Abstract
Insulin is internalized with its cognate receptor into the endosomal apparatus rapidly after binding to hepatocytes. We performed a bioinformatic screen of Golgi/endosome hepatic protein fractions and found that ATIC, which is a rate-limiting enzyme in the de novo purine biosynthesis pathway, and PTPLAD1 are associated with insulin receptor (IR) internalization. The IR interactome (IRGEN) connects ATIC to AMPK within the Golgi/endosome protein network (GEN). Forty-five percent of the IR Golgi/endosome protein network have common heritable variants associated with type 2 diabetes, including ATIC and AMPK. We show that PTPLAD1 and AMPK are rapidly compartmentalized within the plasma membrane (PM) and Golgi/endosome fractions after insulin stimulation and that ATIC later accumulates in the Golgi/endosome fraction. Using an in vitro reconstitution system and siRNA-mediated partial knockdown of ATIC and PTPLAD1 in HEK293 cells, we show that both ATIC and PTPLAD1 affect IR tyrosine phosphorylation and endocytosis. We further show that insulin stimulation and ATIC knockdown readily increase the level of AMPK-Thr172 phosphorylation in IR complexes. We observed that IR internalization was markedly decreased after AMPKα2 knockdown, and treatment with the ATIC substrate AICAR, which is an allosteric activator of AMPK, increased IR endocytosis in cultured cells and in the liver. These results suggest the presence of a signaling mechanism that senses adenylate synthesis, ATP levels, and IR activation states and that acts in regulating IR autophosphorylation and endocytosis.
The insulin receptor (IR)1 (1, 2) activates its tyrosine kinase activity through cross-phosphorylation upon insulin binding, which initiates subsequent signaling and metabolic events (3). The activated complexes are internalized into the endosomal apparatus within seconds following insulin binding (4). Some factors that govern IR endocytosis have been previously described. For instances topogenic sequences in the juxtamembrane region (GPLY and NPEY) are necessary but not sufficient for rapid endocytosis. The IR tyrosine kinase activity is necessary for the autophosphorylation and exposure of the otherwise buried endocytosis sequences (for a review, see (5)). IR autophosphorylation is thought to release a constraint that maintains IR on the microvilli and that allows free, mobile insulin-bound IRs to reach specialized surfaces where endocytosis and signaling are allowed and initiated (6, 7). The interaction with the actin cytoskeleton is an important element as IR compartmentalization at the microvilli on the surface of hepatocytes depends on its coupling with actin (8, 9). IR has also been observed in caveolin-enriched hepatic plasma membrane domains that rapidly associate with the actin cytoskeleton in response to insulin (10). By contrast with the EGF receptor (EGFR, a low recycling receptor), the IR is a rapid recycling receptor in liver and this is important both for insulin signaling and clearance (11, 12). The molecular mechanisms underlying IR regulation remain largely unknown.
Emerging evidence indicates that for coping with cellular and organismal needs the vesicular transport of proteins and lipids to particular regions of the cell, requires plasma membrane and endosome-based signaling devices that would coordinate membrane traffic in response to extracellular signals (13–16). The physiological significance of trafficking and the molecular bases of such coordination have received little attention. In the current work, we characterized an IR protein interaction network in hepatic Golgi/endosomal membranes. We found substantial heterogeneity within the close environment of the internalized IR complexes, with the presence of ATIC, a metabolic enzyme of the de novo purine synthesis pathway.
The de novo purine synthesis pathway includes 10 sequential steps, beginning with phosphoribosyl pyrophosphate and ending with inositol monophosphate (IMP). Purines are the building blocks of DNA and RNA but are also found in adenylates and are thus a source of ATP and cofactors for numerous metabolic and signaling enzymes (17). ATIC, is the last enzyme in this pathway and is a cytosolic enzyme (64 kDa) that includes a transformylase domain (residues 200–593), which transfers a formyl group to the AMP analog AICAR to produce the intermediate formyl-AICAR (FAICAR) and IMP (18). AICAR is an efficient activator of the energy sensor AMPK (19) in a way that is similar to antidiabetic drugs such as metformin (20). ATIC is also a rate-limiting enzyme up-regulated by insulin in C2C12 mouse mesenchymal cells; a 20% decrease in its activity results in a 50% decrease in AICAR-dependent purine synthesis (21). Massive AICAR accumulation and urinary excretion have been observed in children with only 40% of the normal level of ATIC activity (22). We show here the presence of mechanism involving an ATIC protein network and the metabolite AICAR that acts in regulating IR autophosphorylation and endocytosis.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Porcine insulin (I5523) was obtained from Sigma-Aldrich (St. Louis, MO). Mouse receptor-grade EGF was purchased from BD Biosciences (Bedford, MA). The following antibodies were used: antiphosphotyrosine (PY20, Sigma-Aldrich, St. Louis, MO); anti-PTPLAD1 (ab57143, Abcam, Cambridge MA); anti-AMPKα and antiphospho-AMPK (Threonine-172) (#2532 and #2531, Cell Signaling Technology); anti-Akt1/3 and antiphospho-Akt (Ser473) (#9271 and #9272, Cell Signaling Solutions, Lake Placid, NY); and anti-ATIC (#410100, Zymed Laboratories Inc., San Francisco, CA). The anti-Glut 2 antibody, which was directed against the epitope that corresponds to amino acids 32–98 in the human Glut2 extracellular domain (sc-9117: H-67), and the anti-Kif5A (sc-25734: H-75), the EGFR (sc-03) and the IR β-subunit (Sc-711) antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The antibeta actin antibody was obtained from Sigma-Aldrich (St. Louis, MO). Peroxidase-conjugated secondary antibodies were used (1:10,000, Jackson Immuno Research Laboratories, West Grove, PA). The antibody signals were analyzed using an ImageQuant LAS 4000 Imager (GE Healthcare Biosciences). AICA-Riboside was obtained from Calbiochem (Cat # 123040). All other chemicals were of analytical grade and were purchased from Fisher Scientific (Sainte-Foy, Québec) or from Roche Laboratories (Laval, Québec).
Hepatic Fractions
Harlan Sprague-Dawley rats (female 120–140 g b.w.) were purchased from Charles River Ltd. (St. Constant, Québec, Canada) and were maintained under standard laboratory conditions with food and water available ad libitum, except that the food was removed 18 h before the experiments. All animal procedures were approved by the CPA-CRCHUQ (certificate 055-3). The G/E and the PM fractions were prepared as previously described (4, 23). The protein content of each fraction was determined using a modified Bradford method, with bovine serum albumin as the standard (Bio-Rad protein assay, Bio-Rad, Hercules, CA). The G/E fraction was also characterized using an MS-based proteomic analysis of the major proteins (24) and enrichment here against the entire rodent proteome (supplemental Fig. S1).
Database and Network Analyses
A list of proteins (666 unique proteins) from the G/E fraction that we identified in (24) was used to construct the mouse G/E network (GEN), which was generated from functional and physical protein–protein interaction data. The data were obtained using the STRING database (filter set at 0.700) (25). Additional physical interactions were provided by the Biological General Repository for Interaction Data Sets (BioGRID) (26). Protein interactions were visualized on the Cytoscape platform (Version 2.8.2) (27). Self-loops and duplicated edges were removed before the analysis using the “network analyzer” plugin (28). Cellular component grouping was performed following a Gene Ontology analysis using the Biological Networks Gene Ontology tool (BINGO version 2.4) (29). We used a cluster analysis to determine the dense regions of highly interconnected proteins with confident functional relations with the receptor complex (IRGEN) to analyze the environment that was most similar to that of the internalized IR. We used this method with the affinity propagation algorithm implemented in Cytoscape software using clusterMaker plugin (30).
Identification of Internalized IR Partners
The G/E fractions were solubilized and subjected to immunoprecipitation and in-gel analysis procedures using an anti-IR antibody (31).
Liquid Chromatography-Multiple Reaction Monitoring Analysis (LC-MRM)
Peptides (0.5 mg/μl) were analyzed on an ABSciex 5500 QTRAPTM hybrid triple quadrupole/linear ion trap mass spectrometer equipped with an Eksigent nanoLC AS2 cHiPLC nanoflex controlled by Analyst 1.6TM software and with a nanospray ionization source. The quantification value was obtained for each peptide by comparing the peak area of the endogenous peptide with the peak area of the labeled peptide. The data were averaged at the replicate level and the peptide level to obtain the protein quantification values (supplemental Table S1).
Purified Synthetic Peptides
Purified synthetic peptides used for LC-MRM analysis containing [13C6]Lys and [13C6]Arg were obtained from Pierce (Rockford, IL). A solution containing a mix of all the peptides was prepared with concentrations ranging from 0.1 fmol to 20 pmol and used to reconstitute the samples after tryptic digestion for relative quantification (supplemental Table S1).
Endosomal IR Autophosphorylation In Vitro
IR endosomal autophosphorylation was measured as previously reported (4) with the following modifications. Freshly prepared G/E membranes isolated at their IR concentration time-peak (100 μg of protein; 2 min after insulin injection; 1.5 μg/100 g b.w.) were suspended in the cell-free system, maintaining the structural integrity of the endosomes. The IR autophosphorylation reaction was initiated at 37 °C by adding ATP to a final concentration of 1 mm. Where indicated, ATIC (20 ng of protein) that was immunoaffinity-purified from hepatic cytosol (anti-ATIC clone F38P7H9, Sigma-Aldrich, St. Louis, MO) or fresh cytosol (diluted 1/10) was pre-incubated for 15 min on ice before beginning the reaction. After stopping the reaction at the indicated times, membranes were solubilized, IR was immunoprecipitated (insulin Rb (C19): sc-711, Santa Cruz Biotechnology Inc., Santa Cruz, CA), and a phosphotyrosine content analysis was performed using Western blotting. EGF (1.0 μg/100 g b.w.; receptor grade, BD Biosciences, Bedford, MA) was injected where indicated (4). EGFR was immunoprecipitated (EGFR [1005]: sc-03, Santa Cruz Biotechnology) following autophosphorylation and submitted to a Western blot analysis.
siRNA Knockdown in HEK293 Cells
HEK293 cells express IR and allow measurements of insulin-mediated endocytosis (31). We used the following predesigned human sequences to deplete ATIC, PTPLAD1, and AMPKα2 in HEK293 cells:
ATIC: CAGUCUAACUCUGUGUGCUACGCCA, NM_004044.5_stealth_1459; PTPLAD1: GACCCAGAGGCAGGUAAAACAUUACA, NM_016395_stealth_367; and AMPKa2: GAAGUCAUCUCAGGAGAUUGUAUG, NM_006252_stealth_618 (Invitrogen, Life Technologies Corporation, Frederick, MD). Cells were cultivated in DMEM high-glucose medium with 10% fetal bovine serum without antibiotics (Hyclone) and were transfected using LipofectamineTM (Invitrogen) for 48 or 72 h and then subjected to the described analysis. The surface trypsinization assay was performed as previously reported (32), except that the α2β2 IR tetramer was detected by Western blotting under nondenaturing conditions (33) using 5% resolving gels.
RESULTS
Bioinformatics and Proteomic Screens Identify ATIC and PTPLAD1 in Internalized IR Complexes
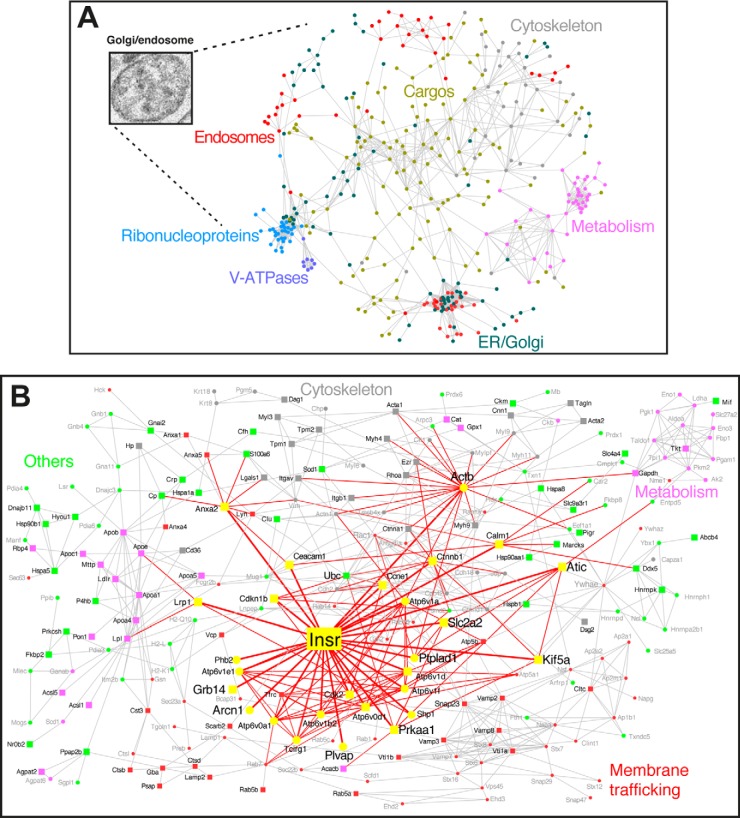
The Golgi/endosome protein interaction network (GEN) was reconstituted from the previous G/E proteome of major proteins (24) and from protein–protein interaction data. This network is characterized by a general vesicle-like topology, with lumenal cargo (lipoproteins, albumin, and proteases) and peripheral membrane elements that are involved in trafficking (SNAREs), cytoskeleton proteins, a lumenal acidification system (vATPase complexes), and other endosome and Golgi elements (Fig. 1A). However, the GEN provides limited information about the interaction partners of IR. Thus, we identified IR interaction partners during insulin-mediated endocytosis using a MS-based screening approach.
Fig. 1.
Construction of the Golgi/endosome network (GEN) and of the IR subnetwork (IRGEN). A, A set of 666 proteins from the G/E fraction (24) were grouped and linked according to their functional and molecular associations; circles correspond to proteins. A link is placed between proteins if a functional or physical association exists, thus forming the GEN. The color corresponds to the cellular component categorization, as calculated by the Gene Ontology plugin Bingo. B, Projection of IR within the GEN (IRGEN). The colors correspond to the functional class to which the associated protein belongs. Yellow represents the IR and the associated proteins (proteins in magnified fonts were identified in the present study). The width of a link is proportional to the distance from IR. Squares correspond to proteins for which the gene was associated with type 2 diabetes. In the IRGEN, 107 genes (45% of the IRGEN) are associated with type 2 diabetes (supplemental Table S4).
Following intravenous insulin injection, IR complexes accumulate in endosomes in the G/E fraction (4). We identified the following five primary proteins that interacted with internalized IR complexes 15 min after insulin stimulation: Grb14, ATIC, PTPLAD1, delta-COP, and PLVAP (Table I, supplemental Fig. S2 and supplemental Table S2). Grb14, is a IR adapter that binds to tyrosine phosphorylation sites in the activation loop (34) and inhibits IR tyrosine kinase activity in cell based assays (35, 36). This adapter protein was previously identified as a common heritable variant in type 2 diabetes (37). The other candidates have not been previously associated with IR. The highest probability identification score was attributed to ATIC, which is a cytosolic protein that has not been previously reported to interact with IR. ATIC forms a complex with its substrate AICAR via the phosphate-binding pocket of the IMP cyclohydrolase (IMPCH) domain (residues 1–599) (38). The functions of PTPLAD1 (also called B-ind1 or PTAD1) are unknown. We recapitulated the presence of ATIC and PTPLAD1 15 min after insulin stimulation and performed quantitative MS with the internalized IR as a reference (LC-MRM; supplemental Fig. S3 and supplemental Table S1). Other identified proteins, such as the nonclathrin coat proteins (delta-COP) and plasmalemma vesicle-associated protein (PLVAP), which belongs to caveolae stromal and fenestral diaphragms (Table I), were not characterized further here.
Table I. Mass spectrometry analysis of internalized IR complexes in the liver. Golgi/endosome fractions were isolated after insulin stimulation. The membranes were solubilized, and IR was immunoprecipitated. The proteins were separated and submitted to LC/MS/MS analysis (supplemental Fig. 2 and supplemental Table S2).
| Protein name | Mass (Dalton) | Accession number | Sequence covered (%) | Peptide (n) | Mascot score | Function |
|---|---|---|---|---|---|---|
| Bifunctional purine biosynthesis protein PURH (ATIC) | 64681 | Q6IN16 | 33 | 26 | 459 | Catalyses the final step of the de novo purine synthesis pathway that produces IMP/AICAR transformylase. |
| Growth factor receptor bound protein 14 (Grb14) | 61181 | O88900 | 17 | 11 | 115 | Interacts with the cytoplasmic domain of the autophosphorylated insulin receptor. |
| Protein-tyrosine phosphatase-like A domain-containing protein 1 (PTPLAD1) | 43132.8 | Q8K2C9 | 9 | 32 | 89.2 | Transmembrane protein in the ER. Putative protein-tyrosine phosphatase. |
| Coatomer subunit delta | 57637 | Q5XJY5 | 8 | 5 | 62 | Reversibly associates with non-clathrin-coated vesicles. |
| Plasmalemma vesicle-associated protein | 50572 | Q9WV78 | 6 | 3 | 49 | Involved in the formation of stromal and fenestral diaphragms of caveolae. |
We included ATIC and PTPLAD1 in the GEN and analyzed their associations. Because AICAR is a potent allosteric AMPK activator (39), AMPK was also included in the analysis. The IR subnetwork (IRGEN) indicated a connection with Kif5A and the microtubule network through AMPK (Prkaa1). Kif5A also possesses two AMPK-dependent phosphorylation sites in the cargo-binding kinesin light chain KCL2, regulating the association of Kif5A with PI3K (40). PTPLAD1 also interacts with the actin network through Rac1 (Fig. 1B and supplemental Table S3). We verified the presence of proteins corresponding to validated type 2 diabetes common variants in the IRGEN to estimate the pathophysiological importance of the IRGEN. We found that 45% of the IRGEN proteins (identified with squares, representing 107 genes) are associated with type 2 diabetes (41–44). A 57% enrichment was calculated for proteins closer to IR (yellow squares), including IR, Grb14, ATIC, AMPK, Kif5A, and GLUT2 (Fig. 1B and supplemental Table S4). This enrichment indicates that these interactions are of mechanistic and pathophysiological importance.
ATIC, PTPLAD1, and AMPK Compartmentalization In Vivo in Response to Insulin
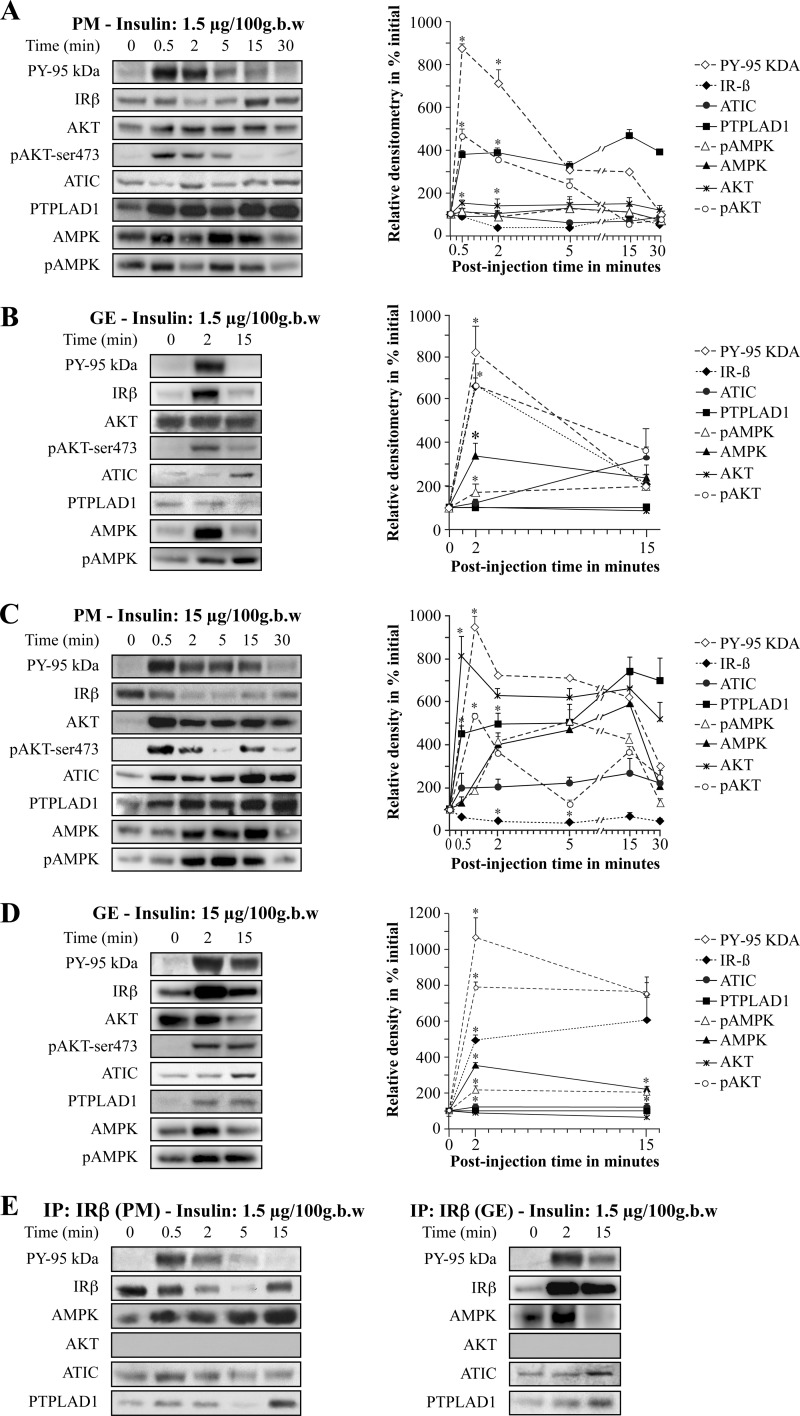
We confirmed the importance of the IRGEN by measuring the responses of ATIC, PTPLAD1 and AMPK to acute insulin stimulation. We conducted these studies with doses of injected insulin, which resulted in 50% rat liver IR occupancy (1.5 μg/100 g body weight [b.w.]) and saturation (15 μg/100 g b.w.) of receptors (4). Rats were killed at different times after insulin injection, PM and G/E cell fractions were prepared and directly submitted to analysis. The time-dependent changes of PM and G/E IR as well as ATIC, PTPLAD1, and AMPK content relative to control fractions were determined. Following insulin injection, there was a rapid and marked increase in G/E IR concentration that peaked at 2–5 min postinjection at about 10-fold-over control for both doses. At 15 min, the G/E IR concentration returned to near control level for the nonsaturating dose of insulin but remained at sixfold above control for the saturating dose. IR autophosphorylation followed a similar profile (Fig. 2B and 2D). There were corresponding rapid decreases in PM receptor concentration with an initial maximal decline by 2–5 min to ∼60–80% of control. At 15 min post-injection, the PM IR concentration returned to control value for the nonsaturating dose. The IR was rapidly (30 s) autophosphorylated in the PM fractions for both doses and then rapidly declined as internalization proceeded (4) (Fig. 2A and 2C).
Fig. 2.
Kinetics of the localization of IR, ATIC, PTPLAD1, and AMPK in the plasma membrane (PM) and endosome (G/E) fractions. Rats were injected with the noted insulin doses via the jugular vein. The PM A, C, and G/E B, D, fractions were prepared from two livers each time. Time 0 corresponds to buffer alone with immediate sacrifice of the animals. Membranes (70 μg proteins) were subjected to immunoblot analysis with the indicated antibody. The relative protein content was determined by expressing all of the densitometric data as a percentage of their levels in the fractions at time 0. The autoradiograms (left) depict data from a typical experiment with each dose of insulin. Each point on the progress curve (right) is the mean ± s.d. of three to five separate experiments (*p < 0.01; two-tailed unpaired Student's t-tests). E, IR was immunoprecipitated from solubilized (200 μg proteins) PM (left) and G/E (right) fractions as described under Methods. The immunoprecipitated proteins were blotted and incubated with antibodies against IR (95 kDa β-subunit), phosphotyrosine (PY-20, IR tyrosine phosphorylation, PY-95 kDa), AMPK, AKT, ATIC, and PTPLAD1.
ATIC was barely detected in both the PM and G/E fractions. There was however, a rapid two- to threefold increase in PM ATIC concentration for the saturating dose (Fig. 2C). The G/E ATIC concentration increased to 1.5- to threefold over control for both doses (Fig. 2B and 2D). This was consistent with the results of the quantitative MS analysis (supplemental Figs. S2 and S3).
PTPLAD1 was also barely detected in the PM and G/E fractions. However, coincident with IR autophosphorylation, there was a rapid and marked increase in PM PTPLAD1 concentration to about five- to sevenfold over control for both doses. A threefold increase in G/E PTPLAD1 concentration was also observed in response to injection with the largest insulin dose (Fig. 2D), which is also consistent with the quantitative MS results (supplemental Fig. S3).
AMPK was detected in both fractions. There was a rapid and marked increase in PM AMPK concentration that peaked at 2–15 min postinjection at about sixfold-over control for the saturating dose. AMPK phosphorylation followed the same profile. (Fig. 2C). Following insulin injection, there was also a rapid and marked increase in G/E AMPK concentration that peaked at 2 min postinjection at about fourfold-over control for both doses. At 15 min, G/E AMPK concentration returned to near control level for both doses. AMPK phosphorylation followed a similar profile but remained at fourfold above control for the saturating dose (Fig. 2B and 2D).
There was a rapid and marked increase in PM Akt concentration and phosphorylation that peaked at 30 s postinjection at about sixfold-over control for both doses. Following insulin injection, there was also a rapid and marked increase in G/E Akt phosphorylation that peaked at 2 min postinjection at about fourfold-over control for both doses. At 15 min, G/E Akt phosphorylation returned to near control level for the lower insulin dose.
The presence of ATIC, PTPLAD1, and AMPK, but not Akt, which is also stimulated by insulin, was confirmed in the IR immunocomplexes prepared following the injection of the 1.5 μg/100 g b.w. dose (Fig. 2E).
Thus, the results showed the presence of an insulin-dependent response for ATIC, PTPLAD1 and AMPK in vivo. They also confirmed their association with the IR.
ATIC Increases IR Autophosphorylation In Vitro
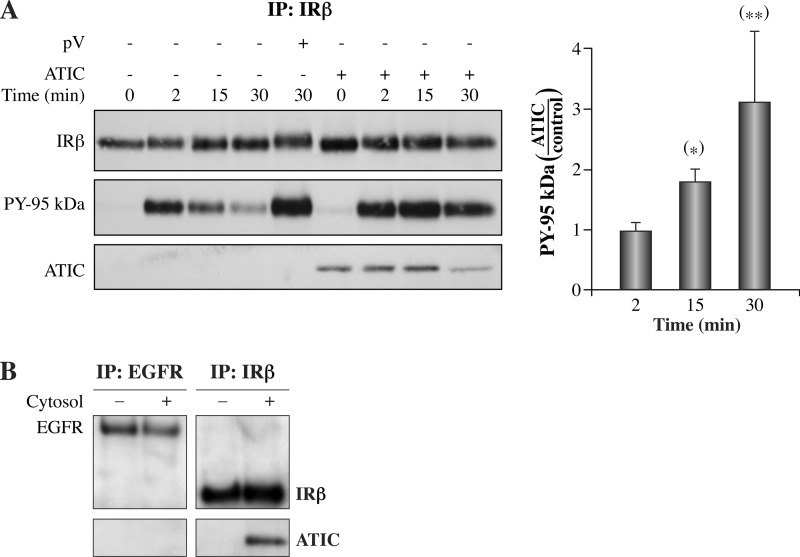
We sought to assess the process of IR autophosphorylation in endosomes directly. Therefore, we used a previously characterized reconstitution system where endosomes isolated 2 min following the injection of insulin were incubated in the presence of purified ATIC. Fig. 3A depicts the time-course of IR autophosphorylation. IR autophosphorylation reached a maximum at 2 min following the addition of ATP and subsequently declined to 60% of the maximal value by 15 min of incubation. In the presence of the protein tyrosine phosphatase (PTP) inhibitor bpV(phen) (pV), IR autophosphorylation was augmented to sixfold above that of control incubations as a result of the inhibition of the endosomal PTP activity (45). In the presence of ATIC, IR autophosphorylation reached a maximum at 2 min following the addition of ATP with no decline by 15 min and 30 min of incubation (Fig. 3A). The IR/ATIC association was recapitulated using fresh cytosol (Fig. 3B). However, ATIC was not detected when IR was replaced with EGFR (Fig. 3B, left; G/E endosomes isolated at the peak of EGFR accumulation, 15 min post-EGF injection, 1.0 μg/100 g b.w. representing approximately the same number of sites) (45). Thus, the results show that IR autophosphorylation is favored by the addition of ATIC.
Fig. 3.
Time course of IR autophosphorylation in isolated endosomes incubated in the presence of purified ATIC. Endosomes (G/E) were prepared at 2 min after insulin injection (1.5 μg/100 g b.w.) or 15 min after EGF injection (1.0 μg/100 g b.w.), corresponding to the time of peak kinase accumulation. The intact membranes were incubated at 37 °C in a buffer mimicking the intracellular milieu in the presence of ATP and in the presence or absence of the PTP inhibitor bpV(phen) (pV) or immunopurified ATIC (100 ng, 5 min preincubation on ice before adding ATP) for the indicated times. A, Left panel: IR immunoprecipitation. IR was detected using an anti-β subunit antibody. IR autophosphorylation (PY 95 kDa) was measured using an antiphosphotyrosine antibody (PY20). The presence of ATIC was verified using an anti-ATIC antibody (portions of the same membrane, detection representing ∼2% or less of the total fraction). Right panel: IR β-subunit tyrosine phosphorylation (PY 95 kDa) densitometry, expressed as the proportion of the control signal (ATIC/minus ATIC). The values shown are the means±s.d. (*p < 0.001; **p < 0.05, n = 5, two-tailed unpaired Student's t-tests). B, The experiments were recapitulated with the presence of cytosol instead of purified ATIC. EGFR or IR-loaded endosomes were incubated for 5 min in the presence or absence of cytosol from fresh liver samples (diluted 1/10, portions of the same membrane; three experiments).
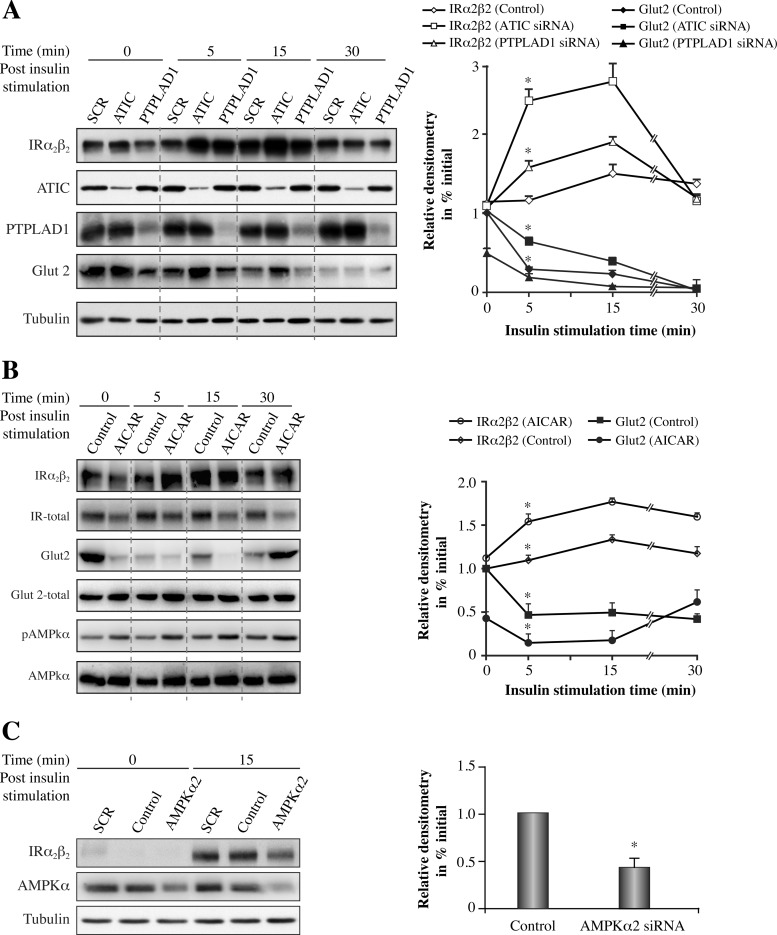
ATIC and PTPLAD1 Knockdown Affects IR Autophosphorylation in HEK293 Cells
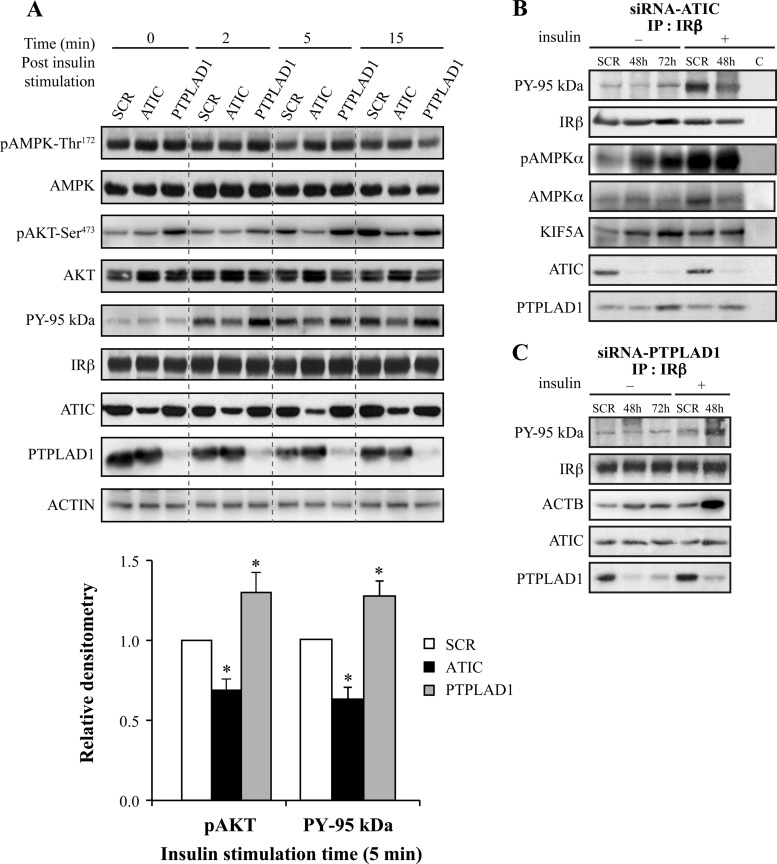
To confirm whether ATIC and the candidate partner PTPLAD1 regulate IR autophosphorylation in a cellular context, we transfected HEK293 cells with siRNA oligonucleotides corresponding to the human ATIC or PTPLAD1 sequences and analyzed IR autophosphorylation. The total ATIC levels were reduced by ∼65% after 48 (Fig. 4A) and 72 h (supplemental Fig. S4) of transfection, and PTPLAD1 levels were reduced by ∼85% (Fig. 4A). The typical flat, square morphology of the cells was unchanged, except after 72 h of transfection with PTPLAD1 siRNA, at which point most of the cells were round and loosely attached to the substratum. ATIC depletion (48 h post-transfection) reduced IR tyrosine phosphorylation by 40% 5 min after insulin stimulation (Fig. 4A and 4B). In contrast, PTPLAD1 depletion increased IR tyrosine phosphorylation (Fig. 4A and C). Akt phosphorylation followed also a similar profile (Fig. 4A). The phosphorylated AMPK−Thr172 readily coprecipitated with IR after ATIC depletion, which was observed before and after insulin stimulation (Fig. 4B). Of further interest, the accumulation of phosphorylated AMPK−Thr172 in whole cell extracts was not detected 48 h after transfection with ATIC siRNA (Fig. 4A) but was readily detected 72 h after transfection and was not fully restored by insulin (supplemental Fig. S4), consistent with AICAR accumulation as a function of time after ATIC depletion (21). Hence, ATIC and PTPLAD1 indeed affect IR tyrosine phosphorylation, then ATIC down-regulation also leads to efficient AMPK−Thr172 phosphorylation and to its rapid compartmentalization in IR complexes. In addition, ATIC depletion also resulted in the presence of KiF5A in IR complexes (Fig. 4B), which is consistent with the link between ATIC and Kif5A, through AMPK, that was suggested by the IRGEN (Fig. 1B). Of further interest, PTPLAD1 depletion and insulin stimulation resulted in the marked presence of actin in the IR complexes (Fig. 4C).
Fig. 4.
ATIC, PTPLAD1, and AMPK bind to IR in cultured cells. ATIC and PTPLAD1 knockdown affects the tyrosine phosphorylation of IR, and ATIC affects the phosphorylation of AMPK. A, HEK293 cells were transfected for 48 or 72 h with ATIC or PTPLAD1 siRNAs. The cells were preincubated in a serum-free medium for 2 h and stimulated for the indicated times with insulin (35 nm). Immunoblots (25 μg protein extracts) show the levels of IR, PY-95 kDa, PTPLAD1, ATIC, AMPK, pAMPK-Thr172, AKT, pAKT-Ser473, and actin in the cells that were transfected with control (SCR: scrambled 48 h), ATIC and PTPLAD1 siRNAs. The salient 5 min time point for pAKT-Ser473 and PY-95Kda (lower panel) is the mean ± s.d. of three experiments (*p < 0.01; two-tailed unpaired Student's t-tests). B, IR immunoprecipitation: After transfection with ATIC siRNA, the cells were incubated in serum-free medium for 2 h and stimulated for 15 min with vehicle (−) or insulin (+, 48 h post-transfection). The tyrosine phosphorylation of the IR β-subunit was measured using an antiphosphotyrosine (PY20) antibody. The presence of AMPK and pAMPK-Thr172 was verified using portions of the same membrane (three experiments). C, IR immunoprecipitations: After transfection with PTPLAD1 siRNA, the cells were incubated in serum-free medium for 2 h and stimulated for 15 min with vehicle (−) or insulin (+, 48 h post-transfection). The tyrosine phosphorylation of the IR β-subunit (PY 95 kDa) and the actin (ACTB) levels were measured using antibeta actin and PY20 antibodies (portions of the same membrane; three experiments).
ATIC, PTPLAD1, and AMPK Knockdown Affects IR Internalization in HEK293 Cells
Next, we assessed the relationship between IR endocytosis and ATIC and PTPLAD1 in HEK293 cells. We used an IR (α2β2) protection assay in which cells were surface-trypsinized before and after insulin-mediated endocytosis (32). α2β2 IR protection peaked 15 min after insulin stimulation and then decreased at 30 min in control cells (Fig. 5A). Both ATIC and PTPLAD1 knockdown increased markedly IR internalization; greater accumulation was observed 5 min after insulin stimulation, and the IR levels then declined to basal levels after 30 min (Fig. 5A). As a control, we blotted for Glut2, which is primarily inside the cells and translocates to the cell surface in an insulin-dependent manner under high glucose conditions (46). Thus, Glut2 was used in the present study as a control to confirm the efficiency of the cell surface protection assay. The results showed that the intracellular Glut2 pool was indeed protected and that this pool rapidly disappeared after insulin stimulation (Fig. 5A). We noted that ATIC depletion delayed the insulin response of Glut2 (Fig. 5A). The opposite effect was observed for PTPLAD1 depletion; a lower Glut2 signal was also observed under basal (no insulin) conditions (Fig. 5A), indicating that ATIC and PTPLAD1 influence the insulin response with an inverse flux from an intracellular compartment to the cell surface. AICAR was reported to affect Glut2 translocation through Akt phosphorylation at its C-terminal Ser-473 by AMPK (47). As the ATIC depletion decreases the insulin-dependent Akt phosphorylation (Fig. 4A), this should explain the effect of ATIC down-regulation on Glut2 translocation. We sought to determine whether AICAR increases IR endocytosis and Glut2 transportation to the cell surface. Coincident with AMPK phosphorylation, a large increase in IR internalization was observed after pre-incubating the cells with AICAR, despite a 50% decrease in AICAR-induced IR expression (Fig. 5B). We cannot explain this AICAR-dependent reduction in IR expression; however, similar effects were previously observed in hepatoma cells (48). Glut2 disappeared after AICAR stimulation, even under basal (time 0) conditions (Fig. 5B).
Fig. 5.
ATIC or PTPLAD1 knockdown in HEK293 cells increases IR internalization, and AMPK knockdown decreases IR internalization. A, HEK293 cells were transfected for 48 h with ATIC or PTPLAD1 siRNA. The cells were preincubated in serum-free medium for 2 h and stimulated with insulin (5 nm). The cells were trypsinized at 4 °C before lysis. The immunoblots (50 μg protein extract) show the IR tetramer (α2β2; 5% resolving gels, no heat, anti-β subunit antibody). PTPLAD1, ATIC, Glut2 (epitope corresponding to amino acids 32–98 in the human Glut2 extracellular domain), and actin were resolved on denaturing gels (resolving gels 7.5%). B, HEK293 cells were preincubated at 37 °C in serum-free medium for 2 h and with AICAR for 15 min before insulin stimulation for the indicated time. The cells were then trypsinized before lysis. The immunoblots (50 μg protein extract) show the IR tetramer (α2β2), PTPLAD1, ATIC, Glut2, and actin. Glut2-total was from control nontrypsinized cells that were submitted to the cell lysis, denaturing electrophoresis and immunoblotting. The values for α2β2 shown in the right panel were corrected according to the total IR signals obtained in the presence of AICAR. C, HEK293 cells were transfected for 48 h with AMPKα2 siRNA. The cells were preincubated in serum-free medium for 2 h and stimulated with insulin. At the indicated time, the cells were trypsinized before lysis. The immunoblots (50 μg protein extract) show the IR tetramer (α2β2). AMPKα and tubulin were resolved on denaturing gels (7.5% resolving gels). The values shown are the means±s.d. (*p < 0.001, n = 3, two-tailed unpaired Student's t-tests).
The AMPKα2 catalytic isoform is the primary contributor to AMPK activation, even during AICAR stimulation, which is not compensated for by the AMPKα1 isoform (for a review, see (49)). Thus, we partially depleted AMPKα2 to determine whether AMPKα2 could inversely affect IR internalization compared with ATIC. The results showed that AMPKα2 knockdown decreased IR internalization (Fig. 5C). Therefore, ATIC, PTPLAD1, and AMPK are effectors of IR internalization in HEK293 cells.
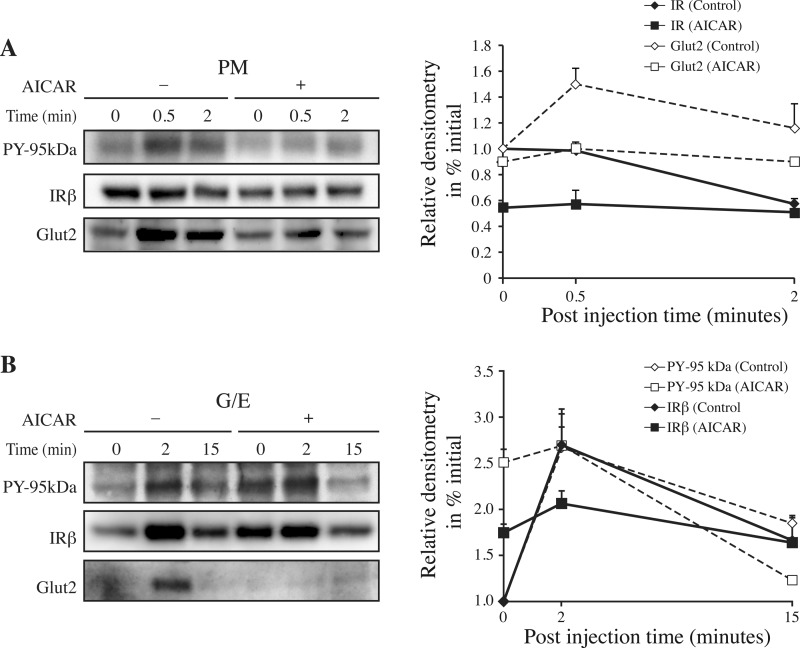
IR Internalization After AICAR Stimulation in the Rat Liver
A key first step in determining the importance of ATIC is to verify whether AICAR has the capacity to affect the kinetics of IR internalization in vivo. Fig. 6 depicts the time course for IR augmentation in the G/E versus PM fractions after the acute administration of AICAR 15 min before insulin injections. Under these conditions, we found that the kinetic of IR endocytosis was perturbed by AICAR with a significant (p < 0.001, n = 3, two-tailed unpaired Student's t-tests) 50% decrease in IR level observed in the PM (Fig. 6A). A coincident increase in IR internalization was observed in the G/E fraction under basal conditions (p < 0.001, n = 3, two-tailed unpaired Student's t-tests) with no further increase 2 min after insulin injection and with a similar area under the curve (Fig. 6B).
Fig. 6.
Time course for IR internalization in liver after AICAR stimulation. A,The plasma membrane (PM), and B, endosome (G/E) fractions were prepared as described in Fig. 2 except that vehicles (−) or AICAR (10 mg/100 g b.w.) (+) were injected via the jugular vein 15 min before the insulin (1.5 μg/100 g b.w.) injections. Portions of the same membranes (70 μg proteins) were subjected to immunoblot analysis (7.5% resolving gels) with the anti-IR-β subunit (IRβ), anti-PY (PY-20, PY-95 kDa), and anti-Glut2 antibodies. The relative contents of the IR and Glut2 proteins, which are shown in the right panels, were determined by expressing the densitometric data as a percentage of their levels the fractions at time 0, without AICAR. Each point on the progress curve is the mean ± s.d. of three separate experiments.
Consistent with the results obtained in HEK293 cells, Glut2 rapidly appeared in the PM fraction after insulin injection (Fig. 6A). However, Glut2 was not present in the G/E fraction (Fig. 6B), supporting the idea that, as for PTPLAD1, Glut2 is in a rapidly insulin-responsive intracellular compartment that is not present in the G/E fraction. Notably, the insulin-dependent appearance of Glut2 along with IR in the G/E fraction, which was almost abolished by AICAR, is consistent with a previously described Glut2 pool that is physically associated with IR in hepatic cells (50) (Fig. 6B). Hence, the results showed that AICAR markedly affects the IR internalization process under acute conditions, as well as the rapid appearance of Glut2 at the cell surface in vivo.
DISCUSSION
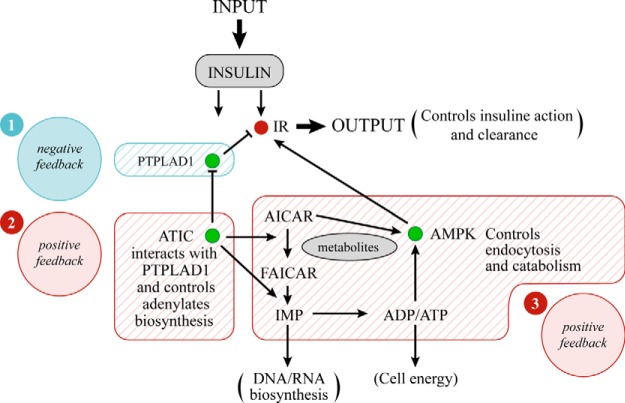
The results reported in this study reveal the presence of an ATIC network that minimally contains proteins PTPLAD1 and AMPK and that regulate IR tyrosine phosphorylation and endocytosis (Fig. 7). The marked increase in IR internalization that was observed at early and later time points after PTPLAD1 knockdown in HEK293 cells is consistent with higher IR tyrosine kinase activity (5, 51). This perturbation may be relevant to local changes in the actin cytoskeleton that indirectly favor IR autophosphorylation (7). In support of this possibility, actin was recruited into IR complexes in an insulin- and PTPLAD1-dependent manner (Fig. 4C). In addition, PTPLAD1 action was also related to Rac1-mediated events (52). Alternatively or in addition, PTPLAD1 may be a bona fide PTP as strongly suggested in Fig. 3A. We were unable to detect any consistent enzymatic activity against the artificial substrate para-nitrophenyl phosphate using bacterially expressed full-length PTPLAD1 or the well-positioned soluble cytosolic loop containing a conserved C(X)5K motif (FRYSFYMLTCIDMDWKVLTWLRY, 257–279). We previously showed however that a IR tyrosine dephosphorylating activity is abolished following the addition of Triton-X100 to isolated endosomes suggesting that an intact membrane microenvironment was critical (45). The topology of PTPLAD1 (a multipass transmembrane protein) is compatible with such a behavior. Also, the rapid insulin-dependent compartmentalization of PTPLAD1 at the cell surface (Fig. 2A and 2C) may explain original observations in rat hepatic cells where IR tyrosine dephosphorylation was shown to occur predominantly in endosomes and not at the cell surface following bpV(phen) administration (53). At this point, the results showed that PTPLAD1 is a protein that is highly responsive to insulin in vivo and in cultured cells. Taguwa et al. reported that PTPLAD1 localizes to endoplasmic reticulum-derived vesicles that display fast movement (54), which may explain the rapid PTPLAD1 accumulation in the PM fraction following insulin stimulation. Thus, similar to Glut2, PTPLAD1 is located in a membrane compartments with flotation properties that differ from those of G/E membranes (devoid of lipoproteins). This suggests that an efficient use of fast-moving insulin-responsive shuttles occurs, with the same insulin input being able to activate IR autophosphorylation and PTPLAD1 compartmentalization, as well as to stimulate glucose entry through Glut2.
Fig. 7.
Architecture of a signaling network that accounts for the relation between the effects of insulin, ATIC, and ATP level. Insulin inputs in a double incoherent mode: IR autophosphorylation plus PTPLAD1 recruitment at the cell surface (1) are converted into robust oscillations in output through the overlay of two positive feedback systems with delay driven by the metabolic enzyme ATIC (2). A local interacting loop counteracting the action of PTPLAD1 (3). Adenylates production. Variations in ATIC levels or in adenylates synthesis or a decline in ATP levels independent of de novo purine biosynthesis increase the AMPK activity and IR endocytosis.
ATIC deficiency also increased insulin-mediated endocytosis (Fig. 5A). This result was the opposite of our original expectation of lower IR tyrosine kinase activity. ATIC is a rate-limiting enzyme, and AICAR accumulates over time under conditions of lower ATIC expression (21, 22). AICAR is a nucleoside that is converted inside the cell to its mono-phosphorylated form, ZMP (5-amino-4-imidazolecarboxamide ribotide), by adenylate kinase; thus, AICAR behaves as an AMP analog that allosterically activates AMPK (19). AMPK activation results in the phosphorylation of the cargo-binding kinesin light chain, KCL2, of Kif5 and in the disruption of Kif5 association with PI3K and Akt (40). The insulin-dependent presence of phosphorylated AMPK and Kif5A in IR complexes (Fig. 4B) argues for the involvement of the AMPK/kinesin/microtubule axes in IR trafficking. PI3K itself is necessary for early and late endocytosis events (55, 56). Kif5 is involved in the Rab5-dependent movement of vesicles toward either the plus or the minus ends of microtubules (56). Thus, compartmentalized AMPK has a wide range of actions during the early and late stages of endocytic trafficking. The mechanism of AMPK localization within the IR environment has yet to be characterized but may be related to the recently described large hydrophobic pocket at the carbohydrate-binding domain/kinase interface (57).
ATIC is the only enzyme involved in the de novo synthesis of purine nucleotides that is bifunctional in every organism studied thus far (18). This apparent evolutionary conservation raised the question of whether a functional advantage to this structural arrangement may exist because substrate channeling is not used by ATIC (58). Another possible advantage of the bifunctionality of ATIC might be more structural in nature. Hence, ATIC exists in a monomer/dimer equilibrium in solution (59). The crystal structure of ATIC has been depicted with a considerable dimer interface for a protein of its size, burying 4929 Angstroms per monomer, of which the IMPCH domain makes up 38% (18, 60). With such a large percentage of the protein contributing to the interface, the physical association of these two activities on the same polypeptide may help to maintain the homodimeric or multimeric organization of the protein.
Insulin-mediated endocytosis is followed by insulin dissociation from IR at the acidic lumenal pH of hepatic endosomes and by free insulin degradation by proteases. A high rate of recycling of the free and tyrosine-dephosphorylated IR is important because the liver is a site for insulin clearance, removing 50–80% of insulin during portal passages (for a review see (12)). A deficiency in insulin-degrading enzyme (IDE), which has also been described as a cell surface protein (61), results in a marked insulin degradation deficit in mouse liver, causing hyperinsulinemia and glucose intolerance (62, 63). By having the capacity in the same time to sense ATP levels and synthesis, and to control IR autophosphorylation and endocytosis, the ATIC node may explain mechanistically the perceived promiscuity that exists between insulin resistance and clearance and thus the difficulty to define the primary mechanism for type 2 diabetes (64, 65).
In conclusion, the connection between IR and ATIC support recent observations indicating that cellular signaling systems are not organized in a linear fashion but rather rely on densely connected protein interaction networks (66). The elevated number of common heritable variants associated with type 2 diabetes in the actual IRGEN (Fig. 1B and supplemental Table S4) favors the idea that the observed confusing genetic heterogeneity (37, 67–69) converges however toward few unanticipated cell biology mechanisms (70, 71).
Supplementary Material
Acknowledgments
We thank Guillaume Diss for his assistance with the bioinformatic analysis. We thank Dr Sabine Elowe for the critical reading of the manuscript.
Footnotes
Author contributions: M.B., S.S.H., and R.L.F. designed research; M.B., G.C., S.F., and I.K. performed research; G.C., S.F., S.S.H., I.K., C.R.L., and R.L.F. analyzed data; M.B., C.R.L., and R.L.F. wrote the paper; R.L.F. director of the research program.
* This work was supported by the National Research Sciences and Engineering Council of Canada (RF, NSERC Discovery Grant: 155751). MBD holds a scholarship from the Foundation of Stars. GCM holds a scholarship from the NSERC. CRL is a FRQ-S Junior II investigator and is funded by CIHR Grants GMX-324265 and GMX-299432. SSH is a FRQ-S junior I investigator.
 This article contains supplemental Figs. S1 to S4 and Table S1 to S4.
This article contains supplemental Figs. S1 to S4 and Table S1 to S4.
1 The abbreviations used are:
- IF
- insulin receptor
- ATIC
- 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase
- PTPLAD1
- protein-tyrosine phosphatase-like A domain-containing protein 1
- IMP
- inositol monophosphate
- AMP
- adenosine monophosphate
- AMPK
- AMP-activated protein kinase
- AICAR
- 5-aminoimidazole-4-carboxamide-1-b-D-ribofuranoside.
REFERENCES
- 1. Hubbard S. R. (2013) Structural biology: insulin meets its receptor. Nature 493, 171–172 [DOI] [PubMed] [Google Scholar]
- 2. Ward C. W., Menting J. G., Lawrence M. C. (2013) The insulin receptor changes conformation in unforeseen ways on ligand binding: sharpening the picture of insulin receptor activation. Bioessays 35, 945–954 [DOI] [PubMed] [Google Scholar]
- 3. Taniguchi C. M., Emanuelli B., Kahn C. R. (2006) Critical nodes in signaling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 [DOI] [PubMed] [Google Scholar]
- 4. Posner B. I., Bergeron J. J. (2014) Assessment of internalization and endosomal signaling: studies with insulin and EGF. Methods Enzymol. 535, 293–307 [DOI] [PubMed] [Google Scholar]
- 5. McClain D. A. (1992) Mechanism and role of insulin receptor endocytosis. Am. J. Med. Sci. 304, 192–201 [DOI] [PubMed] [Google Scholar]
- 6. Carpentier J. L., McClain D. (1995) Insulin receptor kinase activation releases a constraint maintaining the receptor on microvilli. J. Biol. Chem. 270, 5001–5006 [DOI] [PubMed] [Google Scholar]
- 7. Vainio S., Heino S., Mansson J. E., Fredman P., Kuismanen E., Vaarala O., Ikonen E. (2002) Dynamic association of human insulin receptor with lipid rafts in cells lacking caveolae. EMBO Rep. 3, 95–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lange K., Brandt U., Gartzke J., Bergmann J. (1998) Action of insulin on the surface morphology of hepatocytes: role of phosphatidylinositol 3-kinase in insulin-induced shape change of microvilli. Exp. Cell Res. 239, 139–151 [DOI] [PubMed] [Google Scholar]
- 9. Carpentier J. L. (1993) Robert Feulgen Prize Lecture 1993. The journey of the insulin receptor into the cell: from cellular biology to pathophysiology. Histochemistry 100, 169–184 [DOI] [PubMed] [Google Scholar]
- 10. Balbis A., Baquiran G., Mounier C., Posner B. I. (2004) Effect of insulin on caveolin-enriched membrane domains in rat liver. J. Biol. Chem. 279, 39348–39357 [DOI] [PubMed] [Google Scholar]
- 11. Bergeron J. J., Di Guglielmo G. M., Baass P. C., Authier F., Posner B. I. (1995) Endosomes, receptor tyrosine kinase internalization, and signal transduction. Biosci. Rep. 15, 411–418 [DOI] [PubMed] [Google Scholar]
- 12. Duckworth W. C., Bennett R. G., Hamel F. G. (1998) Insulin degradation: progress and potential. Endocr. Rev. 19, 608–624 [DOI] [PubMed] [Google Scholar]
- 13. Di Fiore P. P., De Camilli P. (2001) Endocytosis and signaling. An inseparable partnership. Cell 106, 1–4 [DOI] [PubMed] [Google Scholar]
- 14. Goh L. K., Sorkin A. (2013) Endocytosis of receptor tyrosine kinases. Cold Spring Harb. Perspect. Biol. 5, a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Miaczynska M., Pelkmans L., Zerial M. (2004) Not just a sink: endosomes in control of signal transduction. Curr. Opin. Cell Biol. 16, 400–406 [DOI] [PubMed] [Google Scholar]
- 16. Sallese M., Pulvirenti T., Luini A. (2006) The physiology of membrane transport and endomembrane-based signaling. EMBO J. 25, 2663–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jackson R. C., Harkrader R.J. (1981) Nucleosides and cancer treatment. (Tatersall M.H.N., Fox R.M., eds), pp. 18–31, Academic Press, Sidney, Australia [Google Scholar]
- 18. Beardsley G. P., Rayl E. A., Gunn K., Moroson B. A., Seow H., Anderson K. S., Vergis J., Fleming K., Worland S., Condon B., Davies J. (1998) Structure and functional relationships in human pur H. Adv. Exp. Med. Biol. 431, 221–226 [DOI] [PubMed] [Google Scholar]
- 19. Corton J. M., Gillespie J. G., Hawley S. A., Hardie D. G. (1995) 5-aminoimidazole-4-carboxamide ribonucleoside. A specific method for activating AMP-activated protein kinase in intact cells? Eur. J. Biochem. 229, 558–565 [DOI] [PubMed] [Google Scholar]
- 20. Shaw R. J., Cantley L. C. (2012) Cell biology. Ancient sensor for ancient drug. Science 336, 813–814 [DOI] [PubMed] [Google Scholar]
- 21. Wang W., Fridman A., Blackledge W., Connelly S., Wilson I. A., Pilz R. B., Boss G. R. (2009) The phosphatidylinositol 3-kinase/akt cassette regulates purine nucleotide synthesis. J. Biol. Chem. 284, 3521–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marie S., Heron B., Bitoun P., Timmerman T., Van Den Berghe G., Vincent M. F. (2004) AICA-ribosiduria: a novel, neurologically devastating inborn error of purine biosynthesis caused by mutation of ATIC. Am. J. Hum. Genet. 74, 1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hubbard A. L., Ma A. (1983) Isolation of rat hepatocyte plasma membranes. II. Identification of membrane-associated cytoskeletal proteins. J. Cell Biol. 96, 230–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bilodeau N., Fiset A., Boulanger M. C., Bhardwaj S., Winstall E., Lavoie J. N., Faure R. L. (2010) Proteomic analysis of Src family kinases signaling complexes in Golgi/endosomal fractions using a site-selective antiphosphotyrosine antibody: identification of LRP1-insulin receptor complexes. J. Proteome Res. 9, 708–717 [DOI] [PubMed] [Google Scholar]
- 25. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L. J. (2013) STRING v9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chatr-Aryamontri A., Breitkreutz B. J., Oughtred R., Boucher L., Heinicke S., Chen D., Stark C., Breitkreutz A., Kolas N., O'Donnell L., Reguly T., Nixon J., Ramage L., Winter A., Sellam A., Chang C., Hirschman J., Theesfeld C., Rust J., Livstone M. S., Dolinski K., Tyers M. (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res. 43, D470–D478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saito R., Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Lotia S., Pico A. R., Bader G. D., Ideker T. (2012) A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Doncheva N. T., Assenov Y., Domingues F. S., Albrecht M. (2012) Topological analysis and interactive visualization of biological networks and protein structures. Nat. Protoc. 7, 670–685 [DOI] [PubMed] [Google Scholar]
- 29. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 30. Morris J. H., Apeltsin L., Newman A. M., Baumbach J., Wittkop T., Su G., Bader G. D., Ferrin T. E. (2011) clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics 12, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fiset A., Xu E., Bergeron S., Marette A., Pelletier G., Siminovitch K. A., Olivier M., Beauchemin N., Faure R. L. (2011) Compartmentalized CDK2 is connected with SHP-1 and beta-catenin and regulates insulin internalization. Cell Signal. 23, 911–919 [DOI] [PubMed] [Google Scholar]
- 32. Backer J. M., Kahn C. R., Cahill D. A., Ullrich A., White M. F. (1990) Receptor-mediated internalization of insulin requires a 12-amino acid sequence in the juxtamembrane region of the insulin receptor beta-subunit. J. Biol. Chem. 265, 16450–16454 [PubMed] [Google Scholar]
- 33. Contreres J. O., Faure R., Baquiran G., Bergeron J. J., Posner B. I. (1998) ATP-dependent desensitization of insulin binding and tyrosine kinase activity of the insulin receptor kinase. The role of endosomal acidification. J. Biol. Chem. 273, 22007–22013 [DOI] [PubMed] [Google Scholar]
- 34. Hubbard S. R. (2013) The insulin receptor: both a prototypical and atypical receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol. 5, a008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Béréziat V., Kasus-Jacobi A., Perdereau D., Cariou B., Girard J., Burnol A. F. (2002) Inhibition of insulin receptor catalytic activity by the molecular adapter Grb14. J. Biol. Chem. 277, 4845–4852 [DOI] [PubMed] [Google Scholar]
- 36. Nouaille S., Blanquart C., Zilberfarb V., Boute N., Perdereau D., Roix J., Burnol A. F., Issad T. (2006) Interaction with Grb14 results in site-specific regulation of tyrosine phosphorylation of the insulin receptor. EMBO Rep. 7, 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Visscher P. M., Brown M. A., McCarthy M. I., Yang J. (2012) Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Greasley S. E., Horton P., Ramcharan J., Beardsley G. P., Benkovic S. J., Wilson I. A. (2001) Crystal structure of a bifunctional transformylase and cyclohydrolase enzyme in purine biosynthesis. Nat. Struct. Biol. 8, 402–406 [DOI] [PubMed] [Google Scholar]
- 39. Oakhill J. S., Steel R., Chen Z. P., Scott J. W., Ling N., Tam S., Kemp B. E. (2011) AMPK is a direct adenylate charge-regulated protein kinase. Science 332, 1433–1435 [DOI] [PubMed] [Google Scholar]
- 40. Amato S., Liu X., Zheng B., Cantley L., Rakic P., Man H. Y. (2011) AMP-activated protein kinase regulates neuronal polarization by interfering with PI 3-kinase localization. Science 332, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Becker K. G., Barnes K. C., Bright T. J., Wang S. A. (2004) The genetic association database. Nat. Genet. 36, 431–432 [DOI] [PubMed] [Google Scholar]
- 42. Yu W., Wulf A., Yesupriya A., Clyne M., Khoury M. J., Gwinn M. (2008) HuGE Watch: tracking trends and patterns of published studies of genetic association and human genome epidemiology in near-real time. Eur. J. Hum. Genet. 16, 1155–1158 [DOI] [PubMed] [Google Scholar]
- 43. Bauer-Mehren A., Bundschus M., Rautschka M., Mayer M. A., Sanz F., Furlong L. I. (2011) Gene-disease network analysis reveals functional modules in mendelian, complex, and environmental diseases. PLoS One 6, e20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Faure R., Baquiran G., Bergeron J. J., Posner B. I. (1992) The dephosphorylation of insulin and epidermal growth factor receptors. Role of endosome-associated phosphotyrosine phosphatase (s). J. Biol. Chem. 267, 11215–11221 [PubMed] [Google Scholar]
- 46. Thorens B. (1996) Glucose transporters in the regulation of intestinal, renal, and liver glucose fluxes. Am. J. Physiol. 270, G541–G553 [DOI] [PubMed] [Google Scholar]
- 47. Jiang X., Kenerson H., Aicher L., Miyaoka R., Eary J., Bissler J., Yeung R. S. (2008) The tuberous sclerosis complex regulates trafficking of glucose transporters and glucose uptake. Am. J. Pathol. 172, 1748–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakamaru K., Matsumoto K., Taguchi T., Suefuji M., Murata Y., Igata M., Kawashima J., Kondo T., Motoshima H., Tsuruzoe K., Miyamura N., Toyonaga T., Araki E. (2005) AICAR, an activator of AMP-activated protein kinase, down-regulates the insulin receptor expression in HepG2 cells. Biochem. Biophys. Res. Commun. 328, 449–454 [DOI] [PubMed] [Google Scholar]
- 49. Viollet B., Athea Y., Mounier R., Guigas B., Zarrinpashneh E., Horman S., Lantier L., Hebrard S., Devin-Leclerc J., Beauloye C., Foretz M., Andreelli F., Ventura-Clapier R., Bertrand L. (2009) AMPK: lessons from transgenic and knockout animals. Front Biosci. 14, 19–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leturque A., Brot-Laroche E., Le Gall M. (2009) GLUT2 mutations, translocation, and receptor function in diet sugar managing. Am. J. Physiol. Endocrinol Metab. 296, E985–E992 [DOI] [PubMed] [Google Scholar]
- 51. Carpentier J. L., Paccaud J. P., Gorden P., Rutter W. J., Orci L. (1992) Insulin-induced surface redistribution regulates internalization of the insulin receptor and requires its autophosphorylation. Proc. Natl. Acad. Sci. U.S.A. 89, 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Courilleau D., Chastre E., Sabbah M., Redeuilh G., Atfi A., Mester J. (2000) B-ind1, a novel mediator of Rac1 signaling cloned from sodium butyrate-treated fibroblasts. J. Biol. Chem. 275, 17344–17348 [DOI] [PubMed] [Google Scholar]
- 53. Bevan A. P., Burgess J. W., Drake P. G., Shaver A., Bergeron J. J., Posner B. I. (1995) Selective activation of the rat hepatic endosomal insulin receptor kinase. Role for the endosome in insulin signaling. J. Biol. Chem. 270, 10784–10791 [DOI] [PubMed] [Google Scholar]
- 54. Taguwa S., Kambara H., Omori H., Tani H., Abe T., Mori Y., Suzuki T., Yoshimori T., Moriishi K., Matsuura Y. (2009) Cochaperone activity of human butyrate-induced transcript 1 facilitates hepatitis C virus replication through an Hsp90-dependent pathway. J. Virol. 83, 10427–10436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shpetner H., Joly M., Hartley D., Corvera S. (1996) Potential sites of PI-3 kinase function in the endocytic pathway revealed by the PI-3 kinase inhibitor, wortmannin. J. Cell Biol. 132, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nielsen E., Severin F., Backer J. M., Hyman A. A., Zerial M. (1999) Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1, 376–382 [DOI] [PubMed] [Google Scholar]
- 57. Xiao B., Sanders M. J., Carmena D., Bright N. J., Haire L. F., Underwood E., Patel B. R., Heath R. B., Walker P. A., Hallen S., Giordanetto F., Martin S. R., Carling D., Gamblin S. J. (2013) Structural basis of AMPK regulation by small molecule activators. Nat. Commun. 4, 3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bulock K. G., Beardsley G. P., Anderson K. S. (2002) The kinetic mechanism of the human bifunctional enzyme ATIC (5-amino-4-imidazolecarboxamide ribonucleotide transformylase/inosine 5′-monophosphate cyclohydrolase). A surprising lack of substrate channeling. J. Biol. Chem. 277, 22168–22174 [DOI] [PubMed] [Google Scholar]
- 59. Vergis J. M., Bulock K. G., Fleming K. G., Beardsley G. P. (2001) Human 5-aminoimidazole-4-carboxamide ribonucleotide transformylase/inosine 5′-monophosphate cyclohydrolase. A bifunctional protein requiring dimerization for transformylase activity but not for cyclohydrolase activity. J. Biol. Chem. 276, 7727–7733 [DOI] [PubMed] [Google Scholar]
- 60. Cheong C. G., Wolan D. W., Greasley S. E., Horton P. A., Beardsley G. P., Wilson I. A. (2004) Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates. J. Biol. Chem. 279, 18034–18045 [DOI] [PubMed] [Google Scholar]
- 61. Li Q., Ali M. A., Cohen J. I. (2006) Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell 127, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Farris W., Mansourian S., Chang Y., Lindsley L., Eckman E. A., Frosch M. P., Eckman C. B., Tanzi R. E., Selkoe D. J., Guenette S. (2003) Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc. Natl. Acad. Sci. U.S.A. 100, 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Abdul-Hay S. O., Kang D., McBride M., Li L., Zhao J., Leissring M. A. (2011) Deletion of insulin-degrading enzyme elicits antipodal, age-dependent effects on glucose and insulin tolerance. PLoS One 6, e20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shanik M. H., Xu Y., Skrha J., Dankner R., Zick Y., Roth J. (2008) Insulin resistance and hyperinsulinemia: is hyperinsulinemia the cart or the horse? Diabetes Care 2, S262–S268 [DOI] [PubMed] [Google Scholar]
- 65. Ader M., Stefanovski D., Kim S. P., Richey J. M., Ionut V., Catalano K. J., Hucking K., Ellmerer M., Van Citters G., Hsu I. R., Chiu J. D., Woolcott O. O., Harrison L. N., Zheng D., Lottati M., Kolka C. M., Mooradian V., Dittmann J., Yae S., Liu H., Castro A. V., Kabir M., Bergman R. N. (2014) Variable hepatic insulin clearance with attendant insulinemia is the primary determinant of insulin sensitivity in the normal dog. Obesity 22, 1238–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Levy E. D., Landry C. R., Michnick S. W. (2010) Cell signaling. Signaling through cooperation. Science 328, 983–984 [DOI] [PubMed] [Google Scholar]
- 67. Watanabe R. M. (2010) The genetics of insulin resistance: Where's Waldo? Curr. Diab. Rep. 10, 476–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Pal A., McCarthy M. I. (2013) The genetics of type 2 diabetes and its clinical relevance. Clin. Genet. 83, 297–306 [DOI] [PubMed] [Google Scholar]
- 69. Saxena R., Voight B. F., Lyssenko V., Burtt N. P., de Bakker P. I., Chen H., Roix J. J., Kathiresan S., Hirschhorn J. N., Daly M. J., Hughes T. E., Groop L., Altshuler D., Almgren P., Florez J. C., Meyer J., Ardlie K., Bengtsson Bostrom K., Isomaa B., Lettre G., Lindblad U., Lyon H. N., Melander O., Newton-Cheh C., Nilsson P., Orho-Melander M., Rastam L., Speliotes E. K., Taskinen M. R., Tuomi T., Guiducci C., Berglund A., Carlson J., Gianniny L., Hackett R., Hall L., Holmkvist J., Laurila E., Sjogren M., Sterner M., Surti A., Svensson M., Svensson M., Tewhey R., Blumenstiel B., Parkin M., Defelice M., Barry R., Brodeur W., Camarata J., Chia N., Fava M., Gibbons J., Handsaker B., Healy C., Nguyen K., Gates C., Sougnez C., Gage D., Nizzari M., Gabriel S. B., Chirn G. W., Ma Q., Parikh H., Richardson D., Ricke D., Purcell S. (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316, 1331–1336 [DOI] [PubMed] [Google Scholar]
- 70. Goh K. I., Cusick M. E., Valle D., Childs B., Vidal M., Barabási A. L. (2007) The human disease network. Proc. Natl. Acad. Sci. U.S.A. 104, 8685–8690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sahni N., Yi S., Zhong Q., Jailkhani N., Charloteaux B., Cusick M. E., Vidal M. (2013) Edgotype: a fundamental link between genotype and phenotype. Curr. Opin. Genet. Dev. 23, 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.