Abstract
Ideal nanoparticle (NP)-based drug and vaccine delivery vectors should be free of inherent cytotoxic or immunostimulatory properties. Therefore, determining baseline immune responses to nanomaterials is of utmost importance when designing human therapeutics. We characterized the response of human immune cells to hydrogel NPs fabricated using Particle Replication in Non-wetting Templates (PRINT) technology. We found preferential NP uptake by primary CD14+ monocytes, which was significantly reduced upon PEGylation of the NP surface. Multiplex cytokine analysis of NP treated primary human peripheral blood mononuclear cells (hu-PBMC) suggests that PRINT based hydrogel NPs do not evoke significant inflammatory responses nor do they induce cytotoxicity or complement activation. We furthered these studies using an in vivo humanized mouse model and similarly found preferential NP uptake by human CD14+ monocytes without systemic inflammatory cytokine responses. These studies suggest that PRINT hydrogel particles form a desirable platform for vaccine and drug delivery as they neither induce inflammation nor toxicity.
Keywords: PRINT, Nanoparticles, PEGylation, humanized mice, monocytes, complement, toxicity
Introduction
Biocompatible particles at the nano/micron scale are emerging tools for biological programming with the capacity to induce specific cellular responses in a variety of disease contexts. This has been demonstrated using mouse models of anti-microbial vaccines, cancer immunotherapy, tolerance induction during autoimmunity and siRNA-mediated gene knockdown (1–9). Concomitant with advances in nanotechnology there is a widespread appreciation for ensuring nanoparticle safety and biocompatibility with special consideration for adverse immune responses (10, 11). Therefore, determining how novel drug and vaccine platforms interact with the human immune system is of critical importance.
The particle replication in nonwetting templates (PRINT) process uses soft lithography to generate monodisperse NPs with highly defined size, shape, charge and composition (12, 13). PRINT particles can be engineered to carry a variety of payloads including chemotherapeutics, imaging agents, antigens and adjuvants (14, 15). Additionally, the particle surface can be modified to facilitate antibody or cellular receptor based targeting and the addition of short PEG chains (PEGylation) can reduce NP clearance in mice by preventing serum protein binding and subsequent recognition by phagocytic cells (16, 17).
The nanotechnology field is continually expanding and encompasses a variety of materials, some of which are reported to induce toxicity and/or adverse immune responses, including metals, metal oxides, carbon-nanotubes, liposomes and some polymer based particles (10, 11, 18). A drawback of many therapeutic platforms is the inadvertent induction of host immune responses, which can lead to rapid clearance of the therapeutic and/or be deleterious to host. An additional consideration is the heterogeneity amongst immune responses within the human population. Our recent report showed that macrophages from the blood of healthy donors have differing abilities to ingest NPs that correlated with the M1/M2 activation status of the macrophage, with M2-like cells taking up more NPs (19). We found the same results using various inbred mouse strains with known skewing toward M1 or M2-like phenotypes, indicating that responses to nanomaterials in conventional mouse strains can vary greatly depending on their immunological makeup. More complexity arises when trying to extrapolate results from rodent model systems to therapeutic human applications and suggests that a “one-size-fits-all” approach to nanoscale clinical intervention is too simplistic a model. Furthermore, the fabrication process, choice of material, particle dimensions, route of delivery and dose may differentially affect immune responses. For example, some NP formulations have toxic effects on host cells by inducing cell death through oxidative stress, whereas other particulate compositions of polystyrene and PLGA are sufficient to activate innate immune responses, such as the NLRP3 inflammasome (20, 21). Therefore, a critical first step in development of novel vaccine and drug delivery platforms is to test their baseline compatibility with the human immune system.
One major consideration when developing particulate therapeutics is the potential for activating the NLRP3 inflammasome present in phagocytic cells (22). Inflammasomes are activated by two signals; signal one (also called priming) results in intracellular accumulation of inflammasome subunits and the pro-inflammatory cytokines IL-1β and IL-18. Signal two triggers inflammasome activation and subsequent release of mature IL-1β and IL-18, which rapidly recruit and activate inflammatory cells. If uncontrolled this can lead to systemic inflammation followed by organ failure and even death (23)(24, 25). It is known that particulate irritants such as alum, silica and asbestos are potent activators of the inflammasome and recent work has suggested that particles comprised of titanium oxide, silver, PLG and carbon can also trigger the NLRP3 inflammasome (21, 22, 26–31). The adverse effects of nanotherapeutics depends on many factors, including particle geometry, method of fabrication, dose/route of administration and the tissues/cell types assessed as evidenced by the contrasting results regarding nanoparticle triggering of the NLRP3 inflammasome (21, 32–34). In addition, many studies of particle uptake and toxicity are carried out exclusively in cell lines, most often murinederived, which may not reflect responses by primary human cells (11). The studies in this manuscript test the interaction of human immune cells with PRINT fabricated hydrogel particles using primary human peripheral blood and an in vivo humanized mouse model.
Materials and Methods
Particle Materials
Poly(ethylene glycol) diacrylate (Mn 700) (PEG700DA), 2-aminoetheyl methacrylate hydrochloride (AEM), and Diphenyl (2,4,6-trimethylbenzoyl)-phosphine oxide (TPO) were from Sigma-Aldrich. Tetraethylene glycol monoacrylate (HP4A) was synthesized in-house as previously described (35). Thermo Scientific maleimide-terminated Dylight 650 and Dylight 488, PTFE syringe filters (13mm membrane, 0.220 µm pore size), dimethylformamide (DMF), triethanolamine (TEA), pyridine, sterile water, borate buffer (pH 8.6), Dulbecco’s 1× phosphate buffered saline (DPBS) (pH 7.4), acetic anhydride and methanol were obtained from Fisher Scientific. Methoxy-PEG(5k)-succinimidyl carboxy methyl ester (mPEG5k-SCM) was purchased from Creative PEGWorks. Conventional filters (2 µm) were purchased from Agilent and polyvinyl alcohol (Mw 2000) (PVOH) was purchased from Acros Organics. Liquidia Technologies kindly provided the 80 nm × 320 nm PRINT molds used in these studies.
Fabrication of PRINT HP4A particles
The process of fabricating hydrogel particles has been described previously (12). Briefly, the pre-particle solution of 67.5 wt% HP4A, 20 wt% AEM (functional monomer), 10 wt% PEG700DA (crosslinker), 1 wt% TPO (photo initiator) and 1.5 wt% Dylight 650 or Dylight 488 maleimide was dissolved at 3.5 wt% in methanol and drawn as a thin film using a #3 Mayer rod (R.D. Specialties) onto a roll of corona treated PET using an in-house custom-made roll-to-roll lab line (Liquidia Technologies). The delivery sheet was laminated to the patterned side of the mold, followed by delamination at the nip. Particles were cured by passing the filled mold through a UV-LED and a PVOH harvesting sheet was hot laminated to the filled molds of 80×320 nm rods. Particles were removed from the mold at room temp by splitting the PVOH harvesting sheet from the mold and harvested using 1 mL of water per 5 ft of harvesting sheet. Particle suspensions were passed through a 2 µm filter (Agilent) to remove large particulates. Excess PVOH was removed by centrifugation of NPs and 4 washes with sterile water prior to final resuspension in water.
PEGylation of 80×320 nm HP4A NPs
Free primary amine groups on the HP4A particles were used as chemical handles to react with a methoxy-PEG5K-SCM. Purified 80×320 nm hydrogel particles were exchanged from water to DMF at (1 mg/mL) and were reacted with TEA (100 µL) for 10 min at room temperature on a shaker plate at 1400 rpm. 14 mg of methoxy-PEG5K-SCM was dissolved in DMF and added to the reaction mixture, shaken overnight and then quenched with borate buffer (100 µL). The nanoparticle solution was then centrifuged (21,000 RCF; 15 minutes) and washed 5 times with DMF. Following PEGylation, particles were acetylated with acetic anhydride to quench any unreacted amines and washed once in DMF, once in borate buffer and four times in sterile water.
Print Particle Characterization
Stock particle concentrations were determined using thermogravimetric analysis (TGA; TA Instruments Q5000 TGA). The stock particle solution (20 µL) was pipetted into a tared aluminum sample pan. The sample was heated at 30°C/min to 130°C and held at this temperature for 10 minutes followed by cooling at 30°C/min to 30°C and held for 2 minutes. To account for the mass of any stabilizer remaining in each sample, TGA was performed on a 20 µL aliquot of supernatant from a centrifuged sample of the stock particle solution. A Zetasizer Nano ZS (Malvern Instruments, Ltd.) was used to measure particle size and zeta potential by dynamic light scattering (DLS). NPs were prepared for scanning electron microscopy (SEM) by coating with 1.5 nm of gold-palladium alloy using a Cressington 108 auto sputter coater and imaged using a Hitachi S-4700 scanning electron microscope.
Particle Uptake and Cytokine Secretion by THP-1cells
THP-1 monocytes were seeded at 2×105 cells/well in 96 well plates and incubated with varying doses of NPs for 24 hours or with 10–20 ng/ml LPS (O111:B4 standard preparation, Invivogen) and/or 20 µm nigericin (Invivogen) for the final 40 mins as positive controls for cytokine production. Cell supernatants were collected and assessed for cytokine production by ELISA. Cells were washed 3 times with FACS buffer and assessed for NP uptake using flow cytometry. All studies were prefomed using RPMI media with heat inactivated 10% fetal bovine serum.
Human Peripheral Blood Mononuclear Cell (hu-PBMC) Isolation and Culture
Blood was collected from healthy donors or leukapheresed patients in accordance with the University of North Carolina’s Office of Human Research Ethics (IRB #12-1858 and #05-2860). Donors provided written informed consent and samples were anonymized and de-identified prior to use in the described studies. Peripheral blood mononuclear cell (PBMC) buffy coats were prepared using density gradient centrifugation (Lymphoprep from Cosmo Bio USA). Hu-PBMC were plated at 3×105 cells per well in 96 well plates using AIM V medium (Gibco) supplemented with 1% penicillin/streptomycin, 1% L-glutamine and 10% heat-inactivated ABhuman serum (Invitrogen). After 4 hours of rest, PRINT NP (10µg/ml) were added for 8 hours and supernatant was harvested for ELISA and cells were assessed by flow cytometry.
Enrichment of primary human CD14+ monocytes and NP cytotoxicity analysis
Primary human CD14+ monocytes were enriched from buffy coat using Dynabeads® Untouched™ Human Monocytes Kit (Invitrogen™) according to manufacturer instructions and purity assessed by flow cytometry (>95% pure). Cell cytotoxicity was determined using the luciferase based Toxilight™ bioassay (Lonza) in response to 100, 10 and 1 µg/ml NP concentrations for 12 and 24 hours according the manufacturer’s instructions. ToxiLight™ 100% lysis reagent was used as positive control for the assay. Cell viability analysis using MTT (Life Technologies) was performed on NP treated human peripheral blood buffy coat cells by adding 0.5 mg/ml MTT for 2hr at 37°C followed by formazan solubilization overnight using 5% SDS and 5 mM HCl. Plates were assayed for 550 nm absorbance and a reference wavelength of 690 with values plotted after subtraction of background signal. UV irradiated cells were used as a positive control for cell death in MTT assays.
Analysis of Cytokine Production
Human TNFα, IL-6 and IL-1β secretion by THP-1 cells were analyzed using BD-OPtiea ELISA kits (BD Biosciences) according to the manufacturer’s instructions. Cytokine production by primary hu-PBMC was analyzed using the human 30-Plex Panel (Life Technologies) on the Luminex platform.
Analysis of Complement Activation
Studies of NP mediated complement activation were performed as described with modifications (36). Briefly, samples containing 10 µl human plasma (non-heat inactivated) (Sigma-Aldrich, lot SLBH6826V), 10 µl veronal buffer with Ca2+ and Mg2+ (Complement Technology Inc.), 10 µl HP4A or HP4A-PEG NP in PBS (final concentration 10ug/ml) were incubated for 2 hr or 8 hr at 37°C. Plasma treated with PBS served as negative control and 50 units of cobra venom factor (Complement Technology Inc.) served as positive control for complement activation. Following treatment, samples were boiled for 5 minutes in 1×SDS sample buffer containing DTT, diluted 1:400 and 12 µl was fractionated on a 4%–10% Bis/Tris gel (Invitrogen) followed by transfer to PVDF membrane (Millipore). Blots were probed with polyclonal goat anti-human C3 (Complement Technology Inc.) using a 1:2000 dilution followed by probing with donkey anti-goat IgG- IRDye® 800CW (Li-Core Biosciences) using a 1:20,000 dilution. Membranes were scanned using Li-Core Odyssey Infrared Imaging system (Li-Core Biosciences).
CD14 blockade
The anti-CD14 blocking antibody (clone M5E2 LEAF™ grade, Biolegend) and isotype control (MOPC-173 LEAF™ grade, Biolegend) were tested empirically for the ability to block the CD14 receptor. Primary hu-PBMC were pre-incubated with 50ug/ml anti-CD14 (M5E2) or isotype control for 1 hour followed by NP treatment (10 µg/ml) for 8 hours. Cells were washed 3 times with FACS buffer and NP uptake was assessed by flow cytometry.
Humanized Mouse Experiments
All animal experiments were carried out humanely and followed protocols approved by the University of North Carolina-Chapel Hill Institutional Animal Care and Use Committee (IACUC). A detailed protocol for preparation of NOD.Rag1−/−Il2rg−/− (NRG) humanized mice has previously been published (37–40). In brief, human CD34+ cells were isolated from 15- to 18-week-old fetal liver tissues using a Ficoll gradient (GE Healthcare Bio-science AB) and CD34+ MicroBead Kit from Miltenyi Biotec. One to 3 day old NRG mice were irradiated at 200 rad and injected intra-hepatically with 0.5–1×106 hu-CD34+ cells. Transplanted mice were bled 3–4 months later to assess human immune cell reconstitution by flow cytometry and only mice with >50% chimerism were used in these studies. Humanized mice were injected i.v. with 100µg of 80×320 nm hydrogel particles. Acetylated particles (HP4A) contained Dylight 650, while PEGylated versions of the same particle (HP4A-PEG) contained Dylight 488. Twenty-four hours after particle injections, mice were euthanized for tissue analysis by flow cytometry and quantative realtime PCR (Q-RT PCR).
Flow Cytometry
Antibodies used in hu-PBMC staining; CD3-FITC, CD19-eFluor®450, CD56-PerCP-Cy5, CD14-PE, CD11b-APC-Cy7, and CD11c –PE-Cy7 (eBioscience). Live cells were determined using the Live/Dead Aqua Fixable Dead Stain Kit (Invitrogen). Antibodies used for humanized mouse experiments: CD11c-FITC, CD56-FITC, CD14-PE, CD4-PE-Cy5, CD3-PE-Cy7, CD123-PE-Cy5, CD19-PB, CD11c-APC, CD56-APC, CD45-APC-Cy7 (Biolegend); CD8-PE-TR, CD4-PE-TR, mouse CD45-Pacific Orange (Invitrogen). FACS buffer; 1×PBS (Gibco) + 2% FBS (Sigma). All data were collected using an LSRII (BD Biosciences) or CyAn™ (Beckman Coulter) flow cytometer and analyzed using FlowJo (Tree Star, Inc.).
Quantitative Real-time PCR
RNA was purified from spleens of humanized mice using the RNeasy Plus Minikit (Qiagen) and reverse transcribed to cDNA using M-MLV RT (Invitrogen) and oligo-dT. Human cytokine gene expression was assayed using KiCqStart SYBR Green primers for IL1B, IL6, and TNF (Sigma Aldrich). Primer sequences are as follows: IL1B Fwd – 5’-ACAGTGGCAATGAGG ATGAC-3’, IL1B Rev – 5’-CCATGGCCACAACAACTGA-3’; IL6 Fwd – 5’-GGTACAT CCTCGACGGCATCT -3’, IL6 Rev –5’-GTGCCTCTTTGCTGCTTTCAC -3’; TNF Fwd – 5’-CTCT TCTGCCTGCTGCACTT-3’, TNF Rev – 5’-GGCTACAGGCTTGTCACTC-3’.
Statistical Analysis
Data were evaluated for statistical significance using Analysis of Variance (ANOVA) with the Bonferroni post-test using the GraphPad Prism 5 software.
Results
Characterization of monodisperse and homogenous PRINT nanoparticles
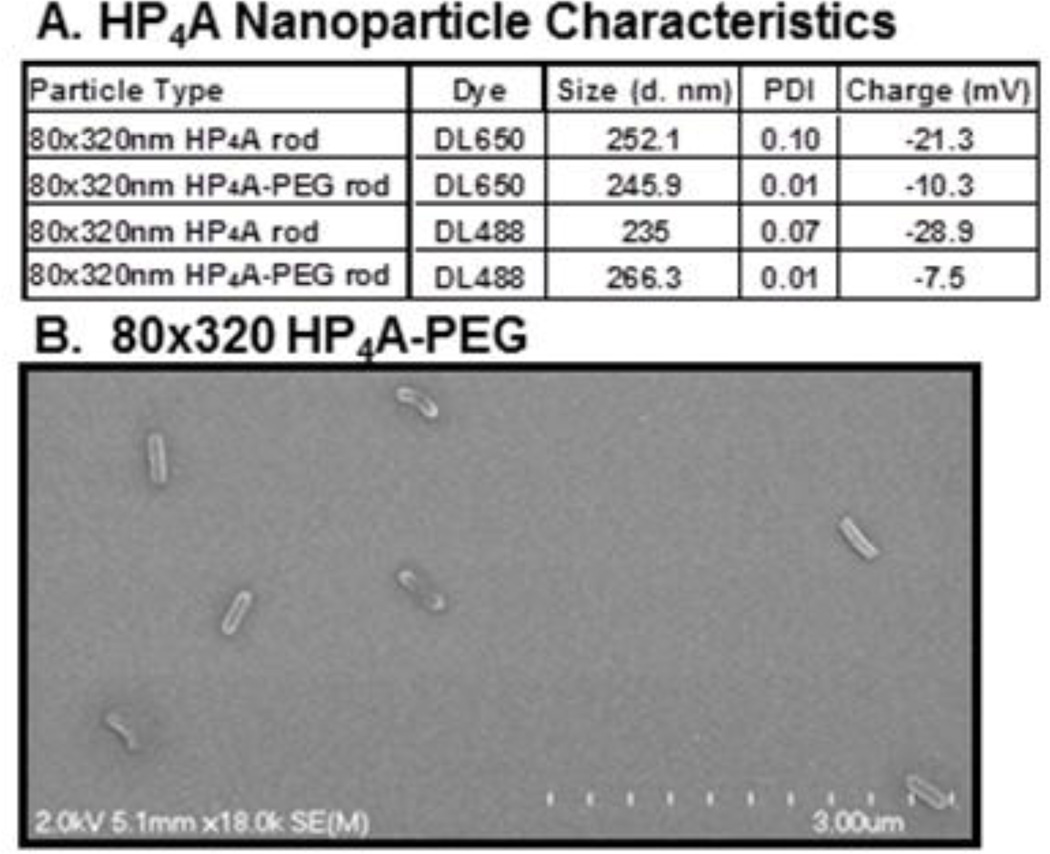
The PRINT fabrication process was used to generate rod shaped 80×320 nm hydrogel particles with a base component of HP4A. Aliquots of the 80×320 nm HP4A-NPs were modified by covalent attachment of short PEG5k chains to yield PEGylated HP4A-NP (HP4A-PEG). Particle uptake was tracked using fluorescent dyes (Dylight 650 or 488) that were covalently incorporated during particle fabrication. NPs were characterized by DLS to measure size, surface charge (zeta potential) and particle homogeneity using the polydispersity index (PDI) (Fig. 1A). All particles were of the expected size and had a negative surface charge ranging from −7.5 to −28.9 mV. The PDI for all nanoparticles was ≤0.1 indicating a high level of homogeneity within each batch which was confirmed using SEM (Fig. 1B).
Figure 1. HP4A-based PRINT NP Characterization.
(A) Size, polydispersity index (PDI) and surface charge of HP4A and HP4A-PEG particles used in these studies. (B) SEM image of 80×320 nm HP4A-PEG rods.
Uptake and immune responses to HP4A particles by the human THP-1 cell line
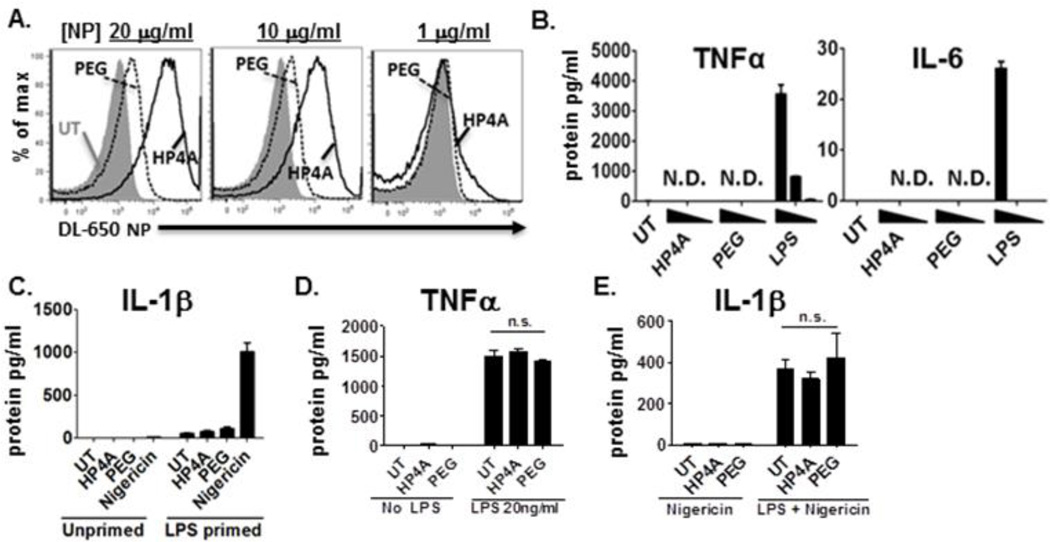
To determine how 80×320 nm HP4A particles interact with human immune cells we incubated the THP-1 monocyte line with fluorescently labeled NP for 24 hours and uptake was measured by flow cytometry. We found that HP4A-NPs were taken up in a dose dependent manner (20, 10, 1 µg/ml) and that PEGylation of the NP surface (HP4A-PEG) dramatically reduced particle uptake (Fig 2A). Supernatants from these cultures had undetectable levels of TNFα and IL-6 by ELISA suggesting that the HP4A based NPs do not elicit inflammatory cytokine responses (Fig. 2B). We further tested whether these NPs induced IL-1β secretion alone or as a second signal following LPS priming. We were unable to detect IL1-β in the supernatant of LPS primed (10 ng/ml) or unprimed THP-1 cells treated with NPs (20µg/ml), whereas nigericin (20 µM, 40 mins), a known activator of the inflammasome, induced robust IL-1β release (Fig. 2C). To determine if pre-treatment with HP4A-based NPs alters cytokine secretion in response to inflammatory cues we incubated THP-1 cells with (10 µg/ml) HP4A or HP4A-PEG NP for 16 hours followed by stimulation with LPS (20 ng/ml) for 4 hours for TNFα production, or LPS (20 ng/ml) for 4 hours and nigericin (20µM) for 40mins for IL-1β production. We detected no alteration in TNFα (Fig. 2D) or IL-1β (Fig. 2E) secretion when cells were pre-treated HP4A-based NPs. Together, these data indicate that HP4A-based NP do not induce, enhance or dampen pro-inflammatory cytokine release or inflammasome activation.
Figure 2. HP4A-based 80×320 nm rod uptake without inflammatory cytokine responses by THP-1 cells.
(A) Dose dependent uptake of HP4A (solid black) and HP4A-PEG (dashed black) NP after 24 hr. Grey fill, untreated cells. (B) ELISA for TNFα and IL-6 secretion 24 hours after NP treatment (20, 10, 1 µg/ml). Pos.cntrl, LPS treatment (100, 10, 1 ng/ml) for 8 hr. (C) ELISA for IL-1β secretion after 24hr NP treatment alone (20 µg/ml) or with LPS (10 ng/ml) added in the final 8 hours. Nigericin (20 µM) was added in the final 40 mins. as pos. cntrl for IL-1β secretion. (D) ELISA for TNFα secretion by THP-1 pre-treated with NP (10 µg/ml) for 16 hr, followed by LPS (20 ng/ml) for 4hr. (E) ELISA for IL-1β secretion by THP-1 pre-treated with NP (10 µg/ml) for 16 hr, followed by LPS treatment (20 ng/ml) for 4hr and nigericin (20 µM) in the final 40 mins. Statistical analysis by 2-way ANOVA. N.D., not detected; n.s., not significant.
Analysis of HP4A NP uptake and toxicity studies using primary human immune cells
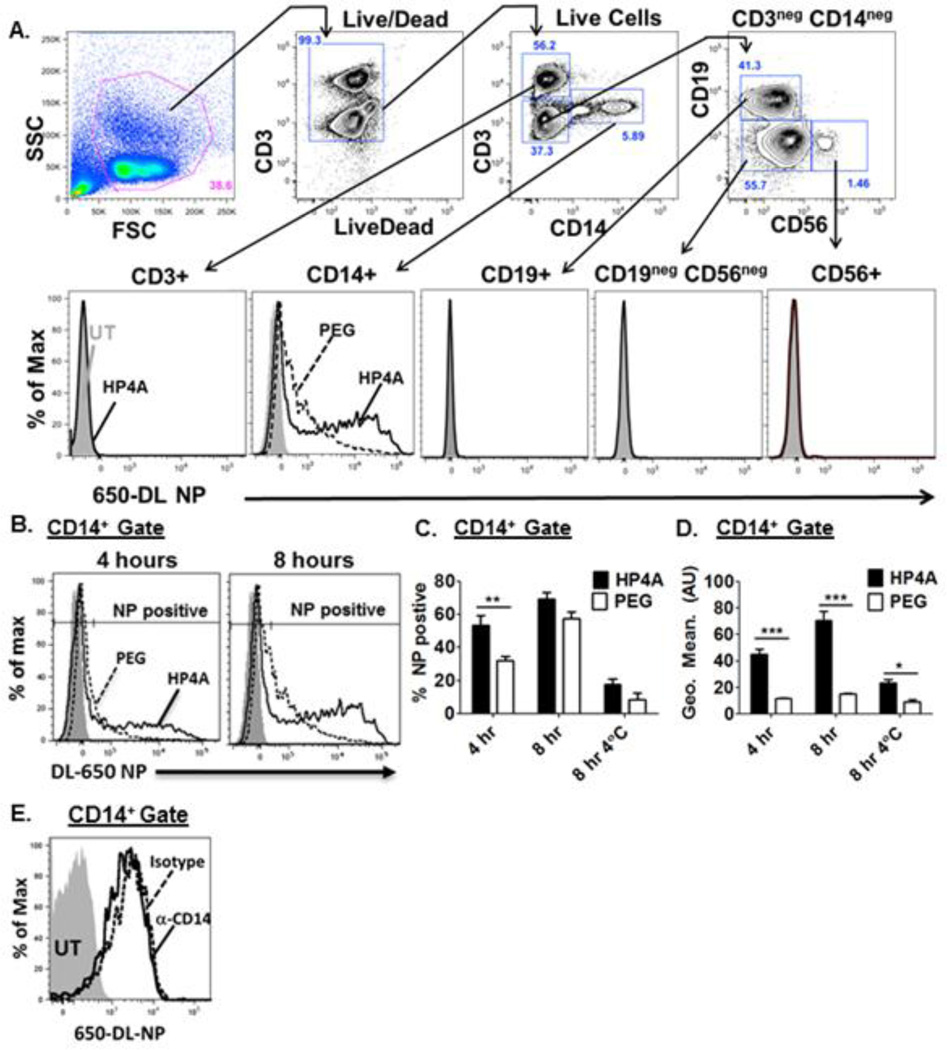
The above data from THP-1 cells suggests that HP4A-based NPs are immunologically inert and we sought to further these studies using primary human cells. Hu-PBMC were obtained from multiple donors and treated ex-vivo with fluorescently labeled HP4A or HP4A-PEG NPs for 8 hours and analyzed for particle uptake using flow cytometry. NPs were exclusively detected in CD14+ monocytes, with undetectable NP uptake in B, T and NK cells (Fig. 3A). PEGylation of the NP surface reduced the frequency of particle uptake at the 4 hour time point; however, by 8 hours similar percentages of cells had taken up detectable levels of NPs (Fig. 3B+C). While the frequency of NP positive cells was similar after 8 hours, the mean fluorescence intensity (MFI) in the CD14+NP+ gate following HP4A-NP treatment was significantly higher than cells treated with HP4A-PEG particles at both 4 hour and 8 hour time points (Fig. 3D), indicating that the actual quantity of particles per cell is reduced with PEGylation (Fig. 3D). These data parallel those obtained in the THP-1 studies and are consistent with other studies showing that PEGylation reduces phagocytosis of drugs and nanomaterials (16, 41). Hu-PMBC were also incubated with NPs at 4°C to distinguish between NP uptake and cell surface binding with the expectation that minimal uptake will occur at low temperatures. We observed a significant difference in the MFI of un-PEGylated NPs compared to PEGylated particles at 4°C to allow NP binding but prevent phagocytosis (Fig. 3D), suggesting that part of the mechanism by which PEGylation reduces uptake may be through reduced NP binding to the cell surface.
Figure 3. HP4A-based 80×320 nm rod uptake by primary human PBMC.
(A) Gating scheme used to analyze HP4A and HP4A-PEG NP uptake by T cells (CD3+), monocytes (CD14+), B cells (CD19+) and NK cells (CD56+) (B) Representative plot of HP 4 A (solid black) and HP4A-PEG (dashed black) NP uptake by CD14 + cells after 4 and 8 hr treatment with NP (10 µg/ml). Grey fill, untreated cells. (C+D) Combined data from 4 donors for frequency of CD14 + NP + cells (C) and mean fluorescence intensity (MFI) of NP signal within the CD14 + NP + gate (D). (E) Uptake of HP 4 A-NP (10 µg/ml) by CD14 + hu-PBMC at 8 hr in the presence of CD14 blocking antibody (50 µg/ml), solid line or isotype control antibody (50 µg/ml), dashed line. Statistical analysis by 2-way ANOVA. *P<0.05, **P<0.01, ***P<0.001 Data in A-D are representative of two independent experiments using a total of 8 donors. Data in E are representative of two independent experiments using a total of 5 donors.
Uptake of HP4A-NPs was exclusively detected in the CD14+ population of hu-PBMC (Fig. 3A), which warranted further investigation as to whether CD14 itself was involved in NP uptake. CD14 has been implicated in binding a variety of ligands with differing characteristics including bacterial lipids (e.g. LPS), viral glycoproteins, double stranded viral RNA and monosodium urate crystals (42–45). To determine if HP4A-NP uptake by CD14+ cells requires sensing through CD14 we assessed NP uptake in the presence of a known CD14 blocking antibody (M2E5) (46). The optimal antibody concentration was empirically determined by incubating THP-1 cells for 1 hour with various doses of M2E5 antibody or an isotype control followed by treatment with LPS and detection of TNFα secretion by ELISA. We found that CD14 blockade reduced THP-1 responses to LPS in a dose dependent manner with a 50µg/ml dose of M2E5 yielding a 90% blockade in CD14 activity (Fig. S1). We therefore used the 50µg/ml dose of M2E5 to determine the effects of CD14 blockade on HP4A-NP uptake by primary hu-PBMC and found that blocking CD14 had no effect on particle uptake compared to isotype control treated cells (Fig. 3E). We conclude that preferential uptake of HP4A-based NP by CD14+ monocytes is likely independent of CD14 function.
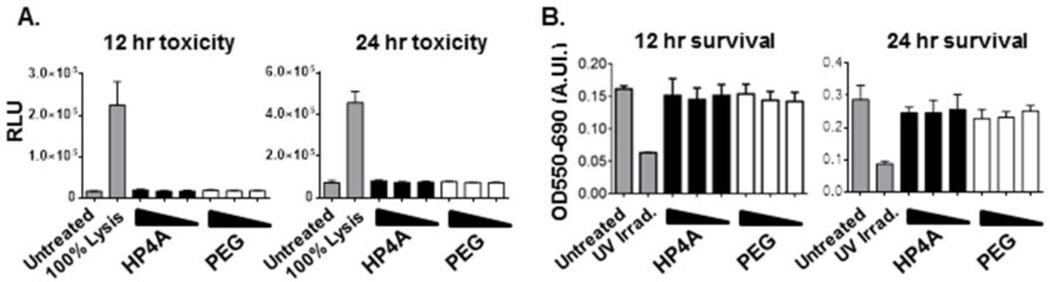
To determine if HP4A-based NPs induce cytotoxicity we treated whole buffy coat or enriched CD14+ monocytes (purity Fig. S2) with 100, 10 and 1 µg/ml doses of HP4A-based NP for 12 and 24 hours and monitored for cytotoxicity using two approaches. The first approach measured adenylate kinase release due to the loss of cell membrane integrity that coincides with cellular damage. We found no increase in adenylate kinase release from CD14 enriched cells treated with NPs compared to untreated cells (Fig. 4A), suggesting that the cell membrane remains uncompromised during NP treatment. Our second measure of cytotoxicity used an MTT-based assay to monitor cell survival following at 12 and 24 hours of NP treatment and found similar survival between untreated and NP treated cells (Fig. 4B). As a positive control for cell death we irradiated cells with UV light, which showed an expected decrease in MTT activity resulting from cytotoxicity (Fig. 4B). Based on these two studies we conclude that PRINT fabricated HP4A hydrogel NPs do not induce cytotoxicity in primary human immune cells.
Figure 4. Undetectable toxicity following treatment of primary hu-PBMC with HP4A-based 80×320 nm rods.
(A) Luciferase based adenylate kinase assay for cell toxicity after treatment of primary human CD14 + cells with 100, 10 and 1 µg/ml HP4A or HP4A-PEG NP at 12 and 24 hr. 100% lysis represents maximal toxicity signal. (B) MTT assay for cell survival after treatment of primary human peripheral blood buffy coat cells with 100, 10 and 1 µg/ml HP4A or HP4A-PEG NP at 12 and 24 hr. UV irradiated cells served as a positive control for cell death. Data in A are representative of two experiments using independently synthesized particles and cells from mixed donors. Data in B are combined results testing 6 independent batches of NP. RLU, relative light units; A.U., arbitrary units. Statistical Analysis by 1-way ANOVA with Tukey’s multiple comparisons test.
Cytokine induction and complement activation in response to HP4A-NP treatment
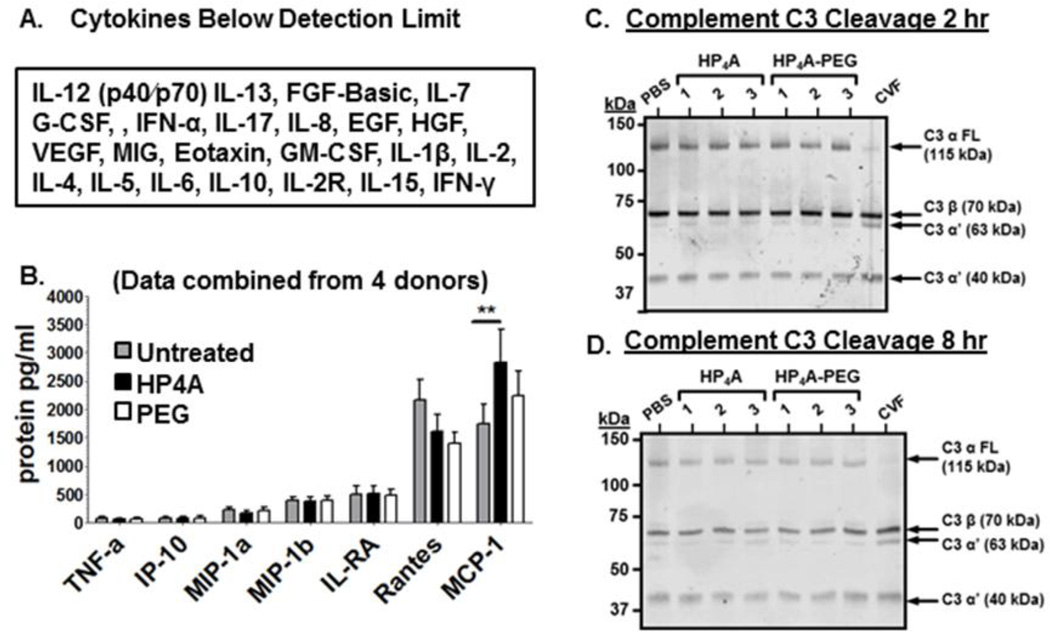
To determine if HP4A-NPs induce inflammatory responses from primary human immune cells we assessed supernatants of total hu-PBMC cultures treated with 10 µg/ml NP for 8 hours using a Luminex based array. Most of the 30 cytokines tested were below the limit of detection (Fig. 5A). Of the cytokines within the detectable range only a modest increase (<40%) in MCP-1 (CCL2) reached statistical significance compared to untreated hu-PBMC (Fig. 5B). Based on the results of this broad cytokine analysis study we conclude that HP4A-based NP do not elicit strong inflammatory cytokine responses from primary human immune cells.
Figure 5. Undetectable cytokine responses and complement activation following treatment of primary hu-PBMC with HP4A-based 80×320 nm rods.
(A) List of cytokines below the limit of detection in hu-PBMC cultures treated with 10 µg/ml HP4A or HP4A-PEG NP. (B) Multiplex cytokine ELISA data for untreated, HP4A or HP4A-PEG NP (10 µg/ml) treated PBMC after 8 hr. (C+D) Human complement C3α activation in plasma at 2 hr (C) and 8 hr (D) following treatment with HP4A and HP4A-PEG NP (10 µg/ml). Positive control; cobra venom factor (CVF) to induce C3α cleavage. Negative control PBS. Numbers indicate independent NP preparations (3 per particle type). Full length C3α (115 kDa). Expected C3α cleavage products (C3α’) 63 kDa and 40 kDa bands. The C3 β-chain (70 kDa) does not undergo cleavage and serves as a loading control. Data in A and B are combined from 4 independent PBMC donors. Statistical Analysis by 2-way ANOVA. **P<0.01.
We also tested whether HP4A-based NPs activate complement as other particle formulations have been shown to activate the complement pathway resulting in enhanced particle clearance and release of inflammatory mediators (36). One of the hallmarks of complement activation is cleavage of the complement C3α chain that results production of smaller fragments with opsonizing and inflammatory activity (47). We used a western blot approach to monitor cleavage of C3α in human plasma flowing incubation with PRINT HP4A-NPs compared to PBS alone or cobra venom factor (CVF) as a positive control for complement activation. Incubating CVF with human plasma results in loss of the full length C3α (115 kDa) and accumulation of the 63 kDa and 40 kDa C3α’ cleavage products after 2 hour and 8 hour treatments compared to PBS (Fig. 5C and D). In contrast, the full length C3α band remains intact when plasma is treated with HP4A and HP4A-PEG NP with no difference in accumulation of C3α’ products compared to PBS controls (Fig. 5C and D). From these data we conclude that PRINT HP4A-NPs are not significant activators of human complement or inflammatory cytokine production.
Uptake and immune responses to HP4A-NPs in humanized mice
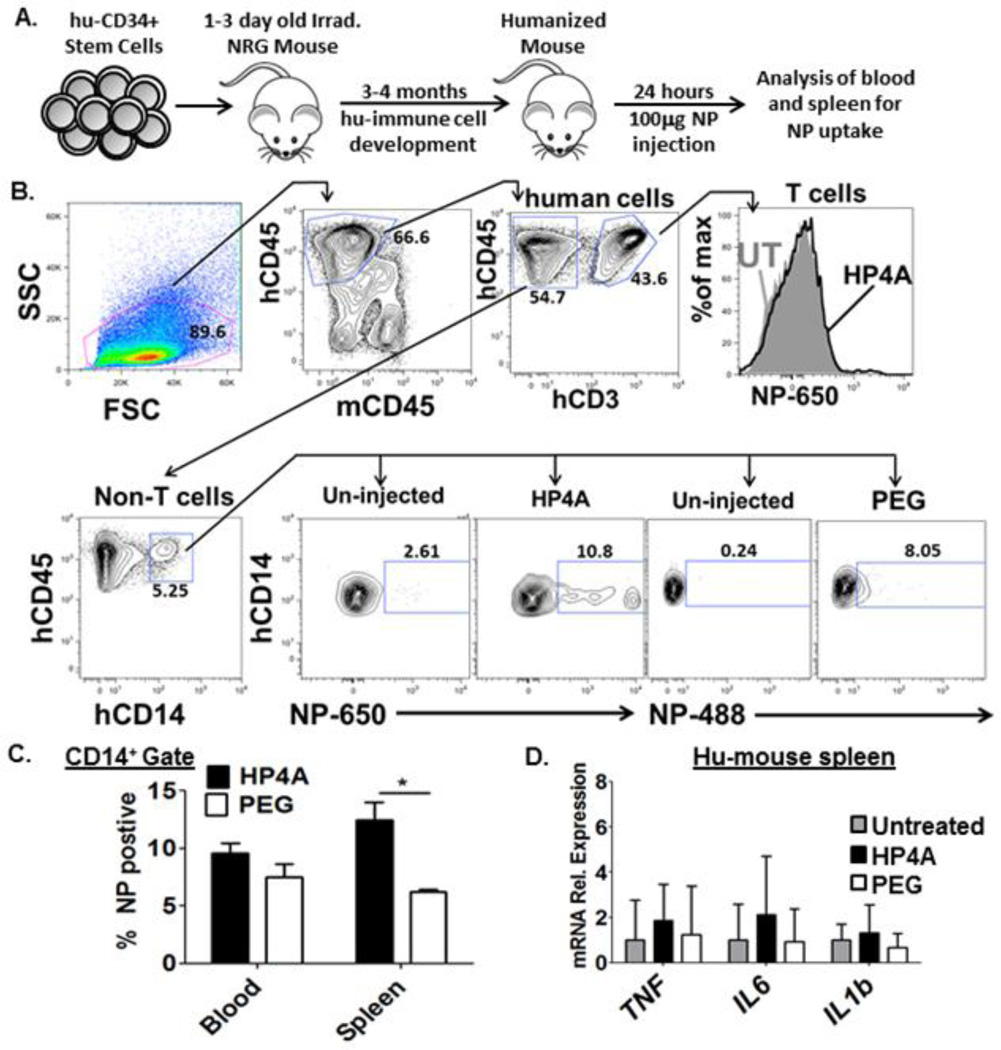
The above data from ex-vivo cultured hu-PBMC suggest that HP4A-NPs are immunologically inert with respect to inflammatory cytokine production, cytotoxicity and complement activation. Whether these findings hold true in vivo would require unobtainable IRB approval for human studies. Thus, to assess how PRINT fabricated HP4A-based NPs interact with human immune cells in vivo we utilized a humanized mouse model (experimental design summarized in Fig. 6A). In this model, human CD34+ hematopoietic stem cells are isolated from fetal liver and injected into irradiated newborn immunodeficient NRG mice. Within 3–4 months the mouse immune system is reconstituted with human immune cells and all mice used in these studies had >50% chimerism. It is worth noting that our previous work identified major differences in the in vivo circulation times of nanoparticles depending on the mouse strain background, specifically the immunologic makeup of these conventional mice (19). Therefore, our efforts to use a humanized mouse model also sought to improve upon this shortcoming of conventional strains by modeling circulating human immune cells in vivo.
Figure 6. HP4A-based 80×320 nm rod uptake and inflammatory cytokine responses using a humanized mouse model.
(A) Schematic of humanized mouse model. CD34+ cells from hu fetal liver are injected into 1–3 day old irradiated immunodeficient NRG (NOD.Rag1−/−Il2rg−/−) mice. 3–4 months post transplant the mice are injected with NP and human immune cells are analyzed for NP uptake. (B) Gating scheme used to analyze HP4A and HP4A-PEG (PEG) NP uptake by human T cells (anti-human CD45+ CD3+) or monocytes (anti-human CD45+CD14+). Gates for NP+ were set based on un-injected controls. (C) Frequency of NP+ cells in human CD14+ gate from blood and spleen of humanized mice 24 hours after 100 µg NP injection. (D) mRNA expression level of human pro-inflammatory cytokines in spleen of untreated and NP treated humanized mice. n = 4 animals per treatment group. Statistical Analysis by 2-way ANOVA. *P<0.05. Data are representative of two independent experiments.
Humanized mice were injected i.v. with 100µg of HP4A or HP4A-PEG NPs and analyzed 24 hours later for particle uptake in the spleen and blood. Similarly to the ex-vivo cultured hu-PBMC we were unable to detect NP uptake in the T, B and NK cell populations in the blood and spleen (Fig. 6B, and data not shown), and CD14+ monocytes were the only cell type with detectable NP compared to un-injected controls (Fig. 6B). There was also a significant difference in frequency of HP4A versus HP4A-PEG NPs uptake by splenic monocytes at 8 hours (Fig. 6C). This in vivo finding that PEGylation of HP4A NP results in decreased uptake is consistent with our observations in the ex vivo hu-PBMC cultures. Lastly, we utilized Q-RT PCR to assess mRNA expression of inflammatory cytokines in the spleens of NP treated humanized mice compared to untreated controls. We did not detect any significant increases in TNF, IL6, or IL1B mRNA expression in mice treated with HP4A- or HP4A-PEG NPs compared to untreated controls (Fig. 6D). These in vivo data combined with our ex vivo primary hu-PBMC culture data indicate that PRINT fabricated HP4A-NPs are preferentially taken up by CD14+ monocytes and do not induce a robust inflammatory response by human immune cells. Together these studies suggest that PRINT hydrogel particles have the capacity for safe vaccine and drug delivery since they do not induce overt inflammatory responses or toxicity of human immune cells.
Discussion
The PRINT fabrication process yields NPs with defined shape and composition making it a leading candidate platform for diagnostic, therapeutic and preventative treatments in human disease. The hydrogel particles used in these studies were fabricated using PRINT and comprised of HP4A based PEG. Select formulations of PEG are F.D.A approved for human use and we sought to test how these particles interact with the human immune system. A variety of particulate molecules (alum, asbestos, silica, cholesterol crystals) can trigger inflammasome activation and release of IL-1β from LPS primed innate immune cells (48). The particulate nature of PRINT derived NPs raised concerns that they may have a similar capacity to activate the inflammasome, especially following LPS priming. We found that HP4A hydrogel particles were readily taken up by human THP-1 cells and primary hu-PBMC without an inflammatory response. We are encouraged by the fact that IL-1β was not secreted in response to HP4A-NPs, even following LPS priming, suggesting that these particles do not induce or enhance inflammasome activation. The 30-Plex cytokine array of supernatant from ex vivo hu-PBMC cultures treated with NP showed only a modest increase in CCL2 secretion (~40%) and while statistically significant, may not be biologically relevant, especially with all other cytokines levels being unchanged compared to untreated controls. Additionally, our in vivo studies using a humanized mouse model also showed no differences in the mRNA level of hallmark inflammatory cytokines (TNF, IL6, IL1B) after NP treatment. Together, these studies support the notion that PRINT based HP4A-NPs are immunologically inert and do not cause cytotoxicity making them a feasible candidate platform for human therapeutics. In addition, given our previously published findings that the murine immune response to nanoparticles is strikingly dependent upon mouse strain background, our current study suggests that humanized mouse models may provide a tractable model for translational studies of nanomedicine as they recapitulate ex-vivo human PBMC studies well (19).
Interestingly, the uptake of HP4A-NPs was exclusively detected in the CD14+ monocytic population of ex vivo cultured hu-PBMC and humanized mice, yet blockade of CD14 did not affect NP uptake and is therefore unlikely to be directly involved in NP recognition. Rather it is likely the inherent phagocytic properties of CD14+ monocytes that contribute to preferential NP uptake over other cell types. Modification of the NP surface by addition of short PEG chains reduced NP uptake by all cells tested, which is consistent with other studies suggesting that PEGylation is a viable means to reduce NP clearance and increase circulation time (49, 50). One hypothesis is that PEGylation reduces complement binding to the NP surface, thereby reducing opsonization and ingestion by phagocytic cells (16). Incubating hu-PBMC with NPs at 4°C, a temperature that should allow NP binding but not uptake, indicated that PEGylated NPs have significantly reduced capacity to bind monocytes compared to un-PEGylated NPs. Our failure to detect complement activation with PRINT fabricated HP4A-based NPs combined with that fact that our NP uptake studies were performed using heat inactivated serum, which is known to eliminate complement activity and reduce opsonization, suggests that our observed enhancement of un-PEGylated NP uptake at 4°C maybe complement independent (51, 52). This does not rule out the possibility that other heat resistant serum proteins bind the NP surface and enhance cellular binding and further studies using serum free conditions are needed to test this hypothesis. However, it is provocative to consider that PEGylation serves as a molecular lubricant that lowers the capacity for immune cells to grip the NP surface in a manner that is independent of serum-mediated opsonization.
Ultimately, the inability of PRINT fabricated HP4A-NPs to induce immune responses by primary human cells is advantageous for future design of human diagnostics and therapeutics. PRINT offers a modular platform in which various agents can be incorporated in a defined manner without concern of unwanted immune activation. The end goal of immune-engineering is to evoke potent and cell specific immune responses. Our studies indicate that PRINT fabricated HP4A-NPs are immunologically inert and passively target a major effector cell population of the human immune system, thereby providing an adjustable scaffold with the capacity to serve a variety of applications related to human health.
Supplementary Material
Acknowledgments
We thank Kristina Riebe of the Duke University Human Vaccine Institute/Regional Biocontainment Laboratory Host Response Monitoring facility for her assistance with multiplex cytokine assays. Select studies were performed in the Regional Biocontainment Laboratory at Duke University, which received partial support for construction from the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH) (grant UC6-AI058607). This work was also supported by NC TRACS 100K1202 (Ting and DeSimone), UCRF UNC Internal Grant (Ting), NMSS Cooperative Center Grant (Ting), NIH 8-VP1-CA174425-04 (DeSimone), NIH UC6-AI058607 (Sempowski), NIH R01AI095097/AI080432 (Su) and NIH U19-AI109784 9 (Ting, DeSimone, Sempowski and Su).
Statements of Funding:
Ting: NC TRACS 100K1202, UCRF UNC Internal Grants and NIH U19AI109784
DeSimone: NIH 8-VP1-CA174425-04 and NIH U19AI109784
Sempowski: NIH UC6-AI058607 and NIH U19AI109784
Su: NIH R01AI095097 and AI080432 and NIH U19AI109784
Robbins: 1F32A1108159-01 and T-32-CA009156-37
Roberts: T-32-GM008719 and T32-AI007273-25
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest:
Joseph M. DeSimone is a founder and maintains a financial interest in Liquidia Technologies. PRINT and Fluorocur are registered trademarks of Liquidia Technologies, Inc.
References
- 1.Dobrovolskaia MA, McNeil SE. Immunological properties of engineered nanomaterials. Nat Nanotechnol. 2007 Aug;2(8):469–478. doi: 10.1038/nnano.2007.223. [DOI] [PubMed] [Google Scholar]
- 2.Bershteyn A, Hanson MC, Crespo MP, Moon JJ, Li AV, Suh H, et al. Robust IgG responses to nanograms of antigen using a biomimetic lipid-coated particle vaccine. J Control Release. 2012 Feb 10;157(3):354–365. doi: 10.1016/j.jconrel.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demento SL, Eisenbarth SC, Foellmer HG, Platt C, Caplan MJ, Mark Saltzman W, et al. Inflammasome-activating nanoparticles as modular systems for optimizing vaccine efficacy. Vaccine. 2009 May 18;27(23):3013–3021. doi: 10.1016/j.vaccine.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, et al. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011 Feb 24;470(7335):543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Getts DR, Martin AJ, McCarthy DP, Terry RL, Hunter ZN, Yap WT, et al. Microparticles bearing encephalitogenic peptides induce T-cell tolerance and ameliorate experimental autoimmune encephalomyelitis. Nat Biotechnol. 2012 Dec;30(12):1217–1224. doi: 10.1038/nbt.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemente-Casares X, Tsai S, Yang Y, Santamaria P. Peptide-MHC-based nanovaccines for the treatment of autoimmunity: A "one size fits all" approach? J Mol Med (Berl) 2011 Aug;89(8):733–742. doi: 10.1007/s00109-011-0757-z. [DOI] [PubMed] [Google Scholar]
- 7.Tsai S, Shameli A, Yamanouchi J, Clemente-Casares X, Wang J, Serra P, et al. Reversal of autoimmunity by boosting memory-like autoregulatory T cells. Immunity. 2010 Apr 23;32(4):568–580. doi: 10.1016/j.immuni.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Hasan W, Chu K, Gullapalli A, Dunn SS, Enlow EM, Luft JC, et al. Delivery of multiple siRNAs using lipid-coated PLGA nanoparticles for treatment of prostate cancer. Nano Lett. 2012 Jan 11;12(1):287–292. doi: 10.1021/nl2035354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010 Apr 15;464(7291):1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naahidi S, Jafari M, Edalat F, Raymond K, Khademhosseini A, Chen P. Biocompatibility of engineered nanoparticles for drug delivery. J Control Release. 2013 Mar 10;166(2):182–194. doi: 10.1016/j.jconrel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Love SA, Maurer-Jones MA, Thompson JW, Lin YS, Haynes CL. Assessing nanoparticle toxicity. Annu Rev Anal Chem (Palo Alto Calif) 2012;5:181–205. doi: 10.1146/annurev-anchem-062011-143134. [DOI] [PubMed] [Google Scholar]
- 12.Rolland JP, Maynor BW, Euliss LE, Exner AE, Denison GM, DeSimone JM. Direct fabrication and harvesting of monodisperse, shape-specific nanobiomaterials. J Am Chem Soc. 2005 Jul 20;127(28):10096–10100. doi: 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- 13.Merkel TJ, Herlihy KP, Nunes J, Orgel RM, Rolland JP, DeSimone JM. Scalable, shape-specific, top-down fabrication methods for the synthesis of engineered colloidal particles. Langmuir. 2010 Aug 17;26(16):13086–13096. doi: 10.1021/la903890h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry JL, Herlihy KP, Napier ME, Desimone JM. PRINT: A novel platform toward shape and size specific nanoparticle theranostics. Acc Chem Res. 2011 Oct 18;44(10):990–998. doi: 10.1021/ar2000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galloway AL, Murphy A, DeSimone JM, Di J, Herrmann JP, Hunter ME, et al. Development of a nanoparticle-based influenza vaccine using the PRINT technology. Nanomedicine. 2013 May;9(4):523–531. doi: 10.1016/j.nano.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 16.Amoozgar Z, Yeo Y. Recent advances in stealth coating of nanoparticle drug delivery systems. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2012 Mar-Apr;4(2):219–233. doi: 10.1002/wnan.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perry JL, Reuter KG, Kai MP, Herlihy KP, Jones SW, Luft JC, et al. PEGylated PRINT nanoparticles: The impact of PEG density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano Lett. 2012 Oct 10;12(10):5304–5310. doi: 10.1021/nl302638g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of toll-like receptor 4 in immune activation. Biomaterials. 2010 Sep;31(26):6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 19.Jones SW, Roberts RA, Robbins GR, Perry JL, Kai MP, Chen K, et al. Nanoparticle clearance is governed by Th1/Th2 immunity and strain background. J Clin Invest. 2013 Jul 1;123(7):3061–3073. doi: 10.1172/JCI66895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manke A, Wang L, Rojanasakul Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res Int. 2013;2013:942916. doi: 10.1155/2013/942916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp FA, Ruane D, Claass B, Creagh E, Harris J, Malyala P, et al. Uptake of particulate vaccine adjuvants by dendritic cells activates the NALP3 inflammasome. Proc Natl Acad Sci U S A. 2009 Jan 20;106(3):870–875. doi: 10.1073/pnas.0804897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tschopp J, Schroder K. NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol. 2010 Mar;10(3):210–215. doi: 10.1038/nri2725. [DOI] [PubMed] [Google Scholar]
- 23.Wen H, Miao EA, Ting JP. Mechanisms of NOD-like receptor-associated inflammasome activation. Immunity. 2013 Sep 19;39(3):432–4341. doi: 10.1016/j.immuni.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 25.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. Into the eye of the cytokine storm. Microbiol Mol Biol Rev. 2012 Mar;76(1):16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yazdi AS, Guarda G, Riteau N, Drexler SK, Tardivel A, Couillin I, et al. Nanoparticles activate the NLR pyrin domain containing 3 (Nlrp3) inflammasome and cause pulmonary inflammation through release of IL-1alpha and IL-1beta. Proc Natl Acad Sci U S A. 2010 Nov 9;107(45):19449–19454. doi: 10.1073/pnas.1008155107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang EJ, Kim S, Kim JS, Choi IH. Inflammasome formation and IL-1beta release by human blood monocytes in response to silver nanoparticles. Biomaterials. 2012 Oct;33(28):6858–6867. doi: 10.1016/j.biomaterials.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 28.Rice-Ficht AC, Arenas-Gamboa AM, Kahl-McDonagh MM, Ficht TA. Polymeric particles in vaccine delivery. Curr Opin Microbiol. 2010 Feb;13(1):106–112. doi: 10.1016/j.mib.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Manke A, Wang L, Rojanasakul Y. Mechanisms of nanoparticle-induced oxidative stress and toxicity. Biomed Res Int. 2013;2013:942916. doi: 10.1155/2013/942916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palomaki J, Valimaki E, Sund J, Vippola M, Clausen PA, Jensen KA, et al. Long, needle-like carbon nanotubes and asbestos activate the NLRP3 inflammasome through a similar mechanism. ACS Nano. 2011 Sep 27;5(9):6861–6870. doi: 10.1021/nn200595c. [DOI] [PubMed] [Google Scholar]
- 31.Yang M, Flavin K, Kopf I, Radics G, Hearnden CH, McManus GJ, et al. Functionalization of carbon nanoparticles modulates inflammatory cell recruitment and NLRP3 inflammasome activation. Small. 2013 Dec 20;9(24):4194–4206. doi: 10.1002/smll.201300481. [DOI] [PubMed] [Google Scholar]
- 32.Neumann S, Burkert K, Kemp R, Rades T, Rod Dunbar P, Hook S. Activation of the NLRP3 inflammasome is not a feature of all particulate vaccine adjuvants. Immunol Cell Biol. 2014 Jul;92(6):535–542. doi: 10.1038/icb.2014.21. [DOI] [PubMed] [Google Scholar]
- 33.Roberts RA, Shen T, Allen IC, Hasan W, DeSimone JM, Ting JP. Analysis of the murine immune response to pulmonary delivery of precisely fabricated nano- and microscale particles. PLoS One. 2013 Apr 12;8(4):e62115. doi: 10.1371/journal.pone.0062115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vaine CA, Patel MK, Zhu J, Lee E, Finberg RW, Hayward RC, et al. Tuning innate immune activation by surface texturing of polymer microparticles: The role of shape in inflammasome activation. J Immunol. 2013 Apr 1;190(7):3525–3532. doi: 10.4049/jimmunol.1200492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzmán J, Iglesias M, Riande E, Compañ V, Andrio A. Synthesis and polymerization of acrylic monomers with hydrophilic long side groups. oxygen transport through water swollen membranes prepared from these polymers. Polymer. 1997;38:5227–5332. [Google Scholar]
- 36.Neun BW, Dobrovolskaia MA. Qualitative analysis of total complement activation by nanoparticles. Methods Mol Biol. 2011;697:237–245. doi: 10.1007/978-1-60327-198-1_25. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Jiang Q, Li G, Jeffrey J, Kovalev GI, Su L. Efficient infection, activation, and impairment of pDCs in the BM and peripheral lymphoid organs during early HIV-1 infection in humanized rag2(−)/(−)gamma C(−)/(−) mice in vivo. Blood. 2011 Jun 9;117(23):6184–6192. doi: 10.1182/blood-2011-01-331173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang Q, Zhang L, Wang R, Jeffrey J, Washburn ML, Brouwer D, et al. FoxP3+CD4+ regulatory T cells play an important role in acute HIV-1 infection in humanized Rag2−/−gammaC−/− mice in vivo. Blood. 2008 Oct 1;112(7):2858–2868. doi: 10.1182/blood-2008-03-145946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bility MT, Zhang L, Washburn ML, Curtis TA, Kovalev GI, Su L. Generation of a humanized mouse model with both human immune system and liver cells to model hepatitis C virus infection and liver immunopathogenesis. Nat Protoc. 2012 Sep;7(9):1608–1617. doi: 10.1038/nprot.2012.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang L, Kovalev GI, Su L. HIV-1 infection and pathogenesis in a novel humanized mouse model. Blood. 2007 Apr 1;109(7):2978–2981. doi: 10.1182/blood-2006-07-033159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasut G, Veronese FM. PEG conjugates in clinical development or use as anticancer agents: An overview. Adv Drug Deliv Rev. 2009 Nov 12;61(13):1177–1188. doi: 10.1016/j.addr.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 42.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ, Mathison JC. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science. 1990 Sep 21;249(4975):1431–1433. doi: 10.1126/science.1698311. [DOI] [PubMed] [Google Scholar]
- 43.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, Tripp RA, et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000 Nov;1(5):398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 44.Lee HK, Dunzendorfer S, Soldau K, Tobias PS. Double-stranded RNA-mediated TLR3 activation is enhanced by CD14. Immunity. 2006 Feb;24(2):153–163. doi: 10.1016/j.immuni.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 45.Scott P, Ma H, Viriyakosol S, Terkeltaub R, Liu-Bryan R. Engagement of CD14 mediates the inflammatory potential of monosodium urate crystals. J Immunol. 2006 Nov 1;177(9):6370–6378. doi: 10.4049/jimmunol.177.9.6370. [DOI] [PubMed] [Google Scholar]
- 46.Power CP, Wang JH, Manning B, Kell MR, Aherne NJ, Wu QD, et al. Bacterial lipoprotein delays apoptosis in human neutrophils through inhibition of caspase-3 activity: Regulatory roles for CD14 and TLR-2. J Immunol. 2004 Oct 15;173(8):5229–5237. doi: 10.4049/jimmunol.173.8.5229. [DOI] [PubMed] [Google Scholar]
- 47.Sahu A, Lambris JD. Structure and biology of complement protein C3, a connecting link between innate and acquired immunity. Immunol Rev. 2001 Apr;180:35–48. doi: 10.1034/j.1600-065x.2001.1800103.x. [DOI] [PubMed] [Google Scholar]
- 48.Gross O, Thomas CJ, Guarda G, Tschopp J. The inflammasome: An integrated view. Immunol Rev. 2011 Sep;243(1):136–151. doi: 10.1111/j.1600-065X.2011.01046.x. [DOI] [PubMed] [Google Scholar]
- 49.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011 Jun;6(4):715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moghimi SM, Hunter AC, Andresen TL. Factors controlling nanoparticle pharmacokinetics: An integrated analysis and perspective. Annu Rev Pharmacol Toxicol. 2012;52:481–503. doi: 10.1146/annurev-pharmtox-010611-134623. [DOI] [PubMed] [Google Scholar]
- 51.Triglia RP, Linscott WD. Titers of nine complement components, conglutinin and C3b-inactivator in adult and fetal bovine sera. Mol Immunol. 1980 Jun;17(6):741–748. doi: 10.1016/0161-5890(80)90144-3. [DOI] [PubMed] [Google Scholar]
- 52.Leijh PC, van den Barselaar MT, van Zwet TL, Daha MR, van Furth R. Requirement of extracellular complement and immunoglobulin for intracellular killing of micro-organisms by human monocytes. J Clin Invest. 1979 Apr;63(4):772–784. doi: 10.1172/JCI109362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.