Abstract
Songbirds represent an important model organism for elucidating molecular mechanisms that link genes with complex behaviors, in part because they have discrete vocal learning circuits that have parallels with those that mediate human speech. We found that ~10% of the genes in the avian genome were regulated by singing, and we found a striking regional diversity of both basal and singing-induced programs in the four key song nuclei of the zebra finch, a vocal learning songbird. The region-enriched patterns were a result of distinct combinations of region-enriched transcription factors (TFs), their binding motifs, and presinging acetylation of histone 3 at lysine 27 (H3K27ac) enhancer activity in the regulatory regions of the associated genes. RNA interference manipulations validated the role of the calcium-response transcription factor (CaRF) in regulating genes preferentially expressed in specific song nuclei in response to singing. Thus, differential combinatorial binding of a small group of activity-regulated TFs and predefined epigenetic enhancer activity influences the anatomical diversity of behaviorally regulated gene networks.
Songbirds offer an important in vivo model system for studying transcriptional programs regulated during behavior. This system consists of interconnected brain nuclei that control production of a learned vocal behavior (singing) with parallels to human speech (1, 2). Four key song nuclei are embedded within three regionally distinct telencephalic brain cell populations: HVC (letter based name), LMAN (lateral magnocellular nucleus of the nidopallium), RA (robust nucleus of the arcopallium), and Area X in the striatum. (Fig. 1A) (3–6). These nuclei are connected in a vocal motor pathway (HVC to RA) and a vocal learning pathway (LMAN and Area X) (7–13). Human functional analogs to these avian brain regions are in the cortex (pallium) and basal ganglia (striatum) (2, 6, 14, 15). This includes song (avian) and speech (human) brain regions that have convergence of differentially expressed genes (15), which suggests that the behavioral and neuroanatomical similarities for the production of learned vocalizations are accompanied by similarities in molecular and genetic mechanisms, such as with FoxP2 (16).
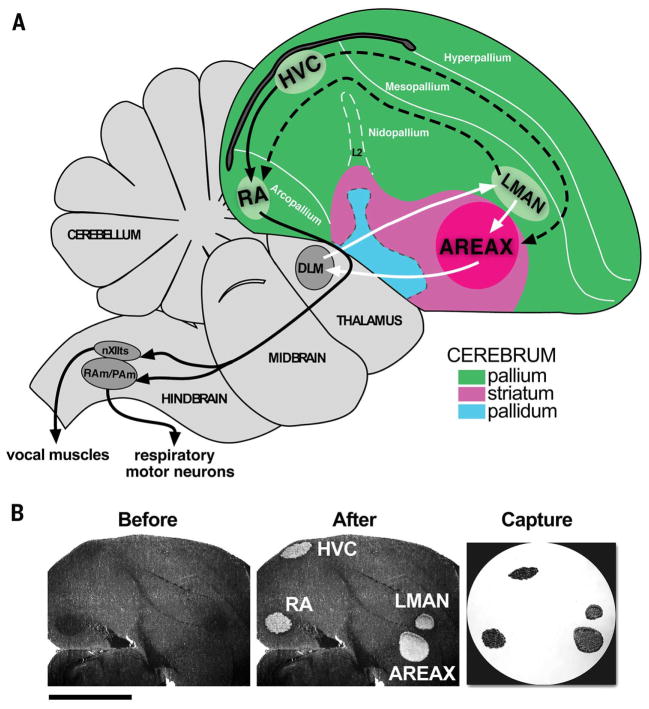
Fig. 1. Song system and laser microdissection.
(A) Sagittal schematic of the zebra finch brain showing positions and some connections of song nuclei. Pallial, striatal, and pallidal regions are distinguished by colors. Black arrows, posterior vocal pathway involved in song production; white arrows, anterior vocal pathway involved in song learning and modulation; dashed arrows, connections between the two pathways. (B) Song nuclei were laser-capture microdissected from males that were either silent or continuously singing for 0.5 hours and 1 hour, and for each hour thereafter up to 7 hours, resulting in more than 200 total microarrays. Shown are images of 10-μm tissue sections before and after laser capture microdissection at 10X magnification. (Before) Following dehydration, song nuclei fiber density appears darker than surrounding tissue. (After) Song nuclei regions are selectively cut out using an infrared laser. (Capture) The cut song nuclei transferred to the cap by the LCM system. For microarray analysis, each of the four song nuclei from each animal was captured separately to individual LCM caps. Dorsal is up; anterior is right. Scale bar, 2 mm.
The neural activity within song nuclei that underlies singing was initially shown to drive induction of two immediate early genes (IEGs), the transcription factors EGR1 and FOS (17–19). Their levels of expression correlate with the amount of singing in a motor-driven and social-context–dependent manner (20–23). Subsequent studies identified an additional 33 genes regulated within song nuclei by singing (24). The identified gene products have a wide range of cellular and biological process functions (24), including from neurogenesis (25, 26) to speech (27, 28). The genes were also found to cluster in a few anatomical and short temporal patterns of expression, although this was determined manually. As a result, we hypothesized that in vivo behaviorally induced gene expression may consist of anatomically and temporally diverse gene expression programs that can be regulated by networks of combinatorial transcription factor complexes or epigenetic chromatin differences (24). Two reports (29, 30) using our oligonucleotide microarrays found many more genes—800 to 2000 gene transcripts—regulated in the song nucleus Area X as a result of singing but could not test this hypothesis because the data were from only one song nucleus and/or one time point.
To test this hypothesis, we profiled baseline and singing-regulated gene expression across time in the four key song nuclei using our songbird gene expression microarray, which we annotated based on recently sequenced avian genomes (15, 31) and the human genome. Combined with genomic transcription factor motif analyses and chromatin immunoprecipitation sequencing (ChIP-seq) detection of active chromatin, we found predominantly diverse networks of simultaneously activated cascades of behaviorally regulated genes across brain regions, which can be explained in part by a combination of transcription factor complexes and epigenetic regulatory activity in the genome.
Results
We analyzed singing-regulated gene expression at a genomic-scale in HVC, LMAN, RA, and Area X of the zebra finch (Fig. 1 and fig. S1). To do so, we recorded moment-to-moment singing behavior of all animals over a 7-hour time course, laser microdissected individual song nuclei from multiple birds at each time point, amplified their mRNA, hybridized the resulting cDNA to our custom-designed 44 K oligonucleotide micro-arrays (table S1), and developed a computational approach that yielded a true positive rate >87%, as verified by in situ hybridization and reverse transcription polymerase chain reaction [fig. S2; tables S2 and S3; supplementary materials sections 1 to 7 (SM1 to SM7)]. This analysis detected 24,498 expressed transcripts among the four song nuclei in silent and/or singing animals (table S4), of which 18,478 (75%) mapped to 9059 Ensembl v60 annotated genes of the zebra finch genome, indicating that at least 50% of the transcribed genome is expressed in the song-control circuit of an adult animal during awake behaving hours.
Distinct baseline gene expression profiles define the song circuit
Using a linear model that we developed to identify differentially expressed transcripts in each brain region and combinations thereof (SM6), we found that of the 24,498 transcripts, ~5167 [21%, representing 3168 genes or ~17% of the genes in the avian genome (29)] were differentially expressed among song nuclei at baseline in silent animals (i.e., before singing began). These 5167 transcripts were organized hierarchically into at least five major region-specific clusters (Fig. 2A and table S5) with different functional enrichments (tables S6 and S7). A striatal song nucleus (Area X) cluster was enriched with noncoding RNAs, G protein–coupled receptors, and synaptic transmission proteins (Fig. 2A, turquoise cluster, and table S6). Cortical-like song nuclei (HVC, LMAN, and RA) were enriched for cell-to-cell signaling membrane-associated, axonal connectivity, and postsynaptic density (PSD) proteins (Fig. 2A, blue cluster, and table S6). The nidopallium song nuclei (HVC and LMAN) were further enriched for another group of cell-cell communication and neural connectivity, membrane-associated proteins (Fig. 2A, yellow cluster, and table S6). The arcopallium song nucleus RA was enriched for another set of neural connectivity proteins and for proteins involved in epilepsy and Alzheimer’s (Fig. 2A, green cluster, and table S6). RA was the only pallial brain region that had a large cluster of genes with a lower level of expression, which was enriched for PSD proteins different from the cortical enrichment (Fig. 2A, brown cluster, and table S6), and LMAN was the only song nucleus that did not have a large enrichment of genes of its own.
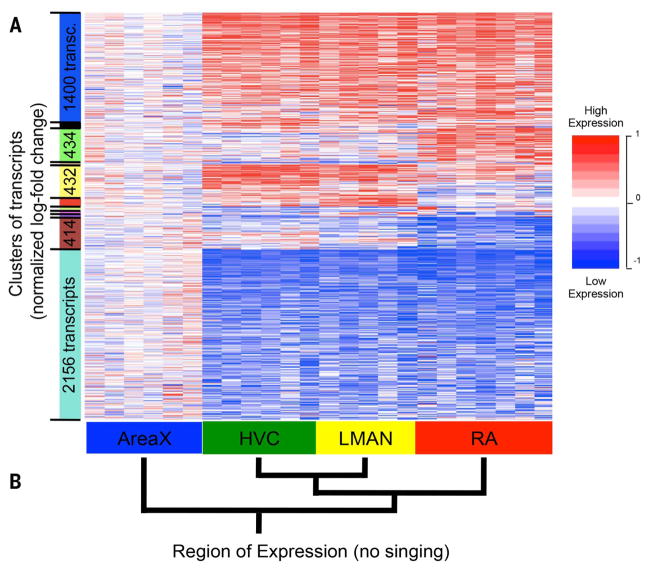
Fig. 2. Region-enriched gene expression at baseline.
(A) A heat map of hierarchically clustered expression profiles of 5167 transcripts (rows) that are differentially expressed across regions at baseline (FDR q < 0.1; see fig. S11 for FDR q < 0.2) in silent birds (red, increases; blue, decreases; white, no change) relative to mean Area X expression (numbers of transcripts not shown for small clusters). Each transcript is normalized to the average value of expression in Area X. Each column is an animal replicate. Detailed results are in table S4. (B) Average linkage hierarchical tree, generated from mean expression in each brain region, representing the molecular expression relationships between regions.
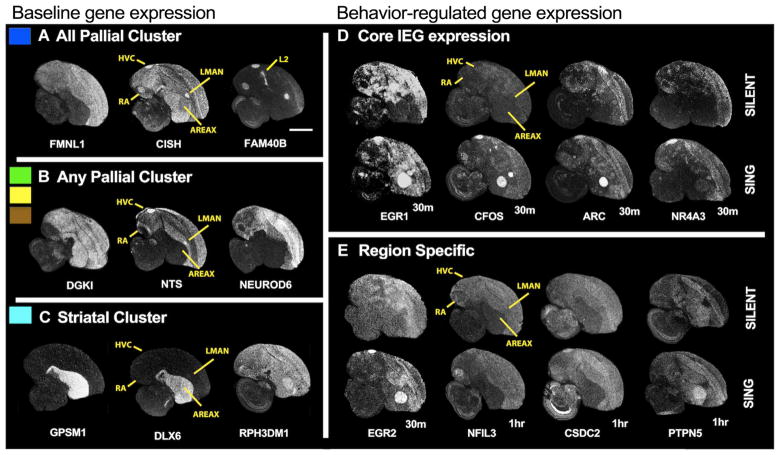
In situ hybridizations of example genes (e.g., some dopamine and glutamate receptors) revealed that most of the song nuclei expression patterns were consistent with the brain subdivisions to which they belonged (Fig. 3, A to C, and table S2) (32–34). However, as seen previously (33, 35, 36), some of the song nuclei had highly differential expression from their surrounding brain divisions (i.e., FMNL1, DGKI, and GPSM1 in Fig. 3, A to C). The most song-nucleus–specific gene was FAM40B (also called STRIP2), a phosphatase that was restricted to cortical-like song nuclei and the primary cortical sensory populations (like auditory area L2 in Fig. 3A).
Fig. 3. In situ hybridizations of baseline and singing-regulated genes.
(A) Genes higher in all pallial song nuclei (RA, HVC, and LMAN) relative to the striatal song nucleus (Area X) at baseline (Fig. 2A, blue clusters). (B) Genes differentially expressed just among the pallial song nuclei (green, yellow, and brown clusters) at baseline. (C) Genes higher in the striatal song nucleus relative to pallial song nuclei (turquoise cluster). (D) Core singing-regulated genes regulated in three to four song nuclei detected by microarrays but detected in all four with diverse levels by in situ hybridization, most peaking at 30 min. (E) Region-enriched singing-regulated genes in one or two song nuclei, with peaks of expression at later time points. Film autoradiograph images are inverted, showing white as labeled mRNA expression of the gene indicated below the image. Dorsal is up; anterior is right. Scale bar, 2 mm.
A dendrogram analysis separated the cortical song nuclei from the striatal and showed a stronger relationship between HVC and LMAN of the nidopallium (Figs. 2B and 1A), consistent with the recently revised understanding of avian brain organization and homologies with mammals (5, 6, 37). These findings show that even before singing starts, the song-learning nuclei have thousands of differentially expressed genes that define specific molecular functions for each [see (15) for characterization of the specializations in song nuclei].
Singing activates both a core and regionally diverse patterns of genes
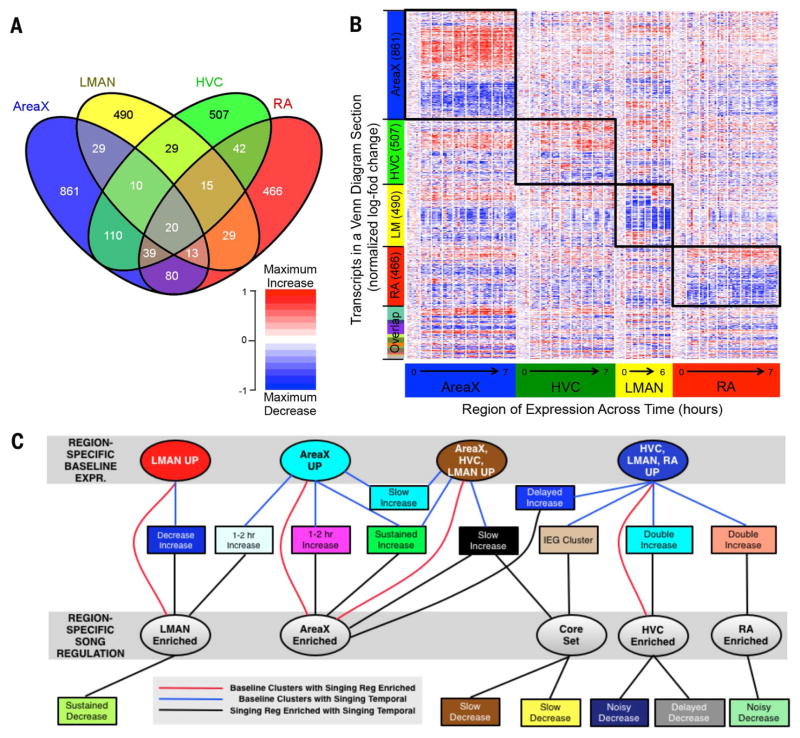
Of the 24,498 transcripts, we found an estimated 2740 (~11%) that were singing-regulated, up or down in time, in one or more song nuclei (Fig. 4, A and B, and table S8). These transcripts mapped to 1833 genes, indicating a conservative estimate of ~10% of the transcribed avian genome that is regulated by singing behavior. Area X had the most regulated transcripts (1162), followed by HVC (772), RA (702), and LMAN (635) (Fig. 4B) (the sum is higher than 2740 because of transcripts expressed in more than one song nucleus). A small number of genes (82) had singing-regulated splice variant differences (table S9), consistent with splice variant differences at baseline among song nuclei for glutamate receptor subunits (33), which can regulate activity-dependent genes in the brain. The vast majority (96%) of the 2740 singing-regulated transcripts were enriched in only one or two song nuclei, and a core set of only about 97 transcripts was regulated in at least three or four (<1.0%) song nuclei; of the latter, only 20 genes were equally regulated in all four song nuclei (Fig. 4, A and B, and table S8, green and yellow).
Fig. 4. Region-enriched gene expression in response to singing.
(A) A four-way Venn diagram showing regional singing-regulated distribution of 2740 transcripts (FDR q < 0.2). (B) Heat map of all 2740 transcripts from the Venn diagram, hierarchically clustered independently in all four song nuclei, then sorted by increased or decreased expression, and level of significance from highest to lowest in the linear model. Each column (170 total) is an animal replicate within a time point, and white lines separate time points. Red, increases; blue, decreases; white, no change relative to 0-hour samples for each song nucleus. Each transcript is normalized so that the maximum increase relative to nonsinging birds in any region is the darkest shade of red for increasing transcripts, and the maximum decrease is the darkest shade of blue for decreasing transcripts. Boxes highlight significant behaviorally regulated enrichment for each region (FDR q < 0.2 for that region). Figure S12 shows a more stringent heat map of region-enriched expression with a similar result. (C) Relationships among clusters of transcripts from the baseline region-enriched (top gray box, from Fig. 2A), singing temporal-enriched (rectangular nodes, from fig. S3, A to D), and singing region-enriched [bottom gray box, from (B)] patterns. Nodes are colored according to their cluster colors in the respective figures. Edges between two nodes correspond to significant overlap between two groups of transcripts (P < 0.001, hyper-geometric test). Nodes are sorted to optimize noncrossing of edges. Detailed results are in table S8.
The core set of 97 transcripts was enriched for known IEGs (38), including membrane depolarization–regulated (Ca2+ responsive) genes identified in cultured hippocampal (39) and cortical neurons (40) and genes induced in the auditory pathway by hearing song (41) (tables S10A and S7). In contrast, the brain region–specific singing-regulated genes had very little overlap with classic IEGs or a list of cell cultured–defined depolarization-induced genes (table S10A). Rather, the striatal Area X singing-regulated genes were enriched for cytoskeletal neural connectivity and neural migration functions, and RA was enriched for mitogen-activated protein kinase pathway transcripts, which control gene expression, differentiation, and cell survival. This suggests that our in vivo analyses are useful for finding region-specific or stimulus-specific genes that may be relevant for the underlying singing behavior.
Similar to the baseline expression, in situ hybridizations revealed that song nuclei expression patterns were consistent with the brain subdivisions to which they belong (Fig. 3, A to C, and table S3), except that the surrounding brain areas in some birds tended to have lower expression, presumably because they sang without much other movement behavior to cause movement-induced gene expression in the surrounding regions (42). We also noted that even among the core early-response genes induced in all song nuclei, expression levels at baseline differed among song nuclei (Fig. 3D). This suggests that there is even greater diversity among the song nuclei singing-regulated genes than simply presence or absence of regulation.
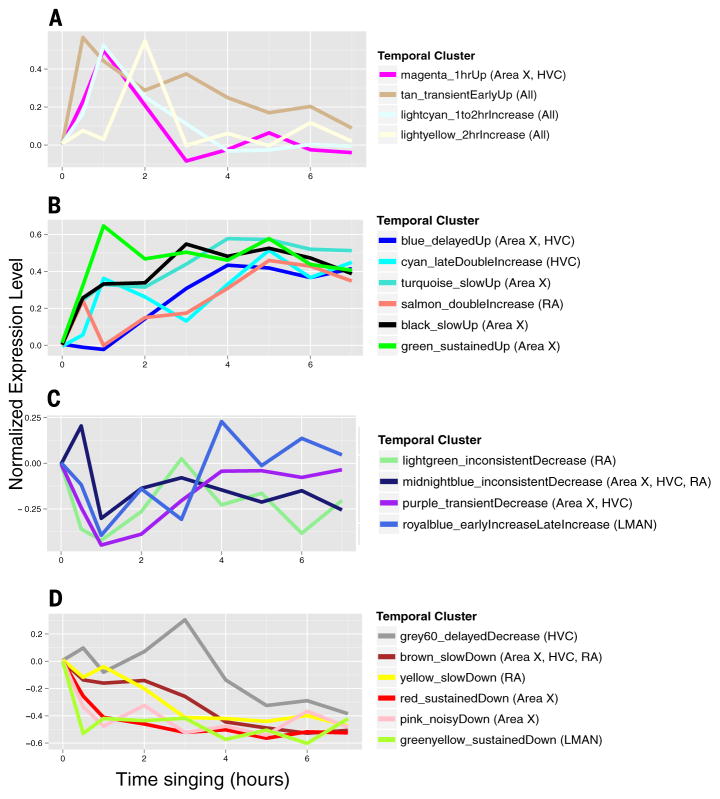
Analysis of the behaviorally regulated gene expression across time, using unsupervised hierarchical clustering (SM8), revealed up to 20 temporal profiles (clusters) among the four song nuclei, including transient or sustained, increased or decreased, early (0.5 to 2 hours) or late (3 to 7 hours), or two peaks of expression (fig. S3, A to D, and table S8). These 20 clusters can be further grouped into four superclusters of temporal profiles: (i) transient early increases, (ii) late-response increases, (iii) transient early decreases, and (iv) late-response decreases (Fig. 5, A to D). Only three of the temporal clusters had relatively comparable representations of genes in all brain regions, all belonging to transient early-increase clusters, including the IEG 0.5 to 1 hour cluster (Fig. 5A; fig. S3, tan cluster; and table S11), which contained a significant proportion (16%) of the core set of 97 transcripts (P < 1 × 10−5, hypergeometric test). For the remaining supertemporal profiles, each song nucleus had a region-enriched set of genes, except the late-response increasing pattern in LMAN (Fig. 5, fig. S3E, and table S11).
Fig. 5. Temporal singing-regulated patterns across time.
(A) Averages of gene expression levels in four temporal clusters of transient early response increases. (B) Averages of six late-response gene cluster increases. (C) Averages of four transient early-response cluster decreases. (D) Averages of six late-response gene cluster decreases. The temporal profiles are normalized such that nonsinging birds have a value of 0 and each gene has a maximum increase or decrease of 1. Each point represents the mean across all gene-brain region combinations for that time point. The 20 colors match the major temporal clusters in fig. S3, A to D.
Functional enrichment analyses showed that the activity-regulated gene expression sets from previous cell culture experiments (table S7) were highly enriched in the early transient IEG temporal cluster expressed in all song nuclei (table S10B). All of the late-increase singing-regulated clusters (Fig. 5B) also had detectable functional enrichments of genes, with Area X+HVC enriched in calcium ion binding and phosphatase proteins (blue temporal cluster); Area X late-increase genes were additionally enriched in chromosome organization, biogenesis (green), activity-dependent late-response genes identified in cultured neurons (40) (turquoise), and ribosomal proteins (black); HVC was additionally enriched in RNA-protein complexes and PSD proteins (cyan); and RA late-increase genes (salmon) were enriched in a different set of calcium ion–binding and ribosomal proteins (table S10B and Fig. 5B). Notably, we did not find any functional enrichment for the remaining transiently increased clusters or any of the decreased clusters, except genes regulated by the serum response transcription factor (SRF) in the slow decreasing cluster of RA (table S10B and Fig. 5D, yellow). These findings show that all song nuclei share a core set of genes with rapid transient up-regulation, but each song nucleus has its own dominant (though partly overlapping) set of other early- and late-responsive behaviorally regulated genes, suggesting cascades of gene regulation specific to each song nucleus with functions that remain to be discovered.
Relationships between differential baseline and differential singing-regulated genes
We next investigated how a small core set of behaviorally regulated transcription factors expressed in most brain regions could regulate a diverse set of downstream genes, with little overlap among regions. We hypothesized that the differential transcriptional state at baseline, before cell stimulation with singing, affects region-enriched singing-regulated expression (43, 44). Three lines of evidence support this hypothesis. First, hypergeometric tests revealed significant overlap between subsets of transcripts from the baseline region-enriched clusters (Fig. 4C, top gray box) with the singing-regulated region-enriched clusters (Fig. 4C, red lines and table S12) and with 10 of the 20 temporal clusters (Fig. 4C, blue and black lines between two gray boxes). If a gene was expressed at higher levels in a region relative to others at baseline before singing, it was also more likely to increase in that region during singing; the converse was not true for the decreasing sets of singing-regulated genes.
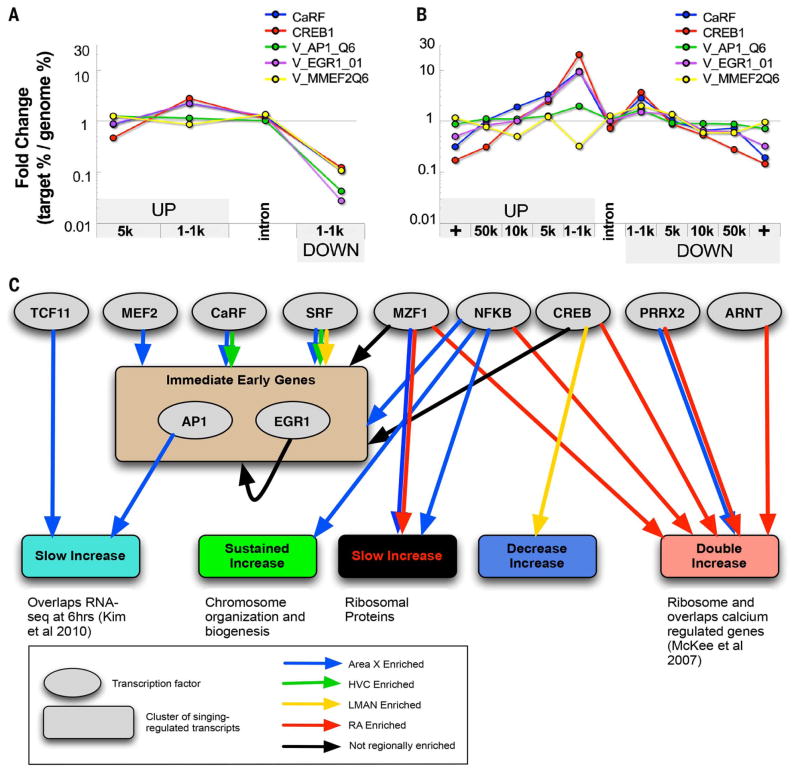
Second, a genome-wide binding site analysis of motifs for transcription factors (SM11) (45, 46) revealed ~100 motifs enriched in regulatory regions (e.g., directly upstream of transcription start sites) of genes in the temporal behaviorally regulated clusters (tables S13 and S14 and Fig. 6, A and B), and these matched genomic locations were also found in mammalian genomes (47, 48). With these motifs, we performed an association analysis between the region-specific and temporal clusters of genes to generate song nuclei–specific transcription factor motif to gene cluster networks (Fig. 6C, simplified network; fig. S4, detailed network; and table S15, edge list) [statistical significance tested with Euclidean distance to randomly generated networks (SM11 and SM12)]. Consistent with the core IEG cluster findings, we found that binding sites for five early-activated transcription factors (EATFs) (MEF2, SRF, NFKB, CREB, and CaRF) that are constitutively expressed at baseline and activated in response to neural activity (38, 49, 50) were significantly overrepresented in the singing-regulated cluster of IEGs expressed in most song nuclei (Fig. 6C and figs. S4 and S5A). In turn, the binding motifs of the singing-regulated AP-1 (bound by a FOS-JUN dimer) and EGR1 IEG transcription factors were also enriched directly upstream of the transcription start sites of many genes in our avian IEG cluster (Fig. 6, A to C). EGR1 can bind to its own promoter and down-regulate itself (51), which is consistent with the transient increase and subsequent decrease of some transcripts in the IEG temporal cluster. Also overrepresented in the IEG cluster was the ARNT motif, which also has the binding motif for the IEG NPAS4.
Fig. 6. Transcription factor binding motifs found in singing-regulated genes.
(A) Location bias of the target window of several motifs relative to its nearby gene when the motif search was confined to the local promoter—i.e., 5 kb upstream and 2 kb downstream of the start of the first nucleotide of the first exon of the gene. Fold change (plotted on the log scale y axis) is the ratio of the percentage of the motif target windows that fell within a particular position category relative to the first exon of a gene (target %) versus the percentage of windows that fall within that position category genome-wide (genome %). (B) Location bias of the motif target window relative to its nearby gene when the motif search was performed over the gene territory—i.e., halfway upstream and halfway downstream to the last or first exon of the nearest nonoverlapping gene. (C) Transcription factor motif-gene cluster network summarized from fig. S4 showing relationships between enriched EATFs (gray circles) and their binding motifs in subsets of genes from the temporal singing-regulated clusters (colored rectangular nodes as in fig. S3, A to D). Edges are colored on the basis of the region-specific expression of the predicted regulatory targets of the TF within each singing-regulated cluster (SM11 and SM12). Detailed results are in table S13 and fig. S4.
Third, consistent with our region-specific clusters, some transcription factors that were differentially expressed in a region or a combination of regions at baseline had binding motifs in genes that were differentially regulated in that region(s) at baseline or during singing. For example, variants of the NFE2L1 and MAF transcription factors that dimerize and bind to the TCF11 motif (52) were higher or lower in Area X relative to the pallial song nuclei at baseline (fig. S6), and the TCF11 binding motif was overrepresented in the slow-increase singing-regulated cluster of genes in Area X (Fig. 6C and figs. S4 and S5B). However, there were many other cases where EATFs and other transcription factors did not exhibit differential regional baseline expression but had binding motifs enriched in clusters of singing-regulated genes specific for a song nucleus. For example, the EATF transcription factors SRF and CaRF, which are not differentially expressed at baseline (table S5), had strong motif associations to singing-regulated genes in Area X and HVC. The MZF1 and PRRX2 transcription factors had associations with different sets of genes in Area X and RA (Fig. 6C and figs. S4 and S5B). Thus, we experimentally tested whether one of these EATFs, CaRF, regulated the predicted region-specific genes (Fig. 7).
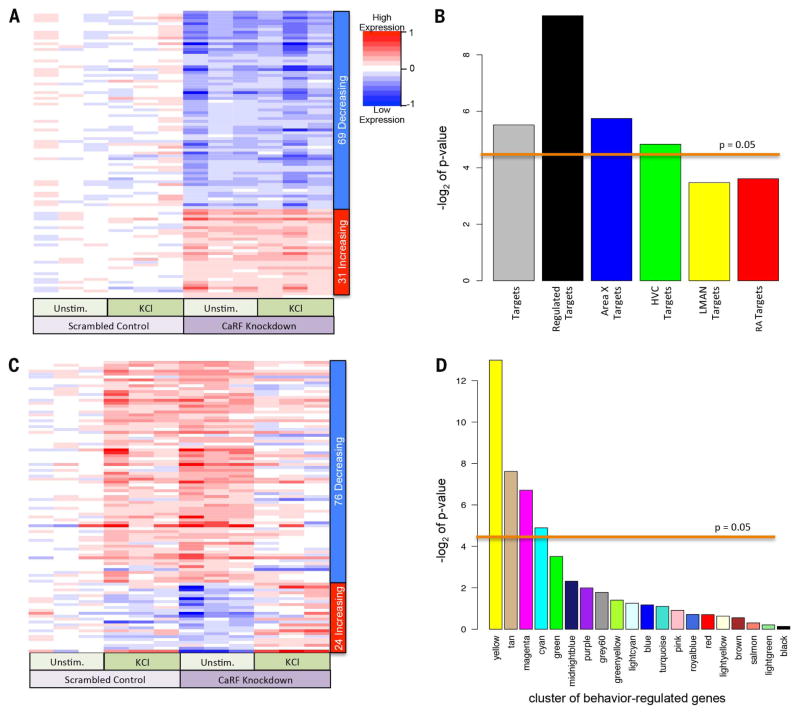
Fig. 7. RNAi knockdown illuminates CaRF binding motif relationships with singing-regulated genes.
(A) Heat map of genes affected by CaRF knockdown independent of membrane depolarization in mouse cultured neurons. Rows represent the 100 transcripts most changed by CaRF RNAi knockdown (P < 0.0014; FDR q < 0.475), sorted according to the t statistic, which takes direction of regulation into account. Each column is an independent sample (n = 3 unstimulated controls; n = 3 KCl depolarized in the presence of either scrambled RNAi or CaRF RNAi knockdown virus). Color intensities (blue to red) represent the log fold change in knockdown cells relative to the mean of the scrambled control conditions. (B) Significance of the enrichment of zebra finch baseline genes (cluster colors according to Fig. 2A) with CaRF promoter motifs in the ranked list of t values for CaRF knockdown–affected genes in mouse cultured neurons. P < 0.05 (above line) is a significant association, Wilcox rank sum statistic over multiple permutations (66). (C) Similar to (A), except for genes that respond differently to KCl activity in the CaRF knockdown cells. Rows represent the 100 transcripts most changed in expression (P < 0.015, factorial test), sorted according to the t statistic. (D) Significance of the enrichment of zebra finch singing-regulated genes (cluster colors according to Fig. 5 and fig. S3), with CaRF promoter motifs in the ranked list of t values for genes differentially regulated by neural activity in mouse cortical neurons during CaRF knockdown versus control. P < 0.05 (above line) is a significant association, Wilcox rank sum statistic over multiple permutations (66).
CaRF is required for regulation of both core and regional expressed sets of genes
We investigated the Ca2+ responsive transcription factor CaRF because the network analyses implicated it in both the regulation of the Ca2+ responsive IEGs that are induced in most song nuclei and some that are regionally enriched in Area X and HVC (Fig. 4C and fig. S6). Because we lacked an established zebra finch neural cell culture method to test CaRF function, we used RNA interference (RNAi) against CaRF in cultured mouse cortical neurons and hybridized labeled cDNA to mouse oligonucleotide micro-arrays representing many of the same genes on our zebra finch oligonucleotide microarray (SM4). We identified a set of genes that showed decreased or increased expression after CaRF knockdown independent of membrane depolarization (Fig. 7A and table S16), and many of these function in calcium signaling pathways (fig. S7 and table S17) (53). This is consistent with the proposed role of CaRF in regulating neuronal gene expression under basal neural activity (48, 54), as both a repressor and activator (48). Importantly, as predicted by our promoter motif analyses in birds, the ranked list of CaRF-regulated genes showed enrichment for singing-regulated genes that had a nearby CaRF binding site (P = 0.0014, Wilcox test) (Fig. 7B). This enrichment was highest in the set of genes regulated in Area X and HVC (Fig. 7B), supporting our network result (Fig. 6C).
CaRF RNAi knockdown also caused genes that were normally up-regulated by membrane depolarization to be suppressed to normal baseline levels and, conversely, genes that were normally down-regulated by membrane depolarization to be up-regulated (Fig. 7C and table S18). This suggests that CaRF is required to buffer activity of these gene promoters under basal conditions such that they can become stimulus-responsive upon membrane depolarization. Importantly, this same set of membrane depolarization- and CaRF-regulated genes significantly overlapped with those that had the CaRF binding site in the singing-regulated genes of the IEG (tan) cluster. They also significantly overlapped with several other clusters that were specifically up-regulated in Area X and HVC (Fig. 7D, magenta and cyan clusters; table S19; and fig. S3E). Genes that showed decreased expression preferentially in RA, but also in other song nuclei (fig. S3, yellow), after 2 to 3 hours of singing (the same amount of time the cultured cells were depolarized) had even greater overlap (Fig. 7D, yellow).
Overall, the findings demonstrate a requirement of the CaRF transcription factor for baseline and activity-dependent regulation of some of the very same genes for which we found CaRF binding motifs that are regulated at baseline and by singing in a region-specific manner, respectively. The calcium signaling and calcium ion–binding genes tended to increase during song production and were affected in the CaRF knockdown experiments, which is evidence of consistent CaRF function across species. We next sought an explanation of how EATFs that are not differentially expressed at baseline could regulate these genes in a region-specific manner.
Epigenetic modifications predefine region specificity of gene regulation
Although transcription factors are the ultimate regulators of gene expression, their ability to bind to sites in the genome is gated by chromatin structural changes. Chromatin regulation by acetylation of histone 3 at lysine 27 (H3K27ac) has been extensively studied and shown to be a strong indicator of active enhancers (55). We thus performed an experiment to identify active transcriptional regulatory regions in the genomes of individual dissected song nuclei (RA and Area X, which showed the largest regional differences) before and after singing, as measured by a genome-wide histone ChIP-seq analysis of H3K27ac (SM14, SM15, and table S20). The active genomic regions can be searched as tracks in the University of California–Santa Cruz (UCSC) browser against the zebra finch genome (56). This analysis also required that we create a more stringent selection of regional, early, and late singing-responsive genes from the respective clusters in RA and Area X (Fig. 5 and fig. S3), using principal components analyses (fig. S8).
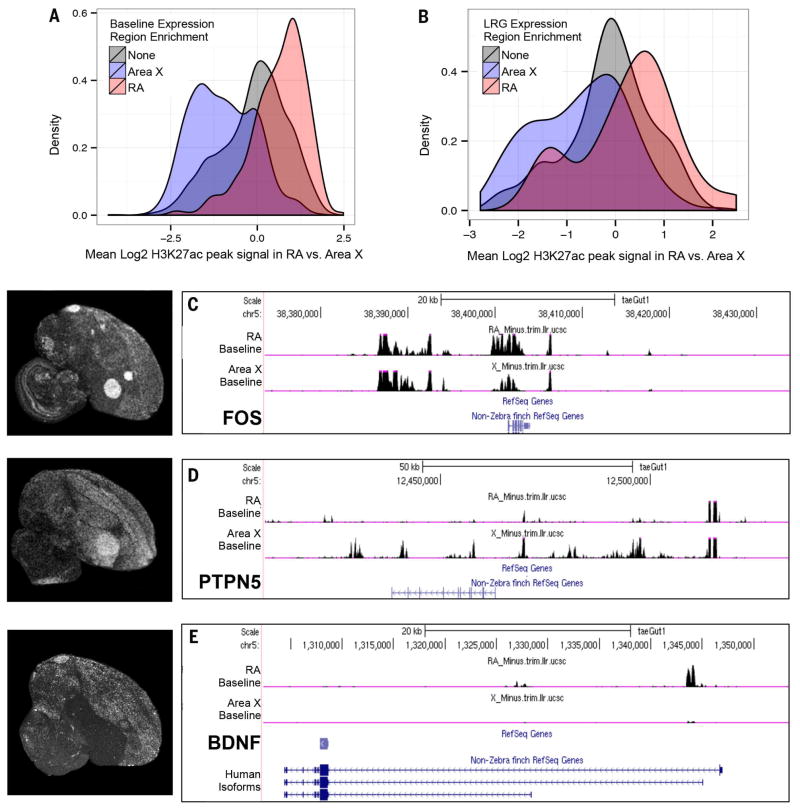
Out of 35,958 peaks, we found 30% (10,749) enriched in Area X and 21% (7673) enriched in RA. Under basal conditions, genes with song nuclei–specific expression patterns had nearby genomic regions that were significantly more likely to be marked by H3K27ac in that brain region (Fig. 8A, blue and red, and table S21) (~1300 genes). Conversely, genes that were expressed similarly in RA and Area X did not show a significant regional bias in the distribution of this chromatin mark (Fig. 8A, gray, and table S21) (~1100 genes examined). Interestingly, when we considered only the set of RA or Area X region-specific genes that were also up-regulated by singing, we found that they were already associated with higher nearby H3K27ac in their preferred brain region before singing (Fig. 6, B, D, and E; fig. S9, A to E; and table S22). There was a strong positive correlation between differences in nearby H3K27ac at baseline and differences in singing-dependent up-regulation of these genes in RA and Area X (R = 0.37, P = 1.6 × 10− 12; Pearson correlation). Conversely, late-response genes that were comparably induced by singing in both RA and Area X showed comparable H3K27ac under basal conditions (Fig. 8B, gray, and table S22). Furthermore, the early-response cluster of genes, which were expressed and induced comparably in both RA and Area X (e.g., FOS), also showed comparable H3K27ac in both brain regions at baseline (Fig. 8C and figs. S9A and S10A). Notably, we did not find any significant difference [e.g., 0 significant peaks; false discovery rate (FDR) threshold < 0.01] in H3K27ac peaks within either song nucleus when we compared ChIP-seq profiles obtained before and after singing (fig. S10A). We detected a weak signal for increased H3K27ac peaks in the Area X down-regulated genes (fig. S10B).
Fig. 8. Region-specific epigenetic signatures predefine behaviorally regulated gene expression.
(A) Density plot of genes differentially expressed at baseline in RA versus Area X and the difference in the level of nearby H3K27ac peaks in the genomes of cells in RA versus Area X. Each H3K27ac peak is mapped to a gene with the nearest transcription start site. For each gene, the changes in all mapped H3K27ac peaks are averaged. The H3K27ac distributions for RA versus Area X enriched genes are significantly different (P = 1.5 × 10−186, t test). (B) Similar plot as in (A) except for differentially expressed late-response singing-regulated genes. The distributions for RA and Area X are also significantly different (P = 1.8 × 10− 5, t test). However, there are two peaks in RA, which suggests that active genomic sites in Area X in the negative peak for RA could be genes that are actively suppressed in Area X. Corresponding data can be found in tables S21 and S22. (C) H3K27ac peaks surrounding a gene induced by singing across all brain regions, FOS; (D) H3K27ac peaks of a gene induced specifically in Area X, PTPN5. (E) H3K27ac peaks of a gene induced at low levels in RA but not detectable in Area X, BDNF. The plots show the log-likelihood ratios of H3K27ac signal in pooled baseline RA and pooled baseline Area X samples versus input DNA around the genomic regions in the zebra finch. The relevant gene models from the UCSC genome browser are shown below. Peaks measure both enhancer and promoter regions. Left of the H3K27ac peaks are in situ hybridization mRNA signal in singing animals. FOS and PTPN5 are shown in Fig. 3, and BDNF is used with permission from (37).
These data suggest that the regional differences in chromatin activity present before singing begins are predictive of differential singing-dependent induction of late-response genes. This hypothesis was further supported by our observation of regional H3K27ac differences at baseline for 50 genes that had equivalent basal expression in RA and Area X but region-specific up-regulation upon singing (table S22, blue and red highlights). An ingenuity pathway analysis on the Area X set of genes out of the 50 mentioned above (table S22, blue, and SM15) revealed that they were enriched for locomotion behavior (P = 0.004; ARNTL, CALB1, FGF14, RCAN2, and RIMS1) and movement-disorder functions (P = 0.004; ARNTL, CALB1, CAPZB, DIRAS2, EEF1A2, ELMO1, FGF14, MTMR2, RPSA, and TMED10), consistent with the function of Area X and the surrounding striatum. There were too few RA-specific genes without baseline differential expression (10 genes) to be tested by pathway analyses. Overall, these findings indicate that region-specific epigenetic chromatin activity at or near transcription factor binding sites for transcription factors expressed in all brain regions could determine which singing or baseline differentially regulated genes are expressed in each brain region.
Discussion
The magnitude of the anatomical diversity of behaviorally regulated genes and their networks in different brain regions of the same circuit was unexpected (24, 29, 30, 41). Our findings suggest two mechanisms that control this diversity: (i) region-enriched transcription factors that regulate region-enriched expression of their target genes and (ii) region-enriched epigenetic marks that determine which genes can be expressed in specific brain regions in both baseline and behaviorally regulated states. The first mechanism is consistent with the hypothesis that interactions between early transcription factors and late-response genes coordinate activity-dependent gene induction associated with behavior (57) but, in this case, in a region-specific manner. The second, epigenetic, mechanism is just beginning to be explored at the level of neural activity (40, 58) and has not been addressed in complex behaviors.
Given our findings and known signaling pathways from experiments in cultured cells (59), we propose the following overall mechanism (see the figure in the print summary, page 1334). Neural activity during the performance of a behavior, such as singing, causes release of neurotransmitters at the synapses between connected cells and activates postsynaptic receptors. These receptors initiate an intracellular signaling response that alters the activity, often through phosphorylation, of constitutively expressed EATFs. The activated EATFs bind or are already bound to the open chromatin of promoters or enhancers of the core IEGs enabled in all brain regions, as measured by H3K27ac, to activate their expression. The IEGs in turn, along with EATFs, bind to recognition regions of open chromatin that have already been primed in a cell type–specific manner, which leads to the induction of region-specific late-response genes. Some transcription factors are already expressed in a region-specific manner and add to the diversity of regulation of the downstream genes. Furthermore, our data show that brain region–specific open enhancers or promoters are already waiting in an active state, ready to do their job at a moment’s notice when the neurons fire to turn on programs of gene expression. Thus, the production of learned behavior modulates an already primed transcriptional and epigenetic network specific to different subregions of the circuit that controls the behavior.
This model may be an explanation for the finding that the IEG and EATF NPAS4, in response to neural activity, activates different sets of genes in cultured excitatory versus inhibitory neurons (60). Likewise, we find that common induction of IEGs across the many different kinds of neurons that comprise all song nuclei is associated with distinct programs of late-response genes, which are likely dependent at least in part on IEG regulation. However, one notable difference between our data and a recent study of activity-dependent enhancers in cultured neuron preparations is that, whereas membrane depolarization was found to further induce H3K27ac at enhancers near activity-regulated genes (58), we find that H3K27ac peaks in vivo in the brain are already enriched near singing-inducible genes under basal conditions and do not show further activation upon singing. It is possible that the neural networks recruited upon singing are sparse enough in the song nuclei that we were unable to detect H3K27ac changes in these cells against the background noise. An alternative possibility is that ongoing neural activity in the brain of an awake behaving animal is sufficient to keep enhancers poised in a fully active state even before execution of a specific behavioral task like singing. In this model, it is regulation of sequence-specific DNA binding of transcription factors that is most important for instructing the level and nature of gene expression, whereas epigenetic marks on chromatin are permissive for expression of the predetermined program.
Our CaRF manipulation experiments help reveal further complexity and potential novel mechanisms of activity-dependent gene regulation in the brain. The increased activity-regulated genes that are reversed in the absence of CaRF in response to membrane depolarization suggest that CaRF may act as a modulating transcription factor for neural activity–dependent regulation of its target genes. In this scenario, it prevents differential expression of its target genes until neural firing increases. When CaRF is removed by knockdown, it can no longer buffer the expression of these genes in the absence of activity; consequently, in the presence of activity, other factors can regulate the genes in a direction opposite of what CaRF would do. The specific mechanisms by which CaRF might achieve this function remain to be determined, but the H3K27ac enhancer activity in CaRF target genes is likely to play a role.
Additional transcriptional anatomical diversity not tested in this study could possibly be generated with differential expression of neurotransmitter membrane receptors at baseline in different brain regions, which could activate different signaling pathways in those neurons during singing (2, 33). Our hypothesis does not explain the down-regulation of some gene clusters where regionally specific transcription factor motifs were not enriched in those genes, and thus their regulation would have to be explained by other mechanisms.
Our findings suggest that each song nucleus has diverse molecular functions and gene networks. Consistent with their dominant roles in song production (7–13) compared to other song nuclei, HVC is specifically enriched with singing-regulated increases in PSD proteins used for cell-to-cell communication and RNA-protein complexes, and RA is enriched with genes in the mitogen-activated protein kinase (MAPK) pathway, such as DUSP1, which is proposed to be involved in neural protection of a brain region that is highly active during behavior performance (61, 62). Consistent with their dominant roles in learning (7–13), LMAN shows greater specificity for the cAMP response element–binding protein (CREB) pathway, a key transcription factor involved in learning and memory (59, 63), and Area X is more enriched with expression of neural connectivity, chromosome organization, and biogenesis genes. In addition, the large overrepresentation of noncoding RNA genes expressed at baseline in Area X indicates that its transcriptional regulatory network may be more extensive than the pallial song nuclei. The larger overrepresentation of neural connectivity and cell signaling genes in the pallial song nuclei indicates greater focus on cell structure and communication.
In terms of memory, a long-held hypothesis is that neural activity will induce an early wave of responsive genes, which in turn regulate a late wave of genes, and that the first wave would act as a molecular switch converting short-term memories into long-term memories (57, 64, 65). If true, singing would be associated with continuous memory consolidation and song fine-tuning, with each nucleus having specific waves of gene regulation for their specific functions. An alternative, not mutually exclusive, proposal states that the activity-dependent waves function as a metabolic mechanism to maintain protein turnover for normal cell homeostasis due to increased protein catabolism that occurs during high activity levels (17). If true, it would be associated with continued repair of the circuit when used. Our transcription factor binding motif analysis suggests that both the early and late transcriptional responses could be driven by some of the same EATFs. This would indicate that the two waves of gene expression may not entirely depend on each other and that they could be used for both memory and homeostasis functions.
In summary, as the mechanisms that define the genome-phenotype relationship, including the diversity of gene expression patterns, begin to be understood, so will the role of individual genes and pathways in learning, maintenance, and production of behavior. Performance of complex behavior involves interaction between neural activity, networks of cells, and networks of genes. Untangling the subtle differences in connected neurons, firing patterns, signaling pathways, and transcription factor activity may lead to a greater understanding of the diversity of the gene expression patterns that we observe here in highly interconnected cells within an intact multicellular organ.
Supplementary Material
Acknowledgments
We thank S. Augustin, J. Coleman, and N. Diaz Nelson for help collecting and processing singing bird samples for microarray analysis; H. Dressman and L.-L. Rowlette at the Duke Institute for Genome Science and Policy for microarray hybridizations; We thank A. Cavanaugh, M. Lawson, and M. Dean for useful comments after careful reading of an earlier version of this manuscript. O.W. was supported by postdoctoral training grants from American Psychological Association fellowship funded by the National Institute of Mental Health grant 5T32MH018882-18 and the National Science Foundation grant 0610337. This work was supported by grants from the National Institute on Deafness and Other Communication Disorders grant R01DC007218 and the Howard Hughes Medical Institute to E.D.J.. Microarray data have been submitted to the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo) under accession no. GSE33365. Annotations are also available in table S1. Raw files of the Chip-seq H3K27ac experiments are available at www.broadinstitute.org/~pfenning/finchWig.
Footnotes
REFERENCES AND NOTES
- 1.Doupe AJ, Kuhl PK. Birdsong and human speech: Common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mooney R, Konishi M. Two distinct inputs to an avian song nucleus activate different glutamate receptor subtypes on individual neurons. Proc Natl Acad Sci USA. 1991;88:4075–4079. doi: 10.1073/pnas.88.10.4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo M, Perkel DJ. A GABAergic, strongly inhibitory projection to a thalamic nucleus in the zebra finch song system. J Neurosci. 1999;19:6700–6711. doi: 10.1523/JNEUROSCI.19-15-06700.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reiner A, et al. Revised nomenclature for avian telencephalon and some related brainstem nuclei. J Comp Neurol. 2004;473:377–414. doi: 10.1002/cne.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarvis ED, et al. Avian brains and a new understanding of vertebrate brain evolution. Nat Rev Neurosci. 2005;6:151–159. doi: 10.1038/nrn1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nottebohm F, Stokes TM, Leonard CM. Central control of song in the canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 8.Bottjer SW, Miesner EA, Arnold AP. Forebrain lesions disrupt development but not maintenance of song in passerine birds. Science. 1984;224:901–903. doi: 10.1126/science.6719123. [DOI] [PubMed] [Google Scholar]
- 9.Scharff C, Nottebohm F. A comparative study of the behavioral deficits following lesions of various parts of the zebra finch song system: Implications for vocal learning. J Neurosci. 1991;11:2896–2913. doi: 10.1523/JNEUROSCI.11-09-02896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams H, Mehta N. Changes in adult zebra finch song require a forebrain nucleus that is not necessary for song production. J Neurobiol. 1999;39:14–28. doi: 10.1002/(SICI)1097-4695(199904)39:1<14::AID-NEU2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 11.Brainard MS, Doupe AJ. Interruption of a basal ganglia-forebrain circuit prevents plasticity of learned vocalizations. Nature. 2000;404:762–766. doi: 10.1038/35008083. [DOI] [PubMed] [Google Scholar]
- 12.Olveczky BP, Andalman AS, Fee MS. Vocal experimentation in the juvenile songbird requires a basal ganglia circuit. PLOS Biol. 2005;3:e153. doi: 10.1371/journal.pbio.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aronov D, Andalman AS, Fee MS. A specialized forebrain circuit for vocal babbling in the juvenile songbird. Science. 2008;320:630–634. doi: 10.1126/science.1155140. [DOI] [PubMed] [Google Scholar]
- 14.Bottjer SW, Johnson F. Circuits, hormones, and learning: Vocal behavior in songbirds. J Neurobiol. 1997;33:602–618. doi: 10.1002/(SICI)1097-4695(19971105)33:5<602::AID-NEU8>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Pfenning AR, et al. Convergent transcriptional specializations in the brains of humans and song learning birds. Science. 2014;346:1256846. doi: 10.1126/science.1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharff C, Petri J. Evo-devo, deep homology and FoxP2: Implications for the evolution of speech and language. Philos Trans R Soc Lond B Biol Sci. 2011;366:2124–2140. doi: 10.1098/rstb.2011.0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci USA. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimpo RR, Doupe AJ. FOS is induced by singing in distinct neuronal populations in a motor network. Neuron. 1997;18:315–325. doi: 10.1016/S0896-6273(00)80271-8. [DOI] [PubMed] [Google Scholar]
- 19.Jin H, Clayton DF. Localized changes in immediate-early gene regulation during sensory and motor learning in zebra finches. Neuron. 1997;19:1049–1059. doi: 10.1016/S0896-6273(00)80396-7. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis ED, Scharff C, Grossman MR, Ramos JA, Nottebohm F. For whom the bird sings: Context-dependent gene expression. Neuron. 1998;21:775–788. doi: 10.1016/S0896-6273(00)80594-2. [DOI] [PubMed] [Google Scholar]
- 21.Whitney O, Soderstrom K, Johnson F. Post-transcriptional regulation of zenk expression associated with zebra finch vocal development. Brain Res Mol Brain Res. 2000;80:279–290. doi: 10.1016/S0169-328X(00)00178-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitney O, Johnson F. Motor-induced transcription but sensory-regulated translation of ZENK in socially interactive songbirds. J Neurobiol. 2005;65:251–259. doi: 10.1002/neu.20187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubikova L, Turner EA, Jarvis ED. The pallial basal ganglia pathway modulates the behaviorally driven gene expression of the motor pathway. Eur J Neurosci. 2007;25:2145–2160. doi: 10.1111/j.1460-9568.2007.05368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wada K, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci USA. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li XC, Jarvis ED, Alvarez-Borda B, Lim DA, Nottebohm F. A relationship between behavior, neurotrophin expression, and new neuron survival. Proc Natl Acad Sci USA. 2000;97:8584–8589. doi: 10.1073/pnas.140222497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombardino AJ, Li X-C, Hertel M, Nottebohm F. Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels. Proc Natl Acad Sci USA. 2005;102:8036–8041. doi: 10.1073/pnas.0503239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teramitsu I, White SA. FoxP2 regulation during undirected singing in adult songbirds. J Neurosci. 2006;26:7390–7394. doi: 10.1523/JNEUROSCI.1662-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller JE, et al. Birdsong decreases protein levels of FoxP2, a molecule required for human speech. J Neurophysiol. 2008;100:2015–2025. doi: 10.1152/jn.90415.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hilliard AT, Miller JE, Fraley ER, Horvath S, White SA. Molecular microcircuitry underlies functional specification in a basal ganglia circuit dedicated to vocal learning. Neuron. 2012;73:537–552. doi: 10.1016/j.neuron.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang G, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haesler S, et al. FoxP2 expression in avian vocal learners and non-learners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wada K, Sakaguchi H, Jarvis ED, Hagiwara M. Differential expression of glutamate receptors in avian neural pathways for learned vocalization. J Comp Neurol. 2004;476:44–64. doi: 10.1002/cne.20201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kubikova L, Wada K, Jarvis ED. Dopamine receptors in a songbird brain. J Comp Neurol. 2010;518:741–769. doi: 10.1002/cne.22255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, et al. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc Natl Acad Sci USA. 2007;104:6834–6839. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lovell PV, Clayton DF, Replogle KL, Mello CV. Birdsong “transcriptomics”: Neurochemical specializations of the oscine song system. PLOS ONE. 2008;3:e3440. doi: 10.1371/journal.pone.0003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jarvis ED, et al. Global view of the functional molecular organization of the avian cerebrum: Mirror images and functional columns. J Comp Neurol. 2013;521:3614–3665. doi: 10.1002/cne.23404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim T-K, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong S, et al. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci USA. 2009;106:11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feenders G, et al. Molecular mapping of movement-associated areas in the avian brain: A motor theory for vocal learning origin. PLOS ONE. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill CS, Treisman R. Growth factors and gene expression: Fresh insights from arrays. Sci STKE. 1999;1999:PE1. doi: 10.1126/stke.1999.3.pe1. [DOI] [PubMed] [Google Scholar]
- 44.Barco A, Alarcon JM, Kandel ER. Expression of constitutively active CREB protein facilitates the late phase of long-term potentiation by enhancing synaptic capture. Cell. 2002;108:689–703. doi: 10.1016/S0092-8674(02)00657-8. [DOI] [PubMed] [Google Scholar]
- 45.Sinha S. On counting position weight matrix matches in a sequence, with application to discriminative motif finding. Bioinformatics. 2006;22:e454–e463. doi: 10.1093/bioinformatics/btl227. [DOI] [PubMed] [Google Scholar]
- 46.Kim J, et al. Functional characterization of transcription factor motifs using cross-species comparison across large evolutionary distances. PLOS Comput Biol. 2010;6:e1000652. doi: 10.1371/journal.pcbi.1000652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfenning AR, Schwartz R, Barth AL. A comparative genomics approach to identifying the plasticity transcriptome. BMC Neurosci. 2007;8:20. doi: 10.1186/1471-2202-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfenning AR, et al. Genome-wide identification of calcium-response factor (CaRF) binding sites predicts a role in regulation of neuronal signaling pathways. PLOS ONE. 2010;5:e10870. doi: 10.1371/journal.pone.0010870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loebrich S, Nedivi E. The function of activity-regulated genes in the nervous system. Physiol Rev. 2009;89:1079–1103. doi: 10.1152/physrev.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao X, Mahendran R, Guy GR, Tan YH. Detection and characterization of cellular EGR-1 binding to its recognition site. J Biol Chem. 1993;268:16949–16957. [PubMed] [Google Scholar]
- 52.Johnsen O, et al. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 1996;24:4289–4297. doi: 10.1093/nar/24.21.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDowell KA, et al. Reduced cortical BDNF expression and aberrant memory in Carf knock-out mice. J Neurosci. 2010;30:7453–7465. doi: 10.1523/JNEUROSCI.3997-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.UCSC Genome Browser. 2014 https://genome.ucsc.edu/cgi-bin/hgTracks?hgS_doOtherUser=submit&hgS_otherUserName=apfenning&hgS_otherUserSessionName=taeGut1_basic.
- 57.Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory—A molecular framework. Nature. 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- 58.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat Neurosci. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:a005744–a005744. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spiegel I, et al. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horita H, Wada K, Rivas MV, Hara E, Jarvis ED. The dusp1 immediate early gene is regulated by natural stimuli predominantly in sensory input neurons. J Comp Neurol. 2010;518:2873–2901. doi: 10.1002/cne.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Horita H, et al. Specialized motor-driven dusp1 expression in the song systems of multiple lineages of vocal learning birds. PLOS ONE. 2012;7:e42173. doi: 10.1371/journal.pone.0042173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakamoto K, Karelina K, Obrietan K. CREB: A multifaceted regulator of neuronal plasticity and protection. J Neurochem. 2011;116:1–9. doi: 10.1111/j.1471-4159.2010.07080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morgan JI, Curran T. Stimulus-transcription coupling in neurons: Role of cellular immediate-early genes. Trends Neurosci. 1989;12:459–462. doi: 10.1016/0166-2236(89)90096-9. [DOI] [PubMed] [Google Scholar]
- 65.Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wettenhall JM, Smyth GK. limmaGUI: A graphical user interface for linear modeling of microarray data. Bioinformatics. 2004;20:3705–3706. doi: 10.1093/bioinformatics/bth449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.