Abstract
Skeletal muscle constitutes 40% of individual body mass and plays vital roles in locomotion and whole-body metabolism. Proteomics of skeletal muscle is challenging because of highly abundant contractile proteins that interfere with detection of regulatory proteins. Using a state-of-the art MS workflow and a strategy to map identifications from the C2C12 cell line model to tissues, we identified a total of 10,218 proteins, including skeletal muscle specific transcription factors like myod1 and myogenin and circadian clock proteins. We obtain absolute abundances for proteins expressed in a muscle cell line and skeletal muscle, which should serve as a valuable resource. Quantitation of protein isoforms of glucose uptake signaling pathways and in glucose and lipid metabolic pathways provides a detailed metabolic map of the cell line compared with tissue. This revealed unexpectedly complex regulation of AMP-activated protein kinase and insulin signaling in muscle tissue at the level of enzyme isoforms.
Skeletal muscle is a highly specialized tissue that plays a fundamental role in locomotion and is indispensable in regulating whole-body carbohydrate metabolism. This tissue alone accounts for about 75% of insulin-stimulated glucose uptake (1). Skeletal muscle exhibits a remarkable plasticity and adapts to a wide range of stimuli such as exercise and nutrient supply (2–4). Metabolism and function of skeletal muscle is severely affected in pathological conditions such as type 2 diabetes (5), neuromuscular disorders (6), cancer cachexia (7), age-related sarcopenia (8), muscular atrophy (9), and muscular dystrophy (10). MS-based proteomics has begun to advance molecular understanding of these muscle diseases (11–13). However, most of these pioneering studies had limited proteome coverage and lacked robust quantitation (14). Significant technological advances over the last decade now allow near exhaustive analysis of cell line proteomes (15–17). These advances have recently enabled the identification of several thousand proteins in muscle tissue preparations (18, 19). However, the challenging dynamic range of protein expression in tissues in general and in muscle in particular has so far prevented measurement of very deep proteomes that would also cover the low abundance, regulatory proteins.
The C2C12 is an immortalized mouse myoblast cell line that can readily be differentiated to myotubes in culture and is commonly used as a model system for investigating molecular, biochemical, or pathological changes of skeletal muscle (20–22). Although these cells express sarcomeric proteins and can develop contractile properties in culture, they lack the 3D structure and specialized muscle functions characteristic of the tissue context. Here we measured a very deep proteome of the C2C12 myotubes and transfer the peptide and protein identifications to a data set obtained under the same conditions from adult mouse skeletal muscle. This strategy allowed us to obtain the largest skeletal muscle proteome so far and to highlight similarities and differences between these two model systems. We further quantitatively mapped the protein isoforms of glucose uptake signaling pathways, key metabolic pathways, and muscle specific transcription factors. Although we choose to present quantitation of these selected proteins and pathways because of their central role in muscle function, our approach could be applied to any protein or pathway as well as expanded to other challenging tissues.
EXPERIMENTAL PROCEDURES
Animal Experiments
All animal experiments were approved by Danish Animal Experimental Inspectorate in compliance with the European Convention for Protection of Vertebrate Animal Used for Scientific Purposes. Fifteen-week-old female C57BL/6 mice were maintained on a 12:12-h light-dark cycle and had free access to standard chow diet. Triceps muscles were surgically removed from the anesthetized mice and quickly frozen in liquid nitrogen followed by storage at −80 °C.
Cell Culture and Differentiation
C2C12 cells (myoblasts) were grown in Eagle's minimum essential medium supplemented with 2 mm l-glutamine and 10% fetal bovine serum plus antibiotics in a humidified atmosphere with 5% CO2 in air. Undifferentiated myoblasts were grown to confluence in normal growth media (day 0). To induce differentiation, the amount of serum in the media was decreased to 2%. Cells were differentiated for 8 days. Growth medium was replaced with fresh medium every 2 days over a period of 8 days. At day 8 post differentiation, we observed less than 5% cell death (Tryphan blue staining). After 8 days, differentiated C2C12 (myotubes) were harvested for proteomics analysis.
Protein Sample Preparation
Triceps muscle and differentiated C2C12 cells were lysed in buffer consisting of 0.1 m Tris-HCl, pH 7.5, 0.1 m DTT, and 4% SDS, and incubated at 95 °C for 5 min. Triceps muscle homogenization was achieved with Ultra Turbax blender (IKA, Staufen, Germany). Lysates were sonicated using a Branson type sonicator and were then clarified by centrifugation at 16,100 × g for 10 min.
Protein Digestion and Peptide Fractionation
Cell lysates were diluted in 8 m urea in 0.1 m Tris-HCl followed by protein digestion with trypsin according to the FASP1 protocol (23). After an over-night digestion peptides were eluted from the filters with 25 mm ammonium bicarbonate buffer. From each sample, 100 μg of peptides were fractionated by isoelectric focusing on an OffGel fractionator (Agilent, Santa Clara, USA) in 12 well formats as described (24). Peptides from each of the 12 fractions were purified on C18 StageTips.
LC-MS Analysis
Analysis was performed in triplicates. Samples were measured using LC-MS instrumentation consisting of an Easy nano-flow HPLC system (Thermo Fisher Scientific, Odense, Denmark) coupled via a nanoelectrospray ion source (Thermo Fisher Scientific, Bremen, Germany) to a Q Exactive mass spectrometer (25). Purified peptides were separated on 50 cm C18 columns (inner diameter 75 μm, 1.8 μm beads, Dr, Maisch GmbH, Germany). Peptides were loaded onto the column with buffer A (0.5% formic acid) and eluted with a 150 min linear gradient from 2–30% buffer B (80% acetonitrile, 0.5% formic acid). After the gradient the column was washed with 90% buffer B and re-equilibrated with buffer A. Mass spectra were acquired in a data-dependent manner, with automatic switching between MS and MS/MS using a top-10 method. MS spectra were acquired in the Orbitrap analyzer with mass range of 300–1750 m/z and 70,000 resolutions at m/z 200. HCD1 peptide fragments acquired at 25 normalized collision energy were analyzed at high resolution in the Orbitrap.
Data Analysis
Raw MS files were analyzed by MaxQuant version 1.3.7.4 (26) (http://www.maxquant.org). MS/MS spectra were searched by the Andromeda search engine (27) against the decoy UniProt-mouse database (Version June 2012, 59,345 entries) supplemented with 262 frequently observed contaminants and forward and forward and reverse sequences. In the main Andromeda search precursor mass and fragment mass were identified with an initial mass tolerance of 6 ppm and 20 ppm, respectively. The search included variable modifications of methionine oxidation and N-terminal acetylation, and fixed modification of carbamidomethyl cysteine. Minimal peptide length was set to seven amino acids and a maximum of two mis-cleavages was allowed. When we checked for the occurrence of demidation, we found it to be negligible (supplemental Fig. S6, supplemental Table S9). We therefore did not include it as a variable modification. The false discovery rate (FDR1) was set to 0.01 for peptide and protein identifications. MS runs from skeletal muscle were analyzed with or without the “match between runs” option. For matching, a retention time window of 30 s was selected. In the case of identified peptides that are all shared between two proteins, these were combined and reported as one protein group. Proteins matching to the reverse database were filtered out. For the proteins that were identified with single peptide, detailed information about the MSMS spectrum, peptide sequence, precursor m/z is provided (supplemental Table S8 and supplemental Fig. S5).
Bioinformatics Analysis
All bioinformatics analysis was performed with the Perseus software (http://www.perseus-framework.org). Categorical annotation was supplied in form of Gene Ontology (GO) biological process (BP), molecular function (MF), and cellular component (CC), as well as participation in a KEGG pathway. All annotations were extracted from UniProt database. Hierarchical clustering and 2D annotation enrichment were based on label-free quantitation of the samples (28). Because of randomness of peptide sampling in shotgun proteomics, quantification will be missing in some samples for each protein. In order to retain sufficiently informative protein expression profiles for bioinformatics analysis, the protein expression data was filtered to have at least two valid abundance values in at least one group (muscle or C2C12). The data was inputed to fill missing abundance values by drawing random numbers from a Gaussian distribution with a standard deviation of 30% in comparison to the standard deviation of measured protein abundances, and one standard deviation downshift of the mean. These parameters have been tuned in order to best simulate the distribution of low abundant proteins. These values are universally applied to nearly all data sets that were generated with the label-free quantitation algorithm. Two sample t test were performed on muscle and C2C12 groups with FDR = 0.05. Hierarchical clustering of significantly different proteins was performed after z-score normalization. We then performed Fisher exact test on particular clusters, testing for enrichment or depletion of any annotation term in the cluster compared with the whole matrix. The specific test used is a two-dimensional version of the nonparametric Mann-Whitney test. Multiple hypothesis testing was controlled by using a Benjamini-Hochberg false discovery rate threshold of 5%. Fisher exact test was performed with an FDR value of 0.04.
2 D Annotation Enrichment
Categorical annotations are supplied in the form of GOCC, GOMF, GOBP, and KEGG. A two-dimensional two-sample test was performed to find the significant differences between the two-dimensional means of the two protein populations (skeletal muscle and C2C12 myotubes). The nonparametric Mann-Whitney test was used. Multiple hypothesis testing was controlled by using a Benjamini-Hochberg FDR threshold of 5%. For categories that are significant, a two-dimensional difference score was calculated by determining the average rank of the protein abundance belonging to the corresponding annotation category. This average rank was then rescaled to the interval between −1 and 1. A value of 1 in one of the dimensions would mean that all members of this category are the largest values in this dimensions, whereas a value of 0 means that the ranks of the members of the category are distributed in same way as the background proteins having no significant bias toward larger or smaller values. For detailed explanation of the rank and scaling of 2D score between 1 to −1, please refer to (29).
Absolute Protein Abundance
Absolute protein abundance (mass) was calculated as described before (30). Briefly, the mass spectrometric signal of an individual protein was divided by the sum of all proteins. Because the resulting values were small, we multiplied them by 1 million and expressed absolute abundance as part-per-million (ppm) (Figs. 5, 6, and supplemental Table I). In other words, the absolute protein abundance described here is the fractional signal of each protein in relation to the total protein signal and hence is expressed in ppm. Quantifiable proteins in the analysis of skeletal muscle and C2C12 myotubes were defined as those identified at least two times in each group. Error bars in Figures 5, 6, and supplemental Fig. S4 denote standard deviation of median. In Table SI, quantifiable proteins were defined as those identified at least two times in skeletal muscle. The absolute abundances that are provided were calculated for the list of the proteins identified after using “match between runs” (Table SI).
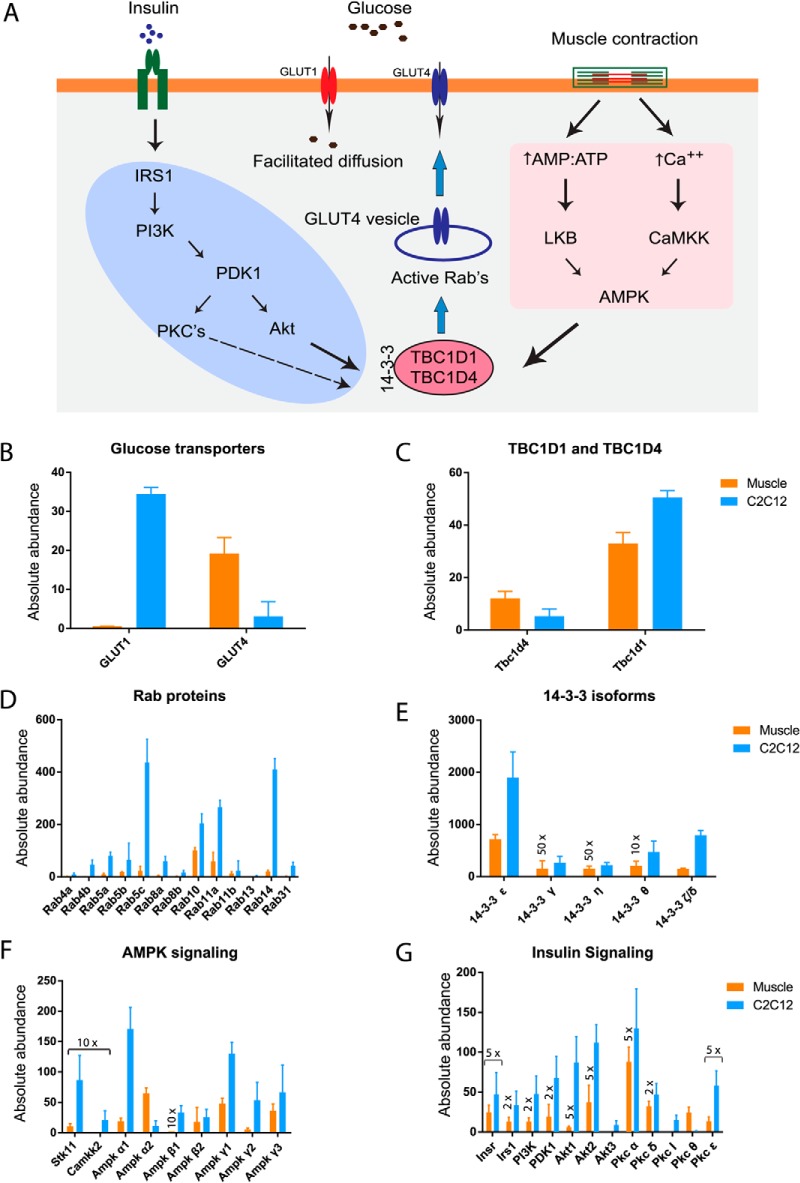
Fig. 5.
Absolute abundance of insulin and AMPK signaling. A, Proteins involved in insulin and AMPK mediated glucose uptake in skeletal muscle (33, 38). Absolute abundance (described under “Experimental Procedures”) of glucose transporter 1 and 4. B, Rab GTPases-activating proteins, C, Rab proteins, D, protein 14–3-3 isoforms, E, members of AMPK, F. and insulin G, signaling. Error bar are standard deviation of median.
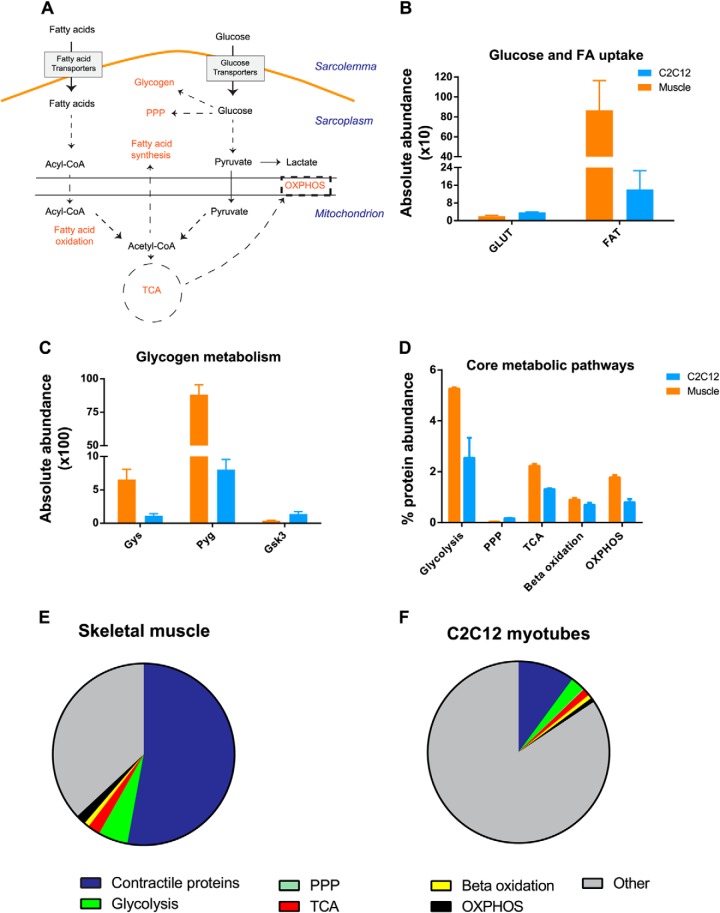
Fig. 6.
A proteomic view of skeletal muscle metabolism. A, Schematic representation of glucose and lipid metabolism in skeletal muscle. B, Summed absolute abundance of glucose transporters (GLUT) and fatty acid transporters (FAT). C, Absolute abundance of glycogen synthase (Gys), glycogen phosphorylase (Pyg) and glycogen synthase kinase 3 (Gsk3). D, Total abundance (%) of metabolic pathways. Total abundance is calculated by summing up absolute abundance of individual enzyme from respective pathways (supplemental Table S6). E and F, Percent contribution abundance of contractile proteins, metabolic pathways and other process in skeletal muscle and C2C12 myotubes. Error bars are standard deviation of median.
Western Blot Analysis
The lysates from the muscle and C2C12 cells were electrophoresed, transferred to nitrocellulose, and probed for selected proteins. This analysis was performed on the very same lysate that were used for MS analysis (n = 2). The following antibodies were used: goat anti-AMPK alpha 2 (Santacruz, Dallas, USA), rabbit anti-HK II (Cell Signaling, Beverly, USA), rabbit anti-GLUT4 (Thermo scientific, Naerum, Denmark), and rabbit anti-GAPDH (Cell signaling, Beverly, USA). The anti-AMPK-a1 antibody (raised in sheep) was kindly provided by Prof. D. G. Hardie (University of Dundee, UK).
RESULTS
Proteome Analysis of Mouse Skeletal Muscle
In previous attempts at very deep proteome coverage of cell lines, we have employed protein level fractionation as well as several proteolytic enzymes (15). However, recent experience in our laboratory suggested that single enzyme digestion and fractionation only at the peptide level, when coupled to advanced MS instrumentation, would be sufficient to achieve great depth of protein identification (31). We therefore developed a streamlined workflow, consisting of cell line or tissue homogenization, filter-aided sample preparation (FASP) (23), followed by peptide separation into twelve fractions according to their isoelectric point as described (24). Peptides were measured by a linear quadrupole-Orbitrap mass analyzer, which achieves sub or low parts per million mass accuracy for peptide ions and their fragments (25). This streamlined method required only 1 day of measurement time for a single analysis each of the proteomes and computational analysis followed by automated computational analysis in the MaxQuant environment (26) (Fig. 1A–1C).
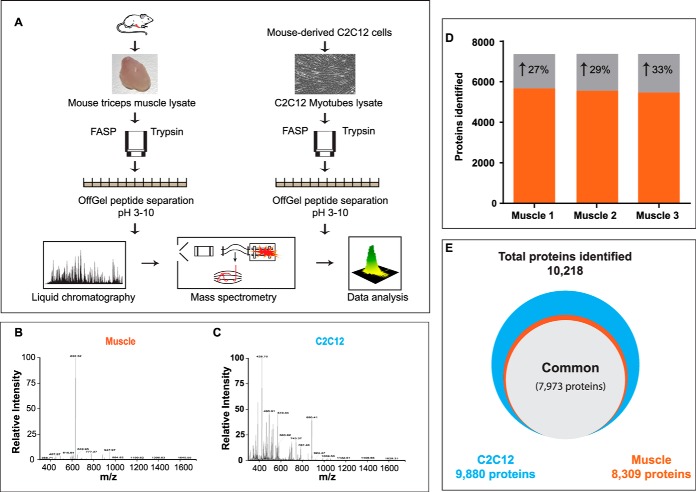
Fig. 1.
Proteome analysis of mouse skeletal muscle. A, Protein lysates (triplicate) from mouse triceps muscle and C2C12 were digested on FASP filter and peptides were separated on an OFFGEL fractionator. Each fraction was analyzed by LCMS on a Q Exactive mass spectrometer. Representative spectrum from skeletal muscle, B, and C2C12 myotubes, C, at MS and MSMS levels. D, Number of protein identification without match between runs (black) and with match between runs option (gray). E, Total number of proteins identified in skeletal muscle and C2C12 cells.
We performed triplicate analyses of triceps muscles from C57BL/6J mice and of differentiated C2C12 cells and analyzed the results together in MaxQuant specifying a false discovery rate of 1% at the peptide and protein level. This identified a total of 10,218 proteins, a vastly larger number than reported in muscle proteome studies so far (Fig. 1D) (supplemental Table S1). A total of 8,309 proteins were identified in skeletal muscle, 9880 proteins in C2C12 myotubes, and 7,973 (78% of the total) proteins were identified in both systems. A mere 338 proteins were exclusively identified in the tissue.
Interestingly, when skeletal muscle samples were analyzed separately, only 5,887 proteins were identified, whereas the corresponding number for myotubes was 9,624 (supplemental Table S2). This large difference is partially because of the complexity of the tissue and its large dynamic range, which makes MS analysis challenging. Peptides from a few highly abundant proteins of skeletal muscle interfere with the identification of lower abundant proteins, thereby reducing the overall number of identifications (Fig. 1B–1C). This is illustrated with a representative example from skeletal muscle: because of one high abundant peptide (originating from myosin), neighboring low abundant peptides are suppressed in comparison and may not be picked for fragmentation (Fig. 1B). This is not the case when analyzing C2C12 myotubes, where a larger number of peptides share approximately equal abundance and can all be fragmented and identified (Fig. 1C).
Combined analysis of mouse skeletal muscle and C2C12 myotubes efficiently mitigated the challenge of dynamic range on protein identification in muscle tissue. MaxQuant aligned the retention times and—based on the accurately determined masses—efficiently transferred the peptide identification from C2C12 myotubes to more complex mouse skeletal muscle (“Match between runs” feature in MaxQuant). This identification, by matching is statistically controlled and high confidence of identification, is further indicated by the fact that 95% of the identified peptides were observed in a tight 0.4 min match time window (supplemental Fig. S1A). Altogether, this approach boosted protein identification by about 30%, which resulted in identification of 8,309 proteins in total in skeletal muscle (Fig. 1D–1E, supplemental Table S2). Most of proteins identified partially or exclusively by matching are of low abundance and include interesting regulatory classes such as skeletal muscle specific transcription factors (myod1, myog, mef2c), nuclear receptors (Smad1, Notch3), and circadian clock proteins (Bmal1, Cry1, Fbxl3) (supplemental Fig. S2A–S2B).
Our experiment did not use isotope labeling but relied on label-free quantitation in the MaxQuant environment (28). We examined reproducibility of this procedure between biological triplicates and found very high correlations within the skeletal muscle and C2C12 groups (0.9–0.96, supplemental Fig. S1B).
Cumulative Protein and Ranked Protein Abundance
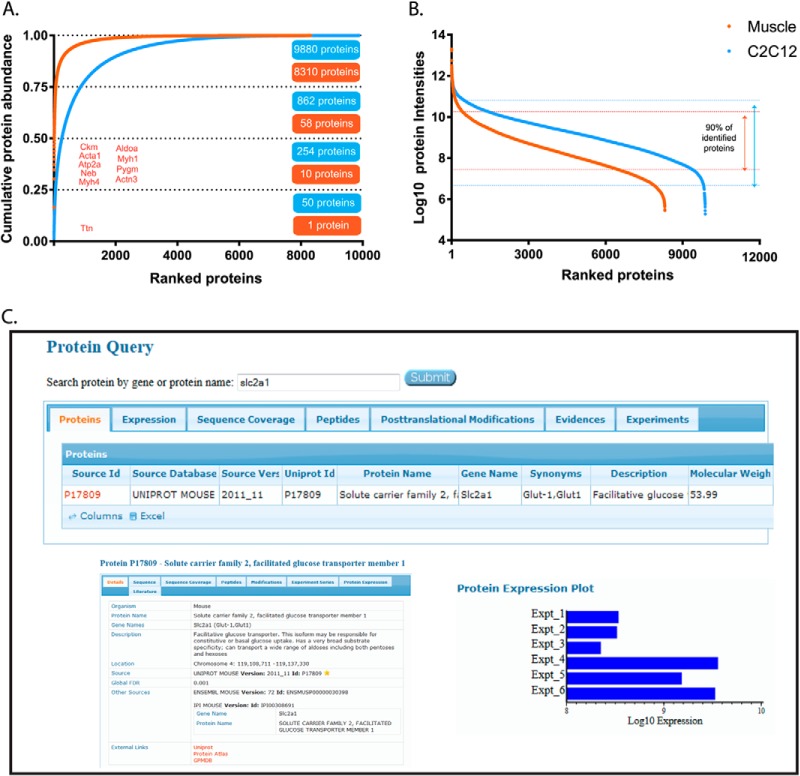
Using summed MS-signals (“abundances”) of the peptides identifying each protein in the MS measurements, we estimated their contribution to the proteome. For protein isoforms, the shared peptides were counted as contributing to each of them. To obtain a global view of the dynamic range of the muscle proteome, the C2C12 myotube proteome and their differences, we plotted the cumulative contribution of each protein to total protein mass (Fig. 2A) as well as the individual, ranked abundance (Fig. 2A, 2B; supplemental Table S3). Combining the abundances of all identified heavy and light myosin isoforms results in 18% of total muscle mass, positioning myosin as the highest abundant protein in skeletal muscle. The giant protein titin (390 kDa) alone accounts for another 16% of total protein mass. The 10 most abundant proteins according to MS measurement were titin, myosin-4, nebulin, calcium ATPase 1, actin α, creatine kinase, α-actinine-3, glycogen phosphorylase, and myosin-1 and together they make up 50% of total protein mass in skeletal muscle. Actin and myosin are the key members of the contractile units (sarcomere) in muscle. The estimated ratio between actin/myosin abundances was ∼1:7. The summed abundance of proteins that are assigned to the contractile machinery by Gene Ontology Cellular Compartment (GOCC) annotation was 53.6% of total protein mass for skeletal muscle. The ranked distribution of all individual proteins revealed a much steeper decline of abundances for the tissue compared with the cell line (Fig. 2B). In skeletal muscle, the lower half of the proteome accounts for a negligible fraction of its total mass (<0.1%). Our data also allows us to determine the amino acid make up of mouse muscle proteome as the weighted contribution from all identified protein sequences. For instance, branched chain amino acids, which are important in protein synthesis, constitute 20.5% of all amino acids weighted by protein abundance, which is similar to values in other mouse tissues (supplemental Fig. S3).
Fig. 2.
Cumulative and ranked protein abundance. A, Cumulative protein mass from highest to lowest abundant proteins. B, Ranked protein abundances from highest to the lowest. C, Screenshot of the MaxQB database. Query protein for glucose transporter slc2a1 (I) returns the search results (II). Selection of protein slc2a1 displays the details on this protein (III). By clicking the expression tab in II, protein expression plots in skeletal muscle (Expt_1, 2, 3) and C2C12 myotubes (Expt_4, 5, 6) are displayed (IV).
Although almost all proteins identified in muscle were also identified in C2C12 myotubes, these two model systems turned out to be quite different on a quantitative level. For instance, the top 10 proteins contribute fivefold less to total proteome mass than they did in muscle tissue (Fig. 2A, 2B).
Our data provides very high confidence identification as well as estimates of the abundance for thousands of skeletal muscle proteins. To make this data conveniently available to the community, it has been deposited in the MaxQB database (http://maxqb.biochem.mpg.de/mxdb) (32). MaxQB has a user-friendly interface that allows querying any protein or gene of interest for the MS-data acquired in the proteomics experiments and it is linked to additional details such as description of protein, chromosome location, ensemble ID, links to other databases such as Uniprot, or Protein Atlas (Fig. 2C). Apart from estimated expression levels within or between proteomes, one can also gather further information such as sequence coverage or post-translational modifications. Such knowledge of the expression pattern of a protein of interest under different circumstances may be helpful in prediction and interpretation of certain phenotypes of skeletal muscle under normal or diseases conditions. Furthermore, expression levels can be compared with those in other data sets including a set of human cell line proteomes as well as an initial atlas of mouse tissue proteomes (18, 31).
Functional Differences between C2C12 Myotubes and Adult Skeletal Muscle
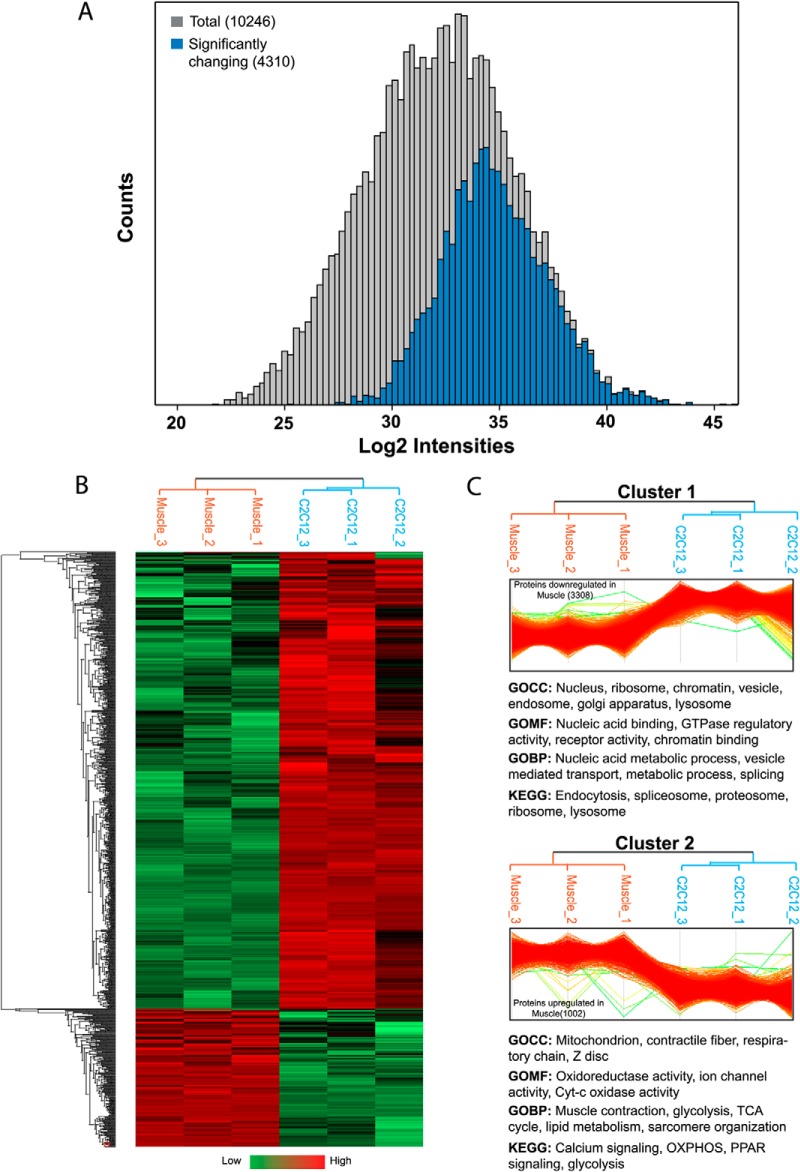
To gain insights into biological differences between the cell line and tissue systems, we analyzed the label-free quantification values of close to 10,000 proteins. Examination of the complete data set by two sample t test extracted 4,310 proteins (44% of total identified) that are significantly different between skeletal muscle and C2C12 myotubes (FDR 0.05, supplemental Table S4). As depicted in Fig. 3A, the majority of them were of high abundance, presumably at least in part because it is more difficult to reach statistical significance for the low expressed proteins. The statistical test will tend to more easily identify statistically significant differences in proteins abundant in one or the other systems. Low abundance proteins can be expressed differently without passing the significance threshold of our test. Note also that we do not claim that proteins that we do not detect are not present, even when we use “match between runs.” Fig. 3B shows the heat map of significantly different proteins, with Cluster 1 representing 3,308 proteins, which were significantly down-regulated and Cluster 2 representing 1,002 proteins, which were significantly up-regulated in skeletal muscle (Fig. 3C). Fischer's exact test for the enrichment of Gene Ontology (GO) protein annotations in the set of significantly changing proteins returned many significantly changing protein categories (FDR 0.05; Fig. 3C). As expected, glycolysis, tricarboxylic acid (TCA) cycle, mitochondrion, contractile proteins, respiratory chain, and calcium signaling were significantly enriched in skeletal muscle (Cluster 2). In fact, members of GOCC category “mitochondrial part” were the most significantly up-regulated proteins in skeletal muscle (p < 10−72) and collectively this category was expressed about twofold higher in muscle. Conversely, cell lines lacked the transverse tubular system (GOCC category “t-tubules”; p < 10−6). Tables of all significantly enriched protein annotations for Cluster 1 and Cluster 2 are presented in supplemental Table S5.
Fig. 3.
Functional differences between C2C12 myotubes and adult skeletal muscle. A, Histogram of total proteins identified in skeletal muscle and C2C12 myotubes (gray) and proteins that are significantly changing between the skeletal muscle and C2C12 myotubes (blue). B, Hierarchical clustering of significantly changing proteins. Cluster 1 (3,308 proteins) represents significantly down-regulated and Cluster 2 (1,002 proteins) significantly up-regulated proteins in skeletal muscle. Annotation categories below the respective clusters are representative examples from significantly enriched categories.
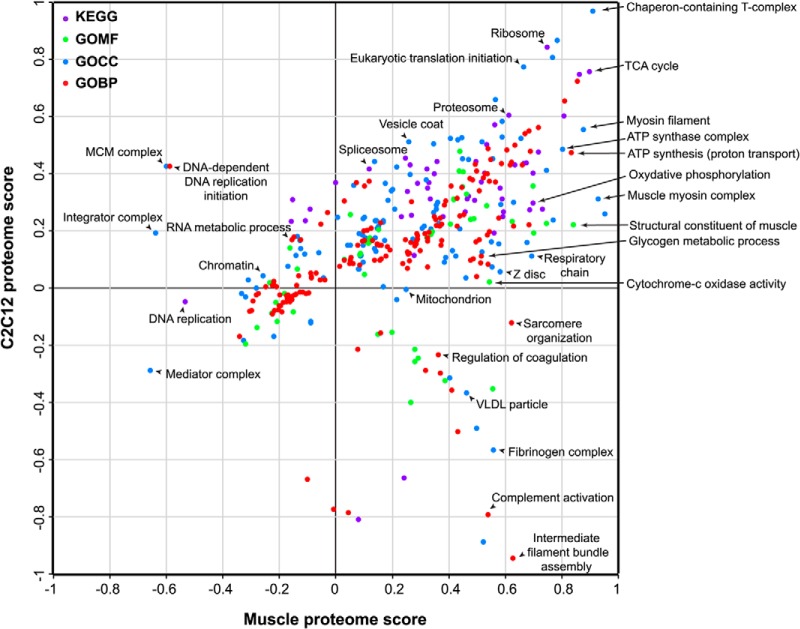
Comparative analysis of two closely related systems highlights their significant differences but not their commonalities. Therefore, we next employed 2D annotation enrichment, which calculates enrichments in each of the systems compared with a background proteome, in this case the mouse proteome (29), and thereby reveals protein categories with significant regulation in the combined space of skeletal muscle and C2C12 myotubes proteome (Fig. 4). Together the differential protein quantification, the cluster analysis, and the 2D analysis allow us to probe functional differences between two different biological systems as reflected in their proteomes.
Fig. 4.
2D annotation distribution. Scatter plot of normalized annotation changes between skeletal muscle and C2C12 myotubes. Calculation of significance is detailed under “Experimental Procedures.” The annotations analyzed were: KEGG (purple), GOMF (green), GOCC (blue), and GOBP (red).
Proteins associated with GO categories like “intermediate filament bundle assembly,” “sarcomere organization,” and “z-disc, myosin filament” were overrepresented in the skeletal muscle tissues compared with the cell line. This was also evident by the percentage contribution of individual sarcomeric proteins to the total proteome (supplemental Fig. S4A). Individual proteins such as titin and myosin were as much as eight times higher in the skeletal muscle tissue whereas SERCA (sarco/endoplasmic reticulum calcium-ATPase), which regulates the calcium homeostasis and muscle contraction, was 10 times higher in the skeletal muscle tissues. Despite these differences, some features are preserved between these systems (diagonal line in Fig. 4). For instance, components of stable protein complexes such as 20S proteasome, large ribosomal subunits, chaperones, protein folding complexes, and proteasome complex maintain similar protein expression in both systems (supplemental Table S6). Proteins of the contractile machinery were of different abundance between the systems; however, the protein abundance ratios were often preserved. For instance, expression ratios between tropomyosin and troponin, which are crucial in the regulation of muscle contraction via calcium binding, were similar between the skeletal muscle tissues and C2C12 (about 1:1.3 in both).
Over-representation of proteins associated with the mini chromosome maintenance (MCM) complex, the integrator complex, the mediator complex, DNA-dependent replication initiation, RNA metabolic processes, and DNA replication in C2C12 myotubes compared with muscle, suggest the presence of a small population of dividing cells in myotubes. Mature skeletal muscle tissue is post-mitotic meaning that myofibers are incapable of further cell division. Muscle cells (myoblasts) differentiated into myotubes are thought to be postmitotic. However, our analysis revealed that even after an 8 day differentiation protocol, C2C12 cells still contain some dividing myoblasts as evidenced by expression of proteins regulating cell division and DNA replication. Our proteomics data also provides a percentage of protein mass attributed to major GO categories in the skeletal muscle tissues and C2C12 myotubes on a global basis (supplemental Fig. S4B–S4C). Proteins associated with various metabolic processes accounts for >30% of the total proteome, indicating the central role of metabolism in skeletal muscles and, by extension, in the whole body. Our analysis also indicates that diverse molecular functions like apoptosis, phosphorylation, proteolysis, translation, endocytosis, and RNA splicing are more pronounced in C2C12 myotubes, highlighting less obvious processes that are rearranged in the cultured muscle cells in contrast to the muscle tissues.
Our analysis also directly reveals the cellular heterogeneity of tissue compared with the cell line. Total protein extract from the skeletal muscle tissues also contain components derived from nerves, capillaries, and other cell types. Their presence was evident from over-represented KEGG categories like “complement and coagulation cascade” (p < 10−8), high expression of proteins like myelin basic protein, and cholinesterase (supplemental Table S4). Over-representation of the proteins categories from the heterogeneous tissue like fibrinogen complex, complements activation, very low density lipoprotein (VLDL) particles, and regulation of coagulation in skeletal muscle was also evident in the 2D annotation distribution. For example, compared with C2C12 myotubes, proteins from coagulation cascade such as fibrinogen and α-2-antiplasmin were several fold higher in skeletal muscles, indicating the presence of a minor contribution from blood cells in skeletal muscles (supplemental Table S1). The abundances of these proteins can be used to estimate the extent of these other cell types in muscle lysate. These results suggest that proteomic studies of skeletal muscles should account for the protein contribution of motor neurons and blood capillaries, the extent of which can easily be gauged by the above analyses.
Glucose Uptake, Insulin, and AMP-activated Protein Kinase (AMPK) Signaling
Skeletal muscle is the major site for glucose disposal. In skeletal muscle, insulin and muscle contraction stimulate translocation of glucose transporter (GLUT4) from the cytoplasm to the plasma membrane by distinct signaling pathways (33, 34). AMP-activated protein kinase (AMPK) is a key player in contraction or exercise-mediated glucose uptake in skeletal muscle (35, 36). Our deep proteome analysis of skeletal muscle enabled us to determine the absolute abundance of glucose transport associated components of insulin and AMPK signaling (Fig. 5). We found dramatically higher levels of GLUT4 than GLUT1 (about 34-fold) underlining the importance of insulin and exercise-like stimuli in the regulated versus basal glucose uptake (Fig. 5B). In contrast, in the C2C12 myotubes, GLUT1 was more highly expressed than GLUT4, which suggests that these cells take up relatively more glucose under nonstimulated conditions (via GLUT1) and that their response to insulin and AMPK mediated glucose uptake (via GLUT4) is compromised.
TBC1D1 and TBC1D4 regulate GLUT4 trafficking via activating Rab proteins. In skeletal muscle expression of TBC1D1 was higher than that of TBC1D4 (Fig. 5C), as has already been reported for a similar muscle type (EDL) consistent with previous studies (37). Out of the 52 identified Rab proteins in our data set, Rab 4a, 4b, 5a, 5b, 5c, 8a, 8b, 10, 11a, 11b, 13, 14, and 31are linked to GLUT4 trafficking (38) (Fig. 5D). Rab5c and Rab14 were the most abundant Rabs in C2C12 myotubes whereas Rab10 and Rab11a were the most abundant in skeletal muscle. Protein 14–3-3 isoforms are also involved in regulation of insulin and AMPK mediated glucose uptake in skeletal muscle (38). Compared with expression levels in the skeletal muscle tissues, all isoforms of protein 14–3-3 were highly expressed in C2C12 myotubes (Fig. 5E). Relative expression between individual isoforms, however, was similar for both systems; for example, isoform ε and ζ/δ are the most abundant and η and γ are the least abundant in both systems.
AMPK exists as a heterotrimeric complex comprising catalytic α, and regulatory β and γ subunits. Each α and β subunit are encoded by distinctive genes (α1, α2 and β1, β2), whereas the γ subunit is encoded by three genes (γ1, γ2, and γ3), giving the possibility to form 12 different heterotrimers (39). Based on absolute abundances of α, β, and γ isoforms, our data indicates that the complexes containing α2β2γ1 are predominant in triceps muscle (Fig. 5F), in line with previous findings in the EDL muscle (40). Remarkably, this MS-based quantification was possible even though the AMPK subunits are relatively low abundance, for instance, we estimate the β1 subunit to be about 70,000-fold less than titin and it ranks in the bottom quarter of the proteome. Among the upstream kinases of AMPK, protein serine threonine kinase 11 (Stk11) appears to be more abundant compared with calcium/calmodulin-dependent protein kinase kinase (CaMKK1), which was identified but did not pass our filter for quantification (supplemental Table S7; Fig. 5F). CaMKK2, another upstream kinase for AMPK was detected in C2C12 cells but not in skeletal muscle, confirming an earlier observation (41).
The absolute abundance of members of the insulin signaling pathways of special interest because of its importance in diabetes and in particular because multiple isoforms of target proteins in insulin signaling drive different metabolic and gene regulatory effects in skeletal muscle (42). Globally all components of the core insulin signaling pathways were highly represented in C2C12 myotubes compared with the skeletal muscle, with the notable exception of protein kinase C θ (Fig. 5G). This negative regulator of insulin signaling (43) was identified with 17 unique peptides in skeletal muscle and we estimate its abundance to be >40 higher than in C2C12 cells (Fig. 5G). Despite this, in both systems, insulin receptor substrate 1 (IRS1) and Akt2, the protein isoforms that primarily drive glucose uptake are much more abundant compared with other isoforms (Fig. 5G). We performed Western blots of selected proteins involved in glucose metabolism. As expected, this followed the same pattern as our MS analysis (supplemental Fig. S5). Together, our results suggest partial conservation of regulatory metabolism for glucose uptake processes between C2C12 myotubes and skeletal muscle. The abundances of members of insulin and AMPK signaling is provided in supplemental Table S7.
A Proteomic View of Skeletal Muscle Metabolism
Glucose and lipids are the major sources of energy in skeletal muscles. Their energy demand is mainly fulfilled by phosphocreatine and ATP produced during glucose and fat oxidation (glycolysis, Krebs cycle, and oxidative phosphorylation). Here we sought to generate a global, systems level map of the proteins involved in glucose and lipid metabolic pathways in skeletal muscle (Fig. 6). Metabolic enzymes were generally very well covered in our MS data. Taking advantage of this fact, we required at least four unique peptides in our analysis of this class of proteins. We first focused on the glucose (GLUT) and fatty acid transporters (FATs), that function in the uptake of glucose and lipids. As expected we quantified GLUT1 and GLUT4, whereas GLUT3 and the liver and blood cell specific GLUT2 were absent in both systems. A wide range of FATs were quantified in both systems—Cluster of differentiation 36 (CD36), Fatty acid binding protein (FABP) 3,4,5, and long chain fatty acid transporter protein 1 and 2. Interestingly, although adipose tissues are the primary storage sites for fatty acids, we found that the combined absolute abundances of FATs were dramatically higher than that of the GLUTs in both skeletal muscle tissues and C2C12 myotubes (Fig. 6B).
Skeletal muscles are the major site of glycogen storage in the body and is readily mobilized during contraction and we were interested in the ratios of the three key enzymes regulating glycogen synthesis (Glycogen synthase (Gys)), glycogen breakdown (Glycogen phosphorylase (Pyg)), and inhibiting glycogen synthase activity (Glycogen synthase kinase 3 (GSK3)). In skeletal muscle, the Pyg to Gys ratio was 1:13, whereas Gys to GSK3 ratio was 1:17 (Fig. 6C). This shows that quantitation of enzymatic levels is readily feasible and could be applied in glycogen turnover studies of resting, exercised, or diabetic muscles. We further quantified the enzymes regulating glycolysis, the Krebs cycle OXPHOS, and pentose phosphate pathways, providing a total proteome overview of muscle function (Fig. 6D). In skeletal muscle, contractile proteins accounted for 53% of total proteome mass. Of the remaining 47%, core metabolic pathways contribute ∼10%, underlining their importance to skeletal muscle function. Total abundances of enzymes involved in glycolytic pathways alone accounted for more than 5% of the muscle proteome. Interestingly, the pentose phosphate pathway (PPP) was strikingly different between C2C12 myotubes and skeletal muscle. The enzymes of oxidative and nonoxidative phase of PPP were 10 and 2.5 times more abundant in C2C12 myotubes, respectively.
DISCUSSION
Recent advances in MS-based proteomics now allow rapid quantification of complete proteome in simple organisms like yeast (44) and near exhaustive proteomes of mammalian cells (15–17). Nevertheless, comprehensive proteomics of complex samples such as tissues in general and skeletal muscle in particular is challenging (18) (reviewed in (14)). This is because the highly abundant sarcomeric proteins including the various isoforms of myosins, troponins, tropomyosins, and associated proteins dramatically increases the dynamic range of the expressed proteome, which extends down to low-abundant proteins such as transcription factors.
In the present study transfer of the identifications from C2C12 myotubes to skeletal muscle also increased protein identification in skeletal muscle by about 30%. Using this method, we identified and quantified many transcriptional regulators that were barely detectable in previous skeletal muscle proteomics studies. Together, we identified over 10,000 proteins in mouse skeletal muscle and C2C12 myotubes, which included skeletal muscle specific transcription factors like myod1 and myogenin and very low abundant circadian clock proteins. Our data represents by far the largest accurately quantified proteome for either skeletal muscle or skeletal muscle cells from any species.
The mouse-derived C2C12 muscle cell line is commonly used as a model system for investigating skeletal muscle. C2C12 myoblasts are differentiated to the myotubes to adjust them more “skeletal muscle like.” Clearly C2C12 myotubes, being an in vitro system, have a number of limitations as a biological system. Although this is generally acknowledged in the field, our proteomic analysis now allows us to pinpoint these differences in an objective way. It should also be noted that C2C12 cell cultures do not contain satellite cells equivalent to adult mouse skeletal muscle tissues. Moreover, the degree and quality of differentiation can vary.
Despite these limitations, we found that at least 8000 proteins were commonly expressed between mouse skeletal muscle and C2C12 myotubes, suggesting similarity of C2C12 cells with mouse skeletal muscle. Quantitative expression profiles showed that 44% of proteome is significantly changing between skeletal muscle and C2C12 cells. The overall picture that emerges from our proteomic and bioinformatic investigation is that C2C12 myotubes do not possess many of the specialized features of skeletal muscle in vivo. Although this might have been expected, functional differences between these models have never been quantified in a global and unbiased way. Our data set now allows biologists to consider advantages and disadvantages of these systems while performing specific biological assays. This may help to manipulate the muscle cells to better reflect the in vivo situation. It also highlights areas in which findings from muscle cell lines can be most readily extrapolated to adult muscle tissue.
In our data set, we accurately quantified every single known member of the insulin and AMPK signaling pathways that are linked to glucose uptake in skeletal muscle (33, 38), which revealed interesting similarities and differences between skeletal muscle and C2C12 myotubes. For instance, given that the absolute abundance of GLUT1 was 60 times higher in C2C12 compared with skeletal muscle, these cells likely take up glucose largely independent of stimulation. This may explain the difficulties in studying GLUT4 mediated glucose uptake upon insulin and AMPK activation in C2C12 myotubes.
The complexity of glucose regulatory signaling is highlighted by the existence of multiple isoforms of target proteins that we found. In human skeletal muscle, based on the intensity of exercise, certain types of AMPK heterotrimers are activated, which potentially dictate muscle preference for either glucose or lipids (45, 46). In our data set, heterotrimers containing α2 AMPK subunits were predominantly present in skeletal muscle whereas C2C12 myotubes predominantly express α1 containing AMPK heterotrimers. These differences might produce a differential effect of AMPK activators in skeletal muscle and C2C12 myotubes. Conversely, our data shows conserved expression patterns of isoform regulating glucose metabolism (IRS1-Akt2) C2C12 myotubes and skeletal muscle, suggesting that C2C12 myotubes can serve as a model system to study glucose metabolism in muscle tissues.
The glycolysis, TCA, and OXPHOS pathways help to maintain steady-state of ATP synthesis flux which is critical for sustained muscle performance (47). Comparative analysis of C2C12 myotubes and skeletal muscle showed very distinct profiles related to glucose utilization and the abundances data of these proteins should be useful in interpreting differential responses to glucose uptake or glycogen synthesis assays in these systems. Although glycolytic enzyme were more than twofold up-regulated in skeletal muscle, enzyme of oxidative PPP were 10-fold higher in C2C12 myotubes. This indicates that C2C12 myotubes use considerable amounts of glucose to produce NADPH and ribose sugars potentially for reductive biosynthesis reaction and nucleic acid synthesis.
Clearly, MS-based quantitation can accurately provide a relationship between individual signaling components and muscle pathology or pathophysiology, for instance in type 2 diabetic muscle (5) and exercise training modulated AMPK signaling (33). This approach can be particularly useful in the context of complex conditions involving skeletal muscle dysfunction, such as type 2 diabetes and the metabolic syndrome and training-induced adaptations in muscle, where many metabolic processes are altered simultaneously.
Data Availability
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000288. The data is also accessible and can be visualized in the MaxQB database (http://maxqb.biochem.mpg.de/mxdb) (29) under “deep muscle proteome.”
Supplementary Material
Acknowledgments
We thank Igor Paron and Korbinian Mayr for assistance in mass spectrometric analysis, and Per Schack Larsen for assistance with Western blotting. We also thank Sean Humphrey and Pablo Garcia-Roves for critical reading of the manuscript and fruitful discussions. We thank Professor Michael Lund Nielsen for providing help with OFFGEL specific modifications.
Footnotes
Author contributions: A.S.D. and M. Mann designed research; A.S.D. and N.N. performed research; A.S.D. and J.C. contributed new reagents or analytic tools; A.S.D., N.N., J.C., and M. Mann analyzed data; A.S.D., M. Murgia, J.T.T., and M. Mann wrote the paper.
* This work was supported by Federation of European Biochemical Societies (FEBS), by the Max-Planck Society for the Advancement of Science, The Novo Nordisk Foundation Center for Basic Metabolic Research Copenhagen, Denmark, and Commission's 7th Framework Programme (grant agreement HEALTH-F4-2008-201648/PROSPECTS).
 This article contains supplemental Figs. S1 to S7 and Tables S1 to S9.
This article contains supplemental Figs. S1 to S7 and Tables S1 to S9.
1 The abbreviations used are:
- FDR
- false discovery rate
- FASP
- filter-aided sample preparation
- HCD
- Higher-energy collisional dissociation.
REFERENCES
- 1. DeFronzo R. A., Gunnarsson R., Bjorkman O., Olsson M., Wahren J. (1985) Effects of insulin on peripheral and splanchnic glucose metabolism in noninsulin-dependent (type II) diabetes mellitus. J. Clin. Invest. 76, 149–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chibalin A. V., Yu M., Ryder J. W., Song X. M., Galuska D., Krook A., Wallberg-Henriksson H., Zierath J. R. (2000) Exercise-induced changes in expression and activity of proteins involved in insulin signal transduction in skeletal muscle: differential effects on insulin-receptor substrates 1 and 2. Proc. Natl. Acad. Sci. U.S.A. 97, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Booth F. W., Thomason D. B. (1991) Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol. Rev. 71, 541–585 [DOI] [PubMed] [Google Scholar]
- 4. Hawley J. A. (2002) Adaptations of skeletal muscle to prolonged, intense endurance training. Clin. Exp. Pharmacol. Physiol. 29, 218–222 [DOI] [PubMed] [Google Scholar]
- 5. DeFronzo R. A., Bonadonna R. C., Ferrannini E. (1992) Pathogenesis of NIDDM. A balanced overview. Diabetes Care 15, 318–368 [DOI] [PubMed] [Google Scholar]
- 6. Lacomis D. (2013) Electrophysiology of neuromuscular disorders in critical illness. Muscle Nerve 47, 452–463 [DOI] [PubMed] [Google Scholar]
- 7. Kadar L., Albertsson M., Areberg J., Landberg T., Mattsson S. (2000) The prognostic value of body protein in patients with lung cancer. Ann. N.Y. Acad. Sci. 904, 584–591 [DOI] [PubMed] [Google Scholar]
- 8. Evans W. J. (1995) What is sarcopenia? J. Gerontol. A Biol. Sci. Med. Sci. 50, 5–8 [DOI] [PubMed] [Google Scholar]
- 9. Mallinson J. E., Murton A. J. (2013) Mechanisms responsible for disuse muscle atrophy: potential role of protein provision and exercise as countermeasures. Nutrition 29, 22–28 [DOI] [PubMed] [Google Scholar]
- 10. Campbell K. P. (1995) Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell 80, 675–679 [DOI] [PubMed] [Google Scholar]
- 11. Hojlund K., Yi Z., Hwang H., Bowen B., Lefort N., Flynn C. R., Langlais P., Weintraub S. T., Mandarino L. J. (2008) Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol. Cell. Proteomics 7, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Doran P., Yi Z., Hwang H., Bowen B., Lefort N., Flynn C. R., Langlais P., Weintraub S. T., Mandarino L. J. (2006) Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics 6, 4610–4621 [DOI] [PubMed] [Google Scholar]
- 13. Parker K. C., Walsh R. J., Salajegheh M., Amato A. A., Krastins B., Sarracino D. A., Greenberg S. A. (2009) Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J. Proteome Res. 8, 3265–3277 [DOI] [PubMed] [Google Scholar]
- 14. Ohlendieck K. (2010) Proteomics of skeletal muscle differentiation, neuromuscular disorders, and fiber aging. Expert Rev. Proteomics 7, 283–296 [DOI] [PubMed] [Google Scholar]
- 15. Nagaraj N., Wisniewski J. R., Geiger T., Cox J., Kircher M., Kelso J., Paabo S., Mann M. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. (2011) The quantitative proteome of a human cell line. Mol. Syst. Biol. 7, 549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altelaar A. F., Munoz J., Heck A. J. (2013) Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet, 14, 35–48 [DOI] [PubMed] [Google Scholar]
- 18. Geiger T., Velic A., Macek B., Lundberg E., Kampf C., Nagaraj N., Uhlen M., Cox J., Mann M. (2013) Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol. Cell. Proteomics 12, 1709–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drexler H. C., Ruhs A., Konzer A., Mendler L., Bruckskotten M., Looso M., Gunther S., Boettger T., Kruger M., Braun T. (2012) On marathons and sprints: an integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers. Mol. Cell. Proteomics 11, M111 010801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsuchiya Y., Hatakeyama H., Emoto N., Wagatsuma F., Matsushita S., Kanzaki M. (2010) Palmitate-induced down-regulation of sortilin and impaired GLUT4 trafficking in C2C12 myotubes. J. Biol. Chem. 285, 34371–34381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharples A. P., Al-Shanti N., Stewart C. E. (2010) C2 and C2C12 murine skeletal myoblast models of atrophic and hypertrophic potential: relevance to disease and aging? J. Cell. Physiol. 225, 240–250 [DOI] [PubMed] [Google Scholar]
- 22. Watanabe T., Sasagawa N., Usuki F., Koike H., Saitoh N., Sorimachi H., Maruyama K., Nakase H., Takagi A., Ishiura S., Suzuki K. (1999) Overexpression of myotonic dystrophy protein kinase in C2C12 myogenic culture involved in the expression of ferritin heavy chain and interleukin-1alpha mRNAs. J. Neurol. Sci. 167, 26–33 [DOI] [PubMed] [Google Scholar]
- 23. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 24. Hubner N. C., Ren S., Mann M. (2008) Peptide separation with immobilized pI strips is an attractive alternative to in-gel protein digestion for proteome analysis. Proteomics 8, 4862–4872 [DOI] [PubMed] [Google Scholar]
- 25. Michalski A., Damoc E., Hauschild J. P., Lange O., Wieghaus A., Makarov A., Nagaraj N., Cox J., Mann M., Horning S. (2011) Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteomics 10, M111 011015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p. p. b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 28. Cox J., Hein M. Y., Luber C. A., Paron I., Nagaraj N., Mann M. (2014) MaxLFQ allows accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction. Mol. Cell. Proteomics 13, 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cox J., Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wisniewski J. R., Ostasiewicz P., Dus K., Zielinska D. F., Gnad F., Mann M. (2012) Extensive quantitative remodeling of the proteome between normal colon tissue and adenocarcinoma. Mol. Syst. Biol. 8, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Geiger T., Wehner A., Schaab C., Cox J., Mann M. (2012) Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteomics, 11, M111 014050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schaab C., Geiger T., Stoehr G., Cox J., Mann M. (2012) Analysis of high accuracy, quantitative proteomics data in the MaxQB database. Mol. Cell. Proteomics 11, M111 014068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deshmukh A. S., Hawley J. A., Zierath J. R. (2008) Exercise-induced phospho-proteins in skeletal muscle. Int. J. Obes. 32, S18–S23 [DOI] [PubMed] [Google Scholar]
- 34. Ploug T., van Deurs B., Ai H., Cushman S. W., Ralston E. (1998) Analysis of GLUT4 distribution in whole skeletal muscle fibers: identification of distinct storage compartments that are recruited by insulin and muscle contractions. J. Cell Biol. 142, 1429–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Egan B., Zierath J. R. (2013) Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 17, 162–184 [DOI] [PubMed] [Google Scholar]
- 36. O'Neill H. M., Maarbjerg S. J., Crane J. D., Jeppesen J., Jorgensen S. B., Schertzer J. D., Shyroka O., Kiens B., van Denderen B. J., Tarnopolsky M. A., Kemp B. E., Richter E. A., Steinberg G. R. (2011) AMP-activated protein kinase (AMPK) beta1beta2 muscle null mice reveal an essential role for AMPK in maintaining mitochondrial content and glucose uptake during exercise. Proc. Natl. Acad. Sci. U.S.A. 108, 16092–16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Szekeres F., Chadt A., Tom R. Z., Deshmukh A. S., Chibalin A. V., Bjornholm M., Al-Hasani H., Zierath J. R. (2012) The Rab-GTPase-activating protein TBC1D1 regulates skeletal muscle glucose metabolism. Am. J. Physiol. Endocrinol. Metab. 303, E524–E533 [DOI] [PubMed] [Google Scholar]
- 38. Sakamoto K., Holman G. D. (2008) Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am. J. Physiol. Endocrinol. Metab. 295, E29–E37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Long Y. C., Zierath J. R. (2006) AMP-activated protein kinase signaling in metabolic regulation. J. Clin. Invest. 116, 1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Treebak J. T., Birk J. B., Hansen B. F., Olsen G. S., Wojtaszewski J. F. (2009) A-769662 activates AMPK beta1-containing complexes but induces glucose uptake through a PI3-kinase-dependent pathway in mouse skeletal muscle. Am. J. Physiol. Cell Physiol. 297, C1041–C1052 [DOI] [PubMed] [Google Scholar]
- 41. Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 42. Bouzakri K., Zachrisson A., Al-Khalili L., Zhang B. B., Koistinen H. A., Krook A., Zierath J. R. (2006) siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 4, 89–96 [DOI] [PubMed] [Google Scholar]
- 43. Li Y., Soos T. J., Li X., Wu J., Degennaro M., Sun X., Littman D. R., Birnbaum M. J., Polakiewicz R. D. (2004) Protein kinase C Theta inhibits insulin signaling by phosphorylating IRS1 at Ser(1101). J. Biol. Chem. 279, 45304–45307 [DOI] [PubMed] [Google Scholar]
- 44. Kulak N. A., Pichler G., Paron I., Nagaraj N., Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 45. Birk J. B., Wojtaszewski J. F. (2006) Predominant alpha2/beta2/gamma3 AMPK activation during exercise in human skeletal muscle. J. Physiol. 577, 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Treebak J. T., Birk J. B., Rose A. J., Kiens B., Richter E. A., Wojtaszewski J. F. (2007) AS160 phosphorylation is associated with activation of alpha2beta2gamma1- but not alpha2beta2gamma3-AMPK trimeric complex in skeletal muscle during exercise in humans. Am. J. Physiol. Endocrinol. Metab. 292, E715–E722 [DOI] [PubMed] [Google Scholar]
- 47. Conley K. E., Kemper W. F., Crowther G. J. (2001) Limits to sustainable muscle performance: interaction between glycolysis and oxidative phosphorylation. J. Exp. Biol. 204, 3189–3194 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD000288. The data is also accessible and can be visualized in the MaxQB database (http://maxqb.biochem.mpg.de/mxdb) (29) under “deep muscle proteome.”