes BF, Keele BF, Hahn BH, Shaw GM. 2010. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J Virol 84:6241–6247. doi: 10.1128/JVI.00077-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog 6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolland M, Edlefsen PT, Larsen BB, Tovanabutra S, Sanders-Buell E, Hertz T, deCamp AC, Carrico C, Menis S, Magaret CA, Ahmed H, Juraska M, Chen L, Konopa P, Nariya S, Stoddard JN, Wong K, Zhao H, Deng W, Maust BS, Bose M, Howell S, Bates A, Lazzaro M, O’Sullivan A, Lei E, Bradfield A, Ibitamuno G, Assawadarachai V, O’Connell RJ, deSouza MS, Nitayaphan S, Rerks-Ngarm S, Robb ML, McLellan JS, Georgiev I, Kwong PD, Carlson JM, Michael NL, Schief WR, Gilbert PB, Mullins JI, Kim JH. 2012. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature 490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaccari M, Keele BF, Bosinger SE, Doster MN, Ma Z-M, Pollara J, Hryniewicz A, Ferrari G, Guan Y, Forthal DN, Venzon D, Fenizia C, Morgan T, Montefiori D, Lifson JD, Miller CJ, Silvestri G, Rosati M, Felber BK, Pavlakis GN, Tartaglia J, Franchini G. 2013. Protection afforded by an HIV vaccine candidate in macaques depends on the dose of SIVmac251 at challenge exposure. J Virol 87:3538–3548. doi: 10.1128/JVI.02863-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roederer M, Keele BF, Schmidt SD, Mason RD, Welles HC, Fischer W, Labranche C, Foulds KE, Louder MK, Yang Z-Y, Todd JP, Buzby AP, Mach LV, Shen L, Seaton KE, Ward BM, Bailer RT, Gottardo R, Gu W, Ferrari G, Alam SM, Denny TN, Montefiori DC, Tomaras GD, Korber BT, Nason MC, Seder RA, Koup RA, Letvin NL, Rao SS, Nabel GJ, Mascola JR. 2014. Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505:502–508. doi: 10.1038/nature12893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sterrett S, Learn GH, Edlefsen PT, Haynes BF, Hahn BH, Shaw GM, Bar KJ. 14 July 2014. Low multiplicity of HIV-1 infection and no vaccine enhancement in VAX003 injection drug users. Open Forum Infect Dis doi: 10.1093/ofid/ofu056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 37.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, Hahn BH, Kappes JC. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J Virol 86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, Hora B, Berg A, Cai F, Hopper J, Denny TN, Ding H, Ochsenbauer C, Kappes JC, Galimidi RP, West AP, Bjorkman PJ, Wilen CB, Doms RW, O’Brien M, Bhardwaj N, Borrow P, Haynes BF, Muldoon M, Theiler JP, Korber B, Shaw GM, Hahn BH. 2013. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A 110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, Aasa-Chapman MM, Pellegrino P, Williams I, Cohen MS, Gao F, Shaw GM, Hahn BH, Ochsenbauer C, Kappes JC, Borrow P. 2013. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ping L-H, Joseph SB, Anderson JA, Abrahams M-R, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar MG, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R, CAPRISA Acute Infection Study and Center for HIV-AIDS Vaccine Immunology Consortium . 2013. Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol 87:7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rolland M, Tovanabutra S, deCamp AC, Frahm N, Gilbert PB, Sanders-Buell E, Heath L, Magaret CA, Bose M, Bradfield A, O’Sullivan A, Crossler J, Jones T, Nau M, Wong K, Zhao H, Raugi DN, Sorensen S, Stoddard JN, Maust BS, Deng W, Hural J, Dubey S, Michael NL, Shiver J, Corey L, Li F, Self SG, Kim J, Buchbinder S, Casimiro DR, Robertson MN, Duerr A, McElrath MJ, McCutchan FE, Mullins JI. 2011. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat Med 17:366–371. doi: 10.1038/nm.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuplin A, Evans DJ, Simmonds P. 2004. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol 85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 43.Kolykhalov AA, Feinstone SM, Rice CM. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol 70:3363–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tellinghuisen TL, Evans MJ, von Hahn T, You S, Rice CM. 2007. Studying hepatitis C virus: making the best of a bad virus. J Virol 81:8853–8867. doi: 10.1128/JVI.00753-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinkert D, Bartenschlager R, Lohmann V. 2005. Quantitative analysis of the hepatitis C virus replication complex. J Virol 79:13594–13605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- 47.Kuntzen T, Timm J, Berical A, Lewis-Ximenez LL, Jones A, Nolan B, Schulze zur Wiesch J, Li B, Schneidewind A, Kim AY, Chung RT, Lauer GM, Allen TM. 2007. Viral sequence evolution in acute hepatitis C virus infection. J Virol 81:11658–11668. doi: 10.1128/JVI.00995-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nevot M, Boesecke C, Parera M, Andrés C, Franco S, Revollo B, Ingiliz P, Tural C, Clotet B, Rockstroh JK, Martinez MA, NEAT Study Group . 2014. Hepatitis C virus NS3/4A quasispecies diversity in acute hepatitis C infection in HIV-1 co-infected patients. J Viral Hepat 21:e19–e28. doi: 10.1111/jvh.12254. [DOI] [PubMed] [Google Scholar]
- 49.Smith JA, Aberle JH, Fleming VM, Ferenci P, Thomson EC, Karayiannis P, McLean AR, Holzmann H, Klenerman P. 2010. Dynamic coinfection with multiple viral subtypes in acute hepatitis C. J Infect Dis 202:1770–1779. doi: 10.1086/657317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 51.Maydt J, Lengauer T. 2006. Recco: recombination analysis using cost optimization. Bioinformatics 22:1064–1071. doi: 10.1093/bioinformatics/btl057. [DOI] [PubMed] [Google Scholar]
- 52.Lohmann V, Körner F, Koch J, Herian U, Theilmann L, Bartenschlager R. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 53.Kumar U, Tuthill T, Thomas HC, Monjardino J. 2000. Sequence, expression and reconstitution of an HCV genome from a British isolate derived from a single blood donation. J Viral Hepat 7:459–465. doi: 10.1046/j.1365-2893.2000.00259.x. [DOI] [PubMed] [Google Scholar]
- 54.Timm J, Neukamm M, Kuntzen T, Kim AY, Chung RT, Brander C, Lauer GM, Walker BD, Allen TM. 2007. Characterization of full-length hepatitis C virus genotype 4 sequences. J Viral Hepat 14:330–337. doi: 10.1111/j.1365-2893.2006.00792.x. [DOI] [PubMed] [Google Scholar]
- 55.Yamada N, Tanihara K, Mizokami M, Ohba K, Takada A, Tsutsumi M, Date T. 1994. Full-length sequence of the genome of hepatitis C virus type 3a: comparative study with different genotypes. J Gen Virol 75:3279–3284. doi: 10.1099/0022-1317-75-11-3279. [DOI] [PubMed] [Google Scholar]
- 56.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 57.Moore PL, Gray ES, Wibmer CK, Bhiman JN, Nonyane M, Sheward DJ, Hermanus T, Bajimaya S, Tumba NL, Abrahams M-R, Lambson BE, Ranchobe N, Ping L, Ngandu N, Abdool Karim Q, Abdool Karim SS, Swanstrom RI, Seaman MS, Williamson C, Morris L. 2012. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med 18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brown RJ, Hudson N, Wilson G, Rehman SU, Jabbari S, Hu K, Tarr AW, Borrow P, Joyce M, Lewis J, Zhu LF, Law M, Kneteman N, Tyrrell DL, McKeating JA, Ball JK. 2012. Hepatitis C virus envelope glycoprotein fitness defines virus population composition following transmission to a new host. J Virol 86:11956–11966. doi: 10.1128/JVI.01079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yi M, Lemon SM. 2003. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol 77:3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka T, Kato N, Cho MJ, Sugiyama K, Shimotohno K. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol 70:3307–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harrison GP, Mayo MS, Hunter E, Lever AM. 1998. Pausing of reverse transcriptase on retroviral RNA templates is influenced by secondary structures both 5′ and 3′ of the catalytic site. Nucleic Acids Res 26:3433–3442. doi: 10.1093/nar/26.14.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quiñones-Mateu ME, Gao Y, Ball SC, Marozsan AJ, Abraha A, Arts EJ. 2002. In vitro intersubtype recombinants of human immunodeficiency virus type 1: comparison to recent and circulating in vivo recombinant forms. J Virol 76:9600–9613. doi: 10.1128/JVI.76.19.9600-9613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lanciault C, Champoux JJ. 2006. Pausing during reverse transcription increases the rate of retroviral recombination. J Virol 80:2483–2494. doi: 10.1128/JVI.80.5.2483-2494.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Riepsamen AH, Gibson T, Rowe J, Chitwood DJ, Subbotin SA, Dowton M. 2011. Poly(T) variation in heteroderid nematode mitochondrial genomes is predominantly an artefact of amplification. J Mol Evol 72:182–192. doi: 10.1007/s00239-010-9414-3. [DOI] [PubMed] [Google Scholar]
- 65.Behrens SE, Tomei L, De Francesco R. 1996. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J 15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 66.Lohmann V, Körner F, Herian U, Bartenschlager R. 1997. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol 71:8416–8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure 7:1417–1426. doi: 10.1016/S0969-2126(00)80031-3. [DOI] [PubMed] [Google Scholar]
- 68.Mosley RT, Edwards TE, Murakami E, Lam AM, Grice RL, Du J, Sofia MJ, Furman PA, Otto MJ. 2012. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J Virol 86:6503–6511. doi: 10.1128/JVI.00386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Odelberg SJ, Weiss RB, Hata A, White R. 1995. Template-switching during DNA synthesis by Thermus aquaticus DNA polymerase I. Nucleic Acids Res 23:2049–2057. doi: 10.1093/nar/23.11.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pfeiffer JK, Topping RS, Shin NH, Telesnitsky A. 1999. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J Virol 73:8441–8447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mader RM, Schmidt WM, Sedivy R, Rizovski B, Braun J, Kalipciyan M, Exner M, Steger GG, Mueller MW. 2001. Reverse transcriptase template switching during reverse transcriptase-polymerase chain reaction: artificial generation of deletions in ribonucleotide reductase mRNA. J Lab Clin Med 137:422–428. doi: 10.1067/mlc.2001.115452. [DOI] [PubMed] [Google Scholar]
- 72.Viguera E, Canceill D, Ehrlich SD. 2001. In vitro replication slippage by DNA polymerases from thermophilic organisms. J Mol Biol 312:323–333. doi: 10.1006/jmbi.2001.4943. [DOI] [PubMed] [Google Scholar]
- 73.Li Y, Korolev S, Waksman G. 1998. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J 17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shinde D, Lai Y, Sun F, Arnheim N. 2003. Taq DNA polymerase slippage mutation rates measured by PCR and quasi-likelihood analysis: (CA/GT)n and (A/T)n microsatellites. Nucleic Acids Res 31:974–980. doi: 10.1093/nar/gkg178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Svarovskaia ES, Cheslock SR, Zhang W-H, Hu W-S, Pathak VK. 2003. Retroviral mutation rates and reverse transcriptase fidelity. Front Biosci 8:d117–d134. doi: 10.2741/957. [DOI] [PubMed] [Google Scholar]
- 76.Das D, Georgiadis MM. 2004. The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure 12:819–829. doi: 10.1016/j.str.2004.02.032. [DOI] [PubMed] [Google Scholar]
- 77.Menéndez-Arias L. 2009. Mutation rates and intrinsic fidelity of retroviral reverse transcriptases. Viruses 1:1137–1165. doi: 10.3390/v1031137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boyer JC, Bebenek K, Kunkel TA. 1992. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci U S A 89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klarmann GJ, Schauber CA, Preston BD. 1993. Template-directed pausing of DNA synthesis by HIV-1 reverse transcriptase during polymerization of HIV-1 sequences in vitro. J Biol Chem 268:9793–9802. [PubMed] [Google Scholar]
- 80.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol 80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Sekiya H, Hijikata M, Shimotohno K. 1992. Characterization of hypervariable regions in the putative envelope protein of hepatitis C virus. Biochem Biophys Res Commun 189:119–127. doi: 10.1016/0006-291X(92)91533-V. [DOI] [PubMed] [Google Scholar]
- 82.Gray RR, Parker J, Lemey P, Salemi M, Katzourakis A, Pybus OG. 2011. The mode and tempo of hepatitis C virus evolution within and among hosts. BMC Evol Biol 11:131. doi: 10.1186/1471-2148-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Friebe P, Bartenschlager R. 2002. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol 76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yanagi M, St. Claire M, Emerson SU, Purcell RH, Bukh J. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci U S A 96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 86.You S, Rice CM. 2008. 3′ RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J Virol 82:184–195. doi: 10.1128/JVI.01796-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ito T, Lai MM. 1997. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol 71:8698–8706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mitchell AM, Stone AEL, Cheng L, Ballinger K, Edwards MG, Stoddard M, Li H, Golden-Mason L, Shaw GM, Khetani S, Rosen HR. 2015. Transmitted/founder hepatitis C viruses induce cell-type- and genotype-specific differences in innate signaling within the liver. mBio 6(1):e02510-14. doi: 10.1128/mBio.02510-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Giugliano S, Kriss M, Golden-Mason L, Dobrinskikh E, Stone AE, Soto-Gutierrez A, Mitchell A, Khetani SR, Yamane D, Stoddard M, Li H, Shaw GM, Edwards MG, Lemon SM, Gale M, Shah VH, Rosen HR 4 November 2014. Hepatitis C virus infection induces autocrine interferon signaling by human liver endothelial cell and release of exosomes, which inhibits viral replication. Gastroenterology doi: 10.1053/j.gastro.2014.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jones CT, Catanese MT, Law LM, Khetani SR, Syder AJ, Ploss A, Oh TS, Schoggins JW, MacDonald MR, Bhatia SN, Rice CM. 2010. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol 28:167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carroll SS, Tomassini JE, Bosserman M, Getty K, Stahlhut MW, Eldrup AB, Bhat B, Hall D, Simcoe AL, LaFemina R, Rutkowski CA, Wolanski B, Yang Z, Migliaccio G, De Francesco R, Kuo LC, MacCoss M, Olsen DB. 2003. Inhibition of hepatitis C virus RNA replication by 2′-modified nucleoside analogs. J Biol Chem 278:11979–11984. doi: 10.1074/jbc.M210914200. [DOI] [PubMed] [Google Scholar]
- 92.Andrus L, Marukian S, Jones CT, Catanese MT, Sheahan TP, Schoggins JW, Barry WT, Dustin LB, Trehan K, Ploss A, Bhatia SN, Rice CM. 2011. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology 54:1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ogawa S, Surapisitchat J, Virtanen C, Ogawa M, Niapour M, Sugamori KS, Wang S, Tamblyn L, Guillemette C, Hoffmann E, Zhao B, Strom S, Laposa RR, Tyndale RF, Grant DM, Keller G. 2013. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development 140:3285–3296. doi: 10.1242/dev.090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee HY, Giorgi EE, Keele BF, Gaschen B, Athreya GS, Salazar-Gonzalez JF, Pham KT, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Hahn BH, Shaw GM, Korber BT, Bhattacharya T, Perelson AS. 2009. Modeling sequence evolution in acute HIV-1 infection. J Theor Biol 261:341–360. doi: 10.1016/j.jtbi.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu J, Keele BF, Li H, Keating S, Norris PJ, Carville A, Mansfield KG, Tomaras GD, Haynes BF, Kolodkin-Gal D, Letvin NL, Hahn BH, Shaw GM, Barouch DH. 2010. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J Virol 84:10406–10412. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cristina J, Colina R. 2006. Evidence of structural genomic region recombination in hepatitis C virus. Virol J 3:53. doi: 10.1186/1743-422X-3-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.González-Candelas F, López-Labrador FX, Bracho MA. 2011. Recombination in hepatitis C virus. Viruses 3:2006–2024. doi: 10.3390/v3102006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Galli A, Bukh J. 2014. Comparative analysis of the molecular mechanisms of recombination in hepatitis C virus. Trends Microbiol 22:354–364. doi: 10.1016/j.tim.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 99.Glynn SA, Wright DJ, Kleinman SH, Hirschkorn D, Tu Y, Heldebrant C, Smith R, Giachetti C, Gallarda J, Busch MP. 2005. Dynamics of viremia in early hepatitis C virus infection. Transfusion 45:994–1002. doi: 10.1111/j.1537-2995.2005.04390.x. [DOI] [PubMed] [Google Scholar]
- 100.Busch MP, Murthy KK, Kleinman SH, Hirschkorn DF, Herring BL, Delwart EL, Racanelli V, Yoon JC, Rehermann B, Alter HJ. 2012. Infectivity in chimpanzees (Pan troglodytes) of plasma collected before HCV RNA detectability by FDA-licensed assays: implications for transfusion safety and HCV infection outcomes. Blood 119:6326–6334. doi: 10.1182/blood-2011-12-393637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sentandreu V, Jiménez-Hernández N, Torres-Puente M, Bracho MA, Valero A, Gosalbes MJ, Ortega E, Moya A, González-Candelas F. 2008. Evidence of recombination in intrapatient populations of hepatitis C virus. PLoS One 3:e3239. doi: 10.1371/journal.pone.0003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barnes E, Folgori A, Capone S, Swadling L, Aston S, Kurioka A, Meyer J, Huddart R, Smith K, Townsend R, Brown A, Antrobus R, Ammendola V, Naddeo M, O’Hara G, Willberg C, Harrison A, Grazioli F, Esposito ML, Siani L, Traboni C, Oo Y, Adams D, Hill A, Colloca S, Nicosia A, Cortese R, Klenerman P. 2012. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci Transl Med 4:115ra111. doi: 10.1126/scitranslmed.3003155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goffard A, Callens N, Bartosch B, Wychowski C, Cosset F-L, Montpellier C, Dubuisson J. 2005. Role of N-linked glycans in the functions of hepatitis C virus envelope glycoproteins. J Virol 79:8400–8409. doi: 10.1128/JVI.79.13.8400-8409.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McCaffrey K, Boo I, Poumbourios P, Drummer HE. 2007. Expression and characterization of a minimal hepatitis C virus glycoprotein E2 core domain that retains CD81 binding. J Virol 81:9584–9590. doi: 10.1128/JVI.02782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goffard A, Dubuisson J. 2003. Glycosylation of hepatitis C virus envelope proteins. Biochimie 85:295–301. doi: 10.1016/S0300-9084(03)00004-X. [DOI] [PubMed] [Google Scholar]
- 106.Helle F, Vieyres G, Elkrief L, Popescu C-I, Wychowski C, Descamps V, Castelain S, Roingeard P, Duverlie G, Dubuisson J. 2010. Role of N-linked glycans in the functions of hepatitis C virus envelope proteins incorporated into infectious virions. J Virol 84:11905–11915. doi: 10.1128/JVI.01548-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schnell G, Loo Y-M, Marcotrigiano J, Gale M. 2012. Uridine composition of the poly-U/UC tract of HCV RNA defines non-self recognition by RIG-I. PLoS Pathog 8:e1002839. doi: 10.1371/journal.ppat.1002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li T, Zhang J. 2000. Determination of the frequency of retroviral recombination between two identical sequences within a provirus. J Virol 74:7646–7650. doi: 10.1128/JVI.74.16.7646-7650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sumpter R, Loo Y-M, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. 2005. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol 79:2689–2699. doi: 10.1128/JVI.79.5.2689-2699.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li K, Chen Z, Kato N, Gale M, Lemon SM. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J Biol Chem 280:16739–16747. doi: 10.1074/jbc.M414139200. [DOI] [PubMed] [Google Scholar]
- 112.Pietschmann T, Zayas M, Meuleman P, Long G, Appel N, Koutsoudakis G, Kallis S, Leroux-Roels G, Lohmann V, Bartenschlager R. 2009. Production of infectious genotype 1b virus particles in cell culture and impairment by replication enhancing mutations. PLoS Pathog 5:e1000475. doi: 10.1371/journal.ppat.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marukian S, Andrus L, Sheahan TP, Jones CT, Charles ED, Ploss A, Rice CM, Dustin LB. 2011. Hepatitis C virus induces interferon-λ and interferon-stimulated genes in primary liver cultures. Hepatology 54:1913–1923. doi: 10.1002/hep.24580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Helle F, Brochot E, Fournier C, Descamps V, Izquierdo L, Hoffmann TW, Morel V, Herpe Y-E, Bengrine A, Belouzard S, Wychowski C, Dubuisson J, Francois C, Regimbeau J-M, Castelain S, Duverlie G. 2013. Permissivity of primary human hepatocytes and different hepatoma cell lines to cell culture adapted hepatitis C virus. PLoS One 8:e70809. doi: 10.1371/journal.pone.0070809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
T/F genomes are representative of three of the major HCV genotypes found globally. The maximum likelihood nucleotide tree is based on the full ORFs with T/F genomes identified in this study highlighted in red. GenBank accession numbers are shown with the names of the strains or clones shown in parentheses. Bootstrap values are included for bootstraps >70% to the left of their respective nodes. The bar represents 0.1% variation at a nucleotide level. Download
Potential N-linked glycan sites are highly conserved between acute T/F lineage sequences and chronic infection sequences. The amino acid (AA) position is based on the H77 AF009606 reference sequence and is indicated on the x axis. The error bars represent the standard error of the mean. Download
Poly(U/UC) tract sequence variation in acute infection subjects. Shown are poly(U/UC) tract sequences for acute infection subjects 10021 (A), 110069 (B), 9055 (C), and 105431 (D). Serial samples are color coded with the corresponding viral kinetics presented on the left. Download
Poly(U/UC) tract sequence variation from chronic infection subjects. Shown are poly(U/UC) tract sequences for chronic infection subjects SLRO5563 (A), WEPA5774 (B), WIMI4025 (C), ARJA6267 (D), JOTO6422 (E), KNPH3730 (F), and ROMI6847 (G). Download
Contributions of E. coli, Taq polymerase, and MMLV reverse transcriptase to poly(U/UC) tract variation. (A) DNA from a molecular clone containing a 41-nucleotide homopoly(U) tract (input) was used to transform E. coli. A total of 60 colonies were sequenced revealing rare deletions and no nucleotide substitutions. (B) Plasmid DNA containing an 87-nucleotide homopoly(U) tract was endpoint diluted and subjected to 45 PCR cycles of amplification, cloned into a plasmid, and transfected into E. coli, and 36 colonies were analyzed, revealing contributions of Taq polymerase slippage to poly(U/UC) length variation. (C) Reverse transcription, single-genome amplification, and subcloning were performed on input synthetic RNA molecules containing a 40-nucleotide homopoly(U) tract, revealing substantial poly(U/UC) length variation—more so than in panel A, B, or D. (D) Reverse transcription, single-genome amplification, and subcloning were performed on input synthetic RNA molecules containing a 30-nucleotide homopoly(U) tract. (E) Electrospray ionization mass spectrometry (ESI-MS) demonstrates that the synthetic RNAs used as the input in panel C are homogeneous in length with the expected molecular weight for RNA oligonucleotide UC1 (predicted, 25,019.4; observed, 25,019.3). (F) The synthetic RNA used as the input in panel D is similarly homogeneous in length and with the expected molecular weight for UC2 (predicted, 25,017.4; observed, 25,017.5). Download
Full-length T/F HCV molecular clones. (A to G) All clones include a 5' T7 transcription site and 3' linearization site for use in generating viral RNA in vitro. Restriction sites used to concatenate the full genomes are labeled in green, and the restriction sites used to introduce the genomes into the vectors are labeled in red. In cases where the terminal restriction site and the RNA runoff sites are different, the runoff site is labeled in blue. (F) The distinctive poly(U/UC) tracts for each molecular clone are listed with the corresponding accession number and clone designation given in parentheses. Download
Sample origin and characteristics of single-genome amplification fragments used for analyses. (a) ZM-SP, Zeptometrix seroconversion panel. (b) SC-SP, SeraCare seroconversion panel. (c) UAB-C, University of Alabama at Birmingham, Center for AIDS Research Network of Integrated Clinical Systems. (d) T/F estimates are based on combined quarter- and half-genome analysis. (e) Percent divergence scores for all regions, except the poly(U/UC) tracts, were determined by DIVEIN multiple pairwise alignment analysis. Needleman-Wunsch pairwise dynamic programming was used for poly(U/UC) comparisons, and the results are presented in Fig. 4 and in Table S2. (f) NA, not applicable.
Poly(UC) tract hypervariation as revealed by Needleman-Wunsch pairwise analysis. Needleman-Wunsch similarity scores are presented as a percentage of the maximum possible score. (a) The short poly(U/UC) fragment denotes the first 45 nucleotides immediately 5’ of the X-tail. (b) The long poly(U/UC) fragment denotes the first 60 nucleotides immediately 5’ of the X-tail. (c) In the case of subjects 105431 and ARJA6267, the poly(U/UC) tract was not sufficiently long for a 60- or 65-nt analysis. NA, not applicable.
Oligonucleotide primers used to amplify and sequence viral genome fragments. Primers used to generate cDNA, amplify genome fragments, and sequence partial or complete genomes are listed with the corresponding AF009606 location where applicable.