Starch contents are highly correlated with the expression levels of a CO2-responsive gene which may regulate coordinated expression of starch synthesis genes.
Abstract
A unique CO2-Responsive CONSTANS, CONSTANS-like, and Time of Chlorophyll a/b Binding Protein1 (CCT) Protein (CRCT) containing a CCT domain but not a zinc finger motif is described, which is up-regulated under elevated CO2 in rice (Oryza sativa). The expression of CRCT showed diurnal oscillation peaked at the end of the light period and was also increased by sugars such as glucose and sucrose. Promoter β-glucuronidase analysis showed that CRCT was highly expressed in the phloem of various tissues such as leaf blade and leaf sheath. Overexpression or RNA interference knockdown of CRCT had no appreciable effect on plant growth and photosynthesis except that tiller angle was significantly increased by the overexpression. More importantly, starch content in leaf sheath, which serves as a temporary storage organ for photoassimilates, was markedly increased in overexpression lines and decreased in knockdown lines. The expressions of several genes related to starch synthesis, such as ADP-glucose pyrophospholylase and α-glucan phospholylase, were significantly changed in transgenic lines and positively correlated with the expression levels of CRCT. Given these observations, we suggest that CRCT is a positive regulator of starch accumulation in vegetative tissues, regulating coordinated expression of starch synthesis genes in response to the levels of photoassimilates.
Global atmospheric CO2 concentration has rapidly increased from the preindustrial level of about 28 Pa to over 39 Pa in 2013 and is projected to increase to 47 to 102 Pa by the end of this century (Meehl, 2007). Consequently, many studies have been made on the effect of elevated CO2 on plant growth and vast amounts of data have been accumulated (Ainsworth and Long, 2005). In general, plants show increased height, larger stem diameter, and increased branching and leaf number in response to elevated CO2 (Ainsworth and Long, 2005; Cheng et al., 2009). On a metabolic level, increases in starch content are typically observed in C3 plants (Ainsworth and Long, 2005). Overall, elevated CO2 positively affects growth and yield by stimulating photosynthesis in most C3 plants.
Rice (Oryza sativa) is one of the most important crops for global food production and also an important model plant for genome analysis in C3 monocots. To understand the molecular basis of plant CO2 response, gene expression profiling using microarrays has been done in a number of plant species including rice (Li et al., 2008; Fukayama et al., 2009, 2011; Leakey et al., 2009; Tallis et al., 2010; Kaplan et al., 2012). These studies showed a variety of responses depending on species and experimental conditions. In rice, we showed that most genes directly involved in CO2 assimilation were down-regulated, whereas genes involved in ribulose 1,5-bisphosphate regeneration and starch synthesis were up-regulated by elevated CO2 both in semiclosed growth chambers (Fukayama et al., 2009) and free-air CO2 enrichment (Fukayama et al., 2011). Up-regulation of genes related to starch synthesis such as ADP-Glc pyrophosphorylase (AGPase) and starch synthase (SS) has been consistently observed and is therefore considered to play a major role in the metabolic adaptation of C3 plants to elevated CO2.
Starch synthesis is a complex and coordinated biochemical reaction catalyzed by multiple enzymes such as AGPase, SS, granule-bound SS (GBSS), α-glucan phosphorylase (Pho), starch-branching enzyme (BE), and starch-debranching enzyme (DBE). Because starch biosynthesis is important for rice grain quality, some transcriptional regulators functioning in seed development have been identified. The APETALA2/Ethylene-Responsive Element-Binding Protein family transcription factor, Rice Starch Regulator1 (RSR1) is involved in the negative regulation of starch synthesis gene expression (Fu and Xue, 2010). It has also been reported that the rice DNA-binding protein OsBP-5 forms a heterodimer with the ethylene-responsive element-binding protein OsEBP-89 to negatively regulate the expression of GBSSI (Zhu et al., 2003). By contrast, FLOURY ENDOSPERM2 (FLO2) which has a tetratricopeptide repeat motif, can positively affect the expression of GBSSI and BEI (She et al., 2010). More recently, a basic Leu zipper transcription factor, OsbZIP58, was shown to bind directly to the promoters of several starch synthesis genes, including AGPase, SS, and GBSS (Wang et al., 2013). Thus, the mechanism of regulation of the expression of starch synthesis genes in rice grain has been determined to some extent, but in vegetative tissues, less is known. In rice, stems, which consist largely of leaf sheath tissue during vegetative growth, are known to serve as temporary storage organs for photoassimilates, mainly as starch. The transiently stored starch is remobilized and translocated to reproductive organs at the grain-filling stage, and its contribution has been estimated to be as much as 30% of grain carbon (Cook and Yoshida, 1972). Moreover, substantial increases in this starch have usually been observed under long-term elevated CO2 treatment, which can lead to significant enhancement of rice yield. This capacity for transient starch storage must be an important component in determining yield as well as CO2 response in rice; however, key regulator molecules controlling expression of starch synthesis genes functioning in these vegetative organs have not been identified.
In general, the effects of elevated CO2 on plant growth and photosynthesis are dependent on nitrogen status (Stitt and Krapp, 1999). Starch accumulation in leaf sheath is also affected by nitrogen status being stimulated by limiting nitrogen and decreased by excess nitrogen application (Hirano et al., 2005). In previous work, we identified many genes of unknown function, which are differentially expressed under elevated CO2 in rice (Fukayama et al., 2009). Reevaluating these data, we found that a unique CONSTANS, CONSTANS-like, and Time of Chlorophyll a/b Binding Protein1 (TOC1; CCT) domain-containing protein gene designated CRCT (for CO2-Responsive CONSTANS, CONSTANS-like, and Time of Chlorophyll a/b Binding Protein1) was notably up-regulated under elevated CO2, and its expression was further enhanced under conditions of low nitrogen supply. In plants, members of CCT domain-containing proteins have been implicated in processes such as photoperiodic flowering (Putterill et al., 1995), regulation of circadian rhythms (Strayer et al., 2000), and light signaling (Kaczorowski and Quail, 2003). Recently, the CCT domain of CONSTANS and TOC1 was found to bind directly to DNA and act as a transcription factor (Tiwari et al., 2010; Gendron et al., 2012). Thus, CRCT could be a candidate regulatory factor controlling the expression of genes related to adaptation to elevated CO2 in rice. In this study, we carried out expression analysis and functional analysis of CRCT using transgenic rice and demonstrate that CRCT can be a positive regulator of starch accumulation regulating the subset of starch synthesis genes in vegetative tissue of rice.
RESULTS
CRCT as a CO2-Responsive Gene
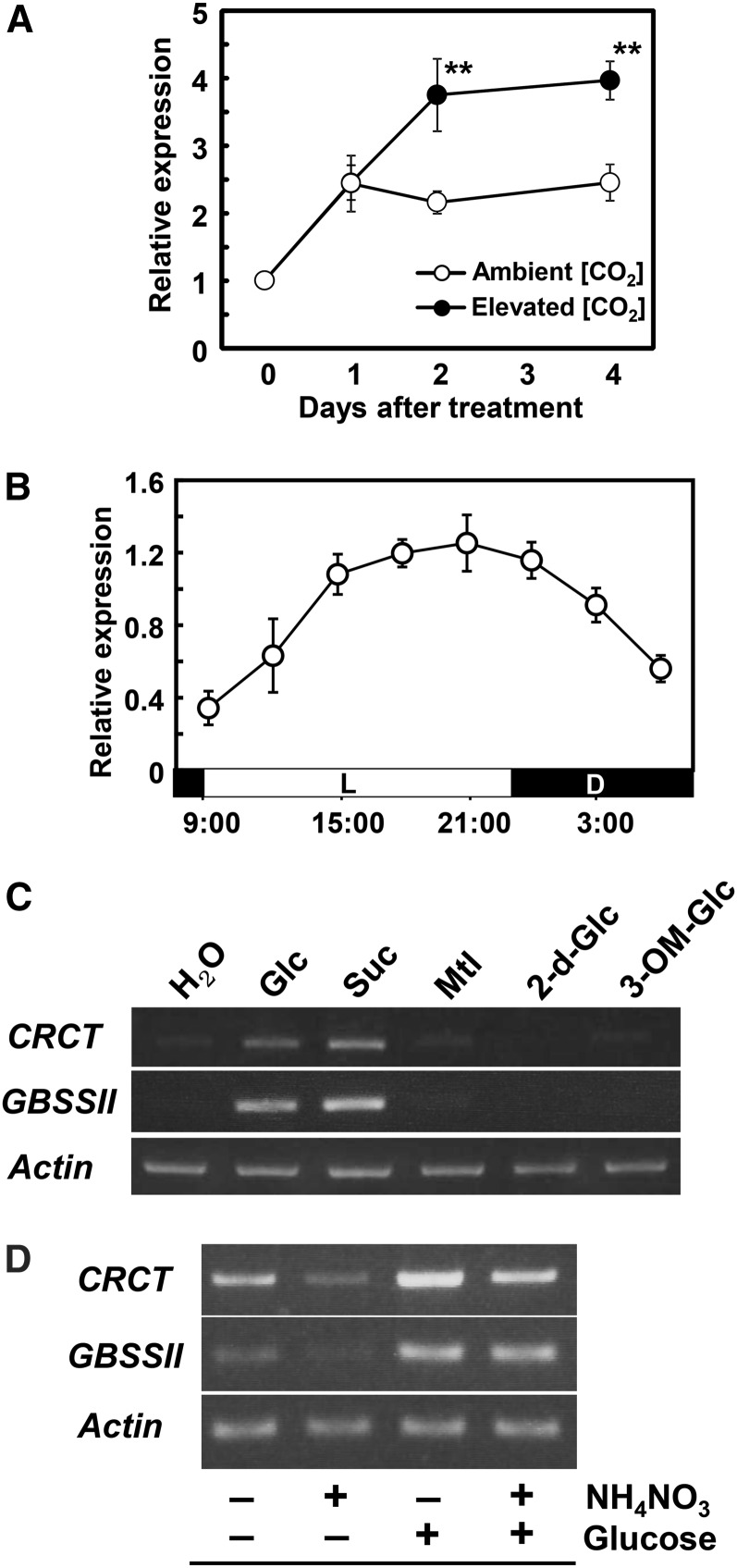
In previous study, we identified many CO2-responsive genes in rice by microarray (Fukayama et al., 2009). Among them, we found that CRCT (Os05g0595300) was markedly up-regulated under elevated CO2. In this study, the effects of elevated CO2 on the expression of CRCT in leaves were analyzed by quantitative reverse transcription (qRT)-PCR (Fig. 1A). The expressions of CRCT were increased during the experiment both in ambient and elevated CO2. This increase in CRCT expression possibly reflects the leaf development because we analyzed upper young leaves, which were undergoing maturation during the experiment. After 2 d, the expression of CRCT in elevated CO2 conditions became significantly higher than in ambient CO2. These results indicate that the expression of CRCT is surely CO2 responsive and this response can occur by relatively short periods of elevated CO2 treatment.
Figure 1.
Expression analysis of CRCT. A, The effects of elevated CO2 on the expression of CRCT. Rice seedlings were grown in a growth chamber until the 4.0 leaf stage and then subjected to CO2 partial pressures of 38 (ambient) or 100 Pa (elevated). Leaf blades above the third leaf were sampled at 11 am. The expression of CRCT relative to Actin was analyzed by qRT-PCR. Data represent mean ± sd of five biological replicates. Asterisks indicate significant difference (P < 0.01) by Student’s t test. The expression of Actin relative to 18S ribosomal RNA in elevated CO2 was not significantly different from that in ambient CO2. B, Diurnal changes in the expression of CRCT. Expanding and uppermost fully expanded leaves at the 4.5 leaf stage were sampled at the designed time of day. The expression of CRCT relative to Actin was analyzed by qRT-PCR. Data represent mean ± sd of five biological replicates. L, Light period; D, dark period. C, Effects of various sugars on the expression of CRCT. Plants were kept in darkness for 2 d, and then the uppermost fully expanded leaves were harvested and floated on 50 mm solutions of Glc, Suc, mannitol (Mtl), 2-deoxyglucose (2-d-Glc), 3-O-methylglucose (3-OM-Glc), or water for 8 h in darkness. D, Effects of Glc and NH4NO3 on the expression of CRCT. Rice seedlings were grown on medium with or without 450 mm Glc and 30 mm NH4NO3. Rice shoots were harvested at 5 d after germination. GBSSII was shown as known sugar-inducible gene (Dian et al., 2003). Actin was used as internal control.
CRCT is located on chromosome 5 and encodes a protein of 308 amino acids. Unlike many other CCT domain-containing proteins such as TOC1, Zinc Finger Protein Expressed in Inflorescence Meristem Like, or CONSTANS, CRCT does not contain a zinc finger domain (Supplemental Fig. S1). Among reported CCT domain-containing proteins, Arabidopsis (Arabidopsis thaliana) gene Activator of Sporamin Promoter::Luciferase2 (ASML2) has some common features in domain structure (that is, no zinc finger domain; Masaki et al., 2005). However, according to the phylogenetic tree of CCT protein sequence (Supplemental Fig. S2), ASML2 is not considered to be orthologous to CRCT. There are two other genes (AT4G27900 and AT5G53420) in Arabidopsis closely related to CRCT that could be orthologous to rice CRCT. Orthologous genes are also found in sorghum (Sorghum bicolor) and grape (Vitis vinifera), suggesting the presence of this gene in a wide range of plants.
Expression Analysis of CRCT
The expression of CRCT in leaves showed diurnal oscillations, with highest expression in the later light period and then gradually decreasing during the dark period from 9 pm to 9 am (Fig. 1B). The effect of sugars on CRCT expression was examined in leaves floating on various sugar solutions. The expression of CRCT was up-regulated by 4- to 24-h Glc treatment, and it was largely dependent on the concentration of Glc (Supplemental Fig. S3). The expression of CRCT was also enhanced by Suc treatments; however, mannitol, 3-O-methylglucose and 2-deoxyglucose (that are not metabolized in plants) did not affect the expression of CRCT (Fig. 1C). The expression of CRCT induced by Suc was slightly higher than that by Glc. These results are mostly consistent with the response of GBSSII to sugar feeding, which was used as a positive control (Dian et al., 2003), though the induction of CRCT was slightly faster than that of GBSSII (Supplemental Fig. S3). Overall, these observations suggest that the expression of CRCT could be regulated by the metabolism of photoassimilates such as Suc. In general, the effects of sugars on plant growth and gene expression vary depending on nitrogen status, such as carbon/nitrogen balance (Stitt and Krapp, 1999). Thus, the effects of nitrogen supplementation on the enhancement of CRCT expression by Glc were analyzed using rice seedlings grown under different concentrations of Glc and ammonium nitrate (Supplemental Fig. S4). Without nitrogen, excess sugar (450 mm Glc) clearly inhibited growth and the accumulation of chlorophyll in rice seedlings. This inhibition of morphogenesis was substantially mitigated by the addition of ammonium nitrate to the medium. Likewise, a stimulatory effect of Glc on CRCT expression was attenuated by ammonium nitrate addition (Fig. 1D). These results suggest that the expression of CRCT is affected by nitrogen status as well as cellular sugar levels.
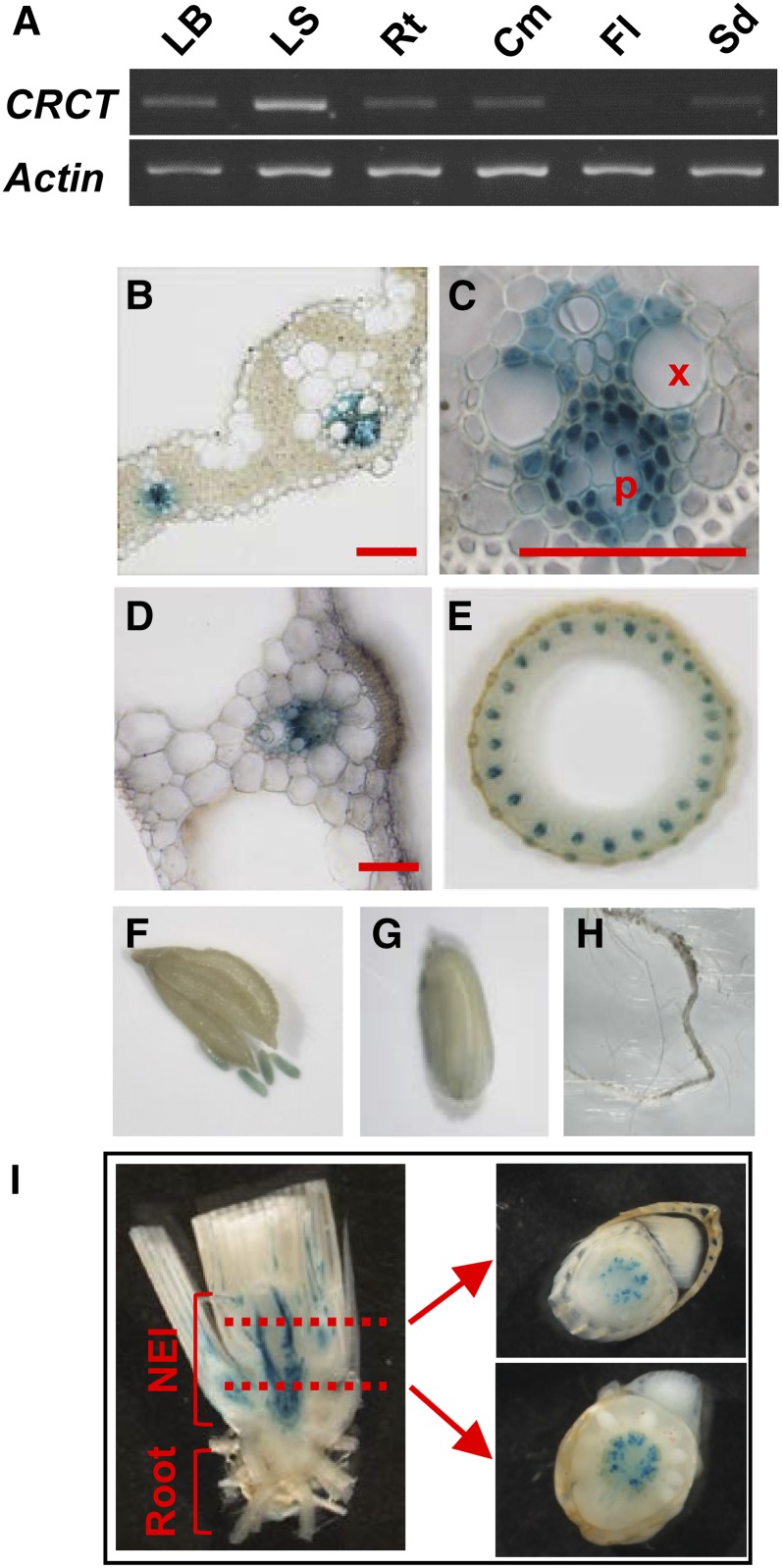
To study the organ specificity of CRCT expression, PCR was carried out in leaf blade, leaf sheath, root, culm, flower, and seed (Fig. 2A). The expression of CRCT was observed in most organs except flower and expression was highest in leaf sheath. Tissue and cellular specificity of CRCT expression was examined by histochemical staining of GUS activity in rice transformed with the CRCT promoter::GUS chimeric gene (Fig. 2, B–I). CRCT expression was mostly found in vascular bundles, particularly around the phloem of the shoot. Expression of CRCT in vascular bundles of the nonelongation portion of main stem and basal portion of leaf sheath was provably high as evidenced by the incubation time required for the GUS reaction in these tissues (2 h), which was much shorter than that required for other tissues (2 d). These findings indicate that CRCT expresses in vascular bundles of a range of organs as observed for other CCT proteins (An et al., 2004; Para et al., 2007).
Figure 2.
Organ and cell specificity of the expression of CRCT. A, Organ specificity of the expression of CRCT by PCR. Leaf blade (LB), leaf sheath (LS), and root (Rt) of the 4.5 leaf stage, culm (Cm) and flower (Fl) just after anthesis, and immature seed (Sd) at 10 d after anthesis were used for analysis. Actin was used as internal control. B to I, Histochemical localization of GUS activity in transgenic rice with CRCT promoter::GUS constructs. Cross section of leaf blade (B) and vascular bundle of leaf blade (C) were observed by light microscope. Cross section of leaf sheath (D), culm (E), flower (F), immature seed (G), and root (H) were observed by stereoscopic microscope. I, Longitudinal (left) and cross (right) sections of stem base. Tissues were stained for 2 d (B–H) or 2 h (I). X, Xylem; P, phloem; NEI, nonelongation internodes. Bars = 0.4 mm.
Effects of Overexpression or Knockdown of CRCT
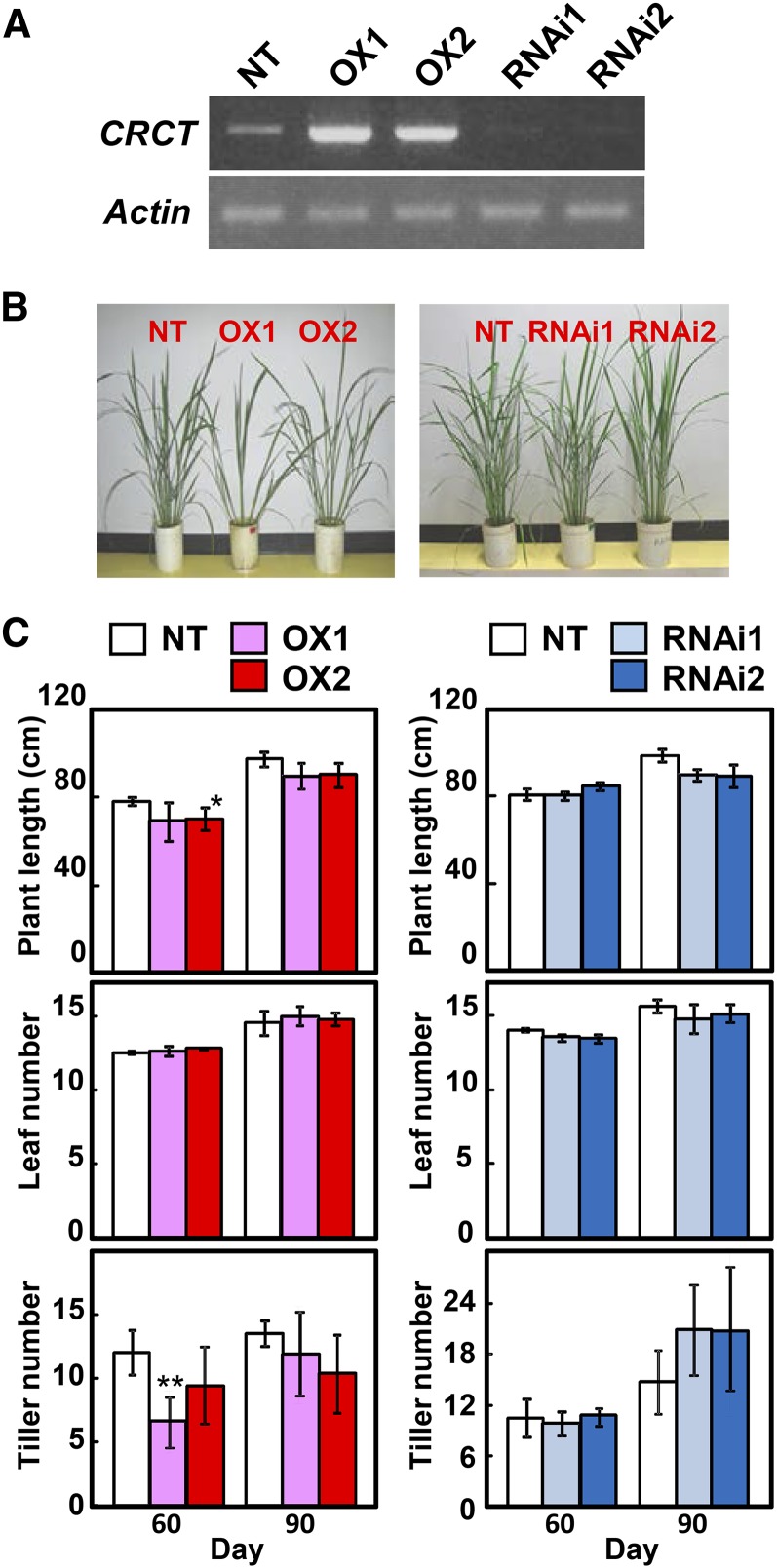
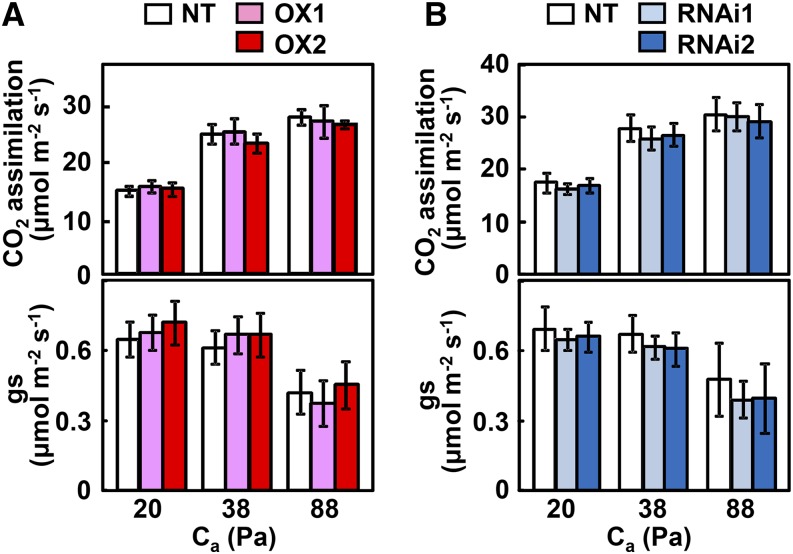
We generated CRCT-overexpressing transgenic lines and RNA interference (RNAi) knockdown transgenic lines to study the effect of CRCT expression level on growth and photosynthesis of rice. Expression of CRCT was significantly increased in the overexpression lines compared with nontransgenic rice (NT), while expression of CRCT was clearly decreased in knockdown lines (Fig. 3A). These transgenic lines showed normal phenotypes with similar growth rates (Fig. 3B). Plant length and leaf number of transgenic lines at 60 d (vegetative stage) and 90 d (reproductive stage) after germination were similar to NT, whereas tiller numbers were slightly decreased in overexpression lines and increased in knockdown lines (Fig. 3C). The photosynthetic rate and stromal conductance of transgenic lines under three different CO2 partial pressures were also similar to NT (Fig. 4). These observations imply that changes in the expression level of CRCT do not cause large-scale effects on plant growth and photosynthesis. However, marked whole-plant morphological differences were observed between NT and overexpression lines in tiller angle (Fig. 5). Tillers were more spreading and tiller angle relative to the main stem was significantly larger in overexpression lines than in NT and knockdown lines. The expression of representative tiller angle-related genes Tiller Angle Control1 (Yu et al., 2007), LAZY1 (Yoshihara and Iino, 2007), and Loose Plant Architecture1 (LPA1; Wu et al., 2013) did not differ between genotypes (Supplemental Fig. S5). Thus, it is proposed that CRCT may function in determining tiller angle by a different mechanism from that reported for these other genes.
Figure 3.
Effects of overexpression or knockdown of CRCT on growth. A, Expression analysis of CRCT in leaf sheath by PCR. Actin was used as internal control. B, Transgenic rice plants at 60 d after sowing. C, Plant growth of CRCT transgenic lines. Plant length, leaf number, and tiller number were determined at 60 and 90 d after sowing. Plants were grown in temperature-controlled greenhouse under natural sunlight. Asterisks indicate significant difference between NT and transgenic rice by Student’s t test (P < 0.01). OX1 and OX2 indicate CRCT overexpression lines, and RNAi1 and RNAi2 indicate CRCT knockdown lines.
Figure 4.
Photosynthetic rate of CRCT transgenic lines. The CO2 assimilation rate and stomatal conductance (gs) of the uppermost fully expanded leaves at vegetative growth stage was measured at a photosynthetic photon flux density of 1,500 µmol m–2 s–1, leaf temperature of 28°C, and CO2 partial pressures (Ca) of 20, 38, and 88 Pa. Data represent mean ± sd of five biological replicates. OX1 and OX2 indicate CRCT overexpression lines, and RNAi1 and RNAi2 indicate CRCT knockdown lines.
Figure 5.
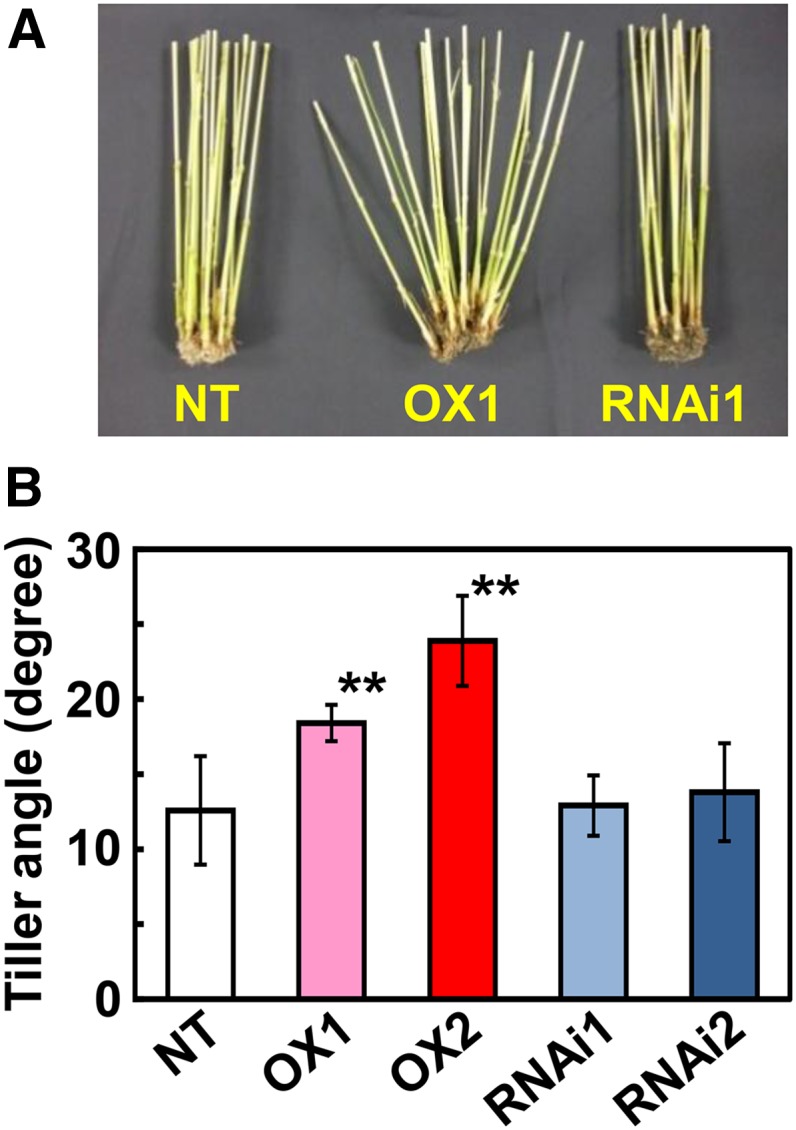
Effects of overexpression or knockdown of CRCT on tiller angle. A, Culm above the base at harvesting stage. B, Tiller angle between the main stem and the tiller emerged from third node at 60 d after sowing. Plants were grown in temperature-controlled greenhouse under natural sunlight. Asterisks indicate significant difference between NT and transgenic rice by Student’s t test (P < 0.01). OX1 and OX2 indicate CRCT overexpression lines, and RNAi1 and RNAi2 indicate CRCT knockdown lines.
CRCT Regulates Starch Accumulation
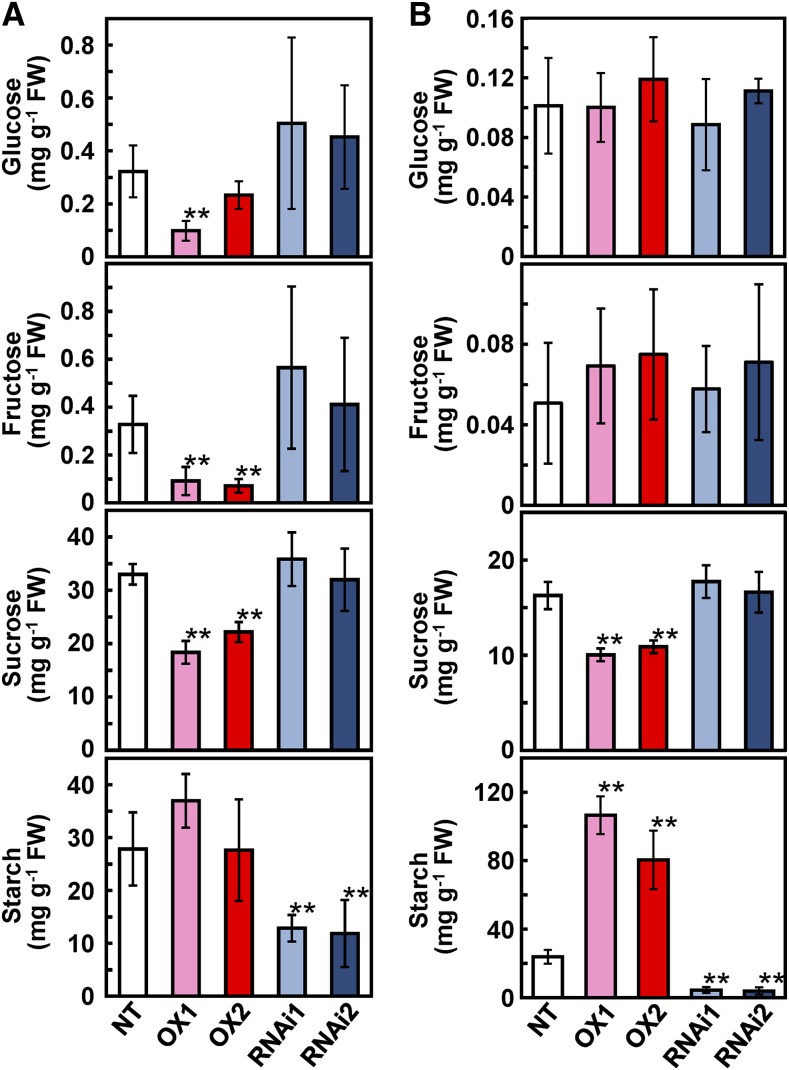
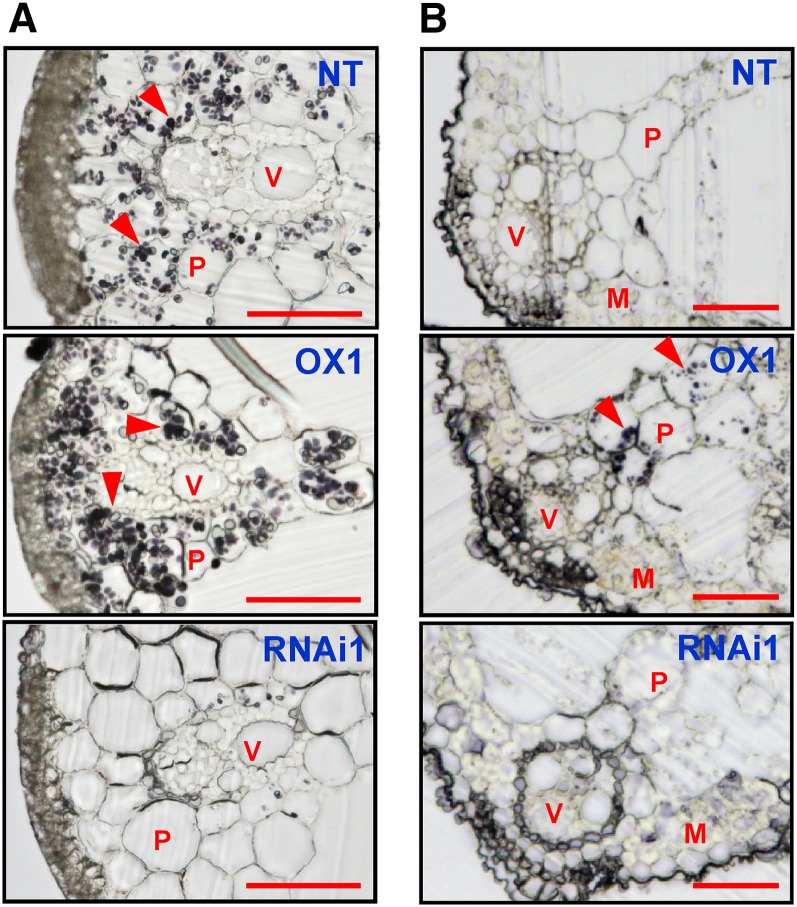
An increase in starch accumulation is a typical response observed in many plants grown under elevated CO2. Thus, we analyzed the levels of soluble sugars and starch in the leaf blade and leaf sheath of transgenic lines (Fig. 6). The Suc content of CRCT overexpression lines was significantly lower than that in NT both in leaf blade and leaf sheath. The contents of Glc and Fru in leaf blade of overexpression lines were also lower than that in NT, whereas its values were quite low in all genotypes compared with Suc. By contrast, it was striking that starch content in leaf sheath was several times higher in overexpression lines compared with NT. In addition, the starch content was significantly lower in knockdown lines than that in NT both in leaf blade and leaf sheath. To confirm the relationship between the content of starch and CRCT, the starch content was also analyzed in additional three overexpression lines and three knockdown lines with different expression level of CRCT (Supplemental Fig. S6). In these lines, the starch content was also correlated with the expression levels of CRCT in leaf sheath. The visualization of starch by iodine staining clearly demonstrated this correlation in leaf sheath and also in culm (Fig. 7, A and B). In leaf sheath, starch is basically accumulated from base to tip with gradation (see the result of NT in Fig. 7A). This observation is consistent with the expression pattern of CRCT, which should be high in basal portion of leaf sheath (Fig. 2I). By contrast, slight changes in iodine staining were observed in leaf blade and glumaceous flower, while no changes were observed in roots. In leaf sheath, the starch was mostly accumulated in the storage parenchyma cell, where starch granules were clearly increased in the overexpression line and almost disappeared in the knockdown line (Fig. 8A). In leaf blade, the starch granule was not observed in the mesophyll cell of all genotypes, whereas it was observed in the parenchyma cell of the overexpression line (Fig. 8B). Above results strongly suggest that changes in the expression of CRCT affect the starch content of vegetative organs.
Figure 6.
Soluble sugars and starch contents in CRCT transgenic lines. A, Soluble sugars and starch contents in leaf blade. B, Soluble sugars and starch contents in leaf sheath. Asterisks indicate significant difference between NT and transgenic rice by Student’s t test (P < 0.01). OX1 and OX2 indicate CRCT overexpression lines, and RNAi1 and RNAi2 indicate CRCT knockdown lines. FW, Fresh weight.
Figure 7.
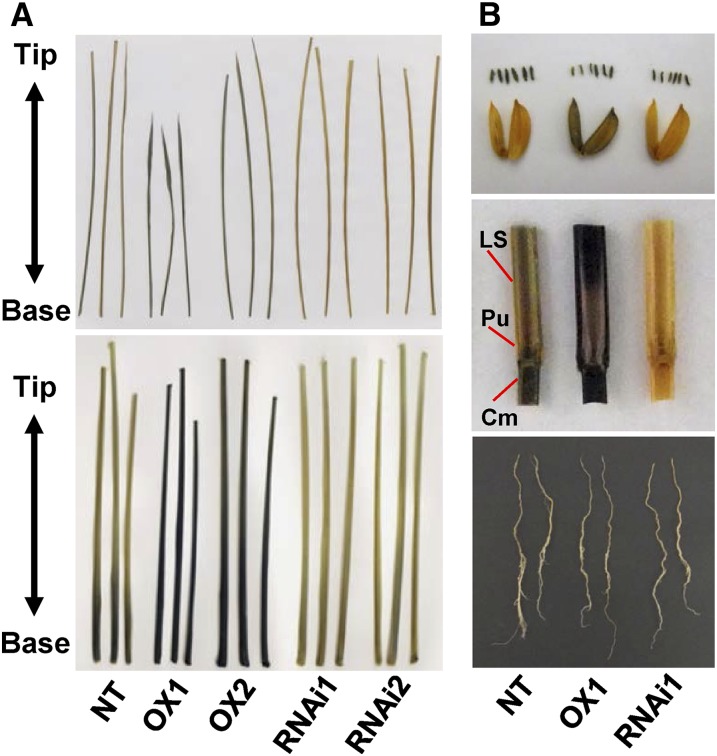
Starch accumulation in CRCT transgenic lines analyzed by iodine staining. A, Starch accumulation of leaf blade (top) and leaf sheath (bottom). Sixth leaf blade and fifth leaf sheath at the 6.5 leaf stage were sampled at the end of the light period and stained by iodine solution. B, Iodine staining of starch in glumaceous flower including anther (top), culm of heading stage (middle), and root at seedling stage (bottom). OX1 and OX2 indicate CRCT overexpression lines, and RNAi1 and RNAi2 indicate CRCT knockdown lines. LS, Leaf sheath; Pu, pluvinus; Cm, culm.
Figure 8.
Starch granule of transverse section of leaf sheath and leaf blade. Transverse sections of middle portion of leaf sheath (A) and leaf blade around the midrib (B) were stained by 1% iodine solution and observed by light microscopy. Starch granule appears dark purple (red arrowhead). OX1 indicates CRCT overexpression line, and RNAi1 indicates CRCT knockdown line. M, Mesophyll cell; P, parenchyma cell (storage parenchyma cell for leaf sheath); V, vascular bundle. Bar = 0.02 mm.
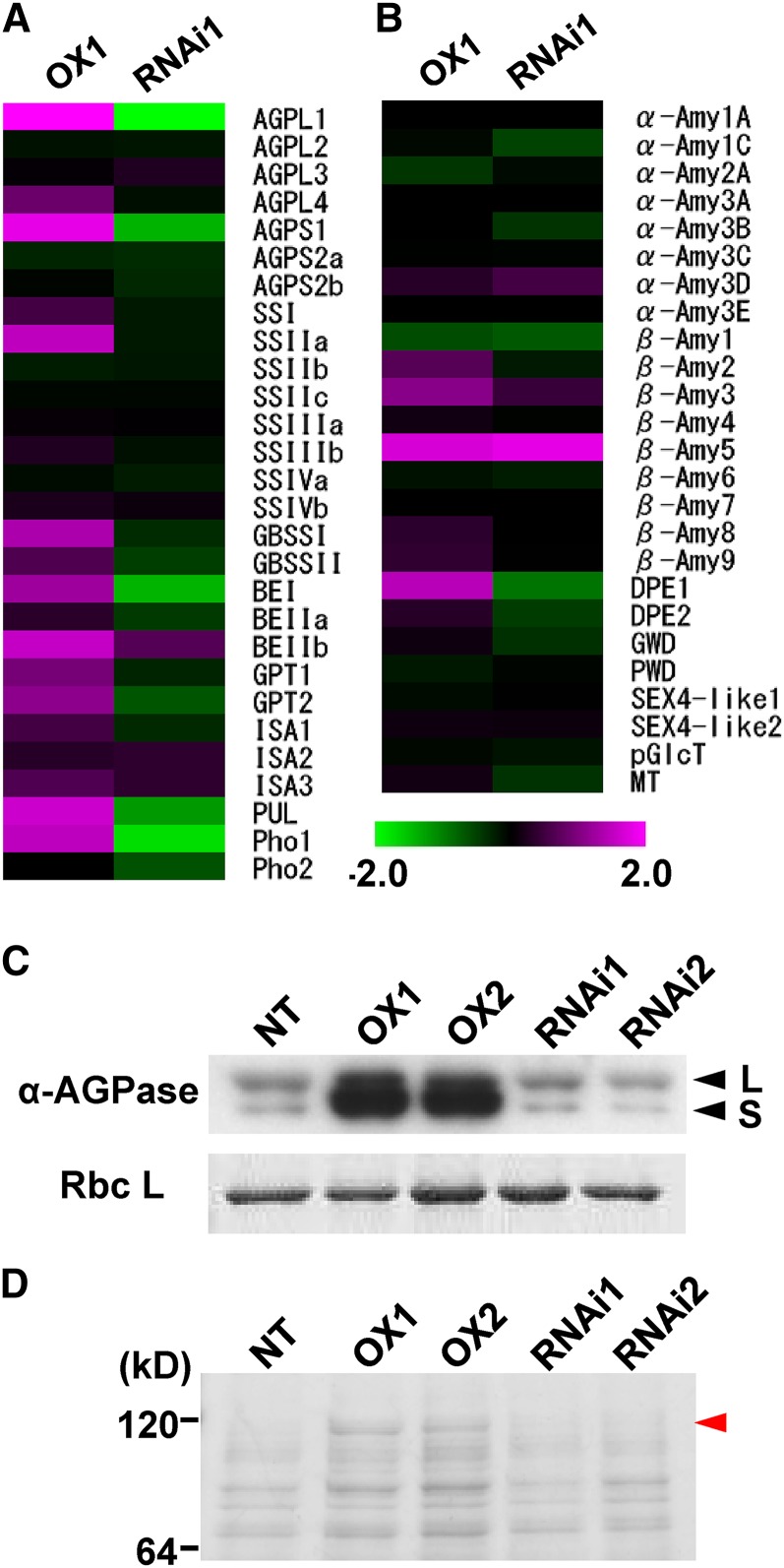
Because the starch content in leaf sheath was correlated with the expression levels of CRCT, the effects of CRCT expression on the transcriptional levels of starch synthesis- and degradation-related genes were analyzed in leaf sheath by microarray (Fig. 9, A and B). Among genes related to starch synthesis, the expression of AGPase L1 (AGPL1), AGPase S1 (AGPS1), BEI, pullulanase (PUL), and Pho1 showed significant differences in both overexpression and knockdown lines compared with NT and were clearly correlated with the expression levels of CRCT. Moreover, these genes appeared on the list of the top 50 down-regulated genes in the knockdown line (Supplemental Tables S1–S4). By contrast, the expression of most genes related to starch degradation was not significantly altered in either overexpression or knockdown transgenic lines. The expression levels of four genes showing significant difference in expression level in transgenic lines and four representative genes highly expressed in leaf sheath (Hirose et al., 2006), BEIIa, SSI, GBSSII, and Glc-6-phosphate translocator2 (GPT2), were also analyzed by qRT-PCR (Supplemental Fig. S7). The expression levels of all selected genes, particularly AGPS1, AGPL1, Pho1, and GPT2 were highly correlated with CRCT expression levels. Furthermore, immunoblot analysis indicated that the expression of AGPL and AGPS proteins were substantially increased in leaf sheath of overexpression lines and were slightly decreased in knockdown lines (Fig. 9C). In addition, we found an unknown band around 120 kD by SDS-PAGE analysis with Coomassie Blue staining, which was markedly increased in leaf sheath by CRCT overexpression (Fig. 9D). This band was identified as Pho1 by liquid chromatography-mass spectrometry analysis and subsequent Mascot search (Supplemental Fig. S8). From these findings, it is postulated that CRCT acts to positively control starch content by regulating the expression levels of a subset of genes related to starch synthesis such as AGPL, AGPS, and Pho1.
Figure 9.
Expression analysis of genes related to starch synthesis and starch degradation in leaf sheath. A, Microarray analysis of genes related to starch synthesis. B, Microarray analysis of genes related to starch degradation. Purple indicates higher, while green represents lower, expression by overexpression (left) and knockdown (right) of CRCT. The scale shows log2 at –2.0 to 2.0. Values are expressed as the mean of four replicates. Heat map view was created using TIGR MeV 4.0 software (Saeed et al., 2003). Data and P values are listed in Supplemental Tables S5 and S6. ISA, Isoamylase (DBE); PUL (DBE); α-Amy, α-amylase; β-Amy, β-amylase; DPE, disproportionating enzyme; GWD, α-glucan water dikinase; PWD, phosphoglucan water dikinase; starch excess4-like (SEX4-like), starch phosphatase-like protein; pGlcT, plastidial Glc translocator; MT, maltose translocator. C, Expression levels of AGPase. Total soluble proteins from leaf sheath were separated by SDS-PAGE. AGPL (L) and AGPS (S) were detected by immunoblotting using an antiserum raised against Arabidopsis AGPase. Rubisco large subunit (RbcL) was detected by Coomassie Blue staining and used as internal control. D, Protein profile of leaf sheath analyzed by SDS-PAGE. Total soluble proteins of leaf sheath were separated by SDS-PAGE and stained by Coomassie Blue. Red arrow head indicates Pho1 identified by liquid chromatography-tandem mass spectrometry after trypsin digestion (for detail, see Supplemental Fig. S8). OX1 and OX2 indicate CRCT overexpression lines, and RNAi1 and RNAi2 indicate CRCT knockdown lines.
DISCUSSION
In this study, we demonstrate that a unique CCT protein, CRCT, could be an important regulator of starch synthesis in vegetative organs of rice. In the process of starch synthesis, AGPase, SS, and BE catalyze the reactions of substrate production, chain elongation, and branching, respectively. In addition to these enzymes, GPT can be an important protein involved in importing the Glc-6-P into the plastid, providing the carbon skeletons for starch synthesis in sink tissues (Flügge et al., 2011). In rice, these proteins related to starch synthesis are encoded by multigene families consisting of 21 genes in total. They can be classified into two groups by their tissue-specific expression patterns: one group is expressed in reproductive organs (highly expressed in endosperm) and the other group is expressed in vegetative organs (expressed in leaf blade and leaf sheath; Hirose et al., 2006). Among these genes, CRCT was shown to control the expression of OsAGPS1, OsAGPL1, GPT2, and BEIIa, all of which are classified as vegetative organ types (Fig. 9; Supplemental Fig. S7). In addition, CRCT also regulates the expression of Pho1, which was recently found to play a crucial role in starch biosynthesis (Dauvillée et al., 2006; Satoh et al., 2008). These observations show that CRCT controls the expression of multiple genes related to starch synthesis, mostly those functioned in vegetative tissues.
The expressions of most genes related to starch synthesis are reported to be controlled by carbon and nutritional status. For example, the expressions of transcripts of AGPase and GBSS have been shown to be increased by Glc or Suc and decreased by nitrate (Scheible et al., 1997; Dian et al., 2003; Akihiro et al., 2005). In this study, the expression of CRCT was increased by sugars such as Suc and reduced by nitrogen (Fig. 1, C and D), suggesting the involvement of CRCT in the carbon/nitrogen response of starch synthesis genes. Moreover, the Suc content was not correlated to the starch content, suggesting the presence of a trade-off relationship between Suc and starch in the transgenic lines (Fig. 6), that is, the starch content was significantly increased in the overexpression lines even though the Suc content was limited. These observations imply that CRCT might act downstream of a regulatory pathway for gene expressions related to starch synthesis, hence controlling the content of starch independent of the levels of substrate sugars in the cell of transgenic rice. Thus, it is likely that the expression levels of CRCT could be an important determinant of starch accumulation in rice.
The expression of CRCT was largely confined to the vascular bundle (Fig. 2), whereas the accumulation of starch occurred in a different cell such as the storage parenchyma cell of the leaf sheath (Fig. 8A). In this study, CRCT was constitutively overexpressed in rice using Actin promoter. However, it is not likely that ectopic expression of CRCT leads to the artifact of the phenotype of our overexpression lines, because knockdown of CRCT expression in vascular tissue also showed significant effect on the starch accumulation of different cells (Figs. 6–8). Thus, it is likely that the observed relation between CRCT and starch accumulation can be true. There are two possible explanations for this spatial discrepancy between CRCT expression and starch accumulation. One possibility is that CRCT can express under some conditions in the storage parenchyma cell. Although we have not found out the condition for CRCT, the expression of APL3 and Impaired Suc-Induction1 (ISI1), which might control the expression of APL3 are usually expressed in vascular tissues, but it can be induced by sugar in the mesophyll cell (Rook et al., 2001, 2006). Another possibility is that CRCT or starch synthesis-related enzymes can be synthesized in vascular tissues and then move to storage parenchyma cell. To verify whether these hypotheses are applicable to CRCT or not, further studies are needed.
In the reproductive organs of rice, some important regulation factors of starch synthesis such as RSR1, FLO2, OsEBP-89, and OsbZIP58 have been identified, and the details of the transcriptional regulation mechanisms for these genes are becoming clearer (Zhu et al., 2003; Fu and Xue, 2010; She et al., 2010; Wang et al., 2013). However, the regulation mechanisms are largely unknown in vegetative tissues. Arabidopsis AGPase genes APL3 and APL4 could be induced by both hexose and Suc in leaves (Crevillén et al., 2005), whereas the induction of APL3 by Suc was more remarkable than that by hexose (Solfanelli et al., 2006). This observation is largely consistent with the expression pattern of CRCT (Fig. 1). Loss of function of ISI1 reduced the expression level of APL3 in leaves (Rook et al., 2006). However, the content of starch was not always correlated with the expression levels of ISI1, suggesting that another regulation factor of starch synthesis genes should be present in plants. Sugar signaling by hexokinase has been suggested to have a central role in the regulation of expression of many photosynthetic genes in plants (Rolland et al., 2001). Another important regulator, Sucrose Nonfermenting Related Kinase1 (SnRK1), targets a wide range of sugar-responsive genes (Baena-González et al., 2007). Both hexokinase and SnRK1 were reported to be involved in the posttranscriptional redox activation of AGPase (Tiessen et al., 2003). However, different from CRCT in this study, it seems unlikely that hexokinase and/or SnRK1 play a major role in regulation of expression of starch synthesis-related genes (Xiao et al., 2000; Baena-González et al., 2007). CRCT clearly affects the expression of starch synthesis genes and the content of starch (Figs. 6, 7, and 9). From these observations, these previously reported factors are not likely to be related to CRCT action. CRCT could be a unique important regulator controlling starch synthesis in vegetative organs possibly through an unknown signal transduction pathway.
One of the few marked morphological differences seen in the CRCT overexpression lines was a wider tiller angle (Fig. 5). Tiller angle can be related to shoot gravitropic response. LAZY1 controls rice shoot gravitropism by regulating polar auxin transport, and loss of function causes a drastic spreading tiller phenotype (Li et al., 2007; Yoshihara and Iino, 2007). Another gene, LPA1, also influences shoot gravitropism by regulating the sedimentation rate of amyloplasts, and loss of function of this gene led to larger tiller angles (Wu et al., 2013). Interestingly, the starch excess mutants are known to be much more sensitive to gravity than the wild type in Arabidopsis (Vitha et al., 2007). Recently, it was also reported in knockout mutants of AGPL1 that the decrease in starch content in leaf sheath and its pulvinus, where graviresponsive bending occurs, significantly increased the tiller angle in rice (Okamura et al., 2013). In our study, the expression levels of CRCT did not influence the expression levels of LAZY1 and LPA1 (Supplemental Fig. S5). However, its overexpression greatly increased the starch content of leaf sheath including the pulvinus (Fig. 7B), implying that the increase in tiller angle observed by CRCT overexpression can be explained by the increased starch content of these tissues. Hence, it is possible that CRCT may influence the morphology of rice indirectly by varying starch content.
From our results obtained in this study, we suggest that CRCT is an important regulatory protein that responds to elevated CO2 and that it displays a diurnal expression pattern that modulates starch levels in vegetative tissues. Moreover, biotechnological application of CRCT could be the production of plants for biofuel feedstock, which accumulate high levels of starch in nonfood parts such as leaf sheath and culm without decreasing grain yield. Most starch excess mutants reported so far result from a defect in some component of starch breakdown or translocation of its products (Weise et al., 2011). The overexpression of CRCT could be an alternative and effective approach for the enhancement of starch content of plants. Further analysis of the molecular function of CRCT and fine-tuning of the expression of CRCT in transgenic rice may contribute to the development of a new generation of crop showing improved yield for food and biofuel.
MATERIALS AND METHODS
Plant Growth Conditions
Rice (Oryza sativa ‘Nipponbare’) plants were grown in soil under natural light conditions in a temperature-controlled greenhouse (28°C day/23°C night). Unless stated otherwise, rice seedlings at the 4.5 leaf stage were transplanted into 1-L pots supplemented with a chemical fertilizer (nitrogen:phosphorus:potassium [8:8:8]) at 0.3 g of nitrogen per pot. At panicle initiation stage, the same chemical fertilizer was applied at a rate of 0.1 g of nitrogen as top dressing. For analysis of the effects of elevated CO2 on the expression of CRCT, rice seedlings were grown in a growth chamber at 30°C day/23°C night with a 14-h photoperiod under illumination at a photon flux density of 500 μmol m–2 s–1. The CO2 treatment (38 and 100 Pa as ambient and elevated CO2, respectively) was started at the 4.0 leaf stage.
Vector Construction and Transformation of Rice
Rice genomic DNA was prepared from young fully expanded leaves using cetyltrimethylammonium bromide as described previously (Morita et al., 2014). To construct the promoter::GUS chimeric gene, 5′ flanking region of CRCT (from –2,096 to +98, numbered from the translation start site) was amplified by PCR using gene-specific primers (Supplemental Table S7) as described previously (Ishikawa et al., 2011a). The amplified DNA was fused upstream of GUS into a binary vector, pBI-Hm (a generous gift from Makoto Matsuoka [Nagoya University]).
Total RNA was isolated from rice leaves using an RNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. First strand complementary DNA (cDNA) was synthesized from total RNA with an oligo(dT)18 primer using PrimeScript II First-Strand cDNA Synthesis Kit (Takara). For overexpression of CRCT, the coding region of CRCT was amplified by reverse transcription-PCR using gene-specific primers (Supplemental Table S7). The amplified cDNA was fused to the rice Actin promoter and cloned into a binary vector pBI-Hm. For knockdown of CRCT, 3′ untranslated region (458 bp) was amplified from the cDNA by PCR and cloned into the RNAi vector pANDA (a generous gift from Ko Shimamoto [Nara Institute of Science and Technology]). These constructs were introduced into calli derived from rice via Agrobacterium tumefaciens-mediated gene transfer. Forty transgenic plants for each construct were regenerated from hygromycin-resistant calli and planted in soil. The expression levels of CRCT in transgenic lines were screened by semiquantitative PCR. Most of transgenic plants showed clear change in the expression of CRCT. Primary transgenic plants exhibiting larger expression changes and segregation ratio of around 1:3 were selected. Among them, plants showing no segregation in the progeny were taken as homozygous with transgenes at one locus. Transgenic plants at T3 or T4 generations were used for analyses.
Reverse Transcription-PCR Analyses
Total RNA was isolated from rice tissues as described above. First strand cDNA was synthesized from 1.2 μg of total RNA with an oligo(dT)18 and a random hexamer as primers using PrimeScript II First-Strand cDNA Synthesis Kit (Takara). Semiquantitative PCR was performed using Quick Taq HS DyeMix (Toyobo) with 5% (v/v) dimethyl sulfoxide and gene-specific primers (Supplemental Table S7). DNA polymerase was first activated at 94°C for 2 min, and PCR was carried out for 20 to 30 cycles of 30 s at 94°C, 30 s at 60°C, and 2 min at 68°C, followed by a final extension step for 4 min at 68°C. qRT-PCR was performed using gene-specific primers (Supplemental Table S7). PCR was carried out using SYBR Premix Ex Taq GC (Takara) and Thermal Cycler Dice TP800 (Takara) according to the manufacturer’s instructions. Expression of the Actin gene (AB047313) was examined as an internal control. The amplification efficiencies of all primer pairs used in this study were greater than 90%. The saturation levels were almost the same in qRT-PCR using these primer pairs.
Histochemical Analysis
Histochemical analysis of GUS activity was performed as described (Matsuoka and Numazawa, 1991). Tissue samples were incubated in buffer containing 50 mm potassium phosphate, 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm 5-bromo-4-chloro-3-indoyl-β-d-GlcA, and 0.1% (v/v) Triton X-100 for 3 to 24 h at 37°C. An ethanol wash was performed to stop the GUS reaction and remove chlorophyll from the tissues.
For staining starch, leaf sheath was incubated in ethanol for 24 h to remove chlorophyll and stained with 1% iodine solution containing 1% (w/v) iodine and 1% (w/v) potassium iodide.
Histological analysis of starch using an optical microscope was performed as described (Ishikawa et al., 2011b). Leaf sheath was fixed in a solution containing 5% (v/v) formaldehyde, 5% (v/v) acetic acid, and 63% (v/v) ethanol. Samples were dehydrated through a graded ethanol series and embedded in Technovit 7100 resin (Kulzer) according to the manufacturer’s instructions. The sections were sliced by microtome and stained with 0.25% (w/v) iodine solution.
Measurement of Photosynthetic Rate
CO2 gas exchange rates of the uppermost fully expanded leaves of the main culm were measured with an open gas exchange system (LI-6400, LI-COR) as described previously (Morita et al., 2014). Measurements were performed at a photosynthetic photon flux density of 1,500 mol m–2 s–1, a leaf temperature of 28°C, and ambient CO2 partial pressures of 20, 38, and 88 Pa.
Determination of Carbohydrate Content
The sixth leaf blade and fifth leaf sheath at the 6.5 leaf stage were sampled at the end of the light period. The sample (100 mg) was ground to fine powder in liquid nitrogen using a mortar and pestle and then homogenized with 80% (v/v) ethanol. The homogenate was centrifuged at 10,000g for 10 min. The supernatant was concentrated using a rotary evaporator and made up to a volume of 5 mL with water. An equal volume of chloroform was added to the solution and mixed. After centrifugation at 10,000g for 10 min, the upper fraction was collected and used for determination of Glc, Fru, and Suc. The pellet obtained from first centrifugation was dried at 60°C and suspended with 0.2 mL of hydrochloric acid and 0.8 mL of dimethyl sulfoxide. The suspension was incubated at 60°C for 30 min and adjusted to pH 4.5 with NaOH. After centrifugation at 10,000g for 10 min, the supernatant was collected and used for measurement of starch. Starch was enzymatically hydrolyzed to Glc by amyloglucosidase and determined by coupled enzyme assay of hexokinase and Glc-6-P dehydrogenase using ENZYTEC Starch kit (code no. E1268, r-biopharm, http://www.r-biopharm.com/) according to the manufacturer’s instructions. Glc, Fru, and Suc were determined by coupled enzyme assay of β-fructosidase, hexokinase, and Glc-6-P dehydrogenase using ENZYTEC d-Glc/d-Fru/Suc kit (code no. E1247, r-biopharm) according to the manufacturer’s instructions.
Microarray Analysis
Microarray analysis was performed as described previously (Fukayama et al., 2009). The fifth leaf sheaths at 6.5 leaf stage were ground to fine powder in liquid nitrogen using a mortar and pestle. Total RNA was isolated from the leaf powder as described above. cDNAs were synthesized from total RNA and labeled with the fluorescent dyes Cyanine 5 and Cyanine 3, respectively. The labeled complementary RNA was hybridized with 44-k Rice Oligo Microarray slides (Agilent Technologies). After hybridization, the signals were detected by a DNA microarray scanner (Agilent Technologies), and spot intensities were digitalized using Feature Extraction software (Agilent Technologies). Four comparative analyses were done for each experiment.
SDS-PAGE and Immunoblotting
Total soluble proteins were extracted from rice tissues in extraction buffer containing 50 mm HEPES-KOH, 10 mm MgCl2, 1 mm EDTA, 5 mm dithiothreitol, 0.01 mm leupeptin, 1 mm phenylmethylsulfonylfluoride, 10% (w/v) glycerol, and 5% (w/v) polyvinylpolypyrrolidone, pH 7.4, with a small amount of quartz sand. The proteins were separated by 12% SDS-PAGE and stained with Coomassie Brilliant Blue R-250 or subjected to immunoblotting using an antiserum raised against synthetic peptide of Arabidopsis (Arabidopsis thaliana) AGPL and AGPS (Agrisera). Immunoreacted bands were visualized with the ECL Select Western Blotting Detection Kit (GE Healthcare) and exposed to x-ray film.
Sequence data of CRCT can be found in the Rice Annotation Project database under accession number Os05g0595300.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene structures of CCT domain-containing proteins.
Supplemental Figure S2. Phylogenetic tree based on motif of amino acid sequence among CCT domain-containing proteins in plants.
Supplemental Figure S3. Effects of Glc on the expression of CRCT.
Supplemental Figure S4. Effects of carbon and nitrogen on growth of rice.
Supplemental Figure S5. Expression of tiller angle-related genes in stem base of transgenic rice.
Supplemental Figure S6. Starch content and expression of CRCT in leaf sheath of different transgenic lines used in Figures 3 to 9.
Supplemental Figure S7. Expression analysis of genes related to starch synthesis by qRT-PCR.
Supplemental Figure S8. Amino acid sequence of Pho1 and detected peptides of unknown band found on SDS-PAGE of soluble protein extracted from leaf sheath of CRCT overexpression line OX1 by liquid chromatography-mass spectrometry analysis.
Supplemental Table S1. Up-regulated gene by the overexpression of CRCT analyzed by microarray.
Supplemental Table S2. Down-regulated gene by the overexpression of CRCT analyzed by microarray.
Supplemental Table S3. Up-regulated gene by the knockdown of CRCT analyzed by microarray.
Supplemental Table S4. Down-regulated gene by the knockdown of CRCT analyzed by microarray.
Supplemental Table S5. Data of expression analysis of starch synthesis-related genes by microarray.
Supplemental Table S6. Data of expression analysis of starch degradation-related genes by microarray.
Supplemental Table S7. The primers used in this study.
Supplementary Material
Acknowledgments
We thank Makoto Matsuoka (Nagoya University) and Ko Shimamoto (Nara Institute of Science and Technology) for generous gifts of vectors, Yoshiaki Nagamura and Ritsuko Motoyama (National Institute of Agrobiological Sciences) for technical support of the rice microarray analysis system, and Ryo Ishikawa and Than Myint Htun (Kobe University) for technical assistance with microscopic observation.
Glossary
- qRT
quantitative reverse transcription
- NT
nontransgenic rice
- cDNA
complementary DNA
Footnotes
This work was supported by the Ministry of Education, Culture, Sport, Science, and Technology of Japan (Scientific Research Grants-in-Aid nos. 20580014 and 22114511 to H.F.).
Articles can be viewed without a subscription.
References
- Ainsworth EA, Long SP (2005) What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol 165: 351–371 [DOI] [PubMed] [Google Scholar]
- Akihiro T, Mizuno K, Fujimura T (2005) Gene expression of ADP-glucose pyrophosphorylase and starch contents in rice cultured cells are cooperatively regulated by sucrose and ABA. Plant Cell Physiol 46: 937–946 [DOI] [PubMed] [Google Scholar]
- An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. (2004) CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131: 3615–3626 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Cheng W, Sakai H, Yagi K, Hasegawa T (2009) Interactions of elevated [CO2] and night temperature on rice growth and yield. Agric For Meteorol 149: 51–58 [Google Scholar]
- Cook JH, Yoshida S (1972) Accumulation of 14C-labelled carbohydrate before flowering and its subsequent redistribution and respiration in the rice plant. Proc Crop Sci Soc 41: 226–234 [Google Scholar]
- Crevillén P, Ventriglia T, Pinto F, Orea A, Mérida A, Romero JM (2005) Differential pattern of expression and sugar regulation of Arabidopsis thaliana ADP-glucose pyrophosphorylase-encoding genes. J Biol Chem 280: 8143–8149 [DOI] [PubMed] [Google Scholar]
- Dauvillée D, Chochois V, Steup M, Haebel S, Eckermann N, Ritte G, Ral JP, Colleoni C, Hicks G, Wattebled F, et al. (2006) Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. Plant J 48: 274–285 [DOI] [PubMed] [Google Scholar]
- Dian W, Jiang H, Chen Q, Liu F, Wu P (2003) Cloning and characterization of the granule-bound starch synthase II gene in rice: gene expression is regulated by the nitrogen level, sugar and circadian rhythm. Planta 218: 261–268 [DOI] [PubMed] [Google Scholar]
- Fu FF, Xue HW (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukayama H, Fukuda T, Masumoto C, Taniguchi Y, Sakai H, Cheng W, Hasegawa T, Miyao M (2009) Rice plant response to long term CO2 enrichment: gene expression profiling. Plant Sci 177: 203–210 [Google Scholar]
- Fukayama H, Sugino M, Fukuda T, Masumoto C, Taniguchi Y, Okada M, Sameshima R, Hatanaka T, Misoo S, Hasegawa T, et al. (2011) Gene expression profiling of rice grown in free air CO2 enrichment (FACE) and elevated soil temperature. Field Crops Res 121: 195–199 [Google Scholar]
- Flügge UI, Häusler RE, Ludewig F, Gierth M (2011) The role of transporters in supplying energy to plant plastids. J Exp Bot 62: 2381–2392 [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA (2012) Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci USA 109: 3167–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Saito Y, Ushimaru H, Michiyama Y (2005) The effect of the amount of nitrogen fertilizer on starch metabolism in leaf sheath of japonica and indica rice varieties during the heading period. Plant Prod Sci 8: 122–130 [Google Scholar]
- Hirose T, Ohdan T, Nakamura Y, Terao T (2006) Expression profiling of genes related to starch synthesis in rice leaf sheaths during the heading period. Physiol Plant 128: 425–435 [Google Scholar]
- Ishikawa C, Hatanaka T, Misoo S, Miyake C, Fukayama H (2011a) Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol 156: 1603–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Ohnishi T, Kinoshita Y, Eiguchi M, Kurata N, Kinoshita T (2011b) Rice interspecies hybrids show precocious or delayed developmental transitions in the endosperm without change to the rate of syncytial nuclear division. Plant J 65: 798–806 [DOI] [PubMed] [Google Scholar]
- Kaczorowski KA, Quail PH (2003) Arabidopsis PSEUDO-RESPONSE REGULATOR7 is a signaling intermediate in phytochrome-regulated seedling deetiolation and phasing of the circadian clock. Plant Cell 15: 2654–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Zhao W, Richards JT, Wheeler RM, Guy CL, Levine LH (2012) Transcriptional and metabolic insights into the differential physiological responses of Arabidopsis to optimal and supraoptimal atmospheric CO2. PLoS ONE 7: e43583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leakey AD, Xu F, Gillespie KM, McGrath JM, Ainsworth EA, Ort DR (2009) Genomic basis for stimulated respiration by plants growing under elevated carbon dioxide. Proc Natl Acad Sci USA 106: 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Ainsworth EA, Leakey ADB, Ulanov A, Lozovaya V, Ort DR, Bohnert HJ (2008) Arabidopsis transcript and metabolite profiles: ecotype-specific responses to open-air elevated [CO2]. Plant Cell Environ 31: 1673–1687 [DOI] [PubMed] [Google Scholar]
- Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J (2007) LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res 17: 402–410 [DOI] [PubMed] [Google Scholar]
- Masaki T, Tsukagoshi H, Mitsui N, Nishii T, Hattori T, Morikami A, Nakamura K (2005) Activation tagging of a gene for a protein with novel class of CCT-domain activates expression of a subset of sugar-inducible genes in Arabidopsis thaliana. Plant J 43: 142–152 [DOI] [PubMed] [Google Scholar]
- Matsuoka M, Numazawa T (1991) Cis-acting elements in the pyruvate, orthophosphate dikinase gene from maize. Mol Gen Genet 228: 143–152 [DOI] [PubMed] [Google Scholar]
- Meehl GA. (2007) Global climate projections. InSolomon S, Qin D, Manning M, Marquis M, Averyt K, Tignor MBB, Miller JHL, Chen Z, eds, Climate Change 2007: The Physical Science Basis. Cambridge University Press, New York, pp 747–845 [Google Scholar]
- Morita K, Hatanaka T, Misoo S, Fukayama H (2014) Unusual small subunit that is not expressed in photosynthetic cells alters the catalytic properties of Rubisco in rice. Plant Physiol 164: 69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M, Hirose T, Hashida Y, Yamagishi T, Ohsugi R, Aoki N (2013) Starch reduction in rice stems due to a lack of OsAGPL1or OsAGPL3 decreases grain yield under low irradiance during ripening and modifies plant architecture. Funct Plant Biol 40: 1137–1146 [DOI] [PubMed] [Google Scholar]
- Para A, Farré EM, Imaizumi T, Pruneda-Paz JL, Harmon FG, Kay SA (2007) PRR3 is a vascular regulator of TOC1 stability in the Arabidopsis circadian clock. Plant Cell 19: 3462–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Rolland F, Winderickx J, Thevelein JM (2001) Glucose-sensing mechanisms in eukaryotic cells. Trends Biochem Sci 26: 310–317 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Baier M, Holman R, May AG, Bevan MW (2006) Impaired sucrose induction1 encodes a conserved plant-specific protein that couples carbohydrate availability to gene expression and plant growth. Plant J 46: 1045–1058 [DOI] [PubMed] [Google Scholar]
- Rook F, Corke F, Card R, Munz G, Smith C, Bevan MW (2001) Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J 26: 421–433 [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34: 374–378 [DOI] [PubMed] [Google Scholar]
- Satoh H, Shibahara K, Tokunaga T, Nishi A, Tasaki M, Hwang SK, Okita TW, Kaneko N, Fujita N, Yoshida M, et al. (2008) Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. Plant Cell 20: 1833–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Gonzalez-Fontes A, Lauerer M, Muller-Rober B, Caboche M, Stitt M (1997) Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell 9: 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She KC, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, et al. (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22: 3280–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt M, Krapp A (1999) The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant Cell Environ 22: 583–621 [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Más P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Tallis MJ, Lin Y, Rogers A, Zhang J, Street NR, Miglietta F, Karnosky DF, De Angelis P, Calfapietra C, Taylor G (2010) The transcriptome of Populus in elevated CO reveals increased anthocyanin biosynthesis during delayed autumnal senescence. New Phytol 186: 415–428 [DOI] [PubMed] [Google Scholar]
- Tiessen A, Prescha K, Branscheid A, Palacios N, McKibbin R, Halford NG, Geigenberger P (2003) Evidence that SNF1-related kinase and hexokinase are involved in separate sugar-signalling pathways modulating post-translational redox activation of ADP-glucose pyrophosphorylase in potato tubers. Plant J 35: 490–500 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Ma SF, McPartland M, Hymus GJ, Adam L, Marion C, et al. (2010) The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol 187: 57–66 [DOI] [PubMed] [Google Scholar]
- Vitha S, Yang M, Sack FD, Kiss JZ (2007) Gravitropism in the starch excess mutant of Arabidopsis thaliana. Am J Bot 94: 590–598 [DOI] [PubMed] [Google Scholar]
- Wang JC, Xu H, Zhu Y, Liu QQ, Cai XL (2013) OsbZIP58, a basic leucine zipper transcription factor, regulates starch biosynthesis in rice endosperm. J Exp Bot 64: 3453–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise SE, van Wijk KJ, Sharkey TD (2011) The role of transitory starch in C3, CAM, and C4 metabolism and opportunities for engineering leaf starch accumulation. J Exp Bot 62: 3109–3118 [DOI] [PubMed] [Google Scholar]
- Wu X, Tang D, Li M, Wang K, Cheng Z (2013) Loose Plant Architecture1, an INDETERMINATE DOMAIN protein involved in shoot gravitropism, regulates plant architecture in rice. Plant Physiol 161: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC (2000) The role of hexokinase in plant sugar signal transduction and growth and development. Plant Mol Biol 44: 451–461 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Iino M (2007) Identification of the gravitropism-related rice gene LAZY1 and elucidation of LAZY1-dependent and -independent gravity signaling pathways. Plant Cell Physiol 48: 678–688 [DOI] [PubMed] [Google Scholar]
- Yu B, Lin Z, Li H, Li X, Li J, Wang Y, Zhang X, Zhu Z, Zhai W, Wang X, et al. (2007) TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J 52: 891–898 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cai XL, Wang ZY, Hong MM (2003) An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. J Biol Chem 278: 47803–47811 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.