Abstract
Ion-electron reaction based fragmentation methods (ExD) in tandem mass spectrometry (MS), such as Electron Capture Dissociation (ECD) and Electron Transfer Dissociation (ETD) represent a powerful tool for biological analysis ExD methods have been used to differentiate the presence of the isoaspartate (isoAsp) from the aspartate (Asp) in peptides and proteins. IsoAsp is a β3-type amino acid that has an additional methylene group in the backbone, forming a Cα-Cβ bond within the polypeptide chain. Cleavage of this bond provides specific fragments that allow differentiation of the isomers. The presence of a Cα-Cβ bond within the backbone is unique to β-amino acids, suggesting a similar application of ExD toward the analysis of peptides containing other β-type amino acids. In the current study, ECD and ETD analysis of several β-amino acid containing peptides was performed. It was found that N-Cβ and Cα-Cβ bond cleavages were rare, providing few c and z• type fragments, which was attributed to the instability of the Cβ radical. Instead, the electron capture resulted primarily in the formation of a• and y fragments, representing an alternative fragmentation pathway, likely initiated by the electron capture at a backbone amide nitrogen protonation site within the beta amino acid residues.
Introduction
Electron capture dissociation (ECD)1-3 and other ion-electron reaction methods such as electron ionization/impact dissociation (EID),4 and electron transfer dissociation (ETD)5 (collectively known as ExD) belong to a group of odd-electron (OE) fragmentation techniques, which also include ultraviolet photodissociation (UVPD)6 and metastable atom-activated dissociation (MAD).7 These methods can generate fragments often unobservable in conventional slow-heating fragmentation techniques such as low energy collisionally activated dissociation (CAD)8 and infrared multiphoton dissociation (IRMPD).9 ECD and ETD are perhaps the most widely used among these OE fragmentation methods. In ECD of polypeptides, a low energy electron is captured by the multiply charged molecular ion either at a protonated backbone carbonyl site according to the Cornell mechanism,1, 10 or in a π orbital of an amide group in the presence of a remote charge according to the Utah-Washington mechanism,11 producing a charge reduced cation radical, with the subsequent backbone N-Cα bond cleavage leading to the formation of c and z• fragments. The radical on z• can induce further rearrangements within the molecule, a free radical cascade,12 producing additional backbone and side chain cleavages both in the proximity of and remote from the initial radical site.13 The fragmentation pattern generated by ECD may vary dramatically depending on the number and location of certain amino acids, modifications, and charges.14-18 A number of experimental and computational studies have been carried out since ECD was first introduced,1, 19 yet the mechanisms remain under discussion, probably because multiple competing pathways are involved.20-31 Nevertheless, the capability of ECD to produce unique fragment ions not obtainable by conventional methods has led to a rapid optimization of ECD and the development of other ion-electron reaction based tandem MS techniques.4, 5, 21 ExD has been broadly applied towards the structural analysis of various types of biomolecules, including proteins, oligosaccharides, oligonucleotides, and others.32-35 Further, ECD and ETD were found to be particularly useful for the characterization of Post Translational Modifications (PTMs) in proteins as they cleave the backbone while preserving the labile groups and non-covalent interactions upon fragmentation.2, 36
Deamidation of asparagine (Asn) and glutamine (Gln) residues and, similarly, isomerization of aspartic (Asp) and glutamic (Glu) acid residues, are two of the most common PTMs found in all proteins, that have been studied recently. These PTMs accumulate with age in long lived proteins and are frequently associated with age related diseases, such as amyloid diseases and cataract formation in the eyes.37-39 Both reactions are non-enzymatic and proceed spontaneously in physiological conditions via formation of a succinimide or glutarimide ring intermediate, followed by rapid hydrolysis. Deamidation introduces an approximately one Dalton mass shift to the protein molecular mass (+0.984 Da) and can be easily identified using mass spectrometry (MS).40 Isomerization does not change the molecular mass of the protein and thus cannot be identified as easily. However, the application of tandem MS methods, in particular, ExD techniques, has shown successful results: isomers of aspartic acid can be unambiguously identified using ECD, ETD, and EID methods;37, 41-48 likewise, identification of γ-glutamic acid by ECD seems to be promising.49
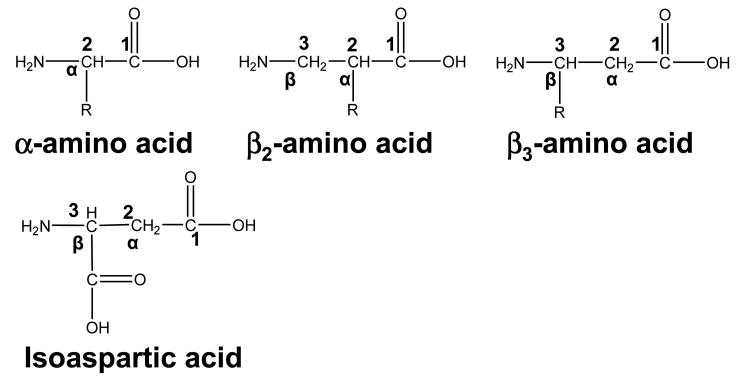
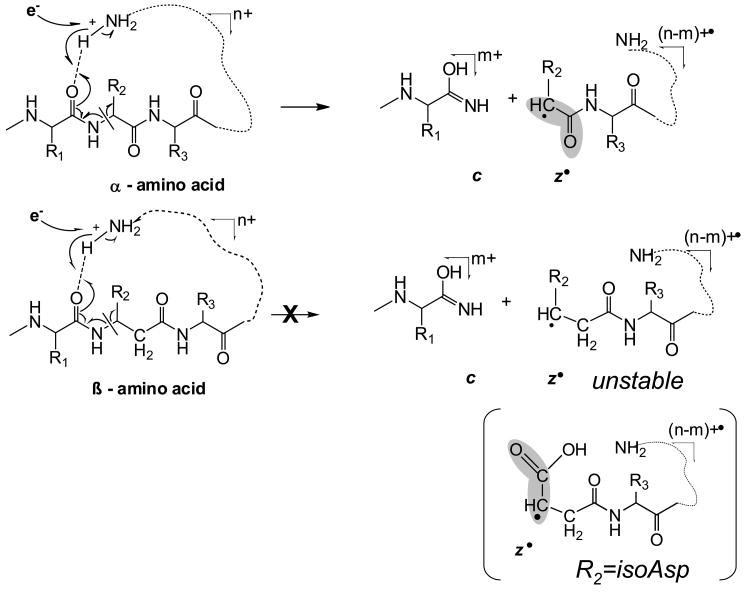
Isoaspartic acid is a β type amino acid that has one extra CH2 group in the polypeptide backbone, and one fewer on the side chain compared to Asp. ExD of isoAsp containing peptides generates additional signature fragment ions (c+57 and z•-57) at the positions of the isomerized residues, allowing differentiation from the non-modified residues (the fragmentation scheme can be found in supplemental material, Figure S1.). These ions are formed by the Cα-Cβ backbone bond cleavage. In peptides consisting solely of α-amino acid residues, there is no such bond within the backbone. Fragments produced upon Cα-Cβ bond rupture are unique to β-amino acid residues and could be used to locate the position of the β-amino acid if observed. The result from the isoAsp experiment suggested a possible extension of the method to characterize peptides containing other β-type amino acids.50-52
Similar to the isoAsp, a β-amino acid has an extra methylene group incorporated between its amino and carboxylate groups compared to its α-analogues. There are two types of β-amino acids: β2 and β3, with the side chain attached to the α and β carbon respectively (Scheme 1). β-amino acids do not normally occur in nature except for β-alanine and β-aspartate (isoAsp); neither do β-peptides, but those can be synthesized.53, 54 It should be noted here, that naturally occurring β-alanine and β-aspartic acid have the same total number of carbons as their α analogues – one more in the backbone and one fewer on the side chain; however, β-amino acids normally used for β-peptide synthesis often have an extra carbon within the backbone, but contain the same side chain as those in α-amino acids, and thus are called β-homo-amino acids (in this study all β-amino acids in synthetic peptides are β-homo-amino acids).
Scheme 1.
β amino acid nomenclature.
β-peptides have been a subject of intense studies that investigated their structural, biological properties, and their interactions involved in peptide folding. β-peptides were found to have richer conformational energy surface with more stable secondary structures.55 They also fold into helices or hairpin-type structures with larger variety than α-peptide secondary structures. β-peptides are very stable against proteolytic degradation and other enzymes in human and various living organisms.56 These features provide great potential for β-peptides in biomedical application as proteolytically stable therapeutics. A fast and accurate MS based method would be of a great utility for analysis of β-peptide structure. In this study the potential of the ExD based tandem MS methods to differentiate β-amino acid containing peptides from their alpha analogues, as well as β2 from β3-type amino acids was investigated. In general, our results are in good agreement with the findings of recent ECD/ETD studies of small β-peptides57 and ε-peptides.58 In the present study, in addition to simple model peptides such as Q06 and substance P, a more complicated system of Puma BH3 peptide analogues of 26 amino residues was investigated. Furthermore, charge state dependence of the fragment appearance was studied. An alternative mechanism of ion-electron reaction induced dissociation of peptides within β-amino acid residues is discussed.
Experimental
Materials
The Q06 β-peptide (β2Vβ2Aβ2Lβ3Vβ3Aβ3L) was kindly provided by Prof. D. Seebach, and Dr J. Gardiner, ETH Zurich, Switzerland. C-terminally amidated Substance P with two amino acids modified to β3-type amino acids (RPKPβQQFFGβLM) was custom synthesized by AnaSpec (San Jose, CA, USA). Puma BH3 pro-apoptotic protein analogue I (βEEQβWAREβIGAβQLRRβMADβDLNAβQYEβRR) and analogue II (βEEQWβAREβIGAQβLRRβMADDβLNAβQYERβR) were kindly provided by the group of Prof. S. Gellman at the University of Wisconsin (Madison, WI, USA), where β indicates β3-type amino acids. Other reagents: non-modified substance P, (2-Aminoethyl)-trimethylammonium chloride hydrochloride (cholamine), triethylamine (TEA), and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA); 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethylaminium hexafluorophosphate (HBTU) from Novabiochem (La Jolla, CA, USA); and 1-Hydroxybenzotriazole (HOBt) from AK Scientific, Inc. (Palo Alto, CA, USA).
Cholamine reaction
Covalent attachment of cholamine to the Q06 β-peptide was performed as previously described.59 Briefly, 0.1 μmol of Q06 β-peptide was treated sequentially with 10 μL of 200 mM HOBt in DMSO, 200 μL of 100 mM cholamine in DMSO containing 200 mM TEA, and 10 μL of 200 mM HBTU in DMSO. The sample was left to react overnight at room temperature, and purified using a ZipTip C18 solid phase micropipette extraction column before MS analysis.
Mass Spectrometry
Most ECD experiments were performed on a custom built qQq-FTICR MS with a nanospray source and a 7 T actively shielded magnet.60, 61 Samples were nanosprayed (50 nL/min, room temperature) at 5 μM concentration in 50:50 MeOH:H2O with 1% formic acid. Ions were isolated in the first quadrupole Q1, accumulated in the second quadrupole Q2, and transmitted into the ICR cell where they were irradiated with electrons emitted from an indirectly heated dispenser cathode (Heatwave, Watsonville, CA, USA) for ion fragmentation. The following ECD and EID parameters were employed: electron irradiation time 35-100 ms, cathode potential -0.2 - 1.2 V (ECD), - 18 V (EID). Acquired spectra were zerofilled twice, internally calibrated, and analyzed manually using BUDA (Boston University Data Analysis, Version 1.4, © 2000 by Peter B. O’Connor). ECD experiments of triply charged Substance P ions were performed on solariX FTICR instrument (Bruker Daltonics, Billerica, MA, USA) with 12 T actively shielded magnet. Electrospray was applied for enhanced production of triply charged ions.
ETD spectra with supplemental activation were acquired on an amaZon Ion Trap instrument (Bruker Daltonics, Billerica, MA, USA) using fluoranthene as the ETD reagent. Peptides were electrosprayed (2 μL/min, glass capillary temperature 220 °C) at 1 μM concentration in 50:50 MeOH:H2O with 1% formic acid using an Apollo II ion source. Data acquired on solariX and amaZon were analyzed using Bruker's ESI Compass DataAnalysis 4.0 software.
Results and Discussion
Q06 beta peptide β2Vβ2Aβ2Lβ3Vβ3Aβ3L
Although the Q06 β-peptide only contains six β-amino acid residues, it can form helical secondary structures.55, 62 This small and relatively simple peptide contains both β2 and β3 type amino acids, making it potentially an ideal system to study for the differentiation of β2 and β3 amino acids. However, only singly charged ions [M+H]+ were detected in all ESI/ECD experiments, either with the nanospray or with the electrospray ionization sources (supplemental figure S2.a), which is likely due to the lack of basic amino acid residues within the peptide sequence. Since ECD and ETD are accompanied by charge neutralization upon the electron capture or transfer, they can not be performed on singly charged ions as the products would be neutral and undetectable. In this case, other fragmentation techniques could be applied that do produce fragments from singly charged ions such as CAD8 or IRMPD.9 Expectedly, IRMPD of the Q06 beta peptide resulted in b and y fragments with no information on the position of the modifications (supplemental figure S2.b).50 Additionally, Electron Ionization/Impact Dissociation (EID) can generate ECD type fragments from singly charged molecular ions,4 as well as Cα-Cβ cleavage for isoaspartic acid.45 However, only b and y fragments were detected in EID mass spectra of the Q06 β-peptide (supplemental figure S2.c).51 The lack of c and z• fragments could be due to the low fragmentation efficiency of EID in this particular experiment. Thus, the well established ECD method would seem to be a better approach, but it requires an increase in the number of charges on the peptide.
Charge increase
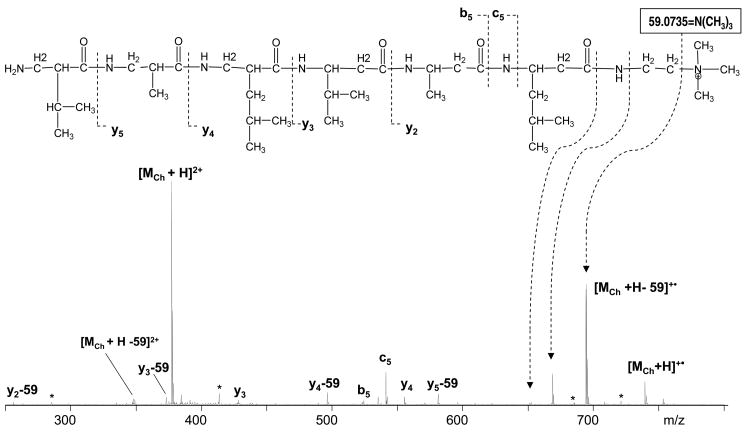
Three different approaches were applied in order to enhance the formation of higher charged molecular ions. These include the addition of nitrobenzyl alcohol or calcium salt into the ESI solution, and the covalent attachment of the cholamine tag to the peptide.50, 59, 63-65 Although doubly charged ions were observed in all three cases, only the cholamine reaction produced sufficient ions for ECD analysis. Inefficiency of other methods could be due to the lack of a preferred calcium ion binding site and potentially low charge stabilization in the peptide. Cholamine reacts with the carboxylic acid and was attached to the peptide at its C-terminus. The second charge could be provided by the protonation of the N-terminal amine. Doubly charged species [M•Ch++H]2+ were readily observed in the ESI MS, isolated, and further subjected to ECD (Figure 1).
Figure 1.
ECD of Q06•cholamine.
Similar experiments of C-terminal charge tag attachment to α-peptides were done by Hunt's group.66 Their result demonstrated increased formation of z ions. In the current study, no z fragments and only one ECD-type fragment, c5, was observed for the Q06 β-peptide. The electron capture at the quaternary ammonium provides an abundant loss of the neutral trimethylamine 59.0735 = N(CH3)3, leaving the radical on the C-terminal methylene group (Figure 1). This is expected to further induce radical initiated reactions and cleavages, such as the loss of ethylene. Although charge neutralization on the quaternary ammonium may not lead to backbone cleavages,67 electron capture at the protonated N-terminal amine of the doubly charged Q06 peptide should result in backbone fragmentations. Further, the recombination energy of the protonated N-terminal amino group is higher than that of the quaternary ammonium as calculated for the singly charged model cations by Jensen et al. (4.3 eV (MeNH3+) vs. 3.1 eV (NMe4+)),68 making it the preferred electron capture site. However, ECD type fragments were lacking in the ECD spectrum of the tagged Q06 peptide, which was dominated by b5 and y fragments which could be merely the result of the residual vibrational excitation.10 Alternatively, the formation of y fragments could be facilitated by the backbone nitrogen protonation or a rearrangement that transfers a hydrogen atom to the backbone nitrogen.2, 15, 58 Upon charge neutralization at the tag site and the subsequent tag loss, the excessive vibrational energy could induce the formation of the b5 ion, similar to that suggested by Cooper et al. in their study of b-ion formation in ECD.14 Nevertheless, it is hard to make definitive conclusions why this unusual fragmentation pattern was observed as two variables were introduced at the same time in the studied system: a cholamine tag and the incorporation of β-amino acids.
ETD of Q06 beta peptide
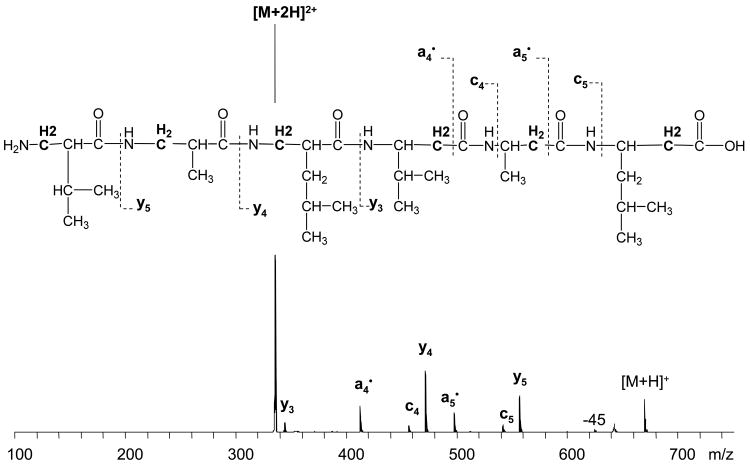
Electron transfer dissociation was further used to analyze the Q06 beta peptide. As opposed to the first ESI experiment, a small peak corresponding to the doubly charged molecular ions, [MQ06+2H]2+, was observed, which could be due to the difference in the ionization sources between two instruments. In particular, the glass capillary in the electrospray source used in the ETD study was heated to 220 °C which could possibly increase the desolvation efficiency and production of higher charged ions compared to the unheated source used in the previous ECD experiment.
ETD of the Q06 beta peptide produced an unusual fragmentation pattern with abundant y and a• fragments, and only two c fragments in low abundance (Figure 2). The assignment of a•5 ion was ambiguous due to the interference from x4 ion [x4 (m/z=497.33), and a•5 (m/z=497.39)], which could not be resolved using the ion trap. The assignment is probably correct, because x ions are not commonly observed in ECD and ETD, and they were not detected for this peptide in ECD experiments. It is not clear how such abundant y fragments could be formed without a C-terminal charge carrier. It might be due to the backbone nitrogen protonation, similar to the cholamine attached Q06 β-peptide ECD results. Likewise, the alternative dissociation pathway seems to be enhanced. Meanwhile, low vibrational energy applied to the charge reduced species to increase dissociation efficiency upon electron transfer (smart decomposition) may also be a reason for enhanced y fragment formation. However, this cannot explain the formation of a• fragment ions as those are radicals species and more likely to be a product of ion-electron reaction. In general, the observed fragmentation pattern for this peptide is not following the usual ETD behavior. ExD of this very simple β-peptide demonstrates a big difference for fragment formation in β-peptides compared to their α-analogues.
Figure 2.
ETD of Q06 β2/β3-beta peptide.
Modified Substance P
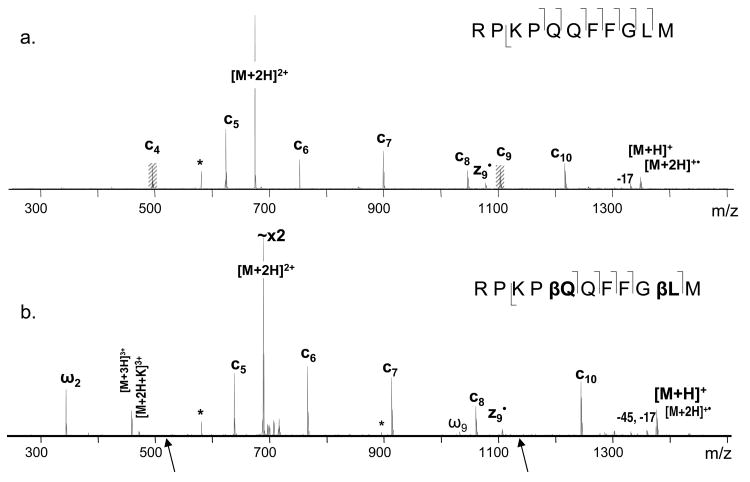
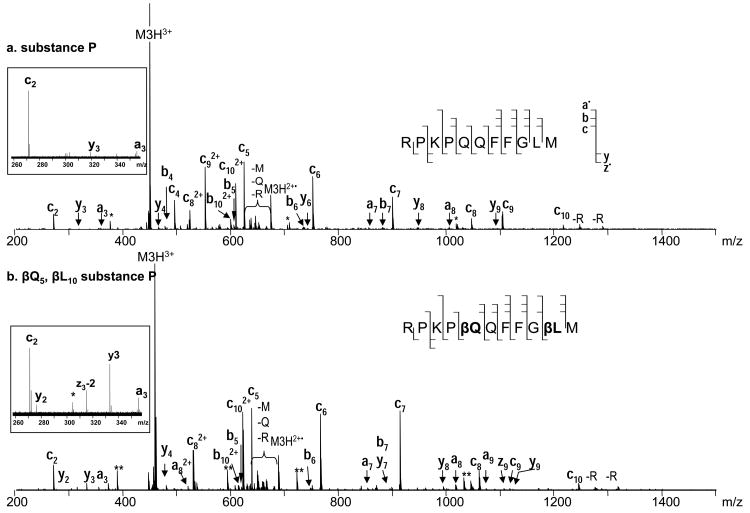
In order to better understand the unusual ExD behavior of β-peptides, ECD experiments were carried out on a well studied system, the substance P peptide, and its variant which was modified at two positions: glutamine 5 and leucine 10 to β3-homo amino acids. Q5 and L10 were specifically selected for modification as they normally provide abundant c4 and c9 fragments (Figure 3.a.), and the expected fragments resulted from the Cα-Cβ bond cleavage (discussed below) would not interfere with other peaks in the spectrum. The introduction of the two extra methylene groups in the backbone increased the molecular mass of the peptide by 28 Da. Fragment ion mass shift at 14 Da per additional methylene group was also observed correspondingly. As was discussed in the introduction, the extra methylene group present in isoAsp leads to a Cα-Cβ backbone bond cleavage and the formation of signature diagnostic fragments. Similar cleavages were expected for the current modifications which would result in formation of c4+85 and c9+70 fragment ions. However, not only were these ions absent, the c4 and c9 fragments also disappeared from the ECD spectrum entirely (Figure 3). One may speculate that introduction of the CH2 group could lead to such a conformational change that would hinder the formation of c4 and c9 fragments; or they may still be formed, but not separated due to the new, possibly tighter hydrogen bonding, although the incorporation of only one extra methylene group is unlikely to create such a big difference in intramolecular hydrogen bonds. As proposed previously,51, 52 it is more likely that the absence of Cα-Cβ bond cleavages resulted from the lack of radical stabilization effect by a sidechain carbonyl group as in isoAsp containing peptides. Further, in α-amino acid residues, the α-carbon radical formed by N-Cα bond cleavage can be resonantly stabilized by the neighboring carbonyl (Scheme 2); in β-amino acid residues, the carbonyl is located further away providing no such stabilization, making the N-Cβ bond cleavage not energetically favorable. Within the isoAsp, however, a carboxylic acid on a side chain is located at the close proximity to the backbone β-carbon atom, which can play a stabilization role for the β-carbon radical and thus ensure both the Cα-Cβ and N-Cβ bond cleavages (supplemental material S1). In agreement with this explanation, in a previous ECD study of γ-glutamic acid containing peptides, N-Cγ, Cα-Cβ, and Cβ-Cγ bond cleavages were all observed.49 Similar stabilizing effect can be provided by the aromatic structure as well, such as that observed in the β-phenylalanine containing peptides, which was recently reported by Hamidane et al.57 supporting the radical stability hypothesis.
Figure 3.
ECD of Substance P: a. unmodified, b. modified at Q5 and L10 to β3 - type amino acids.
Scheme 2.
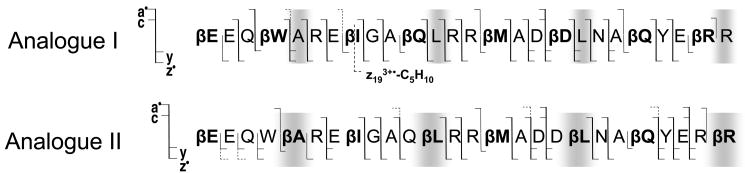
ECD cleavage scheme of Puma BH3 protein analogues I and II.
Puma BH3 protein analogues
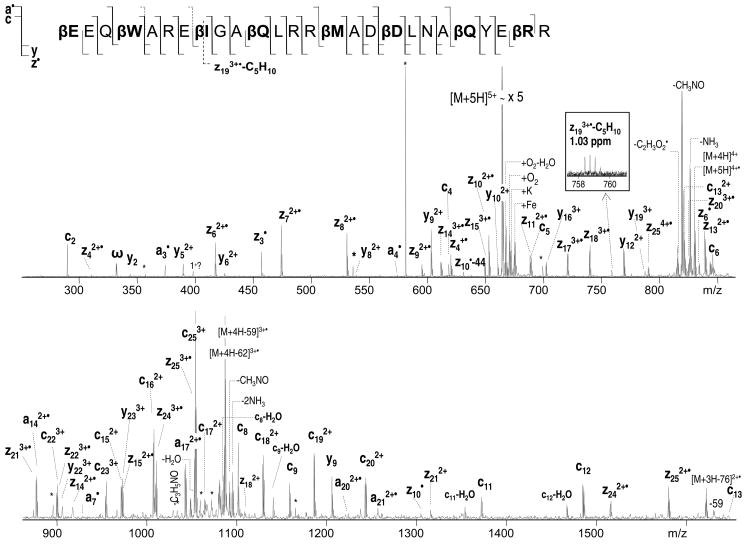
Further ECD analysis was performed on a set of bigger peptides, originally designed to mimic foldamer ligands for the BH3 recognition cleft of the protein Bcl-xL.69 The primary sequence of the two 26-residue α/β-peptide analogues corresponds to a Puma BH3 domain (EEQWAREIGAQLRRMADDLNAQYERR). Both peptide analogues have amino acid residues modified to a β3-homo amino acids after each 2nd or 3rd residue, but not all at the same positions. Such a backbone repeat ααβαααβ allows formation of an α-helix like conformation that helps mimic the original binding behavior of an α-helical domain. For the purpose of the current study, the two peptide analogues provide an excellent system for the direct comparison of fragmentation within α- and β-type amino acids. The ECD spectrum of the 3.3 kDa Puma BH3 protein analogue I (βEEQβWAREβIGAβQLRRβMADβDLNAβQYEβRR) is shown in Figure 4; the ECD spectrum of the analogue II (βEEQWβAREβIGAQβLRRβMADDβLNAβQYERβR) can be seen in supplemental material (S3). The spectra show nearly complete sequence coverage with various types of fragments observed, including a•, y, c, and z• fragments. In agreement with the modified substance P result, no N-Cβ bond cleavages were observed for the β-type amino acid residues, but the corresponding α-amino acid residues all provided such cleavages. For example, c, and z• fragments are observed at the α-Ala5, α-Leu12,20, and α-Arg26 positions (analogue I) but not in the β-Ala5, β-Leu12,20, and β-Arg26 positions (analogue II) (Scheme 3, relevant residues are highlighted). The only N-Cβ bond rupture was detected for β-Asp provided by the z92+ and c172+ fragments (Figure 4). Interestingly, this is a β-homo aspartic acid; i.e. the sidechain carboxylic acid is separated from the backbone β-carbon by one methylene group. Hence, β-Asp side chain carbonyl cannot provide the same stabilization for the Cβ radical as in the native isoAsp, and the z92+ and c172+ fragments were only present at very low abundance.
Figure 4.
ECD of Puma BH3 protein analogue I.
Scheme 3.
ECD mechanism within alpha vs. beta amino acid residues.
As was noted earlier, the Puma BH3 peptides studied here have α-helix like conformations. It is possible that the N-Cβ bonds can be cleaved but protected from dissociation due to strong hydrogen bonding within the α-helix as was suggested previously70 and consistent with the results from ECD mechanistic studies.24, 26 Yet, there were many c and z• fragments present except for those missing within β-residues suggesting that the peptide was relatively unfolded, and thus, the hydrogen bonds should not interfere with the fragment separation. Furthermore, dissociation of the Cα-Cβ bond now seems to be an exception rather than the rule for β-amino acid residues as only one peak representing the Cα-Cβ cleavage of isoleucine was identified provided by the z193+•-C5H10 fragment ion (Figure 4). Indeed, according to Turecek and coworkers71 who studied the β-alanine N-methyl amide model system, dissociation of the Cα-Cβ bond was slow and in competition with other dissociations from the most stable composition with a radical located on the C-terminal amide carbonyl. To conclude, the N-Cβ and Cα-Cβ backbone cleavages are still possible, but do not represent the dominant channel of fragmentation within β-amino acid residues.
Proposed mechanism
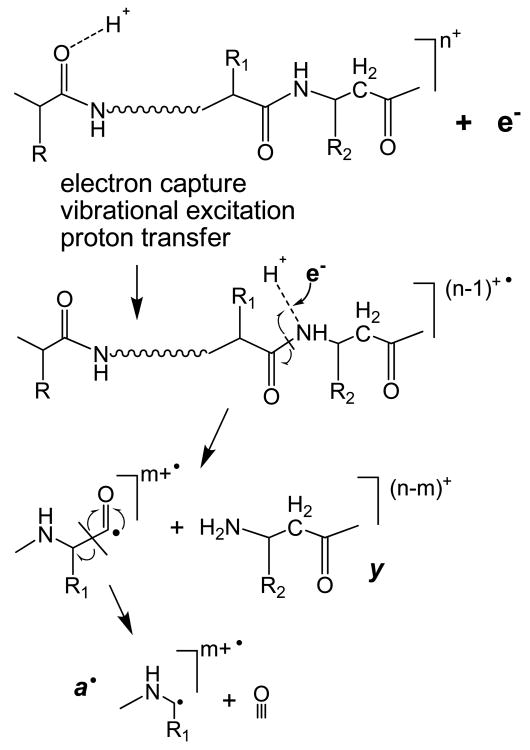
The incorporation of an extra methylene group within the polypeptide backbone increases the flexibility of the molecule. The conformational change is probably not that dramatic, but an increase in internal rotations is expected, resulting in decreased steric hindrance. Thus, the backbone nitrogen may become more exposed for hydrogen bonding. In addition, elongation of the backbone within the β-residue moves the amide nitrogen and the following amide carbonyl apart removing the captodative stabilization effect at the Cβ. Therefore, the N-Cβ bond dissociation becomes a less favorable process, thus shifting dissociation to other fragmentation channels. One such channel may proceed via backbone nitrogen protonation, leading to increased a• and y fragment ion formation (Scheme 4) as was proposed in an early ECD paper.3 The N-Cβ bond rupture may still occur, creating c fragment and unstable intermediate z• fragment that, if formed, will probably undergo further rapid dissociation, and is thus not observed. However, it is more likely, that upon electron capture the radical at the backbone amide hydrogen will induce the homolytic cleavage of the peptide bond and further loss of a CO molecule to produce the more stable a• and y fragment ions. It is interesting to note that, unlike the doubly charged Q06 peptide, the a•/y fragmentation channel was not observed in the ECD spectrum of the doubly charged β-substance P variant.
Scheme 4.
ECD mechanism within β-amino acid residues
A closer look at the Q06 β-peptide and the modified substance P revealed that, besides the nature of the amino acids, the clear difference between the peptides is the presence of basic amino acid residues, which could dictate the sites of protonation and thus the sites of electron capture dissociation. The two charges carried by the substance P peptide would preferably reside at the Arg and Lys sidechains, and are solvated by the carbonyl groups of the peptide. According to the Cornell ECD mechanism,10 upon electron capture at the protonated site, hydrogen migration to various carbonyl groups would result in N-Cα bond cleavages. Due to arginine being a poor hydrogen donor, backbone fragmentations are most likely initiated by electron capture at the protonated N-terminal or lysine sidechain amino group.19, 28, 72 In the case of the β-substance P, this process will result in the formation of unstable Cβ radicals within the βQ and βL residues, which correlates with their disappearance. On the other hand, in the Q06 β-peptide, there are no basic amino acids. As was discussed earlier, the first protonation site would be the N-terminal amine; and the second proton would be mobile within the polypeptide backbone. Thus electron capture would occur at the N-Terminus or on the backbone rather than on the side chain and the electron induced fragmentation of the Q06 peptide would occur via a different mechanism to that of substance P. Introducing the third charge to the substance P peptide should lead to the protonation of an additional site within the molecule, which is likely one of the backbone amide nitrogen or carbonyls (see below), since N-terminal amine protonation seems unlikely because of the strong columbic repulsions by the nearby protonated Lys and Arg residues. To test this hypothesis, triply charged β-substance P was subject to the ECD analysis (Figure 5). Indeed, many a• and y fragments now appeared in the spectra. Interestingly, they often were slightly higher in abundance in the modified substance P variant as shown in the inset (Figure 5). Nonetheless, all of the a• and y fragments were observed in both peptides, except for y2 and a9•, which were exclusively present in the modified substance P variant. The last two fragments are formed due to the cleavages within the β-Leu10 residue. Note that the N-Cβ bond cleavage was not observed within this residue as c9 and z2• fragments were still absent (yet present in the non-modified variant). This is consistent with the previous results and supports the proposed hypothesis for the fragmentation within the β-amino acid residues.
Figure 5.
ECD of triply charged substance P [M+3H]3+: a. unmodified, b. modified at Q5 and L10 to β3-type amino acids.
The role of backbone nitrogen protonation in ECD has been previously discussed in the literature. In the original ECD study, the a• and y ions were proposed to be formed from the backbone nitrogen protonated species.3 Backbone nitrogen protonation was also suggested to play a role in the formation of b ions in ECD. Theoretical investigation by the Uggerud group showed that electron capture by nitrogen protonated N-methyl-acetamide resulted in rapid amide bond dissociation and production of CH3CO and NH2CH2, corresponding to the b• and y ions in peptides.73 Concomitantly, b• ions can further lose CO to form a• ions. In addition, b ions were present in ECD of peptides without basic amino acid residues.16 It was suggested that upon backbone nitrogen protonation, the peptide bond could be cleaved to form ab•/y ion pair, and the subsequent intra-complex hydrogen atom transfer within this long-lived ion pair could lead to the formation of b and y• ions. Backbone amide nitrogen protonation was also invoked to explain the a•, b, and b• ion formation in nitrated peptides that are either acetylated at the N-terminus, or lacking basic amino acid residues.74 In the current study, however, neither b nor y ions observed were radicals. Thus, these b ions were most likely formed via the energetic fragmentation of vibrationally excited even-electron charged reduced species.14
It should be noted that protonation on the backbone nitrogen is not thermodynamically favorable.75-77 For instance, for the model system of N-methylacetamide, the proton affinity of the carbonyl oxygen was calculated to be ∼60 kJ/mol higher than that of the backbone nitrogen.76 This is in agreement with the study of the dipeptide Lys-Gly, where the carbonyl oxygen protonated species was calculated to be ∼45 kJ/mol more stable than the backbone nitrogen protonated species.77 Based on these results, peptide fragmentation via protonation of the backbone nitrogen would seem unlikely. However, theoretical investigations have usually been done on very small model systems, which often do not possess extensive intramolecular interactions such as hydrogen bonding and salt bridges that are expected to play a more important role in the fragmentation of larger peptides. Charge solvation by nearby backbone and sidechain groups could appreciably change the relative stabilities of different protonated species. In β-peptides, the elongation of the backbone may make it better positioned (due to less steric hindrance by the adjacent side chain) for hydrogen bond formation. The addition of an extra methylene group in the backbone can also slightly increases the gas-phase basicity of the amide nitrogen, as the electron withdrawing carbonyl group is replaced by the electron donating alkyl group. Further, electron capture by the precursor ion could increase its internal energy considerably (by several eVs), and less favored protonation sites may become significantly populated as the system relaxes from the initial Rydberg state to low lying electronic states. In other words, the proton may initially reside on the carbonyl oxygen, but could migrate to the backbone amide nitrogen upon excitation. This argument is similar to the one used in the mobile proton model to explain the low energy CAD fragmentation behavior of peptide ions,75, 78 where it was proposed that the proton initially resides on the thermodynamically more stable sites, such as the lysine sidechain or backbone oxygen, and later migrates to the less favorable sites, including the backbone amide nitrogen upon collisional activation to facilitate fragmentations. Finally, for the β-linked peptides studied here, electron capture at the protonated oxygen site cannot lead to “normal” c/z fragmentation due to the radical instability, which may further drive the migration of the proton to the less preferable backbone nitrogen site that could lead to the formation of a• and y ions upon electron capture. In general, the abundance of these unusual ECD fragments was fairly low, as expected from the low population of nitrogen protonated species (Figure 5), but they were nonetheless present in competitive abundance when the primary ECD fragmentation channel was blocked within β-amino acid residues.
Further experiments are needed to test the proposed mechanism. In addition, basicity measurements and theoretical investigations specifically for β-amino acid containing peptides could provide a better understanding on how these a• and y type fragments are formed.
Conclusions
Various peptides containing β-amino acid residues were analyzed in this study. Remarkably, N-Cβ bond cleavages were rare within the β-residues, and Cα-Cβ cleavages were seldom observed providing no evidence of the β-residues in spite of previous results of the isoaspartic acid. Furthermore, no distinct difference was found for the fragmentation within β2 vs. β3-type amino acid residues. Meanwhile, a• and y fragments were often produced at β-residues, particularly for the bigger peptides with α-helical like structures. The lack of z• and c fragments and increased a• and y fragment formation could imply the presence of β-residues in the peptide; however, this is a poor signature, because of the normal appearance of a• and y fragments in ExD spectra of α-peptides, and various other reasons that can contribute to the disappearance of z• and c fragments. Thus, currently, ExD methods cannot be used to reliably differentiate α- from β- or β2 from β3 type amino acids.
The introduction of one extra methylene group into the polypeptide chain destabilizes the Cβ radical formed by the N-Cβ bond rupture making this channel of fragmentation less favorable. Thus the fragmentation occurs via alternative channels. The dominant products appear to be the a• and y fragments, with the exception when the side chain of the β-residue can provide radical stabilization for the formation of the z• and c fragments. It is suggested that appearance of such fragments may require protonation on the backbone amide nitrogen, which is further supported by the charge state-dependent study of modified substance P peptide. The fragmentation mechanism for β-peptides has been proposed via backbone nitrogen protonation similarly to what was originally proposed for the ECD of α-peptides.
The minor ECD pathway of a• and y fragment formations was little studied. Future studies of this fragmentation pathway would help with our understanding of the fragmentation of the β-amino acid containing peptides. Further ExD studies, as well as computational studies and kinetic analysis specifically for the β-amino acids are needed for better characterization of β-peptides.
Supplementary Material
Supplemental figure S1. ECD of isoaspartic acid.
Supplemental figure S2. Q06 beta peptide β2Vβ2Aβ2Lβ3Vβ3Aβ3L: a. ESI, b. IRMPD, c. EID.
Supplemental figure S3. ECD of Puma BH3 protein analogue II.
Acknowledgments
The authors acknowledge Sam Gellman, Lisa Johnson, Dieter Seebach and James Gardiner for providing the samples of β-peptides for this study. We would like to thank Brian Fray and Lloyd Smith for their cholamine reaction protocol. Catherine Costello is appreciated for the opportunity to use the AmaZon and SolariX instruments, Liang Han and Sandrine Bourgoin-Voillard for help with instrument use. This work was supported by the Warwick University chemistry department and Warwick Centre for Analytical Science, and NIH/NCRR-P41 RR10888, NIH/NHLBI-N01HV28178, NIH/NIGMS-R01GM078293, S10 RR025082 grants.
References
- 1.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120(13):3265–3266. [Google Scholar]
- 2.Zubarev RA, Horn DM, Fridriksson EK, Kelleher NL, Kruger NA, Lewis MA, Carpenter BK, McLafferty FW. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72(3):563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 3.Zubarev RA, Kruger NA, Fridriksson EK, Lewis MA, Horn DM, Carpenter BK, McLafferty FW. Electron capture dissociation of gaseous multiply-charged proteins is favored at disulfide bonds and other sites of high hydrogen atom affinity. J Am Chem Soc. 1999;121(12):2857–2862. [Google Scholar]
- 4.Fung YME, Adams CM, Zubarev RA. Electron ionization dissociation of singly and multiply charged peptides. J Am Chem Soc. 2009;131(29):9977–9985. doi: 10.1021/ja8087407. [DOI] [PubMed] [Google Scholar]
- 5.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101(26):9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reilly JP. Ultraviolet photofragmentation of biomolecular ions. Mass Spectrom Rev. 2009;28(3):425–447. doi: 10.1002/mas.20214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook SL, Collin OL, Jackson GP. Metastable atom-activated dissociation mass spectrometry: leucine/isoleucine differentiation and ring cleavage of proline residues. J Mass Spectrom. 2009;44(8):1211–1223. doi: 10.1002/jms.1598. [DOI] [PubMed] [Google Scholar]
- 8.Senko MW, Speir JP, McLafferty FW. Collisional activation of large multiply charged ions using Fourier transform mass spectrometry. Anal Chem. 1994;66(18):2801–2808. doi: 10.1021/ac00090a003. [DOI] [PubMed] [Google Scholar]
- 9.Little DP, Speir JP, Senko MW, O'Connor PB, McLafferty FW. Infrared multiphoton dissociation of large multiply charged ions for biomolecule sequencing. Anal Chem. 1994;66(18):2809–2815. doi: 10.1021/ac00090a004. [DOI] [PubMed] [Google Scholar]
- 10.McLafferty FW, Horn DM, Breuker K, Ge Y, Lewis MA, Cerda B, Zubarev RA, Carpenter BK. Electron capture dissociation of gaseous multiply charged ions by Fourier-transform ion cyclotron resonance. J Am Soc Mass Spectrom. 2001;12(3):245–249. doi: 10.1016/S1044-0305(00)00223-3. [DOI] [PubMed] [Google Scholar]
- 11.Syrstad EA, Turecek F. Toward a general mechanism of electron capture dissociation. J Am Soc Mass Spectrom. 2005;16(2):208–224. doi: 10.1016/j.jasms.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Leymarie N, Costello CE, O'Connor PB. Electron capture dissociation initiates a free radical reaction cascade. J Am Chem Soc. 2003;125(29):8949–8958. doi: 10.1021/ja028831n. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Lin C, Han L, Costello CE, O'Connor PB. Charge remote fragmentation in electron capture and electron transfer dissociation. J Am Soc Mass Spectrom. 2010;21(4):646–656. doi: 10.1016/j.jasms.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper HJ. Investigation of the presence of b ions in electron capture dissociation mass spectra. J Am Soc Mass Spectrom. 2005;16(12):1932–1940. doi: 10.1016/j.jasms.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Tsybin YO, Haselmann KF, Emmett MR, Hendrickson CL, Marshall AG. Charge location directs electron capture dissociation of peptide dications. J Am Soc Mass Spectrom. 2006;17(12):1704–1711. doi: 10.1016/j.jasms.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 16.Liu HC, Hakansson K. Abundant b-type ions produced in electron capture dissociation of peptides without basic amino acid residues. J Am Soc Mass Spectrom. 2007;18:2007–2013. doi: 10.1016/j.jasms.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 17.Li XJ, Cournoyer JJ, Lin C, O'Connor PB. The effect of fixed charge modifications on electron capture dissociation. J Am Soc Mass Spectrom. 2008;19(10):1514–1526. doi: 10.1016/j.jasms.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben Hamidane H, He H, Tsybin OY, Emmett MR, Hendrickson CL, Marshall AG, Tsybin YO. Periodic sequence distribution of product Ion abundances in electron capture dissociation of amphipathic peptides and proteins. J Am Soc Mass Spectrom. 2009;20(6):1182–1192. doi: 10.1016/j.jasms.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 19.Chen XH, Turecek F. The arginine anomaly: Arginine radicals are poor hydrogen atom donors in electron transfer induced dissociations. J Am Chem Soc. 2006;128(38):12520–12530. doi: 10.1021/ja063676o. [DOI] [PubMed] [Google Scholar]
- 20.Zubarev RA, Haselmann KF, Budnik B, Kjeldsen F, Jensen F. Towards an understanding of the mechanism of electron-capture dissociation: a historical perspective and modern ideas. Eur J Mass Spectrom. 2002;8(5):337–349. [Google Scholar]
- 21.Zubarev RA. Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrom Rev. 2003;22(1):57–77. doi: 10.1002/mas.10042. [DOI] [PubMed] [Google Scholar]
- 22.Turecek F, Syrstad EA. Mechanism and energetics of intramolecular hydrogen transfer in amide and peptide radicals and cation-radicals. J Am Chem Soc. 2003;125(11):3353–3369. doi: 10.1021/ja021162t. [DOI] [PubMed] [Google Scholar]
- 23.Turecek F. N-C-alpha bond dissociation energies and kinetics in amide and peptide radicals. Is the dissociation a non-ergodic process? J Am Chem Soc. 2003;125(19):5954–5963. doi: 10.1021/ja021323t. [DOI] [PubMed] [Google Scholar]
- 24.O'Connor PB, Lin C, Cournoyer JJ, Pittman JL, Belyayev M, Budnik BA. Long-lived electron capture dissociation product ions experience radical migration via hydrogen abstraction. J Am Soc Mass Spectrom. 2006;17(4):576–585. doi: 10.1016/j.jasms.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 25.Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. Hydrogen rearrangement to and from radical z fragments in electron capture dissociation of peptides. J Am Soc Mass Spectrom. 2007;18(1):113–120. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Lin C, Cournoyer JJ, O'Connor PB. Probing the gas-phase folding kinetics of peptide ions by IR activated DR-ECD. J Am Soc Mass Spectrom. 2008;19(6):780–789. doi: 10.1016/j.jasms.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turecek F, Chen XH, Hao CT. Where does the electron go? Electron distribution and reactivity of peptide cation radicals formed by electron transfer in the gas phase. J Am Chem Soc. 2008;130(27):8818–8833. doi: 10.1021/ja8019005. [DOI] [PubMed] [Google Scholar]
- 28.Panja S, Nielsen SB, Hvelplund P, Turecek F. Inverse hydrogen migration in arginine-containing peptide ions upon electron transfer. J Am Soc Mass Spectrom. 2008;19(12):1726–1742. doi: 10.1016/j.jasms.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Sohn CH, Chung CK, Yin S, Ramachandran P, Loo JA, Beauchamp JL. Probing the mechanism of electron capture and electron transfer dissociation using tags with variable electron affinity. J Am Chem Soc. 2009;131(15):5444–5459. doi: 10.1021/ja806534r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frison G, van der Rest G, Turecek F, Besson T, Lemaire J, Maitre P, Chamot-Rooke J. Structure of electron-capture dissociation fragments from charge-tagged peptides probed by tunable infrared multiple photon dissociation. J Am Chem Soc. 2008;130(45):14916–+. doi: 10.1021/ja805257v. [DOI] [PubMed] [Google Scholar]
- 31.Simons J. Mechanisms for S-S and N-C-alpha bond cleavage in peptide ECD and ETD mass spectrometry. Chem Phys Lett. 2010;484(4-6):81–95. [Google Scholar]
- 32.Wolff JJ, Laremore TN, Aslam H, Linhardt RJ, Amster IJ. Electron-induced dissociation of glycosaminoglycan tetrasaccharides. J Am Soc Mass Spectrom. 2008;19(10):1449–1458. doi: 10.1016/j.jasms.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper HJ, Hakansson K, Marshall AG. The role of electron capture dissociation in biomolecular analysis. Mass Spectrom Rev. 2005;24(2):201–222. doi: 10.1002/mas.20014. [DOI] [PubMed] [Google Scholar]
- 34.Liang XLR, Liu J, Leblanc Y, Covey T, Ptak AC, Brenna JT, McLuckey SA. Electron transfer dissociation of doubly sodiated glycerophosphocholine lipids. J Am Soc Mass Spectrom. 2007;18(10):1783–1788. doi: 10.1016/j.jasms.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang J, Mo JJ, Adamson JT, Hakansson K. Characterization of oligodeoxynucleotides by electron detachment dissociation Fourier-transform ion cyclotron resonance mass spectrometry. Anal Chem. 2005;77(6):1876–1882. doi: 10.1021/ac048415g. [DOI] [PubMed] [Google Scholar]
- 36.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JEP, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochem Biophys Acta - Proteins and Proteomics. 2006;1764(12):1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson NE, Robinson AB. Molecular clocks: Deamidation of asparaginyl and glutaminyl residues in peptides and proteins. Althouse Press; Cave Junction, OR: 2004. p. 443. [Google Scholar]
- 38.Ritz-Timme S, Collins MJ. Racemization of aspartic acid in human proteins. Ageing Res Rev. 2002;1(1):43–59. doi: 10.1016/s0047-6374(01)00363-3. [DOI] [PubMed] [Google Scholar]
- 39.Lampi KJ, Amyx KK, Ahmann P, Steel EA. Deamidation in human lens beta B2-crystallin destabilizes the dimer. Biochem. 2006;45(10):3146–3153. doi: 10.1021/bi052051k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zabrouskov V, Han XM, Welker E, Zhai HL, Lin C, van Wijk KJ, Scheraga HA, McLafferty FW. Stepwise deamidation of ribonuclease A at five sites determined by top down mass spectrometry. Biochem. 2006;45(3):987–992. doi: 10.1021/bi0517584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan WYK, Chan TWD, O'Connor PB. Electron transfer dissociation with supplemental activation to differentiate aspartic and isoaspartic residues in doubly charged peptide cations. J Am Soc Mass Spectrom. 2010;21(6):1012–1015. doi: 10.1016/j.jasms.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cournoyer JJ, Pittman JL, Ivleva VB, Fallows E, Waskell L, Costello CE, O'Connor PB. Deamidation: Differentiation of aspartyl from isoaspartyl products in peptides by electron capture dissociation. Protein Sci. 2005;14(2):452–463. doi: 10.1110/ps.041062905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cournoyer JJ, Lin C, O'Connor PB. Detecting deamidation products in proteins by electron capture dissociation. Anal Chem. 2006;78(4):1264–1271. doi: 10.1021/ac051691q. [DOI] [PubMed] [Google Scholar]
- 44.Cournoyer JJ, Lin C, Bowman MJ, O'Connor PB. Quantitating the relative abundance of isoaspartyl residues in deamidated proteins by electron capture dissociation. J Am Soc Mass Spectrom. 2007;18(1):48–56. doi: 10.1016/j.jasms.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 45.Sargaeva NP, Lin C, O'Connor PB. Identification of aspartic and isoaspartic acid residues in amyloid beta peptides, Including A beta 1-42, using electron-ion reactions. Anal Chem. 2009;81(23):9778–9786. doi: 10.1021/ac901677t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang HQ, Fung EYM, Zubarev AR, Zubarev RA. Toward proteome-scale identification and quantification of isoaspartyl residues in biological samples. J Proteom Res. 2009;8(10):4615–4621. doi: 10.1021/pr900428m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li XJ, Cournoyer JJ, Lin C, O'Connor PB. Use of O-18 labels to monitor deamidation during protein and peptide sample processing. J Am Soc Mass Spectrom. 2008;19(6):855–864. doi: 10.1016/j.jasms.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Connor PB, Cournoyer JJ, Pitteri SJ, Chrisman PA, McLuckey SA. Differentiation of aspartic and isoaspartic acids using electron transfer dissociation. J Am Soc Mass Spectrom. 2006;17(1):15–19. doi: 10.1016/j.jasms.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 49.Li XJ, Lin C, O'Connor PB. Glutamine deamidation: differentiation of glutamic acid and gamma-glutamic acid in peptides by electron capture dissociation. J Am Soc Mass Spectrom. 2010 doi: 10.1021/ac9028467. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sargaeva NP, Cournoyer JJ, Yao CX, Lin C, O'Connor PB. ECD and IRMPD of amyloid beta protein fragment 1-40 and synthetic beta-linked peptides; The 56th ASMS Conference on Mass Spectrometry and Allied Topics; Denver, CO, USA. 2008. [Google Scholar]
- 51.Sargaeva NP, Yao CX, Lin TY, Cui WD, Aizikov K, Li XJ, Lin C, O'Connor PB. ECD and EID of Amyloid beta and synthetic beta-peptides; The 57th ASMS Conference on Mass Spectrometry and Allied Topics; Philadelphia, PA. 2009. [Google Scholar]
- 52.Sargaeva NP, Lin C, O'Connor PB. Is this a trend or coincidence? Unusual fragmentation of beta - amino acid containing peptides; The 7th Uppsala Conference on Electron Capture and Transfer Dissociation; Nara, Japan. 2009. [Google Scholar]
- 53.Seebach D, Overhand M, Kuhnle FNM, Martinoni B, Oberer L, Hommel U, Widmer H. Beta-peptides: Synthesis by Arndt-Eistert homologation with concomitant peptide coupling. Structure determination by NMR and CD spectroscopy and by X-ray crystallography. Helical secondary structure of a beta-hexapeptide in solution and its stability towards pepsin. Helv Chim Acta. 1996;79(4):913–941. [Google Scholar]
- 54.Appella DH, Christianson LA, Karle IL, Powell DR, Gellman SH. beta-peptide foldamers: Robust Helix formation in a new family of beta-amino acid oligomers. J Am Chem Soc. 1996;118(51):13071–13072. [Google Scholar]
- 55.Seebach D, Matthews JL. Beta-peptides: a surprise at every turn. Chem Commun. 1997;(21):2015–2022. [Google Scholar]
- 56.Seebach D, Beck AK, Bierbaum DJ. The world of beta- and gamma-peptides comprised of homologated proteinogenic amino acids and other components. Chem Biodiver. 2004;1(8):1111–239. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 57.Ben Hamidane H, Vorobyev A, Larregola M, Lukaszuk A, Tourwe D, Lavielle S, Karoyan P, Tsybin YO. Radical stability directs electron capture and transfer dissociation of beta-amino acids in peptides. Chem Eur J. 2010;16(15):4612–4622. doi: 10.1002/chem.200902480. [DOI] [PubMed] [Google Scholar]
- 58.Cooper HJ, Hudgins RR, Marshall AG. Electron capture dissociation Fourier-transform ion cyclotron resonance mass spectrometry of cyclodepsipeptides, branched peptides, and epsilon-peptides. Int J Mass Spectrom. 2004;234(1-3):23–35. [Google Scholar]
- 59.Lamos SM, Shortreed MR, Frey BL, Belshaw PJ, Smith LM. Relative quantification of carboxylic acid metabolites by liquid chromatography - mass spectrometry using isotopic variants of cholamine. Anal Chem. 2007;79(14):5143–5149. doi: 10.1021/ac062416m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jebanathirajah JA, Pittman JL, Thomson BA, Budnik BA, Kaur P, Rape M, Kirschner M, Costello CE, O'Connor PB. Characterization of a new qQq-FTICR mass spectrometer for post-translational modification analysis and top-down tandem mass spectrometry of whole proteins. J Am Soc Mass Spectrom. 2005;16(12):1985–1999. doi: 10.1016/j.jasms.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 61.O'Connor PB, Pittman JL, Thomson BA, Budnik BA, Cournoyer JC, Jebanathirajah J, Lin C, Moyer S, Zhao C. A new hybrid electrospray Fourier transform mass spectrometer: design and performance characteristics. Rapid Commun Mass Spectrom. 2006;20(2):259–266. doi: 10.1002/rcm.2307. [DOI] [PubMed] [Google Scholar]
- 62.Seebach D, Gardiner J. Beta-peptidic peptidomimetics. Acc Chem Res. 2008;41(10):1366–75. doi: 10.1021/ar700263g. [DOI] [PubMed] [Google Scholar]
- 63.Iavarone AT, Jurchen JC, Williams ER. Supercharged protein and peptide ions formed by electrospray ionization. Anal Chem. 2001;73(7):1455–1460. doi: 10.1021/ac001251t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kjeldsen F, Giessing AMB, Ingrell CR, Jensen ON. Peptide sequencing and characterization of post-translational modifications by enhanced ion-charging and liquid chromatography electron-transfer dissociation tandem mass spectrometry. Anal Chem. 2007;79(24):9243–9252. doi: 10.1021/ac701700g. [DOI] [PubMed] [Google Scholar]
- 65.Liu H, Hakansson K. Electron capture dissociation of tyrosine O-sulfated peptides complexed with divalent metal cations. Anal Chem. 2006;78(21):7570–7576. doi: 10.1021/ac061352c. [DOI] [PubMed] [Google Scholar]
- 66.Hunt DF. Innovative mass spectrometry for the study of protein post-translational modifications; The 7th Uppsala Conference on Electron Capture and Transfer Dissociation; Nara, Japan. 2009. [Google Scholar]
- 67.Xia Y, Gunawardena HP, Erickson DE, McLuckey SA. Effects of cation charge-site identity and position on electron-transfer dissociation of polypeptide cations. J Am Chem Soc. 2007;129(40):12232–12243. doi: 10.1021/ja0736764. [DOI] [PubMed] [Google Scholar]
- 68.Jensen CS, Holm AIS, Zettergren H, Overgaard JB, Hvelplund P, Nielsen SB. On the charge partitioning between c and z fragments formed after electron-capture induced dissociation of charge-tagged Lys-Lys and Ala-Lys dipeptide dications. J Am Soc Mass Spectrom. 2009;20(10):1881–1889. doi: 10.1016/j.jasms.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 69.Horne WS, Boersma MD, Windsor MA, Gellman SH. Sequence-based design of alpha/beta-peptide foldamers that mimic BH3 domains. Angew Chem Int Edit. 2008;47(15):2853–2856. doi: 10.1002/anie.200705315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skurski P, Sobczyk M, Jakowski J, Simons J. Possible mechanisms for protecting N-C-alpha bonds in helical peptides from electron-capture (or transfer) dissociation. Int J Mass Spectrom. 2007;265(2-3):197–212. [Google Scholar]
- 71.Yao CX, Syrstad EA, Turecek F. Electron transfer to protonated beta-alanine N-methylamide in the gas phase: An experimental and computational study of dissociation energetics and mechanisms. J Phys Chem A. 2007;111(20):4167–4180. doi: 10.1021/jp0705020. [DOI] [PubMed] [Google Scholar]
- 72.Hayakawa S, Matsubara H, Panja S, Hvelplund P, Nielsen SB, Chen XH, Turecek F. Experimental evidence for an inverse hydrogen migration in arginine radicals. J Am Chem Soc. 2008;130(24):7645–7654. doi: 10.1021/ja800207x. [DOI] [PubMed] [Google Scholar]
- 73.Bakken V, Helgaker T, Uggerud E. Models of fragmentations induced by electron attachment to protonated peptides. Eur J Mass Spectrom. 2004;10(5):625–638. doi: 10.1255/ejms.665. [DOI] [PubMed] [Google Scholar]
- 74.Jones AW, Cooper HJ. Probing the mechanisms of electron capture dissociation mass spectrometry with nitrated peptides. Phys Chem Chem Phys. 12(41):13394–9. doi: 10.1039/c0cp00623h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24(4):508–48. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- 76.Syrstad EA, Stephens DD, Turecek F. Hydrogen atom adducts to the amide bond. Generation and energetics of amide radicals in the gas phase. J Phys Chem A. 2003;107(1):115–126. [Google Scholar]
- 77.Csonka IP, Paizs B, Lendvay G, Suhai S. Proton mobility and main fragmentation pathways of protonated lysylglycine. Rapid Commun Mass Spectrom. 2001;15(16):1457–72. doi: 10.1002/rcm.388. [DOI] [PubMed] [Google Scholar]
- 78.Dongre AR, Jones JL, Somogyi A, Wysocki VH. Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J Am Chem Soc. 1996;118(35):8365–8374. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure S1. ECD of isoaspartic acid.
Supplemental figure S2. Q06 beta peptide β2Vβ2Aβ2Lβ3Vβ3Aβ3L: a. ESI, b. IRMPD, c. EID.
Supplemental figure S3. ECD of Puma BH3 protein analogue II.