Abstract
Ure2, the protein that negatively regulates GATA factor (Gln3, Gat1)-mediated transcription in Saccharomyces cerevisiae, possesses prion-like characteristics. Identification of metabolic and environmental factors that influence prion formation as well as any activities that prions or prion precursors may possess are important to understanding them and developing treatment strategies for the diseases in which they participate. Ure2 exhibits primary sequence and three-dimensional homologies to known glutathione S-transferases. However, multiple attempts over nearly 2 decades to demonstrate Ure2-mediated S-transferase activity have been unsuccessful, leading to the possibility that Ure2 may well not participate in glutathionation reactions. Here we show that Ure2 is required for detoxification of glutathione S-transferase substrates and cellular oxidants. ure2Δ mutants are hypersensitive to cadmium and nickel ions and hydrogen peroxide. They are only slightly hypersensitive to diamide, which is nitrogen source-dependent, and minimally if at all hypersensitive to 1-chloro-2,4-dinitrobenzene, the most commonly used substrate for glutathione S-transferase enzyme assays. Therefore, Ure2 shares not only structural homology with various glutathione S-transferases, but ure2 mutations possess the same phenotypes as mutations in known S. cerevisiae and Schizosaccharomyces pombe glutathione S-transferase genes. These findings are consistent with Ure2 serving as a glutathione S-transferase in S. cerevisiae.
Several neurodegenerative conditions derive from the same pathogenetic mechanism, i.e. a change in protein conformation, polymerization, and plaque formation. These conditions have been called conformational diseases such as Alzheimer’s and the prionoses. Recent studies have demonstrated that a protein associated with such disease, amyloid β-protein, protects neurons from metal-induced oxidative damage (1). Neither the molecular basis for this activity nor how it is affected by environment and neuronal metabolism is yet known. The use of eukaryotic model systems, such as the yeast Saccharomyces cerevisiae, has greatly facilitated our acquisition of information about mammalian proteins, their functions and interactions, and how their synthesis and activities are regulated and integrated. In particular, the genetic study of prions has been facilitated by Wickner’s (2) discovery that S. cerevisiae Ure3 possesses prion-like characteristics. Ure3, the prion form of the nitrogen-regulatory protein Ure2, has been well studied in an attempt to gain further insight into the changes that accompany polymerization and the cellular proteins that impact on that process (3–7, 23). Significant emphasis has been placed on determining whether metabolic activity influences Ure2 → Ure3 conversion (8–13). Ure2 → Ure3 conversion has been reported to decrease in an mks1Δ strain, in a strain expressing the constitutively active dominant Ras2Val19 allele (8), and in strains where the intracellular pool of glutamate is enlarged (11).
Ure2 was originally identified as a mutated genetic locus that permits cells growing with ammonia as nitrogen source to transport the pyrimidine precursor ureidosuccinate; wild type cells are unable to do this (14, 15). The ure2 mutation was subsequently found to possess a pleiotropic phenotype in which transcriptional repression of many genes encoding proteins needed to transport and degrade poor nitrogen sources becomes resistant to nitrogen catabolite repression (NCR),1 i.e. repression no longer occurs in the presence of a good nitrogen source.
When URE2 was cloned and sequenced, it was found to possess homology to known glutathione S-transferases (16). Homology between Ure2 and glutathione S-transferases now extends to the level of its crystal structure (17–19). Two additional structures were determined using crystals in which glutathione or two of its analogues were bound to Ure2 (19). Despite these structural characteristics, multiple attempts to demonstrate that Ure2 catalyzes a glutathione S-transferase reaction have been unsuccessful (7, 16, 20). There are also characteristics of the Ure2 three-dimensional structure that prompt the question of whether it would even be expected to possess S-transferase activity. Most specifically a residue that participates in catalysis by known glutathione S-transferases (i.e. cysteine or histidine in Beta class and tyrosine or serine residues in eukaryotic classes) is not present in Ure2 (17). This residue is critical because it destabilizes the cysteine S–H bond, thereby facilitating formation of the active thiolate anion (GS−). The region of Ure2 that binds glutathione does contain an asparagine, Asn-124, that some, but not all, investigators suggest may be situated at a location and distance that are consistent with permitting it to function in destabilization of this critical S–H bond (19).
Whether or not Ure2 is a glutathione S-transferase is a question that has remained open and tantalizing for 2 decades and is increasingly important to future studies of Ure3 prion formation and the impact of environmental and metabolic influences on it. Therefore, we have investigated the possibility of additional Ure2 functions. To circumvent repeatedly reported problems associated with in vitro enzyme assays, we adopted a more genetic approach, i.e. asking whether ure2Δ mutants exhibit greater sensitivity than isogenic wild type strains to a range of glutathione S-transferase substrates and compounds generating oxidative stress in S. cerevisiae. We show that Ure2 is indeed required for detoxification of glutathione S-transferase substrates and cellular oxidants. Ure2 shares not only structural homology with various glutathione S-transferases, but ure2 mutations exhibit phenotypes similar to those of mutations in known S. cerevisiae and Schizosaccharomyces pombe glutathione S-transferase genes.
MATERIALS AND METHODS
Strains and Media
The S. cerevisiae strains we used were TCY5 (MATα, lys2, ura3, trp1), TCY1 (Matα, lys2, ura3), RR114 (MATα, lys2, ura3, trp1, ure2::TRP1), RR154 (Matα, lys2, ura3, gdh1Δ::hisGURA3-hisG), YHE711 (MATα, ura2, leu2:hisG, [ure-0]), YHE731 (MATα, ura2, leu2:hisG, [URE3] {URE3 cytoduced into YHE711}), STCY32 (Matα, lys2, ura3, trp1, his3::hisG), TIFY3 (MATα, ura2, leu2:hisG, ure2::G418), and BY4741 (Mata, his3 D1, leu2 D0, met1 5D0, ura3 D0). The plasmids were pRA27 (21), pEG202 (22), pRR529 (21), and YEp24 (New England Biolabs, Inc.).
The rich medium was YEPD, and minimal media for plating cells were Difco Yeast Nitrogen Base without amino acids or ammonium sulfate (0.17%) to which was added the indicated nitrogen source at 0.1% if other than 0.5% ammonium sulfate was used. Further additions (added after media were autoclaved and cooled) of metal ions, xenobiotics, etc. are indicated in the figure legends. Our standard auxotrophic supplements were added where necessary. Cells were grown at 30 °C. Although photographs are largely presented of cells at single times and for a single concentration of perturbant, in most cases, we have collected images at multiple times and multiple perturbant concentrations. This approach gives us a better appreciation of what occurs during the experiment and increases our confidence that the images presented here are representative of the effects we report. During this work, we noted wild type strain-to-strain differences in overall sensitivity to various perturbants. However, the patterns of sensitivity in wild type versus mutant cells were always the same. For this reason a wild type control was included on all Petri plates so that wild type versus mutant comparisons could be made directly.
Northern Blot Analyses
TCY5 was grown in Difco Yeast Nitrogen Base (0.17%)-ammonia (0.1%) or -glutamate (0.1%) containing 0.6 mM nickel sulfate to mid-log phase (A600 = 0.5). Cycloheximide (0.01% final concentration) was added to the cells and incubated for 10 min. Cells were then harvested by centrifugation (4 °C), washed in cold lysis buffer (0.5 m NaCl, 0.5 m Tris base (pH 7.5), 0.01 m EDTA) containing 0.005% cycloheximide, and resuspended in cold lysis buffer. An equal volume of acid-washed glass beads was added to the cells along with an equal volume of cold PCI (phenol/chloroform/isoamyl alcohol (25:24:1), and the cells were lysed by vortexing. After two more PCI extractions, an additional extraction with cold chloroform/isoamyl alcohol (24:1) was performed. Total RNA was ethanol-precipitated overnight at −20 °C, pelleted, resuspended in diethylpyrocarbonate-treated water, ethanolprecipitated again, and resuspended as before. RNA concentration was determined spectrophotometrically (A260 nm), and the samples stored at −80 °C until analyzed.
Northern blot analyses were performed as described previously (24) using the PCR-generated probes that were radiolabeled with the Invitrogen RAD Prime DNA Labeling System. The primers used to generate probe DNA were as follows: URE2 (5′-CAAGTGTCGAATCTCTCCAA-3′ and 5′-TCTATCCACGACATTATTCC-3′), GAP1 (12), and H3 (12). Nine micrograms of total RNA were added to each lane for analysis.
RESULTS
Ure2 Is Required for Protection against Heavy Metals
Ure2 possesses clear structural homology with Theta or Beta classes of glutathione S-transferases. Unfortunately attempts in multiple laboratories to demonstrate glutathione S-transferase enzyme activity for Ure2 have been unsuccessful (7, 16, 20). Although negative results are rarely reported in detail, the lack of success might derive from inherent instability in the enzyme, an observed characteristic of glutathione S-transferases, or from performing the assay with substrates that are not the preferred ones for the putative transferase (25–28). The difficulties experienced in attempts to assay an enzyme activity prompted us to take a step backward from in vitro assays and ask more simply whether Ure2 is required for protection of cells against the growth-inhibitory effects of compounds reported to be glutathione S-transferase substrates in other organisms.
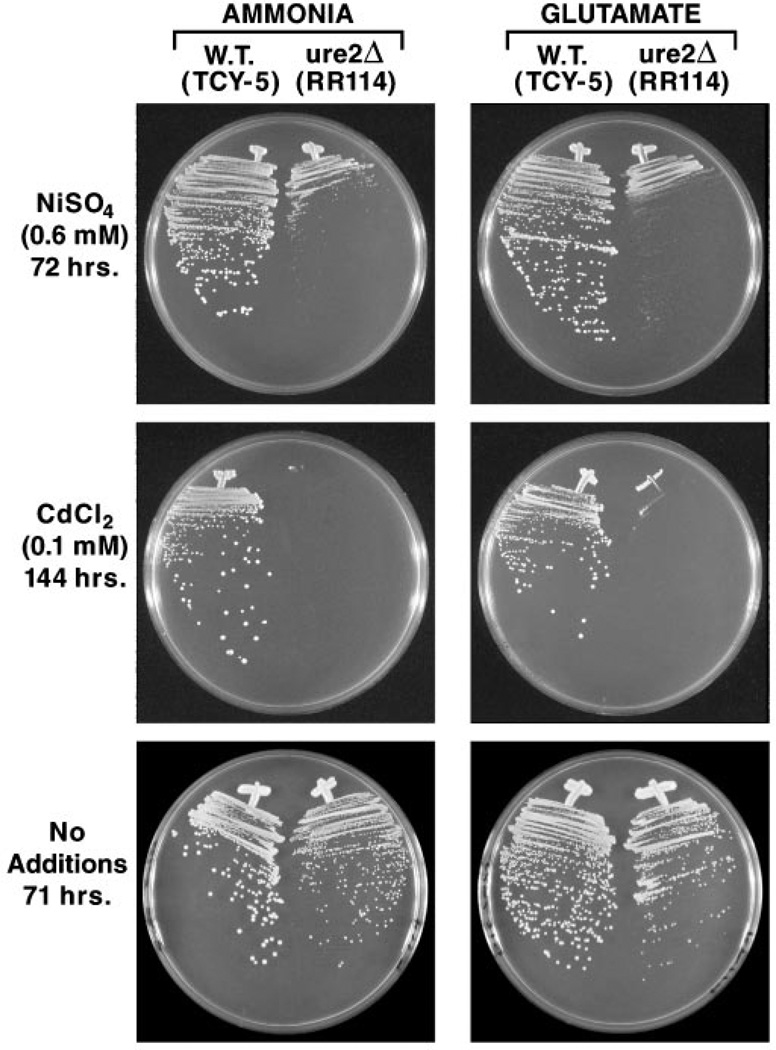
Assays unable to demonstrate Ure2-dependent glutathione S-transferase were performed with the commonly used substrate 1-chloro-2,4-dinitrobenzene (CDNB) (7, 20). Therefore, we first determined whether ure2 mutants might be hypersensitive to previously untested glutathione S-transferase substrates, for example, heavy metal ions such as nickel and cadmium. In wild type S. cerevisiae, cadmium ions are conjugated to glutathione, and the glutathionato-cadmium conjugate is transported into the vacuole or out of the cell (29). Ycf1 is responsible for transport of the conjugate (30), but identity of the enzyme catalyzing conjugation is not known. We compared growth of wild type (TCY-5) and isogenic ure2Δ (RR114) strains in glucose-ammonia or -glutamate medium containing nickel sulfate (0.6 mm) or cadmium chloride (0.1 mm). Both metal ions markedly inhibited growth of the ure2Δ mutant relative to wild type, although at the concentrations used, cadmium ions were far more toxic (note differences in the times of incubation) (Fig. 1). In addition, the ure2 mutant phenotype was much tighter with cadmium than with nickel ions. Equally important, growth was inhibited to roughly the same degree regardless of whether ammonia or glutamate was provided as sole nitrogen source. This is a positive indication that observed sensitivity did not derive indirectly from the influence of the ure2 mutation on NADPH levels in the cell.
Fig. 1. Growth of wild type and ure2Δ cells in the presence and absence of heavy metal ions.
Nitrogen sources and metal ions provided in the medium are indicated. The times of incubation are indicated and were the same for both ammonia and glutamate media. Minimal ammonia or minimal glutamate medium used in the bottom two panels did not contain any added heavy metal ions. W.T., wild type.
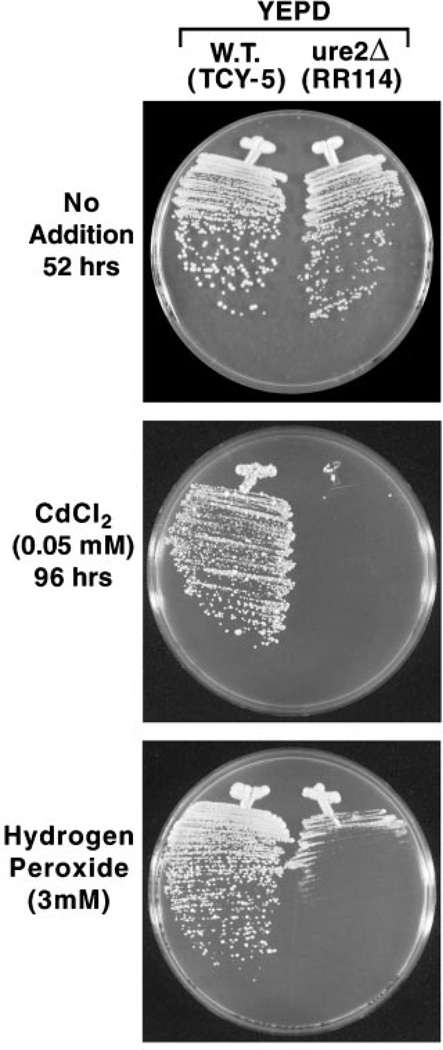
Although growth differences of wild type and ure2Δ strains in Fig. 1 are marked, we were concerned that they might derive trivially from the fact that ure2 mutants in some strain backgrounds grow a bit slower than wild type. Therefore, we asked, did slow growth of the ure2Δ in the presence of nickel or cadmium ions derive from loss of ability to detoxify the metal ions or was it just a manifestation of the fact that ure2 mutants grow slower? To assess these possibilities, we streaked the wild type and ure2Δ strains on the same media used for the metal ions toxicity test but in the absence of the metal ions. As shown in the bottom panels of Fig. 1, the ure2Δ strain does grow a bit slower than wild type. Therefore, ure2Δ hypersensitivity to nickel ions is not as great as depicted in the top two panels but is still present and can be increased if the nickel ion concentration is raised to 0.9 mm (data not shown). For cadmium, the difference in growth is dramatic. In fact, there was no growth of the ure2Δ mutant even after 4 days of incubation on YEPD-cadmium plates (Fig. 2).
Fig. 2. Growth of wild type and ure2Δ mutant cells on YEPD medium in the presence and absence of cadmium chloride or hydrogen peroxide.
W.T., wild type.
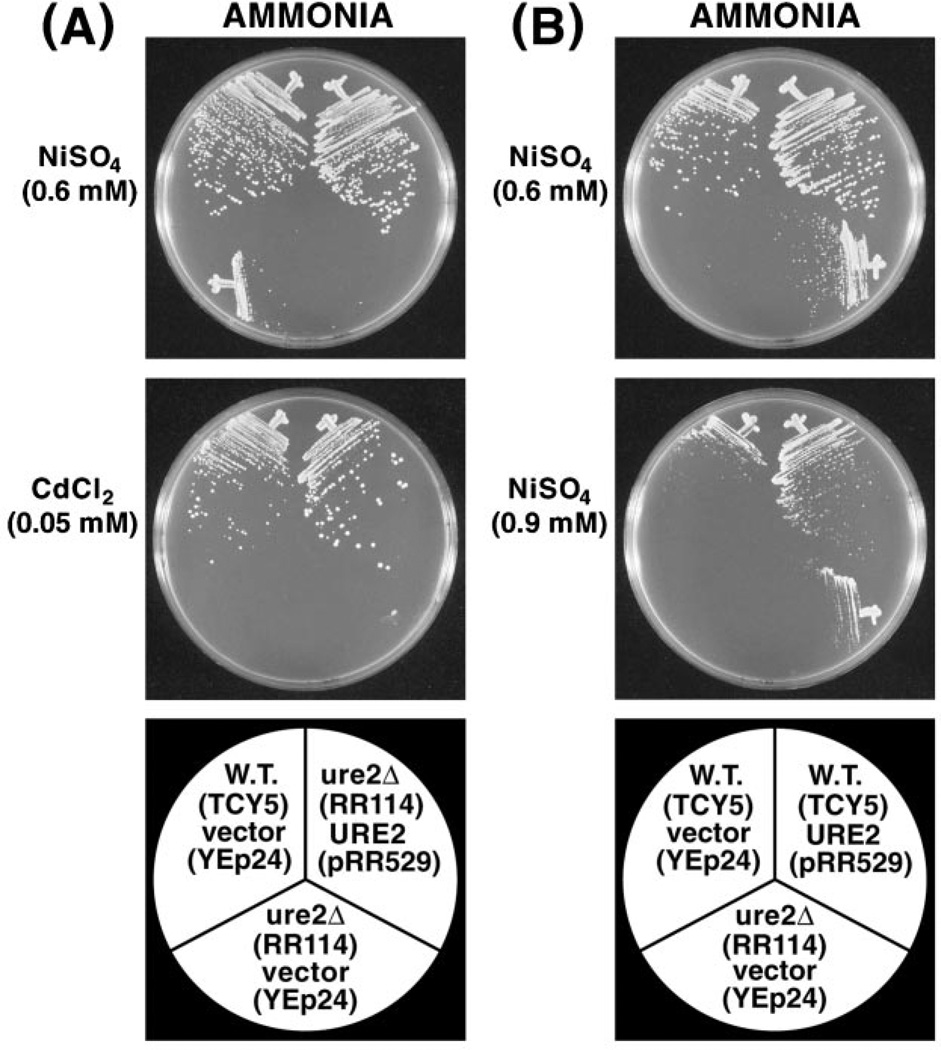
To test whether it was the ure2 mutation in TCY-5 that was specifically responsible for increased metal ion sensitivity, we complemented the mutation by transforming strains RR114 (ure2Δ) with pRR529 (URE2). TCY5 (wild type) transformed with vector YEp24 was the positive control. URE2 pRR529 restored the growth of strain RR114 in the presence of nickel or cadmium ions to wild type levels (Fig. 3A). If Ure2 is itself responsible for wild type detoxification of metal ions and is the limiting entity in metal ion detoxification, one might expect to see increased ability to cope with high levels of environmental metal ions when URE2 is overexpressed. As shown in Fig. 3B, overexpression of URE2 did not generate increased resistance to 0.6 mm nickel sulfate relative to wild type. However, when the nickel ion concentration was increased to 0.9 mm, overexpression of URE2 did result in detectably increased protection relative to the wild type strain transformed with vector YEp24.
Fig. 3. A, complementation of the ure2Δ mutation by plasmid-borne URE2.
Metal ions and nitrogen sources are indicated. B, effect of overexpressing URE2 on the sensitivity of wild type cells to 0.6 and 0.9 mm nickel sulfate. Bottom panels identify the strains and plasmids with which they were transformed. W.T., wild type.
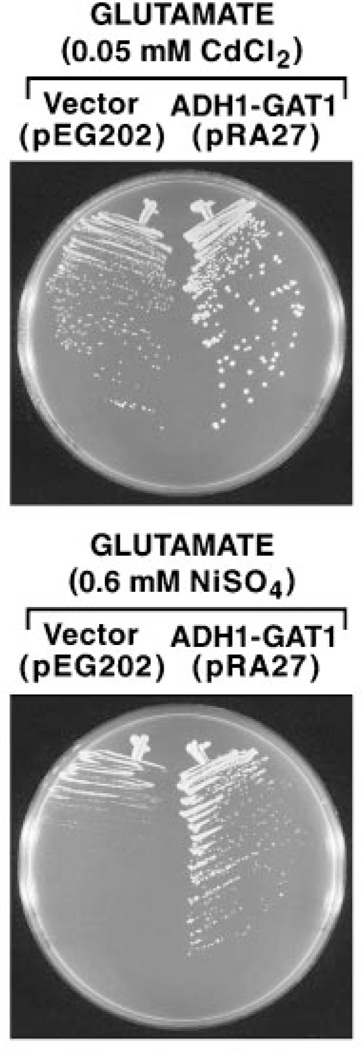
In studies of this nature, one of the most significant problems is to distinguish whether observed effects derive from direct or indirect participation of the gene product being tested. This is an especially serious consideration when that product is known, as Ure2 is, to negatively regulate NCR-sensitive, Gln3- and Gat1-mediated transcription in the presence of a good nitrogen source (31–33). By this reasoning, loss of metal ion detoxification in a ure2Δ mutant might derive indirectly from increased Gln3/Gat1-mediated transcription. To test this possibility, we transformed wild type strain STCY32 with ADH1-GAT1 pRA27 or vector control pEG202 and tested the metal ion sensitivity of the transformants. Overexpression of GAT1 in this way has been previously shown to result in both increased Gat1- and Gln3-mediated transcription and to render that transcription largely resistant to NCR (21, 34). In addition, we tested metal ion sensitivity with glutamate as sole nitrogen source rather than ammonia since glutamate generates much less NCR than ammonia in these strains, which in turn would further increase Gln3/Gat1-mediated gene expression. Rather than increasing metal ion sensitivity, as occurs when URE2 is deleted, overexpression of GAT1 resulted in greater nickel ion resistance than that observed when the wild type strain was transformed with control pEG202 (Fig. 4). The effect of GAT1 overexpression, although visible with cadmium ions, was less striking. When this experiment was repeated in strain BY4741, parent of the consortium-generated set of yeast gene deletion strains, pEG202- and pRA27-containing transformants grew indistinguishably in the presence of cadmium chloride or nickel sulfate (data not shown). These data and those in Fig. 4 argue against the possibility that metal ion hypersensitivity in a ure2Δ mutant results from its only demonstrated function, negatively regulating GATA factor-mediated transcription.
Fig. 4. Effect of overexpressing GAT1 on the sensitivity of wild type cells (STCY32) transformed with control YEp24 or ADH1-GAT1 pRA27 to cadmium and nickel ions.
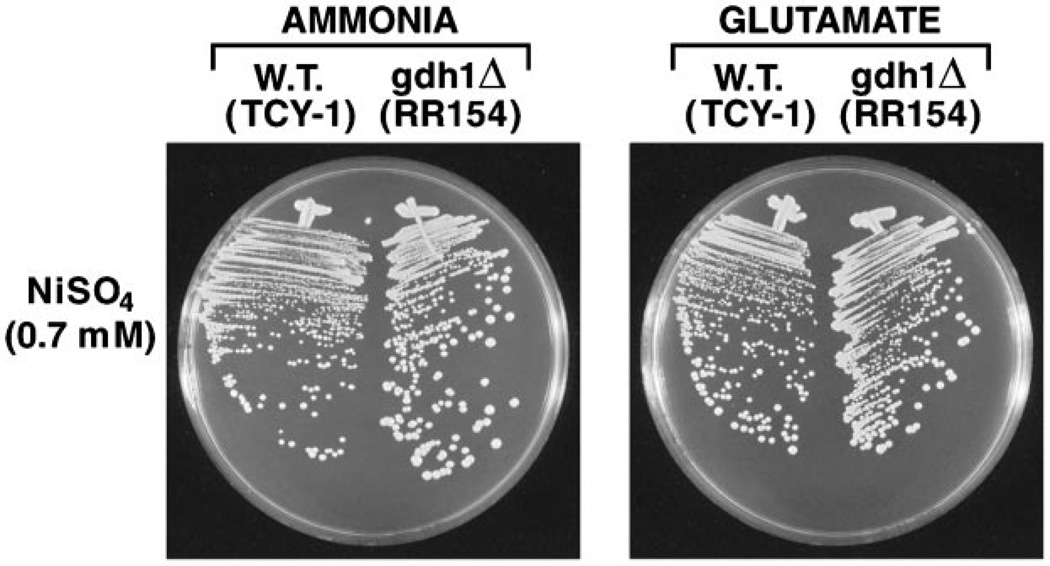
Since Ure2 regulation of Gln3/Gat1-mediated transcription influences expression of GDH1 and GDH2, another conceivable explanation of the Ure2 requirement for metal ion detoxification might be that it alters the level of reducing equivalents (NADPH) required to maintain glutathione in its active, reduced form. Although this explanation was circumstantially addressed in Fig. 1 by comparing metal ion sensitivity with glutamate versus ammonia as nitrogen source, it can be addressed more specifically by comparing metal ion sensitivity of wild type and gdh1Δ mutant cells. In gdh1Δ mutants, ammonia is assimilated without the consumption of NADPH because it occurs through the combined action of the GLN1 (glutamine synthetase) and GLT1 (glutamate synthase, GOGAT) gene products (35). As shown in Fig. 5, wild type and gdh1Δ strains are equally able to detoxify nickel ions. If anything, the gdh1Δ mutant may grow slightly better than wild type.
Fig. 5.
Nickel sulfate sensitivity of wild type and gdh1Δ mutant in minimal ammonia and minimal glutamate media. W.T., wild type.
Ure2 Potential Participation in Detoxification of Organic Xenobiotics and Hydrogen Peroxide
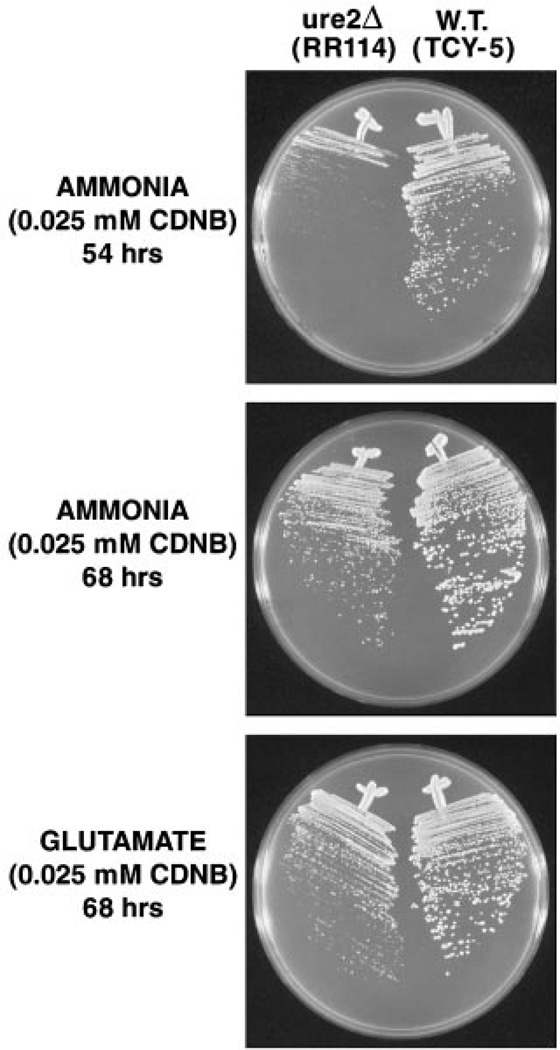
Most glutathione S-transferases exhibit rather broad substrate specificities, although clear substrate preferences exist. Given this characteristic and the fact that all reported in vitro glutathione S-transferase assays of Ure2 were performed using the chromogenic xenobiotic CDNB as the acceptor molecule (7, 20), we compared wild type and ure2Δ strain sensitivity to CDNB. As shown in Fig. 6, wild type colonies are larger than a ure2Δ mutant after 54 h of incubation in the presence of CDNB. At 68 h of incubation, the difference in growth between the two strains, although still apparent, is much less drastic. Similar results were observed with minimal glutamate medium (Fig. 6 and data not shown). However, the growth difference seen at 68 h is not convincingly different from that of wild type and ure2Δ strains growing in the absence of perturbant (Fig. 1, bottom two panels). Therefore, if loss of Ure2 generates CDNB hypersensitivity it is modest at best. This correlates well with the inabilities of multiple investigators, including ourselves, to detect in vitro glutathionation of CDNB.
Fig. 6. Sensitivity of wild type and ure2Δ cells to CDNB over time in minimal ammonia and minimal glutamate media.
W.T., wild type.
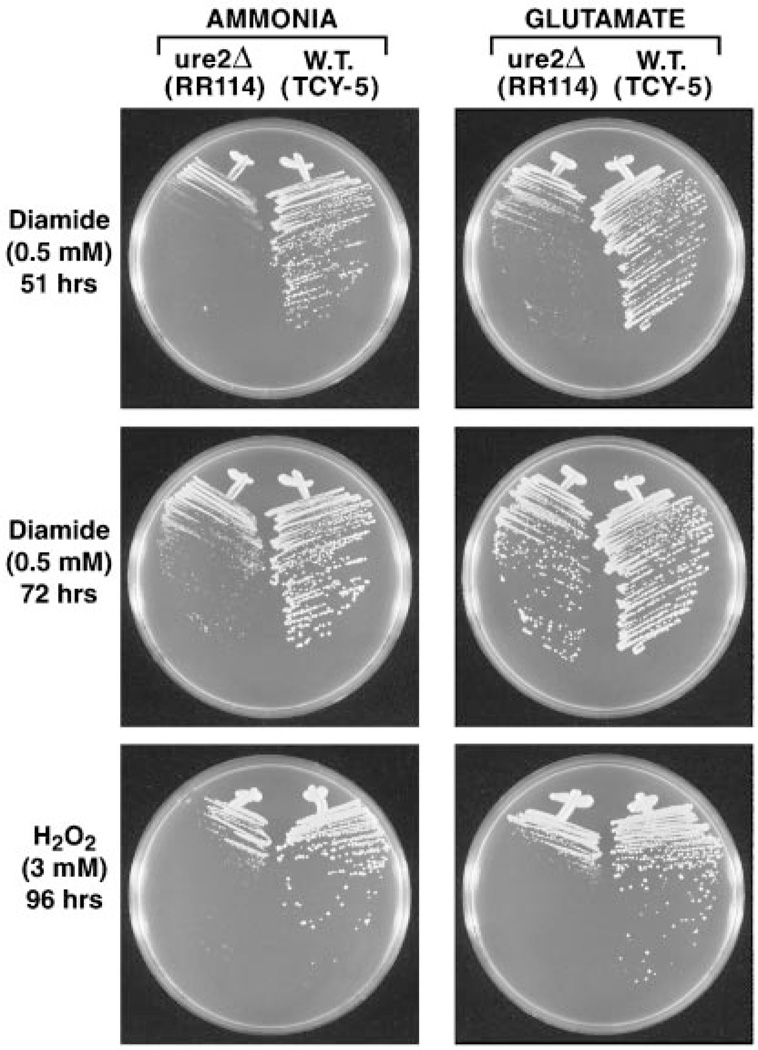
Mutants with defects in glutathione S-transferase genes are often found to exhibit increased sensitivity not only to preferred S-transferase substrates but also to compounds that are not detoxified by direct conjugation to glutathione. Two compounds in this category are hydrogen peroxide and diamide. Hydrogen peroxide is detoxified in two ways: by peroxidation, an activity found in some mammalian Theta-class GSTs and in S. pombe GST3 or by GSH conjugation of cellular products that are oxidized by hydrogen or other peroxides (25, 26, 28). Diamide also oxidizes cellular proteins and other constituents but, in addition, depletes the reduced glutathione pool because it is detoxified via glutathione-dependent reduction (36). ure2Δ colonies grown in the presence of diamide were smaller than wild type after 51 h of incubation in minimal glutamate or minimal ammonia medium (Fig. 7, top two panels). Differences in growth, however, were less marked after 72 h of incubation (Fig. 7, middle two panels). Sensitivity of the ure2Δ to diamide was greater than that observed for CDNB but less than that observed for nickel ions when ammonia was used as nitrogen source. In contrast, when glutamate was used as nitrogen source, differences in growth were not different from those seen in the bottom two panels of Fig. 1. It is important to note that diamide was the only perturbant in which different growth patterns were observed on glutamate versus ammonia medium. This difference correlates with the facts that (i) ammonia as- similation places a greater drain on the NADPH pool than does glutamate and (ii) the only cellular constituent used in detoxifying diamide is NADPH. Therefore, it is conceivable that apparent ure2Δ hypersensitivity to diamide derives indirectly from effects of the mutation on NADPH metabolism or alternatively that hypersensitivity can only be visualized at lower NADPH concentrations.
Fig. 7. Sensitivity of wild type and ure2Δ cells to diamide over time and hydrogen peroxide at 96 h in minimal ammonia and minimal glutamate media.
W.T., wild type.
Mutants lacking glutathione S-transferases have also been reported to become hypersensitive to hydrogen peroxide (25). Therefore, we compared hydrogen peroxide toxicity in wild type and ure2Δ strains. Hydrogen peroxide was more toxic to ure2Δ mutants than were any of the other non-metal ion perturbants (Fig. 7, bottom two panels). In contrast to what occurred with diamide, hypersensitivity to hydrogen peroxide exhibited by the ure2Δ strain was equivalent whether ammonia or glutamate was provided as nitrogen source. Hypersensitivity was also observed in hydrogen peroxide-containing YEPD medium (Fig. 2, bottom panel). In sum, ure2 mutations, although most sensitive to heavy metal ions, exhibit the same pleiotropic hypersensitivity to oxidants, hydrogen peroxide being most toxic, whose detoxification does not involve direct conjugation to glutathione as seen in gst1, gst2, and gst3 mutants of S. pombe (25).
Ure3 Is Capable of Protecting Cells from Heavy Metals
URE3 and ure2 mutants were originally isolated from the same selection (14, 15). Therefore, except for being a bit more leaky, it is not surprising that URE3 exhibits the same phenotype as ure2 mutations. Prior to this work, the only known ure2 phenotype was resistance to NCR (14, 15,31–33). Identification of a new ure2 phenotype prompts the question of whether URE3 strains possess a similar phenotype, as is the case for negative regulation of GATA factor-mediated transcription. The question is pertinent because there is a strong correlation between Ure2-Gln3 and Ure2-Gat1 complex formation and the ability of Ure2 to inhibit NCR-sensitive transcription in the presence of a good nitrogen source, i.e. the Ure2 regulatory activity appears to be associated with a stoichiometric reaction between Gln3 and itself. Since Ure2 in its Ure3 prion form is a polymer, it is not too surprising that it cannot simultaneously interact with itself and Gln3. Indeed portions of the Ure2 molecule that interact with Gln3 also participate in prion formation (37). If we assume that Ure2 does possess glutathione S-transferase activity, it may not be as adversely affected by Ure3 prion formation because this activity is catalytic rather than stoichiometric. Consistent with this possibility, glutathione has been reported to bind to the polymerized form of Ure2 (6).
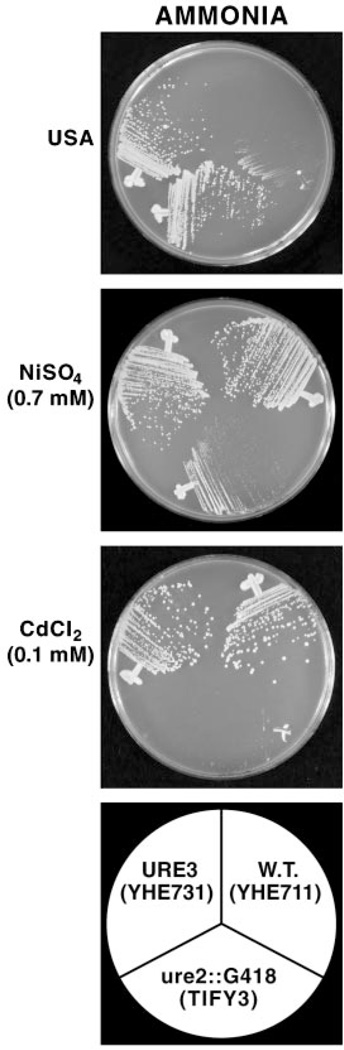
To answer the above question, we used three strains generously provided by Edskes and Wickner: YHE711 (wild type, ure-o), TIFY3 (ure2:G418), and YHE731 (strain YHE711 into which URE3 was cytoduced). Since Ure3 can be lost from some cells during storage in glycerol,2 we streaked out all three strains on YEPD and then scored the phenotypes of multiple isolates on glucose ammonia + ureidosuccinate (USA) medium. Wild type ure-o cells (all strains are ura2, a prerequisite of the plate assay) will not grow in this medium, whereas ure2 and ure3 mutants will. We indeed found that a few of the “ure3” isolates had become identical to the wild type, i.e. no longer able to grow on ammonia + USA medium. We assayed the metal ion sensitivity of seven randomly chosen URE3 clones using wild type and ure2 mutants as controls. As shown in Fig. 8, URE3 and ure2 clones grew similarly in glucose ammonia + USA medium but exhibited opposite phenotypes in the presence of metal ions. The URE3 clones were just as resistant as the wild type to both environmental insults. Note that there was some cross-feeding of the “wild type” (ura2) strain. This did not occur when the streaked cells were more distantly separated.
Fig. 8. Sensitivity of wild type, ure2Δ, and URE3 cells to nickel sulfate and cadmium chloride.
The top panel depicts growth of the three strains in medium supplemented with USA rather than uracil, which was used to supplement media depicted in the lower two panels. W.T., wild type.
Effect of Heavy Metals on URE2 and NCR-sensitive Transcription
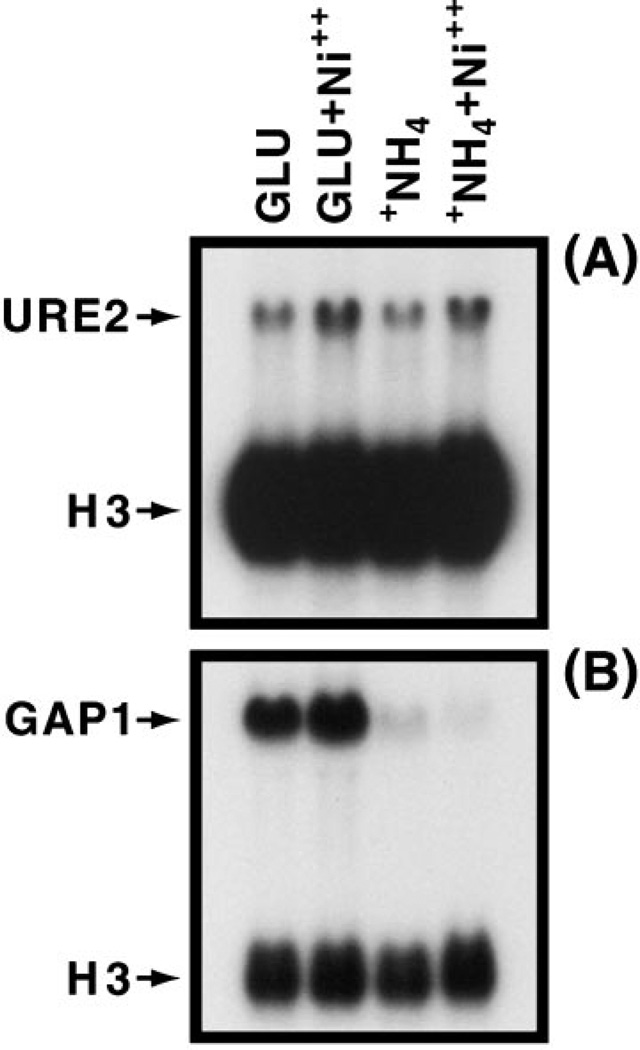
Given Ure2 regulation of NCR-sensitive expression, we determined whether heavy metal ion treatment affects URE2 expression or the ability of Ure2 to regulate GATA factor-mediated transcription. We used 0.6 mm nickel sulfate rather than cadmium chloride as the perturbant here because cadmium ions have such a drastic effect on cell growth. URE2 expression increased about 2-fold in minimal medium containing nickel sulfate (Fig. 9). Similar results were observed whether glutamate or ammonia was used as nitrogen source. Growth in the presence of nickel sulfate had little demonstrable effect on NCR-sensitive gene expression using GAP1 as the NCR-sensitive reporter gene. These data suggest that regulation of URE2 expression is unlikely to be induced at moderate levels of perturbant and that the protection of Ure2 cells from toxic compounds does not demonstrably diminish its ability to regulate NCR-sensitive gene expression.
Fig. 9. Steady state levels of URE2 and GAP1 mRNA in minimal glutamate medium in the presence or absence of 0.6 mm nickel sulfate.
Conditions were as described under “Materials and Methods.”
DISCUSSION
This work identifies a new function for Ure2 in S. cerevisiae, i.e. participation in heavy metal ion and oxidant detoxification. Deletion of URE2 results in hypersensitivity to cadmium ions, hydrogen peroxide, and, to a lesser extent, nickel ions. ure2 mutants are also slightly hypersensitive to diamide, a glutathione cycling reagent, but only when ammonia is used as sole nitrogen source. There is no convincing hypersensitivity to diamide when glutamate is used in place of ammonia, suggesting that hypersensitivity, if it exists, is seen only when NADPH pools are diminished. Our data also offer a possible explanation of negative results encountered in attempts to demonstrate Ure2-mediated glutathione S-transferase activity using CDNB as substrate. We found CDNB to be only slightly more toxic to ure2Δ cells than to wild type, suggesting that Ure2 may play only a peripheral role at best in its detoxification.
Primary sequence and three-dimensional homology between Ure2 and known glutathione S-transferases as well as the observed binding of glutathione and glutathione S-transferase substrates to crystallized Ure2 are consistent with the possibility that URE2 encodes a glutathione S-transferase. Our work provides additional support for that argument, i.e. the demonstration that Ure2 is required for metal ion detoxification and repair or prevention of perturbant-generated cellular damage in vivo. While present evidence argues in favor of Ure2 being a glutathione S-transferase, it does not unequivocally distinguish whether the ure2 mutant phenotype is a direct or indirect effect of the loss of the protein. Our data do, on the other hand, demonstrate that if the effect of Ure2 loss is indirect, it does not occur through the only known function of Ure2, i.e. negative regulation of Gln3 and Gat1. This possibility is eliminated by the observation that increasing Gln3- and Gat1- mediated transcription, the outcome of deleting URE2, does not increase sensitivity to metal ions but rather slightly decreases it or leaves it unaffected depending upon the strain tested.
Deletion of URE2 generates increased sensitivity to multiple compounds including heavy metal ions, strong oxidants (hydrogen peroxide), agents that deplete reduced glutathione pools through oxidation (diamide), and perhaps those depleting the glutathione pools per se through conjugation (CDNB). How- ever, the level of hypersensitivity generated by the ure2Δ varies quantitatively over a very wide range depending upon the perturbant tested. How can Ure2 be a direct participant in protecting cells from all of these compounds when the enzyme mechanisms involved in detoxifying these agents are to varying degrees different? Although this issue cannot be rigorously addressed in the absence of detailed in vitro studies, using purified proteins, varying hypersensitivity to a spectrum of compounds that are detoxified in different ways is a commonly seen phenotype of mutations in known glutathione S-transferase genes. Three glutathione S-transferases (encoded by GST1, GST2, and GST3) have been identified in S. pombe. Single mutants in each of these genes result in increased sensitivity to diamide and hydrogen peroxide. Moreover, overproduction of Gst1, Gst2, or Gst3 increased in vitro activity to catalyze the conjugation of CDNB and glutathione, while overproduction of only Gst3 increased glutathione peroxidase activity using cumene hydroperoxide as substrate (25, 38). Two glutathione S-transferase genes have been reported in S. cerevisiae, GTT1 and GTT2 (20). Both genes were shown to catalyze the glutathione S-transferase reaction using CDNB as substrate. When these investigators similarly assayed Ure2 (20), the results were negative just as they were for others and ourselves (data not shown). Finally, Hynes and his co-workers (39) cloned the gstA gene from Aspergillus nidulans and found its primary protein sequence to be most homologous to Ure2. Among the compounds to which a gstA deletion mutant was hypersensitive were nickel ions, selenium, diamide, and CDNB (39). However, in contrast to S. cerevisiae Ure2, the A. nidulans GstA protein does not participate in the regulation of GATA factor-mediated, nitrogen-responsive transcription.
The fact that URE2 expression increased only 2-fold in the presence of nickel sulfate suggests that cellular Ure2 levels are normally sufficient for it to function effectively in protecting the cell from heavy metal ions and strong oxidants. Consistent with this suggestion is the fact that overexpression of URE2 (Fig. 3) did not result in increased resistance to metal ions until their concentration had been raised substantially. That the regulation of NCR-sensitive GAP1 expression by Ure2 was not demonstrably affected by the presence of metal ions may derive from one or more of several factors. (i) Ure2 levels are sufficient for it to function in both detoxification and GATA factor regulation. (ii) The effects of Ure2 levels on NCR are subtle and hence below detection in the assay we used. (iii) If Ure2 functions both catalytically and stoichiometrically, it would be unlikely for the catalytic function to significantly interfere in the stoichiometric function. (iv) The participation of Ure2 in glutathione S-transferase functions is indirect. Resolving these issues will require a more detailed understanding of the biochemistry of Ure2.
Finally, in addition to identifying a new function for Ure2, this work has identified a phenotype that can be used along with growth in ammonia + USA medium to distinguish wild type, ure2, and URE3 alleles. URE3 strains are as resistant to nickel and cadmium ions as wild type, while ure2 mutants exhibit markedly increased sensitivity to these ions. Both ure2 and URE3 strains, on the other hand, are resistant to NCR, whereas the wild type is not. If Ure2 is a direct participant in enzyme reactions involving glutathione, the shared and distinguishable phenotypes of ure2 and URE3 mutations may derive from the physical requirements of the two processes assayed, one of which requires stoichiometric participation of Ure2, while for the other only catalytic participation is involved. This ability to distinguish Ure2 and Ure3 in vivo will prove highly useful in future studies of relationships between the two forms of this fascinating protein.
Acknowledgments
We thank Drs. H. Edskes and R. Wickner for strains, Dr. Kathleen Cox for many suggestions of pertinent literature, Tim Higgins for preparing the artwork, and the University of Tennessee Yeast Group for suggestions to improve the manuscript.
Footnotes
This work was supported by National Institutes of Health Grant GM-35642.
The abbreviations used are: NCR, nitrogen catabolite repression; CDNB, 1-chloro-2,4-dinitrobenzene; GST, glutathione S-transferase; USA, ureidosuccinate.
H. Edskes, personal communication.
REFERENCES
- 1.Zou K, Gong JS, Yanagisawa K, Michikawa M. J. Neurosci. 2002;22:4833–4841. doi: 10.1523/JNEUROSCI.22-12-04833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner RG. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.Masison DC, Maddelein ML, Wickner RB. Proc. Natl. Acad. Sci. U. S. A. 1997;94:12503–12508. doi: 10.1073/pnas.94.23.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 5.Thual C, Bousset L, Komar AA, Walter S, Buchner J, Cullin C, Melki R. Biochemistry. 2001;40:1764–1773. doi: 10.1021/bi001916l. [DOI] [PubMed] [Google Scholar]
- 6.Bousset L, Thomson NH, Radford SE, Melki R. EMBO J. 2002;21:2903–2911. doi: 10.1093/emboj/cdf303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perrett S, Freeman SJ, Butler PJ, Fersht AR. J. Mol. Biol. 1999;290:331–345. doi: 10.1006/jmbi.1999.2872. [DOI] [PubMed] [Google Scholar]
- 8.Edskes HK, Wickner RB. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6625–6629. doi: 10.1073/pnas.120168697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierce MM, Maddelein ML, Roberts BT, Wickner RB. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13213–13218. doi: 10.1073/pnas.181486098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edskes HK, Hanover JA, Wickner RB. Genetics. 1999;153:585–594. doi: 10.1093/genetics/153.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekito T, Liu Z, Thornton J, Butow RA. Mol. Biol. Cell. 2002;13:795–804. doi: 10.1091/mbc.01-09-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tate JJ, Cox KH, Rai R, Cooper TG. J. Biol. Chem. 2002;277:20477–20482. doi: 10.1074/jbc.M200962200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dilova I, Chen CY, Powers T. Curr. Biol. 2002;12:389–395. doi: 10.1016/s0960-9822(02)00677-2. [DOI] [PubMed] [Google Scholar]
- 14.Lacroute F. J. Bacteriol. 1968;95:824–832. doi: 10.1128/jb.95.3.824-832.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drillien R, Lacroute F. J. Bacteriol. 1972;109:203–208. doi: 10.1128/jb.109.1.203-208.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coschigano PW, Magasanik B. Mol. Cell. Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Umland TC, Taylor KL, Rhee S, Wickner RB, Davies DR. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1459–1464. doi: 10.1073/pnas.041607898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bousset L, Belrhali H, Janin J, Melki R, Morera S. Structure (Camb.) 2001;9:39–46. doi: 10.1016/s0969-2126(00)00553-0. [DOI] [PubMed] [Google Scholar]
- 19.Bousset L, Belrhali H, Melki R, Morera S. Biochemistry. 2001;40:13564–13573. doi: 10.1021/bi011007b. [DOI] [PubMed] [Google Scholar]
- 20.Choi JH, Lou W, Vancura A. J. Biol. Chem. 1998;273:29915–29922. doi: 10.1074/jbc.273.45.29915. [DOI] [PubMed] [Google Scholar]
- 21.Cunningham TS, Andhare R, Cooper TG. J. Biol. Chem. 2000;275:14408–14414. doi: 10.1074/jbc.275.19.14408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham TS, Svetlov VV, Rai R, Smart W, Cooper TG. J. Bacteriol. 1996;178:3470–3479. doi: 10.1128/jb.178.12.3470-3479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriyama H, Edskes HK, Wickner RB. Mol. Cell. Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cox KH, Pinchak AB, Cooper TG. Yeast. 1999;15:703–713. doi: 10.1002/(SICI)1097-0061(19990615)15:8<703::AID-YEA413>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Veal EA, Toonem WM, Jones N, Morgan BA. J. Biol. Chem. 2002;277:35523–35531. doi: 10.1074/jbc.M111548200. [DOI] [PubMed] [Google Scholar]
- 26.Tan KL, Board PG. Biochem. J. 1996;315:727–732. doi: 10.1042/bj3150727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherratt PJ, Pulford DJ, Harrison DJ, Green T, Hayes JD. Biochem. J. 1997;326:837–846. doi: 10.1042/bj3260837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hubatsch I, Ridderstrom M, Mannervik B. Biochem. J. 1998;330:175–179. doi: 10.1042/bj3300175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li ZS, Lu YP, Zhen RG, Szczypka M, Thiele DJ, Rea PA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li ZS, Szczypka M, Lu YP, Thiele DJ, Rea PA. J. Biol. Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman-Bang J. Mol. Biotechnol. 1999;12:35–73. doi: 10.1385/MB:12:1:35. [DOI] [PubMed] [Google Scholar]
- 32.Cooper TG. FEMS Microbiol. Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magasanik B. In: The Molecular and Cellular Biology of the Yeast Saccharomyces—Gene Expression. Jones W, Pringle JR, Broach JR, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 283–318. [Google Scholar]
- 34.Cunningham TS, Rai R, Cooper TG. J. Bacteriol. 2000;182:6584–6591. doi: 10.1128/jb.182.23.6584-6591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avendano A, Deluna A, Olivera H, Valenzuela L, Gonzalez A. J. Bacteriol. 1997;179:5594–5597. doi: 10.1128/jb.179.17.5594-5597.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosower NS, Kosower EM. Methods Enzymol. 1987;143:264–270. doi: 10.1016/0076-6879(87)43050-4. [DOI] [PubMed] [Google Scholar]
- 37.Maddelein ML, Wickner RB. Mol. Cell. Biol. 1999;19:4516–4524. doi: 10.1128/mcb.19.6.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin YH, Park EH, Fuchs JA, Lim CJ. Biochim. Biophys. Acta. 2002;1577:164–170. doi: 10.1016/s0167-4781(02)00422-0. [DOI] [PubMed] [Google Scholar]
- 39.Fraser JA, Davis MA, Hynes MJ. Appl. Environ. Microbiol. 2002;68:2802–2808. doi: 10.1128/AEM.68.6.2802-2808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]