Abstract
Australian antigen, the envelope protein of hepatitis B virus (HBV), was discovered in 1967 as a prevalent serum antigen in hepatitis B patients. Early electron microscopy (EM) studies showed that this antigen was present in 22-nm particles in patient sera, which were believed to be incomplete virus. Complete virus, much less abundant than the 22-nm particles, was finally visualized in 1970. HBV was soon found to infect chimpanzees, gorillas, orangutans, gibbon apes, and, more recently, tree shrews (Tupaia belangeri) and cynomolgus macaques (Macaca fascicularis). This restricted host range placed limits on the kinds of studies that might be performed to better understand the biology and molecular biology of HBV and to develop antiviral therapies to treat chronic infections. About 10 years after the discovery of HBV, this problem was bypassed with the discovery of viruses related to HBV in woodchucks, ground squirrels, and ducks. Although unlikely animal models, their use revealed the key steps in hepadnavirus replication and in the host response to infection, including the fact that the viral nuclear episome is the ultimate target for immune clearance of transient infections and antiviral therapy of chronic infections. Studies with these and other animal models have also suggested interesting clues into the link between chronic HBV infection and hepatocellular carcinoma.
Hepatitis B virus (HBV) has a narrow host range. However, the study of viruses related to HBV in woodchucks, ground squirrels, and ducks has revealed key steps in their replication and in the host response to infection.
Evidence for the existence of a hepatitis B virus (HBV) was obtained in 1967 following the realization that a newly identified human serum antigen, Australia antigen, was produced by a transmissible agent that turned out to be the cause of hepatitis B (Blumberg et al. 1967, 1968). Australia antigen was found in patient sera predominantly in the form of 22-nm particles, which were believed to be incomplete virus (Bayer et al. 1968; Millman et al. 1970). Compared with the 22-nm particles, the larger virion is much less abundant and was finally recognized by electron microscopy (EM) studies in 1970 (Dane et al. 1970). Australia antigen was later found to contain a B-cell epitope present on a region shared by all three of the HBV envelope proteins.
HBV infects and replicates primarily if not exclusively in hepatocytes. A peculiarity of HBV replication is the secretion into the blood of a vast excess of particles made up of the three viral envelope proteins, S, M, and L, but particularly the smallest, S (Heermann et al. 1984). These surface antigen particles (HBsAg) are typically >100-fold more abundant than virus particles (Dane et al. 1970). HBsAg, processed from serum, was the first vaccine to prevent HBV infection. Assays based on HBsAg detection also led to effective screening of blood banks to prevent posttransfusion hepatitis, of which HBV was a major cause (Blumberg 1977).
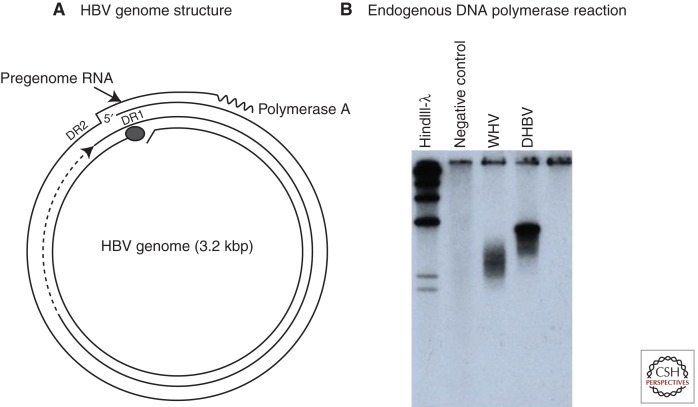
The molecular biology of HBV began with the discovery by Robinson and colleagues that HBV, purified from human serum, had a double-stranded, circular DNA genome and an endogenous DNA polymerase (Kaplan et al. 1973; Robinson et al. 1974; Robinson and Greenman 1974). Summers and colleagues then showed that the genome was partially single-stranded and was held in its circular conformation by a short cohesive overlap between the 5′ ends of the two DNA strands (Summers et al. 1975). One strand of the genome, later determined to be of minus polarity, is always complete; the other, of plus polarity, is incomplete. The endogenous DNA polymerase can partially fill in the gap in the plus strand in an in vitro reaction (Fig. 1A).
Figure 1.
The genome and endogenous polymerase reaction of hepatitis B virus (HBV). (A) The HBV genome is a partially double-stranded DNA, held in a relaxed circular conformation by a short cohesive overlap between the 5′ ends of the two DNA strands (Summers et al. 1975). One strand, later found to be the plus strand, is always incomplete in virus particles, with a gap that may encompass up to 50% of the genome length. The minus strand is always complete. The large circle at the 5′ end of the minus strand represents a covalently attached protein (Gerlich and Robinson 1980) that was later shown to be the viral DNA polymerase/reverse transcriptase (Bartenschlager and Schaller 1988). Pregenomic RNA, the template for viral DNA synthesis, is shown for comparison. DR1 and DR2 are 11-nucleotide direct repeats (12-nucleotide for duck hepatitis B virus [DHBV]) on the pregenome that play essential roles in priming of viral DNA synthesis (see text). (B) HBV and other hepadnaviruses contain the viral DNA polymerase, which can fill in the single-stranded gap in vitro (Summers et al. 1975). The fill-in reaction can be performed by pelleting virus from serum, adding nonionic detergent and radiolabeled deoxynucleotides (dNTPs) to the pellet, and incubating at 37°C. The radiolabeled DNA can be detected by agarose gel electrophoresis and autoradiography, as shown here for DHBV and woodchuck hepatitis virus (WHV). This assay was instrumental in the discovery of WHV, DHBV, and ground squirrel hepatitis virus (GSHV) (Summers et al. 1978; Marion et al. 1980; Mason et al. 1980).
The structure of the genome (Fig. 1A) led to early speculation that HBV might replicate by reverse transcription. A relaxed circular DNA of similar conformation to the HBV genome was believed to be an intermediate in reverse transcription of retrovirus RNA to form linear proviral DNA (Gilboa et al. 1979). However, before this and other ideas about HBV could be studied, it was necessary to develop models of HBV replication that were accessible to routine laboratory analysis.
In the 1970s, cloning of viruses into plasmids was still in its infancy, as was routine transfection of DNA into cells. Studies in the early 1980s showed that cloned viral DNA was infectious when injected into the liver (Will et al. 1982; Seeger et al. 1984; Sprengel et al. 1984). However, a serious barrier to further transfection-based studies was the lack of cell lines that supported hepadnavirus replication from transfected DNA. It was not until 1986 that the HepG2 line of human liver tumor cells (Aden et al. 1979) was found to support HBV replication from cloned viral DNA (Sureau et al. 1986; Sells et al. 1987). In the following 3 years, two other human liver tumor lines, Huh6 and Huh7, and a rat hepatoma cell line were also shown to fulfill this need (Tsurimoto et al. 1987; Yaginuma et al. 1987; Shih et al. 1989). As discussed below, by this time many key steps in hepadnavirus replication had been revealed through the use of animal models and primary hepatocyte cultures. However, these and other cell lines were invaluable for characterizing these steps in greater detail.
DISCOVERY OF ANIMAL MODELS OF HBV INFECTION
To search for animal models of HBV replication, Summers took advantage of the endogenous polymerase reaction of HBV, which repairs the single-stranded gap in plus-strand DNA (Summers et al. 1975). This reaction can be used to screen for new hepatitis B–like viruses, without any knowledge of antigenicity or, in more recent times, DNA sequence. For example, serum samples are centrifuged at high speed. The pellet is then mixed with nonionic detergent to disrupt the virus membrane. Radiolabeled deoxynucleotides are added, and the mixture is incubated at 37°C. If an HBV-like virus is present, its DNA will be labeled by the endogenous DNA polymerase reaction. The DNA can then be released from virus proteins by digestion with a proteinase and detected by agarose gel electrophoresis followed by autoradiography (Fig. 1B) (Summers et al. 1978). Using this approach, new hepadnaviruses were discovered in eastern woodchucks (Summers et al. 1978), domestic ducks (Mason et al. 1980), and Beechey ground squirrels (Marion et al. 1980). These viruses were named woodchuck hepatitis virus (WHV), duck hepatitis B virus (DHBV), and ground squirrel hepatitis virus (GSHV), respectively. Based on similarities and differences in genome organization, nucleotide sequence, and host range specificity, the mammalian isolates were assigned to the genus Orthohepadnavirus, whereas DHBV was designated the prototype Avihepadnavirus. HBV, WHV, and GSHV are considered to be distinct species of Orthohepadnavirus (Fauquet et al. 2005).
In more recent years, new virus species have been added to each genus. These include the Orthohepadnavirus woolly monkey HBV (WMHBV) and the Avihepadnavirus heron hepatitis B virus (HHBV) (Sprengel et al. 1988; Lanford et al. 1998). It is possible that a newly discovered Avihepadnavirus in parakeets and new orthohepadnaviruses found in bats may be designated novel viral species (Drexler et al. 2013; Piasecki et al. 2013).
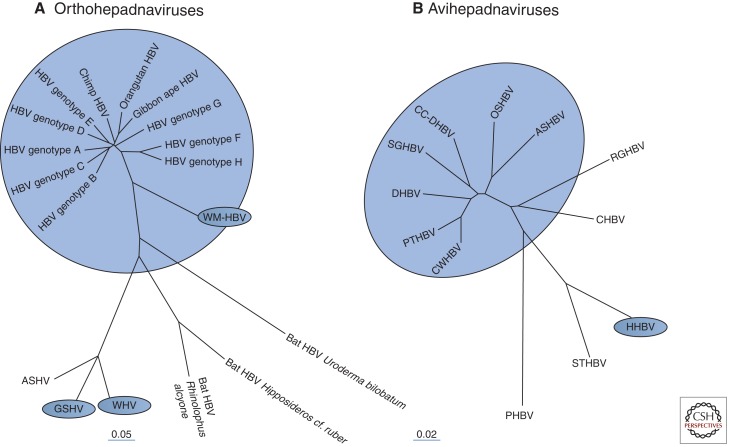
A sampling of ortho- and avihepadnaviruses and their phylogenic relationships are presented in Figure 2. It should be noted that species assignments are currently based on DNA sequence analysis combined with differences in host range. For instance, although WHV and GSHV appear related based on sequence analysis (Fig. 2A), WHV is unable to infect Beechey ground squirrels (although GSHV infects woodchucks) (Seeger et al. 1987, 1991).
Figure 2.
Hepadnavirus phylogeny. (A) The orthohepadnaviruses infect mammals and share similarities in genome sequence and open reading frame location, size, and function. Four species have been defined, with hepatitis B virus (HBV), woolly monkey HBV (WMHBV), ground squirrel hepatitis virus (GSHV), and woodchuck hepatitis virus (WHV) serving as the prototype for each species. As shown, there are at least eight different genotypes of human HBV and additional genotypes in chimpanzees, orangutans, and gibbon apes. WMHBV, WHV, and GSHV are designated as distinct species based on differences in sequence and a unique host range. The arctic squirrel hepatitis virus (ASHV) is most similar to GSHV, but it is not yet known whether it has a host range distinct from that of GSHV or WHV. (B) The Avihepadnavirus isolates to date come mostly from ducks and geese. Viruses that are not from domestic ducks but considered to be of the same species as duck HBV (DHBV) and Chi-tung County (CC)-DHBV isolates from China include isolates from the snow goose (SGHBV), Orinoco sheldgoose (OSHBV), ashy-headed sheldgoose (ASHBV), Puna teal (PTHBV), and Chiloé wigeon (CWHBV) (Guo et al. 2005). DHBV is also found in wild mallards (Cova et al. 1986), from which most domesticated ducks, except Muscovy, were derived. Of the remaining avihepadnaviruses shown here, only heron HBV (HHBV) (Sprengel et al. 1988) is designated a distinct species (Fauquet et al. 2005). Virus isolates from the Ross goose (RGHBV), crane (CHBV) (Prassolov et al. 2003), stork (STHBV) (Pult et al. 2001), and parakeet (PHBV) (Piasecki et al. 2013) remain unassigned.
During the 1970s, the idea that chronic hepatitis B caused primary liver cancer (hepatocellular carcinoma [HCC]) was firmly established by epidemiologic studies performed in areas of Africa and Asia with high levels of chronic HBV infection (Blumberg et al. 1975; Prince et al. 1975; Beasley et al. 1981; Thomas et al. 2015). The search for HBV-like viruses in woodchucks and ducks was, in fact, performed because of reports that woodchucks kept at Penrose Laboratory of the Philadelphia Zoo and domestic ducks resident in China had a high incidence of HCC. A connection between infection and HCC was established in the woodchuck but not in the duck. A link between infection and HCC was subsequently found in the Beechey ground squirrel (Marion et al. 1986). To some extent, the idea that HCC was frequent in ducks may have come about because of the high incidence of secondary amyloid disease of the liver in these birds. This can lead to the outgrowth of regenerative liver nodules, which might have been mistaken, on visual inspection, for tumors (Rigdon 1961; Guo et al. 1996); HCC caused by environmental carcinogens may also have been a factor.
In 1979, the National Institutes of Health (NIH) established a woodchuck colony at Cornell University that was used to explore the connection between WHV infection and HCC and, ultimately, as a model to evaluate antiviral therapies (Tennant and Gerin 2001). The ground squirrel model was also used to study infection and HCC. During the 1980s, the duck model was more widely used than the woodchuck and ground squirrel models to study hepadnavirus replication, probably because of the ready availability of infected ducklings and greater ease of handling. DHBV is common in most domestic duck flocks, often as a chronic infection in 10% or more of the birds. DHBV is generally transmitted in ovo, establishing a largely immunotolerant infection, and is not known to be associated with liver disease in ducks, including secondary amyloidosis (O’Connell et al. 1983; Mason et al. 1987; Uchida et al. 1988).
Like the woodchuck model, the duck model has also been used to evaluate antiviral therapies, primarily but not limited to nucleoside analogs. Therapy with nucleoside analogs to inhibit viral DNA synthesis initially arose as an outgrowth of programs that had already identified inhibitors of human immunodeficiency virus (HIV). Finally, the duck model was also used to study the genetics of DHBV replication, which was made easier by the finding of a chicken cell line, Leghorn male hepatoma (LMH) (Kawaguchi et al. 1987), that supported at least 10-fold-higher levels of virus replication from cloned viral DNA than any of the other transfection systems for HBV or DHBV (Condreay et al. 1990).
THE MOLECULAR BIOLOGY OF HEPADNAVIRUSES AS DEDUCED FROM STUDIES OF SERUM AND LIVER SAMPLES OF DHBV-INFECTED DUCKS AND GSHV-INFECTED GROUND SQUIRRELS
Reverse Transcription
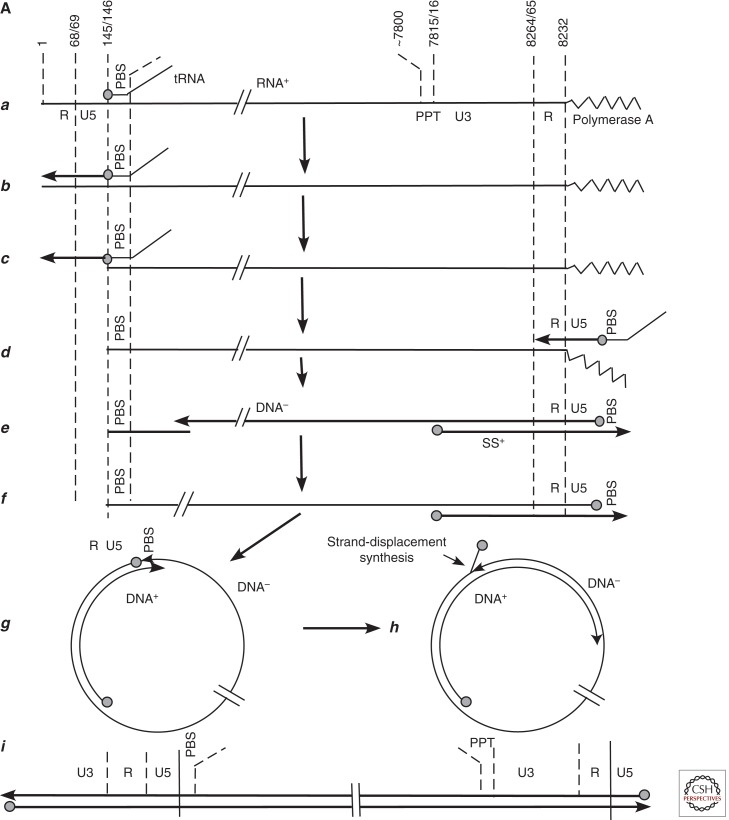
The idea that HBV might be similar to retroviruses came about in the 1970s through discussions between Jesse Summers and John Taylor at the Fox Chase Cancer Center, following publication of the genome structure of HBV (Summers et al. 1975). Taylor’s laboratory was studying retroviral DNA synthesis (Taylor and Illmensee 1975; Sabran et al. 1979). By the late 1970s, it was clear that retrovirus reverse transcription initiated near the 5′ end of the virion RNA and that second-strand synthesis began near the 5′ end of the reverse transcript, as summarized by Gilboa et al. (1979). Thus, it seemed likely that generation of the linear integrated proviral DNA involved formation of circular DNA intermediates. A later version of the model in Gilboa et al. is shown in Figure 3A to highlight the major point, that a proposed intermediate in Moloney murine leukemia virus (MoMLV) provirus formation is similar in structure to the HBV genome.
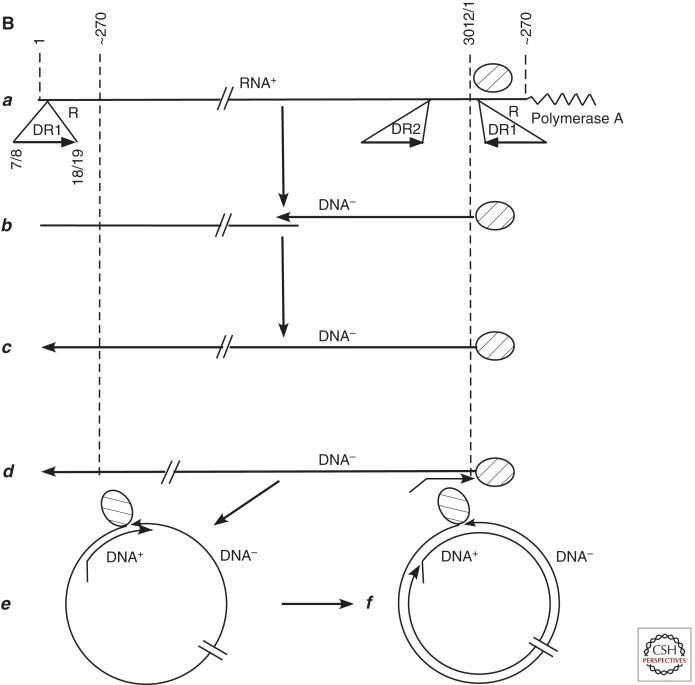
Figure 3.
Comparison of retrovirus and hepadnavirus DNA synthesis pathways. (A) Moloney murine leukemia virus (MoMLV) DNA synthesis. Reverse transcription of MoMLV RNA begins when a cell is infected. The primer for reverse transcription is the 3′ hydroxyl (OH) of tRNAPro, which is annealed near to the 5′ end of the viral RNA genome via hybridization to an 18-nucleotide primer binding site (PBS) (a). Synthesis extends to the 5′ end of the viral RNA. The reverse-transcribed RNA sequences are degraded by the viral RNase H (b,c). At this point, the DNA product can hybridize to the 3′ end of viral RNA through the 68-nucleotide R domain, found at both ends of viral RNA (d). This facilitates reverse transcription of the remainder of the RNA template. Plus-strand synthesis initiates near the 5′ end of the minus strand from an RNA oligonucleotide that is left behind during RNase H degradation of the reverse-transcribed viral RNA (e). (It should be noted that some retroviruses prime plus-strand synthesis from multiple sites, including the polypurine tract [PPT] region as well as upstream sites.) Plus-strand synthesis extends rightward to copy the 3′ 18 nucleotides of the tRNA, recreating the PBS as DNA. Once minus-strand synthesis has made a complementary copy of the PBS (f), the 3′ ends of the nascent plus and minus strands can hybridize to form a circle (g). Minus-strand synthesis extends to the 5′ end of the plus strand, presumably via strand displacement synthesis (h), and plus-strand synthesis extends to the 5′ end of the minus strand to create a DNA with a large terminal redundancy (LTR; U3-R-U5). This terminally redundant linear DNA is the substrate for integration into host DNA. Integrated DNA serves as the provirus template for new viral RNA synthesis. (B) duck hepatitis B virus (DHBV) DNA synthesis. In this early model (Lien et al. 1986), reverse transcription is primed from a tyrosine residue of a protein primer, later shown to map to the terminal protein domain of the viral DNA polymerase (a), and begins within the 3′ copy of DR1. Minus-strand synthesis extends to the 5′ end of the pregenome (b,c). Because it began in R and extended almost to the 5′ end of the RNA template, the nascent minus strand is terminally redundant by 7–8 nucleotides. The pregenome is degraded by RNase H, leaving an 18-nucleotide sequence including the 5′ CAP and first 18 nucleotides of the pregenome (c), including the 12-base repeat, DR1, which is present in the terminal redundancy of the pregenome. This oligoribonucleotide is then translocated from DR1 to DR2, which maps upstream of the 3′ terminal redundancy (d). The oligoribonucleotide hybridizes to DR2 on the minus-strand DNA because DR1 and DR2 are identical in sequence. Plus-strand synthesis initiates at the 3′ end of DR2 and extends to the end of the minus strand. Circularization to continue plus-strand synthesis is apparently facilitated by the 7- to 8-nucleotide terminal redundancy on the minus strand (e,f). Synthesis continues, to produce a partially double-stranded virus genome (with DHBV, most plus strands are nearly full-length, excluding DR2, to which the plus-strand primer remains bound). Approximately 10% of the time, the plus-strand primer is not translocated to DR2, and plus-strand synthesis begins at the upstream copy of DR1, resulting in a linear virus genome (see Fig. 4).
The possibility that a version of the retrovirus model might be correct for hepadnaviruses was supported, first, by the finding that DHBV-infected duck liver contained a large amount of single-stranded viral DNA of minus polarity (Fig. 3A) (Mason et al. 1982). This was also found in subsequent studies of HBV- and GSHV-infected liver (Monjardino et al. 1982; Weiser et al. 1983; Blum et al. 1984; Fowler et al. 1984). To determine whether a reverse transcription model of replication was correct, Summers isolated viral nucleocapsids (cores) from infected duck liver. He then showed that these nucleocapsids contained an endogenous DNA polymerase/reverse transcriptase activity that elongated nascent minus strands in a reaction that was resistant to actinomycin D. In contrast, elongation of plus strands (e.g., as in Fig. 1) was inhibited by actinomycin D. These observations fit nicely with earlier retrovirus studies showing that actinomycin D inhibited DNA-dependent but not RNA-dependent DNA synthesis (Gurgo et al. 1971) and, along with other experiments, indicated that hepadnaviruses replicated via reverse transcription (Summers and Mason 1982). Shortly thereafter, comparison of HBV and retrovirus nucleotide sequences revealed that HBV encodes the reverse transcriptase needed for its replication (Toh et al. 1983).
Many of the steps leading from the discovery of DHBV reverse transcription to a detailed model of viral DNA synthesis were also worked out with virus and liver from animal models.
Priming of Reverse Transcription
Early work addressed the priming of reverse transcription. Gerlich and Robinson (1980) had already shown that a protein was covalently attached to the 5′ end of the minus strand of the HBV genome. At about the same time, it was found that adenovirus DNA synthesis was protein primed (Challberg et al. 1982; Ikeda et al. 1982), suggesting, along with the data from Gerlich and Robinson (1980), that the same might be true for hepadnaviruses. This idea was supported by data showing that a protein was bound to the minus strand of DHBV genomic DNA and to nascent minus-strand DNA in infected duck liver (Molnar-Kimber et al. 1983). In particular, it was found that the smallest minus strands that could be radiolabeled in the endogenous polymerase reaction of virus nucleocapsids, purified from infected liver, were about 20–30 nucleotides and that a protein was covalently attached. Thus, it appeared likely that the protein was the primer of reverse transcription. A protein primer domain was later localized to the amino terminus of the viral DNA polymerase/reverse transcriptase (Bartenschlager and Schaller 1988). In contrast, retrovirus reverse transcription is initiated from a tRNA primer (Fig. 3A).
These early studies with DHBV also showed that the cohesive overlap of DHBV was flanked by 12-base direct repeats that had been previously identified in the DHBV DNA sequence (Mandart et al. 1984). Similar direct repeats are in fact present in all hepadnaviruses, as illustrated for HBV (Fig. 1A). These repeats, known as DR1 (flanking the 5′ end of the minus strand) and DR2 (flanking the 5′ end of the plus strand), have a major role in hepadnaviral DNA synthesis (Seeger et al. 1986).
Priming of Plus-Strand (Second-Strand) DNA Synthesis
It was assumed, at the time that hepadnavirus reverse transcription was discovered, that DHBV and other hepadnaviruses encoded an RNase H because full-length minus-strand DNA was not found as an RNA–DNA hybrid but appeared largely single-stranded. (Indeed, this was later confirmed by DNA sequence comparisons between viral DNA and sequences known to encode RNase H [Khudyakov and Makhov 1989].) This led, by analogy to the retrovirus model (Fig. 3A), to the idea that plus-strand synthesis is primed from an RNA that is created, during degradation of the RNA template, by viral RNase H (Smith et al. 1984) and then remains bound to the complementary minus-strand template for second-strand synthesis. For hepadnaviruses, this would correspond to an oligoribonucleotide just upstream of the 5′ end of plus-strand DNA (Fig. 1A). However, attempts to identify and map the primer revealed two unexpected results. First, although the plus-strand primer was found to be an oligoribonucleotide, as expected, it remained attached to plus-strand DNA even in virus. Second, although the plus-strand primer contained DR2 at its 3′ end, it had an additional 6 nucleotides that did not originate upstream of DR2. Rather, the additional 6 nucleotides mapped upstream of DR1. That is, the plus-strand primer actually came not from the RNA sequence flanking the 5′ end of the plus strand but from the region bracketing the origin of reverse transcription (Lien et al. 1986). Similar findings were reported for GSHV and WHV (Seeger et al. 1986; Will et al. 1987).
Interpretation of these results was possible largely because of the mapping of the ends of the DHBV pregenome, the RNA template for reverse transcription, by Buscher et al. (1985). As shown in Figure 1A, the pregenome was found to be terminally redundant. DR1 is in the terminal redundancy of the pregenome and thus appears twice, with the upstream copy 6 nucleotides downstream from the 5′ cap of the pregenome. The plus-strand primer originates not from DR2 but from the 5′-terminal 18 nucleotides of the pregenome, including the 5′ CAP, and extending through DR1. By inference, the primer was created by RNase H following completion of the minus strand and then translocated to DR2 to allow plus-strand priming. It was later found that the length of the primer is determined by substrate requirements of the RNase H, precluding cleavage any closer than the distance from the 5′ end of the pregenome to the 3′ end of DR1 (Loeb et al. 1991).
A Model of Reverse Transcription Derived from Studies of Virus and Infected Tissues
The first model of hepadnavirus reverse transcription, developed with DHBV and parallel studies with GSHV and HBV, is illustrated in Figure 3B. In summary, reverse transcription initiates from a protein primer, the polymerase, in the 3′ copy of DR1, and extends to the 5′ end of the pregenome. Following completion of the minus strand, a 5′-terminal oligonucleotide of the pregenome, including the CAP and all of DR1, is translocated to DR2, where it can anneal because DR1 and DR2 have the same sequence. Priming of plus-strand synthesis then occurs. Synthesis extends to the 5′ end of the minus strand and then jumps to the 3′ end. This jump is possible because of a terminal redundancy in the minus strand, created because 7–8 nucleotides are reverse transcribed from both the 3′ and 5′ terminal redundancy (R) of the pregenome. This jump creates a relaxed circular DNA for continuation of plus-strand elongation. With DHBV, most of the plus strand is filled in, except for DR2 (Lien et al. 1987). With the orthohepadnaviruses, most virus particles contain a large gap in the plus strand, as illustrated in Figure 1A. As presented in Hu and Seeger (2015), modification of this model came later, with the discovery that a translocation event is also involved in priming of minus-strand synthesis (Wang and Seeger 1992, 1993).
The Hepadnavirus Life Cycle
A second major finding came before the identification of cell lines that supported HBV and DHBV replication from transfected viral DNA. It was known that hepadnavirus DNA replication takes place in the cytoplasm (Burrell et al. 1982; Summers and Mason 1982) and that viral mRNAs were likely transcribed from a unit-length covalently closed circular DNA (cccDNA) (Burrell et al. 1982; Summers and Mason 1982). It was clear that the initial copy of cccDNA was formed from incoming viral DNA during initiation of infection but that cccDNA was later present in multiple copies per cell (Mason et al. 1983; Tagawa et al. 1986). It was not clear, however, whether this cccDNA copy number amplification was semiconservative or occurred through the reverse transcription pathway (Tuttleman et al. 1986b). To address this question, Tuttleman et al. prepared primary duck hepatocyte cultures infected with DHBV and density-labeled newly made cccDNA by addition to the culture medium of bromodeoxyuridine, a thymidine analog (Tuttleman et al. 1986a). They then separated the unlabeled and labeled cccDNAs by ultracentrifugation in CsCl density gradients. They reasoned that if cccDNA was made by semiconservative DNA synthesis, either the plus or minus strand of heavy–light DNA might be labeled. On the other hand, if synthesis was via reverse transcription, then only the plus strand of heavy–light DNA would be labeled. They found the latter result, indicating that cccDNA amplification occurred via the reverse transcription pathway. This, together with earlier results on infection and viral DNA synthesis, led to a simple and straightforward model of the virus life cycle (Fig. 4).
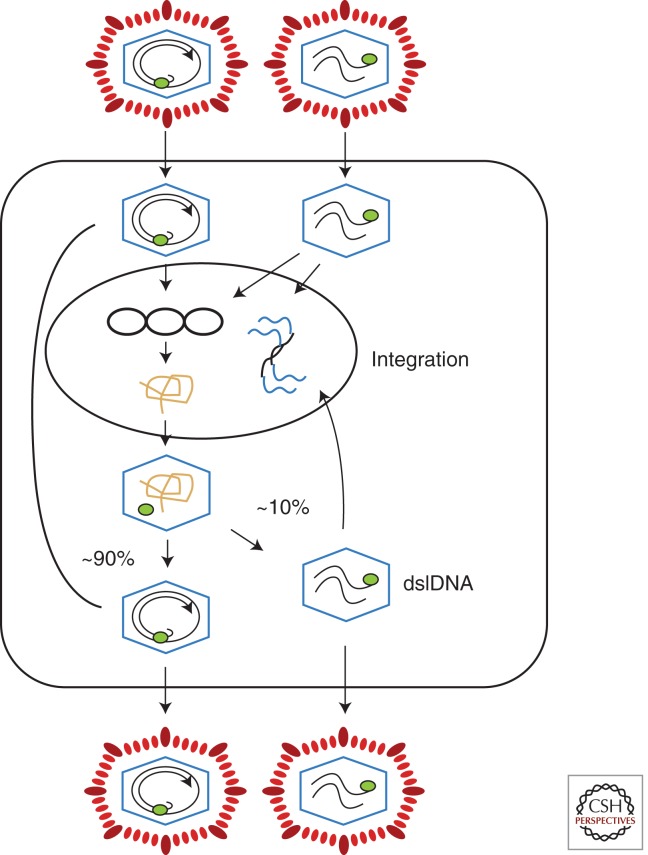
Figure 4.
Hepadnavirus infection. Virus with a relaxed circular (RC) genome is shown on the top right. Upon infection, the DNA is translocated to the nucleus, where the RC DNA genome is converted to covalently closed circular DNA (cccDNA), the template for viral RNA synthesis. When one of the largest viral RNAs, the pregenome, enters the cytoplasm, it can be packaged into viral nucleocapsids along with the viral reverse transcriptase and copied to produce new RC DNA. In the first few days of infection, newly made DNA is transported to the nucleus to amplify cccDNA copy number. As envelope proteins accumulate, this pathway is shut down and the nucleocapsids with RC DNA are enveloped and exported from the cell (Summers et al. 1990; Lenhoff and Summers 1994). Virus with linear DNA (Fig. 3) is also infectious and can form cccDNA via nonhomologous recombination, leading to defective cccDNAs. Linear genomes are also the substrate for integration of viral DNA into host DNA (Gong et al. 1995, 1999; Yang and Summers 1995, 1999). Integration is random on the host genome but appears to occur preferentially near the ends of linear viral DNAs. Some integrants also appear derived from RC DNA that has been linearized by displacement synthesis of plus and minus strands through the cohesive overlap, to create a linear DNA with a large terminal redundancy (Yang et al. 1996). dslDNA, double-stranded linear DNA.
In summary, relaxed circular virion DNA is converted to cccDNA during initiation of infection. Within the next few days, additional relaxed circular viral DNA, newly made in the cytoplasm, is transported to the nucleus to amplify cccDNA copy number. The final cccDNA copy number in vivo probably ranges from 1 to 50 in different hepatocytes, although there have been considerable differences in reports of the exact amounts, perhaps because of different standards used to quantify viral DNA. As later shown, cccDNA copy number amplification in duck hepatocytes is ultimately stopped after a few days by DHBV envelope proteins that have accumulated in the cytoplasm (Summers et al. 1990; Lenhoff and Summers 1994). Subsequently, nucleocapsids with relaxed circular DNA are enveloped and secreted from the infected cell. Shutdown of cccDNA amplification is important because overamplification is toxic to hepatocytes. It is not known whether envelope proteins of orthohepadnaviruses are also responsible for shutting down cccDNA amplification.
By the time the studies described above were completed, most work on hepadnavirus replication was shifting away from animal models to transfection-based studies using liver tumor cell lines (Tsurimoto et al. 1987; Yaginuma et al. 1987; Condreay et al. 1990). Further work with animal models focused primarily on understanding the host response to infection and development of antiviral therapies. A few studies also used animal models to study the molecular biology of infection.
Animal Models and the Role in Infection of Nonstructural Virus Proteins
As discussed in detail in Hu and Seeger (2015), HBV and other orthohepadnaviruses synthesize seven proteins. Core is the subunit of the viral nucleocapsid, Pol encodes the viral DNA polymerase/reverse transcriptase, and Env encodes the three HBV envelope proteins, L, M, and S. DHBV only encodes two envelope proteins, L and S. All of these proteins are found in virions. Two additional, nonvirion proteins are made by HBV, PreCore, and X. PreCore is made by all hepadnaviruses. X is made by all orthohepadnaviruses and by some, but not all, avihepadnaviruses.
PreCore is a variant of the Core protein with a signal peptide at its amino terminus that directs it into a secretory pathway (Takahashi et al. 1983; Ou et al. 1986; Standring et al. 1988). In human serum, it is defined for historic reasons as hepatitis B e antigen (HBeAg). It is widely believed, based on studies in heterologous systems, to suppress the host immune response to infection (Milich et al. 1993; Frelin et al. 2009), although this remains unclear. Whatever its function, it appears to be essential because both ortho- and avihepadnaviruses make this protein and because HBeAg-negative mutants, which often become predominant late in chronic infection, appear to be selected against during virus transmission. Furthermore, it has been shown that e antigen of DHBV and WHV is not required for experimental infection of ducks and woodchucks, respectively (Chang et al. 1987; Chen et al. 1992). However, experiments in animal models did not rule out the possibility that e antigen is needed to establish chronic infections.
Unlike PreCore, the other nonstructural protein of orthohepadnavirus, X, is not found in many avihepadnaviruses, and, even where found, it does not appear necessary for infection (Meier et al. 2003). In contrast, its role in Orthohepadnavirus replication in the liver appears essential, because X-negative WHV is unable to establish a high-titer infection (Chen et al. 1993; Zoulim and Seeger 1994; Zhang et al. 2001). However, despite nearly 30 years of research, it is still unclear whether the role of X in vivo is simply activation of viral mRNA synthesis, as first reported from cell culture studies (Twu and Schloemer 1987; Spandau and Lee 1988), or whether it is much more complex. X has been reported, from cell culture experiments, to activate a variety of host genes, to promote cell cycle progression, to induce apoptosis, to suppress innate immunity, to cause oncogenic transformation, etc. Which of these results have in vivo relevance is unclear. Modification of host gene transcription was not found during the early phase of transient infections of chimpanzees, a natural host of HBV, before the appearance of antiviral cytotoxic T lymphocytes (CTLs) in the liver (Wieland et al. 2004a). Infection by HBV, WHV, or GSHV does not transform hepatocytes. On the other hand, X appears to potentiate chemical carcinogenesis in HBx-transgenic mouse livers (Madden et al. 2001; Zheng et al. 2007). Although these observations in mice are important, it remains uncertain whether the results have a parallel in patients infected with HBV (see Slagle and Bouchard 2015 for more details).
Animal Models and Transient Infections
Chronic hepadnavirus infections are typically lifelong, whereas transient infections are prolonged but still <6–12 months in duration. A peculiar feature of transient infections is the apparent failure to provoke a host response until weeks or months have gone by. Once provoked, the host response is vigorous, typically clearing the infection. In contrast, the immune response in chronic carriers is generally weak and ineffective at virus clearance, although many patients may go through a more vigorous immune clearance phase after decades of infection, leading to a major drop in virus load in the liver (Yim and Lok 2006). Transient infections, although not increasing the risk of HCC, have been studied with the hope that they will provide insights into immunotherapy of chronic infections.
The chimpanzee has recently been used to provide insights into the course and clearance of transient infections. The background for these chimpanzee studies was performed in woodchucks and ducks (Ponzetto et al. 1984; Jilbert et al. 1992; Kajino et al. 1994). Early studies with these models showed that clearance may occur even after infection of the entire hepatocyte population. Clearance in this situation was surprising for several reasons. First, hepatocytes, the major parenchymal cell of the liver, constitute 70% of liver cell mass. Second, cccDNA was believed, even shortly after its discovery, to be stable when new viral DNA synthesis was inhibited (e.g., Hirota et al. 1986). Therefore, virus clearance would appear to require destruction of the entire hepatocyte population to eliminate cccDNA, but it is not obvious how this would occur without destroying the liver and killing the host. However, a study of virus clearance in the woodchuck showed that although clearance involved destruction of at least 70% of the hepatocyte population, the recovered liver was actually populated by hepatocytes that had been infected (Summers et al. 2003). Subsequent studies in the chimpanzee (Guidotti et al. 1999; Wieland et al. 2004b), in transgenic mice (Guidotti and Chisari 1999), and in mouse hepatocyte cultures (Wieland et al. 2005) indicated that antiviral cytokines can induce clearance of replicating DNA from the cytoplasm of infected hepatocytes, which should prevent further cccDNA synthesis (Iannacone and Guidotti 2015).
However, the mechanism of elimination of cccDNA from surviving hepatocytes is still controversial; one view is that cccDNA, like replicating DNA, is destroyed noncytopathically (Wieland et al. 2004b; Murray et al. 2005), whereas another view is that cccDNA cannot survive cell division and is lost when infected hepatocytes divide to replace those killed by antiviral CTLs (Mason et al. 2009a). To address this distinction, Dandri et al. (Lutgehetmann et al. 2010) studied urokinase plasminogen activator-severe combined immunodeficiency disorder (uPA-SCID) mice (Sandgren et al. 1991; Rhim et al. 1995), in which the liver was partially repopulated with HBV-infected hepatocytes from tree shrews. A substantial loss of cccDNA was seen during expansion of the donor hepatocyte population, which appeared consistent with loss of cccDNA at mitosis (Lutgehetmann et al. 2010), supporting the notion that cell division plays an important role in cccDNA loss during resolution of transient infections.
In contrast, a recent study suggests that interferon produced during the antiviral response induces APOBEC, which selectively deaminates cccDNA in infected cells, leading to its nucleolytic degradation (Lucifora et al. 2014). Thus, cytokines, such as interferon, might also have a role in cccDNA elimination. It will be important to know whether this latter result is reproducible in vivo and sufficiently robust to clear more than a small fraction of cccDNA from the infected liver because, as compared with a mechanism involving hepatocyte regeneration, it would have quite distinct implications for development of immune therapies for chronic infections.
Animal Models and Antiviral Therapy
Interferon α was the earliest therapy for HBV to receive Food and Drug Administration (FDA) approval. A disadvantage of interferon α is that it is only effective in curing patients with active hepatitis, presumably because it stimulates the existing immune response to the virus. Most patients do not receive a long-term benefit from this therapy. As a result, significant effort has gone into the development of antiviral therapies that use small molecules to inhibit specific steps in virus replication, including virus uptake (Volz et al. 2013), nucleocapsid assembly (e.g., Wu et al. 2008; Campagna et al. 2013), polymerase activity, and virus release (Noordeen et al. 2013). Of these, only nucleoside analogs, which inhibit viral DNA synthesis, have been FDA-approved for use in humans. Two of the three most studied, lamivudine and tenofovir, were developed initially against HIV, whereas the third, entecavir, was developed primarily for treatment of HBV infection, as it has poor activity against HIV.
A problem with nucleoside analog therapy is that it is not very effective against cccDNA, which maintains the chronic infection. This might be explained by early studies with animal models that showed that cccDNA was stable in infected hepatocytes and suggested that its elimination from the chronically infected liver during nucleoside analog therapy might require killing of infected hepatocytes, generally a slow process (Hirota et al. 1986; Fourel et al. 1994b; Moraleda et al. 1997). On top of this, even if cccDNA is lost during mitosis (i.e., during liver regeneration to replace dying hepatocytes), as suggested by studies in the chimeric mouse model described above, nucleoside analog therapy may be leaky enough in natural hosts to allow some new cccDNA synthesis via the reverse transcription pathway (i.e., viral nucleocapsids containing viral DNA might survive mitosis even if cccDNA did not). This might occur preferentially in dividing hepatocytes (Reaiche-Miller et al. 2013), perhaps because of elevated nucleoside pools that compete with the inhibitor. In any case, it is possible to achieve a reduction in cccDNA levels during long-term therapy with nucleosides, but not its complete elimination. On the positive side, nucleoside analog therapy can reduce disease symptoms associated with chronic hepatitis and also stop, and sometimes even reverse, the progression of fibrosis and cirrhosis. However, although initial results are encouraging (Korba et al. 2004; Lee et al. 2007), nucleoside analog therapy still only reduces the progression to HCC about twofold (Papatheodoris et al. 2010; Zoulim and Durantel 2015).
Animal Models and HCC
Woodchucks infected with WHV at birth have an ∼100% incidence of HCC by 3 years of age, which is mostly attributed to activation of N-myc via insertion of viral DNA into host DNA. This discovery followed the report that B-cell lymphomas in chickens infected by avian leukosis viruses (ALVs) were mostly a result of insertional activation/mutation of c-Myc by the ALV provirus (Hayward et al. 1981). Indeed, an initial effort by some of the same investigators asked whether this model might explain woodchuck HCC, but the results for insertion near woodchuck myc were negative (Ogston et al. 1982). However, later studies (Fourel et al. 1990, 1994a) revealed that this model was correct. Ultimately, about half of woodchuck HCCs result from WHV DNA integration near N-myc2 and another half from integrations at distal loci, win (Fourel et al. 1990, 1994a) and b3n (Bruni et al. 2006). All of these integrations seem to activate N-myc2 expression. In retrospect, the initial study might have failed to detect a woodchuck myc gene because integration had occurred in the win or b3n loci in the tumors that were examined by these investigators.
Interestingly, like the woodchuck, Beechey ground squirrels infected with GSHV also have a high risk of HCC. However, the risk is lower and HCCs are slower to develop (Marion et al. 1986). In addition, HCC is not associated with insertional activation of a myc gene by viral DNA, but instead with amplification of c-Myc (Transy et al. 1992; Hansen et al. 1993). The basis for the differences in HCC incidence and mechanism between the two species is, at least in part, virus-specific. GSHV also infects woodchucks. In this host, chronic GSHV infection is associated with a much slower rate of progression to HCC than is WHV infection (Seeger et al. 1991). However, the viral determinants influencing the rate of HCC development remain elusive. In addition, it is not yet clear how the woodchuck and ground squirrel results apply to human HCC. To date, insertional mutagenesis of a host oncogene does not appear to be a major cause of HCC in HBV patients (however, see Buendia and Neuveut 2015).
Another consideration in emergence of human HCC comes from transgenic mouse studies originally performed by Chisari and colleagues (1989). This group showed that the L protein of HBV is a hepatotoxin when overexpressed. Thus, transgenic mice that overexpress the L protein in hepatocytes develop very severe liver disease, leading to silencing of the transgene and, ultimately, HCC (Chisari et al. 1989). Silencing of the transgene is believed to be associated with clonal repopulation of the liver by hepatocytes that have either lost the transgene or shut down its expression. Clonal hepatocyte repopulation is known in other models of chronic liver disease, including human liver disease, to be a risk factor for HCC (Marongiu et al. 2008; Alison et al. 2009). Although the L protein overexpression model does not appear directly relevant to chronic hepatitis B, the idea of clonal hepatocyte repopulation may apply. For instance, in both woodchucks and chimpanzees, chronic infection appears to be associated with the emergence of foci of hepatocytes, possibly clonally derived, which do not support virus expression, that is, which appear to have escaped the toxic effects of infection caused by the antiviral immune response of the host (Xu et al. 2007; Mason et al. 2009b).
A final issue that needs to be addressed in considering animal models and HCC is viral DNA integration into host DNA. Essentially all human HCCs developing in HBV carriers contain clonally integrated viral DNA. This appears to indicate that the tumors arose by dedifferentiation of hepatocytes that had once been infected, as liver stem cells do not appear to be susceptible to HBV infection. However, this point remains controversial.
What is now clear is that integration is a common by-product of virus infection (Fig. 4). Studies with DHBV by both the Summers and Rogler laboratories showed that linear viral DNAs are the main substrates for integration (Gong et al. 1995, 1999; Yang and Summers 1995, 1999). Linear viral DNAs are created when the plus-strand primer fails to translocate from DR1 to DR2 for plus-strand synthesis, and represent ∼10% of the virion DNA population (Staprans et al. 1991). Interestingly, from the perspective of cccDNA formation, ∼10% of integrants appear to originate not from these linear DNAs but from relaxed circular DNA that had been linearized after entry into the nucleus by strand displacement DNA synthesis through the cohesive overlap region. This would create linear molecules that differed from the linear molecules described above by the presence of a large terminal redundancy (LTR) corresponding to the cohesive overlap domain.
It was also found that molecules with an LTR may give rise to a fraction of cccDNA, possibly via nonhomologous recombination between the LTRs. This results in aberrant cccDNAs. Homologous recombination between the LTRs would, in contrast, give rise to wild cccDNA. It is not yet clear whether this is a normal pathway for cccDNA formation or if, as generally believed, cccDNA is formed directly from relaxed circular DNA (Yang et al. 1996). To date, linear DNAs with an LTR have been inferred but not proven to exist.
Integration appears to occur at random sites on the host genome but near the ends of the linear DNAs. In the woodchuck, there is a clear link between integration site and HCC, but this has not been found in ground squirrels. In rare instances, HBV integration has been found at host oncogenes but, in general, the link is less obvious. This is discussed in detail in Buendia and Neuveut (2015).
SUMMARY
Animal models have made major contributions to our understanding of HBV replication and pathogenesis and, because of limited access to human tissue samples, remain the only reliable tool to understand the link between chronic infection, cirrhosis, and HCC.
Footnotes
Editors: Christoph Seeger and Stephen Locarnini
Additional Perspectives on Hepatitis B and Delta Viruses available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Aden DP, Fogel A, Plotkin S, Damjanov I, Knowles BB 1979. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 282: 615–616. [DOI] [PubMed] [Google Scholar]
- Alison MR, Islam S, Lim S 2009. Stem cells in liver regeneration, fibrosis and cancer: The good, the bad and the ugly. J Pathol 217: 282–298. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R, Schaller H 1988. The amino-terminal domain of the hepadnaviral P-gene encodes the terminal protein (genome-linked protein) believed to prime reverse transcription. EMBO J 7: 4185–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer ME, Blumberg BS, Werner B 1968. Particles associated with Australia antigen in the sera of patients with leukaemia, Down’s syndrome and hepatitis. Nature 218: 1057–1059. [DOI] [PubMed] [Google Scholar]
- Beasley RP, Lin CC, Hwang LY, Chien CS 1981. Hepatocellular carcinoma and hepatitis B virus. Lancet 2: 1129–1133. [DOI] [PubMed] [Google Scholar]
- Blum HE, Haase AT, Harris JD, Walker D, Vyas GN 1984. Asymmetric replication of hepatitis B virus DNA in human liver: Demonstration of cytoplasmic minus strand DNA by blot analysis and in situ hybridization. Virology 139: 87–96. [DOI] [PubMed] [Google Scholar]
- Blumberg BS 1977. Australia antigen and the biology of hepatitis B. Science 197: 17–25. [DOI] [PubMed] [Google Scholar]
- Blumberg BS, Gerstley BJS, Hungerford DA, London WT, Sutnik AI 1967. A serum antigen (Australia antigen) in Down’s syndrome, leukemia and hepatitis. Ann Intern Med 66: 924–931. [DOI] [PubMed] [Google Scholar]
- Blumberg BS, Sutnick AI, London WT 1968. Hepatitis and leukemia: Their relation to Australia antigen. Bull NY Acad Med 44: 1566–1586. [PMC free article] [PubMed] [Google Scholar]
- Blumberg BS, Larouze B, London WT, Werner B, Hesser JE, Millman I, Saimot G, Payet M 1975. The relation of infection with the hepatitis B agent to primary hepatic carcinoma. Am J Pathol 81: 669–682. [PMC free article] [PubMed] [Google Scholar]
- Bruni R, Conti I, Villano U, Giuseppetti R, Palmieri G, Rapicetta M 2006. Lack of WHV integration nearby N-myc2 and in the downstream b3n and win loci in a considerable fraction of liver tumors with activated N-myc2 from naturally infected wild woodchucks. Virology 345: 258–269. [DOI] [PubMed] [Google Scholar]
- *.Buendia M-A, Neuveut C 2015. Hepatocellular carcinoma. Cold Spring Harb Perspect Med 5: a021444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell CJ, Gowans EJ, Jilbert AR, Lake JR, Marmion BP 1982. Hepatitis B virus DNA detection by in situ cytohybridization: Implications for viral replication strategy and pathogenesis of chronic hepatitis. Hepatology 2: 85S–91S. [Google Scholar]
- Buscher M, Reiser W, Will H, Schaller H 1985. Transcripts and the putative RNA pregenome of duck hepatitis B virus: Implications for reverse transcription. Cell 40: 717–724. [DOI] [PubMed] [Google Scholar]
- Campagna MR, Liu F, Mao R, Mills C, Cai D, Guo F, Zhao X, Ye H, Cuconati A, Guo H, et al. 2013. Sulfamoylbenzamide derivatives inhibit the assembly of hepatitis B virus nucleocapsids. J Virol 87: 6931–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg MD, Ostrove JM, Kelly TJ 1982. Initiation of adenovirus DNA replication: Detection of covalent complexes between nucleotide and the 80-kilodalton terminal protein. J Virol 41: 265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Enders G, Sprengel R, Peters N, Varmus HE, Ganem D 1987. Expression of the precore region of an avian hepatitis B virus is not required for viral replication. J Virol 61: 3322–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Kew MC, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH 1992. The precore gene of the woodchuck hepatitis virus genome is not essential for viral replication in the natural host. J Virol 66: 5682–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Kaneko S, Girones R, Anderson RW, Hornbuckle WE, Tennant BC, Cote PJ, Gerin JL, Purcell RH, Miller RH 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol 67: 1218–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD 1989. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell 59: 1145–1156. [DOI] [PubMed] [Google Scholar]
- Condreay LD, Aldrich CE, Coates L, Mason WS, Wu TT 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J Virol 64: 3249–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cova L, Lambert V, Chevallier A, Hantz O, Fourel I, Jacquet C, Pichoud C, Boulay J, Chomel B, Vitvitski L, et al. 1986. Evidence for the presence of duck hepatitis B virus in wild migrating ducks. J Gen Virol 67: 537–547. [DOI] [PubMed] [Google Scholar]
- Dane DS, Cameron CH, Briggs M 1970. Virus-like particles in serum of patients with Australia-antigen-associated hepatitis. Lancet 1: 695–698. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Geipel A, Konig A, Corman VM, van Riel D, Leijten LM, Bremer CM, Rasche A, Cottontail VM, Maganga GD, et al. 2013. Bats carry pathogenic hepadnaviruses antigenically related to hepatitis B virus and capable of infecting human hepatocytes. Proc Natl Acad Sci 110: 16151–16156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauquet CM, Maniloff J, Desselberger U, Ball LA, eds. 2005. Virus taxonomy: VIIIth report of the international committee of taxonomy of viruses, pp. 373–384 Academic, San Diego, CA. [Google Scholar]
- Fourel G, Trepo C, Bougueleret L, Henglein B, Ponzetto A, Tiollais P, Buendia MA 1990. Frequent activation of N-myc genes by hepadnavirus insertion in woodchuck liver tumours. Nature 347: 294–298. [DOI] [PubMed] [Google Scholar]
- Fourel G, Couturier J, Wei Y, Apiou F, Tiollais P, Buendia MA 1994a. Evidence for long-range oncogene activation by hepadnavirus insertion. EMBO J 13: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel I, Cullen JM, Saputelli J, Aldrich CE, Schaffer P, Averett DR, Pugh J, Mason WS 1994b. Evidence that hepatocyte turnover is required for rapid clearance of duck hepatitis B virus during antiviral therapy of chronically infected ducks. J Virol 68: 8321–8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler MJ, Monjardino J, Tsiquaye KN, Zuckerman AJ, Thomas HC 1984. The mechanism of replication of hepatitis B virus: Evidence of asymmetric replication of the two DNA strands. J Med Virol 13: 83–91. [DOI] [PubMed] [Google Scholar]
- Frelin L, Wahlstrom T, Tucker AE, Jones J, Hughes J, Lee BO, Billaud JN, Peters C, Whitacre D, Peterson D, et al. 2009. A mechanism to explain the selection of the hepatitis e antigen-negative mutant during chronic hepatitis B virus infection. J Virol 83: 1379–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich WH, Robinson WS 1980. Hepatitis B virus contains protein attached to the 5′ terminus of its complete DNA strand. Cell 21: 801–809. [DOI] [PubMed] [Google Scholar]
- Gilboa E, Mitra SW, Goff S, Baltimore D 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18: 93–100. [DOI] [PubMed] [Google Scholar]
- Gong SS, Jensen AD, Wang H, Rogler CE 1995. Duck hepatitis B virus integrations in LMH chicken hepatoma cells: Identification and characterization of new episomally derived integrations. J Virol 69: 8102–8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong SS, Jensen AD, Chang CJ, Rogler CE 1999. Double-stranded linear duck hepatitis B virus (DHBV) stably integrates at a higher frequency than wild-type DHBV in LMH chicken hepatoma cells. J Virol 73: 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti LG, Chisari FV 1999. Cytokine-induced viral purging—Role in viral pathogenesis. Curr Opin Microbiol 2: 388–391. [DOI] [PubMed] [Google Scholar]
- Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284: 825–829. [DOI] [PubMed] [Google Scholar]
- Guo JT, Aldrich CE, Mason WS, Pugh JC 1996. Characterization of serum amyloid A protein mRNA expression and secondary amyloidosis in the domestic duck. Proc Natl Acad Sci 93: 14548–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Mason WS, Aldrich CE, Saputelli JR, Miller DS, Jilbert AR, Newbold JE 2005. Identification and classification of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J Virol 79: 2729–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurgo C, Ray RK, Thiry L, Green M 1971. Inhibitors of the RNA and DNA dependent polymerase activities of RNA tumour viruses. Nat New Biol 229: 111–114. [DOI] [PubMed] [Google Scholar]
- Hansen LJ, Tennant BC, Seeger C, Ganem D 1993. Differential activation of myc gene family members in hepatic carcinogenesis by closely related hepatitis B viruses. Mol Cell Biol 13: 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward WS, Neel BG, Astrin SM 1981. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature 290: 475–480. [DOI] [PubMed] [Google Scholar]
- Heermann KH, Goldmann U, Schwartz W, Seyffarth T, Baumgarten H, Gerlich WH 1984. Large surface proteins of hepatitis B virus containing the pre-s sequence. J Virol 52: 396–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Sherker AH, Omata M, Yokosuka O, Okuda K 1986. Effects of adenine arabinoside and corticosteroid on replication of duck hepatitis B virus DNA in the liver. Ann Acad Med Singapore 15: 227–232. [PubMed] [Google Scholar]
- *.Hu J, Seeger C 2015. Hepadnavirus genome replication and persistence. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Iannacone M, Guidotti LG 2015. Mouse models of hepatitis B virus pathogenesis. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda JE, Enomoto T, Hurwitz J 1982. Adenovirus protein-primed initiation of DNA chains in vitro. Proc Natl Acad Sci 79: 2442–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilbert AR, Wu TT, England JM, Hall PM, Carp NZ, O’Connell AP, Mason WS 1992. Rapid resolution of duck hepatitis B virus infections occurs after massive hepatocellular involvement. J Virol 66: 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajino K, Jilbert AR, Saputelli J, Aldrich CE, Cullen J, Mason WS 1994. Woodchuck hepatitis virus infections: Very rapid recovery after a prolonged viremia and infection of virtually every hepatocyte. J Virol 68: 5792–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan PM, Greenman RL, Gerin JL, Purcell RH, Robinson WS 1973. DNA polymerase associated with human hepatitis B antigen. J Virol 12: 995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Nomura K, Hirayama Y, Kitagawa T 1987. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res 47: 4460–4464. [PubMed] [Google Scholar]
- Khudyakov YE, Makhov AM 1989. Prediction of terminal protein and ribonuclease H domains in the gene P product of hepadnaviruses. FEBS Lett 243: 115–118. [DOI] [PubMed] [Google Scholar]
- Korba BE, Cote PJ, Menne S, Toshkov I, Baldwin BH, Wells FV, Tennant BC, Gerin JL 2004. Clevudine therapy with vaccine inhibits progression of chronic hepatitis and delays onset of hepatocellular carcinoma in chronic woodchuck hepatitis virus infection. Antivir Ther 9: 937–952. [PubMed] [Google Scholar]
- Lanford RE, Chavez D, Brasky KM, Burns RB III, Rico-Hesse R 1998. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci 95: 5757–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Eun R, Jang BI, Kim TN 2007. Prevention by lamivudine of hepatocellular carcinoma in patients infected with hepatitis B virus. Gut Liver 1: 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhoff RJ, Summers J 1994. Construction of avian hepadnavirus variants with enhanced replication and cytopathicity in primary hepatocytes. J Virol 68: 5706–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien JM, Aldrich CE, Mason WS 1986. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J Virol 57: 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien JM, Petcu DJ, Aldrich CE, Mason WS 1987. Initiation and termination of duck hepatitis B virus DNA synthesis during virus maturation. J Virol 61: 3832–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb DD, Hirsch RC, Ganem D 1991. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: Implications for other reverse transcribing elements. EMBO J 10: 3533–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucifora J, Xia Y, Reisinger F, Zhang K, Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz T, et al. 2014. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 343: 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutgehetmann M, Volz T, Kopke A, Broja T, Tigges E, Lohse AW, Fuchs E, Murray JM, Petersen J, Dandri M 2010. In vivo proliferation of hepadnavirus-infected hepatocytes induces loss of covalently closed circular DNA in mice. Hepatology 52: 16–24. [DOI] [PubMed] [Google Scholar]
- Madden CR, Finegold MJ, Slagle BL 2001. Hepatitis B virus X protein acts as a tumor promoter in development of diethylnitrosamine-induced preneoplastic lesions. J Virol 75: 3851–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandart E, Kay A, Galibert F 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: Comparison with woodchuck and human hepatitis B virus sequences. J Virol 49: 782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion PL, Oshiro LS, Regnery DC, Scullard GH, Robinson WS 1980. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc Natl Acad Sci 77: 2941–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marion PL, Van DM, Knight SS, Salazar FH, Garcia G, Popper H, Robinson WS 1986. Hepatocellular carcinoma in ground squirrels persistently infected with ground squirrel hepatitis virus. Proc Natl Acad Sci 83: 4543–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marongiu F, Doratiotto S, Montisci S, Pani P, Laconi E 2008. Liver repopulation and carcinogenesis: Two sides of the same coin? Am J Pathol 172: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Seal G, Summers J 1980. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol 36: 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Aldrich C, Summers J, Taylor JM 1982. Asymmetric replication of duck hepatitis B virus DNA in liver cells: Free minus-strand DNA. Proc Natl Acad Sci 79: 3997–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Halpern MS, England JM, Seal G, Egan J, Coates L, Aldrich C, Summers J 1983. Experimental transmission of duck hepatitis B virus. Virology 131: 375–384. [DOI] [PubMed] [Google Scholar]
- Mason WS, Lien J, Petcu DJ, Coates L, London WT, O’Connell A, Aldrich C, Custer RP 1987. In vivo and in vitro studies on duck hepatitis B virus replication. In Hepadna viruses (ed. Robinson WS, et al. ), pp. 3–16 Alan R. Liss, New York. [Google Scholar]
- Mason WS, Xu C, Low HC, Saputelli J, Aldrich CE, Scougall C, Grosse A, Colonno R, Litwin S, Jilbert AR 2009a. The amount of hepatocyte turnover that occurred during resolution of transient hepadnavirus infections was lower when virus replication was inhibited with entecavir. J Virol 83: 1778–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason WS, Low HC, Xu C, Aldrich CE, Scougall CA, Grosse A, Cloustou A, Chavez D, Litwin S, Peri S, et al. 2009b. Detection of clonally expanded heptocytes in chimpanzees with chronic hepatitis B virus infection. J Virol 83: 8396–8408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P, Scougall CA, Will H, Burrell CJ, Jilbert AR 2003. A duck hepatitis B virus strain with a knockout mutation in the putative X ORF shows similar infectivity and in vivo growth characteristics to wild-type virus. Virology 317: 291–298. [DOI] [PubMed] [Google Scholar]
- Milich DR, Jones J, Hughes J, Maruyama T 1993. Role of T-cell tolerance in the persistence of hepatitis B virus infection. J Immunother Emphasis Tumor Immunol 14: 226–233. [DOI] [PubMed] [Google Scholar]
- Millman I, Loeb LA, Bayer ME, Blumberg BS 1970. Australia antigen (a hepatitis-associated antigen): Purification and physical properties. J Exp Med 131: 1190–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar-Kimber KL, Summers J, Taylor JM, Mason WS 1983. Protein covalently bound to minus-strand DNA intermediates of duck hepatitis B virus. J Virol 45: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monjardino J, Fowler MJ, Montano L, Weller I, Tsiquaye KN, Zuckerman AJ, Jones DM, Thomas HC 1982. Analysis of hepatitis virus DNA in the liver and serum of HBe antigen positive chimpanzee carriers. J Med Virol 9: 189–199. [DOI] [PubMed] [Google Scholar]
- Moraleda G, Saputelli J, Aldrich CE, Averett D, Condreay L, Mason WS 1997. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J Virol 71: 9392–9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JM, Wieland SF, Purcell RH, Chisari FV 2005. Dynamics of hepatitis B virus clearance in chimpanzees. Proc Natl Acad Sci 102: 17780–17785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noordeen F, Vaillant A, Jilbert AR 2013. Nucleic acid polymers inhibit duck hepatitis B virus infection in vitro. Antimicrob Agents Chemother 57: 5299–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell AP, Urban MK, London WT 1983. Naturally occurring infection of Pekin duck embryos by duck hepatitis B virus. Proc Natl Acad Sci 80: 1703–1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogston CW, Jonak GJ, Rogler CE, Astrin SM, Summers J 1982. Cloning and structural analysis of integrated woodchuck hepatitis virus sequences from hepatocellular carcinomas of woodchucks. Cell 29: 385–394. [DOI] [PubMed] [Google Scholar]
- Ou JH, Laub O, Rutter WJ 1986. Hepatitis B virus gene function: The precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci 83: 1578–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papatheodoris GV, Lampertico P, Manolakopoulos S, Lok A 2010. Incidence of hepatocellular carcinoma in chronic hepatitis B patients receiving nucleos(t)ide therapy: A systematic review. J Hepatol 53: 348–356. [DOI] [PubMed] [Google Scholar]
- Piasecki T, Harkins GW, Chrzastek K, Julian L, Martin DP, Varsani A 2013. Avihepadnavirus diversity in parrots is comparable to that found amongst all other avian species. Virology 438: 98–105. [DOI] [PubMed] [Google Scholar]
- Ponzetto A, Cote PJ, Ford EC, Purcell RH, Gerin JL 1984. Core antigen and antibody in woodchucks after infection with woodchuck hepatitis virus. J Virol 52: 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prassolov A, Hohenberg H, Kalinina T, Schneider C, Cova L, Krone O, Frolich K, Will H, Sirma H 2003. New hepatitis B virus of cranes that has an unexpected broad host range. J Virol 77: 1964–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince AM, Szmuness W, Michon J, Demaille J, Diebolt G, Linhard J, Quenum C, Sankale M 1975. A case/control study of the association between primary liver cancer and hepatitis B infection in Senegal. Int J Cancer 16: 376–383. [DOI] [PubMed] [Google Scholar]
- Pult I, Netter HJ, Bruns M, Prassolov A, Sirma H, Hohenberg H, Chang SF, Frolich K, Krone O, Kaleta EF, et al. 2001. Identification and analysis of a new hepadnavirus in white storks. Virology 289: 114–128. [DOI] [PubMed] [Google Scholar]
- Reaiche-Miller GY, Thorpe M, Low HC, Qiao Q, Scougall CA, Mason WS, Litwin S, Jilbert AR 2013. Duck hepatitis B virus covalently closed circular DNA appears to survive hepatocyte mitosis in the growing liver. Virology 446: 357–364. [DOI] [PubMed] [Google Scholar]
- Rhim JA, Sandgren EP, Palmiter RD, Brinster RL 1995. Complete reconstitution of mouse liver with xenogeneic hepatocytes. Proc Natl Acad Sci 92: 4942–4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigdon RH 1961. Amyloidosis: Spontaneous occurrence in white Pekin ducks. Am J Pathol 39: 369–378. [PMC free article] [PubMed] [Google Scholar]
- Robinson WS, Greenman RL 1974. DNA polymerase in the core of the human hepatitis B virus candidate. J Virol 13: 1231–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WS, Clayton DA, Greenman RL 1974. DNA of a human hepatitis B virus candidate. J Virol 14: 384–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabran JL, Hsu TW, Yeater C, Kaji A, Mason WS, Taylor JM 1979. Analysis of integrated avian RNA tumor virus DNA in transformed chicken, duck and quail fibroblasts. J Virol 29: 170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandgren EP, Palmiter RD, Heckel JL, Daugherty CC, Brinster RL, Degen JL 1991. Complete hepatic regeneration after somatic deletion of an albumin-plasminogen activator transgene. Cell 66: 245–256. [DOI] [PubMed] [Google Scholar]
- Seeger C, Ganem D, Varmus HE 1984. The cloned genome of ground squirrel hepatitis virus is infectious in the animal. Proc Natl Acad Sci 81: 5849–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Ganem D, Varmus HE 1986. Biochemical and genetic evidence for the hepatitis B virus replication strategy. Science 232: 477–484. [DOI] [PubMed] [Google Scholar]
- Seeger C, Marion PL, Ganem D, Varmus HE 1987. In vitro recombinants of ground squirrel and woodchuck hepatitis viral DNAs produce infectious virus in squirrels. J Virol 61: 3241–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger C, Baldwin B, Hornbuckle WE, Yeager AE, Tennant BC, Cote P, Ferrell L, Ganem D, Varmus HE 1991. Woodchuck hepatitis virus is a more efficient oncogenic agent than ground squirrel hepatitis virus in a common host. J Virol 65: 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chen M, Acs G 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci 84: 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CH, Li LS, Roychoudhury S, Ho MH 1989. In vitro propagation of human hepatitis B virus in a rat hepatoma cell line. Proc Natl Acad Sci 86: 6323–6327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Slagle BL, Bouchard MJ 2015. Regulation of viral gene expression. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JK, Cywinski A, Taylor JM 1984. Specificity of initiation of plus-strand DNA by Rous sarcoma virus. J Virol 52: 314–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spandau DF, Lee C-H 1988. trans-Activation of viral enhancers by the hepatitis B virus X protein. J Virol 62: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel R, Kuhn C, Manso C, Will H 1984. Cloned duck hepatitis B virus DNA is infectious in Pekin ducks. J Virol 52: 932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel R, Kaleta EF, Will H 1988. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol 62: 932–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standring DN, Ou JH, Masiarz FR, Rutter WJ 1988. A signal peptide encoded within the precore region of hepatitis B virus directs the secretion of a heterogeneous population of e antigens in Xenopus oocytes. Proc Natl Acad Sci 85: 8405–8409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staprans S, Loeb DD, Ganem D 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J Virol 65: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Mason WS 1982. Replication of the genome of a hepatitis B–like virus by reverse transcription of an RNA intermediate. Cell 29: 403–415. [DOI] [PubMed] [Google Scholar]
- Summers J, O’Connell A, Millman I 1975. Genome of hepatitis B virus: Restriction enzyme cleavage and structure of DNA extracted from Dane particles. Proc Natl Acad Sci 72: 4597–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Smolec JM, Snyder R 1978. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci 75: 4533–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Smith PM, Horwich AL 1990. Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J Virol 64: 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers J, Jilbert AR, Yang W, Aldrich CE, Saputelli J, Litwin S, Toll E, Mason WS 2003. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci 100: 11652–11659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau C, Romet LJ, Mullins JI, Essex M 1986. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 47: 37–47. [DOI] [PubMed] [Google Scholar]
- Tagawa M, Omata M, Okuda K 1986. Appearance of viral RNA transcripts in the early stage of duck hepatitis B virus infection. Virology 152: 477–482. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Machida A, Funatsu G, Nomura M, Usuda S, Aoyagi S, Tachibana K, Miyamoto H, Imai M, Nakamura T, et al. 1983. Immunochemical structure of hepatitis B e antigen in the serum. J Immunol 130: 2903–2907. [PubMed] [Google Scholar]
- Taylor JM, Illmensee R 1975. Site on the RNA of an avian sarcoma virus at which primer is bound. J Virol 16: 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennant BC, Gerin JL 2001. The woodchuck model of hepatitis B virus infection. ILAR J 42: 89–102. [DOI] [PubMed] [Google Scholar]
- *.Thomas E, Yoneda M, Schiff ER 2015. Viral hepatitis: Past and future of HBV and HDV. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Hayashida H, Miyata T 1983. Sequence homology between retroviral reverse transcriptase and putative polymerases of hepatitis B virus and cauliflower mosaic virus. Nature 305: 827–829. [DOI] [PubMed] [Google Scholar]
- Transy C, Fourel G, Robinson WS, Tiollais P, Marion PL, Buendia MA 1992. Frequent amplification of c-myc in ground squirrel liver tumors associated with past or ongoing infection with a hepadnavirus. Proc Natl Acad Sci 89: 3874–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Fujiyama A, Matsubara K 1987. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc Natl Acad Sci 84: 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuttleman JS, Pourcel C, Summers J 1986a. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47: 451–460. [DOI] [PubMed] [Google Scholar]
- Tuttleman JS, Pugh JC, Summers JW 1986b. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J Virol 58: 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twu JS, Schloemer RH 1987. Transcriptional trans-activating function of hepatitis B virus. J Virol 61: 3448–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida T, Suzuki K, Arii M, Shikata T, Fukuda R, Tao YX 1988. Geographical pathology of duck livers infected with duck hepatitis B virus from Chiba and Shimane in Japan and Shanghai in China. Cancer Res 48: 1319–1325. [PubMed] [Google Scholar]
- Volz T, Allweiss L, MB MB, Warlich M, Lohse AW, Pollok JM, Alexandrov A, Urban S, Petersen J, Lutgehetmann M, et al. 2013. The entry inhibitor Myrcludex-B efficiently blocks intrahepatic virus spreading in humanized mice previously infected with hepatitis B virus. J Hepatol 58: 861–867. [DOI] [PubMed] [Google Scholar]
- Wang GH, Seeger C 1992. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71: 663–670. [DOI] [PubMed] [Google Scholar]
- Wang GH, Seeger C 1993. Novel mechanism for reverse transcription in hepatitis B viruses. J Virol 67: 6507–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser B, Ganem D, Seeger C, Varmus HE 1983. Closed circular viral DNA and asymmetrical heterogeneous forms in livers from animals infected with ground squirrel hepatitis virus. J Virol 48: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland S, Thimme R, Purcell RH, Chisari FV 2004a. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci 101: 6669–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland SF, Spangenberg HC, Thimme R, Purcell RH, Chisari FV 2004b. Expansion and contraction of the hepatitis B virus transcriptional template in infected chimpanzees. Proc Natl Acad Sci 101: 2129–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci 102: 9913–9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will H, Cattaneo R, Koch H, Darai G, Schaller H, Schellekens H, van Eerd P, Deinhardt F 1982. Cloned HBV DNA causes hepatitis in chimpanzees. Nature 299: 740–742. [DOI] [PubMed] [Google Scholar]
- Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengel R, Cattaneo R, Schaller H 1987. Replication strategy of human hepatitis B virus. J Virol 61: 904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Zheng XJ, Yin CC, Jiang D, Zhu L, Liu Y, Wei L, Wang Y, Chen HS 2008. Inhibition of hepatitis B virus replication by Bay 41-4109 and its association with nucleocapsid disassembly. J Chemother 20: 458–467. [DOI] [PubMed] [Google Scholar]
- Xu C, Yemamoto T, Zhou T, Aldrich CE, Frank K, Cullen JM, Jilbert AR, Mason WS 2007. The liver of woodchucks infected with woodchuck hepatitis virus contains foci of virus core antigen negative hepatocytes with both altered and normal morphogy. Virology 359: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaginuma K, Shirakata Y, Kobayashi M, Koike K 1987. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci 84: 2678–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J 1995. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J Virol 69: 4029–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Summers J 1999. Integration of hepadnavirus DNA in infected liver: Evidence from a linear precursor. J Virol 73: 9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Mason WS, Summers J 1996. Covalently closed circular viral DNA formed from two types of linear DNA in woodchuck hepatitis virus-infected liver. J Virol 70: 4567–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim HJ, Lok AS 2006. Natural history of chronic hepatitis B virus infection: What we knew in 1981 and what we know in 2005. Hepatology 43: S173–S181. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Torii N, Hu Z, Jacob J, Liang TJ 2001. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J Clin Invest 108: 1523–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Chen WL, Louie SG, Yen TS, Ou JH 2007. Hepatitis B virus promotes hepatocarcinogenesis in transgenic mice. Hepatology 45: 16–21. [DOI] [PubMed] [Google Scholar]
- *.Zoulim F, Durantel D 2015. Antiviral therapies and prospects for a cure of chronic hepatitis B. Cold Spring Harb Perspect Med 10.1101/cshperspect.a021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoulim F, Seeger C 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol 68: 2026–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]