The mitotic checkpoint protein Spindly is farnesylated in vivo and this modification is required for its interaction with the RZZ complex and its localization to kinetochores.
Abstract
Kinetochore (KT) localization of mitotic checkpoint proteins is essential for their function during mitosis. hSpindly KT localization is dependent on the RZZ complex and hSpindly recruits the dynein–dynactin complex to KTs during mitosis, but the mechanism of hSpindly KT recruitment is unknown. Through domain-mapping studies we characterized the KT localization domain of hSpindly and discovered it undergoes farnesylation at the C-terminal cysteine residue. The N-terminal 293 residues of hSpindly are dispensable for its KT localization. Inhibition of farnesylation using a farnesyl transferase inhibitor (FTI) abrogated hSpindly KT localization without affecting RZZ complex, CENP-E, and CENP-F KT localization. We showed that hSpindly is farnesylated in vivo and farnesylation is essential for its interaction with the RZZ complex and hence KT localization. FTI treatment and hSpindly knockdown displayed the same mitotic phenotypes, indicating that hSpindly is a key FTI target in mitosis. Our data show a novel role of lipidation in targeting a checkpoint protein to KTs through protein–protein interaction.
Introduction
Accurate chromosome segregation during mitosis is essential for the maintenance of genomic stability. The mitotic checkpoint is a molecular mechanism that prevents premature segregation until all chromosomes are bioriented and aligned at the metaphase plate. Mitotic checkpoint proteins were first identified in budding yeast (Hoyt et al., 1991; Li and Murray, 1991; Weiss and Winey, 1996) and are conserved from yeast to human (Chan et al., 2005). Mitotic checkpoint proteins assemble at kinetochores (KTs) during mitosis and include Mad1, Mad2, Bub1, BubR1, Bub3, and Mps1 proteins. The RZZ complex (Roughdeal, ZesteWhite10, and Zwilch) subunits are essential mitotic checkpoint proteins originally identified in flies and are conserved in metazoans (Karess, 2005). The RZZ complex is required for Mad1 and Mad2 KT recruitment, and also recruits hSpindly to KTs (Fang et al., 1998; Buffin et al., 2005; De Antoni et al., 2005; Kops et al., 2005). hSpindly plays a critical role in checkpoint silencing by recruiting the dynein–dynactin motor complex that transports checkpoint proteins, such as Mad1, Mad2, RZZ complex, and hSpindly, from KTs to spindle poles (Howell et al., 2001; Gassmann et al., 2008; Chan et al., 2009; Barisic et al., 2010; Famulski et al., 2011).
hSpindly is a 605-aa protein consisting of two coiled coil domains separated by a conserved 32-aa spindly motif (Griffis et al., 2007; Chan et al., 2009). Spindly was discovered to be a regulator of dynein at KTs during mitosis in Drosophila melanogaster and is also involved in chromosome alignment and mitotic checkpoint silencing in human cells (Griffis et al., 2007; Chan et al., 2009; Barisic et al., 2010; Gassmann et al., 2010). Spindly KT localization is dependent on the RZZ complex because knockdown of Zw10 causes abrogation of Spindly KT localization (Chan et al., 2009; Barisic and Geley, 2011). Knockdown of hSpindly causes chromosome alignment defects, loss of dynein–dynactin KT localization, and prometaphase delay (Gassmann et al., 2008; Chan et al., 2009; Barisic et al., 2010). hSpindly C-terminal residues were previously shown to be important for KT localization and it is speculated that hSpindly undergoes farnesylation; a posttranslational lipid modification (Barisic et al., 2010).
Farnesylation is a type of protein prenylation, where a 15-carbon farnesyl lipid group is transferred onto one or more C-terminal cysteine residues (Zhang and Casey, 1996). A subset of membrane proteins is farnesylated, making the C terminus more hydrophobic, facilitating their membrane binding. A typical farnesylation motif, CAAX, has a C-terminal cysteine that becomes farnesylated, usually followed by two aliphatic amino acids, and the last amino acid is typically methionine, serine, glutamine, or alanine (Sinensky, 2000). It is estimated that >100 proteins undergo farnesylation including two KT proteins, centromere protein (CENP) E and CENP-F (Ashar et al., 2000; Wright and Philips, 2006). RAS family proteins require farnesylation for membrane binding and, because RAS is mutated in a wide variety of cancers, many farnesyl transferase inhibitors (FTIs) have been developed to inhibit Ras farnesylation (Downward, 2003; Karnoub and Weinberg, 2008; Berndt et al., 2011). FTIs efficiently killed tumor cells in culture and in animal models regardless of RAS mutations, suggesting additional unknown farnesylated targets (Nagasu et al., 1995; Sepp-Lorenzino et al., 1995; Crespo et al., 2002). Interestingly, in addition to G1 arrest, FTI-treated tumor cells exhibited prometaphase delay, defective spindle formation, and chromosome misalignments (Ashar et al., 2000; Crespo et al., 2001, 2002). These mitotic defects have been correlated with the inhibition of CENP-E and CENP-F farnesylation (Ashar et al., 2000; Hussein and Taylor, 2002; Schafer-Hales et al., 2007). Studies have shown, however, that FTIs do not affect CENP-E or CENP-F KT localization and it has been hypothesized that mitotic effects of FTIs are caused by unknown targets (Crespo et al., 2001, 2002; Verstraeten et al., 2011). This speculation is further supported by the data that loss of CENP-F function by siRNA in HeLa cells leads to a very brief mitotic delay rather than prometaphase accumulation as seen with FTI treatment (Feng et al., 2006). In addition, siRNA knockdown of CENP-E or microinjection of function-blocking antibodies resulted in unaligned chromosomes at spindle poles in metaphase cells and resembles a metaphase arrest rather than the gross chromosome alignment defect and prometaphase delay observed upon FTI treatment (Schaar et al., 1997; Yao et al., 2000; McEwen et al., 2001; Tanudji et al., 2004). Existence of conflicting data regarding the role of farnesylation in targeting CENP-E and CENP-F proteins to KTs warrants further investigation into how FTIs induce prometaphase delay in tumor cells.
Here we identified hSpindly, a mitotic checkpoint protein, as a novel farnesylation substrate. We defined the hSpindly KT localization domain and reported that farnesylation of hSpindly is required for its interaction with the RZZ complex and its KT localization. Furthermore, we showed that loss of hSpindly KT localization after FTI treatment is likely responsible for the prometaphase delay observed in FTI-treated tumor cells rather than CENP-E and CENP-F as reported previously.
Results
hSpindly KT localization is dependent on its C terminus
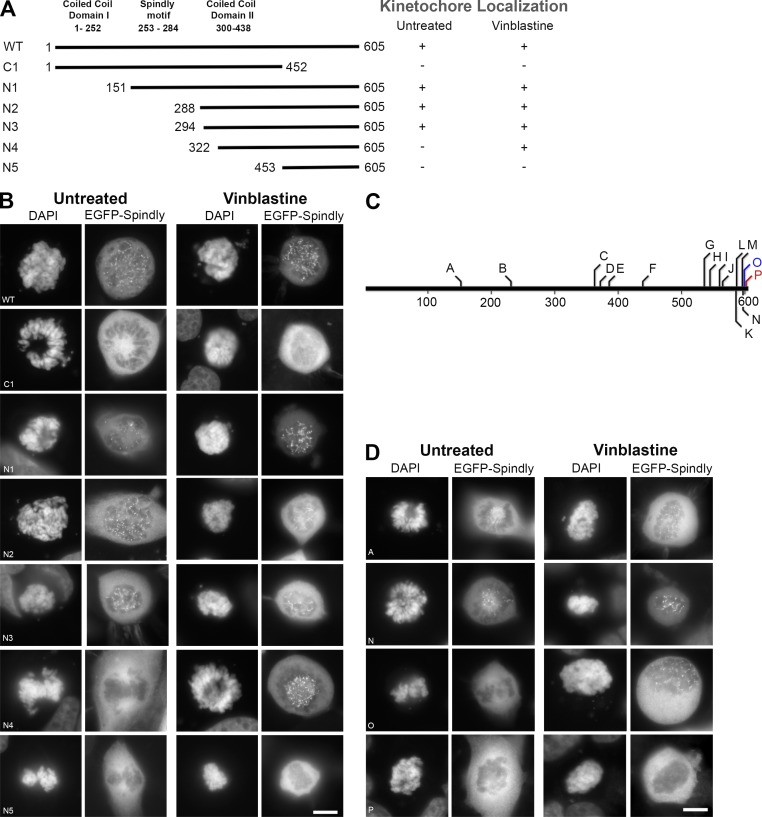
hSpindly localizes to KTs in prophase and prometaphase and accumulates at spindle poles during metaphase in HeLa cells consistent with previous immunofluorescence studies (Fig. S1 A; Chan et al., 2009). EGFP-fused N- and C-terminal truncation constructs of hSpindly were generated and expression of these constructs was confirmed in HEK293T cells by immunoblots (Figs. 1 A and S1 B). Analysis of transfected prometaphase HeLa cells revealed that the 453–605 aa (construct C1) are required for KT localization but the N-terminal 293 aa (construct N3) are dispensable (Fig. 1, A and B). N-terminal deletion construct N5 did not localize and N4 (322-605 aa) localized to KTs only when transfected cells were treated with vinblastine. This microtubule poison disrupts KT–microtubule attachments and maximally enhances checkpoint activity allowing for stringent analysis of KT localization capability of hSpindly mutants (Hoffman et al., 2001; Famulski et al., 2008). The N4 mutant perhaps has very low KT binding affinity (or fast turnover) and is rapidly transported off to spindle poles by dynein (see Discussion for detail).
Figure 1.
hSpindly C-terminal is required for KT localization. (A) A schematic diagram of hSpindly depicting truncation mutants (+, KT localization; −, no KT localization). KT localizing capability of each construct was analyzed under vinblastine treatment, which maximally loads checkpoint proteins on KTs. Amino acid numbers are indicated. (B) HeLa cells were transiently transfected with EGFP-hSpindly fusion constructs (shown in A) and KT localization ability of each construct was analyzed using fluorescence microscopy. DAPI stains chromosomes. Representative images show that the C-terminal 294 to 605 aa of hSpindly are required for KT localization. Bar, 10 µm. (C) A schematic diagram depicting the location of the insertion mutants generated in the hSpindly protein. Constructs shown in red were negative for KT localization and constructs shown in blue localized to KTs only under vinblastine treatment. Site of insertion and inserted residues are shown in Table 1. (1D) HeLa cells transiently transfected with EGFP-hSpindly insertion constructs were analyzed for their KT localization ability. Representative images show that the far C terminus is essential for KT localization. Bar, 10 µm.
To further define the KT localization domain, a transposon-based random insertion mutant library of hSpindly was generated (Table 1). hSpindly mutants containing 5-aa in-frame insertions, spanning 151 to 603 aa residues were screened for KT localization (Fig. 1, C and D; Fig. S1, C and D; and Fig. S2 A). Surprisingly, construct P with an insertion after 603 aa (P603–CGRSP) was the only mutant that completely abrogated KT localization highlighting the importance of the far C-terminal residues (Fig. 1 D). In addition, construct O with an insertion after 600 aa (T600–QLRPH) localized to KTs only when the cells were treated with vinblastine, indicating that residues immediately adjacent to the C terminus are also important (Fig. 1 D; see Discussion for detail).
Table 1.
hSpindly random insertion mutants show that the far C terminus is required for kinetochore localization
| Construct | Insertion site and sequence (amino acid) | Kinetochore localization untreated | Kinetochore localization vinblastine |
| A | M151SAAAM | Positive | Positive |
| B | R230 VCGRS | Positive | Positive |
| C | N364TYAAA | Positive | Positive |
| D | M373NAAAM | Positive | Positive |
| E | G387ECGRS | Positive | Positive |
| F | E441CGRTE | Positive | Positive |
| G | G537CGRIG | Positive | Positive |
| H | S546AAALS | Positive | Positive |
| I | E561CGRTE | Positive | Positive |
| J | T566AAAQT | Positive | Positive |
| K | H587CGRTH | Positive | Positive |
| L | P588MRPHP | Positive | Positive |
| M | T597PVRPH | Positive | Positive |
| N | P598VRPHP | Positive | Positive |
| O | T600QLRPH | Negative | Positive |
| P | P603CGRSP | Negative | Negative |
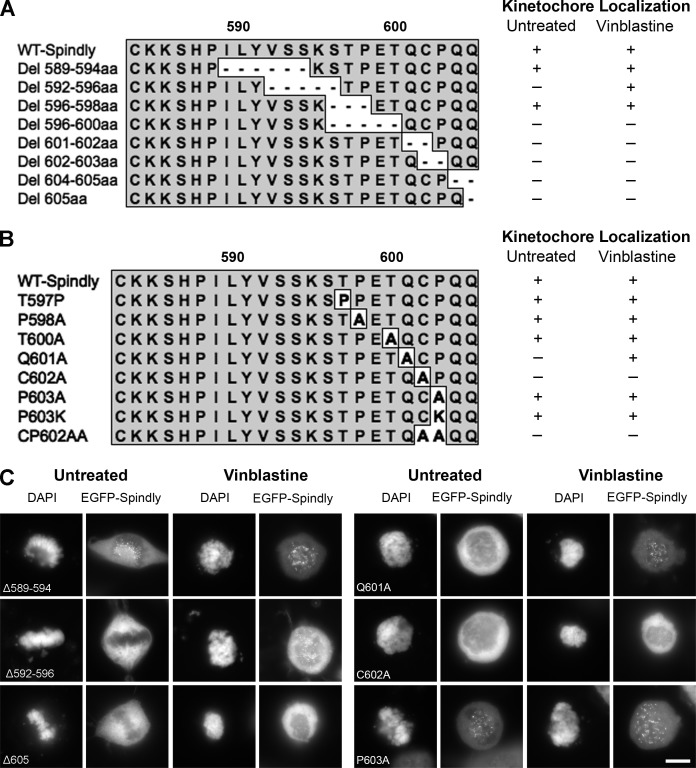
Site-directed mutagenesis was performed to create both deletion and substitution hSpindly mutants to assess the importance of individual C-terminal residues for KT localization. We found that no deletions in the last 10 aa (596–605 aa) were tolerated for KT localization (Fig. 2, A and C; and Fig. S1 F), but all mutants with substitutions in the far C-terminal residues except cysteine (602 aa) localized to KTs (Fig. 2, B and C; and Fig. S1 E). Based on deletion and substitution mutants (Fig. 2 and Fig. S2, B and C), we can conclude that the far C-terminal residues from 596 to 605 aa are critical for KT localization and the cysteine residue (602 aa) is essential for KT localization. Our KT domain mapping data indicate that the N-terminal 293 aa are dispensable for KT targeting and residues 293–322 contribute to high affinity binding.
Figure 2.
Far C-terminal residues of hSpindly are essential for KT localization. (A) Schematic representation of hSpindly C-terminal deletion mutants (589–605 aa). KT localization of each EGFP-tagged mutant was analyzed through fluorescence microscopy (+, KT localization; −, no KT localization). (B) Schematic representation of hSpindly C-terminal substitution mutants (597–605 aa). KT localization of each EGFP-tagged mutant was analyzed through fluorescence microscopy. (C) Representative images of HeLa cells transfected with EGFP-tagged hSpindly deletion or substitution constructs show that the C terminus of hSpindly is essential for its KT localization. Bar, 10 µm
FTI treatment abrogated hSpindly KT localization without affecting the RZZ complex, CENP-E, and CENP-F KT localization
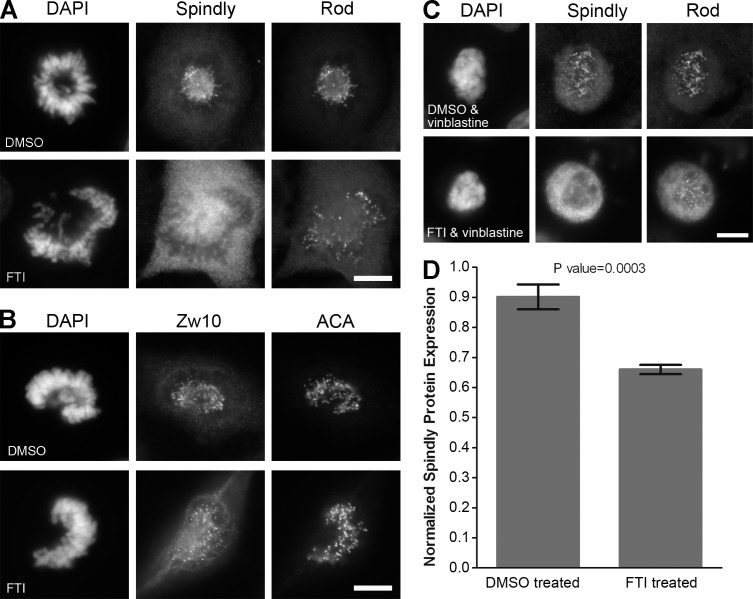
The hSpindly C terminus (CPQQ) is a potential farnesylation site and is highly conserved from zebrafish to humans (Fig. S3 A). To investigate if farnesylation plays a role in targeting hSpindly to KTs, we examined the effect of a previously validated FTI, L-744-832 (Verstraeten et al., 2011), on KT localization of hSpindly along with Zw10 and Rod because the RZZ complex is required for targeting hSpindly to KTs (Chan et al., 2009; Barisic et al., 2010). FTI treatment of HeLa cells led to a complete loss of hSpindly KT localization although Zw10 and Rod localized to KTs normally, indicating that farnesylation is involved in hSpindly KT localization (Fig. 3, A and B). Vinblastine added to FTI treatment also displayed no hSpindly KT localization (Fig. 3 C), indicating that farnesylation is fundamental to hSpindly KT localization and its absence is not a result of rapid dynein-mediated transport. To eliminate the possibility that loss of hSpindly KT localization is a compound-specific effect, we treated HeLa cells with another FTI (FTI-277; Schafer-Hales et al., 2007) and observed identical results (unpublished data). We observed loss of hSpindly KT localization with FTI treatment in breast cancer (MCF7 and T47D) and melanoma cell lines (M2 and MeWo), confirming that the loss of hSpindly KT localization with FTI treatment is not cell line specific (Fig. S3 B). hSpindly protein levels were determined to examine whether the loss of hSpindly KT localization is caused by its degradation in the absence of farnesylation. Immunoblots of FTI- and DMSO-treated HeLa cell lysate showed an ∼25% decrease in hSpindly protein expression with FTI treatment (Fig. 3 D), which does not account for the complete loss of hSpindly KT localization.
Figure 3.
Inhibition of farnesylation abrogates KT localization of hSpindly but not the RZZ complex. (A and B) HeLa cells were treated with 10 µM of L744832 FTI or DMSO for 24 h, fixed, and immunostained for hSpindly, Rod, and Zw10. Complete loss of hSpindly KT localization and normal Rod and Zw10 levels on the KTs were observed in FTI-treated cells. ACA immunostaining labels centromeres and DAPI stains DNA. (C) HeLa cells were treated with FTI or DMSO for 24 h and incubated with 0.5 µM vinblastine for 30 min before harvesting, showing no hSpindly KT localization without affecting Rod KT localization. (D) Normalized hSpindly protein expression from HeLa cells show 25% less expression with FTI treatment as compared with DMSO. n = 3. Error bars are SD from the means. P-value indicated a significant difference. Bars, 10 µm.
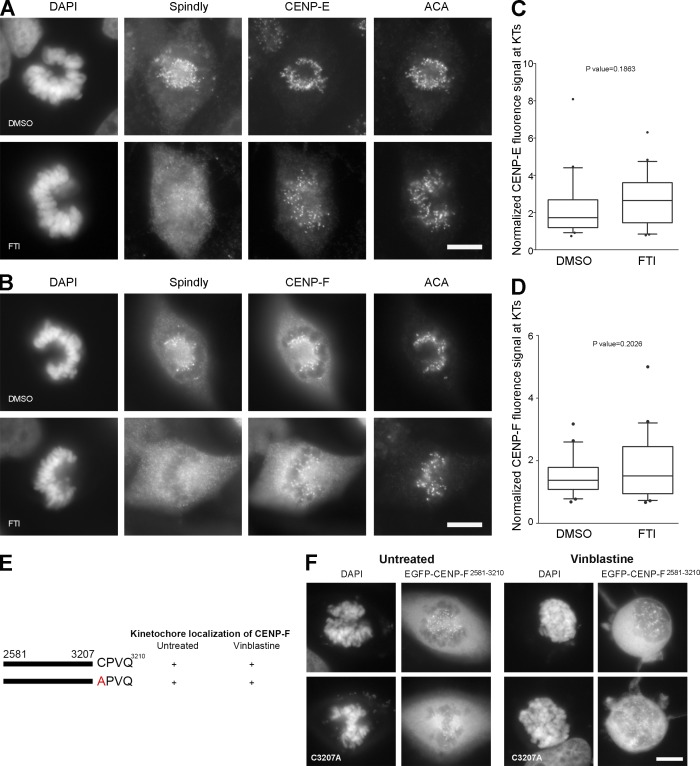
Previous studies have reported conflicting findings regarding whether loss of farnesylation prevents CENP-E and CENP-F KT localization (Ashar et al., 2000; Crespo et al., 2001, 2002; Hussein and Taylor, 2002; Schafer-Hales et al., 2007; Verstraeten et al., 2011). We observed no effect on CENP-E and CENP-F KT localization in HeLa cells with FTI treatment in prometaphase (Fig. 4, A and B) or metaphase (not depicted). Quantitative immunofluorescence microscopy of prometaphase HeLa cells showed no significant difference in CENP-E and CENP-F KT localization with FTI treatment compared with DMSO (Fig. 4, C and D). A cysteine to alanine mutant of CENP-F (C3207A of CAAX motif) was reported to be unable to localize to KTs (Hussein and Taylor, 2002). We cloned the C-terminal 630-aa KT-localizing fragment of CENP-F (2581–3210 aa), found it localized to KTs as expected (Fig. 4, E and F, top; Zhu et al., 1995; Hussein and Taylor, 2002), and generated the CENP-F C3207A mutant (Fig. 4 E and Fig. S1 H). In contrast to the Hussein and Taylor (2002) study, the CENP-F C3207A mutant localized to KTs during mitosis (Fig. 4 F, bottom) but was reproducibly reduced in vinblastine-treated cells for unknown reasons. These results indicate that FTI-mediated mitotic effects cannot be attributed to the loss of CENP-E or CENP-F KT localization.
Figure 4.
Inhibition of farnesylation does not affect CENP-E and CENP-F KT localization. (A and B) HeLa cells were treated with 10 µM of L744832 FTI or DMSO for 24 h, fixed, and immunostained for hSpindly, CENP-E or CENP-F, and ACA. CENP-E and CENP-F localized to KTs in FTI-treated HeLa cells, whereas hSpindly did not. (C and D) Normalized fluorescence signals for CENP-E and CENP-F at KTs with 24-h DMSO or FTI treatment. n = 20. Boxes represent interquartile distributions and whiskers represent 10th and 90th percentiles. FTI-treated cells have slight but not significant increase in CENP-E or CENP-F signal as compared with control DMSO as indicated by p-value. (E) A schematic diagram of hCENP-F depicting the minimal KT localization domain (2581–3210 aa) as previously shown by other studies and the C3207A mutation in the C-terminal farnesylation motif (+, KT localization; −, no KT localization). (F) HeLa cells were transiently transfected with EGFP-hCENP-F fusion constructs (shown in Fig. 3 E) and KT localization ability of each construct was analyzed using fluorescence microscopy. Top panel is EGFP-CENP-F2581-3210 and bottom panel is C3207A mutant of the same fragment. The images show that the farnesylation motif of CENP-F is not required for KT localization. Bars, 10 µm.
hSpindly CPQQ motif can be substituted with other farnesylation motifs
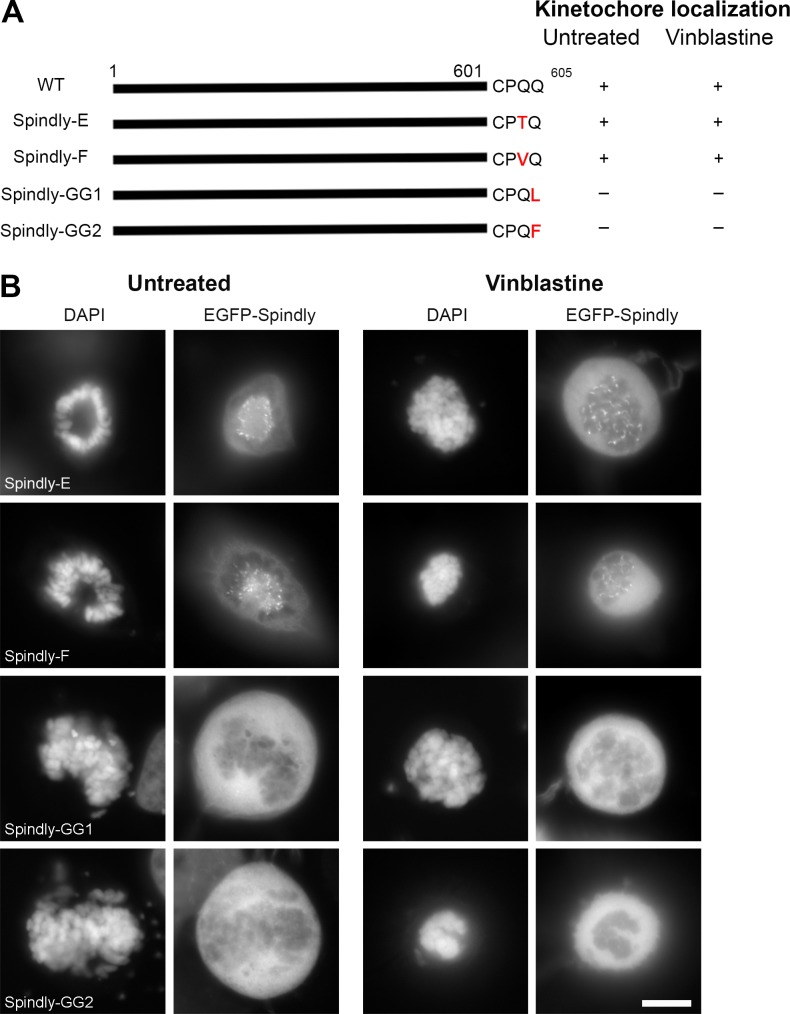
Because the C terminus of hSpindly is required for KT localization through the CAAX farnesylation motif, we hypothesized that any farnesylation motif on the C terminus of hSpindly regardless of the specific residues is sufficient for it to undergo farnesylation and target it to KTs. We replaced the CPQQ residues of hSpindly with the CKTQ (CENP-E) and CKVQ (CENP-F) farnesylation motifs, referred to as Spindly-E and Spindly-F, respectively. Both Spindly-E and -F localized to KTs during prometaphase, supporting our hypothesis that the exact amino acid sequence is not important and the ability to be farnesylated is sufficient for hSpindly KT localization (Fig. 5, A and B; and Fig. S1 G).
Figure 5.
hSpindly farnesylation motif can be substituted with CENP-E or CENP-F farnesylation motif but not a geranylgeranylation motif. (A) A schematic diagram of hSpindly depicting substitution of its farnesylation motif with CENP-E and CENP-F, referred to as Spindly-E and Spindly-F, respectively. The last amino acid of Spindly is changed such that it is a geranylgeranylation motif instead of farnesylation motif referred to as Spindly-GG1 and Spindly-GG2. The KT localization of each construct is shown (+, KT localization; −, non-KT localization). Amino acid numbers are indicated. (B) HeLa cells were transiently transfected with EGFP-hSpindly fusion constructs (shown in A) and KT localization ability of each construct was analyzed using fluorescence microscopy. Representative images show that the farnesylation motif of Spindly can be replaced with the farnesylation motif of known farnesylated proteins CENP-E and CENP-F, but a geranylgeranylation motif cannot target hSpindly to KTs. Bar, 10 µm.
Geranylgeranylation is a prenylation catalyzed by geranylgeranyl transferase enzymes that attaches a 20-carbon geranylgeranyl group to the cysteine of the C-terminal CAAX box (Sinensky, 2000). The nature of the X residue in the CAAX motif determines whether a protein gets farnesylated or geranylgeranylated. Geranylgeranyl transferase enzymes prefer X to be leucine or isoleucine (Zhang and Casey, 1996). However, these rules are not absolute because proteins with phenylalanine at the X position can undergo farnesylation or geranylgeranylation (Carboni et al., 1995). Several studies have demonstrated that when farnesylation is inhibited, KRAS4B and NRAS become geranylgeranylated and are targeted to the membrane, thus retaining their function (Lerner et al., 1997; Rowell et al., 1997; Whyte et al., 1997). Because FTIs inhibit hSpindly KT localization, we predict it cannot inherently be geranylgeranylated to retain its function in the presence of FTIs. We investigated whether a geranylgeranylation motif on the hSpindly C terminus can target it to KTs, assuming it undergoes geranylgeranylation, by replacing the last glutamine of hSpindly with leucine or phenylalanine (referred to as Spindly-GG1 and Spindly-GG2), converting it from a farnesylation substrate to a geranylgeranylation substrate (Fig. 5 A and Fig. S1 G). These constructs did not localize to KTs during mitosis in HeLa cells (Fig. 5 B), demonstrating that a geranylgeranylation motif on hSpindly cannot functionally substitute for the farnesylation motif making hSpindly a potential clinical target for FTIs unlike RAS proteins.
hSpindly is farnesylated on its C-terminal cysteine residue and farnesylation is essential for its interaction with the RZZ complex
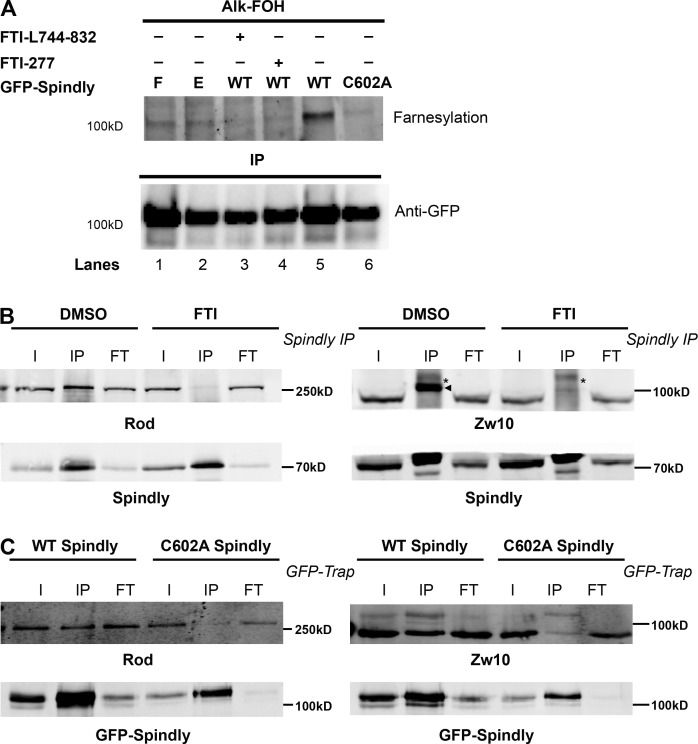
To investigate if hSpindly undergoes farnesylation in vivo, pEGFP-WT Spindly, pEGFP-C602A Spindly (of the CAAX motif), pEGFP-Spindly-E, and pEGFP-Spindly-F transfected HeLa cells were labeled with alkynyl-farnesol, a farnesyl group analogue (Charron et al., 2011), in the presence or absence of FTIs. Farnesylated proteins were detected by performing bioorthogonal ligation (click chemistry) with a fluorescent tag followed by in-gel fluorescence signal detection. Wild-type (WT) Spindly is farnesylated as indicated by the presence of a fluorescent band (Fig. 6 A, lane 5) and farnesylation is inhibited in the presence of FTIs (Fig. 6 A, lanes 3 and 4). Furthermore, Spindly-E and Spindly-F undergo farnesylation as expected because these constructs localize to KTs (Fig. 6 A, lanes 1 and 2). The hSpindly C602A mutant cannot localize to KTs and did not undergo farnesylation (Fig. 6 A, lane 6). These results provide an in vivo validation of hSpindly farnesylation.
Figure 6.
hSpindly is farnesylated in vivo and farnesylation regulates its interaction with the RZZ complex. (A) HeLa cells transiently expressing GFP-WT hSpindly, GFP-C602A hSpindly (of CAAX motif), GFP-hSpindly-E, and GFP-hSpindly-F were metabolically labeled with alkynyl-farnesol (prenylation reporter) and HeLa cells expressing WT GFP-Spindly were either treated with FTI or DMSO. (top) Fluorescence detection of farnesylated immunoprecipitated GFP-tagged hSpindly protein as shown in gel. (bottom) Loading control is shown. WT Spindly, Spindly-E, and Spindly-F exhibit bands indicating farnesylation. Farnesylation of WT Spindly is inhibited in the presence of FTIs (lanes 3 and 4). As expected, C602A mutant of hSpindly is not farnesylated in vivo (lane 6). (B) Immunoprecipitation of hSpindly (70 kD) from mitotic HeLa cell lysates followed by Western blot against Zw10 (89 kD) or Rod (250 kD) RZZ complex subunits. I, input; IP, immunoprecipitated; FT, flowthrough. HeLa cells were treated with either DMSO or 10 µM of FTI-L744832 for 24 h before harvesting and arrested in mitosis with nocodazole treatment. Zw10 is indicated by an arrowhead (IP lanes), and an asterisk denotes a nonspecific band. Inhibition of farnesylation leads to loss of hSpindly and RZZ complex interaction. (C) GFP-Trap from mitotic HeLa cells with endogenous hSpindly knockdown and expressing RNAi resistant GFP-WT hSpindly (98.5 kD) or GFP-C602A hSpindly mutant (98.5 kD). GFP-Trap followed by Western blot against Zw10 (89 kD) or Rod (250 kD) subunits of the RZZ complex. Cells were incubated in nocodazole to accumulate in mitosis before harvesting. Cysteine-to-alanine mutant of GFP-hSpindly leads to loss of hSpindly and RZZ complex interaction.
We have shown that the RZZ complex does not recruit hSpindly to KTs when farnesylation is inhibited (Fig. 3, A and B). hSpindly interacts with the RZZ complex and this interaction is sensitive to the presence of detergent in the lysis buffer (Chan et al., 2009; Barisic et al., 2010), suggesting that farnesylation may regulate hSpindly–RZZ complex interaction. To investigate this, we performed coimmunoprecipitation experiments using HeLa cells treated with either DMSO or FTI for 24 h. Mitotic populations were enriched by double thymidine block and nocodazole treatment. We found that hSpindly associated with Zw10 and Rod (RZZ complex subunits) in DMSO-treated cells as shown previously (Fig. 6 B; Chan et al., 2009; Barisic et al., 2010). This interaction, however, is lost in FTI-treated cells, indicating that farnesylation of hSpindly is required for its interaction with the RZZ complex (Fig. 6 B).
Because the C602A hSpindly mutant is not farnesylated in vivo, we transfected RNAi-resistant GFP-C602A hSpindly and GFP-WT hSpindly into HeLa cells knocked down for endogenous hSpindly (Fig. S4, A and B), immunoprecipitated mitotic cell lysates using GFP Trap, and analyzed for Rod and Zw10 pull-down. The C602A hSpindly mutant showed complete loss of the RZZ complex pull-down, demonstrating that loss of hSpindly farnesylation abrogated the interaction between hSpindly and the RZZ complex subunits (Fig. 6 C). This supports the idea that the presence of a farnesyl group on hSpindly is required for its interaction with the RZZ complex and its subsequent recruitment to KTs.
hSpindly depletion and FTI treatment have similar mitotic phenotype
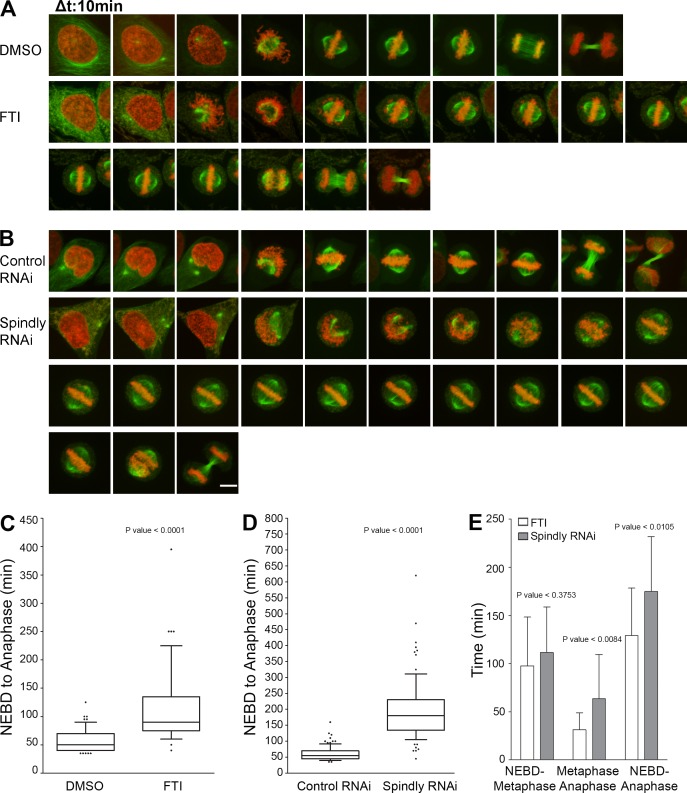
FTI-treated cells are known to accumulate at the G2-M boundary (Ashar et al., 2000; Crespo et al., 2001, 2002). We observed prometaphase accumulation, mostly in the rosette arrangement with FTI treatment as compared with control DMSO-treated cells (Fig. S4 C; Crespo et al., 2001). The percentage of metaphase cells is also significantly reduced, indicating that farnesylation is a critical step for transition from prometaphase to metaphase (Fig. S4 D). The cells in metaphase did not show significant lagging chromosomes at the spindle poles, which is a characteristic phenotype of CENP-E knockdown. To examine the mitotic phenotype of FTI treatment and hSpindly depletion in real time, we performed time-lapse imaging experiments using HeLa cells stably expressing GFP-tubulin and mCherry-histone H2B. FTI-treated and hSpindly-depleted cells spent an increased time in mitosis from nuclear envelope breakdown to anaphase onset (mean of 116 and 180 min for FTI treatment and hSpindly RNAi, respectively, versus a mean of 57 and 55 min for DMSO treatment and scrambled RNAi, respectively; Fig. 7, A–E; and Videos 1–4). hSpindly-depleted and FTI-treated cells exhibited prometaphase delay, as a result of prolonged chromosome alignment, and delayed anaphase onset after metaphase with both delays being more pronounced in hSpindly-depleted cells (Fig. 7, A–E). A fraction of cells with mitotic arrest caused by FTI or hSpindly depletion were observed to undergo cell death or chromatin decondensation (unpublished data). hSpindly-depleted cells spent two times longer transitioning from metaphase to anaphase as compared with FTI-treated cells (Fig. 7 E). There are two possible explanations for the difference. First, in hSpindly-depleted cells there is a lack of both hSpindly protein presence and its KT localization. The fact that hSpindly is present but not localized to KTs in FTI treatment suggests farnesylation is not required for checkpoint silencing. Second, FTIs have a multitude of targets and we cannot rule out the possibility that some other target is compensating for the prolonged prometaphase and anaphase onset (Sebti and Der, 2003; Sebti, 2005). FTI treatment and hSpindly knockdown have the same phenotype but differ in severity, suggesting that the mitotic effects of FTIs are caused by a lack of hSpindly KT localization.
Figure 7.
Phenocopying of farnesylation inhibition with hSpindly knockdown by RNAi. (A) Representative stills from videos of GFP-tubulin and mCherry H2B expressing HeLa cells treated with either DMSO or 10 µM of FTI-L744832 for 24 h before filming. Time is shown in 10-min intervals. (B) Representative stills from videos of GFP-tubulin and mCherry H2B expressing HeLa cells treated with either control (scrambled) RNAi or hSpindly RNAi for 33 h before filming. Time is shown in 10-min intervals. Bar, 10 µm. (C) Box plots comparing the duration of nuclear envelope breakdown (NEBD) to anaphase between FTI- and DMSO-treated cells show significant mitotic delay in FTI-treated cells. Seven experiments were performed with >60 cells in total analyzed for each treatment. Boxes represent interquartile distributions and whiskers represent 10th and 90th percentiles. (D) Box plots comparing the duration of nuclear envelope breakdown to anaphase between control (scrambled) RNAi and hSpindly RNAi–treated cells show significant mitotic delay in hSpindly RNAi–treated cells. Eight experiments were performed with >80 cells in total analyzed for each treatment. Boxes represent interquartile distributions and whiskers represent 10th and 90th percentiles. (E) Duration of nuclear envelope breakdown to metaphase, metaphase to anaphase, and nuclear envelope breakdown to anaphase in FTI- and hSpindly RNAi–treated HeLa cells shows all are increased in Spindly knockdown cells with metaphase-to-anaphase time difference being significant. n = 18 for FTI and n = 22 for hSpindly RNAi. Error bars are SD from the means.
Discussion
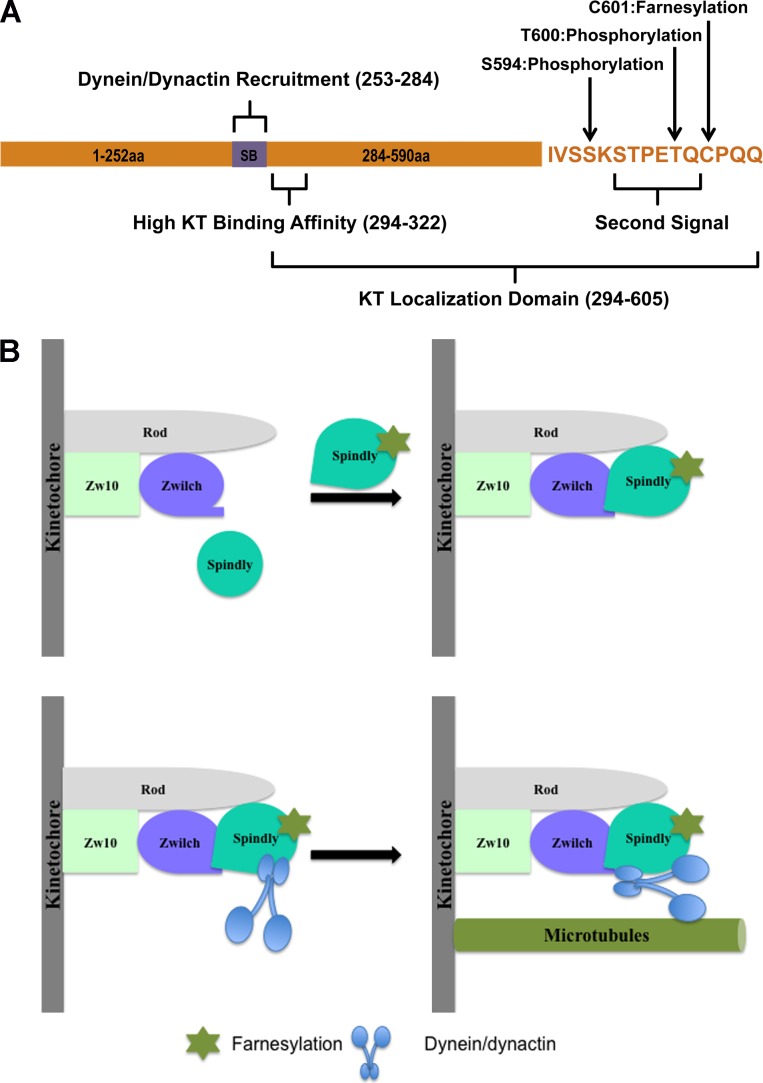
In our current structure function study, we mapped the KT localization domain of hSpindly to its C-terminal 294–605 aa (Fig. 8 A). Furthermore, our data showed that hSpindly undergoes farnesylation, which is essential for hSpindly KT localization. Substitution of the farnesylation motif with a geranylgeranylation motif does not support hSpindly KT localization. We found that FTI treatment does not interfere with KT localization of the RZZ complex, CENP-E, or CENP-F. Additionally, farnesylation plays a pivotal role in the interaction of hSpindly with the RZZ complex providing insight into lipid modification regulating checkpoint protein assembly. Our results showed that FTI treatment and hSpindly knockdown share the same phenotypes, prolonged prometaphase and metaphase with chromosome alignment defects, differing only in severity. Our analysis indicates that hSpindly is likely a key farnesylation target leading to FTI-induced mitotic defects.
Figure 8.
A proposed model of hSpindly KT localization. (A) A schematic representation of hSpindly domains and posttranslational modifications. SB, Spindly box. (B) hSpindly farnesylation induces a conformational change such that it can interact with the RZZ complex subunits Rod and Zwilch, which recruits hSpindly to KTs during mitosis. hSpindly further recruits the dynein–dynactin complex to KTs. Once all the chromosomes are aligned on the metaphase plate, hSpindly and the RZZ complex are transported from the KTs to spindle poles.
KT localization analysis of an extensive hSpindly mutant library consisting of truncation, insertion, deletion, and substitution constructs showed that both the coiled-coil domain II and the C terminus of hSpindly are required for KT localization (Figs. 1 and 2). Although Barisic et al. (2010) has previously shown that the 293–605-aa fragment of hSpindly did not localize to KTs, we found that this specific deletion (N3 construct) does not impair KT localization of hSpindly (Fig. 1). This discrepancy could be the result of overexpression, differences in fusion tags, or the sensitivity of the two assays. Overexpression of coiled-coil proteins often results in aggregation, which can lead to mislocalization of the protein, resulting in a false negative. Immunostaining with anti-FLAG antibody to analyze KT localization by Barisic et al. (2010) compared with GFP fusion constructs may influence the detection sensitivity and our assay is maximized for sensitivity with vinblastine treatment. Barisic et al. (2010) concluded that both coiled-coil domain II and the Spindly box (253–284 aa) contribute to KT localization. In our study, construct N3 (294–605 aa) lacking the Spindly box localized to KTs and we conclude that the Spindly box does not contribute to KT localization. The Spindly box, however, was previously shown to be critical for dynein recruitment (Barisic et al., 2010; Gassmann et al., 2010), suggesting that the hSpindly KT localization domain (294–605 aa) is different from its dynein recruitment domain (Fig. 8 A). Additionally, we found that the N4 construct (322–605 aa) localized to KTs only when cells were treated with vinblastine, suggesting the region between 293 to 322 aa is important for KT binding affinity (Figs. 1 A and 8 A). Deletion of residues in this region might facilitate premature interaction of hSpindly N4 construct with the dynein–dynactin complex, which transports it off KTs to the spindle poles. When KT–microtubule attachment is disrupted by vinblastine, dynein-mediated transport is abolished and the N4 construct is retained on KTs. Thus 294–322 aa of hSpindly is required for high affinity binding to KTs although the precise mechanism warrants further investigation. Insertion and site-directed hSpindly mutants revealed that deletions in residues 596–605 or substitution of the C-terminal cysteine were not tolerated, indicating 596–605 aa are important and the C-terminal cysteine is essential for hSpindly KT localization (Fig. 2). These results are in agreement with Barisic et al. (2010), where deletion mutant 604–605 aa (ΔQQ) and substitution mutant C602S both did not localize to KTs. It also revealed that these residues constitute a potential CAAX farnesylation motif (Fig. 2).
The role of farnesylation in targeting proteins to the membrane is well known. It was first realized with the discovery of CENP-E and CENP-F farnesylation that lipid modifications regulate mitotic protein functions. The hSpindly CPQQ farnesylation motif is conserved in vertebrates but not in D. melanogaster and Caenorhabditis elegans (Fig. S3 A). Inhibition of farnesylation led to complete loss of hSpindly KT localization but had no effect on KT localization of the RZZ complex (Fig. 3). The 25% decrease in hSpindly protein expression upon FTI treatment is not responsible for the absence of hSpindly KT localization and suggests that hSpindly may be slightly more susceptible to degradation in the absence of farnesylation. hSpindly was shown to be degraded when it is not localized to KTs; however, interaction between hSpindly and the RZZ complex and not KT localization stabilizes the hSpindly protein (Chan et al., 2009). Our in vivo labeling experiment results confirmed that hSpindly is farnesylated on the cysteine of its CAAX motif and farnesylation is abolished in the C602A mutant of hSpindly (Fig. 6). These results show that farnesylation is required for hSpindly KT localization.
The presence of the RZZ complex on KTs when farnesylation is inhibited indicates that it is unable to recruit hSpindly. Farnesylation has been reported to regulate protein–protein interactions, two key examples being RAS and Lamin B (Kitten and Nigg, 1991; Kuroda et al., 1993; Porfiri et al., 1994; Bhattacharya et al., 1995; Kisselev et al., 1995; Mical and Monteiro, 1998; Rubio et al., 1999). Farnesylated RAS interacts with and activates Raf-1 kinase; however, if RAS is not farnesylated, it can still interact with Raf-1, but Raf-1 is poorly activated (Luo et al., 1997; Booden et al., 2000). Lamin B localizes to the nuclear membrane and is postulated to bind the lamin B receptor on the membrane. Lamin B cofractionation with lamin B receptor was abolished in cells with lamin B lacking a CAAX farnesylation motif (Mical and Monteiro, 1998). It remains unclear if the farnesyl group binds directly with the interacting protein or if the presence of the farnesyl group induces a conformational change and creates a binding site for its interacting protein. Rubio et al. (1999) proposed that farnesylation induces a conformational change in RAS, creating a binding site for the G protein–responsive phosphoinositide 3-kinase p110γ. Our coimmunoprecipitation and GFP Trap results showed hSpindly farnesylation is a prerequisite for its interaction with the RZZ complex (Fig. 6). Farnesylation remodels the C terminus of a protein from hydrophilic to hydrophobic. Insight into the role of the farnesyl group in binding to a hydrophobic pocket on the RZZ complex could be obtained from the crystal structure of the RZZ complex. The crystal structure of the hZwilch split construct (1–334 and 335–591 aa) and in vitro experiments showed that hZwilch binds to the N terminus of Rod and hZw10 binds to the C terminus of Rod (Civril et al., 2010). Super resolution microscopy mapping has suggested that the C terminus of hSpindly binds Zwilch and/or the N terminus of Rod because they are spatially close to each other (Varma et al., 2013). The main goal of future studies is to identify how farnesylation regulates hSpindly and RZZ complex interaction and which RZZ subunit interacts with hSpindly. Absence of a farnesylation motif in Spindly in D. melanogaster and C. elegans (Fig. S3 A) suggests that farnesylation is not required for its interaction with the RZZ complex in these organisms. We found the hSpindly–RZZ complex interaction mediated through hSpindly farnesylation is essential for its KT recruitment (Fig. 8 B). We propose that farnesylation induces a conformational change in hSpindly such that it can interact with the RZZ complex (Fig. 8 B). Coimmunoprecipitation and GFP Trap experiments showed that nonfarnesylated hSpindly is unable to interact with the RZZ complex subunits Zw10 and Rod. In our model, we predict that the RZZ complex acts as a KT recruitment factor for hSpindly, and Zwilich and/or Rod interact directly with hSpindly based on recent super resolution microscopy findings (Varma et al., 2013; Fig. 8 B). RZZ in complex with hSpindly is recruited to unattached KTs whereby hSpindly recruits the dynein–dynactin complex (Fig. 8 B); although which subunit interacts directly with hSpindly is not clear. Once all chromosomes are bi-oriented, the dynein–dynactin complex transports the checkpoint proteins from the KTs to the spindle poles.
Interestingly, hSpindly constructs O (insertion T600-QLRPH), del592-596aa, and Q601A, all with mutations upstream of the CAAX motif, localized to KTs only when cells were treated with vinblastine (Figs. 1 and 2 and Table 1). These results suggest that the hydrophobicity, charge, and/or posttranslational modifications of the residues upstream of the farnesylation motif might play a role in binding affinity. Insertion mutant O (T600-QLRPH) places a basic residue (H) where there is an acidic residue (E) in WT hSpindly. Del592-596aa replaces hydrophilic serine residues (SSKS) and a basic lysine with hydrophobic residues (PIL). Q601A replaces a polar hydrophilic residue (Q) with an amphiphilic residue (A). These changes likely have further consequences in the 3D structure of the protein, leading to the reduced binding affinity. Our results reflect a system similar to membrane binding in RAS proteins, where the CAAX farnesylation motif requires an upstream second signal, which in HRAS, NRAS, and KRAS4A is an upstream cysteine residue that is palmitoylated (Choy et al., 1999; Apolloni et al., 2000). In KRAS4B, however, the second signal is an upstream lysine-rich polybasic region with a net positive charge of eight (Hancock et al., 1990). Farnesylation and the polybasic region work together with either one alone being insufficient for membrane binding (Ahearn et al., 2012). hSpindly has two residues upstream of the CAAX motif that are phosphorylated: serine594 and threonine600 (Hornbeck et al., 2012). Mutants with a deletion encompassing serine594 (Del592-596aa) and substitution of threonine600 with alanine (T600A) localized to KTs, although Del592-596aa localized to KTs only with vinblastine (Fig. 2). We propose that the region upstream of the hSpindly CAAX motif (hydrophilic and acidic) acts as a second signal for binding affinity as a result of its charge properties because there is no upstream cysteine that can undergo a second lipid modification (Fig. 8 A).
Evidence regarding the effect of FTI on CENP-E and CENP-F KT localization is controversial. Several studies have shown that inhibition of farnesylation does not affect CENP-E and CENP-F KT localization (Ashar et al., 2000; Crespo et al., 2001, 2002; Verstraeten et al., 2011). Ashar et al. (2000) and Verstraeten et al. (2011) treated A549 cells (for 4 d) and human skin fibroblasts (for 3 d) with FTI and did not observe any effect on CENP-E or CENP-F KT localization. Crespo et al. (2001) treated Calu-1 and A549 cells with FTI for 48 h and observed CENP-E and CENP-F KT localization. Schafer-Hales et al. (2007) showed that 16-h FTI treatment in 1A9 ovarian cancer cells disrupts CENP-E and CENP-F KT localization at metaphase but not at prometaphase. Hussein and Taylor (2002) reported the inability of a CENP-F cysteine-to-alanine C3207A mutant (of the CAAX motif) to localize to KTs, indicating that farnesylation is essential for CENP-F KT localization. In the same study, however, FTI treatment did not affect endogenous CENP-F KT localization in consensus with other studies (Crespo et al., 2001; Hussein and Taylor, 2002; Schafer-Hales et al., 2007). In our analysis, inhibition of farnesylation using FTIs did not impair CENP-E or CENP-F KT localization as indicated by quantitative analysis (Fig. 4, A–D). In contrast to Hussein and Taylor (2002), our CENP-F C3207A mutant localized to KTs (Fig. 4, E and F). Thus, the mitotic effects of FTIs cannot be attributed to loss of CENP-E and CENP-F KT localization.
FTIs were originally developed to prevent RAS farnesylation; however, it was soon realized that not all the effects of FTIs were caused by inhibition of RAS farnesylation as RAS can alternatively undergo geranylgeranylation in the presence of FTIs. Additionally, cell lines lacking any RAS mutations responded to FTIs (Sepp-Lorenzino et al., 1995). Prometaphase delay and chromosome alignment defects in FTI-treated cells were previously hypothesized to be caused by CENP-E and CENP-F farnesylation inhibition. One study disputed this hypothesis and suggested the mitotic effects were caused by additional unknown farnesylation targets (Crespo et al., 2001). Our live cell imaging results showed striking similarities between FTI treatment and the hSpindly knockdown phenotype, such as prolonged prometaphase, chromosome alignment defects, and delayed anaphase onset after biorientation, differing only in severity. Unlike KRAS and NRAS, geranylgeranylation does not appear to substitute for farnesylation in hSpindly KT localization, making hSpindly a potentially good target for FTIs as anti-cancer drugs (Fig. 5). Extensive clinical trials using tipifarnib, lonafarnib, BMS-214662, and L-7778123 have mostly demonstrated little efficacy in cancer patients except a subset of patients with hematological malignancies (Berndt et al., 2011). Tipifarnib has shown efficacy against breast cancer in addition to acute myeloid leukemia and chronic myelogenous leukemia (Johnston et al., 2003; Gotlib, 2005). Two studies used a two-gene signature (RASGRP1/APTX ratio) in selection of acute myeloid leukemia patients for receiving tipifarnib treatment (Raponi et al., 2008; Karp et al., 2012). Similarly, lonafarnib has shown efficacy in acute myeloid leukemia and chronic myelogenous leukemia. Recently, FTI therapy showed very promising results in patients with Hutchinson-Gilford Progeria syndrome and a clinical trial is ongoing (Wong and Morse, 2012). The biggest challenge to the development of FTI drugs is the lack of knowledge of their target effectors. Here, we propose hSpindly as a novel FTI mitotic target and potential prognostic marker for using FTIs as a therapeutic treatment.
We therefore propose a novel role of farnesylation in targeting a checkpoint protein, hSpindly, to KTs. Here, we showed that farnesylation of hSpindly regulates its interaction with the RZZ complex and its recruitment to KTs through this interaction. Our data also explains the mitotic effects observed upon FTI treatment. Further research is required to investigate if hSpindly expression is altered in specific cancers.
Materials and methods
Cloning
Full-length hSpindly (NCBI Gene ID: 54908) and 2581–3210 aa of hCENP-F (NCBI Gene ID: 1063) cDNAs were amplified from a human fetal cDNA library (Spring Bioscience) using gene-specific primers and cloned into the pDONR221 vector (Invitrogen). hSpindly N- and C-terminal truncation constructs (Fig. 1 A) were amplified using pDONR221 hSpindly as a template using specific primers. Truncation constructs were cloned into the pDONR221 vector (Invitrogen) using attB recombination linkers of the Gateway Technology cloning system (Invitrogen). Cloning into gateway destination pEGFP (CMV promoter) expression vector (Chan et al., 1998) was performed using the Gateway Technology cloning system.
Mutagenesis and RNAi
The hSpindly random insertion mutant library was generated by transposon-mediated insertion mutagenesis (Mutation Generation System; Finnzymes), which generated mutants of hSpindly protein containing a 5-aa in-frame insertion. Site-directed hSpindly mutants were generated using the QuikChange site-directed mutagenesis kit (Agilent Technologies) using pDONR221 hSpindly as a template. The site of insertion and desired point mutant sequences were confirmed by sequencing using BigDye Terminators v3.1 and an ABI PRISM 310 capillary sequencer (Applied Biosystems). siRNA for Spindly (5′-GAAAGGGUCUCAAACUGAA-3′) or a scrambled control siRNA (5′-UGGUUUACAUGUCGACUAA-3′) from Thermo Fisher Scientific was used. The siRNA-resistant version of hSpindly containing four silent mutations was generated using the QuikChange site-directed mutagenesis kit with the following primer and its antisense (changes shown in bold): 5′-CGTTGCTACAGATGAAGGGATCCCAGACTGAATTTGAGCAGCAGG-3′.
Cell culture and synchronization
HeLa and HEK293 cells were grown as a monolayer in low-glucose DMEM supplemented with 2 mM l-glutamine and 10% (vol/vol) FBS in a humidified incubator at 37°C with 5% CO2. T47D cells were grown in RPMI 1640 supplemented with 2 mM l-glutamine, 10% (vol/vol) FBS, and 1 mM sodium pyruvate. MCF7 cells were grown in low- and high-glucose DMEM in equal volume supplemented with 2 mM l-glutamine, 10% (vol/vol) FBS, and 0.01 mg/ml insulin. M2 and MeWo cells were grown in low-glucose DMEM F12 media supplemented with 2 mM l-glutamine and 10% (vol/vol) FBS. Double thymidine cell synchronization was performed by the addition of 2 mM thymidine to cells for 16 h.
Transient transfection
HeLa cells were seeded and grown to ∼60% confluency on 35-mm dishes. Each dish was transfected with 2 µg of EGFP-Spindly constructs. For each transfection, 2 µg of plasmid DNA was mixed with 100 µl of OPTI-MEM. In a separate tube, 10 µl of polyethylenimine (PEI) was mixed with 100 µl of OPTI-MEM and incubated at room temperature for 5 min. The diluted plasmid DNA and PEI were mixed together and incubated at room temperature for 15 min to allow DNA–PEI complex formation. 2 ml of warmed low-glucose DMEM growth media was added to the plasmid–PEI complex mixture, which was then added to HeLa cell dishes. For hSpindly RNAi transfections, oligofectamine was used according to the manufacturer’s instructions. Cells were incubated with 100 nM RNAi and oligofectamine in OPTI-MEM for 5–6 h followed by the addition of FBS to a final concentration of 10%. Transfection media was replaced with fresh media after 24 h.
For hSpindly in vivo farnesylation analyses, HeLa cells grown to ∼60% confluence in 10-mm-diameter dishes were transiently transfected with 10 µg of the EGFP-Spindly constructs and 40 µl PEI (1 mg/ml) as described in the previous paragraph for ∼39 h. HeLa cells were treated with alkynyl-farnesol (50 µM from 50 mM of stock solution in DMSO) for 4 h (Charron et al., 2011). For coincubation with inhibitors, HeLa cells were pretreated for 1 h with 10 µM of FTI L-744832 (10 mM of stock solution in DMSO; Enzo Life Sciences) or 20 µM of FTI-277 (20 mM of stock solution; Sigma-Aldrich) before alkynyl-farnesol metabolic labeling. Cells were then harvested for immunoprecipitation as previously described (Charron et al., 2011).
Western blotting
For Western blotting, HEK293 cells were seeded at a density of 105 cells/ml in 35-mm-diameter dishes. Cells were transiently transfected with 1–2 µg of EGFP-hSpindly mutant constructs as described under Transient transfection for ∼24 h and processed for Western blotting as described previously (Chan et al., 1998). PageRuler Plus Prestained protein ladder (Fermentas; Thermo Fisher Scientific) was used as a molecular weight marker.
Blots were blocked with Odyssey blocking buffer (LI-COR Biosciences) and probed with IR-800–conjugated mouse monoclonal anti-GFP antibodies (1:10,000 dilution; Rockland Immunochemicals) and rat polyclonal anti-hSpindly (1:1,000 dilution; described below). Rabbit polyclonal anti hRod (N-terminal 809-aa antigen) and rabbit anti-hZw10 (GST full-length hZw10 antigen) were used for labeling the blots (Chan et al., 2000). B512 mouse anti-tubulin antibody (1/3,000; Sigma-Aldrich) was used to label the blot. Secondary antibodies Alexa Fluor 680 anti-rat antibody (1:10,000 dilution; Invitrogen) and Alexa Fluor 680 anti-rabbit antibody (1:10,000 dilution; Invitrogen) were used. Odyssey IR imager system (LI-COR Biosciences) was used to scan the blots.
Fluorescence microscopy
For hSpindly, hZw10, hRod, hCENP-E, and hCENP-F immunofluorescence, HeLa cells were seeded on to 22-mm2 coverslips at a density of 5 × 104 cells/ml in a 35-mm-diameter dish for 36–48 h. One set of coverslips was treated with 10 µM FTI L-744-832 (Enzo Life Sciences) or 20 µM FTI-277 (Sigma-Aldrich) and the other set was treated with same volume of control solvent (DMSO) for 24 h. The cells were fixed with 3.5% (wt/vol) paraformaldehyde in PBS with 10 mM Pipes, pH 6.8, for 7 min; permeabilized in KB (50 mM Tris/HCl, pH 7.4, 150 mM NaCl, and 0.1% BSA) with 0.2% Triton X-100 for 5 min at room temperature (22°C); and rinsed in KB buffer for 5 min at room temperature without Triton X-100. DNA was stained with 0.1 µg/ml DAPI. Rat anti-hSpindly (1:1,000 dilution) and Alexa Fluor 488–conjugated anti-rat (1:1,000 dilution; Molecular Probes) antibodies were used to detect hSpindly. hZw10, hRod, hCENP-E, and hCENP-F were visualized using rat anti-hZw10 sera (1–139-aa antigen), rabbit anti-hRod (N-terminal 809-aa antigen; Chan et al., 2000), rabbit anti-hCENP-E (C-terminal 360-aa antigen; Schaar et al., 1997), and rabbit anti-hCENP-F (8445-end nucleotides; Liao et al., 1995) antibodies. Anticentromere antibody (ACA) sera (1:4,000 dilution) was used to visualize centromeres using human ACA sera (gift from M. Fritzler, University of Calgary, Calgary, Canada). Coverslips were mounted with 1 mg/ml Mowiol 4-88 (EMD Millipore) in phosphate buffer, pH 7.4. Alexa Fluor 488–conjugated anti–rat (1:1,000 dilution; Molecular Probes), Alexa Fluor 555–conjugated anti–rabbit (1:1,000 dilution; Molecular Probes), Alexa Fluor 555–conjugated anti–mouse (1:1,000 dilution; Molecular Probes), and Alexa Fluor 647–conjugated anti–human (1:1,000 dilution; Molecular Probes) secondary antibodies were used to visualize protein localization.
For fluorescence microscopy of GFP-tagged hSpindly constructs, HeLa cells were seeded on to 22-mm2 coverslips at a density of 5 × 104 cells/ml in 35-mm-diameter dishes and grown for 24 h before transfection. Cells were transiently transfected with 1–2 µg of plasmid DNA as described under Transient transfection for 24–36 h. Cells were fixed and permeabilized as described in the previous paragraph. DNA was stained with DAPI (0.1 µg/ml), and coverslips were mounted with Mowiol 4-88.
A microscope (Imager.Z.1; Carl Zeiss) equipped with epifluorescence optics was used to collect the images. Cells were visualized with a 100× Plan-Apochromat objective (Carl Zeiss) with 1.4 NA. Images were captured with a SensiCam (Cooke) charge-coupled device camera (PCO-TECH, Inc.) controlled by Metamorph 7.0 software (Universal Imaging Corp.). Images were processed using Photoshop 7.0 (Adobe). Endogenous Spindly immunofluorescence images (Fig. S1 A) were acquired using a confocal Plan-Aprochromat (Carl Zeiss) with 63× objective (1.4 NA; Carl Zeiss). Data were collected using Zen software 7.0 (Carl Zeiss) and processed using Photoshop 7.0.
Fluorescence quantification
For quantification of fluorescence, fluorescence KT intensities were measured within a 9 × 9-pixel square. Background fluorescence was measured outside the 9 × 9 square using 13 × 13-pixel squares and subtracted (King et al., 2000). For intensity quantifications at KTs, the smaller square encompassed a single KT and results were normalized against ACA signals. 20 KTs were analyzed for each protein.
MBP Spindly and rat anti-hSpindly antibody production
Recombinant full-length MBP-hSpindly fusion protein expression was induced in the Escherichia coli strain JM109 (T7 promoter). Protein expression was induced at mid log phase with 2 mM IPTG at 30°C for 2–3 h. Bacteria were harvested in lysis buffer (50 mM NaPO4, pH 7.6, 0.25 M NaCl, 5 mM MgCl2, 1 mM EGTA, 10 µg AEBSF protease inhibitor, and 0.1 mg/ml lysozyme) and sonicated. The soluble MBP-Spindly fusion protein was bound to an amylose resin (New England Biolabs, Inc.) column washed twice in binding buffer (20 mM Tris-HCl, pH 7.4, 200 mM Nacl, and 1 mM EDTA) following the manufacturer’s directions. The recombinant MBP-Spindly protein was eluted using elution buffer (20 mM Tris-HCl, pH 7.4, 200 mM Nacl, 1 mM EDTA, and 10 mM Maltose), concentrated in PBS (30,000-MW cutoff; Amicon), and used as an antigen for immunization of rats. Immunizations of rats were conducted according to the guidelines set by the Cross Cancer Institute Animal Care Committee and Canadian Council of Animal Care. Antibodies were purified by preabsorbing the rat sera onto an Affigel 15 column (Bio-Rad Laboratories) that was covalently coupled to bacterial lysate containing MBP to remove antibodies against MBP and bacterial proteins. hSpindly antibodies were purified by overnight incubation of preadsorbed serum with an Affigel 15 column that was covalently coupled to MBP-hSpindly fusion protein. The column was washed three times with at least 10 bed volumes of TBS. Antibodies were eluted with elution buffer (0.2 M glycine-HCl, pH 2.5) and immediately neutralized with 1 M Tris, pH 9.0. Protein concentrations of fractions were monitored by absorbance at 280 nm and antibody-containing fractions were pooled, desalted, and concentrated by centrifugation with a protein concentrator (30,000-MW cutoff; Amicon). The antibody concentration was determined and diluted with filter sterile glycerol to a final concentration of 5 µg/ml in 50% glycerol/0.5× PBS. MBP-hZw10 (1–231 aa) recombinant protein was produced, purified, and injected into rats using the same protocol as described for MBP-Spindly protein. Rat anti-hZw10 sera was used for immunofluorescence.
Metabolic labeling, click chemistry reactions, and in-gel fluorescence imaging
HeLa cells were grown and transfected in 10-cm plates as previously described under transient transfection (Charron et al., 2011). Cells were gently washed with PBS to remove dead cells or debris followed by trypsinization. Cells were treated with alkynyl-farnesol (50 µM; 50 mM of stock solution in DMSO) for 4 h using the same volume of solvent (DMSO) in the negative controls (Charron et al., 2011). For coincubation, HeLa cells were pretreated for 1 h with 10 µM L-744832 FTI before alkynyl-farnesol metabolic labeling. The cells were pelleted by centrifuging at 1,000 g for 5 min at RT. Cell pellet was washed twice in ice-cold PBS and pelleted by centrifuging at 1,000 g for 5 min at 4°C. Cells were flash frozen in liquid nitrogen and stored at −80°C. Cell pellets were lysed in ice-cold Brij lysis buffer (1% Brij97, 50 mM triethanolamine, pH 7.4, 150 mM NaCl, 5×EDTA-free Roche protease inhibitor cocktail, and 10 mM PMSF; Charron et al., 2011). Protein concentration was determined by the BCA assay (Thermo Fisher Scientific). EGFP-hSpindly was immunoprecipitated using 1 µl anti-GFP rabbit pAb (Abcam) and 50 µl of packed protein A–agarose beads (Roche) per sample (Charron et al., 2011). Upon incubation at 4°C for 2 h on a nutating mixer (Labnet), the beads were washed (3 × 1 ml) with ice-cold modified RIPA lysis buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 50 mM triethanolamine, pH 7.4, and 150 mM NaCl).
Cu-catalyzed azide-alkyne cycloaddition/click reaction was performed as described previously (Charron et al., 2011). The purified protein bound to beads was suspended in 47 µl of ice-cold PBS, to which 3 µl of freshly premixed click reaction cocktail (azido-rhodamine [100 µM; 10 mM of stock solution in DMSO], tris(2-carboxyethyl)phosphine hydrochloride [TCEP; 1 mM; 50 mM of freshly prepared stock solution in deionized water], tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine [100 µM; 10 mM of stock solution in DMSO], and CuSO4·5H2O (1 mM; 50 mM of freshly prepared stock solution in deionized water]) was added for 1 h at 4°C on a nutating mixer. The beads were washed (3 × 1 ml) with ice-cold modified RIPA lysis buffer, resuspended in 40 µl of loading buffer (27.5 µl [4% SDS, 50 mM triethanolamine, pH 7.4, and 150 mM NaCl], 10 µl 4×SDS-loading buffer [40% glycerol, 200 mM Tris-HCl, pH 6.8, 8% SDS, and 0.4% bromophenol blue], and 2.5 µl of 0.5 M Bond-Breaker TCEP Solution [Thermo Fisher Scientific]), and heated for 5 min at 95°C; and 20 µl of the supernatant was loaded on two separate SDS-PAGE gels (4–20% Criterion Tris-HCl gel [Bio-Rad Laboratories]), one for fluorescence detection and the other for immunoblotting.
For cell lysates, 50 µg of proteins were clicked in 47 µl SDS-buffer (4% SDS, 50 mM triethanolamine, pH 7.4, and 150 mM NaCl) with 3 µl of freshly premixed click reaction cocktail (same as above) for 1 h at room temperature. Proteins were precipitated by adding ice-cold methanol (1 ml), placing at −80°C overnight, and centrifuging at 18,000 g for 10 min at 4°C and discarding the supernatant. The protein pellets were allowed to air dry, resuspended in 50 µl of loading buffer (same as above), and heated for 5 min at 95°C, and 20 µg of protein was loaded on two separate SDS-PAGE gels.
After SDS-PAGE separation, the gel was soaked in destaining solution (40% MeOH, 10% acetic acid, and 50% H2O) overnight at 4°C on an orbital shaker, rehydrated with deionized water, and visualized by scanning the gel on a Typhoon 9400 variable mode imager (excitation of 532 nm, 58-nm filter, and 30-nm band-pass; GE Healthcare).
Immunoprecipitation and GFP Trap
HeLa cells were grown on 10-cm plates and double thymidine treatment was performed. 24 h before harvesting, the cells were treated with either 10 µM of FTI L-744832 FTI or solvent control DMSO. Mitotically arrested HeLa cells (with 0.25 µg/ml nocodazole for 12–16 h) after second thymidine release were resuspended in 20 mM Hepes-KOH, pH 7.5, 10 mM KCl, 1 mM MgCl2, 1 mM EDTA, and 1 mM EGTA, with protease and phosphatase inhibitors (PhosSTOP; Roche), and lysed as previously described (Barisic et al., 2010). 500 µg of lysate was then incubated with the rabbit polyclonal anti-hSpindly (Bethyl Laboratories, Inc.) on a rotator at 4°C for 1 h. The immunoprecipitates were captured with protein A beads, which were blocked overnight with 1% BSA. Western blotting was performed for the identification of Zw10 and Rod pull-down.
For GFP Trap, HeLa cells seeded in the 10-cm plates were transfected with 100 nM hSpindly RNAi for 24 h. After 24 h, the cells were transfected with an RNAi-resistant version of either GFP-WT hSpindly or GFP-C602A mutant hSpindly. Cells were enriched in mitosis by 16-h nocodazole treatment. Mitotic cells were resuspended in buffer as mentioned in the previous paragraph. GFP Trap beads (ChromoTek) were used to pull down hSpindly following the manufacturer’s instructions with the exception of the buffer used.
Live cell imaging
For analysis of mitotic timing, a HeLa cell line stably expressing GFP-tubulin and mCherry H2B was used. Cells seeded in a 35-mm glass-bottom dish (MatTek Corporation or Flourdish) were placed onto a sample stage within an incubator chamber maintained at a temperature of 37°C in an atmosphere of 5% CO2. Cell media was replaced with imaging media (OPTI-MEM; Gibco) supplemented with 2 mM l-glutamine, 10% (vol/vol) FBS, and 14 mM Hepes before imaging. Imaging was performed using a spinning disk confocal on an inverted microscope (Axiovert 200M; Carl Zeiss; with a 40× objective lens and 1.3 NA) equipped with an electron-multiplying charge-coupled device camera (ORCA-FLASH-4.0; Hamamatsu Photonics). Images were collected every 5 min for GFP and Cy3 channel using 100-ms exposure times, for 10–16 h using the velocity software (PerkinElmer). Velocity 6.3.0 software was used to collect and export videos as AVI format using Microsoft video 1 compression. Videos were further converted to mov format with Vegas Pro version 12.0 (Build 394; Sony Creative Software Inc.) using Sorenson 3 compression. Mitotic timing for cells was calculated manually. Still tiff format images from videos were exported using Velocity 6.3.0 software and processed using Photoshop 7.0.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 5.04. P-values were calculated with Student’s t test.
Online supplemental material
Fig. S1 outlines endogenous hSpindly KT localization in prophase and prometaphase, and accumulation at spindle poles in metaphase. Fig. S1 (B–H) shows correct size protein expression for all GFP hSpindly and GFP-CENP-F fusion constructs and their KT localization (with and without vinblastine), which is shown in Fig. S2, demonstrating the importance of the C terminus. Fig. S3 A shows that the hSpindly C terminus is conserved in different species. Fig. S3 B shows that KT localization of hSpindly is inhibited by FTI treatment in breast cancer and melanoma cell lines. Fig. S4 (A and B) shows that expression of hSpindly was knocked down by RNAi transfection. Fig. S4 (C and D) shows HeLa cells treated with FTI accumulate in prometaphase with a reduced percentage of cells in metaphase, revealing a role in the prometaphase-to-metaphase transition. Videos 1–4 show FTI treatment and RNAi knockdown that resulted in prolonged timing for nuclear envelope breakdown to anaphase compared with DMSO or scrambled RNAi controls. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201412085/DC1.
Supplementary Material
Acknowledgments
We thank Chan laboratory members for their helpful discussion and, particularly, Dr. Larissa Vos for reading and editing the manuscript. All microscopy experiments were performed at the Cross Cancer Institute Cell Imaging Facility, University of Alberta.
G.K.T. Chan acknowledges funding by the Alberta Cancer Research Institute (grant number 21594), the Canadian Foundation for Innovation (grant number 9569), the Alberta Heritage Foundation for Medical Research (grant number 200301326), the Natural Sciences and Engineering Research Council of Canada (grant number 355777-08), the Alberta Science and Research Authority (grant number URSI-05-028-SRI), and the Canadian Breast Cancer Foundation–Prairies/NWT. N. Westcott is a Leukemia and Lymphoma Society postdoctoral fellow. H. Hang acknowledges support from National Institutes of Health, National Institute of General Medical Sciences (grant R01 GM087544).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this study:
- ACA
- anticentromere antibody
- CENP
- centromere protein
- FTI
- farnesyl transferase inhibitor
- KT
- kinetochore
- PEI
- polyethylenimine
- WT
- wild type
References
- Ahearn I.M., Haigis K., Bar-Sagi D., and Philips M.R.. 2012. Regulating the regulator: post-translational modification of RAS. Nat. Rev. Mol. Cell Biol. 13:39–51 10.1038/nrm3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolloni A., Prior I.A., Lindsay M., Parton R.G., and Hancock J.F.. 2000. H-ras but not K-ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20:2475–2487 10.1128/MCB.20.7.2475-2487.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashar H.R., James L., Gray K., Carr D., Black S., Armstrong L., Bishop W.R., and Kirschmeier P.. 2000. Farnesyl transferase inhibitors block the farnesylation of CENP-E and CENP-F and alter the association of CENP-E with the microtubules. J. Biol. Chem. 275:30451–30457 10.1074/jbc.M003469200 [DOI] [PubMed] [Google Scholar]
- Barisic M., and Geley S.. 2011. Spindly switch controls anaphase: spindly and RZZ functions in chromosome attachment and mitotic checkpoint control. Cell Cycle. 10:449–456 10.4161/cc.10.3.14759 [DOI] [PubMed] [Google Scholar]
- Barisic M., Sohm B., Mikolcevic P., Wandke C., Rauch V., Ringer T., Hess M., Bonn G., and Geley S.. 2010. Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol. Biol. Cell. 21:1968–1981 10.1091/mbc.E09-04-0356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt N., Hamilton A.D., and Sebti S.M.. 2011. Targeting protein prenylation for cancer therapy. Nat. Rev. Cancer. 11:775–791 10.1038/nrc3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Chen L., Broach J.R., and Powers S.. 1995. Ras membrane targeting is essential for glucose signaling but not for viability in yeast. Proc. Natl. Acad. Sci. USA. 92:2984–2988 10.1073/pnas.92.7.2984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booden M.A., Sakaguchi D.S., and Buss J.E.. 2000. Mutation of Ha-Ras C terminus changes effector pathway utilization. J. Biol. Chem. 275:23559–23568 10.1074/jbc.M001368200 [DOI] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., and Karess R.E.. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Carboni J.M., Yan N., Cox A.D., Bustelo X., Graham S.M., Lynch M.J., Weinmann R., Seizinger B.R., Der C.J., Barbacid M., et al. 1995. Farnesyltransferase inhibitors are inhibitors of Ras but not R-Ras2/TC21, transformation. Oncogene. 10:1905–1913. [PubMed] [Google Scholar]
- Chan G.K., Schaar B.T., and Yen T.J.. 1998. Characterization of the kinetochore binding domain of CENP-E reveals interactions with the kinetochore proteins CENP-F and hBUBR1. J. Cell Biol. 143:49–63 10.1083/jcb.143.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G.K., Jablonski S.A., Starr D.A., Goldberg M.L., and Yen T.J.. 2000. Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat. Cell Biol. 2:944–947 10.1038/35046598 [DOI] [PubMed] [Google Scholar]
- Chan G.K., Liu S.T., and Yen T.J.. 2005. Kinetochore structure and function. Trends Cell Biol. 15:589–598 10.1016/j.tcb.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., and Santamaria A.. 2009. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185:859–874 10.1083/jcb.200812167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G., Tsou L.K., Maguire W., Yount J.S., and Hang H.C.. 2011. Alkynyl-farnesol reporters for detection of protein S-prenylation in cells. Mol. Biosyst. 7:67–73 10.1039/C0MB00183J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy E., Chiu V.K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I.E., and Philips M.R.. 1999. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 98:69–80 10.1016/S0092-8674(00)80607-8 [DOI] [PubMed] [Google Scholar]
- Civril F., Wehenkel A., Giorgi F.M., Santaguida S., Di Fonzo A., Grigorean G., Ciccarelli F.D., and Musacchio A.. 2010. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 18:616–626 10.1016/j.str.2010.02.014 [DOI] [PubMed] [Google Scholar]
- Crespo N.C., Ohkanda J., Yen T.J., Hamilton A.D., and Sebti S.M.. 2001. The farnesyltransferase inhibitor, FTI-2153, blocks bipolar spindle formation and chromosome alignment and causes prometaphase accumulation during mitosis of human lung cancer cells. J. Biol. Chem. 276:16161–16167 10.1074/jbc.M006213200 [DOI] [PubMed] [Google Scholar]
- Crespo N.C., Delarue F., Ohkanda J., Carrico D., Hamilton A.D., and Sebti S.M.. 2002. The farnesyltransferase inhibitor, FTI-2153, inhibits bipolar spindle formation during mitosis independently of transformation and Ras and p53 mutation status. Cell Death Differ. 9:702–709 10.1038/sj.cdd.4401023 [DOI] [PubMed] [Google Scholar]
- De Antoni A., Pearson C.G., Cimini D., Canman J.C., Sala V., Nezi L., Mapelli M., Sironi L., Faretta M., Salmon E.D., and Musacchio A.. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15:214–225 10.1016/j.cub.2005.01.038 [DOI] [PubMed] [Google Scholar]
- Downward J.2003. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 3:11–22 10.1038/nrc969 [DOI] [PubMed] [Google Scholar]
- Famulski J.K., Vos L., Sun X., and Chan G.. 2008. Stable hZW10 kinetochore residency, mediated by hZwint-1 interaction, is essential for the mitotic checkpoint. J. Cell Biol. 180:507–520 10.1083/jcb.200708021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulski J.K., Vos L.J., Rattner J.B., and Chan G.K.. 2011. Dynein/Dynactin-mediated transport of kinetochore components off kinetochores and onto spindle poles induced by nordihydroguaiaretic acid. PLoS ONE. 6:e16494 10.1371/journal.pone.0016494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu H., and Kirschner M.W.. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871–1883 10.1101/gad.12.12.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Huang H., and Yen T.J.. 2006. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 115:320–329 10.1007/s00412-006-0049-5 [DOI] [PubMed] [Google Scholar]
- Gassmann R., Essex A., Hu J.S., Maddox P.S., Motegi F., Sugimoto A., O’Rourke S.M., Bowerman B., McLeod I., Yates J.R. III, et al. 2008. A new mechanism controlling kinetochore–microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 22:2385–2399 10.1101/gad.1687508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R., Holland A.J., Varma D., Wan X., Civril F., Cleveland D.W., Oegema K., Salmon E.D., and Desai A.. 2010. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 24:957–971 10.1101/gad.1886810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotlib J.2005. Farnesyltransferase inhibitor therapy in acute myelogenous leukemia. Curr. Hematol. Rep. 4:77–84. [PubMed] [Google Scholar]
- Griffis E.R., Stuurman N., and Vale R.D.. 2007. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J. Cell Biol. 177:1005–1015 10.1083/jcb.200702062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J.F., Paterson H., and Marshall C.J.. 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 63:133–139 10.1016/0092-8674(90)90294-O [DOI] [PubMed] [Google Scholar]
- Hoffman D.B., Pearson C.G., Yen T.J., Howell B.J., and Salmon E.D.. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 12:1995–2009 10.1091/mbc.12.7.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., and Sullivan M.. 2012. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40:D261–D270 10.1093/nar/gkr1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., and Salmon E.D.. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., and Roberts B.T.. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Hussein D., and Taylor S.S.. 2002. Farnesylation of Cenp-F is required for G2/M progression and degradation after mitosis. J. Cell Sci. 115:3403–3414. [DOI] [PubMed] [Google Scholar]
- Johnston S.R., Hickish T., Ellis P., Houston S., Kelland L., Dowsett M., Salter J., Michiels B., Perez-Ruixo J.J., Palmer P., and Howes A.. 2003. Phase II study of the efficacy and tolerability of two dosing regimens of the farnesyl transferase inhibitor, R115777, in advanced breast cancer. J. Clin. Oncol. 21:2492–2499 10.1200/JCO.2003.10.064 [DOI] [PubMed] [Google Scholar]
- Karess R.2005. Rod–Zw10–Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 15:386–392 10.1016/j.tcb.2005.05.003 [DOI] [PubMed] [Google Scholar]
- Karnoub A.E., and Weinberg R.A.. 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 9:517–531 10.1038/nrm2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp J.E., Vener T.I., Raponi M., Ritchie E.K., Smith B.D., Gore S.D., Morris L.E., Feldman E.J., Greer J.M., Malek S., et al. 2012. Multi-institutional phase 2 clinical and pharmacogenomic trial of tipifarnib plus etoposide for elderly adults with newly diagnosed acute myelogenous leukemia. Blood. 119:55–63 10.1182/blood-2011-08-370825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J.M., Hays T.S., and Nicklas R.B.. 2000. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 151:739–748 10.1083/jcb.151.4.739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev O., Ermolaeva M., and Gautam N.. 1995. Efficient interaction with a receptor requires a specific type of prenyl group on the G protein γ subunit. J. Biol. Chem. 270:25356–25358 10.1074/jbc.270.43.25356 [DOI] [PubMed] [Google Scholar]
- Kitten G.T., and Nigg E.A.. 1991. The CaaX motif is required for isoprenylation, carboxyl methylation, and nuclear membrane association of lamin B2. J. Cell Biol. 113:13–23 10.1083/jcb.113.1.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Kim Y., Weaver B.A., Mao Y., McLeod I., Yates J.R. III, Tagaya M., and Cleveland D.W.. 2005. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J. Cell Biol. 169:49–60 10.1083/jcb.200411118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda Y., Suzuki N., and Kataoka T.. 1993. The effect of posttranslational modifications on the interaction of Ras2 with adenylyl cyclase. Science. 259:683–686 10.1126/science.8430318 [DOI] [PubMed] [Google Scholar]
- Lerner E.C., Zhang T.T., Knowles D.B., Qian Y., Hamilton A.D., and Sebti S.M.. 1997. Inhibition of the prenylation of K-Ras, but not H- or N-Ras, is highly resistant to CAAX peptidomimetics and requires both a farnesyltransferase and a geranylgeranyltransferase I inhibitor in human tumor cell lines. Oncogene. 15:1283–1288 10.1038/sj.onc.1201296 [DOI] [PubMed] [Google Scholar]
- Li R., and Murray A.W.. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531 10.1016/0092-8674(81)90015-5 [DOI] [PubMed] [Google Scholar]
- Liao H., Winkfein R.J., Mack G., Rattner J.B., and Yen T.J.. 1995. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J. Cell Biol. 130:507–518 10.1083/jcb.130.3.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z., Diaz B., Marshall M.S., and Avruch J.. 1997. An intact Raf zinc finger is required for optimal binding to processed Ras and for ras-dependent Raf activation in situ. Mol. Cell. Biol. 17:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen B.F., Chan G.K., Zubrowski B., Savoian M.S., Sauer M.T., and Yen T.J.. 2001. CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell. 12:2776–2789 10.1091/mbc.12.9.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mical T.I., and Monteiro M.J.. 1998. The role of sequences unique to nuclear intermediate filaments in the targeting and assembly of human lamin B: evidence for lack of interaction of lamin B with its putative receptor. J. Cell Sci. 111:3471–3485. [DOI] [PubMed] [Google Scholar]
- Nagasu T., Yoshimatsu K., Rowell C., Lewis M.D., and Garcia A.M.. 1995. Inhibition of human tumor xenograft growth by treatment with the farnesyl transferase inhibitor B956. Cancer Res. 55:5310–5314. [PubMed] [Google Scholar]
- Porfiri E., Evans T., Chardin P., and Hancock J.F.. 1994. Prenylation of Ras proteins is required for efficient hSOS1-promoted guanine nucleotide exchange. J. Biol. Chem. 269:22672–22677. [PubMed] [Google Scholar]
- Raponi M., Lancet J.E., Fan H., Dossey L., Lee G., Gojo I., Feldman E.J., Gotlib J., Morris L.E., Greenberg P.L., et al. 2008. A 2-gene classifier for predicting response to the farnesyltransferase inhibitor tipifarnib in acute myeloid leukemia. Blood. 111:2589–2596 10.1182/blood-2007-09-112730 [DOI] [PubMed] [Google Scholar]
- Rowell C.A., Kowalczyk J.J., Lewis M.D., and Garcia A.M.. 1997. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J. Biol. Chem. 272:14093–14097 10.1074/jbc.272.22.14093 [DOI] [PubMed] [Google Scholar]
- Rubio I., Wittig U., Meyer C., Heinze R., Kadereit D., Waldmann H., Downward J., and Wetzker R.. 1999. Farnesylation of Ras is important for the interaction with phosphoinositide 3-kinase γ. Eur. J. Biochem. 266:70–82 10.1046/j.1432-1327.1999.00815.x [DOI] [PubMed] [Google Scholar]
- Schaar B.T., Chan G.K., Maddox P., Salmon E.D., and Yen T.J.. 1997. CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139:1373–1382 10.1083/jcb.139.6.1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer-Hales K., Iaconelli J., Snyder J.P., Prussia A., Nettles J.H., El-Naggar A., Khuri F.R., Giannakakou P., and Marcus A.I.. 2007. Farnesyl transferase inhibitors impair chromosomal maintenance in cell lines and human tumors by compromising CENP-E and CENP-F function. Mol. Cancer Ther. 6:1317–1328 10.1158/1535-7163.MCT-06-0703 [DOI] [PubMed] [Google Scholar]
- Sebti S.M.2005. Protein farnesylation: implications for normal physiology, malignant transformation, and cancer therapy. Cancer Cell. 7:297–300 10.1016/j.ccr.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Sebti S.M., and Der C.J.. 2003. Opinion: Searching for the elusive targets of farnesyltransferase inhibitors. Nat. Rev. Cancer. 3:945–951 10.1038/nrc1234 [DOI] [PubMed] [Google Scholar]
- Sepp-Lorenzino L., Ma Z., Rands E., Kohl N.E., Gibbs J.B., Oliff A., and Rosen N.. 1995. A peptidomimetic inhibitor of farnesyl:protein transferase blocks the anchorage-dependent and -independent growth of human tumor cell lines. Cancer Res. 55:5302–5309. [PubMed] [Google Scholar]
- Sinensky M.2000. Recent advances in the study of prenylated proteins. Biochim. Biophys. Acta. 1484:93–106 10.1016/S1388-1981(00)00009-3 [DOI] [PubMed] [Google Scholar]
- Tanudji M., Shoemaker J., L’Italien L., Russell L., Chin G., and Schebye X.M.. 2004. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell. 15:3771–3781 10.1091/mbc.E03-07-0482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D., Wan X., Cheerambathur D., Gassmann R., Suzuki A., Lawrimore J., Desai A., and Salmon E.D.. 2013. Spindle assembly checkpoint proteins are positioned close to core microtubule attachment sites at kinetochores. J. Cell Biol. 202:735–746 10.1083/jcb.201304197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten V.L., Peckham L.A., Olive M., Capell B.C., Collins F.S., Nabel E.G., Young S.G., Fong L.G., and Lammerding J.. 2011. Protein farnesylation inhibitors cause donut-shaped cell nuclei attributable to a centrosome separation defect. Proc. Natl. Acad. Sci. USA. 108:4997–5002 10.1073/pnas.1019532108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E., and Winey M.. 1996. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132:111–123 10.1083/jcb.132.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte D.B., Kirschmeier P., Hockenberry T.N., Nunez-Oliva I., James L., Catino J.J., Bishop W.R., and Pai J.K.. 1997. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J. Biol. Chem. 272:14459–14464 10.1074/jbc.272.22.14459 [DOI] [PubMed] [Google Scholar]
- Wong N.S., and Morse M.A.. 2012. Lonafarnib for cancer and progeria. Expert Opin. Investig. Drugs. 21:1043–1055 10.1517/13543784.2012.688950 [DOI] [PubMed] [Google Scholar]
- Wright L.P., and Philips M.R.. 2006. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. J. Lipid Res. 47:883–891 10.1194/jlr.R600004-JLR200 [DOI] [PubMed] [Google Scholar]
- Yao X., Abrieu A., Zheng Y., Sullivan K.F., and Cleveland D.W.. 2000. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat. Cell Biol. 2:484–491 10.1038/35019518 [DOI] [PubMed] [Google Scholar]
- Zhang F.L., and Casey P.J.. 1996. Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65:241–269 10.1146/annurev.bi.65.070196.001325 [DOI] [PubMed] [Google Scholar]
- Zhu X., Chang K.H., He D., Mancini M.A., Brinkley W.R., and Lee W.H.. 1995. The C terminus of mitosin is essential for its nuclear localization, centromere/kinetochore targeting, and dimerization. J. Biol. Chem. 270:19545–19550 10.1074/jbc.270.33.19545 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.