Abstract
Mutations in the TP53 tumor suppressor gene occur in half of all human cancers, indicating its critical importance in inhibiting cancer development. Despite extensive studies, the mechanisms by which mutant p53 enhances tumor progression remain only partially understood. Here, using data from The Cancer Genome Atlas (TCGA), genomic and transcriptomic analyses were performed on 2256 tumors from ten human cancer types. We show that tumors with TP53 mutations have altered gene expression profiles compared to tumors retaining two wildtype TP53 alleles. Among 113 known p53 upregulated target genes identified from cell culture assays, ten were consistently upregulated in at least 8 of 10 cancer types that retain both copies of wildtype TP53. RPS27L, CDKN1A (p21CIP1), and ZMAT3 were significantly upregulated in all ten cancer types retaining wildtype TP53. Using this p53-based expression analysis as a discovery tool, we used cell-based assays to identify five novel p53 target genes from genes consistently upregulated in wildtype p53 cancers. Global gene expression analyses revealed that cell cycle regulatory genes and transcription factors E2F1, MYBL2, and FOXM1 were disproportionately upregulated in many TP53 mutant cancer types. Finally, over 93% of tumors with a TP53 mutation exhibited greatly reduced wildtype p53 messenger expression due to loss of heterozygosity or copy neutral loss of heterozygosity, supporting the concept of p53 as a recessive tumor suppressor. The data indicate that tumors with wildtype TP53 retain some aspects of p53-mediated growth inhibitory signaling through activation of p53 target genes and suppression of cell cycle regulatory genes.
Keywords: p53, TP53, TCGA, nonsense-mediated mRNA decay, MYBL2, FOXM1, E2F1, RPS27L, CDKN1A, ZMAT3
Introduction
The TP53 gene encodes a tumor suppressor protein that functions as a stress responder and cell cycle checkpoint protein that maintains genomic integrity [1]. Loss of its function through genetic alteration is a key event in malignant progression as evidenced by the observation that over half of all human cancers display mutations and deletions of the TP53 gene [2,3]. The recent comprehensive sequencing studies sponsored by The Cancer Genome Atlas (TCGA) consortium confirm the high frequency of TP53 mutations in many sequenced cancers [4–8]. Virtually all of these mutations inactivate the transcriptional functions of p53. As a cell cycle inhibitory transcription factor, p53 upregulates and downregulates hundreds of target genes that enforce its growth inhibitory functions in response to oncogenic or damage-induced stress [9].
Despite many studies, the full functional effects of intact or inactivated TP53 on human cancers remain incompletely understood. The integrated TCGA approach in which large numbers of cancers are examined on multiple high throughput analytical platforms has introduced a rare opportunity to study the relationship between TP53 allele status and other molecular correlates with a higher level of statistical rigor. In this paper we synthesize TP53 sequence data, TP53 allele copy number status, and global gene expression data to better understand how TP53 allele alterations affect transcriptional circuits in ten different human cancer types analyzed by the TCGA network. Comparison of gene expression patterns in wildtype TP53-intact and mutant TP53 cancers shows that wildtype p53 selectively upregulates a subset of known p53 target genes across most cancer types. These types of integrative computational analyses have been used here to experimentally identify five novel p53 target genes. Most tumors with a TP53 mutation show reduction of p53 target gene expression due largely to loss of wildtype p53 RNA expression, through loss of heterozygosity or through copy neutral loss of heterozygosity. These results argue that TP53 behaves as a classic “two hit” tumor suppressor in that absence of wildtype p53 expression is likely a prerequisite for tumor progression. Finally, TP53 allele mutation is also associated with increased expression of cell cycle promoting genes in multiple cancers.
Materials and Methods
Data Procurement
TP53 mutation data, TP53 copy number data, global gene expression data, and individual DNA and RNA sequence reads for each cancer type were obtained from The Cancer Genome Atlas Data Portal, Cancer Genomics Hub (CGHub), Memorial Sloan Kettering Cancer Center sponsored cBio Portal, and TCGA Pan-Cancer SYNAPSE portal. All publicly available TCGA tumor data complies with U.S. law protecting patient confidentiality and other ethical standards. Ten cancer types with sufficient tumor numbers to provide robust statistical power were breast carcinomas (BRCA), colorectal cancers (CRC), glioblastoma (GBM), endometrial carcinomas (UCEC), bladder carcinomas (BLCA), ovarian serous adenocarcinomas (OVCA), acute myelogenous leukemia (LAML), lung adenocarcinoma (LUAD), stomach adenocarcinoma (STAD), and low grade glioma (LGG).
Stratification of tumors by TP53 allele status
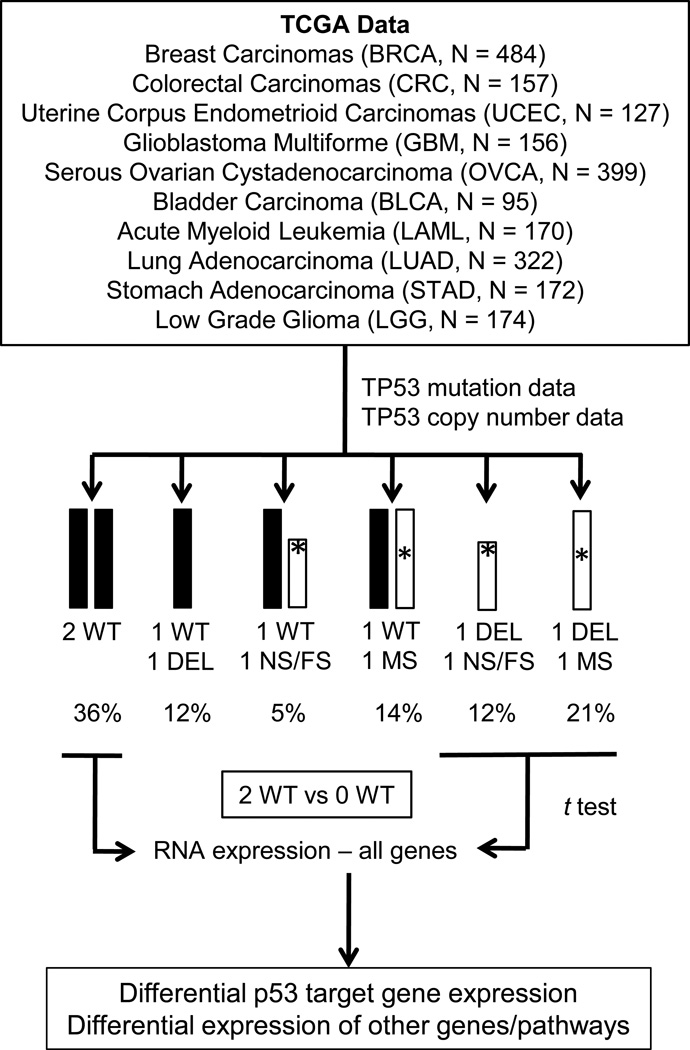
To stratify tumors by TP53 allele status for each cancer type, copy number data was downloaded for each tumor and GISTIC (Genomic Identification of Significant Targets in Cancer) scores for TP53 copy number changes in each tumor were obtained from the Memorial Sloan-Kettering Cancer Center cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal). Integration of TP53 copy number and sequence data resulted in stratification of each tumor into one of six categories: (1) two wildtype TP53 alleles (2 WT), (2) one wildtype TP53 allele and one deleted TP53 allele (WT + DEL), (3) diploid and one nonsense or frameshift or splice site TP53 mutation allele (WT + NS/FS), (4) diploid and one missense or in frame TP53 deletion/insertion mutation (WT + MS), (5) haploid and one nonsense or frameshift or splice site TP53 mutation allele (DEL + NS/FS), and (6) haploid and one missense or in frame TP53 deletion/insertion mutation (DEL + MS) (Figure 1).
Figure 1.
Flow chart showing how TP53 mutation and copy number data was integrated in ten TCGA cancer types to compare gene expression patterns based on six different TP53 allele categories. Percentages of each TP53 allele category across all ten cancers are indicated.
Analysis of global gene expression according to TP53 allele status
Level 3 normalized global RNA expression data files for each tumor were downloaded from The Cancer Genome Atlas Data Portal, the Cancer Genomics Hub (CGHub) and TCGA Pan-Cancer SYNAPSE portal. RNA data for all genes for each tumor were then sorted according to TP53 allele status. For each tumor type, mean gene expression of each gene for each of six TP53 allele status categories was determined. Mean expression for each gene in the 2 WT TP53 category (2 WT) was compared to mean gene expression in the 0 WT TP53 category (DEL + NS/FS and DEL + MS) by determining the ratio of 0 WT / 2 WT expression (Table S1). A two tailed unpaired Student’s t test was used to determine the relative significance of expression for each gene in the 2 WT TP53 category compared to the 0 WT TP53 category. The differentially expressed genes as measured by t test P values were then ranked from most significant to least significant (Table S1).
Analysis of individual DNA and RNA sequence reads
To determine TP53 allele LOH status and TP53 allele RNA expression, we downloaded individual TP53 DNA and RNA sequence reads for four cancer types with sufficient numbers of TP53 mutations from the Cancer Genomics Hub (CGHub). For each tumor we also obtained data for TP53 mutations, TP53 copy number, and tumor purity from the TCGA Pan-Cancer SYNAPSE portal, cBio Portal, and TCGA Data Portal. Each tumor with a somatic TP53 mutation was stratified into one of three categories based on TP53 DNA sequence reads and copy number data after adjustment for tumor purity. Tumors diploid for TP53 copy number showing greater than a 0.65 mutant read fraction were considered copy neutral LOH (“CN LOH”) and those with less than a 0.65 mutant read fraction were categorized “NO LOH”. Tumors with TP53 mutations haploid for TP53 copy number were judged “LOH”. RNA sequence reads were then analyzed and fractions of mutant and wildtype TP53 RNA for each tumor were averaged.
Gene ontology and transcription factor binding site studies
After sorting each tumor for differential gene expression based on TP53 allele status, the top 500 genes for each cancer type expression list in Table S1 was subjected to DAVID (Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov) and GSEA (Gene Set Enrichment Analysis; http://www.broadinstitute.org) gene ontology studies.
p53 target gene lists
p53-induced and p53-repressed target gene lists were assembled from p53 databases (http://p53.iarc.fr, http://p53.free.fr/Database/p53_database.html, http://p53.bii.a-star.edu.sg/abouTP53/targetgene/index.php), reviews, and literature searches. The list in Table S2 (p53-induced genes) is extensive but not exhaustive. Genes were only considered p53 targets if, in addition to being upregulated following wildtype p53 activation, there was functional evidence in the literature (e.g. luciferase reporter assays, chromatin immunoprecipitation assays) that p53 activation of the target gene was direct and not indirect.
Quantitative real-time PCR
HCT116(p53+/+) and HCT116(p53−/−) cells were treated with or without 10 Gy ionizing irradiation and harvested four hours post-irradiation. Total RNA was extracted using TRIzol Reagent (Life Technologies) and reverse transcribed using qScript cDNA SuperMix (Quanta Biosciences) per manufacturer instructions. Real time PCR was performed using Step One Plus Real-Time PCR System (Life Technologies) and Power SYBR Green PCR Master Mix (Life Technologies). Primer sequences are listed in Supplementary Methods. Each sample was tested in replicates and real-time PCR performed twice for each sample and primer pair. ΔΔCt method was used to calculate fold change with respect to p53 wild type cells using GAPDH as endogenous control.
Chromatin immunoprecipitation (ChIP) for candidate p53 target genes
For ChIP, ~16×106 sub-confluent HCT116(p53+/+) cells were gamma irradiated (10 Gy) or mock irradiated. Four hours post radiation cells were fixed, neutralized, washed as described earlier (13) and incubated with buffer #1 (10mM EDTA, 0.5mM EGTA, 10mM HEPES pH 6.5, 0.25% Triton X-100), buffer #2 (1mM EDTA, 0.5MM EGTA, 10mM HEPES pH 6.5, 200mM NaCl) and buffer #3 (10mM EDTA, 50mM Tris-HCl pH 8.1, 0.5% Empigen BB, 1% SDS) for 5 min on ice. Cell lysates were sonicated to average 600–800 bp DNA fragments (S-4000, QSonica LLC). Chromatin samples were diluted with 2mM EDTA, 150mM NaCl, 20mM Tris-Cl pH 8.1, 1% Triton X-100 buffer, 1% kept as input and remaining incubated overnight with 1.5 µg antibodies (Millipore, Cat #17–613, Cat # 05–623), and with Dynabeads Protein G for 90 minutes at 4°C. Immune complexes were washed once with buffer #1 (2mM EDTA, 20mM Tris-Cl pH 8.1, 0.1% SDS, 0.25% Triton X-100) with 100mM NaCl, buffer #1 with 200mM NaCl, buffer #2 (1mM EDTA, 10mM Tris-Cl pH 8.1, 1% Deoxycholate, 0.25% NP-40, 250mM LiCl) and twice with 1mM EDTA, 10mM Tris-Cl, pH 8.0 buffer. Eluted and reverse cross-linked samples were treated with RNase A and Proteinase K followed by phenol-chloroform extraction of DNA. Real-time PCR was performed for P53 responsive elements identified using the P53 FamTaG database (http://p53famtag.ba.itb.cnr.it).
Statistical analyses
Student’s two tailed t test was used to compare RNA expression for 2 WT TP53 tumors versus 0 WT TP53 (DEL + FS/MS, DEL + MS) tumors for each of ten cancer types. Fisher’s exact test was used to determine whether p53-induced target genes were significantly over-represented in the most differentially expressed genes upregulated in 2 WT TP53 tumors for each cancer type.
Results
Upregulation of a subset of p53 target genes in cancers with wildtype TP53
To analyze effects of TP53 allele status on gene expression across multiple cancer types, we integrated TP53 mutation data with TP53 copy number data and global gene expression data in 2256 tumor samples of ten cancer types for which data from all analysis platforms was available (Figure 1). We then segregated tumors of each cancer type into one of six categories: TP53 wildtype and diploid (2 WT), TP53 wildtype and haploid (WT + DEL), TP53 nonsense, frame shift, or splice site mutation and diploid (WT + NS/FS), TP53 missense mutation and diploid (WT + MS), TP53 nonsense/frameshift/splice site mutation allele and haploid (DEL + NS/FS), and TP53 missense mutation allele and haploid (DEL + MS). We then compared mean RNA expression levels for all genes in the two wildtype TP53 allele (2 WT) group versus the tumor groups with zero WT TP53 alleles (DEL + NS/FS, DEL + MS). A two-tailed t test was performed for RNA expression of each gene in the two comparison groups, and all genes were ranked by P value for significance of differential expression (Table S1).
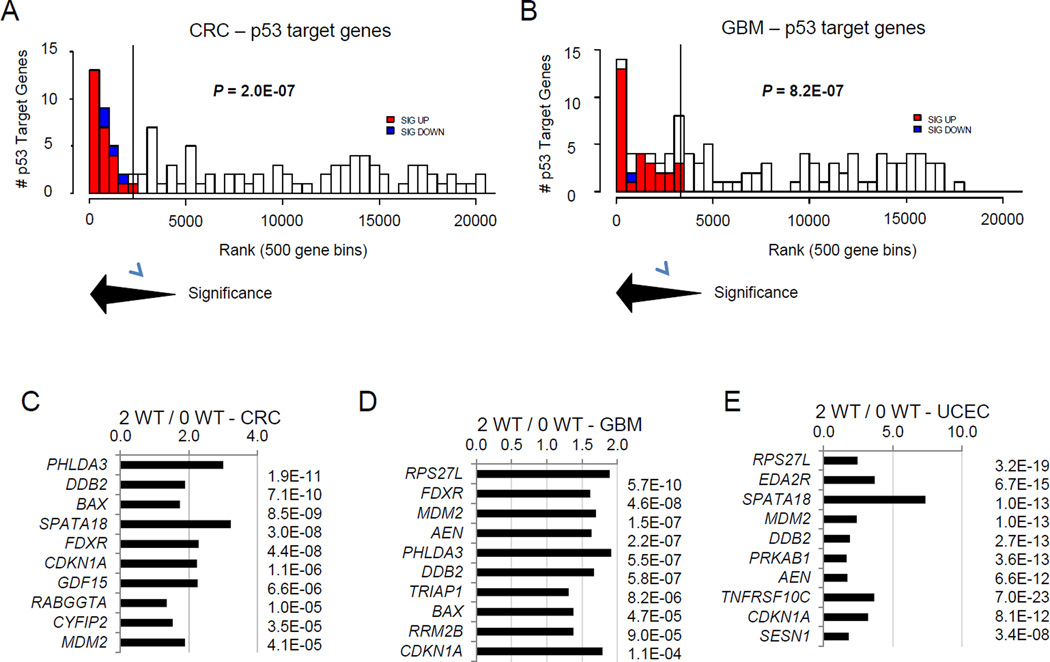
From each list of differentially expressed genes based on TP53 allele status we screened for known p53-induced target genes. We developed from p53 databases [2,3], p53 reviews [9], and literature searches a list of 113 known p53-induced target genes (Table S2). P53-induced target genes were found to be significantly over-represented among the most differentially regulated genes in the 2 WT TP53 tumors versus the 0 WT TP53 tumors (Figure 2A,B, Table S1,S2). Moreover, among the top ten most differentially regulated p53-induced target genes across the ten cancer types, 96 of 100 showed increased expression in the 2 WT TP53 tumors relative to 0 WT TP53 tumors (Figure 2, Figure S1, Table S1). Among 113 known p53-induced targets, only a subset were consistently upregulated in multiple cancer types with 2 WT TP53 alleles (Table 1, Figure 2C–E, Figure S1, Table S2). RPS27L, CDKN1A (p21CIP1), and ZMAT3 were significantly upregulated in 2 WT TP53 tumors of all ten cancer types. The frequently upregulated p53 targets encompass a range of functional types, including cell cycle inhibitors (CDKN1A), apoptosis mediators (TNFRSF10C, AEN, BBC3), p53 regulators (MDM2) and DNA damage responders (DDB2). Some are established p53 targets, yet others (e.g. SPATA18, EDA2R, PHLDA3) have only recently been identified as p53-induced targets and are not well characterized [10,11].
Figure 2.
A subset of p53-induced target genes is consistently upregulated in 2 WT TP53 cancers relative to 0 WT TP53 counterparts. (A) Overrepresentation of upregulated p53-induced target genes among the most differentially regulated genes in colorectal cancers (CRC) with 2 WT TP53 alleles. All genes were ranked in 500 gene bins by significance of differential RNA expression in 2 WT TP53 tumors versus 0 WT TP53 tumors. Numbers of p53-induced target genes are shown in each bin. Significantly upregulated p53 targets are indicated in red and significantly downregulated p53 targets are indicated in blue. A Fischer’s exact test (P = 2.0E-07) showed this over-representation of upregulated p53-induced target genes in the 2 WT TP53 tumors to be highly significant. (B) Over-representation of upregulated p53-induced target genes among the most differentially regulated genes in glioblastomas (GBM) with 2 WT TP53 alleles. Analyses were performed as described for panel A. (C–E) For each of the cancers in C-E, the ten p53-induced target genes most differentially expressed were ranked by significance (P values after t test) and the ratio of mean expression of each gene in 2 WT TP53 tumors relative to 0 WT TP53 tumors is indicated. (C) colorectal carcinomas (CRC). (D) glioblastomas (GBM). (E) Endometrial carcinomas (UCEC).
Table 1.
P53-Induced Target Genes Most Significantly Upregulated in WT TP53 Cancers
| Gene | # Cancers | Significance of Upregulation in 2 WT TP53 vs 0 WT TP53 Tumors (P values) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CRC | BRCA | UCEC | GBM | OVCA | BLCA | LUAD | LAML | STAD | LGG | ||
| RPS27L | 10/10 | 2E-04 | 3E-29 | 3E-19 | 6E-10 | 1E-04 | 1E-02 | 3E-10 | 1E-06 | 2E-10 | 3E-04 |
| TNFRSF10C | 9/10 | 3E-03 | 1E-32 | 7E-12 | 1E-03 | 8E-03 | 5E-04 | 1E-11 | - | 2E-03 | 7E-03 |
| CDKN1A (p21) | 10/10 | 1E-06 | 5E-09 | 8E-12 | 1E-04 | 4E-02 | 1E-02 | 1E-04 | 1E-07 | 5E-02 | 7E-03 |
| ZMAT3 | 10/10 | 8E-05 | 9E-04 | 1E-03 | 9E-03 | 1E-03 | 3E-02 | 2E-03 | 1E-06 | 1E-08 | 3E-10 |
| EDA2R | 9/10 | 2E-03 | - | 7E-15 | 3E-03 | 5E-04 | 3E-02 | 5E-12 | 1E-18 | 4E-03 | 5E-19 |
| DDB2 | 8/10 | 1E-09 | 9E-16 | 2E-13 | 6E-07 | - | 2E-02 | 3E-04 | 5E-05 | 2E-09 | - |
| PHLDA3 | 8/10 | 3E-11 | 6E-11 | 5E-04 | 5E-07 | 2E-02 | - | - | 4E-15 | 1E-02 | 2E-10 |
| SPATA18 | 7/9* | 4E-08 | 9E-14 | 1E-13 | - | * | 1E-04 | 3E-06 | 5E-04 | 2E-05 | - |
| FDXR | 9/10 | 6E-08 | 4E-07 | 2E-04 | 5E-08 | - | 3E-02 | 2E-02 | 6E-05 | 2E-05 | 9E-15 |
| MDM2 | 8/10 | 4E-05 | 2E-04 | 1E-13 | 1E-07 | 3E-02 | - | 1E-02 | 1E-07 | 2E-05 | - |
| TNFRSF10B | 9/10 | 2E-04 | 1E-03 | 5E-03 | 1E-04 | - | 2E-03 | 2E-05 | 2E-03 | 3E-03 | 3E-12 |
| AEN | 7/10 | 8E-05 | - | 7E-12 | 2E-07 | - | 3E-02 | 6E-05 | - | 2E-02 | 3E-12 |
SPATA18 not present in OVCA gene expression platform
CRC - colorectal carcinomas; BRCA - breast carcinomas; UCEC - uterine corpus endometrioid carcinoma; GBM – glioblastoma multiforme; OVCA - ovarian serous cystadenocarcinoma; BLCA – bladder carcinoma; LUAD: lung adenocarcinoma; LAML: acute myeloid leukemia; STAD: stomach adenocarcinoma; LGG: low grade glioma.
P53-based differential expression in cancers as a discovery tool for novel p53 target genes
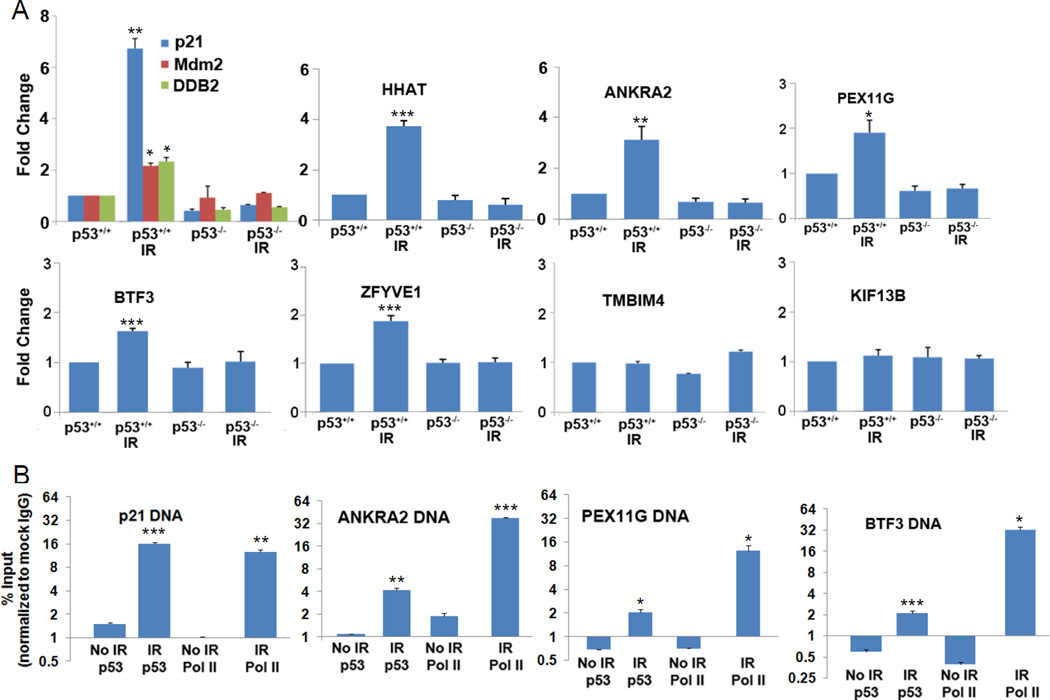
Upregulation of p53 target genes across ten analyzed cancers encouraged us to examine whether other genes consistently upregulated in 2 WT TP53 cancers might be novel p53 target candidates. We screened our p53-based differential expression tables (Table S1) and identified 25 genes not previously known as p53 target genes that were significantly upregulated in at least five of ten cancer types with 2 WT TP53 alleles. To experimentally validate the candidates as bona fide p53 target genes we examined RNA expression of each gene (via qRT-PCR) following ionizing radiation (IR) or mock treatment of isogenic colorectal carcinoma cell lines HCT116(p53+/+) and HCT116(p53−/−). Only those genes that showed IR-induced increases in expression in HCT116(p53+/+) cells and not in HCT116(p53−/−) cells were considered likely p53 targets. Five genes (HHAT, ANKRA2, BTF3, PEX11G, and ZFYVE1) displayed a significant p53-dependent upregulation of expression (Figure 3A). Two of these genes (ANKRA2, and BTF3) are transcription or chromatin-associated proteins and two, ZFYVE1 and PEX11G, are associated with endosome and peroxisome biogenesis, respectively. Of these five genes, ANKRA2, BTF3, and PEX11G, have consensus p53 response elements in their promoter regions, and these sites were used as targets for chromatin immunoprecipitation (ChIP) assays to determine whether p53 could directly bind to these sites following ionizing radiation of HCT116(p53+/+) cells. ChIP assays on all three tested genes using antibodies for p53 or for RNA Polymerase II (Pol II) showed that ionizing radiation resulted in enhanced promoter/chromatin binding by p53 and Pol II, indicating that ANKRA2, BTF3, and PEX11G are direct p53 target genes and actively transcribed post-IR (Figure 3B).
Figure 3.
Experimental validation of novel candidate p53-induced target genes deduced from computational analyses of gene expression patterns in human cancers. (A) Radiation induces p53- dependent upregulation of five candidate p53 targets.Real-time PCR analysis was performed on RNA from HCT116(p53+/+) and HCT116(p53−/−) cells treated with or without IR with primers from 25 candidate genes that were upregulated across five or more different cancer types with two wildtype TP53 alleles. Fold change of indicated genes was normalized to expression ofunirradiated HCT116(p53+/+) samples. GAPDH was used as endogenous control. HHATANKRA2, PEX11G,BTF3, and ZFYVE1were significantly upregulated in HCT116(p53+/+) IR cells. TMBIM4 and KIF13B are representative examples of the 20 p53-unaffected genes. P21 (CDKN1A), MDM2 and DDB2genes in the first panel (known p53- induced target genes) served as positive controls. (B) Chromatin immunoprecipitation (ChIP) analyses confirm some p53 target candidates as direct p53 target genes.Putative p53 targets, ANKRA2, BTF3 and PEX11G were validated by ChIP with p53 antibody and real-time PCR in HCT116(p53+/+) cells treated with or without 10 Gy IR. P21CIP1 served as positive control and mock mouse IgG for each condition (IR/No IR) was used as normalization control. ChIP with Pol II antibody infers increased transcription of p53-induced target genes.Chromatin enrichment is represented as per cent of input chromatin. *P< 0.02; **P< 0.005; ***P< 0.001.
Tumors with a TP53 mutation exhibit loss of p53 target gene expression
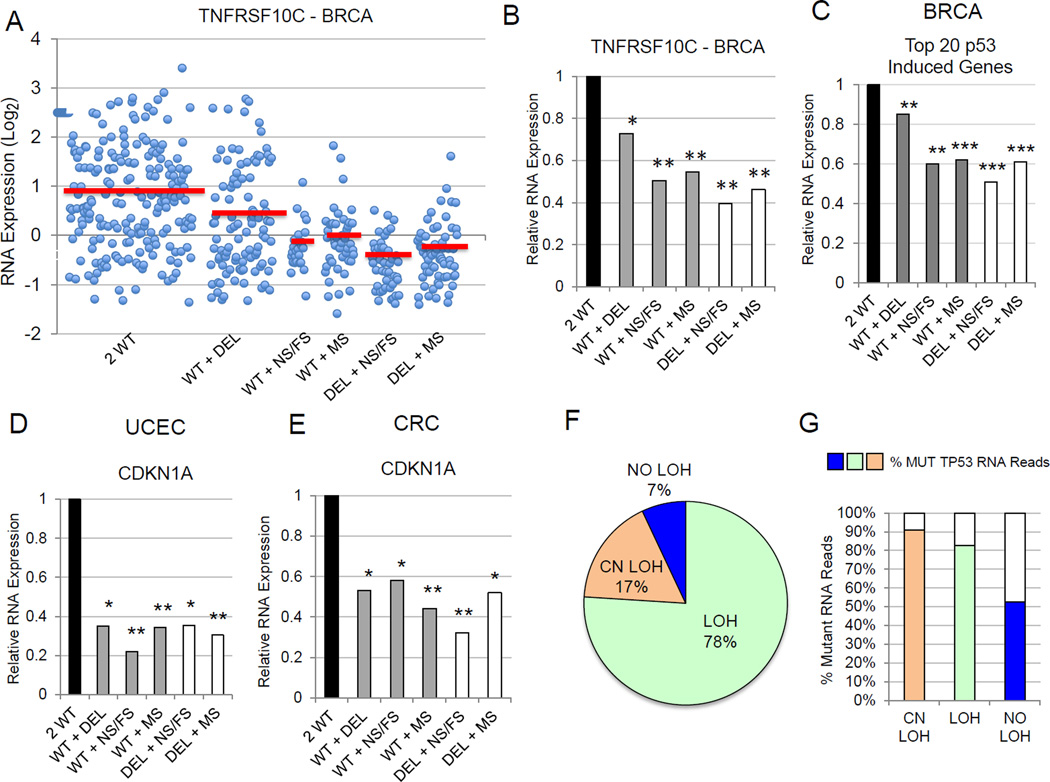
Given the differential p53 target gene expression in the 2 WT and 0 WT TP53 cancers, we examined expression levels of these target genes in tumors with intermediate TP53 genotypes (WT + DEL, WT + NS/FS, WT + MS). Some cancers with a wildtype and deleted copy of TP53 (WT + DEL) exhibited intermediate levels of p53 target expression, while other cancers of this type averaged levels of p53 target expression similar to tumors with no wildtype TP53 alleles (Figure 4A–E). Interestingly, tumors diploid for TP53 and with one mutant TP53 allele (WT + NS/FS, WT + MS) averaged the same low levels of p53 target gene expression as in tumors with TP53 haploidy combined with a mutation (DEL + NS/FS, DEL + MS) (Figure 4A–E). Thus, tumors with a single TP53 mutation generally lose p53 transcriptional function, regardless of whether the tumor is diploid or haploid for TP53.
Figure 4.
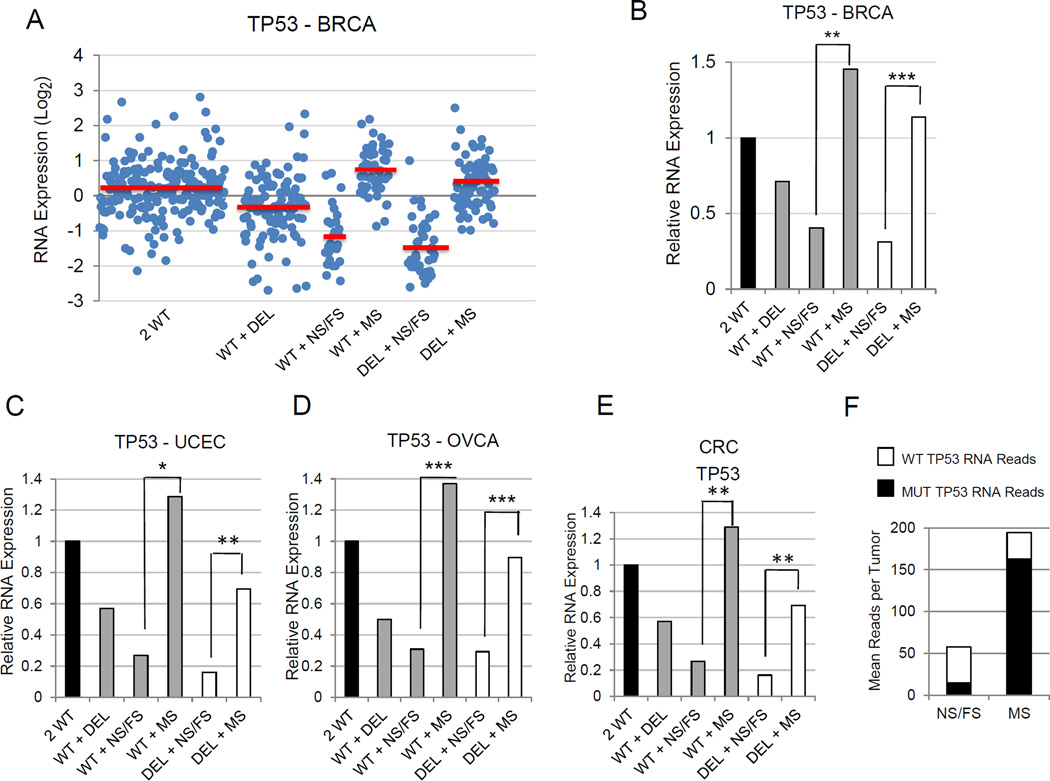
Cancers with TP53 mutations display deficient transcriptional activation of p53 target genes, regardless of the status of the second TP53 allele. (A) Scatter plot showing p53 target gene TNFRSF10C(TRAIL-R3) expression values for each individual tumor in each TP53 allele group of breast carcinomas. The breast carcinomas were stratified into six TP53 allele categories (2 WT, WT + DEL, WT + FS/NS, WT + MS, DEL + NS/FS, DEL + MS) and mean TNFRSF10C expression values for each TP53 allele group are symbolized by red bars. (B) Bar graph simplifying data from panel A showing TNFRSF10CRNA expression levels in breast carcinomas. Mean expression levels were normalized to mean p53 target expression levels in the 2 WT TP53 category (set to a value of 1.0). For panels B-E, asterisks indicate the relative significance of the difference in expression between the designated TP53 category and the 2 WT TP53category. *P < 5E-02; **P < 1E-05; ***P < 1E-25. (C) Bar graph of averaged values of top 20 p53-induced target genes for each TP53 allele category in breast carcinomas relative to values of 2 WT TP53 breast carcinomas. (D,E) Mean CKDN1A (p21CIP1) gene expression in six TP53 allele categories of endometrial cancers (D) and colorectal carcinomas (E). (F) The vast majority of tumors with missense TP53 mutations exhibit either TP53 loss of heterozygosity (LOH) or copy number neutral loss of heterozygosity (CN LOH). For four cancers (BRCA, OVCA, UCEC, CRC) with high TP53 mutation rates, DNA sequence reads for individual tumors were compared for relative frequencies of mutant versus wildtype TP53 alleles. CN LOH status was assigned to tumors with a TP53 mutation that were diploid for TP53 by copy number analysis, but displayed more than a 0.65 mutant TP53 allele fraction based on collected DNA sequence reads after adjustment for tumor purity. Diploid TP53 mutant tumors with less than 0.65 mutant TP53 allele fractions were designated NO LOH. LOH tumors were those tumors with a TP53 mutation but exhibiting a haploid TP53 allele number by copy number analysis. (G) Tumors with missense TP53 mutations and LOH or CN LOH lose expression of wildtype p53 mRNA. Individual tumor mutant and wildtype TP53 RNAseq reads for the four cancer types in (F) were used to determine fractions of mutant TP53 message in each tumor and these were averaged across all tumors with missense TP53 for each LOH category.
The apparent absence of p53 target expression in tumors with a TP53 mutation, regardless of whether the tumor is haploid or diploid for TP53, suggests inactivation of the second TP53 allele in most diploid tumors with a TP53 mutation. We analyzed individual tumor TP53 DNA sequence reads in four cancers to clarify relative fractions of mutant and wildtype TP53 alleles. Haploid tumors with TP53 mutations (DEL + NS/FS and DEL + MS) comprised 76% of all tumors with TP53 mutations and these tumors averaged 82% mutant TP53 reads after adjustment for tumor purity, consistent with loss of heterozygosity (LOH) (Figure 4F). Of the remaining 24% of diploid tumors with TP53 mutations, 17% were categorized as copy neutral LOH (CN LOH). This category was defined as those tumors (WT + NS/FS and WT + MS) that displayed mutant allele read fractions greater than 0.65, after adjustment for tumor purity. The remaining 7% of tumors with TP53 mutations were classified as NO LOH, as these tumors exhibited less than 0.65 mutant allele read fractions (Figure 4F). To confirm loss of wildtype TP53 expression we examined RNAseq reads from WT + MS tumors from four cancer types previously assessed for mutant TP53 allele fractions. Those tumors with TP53 mutations and LOH or CN LOH (93%) consistently showed high percentages (over 80%) of mutant TP53 allele RNA expression (Figure 4G, Figure S2A). Those few tumors (7%) without evidence of TP53 LOH (NO LOH) averaged 50% mutant TP53 allele expression. Thus, across multiple human cancers, tumors with a TP53 mutation are generally associated with loss of wildtype p53 RNA expression, either through simple LOH or copy neutral LOH.
Truncation mutations in TP53 reduce TP53 RNA expression
When p53 RNA expression was stratified according to TP53 allele categories we found that virtually all tumors with a nonsense or frameshift mutation (WT + NS/FS, DEL + NS/FS) had dramatically lower levels of p53 RNA expression (Figure 5A–E). This phenomenon, observed across all ten cancer types, is indicative of nonsense-mediated mRNA decay characteristic of many messenger RNAs with premature translational termination [12]. No differential methylation of the TP53 promoter was observed in any of several examined cancers, eliminating an alternative explanation for reductions in TP53 RNA expression. Direct counting of TP53 RNA sequencing reads in four cancer types confirmed a dramatic reduction in read totals in tumors with a nonsense/frameshift TP53 mutation relative to tumors with a missense TP53 mutation (Figure 5F, Figure S2B). Moreover, a low fraction of the RNA reads from these tumors contained TP53 mutations, consistent with nonsense-mediated mRNA decay. WT TP53 reads appeared not to be affected by nonsense-mediated mRNA decay (Figure 5F).
Figure 5.
TP53 RNA expression is significantly reduced in tumors with TP53 frameshift and nonsense mutations. (A) Scatter plot showing TP53 RNA expression in individual breast carcinomas sorted by TP53 allele status. Mean TP53expression values for each TP53 allele group are indicated by red bars. (B) Bar graph simplifying data from panel A showing TP53RNA expression levels in breast carcinomas segregated by TP53 allele status. Asterisks in panels B-E indicate the relative significance of the difference in expression of the designated TP53 categories. *P < 0.05; **P < 1E-05; ***P < 1E-25. (C–E) Bar graphs showing relative expression of TP53 RNA in endometrial carcinomas (C), ovarian serous adenocarcinomas (D), and colorectal carcinomas (E) by TP53 allele status. (F) Low levels of mutant TP53 RNA levels are found in tumors with nonsense or frameshift TP53 mutations, consistent with nonsense-mediated mRNA decay. Total mutant and wildtype TP53 RNAseq reads were analyzed in individual tumors with TP53 mutations from four different cancers (BRCA, OVCA, UCEC, CRC). The tumors were stratified by those with nonsense or frameshift TP53 mutations and those with missense TP53 mutations. Mean numbers of RNA reads per tumor and relative ratios of wildtype and mutant RNA allele fractions are indicated.
TP53 mutation is associated with upregulation of cell cycle transcriptional programs in some cancers
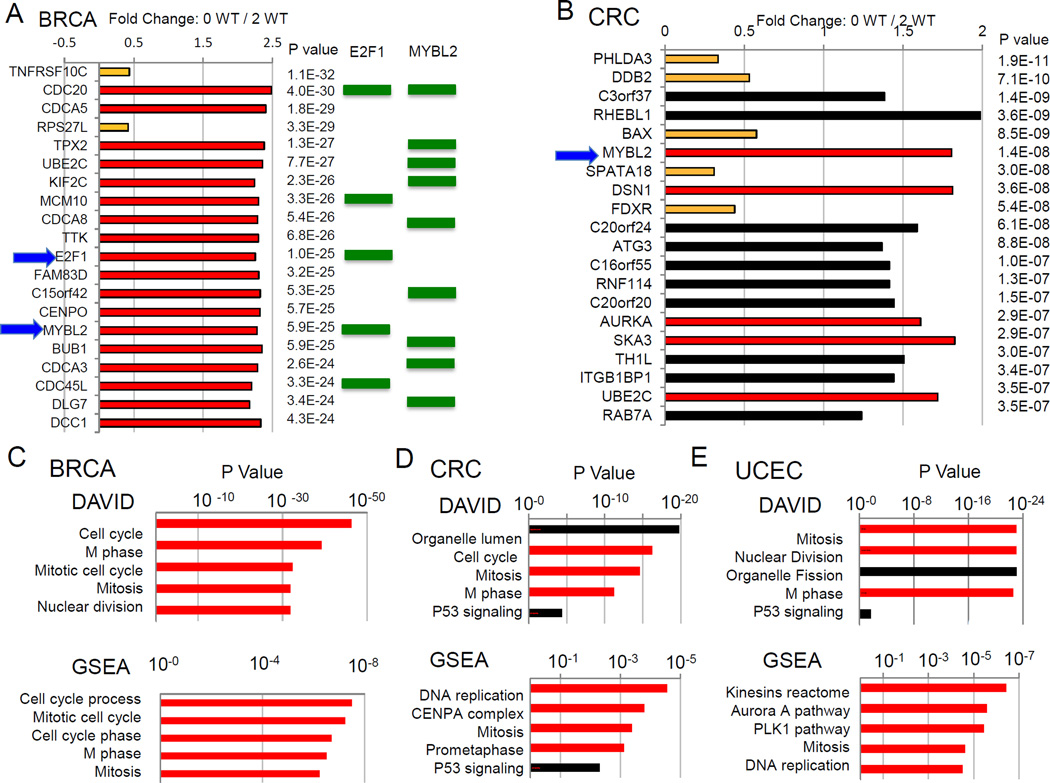
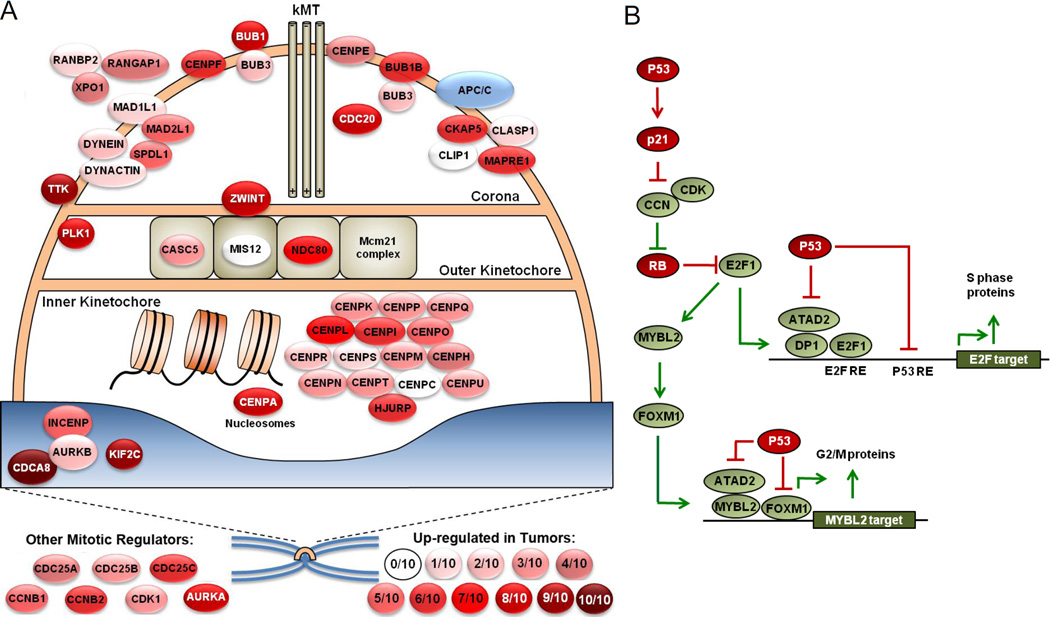
When comparing gene expression in the 2 WT and 0 WT TP53 tumors for each cancer type, most differentially regulated genes were not known p53 target genes (Table S1). Figures 6A and 6B show the top 20 differentially regulated genes in breast cancer and colorectal cancer, respectively. For breast cancer, aside from two p53-induced target genes, the remaining 18 genes are upregulated in the 0 WT TP53 tumors and all are associated with cell cycle regulation, particularly mitosis (Figure 6A). For colorectal cancer, 5 of the top 20 differentially regulated genes were p53-induced target genes and 5 (upregulated in 0 WT TP53 tumors) were cell cycle regulatory in nature (Figure 6B). DAVID and GSEA gene ontology analyses of the top 1000 most differentially regulated genes in breast, colorectal, and endometrial cancers showed that cell cycle regulatory genes were significantly over expressed and over-represented in the tumors with no intact TP53 alleles (Figure 6C–E). Many of the 0 WT TP53 cancers exhibited a recurrent upregulation of CENP and other kinetochore-associated proteins critical for mitotic progression (Figure 7A) [13]. Glioblastomas, ovarian serous adenocarcinomas, lung adenocarcinomas, and stomach adenocarcinomas also displayed significant over-representation of mitotic regulatory genes in 0 WT TP53 tumors (Table S1).
Figure 6.
Cell cycle regulatory genes are over-represented among most differentially regulated genes in wildtype TP53 versus mutant TP53 cancers. (A) Analysis of the 20 most differentially regulated genes in mutant TP53 breast cancers relative to wildtype TP53 breast cancers. Fold differences in expression (ratio of mean gene expression in 0 WT TP53 tumors to mean gene expression in 2 WT TP53 tumors) are indicated by bars and P values for significance of differential expression are indicated. Red bars indicate cell cycle regulatory genes and orange bars indicate p53-induced target genes. Transcription factors E2F1 and MYBL2 are indicated by blue arrows. Each gene directly upregulated by E2F1 or MYBL2 is indicated by a green rectangle.(B) Analysis of the 20 most differentially regulated genes in mutant TP53 colorectal cancers relative to wildtype TP53 colorectal cancers. Results are indicated as in panel A, but black bars indicate genes not associated with p53 (orange) or cell cycle (red) regulation. (CE) Gene ontology analyses of most differentially regulated genes based on TP53 status in breast cancers (C), colorectal cancers (D), and endometrial cancers (E) show a preponderance of cell cycle regulatory pathways. The top 500 most differentially regulated genes between cancers with two wildtype TP53 alleles and those with no wildtype TP53 alleles were analyzed for gene ontology associations by the DAVID and GSEA software programs. The most over-represented gene ontology categories in each analysis are indicated as well as the significance of the association as indicated by P values. Red bars indicated cell cycle-associated gene ontology categories.
Figure 7.
Mutations in TP53 are associated with enhancement of transcriptional programs that drive S phase and mitotic progression. (A) Pictorial diagram showing increased expression of many kinetochore-associated genes in 0 WT TP53 tumors relative to 2 WT TP53 tumors. Diagram of kinetochore adapted from Musacchio and Salmon[35] shows many kinetochore-associated genes and genes that regulated mitosis. Degrees of red shading indicate the number of cancer types (out of ten analyzed) in which that gene is significantly upregulated in 0 WT TP53 tumors relative to 2 WT TP53 tumors. (B) Model showing different mechanisms by which p53 may effect E2F1, MYBL2, and FOXM1 activities in breast cancer.P53 may have direct and indirect suppressive effects on these cell cycle regulatory transcription factors.
Examination of the most differentially expressed cell cycle regulatory genes in breast, colorectal, endometrial, ovarian, and stomach 0 WT TP53 cancers showed that E2F1 and MYBL2 were among the most consistently upregulated transcription factors observed (Figure 6A,B, Table S3). E2F1 is a critical mediator of S phase progression and MYBL2 regulates mitotic progression [14,15]. Similarly, in several 0 WT TP53 cancers, S phase transcriptional regulator E2F2 and mitotic transcriptional regulator FOXM1 were significantly increased in expression (Table S3) [16]. To examine whether loss of p53 associates with activated E2F1/MYBL2/FOXM1 transcriptional programs, we screened for E2F1 response elements in the top 100 genes most upregulated in TP53 breast and colorectal cancers using the CORE_TF transcription factor binding site software program [17] and found that E2F1 response sites were significantly over-represented in these genes compared to promoter regions of 2970 random gene promoters (Figure S3A). Similar findings were shown for E2F1/MYBL2/FOXM1 target gene over-representation in 0 TP53 breast cancers, colorectal cancers and endometrial cancers (Figure S3B,C,D and data not shown).
Discussion
The integrated multi-platform analyses of multiple cancers by The Cancer Genome Atlas research network have provided a rich dataset from which to derive novel insights into the etiology of this disease. This report represents one example of how this data can be mined to better understand the functional role of a single gene (TP53) in intact cancers. Because individual tumors are heterogeneous with respect to many variables, it has been historically difficult to perform the type of study we report here. But the integrated multi-platform approach and large tumor numbers allow novel molecular correlations and extraction of statistically significant patterns that would be obscured by noise associated with smaller datasets.
We show that ten different types of cancers that retain two copies of wildtype TP53 display consistently elevated expression of a subset of known p53-induced target genes. This result suggests that normal p53 transcriptional functions remain partially intact in most 2 WT TP53 tumors and thus p53 likely inhibits some aspects of tumor progression in situ. Only a relatively limited subset of known p53-induced target genes are consistently upregulated across all or most cancer types (Table 1). This should not be surprising since p53 target lists are aggregated from a diverse set of experimental reagents and conditions. Oncogenic signaling represents merely one of many stresses that p53 responds to, so it seems likely that each stress may result in a different subset of p53 targets being activated. Consensus p53-induced targets in tumors don’t fall into any single functional category, and some targets such as the cell cycle inhibitory CDKN1A (p21CIP1) might be expected. Some consensus p53 targets have only recently been discovered and have not been functionally characterized, thus inviting more investigation.
The large datasets generated by the TCGA effort have provided an outstanding discovery tool for identifying new cancer genes and pathways. By filtering for those genes that were consistently upregulated in 2 WT TP53 cancers we were able to validate five new genes as likely p53-induced targets in independent cellular assays. Three of these target genes bind p53 at p53 consensus sites within or near their promoters in a DNA damage-dependent fashion and recruit Pol II, thus confirming their status as p53-induced target genes.
One notable finding was that tumors in all ten cancer types with a TP53 mutation showed similar low levels of p53 target regulation, regardless of TP53 second allele status. Analysis of DNA sequencing reads in these tumors revealed that 76% of tumors displayed TP53 LOH and 17% displayed copy neutral TP53 loss of heterozygosity (CN LOH). CN LOH could occur through uniparental disomy, gene conversion, or mitotic recombination and has been observed in 20% – 80% of human cancers [18–20]. Both CN LOH tumors and LOH tumors exhibited very low expression levels of wildtype TP53 RNA. Only 7% of mutant TP53 tumors were NO LOH. Few tumors with a TP53 mutation retained significant expression of wildtype TP53 RNA. The data confirms that there is a strong selection for elimination of wildtype TP53 RNA expression. Thus, TP53 generally behaves as a classic ‘two hit’ tumor suppressor, as originally proposed by Vogelstein and colleagues [21].
Several cancer types with TP53 mutations displayed significant upregulation of genes that regulate S phase and mitotic progression. We propose that some of the mutant TP53-associated upregulation of cell cycle regulatory genes may be attributed to p53 effects on E2F1/E2F2, MYBL2, and FOXM1, the four most consistently upregulated transcription factors in the majority of the cancer types. This association of p53 mutational status with E2F1, MYBL2 and FOXM1 in breast and ovarian cancers has been noted [5,22]. Our model, shown in Figure 7B, is based partly on known activities of wildtype p53 and partly on inferred activities from literature and TCGA data. E2F1 transcriptionally upregulates MYBL2 and FOXM1 and MYBL2 upregulates FOXM1 to link S phase with M phase progression [23–27]. Wildtype TP53 is known to indirectly suppress E2F1/2 through CDKN1A (p21) which in turn suppresses G1/S cyclin/CDK phosphorylation of RB [28]. P53 may directly suppress E2F1 target genes PLK1, CCNA2, and CDK1, as well as others [29–32]. Many MYBL2 and FOXM1 transcription targets are also repressed by p53 and the FOXM1 gene itself is repressed by p53 [33]. Finally, p53 also suppresses expression of ATAD2, a co-activator for both E2F1 and MYBL2 [34]. Thus, mutation of TP53 in a developing tumor may de-repress these key cell cycle regulatory genes at multiple nodes of regulation, resulting in an enhancement of cell cycle progression.
Supplementary Material
Acknowledgments
We thank Josh Stuart at the TCGA Pan-Cancer effort and The Cancer Genome Atlas Research Network. We are grateful to Bert Vogelstein for provision of HCT116(p53+/+) and HCT116(p53−/−) cells. We thank Alison Roos and Manjula Nakka for technical assistance and Chuck Perou for helpful discussions. This work was supported by grants to D.A.W. and L.A.D. from the National Cancer Institute and National Human Genome Research Institute and the Cancer Prevention Research Institute of Texas.
List of Abbreviations
- CRC
colorectal carcinomas
- BRCA
breast carcinomas
- UCEC
uterine corpus endometrioid carcinoma
- GBM
glioblastoma multiforme
- OVCA
ovarian serous cystadenocarcinoma
- BLCA
bladder carcinoma
- LUAD
lung adenocarcinoma
- LAML
acute myeloid leukemia
- STAD
stomach adenocarcinoma
- LGG
low grade glioma
- TCGA
The Cancer Genome Atlas
- 2 WT
TP53 allele status of tumors with two wildtype TP53 alleles
- WT + DEL
TP53 allele status of tumors with one wildtype and one deleted TP53 allele
- WT + NS/FS
TP53 allele status of tumors with one wildtype and one frameshift or nonsense TP53 mutant allele
- WT + MS
TP53 allele status of tumors with one wildtype and one missense TP53 mutant allele
- DEL + NS/FS
TP53 allele status of tumors with one deleted and one frameshift or nonsense TP53 mutant allele
- DEL + MS
TP53 allele status of tumors with one deleted and one missense TP53 mutant allele
- IR
ionizing radiation
- qRT-PCR
quantitative reverse transcriptase polymerase chain reaction
- ChIP
chromatin immunoprecipitation
- Pol II
RNA polymerase II
- LOH
loss of heterozygosity
- CN LOH
copy number neutral loss of heterozygosity
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- GSEA
Gene Set Enrichment Analysis
Footnotes
There are no conflicts of interest for any of the authors on this manuscript.
Statement of Author Contributions
NP performed experiments shown in Figure 3 and prepared Figures 3 and 7 as well as editing parts of the manuscript. SH and CJC provided statistical guidance and methods used to stratify TP53 mutation categories and expression signatures. TD performed parts of the experiments shown in Figure 3. RS assisted NP and TD in performing Figure 3 experiments and assisted in the compilation of Supplementary Table 1. ES and LX assisted in the extraction and organization of the tumor data from the TCGA-associated databases. RG and DAW provided overall guidance, editorial assistance, and financial support for the work. LAD conceived the described approaches, performed many of the computational tasks, supervised the experiments in Figure 3, and wrote and edited the manuscript.
List of Online Supporting Information
Supplementary Table 1. Most differentially expressed genes in WT TP53 cancers relative to mutant TP53 cancers
Supplementary Table 2.Ranking of p53 target gene expression upregulation in 2 WT TP53 tumors for ten cancers
Supplementary Table 3.Upregulated S Phase and Mitotic Transcription Factors in Cancers Lacking WT p53
Supplementary Figure 1. Upregulated p53-induced target genes across five human cancers
Supplementary Figure 2. Altered RNA expression in tumors with mutant TP53 alleles
Supplementary Figure 3. Absence of p53 is correlated with increased activities of E2F1, MYBL2, and FOXM1
Supplementary Figure Legends
RT-PCR Primer Sequences for Figure 3
References
- 1.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 2.Petitjean A, Achatz MI, Borresen-Dale AL, et al. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 3.Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Advances in cancer research. 2011;110:107–139. doi: 10.1016/B978-0-12-386469-7.00005-0. [DOI] [PubMed] [Google Scholar]
- 4.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riley T, Sontag E, Chen P, et al. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 10.Bornstein C, Brosh R, Molchadsky A, et al. SPATA18, a spermatogenesis-associated gene, is a novel transcriptional target of p53 and p63. Mol Cell Biol. 2011;31:1679–1689. doi: 10.1128/MCB.01072-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawase T, Ohki R, Shibata T, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–550. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson P, Yepiskoposyan H, Metze S, et al. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cellular and molecular life sciences : CMLS. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 14.DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- 15.Efeyan A, Murga M, Martinez-Pastor B, et al. Limited role of murine ATM in oncogene-induced senescence and p53-dependent tumor suppression. PLoS One. 2009;4:e5475. doi: 10.1371/journal.pone.0005475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raychaudhuri P, Park HJ. FoxM1: a master regulator of tumor metastasis. Cancer Res. 2011;71:4329–4333. doi: 10.1158/0008-5472.CAN-11-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hestand MS, van Galen M, Villerius MP, et al. CORE_TF: a user-friendly interface to identify evolutionary conserved transcription factor binding sites in sets of co-regulated genes. BMC Bioinformatics. 2008;9:495. doi: 10.1186/1471-2105-9-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo KC, Bailey D, Burkhardt T, et al. Comprehensive analysis of loss of heterozygosity events in glioblastoma using the 100K SNP mapping arrays and comparison with copy number abnormalities defined by BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2008;47:221–237. doi: 10.1002/gcc.20524. [DOI] [PubMed] [Google Scholar]
- 19.Gondek LP, Tiu R, O'Keefe CL, et al. Chromosomal lesions and uniparental disomy detected by SNP arrays in MDS, MDS/MPD, and MDS-derived AML. Blood. 2008;111:1534–1542. doi: 10.1182/blood-2007-05-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saeki H, Kitao H, Yoshinaga K, et al. Copy-neutral loss of heterozygosity at the p53 locus in carcinogenesis of esophageal squamous cell carcinomas associated with p53 mutations. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:1731–1740. doi: 10.1158/1078-0432.CCR-10-1996. [DOI] [PubMed] [Google Scholar]
- 21.Baker SJ, Fearon ER, Nigro JM, et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 22.Troester MA, Herschkowitz JI, Oh DS, et al. Gene expression patterns associated with p53 status in breast cancer. BMC Cancer. 2006;6:276. doi: 10.1186/1471-2407-6-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. Embo J. 2004;23:4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laoukili J, Kooistra MR, Bras A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 25.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akala OO, Park IK, Qian D, et al. Long-term haematopoietic reconstitution by Trp53−/−p16Ink4a−/−p19Arf−/− multipotent progenitors. Nature. 2008;453:228–232. doi: 10.1038/nature06869. [DOI] [PubMed] [Google Scholar]
- 27.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- 29.Incassati A, Patel D, McCance DJ. Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene. 2006;25:2444–2451. doi: 10.1038/sj.onc.1209276. [DOI] [PubMed] [Google Scholar]
- 30.McKenzie L, King S, Marcar L, et al. p53-dependent repression of polo-like kinase-1 (PLK1) Cell Cycle. 2010;9:4200–4212. doi: 10.4161/cc.9.20.13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scian MJ, Carchman EH, Mohanraj L, et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27:2583–2593. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- 32.Yun J, Chae HD, Choy HE, et al. p53 negatively regulates cdc2 transcription via the CCAAT-binding NF-Y transcription factor. J Biol Chem. 1999;274:29677–29682. doi: 10.1074/jbc.274.42.29677. [DOI] [PubMed] [Google Scholar]
- 33.Barsotti AM, Prives C. Pro-proliferative FoxM1 is a target of p53-mediated repression. Oncogene. 2009;28:4295–4305. doi: 10.1038/onc.2009.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalashnikova EV, Revenko AS, Gemo AT, et al. ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. 2010;70:9402–9412. doi: 10.1158/0008-5472.CAN-10-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.