Abstract
We have evaluated the cloning and functional expression of previously described broad antimicrobial spectrum bacteriocins SRCAM 602, OR-7, E-760, and L-1077, by recombinant Pichia pastoris. Synthetic genes, matching the codon usage of P. pastoris, were designed from the known mature amino acid sequence of these bacteriocins and cloned into the protein expression vector pPICZαA. The recombinant derived plasmids were linearized and transformed into competent P. pastoris X-33, and the presence of integrated plasmids into the transformed cells was confirmed by PCR and sequencing of the inserts. The antimicrobial activity, expected in supernatants of the recombinant P. pastoris producers, was purified using a multistep chromatographic procedure including ammonium sulfate precipitation, desalting by gel filtration, cation exchange-, hydrophobic interaction-, and reverse phase-chromatography (RP-FPLC). However, a measurable antimicrobial activity was only detected after the hydrophobic interaction and RP-FPLC steps of the purified supernatants. MALDI-TOF MS analysis of the antimicrobial fractions eluted from RP-FPLC revealed the existence of peptide fragments of lower and higher molecular mass than expected. MALDI-TOF/TOF MS analysis of selected peptides from eluted RP-FPLC samples with antimicrobial activity indicated the presence of peptide fragments not related to the amino acid sequence of the cloned bacteriocins.
1. Introduction
The antimicrobial peptides (AMPs) are broad spectrum small molecular weight compounds with antagonistic activity against bacteria, viruses, and fungi. Among them, bacteriocins form a widely studied and well-characterized group of ribosomally synthesized peptides produced by bacteria, and those produced by lactic acid bacteria (LAB) attract considerable interest primarily as natural food preservatives but also with great interest in exploring their application as therapeutic antimicrobial agents [1–3]. Most LAB bacteriocins are synthesized as biologically inactive precursors or prepropeptides containing an N-terminal extension that is cleaved off during export to generate their biologically active or mature form. The mature peptides are often cationic, amphiphilic molecules that are generally classified into two main classes: the lantibiotics or class I bacteriocins that consist of modified bacteriocins and the class II or nonmodified bacteriocins [4, 5]. The class II bacteriocins have been further divided into several subgroups, from which the class IIa or pediocin-like bacteriocins show a strong antilisterial activity and the N-terminal consensus sequence YGNGV(X)C [5].
Considering the low amount of AMPs obtained from their direct purification from natural producers and the elevated production costs of chemical synthesis, the biological production of bacteriocins by heterologous microbial hosts may provide an opportunity for their production in large amounts and with higher specific antimicrobial activity [6–8]. Furthermore, the production of bacteriocins by yeasts may have some advantages over bacterial cells regarding specific posttranscriptional and posttranslational modifications [9]. Yeasts are also cost-effective producers with large production and high yields of the desired protein [10]. Pichia pastoris (currently reclassified as Komagataella pastoris) is being used as heterologous producer of bacteriocins because of its ability to produce large amounts of properly folded and biologically active bacteriocins [8, 11, 12]. P. pastoris also produces disulfide-bonded and glycosylated proteins, which are crucial features for functionality [13]. The use of synthetic genes may also constitute a successful approach for heterologous production and functional expression of bacteriocins by recombinant yeasts when the DNA sequence encoding the bacteriocin is not available or difficult to obtain [12]. Furthermore, the use of synthetic genes matching the codon usage of the host microorganism can have a significant impact on gene expression levels and protein folding [14].
Several bacteriocins with broad antimicrobial spectrum have been identified from chicken commensal bacteria including bacteriocin SRCAM 602 produced by Paenibacillus polymyxa [15, 16], bacteriocins OR-7 and L-1077 produced by Lactobacillus salivarius [17, 18], and bacteriocins E-760 and E 50-52 produced by Enterococcus spp. [19, 20]. All these bacteriocins have been reported to be active against Gram-positive and Gram-negative bacteria including Campylobacter spp., reducing Campylobacter colonization in poultry and considered potentially useful towards on-farm control of this foodborne human pathogen [21]. Most animal studies suggest that these bacteriocins considerably reduce C. jejuni colonization in chicken intestine and thus may reduce Campylobacter spp. infections in humans [17, 19, 20]. However, to our knowledge, none of the genes encoding these bacteriocins have been sequenced so far. Accordingly, in this study we report the use of synthetic genes designed from the published amino acid sequence of the mature bacteriocins SRCAM 602, OR-7, E-760, and L-1077 and with adapted codon usage for expression by P. pastoris, their cloning into the protein expression vector pPICZαA, and their expression by recombinant P. pastoris X-33.
2. Materials and Methods
2.1. Microbial Strains and Plasmids
Microbial strains and plasmids used in this study are listed in Table 1. Enterococcus faecium T136 and Pediococcus damnosus CECT4797 were grown in MRS broth (Oxoid Ltd., Basingstoke, UK) at 32°C. P. pastoris X-33 (Invitrogen S.A., Barcelona, Spain) was cultured in YPD medium (Sigma-Aldrich Inc., St. Louis, MO, USA) at 30°C with shaking (200–250 rpm). Escherichia coli JM109 (Promega, WI, USA) was grown in LB broth (Sigma-Aldrich) at 37°C with shaking (250 rpm). Listeria monocytogenes CECT4032 was grown in LB at 37°C and Salmonella typhimurium CECT443 was grown in TSB (Oxoid) at 37°C. Campylobacter jejuni ATCC33560 and C. jejuni NCTC11168 were grown in BHI supplemented with 1% defibrinated horse serum (BD Bioscience, CA, USA) at 37°C in microaerophilic conditions. E. coli O157:H7 was grown in LB at 37°C with shaking (250 rpm). Yersinia ruckeri LMG3279 was grown in TSB at 28°C. Zeocin (Invitrogen) was added when needed at concentrations of 25, 100, or 1000 μg/mL. Strains cited as CECT belong to the Colección Española de Cultivos Tipo (Valencia, Spain), ATCC to the American Type Culture Collection (Rockville, MD, USA), and NCTC to the National Collection of Type Cultures (London, UK).
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Descriptiona | Source and/or referenceb |
|---|---|---|
| Strains | ||
| Enterococcus faecium T136 | Enterocin A and B producer; MPA positive control | DNBTA |
| Pediococcus damnosus CECT4797 | MPA indicator microorganism | CECT |
| Escherichia coli JM109 | Selection of recombinant plasmids | Promega |
| Pichia pastoris X-33 | Yeast producer | Invitrogen Life Technologies |
| Plasmids | ||
| pMA-T | Ampr; carrier of synthetic genes | GeneArt Life Technologies |
| pPICZαA | Zeor; integrative plasmid carrying the secretion signal sequence from the S. cerevisiaeα-factor prepropeptide and functional sites for integration at the 5′ AOX1 locus of P. pastoris X-33 | Invitrogen Life Technologies |
| pMATSRCAM602 | Ampr; pMA-T plasmid carrying the srcam602 synthetic gene with the P. pastoris codon usage | GeneArt Life Technologies |
| pMATOR-7 | Ampr; pMA-T plasmid carrying the or-7 synthetic gene with the P. pastoris codon usage | GeneArt Life Technologies |
| pMATE-760 | Ampr; pMA-T plasmid carrying the e-760 synthetic gene with the P. pastoris codon usage | GeneArt Life Technologies |
| pMATL-1077 | Ampr; pMA-T plasmid carrying the l-1077 synthetic gene with the P. pastoris codon usage | GeneArt Life Technologies |
| pSRCAM602 | pPICZαA derivative with the srcam602 synthetic gene | This work |
| pOR-7 | pPICZαA derivative with the or-7 synthetic gene | This work |
| pE-760 | pPICZαA derivative with the e-760 synthetic gene | This work |
| pL-1077 | pPICZαA derivative with the l-1077 synthetic gene | This work |
aAmpr: ampicillin resistance; Zeor: zeocin resistance.
bDNBTA: Departamento de Nutrición, Bromatología y Tecnología de los Alimentos, Facultad de Veterinaria, Universidad Complutense de Madrid (Madrid, Spain); CECT: Colección Española de Cultivos Tipo Valencia, Spain).
2.2. Basic Genetic Techniques and Enzymes
The published amino acid sequences of mature bacteriocins SRCAM 602, OR-7, E-760, and L-1077 were used as templates to design the nucleotide sequence of the synthetic genes srcam602, or-7, e-760, and l-1077 matching the codon usage of P. pastoris X-33. These synthetic genes contained a 5′-nucleotide appendix including a XhoI restriction site and a 3′-nucleotide appendix including the termination of translation codon (TAA) and the NotI restriction site. All synthetic genes were supplied by GeneArt (Life Technologies, Paisley, UK). DNA restriction enzymes were supplied by New England BioLabs (Ipswich, MA, USA). Ligations were performed with the T4 DNA ligase (Roche Molecular Biochemicals, Mannheim, Germany). E. coli JM109 cells were transformed as described by the supplier. Competent P. pastoris X-33 cells were obtained as recommended by the supplier and electroporation of competent cells was performed as previously described [22]. Electrocompetent cells were transformed with a Gene Pulser and Pulse Controller apparatus (Bio-Rad Laboratories, Hercules, CA, USA).
2.3. PCR Amplification and Nucleotide Sequencing
Oligonucleotide primers were obtained from Sigma-Genosys Ltd. (Cambridge, UK). PCR amplifications were performed in 50 μL reaction mixtures containing 1 μL of purified DNA, 70 pmol of each primer, and 1 U of Platinum Pfx DNA Polymerase (Invitrogen) in a DNA thermal cycler Techgene (Techne, Cambridge, UK). The PCR-generated fragments were purified by a NucleoSpin Extract II Kit (Macherey-Nagel GmbH & Co., Düren, Germany) for cloning and nucleotide sequencing. Nucleotide sequencing of the purified PCR products was performed using the ABI PRISM BigDye Terminator cycle sequence reaction kit and the automatic DNA sequencer ABI PRISM, model 377 (Applied Biosystems, Foster City, CA, USA), at the Unidad de Genómica, Facultad de Ciencias Biológicas, Universidad Complutense de Madrid (UCM), Madrid, Spain.
2.4. Cloning of the srcam602, or-7, e-760, and l-1077 Synthetic Genes in P. pastoris X-33 and Antimicrobial Activity of the Transformants
The primers and inserts used for construction of the recombinant plasmids are listed in Table 2. Derivatives of plasmid pPICZαA were constructed as follows: primers S602-F, S071-F, and SARP-R were used for PCR amplification from plasmids pMATSRCAM602, pMATOR-7, pMATE-760, and pMATL-1077 of nucleotide fragments in frame with the S. cerevisiaeα-factor secretion signal, without the Glu-Ala spacer adjacent to the Kex2 protease cleavage site, fused to the srcam602, or-7, e-760, and l-1077 synthetic genes. Digestion of the above cited fragments with the XhoI-NotI restriction enzymes permitted ligation of the resulting R-SRCAM602, R-OR7, R-E760, and R-L1077 nucleotide fragments of 136-, 181-, 242-, and 167-bp, respectively, into pPICZαA digested with the same enzymes to generate the plasmid-derived vectors pSRCAM602, pOR-7, pE-760, and pL-1077, respectively. Competent E. coli JM109 cells were used for cloning and vector propagation and the resulting transformants were confirmed by PCR amplification and sequencing of the inserts. Subsequently, the SacI-linearized pSRCAM602, pOR-7, pE-760, and pL-1077 vectors were transformed into competent P. pastoris X-33 cells yielding zeocin resistant derivatives on YPD agar supplemented with zeocin (100 and 1,000 μg/mL) and sorbitol (1 M). The presence of the integrated synthetic genes in the transformed yeast cells was confirmed by PCR and DNA sequencing of the inserts.
Table 2.
Primers and PCR products used in this study.
| Primers, PCR products, or bacteriocins | Nucleotide sequence (5′-3′) or description | Amplifications |
|---|---|---|
| Primers | ||
| S602-F | GCCATGAGCTCGAATTCTCGAGAAAAG | R-SRCAM602, R-E760, R-L1077 |
| S071-F | GTCCAGAGCTCGAATTCTCGAGAAAAG | R-SRCAM 602, R-E760, R-L1077, R-OR7 |
| SARP-R | AGGTACCATAAGTTGCGGCCGC | R-OR7 |
| ALFA-F | TACTATTGCCAGCATTGCTGC | pPICZαA amplification fragment including the cloned synthetic gene |
| 3AOX1-R | GCAAATGGCATTCTGACATCC | pPICZαA amplification fragment including the cloned synthetic gene |
| PCR products | ||
| R-SRCAM602 | 136-bp XhoI/NotI fragment containing the α-factor Kex2 signal cleavage fused to the mature synthetic srcam602 gene with the P. pastoris codon usage | |
| R-OR7 | 181-bp XhoI/NotI fragment containing the α-factor Kex2 signal cleavage fused to the mature synthetic or-7 gene with the P. pastoris codon usage | |
| R-E760 | 242-bp XhoI/NotI fragment containing the α-factor Kex2 signal cleavage fused to the mature synthetic e-760 gene with the P. pastoris codon usage | |
| R-L1077 | 167-bp XhoI/NotI fragment containing the α-factor Kex2 signal cleavage fused to the mature synthetic l-1077 gene with the P. pastoris codon usage | |
| Bacteriocins | ||
| BacSRCAM602 (amino acid sequence) | ATYYGNGLYCNKQKHYTWVDWNKASR EIGKITVNGWVQH |
|
| BacSRCAM602 (P. pastoris codon usage) | gctacttactacggtaacggtctttactgtaacaagcagaagcact acacttgggttgactggaacaaggcttccagagagatcggtaag atcactgttaacggttgggttcaaca |
|
| BacOR-7 (amino acid sequence) | KTYYGTNGVHCTKNSLWGKVRLKNMK YDQNTTYMGRLQDILLGWATGAFGKTFH |
|
| BacOR-7 (P. pastoris codon usage) | aagacttactacggaactaacggtgttcactgtactaagaattcctt gtggggtaaggttagattgaagaacatgaagtacgaccagaaca ctacttacatgggtagattgcaggacatcttgttgggttgggctact ggtgctttcggtaagacttttcat |
|
| BacE-760 (amino acid sequence) | NRWYCNSAAGGVGGAAVCGLAGYVGE AKENIAGEVRKGWGMAGGFTHNKACKS FPGSGWASG |
|
| BacE-760 (P. pastoris codon usage) | aacagatggtactgtaactccgctgctggtggtgttggtggtgct gctgtttgtggtttggctggttatgttggtgaggctaaagaaaacat tgctggtgaggttagaaagggttggggtatggctggtggtttcac tcataacaaggcttgtaagtccttcccaggttctggttgggcttctggt |
|
| BacL-1077 (amino acid sequence) | TNYGNGVGVPDAIMAGIIKLIFIFNIRQGY NFGKKAT |
|
| BacL-1077 (P. pastoris codon usage) | actaactacggtaacggtgttggtgttccagacgctattatggctg gtatcatcaagttgatcttcatcttcaacatcagacagggttacaac ttcggtaagaaggctact |
The antimicrobial activity of individual P. pastoris X-33SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-760, and P. pastoris X-33L-1077 was screened by a streak-on-agar test (SOAT). Briefly, the P. pastoris transformants were streaked onto BMMY buffered methanol complex medium (1% yeast, 2% peptone, 100 mM potassium phosphate (pH 6), 1,34% yeast nitrogen base (YNB) without amino acids, 4 × 10−5% biotin, 0.5% methanol) agar and grown at 30°C to induce production of the bacteriocins. After incubation of the plates at 30°C during 24 h, 40 mL of MRS soft-agar containing 105 cfu/mL of the indicator microorganism Pediococcus damnosus CECT4797 was added to the plates that were further incubated at 32°C for 24 h inhibition halos visualization.
2.5. Purification of the Antimicrobial Activity of Supernatants from the Recombinant Yeasts
The antimicrobial activity of supernatants from P. pastoris X-33 and P. pastoris X-33 (pPICZαA) and the recombinant P. pastoris X-33SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-760, and P. pastoris X-33L-1077 was purified using a previously described procedure [12]. Briefly, supernatants from early stationary phase 0.5 L cultures of the recombinant yeasts, grown in BMMY buffered methanol complex medium at 30°C, were precipitated with ammonium sulfate, desalted by gel filtration, and subjected to cation exchange-chromatography, followed by hydrophobic interaction-chromatography and reverse phase-chromatography in a fast protein liquid chromatography system (RP-FPLC) (GE Healthcare, Barcelona, Spain). The antimicrobial activity of the purified fractions was evaluated against Pediococcus damnosus CECT4797 by the microtiter plate assay (MPA).
2.6. Mass Spectrometry Analysis of Purified Supernatants
For determination of the molecular mass of peptides in supernatants of the recombinant yeasts, the eluted purified fractions with antimicrobial activity were subjected to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). Briefly, 1 μL samples were spotted onto a MALDI target plate and allowed to air-dry at room temperature. Then, 0.4 μL of a 3 mg/mL of α-cyano-4-hydroxy-transcinnamic acid matrix (Sigma) in 50% acetonitrile were added to the dried peptide to digest spots and allowed again to air-dry at room temperature. MALDI-TOF MS analyses were performed in a 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Framingham, MA), operated in 1 KV reflector mode. All mass spectra were calibrated externally using a standard peptide mixture (AB Sciex, MA, USA).
To determine the amino acid sequence of the purified peptides, the eluted purified fractions with antimicrobial activity were further subjected to MALDI-TOF/TOF tandem mass spectrometry. The samples were reduced, alkylated, digested with trypsin [23], and analysed in a 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems). The acquisition method for MS analysis was 1 KV reflector positive mode. Peptides from the MS spectra were manually selected for fragmentation analysis. The acquisition method for MS/MS analysis was MS/MS-1 KV in reflector positive mode with CID for fragmentation. The collision gas was atmospheric and the precursor mass window was ±5 Da. The plate model and default calibration were optimized for the MS/MS spectra processing. The parameters used to analyze the data were signal to noise = 20, resolution >6000. For protein identification, the de novo amino acid sequence from the fragmentation spectra of selected peptides was performed using the De Novo tool software (Applied Biosystems), and tentative sequences were manually checked and validated. Homology searches of the deduced amino acid sequences were performed through the NCBI/Blast. All mass spectrometry (MS) analyses were performed at the Unidad de Proteómica, Facultad de Farmacia, Universidad Complutense de Madrid (UCM), Madrid, Spain.
2.7. Antimicrobial Activity of Purified Supernatants from the Recombinant Yeasts
The antimicrobial activity of purified supernatants from the recombinant P. pastoris X-33SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-760, and P. pastoris X-33L-1022 was evaluated against Listeria monocytogenes CECT4032, E. coli O157:H7, Yersinia ruckeri LMG3279, Campylobacter jejuni ATCC33560, and C. jejuni NCTC11168 by using a MPA, as previously described [12].
3. Results
3.1. Cloning of Synthetic Genes Encoding Bacteriocins and Their Expression by Recombinant P. pastoris
The cloning of PCR-amplified fragments from plasmids encoding synthetic genes designed from the published amino acid sequence of the mature bacteriocins SRCAM 602, OR-7, E-760, and L-1077 into the protein expression vector pPICZαA resulted in the recombinant plasmids pSRCAM602, pOR-7, pE-760, and pL-1077 (Table 1). Similarly, transformation of the linearized plasmids into competent P. pastoris X-33 permitted isolation of the P. pastoris X-33SRCAM602 (srcam602), P. pastoris X-33OR-7 (or-7), P. pastoris X-33E-760 (e-760), and P. pastoris X-33L-1077 (l-1077) recombinants. However, none of the recombinant yeasts showed direct antimicrobial activity against P. damnosus CECT4797, even those recombinant yeasts selected for their high zeocin resistance (1,000 μg/mL). Colonies of P. pastoris X-33 and P. pastoris X-33 (pPICZαA) were used as bacteriocin-negative controls to discard the possibility that the antimicrobial activity possibly observed was due to metabolites other than bacteriocins.
3.2. Purification of the Antimicrobial Activity of Supernatants from the Recombinant P. pastoris and Mass Spectrometry Analysis
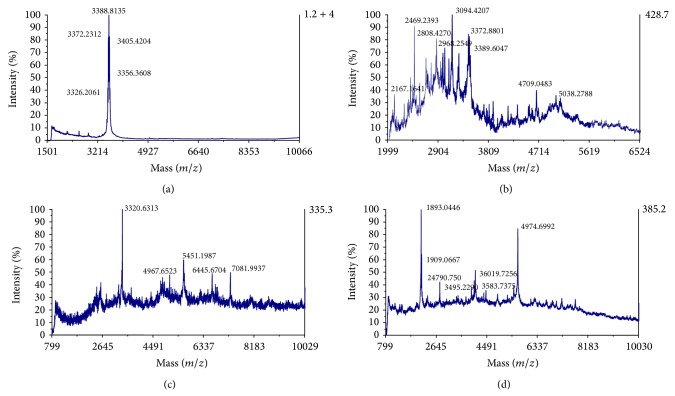
When supernatants from P. pastoris X-33SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-76, and P. pastoris X-33L-1077 were purified by a multistep chromatographic procedure, only eluted fractions after the hydrophobic interaction-chromatography step showed full antimicrobial activity against Pediococcus damnosus CECT4797 as the indicator microorganism (Table 4). MALDI-TOF MS analysis of eluted fractions after the RP-FPLC step showed that supernatants from P. pastoris X-33SRCAM602 showed a major peptide fragment of 3388.8 Da (Figure 1(a)). However, the purified supernatant from P. pastoris X-33OR-7 showed a major peptide fragment of 3094.4 Da and peptide fragments of major and minor molecular mass (Figure 1(b)). Similarly, a large display of peptide fragments of different molecular masses was observed in the purified supernatants from P. pastoris X-33E-760 (Figure 1(c)) and P. pastoris X-33L-1077 (Figure 1(d)). MALDI-TOF MS/MS spectrometry analysis of trypsin-digested peptides from purified supernatants from all recombinant P. pastoris, determined that none of the most probable or de novo amino acid sequence of the evaluated peptide fragments matched the expected amino acid sequence of the bacteriocins SRCAM 602, OR-7, E-760, and L-1077 (Table 5). However, some of the determined peptide fragments were homologous to those observed in proteins from ABC-transporter systems, histidine kinase protein family, peptidase C1 superfamily, and nucleosome binding proteins.
Table 4.
Antimicrobial activity of fractions generated during purification of supernatants from P. pastoris X-SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-760, and P. pastoris X-33L-1077, grown in BMMY with methanol.
| Strain | Antimicrobial activity (BU/mL) of the purified fractionsa | |||||
|---|---|---|---|---|---|---|
| SN | AS | GF | SE | OE | RP-FPLC | |
| P. pastoris X-33SRCAM602 | NA | NA | NA | NA | 12,800 | 1,106,531 |
| P. pastoris X-33OR-7 | NA | NA | 127 | 32 | 1,391 | 10,027 |
| P. pastoris X-33E-760 | NA | NA | NA | NA | 687 | 14,442 |
| P. pastoris X-33L-1077 | NA | NA | 35 | 23 | 2,069 | 13,213 |
Most of the data are mean from two independent determinations in triplicate.
aAntimicrobial activity against Pediococcus damnosus CECT4797 as determined by microtiter plate assay (MPA). BU: bacteriocin units. NA: no activity.
Purification fractions: SN: supernatant; AS: ammonium sulfate precipitation; GF: gel filtration; SE: Sepharose fast flow eluate; OE: Octyl Sepharose eluate; RP-FPLC: reversed-phase eluate.
Figure 1.
Mass spectrometry analysis of purified supernatants from P. pastoris X-33SRCAM602 (a), P. pastoris X-33OR-7 (c), P. pastoris X-33E-760 (b), and P. pastoris X-33L-1077 (d), eluted after RP-FPLC. Numbers indicate the molecular mass in daltons of most of the observed peptide fragments.
Table 5.
Results obtained by MALDI-TOF/TOF MS analysis of the eluted RP-FPLC fractions from purified supernatants of the recombinant P. pastoris X-33 derivatives.
| Bacteriocin | Predicted molecular mass (Da) |
Trypsin-digested precursor (Da) | Amino acid sequence (MS/MS analysis) |
|---|---|---|---|
| SRCAM 602 | 4,630.1 | 926.65 1,500.95 1,614.97 |
SANALRPPT GDKENAAKASSVPAR KTGGNRAVSGAGEIAAR |
| OR-7 | 6,215.1 | Deficient signal | — |
| E-760 | 6,179.8 | 1,166.63 1,579.68 |
VGNPLHGIFGR SLSAYMFFANEQR |
| L-1077 | 4,002.6 | 1,638.76 1,718.73 |
IVGSQAGIGEYLFER LVELSEQELVDCER |
3.3. Antimicrobial Activity of Purified Supernatants from the Recombinant Yeasts
The purified supernatants from recombinant P. pastoris, constructed for expression of the bacteriocins SRCAM 602, OR-7, E-760, and L-1077 showed antimicrobial activity against Pediococcus damnosus CECT4797 but did not display a measurable antimicrobial activity against L. monocytogenes CECT4032, E. coli O157:H7, Y. ruckeri LMG3279, C. jejuni ATCC33560, and C. jejuni NCTC11168, when evaluated by a microtiter plate assay (MPA). The purified supernatants from P. pastoris X-33 and P. pastoris X-33 (pPICZαA) did not show antagonistic activity neither against Pediococcus damnosus CECT4797 nor against any of the indicator bacteria cited above.
4. Discussion
To circumvent the proliferation of emerging pathogenic and antibiotic-resistant bacteria, bacteriocins produced by LAB emerge as natural antimicrobial peptides with potential applications in food preservation, livestock protection, and medical applications [1, 24]. However, the high cost of synthetic bacteriocin synthesis, their low yields, and the production of potential virulence factors from some natural bacterial producers drive the exploration of microbial systems for the biotechnological production of bacteriocins by heterologous LAB and yeasts [6, 8]. Furthermore, the use of synthetic genes may become a useful tool for production and functional expression of bacteriocins by heterologous microbial hosts [12].
The development of heterologous expression systems for bacteriocins may offer a number of advantages over native systems, facilitating the control of bacteriocin gene expression or achieving higher production levels. Although a number of yeast platforms have been used for the production of peptides and proteins, including bacteriocins [10, 11, 25], the use of synthetic genes has been only barely explored for their expression by recombinant yeasts [12]. In this study, the protein expression vector pPICZαA containing an strong and inducible promoter and the Kex2 signal cleavage site for processing of fusion proteins [26] has been used to drive the expression of synthetic genes encoding the mature bacteriocins SRCAM 602, OR-7, E-760, and L-1077, by recombinant P. pastoris X-33 derivatives.
Initial results with P. pastoris X-33SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-760, and P. pastoris X-33L-1077 determined that none of the recombinant yeasts, encoding synthetic genes for expression of the cloned bacteriocins, showed antimicrobial activity when individual colonies were screened by a streak-on-agar test (SOAT). Highly variable yields of secreted proteins have been achieved using the P. pastoris expression system and cases of low secretory yields or complete failure in protein production have also been reported [3, 27, 28]. A number of factors may affect the production of foreign peptides by heterologous yeasts including copy number integration of the expression vectors in the yeast DNA, mRNA stability, errors in mRNA translation, uncoordinated rates of protein synthesis, folding and translocation, and undesired proteolysis of heterologous proteins by resident proteases or by proteases in the extracellular space being secreted, cell-wall associated, or released into the culture medium as a result of cell disruption [29–31]. The use of the P. pastoris expression system for overproduction of peptides and proteins is known to be somewhat hampered by its unpredictable yields of production of heterologous proteins, which is now believed to be caused in part by their varied efficiencies to traffic through the host secretion machinery [3, 28].
The amino acid sequence following the Kex2 secretion signal may also interfere with the secretion of fused peptides or proteins by recombinant P. pastoris. Furthermore, the yields of many recombinant proteins seem to be influenced by the Kex2 P1′ site residue [3]. In this study, the Kex2 P1′ site residues for mature SRCAM 602, OR-7, E-760, and L-1077 were the amino acids A, K, N, and T, respectively (Table 3). However, the cloning in P. pastoris of the bacteriocins enterocin A (EntA) and enterocin P (EntP) with the Kex2 P1′ site residues A and T, respectively, showed an overproduction of both bacteriocins over their natural producers and an intense antimicrobial activity when colonies of the recombinant yeasts were evaluated by the SOAT [11, 22].
Table 3.
Amino acid sequence of cloned bacteriocins and other class IIa bacteriocins.
| Bacteriocin | Amino acid sequence | Number of amino acids |
|---|---|---|
| SRCAM 602 | ATYYGNGLYCNKQKHYTWVDWNKASREIGKITVNGWVQH | 39 |
| OR-7 | KTYYGTNGVHCTKNSLWGKVRLKNMKYDQNTTYMGRLQDILLGW ATGAFGKTFH |
54 |
| E-760 | NRWYCNSAAGGVGGAAVCGLAGYVGEAKENIAGEVRKGWGMAGG FTHNKACKSFPGSGWASG |
62 |
| E 50-52 | TTKNYGNGVCNSVNWCQCGNVWASCNLATGCAAWLCKLA | 39 |
| L-1077 | TNYGNGVGVPDAIMAGIIKLIFIFNIRQGYNFGKKAT | 37 |
| EntP | ATRSYGNGVYCNNSKCWVNWGEAKENIAGIVISGWASGLAGMGH | 44 |
| EntA | TTHSGKYYGNGVYCYKNKCTVDWAKATTCIAGMSIGGFLGGAI PGKC |
47 |
| HirJM79 | ATYYGNGLYCNKEKCWVDWNQAKGEIGKIIVNGWVNHGPWAP RR |
44 |
| SakA | ARSYGNGVYCNNKKCWVNRGEATQSIIGGMISGWASGLAGM | 41 |
Underlined, the class IIa N-terminal consensus sequence YGNGV(X)C.
The use of a multistep chromatographic procedure for purification of the expected antimicrobial activity in supernatants of the recombinant P. pastoris determined that only eluted fractions after the hydrophobic interaction-chromatography step showed full antimicrobial activity (Table 4), probably due to removal of antimicrobial inhibitors, disaggregation of the bacteriocins, or changes in conformation of the bacteriocins in the hydrophobic solvent. The antimicrobial activity of the supernatant from P. pastoris X-33SRCAM602 was much higher than that of the rest of the recombinant P. pastoris. While being interesting, this was not an unexpected observation since purification of the circular bacteriocin garvicin ML, produced by Lactococcus garvieae DCC43, showed a higher antibacterial activity and a broader antimicrobial spectrum as it was increasingly purified [32]. However, MALDI-TOF MS analysis of the purified supernatants from P. pastoris X-33SRCAM602, P. pastoris X-33OR-7, P. pastoris X-33E-760, and P. pastoris X-33L-1077 mostly showed a large display of peptide fragments of different molecular mass than deduced from the calculated molecular mass of the cloned bacteriocins (Figure 1). The different molecular mass of the resulting peptide fragments may suggest the existence of truncated bacteriocins, the interaction of the bacteriocins with unknown biological compounds or the bacteriocins being subjected to posttranslational modifications (PTM) such as phosphorylation, acetylation, methylation, oxidation, formylation, disulfide bond formation, and N-linked and O-linked glycosylation [8, 33]. The presence of cysteine residues in the bacteriocins SRCAM 602, OR-7, and E-760 would permit the formation of disulfide bridges but also permits its oxidation, glutathionylation, and cysteinylation. The absence in all cloned bacteriocins of attachment sites for N-linkages precludes its N-glycosylation, but the presence of threonines and serines makes the bacteriocins sensitive to O-glycosylation [8, 12, 33].
However, MALDI-TOF MS/MS analysis of trypsin-digested peptides from purified supernatants from all recombinant P. pastoris determined that none of the evaluated peptide fragments matched the expected amino acid sequence of the bacteriocins SRCAM 602, OR-7, E-760, and L-1077 (Table 5). From the results obtained it may be suggested that very low yields of secreted and/or purified bacteriocins are obtained after cloning of synthetic genes encoding the bacteriocins SRCAM 602, OR-7, E-760, and L-1077 in P. pastoris. This observation was also not unexpected because bacteriocins cloned into Saccharomyces cerevisiae, P. pastoris, Kluyveromyces lactis, Hansenula polymorpha, and Arxula adeninivorans have been produced with variable success regarding their production, secretion, and functional expression [11]. Furthermore, since one of the main bottlenecks in recombinant protein production is the inability of foreign peptides to reach their native conformation in heterologous yeast hosts, it could also happen that incorrectly folded SRCAM 602, OR-7, E-760, and L-1077 are accumulated in the endoplasmic reticulum (ER) of recombinant P. pastoris, activating the unfolded protein response (UPR) and the ER-associated degradation (ERDA) of the misfolded bacteriocins, leading to persistent ER stress conditions causing much lower efficiencies to traffic through the host secretion machinery, apoptosis, and cell death [8, 28, 34, 35]. In any case, the inconsistent secretory productivity among recombinant proteins has always been a major obstacle for routine application of P. pastoris as an eukaryotic protein expression system in both research and industry [3].
The purified supernatants from the recombinant P. pastoris constructed for expression of the bacteriocins SRCAM 602, OR-7, E-760, and L-1077 showed a measurable antimicrobial activity against Pediococcus damnosus CECT4797 (Table 4), but not against L. monocytogenes CECT4032, E. coli O157:H7, Y. ruckeri LMG3279, C. jejuni ATCC33560, and C. jejuni NCTC11168. One of the remarkable features of bacteriocins is that they are very potent, being active in nanomolar concentrations, thereby surpassing by about 1,000-fold the activity of AMPs produced by eukaryotic cells [36]. One of the major reasons for this extreme potency is that bacteriocins apparently recognize specific receptors on target cells, while the interactions between AMPs and microorganisms are mostly nonspecific. Furthermore, the target receptor for class IIa bacteriocins has been identified as proteins of the sugar transporter mannose phosphotransferase system (Man-PTS), with the most potent receptors being those found in Listeria spp. [37, 38]. However, the very low yields of secreted or purified bacteriocins in supernatants of the recombinant P. pastoris may be responsible for their nondetected antilisterial activity.
It could be also hypothesized that peptide fragments aggregated to or coeluting with the purified bacteriocins may be responsible for the antimicrobial activity of the eluates against the sensitive indicator Pediococcus damnosus CECT4797, but not against any other of the indicator bacteria tested. Many proteins contain encrypted within their primary structure bioactive peptides with antimicrobial activity following hydrolytic release from the native molecule [39, 40]. Incorrect disulfide bond formation, misfolding of the secreted bacteriocin and extensive PTMs were suggested to be responsible for the lower antilisterial activity and the nonmeasurable antimicrobial activity against Gram-negative bacteria of the bacteriocin E 50-52 (BacE50-52), produced by recombinant P. pastoris X-33BE50-52S and K. lactis GG799BE50-52S [12]. This bacteriocin, originally produced by Enterococcus faecium B-30746, was also reported to display a high and broad antimicrobial activity against Gram-positive and Gram-negative bacteria, including Campylobacter spp. [20, 41].
The correct processing, secretion and functional expression of the bacteriocins EntP [22], hiracin JM79 (HirJM79) [42] and EntA [11], produced by recombinant yeasts, contrast with the low biological activity of the sakacin A (SakA) and the chimera EntP/SakA, produced by recombinant P. pastoris and K. lactis producers [8]. Misfolding of SakA and EntP/SakA and induction of the yeasts' UPR may be responsible for apoptosis in recombinant P. pastoris producers of SakA and for extensive PTMs in recombinant P. pastoris and K. lactis, producers of SakA and EntP/SakA [8]. These results, obtained by our research group, also contrast with the low antimicrobial activity against Pediococcus damnosus CECT4797 and the absence of antimicrobial activity against Gram-positive and Gram-negative bacteria of the purified supernatants from recombinant P. pastoris encoding the mature bacteriocins SRCAM 602, OR-7, E-760, and L-1077.
Nevertheless, it should be also worth to notice that bacteriocins SRCAM 602 [16], OR-7 [17], E-760 [19], and L-1077 [18], although being reported as bacteriocins with a broad antimicrobial activity against Gram-positive and Gram-negative microorganisms including Campylobacter spp., have not been fully characterized at their biochemical and genetic level, the genetic identification of their structural and adjacent genes has not been yet reported, and their molecular masses, deduced from their reported amino acid sequences, were not identical to the experimentally determined molecular mass from the purified bacteriocins [16–19]. Furthermore, recent reports suggest that bacteriocin SRCAM 602, previously reported to be produced by Paenibacillus polymyxa NRRL B-30509 and claimed to be responsible for inhibition of C. jejuni, could not be detected in the purified supernatants of the producer strain while the srcam602 structural gene was not found via a PCR-based approach using degenerate nucleotide primers or by genomic sequencing of the bacteriocin producer [43]. Similarly, Bacillus circulans (now Paenibacillus terrae) NRRL B-30644 previously reported to produce SRCAM 1580, a bacteriocin active against C. jejuni [16], has been recently suggested not to produce this bacteriocin and no genetic determinants for its production were shown. Instead the anti-Campylobacter activity of this strain is due to the production of the lipopeptide tridecaptin A1 whereas this strain also produces the novel lantibiotic paenicidin B, active against Gram-positive bacteria [44].
Although the cloning in recombinant yeasts of synthetic genes encoding bacteriocins drives the production, antimicrobial activity, and specific antimicrobial activity of the cloned bacteriocins in the absence of dedicated immunity and secretion proteins [12], these results contrast with the negligible antimicrobial activity against Gram-positive and Gram-negative bacteria of purified supernatants from recombinant P. pastoris encoding the bacteriocins SRCAM 602, OR-7, E-760, and L-1077. Accordingly, since production of bacteriocins from synthetic bacteriocin genes is difficult to predict, further efforts should be performed for a more efficient genetically engineered production and functional expression of other bacteriocin synthetic genes or their chimeras by heterologous producer yeasts. The design of further and novel successful genetic approaches for production and functional expression of bacteriocins by yeasts would facilitate their biotechnological applications as natural antimicrobial agents in food, animal husbandry, and medicine.
Acknowledgments
This work was partially supported by Project AGL2012-34829 from Ministerio de Economía y Competitividad (MINECO) and Project AGL2009-08348 from the Ministerio de Ciencia e Innovación (MICINN), by Grant GR35-10A from the BSCH-UCM and by Grants S2009/AGR-1489 and S2013/ABI-2747 from the Comunidad de Madrid (CAM). S. Arbulu holds a fellowship (FPI) from the MINECO. J. J. Jiménez and L. Gútiez held a fellowship from the MICINN and the Ministerio de Educación y Ciencia (MEC), Spain, respectively.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lohans C. T., Vederas J. C. Development of class IIa bacteriocins as therapeutic agents. International Journal of Microbiology. 2012;2012:13. doi: 10.1155/2012/386410.386410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotter P. D., Ross R. P., Hill C. Bacteriocins-a viable alternative to antibiotics? Nature Reviews Microbiology. 2013;11(2):95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 3.Yang S., Kuang Y., Li H., et al. Enhanced production of recombinant secretory proteins in Pichia pastoris by optimizing Kex2 P1’ site. PLoS ONE. 2013;8(9) doi: 10.1371/journal.pone.0075347.e75347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotter P. D., Hill C., Ross R. P. Food microbiology: bacteriocins: developing innate immunity for food. Nature Reviews Microbiology. 2005;3(10):777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 5.Drider D., Fimland G., Héchard Y., McMullen L. M., Prévost H. The continuing story of class IIa bacteriocins. Microbiology and Molecular Biology Reviews. 2006;70(2):564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borrero J., Jiménez J. J., Gútiez L., Herranz C., Cintas L. M., Hernández P. E. Protein expression vector and secretion signal peptide optimization to drive the production, secretion, and functional expression of the bacteriocin enterocin A in lactic acid bacteria. Journal of Biotechnology. 2011;156(1):76–86. doi: 10.1016/j.jbiotec.2011.07.038. [DOI] [PubMed] [Google Scholar]
- 7.Parachin N. S., Mulder K. C., Viana A. A. B., Dias S. C., Franco O. L. Expression systems for heterologous production of antimicrobial peptides. Peptides. 2012;38(2):446–456. doi: 10.1016/j.peptides.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 8.Jiménez J. J., Borrero J., Diep D. B., et al. Cloning, production, and functional expression of the bacteriocin sakacin A (SakA) and two SakA-derived chimeras in lactic acid bacteria (LAB) and the yeasts Pichia pastoris and Kluyveromyces lactis . Journal of Industrial Microbiology & Biotechnology. 2013;40(9):977–993. doi: 10.1007/s10295-013-1302-6. [DOI] [PubMed] [Google Scholar]
- 9.Gao M., Shi Z. Process control and optimization for heterologous protein production by methylotrophic pichia pastoris. Chinese Journal of Chemical Engineering. 2013;21(2):216–226. doi: 10.1016/S1004-9541(13)60461-9. [DOI] [Google Scholar]
- 10.Ahmad M., Hirz M., Pichler H., Schwab H. Protein expression in Pichia pastoris: recent achievements and perspectives for heterologous protein production. Applied Microbiology and Biotechnology. 2014;98:5301–5317. doi: 10.1007/s00253-014-5732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borrero J., Kunze G., Jiménez J., et al. Cloning, production, and functional expression of the bacteriocin enterocin A, produced by Enterococcus faecium T136, by the Yeasts Pichia pastoris, Kluyveromyces lactis, Hansenula polymorpha, and Arxula adeninivorans . Applied and Environmental Microbiology. 2012;78(16):5956–5961. doi: 10.1128/AEM.00530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiménez J. J., Borrero J., Gútiez L., et al. Use of synthetic genes for cloning, production and functional expression of the bacteriocins enterocin A and bacteriocin E 50-52 by Pichia pastoris and Kluyveromyces lactis . Molecular Biotechnology. 2014;56(6):571–583. doi: 10.1007/s12033-014-9731-7. [DOI] [PubMed] [Google Scholar]
- 13.Darby R. A. J., Cartwright S. P., Dilworth M. V., Bill R. M. Which yeast species shall i choose? Saccharomyces cerevisiae versus Pichia pastoris . Methods in Molecular Biology. 2012;866:11–23. doi: 10.1007/978-1-61779-770-5_2. [DOI] [PubMed] [Google Scholar]
- 14.Öberg F., Sjöhamn J., Conner M. T., Bill R. M., Hedfalk K. Improving recombinant eukaryotic membrane protein yields in Pichia pastoris: the importance of codon optimization and clone selection. Molecular Membrane Biology. 2011;28(6):398–411. doi: 10.3109/09687688.2011.602219. [DOI] [PubMed] [Google Scholar]
- 15.Stern N. J., Svetoch E. A., Eruslanov B. V., et al. Paenibacillus polymyxa purified bacteriocin to control Campylobacter jejuni in chickens. Journal of Food Protection. 2005;68(7):1450–1453. doi: 10.4315/0362-028x-68.7.1450. [DOI] [PubMed] [Google Scholar]
- 16.Svetoch E. A., Stern N. J., Eruslanov B. V., et al. Isolation of Bacillus circulans and Paenibacillus polymyxa strains inhibitory to Campylobacter jejuni and characterization of associated bacteriocins. Journal of Food Protection. 2005;68(1):11–17. doi: 10.4315/0362-028x-68.1.11. [DOI] [PubMed] [Google Scholar]
- 17.Stern N. J., Svetoch E. A., Eruslanov B. V., et al. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrobial Agents and Chemotherapy. 2006;50(9):3111–3116. doi: 10.1128/AAC.00259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svetoch E. A., Eruslanov B. V., Levchuk V. P., et al. Isolation of Lactobacillus salivarius 1077 (NRRL B-50053) and characterization of its bacteriocin, including the antimicrobial activity spectrum. Applied and Environmental Microbiology. 2011;77(8):2749–2754. doi: 10.1128/AEM.02481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Line J. E., Svetoch E. A., Eruslanov B. V., et al. Isolation and purification of enterocin E-760 with broad antimicrobial activity against Gram-positive and Gram-negative bacteria. Antimicrobial Agents and Chemotherapy. 2008;52(3):1094–1100. doi: 10.1128/AAC.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Svetoch E. A., Eruslanov B. V., Perelygin V. V., et al. Diverse antimicrobial killing by Enterococcus faecium E 50-52 bacteriocin. Journal of Agricultural and Food Chemistry. 2008;56(6):1942–1948. doi: 10.1021/jf073284g. [DOI] [PubMed] [Google Scholar]
- 21.Svetoch E. A., Stern N. J. Bacteriocins to control Campylobacter spp. in poultry-a review. Poultry Science. 2010;89(8):1763–1768. doi: 10.3382/ps.2010-00659. [DOI] [PubMed] [Google Scholar]
- 22.Gutiérrez J., Criado R., Martín M., Herranz C., Cintas L. M., Hernández P. E. Production of enterocin P, an antilisterial pediocin-like bacteriocin from Enterococcus faecium P13, in Pichia pastons . Antimicrobial Agents and Chemotherapy. 2005;49(7):3004–3008. doi: 10.1128/AAC.49.7.3004-3008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A., Loboda A., Ens W., Standing K. G. MALDI quadrupole time-of-flight mass spectrometry: a powerful tool for proteomic research. Analytical Chemistry. 2000;72(9):2132–2141. doi: 10.1021/ac9913659. [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization (WHO) Antimicrobial resistance: global report on surveillance 2014. http://www.searo.who.int/thailand/publications/2013/9789241564748/en/
- 25.Böer E., Steinborn G., Kunze G., Gellissen G. Yeast expression platforms. Applied Microbiology and Biotechnology. 2007;77(3):513–523. doi: 10.1007/s00253-007-1209-0. [DOI] [PubMed] [Google Scholar]
- 26.Cregg J. M., Cereghino J. L., Shi J., Higgins D. R. Recombinant protein expression in Pichia pastoris . Applied Biochemistry and Biotechnology—Part B Molecular Biotechnology. 2000;16(1):23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 27.Burrowes O. J., Diamond G., Lee T. C. Recombinant expression of pleurocidin cDNA using the Pichia pastoris expression system. Journal of Biomedicine and Biotechnology. 2005;2005(4):374–384. doi: 10.1155/JBB.2005.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love K. R., Politano T. J., Panagiotou V., Jiang B., Stadheim T. A., Love J. C. Systematic single-cell analysis of pichia pastoris reveals secretory capacity limits productivity. PLoS ONE. 2012;7(6) doi: 10.1371/journal.pone.0037915.e37915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang H. A., Choi E.-S., Hong W.-K., et al. Proteolytic stability of recombinant human serum albumin secreted in the yeast Saccharomyces cerevisiae . Applied Microbiology and Biotechnology. 2000;53(5):575–582. doi: 10.1007/s002530051659. [DOI] [PubMed] [Google Scholar]
- 30.Werten M. W. T., De Wolf F. A. Reduced proteolysis of secreted gelatin and Yps1-mediated α-factor leader processing in a Pichia pastoris kex2 disruptant. Applied and Environmental Microbiology. 2005;71(5):2310–2317. doi: 10.1128/AEM.71.5.2310-2317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ni Z., Zhou X., Sun X., Wang Y., Zhang Y. Decrease of hirudin degradation by deleting the KEX1 gene in recombinant Pichia pastoris . Yeast. 2008;25(1):1–8. doi: 10.1002/yea.1542. [DOI] [PubMed] [Google Scholar]
- 32.Borrero J., Brede D. A., Skaugen M., et al. Characterization of garvicin ML, a novel circular bacteriocin produced by Lactococcus garvieae DCC43, isolated from mallard ducks (Anas platyrhynchos) Applied and Environmental Microbiology. 2011;77(1):369–373. doi: 10.1128/AEM.01173-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y., Jensen O. N. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9(20):4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasser B., Saloheimo M., Rinas U., et al. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microbial Cell Factories. 2008;7, article 11 doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stolz A., Wolf D. H. Endoplasmic reticulum associated protein degradation: a chaperone assisted journey to hell. Biochimica et Biophysica Acta—Molecular Cell Research. 2010;1803(6):694–705. doi: 10.1016/j.bbamcr.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Breukink E., Wiedemann I., van Kraaij C., Kuipers O. P., Sahl H.-G., de Kruijff B. Use of the cell wail precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286(5448):2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 37.Diep D. B., Skaugen M., Salehian Z., Holo H., Nes I. F. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(7):2384–2389. doi: 10.1073/pnas.0608775104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kjos M., Borrero J., Opsata M., et al. Target recognition, resistance, immunity and genome mining of class II bacteriocins from Gram-positive bacteria. Microbiology. 2011;157(12):3256–3267. doi: 10.1099/mic.0.052571-0. [DOI] [PubMed] [Google Scholar]
- 39.Phelan M., Aherne A., FitzGerald R. J., O'Brien N. M. Casein-derived bioactive peptides: biological effects, industrial uses, safety aspects and regulatory status. International Dairy Journal. 2009;19(11):643–654. doi: 10.1016/j.idairyj.2009.06.001. [DOI] [Google Scholar]
- 40.Brand G. D., Magalhães M. T. Q., Tinoco M. L. P., et al. Probing protein sequences as sources for encrypted antimicrobial peptides. PLoS ONE. 2012;7(9) doi: 10.1371/journal.pone.0045848.e45848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Svetoch E. A., Levchuk V. P., Pokhilenko V. D., et al. Inactivating methicillin-resistant Staphylococcus aureus and other pathogens by use of bacteriocins OR-7 and E 50-52. Journal of Clinical Microbiology. 2008;46(11):3863–3865. doi: 10.1128/JCM.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sánchez J., Borrero J., Gómez-Sala B., et al. Cloning and heterologous production of hiracin JM79, a Sec-dependent bacteriocin produced by Enterococcus hirae DCH5, in lactic acid bacteria and Pichia pastoris . Applied and Environmental Microbiology. 2008;74(8):2471–2479. doi: 10.1128/AEM.02559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lohans C. T., Huang Z., Van Belkum M. J., et al. Structural characterization of the highly cyclized lantibiotic paenicidin A via a partial desulfurization/reduction strategy. Journal of the American Chemical Society. 2012;134(48):19540–19543. doi: 10.1021/ja3089229. [DOI] [PubMed] [Google Scholar]
- 44.Lohans C. T., Van Belkum M. J., Cochrane S. A., et al. Biochemical, structural, and genetic characterization of tridecaptin A1, an antagonist of Campylobacter jejuni . ChemBioChem. 2014;15(2):243–249. doi: 10.1002/cbic.201300595. [DOI] [PubMed] [Google Scholar]