Abstract
mRNA structure is important for post-transcriptional regulation, largely because it affects binding of trans-acting factors1. However, little is known about the in vivo structure of full-length mRNAs. Here we present hiCLIP, a high-throughput technique to identify RNA secondary structures interacting with RNA-binding proteins (RBPs) in vivo. Using this technique to investigate RNA structures bound by Staufen 1 (STAU1), we uncover a dominance of intra-molecular RNA duplexes, a depletion of duplexes from coding regions of highly translated mRNAs, an unforeseen prevalence of long-range duplexes in 3′ untranslated regions (UTRs), and a decreased incidence of SNPs in duplex-forming regions. We also discover a duplex spanning 858nts in the 3′ UTR of the X-box binding Protein 1 (XBP1) mRNA that regulates its cytoplasmic splicing and stability. Our study reveals the fundamental role of mRNA secondary structures in gene regulation and introduces hiCLIP as a widely applicable method for discovering novel, especially long-range, RNA duplexes.
RNA secondary structure is emerging as a key determinant of post-transcriptional regulation1-7. However, identifying the base-paired duplexes forming these structures remains challenging on a transcriptomic scale. The recently reported method CLASH (cross-linking, ligation, and sequencing of hybrids) detects RNA duplexes complexed with RBPs8; but several challenges arise from the direct ligation of the two RNA strands forming the duplex9 (Supplementary Note). To this end, we developed hiCLIP (RNA hybrid and individual-nucleotide resolution UV cross-linking and immunoprecipitation). Like CLASH, hiCLIP identifies duplexes by ligating the two RNA strands in the duplex and sequencing the resulting hybrids. However, hiCLIP incorporates an adaptor between the two RNA strands to gain greater control over the ligation reaction and to define duplexes accurately through unambiguous identification of the two arms of hybrid reads (Extended Data Fig. 1a-c). Using hiCLIP, we studied RNA duplexes directly bound by STAU1, a double-stranded RBP that is important for mRNA localization10, stability11,12 and translation13. Despite several investigations into its target RNAs11-18, characteristics of secondary structures bound in vivo remain poorly understood.
We performed hiCLIP from cytoplasmic extracts of Flp-In T-REx 293 cells. To recover a broad spectrum of RNAs and to ensure that only directly bound duplexes are identified, we employed two different RNase concentrations and stringent purification conditions (Fig. 1a, Extended Data Fig. 2a). We obtained 35,358 hybrid reads whose arms could be mapped to non-contiguous segments of RNA transcripts (Extended Data Fig. 1a-c, Supplementary table 1). Hybrid reads comprised approximately 2% of all reads. The remaining non-hybrid reads (1.2 million reads including control library; Supplementary table 1) were equivalent to traditional iCLIP reads and defined STAU1 cross-link sites19 (Extended Data Fig. 1a, Step 6). In contrast, hybrid reads comprised just 0.06% of control experiments omitting the second ligation reaction (Fig. 1b). Despite different RNase concentrations between replicates, there was good correlation in the numbers of reads mapping to each mRNA transcript (r=0.876; Extended Data Fig. 2b).
Figure 1. hiCLIP identifies RNA duplexes bound by STAU1.
a, Autoradiography analysis of the STAU1-RNA complex at different RNase I concentrations or in the absence of cross-linking or STAU1 induction.
b, The proportion of uniquely annotated hybrid reads in the hiCLIP libraries at high and low RNase conditions and from the control in which the second ligation (step 5 in Extended Data Fig. 1a) was omitted.
c, Mapping summary of the arms of hybrid reads.
d, Probability density distributions of minimum free energies of hybridization between the two arms of hybrid reads from mRNAs and long non-coding RNAs, or randomly repositioned sequences. Distributions were compared using the Mann-Whitney U test (n = 6120 for both).
e, Alignment of three newly identified duplexes (hA, hB and hC) that connect distal regions of the human 18S rRNA. The nucleotide position and the nearest annotated helix from the CryoEM structure of the rRNA (Extended Data Fig. 4b) are marked in a different colour for each region.
f, (Top) Proportion of hybrid reads that map to same or different RNA species. (Bottom) For hybrid reads mapping to same mRNA species, proportion in CDS, 3′ UTR, or other (i.e., 5′ UTR or spanning across two regions).
Of the 35,358 hybrid reads, 50% mapped to mRNAs, 21% to rRNAs and the remainder to other RNA species (Fig. 1c). To identify putative STAU1-bound duplexes, we annealed the two arms of hybrid reads to identify the longest predicted double-stranded region (Extended Data Fig. 1d). We assessed the validity of these duplexes first by examining whether hiCLIP identifies the best-characterised STAU1-bound duplex in the 3′ UTR of the ADP-ribosylation factor 1 (ARF1) transcript12,14. Both hybrid reads and STAU1 cross-link sites overlapped with the known duplex, and also revealed additional duplexes in ARF1 3′ UTR (Extended Data Fig. 2c-e).
We also tested the thermodynamic stability of duplexes on a transcriptomic level by comparing the minimum free energy of hybridisation between the two arms of hybrid reads with those of randomly repositioned sequences within the same transcript region. Hybrid reads showed lower energies across all types of RNAs (Fig. 1d, Extended Data Fig. 3a-d). Furthermore, a comparison with the PARS scores (parallel analysis of RNA structure)4 confirmed that hiCLIP duplexes in mRNAs are enriched for double-stranded bases compared with neighbouring regions (Extended Data Fig. 3e).
STAU1 interacts with the ribosome in an RNA-dependent manner13,20. Therefore we compared the distribution of hybrid reads from rRNAs with the human 18S and 28S rRNA structures resolved by cryo-electron microscopy (cryo-EM)21. 78% and 72% of hiCLIP duplexes mapping to the 18S and 28S rRNAs, respectively, agreed with cryo-EM-resolved secondary structures (Extended Data Fig. 4), giving a maximum false discovery rate of 26%. In fact, we propose that many of the non-overlapping hybrid reads are candidate novel duplexes that were missed by cryo-EM; for instance, 8% of hybrid reads in 18S rRNA map to a putative duplex connecting distal regions of the molecule (Fig. 1e). The sequences underlying this newly identified duplex are conserved between yeast and human (Extended Data Fig. 4), suggesting their functional relevance. Thus hiCLIP appears to reveal previously undetected secondary structures or tertiary RNA-RNA contacts that are formed in vivo.
As a final validation, we assessed whether hiCLIP duplexes overlap with STAU1 cross-link sites defined by non-hybrid ‘iCLIP-like’ reads. These cross-link sites were enriched up to 70 nts upstream of hiCLIP duplexes (Extended Data Fig. 3f). Moreover, cross-link sites were more likely to be single-stranded (Extended Data Fig. 3g), suggesting STAU1 cross-links to single-stranded RNA regions located 5′ of the duplexes. This is probably because bases within the duplexes are inaccessible for protein-RNA cross-linking.
Having examined the validity of the hiCLIP data, we investigated general properties of STAU1-bound duplexes in mRNAs and long non-coding RNAs (Extended Data Fig. 5a). 96% of hybrid reads originated from two regions of the same RNA species. Of 4,693 hiCLIP duplexes across 2,964 mRNAs, 3,530 were found in 3′ UTRs, 894 in coding sequences (CDS), and the rest were in 5′ UTRs or spanned two regions of the same mRNA (Fig. 1f). Only 4% of hybrid reads corresponded to inter-molecular duplexes between transcripts of different genes (Extended Data Fig. 1a-c, Fig. 1f). In agreement with previous studies11,22, the proportion of long non-coding RNAs was higher among these inter-molecular hybrids (Extended Data Fig. 5a). Alu repeat elements (Alus) were previously reported as common inter-molecular STAU1 binding sites11; however, we found no enrichment of Alu sequences around the STAU1 cross-link sites. Nevertheless, we detected STAU1-binding to Alus when performing iCLIP on whole-cell extracts, indicating these interactions may occur in the nucleus23 (Extended Data Fig. 6a). We concluded that STAU1 rarely binds duplexes formed by different transcripts in the cytoplasm of 293 cells, and therefore we focused our further analyses on duplexes that have both arms in the same RNA.
Despite efforts to characterise STAU1-bound duplexes 13,15-17, it is still not known whether Staufen proteins recognize duplexes of specific length or sequence. 90% of duplexes were between 5nt to 14nt in length, with a median of 8nt (Extended Data Fig. 5b). We observed a prevalence of G-tracts interspersed by A, or C-tracts interspersed by U (Extended Data Fig. 5c); in other words, each duplex arm generally contained stretches of purines or pyrimidines, but not both (Fig. 2a). Short duplexes displayed higher GC content and long duplexes showed higher AU content, in line with the greater stability of GC base pairs (Fig. 2a). The same sequence characteristics were present at hiCLIP duplexes in the CDS, as well as control duplexes predicted by the RNAfold software24 in 3′ UTRs that did not contain any hiCLIP duplexes (Extended Data Fig. 5d-f). This suggests that STAU1 lacks precise sequence specificity, and instead the asymmetric positioning of purine/pyrimidine tracts is a general property of RNA duplexes in human mRNAs. Importantly as evidence of selection pressure to retain STAU1-bound duplexes, we found decreased occurrence of single nucleotide polymorphisms (SNPs) in hiCLIP duplexes compared with neighbouring 3′ UTR regions (Fig. 2b, Extended Data Fig. 6b-c, Supplementary table 2), whereas there was no similar depletion in control duplexes (Extended Data Fig. 6d).
Figure 2. Properties of STAU1-target RNA duplexes.
a, Heatmap of purine content around the two arms of 2,291 duplexes in 3′ UTRs (only duplexes with distance >80 nucleotides between the two arms are shown in order to allow analysis of loop sequence). Duplexes are oriented so that the arm with the higher purine content is on the left hand side of the plot. Line graphs on both sides show the nucleotide content of each arm.
b, Metaprofile of the normalized occurrences of SNPs in the regions around the two arms of the 2,291 duplexes.
Most past studies of RNA secondary structures were limited to analyses of stem loops; i.e., duplexes with short intervening loops (Extended Data Fig. 1e and Supplementary Note). An important feature of hiCLIP is the ability to detect duplexes regardless of loop length: 57% of hiCLIP duplexes in 3′ UTRs had loop lengths of over 100nts and 20% over 500nts (Fig. 3a, b). Moreover, hiCLIP duplexes in 3′ UTRs had significantly longer loops compared with those in the CDS (Fig. 3b). Many long-range hiCLIP duplexes connected the start and end of the 3′ UTR or of the CDS, so that the stop codon is brought into the vicinity of the start codon or the poly(A) tail (Fig. 3a). Since these regions generally bind proteins controlling mRNA translation or stability25, it is possible that long-range duplexes may affect interactions between these factors.
Figure 3. Insights into the secondary structures of mRNAs.
a, Circos plot illustrating the position of two arms of all duplexes within a standardized mRNA. The mRNA is shown at the circumference of the plot, divided into the 5′ UTR, CDS and 3′ UTR. The left and right arms are positioned relative to the start and end of each segment. The lines connecting the two arms are coloured according to the loop length.
b, Probability density distributions of loop lengths among hiCLIP duplexes in the CDS and 3′ UTRs. Counts of RNA duplexes were weighted by the count of hybrid reads. Distributions were compared using the Mann-Whitney U test (n = 953 reads from 894 unique duplexes for CDS and n = 4447 reads from 3530 unique duplexes for 3′ UTR).
c, Probability density distribution of loop lengths among hiCLIP duplexes in 3′ UTRs that were or were not predicted by RNAfold24. Distributions were compared using the Mann-Whitney U test (n = 1348 and 2182, respectively).
d, Probability density distributions of translational efficiencies among mRNAs with hybrid reads in the CDS or 3′ UTR, compared with all mRNAs. mRNA counts were weighted by the count of hybrid reads. Translational efficiencies were compared using the Mann-Whitney U test (n = 8199 from 8199 mRNAs for all, 885 from 696 mRNAs for CDS, and 4064 from 1986 mRNAs for 3′ UTR).
To evaluate if hiCLIP duplexes could be computationally predicted, we used RNAfold to identify secondary structures in 3′ UTRs by their minimum free energy24. The software predicted just 1,348 of 3,530 hiCLIP duplexes; the remaining 2,182 duplexes generally contain much longer loops with lengths up to 8kb (Fig. 3c). This indicates that current computational and experimental methods that rely on thermodynamic-based folding algorithms miss many duplexes with long loops (Supplementary Note).
Of the 2,964 distinct mRNAs with hiCLIP duplexes, 70% contain at least one duplex in the 3′ UTR, 23% in the CDS, but only 6% contain duplexes both in CDS and 3′ UTR (Extended Data Fig. 7a). We used ribosome profiling to assess how the position of duplexes in these different classes of transcripts relates to their translational efficiencies (Extended Data Fig. 7b-c). Transcripts with hiCLIP duplexes in the CDS were poorly translated compared with all mRNAs, whereas those with 3′ UTR duplexes were highly translated (Fig. 3d). In agreement with previous studies4,5,26, the absence of hiCLIP duplexes from the CDS of highly translated mRNAs indicates that translating ribosomes unwind RNA structures.
Next, we examined the effect of STAU1 on mRNA stability and translational efficiency by analysis of STAU1 knockdown cells with or without inducible expression of siRNA-resistant STAU1 (Extended Data Fig. 7d-e). Transcripts with 3′ UTR duplexes displayed an overall increase in abundance upon STAU1 depletion, but no change in translational efficiency (Extended Data Fig. 7f). This is consistent with the reported function of 3′ UTR-bound STAU1 in mRNA decay12. mRNAs with CDS duplexes displayed the opposite trend, with no change in abundance but increased translational efficiency upon STAU1 depletion (Extended Data Fig. 7f). These results contrast with the reported enhancing effect of STAU1 on translation of structured CDS13; the reason for the discrepancy is unclear, though it might be because the RIPiT-Seq (RNA immunoprecipitation in tandem) method used in the previous study may co-purify ribosomes or other proteins. This might also explain why CDS reads are enriched in STAU1 RIPiT-Seq, whereas 3′ UTR reads are enriched in STAU1 hiCLIP (Fig. 1f).
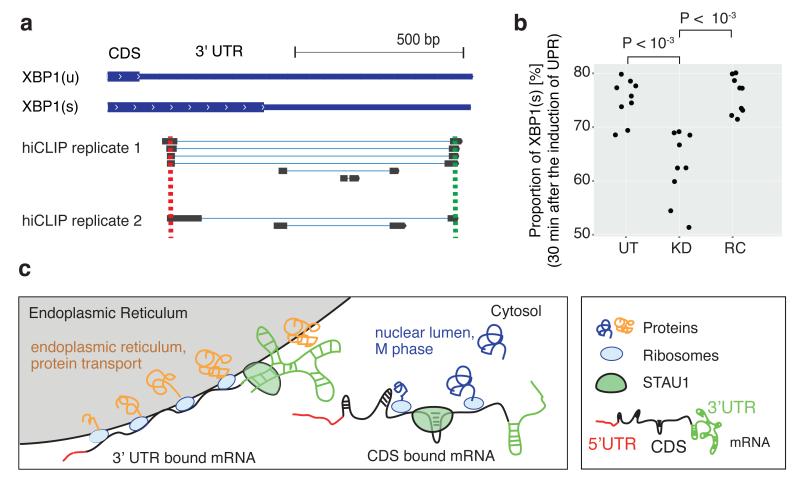
The mRNA encoding XBP1 (also known as endoplasmic reticulum to nucleus signaling 1; ERN1), a central player in ER stress response27, was one of the mRNAs with substantially increased abundance upon STAU1 knockdown (Extended Data Fig. 7f). hiCLIP identified a duplex spanning 858nts in the XBP1 3′ UTR, which is required for efficient splicing of the XBP1 transcript during ER stress (Fig. 4a, b, Extended Data Fig. 8a-d). We produced three reporter constructs to examine the effect of this long-range duplex on the stability of the XBP1 transcript; one containing the XBP1 3′ UTR with the original sequence, one with an AA dinucleotide insertion to disrupt the duplex, and one with an additional insertion of a complementary TT dinucleotide to restore the duplex structure (Extended Data Fig. 8a). We observed that the AA insertion decreased mRNA stability, whereas the complementary TT mutation restored it back to original levels (Extended Data Fig. 8e).
Figure 4. STAU1 regulates cytoplasmic splicing of XBP1 mRNA.
a, Schematic representation of the unspliced XBP1(u) and spliced XBP1(s) mRNA, shown together with hybrid reads that identify RNA duplexes.
b, Proportion of XBP1(s) mRNA 30 min after ER stress induction in untreated (UT), STAU1 knockdown (KD), and rescued (RC) cells. The proportions of XBP1(s) in different conditions were compared using the two-tailed Welch’s t test (n = 9 from 3 independent experiments).
c, Schematic diagram summarizing the functional analyses of STAU1-target mRNAs. 3′ UTR bound mRNAs tend to be highly translated and encode membrane proteins that are translated at the ER (in gray). CDS bound mRNAs tend to be lowly translated and encode proteins that function in the cell cycle M phase. Loops formed by RNA duplexes in the 3′ UTR tend to be longer than in the CDS.
To examine further the functions of proteins encoded by STAU-bound transcripts, we evaluated the enrichment of Gene Ontology terms. Transcripts with duplexes in their 3′ UTRs were enriched for protein trafficking annotations; this suggests that many such transcripts are translated at the rough ER (Fig. 4c, Extended Data Fig. 9, 10, Supplementary table 2), in agreement with the known localization of STAU1 to this compartment28,29. In contrast transcripts with hiCLIP duplexes in the CDS were enriched for annotations relating to nuclear localization and mitosis (Extended Data Fig. 9). This suggests that STAU1 represses translation of mRNAs encoding mitotic proteins, which is consistent with a recent report that STAU1 degradation is required during mitosis for efficient cell cycle progression30.
Our study reveals the unforeseen prevalence of long-range duplexes in 3′ UTRs with importance for mRNA stability, whereas duplexes in the CDS have shorter loops and are mainly found in poorly translated mRNAs. Depletion of SNPs from duplexes suggests a way to improve interpretations of disease-causing mutations in 3′ UTRs. In conclusion, hiCLIP identifies RNA duplexes that form in vivo and thereby opens a new avenue for transcriptome-wide studies of full-length RNA structures.
Online Methods
Analysis pipeline
We developed a computational pipeline for data analysis, which is freely accessible from https://github.com/jernejule/STAU1_hiCLIP. The pipeline is implemented as a package of codes that enables the user to reproduce most plots and results using the sequencing data as input. All sequencing data are available from ArrayExpress (iCLIP and hiCLIP: E-MTAB-2937, mRNA-Seq: E-MTAB-2940, ribosome profiling: E-MTAB-2941).
Plasmids
We produced two Flp-In T-REx 293 cell lines; one expressing FLAG-STAU1 (used for hiCLIP), and the other expressing siRNA-resistant FLAG-STAU1 (used for all functional assays). For the FLAG-STAU1, the sequence of 3× FLAG tag was added to the 3′ end of the STAU1 coding sequence from NM_017454.2, and inserted into the pcDNA5/FRT/TO plasmid (Life Technologies, V6520-20). For the siRNA-resistant FLAG-STAU1, a region of STAU1 CDS (ggactagtaataaagaggatgagtt) was silently mutated to gCacAagCaaCaaGgaAgaCgaAtt (capital letters show the positions of mutations). These mutations were introduced using the si_pr1 (ACGATGCTGCTGCCAAAGCGT), si_pr2 (aaTtcGtcTtcCttGttGct TgtGccattttcatccccagagccagg), si_pr3 (gCacAagCaaCaaGgaAgaCgaAttcaggatgccttatc taagtcatc), si_pr4 (ACGGGGGAGGGGCAAACAAC) primers.
To generate the plasmids used for formaldehyde cross-linking and RNA co-immunoprecipitation assay, reporter constructs were inserted into pcDNA3 FLuc, where the CDS of firefly luciferase (FLuc) was inserted into the pcDNA3 plasmid (Life Technologies). The plasmid containing ARF1 STAU1 binding site (SBS) was a gift from Professor Maquat14. The ARF1 SBS was PCR amplified from this plasmid using C_pr1 (ATTTTTTGGATCCACGCGACCCCCCTCCCTC) and C_pr2 (ATTTTTTCTCGAGGTGCCCATGGGCCTACATCC), and cloned into pcDNA3 FLuc (pcDNA3 FLuc ARF1 SBS). pcDNA3 FLuc ARF1 SBSΔ was generated from pcDNA3 FLuc ARF1 SBS using D_pr1 (GTGCGGCTCGTGGTGTCGTGGTTTGGTCACCG) and D_pr2 (CGGTGACCAAACCACGACACCACGAGCCGCAC). The 3′ UTR of XBP1 was PCR amplified from the total RNA of Flp-In T-REx 293 cells using C_pr3 (ATTTTTTGGATCCTGACCACATATATACCAAGCCCC) and C_pr4 (ATTTTTTCTCGAGGCATTGTACCTTTTAATTGCATGGG), and cloned into pcDNA3 FLuc (pcDNA3 FLuc XBP1). pcDNA3 FLuc XBP1Δ were generated from pcDNA3 FLuc XBP1 using D_pr3 (CTAATGTGGTAGTGAAAATCCTCAGCCCCTCAGAG) and D_pr4 (CTCTGAGGGGCTGAGGATTTTCACTACCACATTAG).
To generate the plasmids used for functional studies of the long-range duplex in the 3′ UTR of XBP1 mRNA, the 3′ UTR of XBP1 was PCR amplified from the total RNA of Flp-In T-REx 293 cells using fX_pr1 (CCGCTCGAGTTCGTTTTGACCACATATATACCAAG) and fX_pr2 (ATAGTTTAGCGGCCGCGATGCT GCATTGTACCTTTTAATTGC), and cloned into psiCHeck2 (Promega, C8021). The XBP1 mut and comp reporter (Fig. 4b) were generated from the XBP1 wt construct using fX_pr3 (GCTAGTGTAGCTTCTGAAAGGTGaaCTTTCTCCATTTATTTAAAACTACCC) and fX_pr4 (GGGTAGTTTTAAATAAATGGAGAAAGttCACCTTTCAGAAGCTACACTAGC) –, and from the XBP1 mut construct using fX_pr5 (CTAATGTGGTAGTGAAAATCGAGGAAGttCACCTCTCAGCCCCTCAGAGAA) and fX_pr6 (TTCTCTGAGGGGCTGAGAGGTGaaCTTCCTCGATTTTCACTACCACATTAG). All cloning was performed with the Phusion® High-Fidelity PCR Master Mix with HF Buffer (NEB, M0531L) according to the manufacturer’s protocol (PCR template plasmid <250 ng).
Antibodies used for western blotting
Anti-GAPDH (14C10) antibody (NEB, 2118) and anti-Staufen 1 antibody (Proteintech, 14225-1-AP) were used for the western blotting analysis.
Cell culture
Flp-In 293 T-REx cells (Life Technologies, R780-07) were cultured in DMEM with 10% FBS, 3 μg/ml Blasticidin S HCl (Life Technologies, A11139-03), 50 μg/ml Zeocin (Life Technologies, R250-01). To produce cell lines, the pcDNA5/FRT/TO plasmid with FLAG-STAU or siRNA-resistant FLAG-STAU1 was co-transfected with pOG44 plasmid into Flp-In 293 T-REx cells. Cells stably expressing these proteins were selected by culturing in DMEM containing 10% FBS, 3 μg/ml Blasticidine S HCl, 200 μg/ml Hygromycine (InvivoGen, ant-hm-5). Absence of mycoplasma contamination was confirmed using the LookOut Mycoplasma PCR Detection Kit (SIGMA, MP0035).
Knockdown (KD) and rescue (RC) of STAU1
We performed all knockdown experiments with siRNA-resistant FLAG-STAU1 Flp-In 293 T-REx cell line under three conditions: untransfected controls (UT), knockdown with an siRNA against STAU1 (KD), and a rescue comprising knockdown combined with inducible expression of siRNA-resistant STAU1 (Extended Data Fig. 7d). 120 pmol of stealth STAU1 siRNA (Life Technology, STAU1HSS110293) was transfected to cells growing on 6 well-dishes with 4.5 μl of RNAiMAX (Life Technologies, 13778-150) using the manufacturer’s reverse-transfection protocol. 24 h after transfection, the medium was replaced with DMEM with 10% FBS and 48 h after the transfection, the cells were collected for the analysis. For untreated control (UT), cells were only treated with RNAiMAX. For rescue experiment (RC), 24 h after transfection of 120 pmol of stealth STAU1 siRNA, the medium was replaced with DMEM with 10% FBS and 100 ng/mL of doxycycline (DOX) to induce expression of siRNA-resistant STAU1, and cells were collected 48 h after transfection.
hiCLIP (Preparation of adaptors)
Adaptor A (/5Phos/rArGrArUrCrGrGrArArGrArGrCrGrGrUrUrCrArG/3ddC/; 5Phos indicates 5′ phosphate, rN indicates the nucleotide is ribonucleotides, and 3ddC indicates 3′-dideoxycytidine) and Adaptor B (/5Phos/rCrUrGrUrArGrGrCrArCrCrArUrArCrArArUrG/3Phos/; 3Phos indicates 3′-phosphate) were ordered as HPLC-purified RNA (Integrated DNA Technologies). They were pre-adenylated using 5′ DNA Adenylation Kit (NEB: E2610) and purified by PAGE with the following protocol. 200 μL of pre-adenylation mix (10 μl 100 μM non adenylated Adaptor A or B, 20 μl 10× 5′ DNA Adenylation Reaction Buffer, 20 μl 1mM ATP, 20 μl Mth RNA ligase, and 130 μl Water) was prepared and incubated at 65°C for 1 hour. 200 μl of TE buffer and 400 μl Acid-Phenol:Chloroform, pH 4.5 (acid-PCI, Life Technologies, AM9722) was added and the mixture was transferred to a 2 ml Phase Lock Gel Heavy tube (VWR, 713-2536). The tube was inverted at least 10 times to mix fully and the phases were separated by 5 min centrifugation at 15,871 ×g at room temperature. The aqueous layer was transferred to a new tube and the isolated RNA was precipitated by mixing 1.5 μl of Linear acrylamide (Life Technologies, AM9510), 40 μl of 3 M sodium acetate pH 5.5, and 1 ml 100% ethanol, and incubated overnight at −20°C. The reaction was centrifuged at 21,800 ×g for 15 min at 4°C. The supernatant was removed and the pellet was washed with 500 μl of 80% ethanol. The pellet was resuspended in 5 μl water. The pre-adenylated linkers were purified by 15% TBE-UREA PAGE (Life Technologies) and gel excision. To extract the adaptors from the gel, 400 μl TE buffer with 1 μl of RNasin (Promega, N2615) was added to excised gel piece, which was crushed into small pieces with a Squisher-Single (Zymo Research, H1001-50). The crushed gel in TE buffer was incubated at 37°C for 1 h at 1100 rpm shaking using the Thermomixer Comfort (Eppendorf, 5355 000.011) or Compact (Eppendorf, 5350 000.013), then placed on dry ice for 2 min, and incubated again at 37°C for 1 h with 1100 rpm shaking. Adaptors were purified using acid-PCI extraction and EtOH precipitation as described above. The adenylated adaptors were used in hiCLIP experiments (and called Adaptor A and Adaptor B respectively).
hiCLIP (UV cross-linking)
STAU1 was induced in FLAG-STAU1 Flp-In 293 T-REx cell line by adding 250 ng / ml of doxycycline to the medium cells growing on a 15 cm dish. After 16 h incubation at 37°C at 5% CO2, cells were washed once by ice-cold PBS, and 10 ml ice-cold PBS was added. Cells were subjected to 150 mJ/cm2 of UV-C (254 nm) irradiations with Stratlinker 2400 on ice, and scraped off. The cells from 5 dishes were transferred to a 50 ml tube and used for single hiCLIP experiment. The cells were pelleted by centrifugation at 514 ×g for 5 min at 4°C, and the supernatant was removed. The pellets were snap-frozen on dry ice and stored at −80 °C until use.
hiCLIP (Preparation of antibody-coupled beads)
Dynabeads covalently coupled to the M2-FLAG antibody were prepared using the Dynabeads Antibody Coupling Kit (Life Technologies, 14311D) following the manufacturer’s protocol (30 μg of antibody / 1 mg of beads). 100 μl antibody-coupled beads were used for a single immunopreciptation experiment. Before immunoprecipitation, beads were washed once with CLIP Lysis Buffer (50 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate), and twice with High Salt Wash (50 mM Tris-HCl, pH 7.4, 1 M NaCl, 1 mM EDTA, 1% Igepal CA-630 (SIGMA, I8896), 0.1% SDS, 0.5% sodium deoxycholate). Beads were left in PGB Cell Lysis buffer (20 m Tris-HCL (pH 7.4), 140 mM NaCl, 5 mM MgCl2, 1% Triton-X 100) until cell lysate was prepared.
hiCLIP (Preparation of the cell lysate and partial RNA digestion)
Frozen cell pellet in a 50 ml tube was resuspended in 30 ml of PGB Cell Lysis Buffer Complete (PGB Cell Lysis Buffer supplemented with 1/1000 1M DTT, 1/1000 anti-RNase (Life Technologies, AM2690), 4/1000 Protease Inhibitor Cocktail Set III, EDTA-Free (MERCK, 539134), 0.1 μl / 1ml TURBO DNase (Life Technologies, AM2238)). The lysate was homogenized by passing twice through a syringe with a 21G needle and cleared by centrifugation at 14,000 ×g for 5 min at 4°C. The supernatant was collected and centrifuged again at 14,000 ×g for 15 min 4°C. The supernatant was collected and 20 ml of PGB Cell Lysis Buffer Complete was added and filtered through a 0.45 μm syringe filter. Unprotected RNA was digested by adding 0.4 U/ml (High RNase condition) or 0.2 U/ml (Low RNase, 2nd round ligation-minus control and STAU1 induction-minus condition) of RNase I (Life Technologies, AM2294) to the lysate, and the lysate was incubated for 5 min at 37°C. After incubation, the tube was transferred to ice for a minimum of 5 min. In order to stop RNase I activity, 20 μl of SUPERaseIn (Life Technologies, AM2694) was added.
hiCLIP (Immunoprecipitation)
PGB Cell Lysis Buffer was removed from the beads. The beads were resuspended in 1 ml of PGB Cell Lysis Buffer and added to the cell lysate. The bead/lysate mix was incubated for 2 ~ 3 h at 4°C while rotating. The beads were collected by centrifugation at 805 ×g for 5 min at 4°C and the supernatant removed. 500 μl of PGB Cell Lysis Buffer was added and the beads were resuspended and transferred to a 1.5 ml tube. The supernatant was discarded using a magnetic stand and the beads were washed twice with PGB Cell Lysis Buffer (the second wash was rotated for at least 1 min in the cold room). To remove completely the residual DNA, the beads were resuspended in 1 ml of PGB Lysis Buffer (containing 1 μl of RNasin, 2 μl of TURBO DNase, 1 μl of SUPERaseIn) and incubated at 37 °C for 3 min at 1100 rpm shaking. The supernatant was discarded and the beads were washed twice with PGB Cell Lysis Buffer (the second wash was rotated for at least 1 min in the cold room). The beads were further washed twice with PNK buffer (20 mM Tris-HCl, pH 7.4 10 mM MgCl2, 0.2% Tween-20) and resuspended in 1 ml PNK Buffer.
hiCLIP (3′ end RNA dephosphorylation and 1st round of adaptor ligation)
5× PNK pH 6.5 buffer (350 mM Tris-HCl (pH 6.5), 50 mM MgCl2, 25 mM DTT) was prepared. The supernatant was discarded and the beads were resuspended in 20 μl of the PNK mix (4 μl 5× PNK pH 6.5 buffer, 0.5 μl T4 PNK (NEB, M0201), 0.5 μl RNasin, 0.5 μl SUPERaseIn, 14.5 μl water) and incubated at 37°C for 20 min with 1100 rpm shaking. The beads were washed once with PNK buffer and twice with PGB Cell Lysis Buffer (the last wash was rotated for at least 5 min in the cold room). Beads were further washed three times with PNK buffer.
We ligated the RNA duplexes bound to STAU1 to an equimolar concentration of RNA adaptors A and B. Assuming that ligation efficiency is 100%, this should lead to 50% of duplexes with Adaptor A ligated to one arm, and Adaptor B to the other arm, which is necessary for the production of hybrid reads by hiCLIP (Extended Data Fig. 1a). 4× ligation buffer was prepared (200 mM Tris-HCl, pH 7.8, 40 mM MgCl2, 40 mM DTT). The supernatant was removed and the beads were resuspended in 20 μl of the 1st round ligation mix (6 μl water, 4 μl 4× ligation buffer, 1 μl T4 RNA Ligase 2, truncated K227Q (NEB, M0351), 0.5 μl RNasin, 0.5 μl SUPERaseIn, 2 μl Adaptor A (10 μM), 2 μl Adaptor B (10 μM), 4 μl PEG400 (SIGMA, 81170)). T4 RNA Ligase 2, truncated K227Q was chosen to selectively ligate pre-adenylated adaptor to RNA. The mixture was incubated overnight at 16°C at 1100 rpm shaking. 500 μl PNK buffer was added and the supernatant was removed. The beads were washed twice with 1 ml PGB Cell Lysis Buffer (both of wash was rotated 5 min at 4°C). The beads were further washed twice with 1 ml PNK buffer. Beads resuspended in 1 ml PNK buffer were transferred to a new tube after the first wash.
hiCLIP (5′ end phosphorylation of RNA, removal of adaptor B’s phosphate blocking, and 2nd round of RNA ligation)
5′ end phosphorylation of RNA and removal of adaptor B’s phosphate blocking were simultaneously performed. The supernatant was removed and 20 μl of PNK mix (1 μl T4 PNK, 2 μl 10mM ATP, 2 μl 10× PNK buffer (NEB), 0.25 μl RNasin, 0.25 μl SUPERaseIn, 14.5 μl water) was added. The reaction was incubated at 37°C for 30 min at 1100 rpm shaking. The beads were washed once with 1 ml PNK buffer, twice with 1 ml PGB Cell Lysis Buffer (the wash was rotated for at least 5 min in cold room), and once with 1 ml PNK buffer.
The supernatant was removed and 20 μl of ligation mix (10 μl 10mM ATP, 72.5 μl water, 10 μl 10× T4 RNA ligase 1 buffer (NEB, B0216S that does not contain ATP), 5 μl T4 RNA Ligase 1 (ssRNA Ligase) (NEB, M0204), 2 μl RNasin, 0.5 μl SUPERaseIn) was added. The reaction was incubated overnight at 16°C at 1100 rpm shaking. After incubation, 800 μl of ice-cold PNK buffer was immediately added, the supernatant was removed, and the tube was transferred to ice. The beads were washed once with High Salt Wash and twice with PNK Buffer.
hiCLIP (Denaturation of cross-linked protein-RNA complexes)
We denatured proteins with urea to ensure that no additional proteins could interact with STAU1 during purification. The protocol for purifying protein-RNA complexes with UREA denaturation was adapted from Kiel et al31. In our preliminary experiments, we found that this UREA denaturation step was required to remove other RNA-binding proteins that tightly interact with STAU1 and are co-purified under standard iCLIP conditions32. Thus, we included these stringent purification conditions in the hiCLIP protocol to ensure that only RNAs directly interacting with STAU1 in vivo were isolated. The supernatant was removed from immunoprecipitated beads and 80 μl of 1.25 × Urea Cracking Buffer (66.6 mM Tris-HCl, pH 7.4, 8M Urea, 1.33% SDS) was added. The reaction was incubated for 3 min at 65 °C at 1100 rpm shaking. The supernatant was collected and 920 μl of ice-cold Tween 20 IP buffer with 1 μl of anti-RNase, 10 μl of Protease Inhibitor Cocktail Set III, EDTA-Free and 1 μl of SUPERaseIn was added. The supernatant was further cleared using a magnetic stand in order to remove any remaining beads.
hiCLIP (Preparation of antibody-attached beads)
100 μl of Dynabeads Protein G (Life Technologies, 10004D) were used per experiment. The beads were washed twice with CLIP Lysis buffer. The beads were resuspended in 100 μl CLIP Lysis buffer with 10 μg M2-FLAG antibody per experiment and incubated at room temperature for 1-2 h with rotation. The beads were washed twice with CLIP Lysis buffer, once with Tween 20 IP Buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% Tween 20, 0.1 mM EDTA)31.
hiCLIP (Stringent purification and radiolabelling of protein-RNA complexes)
The supernatant of denatured protein-RNA complexes was added to 100 μl of antibody-attached beads and incubated at 4 °C for 2h with rotating. The beads were washed twice with High Salt Wash and twice with PNK buffer and left in the final wash. In order to visualize protein-cross-linked RNA complexes, the 5′ end of the RNA was radiolabeled. 200 μl (20%) of beads resuspended in 1 ml PNK buffer was transferred to a new tube and the supernatant was removed. The beads were mixed with hot PNK mix (0.2 μl PNK, 0.4 μl 32P-γ-ATP, 0.4 μl 10× PNK buffer, 3 μl water) and incubated at 37°C for 5 min at 1100 rpm shaking. The supernatant was removed and 20 μl of 1x NuPAGE LDS Sample Buffer (Life Technologies, NP0007) with NuPAGE Sample Reducing Agent (Life Techonologies, NP0004) was added to the beads. The supernatant of the remaining non-radiactive beads (80%) was removed and the radioactively labeled beads (20%) resuspended in 20 μl of NuPAGE buffer were added to the non-radioactive beads.
hiCLIP (SDS-PAGE and transfer of protein-RNA complexes to nitrocellulose)
The supernatant in NuPAGE buffer was incubated at 70°C for 5 min at 1100 rpm shaking. The tube was placed on a magnetic stand to remove the beads, and the supernatant was loaded on the SDS-PAGE gel. The SDS-PAGE was performed using 4-12% NuPAGE Bis-Tris gel (Life Technologies, NP0321BOX) following the manufacturer’s protocol with MOPS running buffer (Life Technologies, NP0001), NuPAGE Antioxidant (Life Technologies, NP0005) was added to the buffer in the upper chamber, and PAGE was performed for 45 min at 180 V. 5 μl of PAGE ruler plus (Thermo Scientific, SM1811) was used as protein size marker.
After the run, the dye front of the gel was removed and discarded since this part contained free radioactive ATP. The protein-RNA complexes from the gel were transferred to a Protran BA85 Nitrocellulose Membrane (Whatman) using the Novex wet transfer apparatus following the manufacturer’s protocol (Life Technologies; transfer for 1 h at 30 V using NuPAGE transfer buffer (Life Technologies, NP0006-1, supplemented with 10% methanol). The membrane was wrapped with saran wrap and exposed to a Carestream Kodak BioMax XAR Film (SIGMA, Z358487) at −80°C for 1 hour and overnight.
hiCLIP (Purification condition test)
To optimize the ideal RNase concentration, purification condition test (Fig. 1a) was performed using the hiCLIP protocol with the following modifications. One third of cells grown on a 15 cm dish were used for each immunoprecipitation experiment. The cells were resuspended in 1 ml of PGB Cell Lysis Buffer Complete. The lysate was cleared with centrifugation. Unprotected RNA was digested by adding 0.05, 0.1, 0.2, 0.4, 2 U/ml (from Low to High RNase condition) RNase I to lysate, or in case of UV-C cross-linking minus control and STAU1 induction-minus control, 0.4 U/ml of RNase I. For this optimization, the steps “3′ end RNA dephosphorylation and 1st round of adaptor ligation” and “5′ end phosphorylation of RNA, removal of adaptor B’s phosphate blocking, and 2nd round of RNA ligation” were skipped to determine the exact molecular weight of the protein that was cross-linked to RNA.
hiCLIP (RNA isolation)
We observed a slower migration of the band after the adaptor ligation (compare Fig. 1a and Extended Data Fig. 2a), showing the linker ligation to cross-linked RNA increases the apparent molecular weight of the protein-RNA complex. The region of the membrane containing the radioactively labeled cross-linked protein-RNA complex was excised using autoradiograph as a mask, and the nitrocellulose piece was transferred to a 1.5 ml tube. 10 μl proteinase K (Roche, 03115828001) in 200 μl PK buffer (100 mM Tris-HCl, pH 7.4, 50 mM NaCl, 10 mM EDTA) was added to the piece of membrane and incubated at 37°C for 20 min at 1100 rpm shaking. 200 μl of PK buffer with 7 M UREA was added and incubated at 37°C for 20 min at 1100 rpm shaking. The supernatant was collected and added together with 400 μl acid-PCI to a 2 ml Phase Lock Gel Heavy tube. The tube was inverted at least 10 times and the phases were separated by centrifugation at 15,871 ×g for 5 min at room temperature. The aqueous layer was transferred to a new tube and isolated RNA was precipitated by mixing 1 μl Linear acrylamide, 40 μl 3 M sodium acetate pH 5.5, and 1 ml 100% ethanol, incubated overnight at −20°C and centrifuged at 21,800 ×g for 15 min at 4°C. The supernatant was removed and the pellet was washed with 500 μl of 80% ethanol. The pellet was resuspended in 6.25 μl of water.
hiCLIP (Reverse transcription and purification of cDNAs)
The RNA duplexes that contain Adaptor A ligated to one arm, and Adaptor B to the other arm, are the template for producing hybrid reads by hiCLIP (Extended Data Fig. 1a). Most cDNAs usually truncate at the position of protein-RNA cross-link site. Our analysis reveals that cross-link generally occurs 5′ of the duplex (Extended Data Fig. 3f). Thus, we expect that the duplexes containing the cross-link site in the arm ligated to the Adaptor B will be well suited for producing hybrid reads, which will contain both arms of the duplex.
The RT mix 1 (1 μl primer Rt#clip (0.5 μM; Rt15clip (/5Phos/NNTATTNNNAGATCGGAAGAGCGTCGTGGATCCTGAACCGC) for the high RNase condition, Rt1clip (/5Phos/NNAACCNNNAGATCGGAAGAGCGTCGTGGATCCTGAACCGC) for the low RNase condition, Rt5clip (/5Phos/NNCGCCNNNAGATCGGAAGAGCGTCGTGGATCCTGAACCGC) for the second round ligation minus control, and Rt12clip (/5Phos/NNGTGGNNNAGATCGGAAGAGCGTCGTGGATCCTGAACCGC) for FLAG-STAU1 minus induction control) and 1 μl dNTP mix (10 mM each)) was added to the resuspended pellet. The primers were annealed to the linkers by incubating at 70°C for 5 min and 25°C for 1 min. RT mix 2 (4 μl 5× RT buffer (Life Technologies), 1 μl 0.1 M DTT, 0.5 μl Superscript III RT (200 U/μl; Life Technologies, 18080-044), 0.5 μl RNasin, 5.75 μl Water) was added and reverse transcription reaction was performed by using the program of 25°C for 5 min, 42°C for 20 min, 50°C for 40 min, 80°C for 5 min, 16°C for hold. The RNA template was hydrolyzed by adding 2.2 μl of 1 M NaOH and incubated at 98°C for 20 min. The reaction was neutralized by adding 25 μl of 1M HEPES-NaOH and 352.8 μl of TE buffer. The cDNA was EtOH-precipitated as described above.
The pellet was resuspended in 6 μl of water and 6 μl of 2× Novex TBE-Urea Sample Buffer (Life Technologies, LC6876) was added. The cDNA was denatured by incubating at 80°C for 5 min and immediately run on 6% TBE-urea gel (Life Technology, EC6865BOX). The region of the gel corresponding to 100 – 300 nts of cDNAs were cut out and transferred to a 1.5 ml tube. In order to extract cDNAs, 400 μl TE buffer was added to the gel piece and the gel was crushed into small pieces with a Squisher-Single. The crushed gel was incubated at 37°C for 1 h at 1100 rpm shaking, then placed on dry ice for 2 min, and incubated again at 37°C for 1 h at 1100 rpm shaking. The extracted cDNA was EtOH-precipitated as described above.
hiCLIP (Circulization and linearization of cDNAs)
The pellet was resuspended in 8 μl ligation mix (6.5 μl water, 0.8 μl 10× CircLigase Buffer II (Epicentre), 0.4 μl 50 mM MnCl2, 0.3 μl CircLigase II (Epicentre, CL9025K)) and incubated for 1 h at 60°C. 30 μl oligo annealing mix (26 μl water, 3 μl 10 × FastDigest Buffer (Thermo Scientific), 1 μl 10 μM Cut_oligo (GTTCAGGATCCACGACGCTCTTCAAAA)) was added. The oligo was annealed to cDNAs with the program of (95°C for 2 min; successive cycles of 20 s, starting from 95°C and decreasing the temperature by 1°C each cycle down to 25°C; and 25°C hold). 2 μl FastDigest BamHI (Thermo Scientific, FD0055) was added and incubated for 30 min at 37°C, and then the enzyme was inactivated by incubating at 80°C for 10 min. cDNA was EtOH-precipitated as described above.
hiCLIP (PCR amplification of cDNA library)
The pellet was resuspended in 11 μl of water. The PCR mix (5 μl cDNA, 2.5 μl primer mix containing P5Solexa (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT) and P3Solexa (CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT) at 10 μM each, 50 μl Accuprime Supermix 1 enzyme (Life Technologies, 12342-010), 42.5 μl water) was prepared. PCR was performed by following program (94°C for 2 min; 24 cycles of 94°C for 15 s, 65°C for 30 s and 68°C for 30 s; 68°C for 3min, and 25°C hold).
Ribosome Profiling and mRNA-Seq (Preparation of cell Lysate)
The published protocols were adjusted33-36. The siRNA-resistant FLAG-STAU1 Flp-In 293 T-REx cell line was grown on a 15 cm dish, and knockdown or rescue was performed as described above by adjusting the volume accordingly. The medium of cells was replaced with DMEM with 10% FBS and 100 μg/ml of Cycloheximide (SIGMA, C4859) and incubated at 37°C for 1 min at 5% CO2. The medium was removed and the dish was moved to ice. The cells were washed once with 10 ml of ice cold PBS with 100 μg/ml of Cycloheximide. PBS was removed and 1 ml of PGB Cell Lysis Buffer with 1 μl of 100 mg/ml Cycloheximide, 10 μl of 100 mM DTT and 12.5 μl of TURBO DNase was added. Cells were scraped off on ice, pipetted up and down, and collected in 1.5 ml tubes. The lysate was homogenized by passing syringe 26G needle twice. The lysate was centrifuged twice at 20,000 ×g for 10 min at 4°C, and 700 μl of the supernatant was collected for ribosome profiling experiment, 250 μl for mRNA-Seq and 50 μl for Western blotting. For mRNA-Seq samples, 250 μl of lysate was directly added to 750 μl of TRIzol LS (Life Technologies, 10296-028), incubated at room temperature for 5 min and stored at −80°C till use.
Ribosome Profiling (Generation of ribosome footprints)
17.5 μl of RNase I was added to 750 μl of the cell lysate and incubated at 23°C for 45 min at 800 rpm shaking. RNase digestion was stopped by adding 35 μl of SUPERaseIn. 600 μl of the lysate was immediately loaded to a pre-cooled centrifuge tube (Beckman, 349622), then 900 μl of 1M ice-cold sucrose cushion buffer (1M sucrose cushion, 20 m Tris-HCl (pH 7.4), 140 mM NaCl, 5 mM MgCl2 and 1 ml of this buffer was supplemented with 1 μl SUPERaseIn, 10 μl 100 mM DTT, 1 μl 100 mg/ml cycloheximide) was added under the cell lysate by placing the tips to the bottom of the tube. The cushion was centrifuged at 70,000 rpm for 4 h at 4°C with TLA-100.3 rotor (Beckman). The supernatant was discarded and the RNA was extracted using Trizol reagent (Life Technologies, 15596-026) following the manufacturer’s protocol except that the sample was incubated overnight at −80°C before centrifugation.
mRNA-Seq (poly(A) RNA selection and fragmentation)
mRNAs were purified from 250 μl of the cell lysate using TRIzol LS following the manufacturer’s protocol except that the sample was incubated overnight at −80°C before centrifugation. Poly(A) RNA was purified using Dynabeads mRNA DIRECT Kit (Life Technologies, 61011) following the manufacturer’s protocol after resuspending RNA pellet to 1 ml of Lysis/Binding Buffer of the kit. RNA was eluted in 20 μl of 10 mM Tris-HCl (pH 8.0).
20 μl of 2× Alkaline fragmentation solution (2 mM EDTA, 10 mM Na2CO3, 90 mM NaHCO3 (pH 9.3)) was added to the eluted RNA and incubated for 20 min at 95°C. Fragmentation was stopped by adding 560 μl ice-cold stop/precipitation solution (final 300 mM NaOAc pH 5.5 and 1 μl of GlycoBlue (Life Technologies, AM9516) per reaction). 600 μl of room-temperature isopropanol was added and incubated at −20°C overnight. The RNA was precipitated by centrifuging at 21,800 ×g for 30 min at 4 °C. The supernatant was discarded and the pellet was washed twice with 500 μl of 75% EtOH and dried at room temperature for 3 min.
Ribosome Profiling and mRNA-Seq (Size selection of fragmented RNAs)
For ribosome footprints, 26-34 nts of RNA was purified using 15% TBE-UREA gel (Life Technologies) as was described by Ingolia et al36, while 50-80 nts of RNA was purified using 10% TBE-UREA gel (Life Technologies) for mRNA-Seq. The RNA was extracted from the gel using the same protocol as hiCLIP cDNA extraction except that 1 μL of RNasin Plus (Promega, N2615) was added to TE buffer and extracted RNA was subjected to acid-PCI extraction before EtOH precipitation.
Ribosome Profiling and mRNA-Seq (3′ end dephosphorylation and adaptor ligation)
The RNA pellet was resuspended in 20 μl of H2O and mixed with PNK master mix (10 μl 5× PNK ph6.5 buffer (custom buffer, described above), 1.25 μl PNK (10 U/μl), 1 μl RNasin, 17.75 μl H2O). The reaction was incubated for 20 min at 37 °C and RNA was purified by acid-PCI extraction and EtOH precipitation as described above.
The RNA was resuspended in 4.5 μl of water and mixed with ligation master mix (4 μl 4 × ligation buffer (custom buffer described above), 1 μl T4 RNA ligase, 0.5 μl RNasin, 1 μl of 100 μM L3-App (/5rApp/AGATCGGAAGAGCGGTTCAG/ddC/; 5rApp indicates 5′ adenylation), 4 μl PEG400, 5 μl water). The reaction was incubated overnight at 16 °C and purified by PCI (UltraPure Phenol:Chloroform:Isoamyl Alcohol (pH 8.05) (Life Technologies; 15593-031); Acid-PCI was used for RNA extraction, while this PCI was used for DNA-RNA hybrid and DNA extraction in this study) extraction and EtOH precipitation as described above. The adaptor ligated RNA fragments corresponding the size of 47 – 54 nts for ribosome profile, while 71-101 nts for mRNA-Seq were PAGE purified as described above using 10% TBE UREA gel.
Ribosome Profiling and mRNA-Seq (Reverse transcription, circulization and linearization of cDNAs, PCR amplification of cDNA library)
Reverse transcription and size selection, circulization and linearization of cDNAs, PCR amplification of cDNA library were performed using the same protocol as hiCLIP except the following modifications. 0.5 μl of 100 μM primer Rt#clip (Rt13clip (/5Phos/NNTCCGNNNAGATCGGAAGAGCGTCGTGGATCCTGAACCGC) for the replicate 1 of UT, KD, and RC conditions, and Rt9clip (/5Phos/NNGCCANNNAGATCGGAAGAGCGTCGTGGATCCTGAACCGC) for the replicate 2 of UT, KD, and RC conditions) was used instead of 0.5 μM for reverse transcription. At cDNA size selection from gel, the sizes of purified cDNAs were 78-88nt for ribosome profiling, and 102-132nt for mRNA-Seq. 68 μl oligo annealing mix and 4 μl FastDigest BamHI were used for linearization. 10-15 cycles of PCR amplifications were used.
High-throughput DNA sequencing
The hiCLIP library was sequenced using MiSeq Reagent Kit v2 (300 cycle) (Illumina, MS-102-2002) and Illumina MiSeq for 196 cycles. Ribosome profiling and mRNA-Seq library was sequenced using Illumina HiSeq for 50 cycles.
iCLIP
STAU1 iCLIP was performed according to the denaturing iCLIP32 protocol as described previously.
Formaldehyde cross-linking and RNA co-immunoprecipitation assay
48h before the immunoprecipitation experiment, 1 μg of plasmid mix (ARF1 WT, ARF1 Δ, XBP1 WT, XBP1 Δ, PRKCSH WT and PRKCSH Δ; equal amount of wt and Δ was mixed) was co-transfected to siRNA-resistant FLAG-STAU1 Flp-In 293 T-REx cell line grown in a 10 cm dish with 10 μl of TurboFect Transfection Reagent (Thermo Scientific, R0531) following the manufacturer’s protocol. PRKCSH WT and Δ did not express, and thus were not reported in this manuscript. 24h before the immunoprecipitation experiment, FLAG tagged STAU1 was induced by adding 250 ng / ml of doxycycline. Cells were cross-linked using 1 % formaldehydes for 10 min and quenched by glycine as described in Niranjanakumari et al37. Whole cell pellet from a 10 cm dish was used for a single immunoprecipitation experiment. Later parts of the protocol were modified in the following way. 100 μl of protein G Dynabeads were washed twice with CLIP Lysis buffer. Beads were resuspended in 100 μl CLIP lysis buffer with 10 μg M2 anti FLAG antibody and incubated for 1-2 h at room temperature. The beads were washed three times with CLIP Lysis buffer. The previously prepared cell pellet was resuspended in 100 μl 6 M UREA cracking buffer and sonicated using a Bioruptor (low amplitude, 5 × of 30 sec on and 30 sec off). 1 ml of T20 IP buffer (with 10 μl of protease inhibitors, 1 μl of anti-RNase) was added to dilute UREA. Lysate was centrifuged two times at 21,800 ×g for 20 at 4°C and the supernatant was collected each time. 35 μl of cleared lysate was used for total RNA extraction by mixing with 205 μl of ChIP elution buffer (100 mM Tris-HCL (pH7.4), 10 mM EDTA, 1% SDS, 200 mM NaCl) and cross-links were reversed as described below.
The cleared cell lysate was mixed with previously prepared beads and incubated for 2 h at 4°C with rotation. The beads were washed twice with high salt buffer (second wash was rotated at 4°C for 1 min). Beads were further washed twice with PNK buffer and resuspended in 240 μl of ChIP elution buffer. Cross-linking was reversed by adding 10 μl of Proteinase K and the mixture was incubated at 42°C for 1 hour, then at 70°C for 45 min. RNA was purified by acid-PCI extraction and EtOH precipitation. To remove residual DNA, RNA was resuspended in 50 μL of TURBO DNase buffer and subjected to DNase digestion and DNase removal using TURBO DNA-free Kit (Life Technologies, AM1907).
Reverse transcription and PCR experiment were performed with SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase (Life Technologies, 12574-026) following the manufacturer’s protocol. 2 μL of RNA prepared above was used for each reaction and following program was used (55 °C for 30 min, 94 °C for 2 min, 40 cycles of (94 °C for 15 sec, 55 °C for 30 sec, and 68 °C for 30 sec), and 68 °C for 5 min). Primers fC_pr1 (GGGCGGAAAGTCCAAATTGT) and fC_pr4 (GATGCACACGGTGACCAAAC) and primers fC_pr1 and fC_pr5 (GGTGATCATTCTCTGAGGGGC) were used for ARF1 and XBP1, respectively. The forward primer annealed to the CDS of FLuc and the reverse primer annealed downstream of the deletion site. Thus, these primer sets only amplified RNAs from the reporter and not endogenous mRNAs, and simultaneously amplified RNAs from wt and Δ constructs, maintaining their ratios. The PCR product was analyzed by QIAxcel system and QIAxcel DNA Screening Kit (QIAGEN, 929004).
Induction of ER stress and XBP1 splicing assay
Knockdown and rescue were performed as described above. The siRNA-resistant FLAG-STAU1 Flp-In 293 T-REx cell line was grown in 6 well plates, and treated with thapsigargin (SIGMA, T9033) by replacing medium with pre-warmed DMEM supplemented with (10% FBS, 300 nM thapsigargin, 100 ng/μl doxycycline (for RC experiment only)). In order to achieve equal thermal transfer for all samples, plates were always placed directly on incubator shelves. The cells were further incubated for 30 min at 5% CO2 to induce ER stress. RNA was purified using RNeasy plus mini kit (QIAGEN, 74136). To avoid any systematic error of experimental handling, we performed three independent experiments by varying the order of each condition. In experiment 1, we induced ER stress and lysed cells by the order of UT, KD and RC conditions, experiment 2 using KD, RC and UT, and experiment 3 using RC, UT, KD.
In order to monitor the cytoplasmic splicing of XBP1, RT-PCR and the analysis were performed using the SuperScript III One-Step RT-PCR System with Platinum Taq DNA Polymerase, X_pr1 (TTACGAGAGAAAACTCATGGC) and X_pr2 (GGGTCCAAGTTGTCCAGAATGC) primers38, and QIAxcel system as described above except that 100 μg of template RNA and 35 cycles of PCR program were used. QIAxcel electropherograms are available at figshare.com/s/d09b7b6e929a11e48bf206ec4bbcf141.
Reporter assay of XBP1 mRNA level
The plasmids (XBP1 wt, XBP1 mut, and XBP1 com) were transfected into siRNA-resistant FLAG-STAU1 Flp-In T-REx 293 cell line using Lipofectamine3000 (Life Technologies, L3000015) according to the manufacturer’s instructions. After 24hrs, RNA was extracted with TRIzol (Life Technologies, 15596018) and Direct-zol™ RNA MiniPrep (Zymo Research, R2052), and reverse-transcribed with RevertAid (Fermentas, #EP0441). Renilla (the 3′ UTR of XBP1 was appended) and Firefly (reference) Luciferases were quantified by qPCR on an Applied Biosystems 7900HT using the ΔΔCt method with default parameters and the PCR conditions for Fast SYBR® Green PCR Master Mix (Life Technologies, 4385612). The oligos Fluc_pr1 (CCACTGTCTAAGGAGGTGGG) and Fluc_pr2 (GGTAATCAGAATGGCGCTGG), and Rluc_pr1 (GCTCATATCGCCTCCTGGAT) and Rluc pr2 (CGTGGCCCACAAAGATGATT) were used to amplify the CDS of Firefly and Renilla, respectively.
Data analysis
Data analysis was performed using R-2.15.139 and Python-2.7.1 (http://www.python.org). The following R (ggplot2 (0.9.3)40, plyr (1.8)41, reshape2 (1.2.2)42) and Bioconductor packages (BSgenome.Hsapiens.UCSC.hg19 (1.3.17)43, GenomicRanges (1.8.13)44, ShortRead (1.14.4)45) were used throughout. latticeExtra_0.6-24, RColorBrewer_1.0-5, Rsamtools_1.8.6, lattice_0.20-10, Biostrings_2.24.1, IRanges_1.14.4 and BiocGenerics_0.2.0 were included in these packages. Other packages used for individual analyses are described in the appropriate sections.
Sequence processing and mapping of reads (hiCLIP, overview)
We used a sequential mapping approach to identify the source RNA. Reads were first mapped to rRNAs and tRNAs (Phase 1). Any remaining unmapped reads were then mapped to the mitochondrial genome and pre-rRNA (Phase 2). The remaining reads were then mapped to representative transcripts for all genes present in ENSEMBL (Phase 3). Finally, the remaining unmapped reads were mapped to the genome (Phase 4). Mapped read counts after each phase are summarized in Supplementary Table 1, and details are described below.
Sequence processing and mapping of reads (hiCLIP, pre-processing of sequence reads, identifying hybrid and non-hybrid reads)
The FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) was used for the basic processing of sequence reads. FASTQ files were converted to FASTA format. Reads were de-multiplexed using the experimental barcode located at positions 3-6 and the random barcode at the positions 1-2 and 7-9.
Hybrid and non-hybrid reads were selected as follows. Our initial analysis showed that 20-30% of reads contained Adaptor B directly followed by Adaptor A. This may have resulted from the imperfect 3′ end phosphorylation of Adaptor B (from the manufacturer’s website). Thus, we first trimmed Adaptor B-Adaptor A sequences in the 3′ end of reads using Cutadapt (http://journal.embnet.org/index.php/embnetjournal/article/view/200; cutadapt -a CTGTAGGCACCATACAATGAGATCGGAAGAGCGGTTCAG -e 0.06 -n 10 -m 186), and the trimmed reads were stored for later analysis. The 60-70% of reads lacking Adaptor B-Adaptor A sequences were trimmed to remove the Adaptor A sequence (cutadapt -a AGATCGGAAGAGCGGTTCAG -e 0.06 -n 10 -O 10 -m 186) and the trimmed reads were used for further analysis. To ensure that we analyzed cDNAs that were fully sequenced, we accepted only those reads in which at least part of the Adaptor A sequence was present before trimming.
After trimming, reads containing one Adaptor B sequence and at least 17nt each of the left and right arms were considered to be hybrids. This leaves a minimum read length of 53 nts for hybrids (minimum 17 nts for the left arm, 19 nts for Adaptor B, and minimum 17 nts for the right arm). The left and right arms were separated and mapped independently. Reads longer than 53 nts lacking Adaptor B sequences were considered to be non-hybrid reads. One mismatch was allowed for the search of Adaptor B sequence. The analysis was performed with the python_Levenshtein-0.10.2 package (https://github.com/miohtama/python-Levenshtein).
Sequence processing and mapping of reads (hiCLIP, Phase 1: mapping to rRNAs and tRNAs)
The processed reads were first mapped to a library of mature rRNAs and tRNAs allowing multiple hits (rRNA: RefSeq id, NR_023363.1, NR_003285.2, NR_003287.2, and NR_003286.2, tRNA: downloaded from Genomic tRNA Database: (http://gtrnadb.ucsc.edu). tRNAs named as Homo_sapiens were selected and CCA was added to the 3′ end of the sequence.). Mapping was performed using Bowtie (v 0.12.746; bowtie -f -p 8 -v 2 -k 1 --best --sam --un).
Sequence processing and mapping of reads (hiCLIP, Phase 2: mapping to mtDNA and pre-rRNA)
Unmapped reads in Phase 1 were aligned against the mitochondrial genome and pre-rRNA allowing multiple hits (Mitochondrial genome: RefSeq id AF347015.1 and pre-rRNA: RefSeq id NR_046235.1; bowtie -f -p 8 -v 2 -k 1 --best --sam --un). Reads mapped in this step were removed from further analyses.
Sequence processing and mapping of reads (hiCLIP, Phase 3: mapping to representative transcripts)
The remaining unmapped reads were aligned to a dataset containing annotated transcript sequences for all genes present in ENSEMBL 67 (http://may2012.archive.ENSEMBL.org/index.html). For genes with multiple transcript isoforms, we selected the longest “protein_coding” transcript as the representative isoform. If all isoforms were non-coding, then the longest non-coding isoform was selected as the representative. Transcript sequences and coordinates were obtained from version hg19 of BSgenome.Hsapiens.UCSC.hg19 (1.3.17)43. Mapping was performed using Bowtie (options -f -p 8 -v 2 -m 1 --best --strata --sam –un). Only uniquely mapping reads were considered for further analysis.
Sequence processing and mapping of reads (hiCLIP, Phase 4: mapping to the genome)
To identify reads from introns and un-annotated transcripts, the remaining unmapped reads were aligned to the human genome (hg19: indexed file downloaded from http://bowtie-bio.sourceforge.net/index.shtml) using Bowtie (options -f -p 8 -v 2 -m 1 --best --strata –sam). Only uniquely mapped reads were considered for further analysis.
Sequence processing and mapping of reads (hiCLIP, reads uniquely mapped to rRNAs)
The complete secondary structures of the 18S, 28S and 5.8S rRNAs were previously determined using cryo-EM data21. To compare the positions of hiCLIP duplexes with the secondary structures, we identified hybrid reads with both arms uniquely mapping to rRNAs in Phase 1 above (bowtie -f -p 8 -m 1 -v 2 --best –sam). The random barcodes were not considered.
Sequence processing and mapping of reads (Ribosome profiling and mRNA-Seq)
Sequence reads from the ribosome profiling and mRNA-Seq experiments were processed and analyzed in a similar way to above with following changes. Between the de-multiplexing and the barcode trimming steps, we performed the Phase 1 and Phase 2 mapping procedures (bowtie -f -p 8 -v 2 --trim5 9 --trim3 15 --un). Sequence reads were trimmed to 35 nts (26 nts of sequence reads + 9 nts of experimental and random barcode). The random barcodes were separated and stored, and the experimental barcodes were trimmed. Phase 3 mapping was performed (bowtie -f -p 8 -v 2 -m 1 --best --strata --sam). After mapping, the positions of the ribosome profiling and mRNA-Seq reads were defined by using the 12th nucleotide of the read as an estimate of the ribosome A site (Extended Data Fig. 6).
Annotation of reads
The reads mapped in Phase 1 were annotated as rRNA or tRNA. Those mapped in Phase 3 and 4 were annotated as mRNA, long non-coding RNA (lncRNA), miRNA or other ncRNA in the following manner. Transcripts annotated as protein_coding in ENSEMBL were categorized as mRNA. Reads mapping to the boundary of regions (e.g. across CDS-3′ UTR junction) were annotated according to the position of the 5′ end nucleotide. miRNAs were annotated as defined in ENSEMBL. Transcripts annotated as (processed_transcript, antisense, lincRNA, sense_intronic, sense_overlapping, 3prime_overlapping_ncrna, pseudogene, transcribed_processed_pseudogene, unprocessed_pseudogene, transcribed_unprocessed_pseudogene) in ENSEMBL and those longer than 200 nts were defined as lncRNAs. Any remaining unannotated transcripts were classified as ‘other ncRNAs’. Reads mapping to the anti-sense of annotated transcripts were classed as intergenic. In the main text, hybrid reads with both arms mapped in Phase 3 were referred to as “mRNA and long non-coding RNA reads”, since vast majority of reads mapped to these RNAs in Phase 3.
Reads mapping in sense to protein-coding transcripts were further annotated as intronic, 5′ UTR, CDS, or 3′ UTR. To enable unique annotation of each read, we defined a single annotation for each fragment of the protein-coding genes by creating collapsed annotation. Transcripts annotated as protein_coding and sharing gene id were collected from ENSEMBL 67. Exons were collapsed and regions of the gene that did not overlap with any exon were annotated as introns. Regions of collapsed exons 5′ to the first start codon in each gene were annotated as 5′ UTR, and those 3′ to the last stop codon in each gene were annotated as 3′ UTR, and remaining exonic regions were annotated as CDS. The collapsed annotation was created by modifying functions from Quantify RNA-Seq data version 0.01 (https://r-forge.r-project.org/scm/viewvc.php/?root=qrnaseq).
Generation of randomly repositioned control sequences
We generated randomly repositioned control sequences by randomly re-positioning each arm of the hybrid reads within the same segments of the gene using ‘sample’ function in R (e.g. if the read mapped to the 3′ UTR of gene A, the control was generated by randomly picking a read of the same length within the 3′ UTR of gene A).
Calculation of folding free energies of hybrid reads
The folding free energies of hybrid reads and randomly repositioned sequences were calculated using the RNAhybrid program47 (options -s 3utr_human -m 1000 -n 1000 -c). The software calculates the minimum free energy of hybridization between two distinct RNA molecules but not of intra-molecular base-pairing. The Mann–Whitney U test and the p-value were calculated using ‘wilcox.test’ function in R.
Probability density distribution plots
The plots in Fig. 1d, and 3b-d were drawn using Kernel density estimate with ‘geom_density’ function from ggplot2 package40 in R.
Selection and processing of hybrid reads used to evaluate the general properties of STAU1-bound RNA duplexes
After analysis of RNA duplexes in rRNAs, we focused all further analyses on the hybrid reads that were mapped in Phase 3 (i.e., mainly mRNAs and ncRNAs). We removed PCR duplicates by keeping only one copy of identical reads containing the same random barcode19. N in random barcode was treated as a wild card.
Enrichment analysis of STAU1 cross-link events around the hiCLIP-identified duplexes
For analysis in Extended Data Fig. 3f, the STAU1 cross-link sites were defined by the start sites of non-hybrid reads. Cross-link events around the two arms of duplexes identified in all transcripts except rRNAs, tRNAs and mtRNAs were compared to those around random transcript positions. Only intra-molecular duplexes were examined. Cross-link sites were not weighted by the cDNA counts.
Identification of intra-molecular RNA duplexes
For all remaining analyses (described in the remaining sections of methods and shown in Fig 3b-3g and Fig. 4), we used only intra-molecular duplexes. ‘Intra-molecular’ duplexes are those in which both arms of hybrid reads mapped to the same RNA species (Extended data Fig. 1b). 2,964 mRNAs were found to contain at least one intra-molecular duplex. We are aware that it is possible that these duplexes are formed by hybridisation of two separate molecules of the same RNA species. We removed PCR duplicates as described above, merged the data for hybrid reads obtained from the high RNase and low RNase conditions, and then retained only hybrid reads where both arms uniquely mapped to the same mRNA (Extended data Fig. 1b). The longest duplex formed by the pair of the arms from a hybrid read was searched by computationally annealing the two arms using the RNAhybrid program47 (options -s 3utr_human -m 1000 -n 1000 -c) and the longest base-paired region was defined as the duplex (Extended data Fig. 1d). If more than two candidate duplex regions tied for length, the one reported first by the RNAhybrid program was chosen. If the right arm of a hybrid read was mapped to the 5′ side of the left arm, the left arm and the right arm were swapped. If the left and right arm of a hybrid read overlapped, the reads were discarded. The resulting hybrid reads were visually examined with the IGV browser (2.1.20)48,49.
Identification of non-redundant intra-molecular RNA duplexes
Some RNA duplexes were identified by more than one hybrid read. In order to extract the non-redundant set of RNA duplexes, we defined the duplexes identified by at least 2 hybrid reads (step 1), and those duplexes identified by single hybrid reads (step 2). Subsequently, the two subsets of the duplexes were merged, and used as the non-redundant set of RNA duplexes. The terminology of this section was described before (http://www.bioconductor.org/help/course-materials/2010/BioC2010/Workflow.pdf).
Step 1. The reads from the left and right arm were pre-filtered separately. If different reads mapped to the same position in transcripts (equal start and end), then they were collapsed (counted as one). The coverage of reads from the left arm of hybrid reads was calculated using the ‘coverage’ function of GenomicRanges44, and the regions covered by more than 1 reads were considered as islands (depth ≥ 2). If the width of an island was less than 9, the island was discarded. Then, hybrid reads with the left arm overlapping with an island were selected, and then the coverage of the right arms was calculated as described above. If the right arm of those hybrid reads formed an island with depth ≥ 2 and width ≥ 9, the two islands formed by the left and right arms were defined as a pair of islands. If the right arm formed more than 1 island with depth ≥ 2 and width ≥ 9, the island with the higher depth was selected. If two islands had the same depth, then the one closer to the left island was selected. If the width of the islands were shorter than 17 nts, the width of the islands was extended to 17 nts. The pair of the islands was treated as the pair of arms from a hybrid read, and the duplex was identified as described above. If more than two candidate duplex regions tied for the length, the one reported first was chosen. The number of hybrid reads overlapping both islands were also calculated and later used to weight the data. This subset of the duplexes were named as confident duplexes, and used to examine the robustness of genome-wide results obtained with only the confident duplexes (Extended Data Fig. 10).
(step 2) We then defined the duplexes identified by a single hybrid read in the following manner. From the hybrid reads where both arms were mapped to the same RNAs, those that did not overlap with the pairs of islands defined above were selected. The duplexes identified by the selected hybrid reads were defined as described above. In a few occasions, two hybrid reads identify the same duplex, but do not pass the filtering of confident duplexes described above because the overlap was shorter than 9nts. Therefore, if identical duplexes were identified by more than 1 hybrid read in this step, we collapsed them as a single duplex.
Subsequently, the set of the RNA duplexes defined by the step 1 and 2 were merged, and the merged data set was defined as the non-redundant set of intra-molecular RNA duplexes.
Generation of randomly repositioned control duplexes
We generated 20 sets of control duplexes by randomizing the positions of the two arms in the transcript region that contained the original hiCLIP duplex. For example, if an arm of the duplex mapped to a 3′ UTR, it was re-positioned within the 3′ UTR of the same transcript, such that the length of the arm was preserved after repositioning.
The STAU1 sequence specificity analysis
We extended the hiCLIP and control duplexes by 10 nucleotides up and downstream (or less if the region between the two arm was shorter than 20 nucleotides). The nucleotide contents were analyzed in the duplexes and in the whole 3′ UTR. The statistical significance of differences in nucleotide content between the hiCLIP duplexes, control duplexes and the whole 3′ UTRs was computed using a binomial test.
To find overrepresented motifs, we used the RSATools suite50 (option –noov -1str). The statistical score (−log(binomial p-values) corrected for multiple testing) of each tetramer in the extended duplexes was computed by comparing the occurrence of motifs to a background model computed from whole 3′ UTRs. The background model consisted of a second order Markov model using the frequencies of di-nucleotides in the 3′ UTRs. The significance threshold was determined by running the same analysis on the control duplexes and using the maximal statistical score found in the analysis (i.e. max(−log10(corrected binomial p-value))=86.6).
The occurrences of each tetramer were mapped using the dna-pattern program in RSATools. Overlapping tetramers were assembled to create matrices using the matrix-from-pattern program in RSATools. The generated matrices were used to scan the duplexes and the control regions using matrix-scan-quick in RSATools with a minimum site weight of 5.2, which corresponds to a p-value of 10−3 (matrix-distrib, RSATools).
We calculated the purine and pyrimidine content of each duplex arm using a 9nt sliding window across a 40nt region up and downstream of the duplex. The two arms of the duplexes are shown in an orientation in which the arm with the higher purine content is on the right (Fig. 2a). The duplexes were then ordered by increasing arm length. The heatmaps were drawn using the gplots R package and the heatmap.2 function.
To check if the results observed for STAU1 targeted duplexes were specific, we picked randomly 1000 non-STAU1 target mRNAs and predicted duplex-formation using the RNAfold software with default parameters24 on their 3′UTRs. Among more than 10000 predicted duplexes, we selected a subset of 3000 duplexes having the same arm length distribution as the one observed in the hiCLIP duplexes. The heatmap was drawn as described above.
Sequence analyses were reproduced on the subset of STAU1-target mRNAs containing the duplexes that were identified by more than one hybrid read to ensure robustness of results (Extended Data Fig. 10a, 10b).
Enrichment analysis of Alu elements at the STAU1 cross-link sites
The enrichment of Alu elements was examined around the STAU1 cross-link sites from the cytoplasmic or total fraction, or from hnRNP C cross-link sites. For this purpose, the enrichment of Alu elements in the non-hybrid reads of STAU1 hiCLIP and in the sequence reads of STAU1 iCLIP and hnRNP C iCLIP were examined. For hnRNP C iCLIP, data from the previous study was used19. The sequence reads of these data sets were trimmed to the length of 26 nts and analyzed using the Repeat Enrichment Estimator server51.
Analysis of single nucleotide polymorphisms (SNPs)
We used the genomic coordinates of hiCLIP duplexes according to ENSEMBL 67 (http://may2012.archive.ENSEMBL.org/index.html) and obtained the list of human SNPs and their genomic coordinates from dbSNP build 138 (ftp.ncbi.nih.gov/snp). We used BEDTools52 to determine SNPs that overlap with duplexes (option intersect). We calculated the frequencies of SNPs in duplexes by dividing the number of SNPs by the number of nucleotides in each duplex. We calculated the background SNP frequency by dividing the number of SNPs in all 3′ UTRs that are STAU1 targets by the total number of nucleotides in these 3′ UTRs. We conducted a simple binomial exact test. The metaprofiles were calculated for the 2,291 3′ UTR-3′ UTR duplexes with loops >80 nucleotides by plotting the normalized count of SNPs over each arm extended by 40 nucleotides from the centre of the left and right arms in each direction. Plots were drawn using the ggplot2 R package and the geom_smooth function. This analysis was reproduced on the subset of STAU1-target mRNAs containing the duplexes that were identified by more than one hybrid read to ensure robustness of results (Extended Data Fig. 10c).
Calculating the length of the loop of hiCLIP duplexes
The loop lengths of hiCLIP duplexes were defined as the distance from the 3′ boundary of the left arm to the 5′ boundary of the right arm (Extended Data Fig. 1e). For the comparison shown in Fig. 3b, the counts of RNA duplexes were weighted by the number of hybrid reads that identified them. Same comparison was reproduced on the subset of STAU1-target mRNAs containing the duplexes that were identified by more than one hybrid read to ensure robustness of results (Extended Data Fig. 10d).
Visualising RNA duplexes in mRNAs with a Circos plot
Circos plots were produced using the Circos53 software. The script and settings are for available from https://github.com/jernejule/STAU1_hiCLIP (options -conf data/HiCLIP_mRNA_norm.conf –pdf).
Computational prediction of the secondary structure of mRNA 3′ UTRs
RNA secondary structures in mRNA 3′ UTRs were computationally predicted using the RNAfold program with default parameters24. Predicted duplexes were extracted using the forgi 0.1 package (http://www.tbi.univie.ac.at/~pkerp/forgi/). hiCLIP duplexes were considered predictable if hybrid read overlapped partially or completely with the predicted duplexes.
mRNA abundance and translational efficiency estimation from mRNA-Seq and ribosome profiling reads
Only reads mapping to mRNAs were considered. For the mRNA abundance analysis all mRNA-Seq reads were used. Translational efficiency was studied for mRNAs with CDS longer than 100 nts and excluding those with HGNC symbol starting with HIST, since histone mRNAs are poorly polyadenylated54; mRNA-Seq and ribosome profiling reads were mapped to trimmed CDS regions (trimmed 30 nts from5′ and 3′ ends because these regions accumulate ribosomes during translation initiation and termination; Extended Data Fig. 6b). The number of reads mapped on each mRNA or trimmed CDS was calculated and a set of mRNAs with sufficient read depth were analyzed (> 30 reads in all 6 replicate experiments). Total library sizes were normalized using DESeq (1.8.3)55 with locfit_1.5-8 and Biobase_2.16.0 and the count of each mRNA was further normalized by a multiplying normalization factor (1000000 / mean value of the total library size of the 6 DESeq normalized libraries).
The translational efficiency of each mRNA was calculated by dividing the number of ribosome profiling reads mapping to the trimmed CDS by the number of mRNA-Seq reads mapping to the trimmed CDS after library normalization. mRNA abundance was defined by dividing normalized count of the mRNA-Seq by the length of the mRNA and multiplying by 1,000 (RPKN: read per kilo base per normalized library). The mRNA abundance and translational efficiency of each condition (UT, KD and RC) was defined as the average of these counts from duplicate experiments. The fold change of translational efficiency or mRNA abundance between conditions was calculated using these values.
Analysis of the relationship between translational efficiency and RNA structures in CDS or 3′ UTR
Since highly expressed mRNAs are expected to have more depth of data, we weighted the translational efficiency of these mRNAs by the number of hybrid reads in the genes (i.e. if mRNA A had 2 hybrid reads, translational efficiency of mRNA A was calculated twice). The weighted translational efficiency was compared with the translational efficiency of all mRNAs that passed our filter by Mann–Whitney U test.
Analysis of the effect of STAU1 depletion on mRNA abundance and translational efficiency
To examine off-target effects of the siRNA, the list of mRNAs was ranked by fold change of mRNA level between untransfected (UT) and knockdown (KD) condition and analyzed by SylArray56 using “Use all available words” option. Top 3 significantly enriched motifs were reported to demonstrate that the primary changes in mRNA stability result from off-target effects of the siRNA (Extended Data Fig. 7e). Therefore, to avoid these off-target effects, all comparisons of ribosome profiling and mRNA-Seq were performed between knockdown (KD) and rescue (RC) conditions.
Gene Ontology analysis
We calculated the enrichment of annotated Gene Ontology terms in hiCLIP duplex containing mRNAs using the David GO tool57. We used the lists of genes for which mRNAs contained duplexes in the 3′ UTR or in the CDS. The list of genes from the whole genome was used as the background. Enrichments with an FDR cut-off of 0.01 were considered statistically significant. Results were visualized using the ReviGO tool58. This analysis was reproduced on the subset of STAU1-target mRNAs containing the duplexes that were identified by more than one hybrid read to ensure robustness of results (Extended Data Fig. 10e).
Extended Data
Extended Data Figure 1. Diagrams explaining the mapping of hybrid reads, duplex assignment and use of terms.
a, Schematic overview of the hiCLIP protocol. (1) Cells are irradiated with UV-C light. (2) After lysis, the unprotected sections of RNAs are digested by RNase I, and the RBP is co-immunoprecipitated with the cross-linked RNA duplex. (3) Two designated adaptors are ligated to both strands of the RNA duplex. Adaptor A (cloning adaptor) has a permanent 3′ block whilst adaptor B (linker adaptor) has a removable 3′ block. (4) 3′ block of adaptor B is removed. (5) The two strands of the RNA duplex are ligated via adaptor B. (6) The RNA hybrid product is then converted into a cDNA library and sequenced as in iCLIP protocol19. The resulting data comprise hybrid and non-hybrid reads. (7) Hybrid reads are selected and adaptors trimmed to define the sequences of left (L) and right (R) arms, which are mapped independently to the transcriptome. b and c, The left arm of hybrid read locates upstream of adaptor B, and the right arm locates downstream of adaptor B. Each arm is mapped independently to transcriptome. If both arms locate into the same gene, then the duplex is considered to be formed by the same RNA. If the arms locate to different genes, then the duplex is formed by two different RNAs. d, A diagram describing how a hybrid read is used to identify an RNA duplex. e, A diagram describing how the loop (intervening sequence) is defined for each RNA duplex.
Extended Data Figure 2. Autoradiography analysis of the STAU1-RNA complex, and analysis of hybrid reads in a known STAU1 mRNA target, ARF1.
a, Autoradiograph of STAU1-RNA complex that was isolated for the hiCLIP experiment. hiCLIP experiments were performed with high and low RNase conditions, and the two controls omitted either the 2nd intermolecular ligation or STAU1 induction. After adaptor ligation, STAU1 cross-linked RNA was radio labeled and the complex was analyzed by denaturing gel electrophoresis and membrane transfer. The size of the band is slightly higher compared to that in Fig. 1a, presumably due to the efficient adaptor ligation that adds to the size of the RNAs (the experiment shown in Fig. 1a didn’t include adaptor ligation). b, Correlation analysis of the non-hybrid read count on each RNA between the replicates of the hiCLIP experiments. c, Schematic representations of ARF1 mRNA and the known STAU1-target RNA duplex, along with the position of STAU1 hybrid reads and cross-link sites identified by non-hybrid reads. The left and right arms of hybrid reads are depicted as black boxes, and lines connect reads originating from the same cDNA. The previously studied STAU1-target RNA duplex12,14 is indicated by green and red boxes. In addition to the known duplex, hybrid reads also identified additional duplexes in the ARF1 3′ UTR. Interestingly, two newly identified duplexes are part of overlapping secondary structures, both of which represent the minimum free energy of folding the local sequence, as predicted by RNAfold24 (shown on the right). This suggests that some regions of the ARF1 3′ UTR may adopt alternative conformations. The overlapping region of the two structures is shaded in blue. d, The constructs of reporters (ARF1 WT and Δ) used for the validation by formaldehyde crosslinking and co-immunoprecipitation experiment are shown. The reporter has firefly luciferase (FLuc) CDS and ARF1 3′ UTR. e, The ratio of ARF1 WT and Δ in total cell lysate fraction or STAU1 co-immunopreciptated fraction were analyzed by RT-PCR using forward primer annealed to CDS of FLuc and reverse primer annealed to downstream of the deletion site. The ratios (log2) of two populations are compared by Welch’s t test (n = 3). The corresponding Qiaxcel electropherograms are available at: figshare.com/s/5f83e88e929b11e4b77106ec4b8d1f61.
Extended Data Figure 3. Hybrid reads identify RNA duplexes.
a, Analysis of hybrid reads in 3′ UTRs demonstrates significantly smaller ensemble folding energy compared to random RNA fragments as calculated by RNAhybrid. The kernel density estimate of ensemble folding energy distribution is plotted, and the ensemble folding energy for hybrid reads and random RNAs is compared by Mann-Whitney U test (n = 4492 for both hybrid reads and random RNAs). b, Similar to a, but the analysis is restricted to hybrid reads in CDS (n = 958). c, Similar to a, but the analysis is restricted to inter-molecular hybrid reads (i.e., hybrid reads whose left arm and right arm originated from different genes; n = 257). d, Similar to a, but the analysis is restricted to hybrid reads in rRNAs (n = 3502). e, Median normalised PARS scores were calculated around center of all mRNA duplexes. PARS scores were obtained from Wan et al4, and positions with 0 values were removed. PARS score represents a ratio between reads starts after cutting with dsRNase (positive) / ssRNase (negative). Assuming that the double-stranded RNase fully digests each duplex, it is expected that the positive values in PARS-seq will be highest at the last nucleotide of each duplex. This might explain why maximum PARS values occur at the positions closer to the 3′ end of duplexes. f, Metaprofiles of the distribution of STAU1 cross-link sites, identified by the start sites of non-hybrid hiCLIP reads (blue), or the randomly repositioned sites (black, mean value of 10 randomizations; gray, standard deviation of the 10 randomizations) around the positions of hiCLIP duplexes. g, Distribution of mRNA median probabilities to be single-stranded from −50 to 100 nucleotides around the cross-link sites.
Extended Data Figure 4. Analysis of duplexes identified by hiCLIP in the secondary structure of human ribosomal RNAs.
a, The position of duplexes identified by hybrid reads in the 28S rRNA secondary structure. 2962 hybrid mapped to 28S rRNA. We first removed the duplexes with only one read, obtaining a final list of 2816 hybrid reads uniquely mapped to 28S rRNA, and 2020 of these reads (72%) identified duplexes that were previously determined with CryoEM structure. These duplexes are marked by blue rectangles, and the number of reads that identify each duplex is marked. 756 hybrid reads (27%) map to different double stranded regions of the 28S rRNA while the remaining 40 hybrid reads (1%) map to single strand regions of the rRNA 28S known structure. The metazoan-specific rRNA expansion segments are indicated by gray shadowing. b, Similar to a, but the position of duplexes identified by hybrid reads in the 18S rRNA secondary structure. 218 hybrid reads uniquely mapped to 18S rRNA, and 170 of these reads identified duplexes that were previously determined with CryoEM structure. Red lines mark the putative newly identified duplexes that are not part of the CryoEM structure, but are complementary and are identified by hybrid reads. The numbers next to the lines shows the number of unique hybrid reads that identify each of these putative duplexes, and the three newly identified duplexes (helices) that are aligned in Fig. 1e are marked as hA, hB and hC. Complementarity of the novel duplex is conserved from yeast to human as seen at: figshare.com/s/47473d24929c11e493f106ec4bbcf141. The rRNA secondary structure is reprinted by permission from Macmillan Publishers Ltd: Nature copyright (2013)21.
Extended Data Figure 5. Analysis of RNA types, stem lengths, and sequence motifs at STAU1-target RNA duplexes.
a, Relative proportions of RNA types that were identified by hybrid reads, where each arm maps to the same (left) or different RNAs (right). b, Distribution of duplex stem lengths for 3′ UTR duplexes. c, Sequence motifs enriched at STAU1-target 3′ UTR duplexes when compared to surrounding regions of same 3′ UTRs (controls). d, Purine content is plotted within each arm of randomly selected 2291 3′ UTR duplexes that were detected using RNAfold24 in mRNAs that don’t contain any STAU1-target hiCLIP duplex in their 3′ UTR, and surrounding sequence up to 40nt on each side. e, Purine content is plotted within each arm of 494 CDS-CDS STAU1-target duplexes and surrounding sequence up to 40nt on each side. f, Boxplots showing the frequencies of consecutive purine tracks in the 3′ UTR STAU1-target duplexes (boxplot on the left) and in the randomly selected 3′ UTR duplexes that were detected using RNAfold in mRNAs that don’t contain any STAU1-target hiCLIP duplex in their 3′ UTR (boxplot on the right).
Extended Data Figure 6. Analysis of repeat elements and SNPs at STAU1-target RNA duplexes.
a, Enrichment analysis of repeat elements. The enrichment was calculated by comparing the proportion of repeat elements in the cytoplasmic STAU1 iCLIP (i.e., hiCLIP non-hybrid reads), STAU1 iCLIP from the total cellular fraction, and hnRNP C iCLIP reads with mRNA-Seq reads using Repeat Enrichment Estimator software. The enrichment was estimated for all repeat element families (e.g. AluJb, AluJo, LTR1, and LTR10A) defined by the Repeat Enrichment Estimator51. The top panels show the enrichment of different types of repeat element; all families of Alu repeat elements are plotted in red, and all remaining repeat elements are plotted in gray. The bottom box plots show the enrichment distribution for all repeat elements excluding Alus, or just for Alus. b, Box plots showing the SNPs frequency in all the 3′ UTRs of the targets (box plot on the left) and inside the duplexes in the 3′ UTRs. A binomial test has been performed to test the statistical significance of the depletion of SNPs inside the duplexes, compared to the total 3′ UTRs (P < 10−04). c, Normalized count profile of SNPs occurrence in the long-range 3′ UTR-3′ UTR hiCLIP duplexes; those that have a loop longer than 500nts. d, Normalized count profile of SNPs occurrence in random control 3′ UTR-3′ UTR duplexes with a loop of at least 80 nucleotides that are identified by RNAfold24 but not by STAU1 hiCLIP.
Extended Data Figure 7. Analysis of mRNA abundance and translational efficiency with mRNA-Seq and ribosome profiling.
a, Venn diagram shows that few mRNAs contain hiCLIP duplexes both in their 3′ UTR and CDS. b, Metagene analysis of ribosome profiling reads. The position + 12 from the 5′ end of the sequence reads was used as the definition of the read positions. The number of reads mapped around start codon or stop codon is shown. The color of dots corresponds to the positions in each codon. The sharp peak at start codon shows our definition of read position well approximated the position of ribosome A site, and trinucleotide periodicity of peak confirms that the reads captured the codon dependent positioning of ribosome. c, Translational efficiency was independent of the mRNA level [RPKN: read per kilo base per normalized library]. All mRNAs that passed our filter are plotted (see Methods, for detail). d, Western blotting analysis for untreated (UT), STAU1 knockdown (KD) and knockdown with rescue (RC) condition for the cell sample used for ribosome profiling and mRNA-Seq library generation. Duplicate experiments were performed for each condition. e, In order to examine the potential off-target effects of siRNA treatment, we analyzed sequence motifs enriched in mRNAs down-regulated in KD condition compared to UT using SylArray56. The plot shows the incremental hypergeometric p-value of the 3 most significantly enriched 7mer motifs in the gene list sorted by the down-regulation level (the leftmost gene is the most down-regulated). The most significantly enriched motif corresponded to the seed sequence of siRNA, indicating that most changes in mRNA abundance between UT and KD corresponded to off-target effects. Therefore, we focused our analyses on the comparison between RC and KD, which had no significant enrichment of such motifs by the SylArray analysis. f, The cumulative fraction of mRNAs relative to their fold change of mRNA abundance or translation efficiency between STAU1 rescue (RC) and knockdown (KD) cells is plotted. The p-value was calculated by Mann–Whitney U test (n = 2269, 752, and 12122 for the RNAs containing the duplexes in their 3′ UTR or CDS or other mRNAs for the analysis of mRNA abundance and n = 1986, 694, and 8199 for the mRNAs containing the duplexes in their 3′ UTR or CDS or other mRNAs for the analysis of translational efficiency).
Extended Data Figure 8. STAU1 regulates the cytoplasmic splicing of XBP1 mRNA.
a, Schematic represents the unspliced (XBP1(u)) and spliced (XBP1(s)) mRNA, together with hybrid reads that identified a long-range STAU1-target RNA structure. The constructs of reporters (XBP1 wt and Δ) used for the validation by formaldehyde cross-linking, co-immunoprecipitation or reporter assay experiment are shown at the bottom. b, STAU1 interacts with XBP1 via the long-range RNA structure. The ratio of XBP1 wt and Δ in total cell lysate fraction or STAU1 co-immunoprecipitated fraction were analyzed by RT-PCR. The ratios (log2) of two populations are compared by two-tailed Welch’s t test (n = 3). The corresponding Qiaxcel electropherograms are available at: figshare.com/s/f0e32272929b11e4a56606ec4b8d1f61. c, Overview of XBP1(u) and XBP1(s) mRNAs. The position of cytoplasmic splicing site is indicated by an arrow. The longer RNA duplex overlaps with the region translated in XBP1(s). d, Thapsigargin induces UPR and cytoplasmic splicing of XBP1. We confirmed that after 30 min of the UPR induction, XBP1 was actively spliced. e, Real-time PCR analysis of reporter mRNA levels containing wt, mut or com XBP1 3′ UTR (as marked in a). Analysis is done in untreated cells (UT), in cells treated with siRNA against STAU1 (KD), or in cells where siRNA-resistant STAU1 is induced with doxycycline to rescue expression of STAU1 in spite of knockdown (RC). Differences in expression are compared by two-tailed Student’s t-Test. The two independent experiments are marked by black and blue colour, and each dot represents replicates performed on separate wells of cells as part of the same experiment. Disruption of duplex destabilises the mRNA, and compensatory mutation restores the stability slightly above the wt level. This may be because the duplex in the ‘com’ reporter is longer by 2 nt compared to ‘wt’. The mechanism whereby the long-range duplex impacts mRNA stability remains to be determined.
Extended Data Figure 9. Gene Ontology analysis and schematic of STAU1 function in 293 cells.
Gene ontology analysis of the genes bound by STAU1 in the 3′ UTR (red) and in the CDS (purple), using the DAVID Gene Ontology Tool57 and visualized using ReviGO58. Node colour indicates the p-value (threshold: p-value < 10−6 - FDR < 0.01), and node size indicates the frequency of the GO term in the GOA database. Each gene is mapped only to the most specific terms that are applicable to it (in each ontology). Highly similar GO terms are linked by edges in the graph, with the edge width depicting the degree of similarity. a, Diagram summarizing the enriched Biological Processes GO terms. 3′ UTR-bound mRNAs tend to encode proteins that function in intracellular transport (in red), whereas CDS-bound mRNAs tend to encode proteins that function in the cell cycle M phase (in blue). b, Diagram summarizing the enriched Cellular Components GO terms in the context of their location in the cell. 3′ UTR-bound mRNAs tend to encode membrane proteins that are translated at the ER (in red), whereas CDS-bound mRNAs tend to encode nuclear proteins (in blue). c, Schematic diagram of the functional analyses of CDS and 3′ UTR bound mRNAs. 3′ UTR-bound mRNAs tend to be highly translated and encode membrane proteins that are translated at the ER (in gray). CDS-bound mRNAs tend to be lowly translated and encode nuclear proteins that function in the cell cycle M phase. Loops formed by RNA duplexes in the 3′ UTR tend to be longer than in the CDS, and 3′ UTRs have higher density of bound duplexes.
Extended Data Figure 10. Analysis of confident duplexes that were identified by >1 hybrid read.
a, Sequence motifs enriched at confident STAU1-target RNA duplexes. b, Visualisation of purine content at confident duplexes (as in Fig. 2a). c, Normalized count profile of SNPs occurrence in confident 3′ UTR-3′ UTR duplexes with a loop of at least 80 nucleotides. d, Comparison of range (the length of loop + duplex) distribution among all duplexes and confident duplexes (those with more than 1 unique hybrid read) located in 3′ UTRs. e, Gene Ontology analysis of mRNAs containing confident STAU1 duplexes in 3′ UTRs (as in Extended Data Figure 9).
Supplementary Material
Acknowledgement
We wish to thank Sander Granneman and Christopher Sibley for discussions on the development of hiCLIP protocol, Kathi Zarnack, Nejc Haberman, Charles Ravarani and Benjamin Lang for assistance with bioinformatic analyses, Dalia Daujotyte and Peter Lukavsky for sharing the STAU1 plasmid and helping in setting up the project, Lynne Maquat for sharing the ARF1 SBS plasmid, the genomic team at the Cancer Research UK Cambridge Institute for Illumina HiSeq sequencing, and Madan Babu Mohan and Ule group members for support and comments on the manuscript. This work was supported by funding from Human Frontier Science Program [RGP0024/2008-C], European Research Council [206726-CLIP and 617837-Translate] and Medical Research Council [U105185858] to J.U., Cancer Research UK and UCL to N.M.L., a Wellcome Trust Joint Investigator Award to N.M.L and J.U. [103760/Z/14/Z], the Nakajima Foundation fellowship and MRC Centenary Early Career Award to Y.S.
Footnotes
Author Information The sequence data and scripts are publically available from ArrayExpress (E-MTAB-2937, E-MTAB-2940, E-MTAB-2941) and https://github.com/jernejule/STAU1_hiCLIP.
The authors declare no competing financial interests.
References
- 1.Wan Y, Kertesz M, Spitale RC, Segal E, Chang HY. Understanding the transcriptome through RNA structure. Nat Rev Genet. 2011;12:641–655. doi: 10.1038/nrg3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 3.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, et al. Global analysis of RNA secondary structure in two metazoans. Cell Rep. 2012;1:69–82. doi: 10.1016/j.celrep.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Goodarzi H, et al. Metastasis-suppressor transcript destabilization through TARBP2 binding of mRNA hairpins. Nature. 2014;513:256–260. doi: 10.1038/nature13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lovci MT, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20:1434–1442. doi: 10.1038/nsmb.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci U S A. 2011;108:10010–10015. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosswendt S, et al. Unambiguous Identification of miRNA:Target Site Interactions by Different Types of Ligation Reactions. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heraud-Farlow JE, Kiebler MA. The multifunctional Staufen proteins: conserved roles from neurogenesis to synaptic plasticity. Trends Neurosci. 2014 doi: 10.1016/j.tins.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 13.Ricci EP, et al. Staufen1 senses overall transcript secondary structure to regulate translation. Nat Struct Mol Biol. 2014;21:26–35. doi: 10.1038/nsmb.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. The EMBO journal. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heraud-Farlow JE, et al. Staufen2 regulates neuronal target RNAs. Cell Rep. 2013;5:1511–1518. doi: 10.1016/j.celrep.2013.11.039. [DOI] [PubMed] [Google Scholar]
- 16.Laver JD, et al. Genome-wide analysis of Staufen-associated mRNAs identifies secondary structures that confer target specificity. Nucleic Acids Res. 2013;41:9438–9460. doi: 10.1093/nar/gkt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lucas S, Oliveros JC, Chagoyen M, Ortin J. Functional signature for the recognition of specific target mRNAs by human Staufen1 protein. Nucleic Acids Res. 2014;42:4516–4526. doi: 10.1093/nar/gku073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeGendre JB, et al. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem. 2013;288:2532–2545. doi: 10.1074/jbc.M112.397349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo M, Duchaine TF, DesGroseillers L. Molecular mapping of the determinants involved in human Staufen-ribosome association. Biochem J. 2002;365:817–824. doi: 10.1042/bj20020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anger AM, et al. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. doi: 10.1038/nature12104. [DOI] [PubMed] [Google Scholar]
- 22.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elbarbary RA, Li W, Tian B, Maquat LE. STAU1 binding 3′ UTR IRAlus complements nuclear retention to protect cells from PKR-mediated translational shutdown. Genes Dev. 2013;27:1495–1510. doi: 10.1101/gad.220962.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorenz R, et al. ViennaRNA Package 2.0. Algorithms for molecular biology : AMB. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends Genet. 2013;29:691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu X, et al. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 28.Marion RM, Fortes P, Beloso A, Dotti C, Ortin J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Molecular and cellular biology. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Molecular and cellular biology. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulay K, et al. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res. 2014;42:7867–7883. doi: 10.1093/nar/gku506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiel JA, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 32.Huppertz I, et al. iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65:274–287. doi: 10.1016/j.ymeth.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Korennykh AV, Behrman SL, Walter P. Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16113–16118. doi: 10.1073/pnas.1010580107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Core Team A language and environment for statistical computing. 2012.
- 40.Wickham H. Ggplot2 : elegant graphics for data analysis. Springer; New York: 2009. [Google Scholar]
- 41.Wickham H. The Split-Apply-Combine Strategy for Data Analysis. J Stat Softw. 2011;40:1–29. [Google Scholar]
- 42.Wickham H. Reshaping data with the reshape package. J Stat Softw. 2007;21:1–20. [Google Scholar]
- 43.The Bioconductor Dev Team BSgenome.Hsapiens.UCSC.hg19: Homo sapiens (Human) full genome (UCSC version hg19)
- 44.Aboyoun P, Pages H, Lawrence M. GenomicRanges: Representation and manipulation of genomic intervals.
- 45.Morgan M, et al. ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25:2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Day DS, Luquette LJ, Park PJ, Kharchenko PV. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol. 2010;11:R69. doi: 10.1186/gb-2010-11-6-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho J, et al. LIN28A Is a Suppressor of ER-Associated Translation in Embryonic Stem Cells. Cell. 2012;151:765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartonicek N, Enright AJ. SylArray: a web server for automated detection of miRNA effects from expression data. Bioinformatics. 2010;26:2900–2901. doi: 10.1093/bioinformatics/btq545. [DOI] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci U S A. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci U S A. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kwok CK, Ding Y, Tang Y, Assmann SM, Bevilacqua PC. Determination of in vivo RNA structure in low-abundance transcripts. Nature communications. 2013;4:2971. doi: 10.1038/ncomms3971. [DOI] [PubMed] [Google Scholar]
- 64.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA-protein complexes. Nat Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J Mol Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 66.Schroeder SJ. Advances in RNA structure prediction from sequence: new tools for generating hypotheses about viral RNA structure-function relationships. J Virol. 2009;83:6326–6334. doi: 10.1128/JVI.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaufmann G, Klein T, Littauer UZ. T4 RNA ligase: substrate chain length requirements. FEBS Lett. 1974;46:271–275. doi: 10.1016/0014-5793(74)80385-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.