Abstract
Picornavirus genomes contain internal ribosomal entry sites (IRESs) that promote end-independent translation initiation. Five structural classes of picornavirus IRES have been identified, but numerous IRESs remain unclassified. Here, previously unrecognized Type IV IRESs were identified in members of three proposed picornavirus genera (Limnipivirus, Pasivirus, Rafivirus) and four recognized genera (Kobuvirus, Megrivirus, Sapelovirus, Parechovirus). These IRESs are ∼230–420 nucleotides long, reflecting heterogeneity outside a common structural core. Closer analysis yielded insights into evolutionary processes that have shaped contemporary IRESs. The presence of related IRESs in diverse genera supports the hypothesis that they are heritable genetic elements that spread by horizontal gene transfer. Recombination likely also accounts for the exchange of some peripheral subdomains, suggesting that IRES evolution involves incremental addition of elements to a pre-existing core. Nucleotide conservation is concentrated in ribosome-binding sites, and at the junction of helical domains, likely to ensure orientation of subdomains in an active conformation.
Keywords: evolution, IRES, picornavirus, recombination, RNA structure
INTRODUCTION
The Picornaviridae currently consist of 46 species grouped into 26 genera (International Committee on Taxonomy of Viruses; http://www.ictvonline.org/), but this number is increasing rapidly with the discovery of new viruses, and at least 10 additional genera have been proposed. Picornaviruses have single-stranded, positive-sense RNA genomes and almost all contain a single large open reading frame (ORF) that is divided into three regions: P1 (encoding the capsid proteins) and P2 and P3 (encoding proteins involved in genome replication). The 5’-untranslated region (5’-UTR) contains elements that function in the initiation of translation by internal ribosomal entry and in positive-sense RNA synthesis. Although the order of conserved genes in picornavirus genomes is invariant, there are regions of significant diversity. Members of a few genera encode a leader (L) protein before P1, various proteins are encoded at the 2A locus in the N-terminal region of P2 (Hughes and Stanway, 2000; Johansson et al., 2002; Honkavuori et al., 2011), and the viral genome-lined protein 3B may be present in one to six copies (Wang et al., 2014). The untranslated regions flanking the ORF contain structural elements with defined functions, and also exhibit considerable variability.
Internal ribosomal entry sites (IRESs) are large, structured RNAs that promote ribosomal entry via non-canonical interactions with components of the translation apparatus, including eukaryotic initiation factors (eIFs) and the ribosomal 40S subunit. Picornavirus IRESs are classified into a few distinct types, each with a common structural core that is maintained by sequence co-variation. Each class of IRES has distinctive factor requirements (Pestova et al., 1996; Lomakin et al., 2000; Pisarev et al., 2004; Yu et al., 2011 Sweeney et al., 2012, 2014). The IRESs in the 5’UTRs of poliovirus (genus Enterovirus) and encephalomyocarditis virus (EMCV; genus Cardiovirus) epitomize Type I and Type II IRESs, respectively. The sole Type III IRES occurs in Hepatitis A virus (genus Hepatovirus)(Brown et al., 1994); Type V IRESs were recently identified in members of the genera Kobuvirus, Salivirus, and Oscivirus (Yu et al., 2011; Sweeney et al., 2012). Type IV IRESs were discovered in Hepatitis C virus (HCV) and Classical swine fever virus (CSFV), members of the genera Hepacivirus and Pestivirus of Flaviviridae, but have subsequently also been identified in several picornavirus genera, including Aquamavirus, Avihepatovirus, Kobuvirus, Megrivirus, Sapelovirus, Senecavirus, Teschovirus, and Tremovirus (Pisarev et al., 2004; Chard et al., 2006a; Hellen and de Breyne, 2007; Kapoor et al., 2008; Reuter et al., 2009; Honkavuori et al., 2011), in the proposed genera Aalivirus (Wang et al., 2014), Anativirus (duck picornavirus TW90A; Hellen and de Breyne, 2007; Tseng and Tsai, 2007), Colbovirus (pigeon picornavirus A (PiPV-A) and PiPV-B; Kofstad and Jonassen, 2011), Kunsagivirus (Boros et al., 2013), Phacovirus (quail picornavirus 1 (QPV-1); Pankovics et al., 2012) and Sakobuvirus (Ng et al., 2014), and in other species, including Bat picornavirus 1 (BPV-1), BPV-2 (Lau et al., 2011) and feline picornavirus 1 (FePV-1)(Lau et al., 2012), that have been provisionally assigned to the genus Sapelovirus.
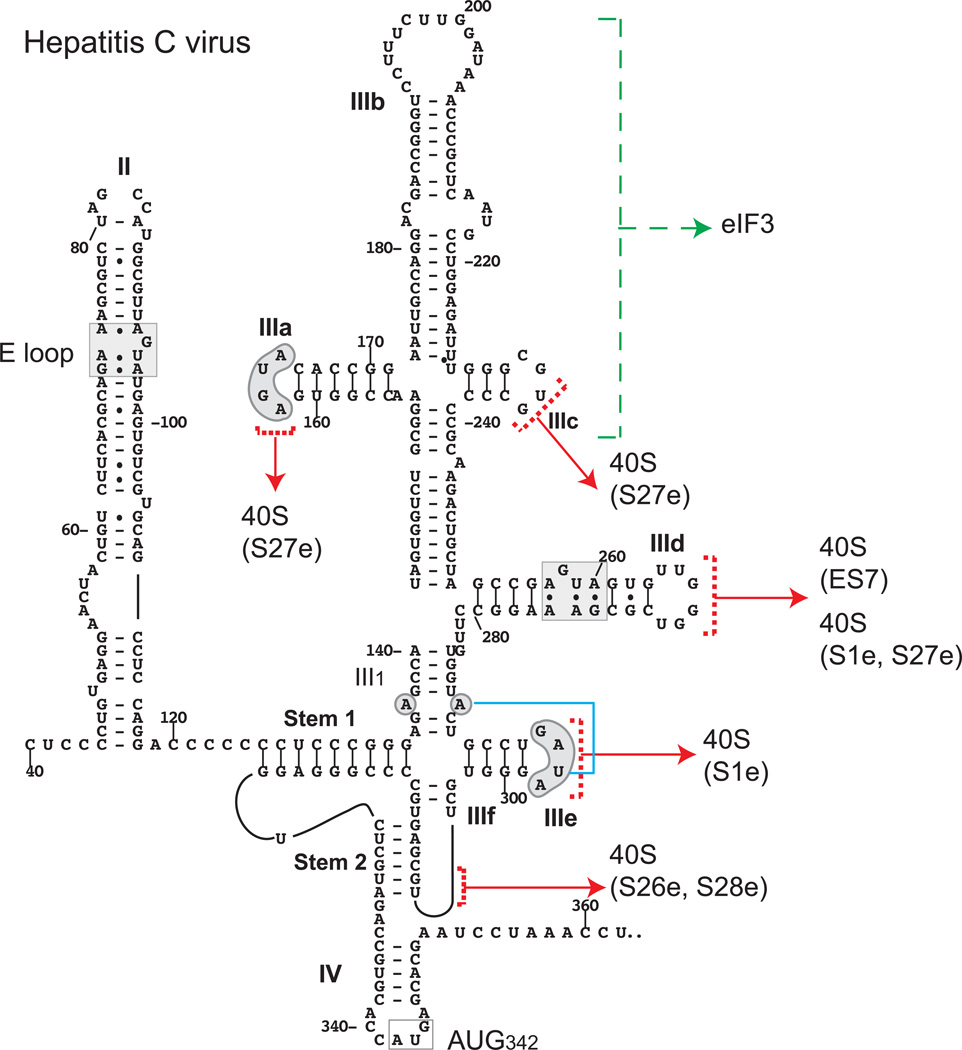
The structural core of Type IV HCV and CSFV IRESs comprises domain II (which is a ∼60–75-nt long, flexible hairpin) and domain III (which consists of branched hairpins IIIa-IIIf and a pseudoknot (PK) that incorporates domain IIIf) (Fig. 1). Pestivirus IRESes contain an additional domain (IIId2) between domains IIId1 and IIIe; the HCV IRES contains a weak additional domain (IV) that harbors the initiation codon (Honda et al., 1996). The picornavirus type IV IRESs identified to date all contain the PK-IIId-IIIe core, including an apical GGG motif in domain IIId and a GA[U/C]A tetraloop in domain IIIe, but apical elements of domain III may be truncated or even absent. Domain II contains an internal loop near its base that enables it to switch between bent and extended conformations, and a ‘loop E’ motif (Lukavsky et al., 2003; Hellen and de Breyne, 2007; Boerneke et al., 2014; Ng et al., 2014; Wang et al., 2014). Very few picornavirus type IV IRESs contain an equivalent of domain IV (Hellen and de Breyne, 2007).
Figure 1. Model of the Type IV IRES of Hepatitis C virus.
The model shows the IRES of HCV genotype 1 (NCBI Reference Sequence: NC_004102.1). Nucleotides are numbered at 20 nt. intervals. The nomenclature of domains is as in [Hellen and de Breyne, 2007], and the initiation codon for the viral polyprotein (AUG342) is boxed and labeled. The ‘loop E’ motif in domain II (Lukavsky et al., 2000) is labeled, and it and sequence motifs in domain III that are conserved in some or all other Type IV IRESs (such as the apical loop in domain IIIa, the conserved apical loops of domains IIId and IIIe and the unpaired purine residue in helix III1) are indicated by grey shading. HCV domain IIId also contains an ‘loop E’ motif (indicated by grey shading), but an equivalent structure does not occur at this position in other Type IV IRESs. The model has been annotated to show interactions of domain III of Type IV IRESs with eIF3 (green arrows) (Sizova et al., 1998; Kieft et al., 2001; Hashem et al., 2013; Sun et al., 2013) and with 18S rRNA ES7 and ribosomal protein S1e, S26e, S27e and s28e components of the 40S ribosomal subunit (red arrows) (Boehringer et al., 2005; Hashem et al., 2013; Matsuda and Mauor, 2014). The tertiary base-pairing interaction between the unpaired purine residue in helix III1 and loop IIIe is indicated by a blue line.
Biochemical studies have established that Type IV IRESs from members of the Flaviviviridae and Picornaviridae use the same mechanism for initiation. The IRES binds to the ribosomal 40S subunit independently of eIFs via sites (Fig. 1) that include the GGG motif in domain IIId, domain IIIe, and elements of the pseudoknot (Pestova et al., 1998; Pestova and Hellen, 1999; Kolupaeva et al., 2000a, b; Kieft et al., 2001; Pisarev et al., 2004; de Breyne et al., 2008; Hellen, 2009; Berry et al., 2010, 2011; Hashem et al., 2013). The apical region of domain III binds eIF3 (Sizova et al., 1998; Hashem et al., 2013; Sun et al., 2013) (Fig. 1), displacing it from its site on the 40S subunit (Hashem et al., 2013), and initiator tRNA is recruited to the peptidyl (P) site of the IRES-bound 40S subunit by eIF2 (Pestova et al., 1998). This mechanism therefore functions without a requirement for the cap-binding complex eIF4F. Domain II induces conformational changes in the 40S subunit, stabilizes binding of mRNA in the ribosomal mRNA-binding cleft, enhances eIF5-mediated hydrolysis of eIF2-bound GTP and promotes joining of a 60S subunit to form an elongation-competent 80S ribosome (Locker et al., 2007; Pestova et al., 2008; Filbin et al., 2013; Yamamoto et al., 2014). These functions of domain II are thought to be related to its ability to switch between bent and extended conformations, possibly in response to interactions with the ribosome (Boerneke et al., 2014). The dependence on domain II differs in different IRESs, and for example, enhances but is not essential for initiation on the CSFV IRES (Kolupaeva et al., 2000a; Fletcher et al., 2002; Friis et al., 2012; Pestova et al., 1998).
The presence of related IRESs in unrelated viruses and adjacent to unrelated coding sequences suggests that they have been ‘captured’ following recombination-mediated lateral gene transfer. A growing number of inter-species and intra-species recombination events involving picornavirus 5’UTRs has been reported, for example leading to exchange of Type 1 IRESs between members of the genus Enterovirus and of Type 2 IRESs between members of the genus Parechovirus (e.g. Zoll et al., 2009; Benschop et al., 2010; McIntyre et al., 2010; Yozwiak et al., 2010). Recombination has also been proposed to account for the juxtaposition of Type V IRESs against unrelated nucleotide sequences in various members of the genera Kobuvirus, Salivirus, and Oscivirus (Sweeney et al., 2012). Further detailed analysis of picornavirus IRESs would likely yield additional insights into processes that have shaped their current structures.
The nature of the IRESs in the 5’UTRs of numerous recently identified picornaviruses representing novel proposed genera have not yet been characterized, and models of several candidate Type IV IRESs are incomplete. We therefore analyzed these sequences, identifying Type IV IRESs in novel members of the genera Kobuvirus, Parechovirus and Sapelovirus, and of the proposed genera Limnipivirus, Pasivirus, Rafivirus and Megrivirus. The unexpectedly wide distribution of Type IV IRESs in the Picornaviridae, and their structural diversity provide insights into the evolution of this important class of noncoding RNA.
METHODS
Sequences
Sequences were analyzed from the following picornaviruses (name followed by Genbank accession number): aalivirus A1 (KJ000696); avian encephalomyelitis virus (AJ225173.1); avian sapelovirus 1 (formerly duck picornavirus TW90A) (AY563023); bat picornavirus 1 strains NC16A and LMH22A (HQ595340 and HQ595341); bat picornavirus 2 strains MH9F and SK17F (HQ595342 and HQ595343); bat picornavirus 3 strains TLC5F and TLC21F (HQ595344 and HQ595345); Bluegill picornavirus (JX134222); California sea lion sapelovirus 1 (JN420368); Caprine kobuvirus strain black goat/12Q108/KOR/2012 (KF793927); Carp picornavirus 1 (KF306267); chicken megrivirus (ChMV) strains chicken/B21-CHV/2012/HUN (KF961186), chicken/CHK-IV-CHV/2013/HUN (KF961187) and chicken/27C/Hong Kong (KF979336); duck hepatitis A virus 1 (DHAV-1)(e.g. KF924552); duck megrivirus (DuMV) strain du/LY/China (KC663628); Fathead minnow picornavirus strains fhm/20/IL/USA/2010, fhm/1/MN/USA/2010, fhm/2/MN/USA/2010 and fhm/09-283/MT/USA/2009 (KF874490, KF183915, KF183916, and KC465953); Feline picornavirus (FePV-1) strains 073F, 127F and 356F, 661F and 1021F (JN572115, JN572116, JN572117, JN572118 and JN572119); Feline sakobuvirus A (KF387721); Ferret kobuvirus strains ferret/MpKoV38/NL/2010, ferret/MpKoV39/NL/2010 and ferret/MpKpV32/NL/2010 (KF006985, KF006986 and KF006987); Ferret parechovirus (FPV) (KF006989); Ia io picornavirus 1 (JQ814852); Kunsagivirus A (KV-A1) strain roller/SZAL6-KuV/2011/HUN (KC935379); Ljungan virus strain 87-012G (EF202833) and strain 174F (AF327921); Manhattan parechovirus isolate MPeV/NYC-A11 (KJ950935); Melegrivirus A (turkey hepatitis virus 1 (THV1)) strains turkey/2993D/CA/USA/c.2009 (HM751199), turkey/0091.1/CA/USA/c.2009 (HQ189775), turkey/B407-THV/2011/HUN (KF961188) and chicken/5C/Hong kong (KF979335); mesivirus 1 (MeV-1) strain pigeon/HK-21/2011/HKN (KC876003) and mesivirus 2 (MeV-2) (pigeon/GALII-5/2011/HUN) (KC811837); Ovine kobuvirus 1 (OKV-1)(Aichivirus B)(GU245693); pasivirus 1 (PaV-A1) strains swine/France/2011 (JQ316470), swine/China/2010 (JX491648), Zsana1/2013/HUN (KM259923), Rom1 (KJ780020), Rom2 (KJ780021) and Rom3 (KJ780022); Pigeon picornavirus B (PiPV-B) (FR727144 and KC560801); Porcine kobuvirus strain swine/S-1-HUN/2007/Hungary (NC_011829) and other isolates (AB624493, GQ249161, GU292559, JN630514, JQ692069, JX177612, JX401523, JX827598, KC204684, KC424638, KC424639, KC424640, KF539763, KF695124, KF814661, KF814662, KF814663, KF814664, KF814665, KF814666, KJ452348); Porcine sapelovirus 1 (formerly, Porcine enterovirus 8) strain V13 (AF406813); Porcine teschovirus 1 strain Teschen-Bozen 654 (AF231767); quail picornavirus 1 (QPV-1) strain quail/HUN/2010 (JN674502); seal aquamavirus (EU142040); Simian sapelovirus 3 (formerly, simian picornavirus type 9) (AY064717) and Tortoise rafivirus A1 (TRaV-1) strain tortoise/UF4/USA/2009 (KJ415177).
Sequence alignment
Viruses containing Type IV IRES-like sequences in their 5’ UTRs were identified using BLAST searches (http://www.ncbi.nlm.nih.gov/BLAST/) of the GenBank database. Searches done with long sequences (>100 nt) used the following parameters: E, 10; word size, 11; reward match/mismatch, 1/−1; gap extension, 0; and gap penalty, 2. IRES sequences were aligned using EMBOSS Matcher (http://www.ebi.ac.uk/Tools/psa/emboss_matcher/nucleotide.html) and protein sequences were aligned using EMBOSS Needle (http://www.ebi.ac.uk/Tools/psa/emboss_needle/) and CLUSTAL W (http://www.ebi.ac.uk/Tools/msa/clustalw2/), using default parameters.
Modeling of secondary and tertiary RNA structures
Secondary structure elements were modeled using complementary posterior decoding (CentroidFold; http://www.ncrna.org/centroidfold) (Sato et al., 2009) and free energy minimization (Mfold (http://mfold.rna.albany.edu/?q_mfold) (Zuker, 2003) and tertiary structures were modeled using pKnotsRG (http://bibiserv.techfak.uni-bielefeld.de/pknotsrg/submission.html) (Reeder et al., 2007) and its successor, pKiss (http://bibiserv2.cebitec.uni-bielefeld.de/pkiss)(Jansssen and Giegerich, 2015), in all instances using default parameters. Models were visualized using Pseudoviewer3 (http://pseudoviewer.inha.ac.kr/)(Byun and Han, 2009). The quality of structural models was assessed by determining their compatibility with sequence variation between virus strains and isolates. The potential of non-Watson-Crick base-pairs to form the ‘loop E’ motif in domain II was evaluated using isostericity matrices (Leontis et al., 2002; Zhong et al., 2006).
RESULTS
Type IV IRESs in the proposed genus Limnipivirus
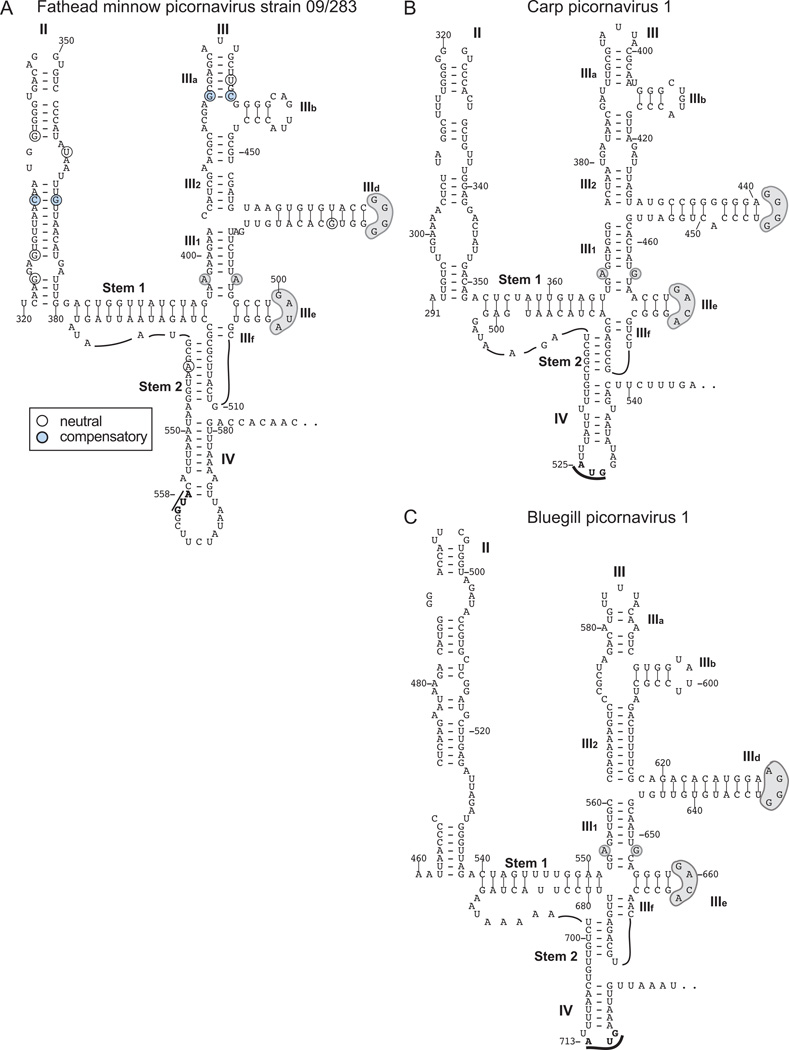
The structures of the IRESs of Bluegill picornavirus (BGPV-1) (Barbknecht et al., 2014), Carp picornavirus 1 (CPV-1)(Lange et al., 2014) and Fathead minnow picornavirus (FHMPV) (Phelps et al., 2014), three species in the proposed genus Limnipivirus, are not known. BLAST searches identified 47–102 nt-long elements with 64–74% identity to the FHMPV 5’UTR in the 5’UTRs of porcine sapelovirus 1, bat picornavirus 2 (BPV-2), Feline sakobuvirus A (SakV-A1) and Ia io picornavirus 1 (IiPV-1). These 5’UTRs contain Type IV IRES structures (Hellen and de Breyne, 2007; Lau et al., 2011, 2012; Ng et al., 2014), and the sequence homology with them maps to domain IIIe and flanking regions. A shorter (11–14nt-long) segment of the FHMPV 5’UTR matched precisely to domain IIIe in the Type IV IRESs of avian encephalomyelitis virus (AEV), duck hepatitis A virus 1 (DHAV-1) and Pigeon picornavirus B (PiPV-B) (Hellen and de Breyne, 2007; Kofstad and Jonassen, 2011) and to segments of the 5’UTRs of California sea lion sapelovirus 1 (Csl SapV-1), Ferret parechovirus (FPV) and Pasivirus 1 (PaV-A1).
These observations suggest that the FHMPV 5’UTR contains a Type IV IRES. We therefore derived models of the 3’-terminal regions of FHMPV, BGPV-1 and CPV-1 5’UTRs using the same or similar methods to those used to model Type V and other Type IV IRESs (Hellen and de Breyne, 2007; Yu et al., 2011; Sweeney et al., 2012). The predicted structure of this part of the FHMPV 5’UTR resembled Type IV IRESs (Fig. 2A) in containing a basal pseudoknot, domains IIId and IIIe and their conserved apical loops. Domain II is similar in size to HCV domain II but has an unrelated sequence. Sequence variation in the available FHMPV sequences is structurally neutral (Fig. 2A). The BGPV-1 and CPV-1 5’UTRs contain Type IV IRESs that are structurally similar to the FHMPV IRES (Figs. 2B, 2C): domain II in each is distinct, but they both have a ‘PK-IIId-IIIe’ core, and a similarly truncated apex of domain III. Members of Limnipivirus thus have Type IV IRESs that are severely truncated relative to e.g. HCV (with domain III in CPV-1 (156nt) being amongst the smallest identified to date), but neverheless contain the key determinants of ribosome binding. These IRESs all have the potential to form weak domain IV-like hairpins (ΔG = −1.5 – −2.0 kcal/mol) that sequester the initiation codon.
Figure 2. Type IV IRESs in members of the proposed genus Limnipivirus.
Models of the structures of the IRESs of fathead minnow picornavirus (FHMPV) strain 09/283 (A), carp picornavirus 1 (CPV-1) (B) and bluegill picornavirus 1 (BGPV-1) (C). The initiation codons for the viral polyproteins are bold and underlined. The nomenclature of domains is as in [Hellen and de Breyne, 2007]. Nucleotides are numbered at 20-nt. intervals. Nucleotide differences between FHMPV strains 09/283 and isolate 20, FHMPV-1 and FHMPV-2 are as indicated in the inset key, and are classified as neutral if they occur at unpaired positions or do not disrupt structure, and as compensatory if substitutions of paired nucleotides lead to formation of a different base-pair. Sequence motifs in domain III of FHMPV, CPV-1 and BGPV-1 IRESs that are conserved in other Type IV IRESs are indicated by grey shading.
Related Type IV IRESs in members of the Parechovirus and proposed Pasivirus genera
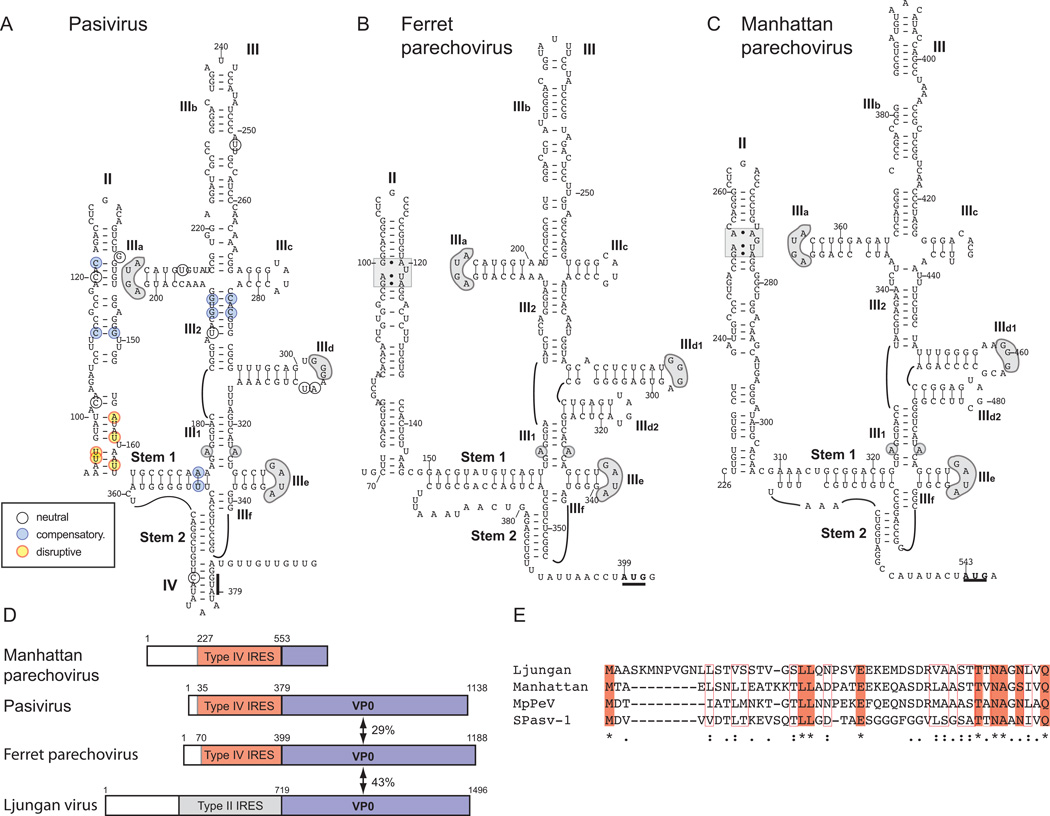
The presence of sequences identical to domain IIIe in picornavirus 5’UTRs that have not been characterized structurally, such as that of pasivirus A (PaV-A1) of the proposed genus Pasivirus (Sauvage et al., 2012; Yu et al., 2013) suggested that they might also contain type IV IRESs. Modeling of the PaV-A1 5’-UTR identified a Type IV IRES, and its predicted structure was strongly supported by the pattern of sequence variation within it (Fig. 3A). Domain II is similar in size to HCV domain II but lacks a ‘loop E’ motif; domain III resembles the HCV equivalent, and like it contains conventional IIIa, IIIb and IIIc domains that form a four-way junction with helix III2, as well as a pseudoknot, domain IIId and domain IIIe. Domain IIIa has an apical ‘AGUA’ loop like that in the HCV IRES, domain IIId has the universal apical GGG motif and domain IIIe has an apical GAUA tetraloop. Domain IV sequesters the initiation codon, as in the HCV IRES.
Figure 3. Related IRESs in Pasivirus A1, Ferret parechovirus and Manhattan parechovirus 5’UTRs.
Models of the structures of the IRESs of (A) Pasivirus A1 (PaV-1) isolate swine/France/2011, (B) Ferret parechovirus (FPV) isolate ferret/MpPeV1/NL and (C) Manhattan parechovirus (MPeV). The initiation codons for the viral polyproteins are bold and underlined, and the nomenclature of domains is as in [Hellen and de Breyne, 2007]. Nucleotide differences between the six current PaV-1 strains are assigned to different classes as indicated in the inset key, and as defined in the legend for Fig. 2. Sequence motifs in domain III of PaV-1, FPV and MPeV IRESs that are conserved in other Type IV IRESs (such as the conserved apical loops of domains IIId and IIIe and the unpaired purine residue in helix III1) are indicated by grey shading, as is an ‘E loop’ motif in domain II of FPV and MPeV IRESs. (D) Schematic representations of the 5’-UTRs and adjacent VP0 coding regions of MPeV, PaV-1 (isolate swine/France/2011), FPV (isolate ferret/MpPeV1/NL) and Ljungan virus strain 174F are divided to show the Type IV IRES in MPeV, PaV-1 and FPV (orange), the Type II IRES in Ljungan virus (grey) and the VP0 coding regions (blue). (E) Sequence alignment of the N-terminal regions of the VP0 proteins of PaV-1, FPV, MPeV and Ljungan virus. Identical amino acid residues are indicated by shading and asterisks, and related amino acid residues are indicated by open boxes and colons.
The entire PaV-A1 IRES is homologous to elements of similar size in the 5’UTRs of Ferret parechovirus (FPV)(59% identity) and Manhattan parechovirus (MPeV) (57% identity). The FPV and MPeV 5’UTRs contain IRESs that are closely related to that of PaV-A1, except that they have ‘E loop’ motifs in domain II, an additional hairpin (domain IIId2), a longer and imperfectly base-paired Stem 1 in the pseudoknot, and in FPV, an extended loop between helices in the pseudoknot (Figs. 3A-3C). Neither IRES forms a stable domain IV. Elements analogous to domain IIId2 occur in the Type IV IRESs of pestiviruses (e.g. CSFV) and Seneca valley virus (a picornavirus in the genus Senecavirus)(Hellen and de Breyne, 2007). The sequence adjacent to FPV, MPeV and PaV-A1 IRESs encodes the VP0 capsid protein (Sauvage et al., 2012; Yu et al., 2013, Smits et al., 2013; Firth et al., 2014), but sequence identity between these moieties is low (∼30%), consistent with assignment of these viruses to different genera, whereas FPV VP0 is more similar (43% identity) to that of Ljungan virus 1 of the genus Parechovirus (Fig. 3D). The presence of unrelated IRESs (type IV IRESs in FPV, MPeV1 and PaV-A1, and a type II IRES in e.g. Ljungan virus and other parechoviruses (Nateri et al., 2000) adjacent to related VP0 sequences suggest that these Type IV IRESs may have been acquired by recombination. The N-terminal truncation of FPV, MPeV and PaV-A1 VP0 relative to VP0 from Ljungan virus (Fig. 3E) suggests that a recombination breakpoint may be located in this region of the genome.
Type IV Pasivirus and Parechovirus IRESs are thus related, and diverge principally in peripheral regions such as domain II, by the presence or absence of domain IIId2, and by whether or not they form domain IV.
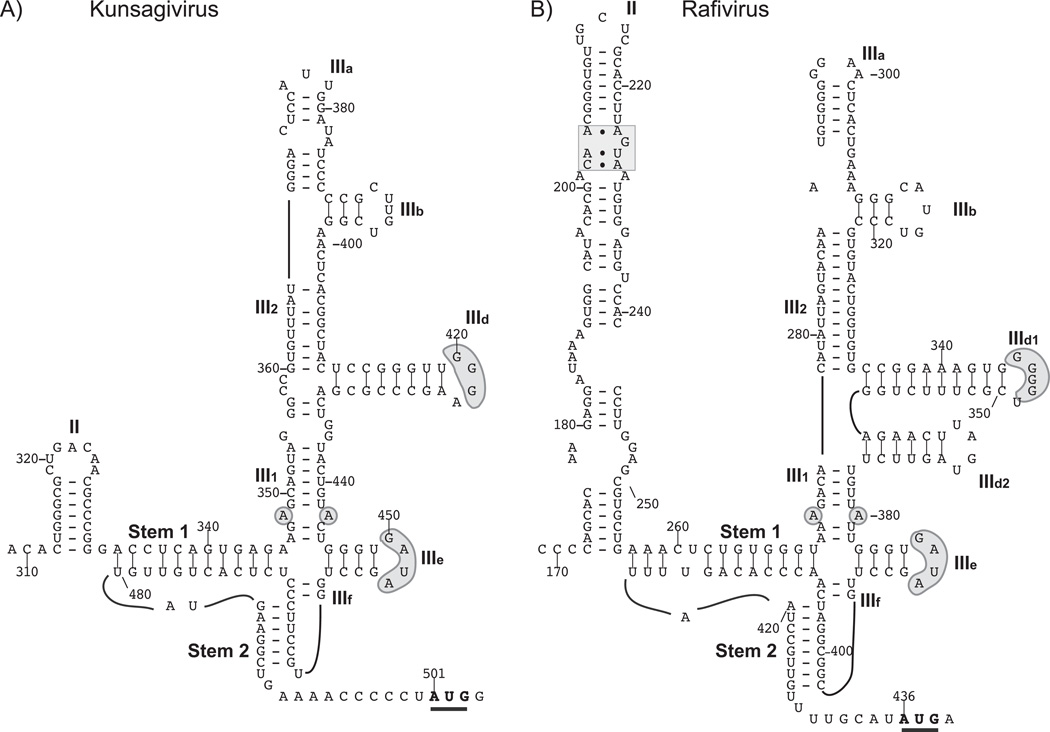
A Type IV IRES in tortoise rafivirus A1 (TRaV-1) in the proposed genus Rafivirus
A fragment of the tortoise rafivirus A1 (TRaV-1) 5’UTR is very similar to segments of Type IV IRESs from avian picornaviruses including nt. 361–408 of the avian sapelovirus 1 5’UTR (73% identity), nt. 441–497 of the quail picornavirus 1 (QPV-1) 5’UTR (73% identity) and nt. 442–481 of the Kunsagivirus A (KV-A) 5’UTR (55% identity). These segments encompass domain IIIe and downstream elements of the pseudoknot. Modeling confirmed the presence of a Type IV IRES in the TRaV-1 5’UTR (Fig. 4B) that closely resembles the KV-A IRES (Fig. 4A), except for an additional domain (IIId2) between domains IIId1 and IIIe and the presence of a conventional rather than a truncated domain II. TRaV-1 domain III (164nt long) is nevertheless very compact and comparable in size to domain III of KV-A (155nt), BGPV-1 (162nt), FHMPV (164nt) and DHV (168nt). As in the Limnipivirus IRESs (Figs. 2A-2C), and the DHV, PTV and KV-A IRESs (Hellen and de Breyne, 2007; Boros et al., 2013), the apex of domain III is truncated and comprises two short hairpins.
Figure 4. Type IV IRESs in members of the proposed genera Kunsagivirus and Rafivirus.
Models of the structures of the IRESs of (A) Kunsagivirus (as proposed in (Boros et al., 2013) and (B) Tortoise rafivirus A1 strain tortoise/UF4/USA/2009. The initiation codons for the viral polyproteins are bold and underlined. The nomenclature of domains is as in [Hellen and de Breyne, 2007]. Nucleotides are numbered at 20nt. intervals and key sequence motifs in domains II and III that are conserved in other Type IV IRESs are indicated by grey shading.
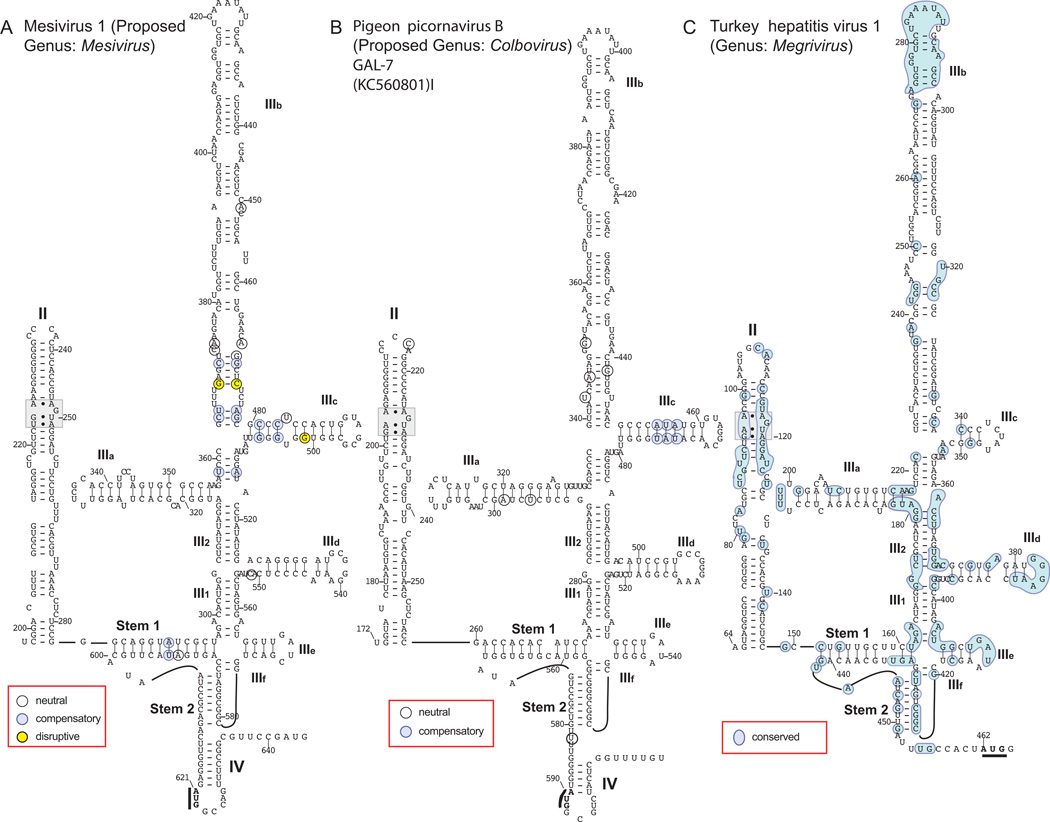
Related Type IV IRESs in members of the genus Megrivirus and the proposed genus Colbovirus
Type IV IRESs have been identified for which models are unavailable or incomplete (a) in pigeon picornavirus B (PiPV-B), of the proposed genus Colbovirus (Kofstad and Jonassen, 2011), (b) in Turkey hepatitis virus 1 (THV1) (Honkavuori et al., 2011; Boros et al., 2014a), of the genus Megrivirus, and (c) in mesiviruses 1 and 2 (Phan et al., 2013), which may be designated either as a new species in the genus Megrivirus or as the prototype of a new genus, Mesivirus. The shorter (620nt-long) 5’UTR of mesivirus 1 is 91% identical to that of mesivirus 2. The sequence differences between them strongly support a model (Fig. 5A) in which domain II (86nt long) contains opposed GAA/AGUA ‘loop E’ motifs, the ‘PK-IIId-IIIe’ core has a canonical size and near-canonical structure, and domain IIId has an apical GGG motif. However, domains IIIa and IIIc branch in a staggered manner from the helix III1-III2-IIIb axis, domain IIIe is exceptional in that its apical loop (GAUC) does not form a GA[U/C]A tetraloop, and domain IIIb (118nt-long) is very elongated in comparison to the equivalent domain in most other Type IV IRESs. Nucleotides downstream of the pseudoknot have the potential to form domain IV.
Figure 5. Type IV IRESs in members of the proposed genera Colbovirus, Megrivirus and Mesivirus.
Models of the structures of the IRESs of (A) Mesivirus 1 strain pigeon/HK-21/2011/HKN (proposed genus Mesivirus), (B) pigeon picornavirus A strain pigeon/Norway/03/603-7/2003 and (C) melegrivirus A (turkey hepatitis virus 1) strain turkey/2993D/CA/USA/c.2009. The initiation codons for the viral polyproteins are bold and underlined. The nomenclature of domains is as in [Hellen and de Breyne, 2007]. Nucleotides are numbered at 20nt. intervals; nucleotide differences between (A) Mesivirus 1 and Mesivirus 2, (B) PiPV-B strains pgeon/Norway/03/641/200 and pigeon/Hungary/GAL-7/2010 are as indicated in the inset keys. Shading of nucleotides in panel (C) indicates nucleotides that are conserved in all ten available sequences of members of the genera Colbovirus, Megrivirus and Mesivirus.
A previous study of the PiPV-B 5’-UTR modeled only the ‘PK-IIId-IIIe’ core and adjacent base of domain II (Kofstad and Jonassen, 2011). A more complete analysis (Fig. 5B) indicates that the base of domain II extends closer to the pseudoknot than previously proposed and contains a ‘loop E’ motif. Domain III closely resembles that of Mesivirus (Fig. 5A) with respect to the pseudoknot and the size and apical loops in domains IIId and IIIe, in the staggered branching of domains IIIa and IIIc, and the elongated nature of domain IIIc (112nt long). In contrast to mesiviruses, PiPV-B domain IIIe is capped by a canonical GAUA tetraloop.
The Type IV IRES in THV1 (Fig. 5C) also adopts a structure very like that proposed here for Mesivirus 1 (Fig. 5A) and elsewhere for the Type IV IRESs in chicken megrivirus (Boros et al., 2014a) and duck megrivirus (Liao et al., 2014). Analysis of the ten current Megrivirus and Mesivirus sequences in which domains II and III are complete or nearly complete indicates that nucleotide substitutions are almost all highly co-variant or structurally neutral, indicating that IRESs in members of the genus Megrivirus maintain a common structure.
Although absolutely conserved nucleotides are dispersed throughout domains II and III in these sequences, they localize to only a few regions. These include the characateristic motifs in domain II (‘loop E’), domain III (GGG, GA[U/C]A tetraloop) and elements of the pseudoknot. Other conserved nucleotides localize (a) to the region surrounding the domain II E-loop, (b) to the base of an internal loop in domain IIIb and the junctions of (b) helices III1, IIIe and IIIf, (d) III1, IIId and III2 and (e) III2 and IIIa. Sequence conservation at these positions suggests that IRES function requires maintenance of the correct spatial orientation of domains. The most strongly conserved sequence is at the apex of domain IIIb: it occurs in all members of the genus Megrivirus, and in the proposed genera Aalivirus, Aquamavirus, Avihepatovirus, Colbovirus and Phacovirus (Kapoor et al., 2008; Kofstad and Jonassen 2011; Pan et al., 2012; Pankovics et al., 2012; Phan et al., 2013; Boros et al., 2014a, b; Liao et al., 2014; Wang et al., 2014). The level of sequence identity of this apical “8”-like structure is remarkable, ranging from perfect identity over 39nt without interruption between THV1 (HM751199) and aalivirus A1 (AalV-1) to identity over a 20nt. core in almost all the rest of these viruses.
Type IV IRESs in members of Megrivirus and Colbovirus genera therefore have a conserved core containing canonical determinants of ribosome binding, but have staggered rather than symmetrically branching domains IIIa and IIIc, and a greatly elongated domain IIIb capped by a highly conserved ‘8-like’ structure that recurs in the Type IV IRESs of diverse avian picornaviruses.
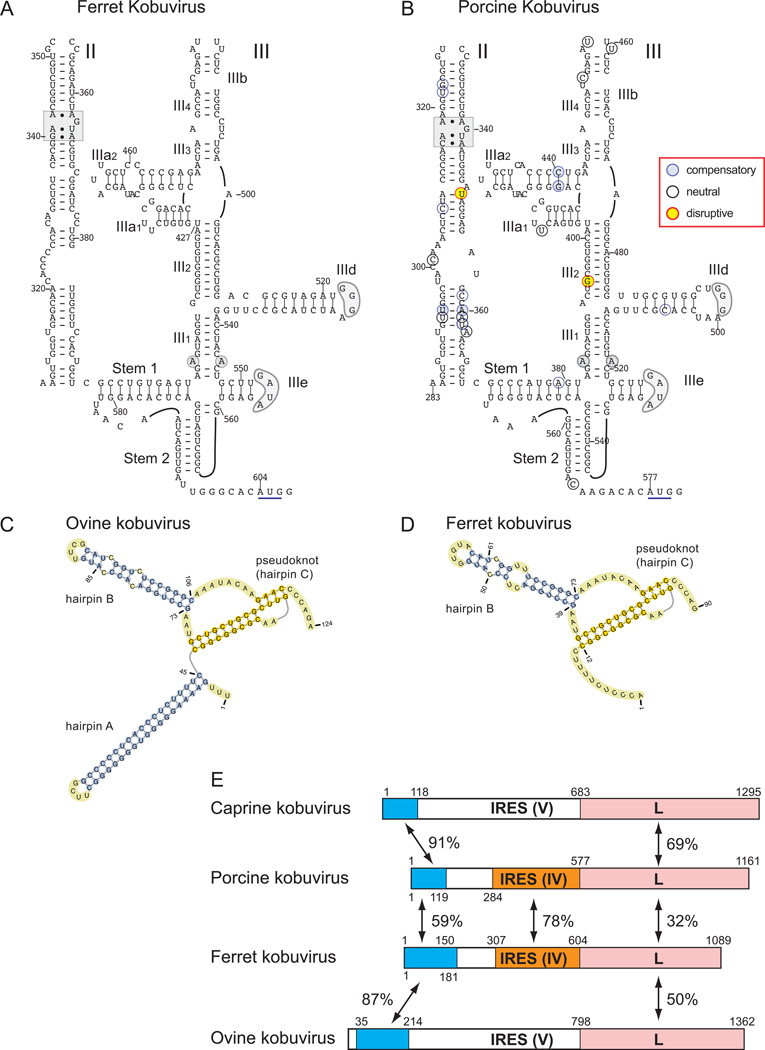
A Type IV IRES in a second member of the genus Kobuvirus
The nt. 368–460 segment of the Turkey hepatitis virus 1 (THV1) strain 2993D IRES (extending from the 5’-border of domain IIId to the initiation codon) also shared 77% identity with a nt.509–603 fragment of the Aichivirus B (ferret kobuvirus isolate MoKoV32; FKV) genome. This region has previously been identified as part of the FKV polyprotein coding region (Smits et al., 2013), even though the encoded amino acid sequence would have no homology with any other picornavirus protein, including the L proteins of other members of the genus Kobuvirus. The structure of the FKV 5’-UTR has not been characterized, and we therefore derived a model of different elements in it.
All kobuviruses have a conserved 5’-terminal structure that consists of a 5’-terminal hairpin ‘A’, followed by a second hairpin (‘B’) protruding from the large downstream loop of a pseudoknot (e.g. Fig. 6C), and with the exception of porcine kobuvirus (PKV), they all have a Type V IRES (Reuter et al., 2009; Sweeney et al., 2012). Sequence alignments suggest that the 5’-terminal region of the FKV 5’-UTR, which has high (87%) sequence identity with the corresponding region of the Ovine kobuvirus (OKV) 5’-UTR (Reuter et al., 2010), is likely missing some nucleotides that in other kobuvirus genomes complete hairpin ‘A’ (Sasaki et al., 2001; Greninger et al., 2009; Sweeney et al., 2012) but it otherwise has the capacity to form a typical kobuvirus-like 5’-terminal structure (Fig. 6D). In addition to their homology with a segment of the THV1 IRES, downstream FKV sequences are also extremely closely related to part of the Aichivirus C (porcine kobuvirus) 5’UTR, which is exceptional amongst members of the genus in in containing a Type IV IRES (Reuter et al., 2009).
FIG 6. Models of predicted secondary and tertiary structures of kobuvirus Type IV IRESs.
The model of the Ferret kobuvirus (FKV) IRES (A) was derived as described in the text and the model of the IRES of PKV-1 strain sw/S-1-HUN/2007 (B) is based on previous proposals (Reuter et al., 2009). Nucleotide differences between the 22 current PKV strains are assigned to different classes as indicated in the inset key, and as defined in the legend for Fig. 2. Domains are labeled II and III; individual helical segments are labeled II1, II2, III1; hairpins are labeled IIIb, IIId, etc. For consistency with the current nomenclature, the additional hairpin element in the FKV and PKV IRESs was named “IIIb2”. Stem 1 and stem 2 are elements of the pseudoknot. AUG triplets marked with a solid black bar represent the translation initiation site for the viral polyprotein. Sequence motifs in domain II and III of FKV and PKV-1 IRESs that are conserved in other Type IV IRES, as indicated by grey shading. (C) Secondary and tertiary 5’-terminal structures of ovine kobuvirus 1 (OKV-1) strain sheep/TB3/HUN/2009 and (D) Ferret kobuvirus, predicted using pKnotsRG. This OKV-1 model has been described elsewhere (Sweeney et al., 2012). Domains A, B, and C are labeled, and nucleotides are numbered in both models. (E) Schematic representations of the 5’ UTRs and adjacent L protein coding region of caprine kobuvirus, porcine kobuvirus, ferret kobuvirus and ovine kobuvirus. 5’-UTRs are divided into domains to show the Type IV IRES in FKV and PKV-1 (orange), the type V IRESs in caprine kobuvirus and ovine kobuvirus and the conserved domain A/B/C/pseudoknot region (blue), which in Aichi virus constitutes the 5’-terminal ori (Sasaki et al., 2001). The L protein region is shaded pink. The ori region, nucleotides forming the Type IV and Type V IRESs and the L protein coding region are labeled; the nucleotides at the borders of conserved regions or defined functional domains are indicated, as are nucleotide sequence identities (ori, IRES) or amino acid sequence identities (L protein). The FKV 5’-UTR is suspected to be incomplete.
The structure of the FKV IRES (Fig. 6A) corresponds almost exactly to the proposed model for the PKV-1 IRES (Reuter et al., 2009)(Fig. 6B). Both contain a conventional domain II, with the only significant difference being the presence in PKV-1 of a variant (AAA/AGUA) of the ‘loop E’ motif. The pseudoknot, domain IIId and domain IIIe of the two IRESs are closely related, and strikingly, the apical region of domain III is also directly analogous in the two IRESs. Both lack domain IIIc, domain IIIb has a conventional length, sequence and structure, but there is an additional hairpin (“IIIa2”) between helix III2 and domain IIIb.
In contrast to the strong sequence and structural similarity between FKV and PKV-1 IRESs, the putative ori sequence upstream of the FKV IRES and the downstream L protein coding sequence are more closely related to sequences in the OKV-1 genome than in the PKV-1 genome, whereas the PKV ori and L sequences are more closely related to those of Caprine kobuvirus (CapKV) than of FKV (Fig. 6E). Whereas the similarities between FKV and PKV-1 IRESs suggest that they had a common origin, an interesting possibility is that the contemporary FKV genome is a mosaic in which a PKV-like type IV IRES has displaced the Type V IRES of an OKV-like genome. Moreover, the greater similarity of FKV to OKV-1 and of PKV-1 to CapKV than to each other in the flanking ori and L -encoding regions suggests that the mosaic nature of FKV and PKV-1 genomes may be due to multiple recombination events.
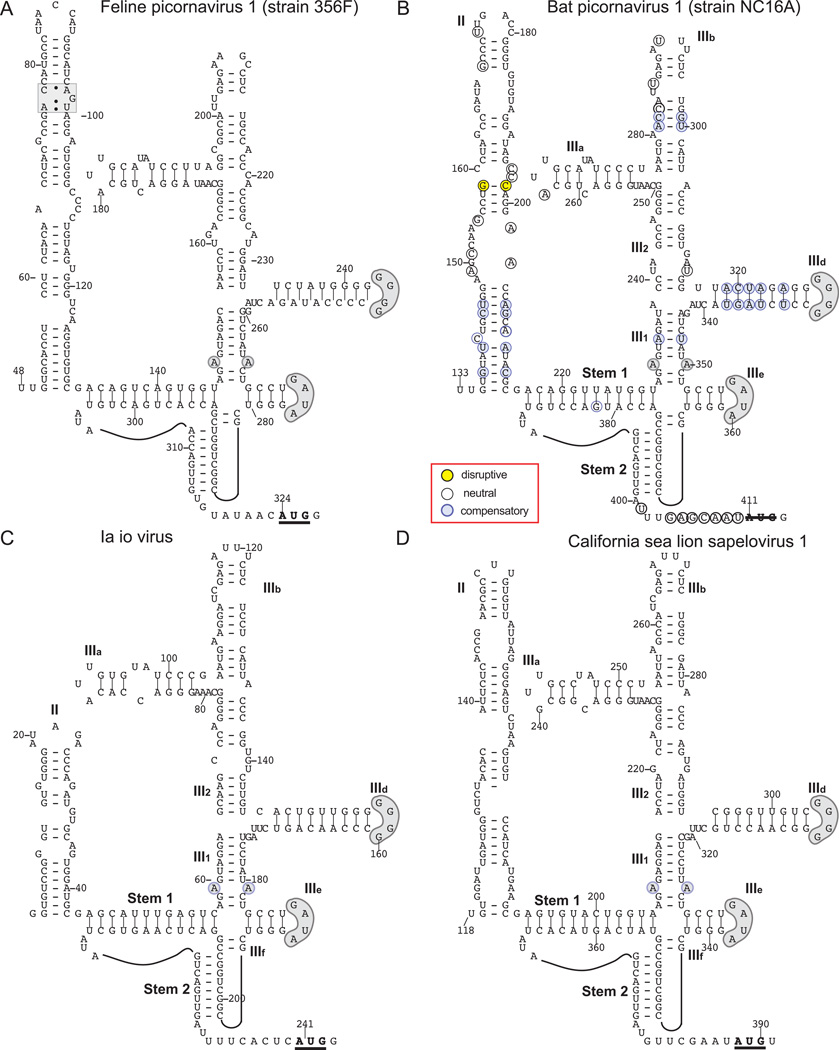
Type IV IRESs in members of the genus Sapelovirus
Sequence homology between the 5’UTR of Ia io picornavirus 1 (IiPV-1) (Wu et al., 2012), provisionally classified as Sapelovirus G in the genus Sapelovirus and the Type IV IRES of FHMPV (see above), as well as with the 5’-UTRs of Feline picornavirus (FePV)-1 (Lau et al., 2012) (provisionally classified as the type member of the species Sapelovirus E), bat picornavirus (BPV)-1 and BPV-2 (Lau et al., 2011) (provisionally classified as the species Sapelovirus D), and Feline sakobuvirus A (SakV-A1) suggested that the IiPV-1 5’-UTR also contains a Type IV IRES. Modeling confirmed that IiPV-1 sequences upstream of the initiation codon form domain III of a Type IV IRES (Fig. 7C) that has significant structural similarities with domain III of the FePV-1 IRES (Fig. 7A; Lau et al., 2012), including the presence of the PK/IIId/IIIe core and the absence of domain IIIc. The 5’-terminal region of the IiPV-1 genome is likely incomplete, so that domain II is undoubtedly larger than the 43nt-long hairpin shown in Fig. 7C.
Figure 7. Type IV IRESs in members of the genus Sapelovirus.
Models of the structures of the IRESs of (A) feline picornavirus strain 356F (as proposed by Lau et al., 2012), (B) bat picornavirus 1 (BPV-1) strain NC16A, (C) Ia io virus (IiPV-1) and (D) California sealion sapelovirus 1 (Csl SapV-1). The initiation codons for the viral polyproteins are underlined and in bold. The nomenclature of domains is as in [Hellen and de Breyne, 2007]. Nucleotides are numbered at 20nt. intervals, and sequence motifs in domains II and III that are conserved in other Type IV IRESs are indicated by grey shading. Nucleotide differences between BPV-1 strains bat/Hong Kong/NC16A/2008 and bat/Hong Kong/LMH22A/2008, and BPV-2 strains bat/Hong Kong/MH9F/2005 and bat/Hong Kong/SK17F/2005 are assigned to different classes as indicated in the inset key (B), and as defined in the legend for Fig. 2.
Despite the extensive sequence similarity between the IiPV-1 5’-UTR and those of BPV-1 and BPV-2 (∼72% identity over the entire length of the current IiPV-1 sequence), the proposed models of IiPV-1 (Fig. 7C) and BPV IRESs (Lau et al., 2011) differ markedly with respect to the sequence and structure of the pseudoknot and the apex of domain III. We therefore re-examined the 5’-UTRs of the currently known strains of BPV-1 and BPV-2, confirmed that each could form a Type IV IRES and derived a consensus structure (Fig. 7B). It differs from the previous model (Lau et al., 2011), but resembled the IiPV-1 IRES Fig. 7C) very closely, and importantly, was strongly supported by the pattern of sequence co-variation.
The IiPV-1 5’-UTR sequence extends only 240nt upstream of the initiation codon, so only domain III will be discussed; like the other sapelovirus IRESs considered here, but in contrast to Sapelovirus B (e.g. simian sapelovirus 3)(Hellen and de Breyne, 2007), the IiPV IRES does not contain a stable domain IV. The sequence from the 5’-border of domain III to the initiation codon shares 68% identity with the Csl SapV-1 5’UTR, which forms a structure (Fig. 7D) that is closely related to those of FePV-1 (Fig. 7A)(Lau et al., 2012), IiPV-1 (Fig. 7C) and BPV-1/BPV-2 (Fig. 7B). In addition to the high level of similarity between these four IRESs from the genus Sapelovirus, IRES domain III and the sequence up to the initiation codon from these four sapeloviruses share 60–73% identity with the equivalent sequences from isolates of porcine teschovirus 1 (PTV) e.g. strain Teschen-Bozen 654.
The Type IV IRESs of Sapelovirus A (e.g. porcine sapelovirus) and Sapelovirus B (e.g. simian sapelovirus 3) contain insertions of 30nt and 46nt, respectively, at positions equivalent to the apical loop of e.g. domain IIIb in the BPV-1 IRES. Excluding these insertions, the structures of Sapelovirus A and Sapelovirus B IRESs (Hellen and de Breyne, 2007) are directly comparable to these Teschovirus and other Sapelovirus Type IV IRESs, sharing 54–66% identity. The extension of the apical region of domain IIIb in Sapelovirus A and Sapelovirus B relative to other sapelovirus Type IV IRESs is reminiscent of the extended nature of domain IIIb in Type IV IRES in the genera Colbovirus, Megrivirus and Phacovirus.
DISCUSSION
We have identified and modeled previously unrecognized Type IV IRESs in members of three proposed novel picornavirus genera (Limnipivirus, Pasivirus, Rafivirus) and in four established picornavirus genera (Kobuvirus, Megrivirus, Sapelovirus, Parechovirus). Other members of the first three of these established genera have previously been reported to contain Type IV IRESs. We also revised existing partial or complete models of Type IV IRESs in BPV (genus Sapelovirus) (Lau et al., 2011) and PiPV-B (genus Colbovirus) (Kofstad and Jonassen, 2011). Type IV IRESs have previously been identified in members of eight recognized and six proposed picornavirus genera (see Introduction): with these additions, they have now been identified in about half of all known picornavirus genera.
Structural conservation and variation
Comparisons between picornavirus Type IV IRESs reveal a remarkable degree of structural heterogeneity, particularly in domain II and the apical region of domain III. The PK-IIId-IIIe core contains some highly conserved elements, but also exhibited significant variability. The apical GGG motif in domain IIId is invariant, consistent with its function in base-pairing with the apical UCCC loop of ES7 of 18S ribosomal RNA (rRNA) (Hashem et al., 2013; Matsuda and Mauro, 2014), and its consequent importance in all Type IV IRESs (Kolupaeva et al., 2000a; Kieft et al., 2001; de Breyne et al., 2007; Easton et al., 2009; Willcocks et al., 2011; Pan et al., 2012). This apical loop occurs in ES7 of 18S rRNA in birds, fish, amphibians and reptiles as well as mammals, and it would therefore be available in ribosomes from them all for binding to Type IV IRESs.
The bulged purine in helix III1 covaries with the pyrimidine at the third position in the GA[U/C]A tetraloop in domain IIIe (Easton et al. 2009), and x-ray crystallography showed that the latter base is flipped out, enabling it to engage in an interhelical Watson-Crick interaction that may stabilize the orientation of the two stems in the pseudoknot (Berry et al., 2011). The same pattern of sequence co-variation was apparent in the Type IV IRESs described here and elsewhere: most contained a bulged adenine helix III1 and a uridine in the tetraloop (Fig. 1), but CPV-1, BGPV-1, seal aquamavirus 1 (SeAV-A1) and QPV-1 instead all contain a bulged guanine in helix III1 and cytosine in the tetraloop (Fig. 2B, 2C; Kapoor et al., 2008; Pankovics et al., 2012). Although domain IIIe, which consists of a 4 bp helix and an apical GA[U/C]A tetraloop, is conserved in almost all Type IV IRESs, Mesiviruses 1 and 2 have a GAUC loop (Fig. 5A) and Aalivirus has a 3 bp helix and 5 nt apical loop (Wang et al., 2014). These variants might nevertheless be able to base-pair with the bulged ‘A’ in helix III1. Positioning of the initiation codon region on the 40S subunit is impaired by substitutions that disrupt this interaction, usually leading to moderate reductions in IRES function that are, however, strongly exacerbated by destabilizing mutations elsewhere in the IRES (Kolupaeva et al., 2000a; Berry et al., 2011). The conservation of this interaction in all Type IV IRESs suggests that its stabilizing influence is important for their structure and function, and may have a buffering role to minimize deleterious effects of mutations elsewhere in the IRES.
The pseudoknot is a characteristic, essential component of Type IV IRESs (Wang et al., 1994; Pestova et al., 1998; Kolupaeva et al., 2000a, Kieft et al., 2001; Fletcher and Jackson, 2002; Chard et al., 2006; Bakhshesh et al., 2008; Berry et al., 2010, 2011; Willcocks et al., 2011). The integrity of pseudoknot stem 1 is critical for binding to the 40S subunit whereas disruption of stem 2 impairs IRES function by abrogating its ability to position the AUG start codon in the ribosomal P site (Pestova et al., 1998; Kolupaeva et al., 2000a; Kieft et al., 2001; Berry et al., 2010, 2011). The sequence variability in stems 1 and 2 (which is predominantly covariant), and differences in the length of this loop in the IRESs identified here are consistent with mutational analyses which showed that insertions in the loop are tolerated, that impairment of IRES function caused by destabilization of the pseudoknot can largely be restored by second-site substitutions that re-establish base-pairing, and that the sequence of the pseudoknot per se is not essential for function (Wang et al., 1994; Kieft et al., 2001; Fletcher and Jackson, 2002; Bakhshesh et al., 2008; Berry et al., 2010).
Several other variable regions in the IRESs described here correspond to non-essential peripheral elements. Domains analogous to domain IIId2 in the FPV, MpeV and TRaV-1 IRESs occur in CSFV and SVV IRESs, where they tolerate significant insertions (Moser et al., 2001) or can be deleted without impairing IRES activity (Friis et al., 2012; Willcocks et al., 2011). Only a subset of the IRESs identified here (BGPV-1, CPV-1, FHMPV, PaV-A1, mesivirus 1 and PiPV-B) contains an equivalent of HCV hairpin domain IV sequestering the initiation codon, and they are all less stable than in HCV.
Domain II has differing sizes and sequences in different hepacivirus and pestivirus Type IV IRESs, and its functional importance varies (Fletcher and Jackson, 2002; Friis et al., 2012). Nevertheless, these IRESs all contain a ‘loop E’ motif, and an asymmetric internal loop near the base (Fig. 1) that is thought to mediate a switch in domain II from a bent to an extended conformation (Boerneke et al., 2014). Cryo-electron microscopy has shown that this switch is accompanied by release of the apical region of HCV domain II from its binding site on the head of the 40S subunit and tentatively, by its interaction with initiator tRNA, thereby adjusting its position to facilitate the transition from initiation to elongation (Yamamoto et al., 2014). A similar asymmetric loop occurs in domain II of almost all picornavirus Type IV IRESs, including those identified here, suggesting that they may undergo a similar induced conformational switch during initiation.
The greatest level of diversity occurs at the apex of domain III. The principal function of this region is to bind to eIF3 (Pestova et al., 1998, Sizova et al., 1998; Hashem et al., 2013; Sun et al., 2013), primarily via interactions with domain IIIb and helix III4, and its frequent expansion beyond the length that is bound by eIF3 suggests that insertions at this locus do not significantly impair IRES function. Some elements in this region are dispensable, such as CSFV domain IIIa (Fletcher and Jackson, 2002). The highly variable nature of this region suggests that it is evolving rapidly: indeed, subdomain IIIb is one of the most variable regions in the HCV IRES (Smith et al., 1995) and comparisons between HCV and other hepaciviruses indicate that its apex is a site of nucleotide insertion (e.g. Burbelo et al., 2012). As discussed below, the presence of a highly conserved element at the apex of domain IIIb in several IRESs may be the result of recombinational insertion of a novel functional element, and the variable nature of domain IIIb may render it permissive for insertion.
Evolution of Type IV IRESs: the role of recombination
Consideration of the sequence and structural diversity of Type IV IRESs suggests different mechanisms that have likely contributed to their evolution. The ability of IRESs to function in heterologous genomes was first demonstrated for the Type IV HCV IRES (Lu and Wimmer, 1996), indicating that they are independent heritable genetic elements that could undergo lateral gene transfer between viral genomes. We previously suggested that the presence of related IRESs in the genomes of unrelated viruses (e.g. Type IV IRESs in members of Flaviviridae and Picornaviridae), and of unrelated IRESs in the genomes of otherwise related viruses (e.g. the Type IV IRES in Aichivirus C (porcine kobuvirus 1) in the genus Kobuvirus) (Reuter et al., 2009) and the Type V IRESs in Aichivirus A (Aichi virus 1, canine kobuvirus 1 and murine kobuvirus 1) and Aichivirus B (bovine kobuvirus 1 and sheep kobuvirus 1) (Sweeney et al., 2012) are suggestive of this phenomenon. This hypothesis is supported by the identification here of a Type IV IRES in another member of the genus Kobuvirus (Aichivirus B; ferret kobuvirus) (Fig. 6A) and of unrelated IRESs in different members of a single genus (Parechovirus), namely Type IV IRESs in FPV and MPeV (Figs. 3B, 3C) but Type II IRESs in e.g. Ljungan virus and human parechovirus (Johansson et al., 2002; Nateri et al., 2000). A complementary observation concerns the genus Parechovirus: whereas the Type IV FPV IRES identified here drives translation of a typical parechovirus polyprotein, ovine and bovine hungaroviruses have parechovirus-like Type II IRESs but encode a Teschovirus-like polyprotein (Reuter et al., 2012).
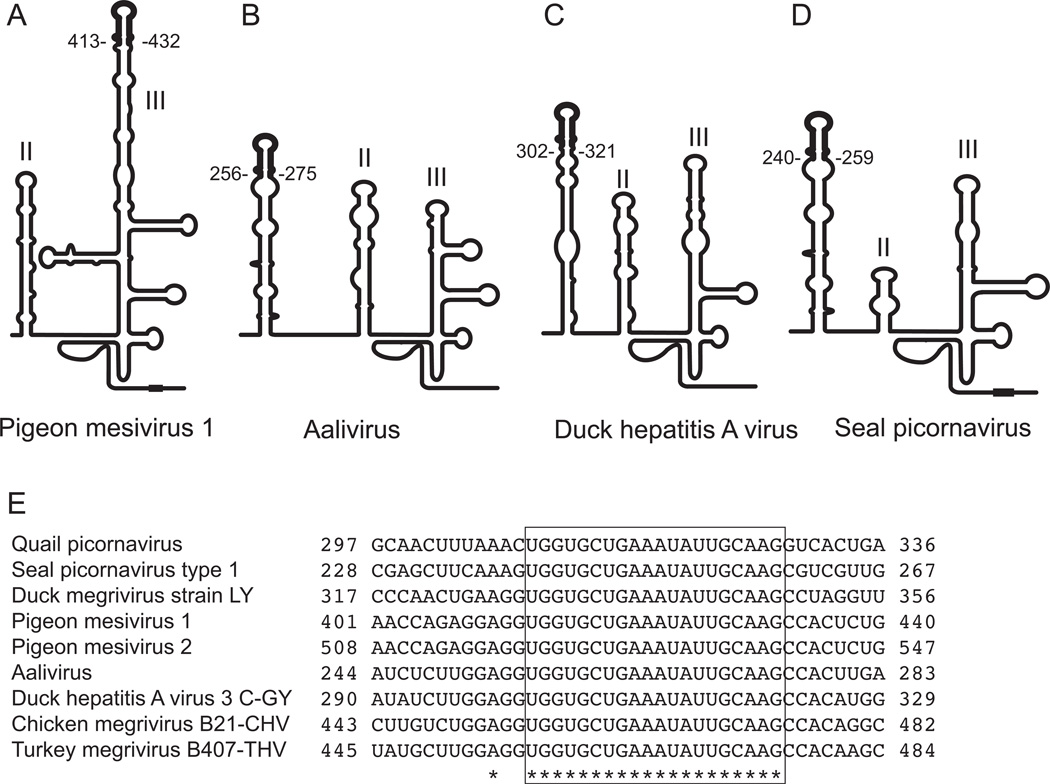
In addition to the exchange of IRESs between viral genomes, recombination could lead to the exchange or even the de novo acquisition of functional domains or subdomains. An event of this type has recently been described in which a 55nt-long translation enhancer element was acquired by Melon necrotic spot virus (family Tombusviridae) from Cucurbit aphid-borne yellow virus (family Luteoviridae) (Miras et al., 2014). A similar process likely accounts for the presence of the 43nt-long stem-loop-2-like motif (s2m) in the 3’-UTRs of members of Astroviridae, Caliciviridae, Coronaviridae and Picornaviridae (Tengs et al., 2013). The HCV IRES retains its activity despite substitution of domain II by analogous elements from other Type IV IRESs (Reusken et al., 2003; Boerneke et al., 2014). Analysis of Type IV IRESs (e.g. Boros et al., 2014b) suggests that a subset of them may also have acquired a subdomain by recombination. The “8-like” motifs that occur at the apex of the greatly elongated (∼110nt-long) domain IIIb in Phacoviruses, Megriviruses and Mesiviruses exhibit a high level of sequence identity (Honkavuori et al., 2011; Pankovics et al., 2012; Phan et al., 2013; Boros et al., 2014a, b). Significantly, exactly the same element occurs outside the core IRES in duck hepatitis virus (e.g. Pan et al., 2012), SeAV-A1 (Kapoor et al., 2008) and Aalivirus (Wang et al., 2014); as noted elsewhere (Kapoor et al., 2008, Boros et al., 2014a, b), it is at the apex of a long hairpin over 50nt upstream of domain II (Fig. 8). The presence of such closely related elements at different locations in diverse picornavirus 5’UTRs is suggestive of their (recent) acquisition by recombination from a common or closely related source. This element augments IRES activity (e.g. Pan et al., 2012) but its precise function has not been elucidated. The acquisition of this conserved element in Type IV IRESs may thus be indicative of an evolutionary process in which peripheral domains are added incrementally to a pre-existing core without perturbing its structure or function.
Figure 8. The variable location of the conserved apical ‘8-like’ motif in divers Type IV picornavirus IRESs.
Schematic representations of Type IV IRESs in (A) pigeon mesivirus 1, (B) Aalivirus, (C) duck hepatitis A virus and (D) Seal picornavirus, based on proposals presented here and elsewhere (Kapoor et al., 2008; Pan et al., 2012; Boros et al., 2014a, b), showing the differing locations of the conserved ‘8-like’ motif. (E) Nucleotide sequenced alignment of the ‘8-like’ motif in these picornaviruses and quail picornavirus, pigeon mesivirus 2 and Duck, Turkey and Chicken megriviruses.
The exchange of IRESs or domains of IRESs between picornaviruses by replication-dependent recombination depends on their ability to co-infect a common host, but whether they can do so has not yet been established for many of the viruses described here, because they have been identified so recently that their host ranges have not yet been determined. The acquisition by IRESs of shorter RNA segments, potentially including the ‘8-like’ motif described above, may not necessarily depend on co-infection of a common host by two distinct viruses, but could instead involve non-homologous replication-dependent recombination with cellular RNA. Transduction of cellular RNA sequences into picornavirus and flavivirus genomes is an established phenomenon (Charini et al., 1994; Becher and Tautz, 2011). As noted below, a further possibility is that IRESs might have acquired novel subdomains by non-replicative recombination with either cellular or viral RNA.
Other IRES subdomains are strikingly variable in terms of size and sequence. In the proposed genera Kunsagivirus, Limnipivirus and Rafivirus, the apical region of domain III is a ∼50nt-long Y-shaped domain (Figs. 2, 4), whereas in Megrivirus and proposed Colbovirus genera, it is ∼220nt long and contains three asymmetrically branching hairpins (Fig. 5). Although an increase in IRES size might be attributable to replicative “stuttering” or non-templated incorporation of nucleotides, picornavirus polymerases are not known to be prone to such errors. However, recent studies have identified mechanisms, including non-homologous, non-replicative recombination (Gmyl et al., 2003; Holmblat et al., 2014) and an imprecise replicative recombinational process (Lowry et al., 2014) that could result in partial duplication of picornavirus genome fragments. Duplicated sequences present in a region of the genome that is not under strong selective pressure could diversify rapidly by nucleotide substitution or by partial deletion (Lowry et al., 2014) and might be particularly permissive for insertion of ‘captured’ domains, such as the “8-like” motif discussed above.
Evolution of Type IV IRESs: the role of nucleotide substitutions
The second principal mechanism that contributes to the evolution of viral IRESs is nucleotide substitution during replication, coupled with selection for retention of functionally important elements. Together, these two processes account for maintenance of IRES structure by sequence covariation coupled with conservation of functionally important nucleotides. As in Type 1 and Type 2 IRESs (Jackson and Kaminski, 1995), nucleotide conservation in Megrivirus and Mesivirus Type IV IRESs (Fig. 5A, 5C) is greatest at sites that interact with components of the translation apparatus (in the case of Type IV IRES, with the 40S subunit) and at the junction of helical elements, likely to maintain the fold of the IRES so that it is competent to present key motifs in orientations and conformations that allow such interactions to be established.
By analogy with rapidly evolving rRNA elements, substitutions at the apex of a helix could promote its invagination, resulting in its division into two smaller helices (Caisova and Melkanian, 2014). This process could account for domain K in Type II IRESs of Ljungan virus (Johansson et al., 2002) and Rosavirus (Phan et al., 2013b) having a Y-shape with a bifurcated apex, rather than the single bent hairpin structure of domain K in the canonical EMCV Type II IRES. An analogous process may account for the appearance of additional branching helical elements in the apical region of domain III in some Type IV IRESs.
Highlights.
Numerous novel Type IV IRES have been identified in diverse picornaviruses
Type IV IRESs retain a common core but still show remarkable structural heterogeneity
Recombinational exchange of functional subdomains may contribute to IRES evolution
Diverse picornavirus genomes may have gained related IRESs by lateral gene transfer
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant AI-51340.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bakhshesh M, Groppelli E, Willcocks MM, Royall E, Belsham GJ, Roberts LO. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J Virol. 2008;82:1993–2003. doi: 10.1128/JVI.01957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbknecht M, Sepsenwol S, Leis E, Tuttle-Lau M, Gaikowski M, Knowles NJ, Lasee B, Hoffman MA. Characterization of a new picornavirus isolated from the freshwater fish Lepomis macrochirus . J. Gen. Virol. 2014;95:601–613. doi: 10.1099/vir.0.061960-0. [DOI] [PubMed] [Google Scholar]
- Becher P, Tautz N. RNA recombination in pestiviruses: cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol. 2011;8 doi: 10.4161/rna.8.2.14514. 216-w24. [DOI] [PubMed] [Google Scholar]
- Benschop KS, de Vries M, Minnaar RP, Stanway G, van der Hoek L, Wolthers KC, Simmonds P. Comprehensive full-length sequence analyses of human parechoviruses: diversity and recombination. J. Gen. Virol. 2010;91:145–154. doi: 10.1099/vir.0.014670-0. [DOI] [PubMed] [Google Scholar]
- Berry KE, Waghray S, Doudna JA. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA. 2010;16:1559–1569. doi: 10.1261/rna.2197210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KE, Waghray S, Mortimer SA, Bai Y, Doudna JA. Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure. 2011;19:1456–1466. doi: 10.1016/j.str.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc. Natl. Acad. Sci. USA. 2014;111:15952–15957. doi: 10.1073/pnas.1414678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros Á, Kiss T, Kiss O, Pankovics P, Kapusinszky B, Delwart E, Reuter G. Genetic characterization of a novel picornavirus distantly related to the marine mammal-infecting aquamaviruses in a long-distance migrant bird species, European Roller (Coracias garrulus) J. Gen. Virol. 2013;94:2029–2035. doi: 10.1099/vir.0.054676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A, Pankovics P, Knowles NJ, Nemes C, Delwart E, Reuter G. Comparative complete genome analysis of chicken and Turkey megriviruses (family picornaviridae): long 3' untranslated regions with a potential second open reading frame and evidence for possible recombination. J. Virol. 2014a;88:6434–6443. doi: 10.1128/JVI.03807-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros A, Pankovics P, Reuter G. Avian picornaviruses: Molecular evolution, genome diversity and unusual genome features of a rapidly expanding group of viruses in birds. Infect. Genet. Evol. 2014b;28C:151–166. doi: 10.1016/j.meegid.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Brown EA, Zajac AJ, Lemon SM. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5’ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 1994;68:1066–1074. doi: 10.1128/jvi.68.2.1066-1074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo PD, Dubovi EJ, Simmonds P, Medina JL, Henriquez JA, Mishra N, Wagner J, Tokarz R, Cullen JM, Iadarola MJ, Rice CM, Lipkin WI, Kapoor A. Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J. Virol. 2012;86:6171–6178. doi: 10.1128/JVI.00250-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byun Y, Han K. Pseudo Viewer3: generating planar drawings of large-scale RNA structures with pseudoknots. Bioinformatics. 2009;25:1435–1437. doi: 10.1093/bioinformatics/btp252. [DOI] [PubMed] [Google Scholar]
- Caisová L, Melkonian M. Evolution of helix formation in the ribosomal Internal Transcribed Spacer 2 (ITS2) and its significance for RNA secondary structures. J. Mol. Evol. 2014;78:324–337. doi: 10.1007/s00239-014-9625-0. [DOI] [PubMed] [Google Scholar]
- Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 2006a;87:927–936. doi: 10.1099/vir.0.81546-0. [DOI] [PubMed] [Google Scholar]
- Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 2006b;80:1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charini WA, Todd S, Gutman GA, Semler BL. Transduction of a human RNA sequence by poliovirus. J. Virol. 1994;68:6547–6552. doi: 10.1128/jvi.68.10.6547-6552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breyne S, Yu Y, Pestova TV, Hellen CU. Factor requirements for translation initiation on the Simian picornavirus internal ribosomal entry site. RNA. 2008;14:367–380. doi: 10.1261/rna.696508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 2013;20:150–158. doi: 10.1038/nsmb.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth C, Bhat M, Firth MA, Williams SH, Frye MJ, Simmonds P, Conte JM, Ng J, Garcia J, Bhuva NP, Lee B, Che X, Quan PL, Lipkin WI. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. MBio. 2014;5:e01933–e01914. doi: 10.1128/mBio.01933-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher SP, Jackson RJ. Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5’ untranslated region important for IRES function. J. Virol. 2002;76:5024–5033. doi: 10.1128/JVI.76.10.5024-5033.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis MB, Rasmussen TB, Belsham GJ. Modulation of translation initiation efficiency in classical swine fever virus. J. Virol. 2012;86:8681–8692. doi: 10.1128/JVI.00346-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmyl AP, Korshenko SA, Belousov EV, Khitrina EV, Agol VI. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA. 2003;9:1221–1231. doi: 10.1261/rna.5111803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Runckel C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, DeRisi JL. The complete genome of klassevirus–a novel picornavirus in pediatric stool. Virol. J. 2009;6:82. doi: 10.1186/1743-422X-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim Biophys Acta. 2009;1789:558–570. doi: 10.1016/j.bbagrm.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmblat B, Jégouic S, Muslin C, Blondel B, Joffret ML, Depeyroux F. Nonhomologous recombination between defective poliovirus and coxsackievirus genomes suggests a new model of genetic plasticity for picornaviruses. Mbio. 2014;5:e01119–e01114. doi: 10.1128/mBio.01119-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda M, Brown EA, Lemon SM. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- Honkavuori KS, Shivaprasad HL, Briese T, Street C, Hirschberg DL, Hutchison SK, Lipkin WI. Novel picornavirus in turkey poults with hepatitis, California, USA. Emerg. Infect. Dis. 2011;17:480–487. doi: 10.3201/eid1703.101410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes PJ, Stanway G. The 2A proteins of three diverse picornaviruses are related to each other and to the H-rev107 family of proteins involved in the control of cell proliferation. J. Gen. Virol. 2000;81:201–207. doi: 10.1099/0022-1317-81-1-201. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Kaminski A. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA. 1995;1:985–1000. [PMC free article] [PubMed] [Google Scholar]
- Janssen S, Giegerich R. The RNA shapes studio. Bioinformatics. 2015;31:423–425. doi: 10.1093/bioinformatics/btu649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson S, Niklasson B, Maizel J, Gorbalenya AE, Lindberg AM. Molecular analysis of three Ljungan virus isolates reveals a new, close-to-root lineage of the Picornaviridae with a cluster of two unrelated 2A proteins. J. Virol. 2002;76:8920–8930. doi: 10.1128/JVI.76.17.8920-8930.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A, Victoria J, Simmonds P, Wang C, Shafer RW, Nims R, Nielsen O, Delwart E. A highly divergent picornavirus in a marine mammal. J. Virol. 2008;82:311–320. doi: 10.1128/JVI.01240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofstad T, Jonassen CM. Screening of feral and wood pigeons for viruses harbouring a conserved mobile viral element: characterization of novel astroviruses and picornaviruses. PLoS One. 2011;6:e25964. doi: 10.1371/journal.pone.0025964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000a;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000b;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange J, Groth M, Fichtner D, Granzow H, Keller B, Walther M, Platzer M, Sauerbrei A, Zell R. Virus isolate from carp: genetic characterization reveals a novel picornavirus with two aphthovirus 2A-like sequences. J Gen Virol. 2014;95:80–90. doi: 10.1099/vir.0.058172-0. [DOI] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Lai KK, Huang Y, Yip CC, Shek CT, Lee P, Lam CS, Chan KH, Yuen KY. Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 2011;85:8819–8828. doi: 10.1128/JVI.02364-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SK, Woo PC, Yip CC, Choi GK, Wu Y, Bai R, Fan RY, Lai KK, Chan KH, Yuen KY. Identification of a novel feline picornavirus from the domestic cat. J Virol. 2012;86:395–405. doi: 10.1128/JVI.06253-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q, Zheng L, Yuan Y, Shi J, Zhang D. Genomic characterization of a novel picornavirus in Pekin ducks. Vet. Microbiol. 2014;172:78–91. doi: 10.1016/j.vetmic.2014.05.002. [DOI] [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CU, Pestova TV. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry K, Woodman A, Cook J, Evans DJ. Recombination in enteroviruses is a biphasic replicative process involving the generation of greater-than genome length 'imprecise' intermediates. PLoS Pathog. 2014;10:e1004191. doi: 10.1371/journal.ppat.1004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu HH, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- Matsuda D, Mauro VP. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc. Natl. Acad. Sci. USA. 2014;111:15385–15389. doi: 10.1073/pnas.1413472111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre CL, McWilliam Leitch EC, Savolainen-Kopra C, Hovi T, Simmonds P. Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J. Virol. 2010;84:10297–10310. doi: 10.1128/JVI.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Sempere RN, Kraft JJ, Miller WA, Aranda MA, Truniger V. Interfamilial recombination between viruses led to acquisition of a novel translation-enhancing RNA element that allows resistance breaking. New Phytol. 2014;202:233–246. doi: 10.1111/nph.12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser C, Bosshart A, Tratschin JD, Hofmann MA. A recombinant classical swine fever virus with a marker insertion in the internal ribosome entry site. Virus Genes. 2001;23:63–68. doi: 10.1023/a:1011135429860. [DOI] [PubMed] [Google Scholar]
- Nateri AS, Hughes PJ, Stanway G. In vivo and in vitro identification of structural and sequence elements of the human parechovirus 5' untranslated region required for internal initiation. J. Virol. 2000;74:6269–6277. doi: 10.1128/jvi.74.14.6269-6277.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TF, Mesquita JR, Nascimento MS, Kondov NO, Wong W, Reuter G, Knowles NJ, Vega E, Esona MD, Deng X, Vinjé J, Delwart E. Feline fecal virome reveals novel and prevalent enteric viruses. Vet. Microbiol. 2014;171:102–111. doi: 10.1016/j.vetmic.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan M, Yang X, Zhou L, Ge X, Guo X, Liu J, Zhang D, Yang H. Duck Hepatitis A virus possesses a distinct type IV internal ribosome entry site element of picornavirus. J. Virol. 2012;86:1129–1144. doi: 10.1128/JVI.00306-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankovics P, Boros A, Reuter G. Novel picornavirus in domesticated common quail (Coturnix coturnix) in Hungary. Arch. Virol. 2012;157:525–530. doi: 10.1007/s00705-011-1192-8. [DOI] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Vo NP, Boros Á, Pankovics P, Reuter G, Li OT, Wang C, Deng X, Poon LL, Delwart E. The viruses of wild pigeon droppings. PLoS One. 2013;8:e72787. doi: 10.1371/journal.pone.0072787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan TG, Vo NP, Simmonds P, Samayoa E, Naccache S, Chiu CY, Delwart E. Rosavirus: the prototype of a proposed new genus of the Picornaviridae family. Virus Genes. 2013b;47:556–558. doi: 10.1007/s11262-013-0968-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps NB, Mor SK, Armien AG, Batts W, Goodwin AE, Hopper L, McCann R, Ng TF, Puzach C, Waltzek TB, Delwart E, Winton J, Goyal SM. Isolation and molecular characterization of a novel picornavirus from baitfish in the USA. PLoS One. 2014;9:e87593. doi: 10.1371/journal.pone.0087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Steffen P, Giegerich R. pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows. Nucleic Acids Res. 2007;35:W320–W324. doi: 10.1093/nar/gkm258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken CB, Dalebout TJ, Eerligh P, Bredenbeek PJ, Spaan WJ. Analysis of hepatitis C virus/classical swine fever virus chimeric 5’NTRs: sequences within the hepatitis C virus IRES are required for viral RNA replication. J Gen Virol. 2003;84:1761–1769. doi: 10.1099/vir.0.19063-0. [DOI] [PubMed] [Google Scholar]
- Reuter G, Boldizsár A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. 2009;154:101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- Reuter G, Boros A, Pankovics P, Egyed L. Kobuvirus in domestic sheep, Hungary. Emerg. Infect. Dis. 2010;16:869–870. doi: 10.3201/eid1605.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Pankovics P, Knowles NJ, Boros Á. Two closely related novel picornaviruses in cattle and sheep in Hungary from 2008 to 2009, proposed as members of a new genus in the family Picornaviridae. J. Virol. 2012;86:13295–13302. doi: 10.1128/JVI.01142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Hamada M, Asai K, Mituyama T. CENTROIDFOLD: a web server for RNA secondary structure prediction. Nucleic Acids Res. 2009;37:W277–W280. doi: 10.1093/nar/gkp367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvage V, Ar Gouilh M, Cheval J, Muth E, Pariente K, Burguiere A, Caro V, Manuguerra JC, Eloit M. A member of a new Picornaviridae genus is shed in pig feces. J. Virol. 2012;86:10036–10046. doi: 10.1128/JVI.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Kusuhara Y, Maeno Y, Kobayashi N, Yamashita T, Sakae K, Takeda N, Taniguchi K. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5' end of the genome. J. Virol. 2001;75:8021–8030. doi: 10.1128/JVI.75.17.8021-8030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Mellor J, Jarvis LM, Davidson F, Kolberg J, Urdea M, Yap PL, Simmonds P. Variation of the hepatitis C virus 5' non-coding region: implications for secondary structure, virus detection and typing. The International HCV Collaborative Study Group. J. Gen. Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- Smits SL, Raj VS, Oduber MD, Schapendonk CM, Bodewes R, Provacia L, Stittelaar KJ, Osterhaus AD, Haagmans BL. Metagenomic analysis of the ferret fecal viral flora. PLoS ONE. 2013;8:E71595. doi: 10.1371/journal.pone.0071595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Querol-Audí J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 2013;41:7512–7521. doi: 10.1093/nar/gkt510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J. 2014;33:76–92. doi: 10.1002/embj.201386124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney TR, Dhote V, Yu Y, Hellen CU. A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J. Virol. 2012;86:1468–1486. doi: 10.1128/JVI.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tengs T, Kristoffersen AB, Bachvaroff TR, Jonassen CM. A mobile genetic element with unknown function found in distantly related viruses. Virol J. 2013;10:132. doi: 10.1186/1743-422X-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CH, Tsai HJ. Sequence analysis of a duck picornavirus isolate indicates that it together with porcine enterovirus type 8 and simian picornavirus type 2 should be assigned to a new picornavirus genus. Virus Res. 2007;129:104–114. doi: 10.1016/j.virusres.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A. A conserved helical element is essential for internal initiation of translation of hepatitis C virus RNA. J. Virol. 1994;68:7301–7307. doi: 10.1128/jvi.68.11.7301-7307.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu N, Wang F, Ning K, Li Y, Zhang D. Genetic characterization of a novel duck-origin picornavirus with six 2A proteins. J. Gen. Virol. 2014;95:1289–1296. doi: 10.1099/vir.0.063313-0. [DOI] [PubMed] [Google Scholar]
- Willcocks MM, Locker N, Gomwalk Z, Royall E, Bakhshesh M, Belsham GJ, Idamakanti N, Burroughs KD, Reddy PS, Hallenbeck PL, Roberts LO. Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. J. Virol. 2011;85:4452–4461. doi: 10.1128/JVI.01107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Ren X, Yang L, Hu Y, Yang J, He G, Zhang J, Dong J, Sun L, Du J, Liu L, Xue Y, Wang J, Yang F, Zhang S, Jin Q. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 2012;86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Bürger J, Mielke T, Spahn CM. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 2014;21:721–727. doi: 10.1038/nsmb.2859. [DOI] [PubMed] [Google Scholar]
- Yozwiak NL, Skewes-Cox P, Gordon A, Saborio S, Kuan G, Balmaseda A, Ganem D, Harris E, DeRisi JL. Human enterovirus 109: a novel interspecies recombinant enterovirus isolated from a case of acute pediatric respiratory illness in Nicaragua. J. Virol. 2010;84:9047–9058. doi: 10.1128/JVI.00698-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JM, Li XY, Ao YY, Li LL, Liu N, Li JS, Duan ZJ. Identification of a novel picornavirus in healthy piglets and seroepidemiological evidence of its presence in humans. PLoS One. 2013;8:e70137. doi: 10.1371/journal.pone.0070137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 2011;30:4423–4436. doi: 10.1038/emboj.2011.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Leontis N, Qian S, Itaya A, Qi Y, Boris-Lawrie K, Ding B. Tertiary structural and functional analyses of a viroid RNA motif by isostericity matrix and mutagenesis reveal its essential role in replication. J. Virol. 2006;80:8566–8581. doi: 10.1128/JVI.00837-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoll J, Galama JM, van Kuppeveld FJ. Identification of potential recombination breakpoints in human parechoviruses. J. Virol. 2009;83:3379–3383. doi: 10.1128/JVI.02529-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]