Abstract
The regulation of flowering time has crucial implications for plant fitness. MicroRNA156 (miR156) represses the floral transition in Arabidopsis thaliana, but the mechanisms regulating its transcription remain unclear. Here, we show that two AGAMOUS-like proteins, AGL15 and AGL18, act as positive regulators of the expression of MIR156. Small RNA northern blot analysis revealed a significant decrease in the levels of mature miR156 in agl15 agl18 double mutants, but not in the single mutants, suggesting that AGL15 and AGL18 co-regulate miR156 expression. Histochemical analysis further indicated that the double mutants showed a reduction in MIR156 promoter strength. The double mutants also showed reduced abundance of pri-miR156a and pri-miR156c, two of the primary transcripts from MIR156 genes. Electrophoretic mobility shift assays demonstrated that AGL15 directly associated with the CArG motifs in the MIR156a/c promoters. AGL18 did not show binding affinity to the CArG motifs, but pull-down and yeast two-hybrid assays showed that AGL18 forms a heterodimer with AGL15. GFP reporter assays and bimolecular fluorescence complementation (BiFC) showed that AGL15 and AGL18 co-localize in the nucleus and confirmed their in vivo interaction. Overexpression of miR156 did not affect the levels of AGL15 and AGL18 transcripts. Taking these data together, we present a model for the transcriptional regulation of MIR156. In this model, AGL15 and AGL18 may form a complex along with other proteins, and bind to the CArG motifs of the promoters of MIR156 to activate the MIR156 expression.
Keywords: AGAMOUS-like 15, AGAMOUS-like 18, CArG motifs, floral transition, miRNA156
INTRODUCTION
In many plant species, flowering marks the transition from vegetative to reproductive growth; induction of flowering at the proper time increases the plant’s reproductive success. In agriculture, the flowering transition crucially determines crop yield, as flowering is mandatory for the production of seeds and fruits in higher plants. By contrast, prolonged vegetative growth and a late floral transition can improve yields in vegetative crops. In Arabidopsis (A. thaliana), extensive studies on the mechanisms that determine the floral transition showed that environmental factors (i.e., temperature and day-length) and intrinsic conditions (i.e., age and gibberellin accumulation) regulate flowering time (Fornara et al., 2010). A better understanding of the factors that govern the floral transition will help in the manipulation of flowering time and the improvement of agricultural practice to increase crop yields (Chung et al., 2014).
MicroRNAs (miRNAs), short non-coding RNAs of 20-24 nucleotides, negatively regulate expression of their target genes (Carrington and Ambros, 2003). Several miRNAs affect flower development and the floral transition in Arabidopsis. For example, transgenic plants overexpressing miR172 flower early and have floral organ defects (Chen, 2004). Also, miR159 and miR319 regulate floral development via interaction with MYB and TCP transcription factors (Rubio-Somoza and Weigel, 2013). In association with phosphate starvation, miR399 has been suggested to regulate flowering time in response to ambient temperature (Lee et al., 2010). The SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 (SPL3) transcription factor promotes the floral transition by activating the expression of FRUITFULL (FUL), LEAFY (LFY), and APETALA1 (AP1) (Yamaguchi et al., 2009). MiR156 negatively regulates SPL3 expression via cleavage of the SPL3 transcripts (Wu and Poethig, 2006); thus overexpression of miR156 prolongs the vegetative phase and delays the floral transition. Lee et al. (2012a) also suggested that, in response to ambient temperature, the miR156-SPL3 module regulates FLOWERING LOCUS T (FT) expression via the direct binding of SPL3 to the GTAC motifs within the FT promoter. Although much is known about miRNA-mediated target gene regulation, the upstream transcriptional regulation of the miRNAs themselves remains relatively unexplored.
MCM1-AGAMOUS-DEFICIENS-SRF (MADS) domain proteins are a family of DNA-binding transcription factors that contain the conserved DNA-binding domains MADS box (56 amino acids) at the N-terminal and a C-terminal extension of approximately 30 amino acids (Shore and Sharrocks, 1995). The SRF core DNA-binding domain selectively binds to a consensus DNA sequence, the C-A/T rich-G (CArG) motif (West et al., 1997) to either activate or repress the expression of target genes. MADS-domain proteins play key roles in regulating developmental processes in eukaryotes (West et al., 1997). Interestingly, plants have more MADS-box gene families, compared to other kingdoms (Jack, 2001). This greater diversity suggests greater specificity of the downstream regulation by selective binding to the target genes (Tang and Perry, 2003). Although different MADS-domain proteins may have a similar binding site, they often show preferential binding (Shore and Sharrocks, 1995). In Arabidopsis, MADS-domain proteins affect several developmental processes, including root growth (Zhang and Forde, 1998), ovary, fruit, and seed coat development (Ferrandiz et al., 2000; and Nesi et al., 2002; Pinyopich et al., 2003), floral organ identity (Jack, 2001), and flowering time determination (Borner et al., 2000; Yoo et al., 2011).
AGAMOUS-like 15 (AGL15), a MADS-domain protein, is expressed during embryo and seed development in both monocot and dicot plants (Perry et al., 1996). AGL15 may function in the regulation of the Arabidopsis MYB transcription factor MYB17 during early development of the inflorescence as well as in seed germination (Zhang et al., 2009). Like many other MADS-domain proteins, AGL15 forms a heterodimer with other proteins (Hill et al., 2008). DNA-protein interactions have also been observed, as AGL15 preferentially binds to CArG motifs in vitro (Tang and Perry, 2003; Wang et al., 2002). AGAMOUS-like 18 (AGL18) functions redundantly with AGL15 in the regulation of flowering time in Arabidopsis. Overexpression of AGL15 and AGL18 in Arabidopsis produced similar phenotypes, including morphological alterations and late flowering time. Moreover, agl15 agl18 double mutants flower early, but the agl15 or agl18 single mutants do not flower early (Adamczyk et al., 2007). As genes in the same family often have overlapping functions, AGL15 and AGL18 likely have functional redundancy and act as co-repressors of floral transition via regulation of multiple flowering time genes (Gu et al., 2013; Fernandez et al, 2014).
MADS-domain proteins also regulate the expression of some miRNAs; for example, the MADS-domain protein SHORT VEGETATIVE PHASE (SVP) binds to a CArG motif in the promoter of MIR172 and negatively regulates its expression (Cho et al., 2012), and the SVP function delays flowering transition (Lee et al., 2010). The AGAMOUS-like 16 (AGL16) transcript is subject to sequence-specific degradation by miR824 in the regulation of stomatal development (Kutter et al., 2007). Although these findings collectively suggest an interaction between MADS-domain proteins and miRNAs, the molecular interactions between MADS-domain proteins and miRNAs, such as between AGL15/AGL18 and miR156 remain unknown. In this study, we demonstrate that AGL15 and AGL18 function as co-regulators of MIR156 expression, as agl15 agl18 double mutants show a significant decrease in miR156 accumulation and weaker expression from the MIR156 promoter. AGL15 bound to the putative CArG motifs present in the MIR156 promoters. AGL15 and AGL18 proteins co-localized in the nucleus and interacted in vitro and in vivo. Collectively, these data suggest that AGL15 and AGL18 may function as direct co-activators of the transcription of MIR156 in the determination of flowering time in Arabidopsis.
MATERIALS AND METHODS
Plant materials
Wild-type Arabidopsis (Col-0), agl15-3, agl15-4, agl18-1, agl15-3 agl18-1, agl15-4 agl18-1 (Adamczyk et al., 2007), and 35S:: MIR156 (Kim et al., 2012) plants were grown on Murashige and Skoog (MS) medium in a growth room under long-day (LD) conditions (16 h light/ 8 dark), at 23°C, with light intensity of 120 μmol m−2s−1. 9-day-old seedlings were sampled, immediately frozen in liquid nitrogen, and stored at −80°C prior to RNA extraction.
Gene expression analysis
Total RNA was isolated from 9-day-old seedlings using Plant RNA Purification Reagent (Invitrogen, USA). The total extracted RNA was pretreated with DNase I (NEB, USA) to eliminate possible DNA contamination, and subsequently subjected to complementary DNA synthesis using the First Strand cDNA Synthesis Kit (Roche Applied Science, USA). To measure the transcript levels of flowering time genes, quantitative real-time RT-PCR (qPCR) was carried out using the Green I Master Mix (Roche Applied Science, USA) with gene-specific primers (Supplementary Table S1). Two reference genes (AT1G13320/AT2G28390) that are stably expressed (Hong et al., 2010) were used for quantification. All reactions were performed with two biological replicates and three technical replicates. Determination of mature miR156 levels was done by miRNA northern hybridization analysis, as described previously (Lee et al., 2010). For histochemical GUS analysis, transgenic plants expressing MIR156::GUS were generated. The promoter region (approximately 2 kb) of MIR156b was amplified by PCR (Supplementary Table S1), and then cloned into the pBI101 vector harboring a GUS reporter gene. The resulting MIR156::GUS construct was introduced into wild-type and double mutant (agl15-3 agl18-1 and agl15-4 agl18-1) plants using a floral dip method (Clough and Bent, 1998).
Recombinant protein production and purification
For production of His-fused proteins, the open reading frame (ORF) of Arabidopsis AGL15 or AGL18 was cloned into the pET21a vector (EMD Biosciences, USA). The resulting construct was introduced into E. coli BL21. Cell pellets were harvested from an overnight culture, resuspended in 1X His-binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl pH 7.9), sonicated, and then centrifuged. The supernatant was collected, and protein purification was performed using the His-Bind Kit (Merck, Germany) according to the manufacturer’s instructions. For GST-fused proteins, the same ORF was cloned into the pGEX-5T vector (GE Healthcare, USA) and introduced into E. coli cells. The subsequent protein production and purification were conducted as previously described (Cho et al., 2012).
Electrophoretic mobility shift assay
Prediction of putative CArG motifs in MIR156a and MIR156c was done using AthaMap (http://www.athamap.de) and PLACE (http://www.dna.affrc.go.jp/PLACE/). The DNA sequences of the predicted CArG motifs were synthesized, with HPLC purification (Cosmogen, Korea). Electrophoretic mobility shift assays (EMSAs) were performed using the LightShift Chemiluminescent EMSA Kit (Thermo Scientific, USA) according to the manufacturer’s instructions. Double-stranded 3′ biotin-labeled oligonucleotides were used as DNA probes (Supplementary Table S1). Purified AGL15-GST or AGL18-GST protein (2 μg) was incubated with the biotin-labeled oligonucleotides at room temperature for 30 min. The reaction samples were electrophoresed in an 8% native polyacrylamide gel, and then transferred onto a nylon membrane. Visualization was done as described in the manufacturer’s instructions (Thermo Scientific, USA).
Yeast 2-hybrid, in vitro GST pull-down, and bimolecular fluorescence complementation (BiFC) analyses
PCR-amplified AGL15 and AGL18 ORFs were cloned into pGADT7 or pGBK7 (Clontech, USA) for the yeast two-hybrid experiment. The yeast 2-hybrid analysis was carried out as described previously (Lee et al., 2012b). The in vitro GST pull-down assay was performed as described by Jang et al. (2009) with minor modifications; purified GST-AGL18 or GST protein was immobilized with glutathione sepharose beads (GE Healthcare, USA), and then incubated with the purified AGL15-His protein. The bead-retained proteins were subsequently eluted, analyzed by 12% SDS-PAGE, and then visualized by Western blot analysis or autoradiography.
For bimolecular fluorescence complementation (BiFC) analysis, the AGL15 and AGL18 ORFs were cloned into pUC-SPYNE/pSPYNE-35S or pUC-SPYCE/pSPYCE-35S (Walter et al., 2004) to generate AGL15-YFPC and AGL18-YFPN constructs. BiFC assays were done in both tobacco (N. benthamiana) epidermal cells and Arabidopsis protoplasts. For transient expression in tobacco plants, Agrobacterium harboring the AGL15-YFPC and AGL18-YFPN constructs was infiltrated into tobacco leaves (Voinnet et al., 2003). Epidermal cells of the transformed tobacco leaves were examined using a Zeiss LSM 510 META confocal microscope (Carl Zeiss, Germany). For transient expression in Arabidopsis mesophyll protoplasts, Arabidopsis protoplasts were prepared from young leaves by macerozyme treatment and transfected with the AGL15-YFPC and AGL18-YFPN constructs as previously described (Yoo et al., 2007). Cellular localization of the fluorescent proteins was examined with a Zeiss LSM 510 META confocal microscope (Carl Zeiss, Germany). Co-expression of the YFP-fused bZIP63 proteins (i.e., bZIP-YFPC/bZIP-YFPN) was used as a positive control (Walter et al., 2004).
Subcellular localization assay
Analysis of subcellular localization was performed using a green fluorescent protein (GFP) reporter assay. The full-length AGL15 or AGL18 ORF was cloned into a Gateway vector harboring the 35S promoter and GFP reporter gene, resulting in 35S::AGL15:GFP and 35S::AGL18:GFP constructs. The 35S::AGL15:GFP and 35S::AGL18:GFP fusion constructs were introduced into Arabidopsis mesophyll protoplasts using a polyethylene glycol (PEG)-calcium method (Yoo et al., 2007). 4′, 6-diamidino-2-phenylindole (DAPI) staining identified the nucleus. Expression of the GFP fusion constructs was monitored by using a Zeiss Leica LSM 510 META confocal microscope (Zeiss, Germany). Tobacco leaves were infiltrated with the constructs and visualized as described above. A 35S::GFP construct was used as a positive control.
RESULTS
Downregulation of miR156 in agl15 agl18 double mutants
To investigate how miR156 expression is regulated by transcription factors, we analyzed the abundance of mature miR156 in agl15 and agl18 single mutants and in agl15 agl18 double mutants, using northern blot analysis (Fig. 1A). Under inductive long day (LD) conditions, neither agl15 nor agl18 single mutants showed a significant reduction of mature miR156 levels, as compared to the wild type plants (Fig. 1A). However, we observed downregulation of miR156 levels in both agl15-3 agl18-1 and agl15-4 agl18-1 double mutants, suggesting that AGL15 and AGL18 act together to positively regulate MIR156 expression.
Fig. 1.
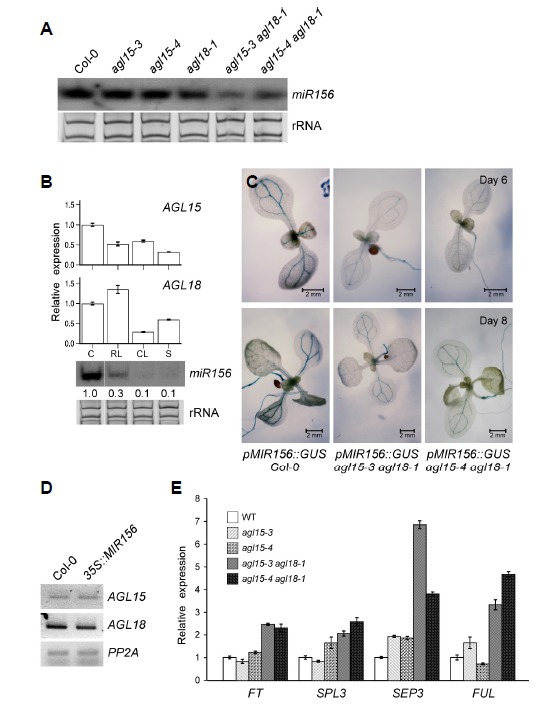
Expression of mature miR156, AGL15, and AGL18. (A) Northern blot analysis of mature miR156 in Col-0, agl15, agl18, and agl15 agl18 double mutants. Ethidium bromide (EtBr)-stained rRNA served as the loading control. (B) Spatial expression patterns of AGL15 and AGL18 compared with those of mature miR156. Y-axis indicates the relative transcript level of AGL15 or AGL18 in the cotyledon (C), rosette leaf (RL), cauline leaf (CL), and stem (S) of mature seedlings. The transcript levels of AGL15 and AGL18 in the cotyledon (C) were set to one. Ethidium bromide (EtBr)-stained rRNA served as the loading control. (C) Histochemical analysis of 6-day-old and 8-day-old seedlings of pMIR156::GUS agl15 agl18 plants. Note that vascular bundle-specific GUS expression was reduced in the agl15 agl18 double mutants. (D) Transcript levels of AGL15 and AGL18 in Col-0 and 35S::MIR156 plants determined by RT-PCR. PP2A was used as an internal control. (E) Transcript levels of FT, SPL3, SEP3, and FUL in Col-0, agl15, and agl15 agl18 double mutants.
We next analyzed the spatial expression patterns of AGL15, AGL18, and miR156. Small RNA northern blots showed that the levels of mature miR156 were high in the cotyledon of mature seedlings (Fig. 1B). Consistent with this observation, reverse transcription quantitative PCR (RT-qPCR) showed that AGL15 and AGL18 transcript levels were also high in the cotyledon. The levels of mature miR156 were low in the rosette leaves, cauline leaves, and stem. AGL15 transcript levels in the same tissues were also lower than in the cotyledon, although AGL18 showed slightly higher transcript levels in the rosette leaves. This result showed that transcript levels of AGL15 and AGL18 were largely correlated with the levels of mature miR156 levels. We then tested pMIR156:GUS reporter activity in agl15 agl18 double mutants. GUS activity was detected in the vascular bundle of the cotyledon, true leaf, and shoot apical region in 6- and 8-day-old wild-type plants (Fig. 1C). Similar GUS patterns were observed in the agl15 agl18 double mutants, but the GUS activity appeared to be lower. This finding further supported the notion that AGL15 and AGL18 regulate MIR156 expression. To further investigate the genetic relationship, we measured the expression levels of AGL15 and AGL18 in 35S::MIR156 transgenic plants. The data showed that expression of AGL15 and AGL18 was not significantly altered in the transgenic lines (Fig. 1D), suggesting that AGL15 and AGL18 likely do not act downstream of miR156 in Arabidopsis. Because agl15 agl18 double mutants showed a decreased level of MIR156, we performed qPCR to measure transcript levels of flowering time genes. We found that FT, SPL3, SEP3, and FUL expression levels significantly increased in agl15-3 agl18-1 and agl15-4 agl18-1 double mutants, compared to their expression in the wild type plants (Fig. 1E). Both double mutant lines showed similar increases in the expression of FT, SPL3, and FUL, whereas in the case of SEP3, the agl15-3 agl18-1 mutants showed higher levels than agl15-4 agl18-1 mutants. By contrast, agl15 mutants did not show significant changes. Taken together, our results suggested that AGL15 and AGL18 positively regulate MIR156 expression to regulate flowering time in Arabidopsis.
Pri-miR156a and pri-miR156c are affected in the agl15 agl18 mutants
The mature miR156 is produced from primary transcripts of miR156 (pri-miR156) from eight different loci (MIR156a-h); therefore, we investigated which locus is mainly affected in the agl15 agl18 double mutants. Absolute quantification of the levels of the primary transcripts revealed that pri-miR156a, pri-miR156c, and pri-miR156d are the main loci that produce miR156 in wild-type plants (data not shown). Thus we measured their transcript levels in wild-type and agl15 agl18 double mutant plants. The expression of pri-miR156a and pri-miR156c were reduced in the double mutants, compared to the wild-type plants (Fig. 2), whereas pri-miR156d levels showed no significant changes in either of the double mutant lines (Fig. 2). This result suggested that AGL15 and AGL18 mainly regulate the transcription of MIR156a and MIR156c. Therefore, we focused on miR156a and miR156c for the subsequent studies.
Fig. 2.
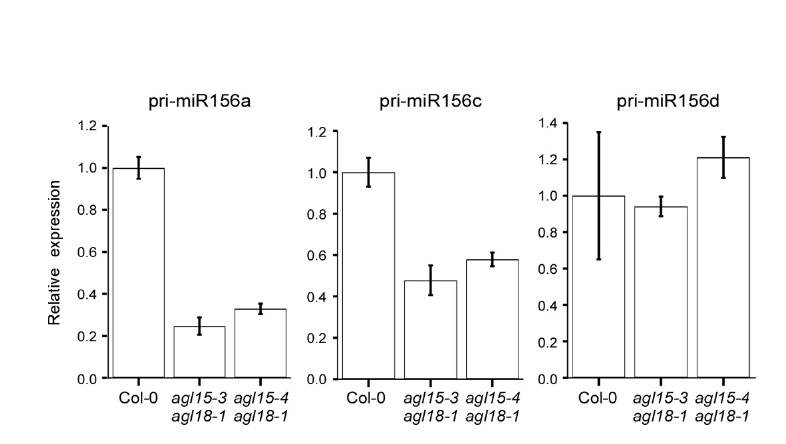
Relative levels of primary transcripts of miR156a, c, and d by qPCR. The levels of each primary transcript in wild-type Col-0 plants were set to one.
AGL15 proteins bind to the CArG motifs in the promoters of MIR156a and MIR156c
Based on the gene expression data (Figs. 1 and 2), we hypothesized that the accumulation of mature miR156 might be caused by the direct interactions of AGL15 and AGL18 with the promoter sequences of MIR156. Using publicly available bioinformatics tools (AthaMap and PLACE) (Hehl and Bülow, 2014; Higo et al., 1998), we identified eight putative CArG motifs in the upstream regions of MIR156a and MIR156c (Figs. 3A and 3B). The sequence of the identified putative CArG motifs, as well as their locations relative to the major transcription start sites (TSSs) (Xie et al., 2005), are shown in Fig. 3C.
Fig. 3.
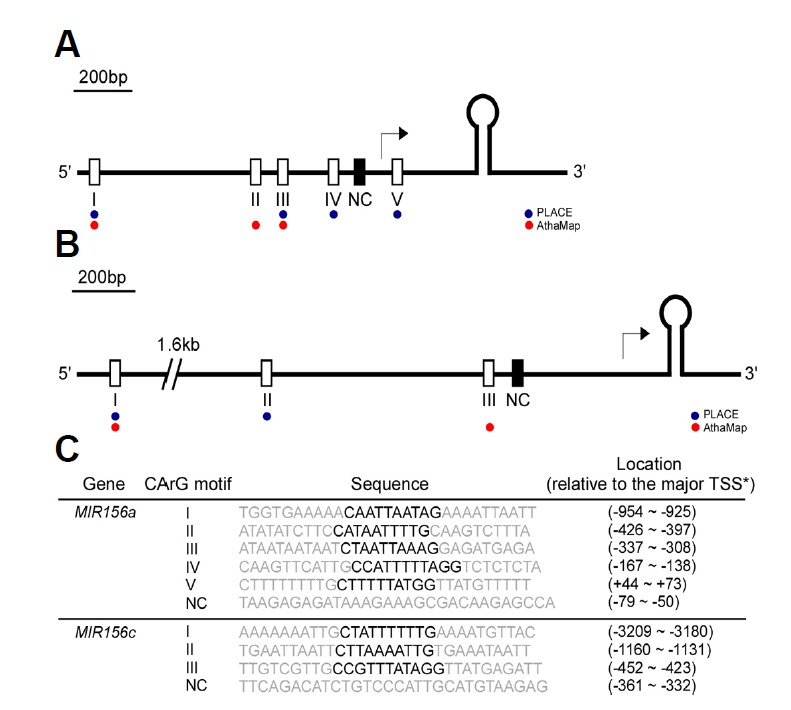
Schematic representation of the locations of CArG motifs within the upstream region of the fold-back structure of miR156a (A) and miR156c (B). The putative CArG motifs predicted by PLACE and AthaMap (Hehl and Bülow, 2014; Higo et al., 1998) are indicated by blue and red dots, respectively. Open and closed boxes indicate putative CArG motifs and negative control (NC), respectively. The major transcription start site (TSS) previously reported (Xie et al., 2005) is denoted with an arrow. (C) Sequence information for the CArG motifs used for probes. Core CArG motifs are marked in bold. TSSs of MIR156a and MIR156c were previously reported (Xie et al., 2005).
To determine whether AGL15 and AGL18 bind to the cis-acting element(s) in the promoters of MIR156a and MIR156c, we first produced GST-tagged, full-length, soluble AGL15-GST (approximately 56 kDa) and AGL18-GST (approximately 55 kDa) (asterisks in Supplementary Fig. S1A). We then purified AGL15-GST and AGL18-GST on glutathione affinity columns. For both recombinant proteins, a single major band eluted from the column at the expected molecular weight (lanes 2 and 4 in Supplementary Fig. S1B).
We used an electrophoretic mobility shift assay (EMSA) to test the possibility that the positive regulation of miR156 expression by AGL15 and AGL18 occurs via direct binding to the cis-acting elements of MIR156a and MIR156c. Purified AGL15-GST and AGL18-GST recombinant proteins and synthesized double-stranded biotin-labeled oligonucleotides of putative CArG motifs were allowed to interact before electrophoresis. AGL15-GST proteins bound to a subset of the CArG motifs in the MIR156a and MIR156c sequences. A shifted band was observed for CArG IV and V of MIR156a and CArG III of MIR156c (Fig. 4A). In contrast, putative CArG motifs I, II, III, and negative control (NC) of MIR156a, and CArG motifs I, II, and NC of MIR156c did not show detectable shifted bands. This suggested that the binding of AGL15-GST protein to the CArG motifs was specific. AGL18-GST produced no apparent band-shifts (Fig. 4B). We further tested the binding of AGL15-GST to the CArG motifs by using unlabeled competitor probes of identical sequence. Our competition assays showed that the addition of unlabeled identical probes greatly reduced the intensity of the shifted bands from CArG motifs IV and V of MIR156a and from CArG motif III of MIR156c (Fig. 4C). Taken together, our results suggest that AGL15 binds to the CArG motifs IV and V of MIR156a and to the CArG motif III of MIR156c to directly regulate the transcription of pri-miR156a and pri-miR156c.
Fig. 4.
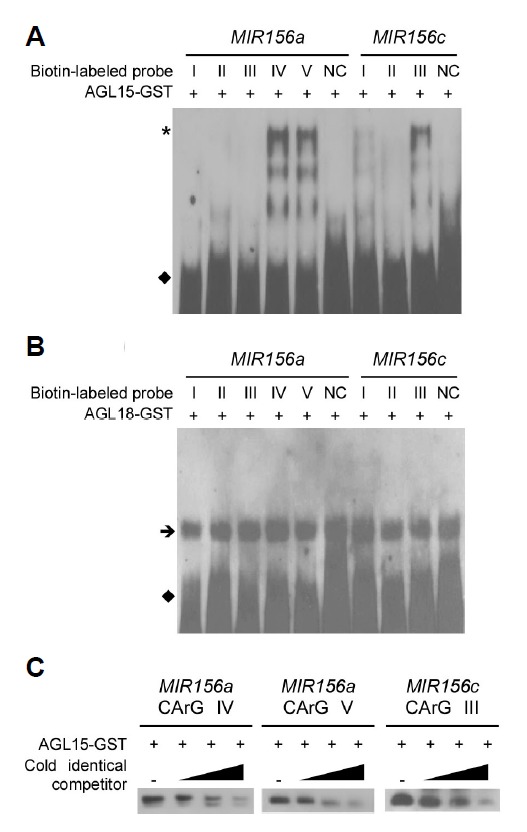
Electrophoretic mobility shift assay (EMSA) with AGL15-GST protein (A) and AGL18-GST protein (B). Shifted bands are indicated with an asterisk. An arrow and closed diamonds indicate non-specific bands and free probes, respectively. (C) Competitor assay of AGL15-GST proteins binding to CArG IV and CArG V motifs (MIR156a) and a CArG III motif (MIR156c). Unlabeled probes of identical sequence were used as competitors at 100X, 500X, and 2500X molar excess. Only shifted bands are shown for simplicity.
Interaction of AGL15 and AGL18
To test the protein-protein interactions between AGL15 and AGL18, we first performed a yeast two-hybrid experiment using pGADT7 and pGBKT7 as bait and prey, respectively. Yeast cells in the two hybrid assay were grown in selective medium lacking leucine and tryptophan (-Leucine -Tryptophan) and assayed for β-galactosidase (β-gal) reporter activity. The result showed that the yeast colonies expressing AGL15 as the prey (i.e., pGAD:AGL15/pGBK:AGL15 and pGAD:AGL15/pGBK18) turned blue in the presence of X-Gal (Fig. 5A; lower panel). When AGL18 was used as the prey (i.e., pGAD:AGL15/ pGBK:AGL18 and pGAD:AGL18/pGBK18), the color development was also observed, but to a lesser extent.
Fig. 5.
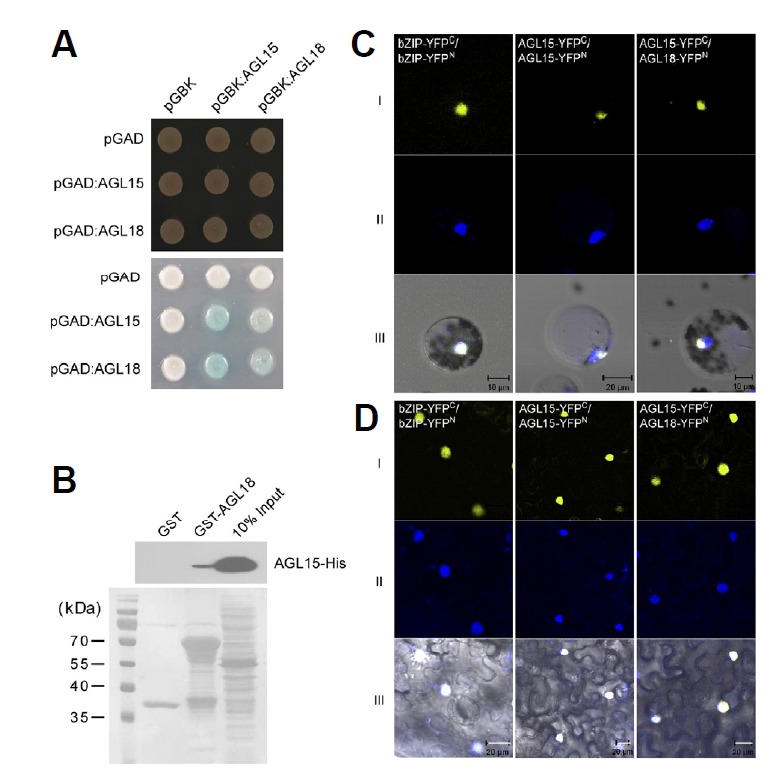
Protein–protein interactions of AGL15 and AGL18 in yeast, in vitro, and in vivo. (A) Protein interactions between AGL15 and AGL18 in yeast two-hybrid analysis. Transformed yeast cells were grown on selective SD/-Leu/-Trp (SD-LT) medium (upper panel) and β-galactosidase assay was performed on SD-LT medium (lower panel). (B) Pull-down assay between AGL15-His and GST-AGL18 proteins. The signals were detected using an anti-His antibody. Proteins stained with Ponceau S are shown below. (C and D) Bimolecular fluorescence complementation using AGL18-YFPN and AGL15-YFPC (right column) (YFPN, N-terminal YFP fragment; YFPC, C-terminal YFP fragment) in Arabidopsis mesophyll protoplasts (C), and tobacco leaves (D). Rows I and II indicate YFP fluorescence and the nucleus stained by 4′, 6-Diamidino-2-phenylindole (DAPI), respectively. Row III indicates the merged image of YFP, DAPI signals, and bright field images. bZIP-YFPN and bZIP-YFPC were used for the positive control (Walter et al., 2004).
To validate the interaction between AGL15 and AGL18, we also conducted in vitro glutathione S-transferase (GST) pull-down assays with recombinant AGL15 and AGL18. We constructed GST-fused AGL18 and His-fused AGL15, expressed these proteins in E. coli, and purified them using glutathione sepharose beads. GST-fused AGL18 and His-fused AGL15 were incubated together, pulled down with glutathione sepharose, and detected by Western blotting. The band signals were detected using anti-His antibody (Fig. 5B). The pull-down assay demonstrated that in vitro synthesized AGL15 interacts with AGL18 (Fig. 5B), but not with GST alone. This result confirmed the heterodimer formation between AGL15 and AGL18 proteins detected in the yeast two-hybrid assay.
In vivo protein-protein interaction
Bimolecular fluorescence complementation (BiFC) assays were conducted to confirm the in vivo AGL15 and AGL18 interaction in Arabidopsis protoplasts. We co-expressed fusion pairs of the C-terminal half of yellow fluorescent protein (YFP) fused to AGL15 (AGL15-YFPC). AGL15 and AGL18 sequences were also fused to the YFP N-terminal half (i.e., AGL15-YFPN and AGL18-YFPN). Co-expression of AGL15-YFPC and AGL18- YFPN resulted in yellow fluorescent signals in Arabidopsis protoplasts (Fig. 5C). The strong YFP fluorescence signals were only observed in the nucleus of the transfected cells, as confirmed by DAPI staining of the nuclei. The BiFC assays also detected self-interaction of AGL15, as the AGL15-YFPC/ AGL15-YFPN transformed cells generated signals. The BiFC experiment was also conducted in tobacco leaves and nucleus-specific signals were observed (Fig. 5D). As a positive control, co-expression of bZIP-YFPC and bZIP-YFPN also produced nuclear signals. These data suggest that the major interaction of AGL15 and AGL18 likely occurs in the nucleus, as expected for transcription factors.
DISCUSSION
The AGAMOUS-like genes AGL15 and AGL18 act together to activate miR156 expression
As AGL15 and AGL18 act redundantly to suppress the floral transition (Adamczyk et al., 2007), we hypothesized that this suppression occurred via activation of miR156, a known floral suppressor (Wu and Poethig, 2006). The down-regulation of miR156 observed only in the double mutants (Fig. 1) indicates that, in addition to their functional redundancy, AGL15 and AGL18 positively co-regulate the expression of miR156. The spatial expression analysis further revealed that the similar expression patterns of AGL15, AGL18, and miR156 are consistent with their possible genetic interactions. The high transcript levels of the three genes in the cotyledon suggest that the cotyledon may be the possible site of their interaction. In the rosette leaf, high expression levels of AGL18 may partially compensate for the low AGL15 expression levels, which result in a decrease in miR156 accumulation. Reduced accumulation of miR156 in cauline leaf and stem are consistent with the reduced AGL15 and AGL18 expression.
The upregulation of floral activator genes (i.e., SPL3, FUL, SEP3, and FT) observed only in the double mutant lines (Fig. 1E) further supports the role of AGL15 and AGL18 as corepressors of the floral transition. MiR156 directly targets SPL3, which is a direct activator of FUL (Yamaguchi et al., 2009), FT, and SEP3 (Lee et al., 2012a); the accumulation of SPL3 transcripts is inversely proportional to the levels of miR156 (Wu and Poethig, 2006). In line with previous reports, our results suggested that upregulation of SPL3 and with its downstream genes (i.e., FUL, FT, and SEP3) is the consequence of the downregulation of miR156 in the double mutant lines. Downregulation of SPL3, FUL, SEP3, and FT can potentially explain the late-flowering phenotype of AGL15 and AGL18 gain-of-function lines (Adamczyk et al., 2007). Histochemical assays showed a reduction of GUS reporter expression in two independent double mutant lines (i.e. agl15-3 agl18-1 and agl15-4 agl18-1). This indicates reduced MIR156 promoter strength in the mutants, implying that AGL15 and AGL18 both affect miR156 expression at the transcriptional level. However, the fact that GUS expression was not completely abolished in the double mutant lines suggested that miR156 expression was partially independent of AGL15 and AGL18 and that other components could affect miR156 expression. The expression levels of both AGL15 and AGL18 were not affected by the overexpression of miR156, suggesting that miR156 does not regulate their expression, and therefore acts downstream of AGL15 and AGL18. The drastic reduction of expression levels of miR156a and miR156c in the double mutant backgrounds implies that their transcriptional regulation directly involves AGL15 and AGL18. The unaltered expression levels of pri-miR156d in agl15 agl18 double mutants is consistent with our observation that pri-miR156d levels were lower than pri-miR156a and pri-miR156c, the two loci that mainly contribute to the production of mature miR156
All together, this evidence strongly suggests that AGL15 and AGL18 act upstream of miR156 to positively co-regulate its transcription. Despite the ambient-temperature responsive nature of miR156 (Lee et al., 2010), neither AGL15 nor AGL18 expression patterns showed a strong correlation with temperature (data not shown). Lee et al. (2007) reported higher expression of SVP, the key factor of the ambient temperature flowering pathway (Lee et al., 2008), in the leaf, whereas AGL15 is expressed more strongly in the cotyledon. This suggests that AGL15 and AGL18 act independently of the ambient temperature pathway, but may be involved in the endogenous aging pathway (Wang et al., 2009).
AGL15 binds to the promoters of MIR156a and MIR156c
Although the members of the MADS-domain protein family interact with similar or identical DNA sequences (Tang and Perry, 2003), our findings showed that AGL15 and AGL18 proteins did not bind to the same CArG motifs. DNA-binding site recognition does not determine the specific physiological role of MADS-domain proteins, as several proteins of the MADS-domain family can bind the same CArG motifs, yet have different developmental roles (Riechmann et al. 1996). Tang and Perry (2003) also reported that AGL15 preferentially binds CArG motifs with a longer A/T-rich core, but we observed that the length of A/T-core of the AGL15-bound motifs was not necessarily longer than those of their counterparts. This implies that factors other than the length of the A/T-rich core (e.g. the sequences flanking the motif) might also contribute to the determination of the preferential binding site(s) for AGL15. Interestingly, in our assays, AGL18 did not seem to directly interact with the promoters of MIR156, as observed in the EMSA experiment (Fig. 4). However, considering that the levels of mature miR156 were downregulated only in agl15 gl18 double mutants, it is possible that AGL18 may have a supporting role in regulation of MIR156. Nevertheless, we still cannot exclude the possibility that AGL18 binds to another factor, which allows it to bind to the MIR156 promoter.
AGL15 forms a heterodimer with AGL18
Our yeast 2-hybrid and in vitro pull-down results indicated that AGL18 interacts with AGL15. In addition, AGL15 also interacted with itself, forming a homodimer (Fig. 5). Co-localization of AGL15 and AGL18 in the nucleus, as observed in the GFP reporter assay (Supplementary Fig. S2), implies that the two proteins likely function at the same site and therefore is consistent with their interaction with each other. The localization of AGL15 in the nucleus is also consistent with a previous report that it is a nuclear protein (Perry et al., 1996). However, we cannot exclude the possiblity that AGL15 and AGL18 may be preferentially localized to the nucleolus, within the nucleus. The in vivo study of the protein-protein interaction further confirmed heterodimer formation between the two proteins. As expected, AGL15 also formed a homodimer in vivo, consistent with a previous study in yeast (Hill et al., 2008). The similar results obtained from tobacco leaves infiltrated with the same constructs provide additional support for the protein-protein interaction. Collectively, our evidence suggests that, although AGL18 does not directly bind to the CArG motifs on MIR156 promoters, it interacts with AGL15, possibly forming a complex that activates MIR156 expression.
Increasing evidence indicates a role for the network of MADS-domain protein members in flowering time control, with individuals contributing more or less to the transition (Yoo et al., 2011). With the fact that neither the phenotype previously reported (Adamczyk et al., 2007) nor the abundance of miR156 of each single mutant in this study resembles those of the double mutants, we suggest that AGL15 and AGL18 function as a complex along with other additional components, which exhibit redundant or additional DNA-binding properties. Therefore, the absence of either AGL15 or AGL18 in the individual single mutant might not be sufficient to cause a major conformational change, and hence the complex’s function as a transcriptional activator is maintained. Further studies are required to identify the other components of the complex.
Based on our findings, we propose a working model (Fig. 6) demonstrating the potential upstream regulation of MIR156 genes by AGL15 and AGL18. These MADS box proteins form a complex, which may include other additional components that directly bind to CArG motifs in the promoters of the MIR156 gene to activate its expression, which subsequently prevents premature floral transition. In binding, AGL15 may act as a molecular surface for the interaction between the complex and MIR156 sequences. Fernandez et al. (2014) reported the direct binding of AGL15 to the FT locus, which contributes to delaying the reproductive phase transition; our evidence additionally suggests that AGL15 and AGL18 delay the floral transition by regulating MIR156, which eventually regulates the expression of multiple flowering genes. Further analysis of the molecular mechanism of the AGL15/18 interaction with the MIR156 sequence will give more information on how flowering time is fine-tuned in Arabidopsis.
Fig. 6.
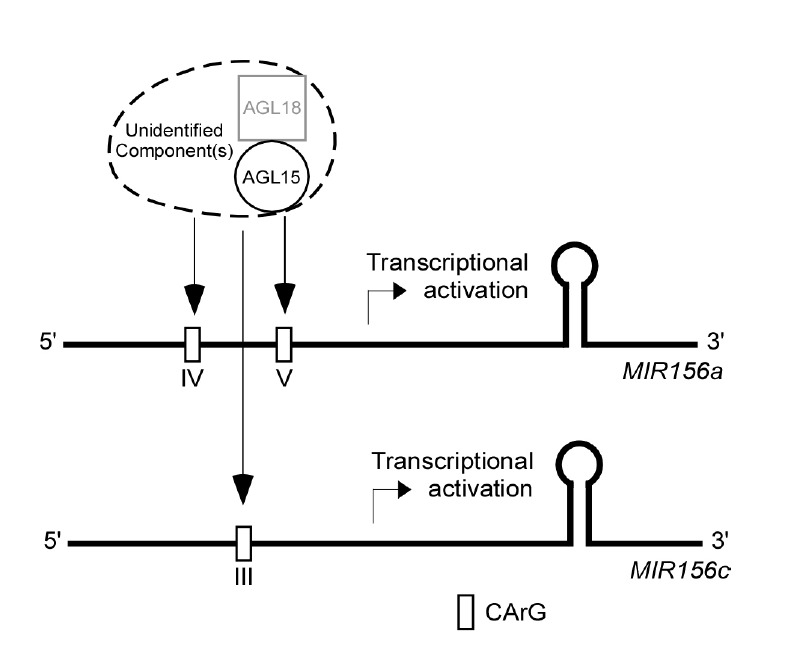
A proposed model for the function of AGL15 and AGL18 in regulating the expression of miR156a and miR156c. AGL15 and AGL18 form a complex, possibly with other unidentified component(s); this complex then binds to the CArG motifs of the MIR156a and MIR156c promoters to activate gene expression.
Acknowledgments
We would like to express our appreciation to Dr. J.H. Lee and Dr. S.Y. Kim for their valuable intellectual and technical support. This work was supported by a National Research Foundation of Korea grant funded by the Korea government (Ministry of Science, ICT, and Future Planning) (2008-0061988) to J.H.A. P.S. was supported by BK21+ program. H.S.R. and W.K. were supported by Korea University.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Adamczyk B.J., Lehti-Shiu M.D., Fernandez D.E. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007;50:1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x. [DOI] [PubMed] [Google Scholar]
- Borner R., Kampmann G., Chandler J., Gleissner R., Wisman E., Apel K., Melzer S. A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J. 2000;24:591–599. doi: 10.1046/j.1365-313x.2000.00906.x. [DOI] [PubMed] [Google Scholar]
- Carrington C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.J., Kim J.J., Lee J.H., Kim W., Jung J., Park C., Ahn J.H. SHORT VEGETATIVE PHASE (SVP) protein negatively regulates miR172 transcription via direct binding to the primiR172a promoter in Arabidopsis. FEBS Lett. 2012;586:2332–2337. doi: 10.1016/j.febslet.2012.05.035. [DOI] [PubMed] [Google Scholar]
- Chung Y., Kwon S.I., Choe S. Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Mol. Cells. 2014;37:795–803. doi: 10.14348/molcells.2014.0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Ferrandiz C., Liljegren S.J., Yanofsky M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- Fernandez D.E., Wang C.T., Zheng Y., Adamczyk B.J., Singhal R., Hall P.K., Perry S.E. The MADS-domain factors AGAMOUS-LIKE15 and AGAMOUS-LIKE18, along with SHORT VEGETATIVE PHASE and AGAMOUS-LIKE24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 2014;165:1591–1603. doi: 10.1104/pp.114.242990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F., Montaigu A., Coupland G. Snapshot: control of flowering in Arabidopsis. Cell. 2010;141:550, e1, 2. doi: 10.1016/j.cell.2010.04.024. [DOI] [PubMed] [Google Scholar]
- Gu X., Wang Y., He Y. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biol. 2013;11:E1001649. doi: 10.1371/journal.pbio.1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hehl R., Bülow L. AthaMap web tools for the analysis of transcriptional and posttranscriptional regulation of gene expression in Arabidopsis thaliana. Methods Mol. Biol. 2014;1158:139–156. doi: 10.1007/978-1-4939-0700-7_9. [DOI] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26:358–359. doi: 10.1093/nar/26.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Wang H., Perry S.E. A transcriptional repression motif in the MADS factor AGL15 is involved in recruitment of histone deacetylase complex components. Plant J. 2008;53:172–185. doi: 10.1111/j.1365-313X.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- Hong S.M., Bahn S.C., Lyu A., Jung H.S., Ahn J.H. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant Cell Physiol. 2010;51:1694–1606. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]
- Jack T. Plant development going MADS. Plant Mol. Biol. 2001;46:515–520. doi: 10.1023/a:1010689126632. [DOI] [PubMed] [Google Scholar]
- Jang Y.H., Park H., Kim S., Lee J.H., Suh M.C., Chung Y.S., Peak K., Kim J. Survey of rice proteins interacting with OsFCA and OsFY proteins which are homologous to the Arabidopsis flowering time proteins, FCA and FY. Plant Cell Physiol. 2009;5:1479–1492. doi: 10.1093/pcp/pcp093. [DOI] [PubMed] [Google Scholar]
- Kutter C., Schob H., Stadler M., Meins F., Si-Ammour A. MicroRNA-mediated regulation of stomatal development in Arabidopsis. Plant Cell. 2007;19:2417–2429. doi: 10.1105/tpc.107.050377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Gene Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Lee J.S., Ahn J.H. Ambient temperature signaling in plants: an emerging field in the regulation of flowering time. J. Plant Biol. 2008;51:321–326. [Google Scholar]
- Lee H., Yoo S.J., Lee J.H., Kim W., Yoo S.K., Fitzgarald H., Carrington J.C., Ahn J.H. Genetic framework for flowering-time regulation by ambient temperature-responsive miRNAs in Arabidopsis. Nucleic Acids Res. 2010;38:3081–3093. doi: 10.1093/nar/gkp1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Kim J.J., Ahn J.H. Role of SEPALLATA3 (SEP3) as a downstream gene of miR156-SPL3-FT circuitry in ambient temperature-responsive flowering. Plant Signal. Behav. 2012a;7:1151–1154. doi: 10.4161/psb.21366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Park S.H., Ahn J.H. Functional conservation and diversification between rice OsMADS22/OsMADS55 and Arabidopsis SVP proteins. Plant Sci. 2012b;185–186:97–104. doi: 10.1016/j.plantsci.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Nesi N., Debeaujon I., Jond C., Stewart A.J., Jenkins G.I., Caboche M., Lepiniec L. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell. 2002;14:2463–2479. doi: 10.1105/tpc.004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.J., Lee J.H., Kim W., Jung H.S., Huijser P., Ahn J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis thaliana. Plant Physiol. 2012;159:461–478. doi: 10.1104/pp.111.192369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry S.E., Nichols K.W., Fernandez D.E. The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell. 1996;8:1977–1989. doi: 10.1105/tpc.8.11.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G.S., Savidge B., Liljegren S.J., Baumann E., Wisman E., Yanofsky M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature. 2003;424:85–88. doi: 10.1038/nature01741. [DOI] [PubMed] [Google Scholar]
- Riechmann J.L., Krizek B.A., Meyerowitz E.M. Dimerization specificity of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. USA. 1996;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio-Somoza I., Weigel D. Coordination of flower maturation by a regulatory circuit of three microRNAs. PLoS Genet. 2013;9:e1003374. doi: 10.1371/journal.pgen.1003374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P., Sharrocks A.D. The MADS-box family of transcription factors. Eur. J. Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- Tang W., Perry S.E. Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J. Biol Chem. 2003;278:28154–28159. doi: 10.1074/jbc.M212976200. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schuutze K., Batistic O., Weckermann K., Nake C., Blazervic D., Grefen C., Schumacher K., Oecking C., et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wang H., Tang W., Zhu C., Perry S.E. A chromatin immunoprecipitation (ChIP) approach to isolate genes regulated by AGL15, a MADS domain protein that preferentially accumulates in embryos. Plant J. 2002;32:831–843. doi: 10.1046/j.1365-313x.2002.01455.x. [DOI] [PubMed] [Google Scholar]
- Wang J., Czech B., Weigel D. MiR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- West A.G., Shore P., Sharrocks A.D. DNA binding by MADS-Box transcription factors: a molecular mechanism for differential DNA bending. Mol. Cell. Biol. 1997;17:2876–2887. doi: 10.1128/mcb.17.5.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Poethig R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133:3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Allen E.A., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. Expression of Arabidopsis MIRNA genes. Plant Physiol. 2005;138:2145–2154. doi: 10.1104/pp.105.062943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi A., Wu M., Yang L., Wu G., Poethig R.S., Wagner D. The MicroRNA-regulated SBP-box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell. 2009;17:268–278. doi: 10.1016/j.devcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.D., Cho Y.H., Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yoo S. K., Wu X., Lee J.S., Ahn J.H. AGAMOUS-LIKE 6 is a floral promoter that negatively regulates the FLC/MAF clade genes and positively regulates FT in Arabidopsis. Plant J. 2011;65:62–76. doi: 10.1111/j.1365-313X.2010.04402.x. [DOI] [PubMed] [Google Scholar]
- Zhang H., Forde B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998;279:407–409. doi: 10.1126/science.279.5349.407. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Cao G., Qu L., Gu H. Characterization of Arabidopsis MYB transcription factor gene AtMYB17 and its possible regulation by LEAFY and AGL15. J. Genet. Genomics. 2009;36:99–107. doi: 10.1016/S1673-8527(08)60096-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


