Abstract
The regulation of plant cell growth and early defense response involves the insolubilization of hydroxyproline-rich glycoproteins (HRGPs), such as extensin, in the primary cell wall. In tomato (Lycopersicon esculentum), insolublization occurs by the formation of tyrosyl-crosslinks catalyzed specifically by the pI 4.6 extensin peroxidase (EP). To date, neither the gene encoding EP nor the protein itself has been identified. Here, we’ve identified tomato EP candidates using both proteomic and bioinformatic approaches. Bioinformatic screening of the tomato genome yielded eight EP candidates, which contained a putative signal sequence and a predicted pI near 4.6. Biochemical fractionation of tomato culture media followed by proteomic detection further refined our list of EP candidates to three, with the lead candidate designated (CG5). To test for EP crosslinking activity, we cloned into a bacterial expression vector the CG5 open-reading frame from tomato cDNA. The CG5 was expressed in E. coli, fractionated from inclusion bodies, and folded in vitro. The peroxidase activity of CG5 was assayed and quantified by ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)) assay. Subsequent extensin crosslinking assays showed that CG5 can covalently crosslink authentic tomato P1 extensin and P3-type extensin analogs in vitro supporting our hypothesis that CG5 encodes a tomato EP.
Keywords: Hydroxyproline-rich glycoproteins, plant cell wall, extensin, peroxidase, protein crosslinking
1. Introduction
1.1 Extensins
Extensins are a major class of the hydroxyproline-rich glycoprotein (HRGP) family that contribute to wall architecture and pathogen defense by forming covalently crosslinked networks (Brysk and Chrispeels, 1972; Epstein and Lamport, 1984; Lamport, 1969, 1977; Mort and Lamport, 1977). Extensins are extensively post-translationally modified. This includes proline hydroxylation to form hydroxyproline (Hyp), Hyp O-glycosylation with arabinosyl oligosaccharides, and serine monogalactosylation. Historically, extensins are defined by the abundance of repetitive Ser-Hyp4 glycomodules that nucleate an extended, polyproline-II conformation (Lamport, 1977; van Holst and Varner, 1984). The Ser-Hyp4 glycomodules alternate with hydrophobic motifs that often contain tyrosine residues involved in covalent crosslinking (Epstein and Lamport, 1983; Fry, 1982; Brady et al., 1996; Brady and Fry, 1997; Brady et al., 1998; Held et al., 2004). Examples include P1- and P3-type extensin repeat motifs of tomato: [Ser-Hyp4-Thr-Hyp-Val-Tyr-Lys]n and [Ser-Hyp4-Ser-Hyp-Ser-Hyp4-Tyr-Tyr-Tyr-Lys]n, respectively (Epstein and Lamport, 1984; Smith et al., 1984; Smith et al., 1986).
1.2 Extensin crosslinking
Extensins are crosslinked both intra- and intermolecularly. Intramolecular crosslinking gives rise to isodityrosine (Idt; Figure 1), whereby the outer two Tyr residues in [-Tyr-X-Tyr-] motifs (where X is usually Tyr, Lys, or Val) are coupled by diphenylether bond formation (Epstein and Lamport, 1984; Fry, 1982). When and how Idt is formed is not currently known. Nonetheless, Idt likely participates in intermolecular crosslinking of extensin monomers in the wall to produce the crosslinking amino acids, pulcherosine (Pul) and di-isodityrosine (di-Idt) (Figure 1; Brady et al., 1996; Brady and Fry, 1997; Brady et al., 1998; Held et al., 2004). It’s hypothesized that the amphiphilic, repetitive nature of extensin monomers likely drives self-assembly, whereby hydrophobic crosslinking motifs position extensin monomers for subsequent covalent crosslinking (Brady et al., 1996; Cannon et al., 2008; Heckman et al., 1988; Kieliszewski and Lamport, 1994).
Figure 1. Intra- and intermolecular extensin crosslinking amino acids.
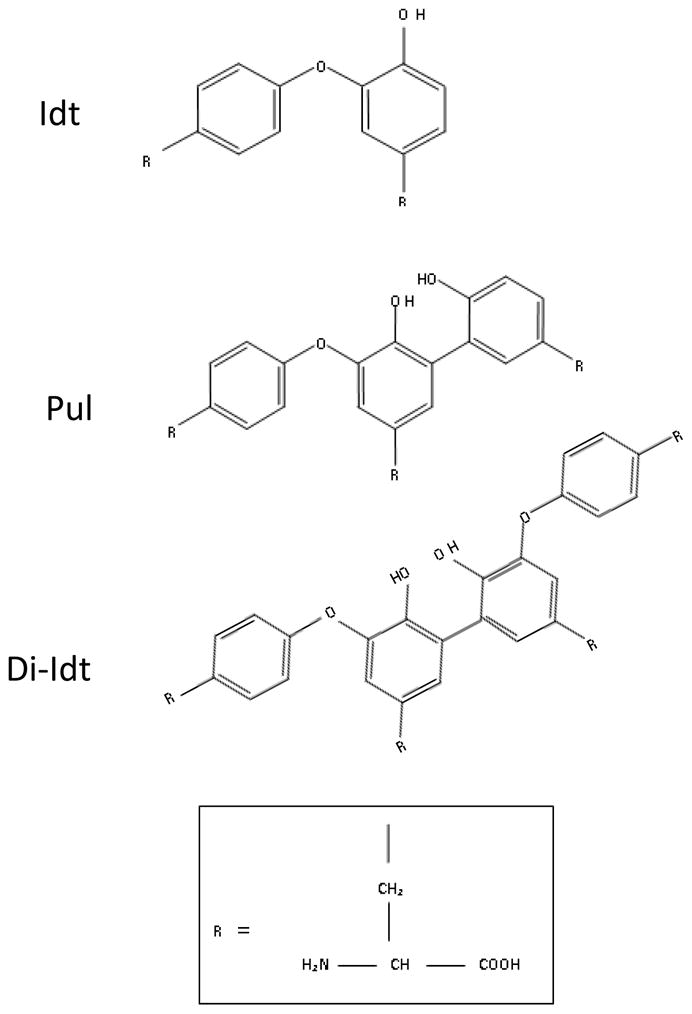
Extensin monomers can be intramolecularly crosslinked through diphenylether linkage of tyrosine side chains as part of (Y-X-Y) crosslinking motifs to form Isodityrosine (Idt) (Lamport, 1977; Epstein and Lamport, 1984; Fry 1982). Idt crosslinks can be intermolecularly coupled to another single Tyr side chain to form Pulcherosine (Pul), or to another Idt to form (di-Idt) (Brady et al. 1996; Brady and Fry 1997; Brady et al. 1998). Both Pul and di-Idt are formed by biphenyl linkages.
1.3 Extensin Peroxidases
Plant peroxidases are involved in a wide range of processes including defense and the hypersensitive response (Bolwell, 1999; Bolwell and Wojtaszek, 1997; O’Brien et al., 2012; Wojtaszek, 1997), hormone catabolism (Lagrimini et al., 1997), suberization (Espelie et al., 1986), salt tolerance (Amaya et al., 1999), and senescence (Abeles et al., 1988) in addition to glycoprotein crosslinking (Everdeen et al., 1988; Fry et al., 1986; Lamport et al., 1980). Formation of the extensin intermolecular crosslinking amino acids, Pul and di-Idt, is likely catalyzed by specific, secreted class III peroxidases, and renders extensin precursors insoluble in the wall. These crosslinking enzymes, designated Extensin Peroxidases (EPs) have been reported in numerous plant systems, including Lupin (LEP1; Price et al., 2003), Grapevine (GvEP1; Jackson et al., 2001), French Bean (FBP1; Wojtaszek et al., 1997), and Tomato (Brownleader et al., 1995; Everdeen et al., 1988; Held et al., 2004; Schnabelrauch et al., 1996). Of these candidates, the pI 4.6 tomato EP is the only acidic EP and it catalyzes the in vitro crosslinking of tomato extensin precursors under physiological conditions (Schnabelrauch et al., 1996).
1.4 Tomato Extensin Peroxidase
The substrate specificity and kinetic properties of an EP has been best characterized in tomato (Lycopersicon esculentum). Schnabelrauch and colleagues (1996) found that the tomato EP had a pI of 4.6, a pH optimum of 5.5, and was highly specific for extensin monomers containing [-Val-Tyr-Lys-] crosslinking motifs, commonly found in P1 extensins (Smith et al., 1984). Subsequently, Held et al. (2004) demonstrated that the tomato EP also rapidly crosslinked P3-type extensins possessing the Idt-motif [-Tyr-Tyr-Tyr-], however the precise identity of the crosslinking enzyme(s) remained unknown. Here we have used a combined approach of bioinformatics and proteomics to identify the pI4.6 tomato EP. The tomato EP gene (CG5) was identified within the tomato genome sequence (solgenomics.net) and subsequently cloned from tomato cDNA. Phylogenetic analysis indicated that the tomato EP was more closely related to GvEP1, than LEP1 or FBP1. The tomato EP was expressed recombinantly in E. coli, purified from inclusion bodies, and folded in vitro in the presence of a heme cofactor. In order to determine if CG5 was indeed an EP, recombinant CG5 was tested for in vitro crosslinking activity using purified tomato P1 extensin and P3 extensin-analog substrates. Extensin crosslinking was observed for recombinant CG5, but not for horseradish peroxidase (HRP). This is the first identification of a pI4.6 extensin peroxidase with confirmation of its biochemical activity in vitro.
2. Results and discussion
Although they account for only about 10% of the primary cell wall, crosslinking extensins are required components for wall assembly, particularly during embryogenesis and germination (Cannon et al., 2008; Hall and Cannon, 2002; Saha et al., 2013), hence the enzyme(s) that catalyze their crosslinking are of interest. Initial work aimed at identifying enzymes involved in tomato extensin crosslinking in vitro was frustrated at the biochemical level due to the requirement for purified peptides to allow sequence analysis by Edman degradation. More recent advances in protein sequence analysis via LC-MS/MS on relatively impure fractions, together with the availability of plant genome databases and facile cloning protocols, enabled us to revisit the tomato EP identification problem.
2.1 A reverse-genomic approach to identify tomato EP candidate genes
Tomato EP is a secreted, heme-containing (class III) peroxidase that has an isoelectric point of 4.6 (Schnabelrauch et al., 1996). To identify candidate genes for encoding the tomato EP, we first collected the sequences of all annotated peroxidase genes from the tomato genome by a keyword search of “peroxidase”. A total of 110 non-redundant peroxidase genes was identified by this approach (Table 1S). This approach was not expected to identify all peroxidase genes in the tomato genome, however, the total number of peroxidase genes agreed well with the total number of peroxidases found in other plant species, (i.e. 73 peroxidase genes in the Arabidopsis (Tognolli et al., 2002) and 138 in Oryzae sativa (Passardi et al., 2004). Next, the protein sequences encoded by these candidates were sorted based on the predicted presence of an N-terminal signal sequence. Proteins that lacked a predicted signal sequence were removed from the candidate list. The remaining candidates were then sorted by the predicted pI of their mature encoded proteins. Eight candidate genes were identified that contained a signal sequence and had a predicted pI within ±0.3 units of 4.6 (Table 1). These candidates were designated candidate genes (CG1-8).
Table 1. Candidate genes for the pI4.6 tomato extensin peroxidase.
Eight candidate genes were identified from 110 annotated peroxidase genes in tomato genome according to Sol Genomics Network (solgenomics.net) by screening for signal sequences and the predicted pI of their mature encoded proteins.
| Candidate gene (CG) | Gene Accession | Predicted pI | Genomic DNA size (bp) | cDNA size (bp) | Predicted MW (kDa) |
|---|---|---|---|---|---|
| 1 | Solyc01g006300 | 4.31 | 2,852 | 1,246 | 35.0 |
| 2 | Solyc02g079510 | 4.37 | 2,393 | 1,272 | 38.6 |
| 3 | Solyc02g079500 | 4.73 | 2,460 | 1,316 | 38.8 |
| 4 | Solyc11g072920 | 4.65 | 1,326 | 984 | 34.8 |
| 5 | Solyc02g094180 | 4.65 | 1,669 | 1,279 | 35.4 |
| 6 | Solyc01g104860 | 4.9 | 1,980 | 1.052 | 34.7 |
| 7 | Solyc10g076190 | 4.84 | 1,142 | 963 | 35.1 |
| 8 | Solyc11g010120 | 4.9 | 2,442 | 1,014 | 37.7 |
2.2 Biochemical fractionation of the pI 4.6 tomato EP and proteomic detection
Schnabelrauch and colleagues (1996) showed that tomato EP could be significantly enriched from cell-suspension culture media by anion-exchange chromatography (Figure S1) and isoelectric focusing (IEF) (Figure S2). In order to further limit the EP candidates identified by our genomic search (Table 1), we used proteomics to identify protein sequences in the pI 4.6 fraction enriched for EP. LC-MS/MS sequencing of tryptic peptides released from the pI 4.6 EP fraction identified three secreted anionic peroxidases, namely CG5 (Solyc02g094180), CG3 (Solyc02g079500), and CG8 (Solyc11g010120) (Table 2). Peptides matching CG5 were the most abundant, in terms of the percentage of total spectra (2.8%) and normalized total spectra (88) (Figure S3A). Peptides matching CG3 and CG8 were each detected in only one of two individual samples, and were both less abundant than CG5 (Figures S3B and S3C), therefore we focused on CG5 for characterization.
Table 2. Annotated tomato peroxidases identified by proteomics.
The pI 4.6 tomato fraction was subjected to proteomic analysis. Listed here are unique peptides matching putative tomato peroxidases. A total of 15 unique peptides matched CG5, while 5 and 2 peptides were identified for CG8 and CG3 respectively. Proteomics of the tomato pI4.6 fraction was performed in duplicate and unique peptides from CG5 were collected from both runs, while peptides for CG8 and CG3 were only detected in one run each.
| Gene Accession # | Protein Identification | Exclusive Unique Peptides |
|---|---|---|
| Solyc02g094180 (CG5) | Anionic Peroxidase 1 | DGLISQASR |
| RDGLISQASR | ||
| DMCPQNVDPTIANMDPATPR | ||
| ELKDMCPQNVDPTIANMDPATPR | ||
| DNLSLAGDGFDTVVK | ||
| DSKDNLSLAGDGFDTVVK | ||
| DVVVLAGGPSYNVELGR | ||
| FSQTFVTIPATLR | ||
| LPEPDFNLIQLNTMFAR | ||
| VAGKLPEPDENLIQLNTMFAR | ||
| LYSFTPSNPVDPSLDPEYAK | ||
| QAVEAQCPGVVSCADILAIATR | ||
| TFDNEYYK | ||
| VGVKTGGQGEIR | ||
| TGGQGEIR | ||
|
| ||
| Solyc11g010120 (CG8) | Peroxidase 17 | EALEQACPGVVSCADLLIIAAR |
| EYVEEYSVDQER | ||
| GFLNSDETLFTNSITR | ||
| LALSNINSLR | ||
| SYEVVDEIK | ||
|
| ||
| Solyc02g079500 (CG3) | Peroxidase | AVVDSAIDAETR |
| LGGQTYSVALGR | ||
2.3 Sequence and phylogenetic analysis of CG5
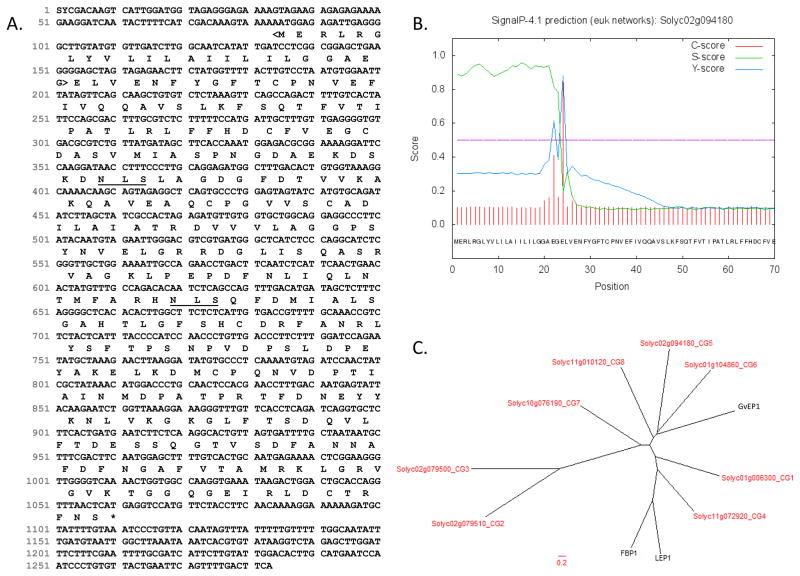
The predicted protein sequence of CG5, deduced from the coding sequence of the CG5 gene in the tomato genome (iTAG 2.3; Figure 2A), is 325 amino acids. The mature CG5 protein, lacking the signal sequence (aa 1–23), is 302aa and has a predicted molecular weight and pI of 35.4 kDa and 4.65, respectively (Table 1; Figure 2B). Unlike LEP1 and FBP1, CG5 does not appear to contain a C-terminal propeptide domain (CTPP). CTPPs are thought to assist trafficking from the trans-Golgi to the vacuole and are removed during protein maturation (Chrispeels and Raikhel, 1992; Nakamura and Matsuoka, 1993; Price et al., 2003; Wojtaszek et al., 1997). Indeed, tomato CG5 retained an intact C-terminus, as peptides were detected by LC-MS/MS from this region (Table 2; VGVKTGGQGEIR). Two predicted N-glycosylation sites were found in CG5 using the NetNGlyc1.0 program (Figure 2A). This sets it apart from LEP1, which has 12 predicted N-glycosylation sites. Whether or not the native CG5 is actually glycosylated remains to be confirmed.
Figure 2. Sequence analyses of the tomato CG5.
(A) Full-length cDNA and predicted protein sequence are from iTAG2.3 (accessed 3/5/14). N-glycosylation sites (underlined) were predicted by NetNGlyc1.0. The N-terminal signal sequence (<bracketed>) and (B) cleavage site were predicted by SignalP 4.0 (Petersen et al. 2011). (C) Phylogenetic analysis of EP proteins and tomato CGs was performed. Predicted protein sequences of tomato CGs were aligned with EP proteins from grapevine (GvEP1; Jackson et al. 2001), lupin (LEP1; Price et al 2004), and French bean (FBP1; Wojtaszek et al. 1997). Multiple sequence alignment was performed by ClustalW using a GONNET weight matrix with a gap penalty of 10 and an extension penalty of 0.05. TreeDyn was used to make the radial dendrogram. Tomato CGs are shown in red.
The phylogenic relationship between the tomato CGs 1–8 (Table 1) and previously characterized EPs was examined by performing a multiple sequence alignment to generate a radial dendrogram (Figure 2C). The dendrogram contains three major clades. Tomato CGs 5, 6 and 8 group in a clade that contains the grapevine peroxidase (GvEP1; Jackson et al., 2001; Pereira et al., 2011), but not LEP1 or FBP1 (Price et al., 2003; Wojtaszek et al., 1997) which instead are more closely related to tomato CG4 and CG1. CGs 2, 3 and 7 group separately in a third clade. Thus CG5 appears to be most closely related to GvEP1 from grapevine.
2.4 Recombinant expression CG5 and in vitro folding
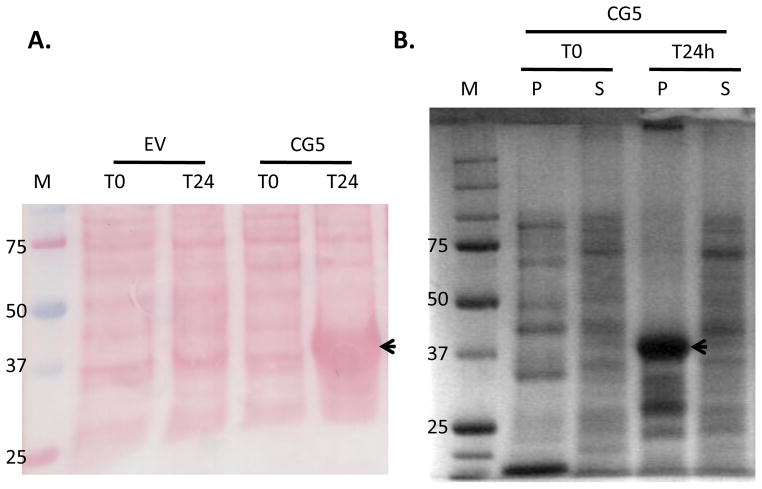
The CG5 ORF was cloned from tomato cDNA into a bacterial expression vector for recombinant expression as a 6xHis-fusion (CG5-6xHis). Protein expression was induced for 24hrs and a protein of expected molecular weight (~37kDa) corresponding to recombinant CG5-6xHis was found in whole cell lysates (Figure 3A). Whole cell lysates were separated into soluble and insoluble (pellet) fractions. The ~37 kDa protein was mainly associated with the insoluble fraction, indicating that it accumulated in inclusion bodies (Figure 3B), as observed with other plant peroxidases (Teilum, 1999; Henriksen et al., 2001). The ~37kDa gel band was excised and submitted for LC-MS/MS sequence analysis. Peptide sequencing confirmed the ~37kDa band was indeed recombinant CG5 (data not shown). Unfolded CG5-6xHis was prepared from inclusion bodies and folded in the presence of a heme cofactor in vitro (Teilum et al., 1999). As a negative control, insoluble proteins from E. coli expressing the empty-vector (EV) construct were prepared after 24 hrs of induction and folded alongside CG5-6xHis, using the same folding procedure.
Figure 3. Expression of recombinant CG5 in E. coli.
(A) E. coli cultures expressing either empty vector (EV) or CG5-6xHis were collected just prior to (T0) and 24 hrs after (T24) induction with IPTG (0.5mM). Whole cell lysates (10μg per lane) were separated by 10% SDS-PAGE, blotted onto nitrocellulose and Ponceau stained. (B) Whole cell lysates of CG5 after 0 (T0) and 24hrs (T24) of induction were separated into soluble (S) and pellet (P) fractions by centrifugation. Protein fractions (10μg) were separated by 10% SDS-PAGE and stained with coomassie. Arrows indicate the expression of CG5 in whole cell lysates (A) and in inclusion bodies (B) 24hrs after induction. M, molecular weight marker (kDa).
2.5 Characterization of the recombinant extensin peroxidase
2,2′-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) is a general substrate for assaying peroxidase activity and has been used previously to quantify EPs in crude eluates (Everdeen et al., 1988; Schnabelrauch et al., 1996). We used ABTS here to assay the activity of folded CG5-6xHis and EV controls. Folding buffer was included as an additional negative control and used to normalize activity measurements. The folded, recombinant CG5 fusion protein samples showed ~3.5 to 6 times more ABTS peroxidase activity than the empty vector (EV) controls (Figure 4). Three individual CG5-6xHis clones were tested, using equal protein contents for all samples, with the exception of the folding buffer control, which lacked protein. These data indicated that CG5-6xHis expressed in E. coli had properly incorporated the heme cofactor and possessed peroxidase activity. The fact that active recombinant protein was obtained from E. coli, suggested that the CG5-6xHis did not require N-glycosylation for peroxidase activity.
Figure 4. Peroxidase activity assay of E. coli expressing CG5-6xHis using ABTS.
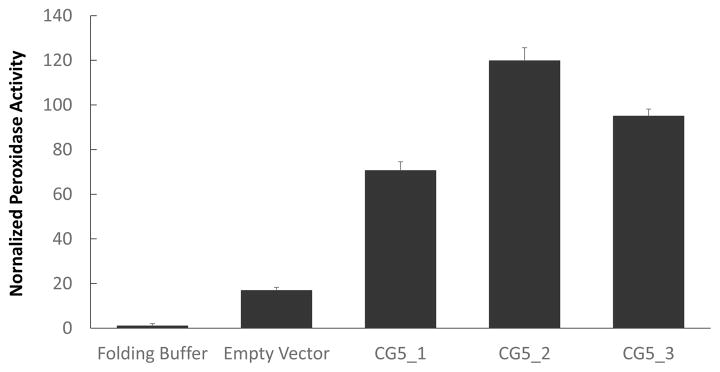
Three individual clones of E. coli expressing CG5-6xHis (CG5_1, CG5_2, and CG5_3) were tested for peroxidase activity using the ABTS assay (Everdeen et al. 1988). Insoluble proteins from E. coli transformed with the empty-vector were assayed as a background control. Activities for each sample were recorded in triplicate, averaged, then normalized to folding buffer-only controls (=1). Error bars represent standard deviations of triplicate measurements.
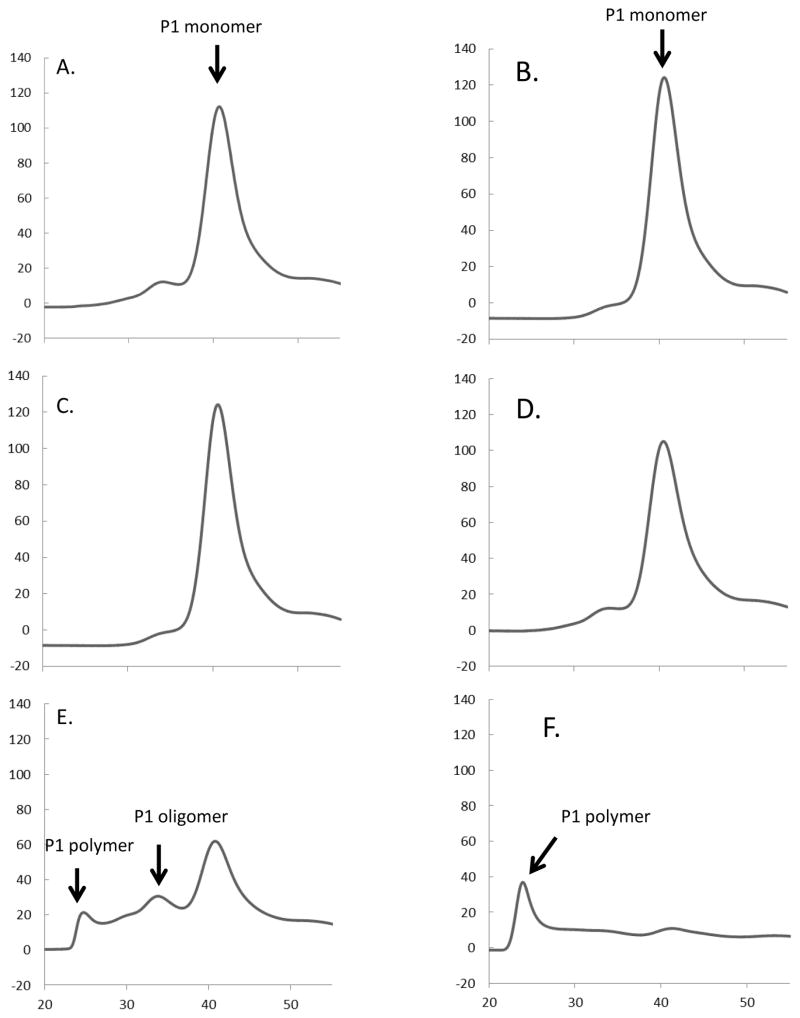
Purified tomato P1 extensin, P3 analog YK20 [-Ser-Hyp4-Ser Hyp-Ser-Hyp4-Tyr-Tyr-Tyr-Lys-]20 and P3 analog FK9 [-Ser-Hyp4-Ser Hyp-Ser-Hyp4-Phe-Phe-Phe-Lys-]9 were used as substrates to examine the extensin crosslinking properties of CG5-6xHis (Schnabelrauch et al., 1996; Smith et al., 1984; Held et al., 2004). Crosslinking reactions were prepared to compare the enzymatic activities of CG5-6xHis with the endogenous tomato pI 4.6 EP, as well as horseradish peroxidase (HRP). HRP is a not an EP and only crosslinks extensin at very high (non-physiological) enzyme concentrations. Equal nkat quantities of each peroxidase, as determined by ABTS activities, were used for all samples (except in the folding buffer control). After 30 min of incubation, both native pI4.6 tomato peroxidase and the recombinant CG5-6xHis polymerized P1 extensin, judging by the size shift assayed by gel-filtration chromatography (Figure 5D and F), while folding buffer, water, EV, and HRP controls did not (Figure 5A–C, E).
Figure 5. In vitro crosslinking assays using tomato P1 extensin.
Extensin crosslinking assays (30min) were performed using tomato P1 extensin substrate to test the activities of folded CG5-6xHis. Control assays included folding solution (A), water (B), corresponding proteins isolated from E. coli transformed with the empty vector-EV (C), and horseradish peroxidase (D). Extensin polymerization was observed when either CG5-6xHis (E) or the tomato pI 4.6 EP (F) was added. Crosslinking is observed by the size shift in P1 monomer by gel permeation chromatography upon forming P1 oligomer and P1 polymer with concomitant decrease in P1 monomer. Extended incubation times leave much of the P1 polymer formed in (E) and (F) insoluble (e.g. not detected by gel-filtration).
Additional reactions were prepared to estimate the crosslinking rates of both enzymes using either authentic P1 or the P3 analog, YK20 as substrate (Held et al., 2004). FK9 substrate was used as a negative control, as it lacks Tyr crosslinking motifs and cannot be crosslinked by EPs (Held et al., 2004). CG5-6xHis and the pI4.6 EP were able to crosslink both P1 and YK20 substrates, but not FK9 (Figure S4) (Held et al., 2004; Schnabelrauch et al., 1996). HRP did not crosslink any of the extensin substrates (Figure S4). The initial rates of these reactions were calculated (Held et al. 2004). Crosslinking rates of CG5-6xHis and tomato EP were comparable for both P1 and YK20 (Figure 6), and both enzymes appear to crosslink the P1 substrate more rapidly than the YK20 substrate. This is agreement with previous reports for the tomato EP with both P1 and YK20 (Held et al. 2004). Together, these data corroborate our conclusion that the CG5-6xHis protein is an EP and is capable of crosslinking the same extensin substrates at rates similar to native tomato EP.
Figure 6. Initial crosslinking rates of Tomato pI4.6 EP and CG5-6xHis.
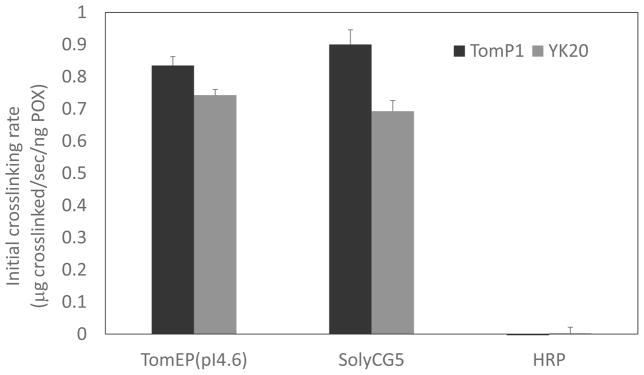
Endogenous tomato P1 and synthetic P3 analog (YK20) extensins were used as substrates for in vitro crosslinking reactions to calculate initial rates (μg of extensin crosslinked/sec/ng peroxidase). Crosslinking reactions were stopped after two-minutes of incubation to facilitate rate calculations for each substrate (Everdeen et al., 1988; Held et al., 2004; Schnabelrauch et al., 1996). Initial rates were calculated as described earlier using the first order rate equation and normalized to the amount of peroxidase (ng POX) per reaction (Everdeen et al. 1988). Error bars represent the standard deviation of the mean crosslinking rates of triplicate reactions. FK9 (not shown) showed no crosslinking whatsoever.
3. Concluding remarks
CG5 catalyzes the crosslinking of extensins at physiological conditions (μg quantities of extensin substrate, μM quantities of peroxide, and fmol quantities of enzyme), which supports the hypothesis that this enzyme is an active EP in muro. The availability of the recombinant CG5 EP and purified extensin substrates/analogs, now allow us to ask more specific questions about extensin network formation and architecture.
How is the extensin network assembled in muro?
Molecular details concerning the assembly and architecture of the extensin network in muro are lacking. Clearly extensin monomers can become covalently crosslinked to each other in the cell wall, most likely through the formation of the crosslinking amino acids, Pul and di-Idt (Brady et al., 1996; Brady and Fry, 1997; Brady et al., 1998). However, we have not yet been able to isolate crosslinked peptides directly from wall preparations to corroborate this hypothesis. This difficulty is likely due to the low abundance of these peptides among cell wall preparations. Using purified extensin and extensin-analog substrates with purified EP, crosslinked polymers can now be produced in vitro and proteolytically cleaved to release these crosslinked peptides for LC-MS/MS identification. Future proteomic studies to identify crosslinked peptides in the wall will benefit by using the in vitro peptides as standards.
What factors influence crosslinking amino acid formation?
Crosslinking of RSH extensin from Arabidopsis results mainly in the formation of Pul, whereas crosslinking of YK20 produces almost exclusively Di-Idt (Cannon et al., 2008; Held et al., 2004). With the in vitro system, we are now able to evaluate the molecular factors that influence the formation of Pul versus di-Idt, using combinations of P1, RSH and/or P3-analog substrates with the purified CG5 EP. These studies will also shed light on whether different extensin types can crosslink to each other (heteropolymeric crosslinking). Current studies have only focuses on the crosslinking of single, purified substrates (homopolymeric crosslinking). Additionally, we do not know when or how intramolecular Idt is formed. Idt is present in YK20 precursors, indicating that it maybe formed prior to wall assembly. However, YK20 precursors are secreted and pass through the wall prior to purification. Therefore it’s possible intramolecular Idt may be formed as YK20 passes through the wall. Additionally, earlier work suggested that Idt may be formed at the wall as well, since monomeric extensins isolated by salt-elution contained relatively little Idt, yet Idt was present in wall crosslinked extensin (Smith et al., 1984). We are now poised for future studies aimed to examine whether the CG5 EP is also involved in forming intramolecular extensin crosslinks.
What makes the CG5 EP especially suited for extensin substrates?
The presence of so many apoplastic peroxidases and extensin types begs questions about the nature of EP:extensin substrate interaction. Specifically, are there multiple EPs and if so, does this reflect the necessity to cross-link different types of extensins/HRGPs? This seems likely as the pI 4.6 EP does not crosslink proline-rich proteins (PRPs), which are HRGPs comprised of repeating Pro-Hyp-Val-Tyr-Lys motifs (Chen and Varner, 1985), yet PRPs are almost certainly part of the crosslinked wall network (Frueauf et al., 2000; Bradley et al., 1992). Structural studies of the purified EP will help identify potential structural aspects which confer high extensin cross-linking activity to these class III peroxidases and the mechanisms of crosslinking amino acid formation.
4. Experimental
4.1 Candidate gene identification
To identify peroxidase genes in tomato, the tomato genome (ITAG2.3) was keyword-searched for “peroxidase” (query date 2/7/2011). A list of 110 unique peroxidase candidate genes (Table S1) was collected from the search. The predicted protein sequences of these candidates were exported from iTAG2.3 and analyzed for the presence of a signal sequence using SignalP3.0 (http://www.cbs.dtu.dk/services/SignalP/) (Petersen et al., 2011) and their predicted pIs were recorded from the tomato genome database at Gramene (http://archive.gramene.org/Solanum_lycopersicum). Candidates that possessed a signal sequence and had a pI of 4.6 ± 0.3 were selected as candidate genes (CGs).
4.2 Phylogenetic analysis of CGs with other EPs
Protein sequences of the tomato CGs (1–8) and three EP candidates (LEP1, GvEP1, and FBP1) were compared by multiple sequence alignment using Clustal W (http://www.genome.jp/tools/clustalw/). The GONNET weight matrix was used with gap penalty of 10 and an extension penalty of 0.05. A radial dendrogram was constructed in TreeDyn using the resulting clustal.dnd file.
4.3 Biochemical fractionation of the pI4.6 EP
The tomato pI 4.6 EP was fractionated by anion-exchange chromatography and isoelectric focusing from tomato culture media essentially as described earlier (Schnabelrauch et al. 1996; Held et al. 2004). Briefly, culture medium (4L) from 30d old tomato cultures was collected by filtration on a sintered glass funnel and concentrated by lyophilization. Dried media was dissolved in water (50ml) and clarified by ultracentrifugation at 250,000 x g for 1h 4°C. Supernatants were dialyzed against 25 mM sodium acetate buffer pH6.0 and then applied onto a DEAE-sepharose column (1.4×10cm) at a flow rate of 1ml/min. A linear gradient to 2M sodium chloride in 25mM sodium acetate buffer pH6.0 was used to elute the acidic fractions. Fractions (1ml) were collected and assayed for peroxidase activity by ABTS assay (Everdeen et al., 1988). ABTS-active fractions were concentrated and desalted by Centricon ultrafiltration (30 kDa MW cutoff) and fractionated by isoelectric focusing (IEF). IEF was performed using a preparative Rotofor apparatus (Bio-Rad) with pH3-pH5 ampholyte according to manufacturer’s instructions. Twenty fractions (2ml each) were collected and assayed for protein concentration by BCA protein assay (Pierce) against bovine serum albumin (BSA) standards. Fractions were also tested for peroxidase activity by ABTS assay. ABTS-active fractions near pH4.6 were collected and used for proteomic analysis.
4.4 Proteomic identification of peroxidases
Proteins (25μg) from the pI 4.6 IEF fraction were separated by SDS-PAGE (10% gel). A section of the gel between 30–40kDa was excised and sent to the Michigan State University Proteomics Facility for proteomics. Proteomic analysis was performed in duplicate. Gel bands were digested in-gel according to Shevchenko et al. (1996) with modifications. Briefly, gel bands were dehydrated using 100% acetonitrile and incubated with 10mM dithiothreitol in 100mM ammonium bicarbonate, pH~8, at 56°C for 45min, dehydrated again and incubated in the dark with 50mM iodoacetamide in 100mM ammonium bicarbonate for 20min. Gel bands were then washed with ammonium bicarbonate and dehydrated again. Sequencing-grade modified typsin was prepared to 0.01μg/μL in 50mM ammonium bicarbonate and ~50μL of this was added to each gel band so that the gel was completely submerged. Bands were then incubated at 37°C overnight. Peptides were extracted from the gel by water bath sonication in a solution of 60%ACN/1%TCA and vacuum dried to ~2μL. Peptides were then re-suspended in 2% acetonitrile/0.1%TFA to 20μL and from this 10μL were automatically injected by a Waters nanoAcquity Sample Manager (www.waters.com) and loaded for 5 minutes onto a Waters Symmetry C18 peptide trap (5μm, 180μm × 20mm) at 4μL/min in 2%ACN/0.1%Formic Acid. The bound peptides were then eluted using a Waters nanoAcquity UPLC (Buffer A = 99.9% Water/0.1% Formic Acid, Buffer B = 99.9% Acetonitrile/0.1% Formic Acid) onto a Waters BH130 C18 column (1.7μm, 150μm × 100mm) and eluted over 35 minutes with a gradient of 10% B to 30% B in 24min, 30%B to 90%B 25min, holding at 90%B for 1min and dropping to 5%B at 26.1min at a constant flow rate of 0.8μl/min.
Eluted peptides were sprayed into a ThermoFisher LTQ-FT Ultra mass spectrometer (www.thermo.com) using a Michrom ADVANCE nanospray source. Survey scans were taken in the FT (25000 resolution determined at m/z 400) and the top five ions in each survey scan were then subjected to automatic low energy collision induced dissociation (CID) in the LTQ. The resulting MS/MS spectra are converted to peak lists using BioWorks Browser, v3.2 (ThermoFisher, www.thermo.com) using the default LTQ-FT parameters and searched against the ITAG tomato protein sequence database, v2.3 (provided by the International Tomato Annotation Group, downloaded from www.solgenomics.net on 5/8/2012), appended with common laboratory contaminants, using the Mascot searching algorithm, v 2.3. The Mascot output was then analyzed using Scaffold, v3.4.7 (www.proteomesoftware.com) to probabilistically validate protein identifications using the ProteinProphet (Nasvizhskii et al., 2003) computer algorithm. Protein assignments validated above the Scaffold 95% confidence filter are considered true.
4.5 Recombinant expression in E. coli, fractionation and folding of CG5
4.5.1 Cloning of CG5 for expression as a 6xHis fusion
First strand cDNA was prepared from total RNA (1μg) isolated from tomato suspension cultured cells using Trizol reagent (Invitrogen). The CG5 open reading frame, lacking the N-terminal signal peptide, was cloned from the cDNA using primers (F: 5′AATCC ATG GAG CTA GTA GAG AAC TTC TAT GGT TTT AC; R: 5′ TTG TCG ACT GAG TTA AAC CTG GTG CAG TCC) and the Pfu Ultra II Fusion HS polymerase (Agilent). The amplified CG5 insert was digested with NcoI-SalI and ligated into the NcoI-XhoI sites of the pET28A vector (EMD Millipore) for expression in E. coli as a C-terminal 6xHis fusion protein. Reading frame was confirmed by DNA sequencing. Intact plasmids for expressing CG5-6xHis and empty-vector (EV) control (lacking insert) were used to transform E. coli strain Rosetta 2 DE3 (EMD Millipore).
4.5.2 Recombinant protein expression and induction
Overnight cultures of CG5-6xHis and EV (10ml) were prepared in LB medium containing 100μg/ml kanamycin and 30μg/ml chloramphenicol and incubated at 37°C overnight. The culture was adjusted to an OD600nm of 0.2 and grown further at 37°C for 1–2hr. Recombinant protein expression was induced with 0.5mM IPTG and cultured at 25°C overnight. Samples from CG5 and EV cultures were collected 0 and 24 hrs after induction to check for protein expression by SDS-PAGE (10%). Proteins in the acrylamide gel were blotted onto a nitrocellulose membrane (Bio-Rad) and stained with Ponceau S (0.1%).
4.5.3 Preparation of inclusion bodies, CG5-6xHis- protein extraction, and in vitro folding
E. coli cultures (50ml) expressing CG5-6xHis were sedimented at 3500rpm at 4°C for 10 min. Cell pellets were resuspended in lysis buffer (20mM Tris-HCl pH 8.0, 2M urea with 1% Triton X-100), ruptured by sonication, and centrifuged at 20,000 x g at 4°C for 10 min. An aliquot of the supernatant (S) was saved for SDS-PAGE analysis and the rest was discarded. The insoluble pellet was again resuspended in 20mM Tris-HCl pH 8.0, 2M urea with 1% Triton X-100, sonicated, and centrifuged. The pellet was then washed three times with 20mM Tris-HCl pH 8.0, 2M urea to remove Triton X-100. Inclusion bodies were then solubilized in 50mM Tris-HCl pH9.2, 8.6 M urea. The suspension was placed on ice in an ultrasound bath for 5min and then centrifuged at 20,000 x g at 4°C for 15min. An aliquot of the solubilized inclusion bodies (P) was saved for SDS-PAGE analysis and the rest was used for folding.
The dissolved CG5-6xHis polypeptide was diluted with folding solution (Teilum et al., 1999) at 4°C giving a final protein concentration of 0.2 mg/ml and final concentrations of 20mM Tris-HCl, pH 9.2, 2.3M urea, 5mM CaCl2, 5μM Hemin (Sigma 51280), 0.25mM reduced glutathione, GSH (Sigma G6529), 0.45mM oxidized glutathione, GSSG (Sigma G6654). The folding process was carried in a closed container in the dark at 4°C overnight.
4.6 In vitro extensin crosslinking assays
4.6.1 Isolation of extensin substrates
P1 extensin substrates were purified from tomato cell suspension cultures, as described earlier (Smith et al., 1984; Schnabelrauch et al., 1996). Synthetic P3-extensin analogs (YK20 and FK9), produced in transgenic tobacco BY2 cells as C-terminal EGFP fusion proteins, were purified as described by Held et al. (2004).
4.6.2 Crosslinking reactions and gel filtration analysis
In vitro crosslinking assays were performed essentially as described earlier (Everdeen et al., 1988; Schnabelrauch et al., 1996; Held et al., 2004). Briefly, extensin stock solutions (10mg/ml) were prepared. Working solutions of extensin substrates were prepared by combining 60 μL of each stock solution with 40 μL of 0.1 M citrate buffer (pH 6.0). Crosslinking reactions were prepared by combining 10 μL of extensin working solution (60 μg), 5 μL peroxidase (0.2–0.4ng), and 5 μL of 0.24 mM H2O2 (60 μM) in a final reaction volume of 20 μL. Peroxidases (tomato pI4.6 EP and CG5-6xHis) were prepared as described above and the horseradish peroxidase was purchased from Sigma (P8125). Enzyme quantities used for these assays were reduced (0.2–0.4ng) compared to Schnabelrauch et al. (1996) and Held et al. (2004) (4–8ng), due to dilution of the protein in folding solution (described above). All peroxidases were quantified by ABTS assay, as described earlier (Everdeen et al., 1988) to ensure equal activity loadings. Timing of the reactions was started upon the addition of H2O2 and the reactions were terminated by the addition of 50μl of 0.05% sodium azide in 200mM sodium phosphate buffer pH 7. Reaction progress and initial crosslinking rates were determined from changes in monomer peak areas after gel-filtration chromatography as described earlier (Everdeen et al., 1988; Held et al., 2004; Schnabelrauch et al., 1996). The column flow rate was adjusted to 0.3 mL/min and protein peaks were monitored at 220 nm.
Supplementary Material
Tomato culture media was separated by DEAE anion-exchange chromatography. Peroxidase activity was monitored by ABTS assay (OD410nm). Fractionation yielded two major peroxidase containing peaks, an acidic (acidic Pox) and a basic (basic Pox) fraction. Previous work has shown that the acidic fraction contains the highest specific activity for crosslinking extensin (Schnabelrauch et al. 1996). The acidic fraction was then further fractionated by IEF.
Acidic peroxidases were further fractionated by IEF using a pH3-5 gradient. Twenty fractions were collected and assayed for total protein and for peroxidase activity via ABTS assay. Peroxidase active fractions are shown and are expressed in terms of their specific activity (ng of peroxidase/mg total protein). The pH4.6 fraction (arrow) was selected for proteomics analysis.
Tryptic digestion of the pI4.6 fraction released peptides, which were sequenced by LC-MS/MS analysis. Peptides sequences matching (A) CG5, (B) CG8, or (C) CG3 are highlighted in yellow. Analyses were performed in duplicate and shown here is the run with the highest percent coverage for each CG protein. Note that peptides derived from CG8 and CG3 were only detected in only one of the two biological replicates, while peptides for CG5 were detected in both trials. Peptides matching CG5 were the most abundant in both trials and had a percent coverage of 51%.
Crosslinking assays (2min) were prepared using P1 and P3-type (YK20 and FK9) extensin analogs to test the substrate specificity of CG5-6xHis. Both CG5-6xHis and the tomato pI4.6 EP were able to form crosslinked P1 (P1XL) and crosslinked YK20 (YK20XL), while HRP did not. FK9 was not crosslinked by any enzyme.
Peroxidase candidates were identified by screening the protein annotations of the tomato genome database (iTAG v2.3; solgenomics.net) by keyword search for “peroxidase”. Candidate genes are listed by their locus tags. Signal sequences were predicted for each peroxidase candidate using the SignalP3.0 program (cbs.dtu.dk). Predicted signal sequences are underlined. The pI of each mature protein (e.g. lacking the signal sequence) was collected for each candidate from the Gramene database. Predicted pIs were subtracted from 4.6 and the absolute values are listed as, delta pI. Candidates were sorted by their delta pI and those having a pI ± 0.3 from 4.6 were selected as CGs (1–8).
Proteomics/bioinformatics identified a tomato extensin peroxidase candidate (CG5)
CG5 was cloned, expressed in E. coli, and folded in vitro.
Recombinant CG5 showed both peroxidase and extensin crosslinking activity
First report of the identification of an anionic extensin peroxidase in tomato
Acknowledgments
We’d like to thank Doug Witten and Curtis Wilkerson (MSU-RTSF) for assistance with proteomics and analysis and Derek TA Lamport (Sussex), Allan Showalter (Ohio University), and Ahmed Faik (Ohio University) for meaningful discussions of our manuscript. This manuscript is dedicated to the memory of GP Bolwell, long-time colleague and friend. Paul’s contributions to the cell wall field were unique, insightful and lasting. This work was supported by a grant from the National Science Foundation (IOS-955569). This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-014575-01 from the National Center for Research Resources, National Institutes of Health.
Footnotes
Dedication: This paper forms part of a special issue of Phytochemistry dedicated to the memory and legacy of Professor (Godfrey) Paul Bolwell, MA DSc (Oxon). (1946–2012), internationally recognised plant biochemist and Regional Editor of Phytochemistry (2004–2012). He is much missed by his friends.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Wen Dong, Email: wd113009@ohio.edu.
Marcia Kieliszewski, Email: kielisze@ohio.edu.
References
- Abeles FB, Dunn LJ, Morgens P, Callahan A, Dinterman RE, Schmidt J. Induction of 33-kD and 60-kD peroxidases during ethylene-induced senescence of cucumber cotyledons. Plant Physiol. 1988;87:609–615. doi: 10.1104/pp.87.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya I, Botella MA, de la Calle M, Medina MI, Heredia A, Bressan RA, Hasegawa PM, Quesada MA, Valpuesta V. Improved germination under osmotic stress of tobacco plants overexpressing a cell wall peroxidase. FEBS Lett. 1999;457:80–84. doi: 10.1016/s0014-5793(99)01011-x. [DOI] [PubMed] [Google Scholar]
- Bolwell GP. Role of active oxygen species and NO in plant defence responses. Curr Opin Plant Biol. 1999;2:287–294. doi: 10.1016/S1369-5266(99)80051-X. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence–a broad perspective. Physiol Mol Plant Path. 1997;51:347–366. [Google Scholar]
- Bradley DJ, Kjellbom P, Lamb CJ. Elicitor-and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- Brady J, Sadler I, Fry S. Di-isodityrosine, a novel tetrametric derivative of tyrosine in plant cell wall proteins: a new potential cross-link. Biochem J. 1996;315:323–327. doi: 10.1042/bj3150323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JD, Fry SC. Formation of di-isodityrosine and loss of isodityrosine in the cell walls of tomato cell-suspension cultures treated with fungal elicitors or H2O2. Plant Physiol. 1997;115:87–92. doi: 10.1104/pp.115.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady JD, Sadler IH, Fry SC. Pulcherosine, an oxidatively coupled trimer of tyrosine in plant cell walls: its role in cross-link formation. Phytochem. 1998;47:349–353. doi: 10.1016/s0031-9422(97)00592-x. [DOI] [PubMed] [Google Scholar]
- Brownleader MD, Ahmed N, Trevan M, Chaplin MF, Dey PM. Purification and partial characterization of tomato extensin peroxidase. Plant Physiol. 1995;109:1115–1123. doi: 10.1104/pp.109.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brysk MM, Chrispeels MJ. Isolation and partial characterization of a hydroxyproline-rich cell wall glycoprotein and its cytoplasmic precursor. Biochim Biophys Acta-Prot Struct. 1972;257:421–432. doi: 10.1016/0005-2795(72)90295-4. [DOI] [PubMed] [Google Scholar]
- Cannon MC, Terneus K, Hall Q, Tan L, Wang Y, Wegenhart BL, Chen L, Lamport DTA, Chen Y, Kieliszewski MJ. Self-assembly of the plant cell wall requires an extensin scaffold. Proc Natl Acad Sci USA. 2008;105:2226–2231. doi: 10.1073/pnas.0711980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Varner JE. An extracellular matrix protein in plants: characterization of a genomic clone for carrot extensin. EMBO J. 1985;4:2145–2151. doi: 10.1002/j.1460-2075.1985.tb03908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels MJ, Raikhel NV. Short peptide domains target proteins to plant vacuoles. Cell. 1992;68:613–616. doi: 10.1016/0092-8674(92)90134-x. [DOI] [PubMed] [Google Scholar]
- Epstein L, Lamport DT. An intramolecular linkage involving isodityrosine in extensin. Phytochem. 1984;23:1241–1246. [Google Scholar]
- Espelie KE, Franceschi VR, Kolattukudy PE. Immunocytochemical localization and time course of appearance of an anionic peroxidase associated with suberization in wound-healing potato tuber tissue. Plant Physiol. 1986;81:487–492. doi: 10.1104/pp.81.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everdeen DS, Kiefer S, Willard JJ, Muldoon EP, Dey PM, Li XB, Lamport DT. Enzymic cross-linkage of monomeric extensin precursors in vitro. Plant Physiol. 1988;87:616–621. doi: 10.1104/pp.87.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueauf JB, Dolata M, Leykam JF, Lloyd EA, Gonzales M, VandenBosch K, Kieliszewski MJ. Peptides isolated from cell walls of Medicago truncatula nodules and uninfected root. Phytochem. 2000;55:429–438. doi: 10.1016/s0031-9422(00)00336-8. [DOI] [PubMed] [Google Scholar]
- Fry SC. Isodityrosine, a new cross-linking amino acid from plant cell-wall glycoprotein. Biochem J. 1982;204:449–455. doi: 10.1042/bj2040449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry SC. Cross-linking of matrix polymers in the growing cell walls of angiosperms. Ann Rev Plant Physiol. 1986;37:165–186. [Google Scholar]
- Hall Q, Cannon MC. The cell wall hydroxyproline-rich glycoprotein RSH is essential for normal embryo development in Arabidopsis. Plant Cell. 2002;14:1161–1172. doi: 10.1105/tpc.010477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckman JW, Terhune BT, Lamport DT. Characterization of native and modified extensin monomers and oligomers by electron microscopy and gel filtration. Plant Physiol. 1988;86:848–856. doi: 10.1104/pp.86.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Tan L, Kamyab A, Hare M, Shpak E, Kieliszewski MJ. Di-isodityrosine is the intermolecular cross-link of isodityrosine-rich extensin analogs cross-linked in vitro. J Biol Chem. 2004;279:55474–55482. doi: 10.1074/jbc.M408396200. [DOI] [PubMed] [Google Scholar]
- Henriksen A, Mirza O, Indiani C, Teilum K, Smulevich G, Welinder KG, Gajhede M. Structure of soybean seed coat peroxidase: A plant peroxidase with unusual stability and haem-apoprotein interactions. Prot Sci. 2001;10:108–115. doi: 10.1110/ps.37301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PA, Galinha CI, Pereira CS, Fortunato A, Soares NC, Amâncio SB, Ricardo CPP. Rapid deposition of extensin during the elicitation of grapevine callus cultures is specifically catalyzed by a 40-kilodalton peroxidase. Plant Physiol. 2001;127:1065–1076. [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ, Lamport DT. Extensin: repetitive motifs, functional sites, post-translational codes, and phylogeny. Plant J. 1994;5:157–172. doi: 10.1046/j.1365-313x.1994.05020157.x. [DOI] [PubMed] [Google Scholar]
- Lagrimini LM, Joly RJ, Dunlap JR, Liu TT. The consequence of peroxidase overexpression in transgenic plants on root growth and development. Plant Mol Biol. 1997;33:887–95. doi: 10.1023/a:1005756713493. [DOI] [PubMed] [Google Scholar]
- Lamport DT. Isolation and partial characterization of hydroxyproline-rich glycopeptides obtained by enzymic degradation of primary cell walls. Biochem. 1969;8:1155–1163. doi: 10.1021/bi00831a049. [DOI] [PubMed] [Google Scholar]
- Lamport DT. Structure, biosynthesis and significance of cell wall glycoproteins. In: Loewus FA, et al., editors. The Structure, Biosynthesis, and Degradation of Wood. Plenum Publishing Corp; New York: 1977. pp. 79–115. [Google Scholar]
- Lamport DTA. Structure and function of plant glycoproteins. In: Stumpf PK, Conn E, editors. The Biochemistry of Plants. Vol. 3. Academic Press; New York: 1980. pp. 501–536. [Google Scholar]
- Lamport DTA, Epstein L. A new model for the primary cell wall: a concatenated extensin-cellulose network. Proc Ann Plant Biochem & Physiol Symp Columbia Missouri. 1983;2:73–83. [Google Scholar]
- Mort AJ, Lamport DT. Anhydrous hydrogen fluoride deglycosylates glycoproteins. Anal Biochem. 1977;82:289–309. doi: 10.1016/0003-2697(77)90165-8. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Matsuoka K. Protein targeting to the vacuole in plant cells. Plant physiology. 1993;101:1–5. doi: 10.1104/pp.101.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Daudi A, Finch P, Butt VS, Whitelegge JP, Souda P, Ausubel FM, Bolwell GP. A peroxidase-dependent apoplastic oxidative burst in cultured Arabidopsis cells functions in MAMP-elicited defense. Plant Physiol. 2012;158:2013–2027. doi: 10.1104/pp.111.190140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passardi F, Longet D, Penel C, Dunand C. The class III peroxidase multigenic family in rice and its evolution in land plants. Phytochem. 2004;65:1879–1893. doi: 10.1016/j.phytochem.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Pereira CS, Ribeiro JM, Vatulescu AD, Findlay K, MacDougall AJ, Jackson PAP. Extensin network formation in Vitis vinifera callus cells is an essential and causal event in rapid and H2O2-induced reduction in primary cell wall hydration. BMC Plant Biol. 2011;11:106. doi: 10.1186/1471-2229-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Price NJ, Pinheiro C, Soares CM, Ashford DA, Ricardo CP, Jackson PA. A biochemical and molecular characterization of LEP1, an extensin peroxidase from lupin. J Biol Chem. 2003;278:41389–41399. doi: 10.1074/jbc.M304519200. [DOI] [PubMed] [Google Scholar]
- Saha P, Ray T, Tang Y, Dutta I, Evangelous NR, Kieliszewski MJ, Chen Y, Cannon MC. Self-rescue of an EXTENSIN mutant reveals alternative gene expression programs and candidate proteins for new cell wall assembly in Arabidopsis. Plant J. 2013;75:104–116. doi: 10.1111/tpj.12204. [DOI] [PubMed] [Google Scholar]
- Schnabelrauch LS, Kieliszewski M, Upham BL, Alizedeh H, Lamport D. Isolation of pl 4.6 extensin peroxidase from tomato cell suspension cultures and identification of Val—Tyr—Lys as putative intermolecular cross-link site. Plant J. 1996;9:477–489. doi: 10.1046/j.1365-313x.1996.09040477.x. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Muldoon EP, Lamport DT. Isolation of extensin precursors by direct elution of intact tomato cell suspension cultures. Phytochem. 1984;23:1233–1239. [Google Scholar]
- Smith JJ, Muldoon EP, Willard JJ, Lamport DT. Tomato extensin precursors P1 and P2 are highly periodic structures. Phytochem. 1986;25:1021–1030. [Google Scholar]
- Teilum K, Ostergaard L, Welinder KG. Disulfide bond formation and folding of plant peroxidases expressed as inclusion body protein in Escherichia coli Thioredoxin Reductase Negative Strains. Protein Express Purif. 1999;15:77–82. doi: 10.1006/prep.1998.0985. [DOI] [PubMed] [Google Scholar]
- Tognolli M, Penel C, Greppin H, Simon P. Analysis and expression of the class III peroxidase large gene family in Arabidopsis thaliana. Gene. 2002;288:129–138. doi: 10.1016/s0378-1119(02)00465-1. [DOI] [PubMed] [Google Scholar]
- van Holst GJ, Varner JE. Reinforced polyproline II conformation in a hydroxyproline-rich cell wall glycoprotein from carrot root. Plant Physiol. 1984;74:247–251. doi: 10.1104/pp.74.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P. Oxidative burst: an early plant response to pathogen infection. Biochem J. 1997;322:681–692. doi: 10.1042/bj3220681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtaszek P, Trethowan J, Bolwell GP. Reconstitution in vitro of the components and conditions required for the oxidative cross-linking of extracellular proteins in French bean (Phaseolus vulgaris L.) FEBS Lett. 1997;405:95–98. doi: 10.1016/s0014-5793(97)00166-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tomato culture media was separated by DEAE anion-exchange chromatography. Peroxidase activity was monitored by ABTS assay (OD410nm). Fractionation yielded two major peroxidase containing peaks, an acidic (acidic Pox) and a basic (basic Pox) fraction. Previous work has shown that the acidic fraction contains the highest specific activity for crosslinking extensin (Schnabelrauch et al. 1996). The acidic fraction was then further fractionated by IEF.
Acidic peroxidases were further fractionated by IEF using a pH3-5 gradient. Twenty fractions were collected and assayed for total protein and for peroxidase activity via ABTS assay. Peroxidase active fractions are shown and are expressed in terms of their specific activity (ng of peroxidase/mg total protein). The pH4.6 fraction (arrow) was selected for proteomics analysis.
Tryptic digestion of the pI4.6 fraction released peptides, which were sequenced by LC-MS/MS analysis. Peptides sequences matching (A) CG5, (B) CG8, or (C) CG3 are highlighted in yellow. Analyses were performed in duplicate and shown here is the run with the highest percent coverage for each CG protein. Note that peptides derived from CG8 and CG3 were only detected in only one of the two biological replicates, while peptides for CG5 were detected in both trials. Peptides matching CG5 were the most abundant in both trials and had a percent coverage of 51%.
Crosslinking assays (2min) were prepared using P1 and P3-type (YK20 and FK9) extensin analogs to test the substrate specificity of CG5-6xHis. Both CG5-6xHis and the tomato pI4.6 EP were able to form crosslinked P1 (P1XL) and crosslinked YK20 (YK20XL), while HRP did not. FK9 was not crosslinked by any enzyme.
Peroxidase candidates were identified by screening the protein annotations of the tomato genome database (iTAG v2.3; solgenomics.net) by keyword search for “peroxidase”. Candidate genes are listed by their locus tags. Signal sequences were predicted for each peroxidase candidate using the SignalP3.0 program (cbs.dtu.dk). Predicted signal sequences are underlined. The pI of each mature protein (e.g. lacking the signal sequence) was collected for each candidate from the Gramene database. Predicted pIs were subtracted from 4.6 and the absolute values are listed as, delta pI. Candidates were sorted by their delta pI and those having a pI ± 0.3 from 4.6 were selected as CGs (1–8).